Abstract
Aims
Sinus venous valve (SVV) and sinoatrial node (SAN) develop together at the sinoatrial junction during embryogenesis. SVV ensures unidirectional cardiac input and SAN generates sinus rhythmic contraction, respectively; both functions are essential for embryonic survival. We aim to reveal the potential role of endocardial NOTCH signalling in SVV and SAN formation.
Methods and results
We specifically deleted Notch1 in the endocardium using an Nfatc1Cre line. This deletion resulted in underdeveloped SVV and SAN, associated with reduced expression of T-box transcription factors, Tbx5 andTbx18, which are essential for the formation of SVV and SAN. The deletion also led to decreased expression of Wnt2 in myocardium of SVV and SAN. WNT2 treatment was able to rescue the growth defect of SVV and SAN resulted from the Notch1 deletion in whole embryo cultures. Furthermore, the Notch1 deletion reduced the expression of Nrg1 in the SVV myocardium and supplement of NRG1 restored the growth of SVV in cultured Notch1 knockout embryos.
Conclusion
Our findings support that endocardial NOTCH1 controls the development of SVV and SAN by coordinating myocardial WNT and NRG1 signalling functions.
Keywords: Sinus venous valve, Sinoatrial node, NOTCH1, WNT2, NRG1
Graphical Abstract
Graphical Abstract.
Translational perspective
Sinus venous valve and sinoatrial node control unidirectional cardiac flow and heart rhythm, respectively, during foetal development. By revealing that NOTCH1 regulates development of sinus venous valve and sinoatrial node through coordinating WNT2 and NRG1 signalling in genetic modified mouse models and whole embryo cultures, this study helps understanding of the potential disease mechanisms in congenital malformation and dysfunction of sinus venous valve and sinoatrial node.
1. Introduction
Sinus venous valve (SVV) and sinoatrial node (SAN) are essential for maintaining unidirectional circulation and sinus rhythmic contraction, respectively.1 SVV and SAN form together at the sinoatrial junction during development.1 In mouse, SVV begins to form at embryonic day (E) 9.5 and is matured by E11.5.2 It prevents the cardiac regurgitation in the early embryos before the formation of functional atrioventricular and semilunar valves.3,4 At the same time of SVV development, a cluster of cardiomyocytes differentiates into specialized primary pacemaker cells that form the SAN at the base of the developing SVV.5,6 SAN cardiomyocytes express HCN4 and generate the pacer action potential that controls the sinus heart rate and rhythm.7
Emerging data from recent studies have underpinned a unique gene regulatory network controlling the development of SVV and SAN, in particular, the differentiation of primary cardiac pacemaker cells.8 Studies by Christoffels et al.6 have shown that SAN progenitors arise from the cardiogenic mesoderm as a genetically distinct subpopulation.9 The proliferation and differentiation of these progenitor cells into SVV/SAN myocardium are regulated by a transcriptional network consisting of the T-box transcription factors, Tbx3,10Tbx5,11 and Tbx18,12 as well as the short stature homeobox transcription factors, Shox and Shox2.2,13,14 Furthermore, ectopic expression of Tbx3 or Tbx18 in the mature quiescent cardiomyocytes reprograms them into the pacemaker-like cells.15,16 In contrast, ectopic expression of Nkx2.5 suppresses the differentiation of SAN myocardium.17 Despite of these findings, the signalling mechanisms that regulate the SVV and SAN transcriptional program remain understudied.
Molecular signalling from the endocardium to the myocardium are essential for myocardial development.18 Among them, the NOTCH1 signalling in the endocardium is a well-studied paracrine signal that regulates myocardial gene expression to promote the proliferation and differentiation of working myocardium during ventricular chamber formation.19 However, whether the NOTCH1 signalling in the endocardium has a role in the development of SVV and SAN myocardium are not addressed in the previous studies, as deletion of Notch1 in the pan-endothelium (including vascular endothelium and endocardium) using the Tie2Cre mouse results in severe vascular and cardiac defects, leading to early embryonic lethality around E9.5–E10.5 before the formation of mature SVV and SAN.19,20
In the present study, we addressed this question by using the Nfatc1Cre mouse to specifically delete Notch1 in the endocardium during heart development.21,22 The endocardial Notch1 knockout embryos died around E11.5 that allowed us to investigate the functions of endocardial NOTCH1 in the development of SVV and SAN between E9.5 and E11.5. Our results show that endocardial NOTCH1 controls the development of SVV and SAN by regulating Wnt2 and Nrg1.
2. Methods
Detailed methods were described in the Supplementary material online.
2.1 Ethics statement
All animal procedures conformed to the Guidelines for the Care and Use of Laboratory Animals published by the National Institution of Health and was approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine.
2.2 Histology, LacZ staining, and immunofluorescence
The pregnant mice were sacrificed by an overdose of isoflurane in a sealed container. Embryos were fixed overnight at 4°C using 4% paraformaldehyde in phosphate-buffered saline, dehydrated through an ethanol gradient, treated with xylene, and embedded in paraffin wax. Embryos were oriented for sagittal sections and cut in 6 µm sections using a Leica microtome. Haematoxylin and eosin (HE), LacZ, and immunofluorescence staining were performed using standard protocols.
2.3 Cell proliferation assays
Cell proliferation was determined using an EdU assay.
2.4 RNA extraction and quantitative PCR
Total RNAs were isolated from pooled heart tissues from E9.5 and E10.5 control and endocardial Notch1 knockout embryos using Trizol (Invitrogen, Thermo Fisher, USA). First strand cDNA was synthesized using the Superscript II Reverse Transcriptase Kit (Invitrogen, Thermo Fisher, USA). Quantitative PCR (qPCR) was performed using the Power SYBR Green PCR Master Mix (ABI, Thermo Fisher, USA). Gene specific primers were used for qPCR (Supplementary material online, Table S1). The expression of Gapdh is used as internal control.
2.5 RNA in situ hybridization
RNA in situ hybridization was performed according to a previously described protocol.21
2.6 Measurement of heartbeat frequency
The heartbeat was measured as described previously.2
2.7 Whole embryo culture
Whole embryo culture was carried out as described previously.21 For rescue experiments, the culture media was supplemented with recombinant NRG1 proteins (1.25 × 10−8 M) (R&D system, USA) or WNT2 conditioned media.21,23 The embryos were cultured in an incubator that contained specialized gas (60% oxygen, 5% CO2, and 35% N2) at 37°C for 24 h. The cultured embryos were subjected to gene expression analysis by whole mount in situ hybridization.
2.8 Statistical analysis
All data were presented as means ± SEM. One-way ANOVA and Tukey’s multiple comparisons test were used for calculating the statistical differences of means among multiple groups. Unpaired Student’s t-test was used for statistical calculation between two groups. The Probability (P) values below 0.05 were considered as significant.
