Preterm birth is the leading cause of neonatal morbidity and mortality worldwide. Multiple etiologies are associated with preterm birth; however, 25% of preterm infants are born to a mother with intra-amniotic infection, most commonly due to invasion of the amniotic cavity by Ureaplasma species. Much research has focused on establishing a link between Ureaplasma species and adverse pregnancy/neonatal outcomes; however, little is known about the taxonomy of and host response against Ureaplasma species. Here, we applied a multifaceted approach, including human samples, in vivo models, and in vitro manipulations, to study the maternal-fetal immunobiology of Ureaplasma infection during pregnancy. Furthermore, we investigated the use of clarithromycin as a treatment for this infection. Our research provides translational knowledge that bolsters scientific understanding of Ureaplasma species as a cause of adverse pregnancy/neonatal outcomes and gives strong evidence for the use of clarithromycin as the recommended treatment for women intra-amniotically infected with Ureaplasma species.
KEYWORDS: Ureaplasma, antibiotics, bacterial burden, intra-amniotic infection, neonate, pregnancy, preterm labor
ABSTRACT
Intra-amniotic infection is strongly associated with adverse pregnancy and neonatal outcomes. Most intra-amniotic infections are due to Ureaplasma species; however, the pathogenic potency of these genital mycoplasmas to induce preterm birth is still controversial. Here, we first laid out a taxonomic characterization of Ureaplasma isolates from women with intra-amniotic infection, which revealed that Ureaplasma parvum is the most common bacterium found in this clinical condition. Next, using animal models, we provided a causal link between intra-amniotic inoculation with Ureaplasma species and preterm birth. Importantly, the intra-amniotic inoculation of Ureaplasma species induced high rates of mortality in both preterm and term neonates. The in vivo potency of U. parvum to induce preterm birth was not associated with known virulence factors. However, term-derived and preterm-derived U. parvum isolates were capable of inducing an intra-amniotic inflammatory response. Both U. parvum isolates invaded several fetal tissues, primarily the fetal lung, and caused fetal inflammatory response syndrome. This bacterium was also detected in the placenta, reproductive tissues, and most severely in the fetal membranes, inducing a local inflammatory response that was replicated in an in vitro model. Importantly, treatment with clarithromycin, a recently recommended yet not widely utilized antibiotic, prevented the adverse pregnancy and neonatal outcomes induced by U. parvum. These findings shed light on the maternal-fetal immunobiology of intra-amniotic infection.
INTRODUCTION
Microbial-associated intra-amniotic inflammation, also known as intra-amniotic infection, is associated with a wide array of adverse pregnancy and neonatal outcomes such as clinical chorioamnionitis (1), funisitis (2), bacteremia (3), and importantly, preterm labor and birth (4)- a syndrome that is the leading cause of neonatal morbidity and mortality worldwide (5, 6). The most frequently identified bacteria in amniotic fluid of women with intra-amniotic infection are Ureaplasma species, followed by Mycoplasma hominis, Gardnerella vaginalis and Streptococcus agalactiae, among others (1, 7–11). Ureaplasma has been classified into two species containing 14 serovars: Ureaplasma parvum (comprised of serovars 1, 3, 6, and 14) and Ureaplasma urealyticum (comprised of serovars 2, 4, 5, and 7 to 13) (12, 13). These two species are distinguishable based on differing genomic characteristics such as 16S rRNA gene sequence identity as well as unique and specific genes such as the multiple banded antigen and urease (14). Both U. parvum and U. urealyticum have been identified in amniotic fluid of women with intra-amniotic infection (13). However, the immune responses triggered by Ureaplasma species in the mother and the offspring have been poorly characterized.
Clinical studies have shown that women intra-amniotically infected with Ureaplasma species exhibit a more intense inflammatory response than those infected with other microorganisms (15). Moreover, the earlier the gestational age, the greater the intensity of the intra-amniotic inflammatory response induced by Ureaplasma species (16). However, not all women intra-amniotically infected with Ureaplasma species deliver preterm (17). This finding is consistent with animal studies in which the intra-amniotic administration of Ureaplasma species does not always lead to preterm birth. Initial studies using an established model of chronically catheterized rhesus macaques (18) showed that intra-amniotic inoculation with U. parvum or Mycoplasma hominis induces preterm labor, intra-amniotic inflammation, inflammation of the placenta (e.g., acute chorioamnionitis), and a systemic fetal inflammatory response (19). However, the ultrasound-guided intra-amniotic injection of U. parvum in rhesus macaques does not induce preterm labor or a severe fetal inflammatory response but causes intra-amniotic and uterine inflammation (20). The difference between the two macaque models could be explained by the distinct methodologies utilized. In mice, surgical intra-amniotic infection with Ureaplasma species does not cause preterm birth (21). Thus, we hypothesized that there is diversity in the Ureaplasma species found in the amniotic cavity of women with intra-amniotic infection, which differentially modulate the host immune responses of the mother and the fetus and may dictate pregnancy outcomes.
In the present study, we isolated and cultivated Ureaplasma species from amniotic fluid of women with intra-amniotic infection and performed 16S rRNA gene and whole-genome sequencing to provide an in-depth characterization of these isolates. The Ureaplasma isolates were then utilized to create a murine model of Ureaplasma-induced intra-amniotic infection to evaluate pregnancy and neonatal outcomes, localize sites of bacterial invasion, and evaluate maternal and fetal immune responses. Finally, we evaluated whether clarithromycin, a clinically relevant antibiotic (22–24), is an effective treatment for intra-amniotic infection caused by Ureaplasma species.
RESULTS
Taxonomic analysis of Ureaplasma isolates from women with intra-amniotic infection.
