Abstract
Expression quantitative trait locus (eQTL) analyses have enabled us to predict the function of disease susceptibility SNPs. However, eQTL for the effector memory T cells (TEM) located in the lamina propria mononuclear cells (LPMCs), which play an important role in Crohn’s disease (CD), are not yet available. Thus, we conducted RNA sequencing and eQTL analyses of TEM cells located in the LPMCs from IBD patients (n = 20). Genome-wide association study (GWAS) was performed using genotyping data of 713 Japanese CD patients and 2,063 controls. We compared the results of GWAS and eQTL of TEM, and also performed a transcriptome-wide association study using eQTL from Genotype Tissue Expression project. By eQTL analyses of TEM, correlations of possible candidates were confirmed in 22,632 pairs and 2,463 genes. Among these candidates, 19 SNPs which showed significant correlation with tenascin-XA (TNXA) expression were significantly associated with CD in GWAS. By TWAS, TNFSF15 (FDR = 1.35e-13) in whole blood, ERV3-1 (FDR = 2.18e-2) in lymphocytes, and ZNF713 (FDR = 3.04e-2) in the sigmoid colon was significantly associated with CD. By conducting integration analyses using GWAS and eQTL data, we confirmed multiple gene transcripts are involved in the development of CD.
Subject terms: Genetic association study, Crohn's disease, Gene expression
Introduction
Inflammatory bowel disease (IBD) is a term for two conditions: Crohn’s disease (CD) and ulcerative colitis. IBD is a multifactorial disease where development of the disease involves both hereditary factors and environmental factors. A genome-wide association study (GWAS) was conducted by various institutions to identify hereditary factors, which revealed that more than 200 regions in the human genome confer susceptibility to IBD1–3. However, many of the polymorphisms that show correlation with the disease are located in non-transcribed regions. Regions that show disease susceptibility due to functional mutation caused by amino acid substitution were limited to regions such as the nucleotide binding oligomerization domain-containing 2 (NOD2), interleukin 23 receptor (IL23R), and autophagy related 16-like 1 (ATG16L1). The GWAS of Japanese CD patients, reported by us in 2019, indicated that the only polymorphism with amino acid substitution among 11 identified disease susceptibility regions was the p.G149R polymorphism located in IL23R4. In cases of polymorphisms with amino acid substitution, it is highly likely that genes with such polymorphisms are involved in the development of IBD. However, the mechanism of IBD development caused by other polymorphisms is unknown, and susceptibility genes remain to be confirmed. It is predicted that the genomic mutations located in these disease susceptibility regions impact the expression of nearby genes and are involved in the development of IBD.
In recent years, many expression quantitative trait locus (eQTL) analyses have been performed with the aim of examining the relationship between comprehensive gene expression in various cell types and the genetic background. These findings have been used to create a database. Within this database, the Genotype Tissue Expression (GTEx) project examined gene polymorphism expression of every human tissue5. Using the eQTL database, it is possible to predict which tissues are affected by gene polymorphisms, which genes are involved, and what is the degree of expression of these genes. Furthermore, it is now possible to examine the relationship between polymorphism and changes in expression, to predict changes in gene expression levels caused by polymorphisms, and to perform a transcriptome-wide association study (TWAS) based on the data6.
By utilizing GWAS and eQTL analysis, polymorphisms that correlate to the development of IBD can be identified and the expression of genes impacted by these polymorphisms can be predicted. Moreover, TWAS enabled us to predict disease susceptibility genes and changes in expression that cause IBD by analyzing each gene unit.
However, each eQTL database is an analysis performed under specific conditions in specific cells. Moreover, racial variations need to be considered. To determine the causes of the development of CD, it is important to consider gene expression and the relationship of gene expression to single-nucleotide polymorphisms (SNPs) in cells that play a role in immunity in the sites of inflammation of the disease (i.e., the intestinal tissues). Although data regarding samples such as the small intestine, large intestine, and whole blood are available from previously described GTEx, data for the immunocompetent cells located in the intestinal sites are not yet available. Thus, the genes involved in IBD and how the expression of such genes is impacted by susceptibility gene polymorphism in Japanese IBD patients remain unknown.
Based on the above, we performed eQTL analyses by collecting CD4+ effector memory T cells (TEM cells) from lamina propria mononuclear cells (LPMCs), the cell type considered to be involved in disease state of Japanese CD patients. Using our results and the eQTL data from previously constructed database for other tissues, disease susceptibility genes involved in the development of CD in the Japanese population were identified.
Results
In LPMC-derived TEM cells, eQTL of 2,463 genes at 22,632 regions were identified
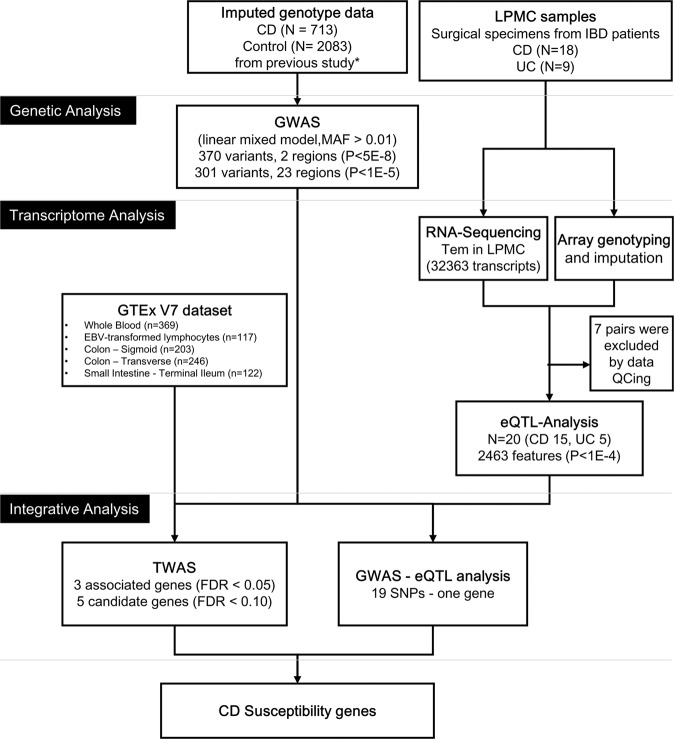
The analysis flow chart is shown in Fig. 1. RNA sequencing performed on TEM cells of 20 IBD patients (15 CD patients, 5 UC patients), which advanced to expression analysis, confirmed expression of 32,363 genes. According to eQTL analyses, 22,632 pairs in 2,463 genes were confirmed to be candidates (p < 1e-04) which showed correlation between gene polymorphism and expression. Among these pairs, 2,000 pairs in 220 genes showed significant (p < 1e-06) correlation (Supplementary Tables S1 and S2).
