Abstract
The genome sequences of 16 Streptomyces strains, showing potential for plant growth-promotion (PGP) activities in rice, sorghum, chickpea and pigeonpea, isolated from herbal vermicompost, have been decoded. The genome assemblies of the 16 Streptomyces strains ranged from 6.8 Mb to 8.31 Mb, with a GC content of 72 to 73%. The extent of sequence similarity (in terms of shared ortholog) in 16 Streptomyces strains showed 70 to 85% common genes to the closest publicly available Streptomyces genomes. It was possible to identify ~1,850 molecular functions across these 16 strains, of which close to 50% were conserved across the genomes of Streptomyces strains, whereas, ~10% were strain specific and the rest were present in various combinations. Genome assemblies of the 16 Streptomyces strains have also provided genes involved in key pathways related to PGP and biocontrol traits such as siderophores, auxin, hydrocyanic acid, chitinase and cellulase. Further, the genome assemblies provided better understanding of genetic similarity among target strains and with the publically available Streptomyces strains.
Subject terms: Applied microbiology, Bacterial genes
Introduction
Plants attract beneficial microbes through their root exudates and such associative or symbiotic microbiomes in-turn induces the plant fitness and immune system via cell-signaling and/or by triggering plant’s physiological process. Microbiome in vicinity of roots plays an important role in plant growth, development and abiotic/biotic stress tolerance1,2. Unraveling of plant-microbiome interaction develops a basis for sustainable strategies in next-generation farming with less input of fertilizers and/or pesticides3. Among the plant-associated microbes, actinobacteria are of particular interest due to their ability to produce a range of secondary metabolites4. Actinobacteria have been found to be associated with biological control of insect pests and pathogens, stress tolerance and growth promotion in plants1,2. They occur in the rhizosphere as well as with in the plants (in the form of endophytes) and have been shown to induce systemic resistance in plants. Among the actinobacteria, Streptomyces is the predominant genus followed by Actinomadura, Microbispora, Micromonospora, Nocardia, Nonomurea, Mycobacterium, Frankia, Actinoplanes, Saccharopolyspora and Verrucosispora and are known for their ubiquitous presence in soil and nutrient cycling; and majorly for the antibiotics and complex secondary metabolite pathways5,6. Streptomyces is the major producer of secondary metabolites (39% of the total metabolites produced by the microbes) including polyene macrolides, actinomycins, aminoglycosides, streptothricins, anthracyclines, cyclopolylactones and quinoxaline-peptides5. The chemical diversity of metabolites produced by Streptomyces ranges from simple lactones to condensed macro-lactones; and simple amino acid derivatives to peptides and high-molecular-weight proteins. A broad range of Streptomyces activities on pharmacological traits have been characterized but the traits related to agriculture have received relatively less consideration. Hence, exploring Streptomyces for agricultural sector becomes an area of active interest in current scenario7,8.
Previously, we had identified 16 candidate Streptomyces strains (through 16 S rDNA sequencing method) and characterized for their plant growth promotion (PGP) traits such as indole acetic acid, siderophore, β-1,3-glucanase, chitinase, hydrocyanic acid and other hydrolytic enzymes9–23 (Table 1). Characterizations of these microbes have provided information on their beneficial traits and were further demonstrated for PGP activities and antagonistic activities (against pathogens of chickpea, pigeonpea and sorghum) in planta9–23. Some of these Streptomyces strains were also demonstrated for their larvicidal activity against Helicoverpa armigera, and other important caterpillar pests such as Spodoptera litura and Chilo partellus15. From these collections, two metabolites with insecticidal activity against H. armigera such as, a diketopiperazine derivative called cyclo (Trp‐Phe), and a novel fatty acid amide derivative called N‐(1‐(2,2‐dimethyl‐5‐undecyl‐1,3‐dioxolan‐4‐yl)‐2‐hydroxyethyl) stearamide were isolated and characterized from the best strains S. griseoplanus SAI‐25 and Streptomyces sp., CAI‐155, respectively21,22. However, the detailed molecular characterization of the above mentioned Streptomyces strains have not been done, so far.
Table 1.
Details of 16 Streptomyces strains and their PGP traits.
| S.no | Strain Number |
Identification by 16S rDNA sequencing |
Gene bank ACC. No. |
In vitro PGP traits | Antifungal activity | Crops | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indole Acetic acid (μg ml−1) |
Sidero phore” |
Hydrocy anic acid# |
Cellu lase! |
Chiti nase! |
β 1,3 glucanase (units) |
FOC | Mp | Rb | Bc | Sr | ||||||
| 1 | CAI-17 | Streptomyces sp. | JQ682619 | 0.34 | 2 | 3 | + | + | 0.66 | + | + | + | + | − | R,S,C | 10,12,17,21 |
| 2 | CAI-21 | Streptomyces sp. | JQ682620 | 1.13 | 1 | 3 | + | − | 0 | + | + | − | − | + | R,S,C,P | 10,11,18,21,23 |
| 3 | CAI-24 | Streptomyces sp. | JN400112 | 5.90 | 3 | 3 | + | + | 0 | + | − | + | + | − | R,S,C | 9,14,16,19,21 |
| 4 | CAI-68 | Streptomyces sp. | JQ682622 | 0.22 | 3 | 3 | + | − | 0.66 | − | − | − | + | − | R,S,C | 10,12,17,21 |
| 5 | CAI-78 | Streptomyces sp. | JQ682623 | 0.95 | 0 | 2 | + | − | 2.92 | + | − | + | + | − | R,S,C | 10,12,17,21 |
| 6 | CAI-85 | Streptomyces sp. | KF770897 | 43.6 | 1 | 2 | + | − | 1.21 | + | + | + | + | − | R,S,C | 13,15,20,21 |
| 7 | CAI-93 | S. fungicidicus | KF742498 | 33.6 | 2 | 2 | + | + | 0 | + | + | + | + | − | R,S,C | 13,20,21 |
| 8 | CAI-121 | Streptomyces sp. | JN400113 | 43.7 | 3 | 2 | + | + | 0 | + | − | − | + | − | R,S,C | 9,14,16,19,21 |
| 9 | CAI-127 | Streptomyces sp. | JN400114 | 3.50 | 4 | 3 | + | + | 0 | + | − | − | + | − | R,S,C | 9,14,16,19,21 |
| 10 | CAI-140 | S. coelicolor | KF742497 | 15.4 | 1.3 | 3 | + | − | 0.353 | + | + | + | + | − | R,S,C | 13,20,21 |
| 11 | CAI-155 | Streptomyces sp. | KF770896 | 12.6 | 2 | 3 | + | + | 0.76 | + | + | + | + | − | R,S,C | 13,15,20,21,22 |
| 12 | KAI-26 | Streptomyces sp. | JQ682624 | 0.40 | 3 | 1 | + | + | 0.35 | + | − | + | + | − | R,S,C | 10,12,17,21 |
| 13 | KAI-27 | Streptomyces sp. | JQ682625 | 0.74 | 1 | 2 | + | + | 0.2 | + | − | + | + | − | R,S,C | 10,12,17,21 |
| 14 | KAI-90 | Streptomyces sp. | JN400116 | 0 | 3 | 3 | + | + | 0 | + | − | + | + | − | R,S,C | 9,14,16,19,21 |
| 15 | KAI-180 | Streptomyces sp. | KF742499 | 30.1 | 0 | 2 | + | + | 0 | + | + | + | + | − | R,S,C | 13,20,21 |
| 16 | MMA-32 | S. roseoviolaceus | JQ682626 | 4.66 | 3 | 2 | + | + | 0 | + | + | + | + | + | R,S,C,P | 10,11,18,21,23 |
Rating scale for siderophore are 0 = no change, 1 = positive, 2 = halo zone of 1 3 mm, 3 = halo zone of 4 6 mm and 4 = halo zone of 7 mm and above. #Ratings scale for HCN production are 0 = no color change; 1 = light reddish brown; 2 = medium reddish brown and 3 = dark reddish brown.! The rating scale for Chitinase and Cellulase are 0 = no change; 1 = 1 6 mm; 2 = halo zone of 7 12 mm; 3 = halo zone of 19 24 mm and 4 = halo zone of 25 30 mm and above. Units One unit of β 1,3 glucanase activity is defined as the amount of enzyme that liberated 1 μmol of glucose h−1 at defined conditions. Foc Fusarium oxysporum f. sp. ciceri; Mp Macrophomina phaseolina; Rb – Rhizoctonia bataticola; Bc Botrytis cinerea; Sr Sclerotium rolfsii; R – Rice, S – Sorghum, C – Chickpea, P – Pigeonpea.
