Summary
Septins form rod-shaped heterooligomeric complexes that assemble into filaments and other higher-order structures such as rings or hourglasses at the cell division site in fungal and animal cells [1–4] to carry out a wide range of functions including cytokinesis and cell morphogenesis. However, the architecture of the septin higher-order assemblies and their control mechanisms, including the regulation by conserved kinases [5, 6], remain largely unknown. In the budding yeast Saccharomyces cerevisiae, the five mitotic septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) localize to the bud neck and form an hourglass before cytokinesis that acts as a scaffold for proteins involved in multiple processes as well as a membrane diffusible barrier between the mother and developing bud [7–9]. The hourglass is remodeled into a double ring that sandwiches the actomyosin ring at the onset of cytokinesis [10–13]. How the septins are assembled into a highly ordered hourglass structure at the division site [13] is largely unexplored. Here we show that the LKB1-like kinase Elm1, which has been implicated in septin organization [14], cell morphogenesis [15], and mitotic exit [16, 17], specifically associates with the septin hourglass during the cell cycle, and controls hourglass assembly and stability, especially for the daughter half, by regulating filament pairing as well as the functionality of its substrate, the septin-binding protein Bni5. This study illustrates how a protein kinase regulates septin architecture at the filament level, and suggests that filament pairing is a highly regulated process during septin assembly and remodeling in vivo.
Keywords: Septin, Elm1 kinase, septin-binding protein Bni5, cytokinesis, morphogenesis, scaffold
Graphical Abstract
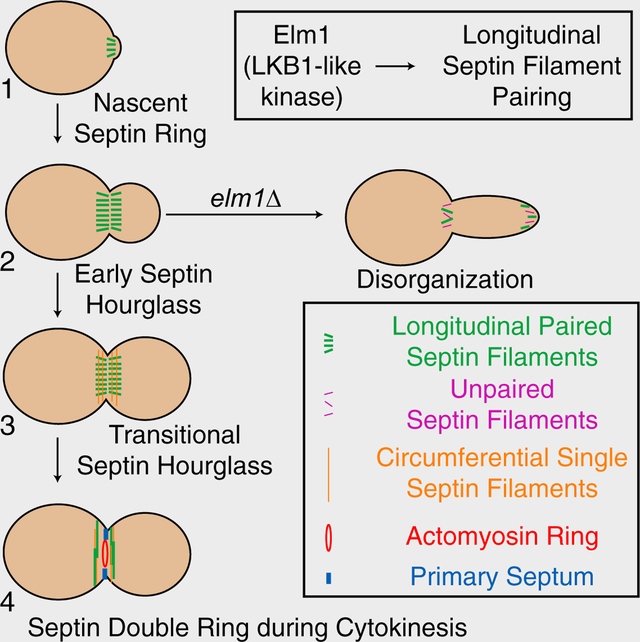
eTOC Blurb:
Protein kinases are known to regulate septin functions, but their impact on septin architecture at the filament level remains unknown. Marquardt et al. show that the LKB1-like kinase Elm1 in budding yeast controls septin hourglass assembly and stability at the cell division site by regulating filament pairing via Bni5 and other targets.
RESULTS AND DISCUSSION
Elm1 preferentially stabilizes the daughter half of the septin hourglass at the cell division site before cytokinesis
Protein kinases regulate septin functions in many different settings. For example, protein kinase A phosphorylates SEPT12 to control septin ring assembly at the annulus of spermatozoa [6, 18]. A number of protein kinases also regulate septin organization at the cell division site (i.e. the bud neck) in budding yeast [5]. However, their precise impact on septin architecture at the filament level remains virtually unknown.
Loss of the LKB1-like kinase Elm1 causes a drastic mislocalization of septins (Cdc3-GFP) to the growing bud tip in nearly 100% of the cells (Figure S1A) [14]. Using time-lapse analysis, we found that assembly of the nascent septin ring prior to bud emergence was largely unperturbed in elm1Δ cells (Figures 1A and 1B). Strikingly, a subset of neck-localized septins began to migrate into the cortex of a growing bud approximately 12–15 min after bud emergence (Figures 1A and 1B, and Video S1), suggesting that the septin hourglass becomes unstable at a specific point of the cell cycle. Kymograph analysis suggested that the daughter half of the hourglass appeared to be preferentially delocalized in elm1Δ cells (Figure S1B). In support of this conclusion, we found that 82% of Bni4, which normally localizes to the mother side of the septin hourglass [19, 20], retained at the bud neck in elm1Δ cells (Figures 1C and 1D). In contrast, Kcc4 and Hsl1, which are normally associated with the daughter side of the hourglass [21, 22], fully migrated to the bud tip of elm1Δ cells (Figures 1C, 1D, S1C, and S1D). These data, together with the observation that Elm1 associated exclusively with the septin hourglass (Figure 1E), suggest that Elm1 plays a critical role in regulating septin hourglass assembly and/or stability.
Figure 1. Elm1 stabilizes the daughter half of the septin hourglass.
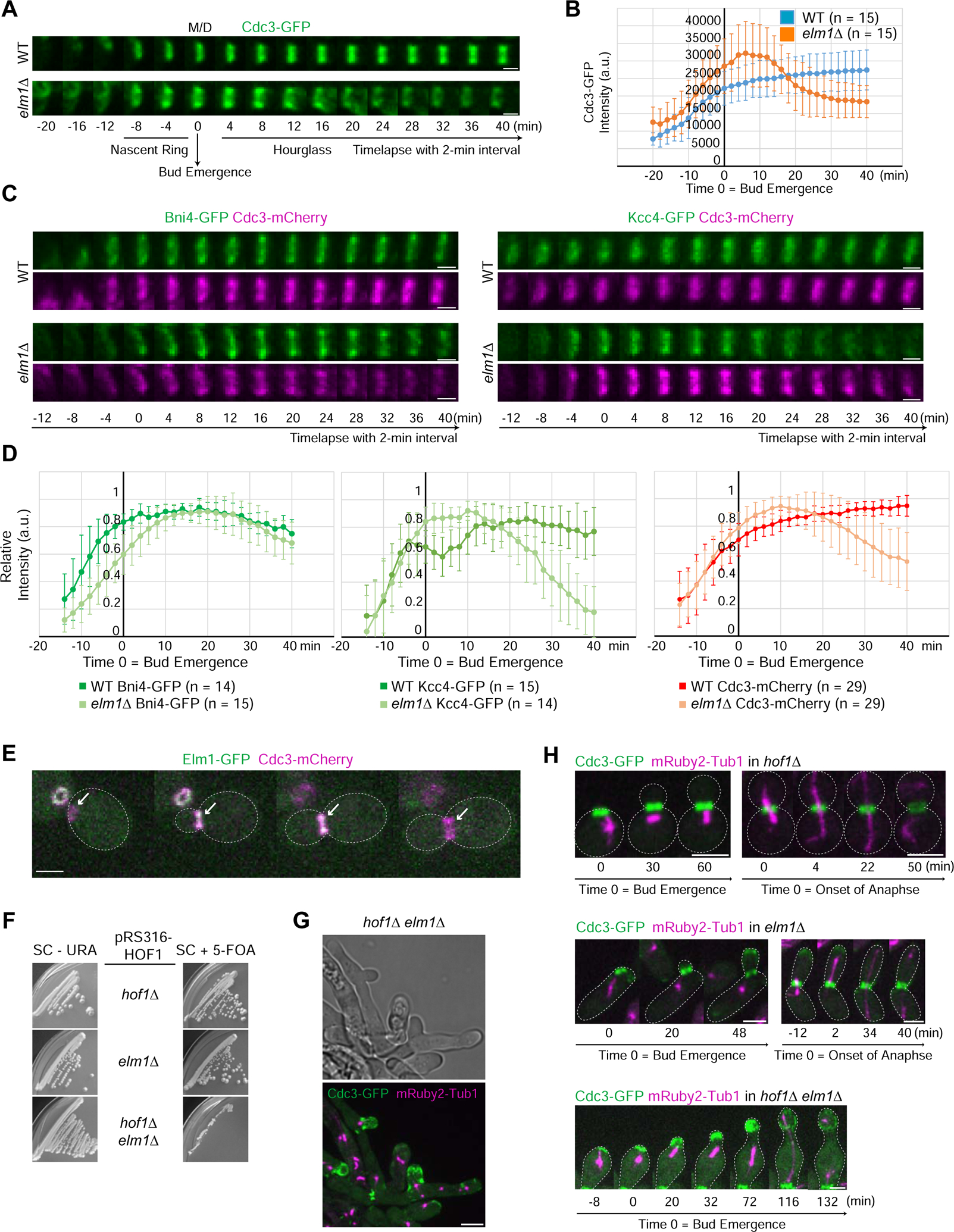
(A) Montages of representative cells of WT (YEF8102) and elm1Δ (YEF8393) strains showing maximum-intensity projections of Cdc3-GFP from 20 min before to 40 min after bud emergence with selected frames from time-lapse series taken with a 2-min interval. For this and all subsequent montages, the mother (M) side is to the left and the daughter (D) side is to the right. T = 0 is bud emergence unless indicated otherwise. Scale bar = 1 μm. See also Figure S1 and Video S1.
(B) Quantification of cells in (1A). Shown is background (outside the cell) subtracted intensity of Cdc3-GFP from the sum projection of given number cells for each strain. The mean is plotted with error bars being the standard deviation. A.U. = arbitrary units.
(C) Montages of representative cells of WT and elm1Δ strains showing maximum-intensity projections of indicated fluorescent protein from 12 min before to 40 min after bud emergence with selected frames from time-lapse series taken with a 2-min interval. Strains used are as follows: YEF8817 (WT BNI4-GFP CDC3-mCherry), YEF8914 (elm1Δ BNI4-GFP CDC3-mCherry), YEF8818 (WT KCC4-GFP CDC3-mCherry), and YEF8915 (elm1Δ KCC4-GFP CDC3-mCherry). Scale bars = 1 μm. See also Figure S1.
