Abstract
The present paper aims to explore the rhizospheric bacterial communities associated with Saccharum arundinaceum grown on organometallic pollutants-rich hazardous distillery sludge. The sequence analysis of 16S rRNA V3–V4 hypervariable region with Illumina MiSeq platform showed 621,897 OTUs derived from rhizospheric and non-rhizospheric distillery sludge samples out of 1,191,014 and 901,757 sequences read, respectively. The major phyla detected in rhizospheric sludge sample were Proteobacteria (50%), Bacteriodetes (33%), Firmicutes (5%) Gemmatimonadetes (2%), Chloroflexi (2%), and Tenericutes (2%). The dominant three genera were detected as Rheinheimera (21%), Sphingobacterium (17%), and Idiomarina (8%). In addition, other minor genera such as uncultured Bacillus (4%), Acidothermus (4%), Bacillus (3%), Pseudomonas (2%), Flavobacterium (2%), uncultured bacterium (2%), Parapedobacter (2%), Alcanivorax (2%), Acholeplasma (2%), Hyphomonas (1%), and Aquamicrobium were also detected (1%) in rhizospheric sludge. Our results suggested that rhizospheric bacterial communities associated with S. arundinaceum were substantially different in richness, diversity, and relative abundance of taxa compared to non-rhizospheric sludge. Further, the comparative organic pollutant analysis from non-rhizospheric and rhizospheric sludge samples through GC–MS analysis revealed the disappearance of few compounds and generation of some compounds as new metabolic products by the activity of rhizospheric bacterial communities. The results of this study will be helpful in understanding the role of rhizospheric bacterial communities responsible for degradation and detoxification of complex organometallic waste and, thus, can help in designing appropriate phytoremediation studies for eco-restoration of polluted sites.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02310-5) contains supplementary material, which is available to authorized users.
Keywords: Proteobacteria, Distillery sludge, Organic pollutants, Illumina MiSeq, Saccharum arundinaceum
Introduction
Distillery industry is one of the major industries in developing countries including India which significantly contributes to the development of economic sector. However, it is also a major contributor of environmental pollution due to the discharge of potentially toxic and hazardous sludge into the natural environment. As per the record of All India Distiller’s Association (AIDA), approximately 397 sugarcane molasses-based distilleries are present in India which are discharging an average of 3.5 × 1013 kL raw effluent per annum (AIDA 2016). Sugarcane molasses-based distilleries also produce large amount of anaerobically digested distillery sludge (ADDS) which contains high concentration of recalcitrant organic pollutants including polymers of aldehyde–amines, nitrogenous heterocyclic compounds of amino-carbohydrate (known as Maillard Reaction Products), melanoidin-like products, and complex organic matter (OM) along with metallic ions (i.e., Cd2+, Cu2+, Mn2+, Fe2+, Pb2+, Ni2+ and Zn2+) and non-metallic ions (i.e., Na+, Cl−, SO−42, PO−43) (Arimi et al. 2014; Hatano et al. 2016; Chandra et al. 2018a, b). The Maillard reaction product which is a major colorant of distillery waste has a strong binding affinity with different heavy metal ions to generate organo-metallic complex compounds (Hatano et al. 2016). Recent reports showed distillery sludge is a source of a high amount of plant-derived long-chain fatty acids, resins, steroids, and organic acids which can act as endocrine-disrupting chemicals (EDCs) (Chandra and Kumar 2017a; Chandra et al. 2018a, b). A continuous disposal of such sludge into the natural environment can result in environmental degradation as well as negative effect on human health (Bharagava and Chandra 2010).
Distillery sludge is limited in freely available carbon–nitrogen sources, but contains large number of complex organic and inorganic compounds. Microorganisms can mineralize the organometallic compounds of ADDS and use as a source energy (Pang et al. 2016; Mukherjee et al. 2017; Kachienga et al. 2018). Thus, comprehensive functional characterization of indigenous bacterial communities living in ADDS can help in the development of sustainable waste management techniques (Chandra and Kumar 2017a, c). Studies suggest how characterization of indigenous bacterial communities is essential for sustainable eco-restoration of polluted sites (Bai et al. 2014; Abed et al. 2018). Use of plants and their rhizospheric microorganisms has been reported for bioremediation of industrial waste-polluted sites (Huaidong et al. 2017; Kumar et al. 2018). Native plant species spontaneously colonized on distillery sludge-dumped sites have been reported as a potential candidate for in situ phytoremediation (Chandra and Kumar 2017b; Chandra et al. 2018b). However, diverse rhizospheric bacterial communities of these plants are still unknown. Functional characterization of rhizospheric bacterial community of native plant growing in distillery waste site can help in better understanding the mechanism of bioremediation. Development of 16S rRNA-based next-generation sequencing (NGS) technology has revolutionized the microbial ecosystem studies and helped in characterization of complex microbial community structure of soil and/or different polluted environment (Das et al. 2017; Sahu et al. 2018; Kumar et al. 2018).
Application of NGS in microbial phylogenetic studies helped in deciphering the functional role of microbial communities for transformation and/or degradation of toxic pollutants at contaminated sites (Bai et al. 2014; Feng et al. 2018). Saccharum arundinaceum, commonly known as hardy sugar cane, is a native grass species to India and has been found actively growing in several distillery sludge disposal sites. Thus, exploring the rhizospheric bacterial communities of S. arundinaceum can connect and correlate how bacterial species are involved in in situ phytoremediation of hazardous organometallic waste.
The present study was aimed to explore and investigate the rhizospheric bacterial community composition (RBCC) of S. arundinaceum which may contribute in discovering potential bacterial species that are capable to remediate melanoidins contaminated site. In the present study, we have used high-throughput sequencing-based metagenomic approach to investigate RBCC associated with S. arundinaceum root grown on distillery sludge-dumped sites. This study will add new information regarding the role of unique rhizospheric bacterial communities in facilitating the growth of S. arundinaceum on hazardous organometallic waste during in situ phytoremediation.
Materials and methods
Chemicals and reagents
Organic solvents such as ethyl acetate and methanol were purchased from Sigma-Aldrich (Saint Louis, MO, USA). Ethyl acetate was used for the extraction of organic pollutants from bulk (non-rhizospheric) and rhizospheric distillery sludge, while methanol was used to dissolve the extracted dried residues for GC–MS analysis. The derivatizing reagents such as TMCS (trimethylchlorosilane), BSTFA [N,O-bis(trimethylsilyl)trifluoroacetamide] and pyridine were procured from Merck (Merck KGaA, Darmstadt, Germany). All the reagents and chemicals used in this study were of high-performance liquid chromatography (HPLC) grade (purity > 99%).
Site description and sampling
The experimental site for plant and distillery sludge sampling was selected in Unnao, Uttar Pradesh, India, situated at the north latitude of 26° 32′ 0″ and east longitude of 80° 30′ 0″, which had been polluted due to extensive and indiscriminate disposal of anaerobically digested sludge. This site is well known for high pollution with organic and inorganic pollutants as previously described by Chandra and Kumar (2017a, b). The climate in this area is classified as humid subtropical dry weather with an average annual rainfall of 850 mm (Rainy season: July–September). Annual temperature of is 25.6 °C, which varies between 15 and 40 °C from winter to summer, respectively. Distillery sludge samples were collected in October 2017 from the two different sludge disposed site located in premises of the M/s Unnao Distilleries & Breweries Limited. The frequent growth of plants, herbs, and grasses on dumped distillery sludge is reported previously (Chandra and Kumar 2017b). The sludge dumping area is dominated mainly by herbs and grasses. However, the dominant growth of perennial grass, S. arundinaceum, on dumped distillery sludge indicates its potentiality for phytoremediation of organometallic pollutants present in distillery waste. Samples of non-rhizospheric and rhizospheric distillery sludge were taken according to a simple randomized schematic strategy (Van Elsas and Smalla 1997). To collect the rhizospheric distillery sludge sample, S. arundinaceum plant root with adhering sludge was transferred into sterilized polyethylene bags and vigorously shaken by hand for 10 min so that loose soil particles were completely removed (Supplementary Fig. S1). The sludge adhering with root hairs was taken in the pre-sterilized 50-mL glass test tube as per the method described by Chen et al. (2010). The height of the S. arundinaceum was found 1.8 m with 1.5 cm in diameter forming large robust clumps. Roots of S. arundinaceum were highly dense and reticulated with root length of 25 cm in dumped sludge. Non-rhizospheric distillery sludge sample was randomly collected in triplicate from a depth of 0–20 cm with a soil auger in pre-sterilized biohazard bags (HiMedia Laboratories Pvt. Ltd., Karnataka, India) from sludge dumping site of anaerobic digester plant located inside the industry’s premises as control. From each sampling site, 150 g of sludge sample was taken. All the collected samples were transported to laboratory in ice-cold condition (~ 4 °C) for further analyses. Samples were divided into two fractions: one fraction of collected non-rhizospheric and rhizospheric distillery sludge samples were stored at − 20 °C for bacterial community analyses and second fraction was stored at 4 °C for physico-chemical characterization. Prior to analysis, triplicate samples of non-rhizospheric or rhizospheric distillery sludge were homogenized to generate one composite sample in each disposed site, respectively. The homogenized sludge samples were designated as DS1 (non-rhizospheric sludge) and DS2 (rhizospheric sludge) samples.