The biosynthetic potential of microorganisms genomes has been greatly underexplored24. For instance, many silent genes referred as cryptic or orphan are often present in the microbial genome pathways. Though, not all cryptic pathways are necessarily silent, some might have given lower rate of metabolite production under specified culturing conditions. This crucial reservoir can be untapped by whole genome sequence (WGS) data. For instance, Streptomyces coelicolor A3 (2) was known to produce four secondary metabolites until the WGS data have revealed the presence of additional 18 biosynthetic gene clusters25. The falling costs of WGS using next-generation sequencing (NGS) technologies has provided opportunity to catalogue genome wide variations present in any organism. Therefore, in the present study de novo genome assemblies of 16 Streptomyces strains have been developed using Illumina sequencing technology. These assemblies have been analyzed to identify the inter-species relationships, relevance of phenotypic and genomic data and additional insights of identified genome locus towards agriculturally important traits. Moreover, the correlation between the genetic makeup of these Streptomyces strains and their metabolites have provided the genes or biochemical pathways associated with phenotypic variability.
Results
De novo assemblies of sixteen Streptomyces strains
De novo assemblies were generated using shotgun sequencing. Each genome was sequenced from ~400 bp insert library to a coverage of ~500X using Illumina HiSeq. 2500. As a result, a total of 17 to 32 million paired-end reads were generated, yielding about 4.5 to 8 Gigabases (Gb) of sequence data per strain (Table 2). The sequencing reads were processed and assembled using de novo assembler SPAdes26, and the contigs with poor support from mapped reads were removed from analysis (Supplementary Fig. S1). As a result, total length of the final assemblies of 16 strains ranged from 6.8 to 8.31 Mb (Table 2), showing consistency with genome size estimates in Streptomyces strains. High congruency was found after mapping paired-end raw reads to the assembled contigs. These assemblies produced between 46 to 659 contigs, depending on the strain, with contig N50 ranging from 37 to 401 kb (Table 2). The average GC content of the Streptomyces strains was 72 to 73%.
Table 2.
Details of genome sequencing and its assembly. The contigs (from de novo assembler SPAdes) having very low read support (<40) were dropped before generation of these statistics.
| Strain no. |
Paired-end reads (in millions) |
Sequence data (in Gigabases) |
Length of assembly (in Mega bases) |
GC composition (in %) |
No. of contigs |
N50 (in Kb) |
L50 (in Kb) |
|---|---|---|---|---|---|---|---|
| CAI-17 | 17.6 | 4.4 | 7.0 | 73 | 188 | 108 | 23 |
| CAI-21 | 22.6 | 5.7 | 7.0 | 73 | 331 | 66 | 30 |
| CAI-24 | 24.6 | 6.1 | 7.7 | 72 | 187 | 118 | 20 |
| CAI-68 | 28.0 | 7.0 | 8.2 | 72 | 74 | 279 | 10 |
| CAI-78 | 27.8 | 6.9 | 7.2 | 73 | 448 | 45 | 47 |
| CAI-85 | 30.0 | 7.5 | 8.3 | 72 | 281 | 148 | 15 |
| CAI-93 | 18.3 | 4.6 | 6.8 | 73 | 97 | 196 | 12 |
| CAI-121 | 27.4 | 6.9 | 8.2 | 72 | 77 | 277 | 10 |
| CAI-127 | 29.4 | 7.4 | 8.2 | 72 | 259 | 108 | 24 |
| CAI-140 | 28.6 | 7.2 | 7.5 | 72 | 335 | 77 | 26 |
| CAI-155 | 22.0 | 5.5 | 7.6 | 72 | 56 | 374 | 8 |
| KAI-26 | 28.5 | 7.1 | 7.7 | 72 | 53 | 367 | 7 |
| KAI-27 | 27.1 | 6.8 | 7.2 | 73 | 659 | 37 | 54 |
| KAI-90 | 32.0 | 8.0 | 8.2 | 73 | 87 | 366 | 10 |
| KAI-180 | 30.0 | 7.5 | 6.8 | 73 | 94 | 182 | 11 |
| MMA-32 | 30.1 | 7.5 | 7.7 | 72 | 46 | 401 | 7 |
Quality assessment of each assembly was performed through, sequence accuracy, gene-space coverage and alignment to protein database. Conserved sets of genes27 were used to estimate gene space content in the 16 de novo assemblies. The results showed an average gene space completeness between 94 to 99% across the 16 de novo assemblies (Table 3). The fraction of entire proteome in de novo assemblies displaying full length alignment (i.e., query coverage of>95%) to the RefSeq proteomes of Streptomyces ranged from 76 to 90% (Table 3).
Table 3.
Assessment of assembly quality.
| Strain no. | Minimal bacterial gene set occurrence (in %) |
Proteins showing BLAST hits with >95% query coverage to RefSeq Streptomyces proteome (in %) |
Protein WITHOUT significant BLAST hits to RefSeq Streptomyces proteome (in %) |
|---|---|---|---|
| CAI-17 | 97 | 90 | 4 |
| CAI-21 | 96 | 89 | 5 |
| CAI-24 | 99 | 87 | 4 |
| CAI-68 | 98 | 84 | 5 |
| CAI-78 | 98 | 85 | 4 |
| CAI-85 | 96 | 76 | 6 |
| CAI-93 | 99 | 87 | 4 |
| CAI-121 | 98 | 84 | 5 |
| CAI-127 | 98 | 84 | 5 |
| CAI-140 | 94 | 83 | 6 |
| CAI-155 | 99 | 87 | 4 |
| KAI-26 | 99 | 86 | 5 |
| KAI-27 | 97 | 85 | 5 |
| KAI-90 | 99 | 80 | 4 |
| KAI-180 | 99 | 85 | 5 |
| MMA-32 | 99 | 86 | 5 |
Annotation and relationships in Streptomyces genomes
Genome assemblies contain 6,001 to 7,455 open reading frames (ORFs); we could assign a putative function through ‘Rapid Annotation using Sub-system Technology (RAST; http://rast.nmpdr.org/) to the encoded proteins for 67 to 73% of these (Table 4); the remaining were either hypothetical proteins or proteins of unknown or doubtful function. About 4–6% of the proteome of 16 strains failed to show any significant homology to the publicly available Streptomyces proteome sequences (Table 3). Curated annotation, involving hierarchical annotation of the genes/proteins, was available only for one-third (30–33%) of them (Table 4).
Table 4.
