Abstract
Purpose
To report a severe phenotype of retinitis pigmentosa associated with novel mutations in CNGB1.
Observations
Six siblings, age range 50–75 years old, were examined using optical coherence tomography and fundus autofluorescene, electroretinogram testing, Goldman visual field testing, and genetic testing using next generation sequencing.
In four affected siblings, two novel compound heterozygous variants in CNGB1 were detected: in exon 26 the missense variant c.2603G > A (p.(Gly868Asp)), and in exon 21, the in-frame 12-bp duplication c.2093_2104dupGCGACCTCATCT (p.(Cys698_lle701dup)). One sibling was unaffected and carried neither of the variants, while another sibling had mild macular degeneration changes and carried the latter variant in heterozygous status. The affected siblings presented with a phenotype showing markedly constricted visual field, flat scotopic and photopic electroretinogram responses and generalized retinal atrophy.
Conclusions and importance
This is the first report of a 12bp in-frame duplication and a missense variant (in compound heterozygous status) in CNGB1, being associated with a severe form of retinitis pigmentosa featuring extensive peripheral and central retinal degeneration. This study expands the molecular genetic basis of CNGB1-related disease.
Keywords: Retinitis pigmentosa, Rod-cone dystrophy, CNGB1, Novel variant
1. Introduction
Retinitis pigmentosa (RP) represents a heterogeneous group of inherited disorders characterized by progressive retinal dystrophy affecting primarily the rod photoreceptors. It affects approximately 1 in 4000 individuals worldwide.1 Typically, RP patients suffer from night blindness, progressive visual field (VF) constriction, and eventually reduction of central vision. The classical fundal picture in RP cases shows mid-peripheral intra-retinal pigment migration, retinal pigment epithelium (RPE) atrophy, attenuated retinal vessels and pale optic disc.
Genes associated with RP encode for proteins involved in the retinal structure and function including phototransduction. Cyclic nucleotide-gated channel beta 1 (CNGB1, OMIM: 600724) encodes the B-subunit of the cyclic nucleotide gated channel (CNGB1a) in the plasma membranes of rods.2 CNGB1 in photoreceptors translate the light mediated changes of the second messenger cyclic Guanosine Monophosphate (cGMP) into voltage signals.3 CNGB1a is important for the trafficking of the channel to the outer segment, its positioning within the cell membrane, and control of the channel's temporal activity. Binding of cGMP in the dark results in transient opening of the CNG channel and rod depolarization. With light-induced activation of the phototransduction, there is a reduction in cGMP levels mediated by the G protein activation of a phosphodiesterase (PDE6). Reduced cGMP leads to closure of the channels and hyperpolarization of the rod. Variants in CNGB1 are uncommon causes of RP, accounting for about 4% of autosomal recessive cases.4,5
In this paper we present a case report of severe RP phenotype associated with 2 novel mutations in the CNGB1 gene.
2. Case report
This is a retrospective case report study. The study adheres to the tenets of the declaration of Helsinki. Ethical approval was obtained from the IRB of KKESH (King Khaled Eye Specialist Hospital). Consent for genetic testing and conduction of the study was obtained from the index patient and his siblings.
A 66-year-old male of Saudi-Syrian origin was referred to KKESH because of poor vision in both eyes and fundal abnormalities. The patient confirmed that both of his parents had no visual problems at all until their death. He has two affected brothers and one affected sister, and eight healthy siblings. The patient complained of night blindness since early childhood (around the age of 6). In his adult life, he experienced gradual constriction of the visual field, which became more noticeable around the age of 30. He had bilateral sequential cataract extraction and intraocular lens implantation at the age of 42 and 43, respectively. A similar history of progressive night blindness and visual field constriction was evident in his affected siblings. Two of his asymptomatic siblings were available for parallel examination.
We performed full ophthalmic examination in the 4 affected and 2 unaffected siblings. Color fundus photos were obtained using Optos fundus camera (Optos 200TX). Fundus autofluorescence (AF) imaging was performed using wide field Optos system (Optos 200TX) with 488 nm wavelength. Retinal structure was analyzed qualitatively with transfoveal horizontal spectral domain optical coherence tomography scans (OCT, Heidelberg Engineering, Inc., Heidelberg, Germany).