(D) Quantification of cells in (1C). Shown is cytoplasmic subtracted intensity of indicated fluorescent protein from the sum projection normalized to the maximum value measured in given number cells for each strain. The mean is plotted with error bars being the standard deviation.
(E) Representative images taken from a time-lapse of strain YEF9305 showing Elm1-GFP localization (green) in relation to septin structures at the bud neck (Cdc3-mCherry; magenta). Gray dotted line is the cell periphery, scale bar = 2 μm.
(F) Growth assay of strains YEF10334 (hof1Δ pRS316-HOF1), YEF10356 (elm1Δ pRS316-HOF1), and YEF10364 (elm1Δ hof1Δ pRS316-HOF1) grown on either SC-URA (left) or SC+5-FOA (right) to select for the loss of the cover plasmid pRS316-HOF1. Plates were grown for 10 days at 25°C.
(G) Representative images of elm1Δ hof1Δ strain grown in YM-1 with CDC3-GFP (septin) in green and mRuby2-TUB1 (tubulin) in magenta. Cells of YEF10364 were pre-grown on SC+5-FOA plates to select for the loss of the cover plasmid pRS316-HOF1. Image is maximum projection. Arrowheads indicate septins retained on the membrane with positive curvature. Scale bar = 5 μm.
(H) Montages of representative cells of indicated strains grown in YM-1 with CDC3-GFP (septin) in green and mRuby2-TUB1 (Tubulin) in magenta. Strains were pre-grown on SC+5-FOA plates to select for the loss of the cover plasmid pRS316-HOF1. Strains used are as follows from top to bottom: YEF10334 (hof1Δ), YEF10356 (elm1Δ), and YEF10364 (elm1Δ hof1Δ). Images are maximum projections. Gray dotted line is the cell periphery, scale bars = 3 μm.
To address why the septin hourglass is preferentially lost at the daughter side in elm1Δ cells, we hypothesized that membrane-associated proteins, which normally localize to the mother side of the hourglass, may stabilize the remaining septins at the bud neck. Deletion of the coding sequence for such a candidate protein, in combination with elm1Δ, would result in a complete loss of septins at the bud neck. Among all the candidate proteins (Syp1, Bud3, Bud4, Bni4, and Hof1) [23–25], we found that only the F-BAR protein Hof1 yielded the expected results. Remarkably, holf1Δ was synthetically sick with elm1Δ (Figure 1F), and the double mutant cells formed extensive, branched chains and elongated buds, indicative of defects in cytokinesis and morphogenesis (Figure 1G). Time-lapse analysis indicated that the double mutant differs from the elm1Δ single mutant in at least two septin hourglass-related phenotypes: 1) the entire septin hourglass was destabilized and moved towards the bud tip in 49% (N =35) cells, which never happened in the elm1Δ single mutant (Figure 1H) and 2) the amount of septins retained at the bud neck appeared to correlate positively with the positiveness of the membrane curvature (Figure 1G, arrowheads). These data imply that the entire septin hourglass is rendered unstable by the deletion of ELM1, but the mother side of the defective hourglass is selectively retained by Hof1, leading to daughter side-specific migration of the hourglass into the bud tip. Thus, Hof1 plays a critical role in the asymmetric stabilization of the septin hourglass before cytokinesis by anchoring the septins to the membrane or by maintaining the membrane curvature.
Surprisingly, the bud tip localization of septins persisted in elm1Δ cells until after the onset of anaphase and completely disappeared by the end of anaphase, as judged by the mitotic spindle geometry (Figure S1E). Importantly, the fluorescent intensity of neck-localized septins increased concurrent with disappearance of the cortical septin signal (Figure S1E; Video S1). This suggests the existence of a mechanism for septin retention at the bud tip before anaphase as well as a mechanism for releasing septins from the bud tip and allowing them to be re-assembled at the bud neck prior to cytokinesis.
Elm1 controls septin hourglass stability via kinase-dependent and -independent mechanisms
The kinase activity of Elm1 is essential for its substrates Hsl1 and Kin4-mediated roles in the morphogenesis and spindle-positioning checkpoint, respectively [16, 17, 26]. To determine whether the kinase activity of Elm1 is also required for septin hourglass stability, we replaced the endogenous gene with a kinase-dead (KD) allele of ELM1, elm1K117R (Elm1KD hereafter) [27]. Surprisingly, only ~70% of the elm1KD cells showed an elongated bud morphology accompanied by mislocalized septins at the bud tip (Figure S2A, yellow arrows). In the remaining 30% of the cells, the bud was round with normal septin localization (Figure S2A, yellow arrowheads). These data suggest that Elm1 may have kinase-dependent and -independent roles in septin organization.
Elm1KD exhibits impaired bud-neck localization [28]. We confirmed this observation using a strain carrying either a GFP- or mApple-tagged elm1KD. Elm1KD was greatly diminished from the bud neck throughout the cell cycle in 100% of elongated cells (Figure S2B), while showing stronger signal in the remaining round cells (Figure 2A, yellow arrow). To test the possibility that the mutant phenotype may be due to a lack of localization, we artificially tethered Elm1KD to the bud neck using the GFP-nanobody/binding peptide (GBP) system [29, 30]. Tagging elm1KD with mApple-GBP at its C-terminus in cells expressing a GFP-tagged core septin (Cdc3 or Cdc10) led to a partial suppression of the elongated-bud and septin-delocalization phenotypes (Figure S2C). Tethering Elm1KD to either terminal septin subunit (Cdc11 or Shs1) led to an even better rescue (Figures 2A, 2B, and S2C). The pattern of rescue was similar in an alternative forced targeting experiment, in which we used an analog-sensitive (AS) allele of ELM1 (elm1AS) [31] to inhibit the kinase activity (Figure S2D). Together, these results demonstrate that the major function of the kinase activity is to target Elm1 to the appropriate position on the septin hourglass. Elm1 can then carry out its role in septin organization largely independent of robust kinase activity. When we repeated the experiment using an N-terminal construct (aa 1–420, known to be cytoplasmic on its own [17]), only the kinase-active Elm11–420 tethered to the septins (via subunit Shs1) was able to rescue the elm1Δ phenotypes, while the kinase-dead Elm11–420 did not exhibit any rescue (Figures S2E and S2F). These results suggest that Elm1 must have substrates at the bud neck that control septin organization, and that the last 220 amino acids are required for a kinase-independent role in septin organization.
Figure 2. The kinase activity and neck localization of Elm1 are critical for its septin function.
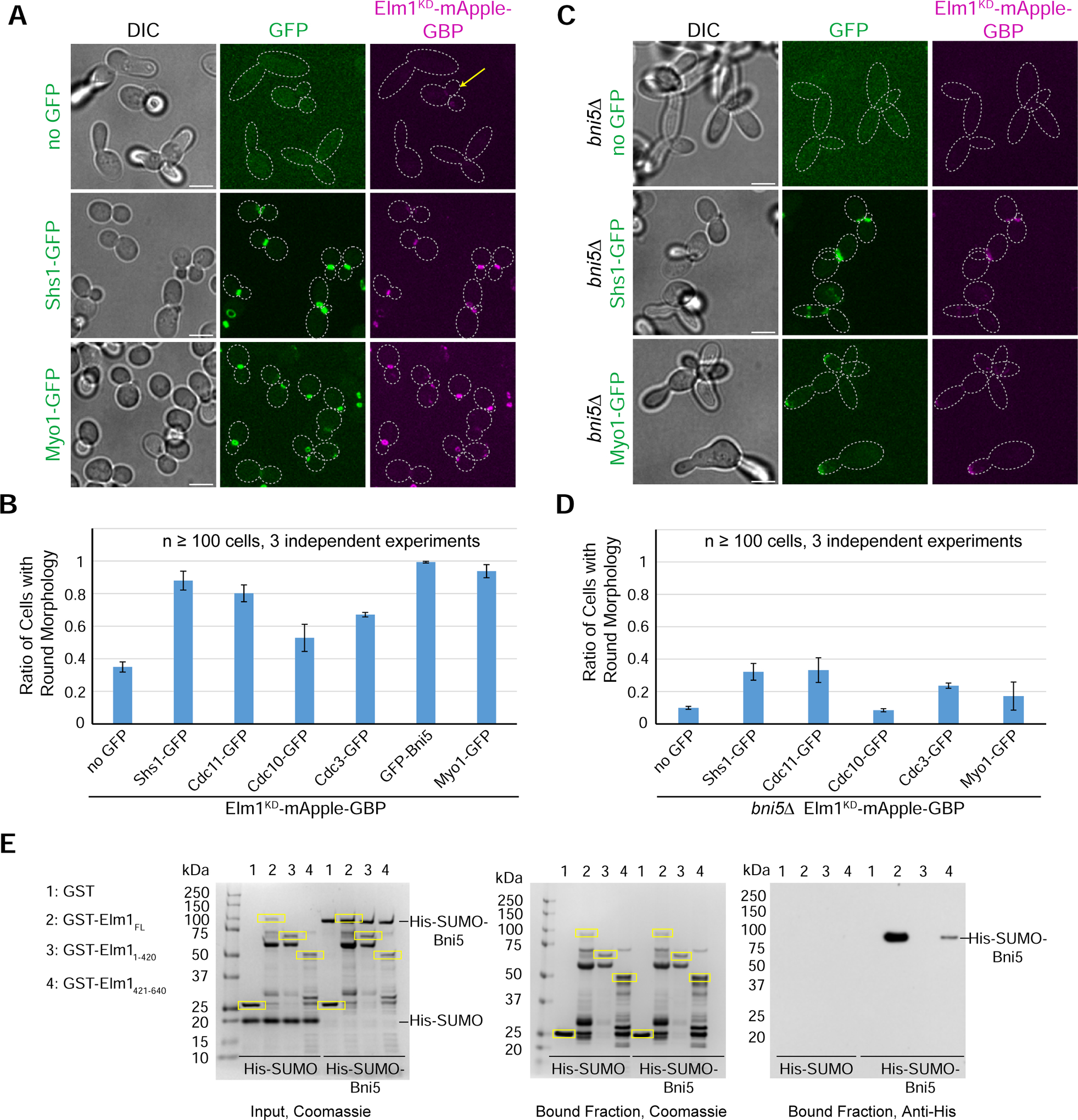
(A) Representative images of indicated strains with Elm1KD-mApple-GBP tethered to various GFP-tagged proteins. Strains used are as follows from top to bottom: YEF9335 (no GFP), YEF9362 (SHS1-GFP), and YEF9370 (MYO1-GFP). Images are maximum projections. Yellow arrow indicates a round cell with Elm1KD-mApple-GBP present at the bud neck. Dotted line is cell periphery. Scale bars = 5 μm. See also Figure S2.
