A micropeptide encoded by MIR155HG modulates antigen presentation and suppresses autoinflammation through binding to HSC70.
Abstract
Many annotated long noncoding RNAs (lncRNAs) harbor predicted short open reading frames (sORFs), but the coding capacities of these sORFs and the functions of the resulting micropeptides remain elusive. Here, we report that human lncRNA MIR155HG encodes a 17–amino acid micropeptide, which we termed miPEP155 (P155). MIR155HG is highly expressed by inflamed antigen-presenting cells, leading to the discovery that P155 interacts with the adenosine 5′-triphosphate binding domain of heat shock cognate protein 70 (HSC70), a chaperone required for antigen trafficking and presentation in dendritic cells (DCs). P155 modulates major histocompatibility complex class II–mediated antigen presentation and T cell priming by disrupting the HSC70-HSP90 machinery. Exogenously injected P155 improves two classical mouse models of DC-driven auto inflammation. Collectively, we demonstrate the endogenous existence of a micropeptide encoded by a transcript annotated as “non-protein coding” and characterize a micropeptide as a regulator of antigen presentation and a suppressor of inflammatory diseases.
INTRODUCTION
Professional antigen-presenting cells (APCs), including dendritic cells (DCs), B cells, and macrophages, internalize exogenous antigens through clathrin-mediated endocytosis and display antigens for CD4+ T cell recognition via endosomal/lysosomal peptide loading to major histocompatibility complex (MHC) class II molecules (1–5). Recent studies revealed that endogenous epitopes derived from cytoplasmic and nuclear proteins also encounter endosomal/lysosomal MHC class II through pathways including chaperone-mediated autophagy (CMA) and macroautophagy (6, 7). In this context, several cytoplasmic chaperones form cascaded machinery that accompanies and transmits self-antigens step by step for MHC class II presentation. As a chaperone, heat shock cognate protein 70 (HSC70) is required for vesicle formation and clathrin uncoating during clathrin-mediated endocytosis (8, 9). Together with other cytoplasmic chaperones such as HSP90, HSC70 transports pentapeptide (KFERQ-like) motif–containing proteins delivered by CMA to the lysosomal membrane, facilitating MHC class II presentation of these antigens (10). Thus, HSC70 is recognized as central for antigen trafficking and MHC class II presentation in APCs. However, the regulatory factors involved in HSC70-mediated antigen processing and presentation cascade are ill-defined. Given the fact that self-antigen presentation drives the development of autoinflammatory disorders, identifying such regulatory factors might benefit the treatment of autoimmune diseases. Small peptides have been shown to be promising therapeutic candidates for treating autoimmune diseases, but choosing a peptide that does not stimulate off-target immune responses is difficult.
Owing to the invention of ribosome profiling (Ribo-seq) and the evolution of in silico approaches, numerous micropeptides predicted to be encoded by small open reading frames (sORFs) within vertebrate long noncoding RNAs (lncRNAs) have been identified (11–14). Whether these micropeptides truly exist and what roles they play in different cell types remain challenging questions, which motivated us to search for functional micropeptides in the first place. We report that human lncRNA MIR155HG is highly expressed in inflamed DCs and encodes a previously unrecognized 17–amino acid micropeptide, which we named miPEP155 (P155). P155 selectively binds to HSC70, modulates MHC class II presentation in DCs, and markedly improves DC-driven autoinflammation in mice. Apart from re-annotation of an lncRNA, our data characterize the cell type–specific role of P155, which is shown to regulate antigen transportation and presentation in APCs. As an endogenously expressed peptide, P155 differs from previous peptide-based treatments targeting autoreactive T cells and proves a strong drug candidate for autoimmune diseases.
RESULTS
MIR155HG encodes an endogenously expressed micropeptide
ORFfinder was used to search for ORFs within human MIR155HG, and a 54–base pair (bp) small ORF was predicted to harbor coding potential among all the sense small ORFs (AUG- and non–AUG-initiated; Fig. 1A and table S1). This ORF (ORF1) is located in the 5′ region of all ORFs identified by ORFfinder and spans the terminal 18 bp in exon 2 and initial 33 bp in exon 3, just 55 bp before pre–miR-155 (fig. S1A). The first ORF at the 5′ region of the sense strand is most likely to be translated (11, 13, 15, 16). To determine whether the start codon of ORF1 is active, we introduced an enhanced green fluorescent protein (EGFP) tag (without its own start codon) into the C terminus of ORF1 to generate an assumed fusion protein using CRISPR/Cas9-mediated homologous recombination in a HEK293T cell line (Fig. 1B). Successful insertion was validated by standard polymerase chain reaction (PCR) with primers on exon 3 and first-generation sequencing (Fig. 1C and fig. S1B), and the HEK293T cells showed normal MIR155HG expression after EGFP knock-in (fig. S1C). Fluorescence imaging showed substantial expression of EGFP-fused protein in the genetically engineered 293T cells, indicating endogenous translation of ORF1 (Fig. 1D). We detected the fusion protein of predicted molecular weight with an anti-EGFP antibody by Western blotting (Fig. 1E). To further confirm the endogenous existence of the 17–amino acid micropeptide encoded by ORF1, we produced an antibody against this peptide. Using chemically synthesized micropeptides as a positive control, the antibody successfully detected the endogenously expressed micropeptide (first enriched by immunoprecipitation), which we termed P155, in a human B cell lymphoma cell line named OCI-LY-1 (fig. S1D) and in human monocyte-derived DCs (moDCs; Fig. 1F). To provide direct evidence of the existence of P155, we performed liquid chromatography–mass spectrometry (LC-MS) on cell-derived immune precipitates, which were generated using P155 antibodies. We detected the amino acid sequence of P155 in OCI-LY-1 cells (Fig. 1G) and in moDCs (fig. S1E) using this approach. Our data demonstrate that human MIR155HG, which is annotated as lncRNA, encodes a 17–amino acid micropeptide.
Fig. 1. Discovery of an endogenously expressed micropeptide encoded by MIR155HG.
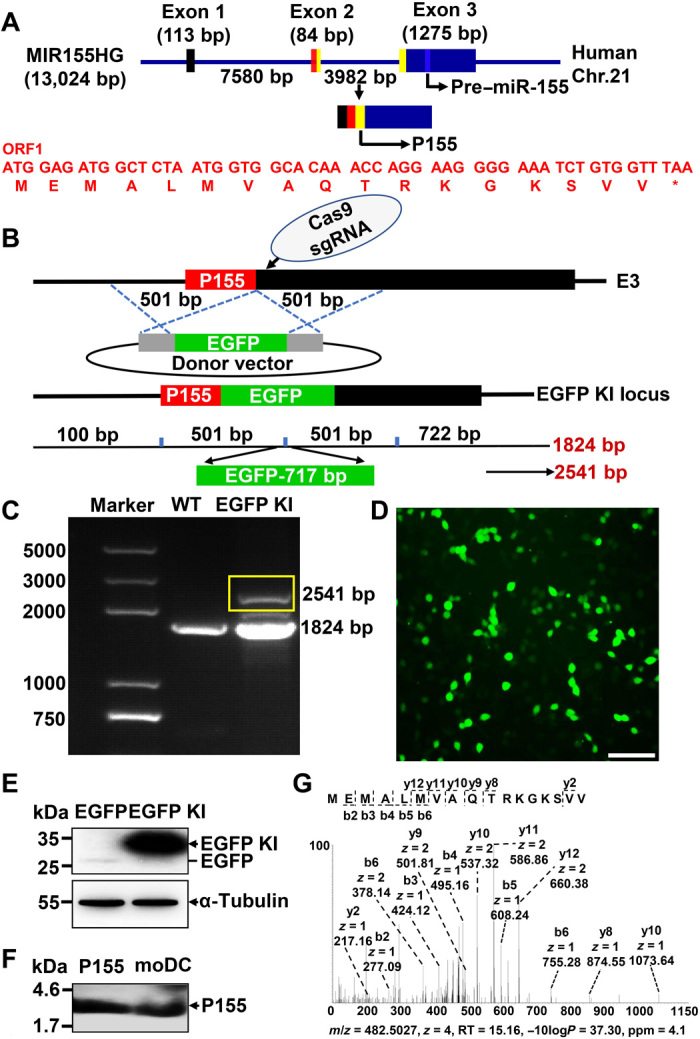
(A) Schematic representation of P155 translation. Human MIR155HG spans 13,024 bp and has three exons. P155 is translated by ORF1 (indicated by yellow boxes), which comprises the end of exon 2 and the head of exon 3. The nucleotide and amino acid sequences of ORF1 are highlighted in red and the pre–miR-155 is indicated by a bluish color. (B) Schematic representation of P155 EGFP knock-in strategy. The EGFP (without its own ATG) was inserted after the last coding codon (GTT-valine) of P155 by CRISPR/Cas9-mediated homologous recombination in HEK293T cells. The front homologous arm is a 501-bp fragment before the termination codon of P155 sequence and the back homologous arm is a 501-bp fragment starting with the P155 termination codon, E3: exon 3. (C) PCR detection of EGFP knock-in efficiency. Target band is indicated by the yellow box. (D) Fluorescence imaging of P155-EGFP fusion protein expression. (E) Immunoblotting verification of P155-EGFP fusion protein in HEK293T cells. Protein lysate of EGFP plasmid–transfected HEK293T cells served as a negative control. The target band is indicated by black arrowheads, and the EGFP location is visible as a black line. (F) Immunoblotting detection of endogenously expressed P155 in human moDCs with P155-specific antibody pre-enrichment. Chemically synthesized P155 served as a positive control, and the target band is indicated by the black arrowheads. (G) LC-MS verification of the P155 endogenous expression in OCI-LY-1 cells with P155-specific antibody pre-enrichment. Scale bar, 100 μm. Data (D to F) are representative of three independent experiments. Photo credit: Liman Niu (Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine).