Physico-chemical characterization of collected samples
Sludge samples were air dried at room temperature, grounded into powder using mortar and pestle, and passed through a 2-mm mesh sieve to get a fine powder. Electrical conductivity (EC) and pH values of distillery sludge samples were measured using Orion conductivity meter (Model-A322, Thermo Scientific, FL, USA) and Orion pH meter (Model-960, Thermo Scientific, FL, USA), respectively. For pH measurement sludge and distilled water was mixed in a ratio of 1:2.5 (weight: volume) (Chandra and Kumar 2017a). Cation exchange capacity (CEC) was quantified using the method as described by Gillman and Sumpter (1986). Total Kjeldahl nitrogen (TKN) and total ammoniacal nitrogen (NH4+-N) were measured by the methods reported by Ju et al. (2007). Total phosphorus (TP) was measured by the colorimetric method (Jackson 1973). Organic matter content was determined using the Walkley and Black procedure (Nelson and Sommers 1982). Total oxygen and hydrogen was estimated by elemental analyzer (EuroVector EA 3000, University of AL-al-Bayt, Jordan). Concentration of Cl−, Na+, PO−43, and SO−42 were estimated according to the method described by Kalra and Maynard (1991). Total phenol concentration in the sludge samples was determined using 4-aminoantipyrine reaction method as described by Ettinger et al. (1951). As outlined, the phenolic material was mixed with 4-aminoantipyrine in the presence of alkaline oxidizing agent, preferable potassium ferricyanide, at high pH, to yield a red chromogenic compound. The absorption of this red solution was measured at 500 nm by UV–Vis spectrophotometer. For heavy metal analyses, distillery sludge samples were first digested with nitric acid–hydrogen peroxide digestion method (3050B) (U.S. Environmental Protection Agency; USEPA 1996). Briefly, 1-g dried sludge sample was placed into a Teflon reaction vessel and digested with 10 mL of the HNO3 (1:1, v/w) at 95 °C for 15 min. After cooling, 5 mL of concentrated HNO3 was added into vessel and reflux at 95 °C until the solution becomes transparent. After cooling the solution, 2 mL of water and 3 mL of 30% H2O2 were added into Teflon vessel. The solution was allowed to evaporate by raising the temperature to 105 °C until the samples were digested and the solution becomes transparent. The acid–peroxide digestate was continually heating at 95 °C without boiling for 2 h. The volume of these samples was reduced to 5 mL by evaporating the acid–peroxide mixture. Thereafter, the samples were allowed to cool to room temperature before the vessel content was filtered with Whatman filter paper and diluted with double deionized water up to 100 mL. Three replicates of every sample were digested, together with the corresponding blanks. Subsequently, the total concentrations of different heavy metals (i.e., Fe, Zn, Cu, Ni, and Mn) in the digested solution were determined by inductively coupled plasma-mass spectrometry (ICP-MS; Agilent 7500Cx, USA). The instrument settings and operational conditions were done in accordance with the manufacturers’ specifications.
Detection and characterization of various organic pollutants
Solid–liquid phase extraction
To investigate the major organic pollutants in collected sludge samples, ethyl acetate was used as an organic solvent for the extraction of organic pollutants under acidic conditions (pH < 2.0) as described by Chandra and Kumar (2017a). The extraction process was repeated three times to ensure complete extraction of organic pollutants from distillery sludge samples. The organic solvent phase was collected from separating funnel in a 50-mL beaker and passed through anhydrous sodium sulfate for removal of water content. Further, it was dried under a stream of nitrogen gas and dried residues were dissolved in methanol (2.0 mL), filtered through 0.22-μm pore size syringe filters (Millipore Ltd., Bedford, USA), and used for GC–MS analysis.
GC–MS analysis
The extracts obtained from rhizospheric and non-rhizospheric distillery sludge were firstly derivatized using TMCS, BSTFA and pyridine as per method described earlier (Chandra and Kumar 2017a) and analyzed by GC–MS (TRACE GC Ultra Gas Chromatographs, Thermo Fisher Scientific, USA) united with a TriPlus autosampler coupled to TSQ Quantum XLS triple quadrupole mass spectrometer (Thermo Scientific, USA). Organic compound separation was carried out in a DB-5MS capillary column (No. 2713S18) by running with helium as the carrier gas at a flow rate of 1.1 mL min−1. The GC oven conditions were as follows: an initial temperature of 65 °C (hold time: 2 min), increased to 230 °C (flow rate of 6 °C min−1) and finally increased up to 290 °C (hold time: 20 min; flow rate of 10 °C min−1) (Chandra and Kumar 2017a). An aliquot (1.0 µl) of derivatized sample was injected in the GC column in the splitless mode. The injector temperature was operated at 250 °C while the mass detector was operated at 300 °C. The MS was operated in full-scan mode (45–800 m z−1) at electron energy of 70 eV with a solvent delay of 7 min. The organic compounds were detected and identified through spectral matching of mass spectra obtained at different retention times with mass spectra of standard compounds mentioned in National Institute of Standard and Technology (version 1.0.0.12, NIST, USA) mass spectra library existing with GC–MS.
Characterisation of bacterial communities of sludge
Sample processing
Roots with adhering sludge were gently shaken in sterile saline solution (0.85 w/v) to remove adhering sludge and centrifuged at 5000 × g for 15 min at 4 °C to concentrate the sludge particles in form of a pellet. The collected pellet was used for DNA extraction, 16S rDNA (variable region: V3–V4) PCR amplification, and Illumina MiSeq sequencing analysis.
Community DNA Extraction and PCR amplification
The rhizospheric bacterial communities of wetland plants are of great interest because of their potential for bioremediation of distillery waste (Chaturvedi et al. 2006; Chandra et al. 2012). The bulk microbial community DNA derived from non-rhizospheric and rhizospheric distillery sludge pellets (0.25 g) was extracted using the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, USA) as per the manufacturer’s instructions. The concentration of extracted community DNA was measured at 260/280 nm and 260/230 nm absorption ratios using NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, USA) as described by Kumbhare et al. (2015). Quality of the extracted genomic DNA was also checked by 1% agarose gel electrophoresis and further purified by gel extraction kit (Qiagen, USA). Isolated genomic DNA was then stored at − 20 °C prior to analysis. For PCR amplification, genomic DNA was diluted up to 1 ng·µL−1 using double distilled water. The universal primer sets 785R (5′-GACTACHVGGGTATCTAATCC-3′) and 341F (5′-CCTAYGGGRBGCASCAG-3′) was used for amplification of hypervariable regions (V3–V4) of 16S rRNA (Wang et al. 2016). PCR reaction mixture was prepared with 3 µL of 10-ng template DNA, 0.5 µL of 785R primer, 0.5 µL of 341F primers and volume was made up to 25 µL with nuclease free water. Amplification of V3–V4 region of 16S rRNA gene was done using following PCR reaction cycle: denaturation at 94 °C for 5 min followed by 35 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min, and a final extension at 72 °C for 7 min. The PCR amplicon was purified using AMPure XP beads (Agencourt Bioscience, USA).
16S rRNA Illumina Miseq sequencing
The MiSeq sequencing procedure was conducted on an Illumina MiSeq platform at the M/s AgriGenome Labs Pvt. Ltd., Kerala, India. The 16S rRNA gene library was prepared in accordance with the Illumina MiSeq platform protocols (https://sapac.illumina.com/systems/sequencing-platforms.html). The sequencing libraries were constructed with NEBNext® Ultra™ DNA II Library Prep Kit for Illumina® (New England Biolabs Inc., Boston, MA, USA), and a Qubit™ 2.0 Fluorometer (Thermo Fisher Scientific, USA) was used to assess the quality of prepared libraries. The constructed libraries were sequenced on an Illumina MiSeq platform using paired-end sequencing method of 2 × 250 bp reads.
Bioinformatics analysis
Rhizospheric bacterial community composition of S. arundinaceum was analyzed based on the raw V3–V4 16S rRNA sequencing data as prepared by Illumina MiSeq platform. Raw sequences obtained through paired-end sequencing were merged using standard procedure described by Magoc and Salzberg (2011). Furthermore, raw sequences were processed using the QIIME (Quantitative Insights Into Microbial Ecology) software package (version 1.7.0) (Caporaso et al. 2010a). Raw sequences were denoised and trimmed to remove barcodes, and primers and then clustered into operational-taxonomic units (OTUs) based on their sequence similarity at 97% using UCLUST program (version 1.2.22q). Phylogenetic relationship with different OTUs was analyzed using PyNAST (Python Nearest Alignment Space Termination) program (version 1.2) against “Core Set” dataset of Greengenes as referencing (Caporaso et al. 2010b). The taxonomic assignment was performed with a ribosomal database project (RDP) classifier (version 2.2) (Cole et al. 2009) against the SILVA OTUs database (version 123) using a confidence threshold of 70% (Quast et al. 2013). Difference in diversity was calculated using Chao1, Shannon-D, and Observed-species indices estimation. The chao1 metric estimates the species richness in environmental sample, while Shannon metric was evaluated to estimate observed OTU abundances, and accounts for both evenness and richness of species. Rarefaction curves were generated based on Shannon, Chao1, and Observed-species indices by QIIME (version 1.7.0) and displayed using R software (version 2.15.3).
Calculation and statistical data analysis
Each experiment was performed in triplicate to avoid any experimental errors. The standard deviation for various physico-chemical parameters of distillery sludge was calculated using IBM SPSS software package (version 22.0; SPSS Inc., USA). Results were represented as mean ± SD value. Statistically significance differences at p < 0.05 among the physico-chemical parameters was assessed using Student’s ‘t’ test.
Results and analysis
Physico-chemical properties of distillery sludge
Physico-chemical characteristics of non-rhizospheric and rhizospheric distillery sludge collected from two different dumping sites are summarized in Table 1. The physicochemical analysis of non-rhizospheric sludge has revealed that it has high EC (4.5 ± 0.00 µS cm−1), NH4+-N (15.70 ± 0.78 mg kg−1), Na+ (39.11 ± 3.21 mg kg−1), Cl− (891.14 ± 15.12 mg kg−1), SO42− (135.07 ± 9.31 mg kg−1), phenol (415.14 ± 1.22 mg kg−1), PO43− (1827.23 ± 7.70 mg kg−1), CEC (63.12 ± 0.47%), TOC (12.21 ± 0.11%), TKN (3.56 ± 0.01%), total hydrogen (3.12 ± 0.01%), and total oxygen (32.16 ± 0.10%) content with pH 8.1. In addition, the non-rhizospheric distillery sludge showed a high concentration of different heavy metals such as Fe (1512 ± 25.28 mg kg−1), Zn (94.25 ± 1.89 mg kg−1), Cu (61.55 ± 0.98 mg kg−1), Cr (17.524 ± 0.00 mg kg−1), Cd (1.012 ± 0.00 mg kg−1), Ni (10.115 ± 0.07 mg kg−1), and Mn (95.15 ± 0.12 mg kg−1). All the values of various physico-chemical parameters were found higher than the permissible limits of industrial discharge (Table 1). However, the sludge obtained from the rhizosphere of S. arundinaceum had lesser concentration of various physico-chemical parameters except for NH4+-N as shown in Table 1.