(B) Quantification of the ratio of round cells in strains used in (2A and S2C). Plotted is the average of 3 independent experiments of n > 100 cells. Error bars are standard deviation. See also Figure S2.
(C) Representative images of indicated bni5Δ strains with Elm1KD-mApple GBP tethered to various GFP-tagged proteins. Strains used are as follows from top to bottom: YEF9594 (no GFP), YEF9645 (SHS1-GFP), and YEF9646 (MYO1-GFP). Images are maximum projections. Dotted line is cell periphery. Scale bars = 5 μm. See also Figure S2.
(D) Quantification of the ratio of round cells in strains used in (2C and S2G). Plotted is the average of 3 independent experiments of n > 100 cells. Error bars are standard deviation. See also Figure S2.
(E) Left: SDS-PAGE stained with Coomassie Blue depicting amounts of each indicated purified protein used as the input for the in vitro binding assay. Middle and Right: In vitro binding assay results for indicated GST-tagged proteins bound to glutathione resin (Coomassie Blue stained, middle) and their ability to pull-down His-SUMO or His-SUMO-Bni5 (immunoblotted with antibody against 6xHis, right). Yellow boxes indicate purified GST-tagged protein. kDa = kiloDalton. This experiment was repeated 4 times with consistent interaction detected for the full-length Elm1, and weak interaction for its C-terminal fragment, and weak or no interaction for its N-terminal fragment.
The tethering experiments with either Elm1KD (Figure 2B) or Elm1AS (Figure S2D) suggest a correlation between the distance to the terminal subunit in a septin complex (Cdc11-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Cdc11 or Shs1-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Shs1) and rescue capacity. The terminal subunits Cdc11 and Shs1 both bind to Bni5 [32, 33], which, in turn, localizes the myosin-II heavy chain Myo1 to the bud neck from bud emergence to the onset of cytokinesis [34]. Strikingly, an N-terminally GFP-tagged Bni5 expressed from its native promoter at its endogenous locus (GFP-Bni5) and Myo1-GFP fully rescued the mApple-GBP-tagged elm1KD or elm1AS phenotype, better than any septin subunit (Figures 2A, 2B, S2C, and S2D), but Myo1-GFP still failed to rescue the phenotypes associated with the elm1 KD1–420 allele (Figures S2E and S2F). These data suggest that the kinase activity and the C-terminal non-kinase domain of Elm1 may act in parallel to control septin organization, most effectively, at the terminal septin subunits.
Bni5 becomes essential for septin hourglass stability in cells with no active Elm1 kinase
Bni5 could mediate the kinase-dependent and/or -independent role of Elm1 in septin organization at the terminal septin subunits, as Bni5 is a potential substrate of Elm1 and may interact with the non-kinase domain [35], which was shown to be critical for its kinase-independent role in septin organization (Figures S2E and S2F). Bni5 also crosslinks paired septin filaments into closer proximity in vitro by interacting with the terminal septin subunits [32, 33, 35, 36]. To test this possibility, we deleted BNI5 in cells expressing the full-length Elm1KD. Strikingly, deletion of BNI5 in elm1KD cells worsened the morphological phenotype and largely abolished the rescue of the elm1KD-GBP phenotypes by GFP-tagged CDC3, CDC10, CDC11 (Figures S2G and 2D), SHS1, and MYO1 (Figures 2C and 2D). These data suggest that the septin-tethered Elm1KD may act through or in parallel with Bni5 to control septin organization. To distinguish these possibilities, we found that the bacterially expressed full-length Elm1 and Bni5 interacted directly in vitro, and this interaction requires its non-kinase C-terminal domain (Figure 2E). In addition, Elm1, but not Elm1KD, displays Bni5-dependent conversion of large septin filament clusters into shorter filaments, suggesting that Elm1 may regulate Bni5 function through its kinase activity [35]. Collectively, these observations suggest that Elm1 could regulate septin organization through its non-kinase domain-mediated binding as well as phosphorylation of Bni5. Because elm1Δ or elm1KD cells display much stronger defects in septin organization and cell morphogenesis than bni5D cells do [36], Elm1 must control septin organization through additional binding partners and/or kinase targets besides Bni5.
Loss of Elm1 causes filament unpairing in the septin hourglass
To understand how Elm1 regulates septin hourglass stability at the filament level, we compared the septin hourglass architecture in WT and elm1Δ cells using platinum-replica electron microscopy (PREM) [13, 37]. In cells synchronized at the small-budded stage by α-factor arrest and release, the early septin hourglass, as indicated by Cdc3-GFP, was apparent as a thick band of fluorescence at the bud neck in WT cells, while the fluorescence signal was split between the bud neck and the bud tip in elm1Δ cells (Figure 3A). At this stage, cells were spheroplasted, unroofed, and the resulting cortices were fixed and processed for PREM. Fragments of early hourglass-like structures were observed in cortices from both WT and elm1Δ cells (Figures 3B and S3A). These fragments were confirmed to contain septin filaments by immunogold labeling of Cdc3-GFP in parallel sample processing (Figure S3B). In most structures (87.6%, N = 73) from WT cells, the filament composition is almost exclusively paired filaments as we reported previously [13], with the remainder being composed of a mixture of bundled paired filaments and scattered single filaments (Figure 3B). Strikingly, in cortices derived from elm1Δ cells, the proportion of structures with predominantly paired septin filaments was dramatically reduced (10.3%, N = 68), and the remaining fraction of structures contained mixtures of paired and single filaments (Figure 3B). The width distribution of filaments in structures from elm1Δ cells had a peak centered at 10 nm (single filaments) while that in WT cells had a broad peak of 15–20 nm (paired filaments) (Figure 3C). Since Bni5 is one of the Elm1 effector pathways as described above, we also analyzed the early-hourglass structure in bni5D cells by PREM (Figures S3C and S3D), and found that the septin architecture resembled those of WT cells (Figure S3D). Thus, Elm1 might regulate filament pairing in septin hourglass independently of or in addition to Bni5.
Figure 3. Elm1 controls paired filament formation in the septin hourglass.
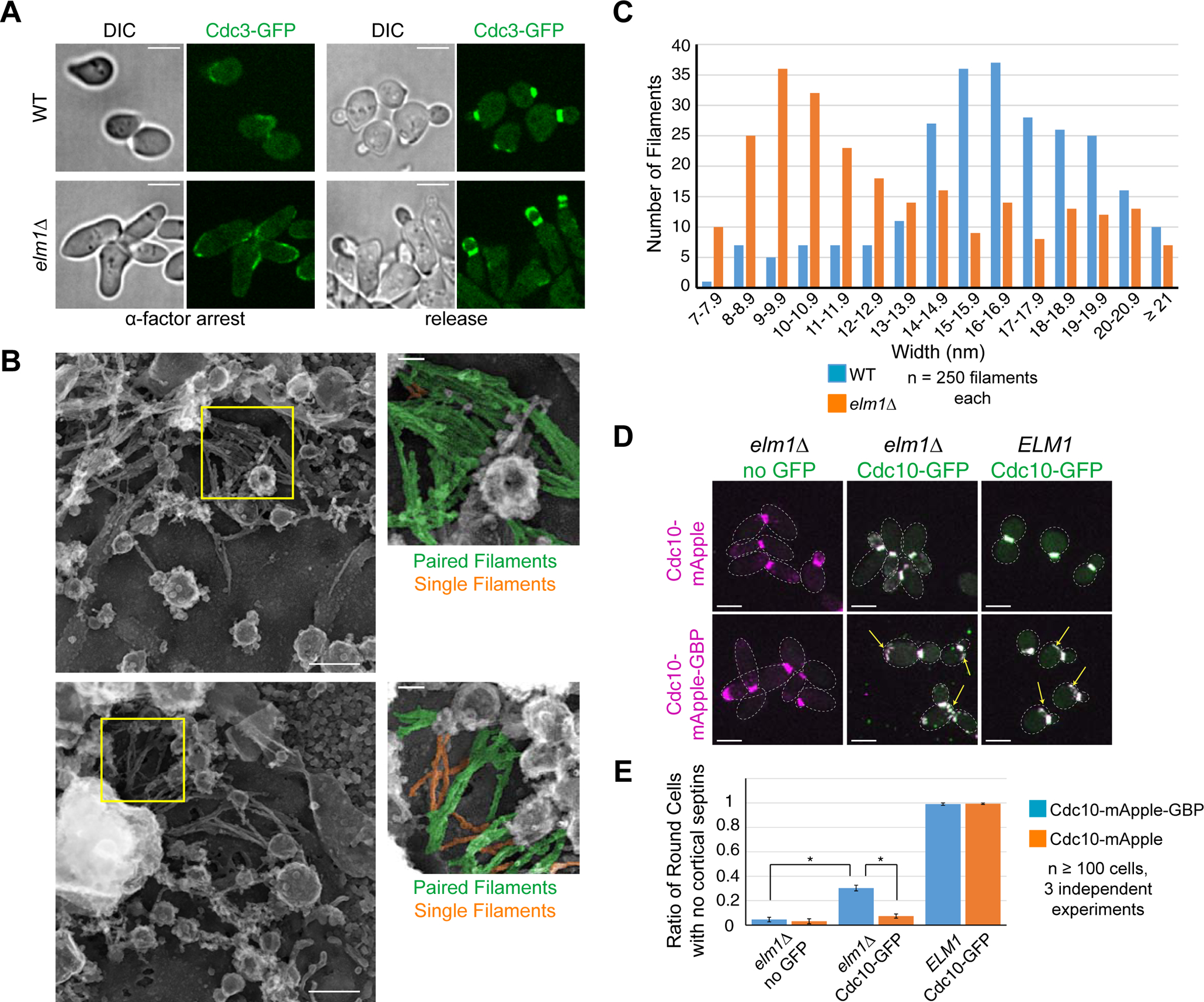
(A) Representative images of cells at 2.5 hours α-factor arrest and after 1.5-hour release for WT (YEF9333) and elm1Δ (YEF9341) with Cdc3-GFP in green. Scale bars = 5 μm.
