Abstract
Colorectal cancer (CRC) has become the second leading cause of new cancer cases and the fifth of cancer deaths in China, and early detection is the most effective way to reduce the incidence and mortality of CRC. A number of methylated DNA biomarkers have been found to associate with CRC and precancerous lesions in stool samples, indicating stool methylated DNA biomarkers are potential tools for CRC early detection. In this study, approximately 5 g of stool specimen was collected from 230 subjects (124 in the training set and 106 in the validation set). Stool DNA was extracted and bisulfite-converted, followed by ColoDefense test, a multiplex qPCR assay, that simultaneously detects methylated SEPT9 (mSEPT9) and methylated SDC2 (mSDC2). Youden index was employed to determine the cut-off value of ColoDefense test for stool specimens. In the training set, the optimized cut-off value of stool ColoDefense test was: mSEPT9 analyzed with 3/3 algorithm and mean mSEPT9 Ct values of <38, or mSDC2 with 2/3 algorithm. Stool ColoDefense test achieved Youden indexes of 79.9 and 57.4% in detecting CRC and advanced adenomas (AA), respectively. Its sensitivities in the training set for AA and CRC were 66.7% (95% CI: 24.1–94.0%) and 89.1% (95% CI: 77.1–95.5%) with a 90.8% (95% CI: 80.3–96.2%) specificity, and AUC was 0.956 (95% CI: 0.924–0.988). In the validation set, its sensitivities for AA and CRC were 66.7% (95% CI: 24.1–94.0%) and 92.3% (95% CI: 78.0–98.0%) with a 93.2% (95% CI: 82.7–97.8%) specificity, and AUC was 0.977 (95% CI: 0.952–1.000). Positive detection rate of stool ColoDefense test has been found to be independent of age, gender, tumor location, and tumor size. In conclusion, stool ColoDefense test demonstrated high sensitivities and specificity for the detection of AA and CRC. Therefore, it has the potential to become a low-cost, convenient, and highly effective tool for CRC early detection.
Keywords: colorectal cancer, stool, mSEPT9, mSDC2, early detection
Introduction
Approximately 2 million new colorectal cancer (CRC) cases and over 881,000 deaths were estimated to occur in 2018 worldwide, accounting for about 1 in 10 new cancer cases and deaths. Overall, CRC ranked the third in terms of incidence but the second in terms of mortality all over the world (Bray et al., 2018). While in China, Over 517,000 new CRC cases and more than 245,000 CRC deaths were estimated for the same year, ranking the second in terms of incidence and the fifth in terms of mortality by the same study (Ferlay et al., 2018). And CRC has been reported to be more common in male aged 60 years in China (Chen et al., 2018).
Early detection is one of the most effective ways to reduce the incidence and mortality of CRC, therefore CRC early screening programs are organized in several countries (Sano et al., 2016). In China, a two-step screening strategy has been recommended for population based CRC screening, the guaiac-based fecal occult blood test (gFOBT) and a quantitative high-risk factor questionnaire as the primary screening, with a full colonoscopy for follow-up (Sano et al., 2016). However, the benefit of gFOBT is limited due to its low sensitivity of 33.3–57.1% (Tinmouth et al., 2015). Colonoscopy, as the gold standard for CRC diagnosis, has been widely used in early CRC screening programs in many countries (Mohammadi et al., 2017). For the past 5 years, CRC incidence has continued to decline by approximately 3% every year in the US due to the increased acceptance of colonoscopy, which allows removal of precancerous lesions (Siegel et al., 2012, 2019). Colonoscopy among US adults elder than 50 years tripled from 21% in 2000 to 60% in 2015 (Siegel et al., 2019). In contrast, a recent population-based CRC screening program in China revealed a relatively low participation rate due to its invasiveness, bothersome bowel preparation and difficult-to-avoid complications (Chen H. et al., 2019).
DNA methylation is known to be abnormal in many cancers (Klutstein et al., 2016). A number of methylated DNA biomarkers have been found to associate with CRC and precancerous lesions in stool or plasma samples, including SEPT9 (Catherine et al., 2008; Lamb and Dhillon, 2017), SDC2 (Barták et al., 2017; Han et al., 2019), SFRP2 (Barták et al., 2017; Li et al., 2019), and TFPI2 (Glöckner et al., 2009), some of which have been developed into commercial kits for CRC early detection (Potter et al., 2014; Lamb and Dhillon, 2017; Li et al., 2019; Zhao et al., 2019). For example, plasma methylated SEPT9 (mSEPT9) test (Epi proColon 2.0 assay) was approved for CRC early detection by FDA in 2016 (Lamb and Dhillon, 2017). Meanwhile, Cologuard, a stool DNA test approved by FDA, also includes two methylated DNA biomarkers (Imperiale et al., 2014), and it has been recommended by the most recent CRC screening guideline in the US (Wolf et al., 2018). We previously demonstrated a multiplex methylated DNA test in plasma, ColoDefense test, with high sensitivity and specificity for CRC early detection. It detects mSEPT9, methylated SDC2 (mSDC2), and an internal control (ACTB) simultaneously in a single reaction (Chen Y. et al., 2019; Zhao et al., 2019). In previous studies, plasma mSEPT9 test showed low sensitivities in detecting early stage CRC and advanced adenomas (AA) (Potter et al., 2014; Timothy et al., 2014). Instead, we have demonstrated that the detection rates of plasma ColoDefense test for AA and early stage CRC were significantly improved by the combination of two biomarkers, mSEPT9 and mSDC2, with high specificity (Zhao et al., 2019). However, plasma ColoDefense test may be a better choice for hospitals, because blood draw is convenient to perform for medical personnel, but requires an appointment in advance which takes more time. While stool samples are convenient to collect at home, it can provide more privacy for those who are concerned, or too busy, or too afraid of a cross-infection (like the Covid-19) to go to the hospital. Therefore, combination of plasma and stool tests can facilitate broader population to participate in the early diagnosis of colorectal cancer. In this study, we evaluated the feasibility of mSEPT9 and mSDC2 in stool specimens as an integrated biomarker for early CRC detection, and optimized its cut-off value.