Retinal function was evaluated with full-field electroretinography (ffERG, Nicolet Biomedical Instruments, Madison, Wisconsin, USA), in dark adapted and light adapted state according to ISCEV standards,6 with a few modifications as follows. Full-field electroretinograms were recorded in a Nicolet analysis system (Nicolet Biomedical Instruments, Madison, Wisconsin, USA), after dark adaptation of subjects for 40 min, dilatation of the pupils with topical cyclopentolate 1% and metaoxedrine 2,5% and topical anaesthesia, with a Burian Allen bipolar contact lens and a ground electrode applied to the forehead. Responses were obtained stimulating with single full-field flash (30 ms) with blue light (0.81 cd-s/m2: rod response) and with white light (10.02 cd-s/m2: combined rod-cone response). Photopic responses were obtained with a background illumination of 3.4 cd-s/m2 in order to saturate the rods.
Molecular genetic testing was performed for the index patient using next-generation sequencing (NGS, performed by Center for Human Genetics Bioscientia, Ingelheim, Germany). 90 genes known to be involved in RP (autosomal dominant, autosomal recessive and X-linked) were investigated.
Genomic DNA was fragmented, and the exons of the analyzed genes as well as the corresponding exon-intron boundaries were enriched using Roche/Nimble Gene sequence capture approach, amplified and sequenced simultaneously by Illumina technology using an Illumina HiSeq1500 system. For 99% of the regions of interest a 20-fold coverage was obtained. NGS data analysis was performed using bioinformatic prediction programs as well as JSI Medical Systems software (version 4.1.2). Identified variants and indels were filtered against external and internal databases and filtered depending on their allele frequency focusing on rare variants with a minor allele frequency (MAF) of 1% or less (ExAC and gnomAD browser, respectively). Nonsense, frameshift and canonical splice site variants were primarily considered likely pathogenic. Assessment of pathogenicity of identified non-synonymous variants was performed using bioinformatic prediction programs (SIFT, PolyPhen-2 HumVar, Mutation Taster, Mutation Assessor, FATHMM, LRT, VEST, CADD). Only those variants were considered likely pathogenic which were predicted probably damaging by the majority of the used algorithms. Variants that have been annotated as common polymorphisms in databases or in the literature were not considered further.
Putatively pathogenic differences between the wild type sequence (human reference genome according to UCSCGenomeBrowser: hg19,GRCh37) and the proband's sequence were validated using polymerase chain reaction (PCR) amplification followed by conventional Sanger sequencing. Sequence data for the CNGB1 gene (OMIM *600724; Locus: Chromosome 16q21) were compared to the reference sequence NM_001297.4, for the ADAMTS18 gene (OMIM *607512; Locus: Chromosome 16q23.1) to the reference sequence NM_199355.2????. Genetic analysis of the 3 affected and 2 unaffected siblings of the index patient was carried out by direct sequencing of the same variants in CNGB1 (and ADAMTS18, respectively.).
3. Results
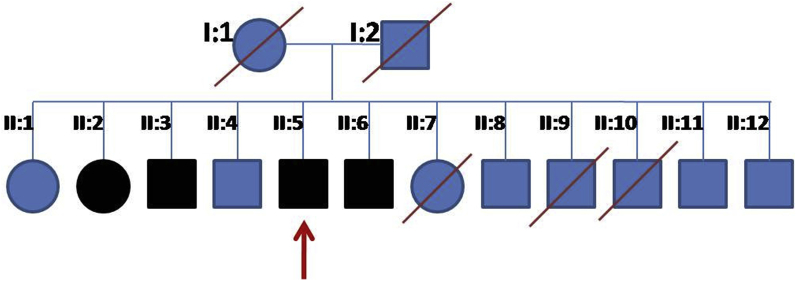
The index patient is a 66-year-old Saudi-Syrian descendant. His family tree is shown in Fig. 1. Molecular genetic testing revealed 2 novel heterozygous variants affecting the CNGB1 gene. The missense variant c.2603G > A leads to the amino acid change p.(Gly868Asp). To the best of our knowledge, this variant has not been described in the literature so far. It has only been found in 1 out of 120726 alleles in the ExAC database (minor allele frequency: 8x10−4%). All eight bioinformatic analysis tools used predict this alteration to be pathogenic. The second CNGB1 variant is an in-frame 12-bp duplication (c.2093_2104dupGCGACCTCATCT)which leads to the duplication of four amino acids (p.(Cys698_lle701dup)). As far as we know, this variant has neither been annotated in databases nor been described in the literature so far. Both variants have been confirmed by conventional Sanger sequencing. All three likewise affected siblings carry both variants. . One of the other siblings (64-year old brother), who had early AMD changes in the macula, carries only the frameshift variant in heterozygous state but not the missense variant. In the other (non-affected) sibling (55-year old brother with normal fundus) none of the variants were detected. Considering the available data and the segregation information, both variants are classified as pathogenic.
