Abstract.
Purpose: Our paper contributes to the burgeoning field of surgical data science. Specifically, multimodal integration of relevant patient data is used to determine who should undergo a complex pancreatic resection. Intraductal papillary mucinous neoplasms (IPMNs) represent cystic precursor lesions of pancreatic cancer with varying risk for malignancy. We combine previously defined individual models of radiomic analysis of diagnostic computed tomography (CT) with protein markers extracted from the cyst fluid to create a unified prediction model to identify high-risk IPMNs. Patients with high-risk IPMN would be sent for resection, whereas patients with low-risk cystic lesions would be spared an invasive procedure.
Approach: Retrospective analysis of prospectively acquired cyst fluid and CT scans was undertaken for this study. A predictive model combining clinical features with a cyst fluid inflammatory marker (CFIM) was applied to patient data. Quantitative imaging (QI) features describing radiomic patterns predictive of risk were extracted from scans. The CFIM model and QI model were combined into a single predictive model. An additional model was created with tumor-associated neutrophils (TANs) assessed by a pathologist at the time of resection.
Results: Thirty-three patients were analyzed (7 high risk and 26 low risk). The CFIM model yielded an area under the curve (AUC) of 0.74. Adding the QI model improved performance with an AUC of 0.88. Combining the CFIM, QI, and TAN models further increased performance to an AUC of 0.98.
Conclusions: Quantitative analysis of routinely acquired CT scans combined with CFIMs provides accurate prediction of risk of pancreatic cancer progression. Although a larger cohort is needed for validation, this model represents a promising tool for preoperative assessment of IPMN.
Keywords: radiomics, cyst fluid, quantitative image analysis, intraductal papillary mucinous neoplasms, pancreas
1. Introduction
Intraductal papillary mucinous neoplasms (IPMNs) of the pancreas are epithelial tumors of mucin-producing cells that arise from pancreatic ducts and may involve the main pancreatic duct (MD-IPMN), branch duct (BD-IPMN), or both (mixed-type IPMN). They have been diagnosed with increasing frequency in recent years due to increased awareness of the entity, increased frequency of imaging, and improved imaging techniques.1,2 These cystic lesions are believed to be precursor lesions to pancreatic cancer and account for 20% to 30% of pancreatic adenocarcinomas, demonstrating a pathway of progression from low-grade to high-grade dysplasia and eventually invasive adenocarcinoma.3–5 However, this progression to malignancy is only seen in a very small percentage of these cysts, and the appropriate management of these cystic lesions represents a growing clinical challenge. Performing opportune resection of high-grade dysplastic lesions before their progression to invasive disease and avoidance of overtreating benign lesions with minimal dysplasia with an invasive surgery is the optimal clinical scenario. One study following patients with nonresected IPMN found that the cumulative probability of developing pancreatic cancer was 5.7% at 5 years and 10.7% at 10 years.6
To date, the clinical management of IPMN relies on several guidelines based on laboratory, endoscopic, cytologic, and imaging findings to properly select patients for surgery. However, their ability to distinguish between IPMNs that represent low-risk disease (low- and intermediate-grade dysplasia) or high-risk disease (high-grade dysplasia or invasive carcinoma) is still not satisfactory.7,8 Of these techniques, the greatest importance is placed on the presence of main pancreatic duct dilatation on preoperative imaging (CT or MRI). Current consensus guidelines recommend the resection of IPMN with a dilated main duct of (MD-IPMN), and of these will have high-grade dysplasia or invasive disease.9–11 In the absence of a dilated duct, resection of BD-IPMN yields a much lower frequency of high-grade or invasive disease, seen in only 10% to 25% of resected lesions.9,12 Even in highly specialized centers, 32% to 38% of patients having MD-IPMN and 73% to 80% having BD-IPMN undergo unnecessary surgery.13 The development of pancreatic cancer in patients with nonresected BD-IPMN has been reported to be between 2.5% and 8.3% over follow-up periods of 4 to 6 years.6,14–16 This compares with a 46% cumulative risk in nonresected MD-IPMN over 5 years.6 With the increasing use of cross-sectional imaging, incidental findings of pancreatic cystic lesions are rising with cysts identified on 2.6% of all CT scans, underscoring the urgent need for biomarkers of pancreatic cancer risk.1,2
Nomograms have recently emerged as a useful prognostic tool capable of providing an individualized risk score for a specific outcome. They have the advantage of inputting multiple variables proven to be prognostic, facilitating the integration of many different clinical information. Our group has investigated and published validated nomograms for the prediction of high-risk disease in patients with IPMN based on predefined clinical and imaging features included in guidelines.17,18 Clinical data used included the presence of specific gastrointestinal (GI) symptoms, weight loss and jaundice, and laboratory results such as carbohydrate antigen (CA) 19-9. Imaging features included cyst size, main duct dilatation, and the presence of mural nodules or solid components.