(B) Representative image of PREM processed sample of WT (YEF9327) and elm1Δ (YEF9334) after α-factor arrest/release. Inset to the right is of area in yellow square. In inset, paired filaments are pseudo-colored green and single filaments are pseudo-colored orange. Scale bar in large image = 200nm, inset = 50nm. See also Figure S3.
(C) Distribution of filament width (in nm) from structures derived from WT or elm1Δ cells. 250 filaments were measured from each strain from a minimum of 6 structures processed from at least two different trials, p = 1.29×10−20. See also Figure S3. P-value is from an unpaired Student’s t-test of width measurements from given number of septin filaments from structures from WT and elm1Δ cells.
(D) Representative images of indicated strains with Cdc10-mApple (top row) or Cdc10-mApple-GBP (bottom row) in magenta and Cdc10-GFP, which is expressed exogenously from a CEN plasmid, in green. Strains used are as follows: YEF10352 (elm1Δ CDC10-mApple no GFP), YEF10358 (elm1Δ CDC10-mApple CDC10-GFP), YEF10357 (ELM1 CDC10-mApple CDC10-GFP), YEF10279 (elm1Δ CDC10-mApple-GBP no GFP), YEF10295 (elm1Δ CDC10-mApple-GBP CDC10-GFP), and YEF10294 (ELM1 CDC10-mApple-GBP CDC10-GFP). Images are maximum projections. Yellow arrow indicates septin ring from previous cell cycle. Dotted line is cell periphery. Scale bars = 5 μm.
(E) Quantification of the ratio of round cells with no cortical septins in strains used in (3D). Plotted is the average of 3 independent experiments of n > 100 cells. Error bars are standard deviation. * indicates p-value < 0.05 by unpaired Student’s t-test between indicated values.
If the defect in filament pairing truly underlies the septin and cell morphology phenotypes in elm1Δ cells, artificial pairing of the septin filaments would rescue these phenotypes. This was the case when adding an exogenous copy of CDC10-GFP to cells expressing CDC10-mApple-GBP in elm1Δ cells. A small but significant proportion of cells (~30%, p-value < 0.05) exhibited normal cell morphology and septin hourglass stability (Figures 3D and 3E). Additionally, the septin rings from the previous cell cycle failed to disassemble in both WT and elm1Δ cells when the septin filaments were artificially tethered together (Figure 3D, arrows). This striking observation suggests that filament unpairing may be required for septin ring disassembly at the cell division site. Thus, our analysis suggests that filament pairing is required for septin hourglass assembly and stability whereas filament unpairing is required for septin ring disassembly at the old cell division site.
Elm1 and the septin Shs1 cooperate to generate an appropriate septin hourglass for its subsequent remodeling into a double ring
The early hourglass consists of paired filaments arranged in parallel to the mother-daughter axis, which develops into a transitional hourglass in anaphase in which the paired filaments are intersected by circumferential single filaments at the spacing of a septin octamer [13, 38]. At the onset of cytokinesis, the transitional hourglass is remodeled into a double ring, consisting of the circumferential single filaments and reorganized circumferential paired filaments, that sandwiches the actomyosin ring (AMR). Since elm1Δ caused profound defects in septin hourglass assembly and stability (Figure S1E and Video S1), we asked whether these defects would impact the hourglass-to-double ring (HDR) transition at the onset of cytokinesis. The HDR transition occurred 6–8 min earlier in elm1Δ than in WT cells (Figure 4A). Perhaps, filament unpairing in elm1Δ cells makes the HDR process more efficient. The septin subunit Shs1 is crucial for the maintenance of the circumferential single filaments in the transitional hourglass [13]. Consistent with this, the septins on average took longer to organize into a double ring in shs1Δ (Figures 4A and 4B). Because Elm1 regulates longitudinal paired filament formation whereas Shs1 regulates the circumferential single filament formation, not surprisingly, shs1Δ elm1Δ cells showed a highly disorganized septin structure during cytokinesis that never resembled a double ring (Figures 4A and 4B). Thus, Elm1 and Shs1 act in concert to generate a proper septin architecture for the HDR transition.
Figure 4. Elm1 and Shs1 promote cytokinesis through their collective control of hourglass assembly and its subsequent remodeling into a double ring.
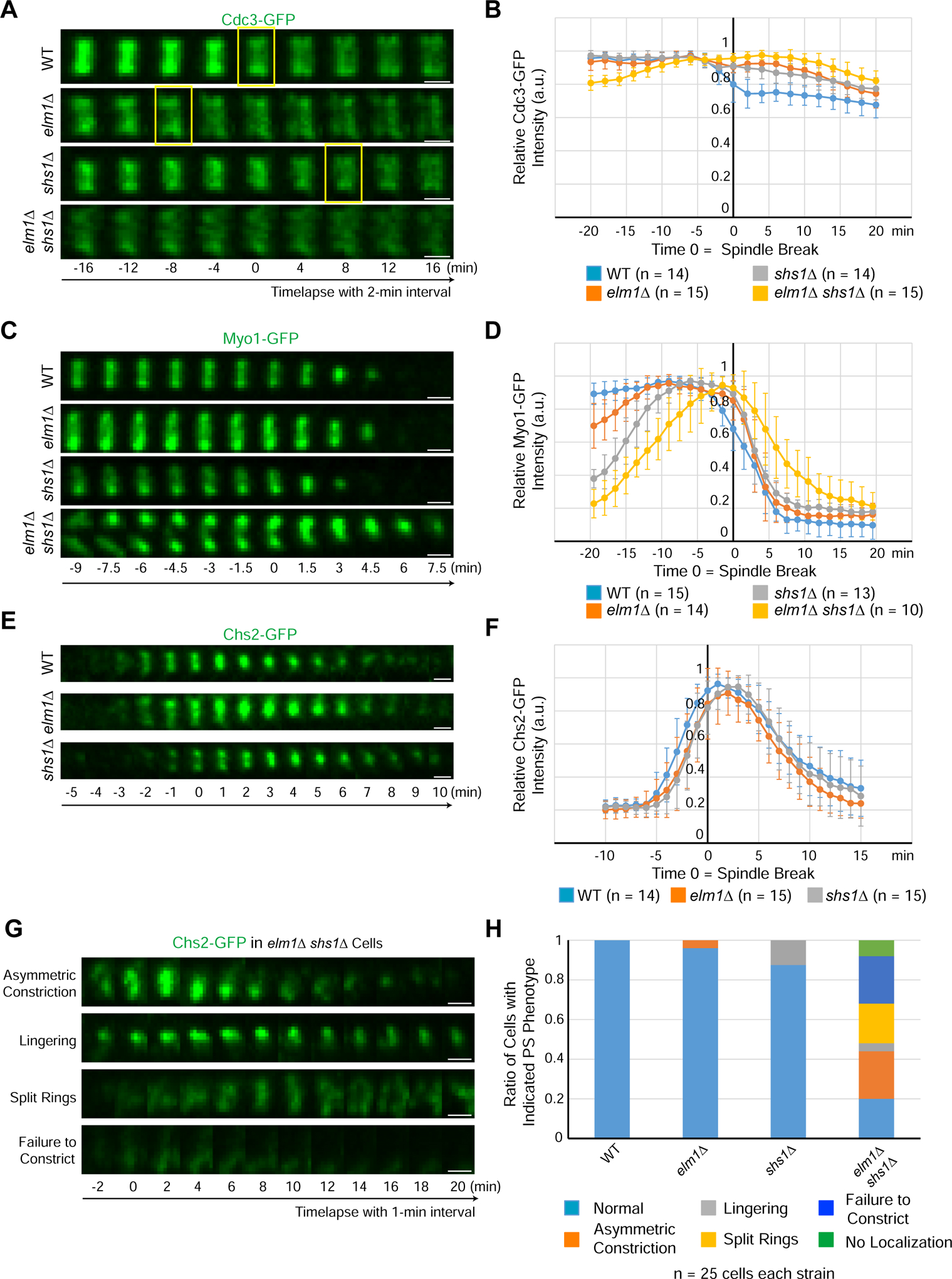
(A) Montages of representative cells of WT (YEF8118), elm1Δ (YEF8195), shs1Δ (YEF8244), and elm1Δ shs1Δ (YEF8246) showing maximum-intensity projections of Cdc3-GFP 16 min before and after T = 0 (spindle break) with selected frames from time-lapse series taken with a 2-min interval. Scale bars = 1 μm, yellow boxes indicate first time point with a distinguishable septin double ring. See also Figure S4.