Annotation of genome assemblies. The prediction of genes and their annotation was done using online RAST server.
| Strain no. | Coding genes |
Genes with annotation (in %) |
Subsystems (Biol. Process) assigned |
Role (Molecular Functions) assigned | Genes with Curated annotation (in %) |
|---|---|---|---|---|---|
| CAI-17 | 6132 | 72 | 421 | 1348 | 33 |
| CAI-21 | 6146 | 71 | 409 | 1262 | 31 |
| CAI-24 | 6615 | 69 | 430 | 1377 | 32 |
| CAI-68 | 7326 | 67 | 440 | 1423 | 31 |
| CAI-78 | 6053 | 72 | 423 | 1352 | 33 |
| CAI-85 | 7455 | 69 | 430 | 1397 | 30 |
| CAI-93 | 6044 | 72 | 413 | 1330 | 33 |
| CAI-121 | 7327 | 67 | 442 | 1427 | 31 |
| CAI-127 | 7299 | 67 | 434 | 1417 | 31 |
| CAI-140 | 6837 | 69 | 412 | 1328 | 30 |
| CAI-155 | 6581 | 69 | 432 | 1369 | 32 |
| KAI-26 | 6744 | 68 | 439 | 1398 | 32 |
| KAI-27 | 6001 | 72 | 417 | 1330 | 33 |
| KAI-90 | 7175 | 73 | 443 | 1439 | 32 |
| KAI-180 | 6050 | 71 | 426 | 1366 | 33 |
| MMA-32 | 6738 | 68 | 439 | 1397 | 32 |
Comparison of genomes based on entire gene set
Gene orthologs across 16 strains were identified using bi-directional best BLAST of peptide sequences along with phylogenetic analysis, as implemented in software package OrthoFinder28. While major fraction of the gene sets formed a total of 9,937 orthogroups, there remained about 3,078 genes from 16 strains unassigned to any of the orthogroups (Fig. 1). Among the orthogroups, over one fourth (28%) of them had member(s) present in all strains, indicating significant inter-genomic variation. Majority of the unassigned genes (~75%) were present in just three strains namely, CAI-85, CAI-140 and KAI-90, indicating the most diverged strains. The entire gene set of these strains were further compared pairwise for presence of common genes (i.e., orthologs present in the pair) and genes unique to each of them. A heatmap of correlation between all pairs of strains showed 3 distinct groups and one singleton (based on inter-nodal distance threshold of 0.2) (Fig. 2A,B). Two of three groups were relatively large (having 6 and 7 strains) compared to the third group having just 2 members (Fig. 2B).
Figure 1.
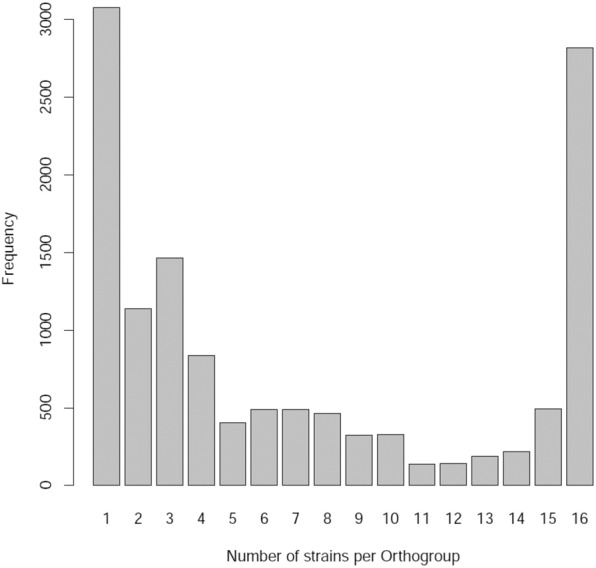
Frequency distribution of size of orthogroup (number of strains having one or more orthologs per orthogroup). The plot also displays genes which remained unassigned to any of the orthogroups (the leftmost bar).
Figure 2.
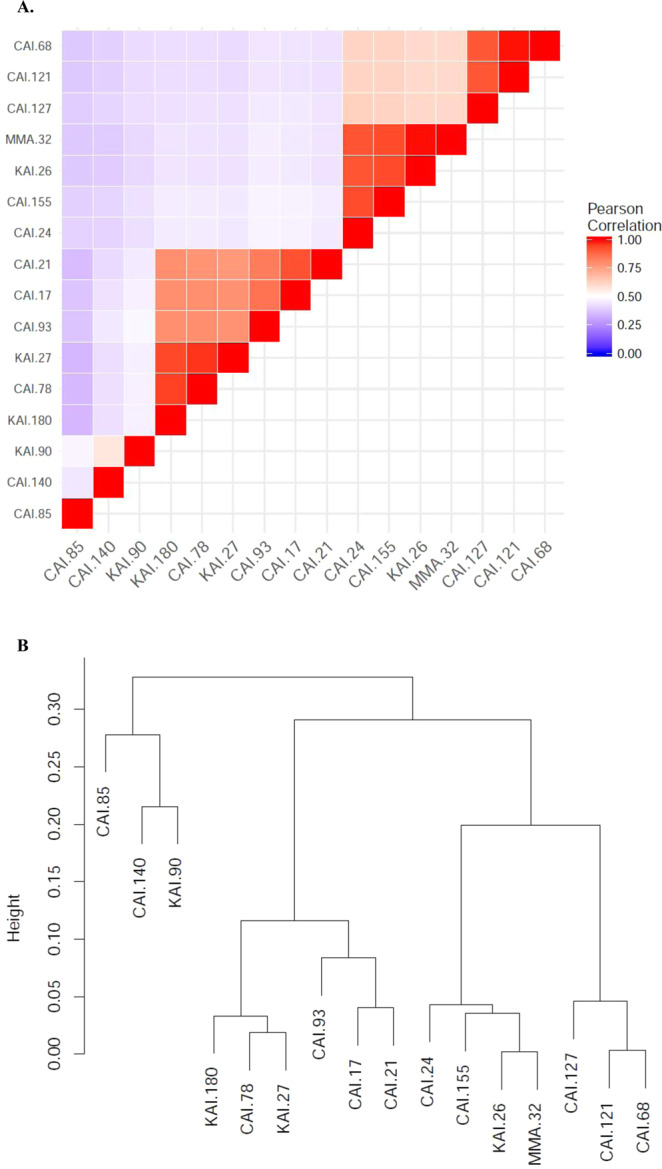
(A) Pearson correlation between all pairs of strains based on entire gene set including both ortholgous and unique genes. (B) Dendrogram based on same data. This pairwise gene-set comparison analysis indicated presence of 3 groups of closely related strains and one singleton.
The de novo assemblies of 16 strains were further compared with the publicly available Streptomyces species genomes/gene-sets available at NCBI/RAST database. These 16 strains were found closer to four public Streptomyces strains, namely, S. griseus NBRC 13350, S. albus J1074, S. avermitilis MA 4680 and S. coelicolor A3(2) (Table 5). Further, we quantified the extent of sequence similarity (in terms of fraction of orthologs) of one of the 16 strains with the closest reference Streptomyces species/strain. This pair-wise comparison have shown 70 to 85% of the genes were found common to the closest publicly available Streptomyces genomes (Table 5).
Table 5.
Closest Streptomyces species/stains, and extent of overlap in terms of fraction of orthologs to total genes.
| Strain no. | Closest public Streptomyces genome |
Orthologs present in corresponding strains (%) |
|---|---|---|
| CAI-17 | Streptomyces albus J1074 | 70 |
| CAI-21 | Streptomyces albus J1074 | 69 |
| CAI-24 | S. griseus NBRC 13350 | 80 |
| CAI-68 | S. griseus NBRC 13350 | 84 |
| CAI-78 | Streptomyces albus J1074 | 70 |
| CAI-85 | S. avermitilis MA-4680 | NA |
| CAI-93 | Streptomyces albus J1074 | 70 |
| CAI-121 | S. griseus NBRC 13350 | 84 |
| CAI-127 | S. griseus NBRC 13350 | 84 |
| CAI-140 | S. coelicolor A3(2) | 75 |
| CAI-155 | S. griseus NBRC 13350 | 80 |
| KAI-26 | S. griseus NBRC 13350 | 80 |
| KAI-27 | Streptomyces albus J1074 | 69 |
| KAI-90 | S. coelicolor A3(2) | 70 |
| KAI-180 | Streptomyces albus J1074 | 70 |
| MMA-32 | S. griseus NBRC 13350 | 80 |
Comparison of genomes based on molecular function of the genes
To understand the large scale functional differences among 16 Streptomyces genomes, the assemblies were also compared using annotated/curated genes. Since multiple genes may perform same molecular-function/role, so comparison was done at the level of molecular-functions/roles across all strains (equivalently, such comparison will be at the level of orthogroup). As a result, we could identify ~1,850 molecular-functions/roles across these genomes (Supplementary Table S1). While ~50% of such molecular-functions/roles were conserved across the genomes of 16 Streptomyces strains, only ~10% were strain specific, and the rest molecular functions/roles were present in various combinations (Supplementary Fig. S2 & Supplementary Table S1).