Table 1.
The physico-chemical characteristics of non-rhizospheric and rhizospheric distillery sludge collected from dumping site located in premises of Unnao Distilleries & Breweries Limited
| S. no. | Parameters | Non-rhizospheric sludge | Rhizospheric sludge | Permissible discharge limit, USEPA (2002) |
|---|---|---|---|---|
| 1. | pH | 8.12 ± 0.00 | 6.40 ± 0.0*** | – |
| 2. | EC | 4.5 ± 0.00 | 3.99 ± 0.01** | – |
| 3. | CEC | 63.12 ± 0.47 | 24.11 ± 1.00*** | – |
| 4. | TOC | 12.21 ± 0.11 | 28.40 ± 1.00*** | – |
| 5. | TKN | 3.56 ± 0.01 | 2.14 ± 0.05*** | – |
| 6. | Ammoniacal nitrogen (NH4+-N) | 15.70 ± 0.78 | 25.19 ± 1.05* | 1 |
| 7. | Total hydrogen | 3.12 ± 0.01 | 1.82 ± 0.05 ± 0.05*** | – |
| 8. | Total oxygen | 32.16 ± 0.10 | 21.23 ± 0.05*** | – |
| 9. | Sodium (Na+) | 39.11 ± 3.21 | 27.27 ± 0.53*** | 200 |
| 10. | Chloride (Cl−) | 891.14 ± 15.12 | 587.14 ± 4.56*** | 1500 |
| 11. | Sulfate (SO42−) | 135.07 ± 9.31 | 119.48 ± 0.92*** | – |
| 12. | Phenol | 415.14 ± 1.22 | 398.55 ± 0.18** | – |
| 13. | Phosphate (PO43−) | 1827.23 ± 7.70 | 1453.24 ± 7.55*** | – |
| 14. | Heavy metals | |||
| A | Iron (Fe) | 1512 ± 25.28 | 952.41 ± 5.08*** | 2.0 |
| B | Zinc (Zn) | 94.25 ± 1.89 | 55.25 ± 1.67*** | 2.0 |
| C | Copper (Cu) | 61.55 ± 0.98 | 35.41 ± 1.02*** | 0.5 |
| D | Chromium (Cr) | 17.524 ± 0.00 | 11.22 ± 0.49*** | 0.05 |
| E | Cadmium (Cd) | 1.012 ± 0.00 | 0.98 ± 0.00*** | 0.01 |
| F | Manganese (Mn) | 95.15 ± 0.12 | 55.12 ± 0.01*** | 0.20 |
| G | Nickel (Ni) | 10.115 ± 0.07 | 6.325 ± 0.10*** | 0.1 |
All values are mean of three replicate ± SD and presented in mg kg−1 except electrical conductivity (µS cm−1), TOC (%), TKN (%), total hydrogen (%), and total oxygen (%); CEC (Cmol), EC electric conductivity, CEC cation exchange capacity, TOC total organic carbon, TKN total Kjeldahl nitrogen. Asterisks indicate significant differences between the non-rhizospheric and rhizospheric distillery sludge; Student’s t test: *non-significant (p < 0.05), **less significant p < 0.01, ***highly significant (p < 0.001)
Characterization of organic pollutants
The comparative GC–MS analysis of ethyl acetate extracted samples obtained from non-rhizospheric and rhizospheric distillery sludge showed the presence of diverse group of organic pollutants. Majority of the identified organic pollutants present in rhizospheric distillery sludge were biodegraded and/or biotransformed through the combined action of plant and their rhizospheric bacterial communities (Fig. 2 and Table 2). The total ion chromatogram (TIC) of non-rhizospheric distillery sludge showed several major peaks at RT 9.07, 10.88, 12.85, 13.26, 13.71, 34.26, 35.45, 36.48, and 36.66 (Supplementary Fig. S2a). These compounds were characterized as acetamide, 2,2,2-trifluoro-n-methyl-(TMS); 2-butanol, tert-butyldimethylsilyl ether; pyridine, 3-trimethylsiloxy; 3-hydroxy-6-methypyridine 1tms; ad 1,4-dimethylpyrrolo(1,2-a) pyrazine; 2-butanone, 4-[2-isopropyl-5-methyl]; 2,5-cyclohexadiene-1,4-dione, 2,6-bis(1,1-dimethylethyl); octadecanoic acid, methyl ester; 17-pentatriacontene. In addition, several minor peaks were also observed at various RT values and their corresponding compounds are listed in Table 2. Moreover, several minor peaks at different RT were also observed but these peaks could not be identified because their respective compounds did not exist in the NIST library. However, in the GC–MS chromatogram of the ethyl acetate extract obtained from the rhizospheric distillery sludge showed disappearance of peaks which are initially present in non-rhizospheric samples along with generation of some new peaks (Supplementary Fig. S2b and Table 3). Presence of new peaks in rhizospheric sample indicates the possible biotransformation and/or biodegradation of toxic organic pollutants that had occurred (carcinogens, mutagens, and EDCs) along with the simultaneous generation of several new compounds as metabolites (Table 3 and Supplementary Fig. S2).
Fig. 2.
Comparison of bacterial communities composition and relative abundance distribution between non-rhizospheric (DS1) and rhizospheric (DS2) distillery sludge at the different taxa a phylum, b class and c genus level which were identified using the RDP classifier and sequences that could not be classified into any known group were designated as unknown. Percent relative abundance refers to cumulative abundance
Table 2.
List of organic compounds detected by GC–MS after derivatization of the ethyl acetate extract derived from non-rhizospheric distillery sludge and their impact in environment
| S. no. | Compounds | RT | Environmental impact | References |
|---|---|---|---|---|
| 1. | Acetamide, 2,2,2-trifluoro-N-methyl-TMS | 9.07 | – | – |
| 2. | 2-Butanol, tert-butyldimethylsilyl ether | 10.88 | – | – |
| 3. | Ethanedioic acid, bis(TMS)ester | 11.11 | – | – |
| 4. | Pyridine, 3-trimethylsiloxy | 12.85 | Neurological effects, renal effects, and irritation of the skin and eye, liver damage | ATSDR (1992) |
| 5. | 3-Hydroxy-6-methypyridine 1TMS | 13.26 | Mutagens | https://pubchem.ncbi.nlm.nih.gov |
| 6. | 1,4-Dimethylpyrrolo(1,2-A)pyrazine | 13.71 | – | |
| 7. | Benzene, 1,3-bis(1,1-dimethylethyl) | 13.98 | Gastrointestinal irritant, reproduction inhibition | OECD (2010) |
| 8. | Docosane | 14.65 | ||
| 9. | 2-Isoropyl-5-methyl-1-heptanol | 15.70 | ||
| 10. | Benzenepropanoic acid, TMS ester | 16.55 | Carcinogenicity and genotoxicity | CEPA (1999) |
| 11. | Decanoic acid, TMS ester | 18.15 | – | |
| 12. | Silane, trimethyl(undecycloxy) | 20.01 | – | – |
| 13. | Benzeneacetic acid, α,4-bis[(TMS)oxy], TMS ester | 20.25 | – | |
| 14. | Emetan, 1′,2-didehydro-6′,7′,10,11-tetramethoxy | 21.88 | – | – |
| 15. | 2,4-Imidazolidinedione, 1-[[(5-nitro-2-firnayl)methylene]amino | 23.00 | – | – |
| 16. | 1-Hexadecanol, 2-methyl | 24.10 | – | – |
| 17. | Hexadecane | 25.61 | Skin problems such as dryness, irritation, and cracking of the skin | Shiri et al. (2015) |
| 18. | 5-Hydroxy-2,2-dimethyl-5,6-bis(2-oxo-1-propyl)-1-cyclohexanone | 26.97 | – | – |
| 19. | Dodecenoic acid, TMS ester | 27.96 | – | – |
| 20. | 2-Propenoic acid, oxybis(methyl-2,1-ethanediyl)ester | 30.37 | – | – |
| 21. | Benzoic acid, 2,5-bis(TMSoxy)-TMS ester | 30.83 | – | – |
| 22. | Dodecanoic acid | 31.16 | – | – |
| 23. | Heptacosane | 32.22 | – | – |
| 24. | 1-Tetradecene, 2-decyl | 32.99 | – | – |
| 25. | 1,2-Propanediol, 3-(octadecycloxy)-diacetate | 33.05 | – | – |
| 26. | β-Sitosterol trimethyl ether | 33.62 | EDC, genotoxic and cytotoxic | USEPA (2012) |
| 27. | 2-Butanone, 4-[2-isopropyl-5-methyl | 34.26 | – | – |
| 28. | Hexadecanoic acid, methyl ester | 34.52 | – | – |
| 29. | 2,5-Cyclohexadiene-1,4-dione, 2,6-bis(1,1-dimethylethyl) | 35.45 | – | – |
| 30. | Octadecanoic acid, methyl ester | 36.48 | EDC, Cytotoxic and genotoxic | USEPA (2012) |
| 31. | 17-Pentatriacontene | 36.66 | – | – |
| 32. | 1,4-Ethano-1,2,3,4-tetrahydroanthracen-3-ol-benzylidene | 38.11 | – | – |
| 33. | Pyrrolo [1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl) | 40.90 | Hemolytic activity | Sanjenbam and Kannabira (2016) |
| 34. | Butyl 11-eicosenoate | 41.47 | – | – |
| 35. | Cholest-8(14)-en-3-one, (5α) | 42.48 | – | – |
| 36. | α-Homocholest-4a-en-3-one | 43.19 | – | – |
| 37. | Stigmasta-5,22-dien-3-ol(3β,22E) | 44.25 | Vasodepressor effect | Barla et al. (2006) |
| 38. | Stigmasterol | 45.20 | EDC | USEPA (2012) |
RT retention time (in min), EDC endocrine-disrupting chemical, TMS trimethylsilyl
Table 3.
