Single-nucleus RNA-seq reveals a dynamic and cell-specific gene program in striatal neurons that contributes to cocaine response.
Abstract
Drugs of abuse elevate dopamine levels in the nucleus accumbens (NAc) and alter transcriptional programs believed to promote long-lasting synaptic and behavioral adaptations. Here, we leveraged single-nucleus RNA-sequencing to generate a comprehensive molecular atlas of cell subtypes in the NAc, defining both sex-specific and cell type–specific responses to acute cocaine experience in a rat model system. Using this transcriptional map, we identified an immediate early gene expression program that is up-regulated following cocaine experience in vivo and dopamine receptor activation in vitro. Multiplexed induction of this gene program with a large-scale CRISPR-dCas9 activation strategy initiated a secondary synapse-centric transcriptional profile, altered striatal physiology in vitro, and enhanced cocaine sensitization in vivo. Together, these results define the transcriptional response to cocaine with cellular precision and demonstrate that drug-responsive gene programs can potentiate both physiological and behavioral adaptations to drugs of abuse.
INTRODUCTION
Nearly 5 million Americans reported cocaine use in 2017, and recent increases in cocaine-related drug overdoses present considerable public health challenges (1). A hallmark trait of drugs of abuse is the acute elevation of dopamine (DA) in the nucleus accumbens (NAc), a central integrator of the reward circuit (2). Abused drugs produce increases in DA that are greater in both concentration and duration than natural rewards (3, 4). Furthermore, this signaling is hypothesized to underlie maladaptive reinforcement following repeated drug use (5). Exposure to drugs of abuse results in widespread transcriptional and epigenetic alterations in the NAc (5), initiating synaptic and behavioral plasticity associated with the transition to drug addiction (5). However, even with well-studied drugs such as cocaine, drug-induced transcriptional responses remain poorly understood. This is, in part, due to the cellular heterogeneity of the NAc, which is a diverse structure containing multiple neuronal and nonneuronal subpopulations and complex neuronal circuitry. In addition, many drugs of abuse engage multiple neurotransmitter systems in the NAc (6, 7), and the specific contributions of DA-induced transcriptional programs to neuronal physiology and behavior are not clear. Furthermore, although drug experience leads to large-scale transcriptional changes in the NAc, previous studies have focused on individual drug-responsive genes instead of coordinated gene programs.
To overcome these challenges, we used high-throughput single-nucleus RNA-sequencing (snRNA-seq) to transcriptionally define cocaine-induced gene expression signatures with single-cell resolution in the NAc of adult male and female rats. We next used a well-characterized striatal culture system to further dissect a core set of immediate early genes (IEGs) robustly induced by DA in discrete neuronal subpopulations. Last, as a proof-of-principle to further understand the molecular and physiological consequences of altering this gene signature, we used a novel multiplexed CRISPR-dCas9 (dead Cas9)–based strategy to induce expression of this DA-responsive gene program. Coordinated induction of targeted genes altered neurophysiological properties in vitro and potentiated behavioral sensitization to cocaine in adult rats in vivo, suggesting that this DA-induced transcriptional signature plays a key role in shaping cellular and physiological adaptations to drugs of abuse.
RESULTS
To map the transcriptional landscape and cocaine response of the NAc with single-cell resolution, we performed snRNA-seq on 15,631 NAc nuclei from both male and female rats after acute exposure to cocaine (Figs. 1A and 2, A to C) using a cocaine dose previously reported to result in enduring synaptic changes in the NAc (8, 9). From a merged dataset containing transcriptomes from all captured cells, unsupervised dimensionality reduction approaches identified discrete NAc neuronal clusters harboring known markers of Drd1 DA receptor–positive and Drd2 DA receptor–positive medium spiny neurons (Drd1-MSNs and Drd2-MSNs, respectively), as well as previously identified (10–14) transcriptionally defined cell classes including somatostatin (Sst)–positive interneurons, microglia, astrocytes, polydendrocytes, and oligodendrocytes (Fig. 1, B to F, and fig. S1). This clustering also revealed potentially novel MSN classes marked by expression of Drd3 (a D2 family DA receptor) and Grm8 (a metabotropic glutamate receptor). While Drd3-MSN and Grm8-MSN clusters were relatively depleted in the classic MSN marker Ppp1r1b (the gene encoding DARPP-32 protein) (15), each cluster exhibited high expression of Foxp2 and Bcl11b, genes strongly linked to MSN differentiation and function (Fig. 1E) (16, 17). Moreover, whereas cell fractions in nearly all clusters were evenly distributed between treatment group (saline and cocaine) and sex, 91% of Drd3-MSNs were found in male animals, indicative of a substantial sex bias in this subpopulation (fig. S2).
Fig. 1. Identification of 16 transcriptionally distinct cell populations in the adult rat NAc.
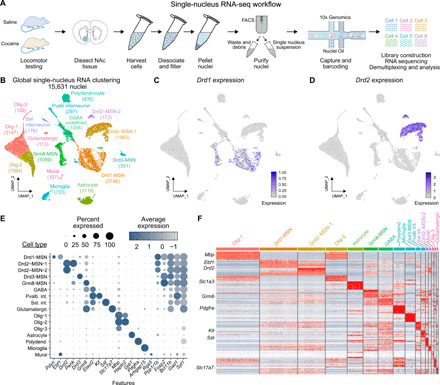
(A) snRNA-seq workflow. Male and female adult rats (n = 4 per sex per treatment) received saline or cocaine (20 mg/kg) (intraperitoneal injection) and underwent locomotor testing before tissue harvesting, NAc dissection (tissue punch area highlighted in blue), nuclei purification, and single-nucleus sequencing on the 10x Genomics platform. (B) Global clustering across experimental treatment and sex for 15,631 individual NAc nuclei identifies all major cell classes of the rat NAc, including MSNs expressing Drd1 and Drd2 mRNA. UMAP, Uniform Manifold Approximation and Projection. (C and D) Enrichment of Drd1 and Drd2 within identified cell types. (E) Dot plot indicating the average expression and percent of cells expressing marker genes of each identified cell type. GABA, γ-aminobutyric acid. (F) Heat map of cell-specific marker genes across all clusters.
Fig. 2. snRNA-seq reveals cell-specific transcriptional response to cocaine.
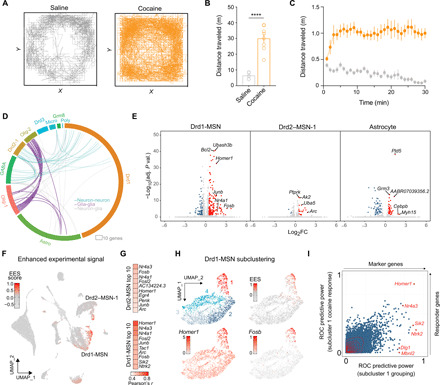
(A) Representative activity trace of saline and cocaine animals over the 30-min open-field test session. (B) Mean total distance traveled is significantly greater following intraperitoneal injection of cocaine (20 mg/kg) compared to saline controls (n = 8 per group, ****P < 0.0001). (C) Locomotor activity is increased by cocaine across all time points. (D) Circos plot of cocaine DEGs by cluster (outer rim) and coherent changes between clusters (internal arcs). Neuron-neuron, glia-glia, and neuron-glia coherent DEGs are reflected in teal, purple, and gray, respectively. (E) Volcano plots showing all DEGs [adjusted P < 0.05, absolute value of log2(fold change) > 0.5] for three distinct cell clusters. X-axis log2(fold change) values represent differences in gene expression between cocaine- and saline-treated groups. (F) Unbiased detection of cocaine-activated cell clusters with Manifold Enhancement of Latent Dimensions pipeline identifies enhanced experimental signal (EES) in distinct Drd1-MSN and Drd2-MSN neuronal clusters. (G) Top 10 gene-EES correlations for Drd1-MSN and Drd2-MSN clusters. (H) Subclustering of Drd1-MSNs highlights specific IEG induction and EES signal in a single subpopulation (subcluster 1). (I) Receiver operating characteristic (ROC) analysis on subcluster 1 identifies “marker” genes (x axis) and “responder” genes that are altered by cocaine.
To identify cocaine-activated cell clusters, we pursued two parallel strategies. First, we performed cluster-specific identification of differentially expressed genes (DEGs) in saline versus cocaine conditions (collapsing across sex; Fig. 2, D and E, and table S1). This analysis revealed robust transcriptional response in Drd1-MSNs, which contained more than twice as many DEGs (232) as any other cluster. Drd1-MSN DEGs were enriched in cyclic adenosine 3′,5′-monophosphate (cAMP) response element–binding protein (CREB) binding motifs and genes involved in mitogen-activated protein kinase (MAPK) pathways, regulation of synaptic signaling, behavior, and cognition (table S2). Drd1-MSN DEGs also exhibited overlap with DEGs arising from Drd2-MSNs (Fig. 2, D to E), suggesting the induction of common transcriptional pathways in these clusters. We next used an unbiased graphical signal processing approach to stratify cellular clusters based on condition-specific gene signatures (18), termed “enhanced experimental signal” (EES) (Fig. 2F and table S3). This method identified two unique neuronal subclusters particularly responsive to cocaine—one from the Drd1-MSN parent cluster and another from Drd2–MSN-1 parent cluster. Both high-EES cell clusters exhibited robust expression of key IEGs (e.g., Fosb, Junb, and Nr4a1; Fig. 2G) in the cocaine condition but little to no expression of these genes in the saline condition (fig. S3). Given that these genes are commonly used markers for neuronal activity (7), these results suggest that cocaine does not alter the activity of the majority of NAc cell types but rather selectively activates discrete ensembles of Drd1-MSNs and Drd2-MSNs. This analysis also revealed correlations between EES and expression of several genes central to neuropeptide signaling in MSNs (19), including the substance P precursor Tac1 (in Drd1-MSNs) and the enkephalin precursor Penk (in Drd2-MSNs; Fig. 2G). These findings are in line with previous evidence that expression of these neuropeptides is elevated in Drd1-MSNs and Drd2-MSNs in the mouse NAc after cocaine exposure (20).
