Streptococcus suis is an important pathogen of pigs but is also transmissible to humans, with potentially fatal consequences. Among 29 serotypes currently recognized, some are clinically and epidemiologically more important than others. This is particularly true for serotypes 2 and 14, which have a large impact on pig production and also on human health. Conventional PCR-based serotyping cannot distinguish between serotype 1/2 and serotype 2 or between serotype 1 and serotype 14.
KEYWORDS: Streptococcus suis, cpsK, BstNI, diagnostics
ABSTRACT
Streptococcus suis is an important pathogen of pigs but is also transmissible to humans, with potentially fatal consequences. Among 29 serotypes currently recognized, some are clinically and epidemiologically more important than others. This is particularly true for serotypes 2 and 14, which have a large impact on pig production and also on human health. Conventional PCR-based serotyping cannot distinguish between serotype 1/2 and serotype 2 or between serotype 1 and serotype 14. Although serotype 1/2 and serotype 2 have a very similar cps locus, they differ in a single-nucleotide substitution at nucleotide position 483 of the cpsK gene. Similarly, serotypes 1 and 14 have a very similar cps locus but also differ in the same nucleotide substitution of the cpsK gene. Fortunately, this cpsK 483G→C/T substitution can be detected by BstNI restriction endonuclease. A PCR-restriction fragment length polymorphism (RFLP) detection method amplifying a fragment of the cpsK gene digested by BstNI restriction endonuclease was developed and tested in reference strains of these serotypes and also in field isolates.
INTRODUCTION
Streptococcus suis is a facultative anaerobic Gram-positive bacterium and is the causative agent of important diseases, particularly in pigs (1, 2), but it can also cause infections in humans and other animal species. The majority of human cases have been identified in Asia (90%), of which Vietnam, Thailand, and China alone accounted for 83.6% of all cases worldwide (3, 4).
Currently, 29 serotypes of S. suis are distinguished, whereas the previously described serotypes 20, 22, 26, 32, 33, and 34 are referred to as S. suis-like (5, 6). Information about the serotype is very important from an epidemiological point of view, as some serotypes have the ability to cause more severe disease than others, and only some serotypes (especially serotypes 2 and 14) cause human disease (4).
Methods traditionally used for serotyping are the agglutination and coagglutination tests using serotype-specific antisera (7, 8). Currently, PCR methods based on the detection of genes encoding capsular polysaccharide (CPS) production of individual serotypes have been developed (5, 9, 10). Unfortunately, PCR-based methods cannot distinguish serotype 2 from serotype 1/2 or serotype 1 from serotype 14 due to the similar gene content of their cps loci (5, 9). However, a single-nucleotide G→C/T substitution at nucleotide position 483 in the cpsK gene (11) causes preferential addition of galactose (in the case of serotypes 2 and 14) or N-acetylgalactosamine residues (in the case of serotypes 1/2 and 1) to the capsular polysaccharide repeating units (12). Identification of this mutation by reading the cpsK sequence or by a recently published method based on high-resolution melting analysis (13) enables specific identification of these serotypes. Here, we present a simple and low-cost method of detecting cpsK gene polymorphism based on PCR-restriction fragment length polymorphism (PCR-RFLP).
MATERIALS AND METHODS
Reference serotype strains were used as controls (14, 15). Another 214 field isolates were recovered from clinically ill pigs on farms in the Czech Republic. S. suis identification from each sample was confirmed by biochemical testing and by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Bruker Daltonik, Bremen, Germany).
All samples were tested serologically using antisera prepared by rabbit immunization with reference S. suis and S. suis-like serotype strains 1 to 34 from the S. suis strain collection at the University of Montreal (Quebec, Canada), according to a methodology originally described for serotyping Actinobacillus pleuropneumoniae (16) and S. suis isolates (8).
All samples were further tested by a multiplex PCR method detecting capsular polysaccharide synthesis genes (10). Each PCR consisted of a particular primer mix and PCR master mix (HotStarTaq Plus mastermix kit, Qiagen, Hilden, Germany). DNA for PCR was isolated by a rapid boiling method (17). PCR products were resolved on 2% agarose gel stained with ethidium bromide and visualized under UV light. Serotypes were determined according to the DNA band position (10). The multilocus sequence type (MLST) was determined according to https://pubmlst.org/ssuis/ (18).