However, recently other important biomarkers not included in those models have emerged as useful for distinguishing high-risk versus low-risk IPMN. Links between inflammation and tumor progression have been described in cancer research, particularly neutrophil infiltration and inflammatory mediators released by these cells promoting tumorigenesis.19 Our group has described an association between tumor-associated neutrophils (TANs) and high-risk IPMNs with most low-risk lesions being negative for the presence of TANs.20,21 However, the presence of TANs can only be determined by pathology on specimen received postresection. A surrogate for the presence of TANs may be found in the presence of cyst fluid inflammatory proteins. Work has been done on analysis of cyst fluid protein markers on resected IPMNs and has identified patterns of cyst fluid protein expression and inflammatory markers between high-risk and low-risk lesions.21–23 Building on this work, cyst fluid inflammatory marker (CFIM) models were created and validated and, when combined with the additional clinical and radiographic information, further increased prediction accuracy for high-risk IPMN.24
More recently, radiomics has emerged in oncologic imaging to describe tumor heterogeneity, which can represent underlying tumor pathophysiology and predict clinical outcomes. A number of studies have evaluated the use of quantitative imaging (QI) in IPMN-risk assessment, demonstrating a potential in the prediction of malignancy or high-risk disease within lesions.25–27 A QI prediction model was created by our group using pretreatment CT scans. Results demonstrated that QI features outperformed the combination of clinical features and qualitative radiographic assessments. Results further improved when the quantitative (radiomic) features were combined with clinical and qualitative radiographic features.28
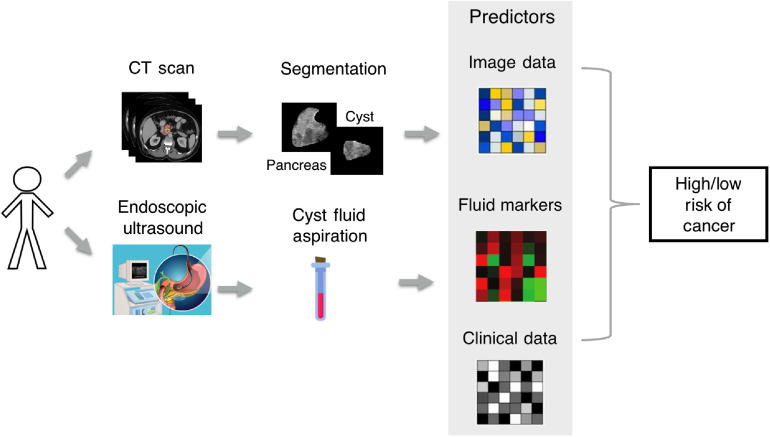
The primary aim of this study is to create a preoperative model for predicting high risk in IPMN using previously defined and validated predictive models created by this group. We hypothesize that combining clinical, qualitative radiographic, QI features, and CFIMs will improve accuracy in the identification of high-risk patients. We propose combining routine diagnostic imaging with cyst fluid aspirate available by endoscopic ultrasound (Fig. 1) as a nonsurgical tool for monitoring IPMN. We also investigate a second model created using pathologic information available postoperatively for comparison.
Fig. 1.
Clinical workflow for noninvasive assessment of risk in IPMN.
2. Methods
2.1. Patient Selection
Following institutional review board approval and using a prospectively maintained institutional database, patients with resected BD-IPMN or MD-IPMN who had cyst fluid available for analysis and preoperative dedicated CT imaging of the pancreas were identified. This resulted in the inclusion of 33 patients in this retrospective study. Patients resected for recurrent IPMN or with concurrent malignancies, such as neuroendocrine tumor or cholangiocarcinoma, were excluded. Demographic and clinical data and laboratory, radiographic, and pathologic features were obtained from the database. GI-specific symptoms as well as a history of jaundice and weight loss were noted. Preoperative laboratory results recorded include serum bilirubin, amylase, CA 19-9, and carcinoembryonic antigen (CEA). Resected lesions were assessed by dedicated GI pathologists. Risk was assigned based on the highest grade of dysplasia noted within the specimen by a pathologist at the time of resection. Patients were classified as “low-risk” in lesions with low-grade or intermediate-grade dysplasia and “high-risk” in lesions with high-grade dysplasia or invasive adenocarcinoma.
2.2. Cyst Fluid Analysis
Cyst fluid samples used in the analysis were obtained at the time of surgery and analyzed on commercially available plates as outlined in detail in a paper published previously by Al Efishat et al.24 Protein concentrations of cyst fluid markers that were used included MMP9, CA72-4, sFASL, and IL-4. The predictive models upon which the cyst fluid nomogram was created used multivariate logistic regression to assess relationships between cyst protein markers and high-risk disease, as assessed and validated on first training and then testing datasets. The data were randomly split into a training and a testing dataset, with no overlap between the two sets. The resultant nomogram was then applied to this patient cohort.24
2.3. Image Acquisition
Standard contrast-enhanced pretreatment CT scans were utilized in the study. Acquisition parameters were as follows. Following the administration of 150 ml iodinated contrast (Omnipaque 300, GE Healthcare, New Jersey), postcontrast CT images were acquired at using a multidetector CT (Lightspeed 16 and VCT, GE Healthcare, Wisconsin). Pitch and table speed were 0.984 to 1.375 and 39.37 to 27.50 mm, respectively, with the remaining variables set to autoMA 220 to 380, noise index 12.5 to 14.0, rotation time 0.7 to 0.8 ms, and scan delay 80 to 85 s. Axial slices from the portal venous phase reconstructed at 2.5 mm intervals were used for analysis.
2.4. Extraction of Quantitative Imaging Features
The cyst region was manually segmented on axial slices using Scout Liver (Pathfinder Technologies Inc., Analogic Corporation, Peabody, Massachusetts) by an expert radiologist. Twelve texture features and one radiographically inspired feature (RiF), which show significant association with IPMN risk in a previous study, were extracted from the segmented cyst region as outlined in a paper published previously by our group, under Attiyeh et al.28 Local binary patterns (LBP) characterize the spatial relationship among neighboring pixels intensity values, where histograms of uniform LBP (ULBP) (LBP2, LBP37, and LBP47), rotation invariant LBP (RI-LBP) (LBP62), and Fourier descriptors extracted from the RI-LBP histogram (LBP107, LBP108, LBP-109, LBP110, LBP111, LBP113, and LBP115) were used in this study.28 Further, angle co-occurrence matrices (ACM) quantify the directional edge patterns of the cyst, with ACM2_5 utilized because it carries both angle and magnitude information. To obtain rotation invariant features, this ACM was calculated in four directions (0 deg, 45 deg, 90 deg, and 135 deg).29 The RiF was developed specifically to represent the appearance of an enhancing mural nodule present within a cyst, which is considered one of the high-risk stigmata in consensus guidelines (when measuring ).11 A list of all features (and their family groups) with brief explanations is provided in Appendix A.
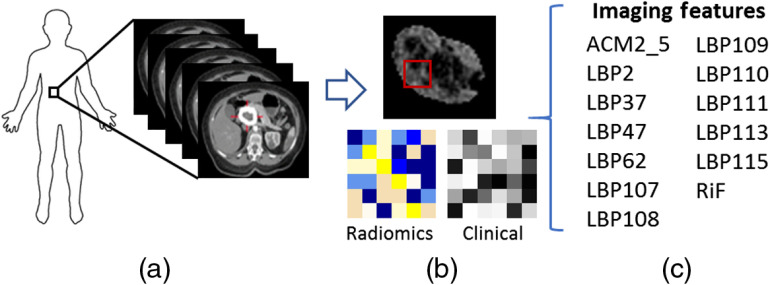
Each feature was extracted from each CT slice of the segmented cyst and averaged to form a single value for a patient as shown in Fig. 2.
Fig. 2.
