Highlights
-
•
Fragment-based drug design (FBDD) is revolutionizing the identification and optimization of new drug candidates.
-
•
Molecular simulations-based approaches are consolidating alongside with experimental and canonical structure-based (SBDD) techniques.
-
•
The implementation of molecular simulations-based techniques affects the entire FBDD pipeline.
Abstract
Fragment-based drug discovery (FBDD) is an innovative approach, progressively more applied in the academic and industrial context, to enhance hit identification for previously considered undruggable biological targets. In particular, FBDD discovers low-molecular-weight (LMW) ligands (<300 Da) able to bind to therapeutically relevant macromolecules in an affinity range from the micromolar (μM) to millimolar (mM). X-ray crystallography (XRC) and nuclear magnetic resonance (NMR) spectroscopy are commonly the methods of choice to obtain 3D information about the bound ligand–protein complex, but this can occasionally be problematic, mainly for early, low-affinity fragments. The recent development of computational fragment-based approaches provides a further strategy for improving the identification of fragment hits. In this review, we summarize the state of the art of molecular dynamics simulations approaches used in FBDD, and discuss limitations and future perspectives for these approaches.
Introduction
Over the past few decades, the advent of high-throughput screening (HTS) methodologies has contributed to revolutionizing the entire drug discovery process, making the identification of new candidates more efficient [1]. Given a relevant pharmaceutical target, thousands of compounds can nowadays be evaluated through robotic screening infrastructures, a number that reaches the impressive value of 177 million screenable molecules if computational approaches are also exploited [2]. Despite the undeniable improvements in the field, the drug-like chemical space magnitude, the dimension of which has recently been approximated to 1063 organic molecules, highlights how even current HTS methodological state of the art is barely able to scratch its surface 3, 4.
An important paradigm change occurred ∼20 years ago with the birth of FBDD, an approach that has become as a robust screening methodology in both the academic and industrial world, allowing the rapid discovery of many clinical candidates and the market approval of two drugs 5, 6. Although there is no unambiguous definition, fragments are small organic molecules usually comprising <20 nonhydrogen atoms, the physicochemical properties of which respect the so-called ‘rule of three’ (RO3) 7, 8. Despite the smaller dimension differentiating the fragment-like chemical space from the drug-sized one, a canonical FBDD campaign in which a few thousand compounds are screened provides better coverage of the chemical diversity compared with a canonical HTS [9]. Given that fragments recognize their molecular targets in an affinity range from μM to mM, their identification only represents the starting point of an iterative medicinal chemistry optimization process 6, 9. Detection of such weak binders depends on the implementation of high-sensitivity biophysical techniques, such as, isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), NMR, and XRC, with only the two latter methodologies able to provide structural information. However, many of these orthogonal techniques, apart from being expensive, have drawbacks that could limit their routine application, making at the same time the parallel implementation of computational methodologies appealing [10].
In silico tools have proven to be crucial in many steps of the FBBD pipeline, such as the identification and characterization of putative binding sites on the target of interest, the fragment screening procedure, and the candidate hit to lead optimization process [11]. However, an accurate and reliable description of the molecular recognition mechanism of the fragment is complicated by the peculiar nature of these low-affinity binders. Fragments usually present transient interactions with their biological targets and are often characterized by a population of different binding modes, rather than just one. Only recently, a series of technological and methodological revolutions have made it possible to reconcile the massive implementation of physics-based molecular simulations approaches, and of molecular dynamics (MD) simulations in particular, with the strict timing characterizing FBDD campaigns.
Here, we provide a general overview of the most innovative methodological implementations of molecular simulations to the field of FBDD, discussing how these approaches can affect all the relevant steps of the drug discovery pipeline flanking faster but less accurate structure-based in silico techniques. We also provide insight into the practical advantages and limitations of each computational protocol.
Hotspots and binding site identification
In a structure-based drug discovery (SBDD) pipeline, the identification and characterization of druggable binding sites represent a key element in determining screening success. Different experimental biophysical approaches, such as XRC or NMR, have highlighted the importance of cosolvent molecules in probing the distribution of hotspots on the surface of proteins and nucleic acids 12, 13. Beginning with this evidence, in silico protocols (e.g., GRID, MCSS, and FTMAP) were developed to exhaustively sample and map putative binding cavities, exploiting a set of chemically diverse LMW molecular probes that configure them as fragments 14, 15, 16.
However, many of these grid-based methodologies lack an adequate description of target conformational flexibility, an aspect that could limit the discovery of cryptic binding pockets, as well as neglect entropic and desolation effects, thus affecting the accuracy and reliability of computational predictions. The recent implementation and validation of molecular simulation-based approaches, such as molecular dynamics (MD) simulations, represented a breakthrough in binding site identification, overcoming some of the aforementioned limitations and, at the same time, guaranteeing competitive computational times.
MD simulations
MD simulations describe the time-dependent evolution of a fully solvated molecular system with an atomistic detail, through the numerical solution of Newton’s second law of motion. The integration is partitioned into the discrete interval (usually 1 or 2 fs)-defined time step, chosen in such a way to guarantee a correct description of the fastest degrees of freedom of the system and efficient calculations [17]. The interatomic forces are approximated using mathematical models based on classical mechanics, define force fields (FF), which have been extensively parameterized to reproduce experimental or quantum-mechanics (QM) data. Therefore, MD is a computational tool with an impressive temporal resolution, able to describe events spanning a range of 12 orders of magnitude among the molecular timescale, from bond vibration (fs) to the folding of small proteins (ms) [18].