P155 interacts with HSC70 in human DCs
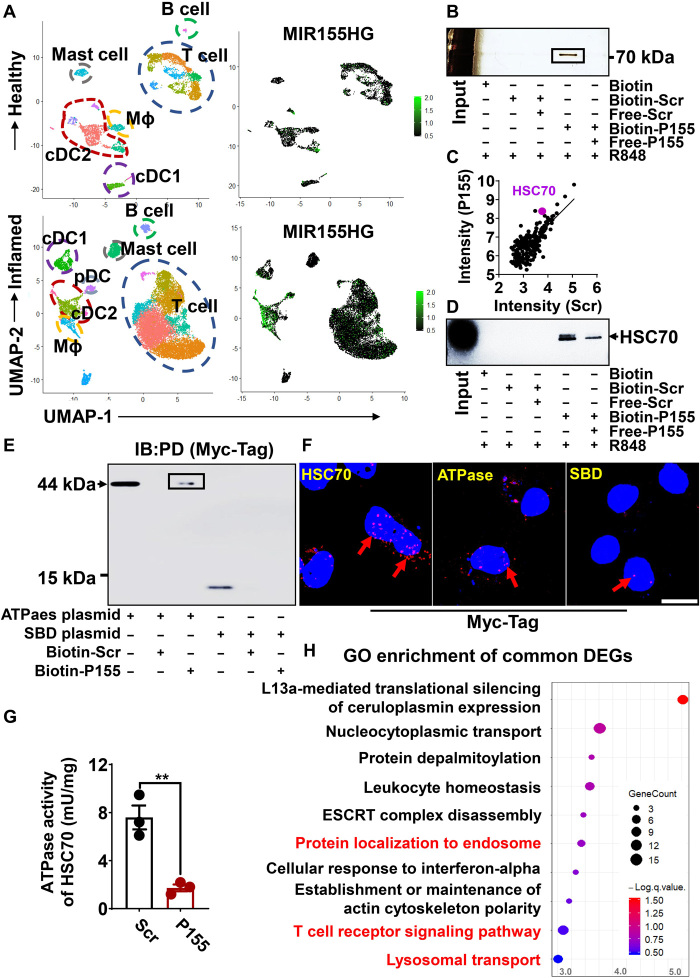
We performed single-cell RNA sequencing (RNA-seq) on CD45+ cells derived from the healthy dermis and inflamed dermis from patients with psoriasis. Unexpectedly, we found that MIR155HG was highly expressed by APCs in inflammation but not at steady state (Fig. 2A). We then sought to investigate whether P155 plays a role in activated DCs harboring the strongest antigen-presenting capacities among professional APCs. To this end, we first showed that fluorescein isothiocyanate (FITC)–labeled synthetic P155 efficiently entered HEK293T cells and colocalized with endogenous P155 in both cytoplasmic and nuclear compartments of the cells (fig. S1F). We then treated human moDCs with biotin-labeled P155 or a scrambled control peptide (Scr) in the presence of a Toll-like receptor (TLR) 7/8 agonist, R848, and then examined the proteins pulled down together with the peptides using SDS–polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining. A ~73-kilodalton (kDa) protein was pulled down by biotin-labeled P155 and could be competed away by free P155, indicating the binding specificity of P155 to this target protein (Fig. 2B and fig. S2A). Using LC-MS and confirmative immunoblotting, we recognized this 73-kDa protein to be HSC70 (Fig. 2, C and D). P155 colocalized finely with HSC70 in wild-type (WT) 293T cells, but its fusion with EGFP impaired such colocalization (fig. S2B).
Fig. 2. P155 interacts with HSC70 in human DCs.
(A) Two-dimensional visualization of the single immune cell (CD45+ cells) transcriptome in the dermis of healthy donors (n = 3) and patients with psoriasis (n = 3). Immune cell compartments are encircled, and feature plots of MIR155HG expression in different subsets are presented. (B) Silver staining of P155 interactive protein in the immunoprecipitants pulled down by streptavidin-agarose from human moDCs pretreated with R848 (1 μg/ml) and biotin-Scr/P155 (25 μM). The black box represents target protein. (C) Scatterplot of representative data for intensity of proteins detected with MS in human moDCs treated with R848 (1 μg/ml) and Biotin-Scr/P155 (25 μM). The dots represent the intensities (log10-transformed) of all proteins identified in the P155 group (y axis) and the Scr group (x axis), and the purple dot represents the protein of interest. (D) Immunoblotting verification of the interaction between HSC70 and P155. The black arrowhead indicates the specific band. (E) Immunoblotting detection of the P155-specific binding domain in the immunoprecipitants pulled down by streptavidin-agarose from biotin-Scr/P155–pretreated HEK293T cells overexpressing Myc-Tag–labeled HSC70 subdomain plasmids. Anti–Myc-Tag antibody was used and the black box indicates the specific banding. IB, immunoblot; PD, pull-down assay. (F) Confocal visualization of PLA signals (red/pink dots) in HEK293T cells overexpressing Myc-Tag–labeled HSC70 or HSC70 subdomain plasmids together with the endogenously expressed P155. Myc-Tag–labeled HSC70 protein served as a positive control, and the red arrows indicate the specific signals. (G) ATPase activity of HSC70 in the presence of P155 or Scr (n = 3). (H) GO pathway enrichment analysis of down-regulated differentially expressed genes (DEGs) of RNA-seq data from THP-1–derived DCs treated with R848 (1 μg/ml) and Scr/P155 (25 μM) compared to Scr-treated controls. Dot color represents the value of −logP; dot size corresponds to the gene counts, and the red words indicate the pathways we are focusing on. Scale bar, 10 μm. Data (B to E and G) are representative of three independent experiments. **P < 0.01, two-tailed Student’s t test (mean ± SEM). Photo credit: Liman Niu (Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine).
HSC70 is composed of two functional subdomains, namely, the adenosine triphosphatase (ATPase) domain and the substrate-binding domain (SBD) (17). We generated HSC70-truncated constructs to further investigate the binding preference of P155 to HSC70 subdomains. Both pull-down assays followed by immunoblotting and proximity ligation assay (PLA) showed that P155 specifically bound to the ATPase domain of HSC70 (Fig. 2, E and F). Moreover, P155 remarkably impaired the ATPase activity of full-length HSC70, which functionally supported P155’s binding specificity (Fig. 2G). Human THP-1 cell line can be induced to up-regulate DC markers including CD80, CD11C, and CD209 (fig. S2C). P155 is bound to HSC70 in THP-1–derived DCs treated with R848 (fig. S2D). RNA-seq data of THP-1–derived DCs treated with P155 or Scr revealed that Gene Ontology (GO) pathways involving vesicle-mediated transport and T cell receptor signaling were enriched in genes down-regulated by P155 treatment, and these functional nodes were related to HSC70 as a protein transport chaperone (Fig. 2H and fig. S2E). Collectively, our data suggest that P155 interacts with the ATPase domain of human HSC70 and probably impairs the function of HSC70 as an antigen transporter in DCs.