3. Results
3.1 Morphogenesis of SVV and SAN and expression of NOTCH1
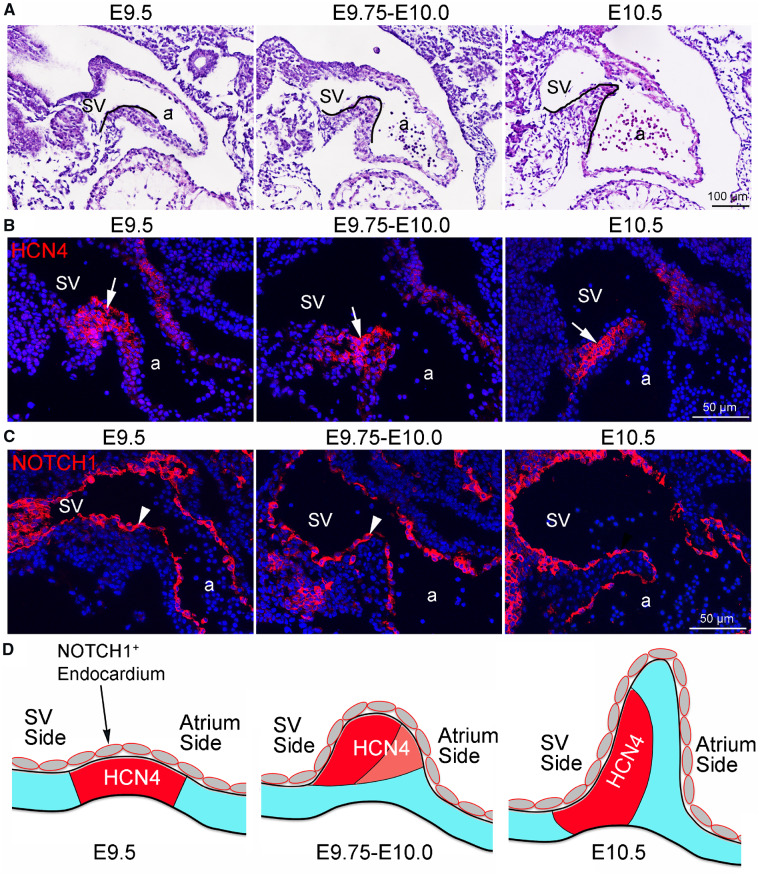
We first examined the morphogenesis of SVV in normal mouse embryos between E9.5 and E10.5 by HE staining. The results showed that the myocardium at the sinoatrial junction started to bend towards the lumen of the heart tube at E9.5, this process continued at E9.75–E10.0, and by E10.5 the SVV structure was clearly formed, which demarks the boundary of sinus venous and atrium (Figure 1A and D). We next examined the SAN development by immunostaining for HCN4, which labels the primary pacemaker cells. The results showed that HCN4 was expressed by the cardiomyocytes at the sinoatrial junction of E9.5 embryos (Figure 1B and D). As the SVV elongated, the HCN4-expressing cardiomyocytes became to be clustered towards the sinus venous side at E9.5–E10.0, by E10.5 the HCN4-expressing cardiomyocytes was further condensed to form the SAN at the base of SVV at the sinus venous side (Figure 1B). We then performed immunostaining for NOTCH1 and found that NOTCH1 was highly expressed in the endocardium of developing SVV and SAN (Figure 1C and D). In addition, by RNA in situ hybridization and immunostaining, we found that the NOTCH ligand Jag1 but not Dll4 was expressed within SVV region (Supplementary material online, Figure S1). Together, these findings suggest that NOTCH1 may be involved in the developmental process of SVV and SAN.
Figure 1.
Morphogenesis of SVV and SAN and the expression of NOTCH1. (A) HE staining shows the morphology of developing SVV (outlined by black lines) in mouse embryos between E9.5 and E10.5. a, atrium; sv, sinus venous. (B) Immunofluorescence shows the expression of HCN4 (arrows) which marks the pacemaker cells of developing SAN. (C) Immunofluorescence shows the expression of NOTCH1 (arrowhead) in the endocardium of developing SVV. (D) Schematics show the morphogenic changes of developing SVV and SAN. At E9.5, the myocardium at the sinoatrial junction begins to bend towards the lumen of the heart tube and expresses HCN4. Between E9.75 and E10.0, the myocardium at the sinoatrial junction continues its protrusion towards the lumen. The HCN4-expressing cardiomyocytes are clustered at the sinus venous side of SVV. By E10.5, the SVV is clearly developed with an elongated tail (or leaflet) at the sinoatrial border. Meanwhile, the SAN head was formed by the HCN4-expressing cardiomyocytes at the base of SVV, with the HCN4 expression extended along the sinus side of the SVV, which is also called SAN tail.
3.2 Deletion of endocardial Notch1 disrupts the development of SVV
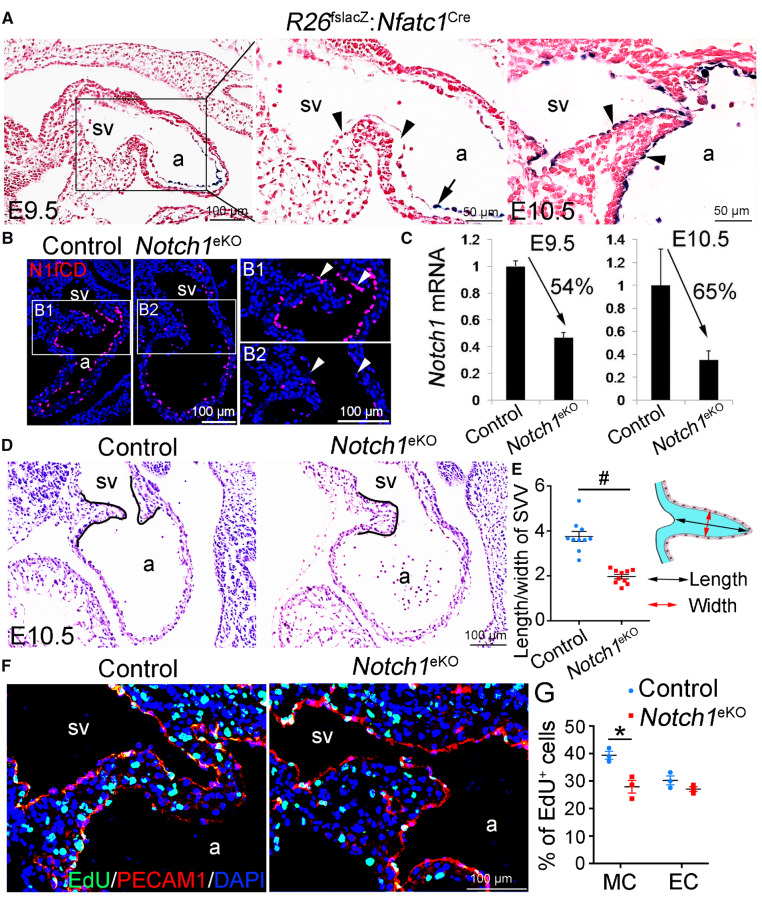
To determine the possible roles of NOTCH1 in the formation of SVV and SAN, we crossed Nfatc1Cremice22 with floxed Notch1 mice24 (Notch1f/f) to generate endocardial Notch1 knockout mice (Notch1f/f: Nfatc1Cre, Notch1eKO thereafter). We first determined the Cre activities in the developing hearts by crossing the Nfatc1Cremouse with R26lacZ reporter line.25 X-gal staining showed that at E9.5 LacZ expression was present in the endocardial cells within the atrium, atrioventricular canal and ventricle, while it was absent in the endocardial cells of the SVV (Figure 2A and Supplementary material online, Figure S2A and B). At E10.5, the LacZ expression was expanded to the endocardial cells of SVV (Figure 2A and Supplementary material online, Figure S2C–E). These results indicated that the Nfatc1Cre started to induce gene recombination in the endocardial cells of SVV around E9.5. Immunofluorescence confirmed that the expression of N1ICD in the endocardial cells of SVV was dramatically reduced in E10.5 Notch1eKO embryos when compared to that in their littermate controls (Figure 2B). Quantitative RT–PCR confirmed that the E9.5 and E10.5 Notch1eKO hearts had significantly reduced levels of Notch1 transcripts (Figure 2C). Additionally, RNA in situ hybridization assays showed that NOTCH target gene Hey1 but not Hey2 was expressed within SVV region of E10.5 control embryos and such expression was reduced in Notch1eKO embryos (Supplementary material online, Figure S3). We noted that the Notch1eKO embryos were grossly normal at E10.5, but they appeared to be underdeveloped and smaller in size at E11.5 and died by E12.5 (Supplementary material online, Figure S4). We therefore analysed the heart histology by HE staining of serial sections crossing the whole heart and found that all the Notch1eKO embryos had more primitive SVV when compared to that in controls (Figure 2D and E and Supplementary material online, Figure S5). In addition, we observed the cushion and trabecular defects that were reported by us previously.21 These observations suggest that endocardial Notch1 is required for the growth of SVV. This notion was confirmed by the EdU labelling that revealed a significant reduction in the proliferation of SVV and atrial cardiomyocytes in the E10.5 Notch1eKO embryos (Figure 2F and G and Supplementary material online, Figure S6). Together, these findings demonstrate that the endocardial Notch1 is essential for the proliferation of SVV cardiomyocytes and the growth of SVV.
