Abstract
Aims
Investigating human heart development and applying this to deviations resulting in disease is incomplete without molecular characterization of the cell types required for normal functioning. We investigated foetal human heart single-cell transcriptomes from mid-gestational healthy and anti-SSA/Ro associated congenital heart block (CHB) samples.
Methods and results
Three healthy foetal human hearts (19th to 22nd week of gestation) and one foetal heart affected by autoimmune-associated CHB (21st week of gestation) were subjected to enzymatic dissociation using the Langendorff preparation to obtain single-cell suspensions followed by 10× Genomics- and Illumina-based single-cell RNA-sequencing (scRNA-seq). In addition to the myocytes, fibroblasts, immune cells, and other minor cell types, previously uncharacterized diverse sub-populations of endothelial cells were identified in the human heart. Differential gene expression analysis revealed increased and heterogeneous interferon responses in varied cell types of the CHB heart compared with the healthy controls. In addition, we also identified matrisome transcripts enriched in CHB stromal cells that potentially contribute to extracellular matrix deposition and subsequent fibrosis.
Conclusion
These data provide an information-rich resource to further our understanding of human heart development, which, as illustrated by comparison to a heart exposed to a maternal autoimmune environment, can be leveraged to provide insight into the pathogenesis of disease.
Keywords: Foetal heart, Congenital heart block, Endothelial cells, Single-cell RNA-seq
Graphical Abstract
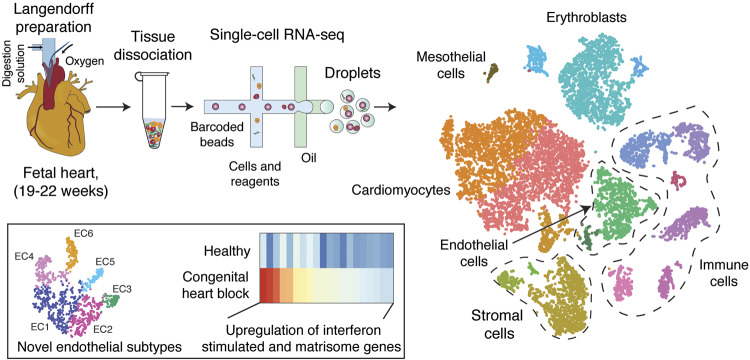
Graphical Abstract.
1. Introduction
The heart, the first organ to function in the embryo, is complex in morphology and composed of diverse cell types that develop during cardiogenesis from the mesoderm. Development of the heart is a finely tuned dynamic process involving cell migration, lineage diversification, proliferation, cell transition, and programmed cell death.1 Our molecular understanding of the developing heart has largely been drawn from studies in animal models and in vitro systems such as human pluripotent stem cell (hPSC) derived cardiomyocytes.2 The cardiac progenitors arise from mesoderm and segregate into two populations that form first (FHF) and second (SHF) heart fields.3 The FHF gives rise to the early cardiac tube that contributes to the left ventricle and parts of the atria whereas the SHF is placed within and at the entry of the developing tube and contributes to the outflow tract, right ventricle, and atria.1 Genetic cell-fate-mapping studies in animal model systems have greatly enhanced the understanding of lineage contribution to diverse cell groups that constitute the heart. Such studies have revealed the epicardium as a major source of cell types that populate the heart.4 However, similar studies of mapping the lineage of cell types in the developing human heart have not been done. In addition, knowledge of cellular composition and gene expression signatures that predict distinct cellular function is extremely crucial for understanding cardiac remodelling, repair, and regeneration.
Single-cell RNA-sequencing (scRNA-seq) provides new and unique opportunities to define the cellular composition and transcriptional heterogeneity in different cell types during development of the human heart.5,6 ScRNA-seq analysis also provides a detailed atlas of ligands and receptors expressed by cell types that can be leveraged to generate a cell–cell communication map of the heart. Such mapping can be used as a reference blueprint for comparing and contrasting diseases affecting human heart development.
Congenital heart block (CHB) is a remarkable foetal disease that occurs de novo in an otherwise normally developing heart during the 18–25th week of human gestation.7 The majority of affected foetuses are exposed to maternal autoantibodies against components of the SSA/Ro and SSB/La ribonucleoprotein complexes via neonatal-Fc-receptor-mediated transplacental passage. The disease carries a significant mortality (17.5%) and most surviving children eventually require permanent pacing.8 Foetal and in utero factors, in addition to maternal autoantibodies, likely contribute to disease since only 2% of anti-SSA/Ro-exposed offspring develop CHB7 and recurrent rates approach 18%.9 Histology of foetuses dying with CHB reveals fibrotic replacement of the atrioventricular node and often a macrophage infiltrate including multinucleated giant cells as the signature lesions.10 Especially given the intracellular location of the candidate antigens, identifying a pathologic link between the putative autoantibodies and tissue damage has been challenging.
This study was initiated to generate an atlas of the human foetal heart to gain insights into cardiogenesis and in doing so to provide understanding of transcriptomic changes in foetal heart cells experiencing the pathologic cascade to heart block. For the former, it should be noted that current approaches to study heart development applying scRNA-seq have relied solely on animal models11,12 or heart-like systems derived from hPSCs13,14 with no direct evaluation of human tissue. To accomplish these goals scRNA-seq analysis of >17 000 cells isolated from three mid-gestational healthy hearts and an anti-SSA/Ro-associated CHB heart, unexposed to any maternal medications, was performed. This study identified several known and previously uncharacterized cell sub-populations in healthy hearts. Moreover, the CHB heart showed diversity in interferon (IFN)-stimulated gene expression across cell types and increased matrisome expression in stromal cells.
2. Methods
2.1 Tissue collection and dissociation
CHB and control hearts were obtained following written informed consent from the New York University (NYU) Institutional Review Board as part of the Research Registry for Neonatal Lupus and the investigation conformed to the principles outlined in the Declaration of Helsinki. Foetal human hearts of the healthy group were obtained at 19, 22, and 22 weeks of gestation. The CHB case was obtained at 21 weeks of gestation. The mother is a 35-year-old Asian anti-SSA/Ro positive female with Sjogren’s syndrome, G1P0. This foetus was found to be bradycardic by auscultation at 20 weeks after a normal echocardiogram at 18 weeks. The echocardiogram at 20 weeks showed 2:1 AV block; shortly thereafter, a subsequent echocardiogram revealed complete AV block with a heart rate of 75 beats/min and no signs of a cardiomyopathy as demonstrated by the absence of endocardial fibroelastosis or evidence of cardiac dilatation or decreased function. The heart was otherwise structurally normal. The mother was on no prior medications, and she declined dexamethasone and IVIG and elected to terminate the pregnancy. The procedure for termination of pregnancy was identical for all cases and involved a standard dilation and evacuation (D & E) with no toxic (KCl or other) injections.
For each specimen, the aorta was cannulated for continuous perfusion of the coronary arteries using a Langendorff preparation with proteolytic enzymes to yield a single-cell suspension of primary human foetal cells as previously described.15 The dissociated cells were collected by centrifugation at 300 g for 5 min and resuspended in 2 mL of phosphate-buffered saline (PBS) maintained at 37°C. To lyse the RBCs, 5 mL of ACK lysing buffer (A104920; Thermo Fisher Scientific) was added and incubated for 4 min at room temperature followed by centrifugation at 300 g for 5 min, supernatant discarded and 5 mL of ice-cold PBS was added. Cells and PBS were gently mixed. Cells were collected by centrifugation at 300 g for 5 min at 4°C and 1 mL of freshly prepared PBS-bovine serum albumin (BSA; 1× PBS and 0.04% BSA) was added. To remove debris, low-speed centrifugation at 200 g for 3 min was performed followed by discarding the supernatant and resuspension of cells in 50 μL of 1× PBS–BSA. Cell viability was assessed using the Trypan blue (1450013; Bio-Rad) exclusion method on a TC20 automated cell counter (Bio-Rad). The time from procurement of the foetal hearts to single-cell capture was <4 h.