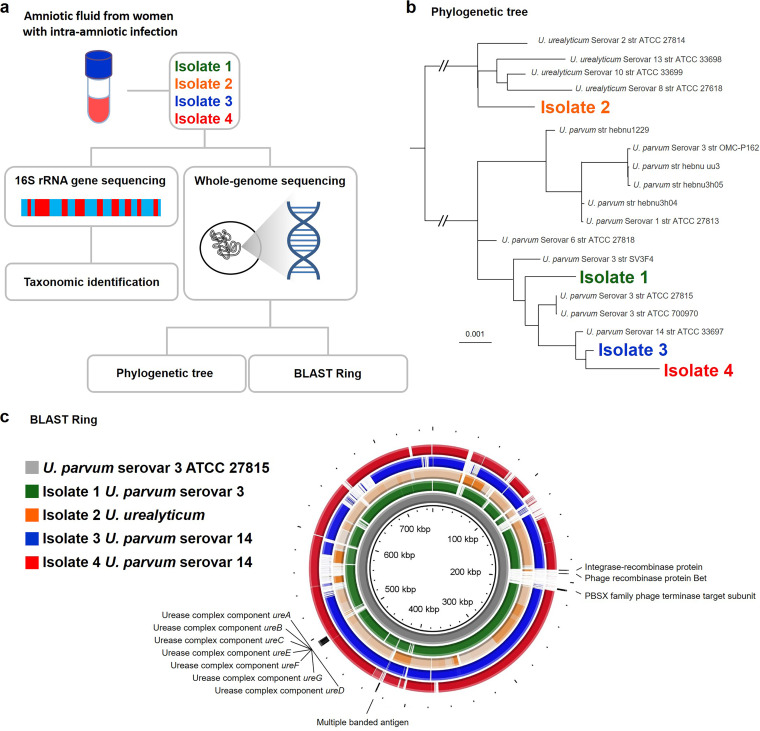
First, we explored whether there were taxonomic differences in the Ureaplasma isolates from women with intra-amniotic infection. DNA was extracted from four Ureaplasma isolates from women with intra-amniotic infection (the clinical and demographic characteristics of these women are shown in Table S1 in the supplemental material) and 16S rRNA gene sequencing was performed (Fig. 1a). The 16S rRNA gene sequences of isolate 1, isolate 3, and isolate 4 were identical to those of U. parvum serovar 3 and serovar 14 (Table 1). Specifically, the trimmed 16S rRNA gene sequences (1,025 bp aligning with nucleotide positions 344101 to 345125 of the genome of U. parvum serovar 3 ATCC 27815, accession no. CP000942.1) for isolates 1, 3, and 4 were identical to U. parvum serovar 3 (ATCC 27815) or serovar 14 (ATCC 33697) (Table 1). The 16S rRNA gene sequence of isolate 2 (1,021 bp) closely matched that of U. urealyticum serovars 2, 8, or 10 (ATCC 27814, ATCC 27618, and ATCC 33699, respectively) (Table 1). Whole-genome sequencing was then performed to further characterize the taxonomic profile of each Ureaplasma isolate (Fig. 1a). The phylogenetic tree based on genomic single nucleotide polymorphism (SNP) data indicated a clear division of two main clusters, with one cluster containing all U. parvum serovars (1, 3, 6, and 14) and the other containing all U. urealyticum serovars (2, 8, 10, and 13) (Fig. 1b). Within the subclusters, isolate 1 grouped with multiple serovar 3 strains, and isolates 3 and 4 grouped with the serovar 14 strain (Fig. 1b). Isolate 2 was not positioned with any specific U. urealyticum serovar (Fig. 1b). Based on these clusters, we determined that the taxonomic identity of isolate 1 corresponded to U. parvum serovar 3, and the taxonomic identities of both isolates 3 and 4 corresponded to U. parvum serovar 14. The taxonomic identity of isolate 2 was determined to correspond with U. urealyticum of an unknown serovar. To further explore the differences between Ureaplasma species isolated from women with intra-amniotic infection, DNA sequence analysis of known virulence genes (urease complex and multiple banded antigen [25]) was performed. No substantial differences were observed between the four isolates (Fig. 1c). Specifically, the similarity of the urease gene complex of the three U. parvum isolates ranged from 99.95 to 99.96% (Fig. 1c). As expected, the similarity of the urease gene complex sequences of the U. urealyticum isolate was only between 90.86 and 90.88% to those of the U. parvum isolates (Fig. 1c). In addition, we evaluated the putative pathogenicity island, a 20-kb region variably present in serovars 1, 3, and 6 that contains potential integrase/recombinase genes with phage orthologs (26), in all isolates. However, this pathogenicity island was not present in any of the genomes of the four isolates (Fig. 1c). Together, these genomic analyses revealed that the taxonomic identity of Ureaplasma species isolated from women with intra-amniotic infection is diverse and yet not distinguishable by known virulence genes. Importantly, U. parvum serovar 14 was present in both a woman who delivered preterm and a woman who delivered at term, suggesting that taxonomy cannot solely account for adverse pregnancy outcomes caused by this genital mycoplasma.
FIG 1.
Taxonomic analysis of Ureaplasma isolates from women with intra-amniotic infection. (a) Experimental design for the 16S rRNA gene sequencing and whole-genome sequencing of bacteria isolated from amniotic fluid of women with intra-amniotic infection (n = 4). (b) Whole-genome single nucleotide polymorphism (SNP) phylogeny of four clinical isolates and 15 publicly available Ureaplasma genomes inferred using whole-genome maximum-likelihood phylogeny under a generalized time-reversible model. The tree was rooted at the midpoint. The scale bar indicates the number of substitutions per nucleotide position. (c) Representation (BLAST Ring) of the four isolate genomes using U. parvum serovar 3 ATCC 27815 (CP000942) as the reference. Known virulence genes of Ureaplasma species are shown.
TABLE 1.