About 10 years ago, Barril and MacKerell and their research groups independently began to investigate the use of mixed-solvent MD simulations (MSMD) to assess the ‘druggability’ of different biological targets, developing MixMD and SILCS protocols, respectively 19, 20. The SILCS approach differs from MixMD in that it uses unphysical organic solvent concentrations close to saturation conditions, an artifact that improves the effectiveness of sampling, but also requires the introduction of a repulsive potential between the probe molecules to avoid their aggregation 21, 22. Both protocols start from a series of nanosecond-long MD trajectories that extrapolate solvent occupancy maps, which can be used not only to characterize the interactivity nature of the binding cavity, but also to improve the accuracy of fragment posing, to rationally drive the optimization process and to estimate fragment binding affinity [23]. In a retrospective study based on 21 protein-protein interactions (PPIs) interfaces with known small molecules inhibitors bound, the performances of MSMD approaches in detecting druggable hotspots were compared with those of traditional, less demanding, grid-based protocols, highlighting a greater accuracy of the former [24]. By contrast, multiple simulations or extensive sampling are required to ensure the convergence of the results, an aspect that might limit their routine implementation. Thus, the CrypticScout platform was recently released on the PlayMolecule webserver to make the set-up, collection, and analysis of MSMD simulations available on a large scale, regardless of the computational infrastructure available to each research group, exploiting the distributed computing power of the GPUGRID project 25, 26.
MSMD simulation performances have often been calibrated on their ability to quantitatively identify experimental known binding sites. However, as recently highlighted by Astex Pharmaceuticals, qualitative and accurate discrimination between ‘warm’ and ‘hot’ spots, reflecting a modest or high fragment affinity, respectively, is even more important [27]. Thus, MD was combined with the grand-canonical Monte Carlo approach (GCMC-MD) by the MacKerell group to assess the druggability profile of the different binding sites. In detail, a multi-step protocol was design to collect via MC posing the configurations of hundreds of drug-like fragments on the target surface, which are then geometrically discretized and ranked, exploiting the previously calculated MSMD occupancy maps [28]. In addition, by exploiting nanosecond timescales, GCMC-MD simulations enhance the sampling of the probe within buried and cryptic pockets, which are otherwise poorly accessible [29]. The identification of these ancillary sites is becoming increasingly important from a pharmaceutical perspective, especially in cases of so-called ‘undruggable’ targets or in the development of allosteric modulators.
Hit fragment identification and characterization
The identification and characterization of putative binding sites represent a fundamental but preparatory step in the complex in silico FBDD pipeline, with the subsequent screening phase having a pivotal role in selecting, among the vast chemical space, new chemotypes for which experimental evaluation should be prioritized. Molecular docking is a relatively fast computational protocol exploited to sample and score ligand binding modes, the application of which has long been debated also in the case of the FBDD. The pioneering work of Shoichet in 2009 laid the foundation for validating docking-based fragment screening, allowing the identification of ten millimolar-range CTX-M β-lactamase inhibitors. Despite many other positive examples, important drawbacks in docking-based approaches have started to emerge, some of which are intrinsic to the methodology, such as a limited consideration of target flexibility or the lack of an accurate treatment of solvent contribution to binding, whereas as others are related to the nature of these low-affinity compounds. Most docking scoring functions have been empirically trained based on potent lead compounds and, thus, concerns have arisen about their ability to distinguish active from nonactive fragments, or native from other low-energy fragment-binding modes [30].
Therefore, the progressive development and validation of more sophisticated physics-based molecular simulation approaches, the most relevant of which are discussed herein, could improve the reliability of in silico predictions in fragment screening.
Nonequilibrium candidate Monte Carlo and molecular dynamic simulations
As early as the 2000s, MD simulations began to be explored as a postprocessing tool to refine and characterize molecular docking-predicted complexes, an approach often identified as ‘post-docking’ [31]. However, it is now clear how the timescale required to extensively sample binding-mode transition and to realistically estimate the distribution of fragment populations massively exceeds μs, thus resulting in computationally expansive simulations. To deal with this problem, Mobley’s research group recently developed a protocol called ‘Binding Modes of Ligands Using Enhanced Sampling’ (BLUES), which improves the sampling of the metastable binding modes of fragments, allowing the simulations to easily escape from local minima of the potential energy surface [32]. The BLUES protocol is based on nonequilibrium candidate Monte Carlo (NCMC), an algorithm that increases the efficacy of configurational sampling concerning classical MD simulation, while providing a higher acceptance rate compared with traditional MC simulation. In detail, BLUES collects a sequence of perturbation steps in which a fragment is alchemically annihilated within its binding site and randomly rotated, followed by propagation steps in which the ligand interactions are restored and the molecular system is relaxed through Langevin MD. In the end, the whole NCMC step is then accepted or rejected based on the nonequilibrium work accumulated during the different perturbation and propagation iterations. The protocol effectiveness was firstly validated on the T4 lysozyme model system, showing an improvement of two orders of magnitude in toluene-binding mode population prediction, in terms of brute-force MD simulations [32]. Subsequently, the pharmaceutically more relevant soluble epoxide hydrolase (SEH) case study was considered, investigating BLUES accuracy in identifying the experimental binding mode of 12 fragments in which structures were solved by Astra Zeneca [33]. Also in this case, the BLUES protocol outperformed the traditional in silico methodologies, recovering the fragment crystallographic binding modes in 86% of cases and providing a reliable estimation of their relative population (whereas docking and MD provided only 7% and 48% of correct predictions, respectively). However, even exploiting this innovative protocol, simulation timescales close to μs are required to ensure the accuracy of results, making BLUES implementation in a real HTS scenario challenging.