Among the unique molecular functions/roles, >95% were limited to just three of the Streptomyces strains namely, CAI-85, CAI-140 and KAI-90 (Supplementary Table S2). These strain-specific molecular functions were involved in biological processes such as, iron acquisition and metabolism, siderophore biosynthesis, phosphate metabolism, auxin biosynthesis, antibiotic resistance and toxin biosynthesis (streptolysin). On the other hand, conserved roles/molecular functions of Streptomyces strains belonged to almost all biological processes listed (Supplementary Table S1).
The remaining molecular functions (~800), other than unique and conserved, were present in multiple Streptomyces strains, that is, in subsets of 16 strains (Supplementary Table S2). There were several instances when same subset of strains was positive for other molecular functions/roles belonging to a common subsystem or biological process. Such behavior was most likely due to arrangement of the genes in an operon. For example, five roles under the subsystem ‘histidine degradation’ were present in same 14 Streptomyces strains, and absent in two strains namely, KAI-27 and CAI-78. Further, we have also estimated the relatedness in Streptomyces strains genomes based on roles/molecular functions. Similar to the observations of comparison of complete gene sets, as mentioned above, these samples formed three groups and a singleton (Supplementary Fig. S3A,B). The smallest group and the singleton, comprised of three strains CAI-85, CAI-140 and KAI-90, was due to the occurrence of almost all unique functions within these three strains.
Biosynthetic gene clusters (BGCs)
Since the Streptomyces genus is known for their ability to produce a variety of secondary metabolites, so the genome assemblies of these strains were predicted for presence of gene clusters involved in biosynthesis of secondary metabolites. While the number of BGCs per strain ranged from 23 to 39, the median number of genes per cluster ranged between 16 to 24, and the median size of BGCs ranged between ~22000 to ~33000 base pairs (Table 6; Fig. 3; Supplementary Table S3). On comparison to one of the closest publicly available genomes namely, S. griseus NBRC 13350, the above quantified properties appeared similar: presence of 40 BGCs, median number of genes per cluster was slightly higher (23), and median size of the BGCs was ~25000 base pairs (Table 6).
Table 6.
Prediction of biosynthetic gene clusters (BGCs) in sixteen Streptomyces genome assemblies, and their comparison with the BGCs from one of the closest species, S. griseus NBRC 13350.
| Strains | Number of biosynthetic clusters predicted | Distribution of number of genes per cluster (1st quartile, median, 3rd quartile) | Distribution of size of clusters (1st quartile, median, 3rd quartile; in base pairs) |
|---|---|---|---|
| CAI-21 | 24 | 13, 21, 28 | 15000, 28000, 42000 |
| CAI-24 | 32 | 13, 20, 40 | 15000, 24000, 41000 |
| CAI-68 | 39 | 13, 21, 35 | 19000, 26000, 47000 |
| CAI-127 | 39 | 11, 21, 31 | 13000, 26000, 39000 |
| CAI-140 | 23 | 11, 16, 25 | 12000, 22000, 31000 |
| CAI-155 | 32 | 16, 23, 40 | 21000, 27000, 43000 |
| KAI-90 | 35 | 10, 19, 32 | 13000, 22000, 40000 |
| CAI-78 | 32 | 7, 18, 23 | 11000, 22000, 40000 |
| CAI-85 | 31 | 11, 22, 38 | 12000, 23000, 49000 |
| CAI-93 | 25 | 9, 20, 38 | 15000, 27000, 44000 |
| CAI-121 | 39 | 13, 21, 35 | 19000, 26000, 47000 |
| KAI-27 | 36 | 7, 17, 22 | 11000, 22000, 38000 |
| KAI-180 | 26 | 6, 15, 27 | 12000, 23000, 32000 |
| MMA-32 | 32 | 17, 24, 42 | 22000, 30000, 48000 |
| KAI-26 | 32 | 18, 24, 42 | 22000, 30000, 49000 |
| S. griseus NBRC 13350 | 40 | 10, 23, 36 | 17000, 25000, 52000 |
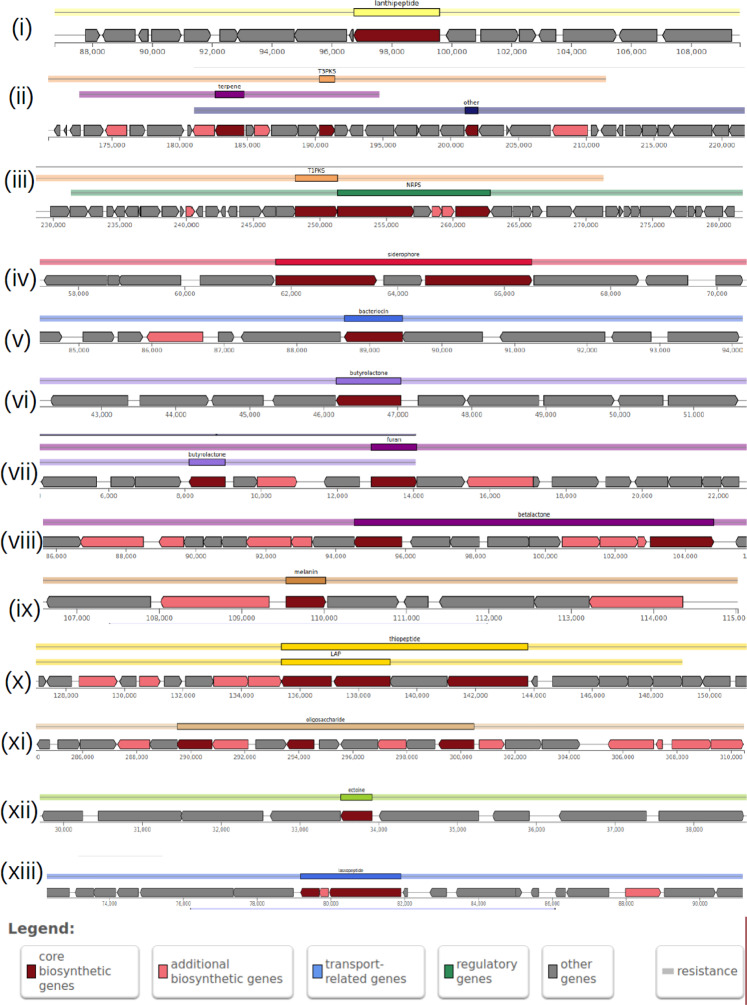
Figure 3.
Graphical representation of key Biosynthetic Gene Clusters (BGCs) in one of the strains having maximum number of BGCs namely, CAI-121. Out of 39 BGCs present, only thirteen non-redundant ones are shown. The annotation of these BGCs are: (i) lantipeptide (ii) T3PKS, tarpene (iii) T1PKS, NRPS (iv) siderophore (v) bacteriocin (vi) butyrolactone (vii) furan (viii) betalactone (ix) melanin (x) thiopeptide, LAP (xi) oligosaccharide (xii) ectoine and (xiii) lassopeptide.
Apart from distribution of BGCs, the conservation of biosynthetic clusters (BC) was also examined in these strains. A genome pair comprising CAI-68 strain and S. griseus NBRC 13350 were selected due to highest percentage of shared genes (Table 5). Out of the 40 experimentally/predicted biosynthetic clusters of S. griseus NBRC 13350 (see methods), while ~40% were found largely intact in CAI-68 assembly, however, ~50% showed structural changes (largely deletions in CAI-68 assembly of size ~4,000 to 1000,000 bp) either at the end or within the BC (Supplementary Table S4). Three such cases of deletions within homologous BC were tested for being observed due to any assembly artifact, however, they all were found genuine (Supplementary Table S4).