List of organic compounds detected by GC–MS after derivatization of the ethyl acetate extract derived from rhizospheric distillery sludge and their impact in environment
| S. no. | Compounds | RT | Environmental impact | References |
|---|---|---|---|---|
| 1. | Benzene, 1-ethyl-3,5-disopropyl | 8.19 | – | – |
| 2. | Butanedioic acid, bis(TMS)ester | 9.01 | – | – |
| 3. | Propanoic acid, 3-[(TMS)oxy], TMS ester | 9.08 | – | – |
| 4. | Heptacosane | 10.25 | – | – |
| 5. | 17-Pentatriacontene | 10.88 | – | – |
| 6. | Ethyl-TMS dipropylmalonate | 11.17 | – | – |
| 7. | Tert-Hexadecanethiol | 12.85 | – | – |
| 8. | 1-Hexadecanol, 2 methyl | 13.72 | – | – |
| 9. | Tricarballylic Acid TMS | 14.65 | Inhibit citric acid cycle in mammals | Russell and Forsberg (1986) |
| 10. | Ethanedioic acid, bis(TMS)ester; | 15.45 | – | – |
| 11. | 2-Monostearin TMS ether | 16.55 | – | – |
| 12. | Octadecanoic acid, 2,3-bis[(TMS)oxy]propyl ester | 17.50 | – | – |
| 13. | Butanedioic acid, bis(TMS) ester | 17.65 | – | – |
| 14. | 2-Monostearin TMS ether | 20.01 | – | – |
| 15. | 2-Methyl-1,3-Propanediol 2TMS | 21.23 | – | – |
| 16. | Pyrrolozine1-one,7-propyl | 21.88 | – | – |
| 17. | Dotriacontane | 22.98 | – | – |
| 18. | Hexacosanoic acid | 24.78 | Mitochondrial dysfunction and Ca2+ deregulation | Hein et al. (2008) |
| 19. | Benzenepropanoic acid, | 27.99 | – | – |
| 20. | Ergosten-3β-ol | 28.10 | – | – |
| 21. | Butane, 2,3-bis(Trimethylsiloxy) | 30.25 | – | – |
| 22. | Undecenoic acid | 30.83 | – | – |
| 23. | 1-Monolinoleoylglycerol TMS ether | 32.25 | – | – |
| 24. | Spirostan-3-one (5α, 20β, 25R) | 33.61 | – | – |
| 25. | Cis-13-docosenoic acid | 33.84 | – | |
| 27. | Hexacosanoic acid | 34.10 | Mitochondrial dysfunction and Ca2+ deregulation | Hein et al. (2008) |
| 28. | Heptadecane, 2,6,10,15-tetramethyl | 34.89 | – | |
| 29. | Benzeneacetic acid, TMS ester | 35.45 | – | |
| 30. | Tetradecanoic acid, TMS ester | 36.06 | Skin and eyes | Burdock and Carabin (2007) |
| 31. | Tetradecanoic acid, ethyl ester | 36.45 | – | |
| 32. | Docosanoic acid, methyl ester | 37.44 | Mitochondrial dysfunction and Ca2+ deregulation | Hein et al. (2008) |
| 33. | Ethyl docosanoate | 38.10 | – | |
| 34. | Octadecanoic acid, 2,3-bis[(TMS)oxy]propyl ester | 38.12 | Acute lung injury | USEPA (2012) |
| 35. | Hexadecane | 39.17 | – | |
| 36. | Stigmasta-5,22-dien-3-ol(3β,22E) | 41.45 | EDC | USEPA (2012) |
| 37. | Stigmasterol | 42.19 | EDC | USEPA (2012) |
RT retention time (in min), EDC endocrine-disrupting chemical, TMS trimethylsilyl
Bacterial community analysis
Diversity analysis and richness of OTUs
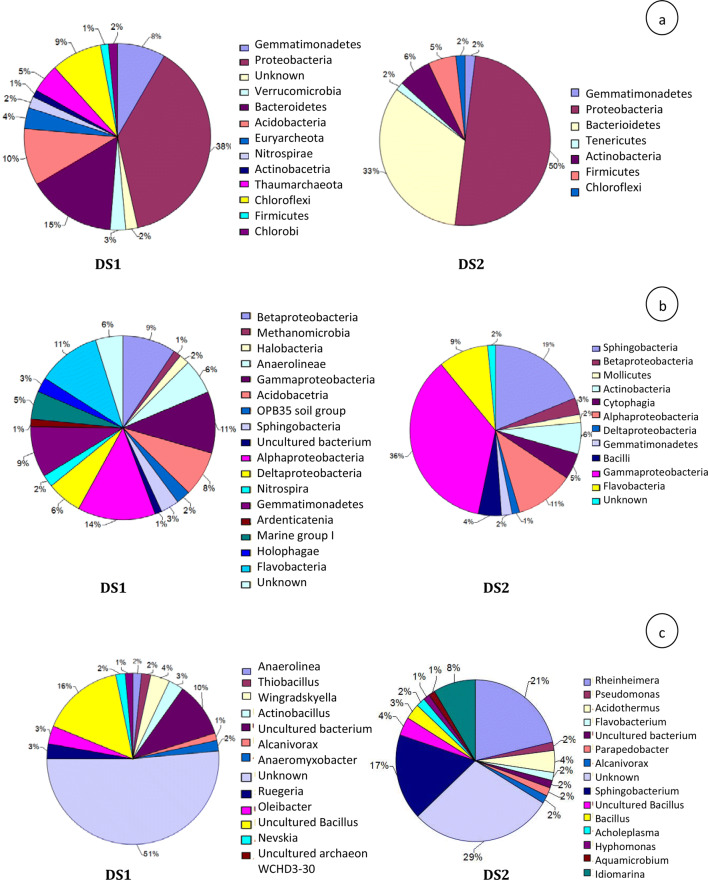
In this study, a total of 1,191,014 and 901,757 high-quality paired reads with an average length of 250 bases were derived from non-rhizospheric and rhizospheric distillery sludge, respectively, using Illumina MiSeq sequencing. The Phred score distribution (≥ Q30) of paired-end reads obtained from non-rhizospheric and rhizospheric distillery was 93.07% and 93.18%, respectively. The average GC content distribution of sequenced reads derived from rhizospheric sludge as well as non-rhizospheric sludge is shown in Supplementary Fig. S3. Sequence read of the non-rhizospheric and rhizospheric distillery sludge had 30–60% GC content (Supplementary Fig. S3a,b). After quality filtering (i.e., conserved region filter, mismatch filter, and chimeras filter), the MiSeq sequencing yielded 1,075,931 and 828,560 pre-processed reads from non-rhizospheric and rhizospheric sludge, respectively. A summary of sequence reads that passed through each filter is provided in Table 4. A total of 621,897 OTUs were identified from 1,904,491 reads. Out of 621,897 OTUs, 539,635 singletons were separated and 82,262 OTUs were selected for further analysis. A total of 82,262 OTUs derived from were generated after clustering at 97% similarity level from 1,904,491 reads (Supplementary Fig. S3c). Additionally, rarefaction curves were produced to determine if the depth of sequencing was indeed suitable to reflect a complete bacterial diversity from the environmental samples. The rarefaction curves demonstrated that OTU abundance was diverse between non-rhizospheric and rhizospheric distillery sludge as shown in Supplementary Fig. S4. Shannon index and Chao1 richness estimator indicated that microbial communities grown in rhizospheric regions are highly rich and diverse compared to non-rhizosphere. All the resulting fragments were then classified into phyla, classes, orders, families, genera, and species (Supplementary Fig. S4). In non-rhizospheric and rhizospheric distillery sludge, the top 10 represented phyla were Proteobacteria (47% and 63%), Acidobacteria (9% and 2%), Bacteroidetes (6% and 10%), Gematimonadetes (7% and 2%), Verrucomicrobia (5% and 2%), unknown (5% and 4%), Firmicutes (3% and 8%), Chloroflexi (3% and 2%), Actinobacteria (2% and 3%), and Nitrospirae (2% and 0.26%) (Fig. 1a). The sequences which did not alignment against the taxonomic database were categorized as unknown. Moreover, the classes other than the top ten results are categorized as other. In addition, an unidentified bacterial phylum (other; 11 and 4%) was also found among top 10 bacterial phyla.
Table 4.
The raw, chimera detection, and quality filter reads derived from distillery sludge samples (a) illumina MiSeq sequencing raw reads, (b) pre-processing of sequence read data and (c) summary of singleton OTUs
| Sample | Total reads | Passed conserved region filter | Passed mismatch filter |
|---|---|---|---|
| (a) | |||
| DS1 | 1,191,014 | 1,191,014 | 1,189,451 |
| DS2 | 901,757 | 901,757 | 900,797 |
| Sample | Consensus read | Chimeric sequences | Pre-processed reads |
|---|---|---|---|
| (b) | |||
| DS1 | 1,189,451 | 113,453 | 1,075,931 |
| DS2 | 900,797 | 72,237 | 828,560 |
| (c) | |||
| Total reads | 1,904,491 | ||
| Total OTUs picked | 621,897 | ||
| Total singleton OTUs | 539,635 | ||
| Total OTUs after singleton removal | 8226 | ||
DS1 non-rhizospheric distillery sludge, DS2 rhizospheric distillery sludge
Fig. 1.