Figure 1.
Analytical flow in this study. CD; Crohn’s disease, UC; ulcerative colitis, LPMC; lamina propria mononuclear cells, GWAS; genome-wide association study, MAF; Minor allele frequency, GTEx; Genotype Tissue Expression, EBV; Epstein–Barr virus, eQTL; expression quantitative trait locus, TWAS; transcriptome-wide association study, FDR; False Discovery Rate.
Twenty-five sites were confirmed as candidates correlated with Japanese CD by GWAS
Manhattan plots were constructed based on the GWAS of CD patients performed using a linear mixed model (Supplementary Fig. S1). Significant correlation was found in 370 SNPs (p < 5e-08). These SNPs were found to be located in two regions, the human leukocyte antigen (HLA) region on chromosome 6 (rs184950714, p = 1.07e-17) and upstream of tumor necrosis factor superfamily member 15 (TNFSF15) (rs55951892, p = 1.76e-23) on chromosome 9. Moreover, 301 SNPs that showed a candidate level of correlation (p < 1e-05) were found in an additional 23 regions (Table 1). Among the SNPs that showed more than a candidate level of correlation, only three polymorphisms, IL23R p.Gly149Arg (p = 4.22e-07), IL27 p.Leu119Pro (p = 3.28e-05), and SULT1A2 p.Asn235Thr (p = 4.38e-05), showed amino acid substitutions (Supplementary Table S3).
Table 1.
Summary of the CD-GWAS results in Japanese patients.
| Chr | Range (bp)* | Top Hit SNP | No. of SNPs** | Genes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| From | To | SNP ID | Position* | A1 | A2 | A2 Frequencies | P-Values | OR (95%CI) | |||
| 1 | 20437634 | 20437764 | rs7515774 | 20437634 | A | T | 0.107 | 1.04E-06 | 1.10 (1.06–1.15) | 2 | PLA2G2D |
| 1 | 67648596 | 67648596 | rs76418789 | 67648596 | G | A | 0.060 | 4.22E-07 | 0.88 (0.84–0.93) | 1 | IL23R (G149R) |
| 1 | 112222702 | 112222702 | rs534888 | 112222702 | C | T | 0.467 | 7.16E-06 | 0.95 (0.92–0.97) | 1 | RAP1A |
| 4 | 38324507 | 38373273 | rs55843528 | 38361416 | G | A | 0.245 | 1.89E-07 | 1.08 (1.05–1.11) | 20 | — |
| 4 | 189893313 | 189893313 | rs12647478 | 189893313 | T | C | 0.391 | 7.58E-06 | 0.92 (0.88–0.95) | 1 | — |
| 5 | 67691469 | 67691469 | rs10068082 | 67691469 | G | A | 0.037 | 1.72E-06 | 1.17 (1.10–1.26) | 1 | PIK3R1 |
| 5 | 158826792 | 158853941 | rs56167332 | 158827769 | C | A | 0.402 | 3.11E-07 | 1.07 (1.04–1.09) | 21 | IL12B |
| 6 | 2957827 | 2966578 | rs79536569 | 2957827 | G | A | 0.017 | 3.50E-06 | 1.23 (1.13–1.35) | 2 | SERPINB6 |
| 6 | 32214010 | 32793981 | rs184950714 | 32636728 | G | A | 0.179 | 1.07E-17 | 1.16 (1.12–1.19) | 452 | (HLA) |
| 7 | 12221801 | 12222116 | rs200319458 | 12221801 | T | C | 0.024 | 8.41E-06 | 1.19 (1.10–1.29) | 2 | TMEM106B |
| 7 | 76948351 | 76948351 | rs4727354 | 76948351 | G | T | 0.051 | 3.31E-06 | 1.14 (1.08–1.20) | 1 | GSAP |
| 8 | 70472493 | 70472493 | rs117742432 | 70472493 | G | A | 0.036 | 6.58E-06 | 0.86 (0.81–0.92) | 1 | SULF1 |
| 8 | 129224694 | 129245849 | rs12678162 | 129224694 | T | C | 0.431 | 3.08E-06 | 1.06 (1.03–1.08) | 4 | PVT1 |
| 9 | 73881874 | 73881874 | rs151258497 | 73881874 | — | C | 0.331 | 4.57E-06 | 0.94 (0.92–0.97) | 1 | TRPM3 |
| 9 | 117480416 | 117697947 | rs55951892 | 117575913 | A | C | 0.490 | 1.76E-23 | 0.89 (0.86–0.91) | 129 | TNFSF15 |