Plant growth-promotion (PGP) and biocontrol traits
De novo genome assemblies of Streptomyces strains were analyzed to identify genes involved in key pathways related to PGP and biocontrol activities. We have analyzed genome assemblies for traits like, biosynthesis and/or release of siderophores, auxin, hydrocyanic acid, chitinase and cellulase.
Siderophores
The phenotyping data have shown that 14 out of 16 Streptomyces strains had siderophore producing capacity in the specified rating scale of 1–4; the strains CAI-78 and KAI-180 did not produce siderophore (Table 1). Further, we have examined genomic data with respect to siderophore production. A total of 31 molecular functions were annotated to be involved in siderophore production (Supplementary Table S5), and majority (19 out of 31) of the molecular functions were conserved across strains (Supplementary Table S5). However, a set of four gene functions namely, Isochorismate synthase, 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase, 2,3-dihydroxybenzoate-AMP ligase and Ferric enterobactin-binding periplasmic protein, were found in high siderophore producing strains (Supplementary Table S6; Table 7). Seven high siderophore scorers (CAI-127, CAI-121, CAI-68, CAI-155, CAI-24, MMA-32 and KAI-26), with mean score of 3, all belonged to a single clade in the phylogenetic tree described earlier (Table 7; Fig. 2B). Out of four genes mentioned above, the three genes were co-located, and were likely to be part of an operon, while the fourth gene (Ferric enterobactin-binding periplasmic protein) was physically apart (Supplementary Table S4; Table 7). One of the strains (KAI-90) from a different clade, however, also showed relatively high siderophore score, but possessed only one of the four genes namely, 2,3-dihydroxybenzoate-AMP ligase. On the other hand, six of the low siderophore scoring (KAI-180, KAI-27, CAI-78, CAI-21, CAI-17 and CAI-93), with mean score of 1, belonged to a distinct clade of the phylogenetic tree, and none of the four gene functions were present in them (Table 7; Fig. 2B). The siderophore scores of these two contrasting clades was significantly different (p-value = 0.00069 for one-sided t.test).
Table 7.
Association of functional composition with the siderophore production. Among 12 molecular functions annotated in RAST for siderophore related, the strains having a set of four molecular functions were high siderophore producers (score 3–4 with an exception of CAI-155). The name of four genes are mentioned in the text. It was also observed that strains showing high siderophore scores belong to same clade in phylogenetic tree (in Figs. 2B and S3B).
| Strain No. | Score for siderophore | Occurrence 4-siderophore related genes | Phylogenetic group the strains belong to |
|---|---|---|---|
| CAI-127 | 4 | All 4 | 7 member clade |
| CAI-121 | 3 | All 4 | - do - |
| CAI-68 | 3 | All 4 | - do - |
| CAI-155 | 2 | All 4 | - do - |
| CAI-24 | 3 | All 4 | - do - |
| MMA-32 | 3 | All 4 | - do - |
| KAI-26 | 3 | All 4 | - do - |
| KAI-180 | — | — | 6 member clade |
| KAI-27 | 1 | — | - do - |
| CAI-78 | — | — | - do - |
| CAI-21 | 1 | — | - do - |
| CAI-17 | 2 | — | - do - |
| CAI-93 | 2 | — | - do - |
| CAI-85 | 1 | — | singleton |
| CAI-140 | 1 | — | 2 member clade |
| KAI-90 | 3 | One of the 4 g | - do - |
Auxin
The phenotyping data have shown that all but one strain (KAI-90) possess the ability of producing auxin (IAA) (Table 1). Four strains were highest producers (CAI-85, CAI-121, CAI-93 and KAI-180; in the range 30–43 µg ml−1), two strains were moderate producers (CAI-140 and CAI-155; in the range 12 and 15 µg ml−1) and the rest strains were least producers (CAI-17, CAI-21, CAI-24, CAI-68, CAI-78, CAI-127, KAI-26, KAI-27 and MMA-32; in the range 0.3–5.9 µg ml−1). In order to correlate the phenotyping data with the genomic composition of strains, we have selected eight genes representing three alternate pathways of auxin biosynthesis for analysis (Table 8). Only one gene, encoding enzyme Indole 3-acetaldehyde dehydrogenase, involved in two auxin biosynthetic pathways (Indole 3-pyruvate and Tryptamine) was present in all 16 strains (Table 8). Another gene namely, Amine/Tyramine oxidase, belonging to Tryptamine pathway was present in all but two strains (Table 8). The highest number (five) of genes involved in auxin biosynthesis was present in CAI-85, and the phenotype data suggested the same strain (CAI-85) to be one of the highest producer of IAA compared to other strains studied (Table 8). Alternatively, homology search of entire protein sequences of 16 strains for orthologs of IAA biosynthesis enzymes reported in literature didn’t add much to the results mentioned above (Supplementary Table S7). Therefore, it can be proposed that the combination of five genes is associated with Auxin biosynthesis at high level in CAI-85.
Table 8.
Orthologs of enzymes involved in auxin (IAA) biosynthetic pathways based on RAST annotation. The data is based on mapping of orthologs to tryptophan metabolic pathway using RAST annotation.
| Pathway | Reaction | Enzyme catalyzing the reaction | No of strains having ortholog | Remarks |
|---|---|---|---|---|
| Tryptophan →Indole 3-pyruvate | EC 2.6.1.99, 2.6.1.27, 1.4.3.2 | |||
| Indole 3-pyruvate | (Aminotransferase) | 0 | ||
| Indole 3- pyruvate→ | EC4.1.1.43, 4.1.1.74 (Indole | |||
| Indole 3- acetaldehyde | pyruvate decarboxylase) | 0 | ||
| Indole 3- acetaldehyde→ | EC 1.2.1.3, 1.2.3.7 (Indole 3- | |||
| Indole acetate | acetaldehyde dehydrogenase) | 12 | ||
| EC 4.1.1.28 (Trip | ||||
| Tryptophan →Tryptamine | decarboxylase) | 1 | Only in CAI-85 | |
| Tryptamine→ Indole 3- | EC 1.4.3.22, 1.4.3.4 | absent in CAI-140 | ||
| Tryptamine | acetaldehyde | (Amine/Tyramine oxidase) | 14 | & KAI-90 |
| Indole 3-acetaldehyde→ | EC 1.2.1.3, 1.2.3.7 (Indole 3- | |||
| Indole acetate | acetaldehyde dehydrogenase) | 16 | ||
| Tryptophan→ Indole 3- | EC 1.13.12.3 (Tryptophan | Present only in | ||
| Indole 3-acetamide | acetamide | mono-oxygenase) | 1 | KAI-90 |
| Indole 3- acetamide→ | Absent in largest | |||
| Indole acetate | EC 3.5.1.4 (IAM hydrolase) | 9 | clade |
Hydro cyanic acid (HCN)
The phenotyping data generated on HCN production have been quantified on the scale of 1–3 (Table 1). All the strains were found to have HCN producing ability with KAI-26 as the least producer with a score of 1 (Table 1). Three genes (hcnA, hcnB and hcnC) corresponding to an operon have been reported in HCN biosynthesis in Pseudomonas fluorescens F11329. These three genes were used for bi-directional best BLAST to search their homologous sequences in 16 Streptomyces strains. We could detect homologous genes only for the hcnC in all the 16 Streptomyces strains (Supplementary Table S8). Nevertheless, a thorough examination of the BLAST results of hcnA, hcnB and hcnC genes indicated that a set of three co-localized genes appeared among top five BLAST hits in ten out of sixteen strains (Supplementary Table S9). The orthologs of hcnA was annotated as similar to sarcosine oxidase alpha subunit, hcnB as putative oxidoreductase in 4 hydroxyproline catabolic gene cluster, and hcnC as D amino acid oxidase (EC 1.4.3.3). Since these gene functions were not directly related to HCN biosynthesis, therefore, based on present results we could not establish a correlation between genotype and phenotyping data. These results indicate the Streptomyces strains use either a different biosynthetic pathway than the one present in Pseudomonas, or they use the above mentioned cluster of three co-localized genes.