To compare serological and multiplex PCR serotyping data, whole-genome sequences of all 35 reference serotypes and an additional 35 field isolates were acquired. The DNA of each sample was isolated by the QIAamp DNA minikit (Qiagen, Hilden, Germany). The sequencing library was prepared with the Nextera XT DNA library preparation kit (Illumina, San Diego, CA) and sequenced on a NextSeq 500 instrument (Illumina, San Diego, CA). Paired-end 2 × 150-bp reads were processed by the Tormes pipeline (19). The serotype of each sample was deduced from the assembled raw genome by comparing cps genes with cps genes corresponding to the known serotypes (5) using the BLAST tool (20). Serotypes 1/2 and 2 and serotypes 1 and 14 were further distinguished according to presence of substitution in nucleotide position 483 of the cpsK gene (11) by reading the cpsK sequence.
All field isolates belonging to serotypes 1, 2, 1/2, or 14 identified by the multiplex PCR method were further tested by both PCR-RFLP identifying substitution on nucleotide 483 of the cpsK gene (Table 1) and by Sanger sequencing (Eurofins, Ebersberg, Germany) of cpsK PCR products. A fragment of the cpsK gene was amplified with forward primer cpsK-F 5′-GTTGCTGGTTATGATAGGGTAG-3′ (11), reverse primer cpsK-R2 5′-AAGCTTCTTTTGCTGTTTGCTC-3′ (this study), and Taq-based PCR PPP mastermix (TopBio, Prague, Czech Republic) in a total volume of 10 μl. Cycling conditions were as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 15 s, 53°C for 30 s, and 72°C for 30 s. The final extension was 72°C for 10 min. A 5-μl aliquot of the resultant 486-bp product was digested with 10 U of BstNI restriction endonuclease (NEB, Ipswich, MA) in a total volume of 20 μl. The resulting products were resolved on 2% agarose gel stained with SimplySafe (EURx Ltd., Gdansk, Poland). In the case of the presence of nucleotide G at nucleotide position 483 of the cpsK gene (serotypes 2 and 14), corresponding to nucleotide 139 of the PCR product, the PCR product could be cleaved by BstNI into two fragments of 139 and 347 bp, respectively (11). In the case of nucleotide C or T at nucleotide position 483 of the cpsK gene (serotypes 1/2 and 1), corresponding to nucleotide 139 of the PCR product (11), the PCR product could not be cleaved, and this resulted in a single 486-bp band (Table 1 and Fig. 1).
TABLE 1.
Schematic representation of the workflow
| Step | Workflow | ||||
|---|---|---|---|---|---|
| 1 | Streptococcus suis multiplex PCR for cps gene typing | ||||
| 2 | Undifferentiated serotype 1/2 or 2 | Undifferentiated serotype 1 or 14 | Other serotypes detected by multiplex PCR | ||
| 3 | PCR-RFLP | PCR-RFLP | |||
| 4 | 486-bp band | 139-bp and 347-bp bands | 486-bp band | 139-bp and 347-bp bands | |
| 5 | Serotype 1/2 | Serotype 2 | Serotype 1 | Serotype 14 | |
FIG 1.
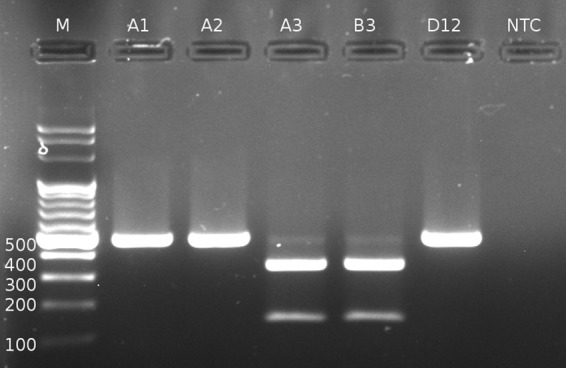
Representative picture of cpsK PCR-RFLP patterns. M, DNA marker, with the number representing fragment length in bp. Samples are indicated as follows: A1, serotype 1/2, 483T; A2, serotype 1, 483T; A3, serotype 2, 483G; B3, serotype 14, 483G; D12, 13/18/Ji/1670 = serotype 1/2, 483C; NTC, no-template control.