Pie chart showing the taxonomic distribution of OTU at different phylogenetic level a phylum, b class, c order, d family, e genus and f species. Analyses were performed separately for rhizospheric and non-rhizospheric distillery sludge. Only top 10 enriched class categories are shown in the figure. The taxonomic classes other than top 10 results are categorized as others. The sequences with very lesser similarity or whose V3–V4 regions do not have any alignment hits against the taxonomic database are categorized as unknown. DS1 non-rhizospheric distillery sludge, DS2 rhizospheric distillery sludge
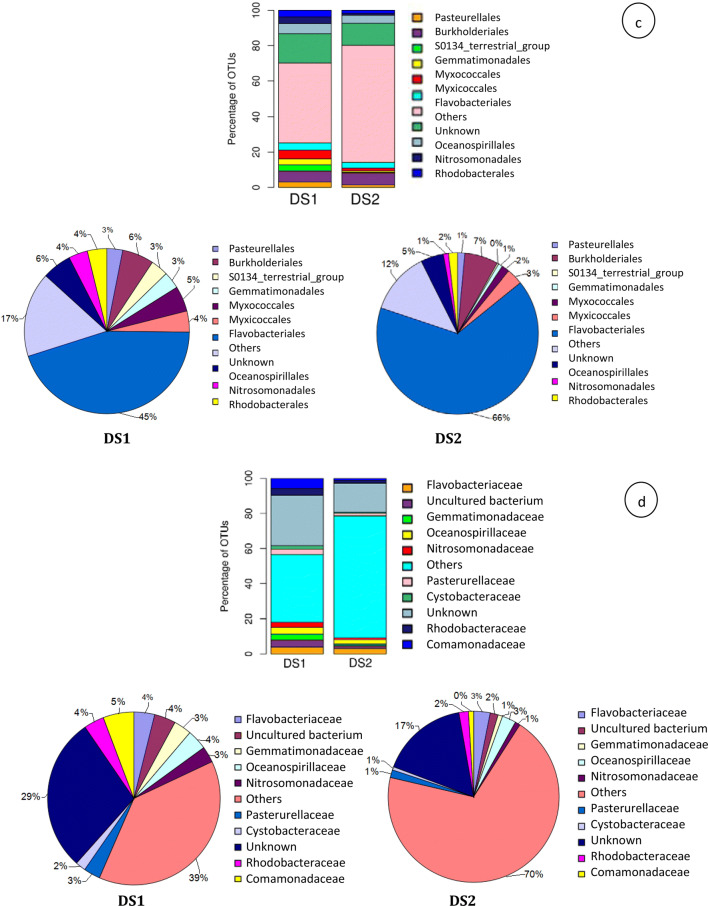
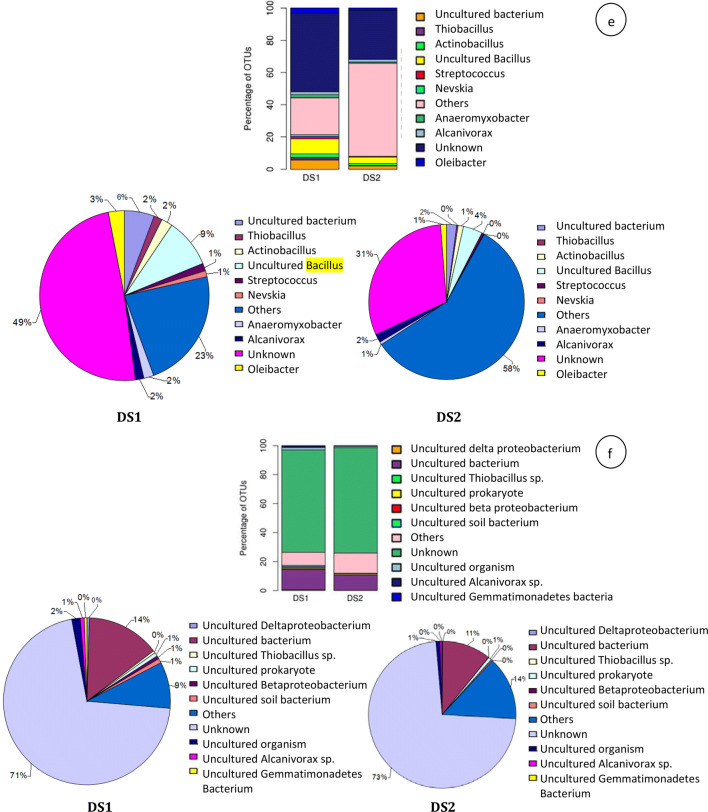
At class level in non-rhizospheric and rhizospheric distillery sludge, top 10 represented classes were Betaproteobacteria (16% and 9%), Gammaproteobacteria (13% and 44%), Deltaproteobacteria (10% and 3%), unknown (9% and 6%), Alphaproteobacteria (7% and 6%), Gemmatimonadetes (7% and 1%) Acidobacteria (6% and 2%), Flavobacteria (4% and 3%), OPB35 soil group (4% and 1%), and Holophage (3% and 1%) (Fig. 1b). Overall, 21% (non-rhizospheric sludge) and 24% (rhizospheric sludge) sequences of the total read were not classified at the class level, suggesting that the identities of these bacteria are unknown. At the order level in non-rhizospheric and rhizospheric distillery sludge, the top ten represented orders were unknown (17% and 12%), Burkholderiales (6% and 7%), Oceanospirillales (6% and 5%), Myxococcales (5% and 2%), Flavobacteriales (4% and 3%), Nitrosomonadales (4% and 1%), Rhodobacterales (4% and 2%), Pasteurellales (3% and 1%), S0134 terrestrial group (3% and 0.31%), and Gemmatimonadales (3% and 1%). Moreover, an unidentified bacterial order (other; 45-66%) was also found among top 10 bacterial orders as shown in Fig. 1c. At the family level in non-rhizospheric and rhizospheric sludge, the top ten represented families were unknown (29% and 16%), Commanadaceae (6% and 1%), Flavobacteraceae (4% and 3%), uncultured bacterium (4% and 1%), Oceanospirillaceae (4% and 3%), Rhodobacteraceae (4% and 2%), Gemmatimonadaceae (3% and 1%), Nitrosomonadaceae (3% and 1%), Pasterurellaceae (3% and 1%), and Cystobacteraceae (2% and 1%) (Fig. 1d). Moreover, an unidentified bacteria family (Others; 38%–70%) was also found among top 10 bacterial families. Further, at the genus level in non-rhizospheric and rhizospheric sludge, the top ten most abundant genera were unknown (49% and 31%), uncultured Bacillus (9% and 4%), uncultured bacterium (6% and 2%), Oleibacter (3% and 1%), Alcanivorax (3% and 2%), Thiobacillus (2% and 0%), Actinobacillus (2% and 1%), Anaeromyxobacter (2% and 1%), Streptococcus (1% and 0%), and Nevskia (1% and 0%). In addition, an unidentified bacterial genus (Others; 23% and 58%) was also found among top 10 bacterial genera (Fig. 1e). Besides, at the species level, the top 10 most abundant bacterial species in non-rhizospheric and rhizospheric sludge were uncultured Deltaproteobacterium (0.49% and 0.09%), uncultured bacterium (14% and 11%), uncultured Thiobacillus sp. (0.45% and 0.06%), uncultured prokaryote (1% and 1%), uncultured Betaproteobacterium (1% and 0.05%), uncultured soil bacterium (1% and 0.28%), unknown (71% and 72%), uncultured organism (2% and 1%), uncultured Alcanivorax sp. (1% and 0.37%) and uncultured Gemmatimonadetes bacterium (0.50% and 0.09%). Moreover, an unidentified bacterial species (Others; 9–14%) were also found among top 10 bacterial species (Fig. 1f).
Bacterial community structure at the phylum, class and genus level
The relative abundance of bacterial communities at the phylum, class and genus levels between the non-rhizospheric and rhizospheric sludge is shown in Fig. 2. The results demonstrated that the rhizobacteria of S. arundinaceum had significantly different community structures and species abundance. In non-rhizospheric sludge, Proteobacteria (38%) represented the most abundant phylum, followed by Chlorobi (15.0%) and Acidobacteria (10%). The other predominant phyla in the non-rhizospheric sludge were Chloroflexi (9%), Gemmatimonadetes (8%), Thaumarchaeota (5%), Euryarchaeota (4%), Verrucomicrobia (3%), and Nitrospirae (2%) (Fig. 2a). Besides the dominant microbes, 1.96% of total sequences could not be assigned to any phylum. In rhizospheric sludge, the bacterial phyla distributions were Proteobacteria (50%), Bacteriodetes (33%), Firmicutes (5%). The other predominant phyla in the rhizospheric sludge were Gemmatimonadetes (2%), Chloroflexi (2%), and Tenericutes (2%). At the class level, the most predominant bacteria in non-rhizospheric sludge were affiliated with Alphaproteobacteria (14%), Flavobaceteria (11%), Gammaproteobacteria (11%), Betaproteobacteria (9%), Gemmatimonadetes (9%), Acidobacteria (8%), Anaerolineae (6%), and Deltaproteobacteria (6%). The other predominant phyla in the non-rhizospheric sludge were Holophage (3%), OPB35 soil group (2%), Halobacteria (2%), Nitrospira (2%), uncultured Bacterium (1%), and Aradenticatenia (1%). On the other hand, rhizospheric sludge represented the most abundant bacteria affiliated with Gammaproteobacteria (36%) followed by Sphingobacteria (19%), Alphaproteobacteria (11%), Flavobacteria (9%), Actinobacteria (6%), Bacilli (5%), Cytophagia (5%), Betaproteobacteria (3%), Mollicutes (2%), and Deltaproteobacteria (1%) (Fig. 2b).