(B) Quantification of cells in (4A). Shown is the mean of the background subtracted intensity of Cdc3-GFP normalized to value at maximum value in given number cells for each strain. Error bars are standard deviation, T = 0 is spindle break.
(C) Montages show maximum-intensity projections of Myo1-GFP 9 min before and 7.5 min after T = 0 (spindle break) with a 1.5-min interval between each picture for representative cells of each WT (YEF8367), elm1Δ (YEF8381), shs1Δ (YEF8502) and shs1Δ elm1Δ (YEF8503) strains. Scale bars = 1 μm. See also Figure S4.
(D) Quantification of cells in (4C). Shown is the mean of the background subtracted intensity of Myo1-GFP normalized to value at maximum value in given number cells for each strain. Error bars are standard deviation, T = 0 is spindle break.
(E) Montages show maximum-intensity projections of Chs2-GFP 5 min before and 10 min after T = 0 (spindle break) with 1-min interval between each picture for representative cells of each WT (YEF8219), elm1Δ (YEF8247), shs1Δ (YEF8911) strains. Scale bars = 1 μm.
(F) Quantification of cells in (4E). Shown is mean of the background subtracted intensity of Chs2-GFP normalized to value at maximum value in given number cells for each strain. Error bars are standard deviation, T = 0 is spindle break.
(G) Montages show maximum-intensity projections of Chs2-GFP 2 min before and 20 min after T = 0 (spindle break) with 2-min interval between each picture taken from a time-lapse series with 1-min interval for representative cells of each shown phenotype in shs1Δ elm1Δ (YEF8912) strain. Scale bars = 1 μm.
(H) Quantification of cells with indicated phenotypes in (4E and 4G). n = 25 cells for each strain. “Asymmetric constriction” is constriction from predominantly one side of the bud neck, “lingering” is persistent localization of signal after constriction, “split rings” is a failure to constrict while separating the single band of signal into two ring-like signals, “failure to condense” is an initial localization of cloudy signal to the bud neck but no clear compaction to a band, and “no localization” is a complete failure to localize any signal even after spindle break.
Proper hourglass-to-double ring transition is critical for the execution of cytokinesis
The septin hourglass is known to scaffold the assembly of the AMR before cytokinesis [39, 40]. After the HDR transition, a septin double ring sandwiches the AMR that constricts and directs the formation of the primary septum (PS) catalyzed by the chitin synthase-II, Chs2 [34, 41]. A proper HDR transition is presumably important for cytokinesis, as septin clearance at the division site is essential for AMR constriction [42]. Because Elm1 and Shs1 act coordinately to enable a proper HDR transition, we monitored their impact on cytokinesis. In agreement with their septin defects, the shs1Δ elm1Δ cells showed a more elongated phenotype as well as more chains of cells, indicative of a defect in cytokinesis and/or cell separation, than the single mutants (Figure S4A). We then analyzed the behaviors of the AMR (indicated by Myo1-GFP) and PS (indicated by Chs2-GFP) in cells deleted for ELM1 and/or SHS1. Strikingly, a majority of the bud-neck Myo1-GFP was found in the growing bud cortex of elm1Δ cells and then returned to the bud neck before cytokinesis (Figure S4B arrow and arrowhead, respectively). In both elm1Δ and shs1Δ single mutants, Myo1-GFP was able to localize to the division site prior to cytokinesis, and constricted with near normal kinetics (Figures 4C and 4D). However, the level of Myo1-GFP at the bud neck during bud formation was significantly lower, especially in shs1Δ cells, than that in WT cells (Figure 4D, before the 12-min time point). In elm1Δ shs1Δ cells, the AMR was much more perturbed during cytokinesis. Cells showed a disorganized, and often incomplete AMR that was highly delayed in constriction initiation, if it constricted at all (Figures 4C and 4D). Thus, the compounding defects found with septins at the division site in the elm1Δ shs1Δ cells gave rise to a much more disorganized and largely nonfunctional AMR. Similarly, PS formation in the elm1Δ and shs1Δ single mutants was largely unaffected (Figures 4E and 4F). However, the double mutant displayed a vast array of defects in both Chs2 localization as well as its symmetric constriction (Figures 4G and 4H). Together, these data indicate that Elm1 and Shs1 likely act in concert to regulate cytokinesis via their direct roles in septin hourglass assembly and remodeling.
In conclusion, a number of protein kinases are known to regulate septin functions in diverse processes in yeast and mammals [5, 6, 43, 44], but their regulatory impact on septin assembly and architecture remains largely unknown. Here, we show that the LKB1-like kinase Elm1 in budding yeast associates specifically with the septin hourglass during the cell cycle, and controls hourglass assembly and stability via both kinase-dependent and -independent mechanisms involving Bni5 and additional targets. Most importantly, this study defines Elm1 as the first regulator of filament pairing in vivo to control the assembly of a specific septin architecture. In mammalian cells, septin filaments are also organized into an hourglass-like shape during furrow ingression [2, 45], which is then remodeled into a double ring-like structure during early midbody stage before abscission [46–48]. It would be of particular interest to determine whether there is an Elm1-like mechanism in mammalian cells that controls septin assembly and/or remodeling during cytokinesis.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for research materials may be directed to and will be fulfilled by the Lead Contact, Erfei Bi (ebi@pennmedicine.upenn.edu).
Materials Availability
Reagents generated in this study will be made available on request.
Data and Code Availability
Data supporting the findings of this study are available within the paper and its Supplemental Information files and from the authors upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The budding yeast Saccharomyces cerevisiae is our experimental model. All yeast strains used in this study are listed in Table S1. Standard culture media and genetic techniques were used [58]. Yeast strains were grown routinely at 25°C in synthetic complete (SC) minimal medium lacking specific amino acid(s) and/or uracil or in rich medium YM-1 [59] or yeast extract/peptone/dextrose (YPD). New strains were constructed either by integrating a plasmid carrying a modified gene at a genomic locus or by transferring a deletion or tagged allele of a gene from a plasmid or from one strain to another via PCR amplification and yeast transformation [52, 53] (see footnotes in Table S1).
METHOD DETAILS
Primers and Plasmids
All primers were purchased from Integrated DNA Technologies and described in Table S2. Sequencing of constructs was performed at the DNA Sequencing Facility, University of Pennsylvania. Plasmids YIp128-CDC3-GFP [49] and YIp128-CDC3-mCherry [50] (integrative, LEU2) carries an N-terminally GFP- or mCherry-tagged CDC3 under the control of its own promoter, respectively. Plasmids pHIS3p::mRuby2-Tub1+3’UTR::URA3, pHIS3p::mRuby2-Tub1+3’UTR::HPH, and pHIS3p:mRuby2-Tub1+3’UTR::HIS3 [51], pFA6a-link-yoEGFP-SpHIS5, pFA6a-link-yoEGFP-CaURA3, and pFA6a-link-yomApple-SpHIS5 [52], pRS316-Hof1 [55], pGFP316-Cdc10 [56], and pFA6a-His3MX6 and pFA6a-TRP1 [53] were described previously. Plasmid pFA6a-URA3-KanMX6 was a generous gift from John Pringle (Stanford University) and is described previously [54]. Plasmid pFA6a-mCherry-URA3 was a generous gift from C. Burd. Plasmid pGEX-4T1 was purchased from GE Healthcare (Cat #: 28-9545-49). Plasmid pET-His6-Sumo-TEV LIC (Addgene # 29659) was a generous gift from Scott Gradia (University of California at Berkeley).
The following plasmids were generated for this study: pFA6a-link-yoEGFP-NatMX6, pFA6a-link-yomApple-GBP-CaURA3, pFA6a-link-yomApple-GBP-SpHIS5, pGEX-4T1-Elm1FL, pGEX-4T1-Elm11–420, pGEX-4T1-Elm1421–640, and pET-His6-Sumo-Bni5. To generate pFA6a-link-yEGFP-NatMX6, the NatMX6 cassette was amplified by PCR using the plasmid pAG25 (Addgene #35121) as the template DNA and the pair of primers pR32-pmeI and pR29-bgIII to flank the NatMX6 cassette with PmeI and BglII sites. The resultant PCR product was then digested with BglII and PmeI and ligated into the plasmid pFA6a-link-yoEGFP-KanMX6 cut with the same restriction enzymes to remove the KanMX6 cassette. The plasmid pRS315-INN1-C2-mApple-GBP-6xHis was constructed by amplifying the GBP-6xHis fragment out of pYA889–1, a generous gift from Yoshinori Ohsumi at the Tokyo Institute of technology [60]. To generate pFA6a-link-yomApple-GBP-CaURA3 and pFA6a-link-yomApple-GBP-SpHIS5, the yomApple-GBP fragment was amplified by PCR using the plasmid pRS315-INN1-C2-mApple-GBP-6xHis as the template DNA and the pair of primers pRS315-F-PacI and GBP-R-AscI-term to flank the yomApple-GBP fragment with PacI and AscI sites. The resultant PCR product was then digested with PacI and AscI and ligated into either plasmid pFA6a-link-yoTagRFP-CaURA3 or pFA6a-link-yoTagRFP-SpHIS5, respectively [52] cut with the same restriction enzymes to remove the yoTagRFP cassette. To generate plasmids pGEX-4T1-Elm1FL, pGEX-4T1-Elm11–420, and pGEX-4T1-Elm1421–640, the chromosomal DNA of the yeast strain YEF473A was used as template DNA for PCR amplification using the pairs of primers Elm1-F-BamHI FL and Elm1-R-XhoI FL, Elm1-F-BamHI FL and Elm1-R-XhoI 1–420, and Elm1-F-BamHI 421–640 and Elm1-R-XhoI FL, respectively, to flank different Elm1 fragments with BamHI and XhoI. These fragments were then digested with BamHI and XhoI and ligated into PGEX-4T1 (GE Healthcare, Chicago, IL, USA) linearized with BamHI and XhoI. To generate pET-His6-Sumo-Bni5, the chromosomal DNA of YEF473A was used as template DNA for PCR amplification using the pair of primers Bni5-F-SspI and Bni5-R-BamHI to flank Bni5 with SspI and BamHI. This fragment was then digested with SspI and BamHI and ligated into pET His6 Sumo TEV LIC (Addgene # 29659) linearized via SspI and BamHI.