P155 modulates antigen presentation via targeting HSC70 in murine DCs
Although the coding sequences of P155 were not found in the mouse genome, we noticed that murine and human HSC70 share 99.85% homology (fig. S3A), and both ATPase and SBD domains of HSC70 are exactly the same in human and mouse. The only different amino acid is serine-579 in house mouse, which does not belong to any functional domain within HSC70. We asked whether P155 interacted with murine HSC70 in the same way observed in human DCs. Using pull-down assays followed by silver staining, we found that P155 selectively bound to HSC70 in bone marrow–derived DCs (BMDCs) treated with R848 (Fig. 3, A to C). We pretreated BMDCs with P155 or Scr and studied antigen trafficking within the endosome-lysosome pathway visualized by confocal microscopy. We found that FITC-labeled soluble ovalbumin (OVA) showed less colocalization with the lysosomes [marked by LAMP2A (lysosomal associated membrane protein 2A); Fig. 3, D and E], but showed comparable colocalization with endosomes (marked by Rab5; fig. S3, B and C) in BMDCs pretreated with P155 compared to BMDCs pretreated with Scr. Together with its “client-protein” HSP90, HSC70 guides antigens to the lysosomal membrane for interactions with LAMP2, which then elicits MHC class II presentation of peptides to the cell membrane (18). A weakened interplay between HSC70 and HSP90 as well as LAMP2A was detected in BMDCs cultured in the presence of P155 compared to BMDCs treated with Scr, which coincided with impaired lysosomal antigen trafficking observed in P155-treated BMDCs (Fig. 3F). We found that P155 treatment remarkably decreased MHC class II expression in BMDCs stimulated with R848, supporting the proposed model that P155 modulates antigen presentation (Fig. 3G). Antigenic peptides transported by the HSC70-HSP90 co-chaperone in APCs are presented by MHC class II to prime CD4+ T cells (19). We loaded OVA into BMDCs and primed CD4+ T cells derived from OT-II mice in the presence of P155 or Scr. As a result, CD4+ T cells primed in the presence of P155 showed a markedly reduced rate of proliferation compared to T cells primed in the presence of Scr (Fig. 3H). Our data suggest that P155 regulates antigen trafficking and membrane presentation of mouse BMDCs through interacting with HSC70 (Fig. 3I).
Fig. 3. P155 modulates antigen presentation via targeting HSC70 in murine DCs.
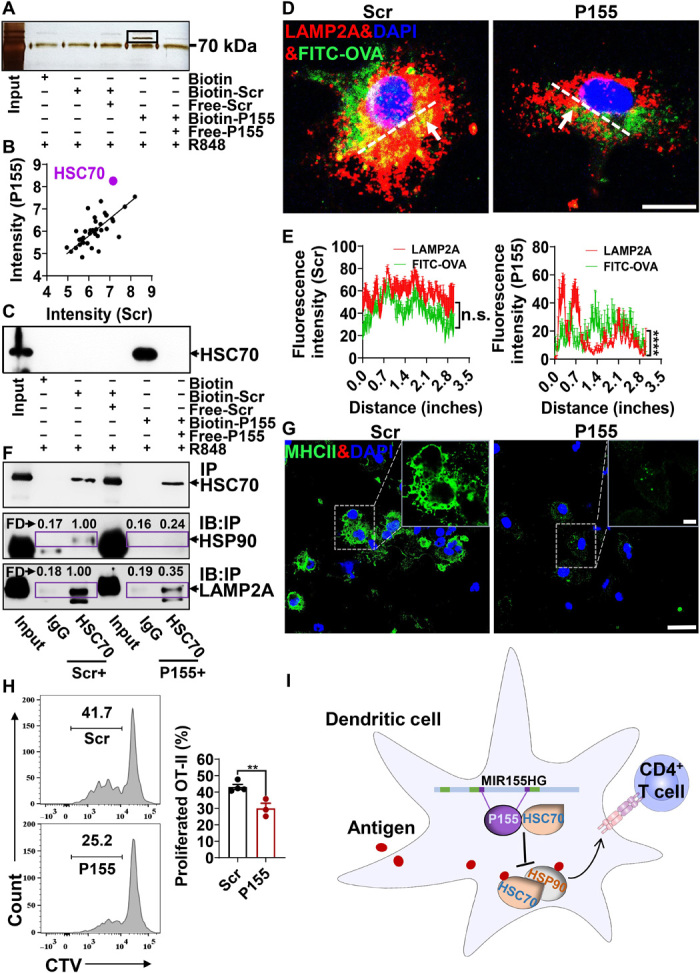
(A) Silver staining of P155 interactive protein in the immunoprecipitants pulled down by streptavidin-agarose from murine BMDCs pretreated with R848 (1 μg/ml) and biotin-Scr/P155 (25 μM). The black box represents target protein. (B) Scatterplot of representative data for intensity of proteins detected with MS in murine BMDCs treated with R848 (1 μg/ml) and Biotin-Scr/P155 (25 μM). The dots represent the intensities (log10-transformed) of all proteins identified in the P155 group (y axis) and the Scr group (x axis), and the purple dot represents the protein of interest. (C) Immunoblotting verification of the interaction between HSC70 and P155. The black arrowhead indicates the target band. (D) Confocal microscopic images of murine BMDCs treated with FITC-OVA with the cells costained with LAMP2A (red); nuclei were stained with DAPI (blue). The white arrows indicate the colocalization of OVA and LAMP2A (yellow), and the white dashed line is the position indicator for the fluorescence colocation analysis. (E) Fluorescence intensity analysis of colocalization of FITC-OVA and LAMP2A in murine BMDCs treated with Scr or P155 (n = 6). ImageJ software was used to analyze the fluorescence colocalization. The white dashed indicator line of statistical analysis centers on the colocation of FITC-OVA and LAMP2A. (F) Immunoblotting detection of HSP90 and LAMP2A expression in the immunoprecipitants pulled down by anti-HSC70 from R848- and Scr/P155-pretreated BMDCs, with immunoprecipitants pulled down by rat IgG from R848- and Scr/P155-pretreated BMDCs serving as the negative controls. The black arrows indicate the target protein. IP, immunoprecipitation; IB, immunoblot. The fold (FD) value represents the protein intensity of HSP90 or LAMP2A and is measured with ImageJ software. (G) Confocal microscopic images of the distribution of MHC class II (green) in R848- and Scr/P155-pretreated murine BMDCs. Nuclei were stained with DAPI (blue). (H) Representative flow cytometry chart (left) and statistics analysis of OT-II T cell proliferation rates (right, indicated by CD4+CD44+Violet− cell percentages) in coculture with BMDCs pretreated with soluble OVA (n = 3 to 4). (I) Schematic diagram of MIR155HG encoding P155 function in regulating DC presentation. Scale bars, 10 μm (D) or 25 μm/5 μm (G). All data are representative of three independent experiments. n.s., not significant; **P < 0.01 and ****P < 0.0001, one-way ANOVA (mean ± SEM). Photo credit: Liman Niu (Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine).