Figure 2.
Deletion of endocardial Notch1 results in defective growth of sinus venous valve. (A) X-gal staining shows the expression of LacZ reporter gene activated by the Nfatc1Cre in the forming SVV of E9.5 and E10.5 embryos. Arrows and arrowheads indicate the endocardial cells in the atrium and SVV, respectively. (B) Immunofluorescence shows the cleaved NOTCH1 (N1ICD) expression in the endocardium (arrowhead) of SVV from the E10.5 control (Notch1f/+) and endocardial Notch1 knockout (Notch1f/f: Nfatc1Cre) embryos. (C) qPCR analysis shows the mRNA level of Notch1 in E9.5 or E10.5 hearts of control and Notch1eKO embryos. The expression of Notch1 was normalized to that of Gapdh. Two to three hearts were pooled as one sample (n = 3/group). (D) HE staining shows histology of SVV (outlined by black lines). (E) Quantification of the ratio of the length/width of SVV of E10.5 control and Notch1eKO embryos (n = 10/group). #P < 0.001. (F) EdU labelling shows the proliferating cells (green) in the SVV. The endocardial cells are marked by PECAM1immunostaining (red). (G) Quantification of proliferation rate in the cardiomyocytes (CM) and endocardial cells (EC) of SVV (n = 3/group). *P < 0.05. Unpaired Student’s t-test was used for statistical calculation. a, atrium; sv, sinus venous.
3.3 Deletion of endocardial Notch1 impairs the formation of SAN
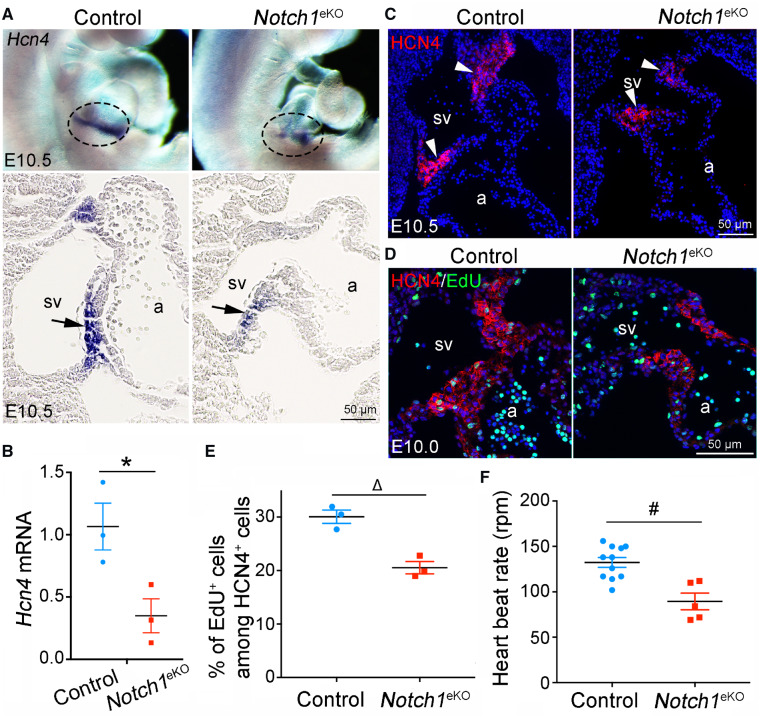
Since SVV and SAN develop concurrently in mouse between E9.5 and E10.5 (Figure 1), we next examined the expression of Hcn4. Whole mount RNA in situ hybridization showed that Hcn4 transcripts marked the forming SAN at the sinoatrial boundary of control embryos at E10.5 (Figure 3A, top panel). In contrast, only a rudimentary SAN with markedly reduced Hcn4 expression was present in the E10.5 Notch1eKO embryos. Sectional analysis of the stained embryos clearly showed that the Hcn4-delineated SAN was present at the base of SVV in the control embryos, while the undersized SAN with much less Hcn4 positive cells was observed in the same region of Notch1eKO embryos (Figure 3A, bottom panel). Consistently, RT–qPCR and immunostaining confirmed a reduced level of HCN4 mRNA and protein respectively in the forming SAN of the E10.5 Notch1eKO embryos (Figure 3B and C). In addition, EdU labelling revealed a reduction in the proliferation of the HCN4-expressing cardiomyocytes in the E10.0 Notch1eKO embryos (Figure 3D and E). SAN generates the primary pacemaker pulse and enables the rhythmic heart contraction. To determine whether the defective SAN affects the heartbeat, we recorded the heartbeat frequencies and the results revealed a significantly reduced heartbeat rate by the E10.5 Notch1eKO hearts when compared to controls (Figure 3F and Supplementary material online, Movies S1 and S2). These results indicate that endocardial Notch1 is also required for the SAN formation and its function.
Figure 3.
Deletion of endocardial Notch1 results in defective development of sinoatrial node. (A) Whole mount RNA in situ hybridization shows that the mRNA level of Hcn4 is markedly reduced in the SAN of E10.5 Notch1eKO embryos when compared to that in control embryos. Note that 3–5 embryos from each genotype were analysed simultaneously. Representative images of whole mount (top panel) or sectional views (bottom panel) show the expression of Hcn4 in the SAN (indicated by circle or arrow) in E10.5 control and Notch1eKO embryos. a, atrium; sv, sinus venous. (B) RT–qPCR analysis of the Hcn4 mRNA expression in E10.5 control and Notch1eKO hearts. The expression of Hcn4 is normalized to that of Gapdh (n = 3/group). *P < 0.05. (C) Immunostaining shows a reduced level of HCN4 protein (arrowhead) in the forming SAN of E10.5 Notch1eKO embryos. (D and E) EdU labelling shows decreased proliferation of the HCN4-expressing cardiomyocytes in the E10.0 Notch1eKO embryos (n = 3/group). ΔP < 0.01. (F) Live recording heart rate indicates a reduced beating rate by the E10.5 Notch1eKO hearts (n = 5) compared with the control hearts (n = 11). #P < 0.001. Unpaired Student’s t-test was used for statistical calculation.