| 10 | 64431973 | 64550071 | rs224136 | 64470675 | C | T | 0.275 | 1.18E-07 | 0.93 (0.90–0.95) | 19 | ZNF365 |
| 11 | 64908062 | 64926722 | rs11227126 | 64908062 | A | T | 0.067 | 1.71E-06 | 1.12 (1.07–1.18) | 3 | SYVN1 |
| 14 | 105695957 | 105695957 | rs117952084 | 105695957 | T | C | 0.021 | 9.73E-06 | 0.83 (0.76–0.90) | 1 | BRF1 |
| 15 | 27156539 | 27156539 | rs781387485 | 27156539 | ACACAA | — | 0.095 | 3.36E-06 | 1.11 (1.06–1.16) | 1 | GABRA5 |
| 16 | 28513068 | 28531287 | rs56354901 | 28523144 | T | C | 0.134 | 2.44E-06 | 1.09 (1.05–1.13) | 3 | NPIPL1, IL27 |
| 19 | 30021446 | 30021446 | rs117223925 | 30021446 | G | A | 0.075 | 6.21E-06 | 0.90 (0.86–0.94) | 1 | VSTM2B |
| 20 | 47804952 | 47804952 | NA | 47804952 | — | CCCGGC | 0.020 | 2.57E-06 | 1.21 (1.12–1.32) | 1 | STAU1 |
| 20 | 51548302 | 51548302 | rs6126698 | 51548302 | A | T | 0.484 | 8.31E-06 | 0.94 (0.91–0.97) | 1 | TSHZ2 |
| 21 | 33326781 | 33326781 | rs2833577 | 33326781 | G | A | 0.286 | 1.23E-06 | 1.07 (1.04–1.11) | 1 | HUNK |
| 22 | 23054614 | 23054614 | rs9623882 | 23054614 | G | A | 0.081 | 1.61E-06 | 1.14 (1.08–1.20) | 1 | GGTLC2 |
*Positions are based on the Genome Reference Consortium human build 37 (GRCh37), **Number of SNPs with p values <1 × 10−5 Chr: Chromosome, OR: Odds ratio, CI: Confidential interval.
Correlation between Japanese CD and TNXA based on GWAS and eQTL results was assessed
Among the candidate polymorphisms identified by the GWAS, 19 SNPs of chromosome 6 showed significant correlation with expression of the tenascin-XA (TNXA) in intestinal TEM cells (rs117433623, PGWAS = 6.34e-09, PeQTL = 3.49e-05) (Fig. 2, Supplementary Figure S2, Supplementary Table S4). Only one SNP showed a genotype of GG; therefore, further analyses were conducted using two groups—CC and G carrier—in which a correlation tendency was also observed (p = 1.60e-03, Wilcoxon rank-sum test).
Figure 2.
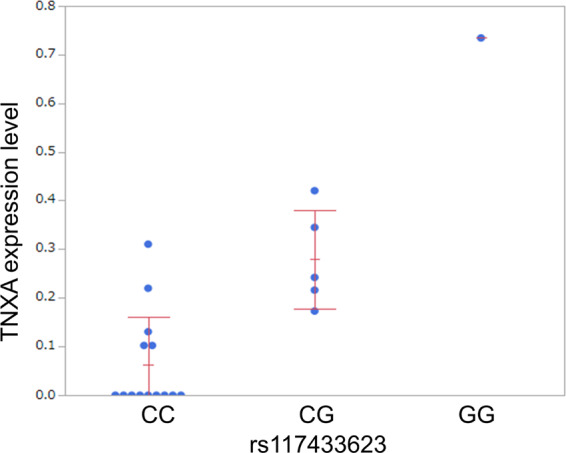
Relationship between rs117433623 and expression of tenascin-XA (TNXA) according to integration analysis of GWAS + eQTL. − Based on eQTL data confirmed in this study, correlation between the expression of 19 SNPs on chromosome 6 in candidate polymorphisms and expression of TNXA in intestinal TEM cells were identified. GWAS; genome-wide association study, eQTL; expression quantitative trait locus.
Six novel genes were identified by TWAS in addition to the previously reported TNFSF15 and RAP1A
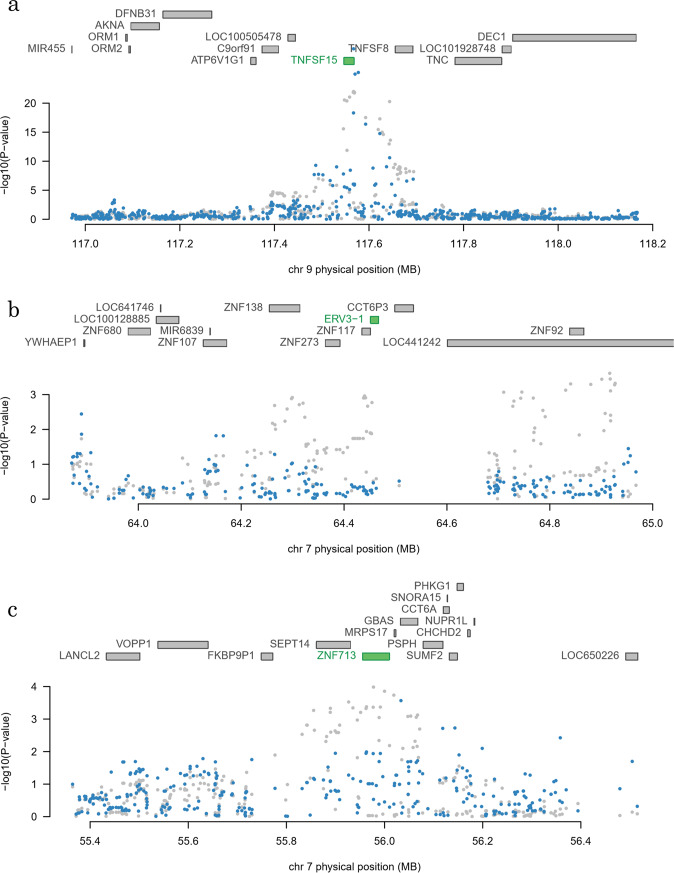
Analyses of HLA regions by TWAS were performed separately from other regions. The relationship of gene expression of multiple genes such as HLA-DQ and HLA-DR with CD was confirmed in all analyzed cell types. Almost all of the correlations were found to be related to re9271170 in the GWAS (Supplementary Table S5). Excluding the HLA region, TNFSF15 (TWAS. p = 2.28e-17, FDR = 1.35e-13) in whole blood, endogenous retrovirus group 3 member 1 (ERV3-1) (TWAS. p = 4.79e-05, FDR = 2.20e-02) in EBV-immortalized lymphocytes, and zinc finger protein 713 (ZNF713) (TWAS. p = 4.41e-05, FDR = 3.03e-02) in the sigmoidal colon showed significant correlation (Table 2). Additionally, apolipoprotein B MRNA editing enzyme catalytic subunit 3 A (APOBEC3A) in whole blood, ras-related protein Rap-1A (RAP1A) in EBV-immortalized