Materials and Methods
Sample Collection
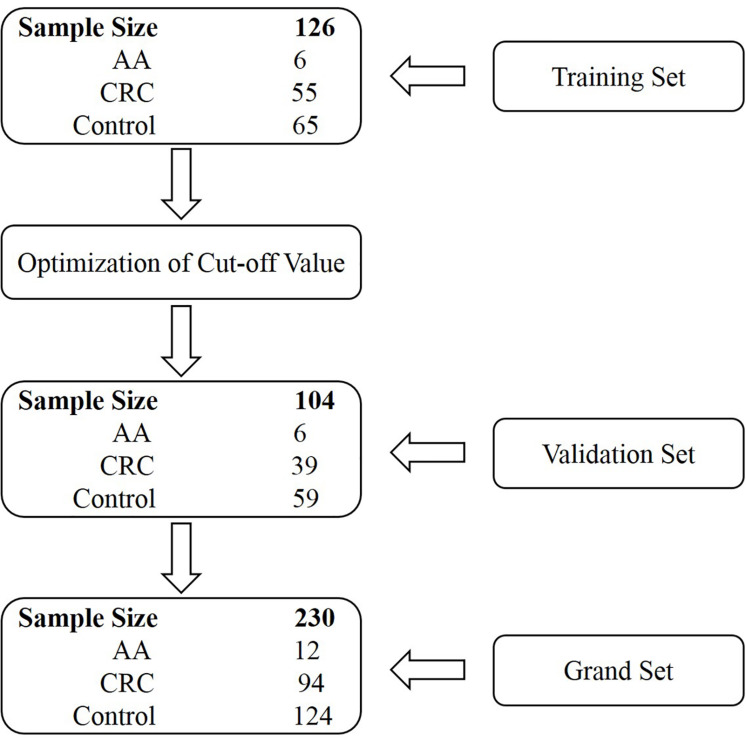
Stool specimens were collected from 230 subjects who underwent colonoscopy at the Affiliated Hospital of Xuzhou Medical University. Participants were prospectively enrolled in two independent cohorts, the training and validation sets. The training set comprised subjects enrolled from July 1, 2018 until February 1, 2019, and the validation set consisted of subjects enrolled from March 1, 2019 until December 1, 2019 (Figure 1). The training set included 55 CRC patients, 6 AA (adenomas measuring ≥1 cm in the greatest dimension or with high-grade dysplasia or with ≥25% villous histologic features) patients and 65 control subjects (colonoscopy negative subjects). The validation set included 39 CRC patients, 6 AA patients, and 59 control subjects (Figure 1 and Supplementary Tables 1, 2). Some CRC and AA patients were tested positive for gFOBT or colonoscopy before being transferred to the Affiliated Hospital of Xuzhou Medical University. All stool samples were collected prior to purgative bowel preparation or colonoscopy. Whole stools were collected in single-use disposable buckets mounted on toilet seats, and approximately 5 g of each stool specimen was transferred into a 50 mL tube containing 25 mL of preservative buffer (Suzhou VersaBio Technologies Co., Ltd., Kunshan, China) to stabilize human genomic DNA in the stool. All stool samples were stored at room temperature for no more than 7 days or −80°C for longer-term storage before usage.
FIGURE 1.
Schematic representation of the study design.
TABLE 2.