2.2 Single-cell capture, library preparation, and sequencing
scRNA-seq libraries were generated using the Chromium Single Cell 3ʹ Reagent Kit V2 (10× Genomics) according to the manufacturer’s instructions. The library was sequenced on Illumina HiSeq 2500 platform as follows: 26 bp (Read1) and 98 bp (Read2). The sequencing was performed to obtain 150–200 million reads (each for Read1 and Read2). The CHB library was sequenced twice to achieve this depth and two sequencing round files were combined for the downstream analysis.
2.3 scRNA-seq data processing
The 10× raw data were processed separately for individual samples using the standard pipeline (Drop-seq core computational protocol V1.2, http://mccarrolllab.com/dropseq/) with the following changes. The Read1 bases 1–16 were tagged with cell barcode ‘XC’, and bases 17–26 were tagged with a unique molecular identifier (UMI) ‘XM’. Reads with low-quality bases were removed, and the 3ʹ-end of Read2 was trimmed to remove poly(A) sequences of length six or more and then aligned to human genome (hg38) reference using the STAR aligner (STAR_2.5.1a), allowing no more than three mismatches. Next, a gene expression matrix was generated for each sample using the ‘MIN_BC_READ_THRESHOLD = 2’ option to retain UMIs with two or more reads.
2.4 Clustering and sub-clustering analysis using Seurat and t-SNE visualization
Clustering, gene expression and cell-type identification were performed independently for healthy and CHB samples using Seurat V2.0. In brief, only those genes that were expressed in more than three cells and cells that expressed more than 100 but less than 5000 genes were retained. Ubiquitously expressed genes such as ribosomal protein-coding (RPS and RPL) and non-coding RNA (MALAT1) genes were removed. We also removed miRNA and snoRNA genes from clustering analysis. Separate Seurat objects were created and then merged by using ‘CreateSeuratObject’ and ‘MergeSeurat’ options followed by scaling data (ScaleData) and finding variable (between 1600 and 2000) genes (FindVariableGenes). Multiple rounds of iteration were performed to identify and remove poor-quality cells. All cells (expect for cardiomyocytes) with >25% mitochondrial content (based on UMIs) were removed as these were considered of poor quality. However, since biologically, cardiomyocytes contain high mitochondrial density, cardiomyocytes with >25% mitochondrial content were not filtered out. Consequently we did still observe a poor-quality cardiomyocyte population of high mitochondrial (all cells with >25%) content but distinctly low numbers of detected genes and transcripts as compared with other cardiomyocyte populations. This poor-quality cardiomyocyte population was removed from the final analysis. Once the poor-quality cells were identified, mitochondrially encoded genes (MT-) were dropped from clustering analysis. For the final round of analysis, clusters were identified using ‘FindClusters’ with PCA parameter ‘dims.use = 1: 20’ and resolution of 0.4. A similar strategy was applied for CHB sample analysis with ‘dims.use = 1:10’ and ‘0.6 resolution’ parameters. With the cell types identified, we performed sub-clustering analysis on single or groups of closely related cell types to identify hidden sub-populations. To identify differentially expressed genes by each cluster, Wilcoxon rank sum test inbuilt in the Seurat package was used with parameters ‘min.pct = 0.25’ and ‘thresh.use = 0.25’. Combination of known lineage markers and genes identified in differential gene expression analysis were then used to generate dotplots.
2.5 Averaged cell-type specific expression profiles and transcription factors enrichment analysis
Cardiomyocytes constituted ∼50% of all cells in the foetal heart and the most abundantly expressed genes in cardiomyocytes showed background ‘noisy’ expression in other cell types. Similarly, haemoglobin subunit and other erythroblast-specific genes showed background expression in other cell types. Such background expression is most likely contributed by ambient RNA in cell suspension due to lysis of cardiomyocytes and erythroblasts. Therefore, for generating cell-type-specific averaged expression profiles, a set of cardiomyocyte- and erythroblast-specific genes were removed from all other cell types whereas only cardiomyocyte-specific genes were removed from erythroblasts and vice versa. After taking averages for each cell type, gene expression was normalized to 10 000 to create a TPM (transcript per million)-like value, followed by transforming to log2(TPM + 1). To identify cell-type-specific transcription factors (TFs), we used a list of published TFs described earlier5,16 and crossed this list with TPM-normalized expression by cell types. For each cell type, only top expressed (max. of 10) TFs and TFs expressed in >10% of cells were considered. For each selected TF, a z-score based on log2(TPM + 1) was calculated, and to find highly cell-type-specific TFs, we applied a threshold of 0.6 to the ratio of the second maximal expression value to the maximal. The only exception for this filtering rule was erythroblasts as they shared many TFs with mast cells.
2.6 Differential gene expression analysis: Healthy vs. CHB
Combined (healthy and CHB) Seurat objects for each of the cell groups were generated by using ‘MergeSeurat’ followed by scaling (ScaleData). A list of the top 60 differentially genes (expressed by >30% of cells) was generated for each cell group using ‘FindAllmarkers’ option. Next, genes with fold change >1.5 log (TPM + 1) were retained for downstream analysis. Only IFN-stimulated genes (ISGs) were plotted as dotplots in Figure 4A, whereas all the differentially expressed genes were plotted in heatmaps (Figure 5).
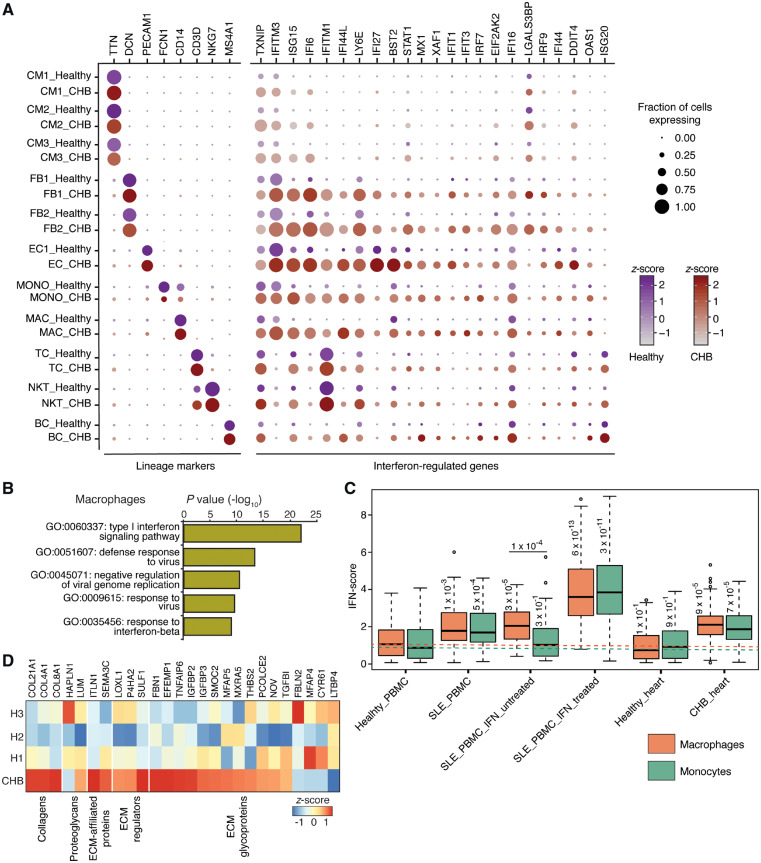
Figure 4.