lymphocytes, nuclear pore complex interacting protein family member B9 (NPIPB9) and immunoglobulin lambda variable 3-29 (IGLV3-29) in the transverse colon, and WD repeat domain 31 (WDR31) in the sigmoidal colon showed possible associations (FDR < 0.10) as candidate genes (Table 2). Some of these genes showed possible associations in other tissues (Supplementary Table S6). Among these genes, correlation of SNPs within the regions of genes such as ERV3-1, RAP1A, ZNF713 was lost when a correction was made using the predicted expression levels, however, multiple SNPs continued to show a strong correlation in TNFSF15 after correction using the predicted expression levels (Fig. 3, Supplementary Figure S3).
Table 2.
Summary of TWAS with the susceptibility genes for CD in Japanese patients (non-HLA genes).
| Tissue | Gene | Chr | GWAS | eQTL | TWAS | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Best SNP | Z-Score | SNP | R2 | Z-Score | Z-Score | P-Value | FDR | |||
| Significantly associated genes (FDR < 0.05) | ||||||||||
| Whole blood | TNFSF15 | 9 | rs4979462 | 9.81 | rs7866342 | 6.47E-02 | 5.12 | −8.48 | 2.28E-17 | 1.35E-13 |
| Blood - EBV-transformed lymphocytes | ERV3-1 | 7 | rs4718244 | −3.67 | rs4718244 | 8.28E-02 | 3.90 | −4.07 | 4.79E-05 | 2.18E-02 |
| Colon – Sigmoid | ZNF713 | 7 | rs6971250 | −3.88 | rs6593287 | 1.06E-01 | 5.31 | 4.08 | 4.41E-05 | 3.04E-02 |
| Candidate genes (FDR < 0.10) | ||||||||||
| Whole blood | APOBEC3A | 22 | rs5750616 | −3.02 | rs4821843 | 2.14E-02 | −4.71 | 3.90 | 9.66E-05 | 6.79E-02 |
| Colon - Transverse | NPIPB9 | 16 | rs4788076 | 4.16 | rs17640009 | 5.35E-02 | −4.60 | −3.92 | 8.74E-05 | 7.66E-02 |
| Colon - Sigmoid | WDR31 | 9 | rs10981725 | −4.38 | rs10817477 | 1.70E-01 | −5.96 | 3.82 | 1.34E-04 | 8.07E-02 |
| Blood - EBV-transformed lymphocytes | RAP1A | 1 | rs2786991 | −4.30 | rs530801 | 9.98E-04 | 3.48 | 3.67 | 2.42E-04 | 9.45E-02 |
| Colon - Transverse | IGLV3-29 | 22 | rs9623882 | 4.80 | rs8140385 | 3.58E-02 | 4.31 | 3.83 | 1.29E-04 | 9.70E-02 |
Chr: Chromosome, GWAS: genome-wide association study, TWAS: transcriptome-wide association study, FDR: false discovery rate
Figure 3.
Correlation plots of SNPs in regions that showed significant correlation by TWAS. Figure shows plots of polymorphism periphery of eight genes (a. TNFSF15, b. ERV3-1, and c. ZNF713) that showed significant correlation by TWAS. Each dot indicates -log10(p-values) before (gray) and after (blue) adjustment by gene (green) that showed correlation. ERV3-1 and ZNF713 lose correlation after adjusting by genes showing correlation, although the correlation of TNFSF15 remains.
Discussion
The novel outcomes of this study were as follows: (1) even though on a small scale, eQTL data of intestinal LPMCs derived from the TEM cells of Japanese IBD patients were constructed for the first time, (2) polymorphisms that showed correlation by GWAS of Japanese CD patients indicated correlation with expression of TNXA in intestinal LPMC-derived TEM cells, (3) TNFSF15 in whole blood and RAP1A in lymphocytes were confirmed to be disease susceptibility genes when using TWAS for the first time in Japanese CD patients, (4) six genes (including 4 candidates) were newly identified to be correlative.