Molecular dynamics simulation and Markov state models
An aspect that needs to be addressed when performing MD simulations is represented by the huge amount of computing time required to sample pharmaceutically relevant events, such as, the molecular recognition of a single fragment with its macromolecular target, from the unbound to the bound state, while ensuring the stability of numerical integration. Alternatively to the use of unphysical fragment concentrations, as previously described with MSMD, dedicated hardware infrastructures have also been engineered to improve the sampling of long-timescale events. A pioneering example is represented by the Anton supercomputer, exploited by the Shaw research group to characterize the recognition of a small library comprising six fragments towards FKBP, a prolyl isomerase protein [34]. Multiple equilibrium simulations reaching the microsecond timescale were collected for each compound, long enough to sample the binding and unbinding molecular events at least a hundred times repeatedly. These trajectories were analyzed to estimate, with a great degree of accuracy with respect to the experimental values, the fragment equilibrium dissociation constants (KD), thus allowing the high confidence ranking of the candidates. This represented the first limited, but promising attempt to perform unbiased MD-based fragment screening [35].
A change in paradigm occurred with the extraction of stochastic information regarding long-timescale events from multiple short simulations, rather than from a single long one [36]. This was achieved by applying Markov state model (MSM) analysis, a framework of a statistical model that discretizes and describes the configurational space sampled by a biomolecular system through, for example, an ensemble of MD simulations. A MSM is constructed by clustering the trajectories into relevant states and then monitoring the transition among each of these states during a specific lag time τ, chosen to ensure memoryless behavior (Markovian) to the system 37, 38. A transition probability matrix approximating the real dynamics of the molecular system is then derived, from which thermodynamic and kinetic quantities can be extracted, as well as phenomena that occur on timescales longer from those sampled by a single simulation. This approach was recently applied by Boehringer Ingelheim against two targets of pharmaceutical interest, neutrophil elastase (NE) and a proline-isomerase domain of FKBP51, which had both been the subject of an FBDD screening [39]. A combination of unbiased MD simulations with MSM analysis was exploited, stressing the methodology performance in reproducing crystallographic binding modes of five molecules. For each fragment–protein complex, an ensemble of 50 μs of simulations was first sampled, and the trajectories were then geometrically clusterized depending on the coordinates of fragment heteroatoms. To improve the accuracy of the results, MSMs were iteratively generated by changing the number of clusters exploited to discretize the simulations and the relative lag time τ, until the most populated state for every parameter combination converged. The procedure ensured not only a reliable fragment-binding mode prediction, but also an estimation of the relative confidence. This work highlighted how the combination of unbiased MD simulation and MSMs significantly improves the posing accuracy concerning molecular docking, correctly anticipating the binding mode of four of the five fragments examined, whereas docking was unable to yield a prediction within 3 Å from the X-ray reference [39].
Although these examples corroborate the methodological accuracy characterizing MD-MSM approaches, it is difficult to reconcile their application to a more traditional drug discovery scenario. For this reason, an MD-based fragment screening of a library containing 129 candidates was recently performed by the De Fabritiis group against a relevant oncological target, the chemokine CXCL12 monomer [40]. In this case, the MSM framework was applied only to perform the analysis of the MD trajectories ensemble, but also to actively drive the sampling, in a protocol-defined adaptive sampling approach. For each fragment–protein complex, a series of short MD simulations [70-nanoseconds (ns) long] was initially collected and exploited to iteratively build MSMs, identifying undersampled regions of the phase space from which to start new simulations 40, 41, 42. Through this adaptive scheme, an average of 45 μs of simulations was collected for each fragment, obtaining 5.85 ms of total MD simulation time if the entire library is considered. This huge amount of data was exploited to build the final MSM, from which both kinetic and thermodynamic information describing ligand binding was extracted. This work represents a first attempt to automate the screening of a small fragment library exploiting molecular simulation, even if the lack of experimental validation does not enable the evaluation of the predictive performances or the accuracy of the methodology [40].
Supervised molecular dynamics simulations
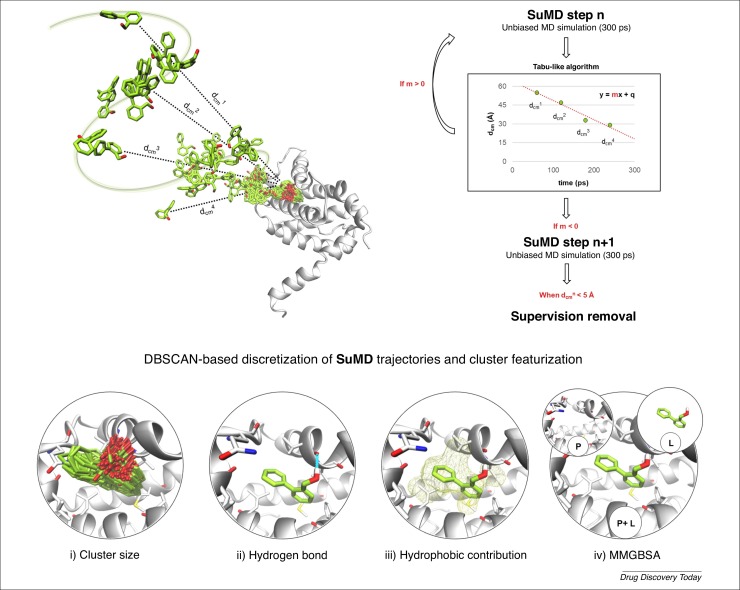
Along with MSMs, other approaches have been implemented to improve the performance of classical molecular simulations for the characterization of long-timescale events. For example, an algorithm called supervised molecular dynamics (SuMD) was developed, which differs from other enhanced sampling approaches because it does not perturb the free energy surface of the system 43, 44, 45. SuMD allows exploration of the entire ligand–receptor recognition pathway, from the unbound to the bound state, in a ns timescale, reducing the computational efforts needed by up to three orders of magnitude. This is achieved by collecting short unbiased MD simulations and monitoring how the protein–ligand distance changes over time (Fig. 1 ). A tabu-like algorithm accepts all the productive steps, simulations in which an approach of the ligand is sampled, rejecting and simulating again from the previous coordinates set those steps describing instead a diffusion of the ligand far from the target. Once the binding site vestibule has been reached, the supervision algorithm is turned off, allowing classic MD simulation to relax the final state. This methodology has proven to be reliable not only in reproducing crystallographic complexes with great geometric accuracy, but also in elucidating the entire recognition pathway for both mature and fragment-like molecules 43, 46, 47.