Live-cell imaging and quantitative analysis
For time-lapse microscopy, cells were grown at 25°C to exponential phase in liquid SC or YM-1 media. For cells grown in SC media, cells were briefly sonicated at 15% power for 5 seconds to declump, concentrated by centrifugation, spotted onto a poly-lysine-coated glass-bottom dish, and then embedded with SC containing agarose [61]. SC medium was added to dish for live imaging. For cells grown in YM-1 media, cells were harvested as described above and spotted onto concanavalin A-coated glass-bottom dish. Supernatant containing non-immobilized cells was removed, and YM-1 medium was added to dish for live imaging. Images were acquired using a Nikon microscope (model Eclipse Ti-U, Tokyo, Japan) equipped with a Nikon 100x/1.49NA oil objective (model CFI Apo TIRF 100x), and a Yokogawa spinning-disk confocal scanner unit (model CSU-X1, Tokyo, Japan). A Photometrics Evolve Delta 512X512 EMCCD Digital Monochrome Camera (Tucson, AZ, USA) was used for image capture. Solid-state lasers for excitation (488 nm for GFP and 561 nm for RFP) were housed in a launch constructed by Spectral Applied Research (model ILE-400, Richmond Hill, Ontario, Canada). The imaging system was controlled by MetaMorph version 7.8.10.0 (Molecular Devices, Downingtown, PA, USA). Images were taken with z-stacks of 13 × 0.7 μm for all the experiments with SC medium and 11 × 0.8 μm or 11 × 1.1 μm for the experiments with YM-1.
For quantification of the fluorescence intensity of targeted protein signal (Bni4-GFP, Cdc3-GFP, Cdc3-mCherry, Hsl1-GFP, Kcc4-GFP, Myo1-GFP, and Chs2-GFP), a sum projection was created and quantification was performed with ImageJ (National Institutes of Health). A polygon was drawn around the region of interest and the integrated density was measured. The fluorescence intensity of the background was subtracted from this measurement (outside the cell except for quantification in Figures 1D and S1D in which cytoplasmic signal was used). For Figure 1B this value was used for data analysis (see below). For Figures 1D, S1D, 4B, 4D, and 4F, the data were normalized to the peak intensity (100%) of the fusion protein signal at the bud neck in the cell. Data were analyzed with Microsoft Excel, and expressed as the mean value ± standard deviation (SD).
For snapshot images, cells were grown at 25°C to exponential phase in liquid YM-1 or SC media. For elm1Δ hof1Δ strain used in Figure 1G, cells were cultured in liquid YM-1 media overnight. For Elm1AS strains used in Figure S2D, cells were treated with 25μM 1NM-PP1 (Santa Cruz Biotechnology, Dallas, TX) or same volume DMSO as control in liquid SC media overnight. Cells were briefly sonicated at 15% power for 5 seconds to declump, concentrated by centrifugation, and spotted onto a slide. Images were taken as described above except with z-stacks of 15 × 0.7 μm for all the experiments. To quantify cell numbers with elongated phenotypes (Figures 2B, 2D, S2D, S2F, and 3E), cells were counted as round if the bud length:width ≤1.2, while they were counted as elongated if >1.2.
Protein purification and in vitro binding assays
BL21 cells transformed with either pGEX-4T1 (GST alone), pGEX-4T1-Elm1FL (GST-Elm1FL), pGEX-4T1-Elm11–420 (GST-Elm11–420), pGEX-4T1-Elm1421–640 (GST-Elm1421–640), pET-His6 Sumo-TEV-LIC (6xHis-SUMO alone) or pET-His6-Sumo-Bni5 (6xHis-SUMO-Bni5) were grown to an OD600 0.6–1.0 before being induced for 3 hours with 0.3mM IPTG (Lab Scientific, Highlands, NJ, USA) at 37°C. Cells were then lysed in either GST lysis buffer (50mM Tris-HCl, pH7.5, 300 mM NaCl, 1.25mM EGTA, 1mM DTT and 0.1%NP-40) or 6xHis-SUMO lysis buffer (50mM Tris-HCl, pH7.5, 300 mM NaCl, 1.25mM EGTA, 1mM DTT, 0.1%NP-40 and 15 mM imidazole) by sonication 10 times for 15 seconds each. The resultant lysates were then centrifuged at 40,000 X g for 30 min at 4°C. The supernatants were then incubated with either Glutathione Sepharose 4B (GE Healthcare) or Complete His-Tag Purification Resin (Roche, Basel, Switzerland), that had been prewashed with TBS, for 1 hour at 4°C. The beads were then washed twice with TBS (50 mM Tris-Cl, pH 7.5. 150 mM NaCl) before being transferred to columns for elution by either GST elution buffer (50mM Tris-HCl, pH8.0, 1 mM DTT, 200 mM NaCl and 10mM reduced glutathione) or 6xHis-SUMO elution buffer (50mM Tris-HCl, pH7.5, 200 mM NaCl and 300 mM imidazole). The eluted proteins were dialyzed in dialysis buffer (100 mM NaCl, 5mM Tris-HCl pH7.5, 1mM DTT) at 4°C overnight. Protein concentrations were determined by standard curve intensity measurements from Coomassie blue-stained BSA of known concentrations.
For in vitro binding, 8 μg of either GST, GST-Elm1FL, GST-Elm11–420, or GST-Elm1421–640 was incubated with 20 μg of either 6xHis-SUMO or 6x-His-SUMO-Bni5 for 1 hour with rotation at 4°C in the presence of Glutathione Sepharose 4B (GE Healthcare) beads in binding buffer (20 mM MOPS, pH7.0, 1 mM EGTA, 100 mM NaCl, 1 mM DTT, 0.1%NP-40). Beads were then washed five times with fresh binding buffer before being extracted with 50 μL of 1X Laemmli Buffer (Bio-Rad Laboratories, Hercules, CA, USA). 15 μL were separated via SDS-PAGE, transferred to PVDF membrane, and blotted with anti-6xHis antibody (1:3000 dilution) followed by HRP-labeled secondary antibody and ECL reagents from the Pierce Fast Western kit (Thermo Scientific, Waltham, MA, USA). The same amount of protein used for the interaction assay was run on a separate gel and stained with Coomassie Blue to serve as the input sample. To check for similar GST pulldown, 15 μL of each in vitro binding reaction was separated via SDS-PAGE and stained with Coomassie Blue.
Cell synchronization, spheroplasting, and unroofing
Cells were prepared for PREM as described previously [13]. Briefly, cells of different strains containing the bar1Δ were cultured in 50 mL YM-1 medium with shaking at 25°C to achieve OD600 = 0.4. 0.5μg/mL of α-factor pheromone (Research Genetics, Huntsville, AL, USA) was added to the culture and further incubated at 25°C to achieve arrest in G1 as unbudded cells. Cells were then washed 3 times with fresh YM-1 media and released into pheromone-free YM-1 for 1.5 hours at 25°C until synchronized cells had small to medium buds.
Cells were washed with 40 mM Tris-EDTA (TE) (pH 8.0), and then incubated in 3 mL TE plus 5μL β-Mercaptoethanol for 15 min at 30°C. After centrifugation, the cell pellet was resuspended in spheroplast buffer (10 mM PIPES, 1.2 M sorbitol, pH 6.5) containing 0.3 mg/mL zymolyase-100T (Amsbio, Oxfordshire, UK) and incubated at 35°C with moderate shaking for 1 hour. Sephroplasts were washed three times with the spheroplast buffer and then mounted to a poly-lysine-coated coverslip. Two coverslips with spheroplasts on top were dipped into the spheroplast buffer and 1X KHMgE buffer (70 mM KCl, 20 mM HEPES, 5 mM MgCl2, 3 mM EGTA) sequentially, and then pressed against each other with the spheroplasts in between the coverslips on a slide with one drop of the 1X KHMgE buffer. The coverslips were lifted and then fixed immediately in 2% glutaraldehyde in 1X KHMgE buffer at room temperature (RT ≈ 23°C) for 20 min.
Immunogold labeling
The coverslips with fixed unroofed spheroplasts were quenched in 2 mg/mL and 5 mg/mL NaBH4 in phosphate-buffered saline (PBS) for 10 min sequentially, blocked in 1% glycine for 10 min, and then washed three times in PBS. The coverslips were blocked in PBS with 5% donkey serum for 30 min. Each coverslip was incubated with the goat polyclonal anti-GFP antibody (1:50 in PBS with 5% donkey serum, ABCAM ab5450) at RT for 1.5 hours. After washing five times with PBS and blocking for 10 min in immunogold-labeling buffer (20 mM Tris-HCl, pH 8.0, 0.5 M Nacl, and 0.05% Tween 20) containing 0.5% donkey serum, the coverslips were incubated with 18-nm Colloidal Gold AffiniPure Donkey Anti-Goat IgG (H+L) (Jackson ImmunoResearch Laboratories, Inc. 705-215-147, 1:5 dilution) in immnogold-labeling buffer containing 5% donkey serum at RT for overnight. After washing five times in immunogold-labeling buffer containing 0.05% donkey serum, the coverslips were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.3.