3.4 Deletion of endocardial Notch1 impairs the expression of Tbx5, Tbx18, and Wnt2
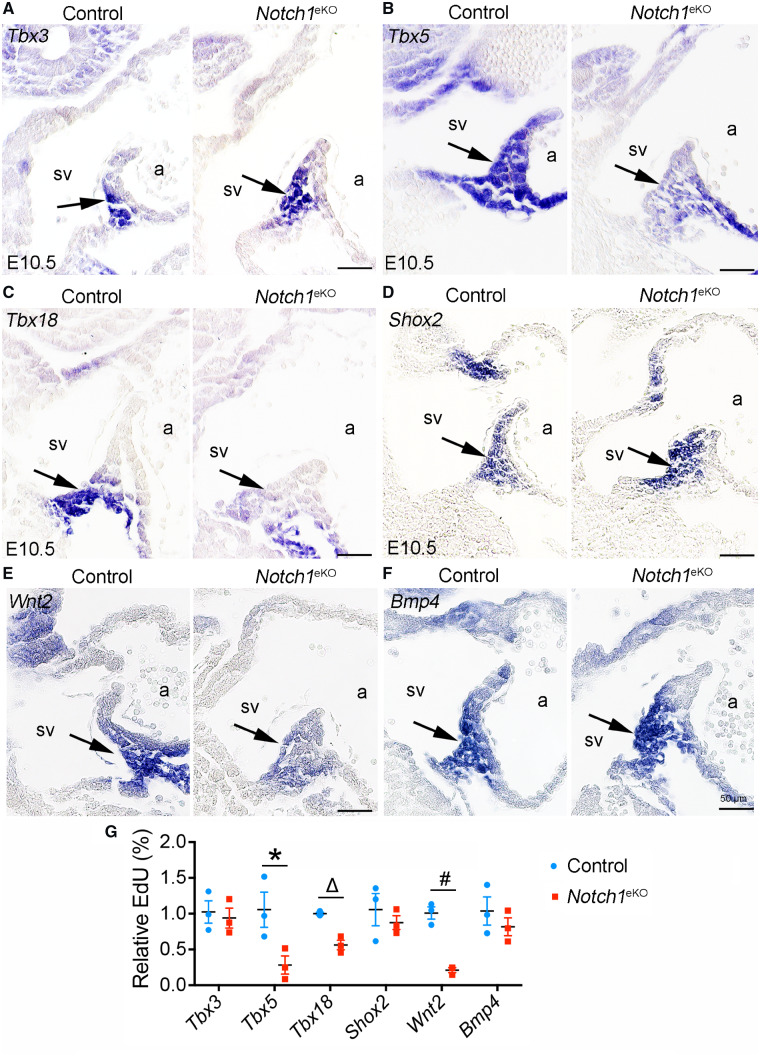
Previous studies have shown that the progenitors arise from the cardiogenic mesoderm to form the SVV and SAN.9 The proliferation and differentiation of these progenitor cells are regulated by T-box transcription factors including Tbx3,10Tbx5,11 and Tbx18.12 We therefore examined the expression of these genes in the E10.5 Notch1eKO embryos by RNA in situ hybridization and qPCR. The results showed that the expression of Tbx3 in SAN was comparable between the control and Notch1eKO embryos (Figure 4A and G). In contrast, the expression of Tbx5 and Tbx18 in the SVV and SAN region were dramatically reduced in E10.5 Notch1eKO embryos (Figure 4B, C, and G). We also examined the expression of Shox2, a key regulator of SVV and SAN development,2,13,14 and found that its expression was not affected in the Notch1eKO embryos (Figure 4D and G). Since Wnt2 and Bmp4 have been implicated in the development of SVV and SAN,26–28 we next examined their expression in the Notch1eKO embryos by RNA in situ hybridization. We found that Wnt2 was predominantly expressed in the SVV and SAN region in control embryos, while such expression was greatly reduced in the Notch1eKO embryos (Figure 4E and G). On the contrary, Bmp4 was expressed at a comparable level in the SVV and SAN region between the control and Notch1eKO embryos (Figure 4F and G). These results suggest that decreased expression of Wnt2 might cause reduced WNT/β-catenin signalling, leading to the SVV and SAN defects in the Notch1eKO embryos.
Figure 4.
Deletion of endocardial Notch1 reduces the expression of Tbx5, Tbx18, and Wnt2. (A–F) RNA in situ hybridization shows the expression of Tbx3 (A), Tbx5 (B), Tbx18 (C), Shox2 (D), Wnt2 (E), and Bmp4 (F) in the E10.5 control and Notch1eKO embryos. The results indicate a reduced expression of Tbx5 and Wnt2 in the forming SAN and SVV, as well as Tbx18 in the forming SAN, of the Notch1eKO embryos. In each experiment, at least three embryos from each genotype were analysed simultaneously for one gene. Representative images are present to show the expression of these genes in SAN/SVV (arrow). a, atrium; sv, sinus venous. (G) RT–qPCR analysis of gene expression in the hearts from E10.5 control and Notch1eKO embryos. The expression of Gapdh is used as internal control (n = 3/group). Unpaired Student’s t-test was used for statistical calculation (n = 3/group). *P < 0.05; ΔP < 0.01; #P < 0.001.
3.5 Endocardial NOTCH1 regulates SVV and SAN formation through myocardial WNT2
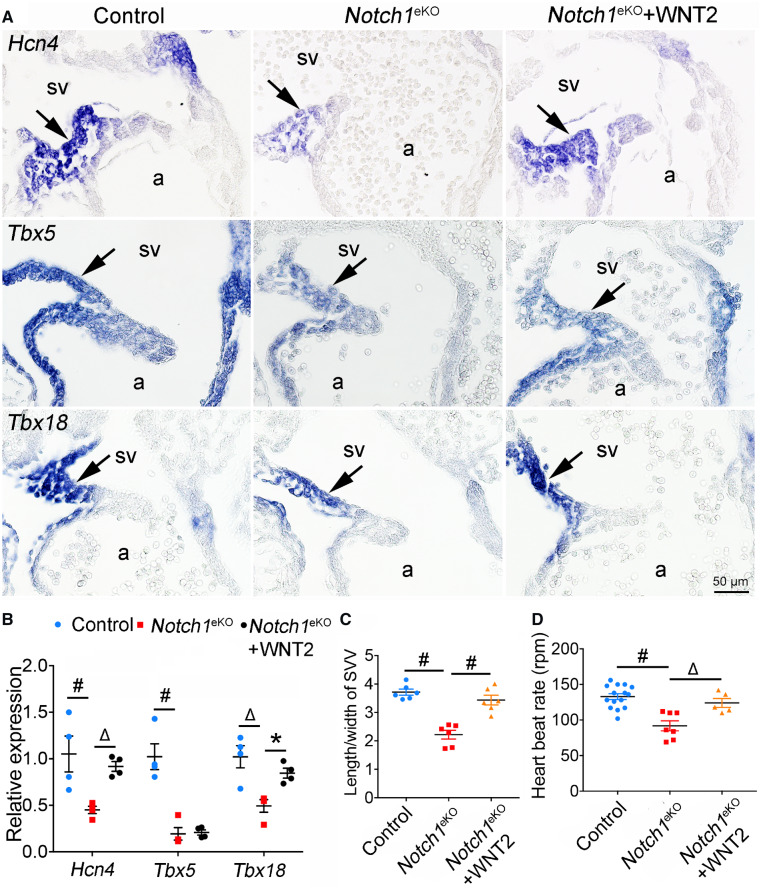
To determine whether myocardial WNT2 acts downstream of the endocardial NOTCH1 signalling to promote the development of SVV and SAN, we performed the rescue experiments by treating the cultured E9.5 Notch1eKO embryos with the recombinant WNT2 protein. The cultured embryos were then subjected to gene expression analysis of Hcn4, Tbx5, and Tbx18 using RNA in situ hybridization and qPCR. The results showed that addition of WNT2 was able to partially restore the expression of Hcn4 and Tbx18 in the Notch1eKO embryos, while it failed to do so for Tbx5 (Figure 5A and B). Of note, the WNT2 treatment also rescued the growth defect of SVV in the Notch1eKO embryos (Figure 5C). In addition, we found that WNT2 treatment restored the heartbeat rate in Notch1eKO embryos (Figure 5D and Supplementary material online, Movies S3–S5). Together, these findings indicate that endocardial NOTCH1 regulates the formation of SVV and SAN, at least partially, through WNT2.