To further investigate novel upstream interactions that may contribute to cocaine response likelihood within a cell type, we restricted our analysis to Drd1-MSNs and repeated dimensionality reduction mapping approaches (Fig. 2H). This analysis identified four unique subclusters of Drd1-MSNs, only one of which (subcluster 1) exhibited a robust transcriptional response to cocaine. Using the four unique subclusters of Drd1-MSNs, we used receiver operator characteristic (ROC) analysis to estimate the success of every individual gene in binary classification of a cell in two steps. First, we identified genes that marked cells found in the cocaine-activated subcluster (subcluster 1) as compared to cells in all other Drd1-MSN subclusters. Genes that were highly expressed in subcluster 1 relative to the other Drd1 subclusters were termed “marker” genes. Next, we identified genes that distinguished cocaine-treated cells from saline-treated cells within this cocaine-activated subcluster (termed “responder” genes). Intriguingly, while genes such as the synaptic protein Homer1 served as both a marker and a responder gene, other genes such as Dlg1 exhibited high predictive power in marking the cocaine-activated subcluster without exhibiting an altered response to cocaine (Fig. 2I and table S5). In contrast, the Drd1 receptor itself exhibited almost no predictive power as a marker gene (ROC power = 0.138). Together, these results highlight a potential contribution of multiple genes in determining which Drd1-MSNs are capable of being activated by cocaine.
Next, to determine whether genetic sex contributed to behavioral or transcriptional responses to cocaine, we split the merged dataset by sex and examined sex-specific locomotor data and transcriptional responses to cocaine. Male and female rats both demonstrated a significant increase in locomotor activity in response to cocaine but did not statistically differ from each other (Fig. 3A). We next recalculated EES scores and EES-gene correlations independently for each sex (Fig. 3, B and C, and table S4). Male and female transcriptional responses were positively correlated in both Drd1-MSNs and Drd2-MSNs, and in both cases, key IEGs associated with high EES scores in our sex-combined analysis also drove EES scores when sex was considered separately (Fig. 3D). In contrast, this analysis revealed unexpected divergence in astrocytes. For both sexes, numerous responsive genes within the astrocyte population exhibited strong correlations with EES (Fig. 3D). However, male and female responses were negatively correlated for this cluster, indicating the presence of distinct, nonoverlapping, and sex-specific astrocytic transcriptional responses to cocaine.
Fig. 3. Contribution of sex to cocaine-mediated locomotor activity and transcriptional responses in specific cell types in adult rat NAc.
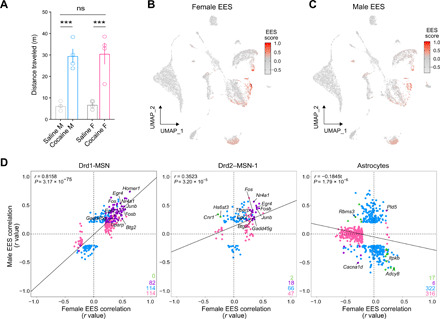
(A) Cocaine experience significantly increased locomotor activity in both male and female rats [20 mg/kg; one-way analysis of variance (ANOVA), ***P < 0.001]; however, Tukey’s post hoc analysis revealed no sex differences in the mean distance traveled in response to saline (n = 4 per sex, P = 0.9988) or acute cocaine (n = 4 per sex, P = 0.9940). ns, not significant. (B and C) UMAPs showing EES generated independently for each sex. (D) EES-gene correlation correspondence plots reveal highly similar cocaine-induced transcriptional responses in Drd1-MSNs and Drd2-MSN clusters. In contrast, cocaine-mediated transcriptional responses in astrocytes were divergent in males and females. EES-gene correlations that reach significance are shown in blue for males and pink for females. EES-gene correlates that occur in the same direction in both sexes are shown in purple, and correlates that diverge by sex are shown in green.
The activation of specific neuronal ensembles observed in Drd1-MSNs and Drd2-MSNs by cocaine may result from circuit-based mechanisms (e.g., differential projections from input structures) or could potentially arise from cell-autonomous mechanisms in response to DA signaling (5, 6, 21–23). However, drugs of abuse target many distinct receptor classes in a variety of neuronal and nonneuronal cell types (9), and these complex drug actions make identification of DA-induced gene expression programs difficult using in vivo models. Therefore, we took advantage of a well-studied and controllable rat primary striatal neuron culture system (24, 25) to enable comprehensive and specific identification of DA-regulated gene expression programs in MSNs. Using this system, we first identified a core signature of the transcriptional response to DA receptor activation by performing deep RNA-seq on bulk striatal neuronal cultures treated with 1 μM DA for 1 hour, a treatment that closely models concentrations and temporal duration of DA increases found in vivo after acute cocaine exposure (2, 4). This approach identified 103 DEGs (Fig. 4, A and B, and table S6) following DA treatment with 97% of DEGs (100 of 103) being up-regulated versus vehicle control samples. Gene Ontology (GO) analysis revealed that genes up-regulated by DA receptor stimulation are primarily associated with transcriptional mechanisms and processes important for neuronal function, such as regulation of synaptic plasticity (Fig. 4C and table S7). Transcription factor motifs significantly enriched in DA-responsive genes included cAMP response elements, which is consistent with previous reports demonstrating a key role for CREB in drug-induced transcriptional changes (26).
Fig. 4. Identification of a cell type–specific DA-mediated transcriptional program.
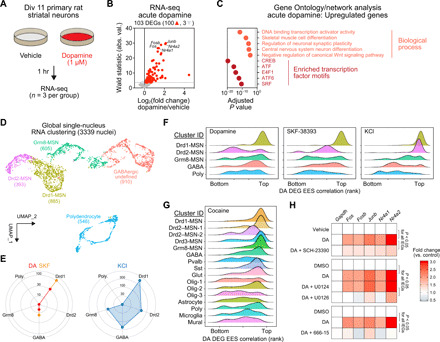
(A) Experimental setup for modeling DA receptor activation in striatal cultures. (B) Volcano plot showing DEGs following 1 hour. DA treatment, including three down-regulated genes (light gray) and 100 up-regulated genes (red). (C) Top five significant biological processes (Gene Ontology) and transcription factor binding site enrichment (motif analysis) for 100 up-regulated DA DEGs. A maximum value was assigned to CREB (adjusted P = 0). (D) snRNA-seq from 3339 nuclei identified five major cell classes in primary striatal neuron cultures. Neurons were treated with DA (50 μM), the DRD1 agonist SKF-38393 (1 μM) or depolarized with 25 mM KCl. (E) DEG number by cell cluster. (F) Ridgeline plots showing density of EES-gene correlation ranks for core DA signature genes [100 up-regulated genes from (B)]. (G) DA signature gene induction in multiple MSN clusters following acute cocaine experience in vivo (clusters from Fig. 1B). (H) Quantitative reverse transcription polymerase chain reaction (RT-qPCR) for representative DA response genes following cotreatment with DRD1 receptor antagonist SCH-23390 (1 μM), MAPK kinase (MEK) inhibitor (U0126, 1 μM), or CREB inhibitor (666-15, 1 μM). P < 0.05 for DA IEGs (n = 7 to 12 per group).
To determine whether this core transcriptional response to DA occurs broadly across MSN subtypes or is limited to specific populations, we performed snRNA-seq from 3339 cultured striatal neurons (mixed from male and female E18 rat brains) in four distinct treatment groups [vehicle, 50 μM DA, 1 μM DRD1 receptor agonist SKF-38393, or 25 mM potassium chloride (KCl) to induce neuronal depolarization]. Cultured neurons were again largely divided into defined Drd1-MSNs, Drd2-MSNs, and Grm8-MSNs (Fig. 4D). Unlike snRNA-seq datasets from the adult NAc, primary striatal neuron cultures were devoid of oligodendrocyte, astrocyte, and microglia markers (because of the embryonic stage of tissue harvested for culture) but did harbor polydendrocytes positive for canonical marker genes Pdgfra and Olig1 (fig. S4). Similar to cocaine experience in vivo, treatment with DA or the DRD1 agonist SKF-38393 predominantly resulted in transcriptional activation of Drd1-MSNs (Fig. 4E). This activation included core DA signature genes identified from bulk RNA-seq (Fos, Fosb, Junb, Nr4a1, and Nr4a2), demonstrating robust induction of this gene program in Drd1-MSNs (fig. S5 and table S8). Moreover, expression of nearly all of these genes was positively correlated with unbiased EES scores for DA and SKF-38393 treatments only in Drd1-MSNs (Fig. 4F and table S9), highlighting the key contribution of this program to overall transcriptional changes in these neurons. In contrast, both Drd1-MSN and Drd2-MSN clusters responded with massive transcriptional alterations following neuronal depolarization with KCl, which was reflected in the total number of DEGs, the fraction of cells activated, and the percentage of reads aligning to DA-responsive genes (Fig. 4E and fig. S5). Together, these results suggest that both Drd1-MSNs and Drd2-MSNs are capable of producing a robust transcriptional response involving the same core gene set, but only Drd1-MSNs exhibit robust transcriptional responses to increased DA neurotransmission in this model. Intriguingly, expression of this core gene set was also positively correlated with EES score in the adult snRNA-seq dataset in multiple MSN clusters following cocaine experience (Fig. 4G), potentially reflective of multimodal (e.g., DA-dependent and DA-independent) contributions to this gene expression profile after cocaine exposure.
Given that DA-responsive genes were preferentially activated in Drd1-MSNs and that these genes are enriched for CREB binding motifs, we hypothesized that responsivity of these genes could be prevented via inhibition of the DRD1 receptor, interference with MAPK signaling cascades linked to CREB activation, or inhibition of CREB itself. To test these hypotheses, we performed quantitative reverse transcription polymerase chain reaction (RT-qPCR) for key DA-responsive genes 1 hour following DA stimulation (1 μM) in the presence of either a DRD1 receptor antagonist (SCH-23390, 1 μM), a MAPK kinase (MEK) inhibitor (U0126, 1 μM), or a CREB inhibitor (666-15, 1 μM). In agreement with our prediction, we found that induction of each DA-responsive gene was inhibited by each of these treatments (Fig. 4H). These results confirm that DA-responsive gene programs, and specifically this core set of IEGs, are DRD1 dependent in this culture model system and demonstrate that these effects require MAPK signaling and classical CREB-mediated transcriptional mechanisms.