(a) Extraction of the tumor from preoperative CT of the pancreas. (b) A segmented tumor with an illustration of the radiomic and clinical features extracted. (c) List of the imaging features extracted from the tumor. ACM, angle co-occurrence matrix; LBP, local binary pattern; and RiF, radiologically inspired feature.
2.5. Prediction Model Building
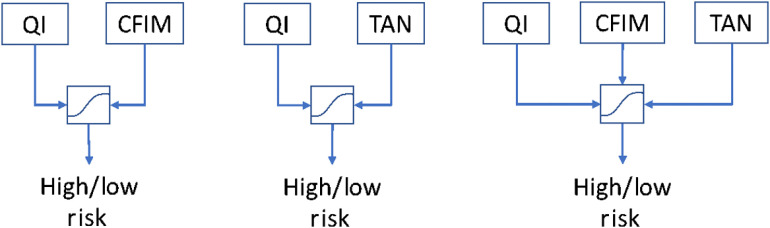
Our QI model, TAN model, and CFIM model were applied to this patient cohort to predict the IPMN risk. The QI model was originally designed based on 103 patients. The model was designed with random forest with 13 selected texture features: RiF, LBP2, LBP37, LBP47, LBP62, LBP107, LBP108, LBP109, LBP110, LBP111, LBP113, LBP115, and ACM2_5. The random forest was developed with 200 trees, with the number of observations per tree as one and the number of variables per random split as the square root of the variables, i.e., 3. A detailed explanation of this model is given in Appendix B. The CFIM model was originally designed based on 149 patients. Three additional prediction models for determining risk were created: (1) a combination of the QI model and CFIM model, (2) a combination of the QI model and TAN model, and (3) a combination of the QI model, CFIM model, and TAN model using logistic regression (Fig. 3). The prediction scores obtained from each model were then used as input to design the combined model using logistic regression. We note that our model was prespecified (no other model was considered a candidate), so sample reuse methods are not needed in this case. The data with which the models were created were separate from the dataset to which the models were then applied.
Fig. 3.
Combinations of models created using logistic regression.
Overall model performance was described using the area under the curve (AUC). Model performance for low-risk and high-risk disease was measured using accuracy, sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV).
2.6. Statistical Confidence Intervals
To reflect the upper and lower ranges of each model’s performance, a 95% confidence interval was calculated for every statistical performance metric. The confidence interval for each metric is calculated as follows:
| (1) |
where the constant represents the fact that 95% of the area of a normal distribution is within 1.96 standard deviations (SD) of the mean, err is the standard error of the metric, and is the number of patients in the observation. The confidence intervals are clipped to values 0 and 1 (or 100%).
3. Results
Thirty-three patients were included in the final analysis, with 21% () classified as high-risk disease (high-grade dysplasia) and the remaining 79% () classified as low-risk (low- or intermediate-grade dysplasia). The cohort was 55% male () and 45% female (), and the median age at resection was 72 years (interquartile range 63 to 76). BD-IPMN was present in 22 patients, and the majority (, 86%) were low-risk disease. MD-IPMN was present in 11 patients, 7 of which had low-risk disease and 4 had high-risk disease.
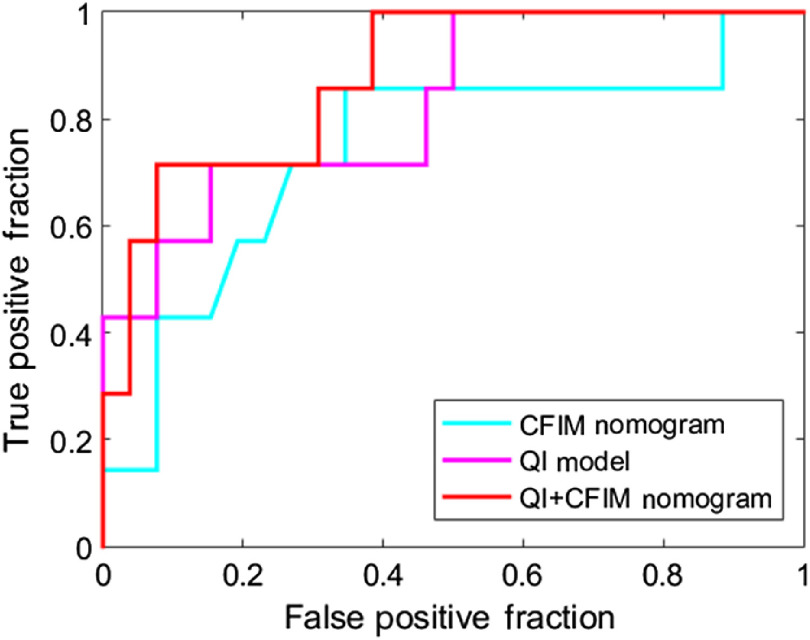
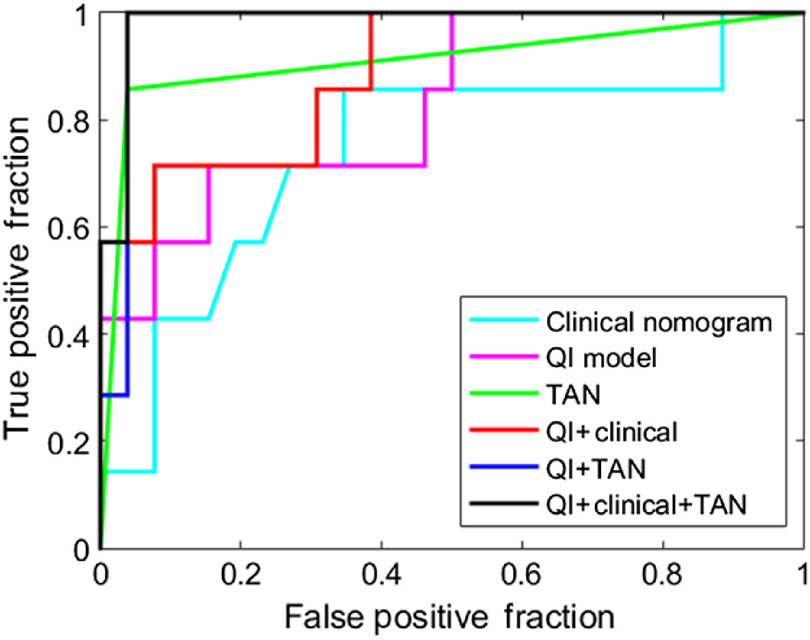
Prediction model results are summarized in Table 1 with a 95% confidence interval to help interpret the results with a likely range of values. The model constructed from the CFIM, QI, and TAN models resulted in an AUC of 0.74, 0.83, and 0.91, respectively. When the QI and CFIM models were combined, the AUC increased to 0.88. The receiver operating characteristic (ROC) curves for these models are shown in Fig. 4. Postoperative models based on the presence of TANs as assessed by a pathologist at the time of resection were also created. The model constructed with TANs, the combined QI and TAN model, and the combined QI, CFIM, and TAN model achieved an AUC of 0.91, 0.97, and 0.98, respectively. The ROC curves for these models are shown in Fig. 5.