Percent similarity of 16S rRNA gene sequences of the isolates to publicly available Ureaplasma type strains
| Isolate | % similarity to U. parvum serovars |
% similarity to U. urealyticum serovars |
||||||
|---|---|---|---|---|---|---|---|---|
| Serovar 1, ATCC 27813 (ASM17137v1) | Serovar 3, ATCC 27815 (ASM1934v1) | Serovar 6, ATCC 27818 (ASM16989v1) | Serovar 14, ATCC 33697 (ASM17135v1) | Serovar 2, ATCC 27814 (ASM17155v1) | Serovar 8, ATCC 27618 (ASM16953v1) | Serovar 10, ATCC 33699 (ASM2126v1) | Serovar 13, ATCC 33698 (ASM17139v1) | |
| 1 | 99.90 | 100.00 | 99.90 | 100.00 | 98.83 | 98.83 | 98.83 | 98.73 |
| 2 | 98.73 | 98.83 | 98.73 | 98.83 | 99.90 | 99.90 | 99.90 | 99.80 |
| 3 | 99.90 | 100.00 | 99.90 | 100.00 | 98.83 | 98.83 | 98.83 | 98.73 |
| 4 | 99.90 | 100.00 | 99.90 | 100.00 | 98.83 | 98.83 | 98.83 | 98.73 |
Clinical and demographic characteristics of women from whose amniotic fluid Ureaplasma species were isolated. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ureaplasma isolates from women with intra-amniotic infection can induce preterm birth and adverse neonatal outcomes.
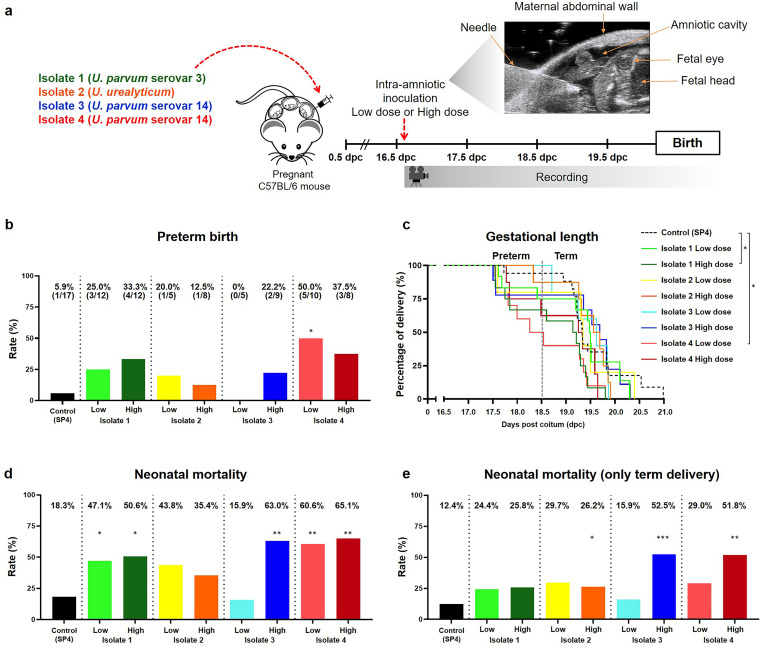
Genomic analysis revealed that there are taxonomic differences in the Ureaplasma isolates obtained from women with intra-amniotic infection. Next, we evaluated whether these four isolates could induce different pregnancy outcomes in mice. Dams were intra-amniotically inoculated under ultrasound guidance in each amniotic sac with a low dose (from ≥1 × 103 to ≤1 × 104 cells/sac) or a high dose (from >1 × 104 to ≤1 × 105 cells/sac) of Ureaplasma species from one of the four clinical isolates and observed until delivery (Fig. 2a). These doses were chosen based on the concentrations of Ureaplasma species recovered from women with intra-amniotic infection. Controls were intra-amniotically injected with sterile 1× phosphate-buffered saline (PBS) or SP4 broth (vehicle control) (see Fig. S1 in the supplemental material). Each clinical Ureaplasma isolate induced a variable rate of preterm birth, with isolate 3 being the least effective at the low dose and isolate 4 showing the greatest potency at the low dose (Fig. 2b). Notably, intra-amniotic inoculation with isolate 4 (low dose) induced the highest rate of preterm birth compared to vehicle controls (Fig. 2b). Survival curve analysis indicated that isolate 1 (high dose) and isolate 4 (low dose) decreased the gestational length compared to vehicle controls (Fig. 2c). No differences in the rates of preterm birth or gestational length were observed between PBS- and vehicle control-injected dams (Fig. S1a and S1b).
FIG 2.
Preterm birth and adverse neonatal outcomes induced by intra-amniotic inoculation with Ureaplasma isolates. (a) Experimental design for the intra-amniotic inoculation with Ureaplasma species isolated from amniotic fluid of women with intra-amniotic infection (positive amniotic fluid culture and an IL-6 concentration of ≥2.6 ng/ml). Dams were intra-amniotically inoculated with Ureaplasma isolates (low dose [1 × 103 to 1 × 104 cells/25 μl] or high dose [1 × 104 to 1 × 105 cells/25 μl], n = 5 to 12 each) or injected with vehicle control (SP4 broth, 25 μl, n = 17) in each amniotic sac under ultrasound guidance at 16.5 days post coitum (dpc) and monitored until delivery. (b) Rates of preterm birth of dams intra-amniotically inoculated with Ureaplasma isolates or injected with control. The P values were determined by Fisher's exact tests. (c) Kaplan-Meier survival curves showing the gestational lengths at delivery of dams intra-amniotically inoculated with Ureaplasma isolates or injected with control. The P values were determined by Gehan-Breslow-Wilcoxon tests. (d) Mortality of neonates born to dams intra-amniotically inoculated with Ureaplasma isolates or injected with control. (e) Mortality of only those neonates born at term to dams intra-amniotically inoculated with Ureaplasma isolates or injected with control. The P values in panels (d) and (e) were determined by unpaired t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Pregnancy outcomes of dams injected with PBS or SP4 broth. (a) Rates of preterm birth of dams intra-amniotically injected with PBS or SP4 broth. The P values were determined by a Fisher's exact test. (b) Kaplan-Meier survival curve showing the gestational ages at delivery of dams intra-amniotically injected with PBS or SP4 broth. Statistical analysis was performed using a Gehan-Breslow-Wilcoxon test. (c) Mortality rates of neonates born to dams intra-amniotically injected with PBS or SP4 broth. Statistical analysis was performed using an unpaired t test. (d) Mortality rates of only those neonates born at term to dams intra-amniotically injected with PBS or SP4 broth. Statistical analysis was performed using an unpaired t test. The results of all statistical analyses were non-significant. Download FIG S1, TIF file, 1.3 MB (1.2MB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Intra-amniotic infection is not only associated with preterm birth but also with adverse neonatal outcomes, even in term neonates (27). Therefore, we also evaluated the mortality rates of neonates born to dams intra-amniotically inoculated with clinical isolates of Ureaplasma species. A significantly elevated rate of mortality was observed for neonates born to dams inoculated with isolate 1 and isolate 4 (high and low doses) as well as the high dose of isolate 3 (Fig. 2d). When considering only neonates that were delivered at term, significantly higher rates of mortality were still observed for those neonates born to dams inoculated with a high dose of Ureaplasma isolates 2, 3, and 4 (Fig. 2e). The rates of neonatal mortality were comparable between PBS and vehicle controls (Fig. S1c and S1d).