RESULTS
Determination of serotype by whole-genome sequencing (WGS) of reference serotypes and 35 field isolates was in accordance with serological and multiplex PCR serotyping. Based on this knowledge, an additional 179 field isolates were tested just by serological and multiplex PCR serotyping. All field isolates identified as serotype 1/2 or 2 and 1 or 14 were further tested by the above-described PCR-RFLP and by Sanger sequencing of cpsK PCR product. Out of 214 field isolates from the Czech Republic, seventeen were of serotype 2 (multilocus sequence type ST1 and ST28), fourteen were of serotype 1/2 (mainly ST28 but also ST1 and ST7), six were of serotype 1 (ST1 and ST11) and two of serotype 14 (ST13 and ST17). WGS data and/or Sanger sequencing of cpsK PCR product confirmed nucleotide G at 483 position of the cpsK gene for serotype 2 and 14 field isolates, while serotype 1/2 and 1 field isolates had C or T at this position (Table 2). All PCR-RFLP results were in 100% accordance with the sequence data.
TABLE 2.
Description of strains examined
| Isolate | Serotype detected bya
: |
cpsK sequenceb | Sequencing methodc | Date of isolation | Species | Age | Tissue | MLSTd | |
|---|---|---|---|---|---|---|---|---|---|
| Multi-PCR | PCR-RFLP | ||||||||
| S1 reference strain | 1 or 14 | 1 | ggtggcctgTaataaac | WGS | 13 | ||||
| S1/2 reference strain | 2 or 1/2 | 1/2 | ggtggcctgTaataaac | WGS | 56 | ||||
| S2 reference strain | 2 or 1/2 | 2 | ggtggcctgGaataaac | WGS | 1 | ||||
| S14 reference strain | 1 or 14 | 14 | ggtggcctgGaataaac | WGS | 6 | ||||
| 3/18/Ji/376 | 2 or 1/2 | 2 | ggtggcctgGaataaac | WGS, S | 16 January 2018 | Pig | 25 days | Lung | 28 |
| 10/18/Ji/830 | 2 or 1/2 | 2 | ggtggcctgGaataaac | WGS, S | 26 January 2018 | Pig | Lung | 28 | |
| 11/18/Ji/830 | 2 or 1/2 | 2 | ggtggcctgGaataaac | WGS, S | 26 January 2018 | Pig | Spleen | 28 | |
| 13/18/Ji/1670 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | WGS, S | 7 February 2018 | Pig | 40 days | Lung | 28 |
| 36/18/Ji/3895 | 2 or 1/2 | 1/2 | ggtggcctgTaataaac | S | 20 March 2018 | Pig | 14 days | Brain | 28 |
| 55/18/Ji/6714 | 1 or 14 | 1 | ggtggcctgTaataaac | S | 9 May 2018 | Pig | 7 days | Brain | 1 |
| 62/18/Ji/8065 | 2 or 1/2 | 1/2 | ggtggcctgTaataaac | S | 6 June 2018 | Pig | 28 days | Spleen | 7 |
| 63/18/Ji/7649 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | S | 6 June 2018 | Pig | Nasal swab | 28 | |
| 74/18/Ji/10270 | 1 or 14 | 1 | ggtggcctgTaataaac | S | 25 June 2018 | Pig | 10 days | Brain | 11 |
| 75/18/Ji/10270 | 1 or 14 | 1 | ggtggcctgTaataaac | S | 22 June 2018 | Pig | 10 days | Joint | 11 |
| 78/18/Ji/12432 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 9 August 2018 | Pig | 75 days | Lung | 1 |
| 94/18/Ji/13557 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 5 September 2018 | Pig | Lung | 28 | |
| 104/18/Ji/15350 | 2 or 1/2 | 1/2 | ggtggcctgTaataaac | S | 10 October 2018 | Pig | 14 days | Lymph node | 1 |
| 105/18/Ji/15351 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | S | 10 October 2018 | Pig | 20 days | Pericardium | 28 |