Table 1.
Prediction model results
| AUC | Accuracy | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|---|
| CFIM nomogram | ||||||
| QI nomogram | ||||||
| TAN nomogram | ||||||
| QI + CFIM | ||||||
| QI + TAN | ||||||
| QI + CFIM + TAN |
Fig. 4.
ROC curves for CFIM, QI, and combined QI and CFIM models for predicting low-risk from high-risk disease.
Fig. 5.
ROC curves for the TAN, combined QI and TAN, and combined CFIM, QI, and TAN models for predicting low-risk from high-risk disease.
4. Discussion
The ability to accurately identify high-risk disease in patients with IPMN remains limited, and those selected for operative intervention undergo surgeries that carry significant morbidity and mortality risks. One study from this institution reported a 2% risk of mortality and 37% rate of major operative complications following resection of IPMN, further underscoring the need for proper patient selection for pancreatic resection. 17 In this study, we aimed to increase the accuracy of preoperative prediction of risk of IPMN progression to cancer, building on previously validated models created by our group. To our knowledge, this is the first study to describe a predictive model combining these variables. The best performing current published model based on clinical data, qualitative radiographic assessment, and analysis of cyst fluid achieved an AUC of 0.74 on these data. When combined with QI (radiomics), the AUC improved to 0.83, demonstrating an increase in accuracy when images are computationally analyzed. Notably, PPV and NPV are high in this combined model, suggesting that low- and high-risk disease can be predicted prior to surgery. The AUC further improved to 0.91 when TANs measured by a pathologist on the resected specimen were included. Interestingly, the addition of TANs improved the sensitivity of all models, suggesting that risk of false negatives can be mitigated at the time of resection with this additional information. The strongest model performance is observed with the combination of QI (radiomics) with any other model. In particular, the combination of QI and cyst fluid analysis provides a high AUC (0.88) and NPV (0.92). To ensure patients at very low risk for progression of pancreatic cancer are spared an aggressive and highly morbid surgery, significant importance is placed on identifying those on whom not to operate.
The International Consensus Guidelines (ICG) were first established in 2006 to assist in the management of IPMN. These guidelines were revised in 2012 (ICG2012), with the most recently revised version published in 2017, which included some minor revisions.9,11,30 These guidelines outline three high-risk stigmata, namely the presence of obstructive jaundice, an enhancing mural nodule , and main duct dilatation . Worrisome features include cyst size , thickened, enhancing cyst walls, enhancing mural nodules , cyst growth rate per 2 years, main duct dilatation between 5 and 9 mm, abrupt change in pancreatic duct with distal pancreatic atrophy, lymphadenopathy, history of pancreatitis, and elevated serum CA 19-9.11 A review into the validity of the ICG2012 guidelines demonstrated a sensitivity of 88% and specificity of 65%.31 Another study found that the accuracy of the ICG2012 guidelines for predicting malignancy in IPMN is 45%.32 In a study using the 2017 guidelines, the presence of all high-risk stigmata and most worrisome features were shown to be associated with malignant IPMN, with the exception of cyst size, presence of thickened, enhancing cyst walls, and increase in cyst size, which did not show an association.33 Another validation study of the 2017 guidelines supported the importance of mural nodules for detection of malignancy but also found an association between malignancy and thickened, enhancing cyst walls. Overall, the diagnostic performance of the 2017 guidelines was found to have improved following these minor revisions.34 However, these findings continue to highlight the challenge in developing a better predictive tool in the determination of high-risk disease.
Serum and cyst fluid protein markers have been investigated to characterize their utility in the differentiation between low- and high-risk disease with studies demonstrating patterns of high cyst fluid protein expression in high-risk disease, the majority of which are inflammatory markers.22,23 Conversely, no CFIMs were found to be overexpressed in low-risk groups. The CFIM predictive model created by this group focused on predicting the degree of dysplasia in IPMN. This group used its experience on previous work identifying overexpression of inflammatory markers in the presence of TANs. TANs or tumor-infiltrating neutrophils are inflammatory cells that are seen in association with a subset of pancreatic neoplasms. Within the IPMN cohort, they are predominantly seen within the histological pancreatobiliary subset, a subset that typically demonstrates high-grade dysplasia and relatively increased risk of progression to invasive adenocarcinoma compared with other IPMN subtypes.20,35 Noncarcinomatous, or low-grade, components of IPMN did not demonstrate TANs. Another study stratified the level of TANs by grades of dysplasia and found high levels of TANs in high-grade lesions versus no TANs on low-risk lesions.21 This has led to work also being done on the tumor microenvironment, the evaluation of cyst fluid protein overexpression, and the presence of inflammatory markers that may act as a surrogate for TANs, which can only be determined on resected samples.21 Higher levels of TANs were seen in 18 CFIMs, of which 16 of these correlated with higher grades of dysplasia, suggesting that CFIMs may be for suitable surrogate markers. Examples of overexpression of CFIM in high-risk lesions included IL-1b, IL-5, and IL-8, with IL-1b shown to be a significant predictor of high-risk disease.22 The predictive model upon which the CFIM nomogram was created used multivariate modeling to choose the two most promising models.24
This study has a number of limitations. First, the cohorts upon which the models have been created consist of patients who underwent resection and pathologically proven IPMN, therefore introducing a selection bias. This also limited our cohort to a smaller sample size. Next, applicability of this cohort to patients with incidentally detected IPMN is difficult. Additional studies are needed in unresected patients undergoing surveillance to validate the models in this population as currently the applicability to patients selected for radiographic surveillance is unknown. Finally, it should be noted that the samples of cyst fluid and TANs were obtained at the time of surgery. Therefore, these samples are prognostic rather than predictive indicators of disease due to the method in which they were obtained. To reflect clinical practice and the potential application of this model, these samples would be obtained at the time of endoscopic ultrasound (EUS) by cyst fluid aspiration and tissue biopsy. Further studies are therefore required to validate cyst fluid aspiration and tissue retrieval by EUS versus at the time of resection.