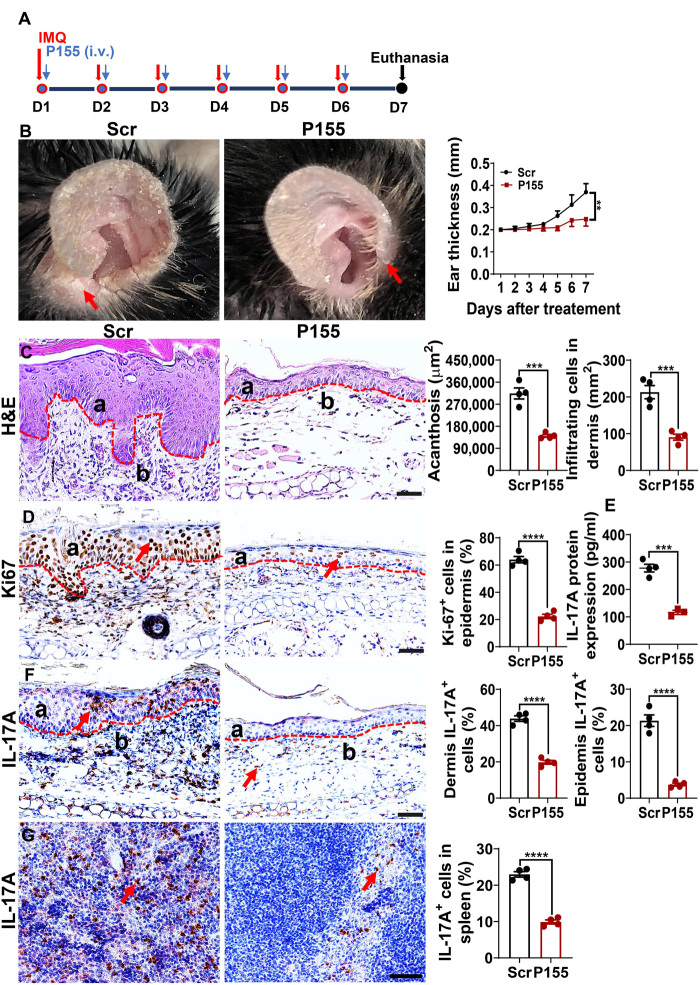
P155 ameliorates psoriasis-like skin inflammation driven by DC activation
Similar to R848, the TLR7/8 agonist imiquimod (IMQ) activates DCs and induces psoriasis-like inflammation when applied to mouse skin. Given that IMQ-induced skin psoriasis-like inflammation relies on MHC class IIhigh conventional DCs (20, 21), we hypothesized that P155 might harbor therapeutic effects in this context. We smeared 5% IMQ cream daily on the ears of C57BL/6 mice for six consecutive days and intravenously injected P155 or Scr at a dosage of 80 μg per mouse per day (Fig. 4A). P155 treatment prevented ear swelling and alleviated inflammatory phenotypes (erythema, thickness, and scaling) compared to the Scr control (Fig. 4B). Histological analysis showed that P155 suppressed epidermal hyperplasia and dermal cellular infiltration (Fig. 4C). Decreased epidermal Ki67+ cell counts further indicated that P155 relieved skin inflammation (Fig. 4D). We found that interleukin-17A (IL-17A), which is pivotal for the formation of psoriasis-like inflammation, was decreased in the IMQ-painted skin homogenate of P155-treated mice compared to the Scr-treated mice (Fig. 4E), but interferon-γ (IFN-γ) and IL-4 were not altered at either protein or mRNA level in P155-treated IMQ-painted skin compared to the Scr controls (fig. S4, A and B). Consistently, the numbers of IL-17A–producing cells in both the epidermis and dermis of the IMQ-induced skin were drastically decreased in P155-treated mice (Fig. 4F). Moreover, splenic IL-17A+ cells also declined with P155 treatment in the IMQ-induced mouse model of psoriasis (Fig. 4G). To rule out direct effects of P155 on T helper 17 (TH17) cell differentiation, we cultured CD4+ T cells under polarizing conditions in the presence of P155 or Scr. We found that P155 did not directly affect TH17 cell differentiation, nor did P155 affect TH1 and T regulatory (Treg) cell differentiation (fig. S4, C to E). To explore whether P155 modulates antigen presentation to skew T cell differentiation, we loaded OVA into mouse BMDCs and cocultured these DCs with OT-II CD4+ T cells for 3 days in the presence of P155 or Scr. The results (fig. S4, F to H) showed that P155 treatment reduced the percentages of T cells differentiated into TH17, TH1, and TH2 cells upon OVA priming. These data indicate that although P155 does not directly monitor T cell differentiation, it modulates antigen presentation to skew T cell polarization statuses. We conclude that P155 alleviates psoriasis-like skin inflammation via regulating DCs but showing marginal direct effects on T cells.
Fig. 4. P155 ameliorates the psoriasis-like skin inflammation in IMQ-induced mouse model.
(A) Schematic diagram of psoriasis-like mouse model induction. (B) Macroscopic views of Scr- or P155-treated IMQ-induced mouse ear (left) and daily measurement of mouse ear thickness (right, n = 4). Scaly plaques are marked by red arrows. (C) Representative images of H&E staining from Scr- or P155-treated IMQ-induced mouse ear sections (left) and statistical analysis for skin acanthosis and infiltrating inflammatory cells in the dermis (right, n = 4). a, epidermis; b, dermis. (D) Representative immunohistochemical staining of Ki67 (left) and quantitation of Ki67+ epidermal cells (right, a) in Scr- or P155-treated IMQ-induced mouse skin sections (n = 4). a, epidermis; red arrows indicate the positive expression of Ki67. (E) ELISA quantification of IL-17A protein expression in supernatants of Scr- or P155-treated IMQ-induced mouse ear homogenates (n = 3 to 4). (F and G) Representative immunohistochemical staining of IL-17A+ cells in Scr- or P155-treated IMQ-induced mouse ear (F, left) and spleen (G, left) sections. a, epidermis; b, dermis. Red arrows indicate the positive staining of IL-17A+ cells. Statistical analysis of IL-17A+ cell percentage in the epidermis, dermis (F, right), and spleen (G, right, n = 4). The dashed line indicates the border between the epidermis and dermis. Scale bars, 50 μm. All data are representative of three independent experiments. **P < 0.01; ***P < 0.001; and ****P < 0.0001, two-way ANOVA (B) and one-way ANOVA (mean ± SEM). Photo credit: Liman Niu (Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine).
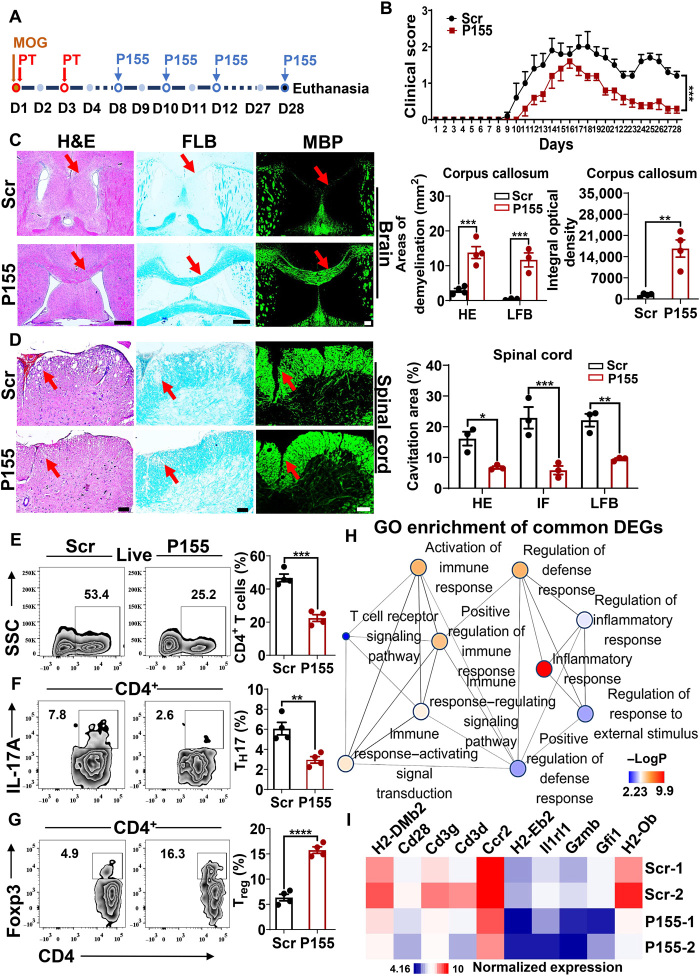
P155 mitigates DC-dependent autoimmunity in the central nervous system
To better define the role of P155 as a modulator of antigen presentation, we took advantage of experimental autoimmune encephalomyelitis (EAE), a classical model of autoimmunity caused by the exposure of myelin-derived autoantigens to DCs. EAE was established and P155 or Scr was injected every other day once clinical manifestations started to appear on day 8 upon immunization (Fig. 5A). P155 treatment resulted in lower clinical scores of EAE compared to the Scr-treated controls, and P155 did not present general toxicity as indicated by untouched body weights (Fig. 5B and fig. S5A). Histological analyses showed mitigated inflammation and alleviated demyelination in the brains and spinal cords of P155-treated mice (Fig. 5, C and D). We examined immune cell infiltration in the brains and spleens of EAE mice treated with P155 or Scr. Notably, P155-treated mice showed reduced CD4+ T cell infiltration in the brain and decreased TH17 cell percentages in CD4+ T cells in the brain and spleen (Fig. 5, E and F, and fig. S5B). Treg cell percentages were increased in CD4+ T cells from inflamed mouse brains and spleens treated with P155 compared to Scr controls (Fig. 5G and fig. S5C). We then investigated gene expression profiles of inflamed spinal cords derived from P155- or Scr-treated EAE mice. GO pathway enrichment of significantly down-regulated genes in the P155 group highlighted the immunomodulatory effects of P155 on inflamed central nervous system tissues (Fig. 5H). Specifically, the reduced expression levels of MHC class II genes (H2-Ob, H2-Eb2, and H2-DMb2) and T cell marker genes (Cd3d, Cd3g, and Cd28) further supported the idea that P155 regulates antigen presentation (Fig. 5I).