Gene expression differences between healthy and CHB heart cell types. (A) Dotplot of lineage markers and differentially expressed IFN-regulated genes (Wilcoxon rank sum test, expressed by >30% of cells of a given cell type, log fold change >1.5 in at least one cell type) between healthy and CHB cell types. (B) Gene ontology (GO) terms enriched in DAVID analysis performed on the top 25 up-regulated genes of CHB macrophages. (C) IFN-score plot of macrophages and monocytes obtained from various sources including healthy and systemic lupus PBMCs in the absence and presence of treatment with exogenous IFN beta. The Healthy_PBMC data was acquired from 10x Genomics (see methods section) and SLE_PBMC data was obtained from Der et al. study48. Both the SLE_PBMC_IFN_untreated and treated datasets were acquired from Kang et al. study17. The Healthy_heart and CHB_heart macrophages and monocytes belong to the present study. Dotted lines highlight median IFN-score of macrophages (orange) and monocytes (green) obtained from PBMCs obtained from healthy donors. Numbers on the boxplots indicate P-values from a Wilcoxon rank sum test. (D) Heatmap showing differential expression and enrichment of matrisome genes between CHB and healthy (H1, H2, and H3) stromal cells (FB and SMC). Only genes with expression >1 TPM and fold change (CHB vs. healthy) of 1.5 TPM were considered for the analysis.
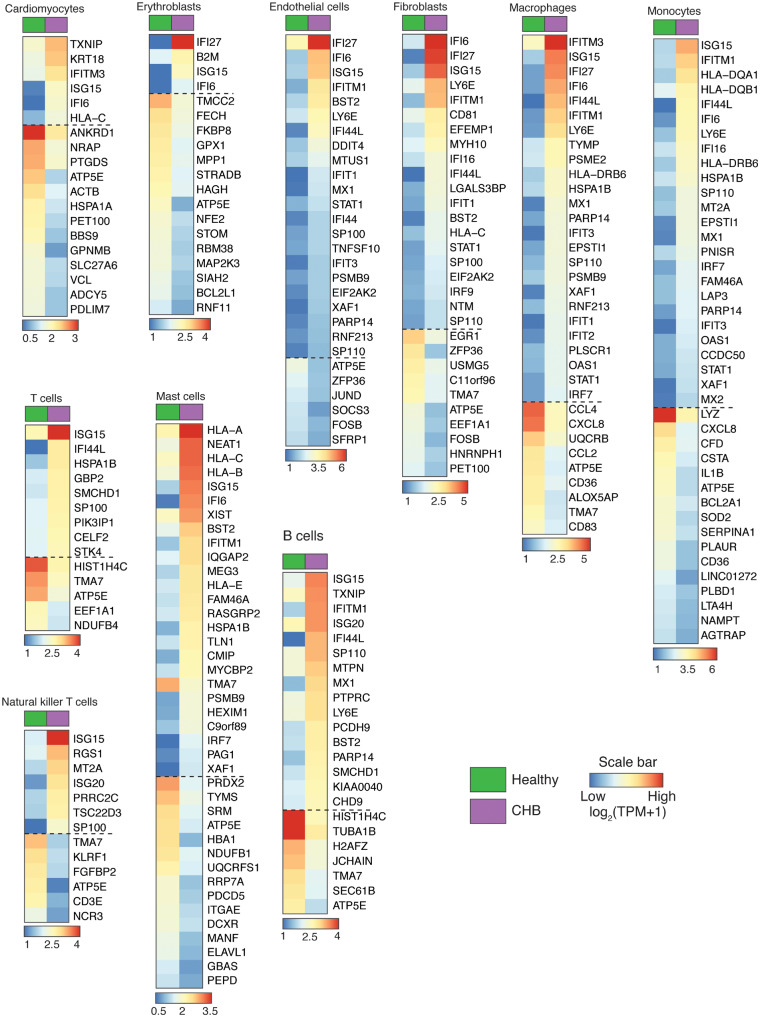
Figure 5.
Gene expression differences between healthy and CHB heart cell types. Heatmap reporting averaged expression level [log2(TPM + 1)] of differentially expressed genes (Wilcoxon rank sum test, >1.5 log fold change, expressed by >30% cells in a given cell type) between healthy and CHB cell types. Note that genes below the dashed line represent a subset of down-regulated genes of CHB vs. healthy cell types.
2.7 Immunostaining of foetal heart tissue
Formalin-fixed, paraffin-embedded sections of human foetal heart were placed onto charged microscope slides (SL6332-4, Avantik SureBond) and adult heart tissue was sectioned onto SHUR/Mark + charged slides (SM-CCU-W). Paraffin was removed by immersing the slides in two changes of Clear-Rite 3 (22-046-341, Richard-Allan, Kalamazoo, MI, USA) for 5 min. Tissue was rehydrated by a series of graded ethanol washes to water. High temperature antigen retrieval was performed in a steamer by immersing the slides in a solution of citrate buffer pH 6.0 (00-5000, Life Technologies, Frederick, MD, USA) for 30 min. Endogenous peroxidase was blocked by incubating the slides in a dual enzyme block (S200389-2, Agilent, Santa Clara, CA, USA). Slides were incubated with a primary antibody to TNNT3 at a 1:200 dilution (HPA037810, Sigma-Aldrich, St. Louis, MO, USA) for 45 min. This was followed by a 30 min incubation with a polymer HRP IgG (PV6119, Leica Biosystems, Pleasanton, CA, USA). The antibody complex was detected with ImmPact DAB (SK-4105, Vector Laboratories, Burlingame, CA, USA) and the slides were counterstained with haematoxylin (7211, Richard-Allen Scientific, Kalamazoo, MI, USA). The stained slides were digitized using an Aperio AT Scanner (Leica Biosystems) and served using Proscia software (Proscia, Philadelphia, PA, USA).
2.8 IFN-score calculations for macrophages and monocytes
Previously published scRNA-seq (10× Genomics) raw data of peripheral blood mononuclear cells (PBMCs) from healthy (https://support.10xgenomics.com/single-cell-gene-expression/datasets), systemic lupus erythematosus (SLE) ‘Der et al’, IFN-untreated, and SLE IFN-treated cells16 were downloaded.17 Graph-based analysis was performed on each of the datasets to identify macrophage and monocyte populations. The averaged expression profiles of macrophage and monocytes were used to generate TPM-like values generated as described above. In addition to the healthy and SLE macrophages and monocytes, we also calculated IFN-scores for the same cell types that we detected in our CHB heart sample that was used in this study. A list of ISGs was obtained as described earlier and crossed with the averaged expression profiles. Only ISGs with >4 TPM in at least one sample were retained. All the TPM for the final list of ISGs were averaged and log2(TPM + 1) was calculated and referred as IFN-score in Figure 4C.
2.9 Matrisome enrichment analysis
Averaged expression profiles of stromal cells [fibroblasts (FBs) and smooth muscle cells (SMCs)] from all individual healthy (H1, H2, and H3) and CHB samples were generated. We used a previously described list of matrisome genes18 for which we derived TPM values from averaged expression profiles. Genes with expression <1 TPM were removed. Next, only genes with >1.5 TPM fold change expression between CHB and combined healthy were retained. For this final list of matrisome genes, z-scores were calculated across all four (three healthy and one CHB) individual samples.