Figure 1.
High-throughput supervised molecular dynamics (HT-SuMD), an automated protocol exploiting molecular simulation to perform fragment screening. The SuMD methodology is summarized with a specific focus on the tabu-like algorithm controlling acceptance or rejection of short unbiased MD simulations, depending on how the distance between the fragment under investigation and the binding site center of mass (dcmn) changes during the trajectory. A density-based clustering algorithm (DBSCAN) clustering algorithm is used to perform a geometrical discretization of SuMD trajectories and identify relevant fragment conformations. Each cluster is then characterized based on four geometric and energetic indicators: (i) cluster size; (ii) hydrogen bond presence; (iii) hydrophobic contribution of binding; and (iv) protein–ligand MMGBSA binding interaction energy. Once all clusters have been characterized, a consensus scoring filter is applied to identify hit fragment molecules.
SuMD application in the context of FBDD was recently investigated, in a screening of a fragment library containing 400 molecules against the target Bcl-xL, an antiapoptotic protein member of the Bcl-2 family [62]. A computational protocol defined as ‘high-throughput supervised molecular dynamics’ (HT-SuMD) was developed to control in a fully automated fashion both the phase of simulation collection and the subsequent analysis of raw data. Given that fragments usually recognize their molecular target through weak and transient interactions, which can determine multiple ligand-binding modes, a specific set of analyses were tailored. The ensemble of SuMD trajectories was geometrically discretized through a density-based clustering algorithm (DBSCAN) to highlight well-populated families of molecule conformations from background noise (Fig. 1) [48]. Each cluster was then characterized based on four geometric or energetic observable (Fig. 1, panel i to iv), which can help to reveal the more stable fragment conformation. Hit fragments were identified by applying a consensus scoring strategy to all the clusters analyzed. The accuracy of the in silico calculation was cross-validated through a comparative but independent NMR study, which highlighted impressive convergence between the hit candidates identified by the two orthogonal methodologies. In particular, all the first-choice hits predicted by HT-SuMD were also confirmed as Bcl-xL binders in the mM range by NMR experiments. To date, this represents the largest fragment screening completely driven by MD simulation reported in the literature, showing how it could be possible to reconcile the use of molecular simulations, even on a large scale, with the tight timing characterizing FBDD. However, HT-SuMD screening protocol requires, contrarily to unbiased MD approaches, a priori knowledge of the binding site localization, thus benefiting from a combined use with methodologies capable of identifying putative binding hotspots.
Steered molecular dynamics simulations
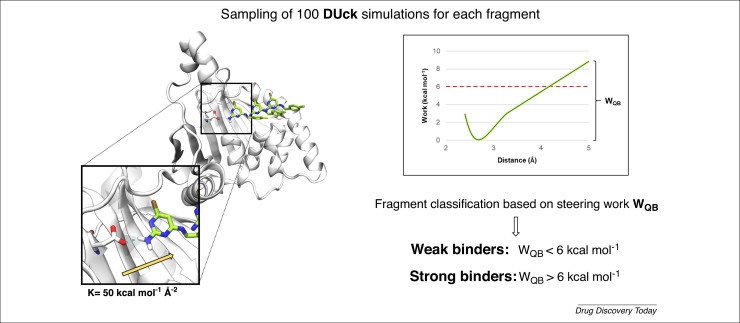
Understanding the molecular determinants underneath protein–fragment structural stability is becoming a crucial step in FBDD. As a consequence, a large-scale analysis was recently performed on 489 high-resolution crystallographic structures, all containing fragments. Remarkably, 92% of the complexes now available are characterized by the presence of at least one intermolecular hydrogen bond and, more importantly, 88% of the hydrogen bonds with the protein target are completely water-shielded [49]. Given that it has been demonstrated how buried hydrogen bond interactions enhance the structural stability of protein–fragment complexes, acting as a kinetic trap, computational approaches have recently been developed to evaluate the energetic contribution of the aforementioned interactions [50]. For example, steered molecular dynamics (SMD) is a technique that takes its inspiration from the experimental methodology atomic force microscopy (AFM), allowing the investigation of force-probe events through the application of an external force vector to the system [51]. A ligand unbinding process can be described by centering the vector on the fragment molecule and then pulling the dissociation process, even though the results might be significantly influenced depending on how the direction and magnitude of the force are chosen. For this specific purpose, a form of SMD called dynamic undocking (DUck) was recently developed. DUck simulations control the application of a force vector on a key hydrogen bond interaction known to stabilize the protein–fragment complex, which is pulled at an approximate distance of 5 Å until the contact breaks [52]. From each nonequilibrium steering process, a property defined as quasi-bound work (WQB) is then calculated, which represents the maximum amount of work characterizing the process of hydrogen bond breaking. Even if it is not possible to establish a direct correlation between the WQB value of a molecule and its binding affinity, which in contrast to the former is an equilibrium property, the Barril research group investigated the application of DUck in a fragment screening campaign. The protocol was applied to a crystallographic set of 41 fragment-like complexes of the cyclin-dependent kinase (CDK2), monitoring the WQB required to break the key hydrogen bond contact with the protein hinge region. As summarized in Fig. 2 , the results showed how WQB can accurately discriminate and classify strong (IC50 < 1 mM) from weak fragment binders (IC50 > 1 mM), obtaining a similar outcome towards a second pharmaceutically relevant target, the BRD4-BD1 bromodomain. DUck methodology was also investigated in a real fragment-screening scenario toward the oncological target heat shock protein 90 kDa (Hsp90), combining docking–undocking simulations. A subset of 139 candidates was selected through conventional molecular docking and subsequently subjected to 100 DUck runs to ensure convergence of the WQB values. Of the 21 fragments predicted as strong binders (WQB > 6 kcal mol−1), eight were confirmed as true Hsp90 binders based on NMR experiments, showing a hit rate value close to 40% and, thus, supporting the implementation of DUck in FBDD pipelines.