Although individual candidate genes in this DA-responsive transcriptional program have been shown to play key roles in drug-induced adaptations (20, 23, 27–30), it is hypothesized that gene expression programs work in concert to exert downstream effects on neuronal function and behavior (31). However, defining the significance of key gene expression programs has remained challenging because of the lack of tools capable of interrogating large-scale polygenic changes. Recent developments in CRISPR-based technology have enabled rapid and multiplexable transcriptional control in the mammalian central nervous system, providing an avenue to investigate how gene programs regulate normal and maladaptive brain states (24, 32). As a proof-of-principle experiment to simultaneously activate multiple genes in this DA-induced gene expression profile, we harnessed a neuron-optimized CRISPR activation (CRISPRa) system (Fig. 5) in which a catalytically dead Cas9 protein (dCas9) is fused to the strong hybrid transcriptional activator VPR (a concatemer of the herpes simplex viral protein VP16, the p65 subunit of nuclear factor κB, and the gammaherpesvirus transactivator Rta) (24). This system allows multiplexed gene programming through the design of single guide RNAs (sgRNAs) that target the promoter region of selected genes. Using this system, we designed sgRNAs to target 16 of the genes most robustly altered by DA receptor activation (including Fos, Fosb, and Nr4a1; Fig. 5A). Following validation of each sgRNA using individual gene targeting in striatal cultures (fig. S6), we engineered custom multiplex sgRNA lentiviruses that contained eight sgRNAs each for a total of 16 unique sgRNAs (Fig. 5, A to C). Expression of these vectors in striatal neurons in tandem with neuron specific, but not subpopulation specific, expression of the dCas9-VPR fusion protein enabled simultaneous induction of all genes targeted (which we termed “Dopaplex”) as compared to a control multiplex virus expressing an sgRNA array targeting the bacterial gene LacZ (Fig. 5D).
Fig. 5. Multiplexed CRISPRa engineering to mimic DA-induced gene expression changes.
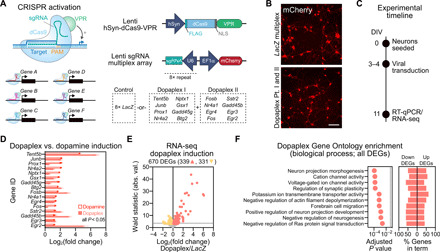
(A) CRISPRa multiplex vector approach for expressing the dCas9-VPR activator fusion and multiplex sgRNAs targeting either the bacterial LacZ gene (nontargeting control) or 16 of the top DA-induced genes (Dopaplex). PAM, protospacer adjacent motif. (B) Live cell imaging reveals successful transduction of the CRISPRa lentiviruses. Scale bar, 50 μm. (C) Experimental timeline for in vitro Dopaplex induction. Primary striatal neurons were generated and transduced with multiplexed CRISPRa constructs at DIV 3 to 4. On DIV11, RNA was extracted and subjected to both RT-qPCR and RNA-seq. (D) dCas9-VPR increases gene expression of Dopaplex genes compared to multiplexed LacZ control (n = 8 per group, unpaired t test, P < 0.05 for all comparisons). Dopamine-induced increases in gene expression of Dopaplex genes from bulk RNA-seq are plotted for comparison. All data are expressed as mean ± SEM. (E) RNA-seq volcano plot showing DEGs in Dopaplex versus LacZ multiplex targeting conditions. Up-regulated genes (salmon, 339 genes) and down-regulated genes (yellow, 331 genes) are indicated, and a standard Wald statistic cutoff corresponding to adjusted P < 0.05 is represented by the horizontal dotted line. (F) Top 10 significant molecular function (GO) for all DEGs (excluding 16 Dopaplex genes) after Dopaplex induction.
Because many of the top DA-responsive genes were transcription factors, we next sought to define the transcriptional consequences of activating this program. Bulk RNA-seq comparisons in primary striatal cultures following constitutive CRISPRa targeting LacZ or Dopaplex identified 670 DEGs, with 339 up-regulated genes and 331 down-regulated genes (Fig. 5E and table S10). After removal of the 16 targeted Dopaplex genes from the gene list, GO analysis revealed that critical neuronal processes were altered in the Dopaplex-targeted group, including genes involved in the regulation of ion channels, projection morphogenesis, and synaptic plasticity (Fig. 5F and table S11). These results demonstrate that selective activation of a limited set of DA-sensitive IEGs can result in large-scale transcriptional consequences, including gene targets that promote neurophysiological alterations.
Exposure to drugs of abuse or alterations in CREB signaling can influence the intrinsic physiological properties of MSNs (6, 23). Therefore, we sought to understand whether Dopaplex induction altered the physiological activity of MSNs using a high-throughput multielectrode array (MEA) system in which striatal neurons are seeded directly on a 768 electrode array divided across 48 culture wells (Fig. 6, A and B). Dopaplex-expressing neurons did not differ from nontargeting LacZ multiplexed control neurons in both the number of spontaneously active neurons and the mean action potential frequency (Fig. 6, C to E). However, action potential burst frequency was increased in Dopaplex-targeted neurons (Fig. 6, F and G), demonstrating that this DA-regulated gene expression program alters MSN firing patterns. To determine whether this electrophysiological phenotype could be mimicked by targeting a single gene within the Dopaplex program, we targeted CRISPRa machinery to either the transcription factor Fosb [which has been frequently implicated in cocaine action (5)], the nucleotidyltransferase Tent5b (the gene with the highest fold change after DA stimulation), or LacZ control. Neurons overexpressing either Fosb or Tent5b alone did not differ from LacZ sgRNA neurons in any physiological measure recorded (active units, firing rate, or burst frequency; Fig. 6H). This suggests that the increase in burst firing following induction of the Dopaplex program is a result of coordinated transcriptional changes, as up-regulation of individual gene targets alone was not sufficient to alter physiological properties of MSNs.
Fig. 6. Dopaplex gene expression program increases bursting activity of MSNs and enhances cocaine locomotor sensitization.
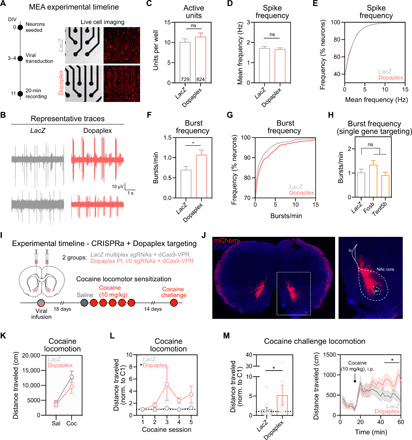
(A) Left: Experimental timeline for viral transduction and MEA recordings. Right: Live cell imaging demonstrating CRISPRa lentivirus expression. (B) Representative single-unit voltage traces. The number of active units per well (C) and action potential frequency (D and E) did not change following Dopaplex targeting (P > 0.275 for both comparisons). (F) Burst frequency is increased in Dopaplex-targeted neurons (n = 729 to 824 neurons per group, *P = 0.0265). (G) Cumulative distribution of burst frequency. (H) Burst frequency was not altered when CRISPRa was targeted to single genes within the Dopaplex gene expression program (n = 356 to 372 neurons per group, P = 0.1012). (I) Experimental timeline for viral transduction and cocaine locomotor sensitization. (J) Representative image of sgRNA viral transduction (mCherry reporter) in the NAc. (K) Absence of baseline locomotion difference between LacZ and Dopaplex-targeted animals following the first cocaine injection (n = 12 to 14 rats per group, P = 0.3474). (L) Over five cocaine administration sessions, Dopaplex-targeted animals exhibited increased locomotion (*P = 0.0464). (M) Cocaine challenge injection revealed enhanced locomotor sensitization in Dopaplex-targeted rats (*P = 0.0270). All data are expressed as means ± SEM.
Our results suggest that DA signaling results in an altered transcriptional profile that may enhance excitability of MSNs, which is consistent with reports that prior drug exposure alters intrinsic physiological properties of MSNs (6). Therefore, we sought to understand how this gene expression program might affect behavioral changes observed after exposure to drugs of abuse. One such behavior is locomotor sensitization, in which DA transmission in the NAc is potentiated as sensitization develops (33, 34). To examine the function of this DA-induced gene expression program on cocaine locomotion sensitization, we stereotaxically infused CRISPRa lentiviral vectors targeting Dopaplex genes (or LacZ control) bilaterally into the NAc core (Fig. 6, I and J, and fig. S7), a region critical for the development and maintenance of sensitization (35). We previously validated robust and neuron-selective expression of this CRISPRa system in vivo within 2 weeks of viral delivery into the NAc, demonstrating that this tool results in elevated protein levels of target genes (24). This manipulation targets all NAc neurons (including Drd1-MSNs and Drd2-MSN populations), which demonstrated partially overlapping transcriptional responses to cocaine in our snRNA-seq dataset (Fig. 2D). Eighteen days after viral infusion, rats began a locomotor sensitization protocol in a novel environment using a subthreshold dose of cocaine (10 mg/kg) that does not reliably produce robust sensitization (36). Animals receiving constitutive CRISPRa constructs targeting Dopaplex genes did not differ from LacZ multiplex controls in baseline locomotion after an injection of saline nor in acute increases in locomotion following the first dose of cocaine (Fig. 6K). However, we observed a significant increase in the development of locomotor sensitization after five cocaine injections in Dopaplex-targeted rats (Fig. 6L). Likewise, this enhancement in locomotor activity was maintained 14 days after the last cocaine pairing by administration of a cocaine challenge dose (Fig. 6M). These findings suggest that induction of a DA-mediated IEG network in the NAc is sufficient to sensitize behavioral responses to a normally subthreshold dose of cocaine. Furthermore, this work provides the first proof-of-principle demonstration that CRISPRa can be used to recapitulate a large-scale coordinated gene expression signature in vivo.
DISCUSSION
Substance use disorder is characterized by long-lasting molecular and physiological changes in the mesocorticolimbic reward system. A unifying feature of all drugs of abuse is their ability to increase DA signaling within the brain regions that comprise this system, including the NAc. Our present understanding of dopaminergic transcriptional control in reward circuitry is largely based on next-generation sequencing approaches conducted with bulk tissue homogenate. While this work has provided useful information regarding global drug-related changes in gene expression, the neurocircuitry that underlies maladaptive reward seeking contains a great deal of cellular diversity. Traditional RNA-seq approaches fail to capture the unique landscape of these distinct cell populations and at best represent aggregate changes in gene expression. Here, we used snRNA-seq to capture the heterogeneity of the NAc and define the transcriptional architecture of this region. Unbiased dimensionality reduction approaches revealed 16 transcriptionally defined populations of cells in male and female rat NAc and allowed for comprehensive examination of cocaine-mediated transcriptional states between these distinct populations. We further supplemented this work using high-depth bulk RNA-seq and snRNA-seq of well-defined striatal neuronal cultures to dissect DA-dependent gene expression programs in MSNs. This combinatorial approach allowed us to identify a core set of IEGs robustly induced by DA in a small subset of MSNs.