Fig. 1.
The family pedigree of a 66-year-old male with severe RP associated with a heterozygous missense variant c.2603G > A (p.Gly868Asp) in exon 26 of CNGB1 and a heterozygous in-frame 12-bp duplication c.2093_2104dupGCGACCTCATCT (p.Cys698_lle701dup) in exon 21of CNGB1variants.
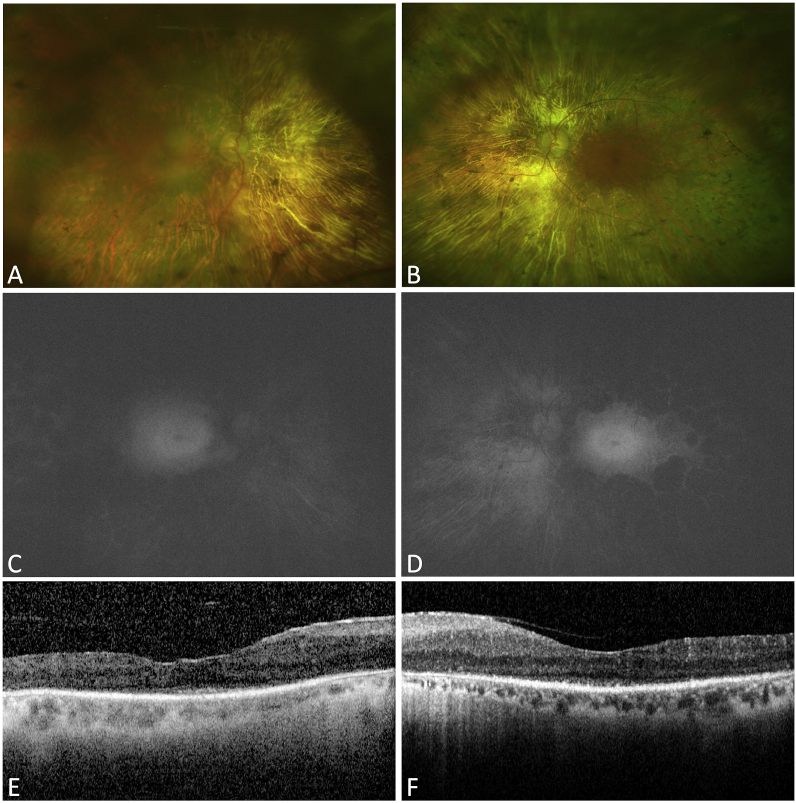
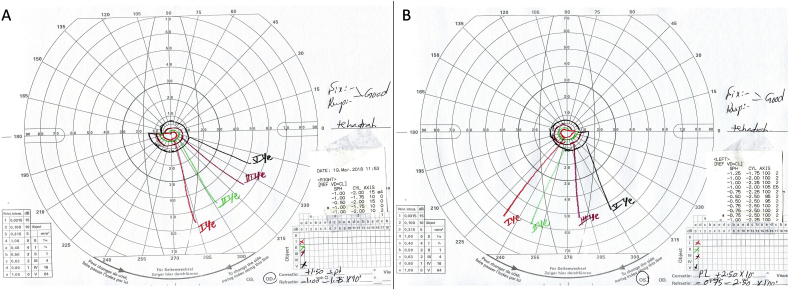
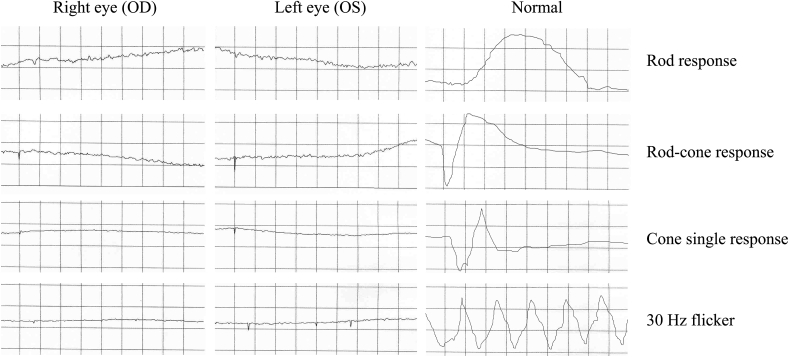
Visual acuity was 20/100 in the right eye and 20/80 in the left eye. Fundal examination showed bilateral widespread RPE atrophy with attenuated retinal vessels and pale optic discs (Fig. 2A and B). AF imaging (Fig. 2C and D) showed a small preserved central island in both eyes (about 3 disc diameters) with high AF signal and loss of AF signal outside that area. OCT in both eyes (Fig. 2E and F) demonstrates central preservation of outer retina and loss of ellipsoide zone outside that. Kinetic Goldmann VF testing showed a constricted VF down to 10° in both eyes with size IV target (Fig. 3). FfERG testing showed flat photopic and scotopic responses in both eyes (Fig. 4).
Fig. 2.
Imaging in a 66-year-old male with severe RP associated with a heterozygous missense variant c.2603G > A (p.Gly868Asp) in exon 26 of CNGB1 and a heterozygous in-frame 12-bp duplication c.2093_2104dupGCGACCTCATCT (p.Cys698_lle701dup) in exon 21of CNGB1variants. A&B: Color fundus photos of the right and left eye. Both eyes show extensive RPE atrophy, intra-retinal pigment migration, attenuated retinal vessels, and pale optic discs. C&D: AF imaging of the right and