The eQTL constructed in this study, albeit at a very small scale, was limited to intestinal LPMC-derived TEM cells of Japanese IBD patients and has not previously been reported. The reason why we analyzed eQTL in TEM cells was TEM is considered to be strongly associated with IBD pathogenesis. For example, colitis can be induced in immunodeficient mice by transferring naïve T cells7, strategies blocking T-cell function are useful for attenuating mucosal inflammation in mice with experimental colitis8, and IBD is frequently associated with other T-cell mediated diseases (i.e., psoriasis and multiple sclerosis)9,10. Based on the integration analysis of this eQTL data and the GWAS, new polymorphisms involved in the development of CD in the Japanese population that correlated to the expression of TNXA were identified. TNXA is considered a pseudogene which is not capable of producing functional protein. Therefore, it is unclear whether the gene is involved in the disease state, and if so, how it is involved. However, a report has suggested that TNXA is a serum protein characteristic of stricturing CD11. Thus, combined with this report, it is possible that TNXA actually codes for a protein with unknown function. In addition, it may be involved in the development of the specific disease phenotype of CD. However, there is currently insufficient data to conclude that an increased level of TNXA in the serum of CD patients is involved in the development of CD. Polymorphisms that showed correlation with CD may have two functions: one may involve the development of CD via other functions and the other may involve the expression of TNXA that does not code for functional protein. Hence, the expression of TNXA may function as a marker of polymorphism in the gene. Future studies should consider the function of TNXA using models (i.e., mice) in addition to the conformation of a TNXA expression level in intestinal sites in Japanese CD patients. Additionally, the association of TNXA gene with CD causality was shown indirectly by connecting the results of GWAS and eQTL. To confirm this association, additional analysis such as Mendelian randomization analysis with a larger eQTL data set of Tem from LPMC in the Japanese population should be performed.
In this study, TWAS was first conducted on Japanese CD patients with the use of previously reported eQTL data. A verified correlation around the periphery of TNFSF15 may indicate disease susceptibility via TNFSF15 expression in whole blood according to TWAS. In recent years, many statistical correlations of polymorphisms with unknown function have been identified because genome-wide studies has become available due to low-cost genome analysis technology. However, it is difficult to analyze the expression of genes of various tissue samples in terms of sample collection cost. On the other hand, eQTL databases of various cell types have been constructed and such databases have become freely available. TWAS is one approach that can be used to solve the limitations of GWAS by analyzing such databases integrally and is an analytical method that can be used to identify new disease susceptibility genes. Those regions sometimes contain multiple genes; however, correlation with each gene can be identified by TWAS due to the analysis of a gene unit. The correlation of the TNFSF15 periphery identified by GWAS was found to exist in the region stretching from TNFSF15 to TNFSF8; however, the whole region was indicated to be involved in TNFSF15 expression and to correlate with CD, according to TWAS.
TNFSF15 is a cytokine gene belonging to the TNF family (also called TNF-like ligand 1 A (TL1A)) and is known to show increased expression at intestinal CD sites12. TNFSF15 is mainly secreted from monocytic cells, such as macrophage and dendric cells, and is thought to promote Th1 and Th17 cell activities, leading to CD development13. Multiple studies have reported that TNFSF15 polymorphisms involve gene expression14,15. TWAS results in this study agree with these reports. Therefore, the usefulness of TWAS is supported by analyses using independent databases such as TWAS.
The TWAS method used in this study confirmed multiple novel candidate genes in addition to TNFSF15 and RAP1A4,16,17. APOBEC3A (cytidine deaminase) targets single-stranded DNA and functions as a restriction factor in retrovirus replication. It has been previously reported that this gene is involved in cell cycle arrest caused by DNA damage and oxidative stress18. Polymorphisms located relatively close to the gene are reported to correlate to IBD in the Western population; however, involvement of the genes in IBD has not been indicated. Therefore, this study showed such a correlation for the first time.
ERV3-1 is a gene found in endogenous retroviruses; however, the relationship of ERV3-1 with IBD has not been reported previously. The function of both NPIPB9 and IGLV3-29 is also unknown. One study has reported that changes in the expression level of ZNF713 due to mutation in the gene are involved in autism spectrum disorder19; however, the function of ZNF713 and its relationship with IBD are unknown.
WDR31 is a member of the family of WD40 repeat proteins. WD40 repeat proteins belong to a large family observed among all eukaryotes and are involved in various functions, including signal transduction, regulation of transcription, regulation of cell cycle, autophagy, and apoptosis. It is plausible that changes in the expression of members belonging to this family of genes would relate to disease. In fact, WDR30 is also known as ATG16L1, which is a disease susceptibility gene in Western CD patients and is involved in autophagy20. However, the function of WDR31, which showed correlation in this study, is currently unknown, and no relationship with IBD has been reported. Many of these novel candidate genes have unknown functions and unknown relationships with IBD; however, future functional analyses may provide this information. And these associations were only observed in colon, it will be interesting to see associations of these genes with each clinical sub-phenotype (i.e. disease locations) of CD. Further analyses using additional sample set will be needed.
This study showed that multiple correlations could be confirmed with the use of TWAS. Correlation shown by GWAS at the regions of some genes such as ERV3-1 can be lost when a predicted expression level of ERV3-1 was taken into consideration. Therefore, it was indicated that correlation in the region is due to changes in the expression level of ERV3-1. However, correlation of some SNPs in genes such as TNFSF15 does not diminish when a predicted expression level of the genes is taken into consideration; thus, it has been confirmed that some SNPs have correlation regardless of predicted gene expression levels. In fact, it has been demonstrated previously that there are two independent correlations in this region21, the result of which are consistent with those found this study. However, how the polymorphisms that showed independent correlations are involved in the disease is unknown. The referenced eQTL data are from the Western population, and there may be vastly distinctive Asian-specific eQTL data. Further research is necessary.