Figure 5.
Endocardial NOTCH1 regulates the formation of SVV and SAN through WNT2. (A) E9.5 control and Notch1eKO embryos were cultured with or without the WNT2 treatment for 24 h. RNA in situ hybridization shows the expression of Hcn4, Tbx5, and Tbx18 in SAN (arrow) of cultured embryos. In each experiment, 3–5 embryos from each group were analysed simultaneously. (B) RT–qPCR analysis of the expression of Hcn4, Tbx5, and Tbx18 in the hearts from cultured embryos. The expression of Gapdh is used as internal control (n = 4/group). (C) Quantification of the length/width ratio of SVV in cultured embryos indicates the rescued SVV growth in the Notch1eKO embryos by WNT2 (n = 6/group). #P < 0.001. (D) Live recording heart rate of cultured embryos. Control (n = 15); Notch1eKO (n = 7); Notch1eKO + WNT2 (n = 5). Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. a, atrium; sv, sinus venous. *P < 0.05; ΔP < 0.01; #P < 0.001.
3.6 WNT/β-catenin signalling is essential for SVV growth
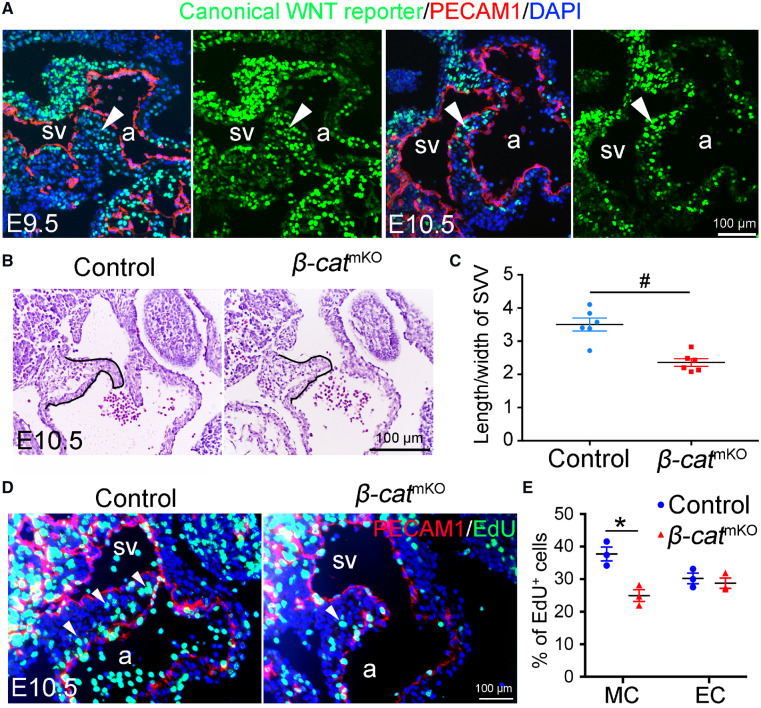
Since WNT2 is a ligand for activating canonical WNT (WNT/β-catenin) signaling,29 we then sought to determine whether the WNT/β-catenin signalling is required for the formation of SVV and SAN. We first traced WNT/β-catenin activities in the developing SVV by using a canonical WNT signalling reporter mouse model in which the expression of GFP was directed by of six tandem TCF/LEF binding sites.30 The results showed that the WNT/β-catenin activities were exclusively present in the SVV myocardium and rare in the SVV endocardium (Figure 6A). This finding suggests that the myocardial WNT/β-catenin activities might be involved in the development of SVV and SAN. To test this idea, we generated myocardial β-catenin knockout mice (β-cateninf/f: TntCre, β-catmKO thereafter) by crossing the floxed β-catenin mice31 with the TntCre mice.32 Histological analysis by HE staining showed that β-catmKO embryos had primitive SVV (Figure 6B and C, and Supplementary material online, Figure S7) that was similar to the phenotypes observed in the Notch1eKO embryos (Figure 2D and E). Consistent with the reduced size of SVV, EdU labelling revealed a significantly decreased cell proliferation in the SVV cardiomyocytes of β-catmKO embryos (Figure 6D and E). RNA in situ hybridization and qPCR were then performed to determine the expression of Hcn4 and the results showed that the deletion of β-catenin in the myocardium had no effect on the expression of Hcn4 (Supplementary material online, Figure S8Aand B). Consistently, β-catmKO embryos had a normal heartbeat rate (Supplementary material online, Figure S8C). Together, these results indicate that the WNT/β-catenin signalling in the myocardium is required for the growth of SVV, but not for the formation of SAN.
Figure 6.
Deletion of myocardial β-catenin impairs the growth of SVV. (A) A canonical WNT signalling reporter line shows the canonical WNT activities (arrowhead) in the SVV/SAN region of E9.5 or E10.5 hearts. Endocardial cells were marked by immunostaining for PECAM1 (red). (B) HE staining shows the histology of SVV (outlined by black lines) in E10.5 control and myocardial β-catenin knockout (β-catmKO) embryos. (C) Quantification of the ratio of the length/width of SVV in the E10.5 control and β-catmKO embryos (n = 6/group). #P < 0.001. (D) EdU labelling shows the proliferating cells (green) in SVV, while endocardial cells are marked by PECAM1immunostaining (red). (E) Quantification of cell proliferation rate in the cardiomyocytes (CM) and endocardial cells (EC) within the SVV of E10.5 control and β-catmKO embryos (n = 3/group). *P < 0.05. Unpaired Student’s t-test was used for statistical calculation. a, atrium; sv, sinus venous.