Figure 2.
Dynamic undocking (DUck) is a steered molecular dynamics (SMD)-based protocol in which a fragment unbinding pathway is sampled by pulling a key stabilizing hydrogen bond interaction through the application of a directional force vector. The maximum amount of steering work required for the contact rupture is exploited as a nonequilibrium property differentiating strong from weak fragment binding molecules.
Hit to lead fragment optimization
Once a low-affinity hit fragment has been identified, a multistep medicinal chemistry optimization process begins to improve candidate pharmacodynamic and pharmacokinetic properties. For this purpose, a multitude of structure-based in silico protocols have been developed over the past few decades. Fragment maturation could be driven by exploiting core positional restraints, pharmacophoric models, grid-based approaches, or statistics/active learning techniques [53]. However, exploration of a focused region of the chemical space, starting from an initial fragment seed, is only useful if combined with in silico methodologies able to anticipate the binding affinity of the candidate, thus guiding research decisions and prioritizing the synthesis of the most promising lead compounds. From this perspective, molecular simulation-based binding free energy calculations are becoming a gold standard in hit-to-lead optimization pipelines.
Binding free energy calculations
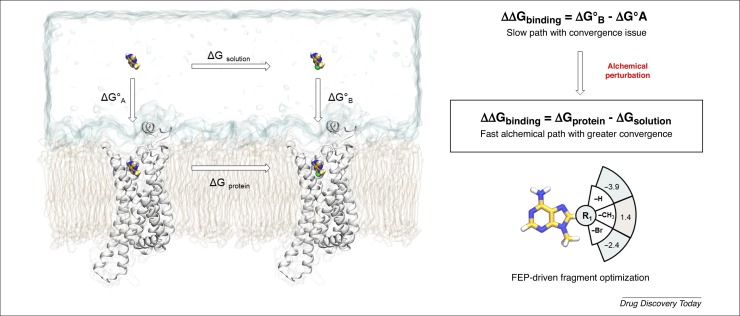
Calculation of absolute binding affinity (ΔG), defined as the free energy difference between a ligand in its bound and unbound state, still remains challenging. The impossibility of describing the molecular system energetics with a quantum mechanics level of accuracy, along with inadequate sampling of the system configurations, affect the prediction reliability [54]. The calculation of relative binding free energy (ΔΔG) instead, defined as the difference in free energy characterizing a series of congener compounds, is becoming increasingly rigorous and efficient. Among the different approaches based on statistical mechanics, free energy perturbation (FEP) methods currently represent the state of the art for ΔΔG predictions. Instead of computing free energy changes as the difference between two absolute ΔG values, FEP exploits a nonphysical thermodynamic cycle in which a ligand is perturbed through an alchemical transformation into another, both in the aqueous solution and within its protein binding site (Fig. 3 ). In recent retrospective work by Schrödinger, the application of FEP protocols to the FBDD field was extensively validated, investigating more than 90 fragment molecules recognizing eight pharmaceutically relevant targets [55]. Results showed a good correlation between the experimental changes in binding affinity and the predicted ΔΔG values, with an R2 value of 0.65 and a root mean square error (RMSE) of 1.14 kcal/mol. Moreover, in 89% of the predictions, the FEP protocol correctly anticipated the sign of ΔΔG, thus allowing the researchers to discern the putative effect of the chemical modification, that is, whether it improved the candidate affinity. These results become even more interesting when comparing FEP predictive performances with those of an empirical scoring function (Glide SP) or with an FF-based scoring method (MMGBSA), demonstrating how the first approach consistently outperforms the latter two. The Carlsson group evaluated the performance of FEP in a fragment optimization pipeline applying the protocol to A2A adenosine receptor (A2AAR), one of the most investigated G-protein-coupled receptors (GPCRs) [56]. The relative binding affinity of a series of 23 adenine compounds, which explore two different cavities of the A2AAR orthosteric binding site, was evaluated, also highlighting a strong correlation between FEP prediction and experimental ΔΔG, with an R2 value of 0.78. Apart from the encouraging evaluation of FEP performance and accuracy, some crucial issues could impact this protocol and, therefore, must be taken into consideration in an FBDD project. Given that a typical fragment optimization usually results in a modest perturbation of ligand-binding affinity (∼1 kcal/mol), the uncertainty that it is necessary to handle in free energy calculations could be comparable or, in some case greater, to the thermodynamic quantity of interest [54]. Furthermore, despite the continuous improvements in FF, an inaccurate parameterization of the ligand under investigation, especially as in terms of the torsional energy profile, can negatively influence the reliability of the prediction.
Figure 3.
Relative binding free energy calculation as a valuable tool for driving fragment optimization campaigns. On the left side of the panel, the traditional thermodynamics cycle for ΔΔG calculation exploiting molecular simulation is depicted. The physical path (vertical arrows) describing the absolute free energy of binding (ΔG°) are affected by convergence because of the massive system perturbation sampled, making the calculation inefficient. On the right side, the nonphysical alchemical path (horizontal arrow) is shown, in which a ligand is perturbed into another both in the bound and unbound state, providing greater convergence. Abbreviation: FEP, free energy perturbation.