Positive detection rates of mSEPT9 and mSDC2 with 1/3, 2/3, and 3/3 algorithms for AA, CRC, and control groups.
| Algorithm | Group | Subject number (n) |
Positive detection rate (95% CI:%) |
|
| mSEPT9 | mSDC2 | |||
| 1/3 | AA | 6 | 100.0 (51.7–100.0) | 50.0 (13.9–86.1) |
| CRC | 55 | 98.2 (89.0–99.9) | 90.9 (79.3–96.6) | |
| Control | 65 | 53.8 (41.1–66.1) | 26.2 (16.4–38.8) | |
| 2/3 | AA | 6 | 100.0 (51.7–100.0) | 33.3 (6.0–75.9) |
| CRC | 55 | 94.5 (83.9–98.6) | 83.6 (70.7–91.8) | |
| Control | 65 | 41.5 (29.7–54.4) | 6.2 (2.0–15.8) | |
| 3/3 | AA | 6 | 83.3 (36.5–99.1) | 33.3 (6.0–75.9) |
| CRC | 55 | 85.5 (72.8–93.1) | 78.2 (64.6–87.8) | |
| Control | 65 | 23.1 (13.9–35.5) | 0.0 (0.0–7.0) | |
This study was approved by the Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYFY2018-KL081), and the informed consent was obtained for all participating patients and control subjects.
DNA Extraction, Bisulfite Treatment, and Quantitative Real-Time PCR
All stool samples were thawed for about 30 min at 15–30°C and subsequently homogenized for 1 min with a shaking device. After homogenization, each stool sample was centrifuged for 20 min at 10,000 g. One hundred and fifty microliters supernatants were removed for human genomic DNA extraction with a stool DNA extraction kit (Suzhou VersaBio Technologies Co., Ltd.). Briefly, each supernatant was added with 500 μL fresh preservative buffer and centrifuged at 20,000 g for 3 min. The resulting supernatant was then transferred to a new tube, and 600 μL lysis buffer and 20 μL proteinase K solution were added to each sample followed by incubation at 70°C for 10 min. Next, each sample was added with 600 μL absolute ethanol and then loaded onto a spin column. After two washing steps, the column was dried and DNA was eluted with 100 μL elution buffer. Bisulfite conversion of purified DNA and purification of the converted products were performed with a fast bisulfite conversion kit (Suzhou VersaBio Technologies Co., Ltd.). Briefly, 150 μL conversion buffer and 25 μL protection buffer were added to 100 μL purified DNA solution, and followed by incubation at 80°C for 45 min. Next, 1 mL wash buffer A was added to each sample and loaded onto a spin column. After two washing steps, the column was air dried and DNA was eluted with 100 μL elution buffer.
Purified and converted DNA from the above steps was tested by ColoDefense test developed by Suzhou VersaBio Technologies Co., Ltd. Three PCR replicates were performed for each sample. The total volume of the ColoDefense test was 30 μL including 15 μL template and 15 μL PCR master mix. qPCR was analyzed on LC480-II thermal cycler (Roche Diagnostics, Basel, Switzerland) using the following cycling conditions: activation at 95°C for 30 min, 50 cycles of 95°C for 10 s, 56°C for 30 s, and final cooling to 40°C for 30 s (Zhao et al., 2019).
Data Analysis
ACTB was used as the internal control gene for valid sample collection and processing to avoid false negatives. The result for a stool sample was considered “invalid” if ACTB Ct was greater than 41.0, and mSEPT9 and mSDC2 were considered “detected” if their Ct values were less than 45.0 and 50.0, respectively. As ColoDefense test is a multiplex qPCR reaction run in triplicates and therefore returns with several possible results depending on different scoring algorithms (1/3, 2/3, or 3/3 for each target). According to this principle, the results of ColoDefense test were analyzed with different algorithms to determine the optimal algorithm (Tables 2, 3). Youden index (sensitivity + specificity - 1) was employed to determine cutoff values, where specificity equals to 1 minus positive detection rate of control, and sensitivity equals to positive detection rate of CRC or AA. Therefore, Youden index of CRC or AA equals to positive detection rate of CRC or AA minus positive detection rate of control.
TABLE 3.
Positive detection rates of ColoDefense test with different algorithm combinations for mSEPT9 and mSDC2 in detecting AA, CRC, and control.
|
Algorithm |
Group | Subject number (n) | Positive detection rate (95% CI:%) | Youden index (%) | |
| mSEPT9 | mSDC2 | ||||
| 3/3 | 2/3 | AA | 6 | 83.3 (36.5–99.1) | 54.1 |
| CRC | 55 | 94.5 (83.9–98.6) | 65.3 | ||
| Control | 65 | 29.2 (18.9–42.0) | N/A | ||
| 3/3 | 3/3 | AA | 6 | 66.7 (24.1–94.0) | 42.1 |
| CRC | 55 | 90.9 (3.4–20.7) | 66.3 | ||
| Control | 65 | 24.6 (15.1–37.1) | N/A | ||
| 3/3 and mean Ct <40 | 2/3 | AA | 6 | 83.3 (36.5–99.1) | 60.3 |
| CRC | 55 | 90.9 (3.4–20.7) | 67.8 | ||
| Control | 65 | 23.1 (13.9–35.5) | N/A | ||
| 3/3 and mean Ct <40 | 3/3 | AA | 6 | 66.7 (24.1–94.0) | 48.2 |
| CRC | 55 | 87.3 (74.9–94.3) | 68.8 | ||
| Control | 65 | 18.5 (10.3–30.4) | N/A | ||
| 3/3 and mean Ct <39 | 2/3 | AA | 6 | 83.3 (36.5–99.1) | 71.0 |
| CRC | 55 | 89.1 (77.1–95.5) | 76.8 | ||
| Control | 65 | 12.3 (5.8–23.4) | N/A | ||
| 3/3 and mean Ct <39 | 3/3 | AA | 6 | 66.7 (24.1–94.0) | 59.0 |
| CRC | 55 | 87.3 (74.9–94.3) | 79.6 | ||
| Control | 65 | 7.7 (2.9–17.8) | N/A | ||
| 3/3 and mean Ct <38 | 2/3 | AA | 6 | 66.7 (24.1–94.0) | 57.4 |
| CRC | 55 | 89.1 (77.1–95.5) | 79.9 | ||
| Control | 65 | 9.2 (3.8–19.7) | N/A | ||
| 3/3 and mean Ct <38 | 3/3 | AA | 6 | 50.0 (13.9–86.1) | 43.8 |
| CRC | 55 | 87.3 (74.9–94.3) | 81.1 | ||
| Control | 65 | 6.2 (2.0–15.8) | N/A | ||
N/A, not applicable.