3.7 Endocardial NOTCH1 regulates the elongation of SVV through myocardial NRG1
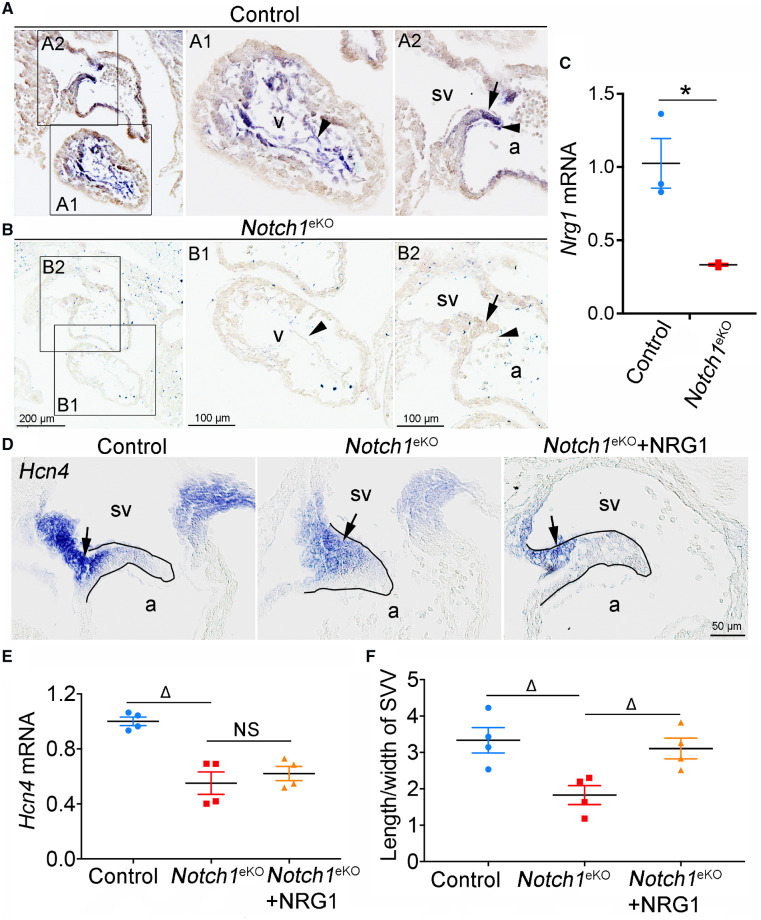
Nrg1 is specifically expressed in the endocardium and known to mediate the endocardial NOTCH signalling to regulate myocardial gene expression during cardiac chamber formation.11,19,33 To determine whether Nrg1 plays a similar role in the development of SVV and SAN, we examined its expression in the Notch1eKO embryos by RNA in situ hybridization and qPCR. We found that the expression of Nrg1 in the ventricular endocardium was dramatically reduced in the Notch1eKO embryos compared to that of controls (Figure 7A–C). Unexpectedly, we noted that Nrg1 was expressed in both endocardium and myocardium of SVV tail in the control embryos and such expression was markedly reduced in the Notch1eKO embryos (Figure 7A–C). Consistent with its expression pattern, addition of NRG1 in the cultured E9.5 Notch1eKO embryos for 24 h was able to rescue the SVV growth defect (Figure 7D and F), but not the expression of Hcn4 and Wnt2 in the SVV head (Figure 7D and E and Supplementary material online, Figure S9). These findings suggest that NRG1 acts downstream of endocardial NOTCH1 to promote the SVV growth.
Figure 7.
Endocardial NOTCH1 regulates the SVV formation through myocardial NRG1. (A and B), RNA in situ hybridization shows the expression of Nrg1 in E10.5 control and Notch1eKO embryos. The high magnification of ventricle (A1 and B1) and SVV (A2 and B2) regions is showed on the right. The arrowhead and arrow indicate the endocardium and myocardium respectively (n = 3/group). (C) RT–qPCR analysis of the Nrg1 mRNA expression in E10.5 control and Notch1eKO hearts. The expression of Nrg1 was normalized to that of Gapdh (n = 3/group). (D) RNA in situ hybridization shows the expression of Hcn4 (arrow) in the cultured E9.5 control and Notch1eKO embryos with or without the NRG1 treatment for 24 h. a, atrium; sv, sinus venous. (E) RT–qPCR analysis of the Hcn4 mRNA expression. The expression of Hcn4 is normalized to that of Gapdh (n = 4/group). (F) Quantification of the ratio of the length/width of SVV in cultured embryos indicates the rescued SVV growth in the Notch1eKO embryos by NRG1 (n = 4/group). Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. *P < 0.05, ΔP < 0.01.
4. Discussion
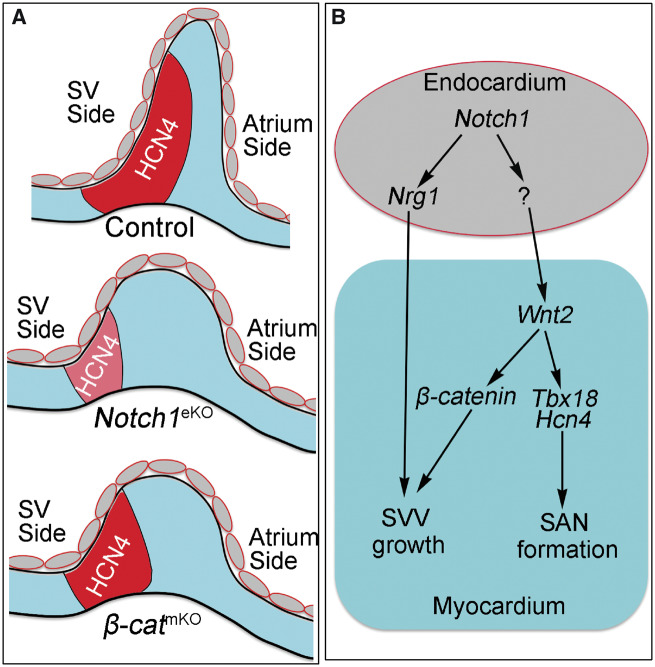
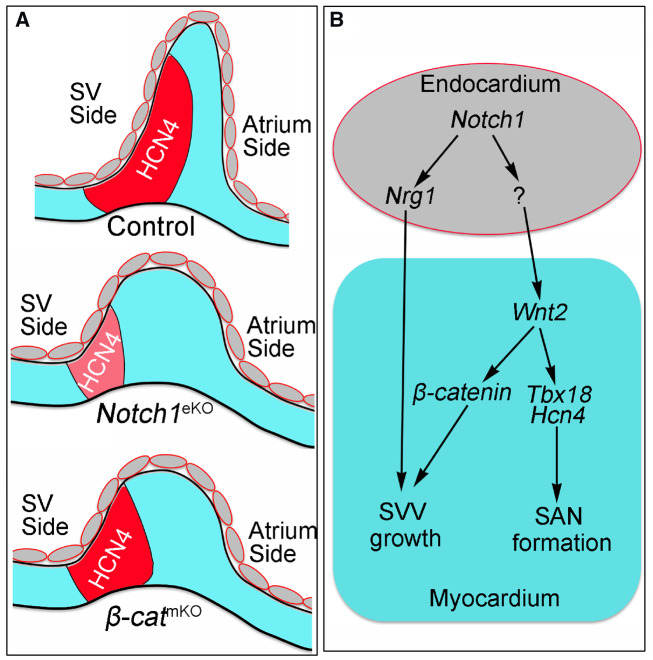
NOTCH signalling mediates signal communications between neighbouring cells and play multiple roles during heart development.34 NOTCH signalling critically regulates early cardiomyocyte differentiation,35,36 trabeculation,19,37 heart valve development,21,38 atrioventricular node formation,39 and coronary artery development.40,41 Here we present the first genetic evidence that demonstrates a previous unknown role of endocardial NOTCH1 in regulation of SVV and SAN development (Figure 8A). Collectively, our data support that endocardial Notch1 controls the formation of SVV and SAN through regulating Wnt2 and Nrg1 (Figure 8B).
Figure 8.