5. Impact on Interventional and Surgical Data Science
There is no reliable method of discerning between low-risk and high-risk IPMNs, despite current laboratory, endoscopic, cytologic, and imaging technologies. The 2012 ICG recommended resection for MD-IPMN patients and observation for the majority of BP-IPMN. However, with of MD-IPMN resected patients high-risk and 10% to 15% of BP-IPMN resected patients high-risk, this method of determining patients for resection is remarkably inaccurate. Therefore, the development of preoperative models able to discern high-grade dysplasia in IPMN patients is necessary. Identifying high-risk patients with precision allows for more definitive treatment options and decision-making.17,18 Low-risk patients would be presented with nonsurgical treatment options (typically MRI surveillance), avoiding a potentially morbid and life-threatening operation, while high-risk patients would undergo resection, possibly prior to the formation of invasive disease.17,18
This research hypothesized that multimodal models of quantitative analysis and cyst fluid marker analysis would offer more robustness than models rendered from singular categories of quantitative or cyst fluid analysis. The combination models developed in this study show great promise in providing accurate answers of risk level for IPMN patients. The addition of the QI model enhanced the statistical results overall of the CFIM and TAN models, while the combination of all modalities yielded the best results.
6. Conclusion
For patients in whom IPMN are identified, we present a model that combines quantitative analysis of routinely acquired CT scans (radiomics) with analysis of cyst fluid markers for the prediction of progression to pancreatic cancer. These models can aid the surgeon in optimally selecting patients for pancreatic resection.
7. Appendix A: List of Texture Features (13 Used in the Model)
The specific characteristics of the local binary patterns (LBP) used in the model are described in Table 2. The angle co-occurrence matrices (ACM) used in the model are listed with brief explanations in Table 3.
Table 2.
LBP (128 features).36
| Feature description | Corresponding features |
|---|---|
| 1. 59 unique output levels of ULBP | LBP2, LBP37, LBP47 |
| 2. 10 unique output levels of RI-ULBP | LBP62 |
| 3. SD, skewness, kurtosis, and entropy of ULBP histogram | n/a |
| 4. SD, skewness, kurtosis, and entropy of RI-ULBP histogram | n/a |
| 5. Mean, SD, skewness, kurtosis, and entropy of LBP histogram | n/a |
| 6. SD, skewness, kurtosis, and entropy of efficient RI-LBP histogram | n/a |
| 7. SD, skewness, kurtosis, and entropy of rotated LBP histogram | n/a |
| 8. 38 Fourier descriptors of RI-ULBP histogram | LBP107, LBP108, LBP-109, LBP110, LBP111, LBP113, and LBP115 |
Table 3.
ACM1 and ACM2 (total 38 features, 19 from each).37
| Feature description | Corresponding features |
|---|---|
| 1. Energy | n/a |
| 2. Contrast | n/a |
| 3. Correlation | n/a |
| 4. Sum of squares | n/a |
| 5. Inverse difference moment | ACM2_5 |
| 6. Sum average | n/a |
| 7. Sum variance | n/a |
| 8. Entropy | n/a |
| 9. Difference variance | n/a |
| 10. Sum entropy | n/a |
| 11. Difference entropy | n/a |
| 12. Information-theoretic measures of correlation 1 | n/a |
| 13. Information-theoretic measures of correlation 2 | n/a |
| 14. Maximal correlation coefficient | n/a |
| 15. Inertia | n/a |
| 16. Cluster shade | n/a |
| 17. Cluster prominence | n/a |
| 18. Renyi entropy | n/a |
| 19. Tsallis entropy | n/a |
7.1. RiF (Filled Largest Connected Component)
The RiF is based on radiographic observations that relate to high-risk disease.29 Adaptive thresholding was used to capture high-intensity pixels, which represent the solid component in a cyst, and low-intensity pixels, which represent hypoattenuated dilation in parenchyma regions. The cyst or pancreas regions were initially smoothed to remove minor fluctuations with an averaging filter of size . The filtered image was then thresholded to discover the high-/low-intensity pixels of cyst/pancreas. To compute the threshold, all pixels within the region under consideration (pancreas/cyst) were sorted in ascending order of intensity, and the threshold was selected such that 90% of the total pixels were less than the intensity. The threshold is computed as
| (2) |
where , with c and p representing cyst and pancreas, respectively. is the total number of pixels in the segmented region , and is the thresholded image. and represent the segmented cyst and pancreas regions, respectively. Pixels greater/less than the threshold are noted as enhanced pixels for cyst/pancreas region, respectively. We defined boundary and nonboundary regions of cyst/pancreas for extracting the RiFs based on the radiologist’s observations regarding the appearance of high-risk IPMN. The boundary width was selected empirically as three pixels to quantify the enhanced wall appearing as a thin ring-like structure. and are the number of boundary pixels and nonboundary pixels of , respectively, where boundary and nonboundary regions are mutually exclusive.
The largest enhanced area obtained by filling the largest enhanced component within the nonboundary region is referred to as the filled largest connected component (FLCC). contains number of connected components ; after filling the holes inside, then is the largest region representing FLCC. FLCCF is the ratio of area of FLCC to the area of the nonboundary region:
| (3) |
8. Appendix B: Random Forest Model and Parameters
Random forest combines the results of many bootstrapped decision trees to reduce overfitting and improve generalization. The random forest for the QI model was developed in MATLAB using the TreeBagger class. The parameters for this algorithm are given in Table 4. The tree depth is not an explicit parameter offered in MATLAB’s decision tree algorithms, but it is implicitly derived from the MinLeafSize and MinParentSize parameters. The number of features () selected per random split (NumPredictorsToSample) is calculated by taking the square root of the number of features ().
Table 4.