Statistical analysis was performed using IBM SPSS for Windows, Version 22.0, and t-test was used for comparison between two testing subjects at the significance level of p <0.05. Receiver operating characteristic (ROC) curves were plotted using the mean Ct values from CRC and control subjects. Because mSEPT9 and mSDC2 were not detected from most control subjects by the qPCR reaction, we set the corresponding Ct values to 50.0 (the maximal number of PCR cycles) for such samples to plot the curves (Wu et al., 2016).
Results
Two hundred and thirty participants in total were originally enrolled in our study, whose baseline characteristics are shown in Table 1. The training set included 126 subjects, among which 55 were CRC patients including one stage 0, 11 stage I, 15 stage II, 18 stage III, four stage IV patients and six patients of unknown stage. For the validation set, there were 39 CRC patients including nine stage I, eight stage II, 16 stage III, three stage IV patients and three patients of unknown stage (Supplementary Tables 1, 2).
TABLE 1.
Subjects characteristics.
|
Training set |
Validation set |
|||
| Age (years) | Min-Max | Mean | Min-Max | Mean |
| AA | 47–75 | 62.2 | 46–67 | 56.2 |
| CRC | 35–86 | 60.0 | 27–83 | 59.0 |
| Control | 24–69 | 45.2 | 24–83 | 47.9 |
| Gender [N (%)] | Male | Female | Male | Female |
| AA | 5 (83.3) | 1 (16.7) | 3 (50.0) | 3 (50.0) |
| CRC | 32 (58.2) | 23 (41.8) | 21 (53.8) | 18 (46.2) |
| Control | 31 (47.7) | 34 (52.3) | 31 (52.5) | 28 (47.4) |
The cut-off value of ColoDefense test for plasma specimens is the detection of mSEPT9 in 1 or mSDC2 in 2 out of 3 replicate PCR reactions (Zhao et al., 2019). However, mSEPT9 showed low specificity with 1/3 algorithm for stool specimens (Table 2). Therefore, we used a two-step approach to optimize the cut-off value of ColoDefense test for stool specimens. First, we analyzed the performance of mSEPT9 and mSDC2 with different algorithms (Table 2). Similar to the results for plasma specimens, mSDC2 showed relatively high sensitivity (83.6%) and specificity (93.8%) in detecting CRC with 2/3 algorithm. However, with 1/3 algorithm, mSDC2 resulted in higher sensitivity in detecting both AA and CRC but a lower specificity. In contrast, with 3/3 algorithm, mSDC2 showed a 100% specificity and a 78.2% sensitivity, even better than its performance for plasma specimens. As for mSEPT9, it showed very high sensitivities and low specificities in detecting both AA and CRC with 1/3, 2/3 and 3/3 algorithms.
While the combination of mSEPT9 and mSDC2 could achieve higher sensitivity than mSEPT9 or mSDC2 alone, it would decrease specificity. Therefore, in the second step, we gradually tightened the Ct requirement for mSEPT9, and the resulting mSEPT9 readouts were combined with mSDC2 in 2/3 and 3/3 algorithms to choose the optimal cut-off value. As shown in Table 3, mSEPT9 with 3/3 algorithm and a mean mSEPT9 Ct value of less than 38 combined with mSDC2 with 2/3 algorithm achieved the relatively high Youden index for detecting CRC (79.9%) and AA (57.4%), showed a balance of sensitivities and specificity. Therefore, all subsequent data were analyzed with these criteria.