Although a number of studies have previously shown that discrete ensembles of MSNs are activated as a consequence of cocaine exposure (7, 20, 37), the present findings expand on this body of work in several important ways. For example, while our data are congruent with previously reported IEG changes (e.g., Fosb, Egr1, and Nr4a1) in response to drug experience (7, 20, 38), our results extend this work by revealing hundreds of other cocaine-induced gene changes in transcriptionally defined populations, many of which have not been previously explored in the context of addiction. Cell type–specific transcriptional profiling with snRNA-seq revealed that cocaine activates a subset of Drd1-MSNs and Drd2-MSNs, but this effect was more robust in the Drd1-MSN population—both in terms of number of cells activated and the total number of gene expression changes. Furthermore, ROC analysis performed on a specific subpopulation of Drd1-MSNs that exhibited elevated responses to cocaine suggests that certain gene markers may hold predictive power for which cells are most likely to be transcriptionally altered by cocaine. However, additional studies are required to validate this intriguing possibility. Although our focus here was predominantly on striatal MSN populations most robustly altered by cocaine exposure, we have created a public web browser where the datasets produced by these experiments can be viewed (available via day-lab.org/resources). We hope this will serve as a rich, user-friendly resource for other investigators to explore cocaine- and DA-regulated transcriptional changes in any of the clusters identified.
Critically, snRNA-seq approaches used here allow for comparison of transcriptional responses to acute cocaine in each of the unique cell populations found in the NAc, a previously unachieved level of cellular resolution for cocaine-mediated gene regulation in this region. This is particularly important, as transcriptional alterations observed from bulk transcriptional profiling only provide a representative average of RNA changes in a region of interest, and this metric can be heavily biased by only a small percentage of cells (39). While strategies that focus on transcriptional profiling of Fos+ cells isolated after cocaine experience can help to identify other genes enriched in activated cells, these approaches do not allow for direct comparisons between similarly activated yet distinct cell types (7). In addition, these techniques rely on the presence of mature FOS protein, limiting the transcriptional profiling to time points after the first wave of IEGs as characterized in this study. Furthermore, transcript levels within similar cellular phenotypes can also vary by several orders of magnitude (40). In addition to enabling comparison of gene expression patterns between unique neuronal subpopulations within the NAc, this study also revealed a robust transcriptional response to cocaine in astrocytes, which appears to diverge based on sex. While previous research has identified astrocytes as an addiction-relevant cell population (41), to our knowledge, no studies have conducted selective in-depth sequencing of an astrocyte population in response to cocaine. Therefore, this work also expands on previous investigations of cocaine-induced neuroadaptations by providing new insights into relevant cell type–specific gene targets for substance abuse intervention strategies. An additional strength of the present study is the incorporation of both male and female rats in snRNA-seq experiments. This is an improvement upon most of the studies characterizing transcriptional responses to cocaine, which have historically been conducted exclusively in male animals.
Single-nucleus capture and RNA-seq approaches used here identified 16 transcriptionally distinct clusters from 15,631 male and female rat NAc nuclei, defining the organizational principles of transcriptional heterogeneity in the NAc. These observations are largely consistent with recently published single-cell sequencing datasets from the mouse striatum. In agreement with other single-cell sequencing studies, we identified two major MSN populations defined by distinct patterns of DA-receptor expression (Drd1-MSNs and Drd2-MSNs, respectively), each of which contained classic MSN markers including Bcl11b and Ppp1r1b (10–13). Similar to other reports, we found that substance P precursor Tac1 was enriched predominately in Drd1-MSNs, whereas expression of opioid neuropeptide Penk overlapped primarily with Drd2-MSNs (13, 14). We also observed several classes of interneurons expressing the GABAergic enzyme Gad1, as well as physiologically dissociable interneuron classes expressing Sst and Pvalb recently described in the mouse dorsal striatum (12). Although we did observe sparse expression of cholinergic interneuron marker Chat within our GABAergic undefined cluster, this signal was not observed in most of the cells in this population nor was it robust enough to yield its own discrete cluster. Unbiased clustering also revealed smaller classes of NAc MSNs defined by enrichment in the D2-family receptor Drd3 or the metabotropic glutamate receptor Grm8, populations not previously reported in mouse single-cell RNA-seq papers that focused largely on the dorsal striatum. Although each of these clusters exhibited limited expression of Ppp1r1b, commonly found in Drd1-MSNs and Drd2-MSN populations, they expressed high levels of other common MSN markers Foxp2 and Bcl11b. Last, neuronal populations in the NAc were accompanied by a variety of nonneuronal cell populations, including oligodendrocytes, astrocytes, polydendrocytes, microglia, and mural cells, each containing population-specific gene markers similar to those previously described in single-cell datasets originating from the adult mouse brain (10, 11).
Following chronic drug administration, drug-induced dopaminergic signaling in the NAc is potentiated (33, 34), providing multiple opportunities for induction of long-lasting changes in gene expression that are observed after repeated drug use (5, 42). Traditional approaches focusing on single candidate genes have been critical in understanding some of the fundamental transcriptional changes that occur following exposure to drugs of abuse (20, 23, 27–30). However, activity-dependent transcriptional events that occur after drug use produce coordinated changes that may be distinct from isolated changes in the expression of a single gene. Recent advances in next-generation sequencing have provided tremendous insight into the large-scale genome-wide transcriptional changes mediated by DA signaling after drug use. However, approaches that allow for selective and simultaneous regulation of multiple gene targets to mimic gene expression patterns have been largely absent. Here, we used a large-scale CRISPR-dCas9 activation strategy to recreate the gene signature program identified in MSNs by bulk and snRNA-seq datasets. This work provides the first proof-of-principle evidence that CRISPRa can be used for simultaneous and selective regulation of a gene expression signature in vivo. An important consideration of the present work is that we chose to perform viral manipulations in a neuron-specific, but not subpopulation-specific, manner because of our observation that cocaine strongly induced expression of many of these IEGs in both Drd1-MSNs and Drd2-MSN cell types. An important future direction for this line of research will involve multiplexed CRISPRa manipulations within distinct Drd1-MSNs and Drd2-MSN subpopulations. Although transgenic Drd1-Cre and Drd2-Cre rat lines and Cre-dependent CRISPR gRNA strategies have previously been described (43), these strategies are not currently compatible with the multiplexed guide vector used for CRISPRa approaches described in this study. Inducible and cell type–specific strategies compatible with multiplexed CRISPR regulation will be critical for unraveling how temporal- and population-specific regulation of this core gene program affects striatal physiology- and reward-related behaviors in vivo. In addition to Drd1-MSNs and Drd2-specific manipulations, an inducible CRISPRa system could be modified to allow for exploration of gene programs in nonneuronal cell types that are also transcriptionally responsive to cocaine, such as astrocytes.
The DA-induced gene expression program identified here consisted largely of IEGs. This is likely a reflection of altered neuronal activity in response to DA but does not indicate that this gene program is exclusively regulated by DA. Several of these IEGs (Fos, Fosb, Junb, and Nr4a1) were also induced by KCl depolarization in our striatal snRNA-seq datasets. Given that induction of several of these plasticity-associated genes is a shared feature of learning and memory research, it will be important for future work to delineate whether similar gene expression profiles emerge in other brain regions in response to alternate stimuli or experiences. For example, several IEGs in the Dopaplex construct (e.g., Fos, Egr, and Nr4a family genes) are markers of cellular ensemble activation during fear conditioning and episodic memory paradigms (44), and these genes have been shown to be correlated with learning in behavioral tasks (45, 46). Given the versatile and modular nature of the CRISPRa approach described here, it does not escape our attention that this system could similarly be applied to dissect the role of any gene program in mediating other types of behavior, memory formation, or neuropsychiatric disorders.
In addition to the robust induction of IEGs observed in Drd1-MSNs and Drd2-MSNs, we also observed concomitant increases in neuropeptide expression in each of these subpopulations in response to cocaine. These findings are in line with previous evidence that Tac1 and Pdyn are elevated in Drd1-MSNs and that Penk expression is elevated in Drd2-MSNs in the mouse NAc after cocaine exposure (20). Similarly, others have demonstrated that Penk expression is mediated by cocaine experience in rodents (47), and Penk knockout mice demonstrate reduced motivation to seek a cocaine reward (48). This work may be translationally relevant, as polymorphisms in the Penk gene have previously been implicated in substance use disorder in humans (49). Overall, our results add to the growing body of literature, suggesting that opioid neuropeptide systems affect DA transmission in reward neurocircuitry (50, 51) and may play a critical role in psychostimulant abuse.
While the present work provides insight into how cellular diversity contributes to transcriptional responses after an initial cocaine experience, repeated exposure to drugs of abuse promotes neurophysiological adaptations that are thought to drive compulsive drug seeking long after cessation of use. Hence, it will be critical for future studies to expand on this work by examining the transcriptional consequence of repeated or self-administered drug use at the single-cell level, as well as understanding how these changes are maintained within different cell populations over longer periods of time and as a result of volitional drug experience. Likewise, although drug use can alter chromatin accessibility in reward-relevant brain regions, little is known about how these changes are dynamically regulated and sustained in specific cell populations within reward circuitry. Therefore, mapping chromatin modifications in distinct cell populations in parallel with transcriptional changes will reveal essential information about how these processes act in concert to shape functional properties of individual cells within reward regions. Collectively, this work will provide a necessary level of resolution for improved therapeutic strategies aimed at reversing gene expression programs in defined cell populations that perpetuate maladaptive states.