Collectively, these results indicate that intra-amniotic inoculation with clinically isolated Ureaplasma species induces variable rates of preterm birth and neonatal mortality. Importantly, U. parvum serovar 14 isolated from a woman who delivered preterm (isolate 4) induced higher rates of preterm birth in mice than that from a woman who delivered at term (isolate 3), yet both isolates were associated with intra-amniotic inflammation (an interleukin-6 [IL-6] concentration of ≥2.6 ng/ml [28]) and acute maternal and fetal inflammatory responses in the placenta (Table S1). These data indicate that, although U. parvum serovar 14 displays variable in vivo potency to induce preterm birth, this bacterium consistently causes damage to the fetus and at the maternal-fetal interface (e.g., chorioamniotic membranes and decidua). Therefore, we next investigated the host immune responses triggered by both isolates of U. parvum serovar 14 in the amniotic cavity, in the fetus, and at the maternal-fetal interface.
Intra-amniotic inoculation with U. parvum serovar 14 induces an intra-amniotic inflammatory response but not a maternal inflammatory response.
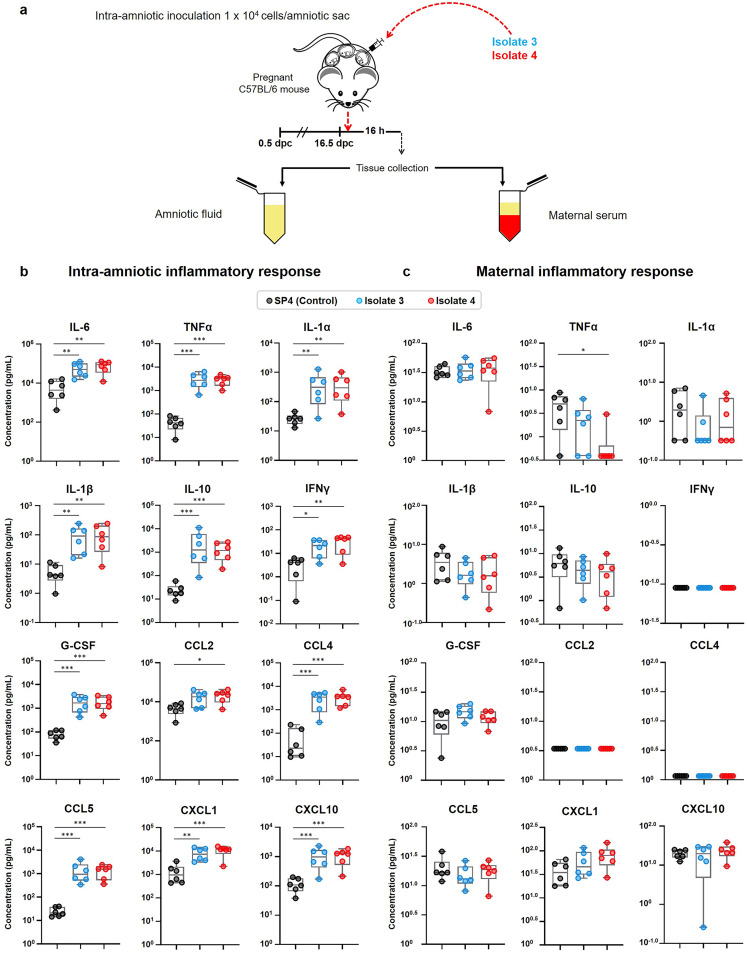
It is well documented that women with Ureaplasma-associated intra-amniotic infection display a severe intra-amniotic inflammatory response (15). Therefore, we next evaluated whether intra-amniotic inoculation with low doses of isolates 3 and 4 (both U. parvum serovar 14) induced inflammatory responses in the amniotic cavity. Given that the intra-amniotic inflammatory response in women with Ureaplasma-associated intra-amniotic infection is subclinical in nature (29), we also determined the inflammatory response in the maternal circulation. Dams were intra-amniotically inoculated with either isolate 3 or isolate 4 of U. parvum serovar 14, and both amniotic fluid and maternal serum were collected (Fig. 3a). The amniotic fluid concentrations of IL-6, a cytokine used for the clinical diagnosis of intra-amniotic inflammation (28), were increased in dams inoculated with isolate 3 and isolate 4 compared to vehicle controls (Fig. 3b). Other cytokines upregulated in women with intra-amniotic infection (30) such as tumor necrosis factor alpha (TNFα), IL-1α, IL-1β, IL-10, gamma interferon (IFNγ), granulocyte colony-stimulating factor (G-CSF), CCL2, CCL4, CCL5, CXCL1, and CXCL10 were also increased in amniotic fluid from dams inoculated with isolate 3 or isolate 4 compared to vehicle controls (Fig. 3b). No differences were observed between isolate 3 and isolate 4 (Fig. 3b). Conversely, when we evaluated the maternal systemic inflammatory response, none of the cytokines were increased upon inoculation with isolate 3 or isolate 4 compared to vehicle controls (Fig. 3c). Cytokine concentrations in amniotic fluid and sera of dams injected with PBS and vehicle control are shown in Fig. S2 and S3.