Limitations in this study regarding eQTL are as follows: (1) the sample size was small, (2) only IBD patients who required surgery were studied, and mild IBD patients who did not require surgery were not included in this study, (3) there were differences in inflammation sites and degree of inflammation in surgical specimens, and (4) there were difference in drugs administered before surgery (individual results may be affected by such drugs). Limitations of TWAS are that (6) referenced gene expression data are from a different ethnic group and (7) evaluation of genes induced under specific conditions was not possible. To increase the number of subjects and reduce the effect of medications or severity issue, analyses of biopsy samples at the initial endoscopy will be informative. However, we aimed to establish eQTL dataset of specific cell population in this study, we analyzed surgical specimens. The most serious limitation of our study was we could only see eQTLs of TEM cells in Japanese patients with IBD, because the number of LPMCs, which could be isolated from surgical specimens, was still too few to analyze several immunocompetent cells. The increasing number of samples and cell species of immunocompetent cells and/or adopting new technologies (i.e. single cell analysis) may show more certainly eQTL, although this is a subject for future analysis. However, this study included a functional approach utilizing data regarding function of polymorphisms in addition to existing GWAS, which simply examines whether SNPs are involved in the development of the disease. Factors related to the development of the disease at a gene level in a specific tissue could be predicted. Moreover, the results obtained in this study included genes (TNFS15 and RAP1A) that have shown correlation by functional analyses as candidate genes and thus the usefulness of this approach was shown. Integration analyses using GWAS and eQTL data are considered useful not only for the analysis of disease susceptibility genes but also for analyzing disease-modifying genes that determine the disease state and pharmacogenomics, which involves analysis of drug efficacy and adverse effects. Future analyses are anticipated.
In conclusion, by conducting integration analyses using information regarding polymorphism and transcriptome-related analysis data, we confirmed multiple gene transcripts involved in the development of CD in the Japanese population. The study also indicated that expression of TNFSF15 in blood cells was likely to be involved in the development of CD in the Japanese population.
Materials and Methods
In this study, analyses were processed using the following two approaches to accomplish our objective. First, eQTL analyses were conducted on intestinal TEM cells of Japanese CD patients and disease susceptibility genes were predicted by projecting the function of disease susceptibility polymorphisms in these patients. Second, a TWAS was conducted using data from the existing eQTL database and the GWAS results to analyze the susceptibility genes of Japanese CD patients.
Subjects
For TEM transcriptome analyses, cells were isolated from 18 patients who were in an active phase of CD and nine patients who were in an active phase of UC from a cohort of IBD patients hospitalized in Tohoku University Hospital between July 2015 and July 2018. The studied cohort underwent surgery that involved intestinal resection and consented to research including genetic analysis. The subjects for GWAS were 713 Japanese CD patients who regularly visited either Tohoku University Hospital (379 patients) or Kyushu University Hospital (334 patients) and could be analyzed by previous GWAS of Crohn’s disease4. A total of 2,063 healthy individuals who resided in Tohoku (1,621 individuals) or Kyushu (462 individuals) were also studied as controls22. Diagnosis was performed according to the diagnostic criteria proposed by the Japanese Ministry of Health, Labor and Welfare23, based on clinical symptoms and endoscopic, X-ray, and tissue findings. All subjects were Japanese.
This study was conducted after receiving written consent from subjects and approval from the ethics committee of the School of Medicine at Tohoku University (2017-1-253, 2019-1-161). All methods in this study were performed in accordance with ethical guidelines for medical and health research involving human subjects established by the Ministry of Health, Labour and Welfare in Japan. The demographic profiles of the subjects are shown in Table 3.
Table 3.
Patient characteristics.
| Sample | Disease | Age | Sex | Disease Location | Sampling site | Medication (active intervention) |
|---|---|---|---|---|---|---|
| IBD1 | CD | 36 | M | ileum | ileum | 5ASA |
| IBD2 | CD | 18 | M | ileum | ileum | ADA |
| IBD3 | CD | 27 | M | ileum | ileum | None |
| IBD4 | CD | 21 | M | ileocolon | ileum | 5ASA, UST, AZA |
| IBD5 | CD | 35 | M | ileocolon | ileum | 5ASA, IFX |
| IBD6 | CD | 40 | M | ileocolon | ileum | 5ASA |
| IBD7 | CD | 48 | F | ileocolon | ileum | 5ASA, ADA, AZA |
| IBD8 | CD | 26 | M | ileocolon | ileum | 5ASA |
| IBD9 | CD | 58 | M | ileocolon | ileum | 5ASA |
| IBD10 | CD | 40 | M | ileocolon | ileum | 5ASA, IFX |
| IBD11 | CD | 28 | M | ileocolon | colon | 5ASA, IFX, AZA |
| IBD12 | CD | 40 | M | ileocolon | colon | 5ASA, IFX |
| IBD13 | CD | 19 | M | ileocolon | colon | IFX, 6MP |
| IBD14 | CD | 48 | M | ileocolon | colon | 5ASA, ADA, AZA |
| IBD15 | CD | 42 | M | ileocolon | colon | 5ASA |
| IBD16 | UC | 65 | F | pancolitis | colon | 5ASA, PSL, ADA |
| IBD17 | UC | 26 | M | pancolitis | colon | PSL, IFX |
| IBD18 | UC | 75 | F | pancolitis | colon | 5ASA, PSL |
| IBD19 | UC | 49 | F | pancolitis | colon | 5ASA, PSL, AZA |
| IBD20 | UC | 65 | M | pancolitis | colon | 5ASA, PSL, Tac |
CD: Crohn’s disease, UC: ulcerative colitis, 5ASA: 5 aminosalicylic acid, IFX: infliximab, ADA: adalimumab, UST: ustekinumab, AZA: azathiopurine, 6MP: 6 mercaptopurine, PSL: prednisolone, Tac: Tacrolimus
Isolation of LPMCs
LPMCs were isolated from inflammation sites surgically resected from the small intestine or the large intestine according to the method described by Fiocchi et al.24,25. In brief, a resected specimen was cut lengthwise, and feces were removed by washing the intestine in Hank’s balanced salt solution (HBSS) (Wako, Osaka, Japan). The specimen was then cut into 2–3 cm × 10 cm sections. The sections were then washed in HBSS containing 0.15% dithiothreitol (Wako) for 30 minutes with shaking. The specimens were then washed in HBSS containing 1 mM ethylenediaminetetraacetic acid (Wako) for 90 minutes with shaking. This wash was repeated until the epithelial layer was completely removed. After removing the epithelial layer completely, the specimens were washed again in HBSS with shaking and the washed specimens were finely divided into 5-mm sections. The specimens were then digested in HBSS containing 1 mg/ml collagenase-3 (Worthington Biochemical Corporation, Lakewood, USA) and DNase I (Roche, Basel, Switzerland) at 37 °C for 8–10 hours. The digested specimens were then passed through a 100 μm cell strainer (BD Biosciences, Franklin Lake, USA) and the cell suspension was recovered. The suspension was centrifuged at 700 × g and the cell pellet was resuspended in HBSS. The suspension was overlaid on Ficoll–Hypaque (GE Healthcare, Little Chalfont, UK) and centrifuged for 20 minutes at 1,000 × g. LPMC cells located at the interface between HBSS and Ficoll–Hypaque were recovered.