Schematic summary of the roles of endocardial Notch1 in SVV and SAN formation. (A) A carton shows the normal development of SVV and SAN in the E10.5 control Notch1eKO or β-catmKO embryos. Red area marks the HCN4-expressing SAN (head and tail). The expression of HCN4 is reduced in the Notch1eKO embryos, but not affected in the β-catmKO embryos when compared to that in controls, whereas the elongation of SVV is affected in both mutant embryos. (B) A schematic working model shows the possible signalling mechanisms through which endocardial Notch1 regulates the SVV growth and SAN formation. Endocardial Notch1 promotes the SVV growth through regulating Nrg1 expression in the endocardium. Endocardial Notch1 also regulates myocardial Wnt2 to promote the SVV growth through β-catenin, as well as supports the SAN formation and maturation through regulating Tbx18.16 The downstream mediators of Notch1 in the endocardium that regulate Wnt2 expression in myocardium are unknown and future studies are needed to identify them.
We specifically deleted Notch1 in the endocardium, without affecting the vascular endothelium.21 The endocardial Notch1 knockout embryos died around E11.5, thereby allowing us to study the functions of Notch1 in heart development between E9.5 and E11.5. We found that loss of Notch1 results in the primitive SVV that may cause blood regurgitation from the atrium to the sinus venous. In addition, loss of Notch1 leads to markedly reduced expression of Hcn4, a functional marker specific for SAN that generate the primary pacemaker pulse.7,42 The reduced heartbeat rate in the endocardial Notch1 knockout embryos further suggests a defective SAN function. Together, the primitive SVV and defective SAN function may contribute to the embryonic lethality of the endocardial Notch1 knockout embryos, in addition to the chamber and endocardial cushion defects.21
Previous studies have uncovered that the development of SAN is achieved through highly localized suppression of the working cardiomyocyte differentiation by a transcriptional network consisting of Tbx3, Tbx5, Tbx18, and Shox2.6,9,14,43Tbx18, initially expressed in a distinct precursor cell population within the septum transversum for the sinus venous horn myocardium, is required for the specification, differentiation, and localization of these cells into the myocardium to form the SAN head.9,12,44 In addition, Tbx3 and Shox2 are essential for the establishment of SAN gene program and the differentiation of SAN cardiomyocytes.2,10,12–14,45 Our data showed that deletion of Notch1 significantly reduced the expression of Tbx18 but had no effect on the expression of Tbx3 and Shox2. These findings suggest that endocardial NOTCH1 might recruit the TBX18-expressing cardiomyocytes and/or promote their proliferation during the formation of SAN. Thus, the reduced expression of Hcn4 in the endocardial Notch1 knockout embryos represents the underdeveloped SAN structure, not an early differentiation defect. It is worth to discuss that, while previous genetic studies have revealed a link between Tbx5, Shox2, and Bmp4 in the developing SAN where Tbx5 regulates Bmp4 expression through Shox2,11 our data show that the endocardial Notch1 knockout embryos had reduced expression of Tbx5 in the developing SVV and SAN, whereas the expression levels of Shox2 and Bmp4 are not changed. One explanation for this discrepancy between our and the previous study is that the reduced Tbx5 expression in the endocardial Notch1 knockout embryos is less dramatic and thus not sufficient to have an impact on the Shox2 expression.
To explore the molecular mechanisms by which the endocardial NOTCH1 promotes myocardial cell proliferation and SVV growth, we examined the expression of candidate genes involved in the development of SVV and SAN. Wnt2 has been reported to induce the WNT/β-catenin signalling that promotes cell proliferation and cardiac inflow tract development.29 Interestingly, we found that the expression of Wnt2 in the SVV and SAN myocardium was significantly reduced in the Notch1 knockout embryos. In addition, we observed a high level of WNT/β-catenin signalling in the SVV and SAN myocardium of the transgenic mice that specifically report the canonical WNT signalling. These observations suggest that Wnt2 might be involved in the formation of SVV and SAN by activating the WNT/β-catenin signalling. Indeed, our rescue experiments showed that recombinant WNT2 could rescue the SVV growth defect and partially restore the expression of Hcn4 in the cultured Notch1eKO embryos. Furthermore, we showed that disruption of β-catenin in the myocardium recapitulated the SVV growth defect resulted from the deletion of Notch1 in the endocardium. However, loss of β-catenin had no effect on the expression of Hcn4. Consistent with these observations, Norden et al.27 have shown that the WNT/β-catenin signalling is required to maintain the proliferation of the Tbx18-expressing mesenchymal progenitors, but not for the later formation of SAN. Taken together, these observations support that WNT2 regulates the development of SVV through a β-catenin dependent manner, while it modulates the SAN formation independent of β-catenin.
One limitation of our study is that we do not know the downstream factors that mediate the endocardial NOTCH1 signalling to induce the expression of Wnt2 in the myocardium. One such candidate would be Nrg1, since previous studies have shown that Nrg1 works downstream of NOTCH1 signalling in the endocardium to regulate myocardial development including the central cardiac conduction system.19,33,46,47 We found Nrg1 expression in both endocardium and myocardium of SVV in control embryos and this expression was markedly reduced in Notch1eKO embryos. Furthermore, addition of NRG1 was able to rescue the SVV growth defect resulted from the endocardial Notch1 deletion, suggesting that NRG1 acts downstream of endocardial NOTCH1 signalling to promote SVV growth. This finding is consistent with the requirement of NOTCH1-NRG1 signalling in the ventricular endocardium for trabeculation,19,33 suggesting a common NOTCH1-NRG1 mechanism regulating SVV elongation and trabecular protrusion. In contrast, exogenous NRG1 failed to restore the expression of Hcn4 and Wnt2, indicating that the formation of SAN and the expression of Wnt2 expression are not dependent on the endocardial NOTCH-NRG1 signalling. Future studies are required to identify other endocardial factors that relay the NOTCH signalling to the SVV and SAN myocardium.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Supplementary Material
Acknowledgement
We thank Dr Diana Klein for providing WNT2 conditioned media.
Conflict of interest: none declared.
Funding
This work was supported by grants from the American Heart Association (13POST16970056 to Y.W.); and the National Heart, Lung, and Blood Institute (R01HL111770, R01HL116997, R01HL133120 to B.Z.).