left eye showing a small preserved island of about 3 disc diameters with loss of signal outside that island. E&F: OCT horizontal cut through the fovea of the right and left eye showing a small central zone of preserved inner segment ellipsoid (ISe) (the area between the arrows) and loss of the ISe outside that area. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Goldman visual field test in of a 66-year-old male with severe RP associated with a heterozygous missense variant c.2603G > A (p.Gly868Asp) in exon 26 of CNGB1 and a heterozygous in-frame 12-bp duplication c.2093_2104dupGCGACCTCATCT (p.Cys698_lle701dup) in exon 21of CNGB1variants. Note the severe constriction of the VF down to 10° with size IV object.
Fig. 4.
ffERG results in of a 66-year-old male with severe RP associated with a heterozygous missense variant c.2603G > A (p.Gly868Asp) in exon 26 of CNGB1 and a heterozygous in-frame 12-bp duplication c.2093_2104dupGCGACCTCATCT (p.Cys698_lle701dup) in exon 21of CNGB1variants. OD = Right eye. OS = Left eye. N = Normal age matched result.
All affected siblings show variable constricted visual field (≤20° with the largest object V:4) and non-recordable ERGs. OCT demonstrated very limited preservation of the photoreceptor layer and the absence of this layer outside a small central zone. Among the 2 unaffected siblings, one had normal fundi on examination and imaging The other one, who carried only one of the 2 compound heterozygous CNGB1 variants, shows changes/degeneration around the macula with normal retinal periphery.