3. Results
3.1 Strategy for scRNA-seq, clustering analysis, and cell-type identification
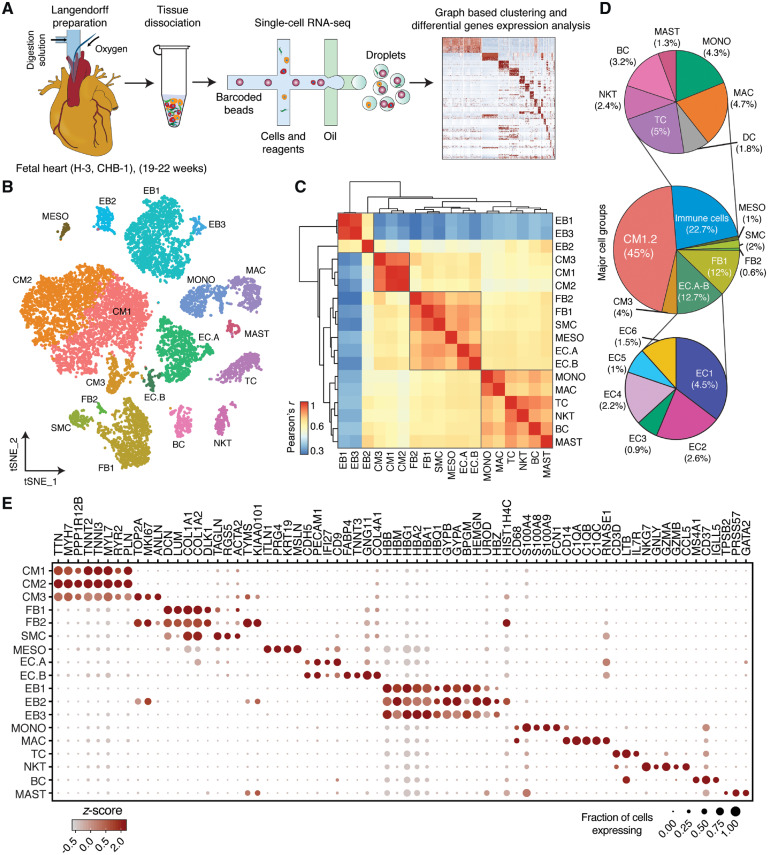
Freshly dissociated cells from three healthy foetal human hearts were subjected to 10× Genomics-19 and Illumina-based scRNA-seq; Figure 1A, Supplementary material online, Table S1), along with one foetal heart with anti-SSA/Ro associated CHB. Bioinformatic analysis identified 12 461 and 5286 high-quality scRNA-seq profiles originating from the healthy and CHB samples, respectively, with average read depth of 42 000/cell. The data of healthy samples were combined and analysed by graph-based clustering as described earlier keeping track of the sample identity for each cell barcode (H1, H2, and H3)20 (Figure 1B, Supplementary material online, Figure S1A, B). Subsequent gene expression analysis and the presence of established cell lineage markers differentiated 18 cell types referred to as cardiomyocytes (CM1 to 3), fibroblasts (FB1 and 2), SMCs, mesothelial cells (MESO), endothelial cells (EC.A and EC.B), erythroblasts (EB1 to 3), monocytes (MONO), macrophages (MAC), T cells (TC), natural killer cells (NKT), B cells (BC), and mast cells (MAST). Pearson correlation of averaged expression profiles of healthy foetal heart cells provided a matrix of cell-type similarity. FB1 and 2, SMC, EC.A, EC.B, and MESO clustered together, suggesting a common origin (Figure 1C). Cardiomyocytes have been debated to originate from MESO4 but CM1-3 formed their own distinct subgroup. Immune-cell types and erythroblasts EB1-3 also clustered separately. With respect to the cell-type composition of these dissociated foetal hearts, CMs accounted for nearly half of the total cell composition, followed by ECs and FBs at similar proportions adding up to nearly a quarter of the cells (Figure 1D, Supplementary material online, Figure S1C). Immune cells dominated in the remaining quarter. EBs were excluded from calculating relative cell proportions, because the heart was perfused and resulting cell suspensions were also treated to minimize EB contributions. In addition, the majority (more than 90%) of EBs was contributed by one (H3) sample (Supplementary material online, Figure S1A).
Figure 1.
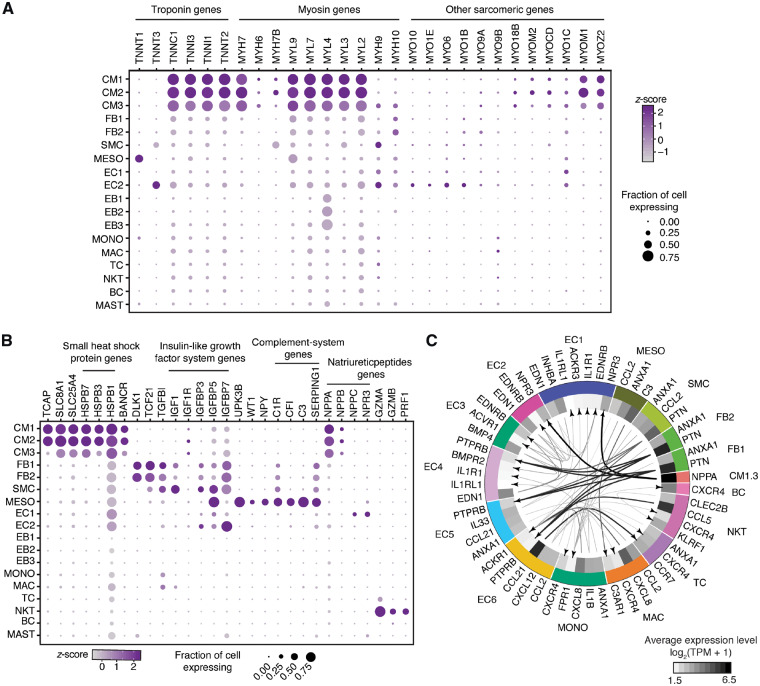
Identification of cell types in human foetal heart. (A) Workflow for single-cell transcriptome profiling of foetal hearts. Numbers in parentheses indicate the number of healthy (H) and disease (CHB) samples analysed. (B) Clustering for 12 461 cells from three healthy samples based on established lineage markers and visualized using t-SNE. (C) Pearson’s correlation coefficient (r, for values see colour scale) comparing averaged gene expression of each cell type. (D) Averaged proportion of all cell types (except EBs) contributing to foetal human hearts. Middle panel indicates major cell groups with subgroups of immune cells (top panel) and endothelial cells (bottom panel) and number in parenthesis indicates percentage contribution to foetal heart cells. (E) Dot plots showing expression of known lineage markers and co-expressed lineage-specific genes.
3.2 Cell types of healthy foetal hearts
3.2.1 Cardiomyocytes
Resolving gene expression to cellular levels provides unique insights into cellular processes active in each cell type (Figures 1E and 3A). CMs were characterized by parallel expression of myosin light (MYL2, MYL3, MYL4, MYL7, and MYL9) and heavy chains (MYH7, MYH7B, and MYH6) as well as troponins (TNNT2, TNNI1, TNNI3, and TNNC1), together constituting the contractile system.21 The most distinctive feature between CM subtypes was a cluster of proliferation markers including TOP2A and MKI67 expressed at high levels in CM3, which were also defined by the absence of muscle assembly regulating factor TCAP (Figure 3B). All CMs expressed the Na+/Ca2+ exchanger-1 (SLC8A1) important for Ca2+ homeostasis and contraction and relaxation of the heart;22 however, the parallel expression of SLC25A4, an adenine nucleotide translocator-1, whose complete absence causes cardiomyopathy,23 has not been noted previously. A subset of small-heat-shock-protein genes, HSPB7 and HSPB3, was specifically expressed in CMs, implicated in CM-specific protein folding,24 whereas the similarly abundant HSPB1 was expressed ubiquitously. The spliced lncRNA BANCR was also uniquely expressed by CMs although it was originally reported for its involvement in cancer cell migration and epithelial–mesenchymal transition (EMT).25
Figure 3.