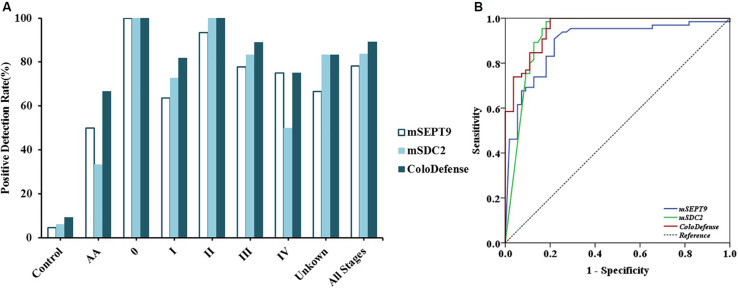
Out of 126 stool samples in the training set, mSEPT9 was detected in 4.6% of control, 50.0% of AA, 100.0% of stage 0 CRC (1/1), 63.6% of stage I CRC (7/11), 93.3% of stage II CRC (14/15), 77.8% of stage III CRC (14/18), 75.0% of stage IV CRC (3/4), and 66.7% of unknown stage CRC (4/6) samples (Figure 2A). mSDC2 was detected in 6.2% of control, 33.3% of AA, 100.0% of stage 0 CRC (1/1), 72.7% of stage I CRC (8/11), 100.0% of stage II CRC (15/15), 83.3% of stage III CRC (15/18), 50.0% of stage IV CRC (2/4), and 83.3% of unknown stage CRC (5/6) samples. In contrast, with ColoDefense test, the positive detection rates improved to 66.7% (4/6) for AA, 100.0% (1/1) for stage 0 CRC, 81.8% for stage I CRC (9/11), 100.0% for stage II CRC (15/15), 88.9% for stage III CRC (16/18), 75.0% for stage IV CRC (3/4), and 83.3% of unknown stage CRC (5/6), and the positive detection rate for control was 9.2% (refer to specificity was 90.8%) (Supplementary Table 3). AUC for mSEPT9 alone in detecting CRC was 0.892 (95% CI: 0.831–0.953), and AUC for mSDC2 alone in detecting CRC was 0.930 (95% CI: 0.877–0.983) (Figure 2B). In contrast, ColoDefense test improved AUC to 0.956 (95% CI: 0.924–0.988).
FIGURE 2.
Performance of ColoDefense test in detecting control, AA and CRC across stages 0–IV in the training set. (A) Positive detection rates for control and all stages of CRC. (B) ROC curves for ColoDefense test in detecting CRC.
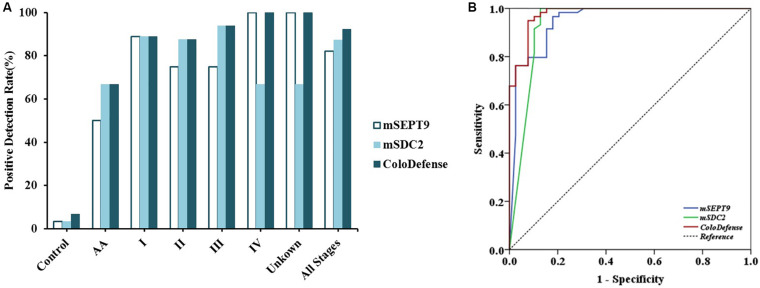
To validate the performance of the cut-off value of ColoDefense for stool samples, we enrolled an independent cohort of 104 subjects as a validation set. In the validation set, mSEPT9 showed 50.0% (95% CI: 13.9–86.1%) and 82.1% (95% CI: 65.9–91.9%) sensitivities and 96.6% (95% CI: 87.3–99.4%) specificity for detecting AA and CRC, and mSDC2 showed 66.7% (95% CI: 24.1–94.0%) and 87.2% (95% CI: 71.8–95.2%) sensitivities and 96.6% (95% CI: 87.3–99.4%) specificity for detecting AA and CRC (Figure 3A). In contrast, ColoDefense test improved the sensitivities to 66.7% (95% CI: 24.1–94.0%) for AA and 92.3% (95% CI: 78.0–98.0%) for CRC with a specificity of 93.2% (95% CI: 82.7–97.8%) (Supplementary Table 3). AUC for mSEPT9 alone in detecting CRC was 0.948 (95% CI: 0.901–0.995), and AUC for mSDC2 alone in detecting CRC was 0.937 (95% CI: 0.875–0.999), whereas ColoDefense test improved the AUC to 0.977 (95% CI: 0.952–1.000) (Figure 3B).
FIGURE 3.
Performance of ColoDefense test in detecting control, AA and CRC across stages I–IV in the validation set. (A) Positive detection rates for control and all stages of CRC. (B) ROC curves for ColoDefense test in detecting CRC.
Furthermore, there was no significant difference for the positive detection rates of mSEPT9 alone, mSDC2 alone or ColoDefense test between different stage, age groups or genders (p > 0.05, Table 4). The positive detection rates of mSEPT9 showed significant difference among different tumor locations (p < 0.05), and the positive detection rates of mSDC2 seemed to increase with the increase of tumor sizes (p < 0.05). However, when mSEPT9 with mSDC2 were combined as ColoDefense test, no significant difference was apparent in detecting CRC among different locations or tumor sizes.
TABLE 4.