Chitinase
As per the phenotypic results, chitinase production was observed in 11 strains such as, CAI-17, CAI-24, CAI-93, CAI-121, CAI-127, CAI-155, KAI-26, KAI-27, KAI-90, KAI-180 and MMA-32 (Table 1). The strains CAI-21, CAI-68, CAI-78, CAI-85 and CAI-140 were devoid the chitinase producing traits (Table 1). There were eleven gene functions mapped to Chitin and N-acetylglucosamine utilization subsystem. Nine out of eleven gene functions were present in all 16 strains (Supplementary Table S10). The two gene functions which were present in a subset of strains encoded for ‘Chitodextrinase precursor’ (EC 3.2.1.14) and ‘N-Acetyl- glucosamine ABC transport system, permease protein 2’.
Cellulase
Phenotyping for cellulase activity showed all strains were positive (Table 1). Cellulase activity involves two enzymes (Endoglucanase (EC 3.2.1.4)) and Beta-glucosidase (EC 3.2.1.21)). Therefore, we searched the genome sequence data for their homologous sequences in 16 studied strains. Interestingly we have identified both the enzymes homologues in all 16 strains (Table 9).
Table 9.
Occurrence of enzymes involved in cellulase activity. The codes 0 and 1 indicates presence and absence of orthologs of the enzyme in a given strain. In three strains RAST couldn’t find an ortholog which was however predicted by Orthofinder (highlighted with blue).
| Strain no | Endoglucanase (EC 3.2.1.4) | Beta-glucosidase (EC 3.2.1.21) | Phenotype |
|---|---|---|---|
| CAI-17 | 1 | 1 | + |
| CAI-21 | 1 | 1 | + |
| CAI-24 | 1 | 1 | + |
| CAI-68 | 1 | 1 | + |
| CAI-78 | 1 | 1 | + |
| CAI-85 | 1 | 1 | + |
| CAI-93 | 1 | 1 | + |
| CAI-121 | 1 | 1 | + |
| CAI-127 | 1 | 1 | + |
| CAI-140 | 1 | 1 | + |
| CAI-155 | 1 | 1 | + |
| KAI-26 | 1 | 1 | + |
| KAI-180 | 1 | 1 | + |
| KAI-27 | 1 | 1 | + |
| KAI-90 | 1 | 1 | + |
| MMA-32 | 1 | 1 | + |
Discussion
Relatively less number of actinobacterial genera relevant to agriculture have been studied at the whole genome level as compared to clinically-important genera e.g. Mycobacterium, Propionibacterium, etc. Hence, in the present study we have developed de novo assemblies for 16 Streptomyces strains, which were phenotypically characterized for their PGP, antagonistic and larvicidal (including one metabolite with insecticidal) activities against pathogens and insect pests of chickpea, pigeonpea and sorghum in planta15,21,22 (Table 1). De novo assemblies of selected 16 Streptomyces strains led to better understanding of the molecular mechanisms of their PGP/antagonistic/entomopathogenic functions and provided opportunities to discover more secondary metabolites.
The phylogenetic analysis conducted in the present study has demonstrated a much more accurate view of the species/strain phylogeny in Streptomyces that reflects different parts of the genome. Comparative analysis of gene annotations across the Streptomyces strains revealed many apparent lineage-specific gene families that might have emerged in the common ancestor of Streptomyces clade. The selected 16 Streptomyces strains have also shown to produce hydrolytic enzymes/harmones (as PGP and biocontrol traits) such as siderophore, indole acetic acid, hydrocyanic acid, chitinase, cellulase, protease, lipase and β-1,3-glucanase under in vitro conditions (Table 1). A number of genes/gene functions have been found associated with above mentioned traits in the present study. For instance, a set of four gene functions (Isochorismate synthase, 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase, 2,3-dihydroxybenzoate-AMP ligase and Ferric enterobactin-binding periplasmic protein) were found in high siderophore producing strains such as CAI-127, CAI-121, CAI-68, CAI-155, CAI-24, MMA-32 and KAI-26. Similarly30 reported five gene clusters for siderophore biosynthesis in Streptomyces sp.31 reported another siderophore gene cluster, 2,3-dihydroxybenzoate, for the first time in Streptomyces sp. ATCC 700974, which was also observed in this study. Siderophores are iron chelators secreted by bacteria, fungi and plants for their uptake, occur in several chemical forms: enterobactin (in Escherichia coli), aerobactin (in Aerobacter aerogenes), anguibactin, pyochelin and rhodotorulic acid (in Rhodotorula pilimanae) and ferrichrome (in Ustilago maydis)32. Siderophores also forms stable complexes with heavy metals such as U, Np, Al, Cu, Cd, Ga, Zn and Pb and increases the soluble metal concentrations23. This process helps to alleviate the heavy metal stresses in soils. Functional characterization (gene expression based) of this phenotype was earlier reported for all strains13,15,17,19. In brief, all the strains were grown separately in Bennett’s broth at 28 °C for 72 h in laboratory conditions and RNA was extracted to perform quantitative real time- PCR (qRT-PCR). Varying level of expression of siderophore synthetase (conserved across the strains) was observed, and the expression level correlated well with the siderophore production level, with exception in three cases13,15,17,19 (Supplementary Table S11). The expression results indicates that in addition to genetic makeup, this PGP trait may also be regulated at the transcriptional level.
Another PGP agent or plant growth hormone, i.e. Auxins, were also searched in the present data. Auxin producing bacteria are known to stimulate seed germination, root formation and root proliferation, thereby providing the host plant greater access to water and soil nutrients33,34. Several pathways exist in bacteria for auxin biosynthesis and in few of the cases tryptophan has been used as a precursor. In the present study, we have identified five genes associated with auxin biosynthesis and the pathway involving indole-3-pyruvate. Genes involved in another pathway for auxin biosynthesis i.e. pathway involving tryptamine were also searched in the present data. In tryptamine pathway three enzymes would be required and two of them are unique to it. Only one strain (CAI-85) in the present study had all the enzymes, whereas in at least 14 strains, enzymes catalyzing last two reactions were present. The third pathway involved in auxin biosynthesis i.e. indole-3- acetamide as a precursor and two enzymes were looked for their presence in the targeted strains. Sequence data showed that in just one strain (KAI-90) both the enzymes were present, and in about 9 strains only the enzyme catalyzing second reaction was present (Supplementary Table S7). Similar to present findings, Streptomyces sp. such as S. violaceus, S. scabies, S. griseus, S. exfoliates, S. coelicolor and S. lividans synthesize IAA in the presence of tryptophan via indole-3-acetamide pathway35,36. In addition to indoleacetamide hydrolase pathway nitrilase pathway has also reported in Streptomyces sp. (GKU 895)37. Like siderophore, function characterization of IAA pathway by qPCR was earlier reported for almost all strains, showing correlated expression with the IAA production level13,15,17,19. While minimal gene expression was reported among the low IAA producers, as high as 24 fold up-regulation was found among the high IAA producers, with an exception of strain CAI-6813,15,17,19 (Supplementary Table S12). This behavior is indicative of regulation (of IAA production pathway) at transcriptional level.
Similar to above mentioned PGP or biocontrol agents, in the present study, few genes or molecular functions involved in HCN, chitinase and cellulase were also discovered in Streptomyces strains following candidate based approach. However, in the present study we could not establish association of genes with HCN, chitinase and cellulase synthesis. A whole genome transcriptome analysis in future together with de novo assemblies generated in the present study may help in identification of the candidate genes responsible for HCN, chitinase and cellulase.
Conclusion
The present study has developed complete genome assemblies for 16 Streptomyces strains and provided better understanding of genomic composition, genetic similarity and information on genes associated with favorable traits. However, identified favorable traits associated genes in the present study needs to be validated through construction of knock-outs or gene expression analysis in future studies. This can be considered as limitation of the present study and an opportunity for the upcoming studies. Moreover, we anticipate advancements made in the present study will provide opportunities for genome mining particularly of biosynthetic gene clusters from these and other micro-organisms, cloning of target genes, heterologous expression etc.