FIG 3.
Intra-amniotic and maternal systemic inflammatory responses after intra-amniotic inoculation with U. parvum serovar 14. (a) Experimental design for the evaluation of intra-amniotic and maternal systemic inflammatory responses after intra-amniotic inoculation with two clinical isolates of U. parvum serovar 14. Dams were intra-amniotically inoculated with U. parvum isolate 3 (1.0 × 104 cells/25 μl) or U. parvum isolate 4 (1.0 × 104 cells/25 μl), or injected with vehicle control (SP4 broth, 25 μl) at 16.5 days post coitum (dpc) (n = 6 each). Amniotic fluid and maternal serum samples were collected 16 h after intra-amniotic inoculation for cytokine/chemokine multiplex analysis. (b and c) Cytokine/chemokine concentrations in amniotic fluid (b) and maternal serum (c). The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. The P values were determined by one-way analysis of variance (ANOVA), followed by Tukey’s tests to determine differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Cytokine and chemokine concentrations in amniotic fluid from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. P values were determined by using unpaired t tests. *, P < 0.05; **, P < 0.01. Download FIG S2, TIF file, 0.1 MB (102.6KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cytokine and chemokine concentrations in maternal serum from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. Statistical analysis was performed by using unpaired t tests. The results of all statistical analyses were non-significant. Download FIG S3, TIF file, 0.1 MB (96.8KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Together, these results show that intra-amniotic inoculation with either isolate of U. parvum serovar 14 induces a similar intra-amniotic inflammatory response in the absence of a maternal systemic inflammatory response. This finding indicated that a deeper characterization of the immunological effects of U. parvum serovar 14 on the fetus was required.
Ureaplasma parvum serovar 14 invades the fetal organs, inducing fetal inflammatory response syndrome.
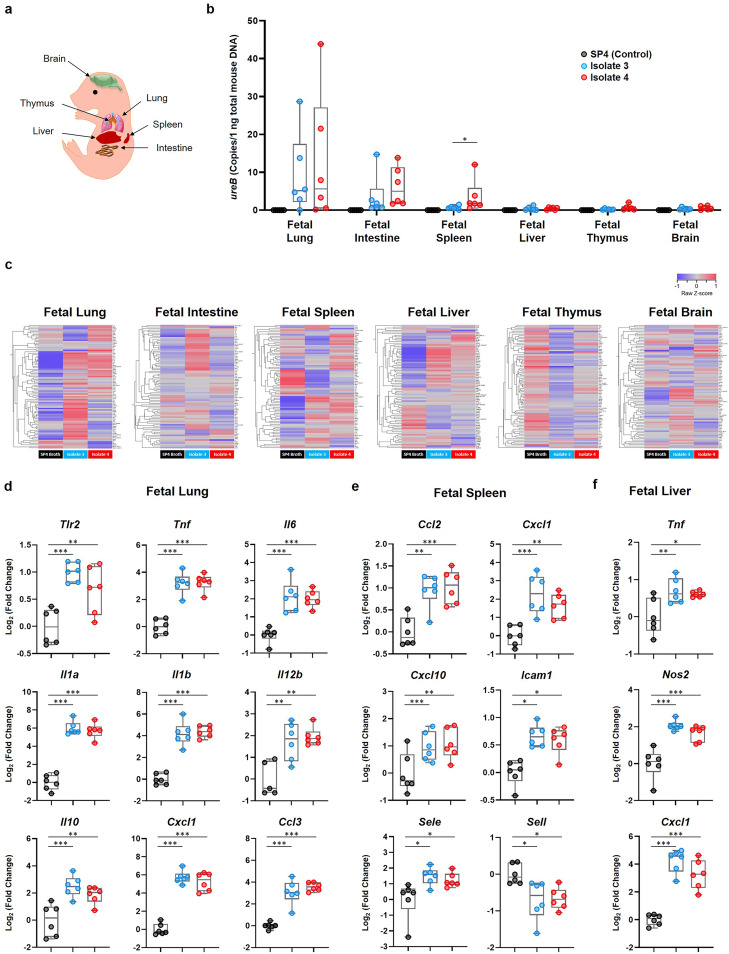
To further explore the effects of U. parvum serovar 14 on the fetus, we first evaluated whether isolate 3 and isolate 4 can be detected in the fetal organs. The fetal lung, intestine, spleen, liver, thymus, and brain were collected from pregnant dams intra-amniotically inoculated with U. parvum isolate 3 or isolate 4 (Fig. 4a), and the bacterial burden was quantified by real-time quantitative PCR (qPCR) of the ureB gene. Both isolates of U. parvum were most abundant in the fetal lung, followed by the fetal intestine and fetal spleen (Fig. 4b). Interestingly, the abundance of isolate 4 in the fetal spleen was higher than that of isolate 3 (Fig. 4b). There was a detectable yet minimal bacterial burden in the fetal liver, thymus, and brain (Fig. 4b). Fetal organs from PBS- and vehicle control-injected dams had no detectable ureB gene copies (Fig. 4b; see also Fig. S4a in the supplemental material). These results show that when U. parvum serovar 14 invades the amniotic cavity, the primary sites of infection are the fetal lung, fetal intestine, and fetal spleen.
FIG 4.