Among the assigned taxa at genus level in non-rhizospheric sludge, the most abundant genera were affiliated with unknown (51%), followed by uncultured Bacillus (16%), uncultured bacterium (10%), Winogradskyella (4%), Actinobacillus (3%), Oleibacter (3%), Ruegeria (3%), Anaerolinea (2%), Thiobacillus (2%), Anaeromyxobacter (2%), Nevskia (2%), and Alcanivorax (1%). In rhizospheric sludge, the most predominant bacteria were affiliated with Rheinheimera (21%) followed by Sphingobacterium (17%), Idiomarina (8%). Acidothermus (4%), Pseudomonas (2%), Flavobacterium (2%), uncultured bacterium (2%), Parapedobacter (2%), Alcanivorax (2%), uncultured Bacillus (4%), Bacillus (3%), Acholeplasma (2%), Hyphomonas (1%), and Aquamicrobium (1%) as shown in Fig. 2c. The presence of uncultured Bacillus sp. in spent wash and post methanated distillery sludge has been reported earlier using restriction fragment length polymorphism approach to explore their role during in situ remediation of such waste (Chandra and Kumar (2017a, b). In this study, Alcanivorax was the secondary predominant genera found in non-rhizospheric and rhizospheric distillery sludge samples. Alcanivorax accounts for 1.07% sequence obtained from DS1, while it occupied 1.25% in DS2. Hence, our result indicated that these genera could be well adapted to grow in highly stressed environmental conditions. It was observed that 29.75 and 22.90% sequences of the rhizospheric and non-rhizospheric distillery sludge were not assigned to any genera. These unassigned sequences might be novel microbes were existing in sludge that needed to be further investigation. Principal component analysis (PCA) based on the relative abundance of all detected bacterial genera explained 76.50% and 23.50% of the total variance by two components, respectively (Fig. 3). In non-rhizospheric and rhizospheric sludge, it was shown that 12.03 and 2.92 of the total sequence belonged to uncultured Bacillus (Fig. 4). The findings of this study suggest a significant difference in relative abundance of bacterial phyla between non-rhizospheric and rhizospheric distillery sludge samples. The metagenomics analysis also suggested that the identified bacterial phyla that were enriched at plant growing site play a major role in in situ phytoremediation of organometallic rich distillery sludge.
Fig. 3.

Principal components analyses of relative abundance of all detected bacterial genera of the two studied distillery sludge samples
Fig. 4.
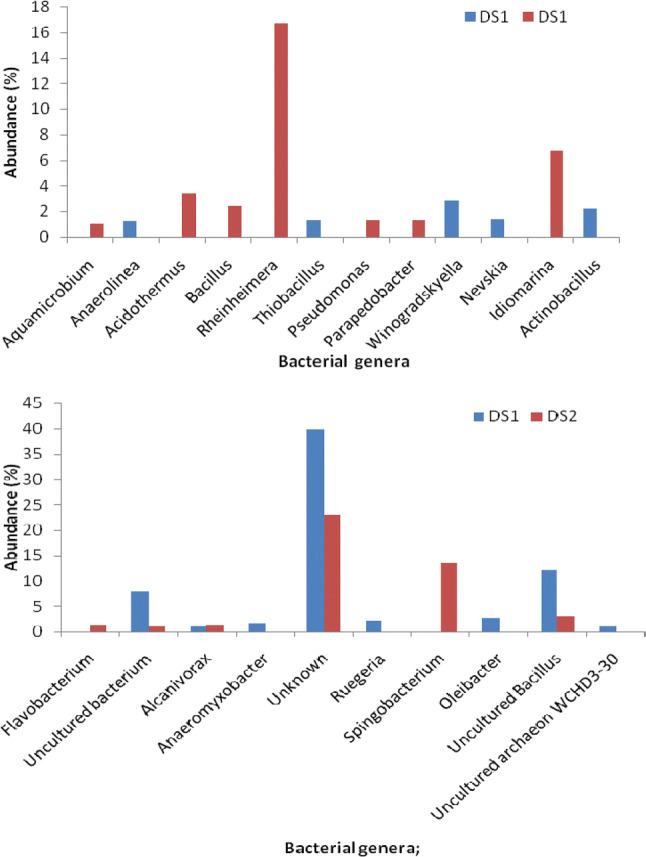
Major identified bacterial genera (sequence percentage above 1%) grown in the non-rhizospheric (DS1) and rhizospheric (DS2) distillery sludge
Discussion
In this study, distillery sludge was found to be alkaline (pH 8.1) in nature. The alkalinity of distillery sludge arises due to the presence of bicarbonates, carbonates, hydroxides, calcium and magnesium ions, which are used in distilleries for adjustment of pH of diluted sugarcane molasses during fermentation process. In addition, the high level of EC in distillery sludge indicated the presence of diverse anions and cations such as Cl−, Na+, PO43−, SO42−, NH4+. Moreover, the higher concentration of heavy metal in distillery sludge could be due to the corrosive effect of fermented molasses and distillation process of metallic ions in sugarcane molasses and further addition in the raw effluent. The presence of a high level of inorganic and organic parameters in non-rhizospheric distillery sludge is also concordant with the earlier reports (Bharagava and Chandra 2010; Chandra and Kumar 2017b). On the other hand, the sludge obtained from the rhizosphere of S. arundinaceum, an important perennial grass of Poaceae family, native to India and distributed to tropical countries of South Asia and Southeast Asia, had lower values of various physico-chemical parameters as shown in Table 1. The result of physico-chemical analyses of rhizospheric sludge shows that pH of distillery sludge was found alkaline in nature (pH 8.12) which slowly changed into acidic (pH 6.4); this might be due to the interaction of microbial communities with organometallic waste in root zone, where root exudates are released from plant in the form of amino acids, and organic acids as metal chelators. These metal chelators decrease the pH of distillery sludge and facilitate the growth of rhizospheric microbial communities. Overall, the reduction of all physico-chemical parameters in rhizospheric distillery sludge is a combined phenomenon of S. arundinaceum and rhizosphere bacterial communities grow in vicinity of root with the support of plant. Bacterial communities degrade and detoxify organic and inorganic pollutants in rhizosphere and facilitate the plant growth in highly polluted environment. Consequently, all the values of various physico-chemical parameters were reduced in rhizospheric distillery sludge (Table 1).
GC–MS is a powerful sophisticated technique for characterization of organic pollutants in the environmental sample. In this study, the majority of detected organic pollutants in distillery sludge a threat to human health and environment due to their toxic nature (USEPA 2012). These toxic compounds are derived from biochemical processes at different stages of sugar manufacturing, ethanol production, and anaerobic digestion of raw effluent and constituted the major components of sludge. Similar organic compounds have also been reported by the earlier workers in case of anaerobically digested distillery sludge (Chandra et al. 2018b). However, luxuriant growth of S. arundinaceum on dumped site indicates the capabilities of their rhizospheric bacterial communities which might be degraded and detoxifying the complex organic pollutants of distillery sludge during in situ phytoremediation.
Next-generation sequencing technology provides a platform for exploring the composition and diversity of the bacterial communities in environmental samples. Using Illumina (MiSeq) sequencing of the V3–V4 hypervariable region of bacterial 16S rRNA gene analysis, we found conspicuous differences in rhizospheric and non-rhizospheric bacterial community structure derived from distillery sludge. In the present study, rarefaction curves confirmed that OTU abundance was diverse in rhizospheric sludge compared to non-rhizospheric as shown in Supplementary Fig. 4. For instance, we have calculated Chao1 species richness estimator and Shannon diversity indices using the rarefaction sampling to estimate the alpha diversity in rhizospheric and non-rhizospheric distillery sludge sample. The Chao1 and Shannon-D indices both indicated that the bacterial communities in rhizospheric and non-rhizospheric sludge samples were more diverse. The diversity and composition of rhizosphere microbial communities differ between various plant species (Marschner et al. 2004) and growth season (Marschner et al. 2002). Besides, the microbial diversity exhibited variations depends on the physico-chemical properties of soils, pollution level, plant root exudates, and environmental conditions (Erguven et al. 2016; Huang et al. 2014).
The composition and diversity of microbial communities in the substrate also play a critical role in pollutant removal. The results of this study revealed that rhizosphere sludge showed higher bacterial diversity and more unique OTUs in contrast to non-rhizospheric sludge at organometallic-rich polluted sites. As a result of OTUs, the microbial taxonomic distributions in this study displayed top 10 phyla Proteobacteria, Acidobacteria, Bacteroidetes, Gemmatimonadetes, Verrucomicrobia, Firmicutes, Actinobacteria, Clororoflexi, and Nitrospirae. The abundance of Proteobacteria can be correlated with their ability to survive in organometallic rich stressed environments. Classification at phylum level revealed that Proteobacteria was the most abundant phylum in all sludge samples with the class Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, Alphaproteobacteria, and Gemmatimonadetes. They gradually increased by 50% in rhizospheric distillery sludge. Members of Gammaproteobacteria and Betaproteobacteria are known to be highly versatile for their bioremediation and biodegradation ability (Shah et al. 2013; Kostka et al. 2011). Members of the phylum Proteobacteria are believed to be involved in the elimination and/or reduction of organic and inorganic pollutants such as phenolic compounds, phosphorus, nitrogen, and heavy metals. Similar results were reported in previous studies, which found that Proteobacteria was the dominant bacterial community grown in the rhizospheric zone of P. cummunis during treatment two-stage sequential treatment of anaerobically digested distillery effluent (Chandra et al. 2012). Phyla Acidobacteria and Bacterioidetes turned dominant in rhizospheric sludge sample. Phylum Acidobacteria is one of the five phyla, including Planctomycetes, Chloroflexi, Gemmatimonadetes, and Verrucomicrobia, which have been proven to be poorly characterized and hardly cultivated in the laboratory due to their slow-growth properties that lead towards dormancy (Janssen et al. 2002; Bergmann et al. 2011). While at the species level, the top ten most abundant species were uncultured Deltaproteobacterium, uncultured Bacterium, Uncultured Thiobacillus sp., uncultured Betaproteobacterium, uncultured soil bacterium, unknown, uncultured organism, uncultured Alcanivorax sp. and uncultured Gemmatimonadetes bacterium.