Materials and Methods
PGP strains
Sixteen strains of Streptomyces (CAI-17, CAI-21, CAI-24, CAI-68, CAI-78, CAI-85, CAI-93, CAI-121, CAI-127, CAI-140, CAI-155, KAI-26, KAI-27, KAI-90, KAI-180 and MMA-32) isolated previously from various herbal vermicompost and reported as potential for the PGP in chickpea, pigeonpea, rice and sorghum and biocontrol of important pathogens of chickpea and sorghum (Table 1)9–23 were further studied.
Isolation of DNA
DNA was isolated as per the protocols of38. In brief, Streptomyces strains were inoculated in starch casein broth (SCB) and incubated for 5 days at 28 °C. At the end of incubation, the cultures were centrifuged at 8,000 g for 10 min at 4 °C and the cells washed twice with STE buffer (0.3 M sucrose, 25 mM Tris/HCl and 25 mM Na2EDTA, pH 8.0). One g of the pellet was re suspended in 8.55 ml STE buffer and 950 µl lysozyme (20 mg/ml STE buffer) and incubated for 20–30 min at 30 °C. This was followed by addition of 500 µl of 10% SDS (w/v) and 50 µl of protease (20 mg/ml) and the mixture was held at 37 °C for 1 h. At the end of incubation, 1.8 ml 5 M NaCl was added with gentle mixing to avoid shearing the DNA and 1.5 ml 10% (w/v) CTAB in 0.7 M NaCl (CTAB/NaCl solution) and incubated for 20 min at 65 °C. After the addition of CTAB, all the steps were carried out at room temperature. The lysate was extracted twice with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, by vol) and centrifuged at 12,000 g for 10 min. The aqueous phase was finally extracted with chloroform/isoamyl alcohol (24:1, by vol) and transferred to a fresh tube. This was followed by addition of 600 µl of propan 2 ol and DNA spooled out after 10 min. Alternatively, it was recovered by centrifugation at 12,000 g for 10 min. The pellet was washed twice with 70% (v/v) ethanol, vacuum dried and dissolved in 2 ml TE buffer (10 mM Tris/HCl and 1 mM EDTA, pH 8.0). RNaseA (50 mg/ml) was added with incubation at 37 °C for 2 h. The sample was again extracted with phenol as described above. DNA was re precipitated from the aqueous phase with addition of 100 µl of 3 M sodium acetate (pH 5.3) and 600 µl of propan 2 ol. The DNA pellet was washed with 70% (v/v) ethanol, dried and dissolved in TE buffer.
Sequencing, assembly and annotation
The genomic DNA was sequenced to generate paired end reads using Hiseq. 2500 platform with a target of ~500X coverage. The reads were assembled using de novo assembler SPAdes version 3.10.126. The contigs were filtered for minimum size (500 bp) and minimum read support (40). To evaluate the integrity of samples, 5–10 longest contigs were compared using BLAST (version 2.4.0+) against reference genomes of bacteria (source: ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/bacteria/), and consistency in term of target sequence was examined for the top hits (upto two). Hits to targets other than genus Streptomyces, would be indicative of issues with the assembly and/or contamination, but none was observed in the 16 assemblies.
To annotate the contigs, the sequence data were uploaded to Rapid Annotation using Subsystem Technology (RAST) online server (http://rast.nmpdr.org/rast.cgi)39. The RAST server predicted genes, translation of protein coding genes, and their annotation. Whether the proteome of each sample covers the minimal bacterial proteins27, the protein sequence of 339 such genes were first obtained from uniprot database (www.uniprot.org) such that the sequences of majority were form Streptomyces species present in uniprot database. Homology search of database sequences against proteome of each of the sample was performed using NCBI-BLASTP, and hits with Evalue <1E-05 were considered significant. 24 of the 339 query sequences didn’t get a hit in any of the 16 strains, and were dropped from the list of minimal bacterial gene set.
Besides, for examining quality of gene/protein sequences, a set comprising 32 RefSeq proteomes of publicly available Streptomyces genus (https://www.ncbi.nlm.nih.gov/assembly?LinkName=genome_assembly&from_uid=13511), was used as a database against each strain using NCBI-BLASTP. Sequences without a hit or hit with Evalue > =1E-03 were removed. Further, for a significant hit, if the alignment length covered 95% or more of length of query sequence, then it indicated assembly of full gene (i.e., full-length alignment).
Comparison of genomes using gene sets
To compare the genomes at gene level, two approaches were implemented. In first approach, entire protein sequences of all strains were subject to prediction of orthogroups using default parameters of OrthoFinder tool28. In second approach, the genes which were successfully annotated using Subsystem technology were grouped based on molecular function (role). This was equivalent to orthogroups predicted by OrthoFinder. For each molecular function, if one or more genes were present identified in a strain, then assigned a binary code of 1, otherwise 0 (Supplementary Tables S1–S3). To get a summary, these binary codes were summed to report number of strains having a molecular function (role) per orthogroups.
Prediction of biosynthetic gene clusters (BGCs) and their conservation
The BGCs were predicted using standalone version of antiSmash-v5.040 with following parameters:–minimal,–genefinding-tool none. For finding conservation of BGCs, those identified by IMG database in S. griseus NBRC 13350 genome were obtained (https://img.jgi.doe.gov/)41, and nucleotide sequences of BGC regions were extracted from the complete genome downloaded from NCBI-RefSeq database (ID: GCF_000010605.1_ASM1060v1). Homology of these sequences were searched against CAI-68 genome assembly using BLASTN (pvalue cutoff: 1E-150), and the high scoring pairs were arranged in the increasing order of genomic position to figure out any structural variations (i.e., large insertions, deletions and translocations). Three BGC cases where deletions were observed in CAI-68 genome with respect to S. griseus NBRC 13350 genome, their sequences extracted and were BLASTed online against the NCBI complete genome database, followed by evaluation of the query matched with any of the database sequences without any break (that is, no fragmented match).
Discovery of candidate genes for PGP/biocontrol properties
To discover the genes underlying various PGP or biocontrol traits, multiple approaches were implemented. If the pathway or process was characterized in curated annotation of RAST or its pathways, then the genomic information was directly compared to the phenotype. If incomplete/nil data was found from RAST annotation, then KEGG pathway database (http://www.genome.jp/kegg/kegg2.html) was examined followed by searching orthologs of the KEGG enzymes in the peptide data through bi-directional Best BLAST. In case even KEGG database didn’t have any information, the literature was searched for genetic/genomic studies about that process. Only literature on bacterial species, in particular, in actinomycetes group were preferred. These analysis were often complemented by exploiting the orthogroups predicted by OrthoFinder.
Supplementary information
Acknowledgements
This work was undertaken as part of the CGIAR Research Program on Grain Legumes and Dry Land Cereals (GLDC). ICRISAT is a member of CGIAR Consortium. This study was also partly supported by DBT’s Ramalingaswami grant and author Vivek Thakur is grateful to this. We also thank Mr. PVS Prasad for technical assistance.
Author contributions
G.S., V.T., R.K.S. and R.K.V. conceived, designed, supervised the study and finalized the manuscript. G.S., R.K.S., S.V., V.K. and A.C. generated the data. V.T., S.P. and A.R. along with R.K.V., G.S., R.K.S., analyzed the data. All authors read, and approved the manuscript.
Data availability
The sequencing data generated in this study has been submitted at National Centre for Biotechnology Information (NCBI) under the Bioproject ID PRJNA510915.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gopalakrishnan Subramaniam, Vivek Thakur and Rachit K. Saxena.
Contributor Information
Gopalakrishnan Subramaniam, Email: s.gopalakrishnan@cgiar.org.
Vivek Thakur, Email: vivek22@gmail.com.