| 106/18/Ji/15351 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | S | 10 October 2018 | Pig | 20 days | Spleen | 28 |
| 107/18/Ji/15351 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | S | 10 October 2018 | Pig | 20 days | Lung | 28 |
| 119/18/Ji/16786 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | S | 8 November 2018 | Pig | 20 days | Joint | 28 |
| 120/18/Ji/16789 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | S | 8 November 2018 | Pig | 20 days | Lung | 28 |
| 124/18/Ji/18103 | 2 or 1/2 | 1/2 | ggtggcctgTaataaac | S | 5 December 2018 | Pig | 20 days | Nasal Swab | 28 |
| 125/18/Ji/18566 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 13 December 2018 | Pig | Weaned piglet | Pericardium | 1 |
| 126/18/Ji/18567 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 13 December 2018 | Pig | Weaned piglet | Brain | 1 |
| 131/18/Ji/18762 | 1 or 14 | 1 | ggtggcctgTaataaac | S | 19 December 2018 | Pig | 20 days | Brain | 1 |
| 132/18/Ji/18762 | 1 or 14 | 1 | ggtggcctgTaataaac | S | 19 December 2018 | Pig | 20 days | Lung | 1 |
| 141/19/JI/1352 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 22 January 2019 | Pig | 68 days | Lymph node | 1 |
| 142/19/JI/1353 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 22 January 2019 | Dog | Brain | 1 | |
| 143/19/JI/1352 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 22 January 2019 | Pig | 68 days | Brain | 1 |
| 144/19/JI/1625 | 1 or 14 | 14 | ggtggcctgGaataaac | S | 24 January 2019 | Pig | Fattening pig | Heart | 13 |
| 145/19/JI/1634 | 1 or 14 | 14 | ggtggcctgGaataaac | S | 25 January 2019 | Pig | Weaned piglet | Brain | 17 |
| 149/19/JI/2993 | 1 or 14 | 1 | ggtggcctgTaataaac | S | 29 January 2019 | Pig | 19 days | Lymph node | 1 |
| 156/19/JI/3589 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 19 February 2019 | Pig | 8 weeks | Lung | 28 |
| 159/19/JI/3959 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 28 February2019 | Pig | Lung | 28 | |
| 163/19/JI/4739 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 11 March 2019 | Pig | Fattening pig | Liver | 28 |
| 187/19/JI/10440 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 19 June 2019 | Pig | Fattening pig | Brain | 1 |
| 188/19/JI/10440 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 21 June 2019 | Pig | Fattening pig | Intestine | 1 |
| 189/19/JI/10440 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 24 June 2019 | Pig | Fattening pig | Lymph node | 1 |
| 203/19/JI/11813 | 2 or 1/2 | 1/2 | ggtggcctgCaataaac | S | 18 July 2019 | Pig | Weaned piglet | Lung | 28 |
| 211/19/JI/14336 | 2 or 1/2 | 1/2 | ggtggcctgTaataaac | S | 28 August 2019 | Chicken | 1 day | Yolk sac | 28 |
| 213/19/JI/14852 | 2 or 1/2 | 1/2 | ggtggcctgTaataaac | S | 4 September 2019 | Pig | 40 days | Heart | 28 |
| 214/19/JI/14881 | 2 or 1/2 | 2 | ggtggcctgGaataaac | S | 4 September 2019 | Pig | 55 days | Lung | 28 |
Multi-PCR, multiplex PCR for detecting capsular polysaccharide synthesis genes (10).
Nucleotide position 483 of the cpsK gene is indicated in uppercase, and the BstNI recognition site is underlined.
WGS, whole-genome sequence; S, Sanger sequence of cpsK PCR product.
MLST, multilocus sequence type.