Dot plots showing expression of selected genes. (A) Troponin, myosin and other sarcomeric genes expression in healthy heart cell types. (B) Expression of other important genes in major cell types of foetal heart. (C) Putative signalling within foetal heart cell types with size of the arrow stem proportionate to expression levels of the ligands. All the arrows are pointing to the receptors. Only ligands with >5 TPM and receptors with >1.8 TPM average expression were used for interaction analysis display. MDK–PTPRB interactions were excluded, and provided as a separate figure (Supplementary material online, Figure S2B).
3.2.2 Immune cells
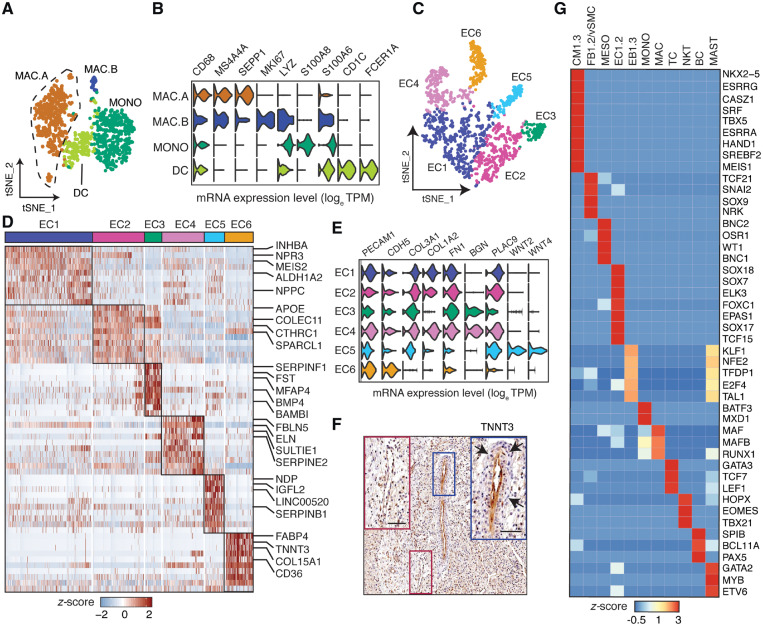
Several major populations of lymphoid cell types were identified in the normal foetal heart, including TCs differentiated by LTB, and IL7R from cytotoxic NKTs expressing NKG7, GNLY, GZMA, GZMB, and PRF1, and BCs uniquely expressing MS4A1 and IGLL5. The myeloid lineage cell types included MASTs specific for TPSB2 and GATA2 expression, and EBs identified by haemoglobin expression, including a proliferating EB2 sub-population. The myeloid origin MONO and MAC cell populations identified by first-level graph-based analysis separated into four distinct cell clusters at second-level analysis (Figure 2A, B). The macrophages separated into non-proliferating (MAC.A) and proliferating (MAC.B) subtypes both referred to as alternatively activated M2 macrophage based on co-expressing MS4A4A and SEPP1. In addition to CD68, MONO abundantly expressed LYZ, S100A8, and S100A6. Finally, a small group of dendritic cells (DCs) of the CD1C+ subtype was uncovered.
Figure 2.
Sub-clustering and TF enrichment analysis of foetal heart cell types. (A) t-SNE visualization of 1093 myeloid-derived cells as four subgroups. (B) Violin plots showing expression for different myeloid-derived cell lineage markers and other co-expressed genes. (C) t-SNE visualization of 1263 EC cells sub-clustered into six subgroups. (D) Differentially expressed genes of EC sub-populations using Wilcoxon rank sum test. (E) Violin plots for EC lineage markers. (F) Immunohistochemical staining of a foetal human heart for TNNT3. Blue inset: EC6 endothelium and red inset: non-EC6 endothelium. Arrows indicate perivascular density surrounding the TNNT3-positive endothelium. Scale bar, 50 μm. (G) TF enrichment analysis showing the most abundant (maximum of 10) and specific TFs of major cell groups.
3.2.3 Diversity of ECs
ECs separated into EC.A and EC.B populations by first-level analysis, but separated further into six distinct EC sub-populations upon second-level analysis (EC1-6; Figure 2C, D). ECs shared expression of the canonical markers PECAM1 and CDH5 (Figure 2E). In addition, several ECs expressed genes such as COL3A1, COL1A2, FN1, and BGN usually expressed by fibroblasts indicating endothelial-mesenchymal transition (EndMT) required for the formation of atrioventricular cushion and valves.26 Natriuretic peptide hormone NPPC was exclusively expressed by EC1 and EC5, whereas NPPA and NPPB were expressed uniquely by CMs (Figure 3B, Supplementary material online, Figure S2). Natriuretic peptides are implicated in a wide range of functions such as control of blood pressure and volume, protection against atherosclerosis, myocardial ischaemia, and cardiac remodelling. Impairment or abnormality in the natriuretic peptide system leads to cardiovascular complications. The natriuretic peptide receptor NPR3 was predominantly expressed by EC1 and EC2 with NPR1 sparsely expressed by all ECs. NPR3 is preferentially expressed in mouse atrial and ventral endocardium suggesting that EC1 and EC2 represent endocardial ECs. EC1 also expressed MEIS2, mutations of which cause septal defects and aortic coarctation in humans27 (Figure 2D). EC2 and EC3 highly expressed APOE, an important secreted lipid transporter. EC3 also expressed SERPINF1, a neurotrophic agent and inhibitor of angiogenesis, as well as PDGFRA28 and BMP4,29 both crucial for outflow tract development as shown in the mouse. EC4s are defined by elastin ELN and elastin-binding component FBLN5 expression, both of which are necessary for organizing elastic fibres and organ remodelling during development, as well as expression of RSPO3 promoting proliferation and sprouting in human ECs.30 ECs were also the only source of nitric oxide (NO) synthase (NOS) in the foetal hearts with NOS1 expressed by EC4 and NOS3 by EC1 and to a lesser extent also by EC2-6. The pathway is crucial for cardiovascular functions through regulation of Ca2+ fluxes and homeostasis, sarcomeric protein Ca2+ sensitivity, and mitochondrial respiration.31WNT2, WNT4, and NDP involved in WNT signalling were abundantly and specifically expressed in EC5 along with CD44 and SELL previously implicated in lymphocyte migration suggesting EC5 is a migratory endothelial cell type in the developing heart. LncRNA LINC00520 was also uniquely expressed by EC5. EC6 uniquely expressed genes involved in fatty acid (FA) binding and transport including CD36, FABP4, and FABP5, thereby providing the basis for the FA-biased energy metabolism that contributes up to 70% of cardiac ATP (Supplementary material online, Figure S3). Unexpectedly, fast skeletal muscle troponin TNNT3 is specifically expressed in EC6 in the foetal heart, which was further confirmed by immunohistochemistry (Figures 2D–F and 3A). Furthermore, blood vessels formed by EC6 are characterized also by the presence of perivascular cells surrounding them. This observation is supported by the specific co-expression of PDGFB in EC6 implicated in recruitment of these perivascular cells.