This study exposed that phylum Proteobacteria was dominant in both non-rhizospheric and rhizospheric distillery sludge sample and remained as high throughout among the samples. This indicated that Proteobacteria phylum may have a key role in degradation and detoxification of organic and inorganic pollutants, respectively, in both non-rhizospheric and rhizospheric distillery sludge. In our study, Bacteriodetes was the second-highest represented phylum found in rhizospheric distillery sludge. Members of Bacteroidetes phylum have colonized several different ecological niches where they demonstrate diverse biological functions. In particular, they are well-known degraders of various organic compounds present in distillery waste due to their hydrolytic capacities (Acharya et al. 2011). In this study, a third phylum Firmicutes was also detected from rhizospheric distillery sludge. The presence of Firmicutes is quite rare in natural samples. However, lignocellulosic materials like sugarcane bagasse and sugarcane molasses represent an abundant source of organic and inorganic materials that can be used as a sole source of carbon and nitrogen for growing bacteria (Womersley 2006). Thus, these results indicated that Firmicutes might have diverse metabolic capabilities, as well as the ability to use various sugar and organic compounds as a sole carbon and nitrogen source. Hence, this ability creates a specific niche of these grown bacterial communities that may lead to in situ phytoremediation of toxic pollutants. These findings were consistent with a lot of studies that reported Firmicutes as the dominant phylum during bioremediation of sugarcane molasses-based distillery waste (Chaturvedi et al. 2006; Sharmin et al. 2013). The class of Alphaproteobacteria appeared as the most abundant within the group Proteobacteria in non-rhizospheric sludge, but lesser numbers were found in rhizospheric distillery sludge. However, Gammaproteobacteria grows dominantly in the rhizospheric sludge. Gammaproteobacteria had been extensively present in various environmental conditions and play a major role in the degradation and denitrification of OM. This finding was similar to the results of previous studies, which revealed that the Gammaproteobacteria was the predominant one within Proteobacteria in the rhizosphere of Phragmites communis during treatment of anaerobically treated distillery effluent in two-stage sequential treatment system (Chandra et al. 2012). The genus Rheinheimera, a branch of Gammaproteobacteria, has been detected in our study. Species of Rheinheimera are capable to grow on and to degrade rapidly the more easily degradable OM. Besides, the second most common genus detected in our study was Sphingobacterium belonging to the phylum Bacteroidetes. Moreover, other genera identified from rhizospheric distillery sludge samples representing the Proteobacteria phylum were Idiomarina, Acidothermus, Pseudomonas, Flavobacterium, Parapedobacter, Alcanivorax, Bacillus, Acholeplasma, Hyphomonas, and Aquamicrobium. Pseudomonas and Bacillus are well-known genera in various ways due to their physiological and versatile metabolic as well as plant growth-promoting activities (Glick 2012, 2014). As noted by Ghosh et al. (2004) and Kaushik et al. (2010) Pseudomonas and Bacillus communities play a vital role in biodegradation of organic pollutants present in distillery effluent. Previously, Chaturvedi et al. (2006) characterized Bacillus spp., Pseudomonas spp. and Achromobacter spp. as potential rhizospheric bacterial communities of Phragmites australis L. responsible for bioremediation of distillery effluent from a contaminated site. Apart from this, genera such as Idiomarina, Acidothermus, Parapedobacter, Hyphomonas, Acholeplasma, and Aquamicrobium were found in abundance from rhizospheric sludge which depicts their significant role in the degradation of sludge pollutants. The above findings suggest significant variance for the relative abundance of bacterial phyla between non-rhizospheric and rhizospheric distillery sludge samples. The higher bacterial diversity in rhizospheric sludge could be due to the presence of a high level of organic root exudates, which provide appropriate ecological niches for survival and bacterial growth of bacterial communities and lead to an increased number of microbes (Buee et al. 2009). These bacterial communities help in the survival and growth of the plants at the polluted site.
Conclusion
This study has concluded that rhizospheric as well as non-rhizospheric distillery sludge contains several toxic organometallic complex compounds, some of them are listed among EDCs. Further, the luxuriant growth of S. arundinaceum showed the potential of plant for phytoremediation of disposed distillery sludge which gives an evidence of mineralisation of organometallic complex pollutants. In addition, metagenomic analysis of rhizospheric sludge of S. arundinaceum revealed the presence of Rheinheimera (21%), Sphingobacterium (17%), Idiomarina (8%). Acidothermus (4%), uncultured Bacillus (4%), Bacillus (3%), Pseudomonas (2%), Flavobacterium (2%), uncultured bacterium (2%), Parapedobacter (2%), Alcanivorax (2%), Acholeplasma (2%), Hyphomonas (1%), and Aquamicrobium (1%) as dominant taxa compared with non-rhizospheric distillery sludge. The taxonomic information confirmed that the diversity of rhizospheric bacterial communities of S. arundinaceum quite different from the non-rhizospheric sludge communities. These bacteria are likely to contribute to the biodegradation and/or biotransformation of organic pollutants exhibiting in distillery sludge. Furthermore, the comparative GC–MS analysis also supported that the majority of organic pollutants detected in non-rhizospheric sludge were eliminated from rhizospheric sludge samples and few compounds were generated as metabolic products by the combined action of plant and bacterial communities. Future work should concentrate to reveal the specific role of S. arundinaceum and associated rhizo and/or endophytic bacteria under polluted environment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We would like to thank M/s AgriGenome Labs Pvt Ltd., Kerala, India for the 16S rDNA Illumina Miseq analysis. The authors gratefully acknowledge the financial assistance received from University Grants Commission (UGC), New Delhi, in form of Rajiv Gandhi National Fellowship (Letter No. F1-17.1/2012-13/RGNF-2012-13-SC-UTT-30458) to Vineet Kumar and Department of Biotechnology (DBT), Govt. of India as Grant-in-Aid Project (Letter No. BT/PR13922/BCE/8/1129/2015) to Ram Chandra. The authors gratefully acknowledge the anonymous reviewers and executive editor-in-chief for their valuable comments and suggestion on the earlier version of this paper. The authors would like to acknowledge the English Language Editing help from Dr. Saurav Das, University of Nebraska at Lincoln, United States.
Author contributions
VK and RC designed the study, carried out sampling and performed the experiments and analyzed data. VK wrote the first manuscript draft, which was then revised by RC. RC contributed to the development of the idea for this manuscript and contributed text to the final version, and edited the manuscript. All authors gave approval to the final version.
Compliance with ethical standards
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical statement
We confirm that this research study does not include either vertebrates or higher invertebrates.
References
- Abed RMM, Al-Kharusi S, Gkorezis P, Prigent S, Headley T. Bacterial communities in the rhizosphere of Phragmites australis from an oil-polluted wetland. Arch Agron Soil Sci. 2018;64(3):360–370. [Google Scholar]
- Acharya BK, Pathak H, Mohana S, Shouche Y, Singh V, Madamwar D. Kinetic modelling and microbial community assessment of anaerobic biphasic fixed film bioreactor treating distillery spent wash. Water Res. 2011;45:4248–4259. doi: 10.1016/j.watres.2011.05.048. [DOI] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR) Toxicological profile for pyridine. Atlanta: U.S. Department of Health and Human Services, Public Health Service; 1992. [PubMed] [Google Scholar]
- All India Distillers’ Associations (AIDA) (2016) Annual report. New Delhi. http://www.aidaindia.org/annual-report.html
- Arimi MM, Zhang Y, Goetz G, Kiriamiti K, Geissen SU. Antimicrobial colorants in molasses distillery wastewater and their removal technologies. Int Biodeter Biodegr. 2014;87:34–43. [Google Scholar]
- Bai Y, Liang J, Liu R, Hu C, Qu J. Metagenomic analysis reveals microbial diversity and function in the rhizosphere soil of a constructed wetland. Environ Technol. 2014;35(20):2521–2527. doi: 10.1080/09593330.2014.911361. [DOI] [PubMed] [Google Scholar]
- Barla A, Birman H, Oksuz K. Secondary metabolites from € Euphor- bia helioscopia and their vasodepressor activity. Turk J Chem. 2006;30:325–332. [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharagava R, Chandra R. Effect of bacteria treated and untreated post-methanated distillery effluent (PMDE) on seed germination, seedling growth and amylase activity in Phaseolus mungo L. J Hazard Mater. 2010;180:730–734. doi: 10.1016/j.jhazmat.2010.04.100. [DOI] [PubMed] [Google Scholar]
- Buee M, De Boer W, Martin F, Van Overbeek L, Jurkevitch E. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil. 2009;321:189–212. [Google Scholar]
- Burdock GA, Carabin IG. Safety assessment of myristic acid as a food ingredient. Food Chem Toxicol. 2007;45(4):517–529. doi: 10.1016/j.fct.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, De-Santis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R, Kumar V. Detection of androgenic-mutagenic compounds and potential autochthonous bacterial communities during in situ bioremediation of post methanated distillery sludge. Front Microbiol. 2017;8:88. doi: 10.3389/fmicb.2017.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R, Kumar V. Phytoextraction of heavy metals by potential native plants and their microscopic observation of root growing on stabilised distillery sludge as a prospective tool for in situ phytoremediation of industrial waste. Environ Sci Pollut Res. 2017;24:2605–2619. doi: 10.1007/s11356-016-8022-1. [DOI] [PubMed] [Google Scholar]