As the aforementioned studies have highlighted, the precise knowledge of fragment-binding modes represents a crucial aspect for FEP calculation correctness, with the most significant ΔΔG deviations described for those fragments characterized by an obscure modality of recognition. Moreover, in an FEP protocol, it is usually assumed that a fragment maintains its original conformation even after a chemical perturbation; this does not always correspond to reality and both ligand and protein, along with the water hydration network organization, can be significantly altered. In all these cases, an incorrect ΔΔG prediction will be obtained unless the FEP simulation allows sampling of an interconversion of the binding mode [57]. Lastly, particular attention must be paid before performing FEP calculations of fragments that could undergo tautomeric or ionization equilibrium, a not so infrequent event, because the interconversion cannot be sampled through MD simulations, thus altering the molecular recognition free energy profile [54].
Concluding remarks and perspectives
Starting from pioneering research by Karplus and McCammon in 1977, the year in which the first all-atom MD simulation of a protein was performed, molecular simulations have begun to acquire increasing importance among the scientific community [58]. The progressive optimization of MD algorithms, together with the advent of GPU architectures, has contributed to turning molecular simulations in a computational microscope characterized by an impressive spatiotemporal resolution. Therefore, physics-based simulations have started to accompany classic structure-based computational approaches in drug discovery pipelines and, recently, have also found application in the field of FBDD. This review offers an overview of the latest implementations of molecular simulations in a typical FBDD campaign, highlighting how these approaches can impact many crucial steps, from the identification of druggable binding sites, the screening of fragments libraries, to subsequent hit to lead optimization phase (Fig. 4 ). Many methodological applications have been described herein, comparing both their applicability domain as well as the simulation timescales required. Molecular simulation-based approaches efficiently capture the highly dynamic nature of low-affinity fragments and, contrary to many structural biology techniques, also unveil vital information about the lowest populated conformational states. This knowledge could be particularly important to rationally drive the fragment maturation process, suggesting different linking or growing directions. A further emerging aspect is the better ranking ability of fragment compounds by using an MD-based strategy compared with conventional rigid techniques. Again, the possibility of investigating both protein flexibility and desolvation effect will have a significant impact on improving the fragment-screening procedure, both in terms of the initial problem as well as the characterization of thermodynamics and kinetics.
Figure 4.
Summary of a canonical in silico fragment-based drug discovery (FBDD) pipeline, highlighting the applicability of different molecular simulation-based approaches. For each computational methodology, the simulation timescale that is required is reported, as well as the main (black dot) or secondary (white dots) applications. The table highlights the differences between approaches that can provide cross-cutting support to many FBDD phases (e.g., Markov state models; MSMs) from those that, because of their specificity and complexity, have a more focused use (e.g., free energy perturbation; FEP). Abbreviations: HT-SuMD, high-throughput supervised molecular dynamics; MSMD, mixed-solvent molecular dynamics; NCMC, nonequilibrium candidate Monte Carlo; SMD, steered molecular dynamics.
The implementation of molecular simulations in the field of FBDD is becoming a consolidated practice. As evidence of this, following the contemporary pandemic condition caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spread, a timely fragment screening against the Main protease (Mpro) of the virus has made tens of crystallographic structures available to the scientific community [63]. The massive distributed computing power provided by the Folding@home project is currently being exploited to in silico screen tens of thousands of promising inhibitors, using the state of the art of binding free energy calculation to prioritize the synthesis and the experimental validation of promising candidates, hopefully accelerating the discovering of new therapeutics 59, 60.
Despite the undeniable methodological improvements and success described so far, the predictive accuracy of these methodologies remains an issue. Continuous optimization of the FF parameters is desirable, for example through the implementation of polarization effects, as is the necessary improvement of the efficiency with which the configurational space of the molecular system of interest is sampled [61]. In light of these issues and considering the continuous and exponential improvements in computational performance, ever greater implementation of molecular simulations in the FBDD field can be envisioned, accelerating and making more efficient the entire process of rational drug discovery.
Acknowledgments
The M.M.S. lab is very grateful to the Chemical Computing Group, NVIDIA Corporation, OpenEye, and Acellera for scientific and technical partnership. This research was financially supported by MIUR (PRIN2017, n.2017MT3993).