3.2.4 Cardiac fibroblasts and smooth muscle cells
Cardiac FBs that play a crucial role in organogenesis by depositing and maintaining extracellular matrix (ECM) proteins32 shared the expression of abundant collagens and collagen-binding protein COL1A1, COL1A2, and TGFBI with SMCs (Figures 1E and 3B). Although the FB population separated into proliferating (FB2) and non-proliferating sub-populations (FB1), SMCs did not. FBs and SMCs were discriminated by expression of DCN, DLK1, LUM, and TAGLN, RGS5, ACTA2, respectively. Insulin-like growth factor (IGF) signalling is important for heart development,33 with the receptor IGF1R being mainly expressed by CMs, whereas secreted IGF1 was highly expressed by SMCs and moderately by FBs, and an even wider range of cell types expressed the secreted modulators of IGF signalling proteins including IGFBP3, IGFBP5, and IGFBP7 (Figure 3B).
3.2.5 Mesothelial cells
The epicardium is composed by a layer of MESO, which specifically expressed UPK3B, MSLN, WT1,4 as well as NPY (Figure 3B). Neuropeptide Y (NPY) is expressed in neurons but is also involved in cardiac remodelling, angiogenesis, and vasoconstriction.34 It was, therefore, surprising to discover its expression in MESO. Another peculiarity of MESO was the unique expression of slow muscle troponin TNNT1 and a wide range of complement immune system genes such as C1R, CFI, C3, and SERPING1 (Figure 3B).
3.3 TFs enrichment analysis
We performed TF enrichment analysis to annotate the highest expressed TFs in each cell type and identified at least one cell-type-specific TF able to complement lineage assignment. In addition to the classical cardiomyocyte TF NKX2-5, CMs also expressed ESRRG, CASZ1, and SRF (Figure 2G). Stromal cells (FBs and SMCs) preferentially expressed TCF21 and SNAI2, known mesenchymal markers, among other TFs. MESOs are defined by enriched expression of WT1, BNC1, BNC2, and OSR1. ECs transcribed a range of SOX family TFs including SOX7, SOX17, and SOX18. Immune cell types MONO, MAC, TC, NKT, BC, and MAST preferentially expressed immune-cell-specific TFs BATF3, MAF, GATA3, EOMES, BCL11A, and GATA2, respectively. Although, EBs expressed the erythropoiesis regulator KLF1, interestingly, MAST also expressed TFs enriched in EBs suggesting a shared developmental trajectory.
3.4 Potential ligand-receptor interactions in foetal heart
To investigate intercellular communication in the healthy foetal heart, we visualized averaged expression of the most abundant ligands (>5 TPM) and their cognate receptors (>1.8 TPM) from each cell type (Figure 3C). NPPA that regulates cardiovascular homeostasis through natriuresis, diuresis, and vasodilation showed the highest expression in CMs communicating with EC1 and EC2 via NPR3 receptor. MDK (midkine), the deficiency of which causes heart failure in mice, was the ubiquitously and highly expressed ligand that interacts with PTPRB receptor expressed by several cell types (Supplementary material online, Figure S2B). In addition, PTN, an angiogenic as well as MDK-related factor, was abundantly expressed by FBs and potentially interacts with its EC-specific receptor (EC4-6) PTPRB to regulate vasculogenesis in the developing heart. CCL21 was the most abundantly expressed cytokine in EC6 with receptor CCR7 being expressed on TCs. Other cytokines with high-expression levels were CCL5 from NKT and CXCL8 from MAC and MONO interacting with ACKR1 receptor expressed by EC6.
3.5 CHB specimen analysis
In the CHB foetal heart similar major cell types were obtained compared with each healthy foetal heart, although MESO, EC5, SMC, and DC were not captured, most likely because they represented rare cell populations (<1.3%) even in the healthy heart. To provide insights into the pathogenesis of CHB and the cellular mechanisms by which exposure to maternal anti-SSA/Ro antibodies compromises atrioventricular function in an otherwise normally developing heart in the second trimester of gestation, we investigated differentially expressed genes (Wilcoxon rank sum test) between cell types of healthy and CHB samples (Figures 4A and 5). Overall, all CHB cell types showed overexpression of ISGs including TXNIP, IFITM3, ISG15, IFI6, and IFI16, and ‘interferon signalling’ was also identified as the most up-regulated biological process according to pathway analysis (Figure 4B). However, varying degrees of ISG up-regulation were also apparent among cell types. Specifically, the CHB CM population showed the least up-regulation of ISGs whereas FB1 and 2, EC, MONO, and MAC displayed the greatest up-regulation. The expression of individual ISGs also differed by cell type, e.g. ISG20 was preferentially induced in lymphoid origin cell types.
3.6 IFN-score of macrophages and monocytes in CHB and SLE
Systemic IFN responses are observed in other autoimmune diseases including SLE, which has recently been studied by scRNA-seq.35 For comparison to CHB, we calculated an IFN response score for MAC and MONO from the CHB and healthy sample (this study) and SLE patients as well as from an in vitro IFN-stimulated PBMC sample17 (Figure 4C). As expected, the in vitro IFN-stimulated cells displayed the highest IFN response. Nevertheless, the CHB macrophages and monocytes showed greater IFN responses than cells isolated from the SLE patients.
3.7 Enrichment of fibrotic gene signature in CHB
The signature histologic feature of CHB is fibrotic replacement of the atrioventricular node.10 Accordingly, to address the contribution of ECM components to tissue injury, we performed differential expression analysis of matrisome genes18 on stromal cells (FB and SMC). Most of the genes that belong to different categories of matrisome including COL21A1, COL4A1, and COL8A1 were highly enriched in the CHB sample compared with all three healthy hearts (Figure 4D) supporting stromal cells as the potential source of ECM thereby leading to fibrosis.
4. Discussion
In summary, our transcriptome profiles provide insights into the human foetal heart at single-cell resolution. We show an unanticipated diversity of cardiac cell types and gene expression patterns including novel EC sub-populations highlighting the complexity of the vasculature in the developing human heart. In addition, using ligand-receptor enrichment analysis, we also identified potential cell–cell communication in the niche of the developing human heart. Cardiomyocytes that formed the major cell population in foetal heart samples revealed proliferative and non-proliferative states. However, they failed to group into region-specific (atrial or ventricular) or any other subtype-specific cardiomyocytes. Applying the recent finding of AV-nodal-cell-specific expression of the GJD3 gene in a murine model36 we identified that of approximately 5000 cardiomyocytes that were captured, only four GJD3+ cells likely represent AV-nodal cells. (Supplementary material online, Figure S2C). The fraction of AV node cells in the overall heart is expected to be low and fluorescent labelling and microdissection or cell sorting may be necessary to increase their proportion.
Having generated an atlas of the human second trimester foetal heart, we leveraged a truly unique opportunity to compare cell expression levels in a CHB heart in the absence of exposure to any maternal medications, which could have influenced the transcriptome. To date, delineating the pathogenesis of CHB has relied on in vitro models of human foetal cardiomyocytes,15,37,38 bulk mRNA sequencing,39 and immunohistochemistry.40 The comparative cell-type-specific expression analysis performed herein revealed a previously unexplored, wide, and dynamic spectrum of IFN response in different cardiac cell types isolated from a human heart with CHB. The profound IFN response confirms and extends previous work, which employed bulk sequencing39,41 and did not address the foetal myocardiocytes per se. Although expression of ISGs in these cells was less than that observed in the fibroblast and macrophages populations, the scRNA-seq data attest to the profound bystander effect of an IFN-rich environment present in the CHB heart. It remains speculative whether the source of IFN generates from the maternal or foetal circulation, but it has been reported that mothers of CHB children have significantly higher levels of circulating IFNα than anti-SSA/Ro-exposed healthy children, which the authors speculate influences the development of cardiac disease via maternal transplacental passage.42 Pertinent to therapy is the observation that hydroxchloroquine, which inhibits TLR signalling and likely downstream IFN responses,43,44 has been shown to decrease both the primary and secondary rate of CHB.45–47 In addition, we also identified a set of genes that potentially contribute to the characteristic fibrosis of CHB via ECM deposition. Both of these findings provide potentially targetable candidates for treatment of a disease that is irreversible with current anti-inflammatory approaches.