Bacterial burden and inflammatory gene expression in the fetal organs upon inoculation with U. parvum serovar 14. Dams were intra-amniotically inoculated with U. parvum isolate 3 (1.0 × 104 cells/25 μl) or U. parvum isolate 4 (1.0 × 104 cells/25 μl), or injected with vehicle control (SP4 broth, 25 μl) at 16.5 days post coitum (dpc) (n = 6 each). Tissues were collected 16 h after intra-amniotic inoculation. (a) Spatial localization of the murine fetal organs included in this experiment. (b) Copy numbers of the ureB gene in the fetal organs. (c) Heatmap visualization of inflammatory gene expression in the fetal organs. (d to f) Expression of specific inflammatory genes in the fetal lung (d), spleen (e), and liver (f). The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. The P values were determined by Mann-Whitney U tests (b) or one-way analysis of variance (ANOVA), followed by Tukey’s tests to determine differences between groups (d to f). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Bacterial burden in the fetal, maternal, reproductive, and gestational tissues from dams intra-amniotically injected with PBS. (a) Copy numbers of the ureB gene in the fetal organs. (b) Copy numbers of the ureB gene in the maternal, reproductive, and gestational tissues. Download FIG S4, TIF file, 0.1 MB (53.3KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we evaluated the expression of several inflammatory mediators in the fetal organs upon infection with U. parvum serovar 14. Intra-amniotic inoculation with either isolate of U. parvum serovar 14 induced the upregulation of multiple inflammatory genes in the fetal lung, spleen, and liver, with the number of upregulated genes in the fetal lung being highest among the fetal tissues (Fig. 4c). Specifically, the expression of Tlr2, Tnf, Il6, Il1a, Il1b, Il12b, Il10, Cxcl1, and Ccl3 was significantly upregulated in the fetal lung from dams intra-amniotically inoculated with U. parvum serovar 14 (Fig. 4d). The fetal intestine contained the second greatest bacterial burden among the fetal organs (Fig. 4b); however, only the expression of Il1b was significantly upregulated in this fetal organ from dams inoculated with either U. parvum isolate (Fig. S5a). In the fetal spleen, the relative expression of Ccl2, Cxcl1, Cxcl10, Icam1, Sele, and Sell was significantly altered in response to either U. parvum isolate (Fig. 4e). Although the bacterial burden of the fetal liver was low (Fig. 4b), the relative expression of Tnf, Nos2, and Cxcl1 was upregulated in response to isolate 3 or isolate 4 (Fig. 4f). No differences in the expression of the above-mentioned inflammatory genes were observed between fetal organs from PBS- or vehicle control-injected dams (Fig. S5b and S6a to c). Together, these findings indicate that a primary site of U. parvum serovar 14 invasion in the fetus is the lung, in which a significant bacterial burden and upregulation of inflammatory mediators take place.
Inflammatory gene expression in the fetal intestine from dams intra-amniotically inoculated with U. parvum or injected with SP4 broth or PBS. (a) Expression of Il1b in the fetal intestine from dams intra-amniotically inoculated with U. parvum serovar 14 isolate 3 or U. parvum serovar 14 isolate 4, or injected with SP4 broth (control). (b) Expression of Il1b in the fetal intestine from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. Statistical analysis was performed using one-way analysis of variance (ANOVA), followed by a Tukey’s test, to determine differences between groups (a), or an unpaired t test (b). **, P < 0.01; ***, P < 0.001. Download FIG S5, TIF file, 0.04 MB (45.7KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inflammatory gene expression in the fetal organs from dams intra-amniotically injected with PBS or SP4 broth. (a to c) Expression of specific inflammatory genes in the fetal lung (a), fetal spleen (b), and fetal liver (c) from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. Statistical analysis was performed by using unpaired t tests. The results of all statistical analyses were non-significant. Download FIG S6, TIF file, 0.1 MB (149.4KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
U. parvum serovar 14 is detected in the reproductive tissues and induces a local inflammatory response.
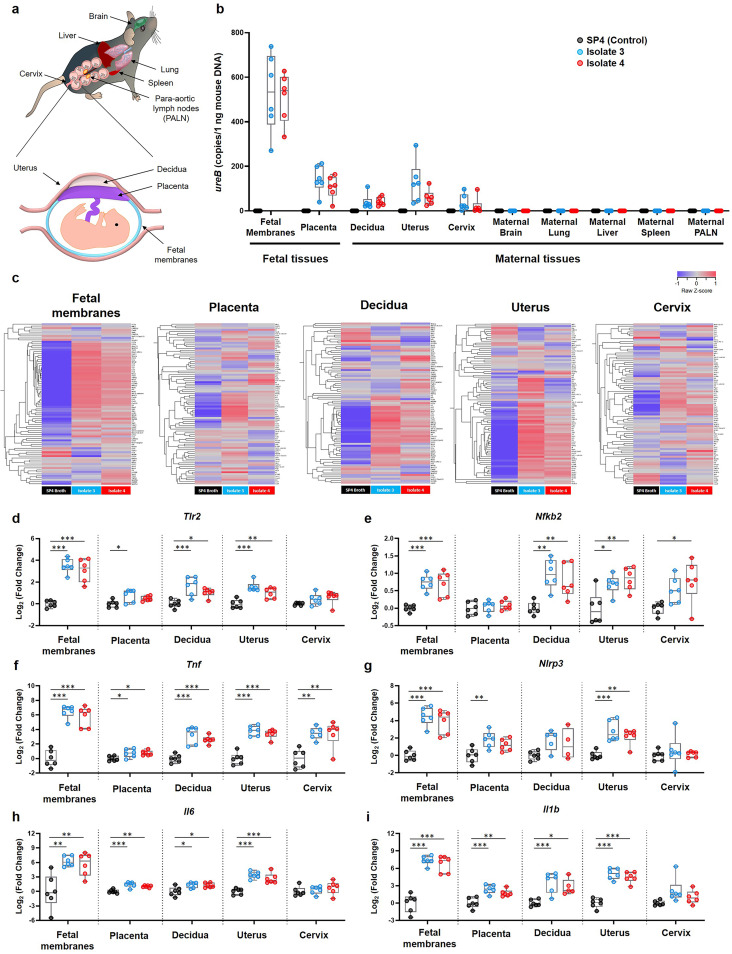
Similar to the fetus, microbes in the amniotic cavity can also invade the surrounding tissues such as the chorioamniotic membranes (also known as fetal membranes) and placenta (31, 32). Thus, we proceeded to evaluate whether intra-amniotic U. parvum serovar 14 can be detected in the reproductive tissues surrounding the amniotic cavity as well as the maternal organs. The fetal membranes, placenta, decidua, uterus, cervix, maternal brain, maternal lung, maternal liver, maternal spleen, and the maternal para-aortic lymph nodes (PALN) were collected from pregnant dams intra-amniotically inoculated with U. parvum isolate 3 or isolate 4 (Fig. 5a), and the bacterial burden was quantified by qPCR of the ureB gene. The greatest bacterial burden was found in the fetal membranes, distantly followed by the placenta, decidua, uterus, and cervix compared to vehicle controls (Fig. 5b). There was no detectable bacterial burden in the maternal brain, lung, liver, spleen, and PALN (Fig. 5b). Reproductive and maternal organs from PBS- and vehicle control-injected dams had no detectable ureB gene copies (Fig. 5b; see also Fig. S4b in the supplemental material). These results show that intra-amniotic U. parvum serovar 14 disseminates into the surrounding tissues (e.g., fetal membranes and placenta) and also reaches the maternal-fetal interface (e.g., decidua) and reproductive tissues (e.g., uterus and cervix).