- Chandra R, Kumar V. Detection of Bacillus and Stenotrophomonas species growing in an organic acid and endocrine-disrupting chemicals rich environment of distillery spent wash and its phytotoxicity. Environ Monit Assess. 2017;189:26. doi: 10.1007/s10661-016-5746-9. [DOI] [PubMed] [Google Scholar]
- Chandra R, Bharagava RN, Kapley A, Purohit HJ. Characterization of Phragmites cummunis rhizosphere bacterial communities and metabolic products during the two stage sequential treatment of post methanated distillery effluent by bacteria and wetland plants. Bioresour Technol. 2012;103:78–86. doi: 10.1016/j.biortech.2011.09.132. [DOI] [PubMed] [Google Scholar]
- Chandra R, Kumar V, Tripathi S. Evaluation of molasses-melanoidin decolourisation by potential bacterial consortium discharged in distillery effluent. 3 Biotech. 2018;8:187. doi: 10.1007/s13205-018-1205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R, Kumar V, Tripathi S, Sharma P. Heavy metal phytoextraction potential of native weeds and grasses from endocrine-disrupting chemicals rich complex distillery sludge and their histological observations during in situ phytoremediation. Ecol Eng. 2018;111:143–156. [Google Scholar]
- Chaturvedi S, Chandra R, Rai V. Isolation and characterization of Phragmites australis (L.) rhizosphere bacteria from contaminated site for bioremediation of colored distillery effluent. Ecol Eng. 2006;27:202–207. [Google Scholar]
- Chen X, Sun X, Bi S, Lu G. Fungi diversity of ginseng rhizospheric soil in northeastern China. Agric Sci Technol. 2010;11:132–136. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Bora SS, Yadav RNS, Barooah M. A metagenomic approach to decipher the indigenous microbial communities of arsenic contaminated groundwater of Assam. Genom data. 2017;12:89–96. doi: 10.1016/j.gdata.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger M, Ruchhoft C, Lishka R. Sensitive 4-aminoantipyrine method for phenolic compounds. Anal Chem. 1951;23(12):1783–1788. [Google Scholar]
- Erguven GO, Bayhan H, Demir G, Ikizoglu B, Kanat G. Monitoring aclonifen remediation in soil with a laboratory scale research. J Chem. 2016;2016:5059049. doi: 10.1155/2016/5059049. [DOI] [Google Scholar]
- Feng G, Xie T, Wang X, Bai J, Tang L, Zhao H, Wei W, Wang M, Zhao Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol. 2018;18:11. doi: 10.1186/s12866-018-1152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Verma SC, Mengoni A, Tripathi AK. Enrichment and identification of bacteria capable of reducing chemical oxygen demand of anaerobically treated molasses spent wash. J Appl Microbiol. 2004;96:1278–1286. doi: 10.1111/j.1365-2672.2004.02289.x. [DOI] [PubMed] [Google Scholar]
- Gillman GP, Sumpter EA. Modification to the compulsive exchange method for measuring exchange characteristics of soils. Aust J Soil Res. 1986;24(1):61–66. [Google Scholar]
- Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Hatano K, Kanazawa K, Tomura H, Yamatsu T, Tsunoda K, Kubota K. Molasses melanoidin promotes copper uptake for radish sprouts: the potential for an accelerator of phytoextraction. Environ Sci Pollut Res. 2016;23:17656–17663. doi: 10.1007/s11356-016-6904-x. [DOI] [PubMed] [Google Scholar]
- Hein S, Schönfeld P, Kahlert S, Reise G. Toxic 750 effects of X-linked adrenoleukodystrophy associated, very long chain fatty acids on glial cells and neurons from rat hippocampus in culture. Hum Mol Genet. 2008;17(12):1750–1761. doi: 10.1093/hmg/ddn066. [DOI] [PubMed] [Google Scholar]
- Huaidong HE, Waichin LI, Riqing YU, Zhihong YE. Illumina-based analysis of bulk and rhizosphere soil bacterial communities in paddy fields under mixed heavy metal contamination. Pedosphere. 2017;27(3):569–578. [Google Scholar]
- Huang XF, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany. 2014;92:267–275. [Google Scholar]
- Jackson ML. Soil chemical analysis. New Delhi: Prentice Hall; 1973. [Google Scholar]
- Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol. 2002;68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju XT, Kou CL, Christie P, Dou ZX, Zhang FS. Changes in the soil environment from excessive applicationof fertilizers and manures to two contrasting intensive cropping systems on the North China Plain. Environ Pollut. 2007;14:497–506. doi: 10.1016/j.envpol.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Kachienga L, Jitendra K, Momba M. Metagenomic profiling for assessing microbial diversity and microbial adaptation to degradation of hydrocarbons in two South African petroleum contaminated water aquifers. Sci Rep. 2018;8:7564. doi: 10.1038/s41598-018-25961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra YP, Maynard DG. Methods manual for forest soil and plant analysis, information report NOR-X-319 Forestry Canada. Edmonton: Northwest region, Northern Forest Centre; 1991. [Google Scholar]
- Kaushik G, Gopal M, Thakur IS. Evaluation of performance and community dynamics of microorganisms during treatment of distillery spent wash in a three stage bioreactor. Bioresour Technol. 2010;101:4296–4305. doi: 10.1016/j.biortech.2010.01.046. [DOI] [PubMed] [Google Scholar]
- Kostka JE, Prakash O, Overholt WA, et al. Hydrocarbon degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl Environ Microbiol. 2011;77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, AlMomin S, Al-Aqeel H, Al-Salameen F, Nair S, Shajan A. Metagenomic analysis of rhizosphere microflora of oil-contaminated soil planted with barley and alfalfa. PLoS One. 2018 doi: 10.1371/journal.pone.0202127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhare SV, Dhotre DP, Dhar SK, et al. Insights into diversity and imputed metabolic potential of bacterial communities in the continental shelf of Agatti Island. PLoS One. 2015;10(6):e0129864. doi: 10.1371/journal.pone.0129864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P, Neumann G, Kania A, et al. Spatial and temporal dynamics of the microbial community structure in the rhizosphere of cluster roots of white lupin (Lupinus albus L.) Plant Soil. 2002;246:167–174. [Google Scholar]
- Marschner P, Crowley D, Yang CH. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil. 2004;261:199–208. [Google Scholar]
- Mukherjee A, Chettri B, Langpoklakpam JS, Basak P, Prasad A, Mukherjee AK, Bhattacharyya M, Singh AK, Chattopadhyay D. Bioinformatic approaches including predictive metagenomic profiling reveal characteristics of bacterial response to petroleum hydrocarbon contamination in diverse environments. Sci Rep. 2017;7:1108. doi: 10.1038/s41598-017-01126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DW, Sommers LE. Total carbon, organic carbon and organic matter. In: Page L, editor. Methods of soil analysis. Part 2: agronomy 9. Madison: American Society of Agronomy; 1982. pp. 279–539. [Google Scholar]
- OECD SIDS (2010) SIDS Initial Assessment Report FOR 13TH SIAM
- Pang S, Zhang S, Lv XY, et al. Characterization of bacterial community in biofilm and sediments of wetlands dominated by aquatic macrophytes. Ecol Eng. 2016;97:242–250. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Gloeckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JB, Forsberg N. Production of tricarballylic acid by rumen microorganisms and its potential toxicity in ruminant tissue metabolism. Br J Nutr. 1986;56(1):153–162. doi: 10.1079/bjn19860095. [DOI] [PubMed] [Google Scholar]
- Sahu N, Deori M, Ghosh I. Metagenomics study of contaminated sediments from the Yamuna River at Kalindi Kunj, Delhi, India. Genome Announc. 2018;6:e01379–17. doi: 10.1128/genomeA.01379-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjenbam P, Kannabira K. Bioactivity of Pyrrolo[870 1,2-A]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)-extracted from Streptomyces sp VITPK9 isolated from the Salt Spring Habitat of Manipur, India. Asian J Pharmace. 2016;10(4):26. [Google Scholar]
- Shah V, Zakrzewski M, Wibberg D, Eikmeyer D, Schluter A, Madamwar D. Taxonomic profiling and metagenome analysis of a microbial community from a habitat contaminated with industrial discharges. Microb Ecol. 2013;66:533–550. doi: 10.1007/s00248-013-0244-x. [DOI] [PubMed] [Google Scholar]
- Sharmin F, Wakelin S, Huygens F, Hargreaves M. Firmicutes dominate the bacterial taxa within sugar-cane processing plants. Sci Rep. 2013 doi: 10.1038/srep03107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiri Z, Kermanshahi RK, Soudi MR, Farajzadeh D. Isolation and characterization of an n-hexadecane degrading Acinetobacter baumannii KSS1060 from a petrochemical wastewater treatment plant. Int J Environ Sci Technol. 2015;12:455–464. [Google Scholar]
- United States Environmental Protection Act (USEPA) (1996) Acid digestion of sediments, sludges, and soils. https://www.epa.gov/sites/production/files/2015-06/documents/epa-3050b.pdf
- United States Environmental Protection Act (USEPA) The environment protection rules, 3A, schedule-II, III. Cincinnati: U.S. Environmental Protection Agency, Ofce of research and development; 2002. [Google Scholar]
- USEPA (2012) U.S. Environmental protection agency endocrine disruptor screening program universe of chemicals and general validation principles. https://www.epa.gov/sites/production/files/2015-07/documents/edsp_chemical_universe_and_general_validations_white_paper_11_12.pdf
- Van Elsas JD, Smalla K. Methods for sampling soil microbes. In: Hurst CJ, Knudsen GR, McInerney MJ, Stetzenbach LD, Walter MV, editors. Manual of environmental microbiology. Washington, DC: American Society of Microbiology Press; 1997. pp. 383–390. [Google Scholar]
- Wang P, Zhang H, Zuo J, Zhao D, Zou X, Zhu Z, Jeelani N, Leng X, An S. A hardy plant facilitates nitrogen removal via microbial communities in subsurface flow constructed wetlands in winter. Sci Rep. 2016;6:33600. doi: 10.1038/srep33600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womersley J. Guideline: managing impacts from the bulk storage of bagasse. Queensland: Department of Environment and Resource Management Publication; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.