It is readily acknowledged that there are limitations to the work presented. Specialized cardiomyocytes that form the sinoatrial and atrioventricular nodes were with extremely rare exception not identified in normal or the CHB heart. These and other rare cell populations may be absent not only because of their assumed low abundance but also because of their sensitivity to tissue digestion, unusual size, or short-term viability upon dissociation. It is also conceivable that the disease impacted the specialized cardiomyocytes. Although only one CHB heart was analysed, it is important to point out that it is highly unlikely to have another CHB heart donated in proximity to a major research facility, unexposed to maternal immunosuppression, and electively terminated with early disease. The agnostic cell dissociation approach allowed for numerous cell types to be evaluated with no a priori biases.
In summary, providing a high-resolution cellular atlas has fundamental implications for investigating human heart development and congenital heart diseases. This foundation will guide developing novel regenerative strategies for treatment of myocardial infarction. Our study that catalogues the cell-type-specific gene programmes will also help to evaluate results previously obtained using human heart tissue culture and differentiation models.
Data and materials availability
All processed data are available in the main text or the Supplementary material online. All additional data needed to evaluate the conclusions in the paper are available under BioProject ID PRJNA576243.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
J.B., T.T., R.C., and H.S. conceived the study. R.C. performed the sample tissue dissociations. H.S. performed single-cell suspension clean-up and sample preparation for single-cell experiments. H.S. and P.M. performed all the computational analysis. M.H. carried out the immunohistochemical staining experiments. H.S., T.T., R.C., and J.B., prepared and wrote the manuscript.
Supplementary Material
Acknowledgements
We thank members of the Rockefeller University Genomics Center for expert support. We acknowledge Karen Fox-Talbot (Johns Hopkins Medicine, MD, USA) for the expert support on immunostaining experiments. We also thank Tuschl lab members and M. Yamaji (Cincinnati Children’s Hospital Medical Center, OH, USA) for their feedback on the manuscript. The authors thank Arlene Hurley NP and the Research Facilitation Office staff for regulatory and administrative assistance.
Conflict of interest: none declared.
Funding
This work was supported by R37 AR042455, R37 AR042455-21S1, R37 AR042455-21S2 (J.P.B.), N01 AR-4-2220 (J.P.B.), R03 HD069986 (J.P.B.), and R01 HD079951-01A1 (J.P.B.). Supported in part by grant # UL1TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) programme.
Translational perspective
This study provides a single cell transcriptomic atlas of cells obtained from healthy second trimester fetal hearts to further understand human heart development and impart insight into autoimmune associated congenital heart block. In addition to myocytes and fibroblasts, previously uncharacterized subpopulations of endothelial cells were identified. Leveraging an unprecedented opportunity, healthy heart transcriptomes were compared to an age matched anti-SSA/Ro exposed fetal heart with third degree block in which no maternal medications were taken. Differential gene expression analysis revealed a remarkable interferon response in many cell types of the diseased heart. In addition, matrisome transcripts were enriched in the stromal cells likely contributing to the extracellular matrix deposition and thereby fibrosis, a signature lesion of heart block. Thus, targeting the interferon pathway merits therapeutic consideration.
References
- 1. Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell 2006;126:1037–1048. [DOI] [PubMed] [Google Scholar]
- 2. Meilhac SM, Buckingham ME.. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol 2018;15:705–724. [DOI] [PubMed] [Google Scholar]
- 3. Ivanovitch K, Esteban I, Torres M.. Growth and morphogenesis during early heart development in amniotes. J Cardiovasc Dev Dis 2017;4:20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao J, Poss KD.. The epicardium as a hub for heart regeneration. Nat Rev Cardiol 2018;15:631–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, Williams Z.. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv 2018;4:eaau4788.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Domínguez Á, Pinho S, Akhmetzyanova I, Gao J, Witkowski M, Guillamot M, Gutkin MC, Zhang Y, Marier C, Diefenbach C, Kousteni S, Heguy A, Zhong H, Fooksman DR, Butler JM, Economides A, Frenette PS, Adams RH, Satija R, Tsirigos A, Aifantis I.. The bone marrow microenvironment at single-cell resolution. Nature 2019;569:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brito-Zerón P, Izmirly PM, Ramos-Casals M, Buyon JP, Khamashta MA.. The clinical spectrum of autoimmune congenital heart block. Nat Rev Rheumatol 2015;11:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Izmirly PM, Saxena A, Kim MY, Wang D, Sahl SK, Llanos C, Friedman D, Buyon JP.. Maternal and fetal factors associated with mortality and morbidity in a multi-racial/ethnic registry of anti-SSA/Ro-associated cardiac neonatal lupus. Circulation 2011;124:1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Llanos C, Izmirly PM, Katholi M, Clancy RM, Friedman DM, Kim MY, Buyon JP.. Recurrence rates of cardiac manifestations associated with neonatal lupus and maternal/fetal risk factors. Arthritis Rheum 2009;60:3091–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Llanos C, Friedman DMSaxena A, Izmirly PM, Tseng C-E, Dische R, Abellar RG, Halushka M, Clancy RM, Buyon JP.. Anatomical and pathological findings in hearts from fetuses and infants with cardiac manifestations of neonatal lupus. Rheumatology (Oxford) 2012;51:1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sànchez-Dànes A, Moignard V, Dubois C, Paulissen C, Kinston S, Göttgens B, Blanpain C.. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 2018;359:1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR.. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep 2018;22:600–610. [DOI] [PubMed] [Google Scholar]
- 13. Friedman CE, Nguyen Q, Lukowski SW, Helfer A, Chiu HS, Miklas J, Levy S, Suo S, Han J-DJ, Osteil P, Peng G, Jing N, Baillie GJ, Senabouth A, Christ AN, Bruxner TJ, Murry CE, Wong ES, Ding J, Wang Y, Hudson J, Ruohola-Baker H, Bar-Joseph Z, Tam PPL, Powell JE, Palpant NJ.. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell 2018;23:586–598.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Churko JM, Garg P, Treutlein B, Venkatasubramanian M, Wu H, Lee J, Wessells QN, Chen S-Y, Chen W-Y, Chetal K, Mantalas G, Neff N, Jabart E, Sharma A, Nolan GP, Salomonis N, Wu JC.. Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat Commun 2018;9:4906.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clancy RM, Askanase AD, Kapur RP, Chiopelas E, Azar N, Miranda-Carus ME, Buyon JP.. Transdifferentiation of cardiac fibroblasts, a fetal factor in anti-SSA/Ro-SSB/La antibody-mediated congenital heart block. J Immunol 2002;169:2156–2163. [DOI] [PubMed] [Google Scholar]
- 16. Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM.. A census of human transcription factors: function, expression and evolution. Nat Rev Genet 2009;10:252–263. [DOI] [PubMed] [Google Scholar]