Fig. 5. P155 mitigates DC-dependent autoimmunity in the central nervous system.
(A) Schematic diagram of MOG-induced EAE. (B) Clinical score of Scr- or P155-treated EAE mice (n = 5). (C and D) Representative images of H&E, LFB, MBP staining, and statistical analysis for Scr- or P155-treated EAE mouse brain (C, left and right) and spinal cord sections (D, left and right). n = 3 to 4, red arrows indicate the demyelination. (E to G) Flow cytometric analysis of CD4+ T cell (E), TH17 cell (F), and Treg cell (G) percentages gated on CD4+ T cells from Scr- or P155-treated EAE mouse brain on day 15 after immunization (n = 4). (H) Pathway enrichment analysis of down-regulated DEGs in Scr- or P155-treated EAE mouse spinal cord on day 15 after immunization (n = 2). Color key represents the value of −logP; the size of the dot corresponds to the gene counts. (I) Heat map of selected genes based on RNA-seq data from the spinal cord of Scr- or P155-treated EAE mice on day 15 after immunization (P < 0.05, Log2FC < −1, n = 2). Color key represents the normalized expression of genes. Scale bars, 200 μm (C) or 100 μm (D). Data (C to G) are representative of three independent experiments. **P < 0.01; ***P < 0.001; and ****P < 0.0001, two-way ANOVA (B) and one-way ANOVA (mean ± SEM). Photo credit: Liman Niu (Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine).
DISCUSSION
Originally identified as being highly expressed by B-cell lymphomas, MIR155HG has long been regarded as an lncRNA serving exclusively to produce miR-155, which has become one of the best-characterized microRNAs (miRNAs) in mammals (22). MIR155HG and miR-155 expression levels are greatly increased in macrophages and DCs upon TLR activation (23–25). Normal DC functions were abrogated in miR-155–deficient mice, highlighting the roles of MIR155HG and miR-155 in mediating inflammation and immune responses (26). However, the up-regulation of MIR155HG does not always result in miR-155 induction (27). Meanwhile, measurable MIR155HG ncRNA levels were detected in the cytoplasm, denying its destiny to serve as a miRNA precursor (28). Using the EGFP knock-in reporter, antibody detection, and, most importantly, LC-MS, in the current study, we explicitly demonstrated the endogenous existence of P155, a micropeptide coded by MIR155HG. Furthermore, we proposed a bidirectional fate of MIR155HG such that MIR155HG can either be processed to generate a pre-miRNA in the nucleus or be transported to the cytoplasm to encounter ribosomes. An interesting idea brought by our study is such a fate transformation of a mammalian transcript, which should incent the reevaluation of the oncogenic or proinflammatory roles of MIR155HG in different cell types and diseases.
Our study characterized the functions of the newly discovered P155, and we identified the peptide chaperone HSC70 as an interacting protein of P155 in both human and mouse DCs. Although HSC70 often appears as an artifact in MS assays (29), we did not find HSC70 among proteins pulled down with the Scr control peptide. P155 did not bind to the general peptide-binding region, namely, the SBD domain of HSC70, but selectively bound to the ATPase region of HSC70 and suppressed the ATPase activity of full-length HSC70, which is critical for the HSC70-mediated chaperon cycle of peptide transportation (30). Given that HSC70 is central for antigen trafficking in DCs, we found that P155 modulated lysosomal localization of antigens and subsequent antigen presentation by MHC class II to CD4+ T cells. Meanwhile, we found that P155 also suppressed MHC II gene expression in DCs, as indicated by the data from RNA-seq and immunofluorescence, suggesting that P155 might act through an alternative path other than binding to HSC70 to regulate antigen presentation. Thus, we identified a potential anti-inflammatory role of P155, suggesting that both the production mode and the functionality of P155 act against miR-155. These findings advance our understanding of the regulatory mechanisms of antigen presentation and miRNA function implementation, but the intrinsic roles of P155 need further elucidation with genetic approaches.
Intravenously delivered synthetic P155 harbors striking therapeutic effects on autoinflammatory conditions driven by MHC class IIhigh DCs and CD4+ T cells. Displaying unparalleled advantages including ready synthesis, quick optimization, rapid clearance, and limited side effects, peptide drugs hold great promise. However, their delivery might result in unwanted immune responses facilitated by antigen-reactive T cells primed by MHC presentation (31, 32). Shown to be a micropeptide that regulates antigen presentation, P155 is unlikely to face immune-boosting limitations and presents itself as a good peptide drug candidate for the treatment of autoimmune diseases.
MATERIALS AND METHODS
Mice
C57BL/6J mice were purchased from LINGCHANG Biotech, Shanghai, China. C57BL/6-Tg (Tcra Tcrb) 425Cbn/J (OT-II) mice were a gift from F. Ginhoux. All mice were kept in specific pathogen–free facilities following the criteria of the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals with the approval (SYXK-2003-0026) of the Scientific Investigation Board of Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Human subjects
Human peripheral blood mononuclear cells (PBMCs) were obtained from blood buffy coats of healthy donors provided by SAILY BIO; all of the individuals provided informed consent. The study was performed in accordance with the principles of the Declaration of Helsinki and approved by the Research Ethics Board of Shanghai Tenth People’s Hospital (no. 2018121901).
Constructs
pcDNA3.1 was used to generate the expression constructs of HSC70-SBD and HSC70-ATPase with a C-Myc-Tag for protein interaction studies. pSpCas9(BB)-2A-Puro (PX459) V2.0v2 was used for the expression constructs of P155 guide RNA (gRNA). PUC57 was used to generate the expression constructs of EGFP homologous recombinant plasmid (all from GenScript).
Induction of IMQ-induced psoriasis-like mouse model
Male C57BL/6J mice were subjected to a daily topical dose of 25 mg of IMQ cream (5%) (MedShine) on the ear for six consecutive days. Skin thickness and body weight were measured on the indicated days, and mice were treated with P155 (80 μg per mouse; GenScript) or Scr (80 μg per mouse; GenScript) daily via tail vein injection. All procedures were approved and supervised by the Shanghai Jiao Tong University School of Medicine Animal Care and Use Committee.
EAE induction and treatment
EAE was induced by complete Freund’s adjuvant (CFA; Sigma-Aldrich)–myelin oligodendrocyte glycoprotein (MOG35–55) peptide subcutaneous immunization (China Peptides Biotechnology). Briefly, a 200-μl mixture containing 300 μg of MOG35–55 peptide emulsified in CFA was subcutaneously injected into the mouse tail base on the first day, and 200 ng of pertussis toxin (Calbiochem) was injected intravenously on days 0 and 2 after immunization. Mice were scored daily according to the following scale: 0, no symptoms; 0.5, partially limp tail; 1, completely limp tail; 1.5, impaired righting reflex; 2, hindlimb paresis; 2.5, hindlimb paralysis; 3, forelimb weakness; 4, complete paralysis; and 5, moribund or death. P155 or Scr treatment was carried out every other day from the 8th day of immunization.