FIG 5.
Bacterial burden and inflammatory gene expression in reproductive and gestational tissues upon inoculation with U. parvum serovar 14. Dams were intra-amniotically inoculated with U. parvum isolate 3 (1.0 × 104 cells/25 μl) or U. parvum isolate 4 (1.0 × 104 cells/25 μl), or injected with vehicle control (SP4 broth, 25 μl) at 16.5 days post coitum (dpc) (n = 6 each). Tissues were collected 16 h after intra-amniotic inoculation. (a) Spatial localization of the murine tissues collected in this experiment. (b) Copy numbers of the ureB gene in maternal, fetal, and reproductive tissues. (c) Heatmap visualization of inflammatory gene expression in the fetal membranes, placenta, decidua, uterus, and cervix. (d to i) Expression of Tlr2 (d), Nfkb2 (e), Tnf (f), Nlrp3 (g), Il6 (h), and Il1b (i) in the fetal membranes, placenta, decidua, uterus, and cervix. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. The P values were determined by Mann-Whitney U tests (b) or one-way analysis of variance (ANOVA), followed by Tukey’s tests to determine differences between groups (d to i). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
U. parvum serovar 14 was detected only in the gestational and reproductive tissues; therefore, we next evaluated the expression of inflammatory mediators in these compartments. The fetal membranes, placenta, decidua, uterus, and cervix each displayed consistent upregulation of inflammatory mediators upon intra-amniotic infection with isolate 3 or isolate 4 compared to vehicle controls (Fig. 5c). Interestingly, the fetal membranes and uterus had the greatest upregulation of inflammatory genes, whereas the placenta displayed a weaker response (Fig. 5c).
We then compared the relative expression of specific inflammatory genes (Tlr2, Nfkb2, Tnf, Nlrp3, Il6, and Il1b) in the gestational and reproductive tissues upon inoculation with U. parvum isolate 3 or isolate 4. Strikingly, the fetal membranes displayed the highest expression of inflammatory genes (Fig. 5d and f to i), which corresponded to the high bacterial burden detected in these tissues (Fig. 5b). Specifically, the expression of Tlr2 was upregulated in the fetal membranes, decidua, uterus, and—to a lesser degree—in the placenta in response to both isolates (Fig. 5d). The expression of the transcription factor Nfkb2 was upregulated in every tissue, except for the placenta, in response to both isolates (Fig. 5e). The expression of Tnf was upregulated in all tissues in response to both isolates (Fig. 5f), whereas the expression of Nlrp3 (the sensor molecule of the NLRP3 inflammasome [33]) was upregulated only in the fetal membranes, placenta, and uterus (Fig. 5g). Consistently, the expression of Il6 and Il1b increased in response to both isolates in the fetal membranes, placenta, decidua, and uterus but not in the cervix (Fig. 5h and i). The expression of Tlr2, Nfkb2, Tnf, Nlrp3, Il6, and Il1b in PBS- and vehicle control-injected dams is shown in Fig. S7. These results show that intra-amniotic inoculation with U. parvum serovar 14 results in the upregulation of several inflammatory mediators in the gestational and reproductive tissues.
Inflammatory gene expression in the reproductive and gestational tissues from dams intra-amniotically injected with PBS or SP4 broth. The expression of specific inflammatory genes in the fetal membranes, placenta, decidua, uterus, and cervix of dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. P values were determined by using unpaired t tests. *, P < 0.05. Download FIG S7, TIF file, 0.1 MB (118.2KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Does U. parvum serovar 14 induce the upregulation of inflammatory mediators in human amniocytes in vitro?