Isolation of TEM cells and extraction of DNA/RNA
CD4+ T cells were isolated by negative selection from isolated LPMCs using an Easy Sep Magnet (STEMCELL Technology, Vancouver, Canada) and an Easy Sep Human CD4+ T cell Enrichment kit (STEMCELL Technology). Furthermore, the isolated CD4 positive T cells were stained with anti-CD3-FITC, anti-CD4-PE, anti- CD45RO-APC, anti-CD197 (CCR7) -BV421, and 7ADD-Cell Viability Solution (BD Biosciences), followed by isolation of TEM cells using a FACS Aria II cell sorter (BD Biosciences). Sorting efficiency was consistently over 98%. These TEM cells may include a few regulatory T Cells. However, to keep the number of cells to perform RNA sequencing, we used these samples as TEM cells. DNA and total RNA were extracted from isolated TEM cells using an AllPrep DNA/RNA mini kit (QIAGEN, Hilden, Germany).
Genotyping
Transcriptome analysis of subjects by Japonica array V1 (Thermo-Fisher Scientific Inc., Waltham, MA) was contracted to Toshiba Inc. (Tokyo, Japan)26. Affymetrix Power Tools software (Thermo-Fisher Scientific Inc.) was used for genotyping. For genotyping of SNPs that could not be typed by the array, IMPUTE2 (Version 2.3.2) (Center for Statistical Genetics, University of Michigan, USA) was used for performing imputation with the genome reference panel of people from the Tohoku region (2KJPN)27,28. For genotyping data for the GWAS, data which had undergone analyses by Japonica array VI, imputation by the 1KJPN panel, and quality control (QC) by previous studies were used4.
Transcriptome and eQTL analyses
For the total RNA collected from the intestines of 27 IBD patients (18 CD patients, 9 UC patients), QC, library construction, and transcriptome analysis by RNA sequencing were contracted to Macrogen Inc, Japan. QC was performed using TapeStation HighSensitivity RNA ScreenTape (Agilent Technologies, Santa Clara, USA), where the standard was set as RNA integrity number >7. RNA amplification, was performed using SMART Seq V4 Ultra Low Input RNA Kit (Takara Bio, Kusatsu, Japan), following the manufacturer’s protocol. TruSeq Stranded mRNA Library Prep (Illumina, San Diego, USA) was used for library construction. NovaSeq. 6000 (Illumina) was used for RNA sequencing. Processes from alignment to post-treatment of FASTQ data obtained by RNA sequencing was performed using STAR29 and Picard software (http://broadinstitute.github.io/picard/) according to TOPMed RNA-seq pipeline guidelines using the supercomputer system at Tohoku University’ medical-megabank institute. The consistency of RNA/DNA samples was confirmed by comparing RNA sequence data and genotype of genomic DNA. Samples with insufficient data or a low number of reads were excluded, which resulted in 15 active-phase CD patients and five active-phase ulcerative colitis patients for expression analysis. The number of reads of each transcript was calculated using the featureCounts (Ver 1.6.4)30 and were standardized against entire transcripts using edgeR (Ver 3.20.9)31. eQTL analysis and standardization at the gene level were performed using FastQTL (Ver 2.184) with–normal option32.
GWAS
The GWAS data were analyzed with a linear mixed model. Genome-wide Complex Trait Analysis software (Ver 1.91.7b1) was used for the analysis33, where 7,424,691 polymorphisms with minor allele frequencies of over 0.5% were analyzed among the 16,919,636 polymorphisms input.
TWAS
FUSION software was used for the TWAS6. The data for analysis consisted of RNA sequence data from whole blood, Epstein–Barr virus (EBV)-immortalized B cells, the transverse colon, the sigmoidal colon, and the small intestine (the ileum terminal), as these tissues are considered, among GTEx V7 data released in GTEx5, to be highly related to IBD.
Statistical analysis
In eQTL analysis, samples showing p values less than 1e-06 were considered significant correlation. p values under 1e-04 were considered as candidates and were used for further analyses. In GWAS, polymorphisms showing p < 5e-08 in the linear mixed model were considered to be significant and those with p values under 1e-05 were considered to be candidates. Polymorphisms showing correlation within 500 kbps upstream and downstream of the polymorphism were considered cases of correlation at the same region. For TWAS, the genes with false discovery rate (FDR) of <0.05 were considered to be susceptibility genes, and genes with FDR < 0.10 were considered to be candidates. Data obtained from each analysis was further analyzed using R software (Ver 3.4.4). Supplementary Table S7 shows the eQTL data set analyzed in this study.
Supplementary information
Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers JP15H04805, 18K07929 and in part by the Tohoku Medical Megabank Project (Special Account for reconstruction from the Great East Japan Earthquake). SNP genotyping was supported in part by the Center of Innovation Program from the Japan Science and Technology Agency, JST. Computational resources were provided in part by the ToMMo supercomputer system.