T cell differentiation
Splenocytes were harvested from 7-week-old mice, and naïve CD4+ T cells (CD4+CD25−CD62Lhigh) were enriched with the Naïve CD4+ Isolation Kit (STEMCELL Technologies). T cells were cultured with RPMI 1640 culture medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 1% Antibiotic-Antimycotic, 10 mM Hepes, and 50 μM β-mercaptoethanol (all from Gibco). The naïve T cells were activated with plate-bound anti-CD3 (5 mg/ml; BD Biosciences) and soluble anti-CD28 (2 mg/ml; BD Biosciences). The Treg cell differentiation condition was achieved by adding transforming growth factor–β1 (TGF-β1) (5 ng/ml; R&D Systems). The TH17 cell differentiation condition was achieved by adding TGF-β1 (10 ng/ml, R&D Systems), anti–IFN-γ (10 μg/ml; BioLegend), anti–IL-4 (5 μg/ml; BioLegend), IL-23A (20 ng/ml; R&D Systems), and IL-6 (10 ng/ml; R&D Systems). The TH1 cell differentiation condition was achieved by adding IL-12 (20 ng/ml; R&D Systems) and anti–IL-4 (10 μg/ml; BioLegend). All T cell subsets were cultured for 3 days and detected using flow cytometry.
Antibodies and flow cytometry
Cytokines, transcriptional factors, and surface markers were detected using flow cytometry with FACSCanto II or LSRFortessa (BD Biosciences) and analyzed with FlowJo (Version). To detect the intracellular expression of IL-17A, IFN-γ, IL-4, and Foxp3 in CD4+ T cells, cells were first treated with ionomycin (750 ng/ml; Sigma-Aldrich), phorbol 12-myristate 13-acetate (PMA; 50 ng/ml; Sigma-Aldrich), and GolgiPlug (BD Biosciences) for 4 hours at 37°C. Cells were fixed and permeabilized using the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences) or Foxp3 Transcription Factor Staining Buffer Set (eBioscience) and stained with the corresponding fluorescent antibodies. For flow cytometry, mouse monoclonal antibodies (mAbs) against CD4 (clone GK1.5), CD44 (clone IM7), CD25 (clone PC61.5), IL-17A (clone eBio17B7), IFN-γ (clone XMG1.2), IL-4 (clone 11B11), and Foxp3 (clone FJK-16s) were purchased from eBioscience. To detect the induction of THP-1–derived DCs, antibodies of CD80, CD11C, and CD209 were used for the flow cytometry.
CRISPR/Cas9-mediated editing and cell sorting
gRNA sequences were obtained using the ATUM gRNA design tool. HEK293T cells were cotransfected with the EGFP knock-in plasmid and single gRNA (sgRNA) plasmid (GenScript) for 2 days. DNA of cotransfected HEK293T cells was extracted using the TIANamp Genomic DNA Kit (TianGen), PCR was performed on genomic DNA, and the EGFP fusion protein was detected with immunofluorescence. Cotransfected HEK293T cells were collected for single-cell suspension. After washing with 1 × MACS (magnetic-activated cell sorting) buffer twice, the cells were sorted using the FITC channel to select EGFP-integrated cells on a BD FACSAria III instrument (BD Biosciences).
Single-cell RNA-seq
Fresh skin was placed in saline at 4°C until further processing. Single-cell suspensions were generated by enzyme digestion and subjected to fluorescence-activated cell sorting to exclude doublets, debris, and 4′,6-diamidino-2-phenylindole (DAPI)–positive dead cells. Sorted cells were centrifuged and resuspended in 0.04% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Chromium Single Cell 3′ v3 (10× Genomics) library preparation was conducted by the Sequencing Core at the Shanghai Institute of Immunology, according to the manufacturer’s instructions. The resulting libraries were sequenced with an Illumina HiSeq 4000 platform. Raw data were processed using Cell Ranger (version 3.0) and further filtered, processed, and analyzed using Seurat package.
Antigen presentation assays
OT-II mice were euthanatized and splenic CD4+ T cells were isolated and enriched by CD4 microbeads (Miltenyi Biotec) and labeled with Cell Trace Violet (CTV; Invitrogen) for 15 min at 37°C. BMDCs were loaded with 50 μg/ml of soluble OVA (Worthington) for 2 hours and washed with preheated RPMI 1640 culture medium twice. Then, treated BMDCs and CTV-labeled OT-II T cells were cocultured for 3 days at a ratio of 1:3. P155 or Scr (25 μM) was added to the corresponding group every day. For the polarizing direction of OT-II T cells induced upon OVA-loaded BMDCs, we harvested the CD4+ T cells and cocultured these cells with OVA (50 μg/ml)–loaded BMDCs for 3 days in the presence of P155 or Scr (25 μM). In vitro presentation assays were detected by flow cytometry using APC/BV510-labeled anti-CD44 and anti-CD4 antibody.
Histological analysis and immunohistochemistry
On day 7 following IMQ induction, mouse ears were harvested, fixed in 4% paraformaldehyde (PFA), and embedded in paraffin. Tissue sections (5 μm) were stained with hematoxylin and eosin (H&E) and Luxol fast blue (LFB). To measure acanthosis, the epidermis was outlined and measured with the lasso tool in Adobe Photoshop CC. The relative area of the epidermis was calculated using the following formula: 0.5119 × area pixels/(horizontal resolution × vertical resolution). To count dermal infiltrating cells, three areas in four sections of each sample were randomly taken, in which the number of infiltrating cells was calculated. For immunohistochemistry, anti-rabbit Ki67 Ab (1:1000; abcam) and goat polyclonal immunoglobulin G (IgG) anti–IL-17A Ab (1:100; Santa Cruz Biotechnology) were incubated overnight, and donkey anti-goat IgG-HRP (1:200; Santa Cruz Biotechnology) and rabbit enhanced polymer detection system (PV9000; ZSGB Bio) were used. For the EAE model, brain and spinal cord from P155- or Scr-treated mice were harvested and treated as above for H&E staining. All images were acquired using an Olympus IX51 microscope.
Murine BMDCs, human moDCs, and THP-1–derived DC induction in vitro
For mouse BMDC induction, bone marrow cells from C57BL/6J mice were cultured in RPMI 1640 culture medium containing 10% FBS, 1% Antibiotic-Antimycotic, 10 mM Hepes, 50 μM β-mercaptoethanol, 2 mM l-glutamine (all from Gibco), granulocyte-macrophage colony-stimulating factor (GM-CSF) (20 ng/ml), and IL-4 (10 ng/ml; PeproTech). The culture medium was partially replaced every 2 days, and the suspended cells were harvested on day 6. For human moDCs induction, the PBMCs were prepared using Ficoll gradient centrifugation (STEMCELL Technologies) and cultured for 6 days in RPMI 1640 culture medium supplemented with GM-CSF (100 ng/ml) and IL-4 (30 ng/ml; R&D Systems). Fresh culture medium was replaced every 3 days, and the suspended cells were harvested on day 6. For the induction of THP-1–derived DCs, THP-1 cells were cultured for 6 days in RPMI 1640 culture medium supplemented with GM-CSF (100 ng/ml) and IL-4 (30 ng/ml; R&D Systems), fresh culture medium was added every 2 days, and the suspended cells were harvested on day 6.
RNA-seq and transcriptome analysis
P155- or Scr-treated EAE mice were euthanized 15 days after disease induction, and mononuclear cells of the spinal cord were isolated using Percoll gradient centrifugation (GE Healthcare). For THP-1–derived DCs, cells were treated with P155 or Scr for 2 hours and then treated with R848 (1 μg/ml) for another 2 hours. All samples were washed twice with PBS, and total RNA was extracted with RNAiso Plus (Takara Bio) and purified with magnetic oligo (dT) beads after denaturation. Purified mRNA samples were reverse-transcribed into fragmented DNA samples and adenylated at the 3′ ends. Adaptors were ligated to construct a library. DNA was quantified by Qubit (Invitrogen). After cBot cluster generation, DNA samples were then sequenced by an Illumina HiSeq X Ten SBS instrument from Genergy Bio (Shanghai). Raw data were converted into FASTQ format, and transcript per million fragments mapped (fragments per kilobase) was calculated and log2-transformed with Cuffnorm. Differential gene transcripts were analyzed with DESeq and enriched for the GO/Kyoto Encyclopedia of Genes and Genomes pathway.