In summary, our results provide single-cell resolution of cocaine-induced transcriptional regulation by identifying specific MSN subpopulations altered by cocaine experience, defining a DA-responsive gene program that alters neuronal function and behavioral responses to cocaine. Together, these results allow comprehensive exploration of NAc transcriptional programs in an addiction-relevant model species and identify novel target cells and genes for perturbation in the context of dopaminergic signaling. Given that addiction is a polygenic disorder that involves a complex interplay between genetic background and experience-dependent cellular responses, the use of multiplexed gene regulation and cell-specific manipulations may enable a more complete understanding of how initial drug reinforcement transitions to long-term substance abuse disorders.
MATERIALS AND METHODS
Animals
All experiments were performed in accordance with the University of Alabama at Birmingham Institutional Animal Care and Use Committee. Sprague-Dawley timed-pregnant dams and 90- to 120-day-old male or female rats were purchased from Charles River Laboratories. Dams were individually housed until embryonic day 18 (E18) for cell culture harvest. Male or female adult rats were cohoused in pairs in plastic filtered cages with nesting enrichment in an Association for Assessment and Accreditation of Laboratory Animal Care–approved animal care facility maintained between 23° and 24°C on a 12-hour light/12-hour dark cycle with ad libitum food (Lab Diet Irradiated rat chow) and water. Bedding and enrichment were changed weekly by animal resources program staff. Animals were randomly assigned to experimental groups.
Drugs
Cocaine hydrochloride was dissolved in sterile 0.9% sodium chloride. Drugs were purchased from Sigma-Aldrich (St. Louis, MO) and intraperitoneally injected at a dose of 20 or 10 mg/kg for locomotor testing and locomotor sensitization, respectively. Cocaine solution was made fresh immediately before behavioral testing and protected from light. For in vitro experiments, drugs were diluted in Neurobasal medium (Invitrogen) immediately before treating cell culture plates. DA hydrochloride (Sigma-Aldrich, H8502-5G) was dissolved in Neurobasal medium, and cells were treated at a dose of 1 μM for RT-qPCR experiments or 50 μM for snRNA-seq experiments. R(+)–SCH-23390 hydrochloride (Sigma-Aldrich, D054-5MG) and R(+)–SKF-38393 hydrochloride (Sigma-Aldrich, S101-5MG) were dissolved in sterile Milli-Q water, and cells were treated at a dose of 1 μM. For MEK inhibitor experiments, U0124 (1 μM; Millipore, 662006-1MG) and U0126 (1 μM; Millipore, 662005-1MG) were dissolved in dimethyl sulfoxide (DMSO) (Invitrogen, D12345). The CREB inhibitor 666-15 (1 μM; Tocris, 5661), also called 3-(3-aminopropoxy)-N-[2-[[3-[[(4-chloro-2-hydroxyphenyl)amino]carbonyl]-2-naphthalenyl]oxy]ethyl]-2-naphthalenecarboxamine hydrochloride, was dissolved in DMSO. KCl (Fisher Scientific, P330-500) was dissolved in Neurobasal media, and cells were treated with a 25 mM dose.
Neuronal cell cultures
Primary rat neuronal cultures were generated from E18 rat striatal tissue as described previously (24, 25). Briefly, cell culture plates (Denville Scientific Inc.) were coated overnight with poly-l-lysine (50 μg/ml; Sigma-Aldrich) supplemented with laminin (7.5 μg/ml; Sigma-Aldrich) and rinsed with diH2O. MEAs (Axion BioSystems) were coated with polyethyleneimine (Sigma-Aldrich). Dissected striatal tissue was incubated with papain (Worthington, LK003178) for 25 min at 37°C. After rinsing in complete Neurobasal media (supplemented with B27 and l-glutamine, Invitrogen), a single-cell suspension was prepared by sequential trituration through large to small fire-polished Pasteur pipettes and filtered through a 100-μm cell strainer (Fisher Scientific). Cells were pelleted, resuspended in fresh media, counted, and seeded to a density of 125,000 cells per well on 24-well culture plates (65,000 cells/cm2) or 30,000 cells per well on 48-well MEA plates. Cells were grown in complete Neurobasal media for 11 days in vitro (DIV 11) in a humidified CO2 (5%) incubator at 37°C with half media changes at DIV 1, 4 to 5, and 8 to 9. MEAs received a one-half media change to BrainPhys (STEMCELL Technologies Inc.) with SM1 and l-glutamine supplements starting on DIV 4 to 5 and continued every 3 to 4 days. All MEA media were supplemented with penicillin-streptomycin.
RNA extraction and RT-qPCR
Total RNA was extracted (RNeasy Mini kit, Qiagen) and reverse-transcribed (iScript cDNA Synthesis Kit, Bio-Rad). Complementary DNA (cDNA) was subject to RT-qPCR for genes of interest, as described previously (25). A list of PCR primer sequences is provided in table S12.
Tissue collection from adult NAc
One hour after intraperitoneal injection of either saline vehicle or cocaine (20 mg/kg), rats were euthanized by live decapitation, and the brain was rapidly removed and blocked into 1-mm-thick coronal sections in ice-cold Hibernate A media (Thermo Fisher Scientific, A1247501) supplemented with B27 and GlutaMAX (Life Technologies, 35050-061). Tissue punches, 3 mm in diameter, were then centered around the NAc core and shell for tissue collection [~+1.68 anterior-posterior (AP) from bregma; n = 4 rats per treatment per sex], transferred to ice-cold centrifuge tubes, and rapidly frozen on dry ice. Tissue was stored at −80°C until the day of cDNA library preparation.
Single-nuclei dissociation
Frozen tissue from adult NAc
Frozen tissue punches were thawed slowly on ice before being chopped by scalpel 100 times in two orthogonal directions. Tissue from four animals in the same treatment condition was then combined and transferred to 5 ml of chilled (4°C) lysis buffer [10 mM tris-HCl, 10 mM NaCl, 3 mM MgCl2 (Thermo Fisher Scientific, BP214-500), and 0.1% Igepal in nuclease-free water (Sigma-Aldrich, 18896-50ML)] for 15 min, mixing the tissue by inversion every 2 min. Lysis was quenched after 15 min with 5 ml of complete Hibernate A (Thermo Fisher Scientific, A1247501) supplemented with B27, GlutaMAX (Life Technologies, 35050-061), and NxGen ribonuclease (RNase) inhibitor (0.2 U/μl; Lucigen, 30281-2). Tissue was then triturated by fire-polished Pasteur pipette (two pipettes of decreasing diameter; 8 to 10 passes per pipette) for 35 min and passed through a 40-μm prewet filter. The samples were then pelleted at 500 relative centrifugal force (rcf) for 10 min at 4°C followed by a wash in 10 ml of nuclei wash and resuspension buffer [1× phosphate-buffered saline (PBS), 1% bovine serum albumin (BSA), and NxGen RNase inhibitor (0.2 U/μl; Lucigen, 30281-2)]. Supernatant was then removed, and the pellet was gently resuspended in a total volume of 800 μl before 7-aminoactinomycin D (Thermo Fisher Scientific, 00-6993-50) staining and fluorescence-activated cell sorting (FACS) to further purify the nuclei for sequencing (BD FACS Aria II, 70-μm nozzle, BD Biosciences). Immediately after FACS, nuclei were washed a final time at 200 rcf in 10 ml of supplemented Hibernate A containing 1% BSA for 10 min at 4°C to remove any remaining fine debris and nascent RNA. Nuclei were then brought to a concentration of 1700 nuclei/μl. Last, an average total of 2400 nuclei pooled from four rats per sex and treatment group were loaded into a single well of the Chromium Single Cell B Chip (10x Genomics, catalog no. 1000074) using four of the eight available wells.
Striatal cell culture dissociation
On DIV 11, striatal primary cells cultured on 12-well plates (250 K per well) were treated with Neurobasal vehicle, 50 μM DA, 1 μM SKF-38393, or 25 mM KCl and placed back into the incubator for 1 hour. Cells were then washed with 1 ml of PBS per well, followed by a 5-min incubation period in 600 μl of lysis buffer. Cells were then gently pipette mixed five times with a P1000 before being transferred to a 1.5-ml tube and centrifuged at 500 rcf for 5 min at 4°C. Supernatant was then removed, and the pellet was resuspended in 100 μl of nuclei wash buffer. Tubes for each treatment group were then combined, gently pipette-mixed five times, and passed through a 40 μM filter before FACS to further purify nuclei for sequencing. After FACS, 2400 nuclei per treatment condition were loaded into individual wells of the Chromium Single Cell B Chip (10x Genomics, catalog no. 1000074) using four of the eight available wells. Chromium single-nuclei capture and barcoding were completed on the Chromium Single Cell Controller according to manufacturer’s instructions.
snRNA-seq and analysis
Libraries were constructed according to manufacturer’s instructions using the Chromium Single Cell 3′ Library Construction Kit (10x Genomics, catalog no.1000092), which uses version 3 chemistry for gene expression. Nuclei (15,676) from adult rat NAc and nuclei (3600) from cultured primary rat striatal neurons were sequenced on the Illumina NextSeq 500 at the UAB Heflin Genomics Core to a depth of ~38,000 reads per nuclei and ~147,201 reads per nuclei, respectively. A Cell Ranger (v3.0.2) reference package was generated from the Ensembl Rn6 rat genome with a modified gene transfer format (GTF) file (version 95) to create a custom pre-mRNA package, which ensured alignment to unspliced pre-mRNAs and mature mRNAs. Cell Ranger filtered outputs were analyzed with Seurat v3.0.2 using R v3.6.0 (52, 53). Nuclei containing <200 features and >5% of reads mapping to the mitochondrial genome were removed from further analysis. Molecular count data from each GEM (gel beads in emulsion) well were then log normalized with a scaling factor of 10,000. Following normalization and before dimensionality reduction, all GEM wells were integrated with FindIntegrationAnchors() and IntegrateData() using 17 principal components (PCs) for the adult dataset and 10 PCs for the culture dataset. Dimensionality reduction is dependent on two main parameters, number of PCs, and the resolution value. To determine the appropriate values, uniform manifold approximation and projection (UMAPs) were generated for every combination of 15 different PCs (2 to 30) and 20 different resolution values (0.1 to 2.0) using the standard Louvain algorithm. To produce UMAPs that capture previously defined cell types, we used 17 PCs with a resolution value of 0.2 for the adult dataset and 10 PCs with a 0.1 resolution value for the culture dataset. To validate that the clusters are specific and consist of known cell types, the log-normalized expression value of the top two to six genes from each striatal cell type, as identified by publicly available single-cell sequencing databases [e.g., DropViz.org (10) and mousebrain.org (11)], found within each dataset was overlaid on the UMAP using FeaturePlot(). To identify stimulation-dependent DEGs within each cell type, a Wilcoxon ranked-sum test was performed on the log-normalized corrected unique molecular identifier counts using custom R scripts based on Seurat source code. P values were then adjusted using a Bonferroni correction based on the total number of genes identified within the dataset. Biological process GO and transcription factor motif enrichment analysis for DEGs from specific cell clusters were performed in WEB-based Gene Set Analysis Toolkit (WebGestalt) (54) using all protein coding genes as a reference set. Correction for multiple testing was performed using Benjamani-Hochberg adjustment, with statistical significance assessed at false discovery rate (FDR) < 0.05.