Digestion of the cpsK PCR product of reference strains and isolate number 13/18/Ji/1670 with BstNI and subsequent electrophoresis on agarose gel led to the presence of 486-bp undigested PCR product in the case of serotypes 1/2 and 1 (samples were as follows: A1, serotype 1/2, 483T; A2, serotype 1, 483T; D12, 13/18/Ji/1670 = serotype 1/2, 483C) and two bands of 139 bp and 347 bp in the case of serotype 2 and 14 (samples were as follows: A3, serotype 2, 483G; B3, serotype 14, 483G) (Fig. 1).
DISCUSSION
Identification of S. suis serotypes by serological methods is still considered the standard, but the development of multiplex PCR typing methods detecting particular genes in the cps locus of the known S. suis and S. suis-like serotypes made serotyping more convenient and enabled resolution of dubious results occasionally obtained by serological typing (12). Unfortunately, serotypes 1/2 and 2 share highly similar genes in the cps cluster. The same applies to serotypes 1 and 14, and thus detection of the serotype-specific cps gene cannot distinguish these serotypes (5, 9). However, the discovery of a causal 483G→C/T mutation in the cpsK gene (11) enables clear identification of these serotypes.
Detection of cpsK 483G→C/T gene polymorphism is possible by Sanger sequencing of cpsK PCR product or by whole-genome sequencing. Although this approach is accurate, it is time consuming and probably not available for all diagnostic laboratories. Recently, a method based on high-resolution melting analysis of cpsK PCR product was reported (13). This method could be quick and reliable in laboratories equipped with a quantitative PCR (qPCR) apparatus. Here, we describe a PCR-RFLP-based method detecting cpsK 483G→C/T substitution by BstNI restriction endonuclease. The presence of one 486-bp band, in the case of serotypes 1/2 and 1, or of two bands of 139 bp and 347 bp, in the case of serotype 2 and 14, can easily be resolved by agarose gel electrophoresis. PCR-RFLP results were 100% consistent with WGS and/or Sanger sequences of the tested 43 strains of serotype 1/2, 1, 2, and 14. As additional information in this report, although we found no exclusive association between a particular serotype and sequence type (ST), some ST data are interesting. As expected for European strains, potentially zoonotic ST1 serotype 2 strains were identified (3). However, the finding of serotype 1/2 ST1 or ST7 strains was unexpected. A serotype 2 ST7 strain was responsible for the deadly outbreak in humans in China in 2005 (21). More recently, serotype 14 ST7 strains were also described in China (22). This is the first report on serotype 1/2 ST7 strains, and the zoonotic potential of such strains should be further studied.
In conclusion, the PCR-RFLP method described here is relatively inexpensive, rapid, and simple but still highly reliable, and it requires just basic laboratory equipment.
ACKNOWLEDGMENTS
This study was supported by the Ministry of Agriculture of the Czech Republic by grants NAZV QK1810193 and RO0518.
We thank Eva Audova and Romana Ondriasova for their technical assistance. We also thank Ludmila Faldikova for proofreading the manuscript.