Critical point drying and platinum coating
Unroofed cortices were dried and coated for PREM as described previously [13, 62]. The coverslips with fixed unroofed spheroplasts were treated in 0.1% tannic acid in water for 20 min. After washing three times with water, they were stained in 0.2% uranyl acetate in water for 20 min, which was followed by sequential incubation in 10, 20, 40, 60, 80, 100% ethanol and then critical point-dried with Samdri PVT-3D (Tousimis) CPD. Platinum was rotary-shadowed to the samples at 45° angle to form ~2-nm layer and carbon was then rotary-shadowed at 90° angle to form a 3.5–4 nm layer.
PREM imaging and analysis
After detaching the glass coverslips with hydrofluoric acid, coated samples were mounted on EM grids and imaged using a JEM 1011 transmission electron microscope (JEOL USA, Peabody, MA) operated at 100 kV. Images were captured with an ORIUS 832.10W charge-coupled device (CCD) camera (Gatan, Warrendale, PA), and were presented in inverted contrast. Structures of interest were color-labeled using the Adobe Photoshop. To determine the filament composition of hourglass fragments, filaments between 7–12 nm in width were considered to be single septin filaments while those between 14–20 nm in width were considered to be paired septin filaments [13]. More than 5 single septin filaments in a given structure was designated as a mixture of single and paired. The entire procedure of synchronizing, speroplasting, unroofing, and PREM processing was repeated four times to attain the number of structures reported as counted.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean value ± SD throughout this study. All p-values shown are the results of an unpaired Student’s T-test assuming unequal variances from the datasets from WT and elm1Δ cells.
Supplementary Material
Video S1. Deletion of ELM1 causes destabilization of the septin hourglass preferentially at the daughter side from bud emergence to late anaphase. Related to Figures 1 and S1. Top : Time-lapse analysis of Cdc3-GFP localization (shown in green) in relation to bud emergence and the mitotic spindle (shown in magenta) during the cell cycle by spinning-disk microscopy in WT (YEF8102) cells. Time 0 = Bud emergence. Time interval = 2 min. Scale bar = 2 μm. Imaging was performed on cells cultured at 25°C. Bottom: Time-lapse analysis of Cdc3-GFP localization (shown in green) in relation to bud emergence and the mitotic spindle (shown in magenta) during the cell cycle by spinning-disk microscopy in elm1Δ (YEF8393) cells. Time 0 = Bud emergence. Time interval = 2 min. Scale bar = 2 μm. Imaging was performed on cells cultured at 25°C.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GST antibody (3G10/1B3) 1:3,000 dilution | Abcam | Cat #: ab92; RRID: AB_307067 |
| Anti-6x His antibody (HIS.H8) 1:3,000 dilution | Abcam | Cat #: ab18184; RRID: AB_444306 |
| Anti-GFP antibody 1:50 dilution | Abcam | Cat#: ab5450; RRID: AB_304897 |
| 18 nm Colloidal Gold AffiniPure Donkey Anti-Goat IgG (H+L) 1:5 dilution | Jackson ImmunoResearch | Cat#: 705-215-147; RRID: AB_2340419 |
| Bacterial and Virus Strains | ||
| E. coli strain DH5α | Invitrogen | Cat #: 18258012 |
| E. coli strain One-Shot BL21 | Invitrogen | Cat #: C600003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 5-fluoro-orotic acid | Formedium, Hunstanton, UK | Cat #: 5FOA10 |
| G-418 Disulphate | Formedium, Hunstanton, UK | Cat #: G4185 |
| 1NM-PP1 | Santa Cruz | Cat #: sc-203214 |
| alpha-factor (WHWLQLKPGQPMY) | Research Genetics | Synthesized peptide |
| IPTG | Lab Scientific | Cat #: I-555 |
| Glutathione Sepharose 4B | GE Healthcare | Cat #:17-0756-01 |
| Complete His-Tag Purification Resin | Roche | Cat #: 05893682001 |
| Imidazole | Acros, Geel, Belgium | Cat #: 301872500 |
| L-Glutathione, reduced | Sigma-Aldrich | Cat #: G4251 |
| Restore Western Blot Stripping Buffer | Thermo Scientific | Cat. # 21059 |
| Zymolyase 100T | Amsbio, Oxfordshire, UK | Cat#: 120493-1 |
| Glutaraldehyde | Polyscience, Pennsylvania, USA | Cat#: 01909-10 |
| Tannic Acid | Mallinckrodt, Kentucky, USA | Cat#:1764 |
| Uranyl Acetate | Structure Probe, Pennsylvania, USA | Cat#:02624 |
| Critical Commercial Assays | ||
| Pierce Fast Western Blotting Kit, Supersignal West Pico,Mouse | Thermo Scientific | Cat #: 35060 |
| Quick-Fusion Cloning Kit | BiMake | Cat #: B22612 |
| Experimental Models: Organisms/Strains | ||
| Saccharomyces cerevisiae strains, see Table S1 | This paper | N/A |
| Oligonucleotides | ||
| Primers, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: YIp128-CDC3-GFP | [49] | N/A |
| Plasmid: YIp128-CDC3-mCherry | [50] | N/A |
| Plasmid: pHIS3p: mRuby2-Tub1+3’UTR::HIS3 | [51] | N/A |
| Plasmid: pHIS3p: mRuby2-Tub1+3’UTR::URA3 | [51] | N/A |
| Plasmid: pHIS3p: mRuby2-Tub1+3’UTR::HPH | [51] | N/A |
| Plasmid: pFA6a-mCherry-URA3 | C. Burd | N/A |
| Plasmid: pFA6a- link-yoEGFP-SpHIS5 | [52] | Addgene Plasmid #: 44836 |
| Plasmid: pFA6a- link-yoEGFP-CaURA3 | [52] | Addgene Plasmid #: 44872 |
| Plasmid: pFA6a-link-yomApple-SpHIS5 | [52] | Addgene Plasmid #: 44844 |
| Plasmid: pFA6a- link-yoEGFP-NatMX6 | This Paper | N/A |
| Plasmid: pFA6a- link-yomApple-GBP-CaURA3 | This Paper | N/A |
| Plasmid: pFA6a-TRP1 | [53] | Addgene Plasmid #: 41595 |
| Plasmid: pFA6a-HIS3MX6 | [53] | Addgene Plasmid #: 41596 |
| Plasmid: pFA6a-URA3-KanMX6 | [54] | Addgene Plasmid #: 53194 |
| Plasmid: pRS316-HOF1 | [55] | N/A |
| Plasmid: pGFP316-CDC10 | [56] | N/A |
| Plasmid: pGEX-4T1 | GE Healthcare | Cat #: 28-9545-49 |
| Plasmid: pGEX-4T1-Elm1FL | This Paper | N/A |
| Plasmid: pGEX-4T1-Elm11–420 | This Paper | N/A |
| Plasmid: pGEX-4T1-Elm1421–640 | This Paper | N/A |
| Plasmid: pET His6 Sumo TEV LIC | Scott Gradia | Addgene Plasmid #: 29659 |
| Plasmid: pET-His6-Sumo-Bni5 | This Paper | N/A |
| Software and Algorithms | ||
| MetaMorph version 7.8.10.0 | Molecular Devices | N/A |
| Fiji | [57] | https://imagej.nih.gov/ij/ |
| Other | ||
| Amicon Ultra-Ultracel-10K (10,000 MWCO) | Millipore | Cat #: UFC801024 |
Highlights:
Elm1 is specifically associated with the septin hourglass at the division site
Deletion of ELM1 causes filament unpairing in septin hourglass
Elm1 controls septin hourglass assembly and stability via Bni5 and other targets
Elm1 and Shs1 impact cytokinesis via their roles in septin assembly and remodeling
ACKNOWLEDGEMENTS
The authors would like to thank Kangji Wang and the members of the Svitkina lab for help with PREM, the members of the Bi lab for critical discussions of data interpretation and figure design, Michael McMurray for suggesting the experiment on forced filament pairing using Cdc10 GFP-GBP, Satoshi Okada in the lab of Takashi Ito and Masayuki Onishi in the lab of John Pringle for generously sharing plasmids, Doug Kellogg for generously sharing yeast strains, and the former postdoc Yogini Bhavsar and undergraduates Yoorim Oh and Alaina Hunt for help in yeast strain construction. This work is supported by the National Institutes of Health grants GM116876 (to E.B.) and GM095977 (to T.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATIONS OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at
REFERENCES
- 1.Byers B, and Goetsch L (1976). A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol 69, 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estey MP, Di Ciano-Oliveira C, Froese CD, Bejide MT, and Trimble WS (2010). Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol 191, 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridges AA, and Gladfelter AS (2014). Fungal pathogens are platforms for discovering novel and conserved septin properties. Curr. Opin. Microbiol 20, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marquardt J, Chen X, and Bi E (2019). Architecture, remodeling, and functions of the septin cytoskeleton. Cytoskeleton (Hoboken) 76, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez AM, Finnigan GC, Roelants FM, and Thorner J (2016). Septin-associated protein kinases in the yeast Saccharomyces cerevisiae. Front. Cell Dev. Biol 4, 119–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin CH, Shen YR, Wang HY, Chiang CW, Wang CY, and Kuo PL (2019). Regulation of septin phosphorylation: SEPT12 phosphorylation in sperm septin assembly. Cytoskeleton (Hoboken) 76, 137–142. [DOI] [PubMed] [Google Scholar]
- 7.Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, and Pringle JR (1996). The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol 8, 106–119. [DOI] [PubMed] [Google Scholar]
- 8.Barral Y, Mermall V, Mooseker MS, and Snyder M (2000). Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5, 841–851. [DOI] [PubMed] [Google Scholar]
- 9.Takizawa PA, DeRisi JL, Wilhelm JE, and Vale RD (2000). Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341–344. [DOI] [PubMed] [Google Scholar]
- 10.Kim HB, Haarer BK, and Pringle JR (1991). Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol 112, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cid VJ, Adamikova L, Sanchez M, Molina M, and Nombela C (2001). Cell cycle control of septin ring dynamics in the budding yeast. Microbiology 147, 1437–1450. [DOI] [PubMed] [Google Scholar]
- 12.Lippincott J, Shannon KB, Shou W, Deshaies RJ, and Li R (2001). The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci 114, 1379–1386. [DOI] [PubMed] [Google Scholar]
- 13.Ong K, Wloka C, Okada S, Svitkina T, and Bi E (2014). Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun 5, 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouquin N, Barral Y, Courbeyrette R, Blondel M, Snyder M, and Mann C (2000). Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci 113, 1435–1445. [DOI] [PubMed] [Google Scholar]