EES was calculated using the Manifold Enhancement of Latent Dimensions (version 0.0) (18) package on a cell-state graph generated using 100 PC analysis (PCA) dimensions calculated with edge weights between cells generated through the graph-tools library with a knn = 9 and a decay = 10 using Python v3.7.3. For main analyses, lowly expressed genes [genes not expressed in at least 10 cells (adult NAc dataset) or 5 cells (primary striatal culture dataset)] were removed before EES analysis. For sex-separated analysis from adult NAc datasets, all genes were used for EES calculations. The saline and cocaine experimental labels in the cell state graph were set with a smoothing parameter of beta = 1. The Markov affinity–based graph imputation of cells (v1.5.5) (55) package was used to impute gene expression using default settings on the cell-state graph. To determine genes that vary with the EES, pairwise Pearson correlation r values were calculated between each gene and the EES by cluster using Single Cell PREParation (SCPREP) (v0.12.1). To generate the ridgeline plots, pairwise EES-gene Pearson correlation r values were calculated by cluster and ranked. Ridgeline plots reflect the ranks of 100 up-regulated DEGs identified in the DA bulk RNA-seq experiment. The CIRCOS plot was generated using CIRCOS version 0.69 in Perl v5.18.4. The size of the outer rings represents the number of DEGs within each cluster. Genes were positioned by adjusted P values, and arcs were then drawn between genes that changed in the same direction in pairwise clusters and color coded to indicate neuron-neuron, glia-glia, and neuron-glia matches. ROC analysis was performed with a subset of snRNA-seq data from the parent Drd1-MSN cluster in Seurat v3.0.2. Dimensionality reduction was performed with 17 PCs and resolution of 0.2. For ROC evaluation, a binary classifier was built with each gene using FindMarkers(test.use = “roc”, assay = “RNA”, slot = “data”, logfc.threshold = 0, min.pct = 0.1). R and Python code for snRNA-seq analysis is available via GitLab (https://gitlab.rc.uab.edu/day-lab).
CRISPR-dCas9 construct design
CRISPRa experiments used lentivirus compatible plasmid constructs we previously optimized for robust neuronal expression (24). Gene-specific sgRNA targets were designed using online tools provided by the Zhang laboratory at Massachusetts Institute of Technology (crispr.mit.edu) and CHOPCHOP (http://chopchop.cbu.uib.no/). To ensure specificity, all CRISPR RNA (crRNA) sequences were analyzed with National Center for Biotechnology Information’s Basic Local Alignment Search Tool. A list of the target sequences is provided in table S12. crRNAs were annealed and ligated into the sgRNA scaffold using the BbsI cut sites. Plasmids were sequence-verified with Sanger sequencing. Selected CRISPRa target genes were the genes with the highest confidence changes in gene expression following DA treatment. We applied a log2 fold change cutoff of >1.2 on the bulk DA RNA-seq dataset, resulting in 23 potential target genes (table S6). We first designed sgRNAs to target each gene individually (fig. S6A), excluding three genes due to significant predicted off-target localization of CRISPRa sgRNAs. At DIV 3 to 4, primary striatal neurons were transduced with lentiviruses expressing sgRNAs targeting either a negative control sequence (the bacterial LacZ gene) or one of the high confidence DA-induced genes (fig. S6A). Compared to the nontargeting control, we measured significant induction of 17 of the high confidence genes. To determine whether these genes could be induced simultaneously, we pooled all 17 sgRNA viruses (fig. S6B) and observed that all but one gene (Fosl2) was induced by this approach. Therefore, we excluded Fosl2 from the multiplex CRISPRa targeting experiments. To deliver sgRNAs for each of these targets in a more efficient way, we generated new multiplex sgRNA lentiviruses that contained eight sgRNA sequences each (driven by distinct U6 promoters) for a total of 16 unique sgRNAs between two viruses (Fig. 5A). The bacterial LacZ gene target was again used as a sgRNA nontargeting control by constructing an 8× LacZ sgRNA array. All multiplexed sgRNA arrays were created by first amplifying individual U6-sgRNA constructs with unique overhangs. Individual U6-sgRNA constructs were then assembled into a custom lentivirus backbone using an eight-insert Golden Gate Assembly reaction. For multiplexing sgRNAs, individual sgRNAs targeting either LacZ or the DA-induced genes were inserted by PCR amplification and Golden Gate Assembly into a custom-made destination plasmid coexpressing mCherry with unique BsmBI sites. All lentiviruses were produced as described previously (24), and titers were >1 × 1011 genome copies (GC)/ml for in vitro experiments and >1 × 1012 GC/ml for in vivo experiments. Titers were determined with the Lenti-X RT-qPCR Titer Kit (Clonetech).
Lentivirus production
Viruses were produced in a sterile environment subject to BSL-2 safety by transfecting human embryonic kidney–293T cells (American Type Culture Collection, CRL-3216) with the specified CRISPR plasmid, the psPAX2 packaging plasmid, and the pCMV-VSV-G envelope plasmid (Addgene 12260 and 8454) with FuGENE HD (Promega) for 40 to 48 hours in supplemented UltraCULTURE media (l-glutamine, sodium pyruvate, and sodium bicarbonate) in a T225 culture flask. Supernatant was passed through a 0.45-μm filter and centrifuged at 106,883 rcf for 1 hour and 45 min at 4°C. The viral pellet was resuspended in 1/100th (in vitro) or 1/1000th (in vivo) supernatant volume of sterile PBS and stored at −80°C. Physical viral titer was determined using the Lenti-X qRT-PCR Titration Kit (Takara), and only viruses greater than 1 × 1011 GC/ml were used. Viruses were stored in sterile PBS at −80°C in single-use aliquots. For smaller-scale virus preparation, each sgRNA plasmid was transfected in a 12-well culture plate as described above. After 40 to 48 hours, lentiviruses were concentrated with Lenti-X concentrator (Takara), resuspended in sterile PBS, and used immediately.
MEA recordings
Single-neuron electrophysiological activity was recorded using an Axion Maestro recording system (Axion Biosystems). E18 rat primary striatal neurons were seeded in 48-well MEAs at 30,000 cells per well, as described above. Each MEA well contains 16 extracellular recording electrodes and a ground electrode. Neurons were transduced with CRISPRa constructs on DIV 3 to 4, and MEA recordings were performed at DIV 11 while connected to a temperature- and CO2-controlled headstage (monitored at 37°C and 5% CO2). Electrical activity was measured by an interface board at 12.5 kHz, digitized, and transmitted to an external computer for data acquisition and analysis in Axion Navigator software (Axion Biosystems). All data were filtered using dual 0.01 Hz (high pass) and 5000 Hz (low pass) Butterworth filters. Action potential thresholds were set automatically using an adaptive threshold for each electrode (>6 SDs from the electrode’s mean signal). Neuronal waveforms collected in Axion Navigator were exported to Offline Sorter (Plexon) for sorting of distinct waveforms corresponding to multiple units on one electrode channel and confirmation of waveform isolation using PCA, interspike intervals, and auto- or cross-correlograms. Further analysis of burst activity and firing rate was performed in NeuroExplorer (v.5.0).
Bulk RNA-seq
Bulk RNA-seq was carried out at Hudson Alpha Genome Services Laboratory or the Heflin Center for Genomic Science Genomics Core Laboratories at the University of Alabama at Birmingham. RNA was extracted, purified (RNeasy, Qiagen), and deoxyribonuclease-treated for three biological replicates per experimental condition. One microgram of total RNA underwent quality control (Bioanalyzer) and was prepared for directional RNA-seq using NEBNext reagents (New England Biolabs) or the SureSelect Strand Specific RNA Library Prep Kit (Agilent Technologies) according to the manufacturer’s recommendations. PolyA+ RNA libraries underwent sequencing (50– to 75–base pair paired-end directional reads; ∼25 M reads per sample) on an Illumina sequencing platform (HiSeq2000 or NextSeq500).
Bulk RNA-seq data analysis
Paired-end FASTQ files were uploaded to the University of Alabama at Birmingham’s High Performance Computer cluster for custom bioinformatics analysis using a pipeline built with Snakemake (v5.1.4) (56). Read quality, length, and composition were assessed using FastQC before trimming low quality bases (Phred < 20) and Illumina adapters (Trim_Galore!, v04.5). Splice-aware alignment to the Rn6 Ensembl genome assembly (v90) was performed with spliced transcripts alignment to a reference (STAR) v2.6.0c. (57). An average of 88.4% of reads was uniquely mapped. Binary alignment map files were merged and indexed with SAMtools (v1.6). Gene-level counts were generated using the featureCounts (58) function in the Rsubread package (v1.26.1) in R (v3.4.1), with custom options (isGTFAnnotationFile = TRUE, useMetaFeatures = TRUE, isPairedEnd = TRUE, requireBothEndsMapped = TRUE, strandSpecific = 2, and autosort = TRUE). DESeq2 (v 1.16.1) (59) in R was used to perform count normalization and differential gene expression analysis with the application of Benjamini-Hochberg FDR for adjusted P values. DEGs were designated whether they passed a P < 0.05 adjusted P value cutoff and contained baseMean of >39.
GO analysis was conducted on DA (compared to vehicle control) and CRISPRa-induced Dopaplex gene expression program (compared to LacZ sgRNA control) using the ClueGO application in Cytoscape. Overrepresentation enrichment analysis was performed using nonredundant terms in biological process using the protein-coding rat genome as a reference set. Enrichment analysis applied Benjamini-Hochberg correction for multiple comparisons and required a minimum of three genes per enriched GO term category. The transcription factor binding prediction used the WebGestalt (54).