Results of ColoDefense test in detecting CRC for different stage, age groups, genders, tumor locations, and tumor sizes.
| mSEPT9 positive detection rate (%) | p-value | mSDC2 positive detection rate (%) | p-value | ColoDefense positive detection rate (%) | p-value | |
| Stage | ||||||
| I–II (n = 44) | 81.8(36/44) | 0.664 | 88.6(39/44) | 0.450 | 90.9(40/44) | 0.916 |
| III–IV (n = 41) | 78.1(32/41) | 82.9(34/41) | 90.2(37/41) | |||
| N/A (n = 9) | 77.8(7/9) | 77.8(7/9) | 88.9(8/9) | |||
| Age | ||||||
| <60 (n = 48) | 79.2(38/48) | 0.878 | 85.4(41/48) | 0.931 | 93.8(45/48) | 0.263 |
| ≥60 (n = 46) | 80.4(37/46) | 84.8(39/46) | 87.0(40/46) | |||
| Gender | ||||||
| Male (n = 53) | 86.8(46/53) | 0.054 | 86.8(46/53) | 0.602 | 90.6(48/53) | 0.985 |
| Female (n = 41) | 70.7(29/41) | 82.9(34/41) | 90.2(37/41) | |||
| Location | ||||||
| Proximal (n = 46) | 89.1(41/46) | 0.031* | 84.8(39/46) | 0.113 | 93.5(43/46) | 0.361 |
| Distal (n = 41) | 70.7(29/41) | 87.8(29/41) | 87.8(36/41) | |||
| N/A (n = 7) | 71.4(5/7) | 71.4(5/7) | 85.7(6/7) | |||
| Size | ||||||
| <3 cm (n = 11) | 81.8(9/11) | 0.870a | 54.6(6/11) | 0.003**a | 81.8(9/11) | 0.324a |
| 3–6 cm (n = 59) | 79.7(47/59) | 0.719b | 89.8(53/59) | 0.026*b | 91.5(54/59) | 0.202b |
| >6 cm (n = 8) | 75.0(6/8) | 0.761c | 100.0(8/8) | 0.345c | 100.0(8/8) | 0.392c |
| N/A (n = 16) | 81.3(13/16) | 81.3(13/16) | 87.5(14/16) | |||
*p < 0.05; **p < 0.01. N/A, not applicable. ap-value between <3 cm and 3–6 cm. bp-value between <3 cm and >6 cm. cp-value between 3–6 cm and >6 cm.
Discussion
As one of the most common cancers globally, CRC caused millions of new cases and deaths in 2018 (Bray et al., 2018). Longstanding and early screening program is the most effective way to reduce the incidence and mortality of CRC. Up to know, several strategies have been demonstrated to be effective for CRC early screening, including colonoscopy, fecal immunochemical test, and high-sensitivity gFOBT (Wolf et al., 2018; Siegel et al., 2019). In 2018, multitarget stool DNA test was recommended as a new CRC screening method in the updated guideline from American Cancer Society (ACS) for adults elder than 45 years with an average risk of CRC (Wolf et al., 2018). In this study, we optimized the cut-off value of ColoDefense test for stool samples and evaluated the feasibility of stool ColoDefense test for CRC early detection.
In our previous study, we demonstrated that ColoDefense test detected 47.8% AA, 87.1% early stage (0–II) CRC and 88.9% stage I–IV CRC, with a specificity of 92.8% for plasma samples (Zhao et al., 2019). And plasma ColoDefense test showed better performance than mSEPT9 or mSDC2 alone in detecting AA and CRC. In the present study, we attempted to determine whether stool ColoDefense test was a viable option for CRC early detection. The results showed that the sensitivities of stool ColoDefense test in detecting AA, early stage (0–II) CRC and stage I–IV CRC were 66.7% (8/12), 90.9% (40/44), and 90.4% (85/94), respectively, with a specificity of 91.9%. Compared with plasma ColoDefense test, stool ColoDefense test showed better performance in detection AA (66.7 vs. 47.8%) and stage I CRC (85.0 vs. 80.0%). This was likely because DNA in stool samples originates directly from the cells of the intestinal wall where precancerous and tumor tissues are located, while ctDNA in plasma has passed through various barriers within the body to reach the circulation. Therefore, the higher DNA abundance in stool samples than that in plasma resulted in higher sensitivities in AA and early stage CRC detection, which is also consistent with the different cut-off values between stool and plasma ColoDefense test. Another advantage of stool ColoDefense test is that sample can be collected at home and sent to laboratories for analysis, a convenience desirable for those people who are concerned with privacy or too busy to go to the hospital. On the contrary, plasma ColoDefense test may be the better choice for hospitals and other medical institutions. Blood draw is more convenient for medical personnel to perform, and the pretreatment of blood samples is easier than that of stool samples.
In previous studies, several stool methylated DNA biomarkers have been investigated for CRC early detection. Glöckner et al. (2009) developed a methylation-specific PCR (MSP) assay for methylated TFPI2, which could detect stage I to III CRC patients with a sensitivity of 76–89% and a specificity of 79–93% with stool samples. Tang et al. (2011) reported a MSP assay for methylated SFRP2, which could detect 84.0% CRC with a specificity of 54.0% with stool DNA. Both methylated TFPI2 and SFRP2 tests employed agarose gel electrophoresis-based MSP approach, not suitable for clinical application.