4. Discussion
In the reported case here, severe RP phenotype was evident with a tunnel VF. Clinically, the small central island on AF probably corresponds to the remaining functional area at the macular centre. The flat ffERG responses fits with the severe loss of photoreceptor function and visual field restriction. The proband is 66 year old which could, at least partially, explain the severity of the disease at that point. However, the effect of the patient's genotype cannot be excluded. The 3 affected siblings showed similar phenotype as well. It has been found that CNGB1 related RP is generally not an aggressive form of the disease with well-preserved central vision till the fourth decade.7,8 Hull et al. identified a previously reported missense variant (p.[N986I]) and 7 variants not previously reported in disease including 4 nonsense (p.[(Q88*], p.[Q222*], p.[Q318*], and p.[R729*]), 2 frameshift (p.[A1048fs*13], p.[L849Afs*3]), and a splice site variant (c.761 + 2T > A).7 On the other hand, we speculate that the generalized severe phenotype observed here may resulted from the complex novel 12-bp duplication c.2093_2104dupGCGACCTCATCT, possibly leading to RP rather through protein misfolding or aggregation than (only) CNG channel dysfunction. CNGB1 is important for the trafficking of the channel to the outer segment, its positioning within the cell membrane, and control of the channel's temporal activity.
In a recent study, CNGB1-deficient RP patients and mouse and dog models had a phenotype characterized by early loss of rod function and slow rod loss with a secondary decline in cone function. Imaging showed promise for evaluating RP progression in human patients, and gene augmentation using adeno-associated virus vectors robustly sustained the rescue of rod function and preserved retinal structure in a dog model. An early loss of rod function in CNGB1-deficient patients and a wide window for therapeutic intervention was demonstrated.8
5. Conclusions
The amount of available literature on CNGB1 related RP is limited. Accordingly, this case adds useful information to the available literature. The 2 novel variants found in this case expands our knowledge regarding the molecular genetics of this category of RP. The severe phenotype reported here helps to widen the spectrum of RP related to CNGB1 mutations.
Patients’ consent
Written consent to publish case details has been obtained from each patient.
Funding
No funding was received for this work.
Declaration of competing interestCOI
The authors have no conflict of interest to declare.
Authorship
All authors attest that they meet the current ICMJE criteria for authorship.
Acknowledgements
None.
References
- 1.Hartong D.T., Berson E.L., Dryja T.P. Retinitis pigmentosa. Lancet. 2006 Nov 18;368(9549):1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 2.Zheng J., Trudeau M.C., Zagotta W.N. Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron. 2002 Dec 5;36(5):891–896. doi: 10.1016/s0896-6273(02)01099-1. [DOI] [PubMed] [Google Scholar]
- 3.Kaupp U.B., Seifert R. Cyclic nucleotide-gated ion channels. Physiol Rev. 2002 Jul;82(3):769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 4.Bocquet B., Marzouka N.A., Hebrard M. Homozygosity mapping in autosomal recessive retinitis pigmentosa families detects novel mutations. Mol Vis. 2013 Dec 8;19:2487–2500. [PMC free article] [PubMed] [Google Scholar]
- 5.Bareil C., Hamel C.P., Delague V., Arnaud B., Demaille J., Claustres M. Segregation of a mutation in CNGB1 encoding the beta-subunit of the rod cGMP-gated channel in a family with autosomal recessive retinitis pigmentosa. Hum Genet. 2001 Apr;108(4):328–334. doi: 10.1007/s004390100496. [DOI] [PubMed] [Google Scholar]
- 6.Marmor M.F., Zrenner E. Standard for clinical electroretinography. Doc Ophthalmol. 1995;97:143–156. doi: 10.1023/a:1002016531591. [DOI] [PubMed] [Google Scholar]
- 7.Hull S., Attanasio M., Arno G. Clinical Characterization of CNGB1-Related Autosomal Recessive Retinitis Pigmentosa. JAMA Ophthalmol. 2017 Feb;135(2):137–144. doi: 10.1001/jamaophthalmol.2016.5213. [DOI] [PubMed] [Google Scholar]
- 8.Petersen-Jones S.M., Occelli L.M., Winkler P.A. Patients and animal models of CNGβ1-deficient retinitis pigmentosa support gene augmentation approach. J Clin Invest. 2018 Jan 2;128(1):190–206. doi: 10.1172/JCI95161. [DOI] [PMC free article] [PubMed] [Google Scholar]