Stereotaxic surgery
Naïve adult Sprague-Dawley rats were anaesthetized with 4% isoflurane and secured in a stereotaxic apparatus (Kopf Instruments). During surgical procedures, an anesthetic plane was maintained with 1 to 2.5% isoflurane. Under aseptic conditions, guide holes were drilled using Paxinos and Watson (60) stereotaxic coordinates (all coordinates in respect to bregma: AP, +1.6 mm; mediolateral, ±1.4 mm; dorsoventral, −7.0) to target the NAc core. All infusions were made using a gastight 30-gauge stainless steel injection needle (Hamilton Syringes) that extended into the infusion site. Bilateral lentivirus microinfusions of (1.5-μl total volume per hemisphere) were made using a syringe pump (Harvard Apparatus) at a rate of 0.25 μl/min. Injection needles remained in place for 10 min following infusion to allow for diffusion. Rats were infused bilaterally with either 1.5 μl of total lentivirus mix composed of 0.75 μl of sgRNA and 0.75 μl of dCas9-VPR viruses in sterile PBS. After infusions, guide holes were covered with sterile bone wax, and surgical incision sites were closed with nylon sutures. Animals received buprenorphine and carprofen for pain management and topical bacitracin to prevent infection at the incision site.
Acute cocaine locomotion testing
For basic locomotor testing, naïve male (n = 8) and female (n = 8) animals were given a single intraperitoneal injection of either cocaine (20 mg/kg) or saline immediately before being placed in the activity chamber for 30 min of testing. Testing took place over two consecutive days between 2:00 and 5:30 p.m., and sessions were counterbalanced across time by sex and treatment condition. The same female experimenter conducted locomotor testing and was present in the room during testing. Behavior chambers consisted of 43-cm by 43-cm Plexiglass locomotor activity chambers (Med Associates Inc., St. Albans, VT) with opaque white wall covering and an open top. Each chamber included a 48-channel X-Y infrared array (Med Associates Inc., St. Albans, VT) that was used to measure distance traveled in conjunction with Activity Monitor software (Med Associates Inc., St. Albans, VT). Test chambers were cleaned with 0.0156% chlorhexidine and 70% ethanol at the beginning and end of each testing day and cleaned with 70% ethanol in between trials. Animals were transported to the behavioral testing core 30 min before testing. Only the two animals currently undergoing locomotor testing were kept in the testing room; other animals were kept in an adjacent room. Behavior sessions were conducted during the light cycle, and the overhead lights and white noise generator in the behavior room remained on.
Cocaine locomotor sensitization
After 2 weeks to allow for recovery and viral expression, animals were habituated to handling for a minimum of 4 days before locomotion testing began. Locomotor sensitization experiment sessions were 1 hour, with a 15-min habituation time before an intraperitoneal injection of either saline (day 1) or cocaine (10 mg/kg; days 3, 5, 7, 9, 11, and 25).
Immunohistochemistry
Immunohistochemistry was performed as described previously (24). Adult male rats were transcardially perfused with formalin (1:10 dilution in PBS; Fisher Scientific) 24 to 48 hours after the cocaine challenge. Brains were removed and postfixed for 48 hours in formalin then sliced at 50 μm using a vibratome. Tissue slices were permeabilized with 0.25% Triton X-100 in PBS then blocked for 1 hour at room temperature with blocking buffer (1× PBS with 10% Thermo Blocker BSA and 1% goat serum). Expression of mCherry was examined by incubation with an anti-mCherry primary antibody (1:500 in PBS with 10% Thermo Blocker BSA and 1% goat serum; Abcam, catalog no. ab183628) overnight at 4°C. Slices were washed three times with PBS and incubated for 1 hour at room temperature with a fluorescent secondary antibody (1:500; Alexa Fluor 546 goat anti-rabbit, Thermo Fisher Scientific, catalog no. A-11010). Slices were washed three times with PBS and mounted onto microscope slides with ProLong Gold Antifade medium (Invitrogen) containing 4,6-diamidino-2-phenylindole stain as a marker for cell nuclei. Images (4×) of the viral infusion site were taken on a Nikon TiS inverted fluorescent microscope, and viral placement was mapped onto to coronal diagrams adapted from Paxinos and Watson (60) (fig. S7). Animals with strong viral expression in at least one hemisphere were included in behavioral studies, and one animal was excluded due to targeting.
Statistical analysis
Sample sizes were calculated using a freely available calculator [Lenth, R. V. (2006-9)]. Java Applets for Power and Sample Size [Computer software] were available at www.stat.uiowa.edu/~rlenth/Power. Correspondence between the DA and SKF datasets was determined with a linear regression. Transcriptional differences from RT-qPCR experiments were compared with an unpaired t test with Welch’s correction or one-way analysis of variance (ANOVA) with Dunnett’s or Tukey’s post hoc tests where appropriate. MEA data were compared with Mann-Whitney U tests or a one-way ANOVA. Sensitization data were compared with either an unpaired t test with Welch’s correction or two-way ANOVA. Statistical significance was designated at α = 0.05 for all analyses. Statistical and graphical analyses were performed with Prism software (GraphPad). Statistical assumptions (e.g., normality and homogeneity for parametric tests) were formally tested and examined via boxplots.
Data availability
Sequencing data that support the findings of this study are available in Gene Expression Omnibus (GSE137568, GSE137658, GSE137759, and GSE137763). All relevant data that support the findings of this study are available by request from the corresponding author (J.J.D.). With the exception of multiplexed or gene-specific sgRNA plasmids, all CRISPRa constructs are available in the Addgene plasmid repository.
Supplementary Material
Acknowledgments
Funding: This work was supported by NIH grants DA039650, DA034681, and MH114990 (to J.J.D.); DA042514 (to K.E.S.); GM008361 (to M.E.Z. and C.G.D); GM008111 (to M.E.Z.); and DA041778 (to F.A.S.). L.I. was supported by the Civitan International Research Center at UAB. Additional assistance to J.J.D. was provided by the UAB Pittman Scholars Program. We thank S. Black for assistance with stereotaxic surgery and all current and former Day laboratory members for assistance and support. We specifically thank M. G. Karlsson and S. Bach for their contributions to establishment of single-cell RNA sequencing platforms in the laboratory. We thank the UAB Flow Cytometry Core for assistance with 10× snRNA-seq and FACS and the UAB Heflin Genomics Core facility for assistance with sequencing. Author contributions: J.J.D., J.J.T., C.G.D., M.E.Z., and R.A.P. conceived of snRNA-seq experiments. M.E.Z. and J.J.T. performed behavioral assays and generated tissue for snRNA-seq with assistance from R.A.P. and C.G.D. C.G.D. and J.J.T developed the single-nuclei dissociation protocol with assistance from R.A.P., M.E.Z., and the UAB Flow Cytometry Core. R.A.P., C.G.D., and J.J.D. performed statistical and graphical analysis from snRNA-seq datasets with assistance from L.I. and J.J.T. M.E.Z. performed DRD1 antagonist, MEK inhibitor, and CREB inhibitor experiments. K.E.S. and J.J.D. conceived of the CRISPR-dCas9 multiplexing experiments. K.E.S., L.I., R.A.P., and J.J.D. performed statistical and graphical analysis of bulk RNA-seq datasets. K.E.S. designed, cloned, and validated custom sgRNA arrays and performed electrophysiological and behavioral assays for CRISPR-dCas9 multiplexing with assistance from A.J.B., S.T., F.A.S., and N.A.G. All projects were supervised by J.J.D. K.E.S., J.J.T., and J.J.D. wrote the main text of the manuscript. K.E.S., C.G.D., J.J.T., R.A.P., M.E.Z., and J.J.D. wrote the Materials and Methods section of the manuscript. All authors have approved the final version of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/26/eaba4221/DC1
REFERENCES AND NOTES
- 1.Centers for Disease Control and Prevention, 2018 Annual Surveillance Report of Drug-Related Risks and Outcomes — United States. (2018).