Oh et al. (2017) recently reported a stool mSDC2 assay based on a nested-PCR method (LTE-qMSP) for early CRC screening, which detected 90.0% CRC and 33.3% small polyps with a specificity of 90.9%. Niu et al. (2017) also published a qPCR based mSDC2 assay in 2017, which detected 81.1% of CRC and 58.2% of >1 cm adenomas with a specificity of 93.3%. Both results are comparable to the results of mSDC2 alone in our study, indicating that mSDC2 has a high potential as an early diagnostic biomarker for CRC. However, the results in our study indicated that mSDC2 alone may result in more false negatives (Figures 2, 3) due to its different sensitivities for tumors of different sizes (Table 4). In contrast, the combination of mSDC2 and mSEPT9 for stool DNA test led to higher sensitivities for all CRC stages (Figure 3) and the best balance of positive detection rates for different age groups, genders, tumor locations, and tumor sizes (Table 4), which was also demonstrated for plasma ColoDefense test (Zhao et al., 2019).
Cologuard, a stool DNA test approved by FDA for CRC screening in 2014, employs multiple biomarkers including two methylated genes (BMP3 and NDRG4), 7 KRAS mutation sites and an immunochemical assay for human hemoglobin. It detected CRC and AA with 92.3 and 42.4% sensitivities, respectively, and a specificity of 86.7% (Imperiale et al., 2014). As the first FDA approved test for CRC screening using multiple biomarkers demonstrating impressive sensitivity and specificity, Cologuard has been recommended by ACS as an option for CRC screening (Wolf et al., 2018). Compared with Cologuard, stool ColoDefense test only requires approximately 5 g stool for each subject and detects two methylation biomarkers and an internal control gene in one multiplex qPCR reaction, resulting in significantly lower cost and much simpler procedure. Moreover, stool ColoDefense test achieved a higher sensitivity for AA (66.7 vs. 42.4%) and a similar sensitivity of CRC (90.4 vs. 92.3%) with a high specificity of 91.9%. Therefore, stool ColoDefense test may be a superb alternative for CRC screening in developing countries such as China.
However, there are a few limitations in this study. For example, the numbers of AA samples, stage IV CRC samples, and the total number of clinical samples examined were relatively small, which could lead to fluctuations of estimates of performance. This can be improved by increasing the number of enrolled patients in future studies. In this study, we analyzed performance of stool samples in a training and a validation set as a prospective study, but more validation studies in multiple clinical centers as well as a large prospective study within a population screening program should be carried out in the future. In addition, as non-advanced colon polyps were prevalent in 7.4–52.5% of patients undergoing screening colonoscopy (Corley et al., 2014), the performance of ColoDefense test on non-advanced colon polyps should also be examined in further studies.
Conclusion
Stool ColoDefense test demonstrated high sensitivities for AA and CRC detection, which were higher than those of either mSEPT9 or mSDC2 alone. Therefore, stool ColoDefense test has the potential to become a powerful, convenient and highly effective screening tool for CRC early detection.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYFY2018-KL081). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
GZ, XL, and YM performed the statistical analyses and drafted the manuscript. YL, HL, SL, YZ, and JM participated in sample collection and data analysis. GZ, SX, SF, and MZ conceived of the study and participated in the design and coordination of the study. All authors read and approved the final manuscript.
Conflict of Interest
GZ and SX were employees of Suzhou VersaBio Technologies Co., Ltd. SX was the shareholder of Suzhou VersaBio Technologies Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the grants from the Suzhou Technology Entrepreneur Angel Project (Grant No. CYTS2018051), Key Technologies R&D Program for Social Development of Jiangsu Province (Grant No. BE2019688), Kunshan Leading Talent Project (Grant No. 00311), and Key Technologies R&D Program for Social Development of Xuzhou (Grant No. KC17184).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00643/full#supplementary-material
References
- Barták B. K., Kalmár A., Péterfia B., Patai ÁV., Galamb O., Valcz G., et al. (2017). Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics 12 751–763. 10.1080/15592294.2017.1356957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Catherine L. D., Fabian M., Theo D., Reimo T., Juergen D., Matthias S., et al. (2008). DNA methylation biomarkers for blood-based colorectal cancer screening. Clin. Chem. 54 414–423. [DOI] [PubMed] [Google Scholar]