Immunofluorescence microscopy
Murine BMDCs were seeded into a 48-well plate with glass coverslips overnight and then incubated with P155 or Scr (10 μM) for 3 hours under R848 (1 μg/ml) stimulation. Cells were washed with PBS twice and fixed with 4% PFA for 10 min. Glass coverslips were transferred to microscope slides, adhered, and washed with PBS twice, and then stained with an FITC anti-mouse MHC class II antibody (1:200; BioLegend) overnight at 4°C in a semi-humid chamber after blocking with 1% BSA in PBS for 1 hour at room temperature. The slides were rinsed three times for 5 min each with PBS, mounted with DAPI (1:1000; Thermo Fisher Scientific), and sealed. For protein trafficking detection, BMDCs were incubated with FITC-labeled OVA (1:10,000; NobleRyder) for 10 min at 37°C. Cells were washed with PBS twice and fixed with 4% PFA for 10 min. Blocking was performed with 1% BSA in PBS for 30 min at room temperature, and anti-rabbit Rab5 antibody (1:500; abcam) and anti-rabbit LAMP2A antibody (1:500; abcam) were incubated overnight, respectively. Anti-rabbit IgG Fab2 Alexa Fluor 647 Molecular Probes (1:1000; Thermo Fisher Scientific) were incubated for 1 hour at room temperature, and DAPI (1:1000; BD Biosciences) was used for nuclear staining and mounting. For EAE brain and spinal cord tissues, sections were stained with a rat mAb against myelin basic protein (MBP) (1:1000; abcam) and later labeled with Alexa Fluor 488 donkey anti-rabbit IgG (H+L) (1:200; Invitrogen) for 1 hour at room temperature; other steps were as described above. For the colocalization of exogenous and endogenous P155 detection, we treated HEK293T cells with FITC-labeled P155 and stained the cells with the anti-rabbit P155 antibody (1:200; HuaBio) overnight, and Alexa Fluor 594 donkey anti-rabbit IgG (H+L) (1:1000; Thermo Fisher Scientific) was used. To detect the colocalization of P155 and HSC70 in WT 293T and EGFP-KI 293T cells, anti-rabbit P155 antibody (1:200; HuaBio) and anti-rat HSC70 (1:400; abcam) were incubated overnight, and Alexa Fluor 555/488 goat anti-rat IgG (H+L) and Alexa Fluor 594 donkey anti-rabbit IgG (H+L) were used (1:200; Thermo Fisher Scientific); other steps were as described above. Images were acquired with an Olympus IX51 microscope or a Leica TCS SP8 laser scanning confocal microscope.
Western blotting
Immunoprecipitation samples harvested from human moDCs, THP-1–derived DCs, or murine BMDCs were separated using SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. After three times washing, the membrane was incubated with HRP-labeled anti-rabbit IgG (H+L) for 1 hour at room temperature (1:1000; Beyotime) and developed with an enhanced chemiluminescence detection kit (1:1; YESEN). For EGFP detection, an anti-EGFP antibody was used (1:1000; CST). For the combination of P155 with HSC70 subdomain detection, a Myc-Tag mouse mAb (1:1000; CST) and HRP-labeled anti-mouse IgG (H+L) were used. For P155 detection, lysis was conducted using Tricine–SDS-PAGE and transferred to a PVDF membrane at 150 mA for 30 min and then fixed with 1% glutaraldehyde solution (Servicebio) for 1 hour. An anti-P155 antibody was used (1:400; HuaBio), and all other steps were the same as described above.
Silver staining and LC-MS
The samples of human moDCs or murine BMDCs harvested from immunoprecipitation were separated with SDS-PAGE and stained with Pierce Silver Stain Kit (Thermo Fisher Scientific) according to the instruction manual. The specific strip was cut for LC-MS. All MS was performed on a QE-Plus mass spectrometer connected to an Easy-nLC2000 via an Easy Spray (Thermo Fisher Scientific). Spectra were analyzed using PEAKS 8.0 (Bioinformatics Solutions).
Immunoprecipitation
Cultured OCI-LY-1 cells (10 million cells per dish) and human moDCs were lysed in immunoprecipitation assay buffer supplemented with a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). The extracted protein was incubated with a rabbit anti-P155 antibody (40 μg/ml; HuaBio) overnight at 4°C with rotation and then transferred to an Amicon Ultra Centrifugal Filters (Merck) to segregate ingredients ≤10 kDa. Harvests were then coincubated with pre-washed Protein A/G Magnetic Beads for immunoprecipitation (1:10; Bimake) for 4 hours at 4°C with rotation. The mixture was washed three times and eluted with 0.2 M glycine (pH 2.6) before MS. To explore the effects of P155 on HSC70-HSP90 machinery, the immunoprecipitants were pulled down with anti-HSC70 antibody (1:250; abcam) from R848- and Scr/P155-pretreated BMDCs and stored for using.
Pull-down assays
To detect the interaction protein with Biotin-Scr/P155, human moDCs, THP-1–derived DCs, or murine BMDCs were seeded at 1 million cells per plate and incubated with 25 μM Biotin, Biotin-P155, Biotin-Scr, or P155/Scr-treated Biotin-Scr/P155 (GenScript). After 2 hours of incubation, R848 (1 μg/ml; Sigma-Aldrich) was added to all samples and incubated for another 1.5 hours. Cells were lysed on ice with modified lysis buffer (1% NP-40, 150 mM NaCl, 4 mM EDTA-2Na, and 50 mM Hepes, pH 7.5) with a protease and phosphatase inhibitor cocktail (1:100; Thermo Fisher Scientific) for 20 min and centrifuged for another 20 min at 12,000g at 4°C, and cleared lysates were harvested. The streptavidin-agarose from Streptomyces avidinii (20 μl per sample; Sigma-Aldrich) was washed six times with 0.1% Triton PBS and coincubated with protein lysates for 4 hours at 4°C with rotation. The coincubated beads were then washed six times with lysis buffer and resuspended with 20 μl of lysis buffer and 5 μl of loading buffer. The mixture was boiled for 5 min at 100°C and harvested by centrifugation. To detect the binding domain of P155 with HSC70, HEK293T cells were seeded on a 6-cm dish and transfected with plasmids overexpressing C-Myc-Tag–labeled HSC70-SBD and HSC70-ATPase for 48 hours. Cells were lysed in ProteinExt Mammalian Total Protein Extraction Buffer (TransGen Biotech) with a protease and phosphatase inhibitor cocktail (1:100; Thermo Fisher Scientific) for 20 min, and the extracted protein was incubated with 25 μM Biotin-P155 or Scr for 2 hours at 4°C with rotation; other steps were the same as described above in the experiment of interaction protein detection.
Enzyme-linked immunosorbent assay
Total protein extracted from P155- or Scr-treated IMQ mouse ears was quantified with the BCA kit (Thermo Fisher Scientific). Protein expression of IL-17A, IFN-γ, and IL-4 was measured using mouse IL-17A (BioLegend) enzyme-linked immunosorbent assay (ELISA) Ready-SET-Go kit, mouse IFN-γ, and IL-4 quantikine ELISA kit (R&D Systems) according to the instructions.
Quantitative PCR
For the mRNA expression of MIR155HG detection, total RNA was extracted from NIH3T3, WT 293T, and EGFP-KI 293T cells using an RNAiso reagent (Takara Bio) in accordance with the manufacturer’s instructions. Complementary DNAs (cDNAs) were synthesized by HiScript qRT Super Mix (Vazyme Bio). Quantitative PCR (qPCR) was carried out with the Hieff qPCR SYBR Green Master Mix (Low Rox Plus) (Yeasen Bio) using primers recognizing human MIR155HG but not mouse mir155hg with ViiA7 Real-Time PCR instrument (Applied Biosystems). The relative expression of target genes was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). For the detection of IFN-γ and IL-4 mRNA expression, total RNA was extracted from P155- or Scr-treated IMQ mouse ears, and primers on IFN-γ and IL-4 were used; other steps were the same as described above.
Proximity ligation assay
HEK293T cells were seeded on a 48-well plate with glass coverslips and transfected with plasmids overexpressing Myc-Tag–labeled HSC70, HSC70-SBD, and HSC70-ATPase for 24 hours. Cells were fixed and permeabilized with 4% PFA for 10 min at room temperature. A proximity ligation assay (PLA) was performed using Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, cells were blocked with blocking solution for 30 min at 37°C, and primary antibodies against P155 (1:400) and Myc-Tag (1:5000; CST) were incubated overnight at 4°C. PLA probe anti-mouse plus and PLA probe anti-rabbit minus were incubated for 1 h at 37°C. Ligation and amplification were performed using Detection Reagents Red. Duolink mounting medium with DAPI was used for nuclear staining and mounting.