Random forest parameters.
| Parameter | Value |
|---|---|
| NumTrees | 200 |
| NumObservations | 1 |
| NumPredictorsToSample | 3 |
| MinLeafSize | 2 |
| MinParentSize | 4 |
| SplitCriterion | Deviance |
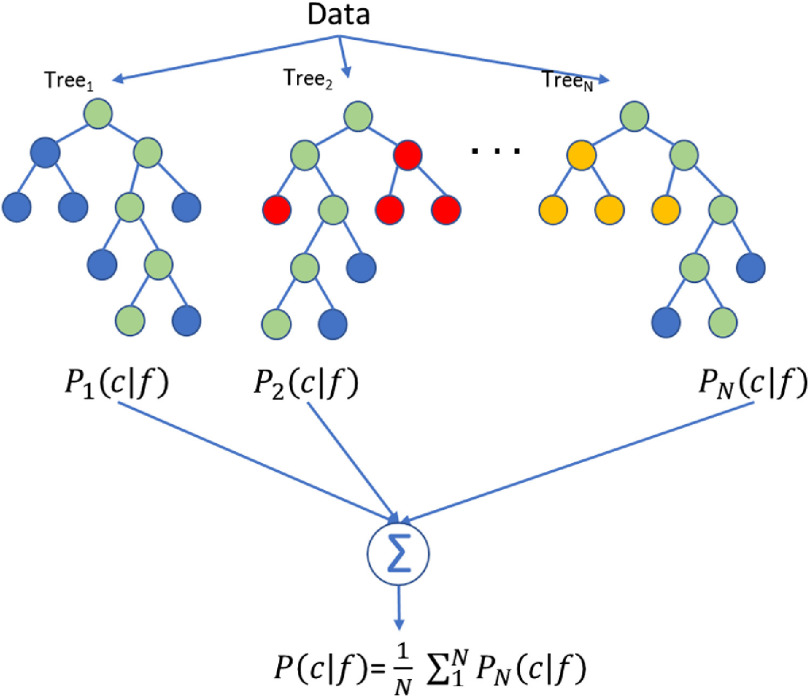
A visual representation of the random forest model is shown in Fig. 6. Each tree calculates a conditional probability of a class given features . These probabilities are averaged to give the final conditional probability of the random forest. In this figure, the green nodes are the selected nodes at each branch.
Fig. 6.
A visual representation of the random forest model for the QI model.
Acknowledgments
This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748, the American Association for Cancer Research, and the Pancreatic Cancer Action Network.
Biographies
Kate A. Harrington is a fellowship-trained diagnostic radiologist specializing in oncologic body imaging, with special interest in imaging of hepatopancreatobiliary tumors. She is a member of the Disease Management Team for liver, biliary and pancreatic tumors and endeavors to offer high-quality, subspecialized radiology, working closely with surgeons and oncologists as part of the multidisciplinary network of patient care provided at Memorial Sloan Kettering Cancer Center. Her research interests are centered on using novel imaging techniques.
Travis L. Williams received his PhD in electrical engineering from North Carolina A&T State University in 2017. His dissertation, “Advanced Image Classification using Deep Neural Networks,” combined wavelet theory and deep learning to increase the accuracy and efficiency of deep learning algorithms. Currently a Research Scholar under Dr. Amber Simpson, his research applies machine learning to classifying tumors in the pancreas and liver as cancerous, benign, predicting survival, etc.
Jayasree Chakraborty research focuses on the early detection, diagnosis, and prognostication of pancreatic and liver cancers via quantitative analysis of clinical CT imaging using machine learning. Previously, she developed computer-aided methods for the early detection of breast cancer. She served as an assistant professor at National Institute of Technology Silchar, India, from March 2013 to May 2015, after receiving her PhD in electronics and electrical communication engineering from Indian Institute of Technology Kharagpur in 2013.
Mithat Gonen has been serving as chief of the Biostatistics Service for Memorial Sloan Kettering since 2015. His translational and clinical collaborations focus on surgical treatment of gastrointestinal and hepatobiliary cancers, response evaluation and imaging endpoints for clinical trials and biomarker development. Most of his methodological research originates from these collaborations, including building, assessing and comparing prognostic and predictive models, design and analysis of clinical trials and imaging studies, as well as Bayesian methods.
Olca Basturk is a pathologist trained in oncologic and gastrointestinal pathology. Her main interest is pancreaticobiliary diseases, and her research has particularly focused on pancreas and gallbladder neoplasms. Her career goals are to investigate these tumors from clinical, morphologic, and molecular perspectives such as exploring the pathologic factors that may predict the behavior of these neoplasms or genetic factors in their pathogenesis.
Vinod P. Balachandran is a surgical oncologist specializing in treating benign and cancerous diseases of the pancreas, bile duct, gallbladder, and liver. He researches new ways to use the immune system to fight pancreatic cancer. By studying T cells in sarcoma, he discovered imatinib activates T cells to attack cancers. He combined imatinib with an immunotherapy to increase T cell tumor killing. These findings changed our understanding of how imatinib and other kinase inhibitors destroy tumors.
T. Peter Kingham is the international surgical oncology fellowship director and the assistant director for the Hepatopancreatobiliary (HPB) fellowship at Memorial Sloan Kettering. His primary research interest is determining how to improve cancer care for patients in low- and middle-income countries (LMIC). He is co-PI on multiple prospective studies on colorectal and breast cancer in Nigeria, co-founded the African Research Group for Oncology (ARGO), and is president and cofounder of Surgeons OverSeas (SOS).
Michael I. D’Angelica is an assistant attending in the Department of Surgery, hepatopancreatobiliary service at Memorial Sloan Kettering. He is a board-certified surgical oncologist with expertise in treating cancer of the liver, bile ducts, gall bladder, and pancreas. He works with a team that has developed and uses many surgical and non-surgical techniques that have shown remarkable success treating these cancers. Sloan-Kettering’s multi-disciplinary methods of treatment offer patients a good chance of long-term survival and cure.
William R. Jarnagin has served as chief of the hepatopancreatobiliary service since 2008 at Memorial Sloan Kettering. His research has focused on genomics, novel therapies and biomarkers of treatment response in patients with biliary cancer, intraoperative navigation, and intraoperative management during major liver and pancreas resection. He has authored or co-authored over 250 articles in peer-reviewed journals, over 60 chapters or invited reviews, and has coedited three textbooks.
Jeffrey A. Drebin is chair of the Department of Surgery at Memorial Sloan Kettering. He oversees a team that is world-renowned for its innovation and expertise in cancer surgery. In addition to his administrative duties, he is also a surgeon and scientist. He specializes in treating people with pancreatic cancer, gallbladder and bile duct cancers, liver cancer, and stomach cancer.