- Chen H., Li N., Ren J., Feng X., Lyu Z., Wei L., et al. (2019). Participation and yield of a population-based colorectal cancer screening programme in China. Gut 68 1450–1457. 10.1136/gutjnl-2018-317124 [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang Z., Zhao G., Sun C., Ma Y., Zhang L., et al. (2019). Performance of a novel blood-based early colorectal cancer screening assay in remaining serum after the blood biochemical test. Dis. Mark. 2019:5232780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Sun K., Zheng R., Zeng H., Zhang S., Xia C., et al. (2018). Cancer incidence and mortality in China, 2014. Chin. J. Cancer Res. 30 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley D. A., Jensen C. D., Marks A. R., Zhao W. K., Lee J. K., Doubeni C. A., et al. (2014). Adenoma detection rate and risk of colorectal cancer and death. N. Engl. J. Med. 370 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Colombet M., Soerjomataram I. (2018). Global and Regional Estimates of the Incidence and Mortality for 38 Cancers: GLOBOCAN 2018. Lyon: International Agency for Research on Cancer. [Google Scholar]
- Glöckner S. C., Dhir M., Yi J. M., Mcgarvey K. E., Van Neste L., Louwagie J., et al. (2009). Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 69 4691–4699. 10.1158/0008-5472.can-08-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. D., Oh T. J., Chung T.-H., Jang H. W., Kim Y. N., An S., et al. (2019). Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin. Epigenet. 11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale T. F., Ransohoff D. F., Itzkowitz S. H., Levin T. R., Lavin P., Lidgard G. P., et al. (2014). Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 370 1287–1297. [DOI] [PubMed] [Google Scholar]
- Klutstein M., Nejman D., Greenfield R., Cedar H. (2016). DNA methylation in cancer and aging. Cancer Res. 76 3446–3450. 10.1158/0008-5472.can-15-3278 [DOI] [PubMed] [Google Scholar]
- Lamb Y. N., Dhillon S. (2017). Epi proColon(®) 2.0 CE: a blood-based screening test for colorectal cancer. Mol. Diagnosis Ther. 21 1–8. [DOI] [PubMed] [Google Scholar]
- Li H., Wang Z., Zhao G., Ma Y., Chen Y., Xue Q., et al. (2019). Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int. J. Biol. Mark. 34 54–59. 10.1177/1724600818820536 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., Jafari S. M., Mohaghegh M. A., Ghasemi A., Momenzadeh N. (2017). Comparison of SFRP2 promoter methylation in stool sample and cfDNA regarding patients with colorectal cancer. Comp. Clin. Pathol. 27 155–159. 10.1007/s00580-017-2568-2 [DOI] [Google Scholar]
- Niu F., Wen J., Fu X., Li C., Zhao R., Wu S., et al. (2017). Stool DNA Test of methylated syndecan-2 for the early detection of colorectal neoplasia. Cancer Epidemiol. Biomark. Prevent. 26 1411–1419. 10.1158/1055-9965.epi-17-0153 [DOI] [PubMed] [Google Scholar]
- Oh T. J., Oh H. I., Yang Y. S., Jeong D., Kim C., Kang H. W., et al. (2017). Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin. Epigenet. 9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter N. T., Hurban P., White M. N., Whitlock K. D., Lofton-Day C. E., Tetzner R., et al. (2014). Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 60 1183–1191. 10.1373/clinchem.2013.221044 [DOI] [PubMed] [Google Scholar]
- Sano Y., Byeon J. S., Li X. B., Wong M. C., Chiu H. M., Rerknimitr R., et al. (2016). Colorectal cancer screening of the general population in East Asia. Digest. Endoscopy 28 243–249. 10.1111/den.12579 [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2019). Cancer statistitistics, 2019. CACancer J. Clin. 69 7–34. [Google Scholar]
- Siegel R. L., Ward E. M., Jemal A. (2012). Trends in colorectal cancer incidence rates in the United States by tumor location and stage, 1992–2008. Cancer Epidemiol. Prevent. Biomark. 21 411–416. 10.1158/1055-9965.epi-11-1020 [DOI] [PubMed] [Google Scholar]
- Tang D., Liu J., Wang D. R., Yu H. F., Li Y. K., Zhang J. Q. (2011). Diagnostic and prognostic value of the methylation status of secreted frizzledrelated protein 2 in colorectal cancer. Clin. Investigat. Med. 34 88–95. [DOI] [PubMed] [Google Scholar]
- Timothy R. C., Wandell M., Lofton-Day C., Mongin S. J., Burger M., Payne S. R., et al. (2014). Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 63 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinmouth J., Lansdorp-Vogelaar I., Allison J. E. (2015). Faecal immunochemical tests versus guaiac faecal occult blood tests: what clinicians and colorectal cancer screening programme organisers need to know. Gut 64 1327–1337. 10.1136/gutjnl-2014-308074 [DOI] [PubMed] [Google Scholar]
- Wolf A. M., Fontham E. T., Church T. R., Flowers C. R., Guerra C. E., Lamonte S. J., et al. (2018). Colorectal cancer screening for average-risk adults: 2018 guideline update from the American cancer society. CA Cancer J. Clin. 68 250–281. 10.3322/caac.21457 [DOI] [PubMed] [Google Scholar]
- Wu D., Zhou G., Jin P., Zhu J., Li S., Wu Q., et al. (2016). Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J. Mol. Diagnost. 18 535–545. 10.1016/j.jmoldx.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Zhao G., Li H., Yang Z., Wang Z., Xu M., Xiong S., et al. (2019). Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 8 5619–5628. 10.1002/cam4.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.