- 2.Di Chiara G., Imperato A., Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc. Natl. Acad. Sci. U.S.A. 85, 5274–5278 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day J. J., Roitman M. F., Wightman R. M., Carelli R. M., Associative learning mediates dynamic shifts in dopamine signaling in the nucleus accumbens. Nat. Neurosci. 10, 1020–1028 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Stuber G. D., Roitman M. F., Phillips P. E., Carelli R. M., Wightman R. M., Rapid dopamine signaling in the nucleus accumbens during contingent and noncontingent cocaine administration. Neuropsychopharmacology 30, 853–863 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Nestler E. J., Luscher C., The Molecular Basis of Drug Addiction: Linking Epigenetic to Synaptic and Circuit Mechanisms. Neuron 102, 48–59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacAskill A. F., Cassel J. M., Carter A. G., Cocaine exposure reorganizes cell type- and input-specific connectivity in the nucleus accumbens. Nat. Neurosci. 17, 1198–1207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guez-Barber D., Fanous S., Golden S. A., Schrama R., Koya E., Stern A. L., Bossert J. M., Harvey B. K., Picciotto M. R., Hope B. T., FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. J. Neurosci. 31, 4251–4259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pascoli V., Turiault M., Luscher C., Reversal of cocaine-evoked synaptic potentiation resets drug-induced adaptive behaviour. Nature 481, 71–75 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Korpi E. R., den Hollander B., Farooq U., Vashchinkina E., Rajkumar R., Nutt D. J., Hyytiä P., Dawe G. S., Mechanisms of Action and Persistent Neuroplasticity by Drugs of Abuse. Pharmacol. Rev. 67, 872–1004 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Saunders A., Macosko E. Z., Wysoker A., Goldman M., Krienen F. M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S., Goeva A., Nemesh J., Kamitaki N., Brumbaugh S., Kulp D., McCarroll S. A., Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L. E., Manno G. L., Codeluppi S., Furlan A., Lee K., Skene N., Harris K. D., Hjerling-Leffler J., Arenas E., Ernfors P., Marklund U., Linnarsson S., Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Manchado A. B., Gonzales C. B., Zeisel A., Munguba H., Bekkouche B., Skene N. G., Lönnerberg P., Ryge J., Harris K. D., Linnarsson S., Hjerling-Leffler J., Diversity of Interneurons in the Dorsal Striatum Revealed by Single-Cell RNA Sequencing and PatchSeq. Cell Rep. 24, 2179–2190.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gokce O., Stanley G. M., Treutlein B., Neff N. F., Camp J. G., Malenka R. C., Rothwell P. E., Fuccillo M. V., Südhof T. C., Quake S. R., Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 16, 1126–1137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley G., Gokce O., Malenka R. C., Sudhof T. C., Quake S. R., Continuous and Discrete Neuron Types of the Adult Murine Striatum. Neuron 105, 688–699.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Walaas S. I., Greengard P., DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J. Neurosci. 4, 84–98 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hachigian L. J., Carmona V., Fenster R. J., Kulicke R., Heilbut A., Sittler A., de Almeida L. P., Mesirov J. P., Gao F., Kolaczyk E. D., Heiman M., Control of Huntington's Disease-Associated Phenotypes by the Striatum-Enriched Transcription Factor Foxp2. Cell Rep. 21, 2688–2695 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Arlotta P., Molyneaux B. J., Jabaudon D., Yoshida Y., Macklis J. D., Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J. Neurosci. 28, 622–632 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burkhardt D. B., Stanley J. S. III, Pertigoto A. L., Gigante S. A., Herold K. C., Wolf G., Giraldez A., van Dijk D., Krishnaswamy S., Enhancing experimental signals in single-cell RNA-sequencing data using graph signal processing. bioRxiv, 532846 (2019). [Google Scholar]
- 19.Castro D. C., Bruchas M. R., A Motivational and Neuropeptidergic Hub: Anatomical and Functional Diversity within the Nucleus Accumbens Shell. Neuron 102, 529–552 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandra R., Francis T. C., Konkalmatt P., Amgalan A., Gancarz A. M., Dietz D. M., Lobo M. K., Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. J. Neurosci. 35, 7927–7937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiang H. L., Epp J. R., van den Oever M. C., Yan C., Rashid A. J., Insel N., Ye L., Niibori Y., Deisseroth K., Frankland P. W., Josselyn S. A., Manipulating a "cocaine engram" in mice. J. Neurosci. 34, 14115–14127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trouche S., Perestenko P. V., van de Ven G. M., Bratley C. T., McNamara C. G., Campo-Urriza N., Black S. L., Reijmers L. G., Dupret D., Recoding a cocaine-place memory engram to a neutral engram in the hippocampus. Nat. Neurosci. 19, 564–567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Y., Green T., Saal D., Marie H., Neve R., Nestler E. J., Malenka R. C., CREB modulates excitability of nucleus accumbens neurons. Nat. Neurosci. 9, 475–477 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Savell K. E., Bach S. V., Zipperly M. E., Revanna J. S., Goska N. A., Tuscher J. J., Duke C. G., Sultan F. A., Burke J. N., Williams D., Ianov L., Day J. J., A Neuron-Optimized CRISPR/dCas9 Activation System for Robust and Specific Gene Regulation. eNeuro 6, ENEURO.0495-18.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savell K. E., Gallus N. V. N., Simon R. C., Brown J. A., Revanna J. S., Osborn M. K., Song E. Y., O’Malley J. J., Stackhouse C. T., Norvil A., Gowher H., Sweatt J. D., Day J. J., Extra-coding RNAs regulate neuronal DNA methylation dynamics. Nat. Commun. 7, 12091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlezon W. A. Jr., Thome J., Olson V. G., Lane-Ladd S. B., Brodkin E. S., Hiroi N., Duman R. S., Neve R. L., Nestler E. J., Regulation of cocaine reward by CREB. Science 282, 2272–2275 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Grueter B. A., Robison A. J., Neve R. L., Nestler E. J., Malenka R. C., ∆FosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc. Natl. Acad. Sci. U.S.A. 110, 1923–1928 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson E. B., Graham D. L., Arzaga R. R., Buzin N., Webb J., Green T. A., Bass C. E., Neve R. L., Terwilliger E. F., Nestler E. J., Self D. W., Overexpression of CREB in the nucleus accumbens shell increases cocaine reinforcement in self-administering rats. J. Neurosci. 31, 16447–16457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu M., c-Fos is an intracellular regulator of cocaine-induced long-term changes. Ann. N. Y. Acad. Sci. 1139, 1–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Zhang L., Jiao H., Zhang Q., Zhang D., Lou D., Katz J. L., Xu M., c-Fos facilitates the acquisition and extinction of cocaine-induced persistent changes. J. Neurosci. 26, 13287–13296 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pope S. D., Medzhitov R., Emerging Principles of Gene Expression Programs and Their Regulation. Mol. Cell 71, 389–397 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Savell K. E., Day J. J., Applications of CRISPR/Cas9 in the Mammalian Central Nervous System. Yale J. Biol. Med. 90, 567–581 (2017). [PMC free article] [PubMed] [Google Scholar]
- 33.Addy N. A., Daberkow D. P., Ford J. N., Garris P. A., Wightman R. M., Sensitization of rapid dopamine signaling in the nucleus accumbens core and shell after repeated cocaine in rats. J. Neurophysiol. 104, 922–931 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalivas P. W., Duffy P., Time course of extracellular dopamine and behavioral sensitization to cocaine. I. Dopamine axon terminals. J. Neurosci. 13, 266–275 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cadoni C., Solinas M., Di Chiara G., Psychostimulant sensitization: differential changes in accumbal shell and core dopamine. Eur. J. Pharmacol. 388, 69–76 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Gulley J. M., Hoover B. R., Larson G. A., Zahniser N. R., Individual differences in cocaine-induced locomotor activity in rats: behavioral characteristics, cocaine pharmacokinetics, and the dopamine transporter. Neuropsychopharmacology 28, 2089–2101 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Bertran-Gonzalez J., Bosch C., Maroteaux M., Matamales M., Hervé D., Valjent E., Girault J.-A., Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. J. Neurosci. 28, 5671–5685 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpenter M. D., Hu Q., Bond A. M., Lombroso S. I., Czarnecki K. S., Lim C. J., Song H., Wimmer M. E., Pierce R. C., Heller E. A., Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nat. Commun. 11, 504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bengtsson M., Stahlberg A., Rorsman P., Kubista M., Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 15, 1388–1392 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raj A., Peskin C. S., Tranchina D., Vargas D. Y., Tyagi S., Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beardsley P. M., Hauser K. F., Glial modulators as potential treatments of psychostimulant abuse. Adv. Pharmacol. 69, 1–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker D. M., Cates H. M., Loh Y.-H. E., Purushothaman I., Ramakrishnan A., Cahill K. M., Lardner C. K., Godino A., Kronman H. G., Rabkin J., Lorsch Z. S., Mews P., Doyle M. A., Feng J., Labonté B., Koo J. W., Bagot R. C., Logan R. W., Seney M. L., Calipari E. S., Shen L., Nestler E. J., Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain's Reward Circuitry. Biol. Psychiatry 84, 867–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Back S., Necarsulmer J., Whitaker L. R., Coke L. M., Koivula P., Heathward E. J., Fortuno L. V., Zhang Y., Yeh C. G., Baldwin H. A., Spencer M. D., Mejias-Aponte C. A., Pickel J., Hoffman A. F., Spivak C. E., Lupica C. R., Underhill S. M., Amara S. G., Domanskyi A., Anttila J. E., Airavaara M., Hope B. T., Kent Hamra F., Richie C. T., Harvey B. K., Neuron-Specific Genome Modification in the Adult Rat Brain Using CRISPR-Cas9 Transgenic Rats. Neuron 102, 105–119.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallo F. T., Katche C., Morici J. F., Medina J. H., Weisstaub N. V., Immediate Early Genes, Memory and Psychiatric Disorders: Focus on c-Fos, Egr1 and Arc. Front. Behav. Neurosci. 12, 79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maleeva N. E., Ivolgina G. L., Anokhin K. V., Limborskaia S. A., Analysis of the expression of the c-fos proto-oncogene in the rat cerebral cortex during learning. Genetika 25, 1119–1121 (1989). [PubMed] [Google Scholar]
- 46.Day J. J., Childs D., Guzman-Karlsson M. C., Kibe M., Moulden J., Song E., Tahir A., Sweatt J. D., DNA methylation regulates associative reward learning. Nat. Neurosci. 16, 1445–1452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Przewlocka B., Lason W., Adaptive changes in the proenkephalin and D2 dopamine receptor mRNA expression after chronic cocaine in the nucleus accumbens and striatum of the rat. Eur. Neuropsychopharmacol. 5, 465–469 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Gutierrez-Cuesta J., Burokas A., Mancino S., Kummer S., Martín-García E., Maldonado R., Effects of genetic deletion of endogenous opioid system components on the reinstatement of cocaine-seeking behavior in mice. Neuropsychopharmacology 39, 2974–2988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moeller S. J., Beebe-Wang N., Schneider K. E., Konova A. B., Parvaz M. A., Alia-Klein N., Hurd Y. L., Goldstein R. Z., Effects of an opioid (proenkephalin) polymorphism on neural response to errors in health and cocaine use disorder. Behav. Brain Res. 293, 18–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levran O., Yuferov V., Kreek M. J., The genetics of the opioid system and specific drug addictions. Hum. Genet. 131, 823–842 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trifilieff P., Martinez D., Kappa-opioid receptor signaling in the striatum as a potential modulator of dopamine transmission in cocaine dependence. Front. Psychol. 4, 44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J., Vasaikar S., Shi Z., Greer M., Zhang B., WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A. J., Burdziak C., Moon K. R., Chaffer C. L., Pattabiraman D., Bierie B., Mazutis L., Wolf G., Krishnaswamy S., Pe’er D., Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koster J., Rahmann S., Snakemake-a scalable bioinformatics workflow engine. Bioinformatics 34, 3600 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y., Smyth G. K., Shi W., featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.G. Paxinos, C. Watson, The Rat Brain in Stereotaxic Coordinates (El Sevier, New York, ed. 5, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/26/eaba4221/DC1
Data Availability Statement
Sequencing data that support the findings of this study are available in Gene Expression Omnibus (GSE137568, GSE137658, GSE137759, and GSE137763). All relevant data that support the findings of this study are available by request from the corresponding author (J.J.D.). With the exception of multiplexed or gene-specific sgRNA plasmids, all CRISPRa constructs are available in the Addgene plasmid repository.