REFERENCES
- 1.Power SB. 1978. Streptococcus suis type 2 infection in pigs. Vet Res 102:215–216. [DOI] [PubMed] [Google Scholar]
- 2.Gottschalk M. 2014. Streptococcus suis infection. Merck veterinary manual. Merck & Co, Kenilworth, NJ. [Google Scholar]
- 3.Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. 2014. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microb Infect 3:e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutkiewicz J, Sroka J, Zając V, Wasiński B, Cisak E, Sawczyn A, Kloc A, Wójcik-Fatla A. 2017. Streptococcus suis: a re-emerging pathogen associated with occupational exposure to pigs or pork products. Part I—Epidemiology. Ann Agric Environ Med 24:683–695. doi: 10.26444/aaem/79813. [DOI] [PubMed] [Google Scholar]
- 5.Okura M, Lachance C, Osaki M, Sekizaki T, Maruyama F, Nozawa T, Nakagawa I, Hamada S, Rossignol C, Gottschalk M, Takamatsu D. 2014. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J Clin Microbiol 52:1714–1719. doi: 10.1128/JCM.03411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okura M, Osaki M, Nomoto R, Arai S, Osawa R, Sekizaki T, Takamatsu D. 2016. Current taxonomical situation of Streptococcus suis. Pathogens 5:45. doi: 10.3390/pathogens5030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins R, Gottschalk M. 1990. An update on Streptococcus suis identification. J Vet Diagn Invest 2:249–252. doi: 10.1177/104063879000200324. [DOI] [PubMed] [Google Scholar]
- 8.Gottschalk M, Higgins R, Boudreau M. 1993. Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J Clin Microbiol 31:2192–2194. doi: 10.1128/JCM.31.8.2192-2194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Zheng H, Gottschalk M, Bai X, Lan R, Ji S, Liu H, Xu J. 2013. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS One 8:e72070. doi: 10.1371/journal.pone.0072070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerdsin A, Akeda Y, Hatrongjit R, Detchawna U, Sekizaki T, Hamada S, Gottschalk M, Oishi K. 2014. Streptococcus suis serotyping by a new multiplex PCR. J Med Microbiol 63:824–830. doi: 10.1099/jmm.0.069757-0. [DOI] [PubMed] [Google Scholar]
- 11.Athey TB, Teatero S, Lacouture S, Takamatsu D, Gottschalk M, Fittipaldi N. 2016. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol 16:162. doi: 10.1186/s12866-016-0782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy D, Athey TBT, Auger JP, Goyette-Desjardins G, Van Calsteren MR, Takamatsu D, Okura M, Teatero S, Alcorlo M, Hermoso JA, Segura M, Gottschalk M, Fittipaldi N. 2017. A single amino acid polymorphism in the glycosyltransferase CpsK defines four Streptococcus suis serotypes. Sci Rep 7:4066. doi: 10.1038/s41598-017-04403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherrer S, Rademacher F, Spoerry Serrano N, Schrenzel J, Gottschalk M, Stephan R, Landolt P. 2020. Rapid high resolution melting assay to differentiate Streptococcus suis serotypes 2, 1/2, 1, and 14. Microbiologyopen 9:e995. doi: 10.1002/mbo3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perch B, Pedersen KB, Henrichsen J. 1983. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J Clin Microbiol 17:993–996. doi: 10.1128/JCM.17.6.993-996.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottschalk M, Higgins R, Jacques M, Mittal KR, Henrichsen J. 1989. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol 27:2633–2635. doi: 10.1128/JCM.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittal KR, Higgins R, Lariviere S. 1983. Identification and serotyping of Haemophilus pleuropneumoniae by coaglutination test. J Clin Microbiol 18:1351–1353. doi: 10.1128/JCM.18.6.1351-1354.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aslam M, Hogan J, Smith KL. 2003. Development of a PCR-based assay to detect Shiga toxin-producing Escherichia coli, Listeria monocytogenes, and Salmonella in milk. Food Microbiol 20:345–350. doi: 10.1016/S0740-0020(02)00121-1. [DOI] [Google Scholar]
- 18.Jolley KA, Bray JE, Maiden M. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quijada NM, Rodríguez-Lázaro D, Eiros JM, Hernández M. 2019. TORMES: an automated pipeline for whole bacterial genome analysis. Bioinformatics 35:4207–4212. doi: 10.1093/bioinformatics/btz220. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Ye C, Bai X, Zhang J, Jing H, Zheng H, Du H, Cui Z, Zhang S, Jin D, Xu Y, Xiong Y, Zhao A, Luo X, Sun Q, Gottschalk M, Xu J. 2008. Spread of Streptococcus suis sequence type 7, China. Emerg Infect Dis 14:787–791. doi: 10.3201/eid1405.070437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M, Du P, Wang J, Lan R, Huang J, Luo M, Jiang Y, Zeng J, Quan Y, Shi Z, Zheng H. 2019. Genomic epidemiology of Streptococcus suis sequence type 7 sporadic infections in the Guangxi Zhuang Autonomous Region of China. Pathogens 8:187. doi: 10.3390/pathogens8040187. [DOI] [PMC free article] [PubMed] [Google Scholar]