Richard K. Do is a radiologist whose expertise is in imaging for liver cancer, bile duct cancer, and pancreatic cancer. His research interests are in advanced CT and MRI techniques for hepatopancreatobiliary tumors. He works with computer scientists to investigate the potential of computer-based medical image analysis to predict tumor pathology, genetics, and treatment response. He is active in the American College of Radiology, working to develop new guidelines for treatment response assessment for liver tumors.
Peter J. Allen is board certified in surgery and specializes in treating cancerous and precancerous conditions of the pancreas, liver, bile duct, and stomach. His research focuses on developing nonsurgical methods for diagnosing pancreatic cancer, including the identification of novel biomarkers and imaging modalities for patients with precancerous conditions of the pancreas. He has led multiple prospective trials, and his research has been funded by the NIH since 2009.
Amber L. Simpson is an associate professor of computer science and biomedical and molecular sciences specializing in medical image analysis and computer-aided surgery. Her lab focuses on developing novel computing strategies for precision oncology. She joined Queen’s University faculty in 2019, after four years as Memorial Sloan Kettering faculty and three years as a Biomedical Engineering Research assistant professor at Vanderbilt University. She received her PhD in Computer Science from Queen’s University in Kingston, Ontario, Canada.
Disclosures
K. A. Harrington, T. L. Williams, S. A. Lawrence, J. Chakraborty, M. A. Al Efishat, M. A. Attiyeh, G. Askan, Y. Chou, A. Pulvirenti, C. A. McIntyre, M. Gonen, O. Basturk, V. P. Balachandran, T. P. Kingham, M. I. D’Angelica, W. R. Jarnagin, J. A. Drebin, R. K. Do, A. L. Simpson, and P. J. Allen have no disclosures to declare.
Contributor Information
Kate A. Harrington, Email: harrink1@mskcc.org.
Travis L. Williams, Email: williat4@mskcc.org.
Sharon A. Lawrence, Email: sl1203@rwjms.rutgers.edu.
Jayasree Chakraborty, Email: chakrabj@mskcc.org.
Mohammad A. Al Efishat, Email: malefis1@jhmi.edu.
Marc A. Attiyeh, Email: marcattiyeh@gmail.com.
Gokce Askan, Email: gokceaskan@gmail.com.
Yuting Chou, Email: chouy@mskcc.org.
Alessandra Pulvirenti, Email: ale.pulvirenti@gmail.com.
Caitlin A. McIntyre, Email: mcintyrc@mskcc.org.
Mithat Gonen, Email: gonenm@mskcc.org.
Olca Basturk, Email: basturko@mskcc.org.
Vinod P. Balachandran, Email: balachav@mskcc.org.
T. Peter Kingham, Email: kinghamp@mskcc.org.
Michael I. D’Angelica, Email: dangelim@mskcc.org.
William R. Jarnagin, Email: jarnagiw@mskcc.org.
Jeffrey A. Drebin, Email: drebinj@mskcc.org.
Richard K. Do, Email: DoK@mskcc.org.
Peter J. Allen, Email: peter.allen@duke.edu.
Amber L. Simpson, Email: amber.simpson@queensu.ca.
References
- 1.Laffan T. A., et al. , “Prevalence of unsuspected pancreatic cysts on MDCT,” AJR Am. J. Roentgenol. 191(3), 802–807 (2008). 10.2214/AJR.07.3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X. M., et al. , “Pancreatic cysts: depiction on single-shot fast spin-echo MR images,” Radiology 223(2), 547–553 (2002). 10.1148/radiol.2232010815 [DOI] [PubMed] [Google Scholar]
- 3.Maitra A., et al. , “Precursors to invasive pancreatic cancer,” Adv. Anat. Pathol. 12(2), 81–91 (2005). 10.1097/01.pap.0000155055.14238.25 [DOI] [PubMed] [Google Scholar]
- 4.Matthaei H., Dal Molin M., Maitra A., “Identification and analysis of precursors to invasive pancreatic cancer,” in Pancreatic Cancer: Methods and Protocols, (Methods in Molecular Medicine), Su G. H., Ed., Vol. 980, pp. 1–12, Humana Press, Totowa, New Jersey: (2013). [DOI] [PubMed] [Google Scholar]
- 5.Basturk O., et al. , “A revised classification system and recommendations from the Baltimore Consensus Meeting for neoplastic precursor lesions in the pancreas,” Am. J. Surg. Pathol. 39(12), 1730–1741 (2015). 10.1097/PAS.0000000000000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagata N., et al. , “Development of pancreatic cancer, disease-specific mortality, and all-cause mortality in patients with nonresected IPMNs: a long-term cohort study,” Radiology 278(1), 125–134 (2016). 10.1148/radiol.2015150131 [DOI] [PubMed] [Google Scholar]
- 7.Maker A. V., et al. , “Cytology from pancreatic cysts has marginal utility in surgical decision-making,” Ann. Surg. Oncol. 15(11), 3187–3192 (2008). 10.1245/s10434-008-0110-0 [DOI] [PubMed] [Google Scholar]
- 8.Heckler M., et al. , “The Sendai and Fukuoka consensus criteria for the management of branch duct IPMN—a meta-analysis on their accuracy,” Pancreatology 17(2), 255–262 (2017). 10.1016/j.pan.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 9.Tanaka M., et al. , “International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas,” Pancreatology 12(3), 183–197 (2012). 10.1016/j.pan.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 10.Marchegiani G., et al. , “IPMN involving the main pancreatic duct: biology, epidemiology, and long-term outcomes following resection,” Ann. Surg. 261(5), 976–983 (2015). 10.1097/SLA.0000000000000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka M., et al. , “Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas,” Pancreatology 17(5), 738–753 (2017). 10.1016/j.pan.2017.07.007 [DOI] [PubMed] [Google Scholar]
- 12.Allen P. J., “The management of intraductal papillary mucinous neoplasms of the pancreas,” Surg. Oncol. Clin. N. Am. 19(2), 297–310 (2010). 10.1016/j.soc.2009.11.002 [DOI] [PubMed] [Google Scholar]
- 13.Pulvirenti A., et al. , “Intraductal papillary mucinous neoplasms,” Ann. Surg. (2019). 10.1097/SLA.0000000000003703 [DOI] [Google Scholar]
- 14.Tanno S., et al. , “Pancreatic ductal adenocarcinomas in long-term follow-up patients with branch duct intraductal papillary mucinous neoplasms,” Pancreas 39(1), 36–40 (2010). 10.1097/MPA.0b013e3181b91cd0 [DOI] [PubMed] [Google Scholar]