References
- 1. van Weerd JH, Christoffels VM.. The formation and function of the cardiac conduction system. Development 2016;143:197–210. [DOI] [PubMed] [Google Scholar]
- 2. Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, Chen Y.. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev Biol 2009;327:376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pennisi DJ, Rentschler S, Gourdie RG, Fishman GI, Mikawa T.. Induction and patterning of the cardiac conduction system. Int J Dev Biol 2002;46:765–775. [PubMed] [Google Scholar]
- 4. Gomez Stallons MV, Wirrig-Schwendeman EE, Fang M, Cheek JD, Alfieri CM, Hinton RB, Yutzey KE, Molecular mechanisms of heart valve development and disease In Nakanishi T, Markwald RR, Baldwin HS, Keller BB, Srivastava D, Yamagishi H (eds). Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology. Tokyo: Springer; 2016. p145–151. [Google Scholar]
- 5. Blom NA, Gittenberger-de Groot AC, DeRuiter MC, Poelmann RE, Mentink MM, Ottenkamp J.. Development of the cardiac conduction tissue in human embryos using HNK-1 antigen expression: possible relevance for understanding of abnormal atrial automaticity. Circulation 1999;99:800–806. [DOI] [PubMed] [Google Scholar]
- 6. Christoffels VM, Smits GJ, Kispert A, Moorman AF.. Development of the pacemaker tissues of the heart. Circ Res 2010;106:240–254. [DOI] [PubMed] [Google Scholar]
- 7. Stieber J, Herrmann S, Feil S, Loster J, Feil R, Biel M, Hofmann F, Ludwig A.. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A 2003;100:15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Munshi NV. Gene regulatory networks in cardiac conduction system development. Circ Res 2012;110:1525–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mommersteeg MT, Dominguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF, Christoffels VM.. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res 2010;87:92–101. [DOI] [PubMed] [Google Scholar]
- 10. Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM.. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev 2007;21:1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H, Rappold G.. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum Mol Genet 2010;19:4625–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM.. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res 2009;104:388–397. [DOI] [PubMed] [Google Scholar]
- 13. Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Schöler H, Feitsma H, Rottbauer W, Blum M, Meijlink F, Rappold G, Gittenberger-de Groot AC.. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 2007;115:1830–1838. [DOI] [PubMed] [Google Scholar]
- 14. Liu H, Espinoza-Lewis RA, Chen C, Hu X, Zhang Y, Chen Y.. The role of Shox2 in SAN development and function. Pediatr Cardiol 2012;33:882–889. [DOI] [PubMed] [Google Scholar]
- 15. Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, Tan HL, Moorman AF T, Hoen PA, Christoffels VM.. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res 2012;94:439–449. [DOI] [PubMed] [Google Scholar]
- 16. Kapoor N, Liang W, Marban E, Cho HC.. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol 2013;31:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Espinoza-Lewis RA, Liu H, Sun C, Chen C, Jiao K, Chen Y.. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev Biol 2011;356:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacGrogan D, Munch J, de la Pompa JL.. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat Rev Cardiol 2018;15:685.. [DOI] [PubMed] [Google Scholar]
- 19. Grego-Bessa J, Luna-Zurita L, del Monte G, Bolós V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama Y-S, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Pérez-Pomares JM, de la Pompa JL.. Notch signaling is essential for ventricular chamber development. Dev Cell 2007;12:415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK.. Essential role of endothelial Notch1 in angiogenesis. Circulation 2005;111:1826–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y, Wu B, Chamberlain AA, Lui W, Koirala P, Susztak K, Klein D, Taylor V, Zhou B.. Endocardial to myocardial notch-wnt-bmp axis regulates early heart valve development. PLoS One 2013;8:e60244.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez RA, Markwald RR, O’Rourke BP, Sharp DJ, Zheng D, Lenz J, Baldwin HS, Chang C-P, Zhou B.. Endocardial cells form the coronary arteries by angiogenesis through myocardial–endocardial VEGF signaling. Cell 2012;151:1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klein D, Demory A, Peyre F, Kroll J, Augustin HG, Helfrich W, Kzhyshkowska J, Schledzewski K, Arnold B, Goerdt S.. Wnt2 acts as a cell type-specific, autocrine growth factor in rat hepatic sinusoidal endothelial cells cross-stimulating the VEGF pathway. Hepatology 2008;47:1018–1031. [DOI] [PubMed] [Google Scholar]
- 24. Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J.. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol 2004;269:81–94. [DOI] [PubMed] [Google Scholar]
- 25. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999;21:70–71. [DOI] [PubMed] [Google Scholar]
- 26. Schlueter J, Manner J, Brand T.. BMP is an important regulator of proepicardial identity in the chick embryo. Dev Biol 2006;295:546–558. [DOI] [PubMed] [Google Scholar]
- 27. Norden J, Greulich F, Rudat C, Taketo MM, Kispert A.. Wnt/beta-catenin signaling maintains the mesenchymal precursor pool for murine sinus horn formation. Circ Res 2011;109:e42–e50. [DOI] [PubMed] [Google Scholar]
- 28. Norden J, Kispert A.. Wnt/Ctnnb1 signaling and the mesenchymal precursor pools of the heart. Trends Cardiovasc Med 2012;22:118–122. [DOI] [PubMed] [Google Scholar]
- 29. Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, Morrisey EE.. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev Cell 2010;18:275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, Hadjantonakis AK.. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC Dev Biol 2010;10:121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R.. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 2001;128:1253–1264. [DOI] [PubMed] [Google Scholar]
- 32. Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL.. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev 2003;17:2362–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai D, Liu X, Forrai A, Wolstein O, Michalicek J, Ahmed I, Garratt AN, Birchmeier C, Zhou M, Hartley L, Robb L, Feneley MP, Fatkin D, Harvey RP.. Neuregulin 1 sustains the gene regulatory network in both trabecular and nontrabecular myocardium. Circ Res 2010;107:715–727. [DOI] [PubMed] [Google Scholar]
- 34. MacGrogan D, Nus M, de la Pompa JL.. Notch signaling in cardiac development and disease. Curr Top Dev Biol 2010;92:333–365. [DOI] [PubMed] [Google Scholar]
- 35. Rones MS, McLaughlin KA, Raffin M, Mercola M.. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development 2000;127:3865–3876. [DOI] [PubMed] [Google Scholar]
- 36. Schroeder T, Fraser ST, Ogawa M, Nishikawa S, Oka C, Bornkamm GW, Nishikawa S, Honjo T, Just U.. Recombination signal sequence-binding protein Jkappa alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc Natl Acad Sci U S A 2003;100:4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Del Monte-Nieto G, Ramialison M, Adam AAS, Wu B, Aharonov A, D'Uva G, Bourke LM, Pitulescu ME, Chen H, de la Pompa JL, Shou W, Adams RH, Harten SK, Tzahor E, Zhou B, Harvey RP.. Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature 2018;557:439–445. [DOI] [PubMed] [Google Scholar]
- 38. Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL.. Notch promotes epithelial–mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 2004;18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rentschler S, Harris BS, Kuznekoff L, Jain R, Manderfield L, Lu MM, Morley GE, Patel VV, Epstein JA.. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest 2011;121:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y, Wu B, Lu P, Zhang D, Wu B, Varshney S, Del Monte-Nieto G, Zhuang Z, Charafeddine R, Kramer AH, Sibinga NE, Frangogiannis NG, Kitsis RN, Adams RH, Alitalo K, Sharp DJ, Harvey RP, Stanley P, Zhou B.. Uncontrolled angiogenic precursor expansion causes coronary artery anomalies in mice lacking Pofut1. Nat Commun 2017;8:578.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. del Monte G, Casanova JC, Guadix JA, MacGrogan D, Burch JB, Perez-Pomares JM, de la Pompa JL.. Differential Notch signaling in the epicardium is required for cardiac inflow development and coronary vessel morphogenesis. Circ Res 2011;108:824–836. [DOI] [PubMed] [Google Scholar]
- 42. Baruscotti M, Bucchi A, Viscomi C, Mandelli G, Consalez G, Gnecchi-Rusconi T, Montano N, Casali KR, Micheloni S, Barbuti A, DiFrancesco D.. Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4. Proc Natl Acad Sci U S A 2011;108:1705–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu H, Chen CH, Espinoza-Lewis RA, Jiao Z, Sheu I, Hu X, Lin M, Zhang Y, Chen Y.. Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J Biol Chem 2011;286:17029–17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, Kispert A.. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res 2006;98:1555–1563. [DOI] [PubMed] [Google Scholar]
- 45. Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM.. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res 2004;62:489–499. [DOI] [PubMed] [Google Scholar]
- 46. Rentschler S, Zander J, Meyers K, France D, Levine R, Porter G, Rivkees SA, Morley GE, Fishman GI.. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A 2002;99:10464–10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA.. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 2006;133:1125–1132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.