References
- 1.MacArron R. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 2.Lyu J. Ultra-large library docking for discovering new chemotypes. Nature. 2019;566:224–229. doi: 10.1038/s41586-019-0917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reymond J.L. The Chemical Space Project. ACC Chem. Res. 2015;48:722–730. doi: 10.1021/ar500432k. [DOI] [PubMed] [Google Scholar]
- 4.Hall R.J. Efficient exploration of chemical space by fragment-based screening. Prog. Biophys. Mol. Biol. 2014;116:82–91. doi: 10.1016/j.pbiomolbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Jacquemard C., Kellenberger E. A bright future for fragment-based drug discovery: what does it hold? Expert Opin. Drug. Discov. 2019;14:413–416. doi: 10.1080/17460441.2019.1583643. [DOI] [PubMed] [Google Scholar]
- 6.Hajduk P.J., Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 7.Congreve M. A ‘Rule of Three’ for fragment-based lead discovery? Drug Discov. Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 8.Jhoti H. The ‘rule of three’ for fragment-based drug discovery: where are we now? Nat. Rev. Drug Discov. 2013;12:644. doi: 10.1038/nrd3926-c1. [DOI] [PubMed] [Google Scholar]
- 9.Erlanson D.A. Twenty years on: the impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 10.Zoete V. Docking, virtual high throughput screening and in silico fragment-based drug design. J. Cell. Mol. Med. 2009;13:238–248. doi: 10.1111/j.1582-4934.2008.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortier J. Computational tools for in silico fragment-based drug design. Curr. Top. Med. Chem. 2012;12:1935–1943. doi: 10.2174/156802612804547371. [DOI] [PubMed] [Google Scholar]
- 12.Mattos C. Multiple solvent crystal structures: probing binding sites, plasticity and hydration. J. Mol. Biol. 2006;357:1471–1482. doi: 10.1016/j.jmb.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 13.Liepinsh E., Otting G. Organic solvents identify specific ligand binding sites on protein surfaces. Nat. Biotechnol. 1997;15:264–268. doi: 10.1038/nbt0397-264. [DOI] [PubMed] [Google Scholar]
- 14.Goodford P.J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 15.Miranker A., Karplus M. Functionality maps of binding sites: a multiple copy simultaneous search method. Proteins Struct. Funct. Genet. 1991;11:29–34. doi: 10.1002/prot.340110104. [DOI] [PubMed] [Google Scholar]
- 16.Ho Ngan C. FTMAP: extended protein mapping with user-selected probe molecules. Nucleic Acids Res. 2012;40:W271–W275. doi: 10.1093/nar/gks441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Vivo M. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 2016;59:4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- 18.Dror R.O. Perspectives on: molecular dynamics and computational methods Exploring atomic resolution physiology on a femtosecond to millisecond timescale using molecular dynamics simulations. J. Gen. Physiol. 2010;135:555–562. doi: 10.1085/jgp.200910373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Garcia D., Barril X. Molecular simulations with solvent competition quantify water displaceability and provide accurate interaction maps of protein binding sites. J. Med. Chem. 2014;57:8530–8539. doi: 10.1021/jm5010418. [DOI] [PubMed] [Google Scholar]
- 20.Guvench O., MacKerell A.D. Computational fragment-based binding site identification by ligand competitive saturation. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faller C.E. Site identification by ligand competitive saturation (silcs) simulations for fragment-based drug design. Methods Mol. Biol. 2015;1289:75–87. doi: 10.1007/978-1-4939-2486-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Defelipe L.A. Solvents to fragments to drugs: MD applications in drug design. Molecules. 2018;23:3269. doi: 10.3390/molecules23123269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arcon J.P. Molecular dynamics in mixed solvents reveals protein-ligand interactions, improves docking, and allows accurate binding free energy predictions. J. Chem. Inf. Model. 2017;57:846–863. doi: 10.1021/acs.jcim.6b00678. [DOI] [PubMed] [Google Scholar]
- 24.Ghanakota P. Large-scale validation of mixed-solvent simulations to assess hotspots at protein–protein interaction interfaces. J. Chem. Inf. Model. 2018;58:784–793. doi: 10.1021/acs.jcim.7b00487. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Rosell G. PlayMolecule CrypticScout: predicting protein cryptic sites using mixed-solvent molecular simulations. J. Chem. Inf. Model. 2020;2020:2314–2324. doi: 10.1021/acs.jcim.9b01209. [DOI] [PubMed] [Google Scholar]
- 26.Buch I. High-throughput all-atom molecular dynamics simulations using distributed computing. J. Chem. Inf. Model. 2010;50:397–403. doi: 10.1021/ci900455r. [DOI] [PubMed] [Google Scholar]
- 27.Rathi P.C. Predicting ‘hot’ and ‘warm’ spots for fragment binding. J. Med. Chem. 2017;60:4036–4046. doi: 10.1021/acs.jmedchem.7b00366. [DOI] [PubMed] [Google Scholar]
- 28.MacKerell A.D. Identification and characterization of fragment binding sites for allosteric ligand design using the site identification by ligand competitive saturation hotspots approach (SILCS-Hotspots) Biochim. Biophys. Acta. 2020;1864:129519. doi: 10.1016/j.bbagen.2020.129519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ustach V.D. Optimization and Evaluation of site-identification by ligand competitive saturation (SILCS) as a tool for target-based ligand optimization. J. Chem. Inf. Model. 2019;59:3018–3035. doi: 10.1021/acs.jcim.9b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdonk M.L. Docking performance of fragments and druglike compounds. J. Med. Chem. 2011;54:5422–5431. doi: 10.1021/jm200558u. [DOI] [PubMed] [Google Scholar]
- 31.Alonso H. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 2006;26:531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 32.Gill S.C. Binding modes of ligands using enhanced sampling (BLUES): rapid decorrelation of ligand binding modes via nonequilibrium candidate Monte Carlo. J. Phys. Chem. B. 2018;122:5579–5598. doi: 10.1021/acs.jpcb.7b11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim N.M. Fragment pose prediction using non-equilibrium candidate Monte Carlo and molecular dynamics simulations. J. Chem. Theory Comput. 2020;16:2778–2794. doi: 10.1021/acs.jctc.9b01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw D.E. Millisecond-scale molecular dynamics simulations on Anton. Proc. Conference on High Performance Computing, Networking, Storage And Analysis; Association for Computing Machinery, New York; 2009. pp. 1–11. [Google Scholar]