- 15.Tada M., et al. , “Pancreatic cancer in patients with pancreatic cystic lesions: a prospective study in 197 patients,” Clin. Gastroenterol. Hepatol. 4(10), 1265–1270 (2006). 10.1016/j.cgh.2006.07.013 [DOI] [PubMed] [Google Scholar]
- 16.Uehara H., et al. , “Development of ductal carcinoma of the pancreas during follow-up of branch duct intraductal papillary mucinous neoplasm of the pancreas,” Gut 57(11), 1561–1565 (2008). 10.1136/gut.2007.145631 [DOI] [PubMed] [Google Scholar]
- 17.Attiyeh M. A., et al. , “Development and validation of a multi-institutional preoperative nomogram for predicting grade of dysplasia in intraductal papillary mucinous neoplasms (IPMNs) of the pancreas: a report from the pancreatic surgery consortium,” Ann. Surg. 267(1), 157–163 (2018). 10.1097/SLA.0000000000002015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Correa-Gallego C., et al. , “Predicting dysplasia and invasive carcinoma in intraductal papillary mucinous neoplasms of the pancreas: development of a preoperative nomogram,” Ann. Surg. Oncol. 20(13), 4348–4355 (2013). 10.1245/s10434-013-3207-z [DOI] [PubMed] [Google Scholar]
- 19.Steele C. W., et al. , “Exploiting inflammation for therapeutic gain in pancreatic cancer,” Br. J. Cancer 108(5), 997–1003 (2013). 10.1038/bjc.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid M. D., et al. , “Tumor-infiltrating neutrophils in pancreatic neoplasia,” Mod. Pathol. 24(12), 1612–1619 (2011). 10.1038/modpathol.2011.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadot E., et al. , “Tumor-associated neutrophils and malignant progression in intraductal papillary mucinous neoplasms: an opportunity for identification of high-risk disease,” Ann. Surg. 262(6), 1102–1107 (2015). 10.1097/SLA.0000000000001044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maker A. V., et al. , “Cyst fluid interleukin-1beta (IL1beta) levels predict the risk of carcinoma in intraductal papillary mucinous neoplasms of the pancreas,” Clin. Cancer Res. 17(6), 1502–1508 (2011). 10.1158/1078-0432.CCR-10-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maker A. V., et al. , “Pancreatic cyst fluid and serum mucin levels predict dysplasia in intraductal papillary mucinous neoplasms of the pancreas,” Ann. Surg. Oncol. 18(1), 199–206 (2011). 10.1245/s10434-010-1225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al Efishat M. A., et al. , “Multi-institutional validation study of pancreatic cyst fluid protein analysis for prediction of high-risk intraductal papillary mucinous neoplasms of the pancreas,” Ann. Surg. 268(2), 340–347 (2018). 10.1097/SLA.0000000000002421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanania A. N., et al. , “Quantitative imaging to evaluate malignant potential of IPMNs,” Oncotarget 7(52), 85776–85784 (2016). 10.18632/oncotarget.11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Permuth J. B., et al. , “Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms,” Oncotarget 7(52), 85785–85797 (2016). 10.18632/oncotarget.11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman D. H., et al. , “Utility of whole-lesion ADC histogram metrics for assessing the malignant potential of pancreatic intraductal papillary mucinous neoplasms (IPMNs),” Abdom. Radiol. 42(4), 1222–1228 (2017). 10.1007/s00261-016-1001-7 [DOI] [PubMed] [Google Scholar]
- 28.Attiyeh M. A., et al. , “Preoperative risk prediction for intraductal papillary mucinous neoplasms by quantitative CT image analysis,” HPB 21(2), 212–218 (2019). 10.1016/j.hpb.2018.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty J., et al. , “CT radiomics to predict high-risk intraductal papillary mucinous neoplasms of the pancreas,” Med. Phys. 45(11), 5019–5029 (2018). 10.1002/mp.13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M., et al. , “International consensus guidelines for management of intraductal papillary mucinous neoplasms and mucinous cystic neoplasms of the pancreas,” Pancreatology 6(1–2), 17–32 (2006). 10.1159/000090023 [DOI] [PubMed] [Google Scholar]
- 31.Watanabe Y., et al. , “Validity of the management strategy for intraductal papillary mucinous neoplasm advocated by the international consensus guidelines 2012: a retrospective review,” Surg. Today 46(9), 1045–1052 (2016). 10.1007/s00595-015-1292-2 [DOI] [PubMed] [Google Scholar]
- 32.Yamada S., et al. , “Comparison of the international consensus guidelines for predicting malignancy in intraductal papillary mucinous neoplasms,” Surgery 159(3), 878–884 (2016). 10.1016/j.surg.2015.08.042 [DOI] [PubMed] [Google Scholar]
- 33.Lee J. E., et al. , “Determining malignant potential of intraductal papillary mucinous neoplasm of the pancreas: CT versus MRI by using revised 2017 International Consensus Guidelines,” Radiology 293, 190144 (2019). 10.1148/radiol.2019190144 [DOI] [PubMed] [Google Scholar]
- 34.Kang J. S., et al. , “Clinical validation of the 2017 international consensus guidelines on intraductal papillary mucinous neoplasm of the pancreas,” Ann. Surg. Treat. Res. 97(2), 58–64 (2019). 10.4174/astr.2019.97.2.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furukawa T., et al. , “Prognostic relevance of morphological types of intraductal papillary mucinous neoplasms of the pancreas,” Gut 60(4), 509–516 (2011). 10.1136/gut.2010.210567 [DOI] [PubMed] [Google Scholar]
- 36.Ojala T., Pietikäinen M., Harwood D., “A comparative study of texture measures with classification based on feature distributions,” Pattern Recognit. 29(1), 51–59 (1996). 10.1016/0031-3203(95)00067-4 [DOI] [Google Scholar]
- 37.Chakraborty J., et al. , “Statistical measures of orientation of texture for the detection of architectural distortion in prior mammograms of interval-cancer,” J. Electron. Imaging 21(3), 033010 (2012). 10.1117/1.JEI.21.3.033010 [DOI] [Google Scholar]