Measurements of HSC70 ATPase activity
ATPase activity was measured using the ATPase Activity Assay Kit (BioVision). To determine the HSC70 ATPase activity, the recombinant protein of human heat shock 70-kDa protein 8 (HSPA8) transcript variant 1 (OriGene) was prepared in assay buffer at a final concentration of 0.2 μM with P155 or Scr (25 μM) added to the solution. An aliquot (100 μl) of this mixture was added into each well of a 96-well plate and incubated for 30 min at room temperature. All subsequent procedures followed the instructions of the assay kit and the absorbance was determined at 650 nm on a Multiskan GO (Thermo Fisher Scientific).
Statistics
All data were expressed as means ± SEM, and statistical analysis was performed using GraphPad Prism 7. Student’s t test, one-way analysis of variance (ANOVA), or two-way ANOVA was performed to determine statistical significance.
Supplementary Material
Acknowledgments
We thank C. Gao, M. Zhang, L. Ding, and L. Chen at the Sequencing Core of Shanghai Institute of Immunology. We also thank L. Xia and L. Meng at the Proteomics Core of College of Basic Medical Sciences, SJTU-SM. Funding: This work was supported by grants from the National Natural Science Foundation of China (nos. 81725018, 31570922, 81703118, and 81803123), the Shanghai Collaborative Innovation Center for Translational Medicine (TM201925), and the Innovative Research Team of High-Level Local Universities in Shanghai. Author Contributions: L.N. and Honglin Wang designed the study and prepared the manuscript. L.N. conducted the experiments and analyzed the data. F.L. and Y.S. reviewed and edited the manuscript. Honglin Wang supervised the research. L.S., X.C., Z.L., H.Z., Z.W., Hong Wang, J.B., Q.Y., J.Z., L.C., D.P., Z.X., Y.G., S.T., and L.F. helped with experimental details. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the corresponding author. RNA-Seq datasets were deposited in GEO under accession numbers GSE142710 and GSE142711.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz2059/DC1
REFERENCES AND NOTES
- 1.Irla M., Küpfer N., Suter T., Lissilaa R., Benkhoucha M., Skupsky J., Lalive P. H., Fontana A., Reith W., Hugues S., MHC class II–restricted antigen presentation by plasmacytoid dendritic cells inhibits T cell–mediated autoimmunity. J. Exp. Med. 207, 1891–1905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuseff M.-I., Pierobon P., Reversat A., Lennon-Duménil A.-M., How B cells capture, process and present antigens: A crucial role for cell polarity. Nat. Rev. Immunol. 13, 475–486 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Roche P. A., Furuta K., The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuseff M.-I., Reversat A., Lankar D., Diaz J., Fanget I., Pierobon P., Randrian V., Larochette N., Vascotto F., Desdouets C., Jauffred B., Bellaiche Y., Gasman S., Darchen F., Desnos C., Lennon-Duménil A.-M., Polarized secretion of lysosomes at the B cell synapse couples antigen extraction to processing and presentation. Immunity 35, 361–374 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Nagy L., Szanto A., Szatmari I., Széles L., Nuclear hormone receptors enable macrophages and dendritic cells to sense their lipid environment and shape their immune response. Physiol. Rev. 92, 739–789 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Germic N., Frangez Z., Yousefi S., Simon H.-U., Regulation of the innate immune system by autophagy: Monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ. 26, 715–727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushik S., Cuervo A. M., The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu A., Shibata Y., Shah B., Calamini B., Lo D. C., Morimoto R. I., Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc. Natl. Acad. Sci. U.S.A. 111, E1481–E1490 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon H. T., Boucrot E., Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Navarro J. A., Cuervo A. M., Dietary lipids and aging compromise chaperone-mediated autophagy by similar mechanisms. Autophagy 8, 1152–1154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K. I., Clohessy J. G., Pandolfi P. P., mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Ransohoff J. D., Wei Y., Khavari P. A., The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slavoff S. A., Mitchell A. J., Schwaid A. G., Cabili M. N., Ma J., Levin J. Z., Karger A. D., Budnik B. A., Rinn J. L., Saghatelian A., Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plaza S., Menschaert G., Payre F., In search of lost small peptides. Annu. Rev. Cell Dev. Biol. 33, 391–416 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Lauressergues D., Couzigou J.-M., Clemente H. S., Martinez Y., Dunand C., Bécard G., Combier J.-P., Primary transcripts of microRNAs encode regulatory peptides. Nature 520, 90–93 (2015). [DOI] [PubMed] [Google Scholar]
- 16.D’Lima N. G., Ma J., Winkler L., Chu Q., Loh K. H., Corpuz E. O., Budnik B. A., Lykke-Andersen J., Saghatelian A., Slavoff S. A., A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 13, 174–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauch J. N., Zuiderweg E. R., Gestwicki J. E., Non-canonical interactions between heat shock cognate protein 70 (Hsc70) and Bcl2-associated anthanogene (BAG) co-chaperones are important for client release. J. Biol. Chem. 291, 19848–19857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou D., Li P., Lin Y., Lott J. M., Hislop A. D., Canaday D. H., Brutkiewicz R. R., Blum J. S., Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity 22, 571–581 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Deffit S. N., Blum J. S., A central role for HSC70 in regulating antigen trafficking and MHC class II presentation. Mol. Immunol. 68, 85–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T.-G., Kim S. H., Park J., Choi W., Sohn M., Na H. Y., Lee M., Lee J. W., Kim S. M., Kim D.-Y., Kim H.-P., Choi J.-H., Park C. G., Lee M.-G., Skin-specific CD301b+ dermal dendritic cells drive IL-17–mediated psoriasis-like immune response in mice. J. Invest. Dermatol. 138, 844–853 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Wohn C., Ober-Blöbaum J. L., Haak S., Pantelyushin S., Cheong C., Zahner S. P., Onderwater S., Kant M., Weighardt H., Holzmann B., Reizis B., Becher B., Prens E. P., Clausen B. E., Langerinneg conventional dendritic cells produce IL-23 to drive psoriatic plaque formation in mice. Proc. Natl. Acad. Sci. U.S.A. 110, 10723–10728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tam W., Ben-Yehuda D., Hayward W. S., bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell Biol. 17, 1490–1502 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceppi M., Pereira P. M., Dunand-Sauthier I., Barras E., Reith W., Santos M. A., Pierre P., MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2735–2740 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D., MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanczyk J., Pedrioli D. M. L., Brentano F., Sanchez-Pernaute O., Kolling C., Gay R. E., Detmar M., Gay S., Kyburz D., Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 58, 1001–1009 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A., Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kluiver J., van den Berg A., de Jong D., Blokzijl T., Harms G., Bouwman E., Jacobs S., Poppema S., Kroesen B.-J., Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene 26, 3769–3776 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Eis P. S., Tam W., Sun L., Chadburn A., Li Z., Gomez M. F., Lund E., Dahlberg J. E., Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. U.S.A. 102, 3627–3632 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellacheruvu D., Wright Z., Couzens A. L., Lambert J.-P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J. R., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A.-C., Nesvizhskii A. I., The CRAPome: A contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 10, 730–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabrera Y., Dublang L., Fernández-Higuero J. A., Albesa-Jové D., Lucas M., Viguera A. R., Guerin M. E., Vilar J. M. G., Muga A., Moro F., Regulation of human Hsc70 ATPase and chaperone activities by Apg2: Role of the acidic subdomain. J. Mol. Biol. 431, 444–461 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Fosgerau K., Hoffmann T., Peptide therapeutics: Current status and future directions. Drug Discov. Today 20, 122–128 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Martinez N. R., Augstein P., Moustakas A. K., Papadopoulos G. K., Gregori S., Adorini L., Jackson D. C., Harrison L. C., Disabling an integral CTL epitope allows suppression of autoimmune diabetes by intranasal proinsulin peptide. J. Clin. Invest. 111, 1365–1371 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz2059/DC1