Rajeev K. Varshney, Email: r.k.varshney@cgiar.org
Supplementary information
is available for this paper at 10.1038/s41598-020-67153-9.
References
- 1.Pérez-Montano F, et al. Plant growth-promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014;169:325–336. doi: 10.1016/j.micres.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Gopalakrishnan, S., Sathya, A. & Vijayabharathi, R. Plant Growth-Promoting Actinobacteria: A New Avenue for Enhancing the Productivity & Soil Fertility of Grain Legumes, Springer Singapore, ISBN 978-981-10-0705-7, (2016).
- 3.Vacheron J, et al. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behie SW, Bonet B, Zacharia VM, McClung DJ, Traxler MF. Molecules to Ecosystems: Actinomycete Natural Products in situ. Front. Microbiol. 2017;7:2149. doi: 10.3389/fmicb.2016.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012;65:385–395. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- 6.Martinez Hidalgo P, Olivares J, Delgado A, Bedmar E, Martínez Molina E. Endophytic Micromonospora from Medicago sativa are apparently not able to fix atmospheric nitrogen. Soil Biol. Biochem. 2014;74:201–203. doi: 10.1016/j.soilbio.2014.03.011. [DOI] [Google Scholar]
- 7.Tarkka, M. T. & Hampp, R. Secondary metabolites of soil Streptomycetes in biotic interactions. In: Karlovsky P, editor. Secondary Metabolites in Soil Ecology. 14. Berlin, Germany: Springer, Soil Biology Series. pp 107–118 (2008).
- 8.Sathya A, Vijayabharathi R, Gopalakrishnan S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3Biotech. 2017;7:102. doi: 10.1007/s13205-017-0736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gopalakrishnan S, et al. Evaluation of actinomycete isolates obtained from herbal vermicompost for biological control of Fusarium wilt of chickpea. Crop Prot. 2011;30:1070–1078. doi: 10.1016/j.cropro.2011.03.006. [DOI] [Google Scholar]
- 10.Gopalakrishnan S, et al. Biocontrol of charcoal rot of sorghum by actinomycetes isolated from herbal vermicompost. Afric J. Biotechnol. 2011;10:18142–18152. [Google Scholar]
- 11.Gopalakrishnan S, et al. Plant growth-promoting traits of biocontrol potential Streptomyces isolated from herbal vermicompost. Biocont Sci. Technol. 2012;22:1199–1210. doi: 10.1080/09583157.2012.719151. [DOI] [Google Scholar]
- 12.Gopalakrishnan S, et al. Evaluation of Streptomyces spp. for their Plant growth-promoting traits in rice. Can. J. Microbiol. 2013;59:534–539. doi: 10.1139/cjm-2013-0287. [DOI] [PubMed] [Google Scholar]
- 13.Gopalakrishnan S, Srinivas V, Prakash B, Vijayabharathi R, Rupela O. Evaluation of Streptomyces strains isolated from herbal vermicompost for their plant growth promotion traits in rice. Microbiol. Res. 2014;169:40–48. doi: 10.1016/j.micres.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Gopalakrishnan S, Srinivas V, Vidya MS, Rathore A. Plant growth-promoting activities of Streptomyces spp. in sorghum and rice. Springer Plus. 2013;2:1–8. doi: 10.1186/2193-1801-2-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vijayabharathi R, et al. Biological activity of entomopathogenic actinomycetes against lepidopteran insects (Noctuidae: Lepidoptera) Can. J. Plant Sci. 2014;94:759–769. doi: 10.4141/cjps2013-298. [DOI] [Google Scholar]
- 16.Gopalakrishnan S, Srinivas V, Prakash B, Sathya A, Vijayabharathi R. Plant growth-promoting traits of Pseudomonas geniculata isolated from chickpea nodules. 3Biotech. 2015;5:653–661. doi: 10.1007/s13205-014-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gopalakrishnan S, et al. The extent of grain yield and plant growth enhancement by plant growth-promoting broad spectrum Streptomyces sp. in chickpea. SpringerPlus. 2015;4:31. doi: 10.1186/s40064-015-0811-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopalakrishnan S, et al. Evaluation of broad spectrum Streptomyces sp. for plant growth-promotion traits in chickpea. Philipp. Agric. Scientist. 2015;98:270–278. [Google Scholar]
- 19.Gopalakrishnan S, et al. Evaluation of Streptomyces sp. obtained from herbal vermicompost for broad spectrum of plant growth promoting activities in chickpea. Org. Agric. 2015;5:123–133. doi: 10.1007/s13165-015-0099-1. [DOI] [Google Scholar]
- 20.Gopalakrishnan S, Srinivas V, Alekhya G, Prakash B. Effect of plant growth-promoting Streptomyces sp. on growth promotion and grain yield in chickpea (Cicer arietinum L.) 3Biotech. 2015;5:799–806. doi: 10.1007/s13205-015-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sathya A, Vijayabharathi R, Srinivas V, Gopalakrishnan S. Plant growth-promoting actinobacteria on chickpea seed mineral density: An upcoming complementary tool for sustainable biofortification strategy. 3 Biotech. 2016;6:138. doi: 10.1007/s13205-016-0458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopalakrishnan S, et al. Insecticidal activity of a novel fatty acid amide derivative from Streptomyces species against Helicoverpa armigera. Nat. Prod. Res. 2016;30:2760–2769. doi: 10.1080/14786419.2016.1154055. [DOI] [PubMed] [Google Scholar]
- 23.Gopalakrishnan S, Vadlamudi S, Kumar CVS. Plant growth-promoting traits of Streptomyces sp. in pigeonpea. Leg. Perspec. 2016;11:43–44. [Google Scholar]
- 24.Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomole. Chem. 2009;7(9):1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 25.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 26.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye YN, et al. A novel proposal of a simplified bacterial gene set and the neo-construction of a general minimized metabolic network. Scientific Reports. 2016;6:35082. doi: 10.1038/srep35082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome. Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laville J, et al. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 1998;180:3187–3196. doi: 10.1128/JB.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. PNAS. 2001;98(21):12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patzer SI, Braun V. Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 2010;192:426–435. doi: 10.1128/JB.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajkumar M, Ae N, Prasad MNV, Freitas H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010;28:142–149. doi: 10.1016/j.tibtech.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Ahemad M, Kibret M. Mechanisms and applications of plant growth-promoting rhizobacteria: Current perspective. J. King Saud Univer. 2014;26:1–20. doi: 10.1016/j.jksus.2013.05.001. [DOI] [Google Scholar]
- 35.Manulis S, Shafrir H, Epstein E, Lichter A, Barash I. Biosynthesis of indole-3-acetic acid via the indole-3-acetamide pathway in Streptomyces spp. Microbiol. 1994;140:1045–1050. doi: 10.1099/13500872-140-5-1045. [DOI] [PubMed] [Google Scholar]
- 36.Lin L, Xu X. Indole-3-acetic acid production by endophytic Streptomyces sp. En-1 isolated from medicinal plants. Curr. Microbiol. 2013;67:209–217. doi: 10.1007/s00284-013-0348-z. [DOI] [PubMed] [Google Scholar]
- 37.Kruasuwan W, Salih TS, Brozio S, Hoskisson PA, Thamchaipenet A. Draft genome sequence of plant growth-promoting endophytic Streptomyces sp. GKU 895 isolated from the roots of sugarcane. Genome. Announc. 2017;5(19):e00358–17. doi: 10.1128/genomeA.00358-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi G, Rawal SK. Simple and efficient protocol for isolation of high molecular weight DNA from Streptomyces aureofaciens. Biotech Techniques. 1998;12:629–631. doi: 10.1023/A:1008836214495. [DOI] [Google Scholar]
- 39.Overbeek, R. et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. (Database issue):D206–214 (2014). [DOI] [PMC free article] [PubMed]
- 40.Blin K, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen IA, et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019;47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this study has been submitted at National Centre for Biotechnology Information (NCBI) under the Bioproject ID PRJNA510915.