- 35.Pan A.C. Quantitative characterization of the binding and unbinding of millimolar drug fragments with molecular dynamics simulations. J. Chem. Theory Comput. 2017;13:3372–3377. doi: 10.1021/acs.jctc.7b00172. [DOI] [PubMed] [Google Scholar]
- 36.Chodera J.D., Noé F. Markov state models of biomolecular conformational dynamics. Curr. Opin. Struct. Biol. 2014;25:135–144. doi: 10.1016/j.sbi.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Husic B.E., Pande V.S. Markov state models: from an art to a science. J. Am. Chem. Soc. 2018;140:2386–2396. doi: 10.1021/jacs.7b12191. [DOI] [PubMed] [Google Scholar]
- 38.Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front. Pharmacol. 2018;9:923. doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linker S.M. Fragment binding pose predictions using unbiased simulations and Markov-state models. J. Chem. Theory Comput. 2019;15:4974–4981. doi: 10.1021/acs.jctc.9b00069. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Rosell G. Molecular-simulation-driven fragment screening for the discovery of new CXCL12 inhibitors. J. Chem. Inf. Model. 2018;58:683–691. doi: 10.1021/acs.jcim.7b00625. [DOI] [PubMed] [Google Scholar]
- 41.Doerr S. HTMD: High-throughput molecular dynamics for molecular discovery. J. Chem. Theory Comput. 2016;12:1845–1852. doi: 10.1021/acs.jctc.6b00049. [DOI] [PubMed] [Google Scholar]
- 42.Doerr S., De Fabritiis G. On-the-fly learning and sampling of ligand binding by high-throughput molecular simulations. J. Chem. Theory Comput. 2014;10:2064–2069. doi: 10.1021/ct400919u. [DOI] [PubMed] [Google Scholar]
- 43.Sabbadin D., Moro S. Supervised molecular dynamics (SuMD) as a helpful tool to depict GPCR–ligand recognition pathway in a nanosecond time scale. J. Chem. Inf. Model. 2014;54:372–376. doi: 10.1021/ci400766b. [DOI] [PubMed] [Google Scholar]
- 44.Sabbadin D. Exploring the recognition pathway at the human A 2A adenosine receptor of the endogenous agonist adenosine using supervised molecular dynamics simulations. Medchemcomm. 2015;6:1081–1085. [Google Scholar]
- 45.Salmaso V. Exploring protein-peptide recognition pathways using a supervised molecular dynamics approach. Structure. 2017;25:655–662. doi: 10.1016/j.str.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Bissaro M. Targeting protein kinase CK1δ with riluzole: could it be one of the possible missing bricks to interpret its effect in the treatment of ALS from a molecular point of view? ChemMedChem. 2018;13:2601–2605. doi: 10.1002/cmdc.201800632. [DOI] [PubMed] [Google Scholar]
- 47.Deganutti G., Moro S. Supporting the identification of novel fragment-based positive allosteric modulators using a supervised molecular dynamics approach: a retrospective analysis considering the human A2A adenosine receptor as a key example. Molecules. 2017;22:818. doi: 10.3390/molecules22050818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ester M. A density-based algorithm for discovering clusters in large spatial databases with noise. Kdd. 1996;96:226–231. [Google Scholar]
- 49.Giordanetto F. Fragment hits: what do they look like and how do they bind? J. Med. Chem. 2019;62:3381–3394. doi: 10.1021/acs.jmedchem.8b01855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidtke P. Shielded hydrogen bonds as structural determinants of binding kinetics: Application in drug design. J. Am. Chem. Soc. 2011;133:18903–18910. doi: 10.1021/ja207494u. [DOI] [PubMed] [Google Scholar]
- 51.Do P.C. Steered molecular dynamics simulation in rational drug design. J. Chem. Inf. Model. 2018;58:1473–1482. doi: 10.1021/acs.jcim.8b00261. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz-Carmona S. Dynamic undocking and the quasi-bound state as tools for drug discovery. Nat. Chem. 2017;9:201–206. doi: 10.1038/nchem.2660. [DOI] [PubMed] [Google Scholar]
- 53.de Souza Neto L.R. In silico strategies to support fragment-to-lead optimization in drug discovery. SILC Chem. 2020;8:93. doi: 10.3389/fchem.2020.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cournia Z. Relative binding free energy calculations in drug discovery: recent advances and practical considerations. J. Chem. Inf. Model. 2017;57:2911–2937. doi: 10.1021/acs.jcim.7b00564. [DOI] [PubMed] [Google Scholar]
- 55.Steinbrecher T.B. Accurate binding free energy predictions in fragment optimization. J. Chem. Inf. Model. 2015;55:2411–2420. doi: 10.1021/acs.jcim.5b00538. [DOI] [PubMed] [Google Scholar]
- 56.Matricon P. Fragment optimization for GPCRs by molecular dynamics free energy calculations: Probing druggable subpockets of the A2A adenosine receptor binding site. Sci. Rep. 2017;7:6398. doi: 10.1038/s41598-017-04905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mobley D.L., Klimovich P.V. Perspective: alchemical free energy calculations for drug discovery. J. Chem. Phys. 2012;137 doi: 10.1063/1.4769292. XXX–YYY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCammon J.A. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 59.Larson S.M. Folding@Home and Genome@Home: using distributed computing to tackle previously intractable problems in computational biology. ArXiv. 2009;2009 0901.0866. [Google Scholar]
- 60.Voelz V. 2020. New COVID-19 Small Molecule Screening Simulations Are Running on Full Folding@home! Folding@home. [Google Scholar]
- 61.Jing Z. Polarizable force fields for biomolecular simulations: recent advances and applications. Annu. Rev. Biophys. 2019;48:371–394. doi: 10.1146/annurev-biophys-070317-033349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferrari, F. Bissaro, M. Fabbian, S. de Almeida Roger, J. Mammi, S. and Moro, S. et al. (2020) HT-SuMD: making molecular dynamics simulations suitable for fragment-based screening. A comparative study with NMR. 10.26434/CHEMRXIV.12582662.V1. [DOI] [PMC free article] [PubMed]
- 63.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. BioRxiv. 2020 doi: 10.1101/2020.05.27.118117. 2020.05.27.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]