- 17. Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ.. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 2018;36:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hynes RO, Naba A.. Overview of the matrisome–an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol 2012;4:a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH.. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA.. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordon AM, Homsher E, Regnier M.. Regulation of contraction in striated muscle. Physiol Rev 2000;80:853–924. [DOI] [PubMed] [Google Scholar]
- 22. Philipson KD, Nicoll DA.. Sodium-calcium exchange: a molecular perspective. Annu Rev Physiol 2000;62:111–133. [DOI] [PubMed] [Google Scholar]
- 23. Strauss KA, DuBiner L, Simon M, Zaragoza M, Sengupta PP, Li P, Narula N, Dreike S, Platt J, Procaccio V, Ortiz-González XR, Puffenberger EG, Kelley RI, Morton DH, Narula J, Wallace DC.. Severity of cardiomyopathy associated with adenine nucleotide translocator-1 deficiency correlates with mtDNA haplogroup. Proc Natl Acad Sci USA 2013;110:3453–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kötter S, Unger A, Hamdani N, Lang P, Vorgerd M, Nagel-Steger L, Linke WA.. Human myocytes are protected from titin aggregation-induced stiffening by small heat shock proteins. J Cell Biol 2014;204:187–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y.. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett 2014;8:869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Armstrong EJ, Bischoff J.. Heart valve development: endothelial cell signaling and differentiation. Circ Res 2004;95:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Louw JJ, Corveleyn A, Jia Y, Hens G, Gewillig M, Devriendt K.. MEIS2 involvement in cardiac development, cleft palate, and intellectual disability. Am J Med Genet A 2015;167A:1142–1146. [DOI] [PubMed] [Google Scholar]
- 28. Bloomekatz J, Singh R, Prall OW, Dunn AC, Vaughan M, Loo C-S, Harvey RP, Yelon D.. Platelet-derived growth factor (PDGF) signaling directs cardiomyocyte movement toward the midline during heart tube assembly. Elife 2017;6:40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu W, Selever J, Wang D, Lu M-F, Moses KA, Schwartz RJ, Martin JF.. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc Natl Acad Sci USA 2004;101:4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scholz B, Korn C, Wojtarowicz J, Mogler C, Augustin I, Boutros M, Niehrs C, Augustin HG.. Endothelial RSPO3 controls vascular stability and pruning through non-canonical WNT/Ca(2+)/NFAT signaling. Dev Cell 2016;36:79–93. [DOI] [PubMed] [Google Scholar]
- 31. Farah C, Michel LYM, Balligand J-L.. Nitric oxide signalling in cardiovascular health and disease. Nat Rev Cardiol 2018;15:292–316. [DOI] [PubMed] [Google Scholar]
- 32. Tallquist MD, Molkentin JD.. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol 2017;14:484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN.. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal 2011;4:ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan CMJ, Green P, Tapoulal N, Lewandowski AJ, Leeson P, Herring N.. The role of neuropeptide Y in cardiovascular health and disease. Front Physiol 2018;9:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crow MK, Olferiev M, Kirou KA.. Type I interferons in autoimmune disease. Annu Rev Pathol 2019;14:369–393. [DOI] [PubMed] [Google Scholar]
- 36. Bhattacharyya S, Duan J, Wang L, Li B, Bhakta M, Fernandez-Perez A, Hon GC, Munshi NV.. Using Gjd3-CreEGFP mice to examine atrioventricular node morphology and composition. Sci Rep 2019;9:2106–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miranda-Carús ME, Askanase AD, Clancy RM, Di Donato F, Chou TM, Libera MR, Chan EK, Buyon JP.. Anti-SSA/Ro and anti-SSB/La autoantibodies bind the surface of apoptotic fetal cardiocytes and promote secretion of TNF-alpha by macrophages. J Immunol 2000;165:5345–5351. [DOI] [PubMed] [Google Scholar]
- 38. Clancy RM, Neufing PJ, Zheng P, O'Mahony M, Nimmerjahn F, Gordon TP, Buyon JP.. Impaired clearance of apoptotic cardiocytes is linked to anti-SSA/Ro and -SSB/La antibodies in the pathogenesis of congenital heart block. J Clin Invest 2006;116:2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clancy RM, Halushka M, Rasmussen SE, Lhakhang T, Chang M, Buyon JP.. Siglec-1 macrophages and the contribution of IFN to the development of autoimmune congenital heart block. J Immunol 2019;202:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clancy RM, Kapur RP, Molad Y, Askanase AD, Buyon JP.. Immunohistologic evidence supports apoptosis, IgG deposition, and novel macrophage/fibroblast crosstalk in the pathologic cascade leading to congenital heart block. Arthritis Rheum 2004;50:173–182. [DOI] [PubMed] [Google Scholar]
- 41. Clancy RM, Markham AJ, Jackson T, Rasmussen SE, Blumenberg M, Buyon JP.. Cardiac fibroblast transcriptome analyses support a role for interferogenic, profibrotic, and inflammatory genes in anti-SSA/Ro-associated congenital heart block. Am J Physiol Heart Circ Physiol 2017;313:H631–H640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lisney AR, Szelinski F, Reiter K, Burmester GR, Rose T, Dörner T.. High maternal expression of SIGLEC1 on monocytes as a surrogate marker of a type I interferon signature is a risk factor for the development of autoimmune congenital heart block. Ann Rheum Dis 2017;76:1476–1480. [DOI] [PubMed] [Google Scholar]
- 43. Kuznik A, Bencina M, Svajger U, Jeras M, Rozman B, Jerala R.. Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J Immunol American Association of Immunologists 2011;186:4794–4804. [DOI] [PubMed] [Google Scholar]
- 44. Lamphier MZheng W, Latz E, Spyvee M, Hansen H, Rose J, Genest M, Yang H, Shaffer C, Zhao Y, Shen Y, Liu C, Liu D, Mempel TR, Rowbottom C, Chow J, Twine NC, Yu M, Gusovsky F, Ishizaka ST.. Novel small molecule inhibitors of TLR7 and TLR9: mechanism of action and efficacy in vivo. Mol Pharmacol 2014;85:429–440. [DOI] [PubMed] [Google Scholar]
- 45. Izmirly PM, Kim MY, Llanos C, Le PU, Guerra MM, Askanase AD, Salmon JE, Buyon JP.. Evaluation of the risk of anti-SSA/Ro-SSB/La antibody-associated cardiac manifestations of neonatal lupus in fetuses of mothers with systemic lupus erythematosus exposed to hydroxychloroquine. Ann Rheum Dis 2010;69:1827–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Izmirly PM, Costedoat-Chalumeau N, Pisoni CN, Khamashta MA, Kim MY, Saxena A, Friedman D, Llanos C, Piette J-C, Buyon JP.. Maternal use of hydroxychloroquine is associated with a reduced risk of recurrent anti-SSA/Ro-antibody-associated cardiac manifestations of neonatal lupus. Circulation 2012;126:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barsalou J, Jaeggi E, Laskin CA, Brown P, Tian SY, Hamilton RM, Silverman ED.. Prenatal exposure to antimalarials decreases the risk of cardiac but not non-cardiac neonatal lupus: a single-centre cohort study. Rheumatology (Oxford) 2017;56:1552–1559. [DOI] [PubMed] [Google Scholar]
- 48. Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabathou SIzmirly P, Clancy R, Belmont HM, Koenigsberg M, Mokrzycki M, Rominieki H, Graham JA, Rocca JP, Bornkamp N, Jordan N, Schulte E, Wu M, Pullman J, Slowikowski K, Raychaudhuri S, Guthridge J, James J, Buyon J, Tuschl T, Putterman C, Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium.. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol Nature Publishing Group 2019;20:915–927. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.