Author contributions
Y. Kakuta, R.I., M. Nagasaki and Y. Kinouchi designed the study. R.I., Takeo Naito, Y. Kawai, K.T., A.A. and Y. Kakuta acquired data. A.H., J.U., Y.F., T.T., T. Nakano, Y.I., R.I., D.O., R.M., M.K., H.S., Y. Kanazawa, T.K., M. Nakamura, K.W., Takeshi Naito, M.U., T.M. and M.E. recruited patients. R.I., Y. Kakuta, Y. Kawai and M. Nagasaki analysed data. Y. Kakuta, R.I., Y. Kinouchi, A.M., drafted the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yoichi Kakuta and Ryo Ichikawa.
Supplementary information
is available for this paper at 10.1038/s41598-020-66951-5.
References
- 1.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang H, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kakuta Y, et al. A Genome-wide Association Study Identifying RAP1A as a Novel Susceptibility Gene for Crohn’s Disease in Japanese Individuals. J. Crohns Colitis. 2019;13:648–658. doi: 10.1093/ecco-jcc/jjy197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carithers LJ, et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusev A, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powrie F, Coffman RL, Correa-Oliveira R. Transfer of CD4+ T cells to C.B-17 SCID mice: a model to study Th1 and Th2 cell differentiation and regulation in vivo. Res Immunol. 1994;145:347–353. doi: 10.1016/S0923-2494(94)80198-3. [DOI] [PubMed] [Google Scholar]
- 8.Monteleone G, Caprioli F. T-cell-directed therapies in inflammatory bowel diseases. Clin Sci (Lond) 2010;118:707–715. doi: 10.1042/CS20100027. [DOI] [PubMed] [Google Scholar]
- 9.Fiorino G, Omodei PD. Psoriasis and Inflammatory Bowel Disease: Two Sides of the Same Coin? J Crohns Colitis. 2015;9:697–698. doi: 10.1093/ecco-jcc/jjv110. [DOI] [PubMed] [Google Scholar]
- 10.Sonnenberg A, Ajdacic-Gross V. Similar birth-cohort patterns in Crohn’s disease and multiple sclerosis. Mult Scler. 2018;24:140–149. doi: 10.1177/1352458517691620. [DOI] [PubMed] [Google Scholar]
- 11.Townsend P, et al. Serum Proteome Profiles in Stricturing Crohn’s Disease: A Pilot Study. Inflamm Bowel Dis. 2015;21:1935–1941. doi: 10.1097/MIB.0000000000000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bamias G, et al. Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. J Immunol. 2003;171:4868–4874. doi: 10.4049/jimmunol.171.9.4868. [DOI] [PubMed] [Google Scholar]
- 13.Takedatsu H, et al. TL1A (TNFSF15) regulates the development of chronic colitis by modulating both T-helper 1 and T-helper 17 activation. Gastroenterology. 2008;135:552–567. doi: 10.1053/j.gastro.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakuta Y, et al. TNFSF15 transcripts from risk haplotype for Crohn’s disease are overexpressed in stimulated T cells. Hum Mol Genet. 2009;18:1089–1098. doi: 10.1093/hmg/ddp005. [DOI] [PubMed] [Google Scholar]
- 15.Michelsen KS, et al. IBD-associated TL1A gene (TNFSF15) haplotypes determine increased expression of TL1A protein. PLoS One. 2009;4:e4719. doi: 10.1371/journal.pone.0004719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakuta Y, Kinouchi Y, Negoro K, Takahashi S, Shimosegawa T. Association study of TNFSF15 polymorphisms in Japanese patients with inflammatory bowel disease. Gut. 2006;55:1527–1528. doi: 10.1136/gut.2006.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamazaki K, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14:3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 18.Niocel M, Appourchaux R, Nguyen XN, Delpeuch M, Cimarelli A. The DNA damage induced by the Cytosine Deaminase APOBEC3A Leads to the production of ROS. Sci Rep. 2019;9:4714. doi: 10.1038/s41598-019-40941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metsu S, et al. A CGG-repeat expansion mutation in ZNF713 causes FRA7A: association with autistic spectrum disorder in two families. Human mutation. 2014;35:1295–1300. doi: 10.1002/humu.22683. [DOI] [PubMed] [Google Scholar]
- 20.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 21.Kakuta, Y. et al. Rare Variants of <em>TNFSF15</em> Are Significantly Associated With Crohn’s Disease in Non-Jewish Caucasian Independent of the Known Common Susceptibility SNPs. Gastroenterology 144, S–466, 10.1016/S0016-5085(13)61723-0 (2013).
- 22.Kuriyama S, et al. The Tohoku Medical Megabank Project: Design and Mission. J Epidemiol. 2016;26:493–511. doi: 10.2188/jea.JE20150268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsui T.H. F., Hisabe T. Proposed diagnostic criteria for Crohn’s disease. Annual reports of the research group of intractable inflammatory bowel disease granted by the Minitry of Health, Labour, and Welfare of Japan., 52–54 (2011).
- 24.Fiocchi A, et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ J. 2015;8:4. doi: 10.1186/s40413-015-0055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiocchi, C. & Youngman, K. R. Isolation of human intestinal mucosal mononuclear cells. Curr Protoc Immunol Chapter 7, Unit 7 30, 10.1002/0471142735.im0730s19 (2001). [DOI] [PubMed]
- 26.Kawai Y, et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J Hum Genet. 2015;60:581–587. doi: 10.1038/jhg.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagasaki M, et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018. doi: 10.1038/ncomms9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi-Kabata Y, et al. iJGVD: an integrative Japanese genome variation database based on whole-genome sequencing. Human genome variation. 2015;2:15050. doi: 10.1038/hgv.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.