- 15.Kang H, Tsygankov D, and Lew DJ (2016). Sensing a bud in the yeast morphogenesis checkpoint: a role for Elm1. Mol. Biol. Cell 27, 1764–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caydasi AK, Kurtulmus B, Orrico MI, Hofmann A, Ibrahim B, and Pereira G (2010). Elm1 kinase activates the spindle position checkpoint kinase Kin4. J Cell Biol 190, 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore JK, Chudalayandi P, Heil-Chapdelaine RA, and Cooper JA (2010). The spindle position checkpoint is coordinated by the Elm1 kinase. J. Cell Biol 191, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen YR, Wang HY, Kuo YC, Shih SC, Hsu CH, Chen YR, Wu SR, Wang CY, and Kuo PL (2017). SEPT12 phosphorylation results in loss of the septin ring/sperm annulus, defective sperm motility and poor male fertility. PLoS Genet 13, e1006631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeMarini DJ, Adams AEM, Fares H, De Virgilio C, Valle G, Chuang JS, and Pringle JR (1997). A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol 139, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozubowski L, Panek H, Rosenthal A, Bloecher A, DeMarini DJ, and Tatchell K (2003). A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 14, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, and Lew DJ (2000). Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozubowski L, Larson JR, and Tatchell K (2005). Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 3455–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladfelter AS, Pringle JR, and Lew DJ (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol 4, 681–689. [DOI] [PubMed] [Google Scholar]
- 24.Qiu W, Neo SP, Yu X, and Cai M (2008). A novel septin-associated protein, Syp1p, is required for normal cell cycle-dependent septin cytoskeleton dynamics in yeast. Genetics 180, 1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurray MA, and Thorner J (2009). Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szkotnicki L, Crutchley JM, Zyla TR, Bardes ES, and Lew DJ (2008). The checkpoint kinase Hsl1p is activated by Elm1p-dependent phosphorylation. Mol. Biol. Cell 19, 4675–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blacketer MJ, Koehler CM, Coats SG, Myers AM, and Madaule P (1993). Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol. Cell. Biol 13, 5567–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas CL, Blacketer MJ, Edgington NP, and Myers AM (2003). Assembly interdependence among the S. cerevisiae bud neck ring proteins Elm1p, Hsl1p and Cdc12p. Yeast 20, 813–826. [DOI] [PubMed] [Google Scholar]
- 29.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, et al. (2006). Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889. [DOI] [PubMed] [Google Scholar]
- 30.Kubala MH, Kovtun O, Alexandrov K, and Collins BM (2010). Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci 19, 2389–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sreenivasan A, Bishop AC, Shokat KM, and Kellogg DR (2003). Specific inhibition of Elm1 kinase activity reveals functions required for early G1 events. Mol. Cell. Biol 23, 6327–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finnigan GC, Booth EA, Duvalyan A, Liao EN, and Thorner J (2015). The Carboxy-Terminal Tails of Septins Cdc11 and Shs1 Recruit Myosin-II Binding Factor Bni5 to the Bud Neck in Saccharomyces cerevisiae. Genetics 200, 843–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Booth EA, Sterling SM, Dovala D, Nogales E, and Thorner J (2016). Effects of Bni5 Binding on Septin Filament Organization. J. Mol. Biol 428, 4962–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, and Bi E (2010). Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin-II. J. Cell Biol 191, 1333–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patasi C, Godocikova J, Michlikova S, Nie Y, Kacerikova R, Kvalova K, Raunser S, and Farkasovsky M (2015). The role of Bni5 in the regulation of septin higher-order structure formation. Biol. Chem 396, 1325–1337. [DOI] [PubMed] [Google Scholar]
- 36.Lee PR, Song S, Ro HS, Park CJ, Lippincott J, Li R, Pringle JR, De Virgilio C, Longtine MS, and Lee KS (2002). Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol 22, 6906–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svitkina T (2009). Imaging cytoskeleton components by electron microscopy. Methods Mol. Biol 586, 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Wang K, Svitkina T, and Bi E (2020). Critical roles of a RhoGEF-Anillin module in septin architectural remodeling during cytokinesis. Curr. Biol doi: 10.1016/j.cub.2020.02.023. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, and Pringle JR (1998). Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol 142, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider C, Grois J, Renz C, Gronemeyer T, and Johnsson N (2013). Septin rings act as a template for myosin higher-order structures and inhibit redundant polarity establishment. J. Cell Sci 126, 3390–3400. [DOI] [PubMed] [Google Scholar]
- 41.Sburlati A, and Cabib E (1986). Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J. Biol. Chem 261, 15147–15152. [PubMed] [Google Scholar]
- 42.Tamborrini D, Juanes MA, Ibanes S, Rancati G, and Piatti S (2018). Recruitment of the mitotic exit network to yeast centrosomes couples septin displacement to actomyosin constriction. Nat. Commun 9, 4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brand F, Schumacher S, Kant S, Menon MB, Simon R, Turgeon B, Britsch S, Meloche S, Gaestel M, and Kotlyarov A (2012). The extracellular signal-regulated kinase 3 (mitogen-activated protein kinase 6 [MAPK6])-MAPK-activated protein kinase 5 signaling complex regulates septin function and dendrite morphology. Mol. Cell. Biol 32, 2467–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yadav S, Oses-Prieto JA, Peters CJ, Zhou J, Pleasure SJ, Burlingame AL, Jan LY, and Jan YN (2017). TAOK2 kinase mediates PSD95 stability and dendritic spine maturation through Septin7 phosphorylation. Neuron 93, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinoshita M, Kumar S, Mizoguchi A, Ide C, Kinoshita A, Haraguchi T, Hiraoka Y, and Noda M (1997). Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev 11, 1535–1547. [DOI] [PubMed] [Google Scholar]
- 46.Renshaw MJ, Liu J, Lavoie BD, and Wilde A (2014). Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol 4, 130190–130190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Wloka C, and Bi E (2019). Non-muscle myosin-II is required for the generation of a constriction site for subsequent abscission. iScience 13, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karasmanis EP, Hwang D, Nakos K, Bowen JR, Angelis D, and Spiliotis ET (2019). A septin double ring controls the spatiotemporal organization of the ESCRT machinery in cytokinetic abscission. Curr Biol 29, 2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caviston JP, Longtine M, Pringle JR, and Bi E (2003). The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao XD, Sperber LM, Kane SA, Tong Z, Hin Yan Tong A, Boone C, and Bi E (2007). Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol. Biol. Cell 18, 2542–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markus SM, Omer S, Baranowski K, and Lee WL (2015). Improved plasmids for fluorescent protein tagging of microtubules in Saccharomyces cerevisiae. Traffic 16, 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee S, Lim WA, and Thorn KS (2013). Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One 8, e67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Longtine MS, McKenzie A III,, DeMarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 54.Onishi M, Ko N, Nishihama R, and Pringle JR (2013). Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J. Cell Biol 202, 311–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallen EA, Caviston J, and Bi E (2000). Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwase M, and Toh-e A (2001). Nis1 encoded by YNL078W: a new neck protein of Saccharomyces cerevisiae. Genes Genet. Syst 76, 335–343. [DOI] [PubMed] [Google Scholar]
- 57.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guthrie C, and Fink GR (1991). Guide to Yeast Genetics and Molecular Biology. Methods Enzymol Vol 194, 933 pp. [PubMed] [Google Scholar]
- 59.Lillie SH, and Pringle JR (1980). Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol 143, 1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, and Ohsumi Y (2013). Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol 203, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okada S, Wloka C, and Bi E (2017). Analysis of protein dynamics during cytokinesis in budding yeast. Methods Cell Biol 137, 25–45. [DOI] [PubMed] [Google Scholar]
- 62.Svitkina T (2016). Imaging cytoskeleton components by electron microscopy. Methods Mol. Biol 1365, 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Deletion of ELM1 causes destabilization of the septin hourglass preferentially at the daughter side from bud emergence to late anaphase. Related to Figures 1 and S1. Top : Time-lapse analysis of Cdc3-GFP localization (shown in green) in relation to bud emergence and the mitotic spindle (shown in magenta) during the cell cycle by spinning-disk microscopy in WT (YEF8102) cells. Time 0 = Bud emergence. Time interval = 2 min. Scale bar = 2 μm. Imaging was performed on cells cultured at 25°C. Bottom: Time-lapse analysis of Cdc3-GFP localization (shown in green) in relation to bud emergence and the mitotic spindle (shown in magenta) during the cell cycle by spinning-disk microscopy in elm1Δ (YEF8393) cells. Time 0 = Bud emergence. Time interval = 2 min. Scale bar = 2 μm. Imaging was performed on cells cultured at 25°C.
Data Availability Statement
Data supporting the findings of this study are available within the paper and its Supplemental Information files and from the authors upon request.