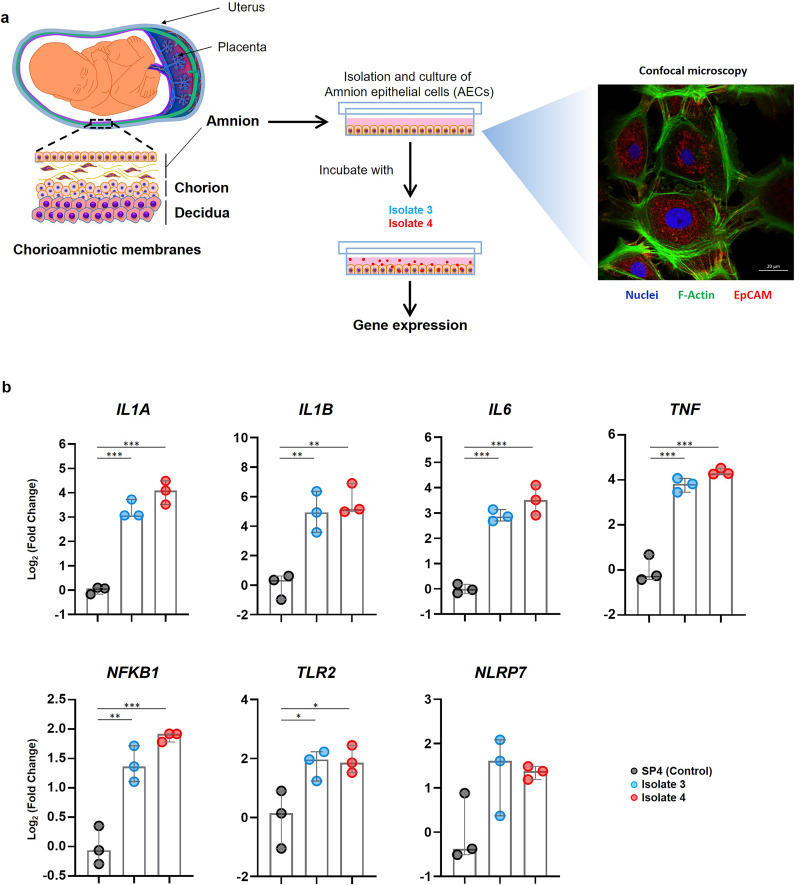
Given that the fetal membranes were the primary site of U. parvum serovar 14 infection resulting in the upregulation of inflammatory mediators, we investigated whether this bacterium could induce similar immune responses in the inner layer (i.e., amnion epithelial cells [AECs]) of the chorioamniotic membranes, which faces the amniotic cavity. AECs were isolated from the placentas of normal term deliveries and incubated with U. parvum isolate 3 or isolate 4 (Fig. 6a). A representative confocal microscopy image of primary cultured AECs is shown in Fig. 6a. Consistent with our murine findings (Fig. 5c to i), AECs incubated with either of the U. parvum serovar 14 isolates had significantly higher relative expression of the inflammatory genes IL1A, IL1B, IL6, TNF, NFKB1, and TLR2 compared to the vehicle control (Fig. 6b). The expression of NLRP7, an inflammasome sensor molecule upregulated upon infection with Ureaplasma species (34), tended to increase as well, although this did not reach statistical significance (Fig. 6b). The expression of NLRP3 was not upregulated in response to either U. parvum isolate (Fig. S8a). The expression of IL1A, IL1B, IL6, TNF, NFKB1, TLR2, NLRP7, and NLRP3 in AECs incubated with PBS or vehicle control is shown in Fig. S8b. These data indicate that U. parvum serovar 14 upregulates the expression of several inflammatory mediators in human amniocytes, resembling the inflammatory milieu observed in the fetal membranes of animals intra-amniotically inoculated with this bacterium.
FIG 6.
Incubation of U. parvum serovar 14 and primary human amniocytes. (a) Experimental design (left to right). Primary human amnion epithelial cells (AECs) (n = 3) were isolated from term human chorioamniotic membranes and incubated with U. parvum isolate 3 (1.3 × 105 cells), U. parvum isolate 4 (1.3 × 105 cells), or vehicle control (SP4 broth) for 24 h. The AECs were then collected to determine gene expression. A representative confocal microscopy image of the immunofluorescence staining of AECs to show the expression of EpCAM and F-Actin is displayed. (b) Expression of IL1A, IL1B, IL6, TNF, NFKB1, TLR2, and NLRP7 in AECs incubated with U. parvum isolates. The data are shown as medians with minimum and maximum ranges. The P values were determined by one-way analysis of variance (ANOVA), followed by Tukey’s tests to determine differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Expression of specific inflammatory genes in primary human amniocytes incubated with U. parvum serovar 14, PBS, or SP4 broth. (a) Expression of NLRP3 in primary human amnion epithelial cells incubated with U. parvum isolate 3, isolate 4, or SP4 broth. (b) Expression of inflammatory genes in primary human amnion epithelial cells incubated with PBS or SP4 broth. The data are shown as medians with minimum and maximum ranges. P values were determined by one-way analysis of variance (ANOVA), followed by a Tukey’s test, to determine differences between groups (a), or unpaired t tests (b). **, P < 0.01; ***, P < 0.001. Download FIG S8, TIF file, 0.1 MB (130.2KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
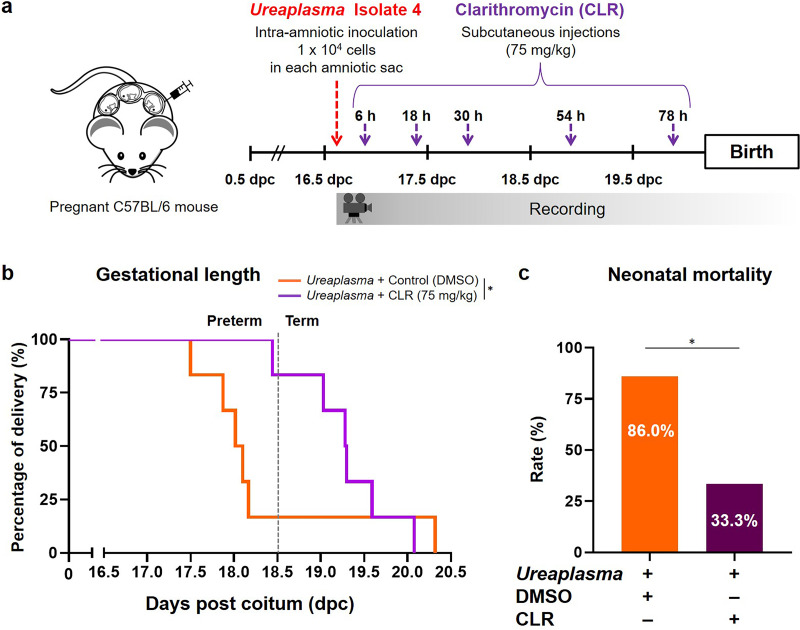
Treatment with clarithromycin reduces adverse pregnancy and neonatal outcomes induced by U. parvum serovar 14.
Recent clinical studies have shown that the administration of antibiotics improves perinatal outcomes in women with intra-amniotic infection/inflammation (22–24). Three antibiotics were administered in these studies to achieve broad coverage, but among these three only clarithromycin (CLR) is effective against Ureaplasma species (35). Therefore, we tested whether treatment with this antibiotic could prevent adverse pregnancy and neonatal outcomes in dams intra-amniotically inoculated with U. parvum serovar 14 (Fig. 7a). We verified the efficacy of CLR against intra-amniotic infection with isolate 4 (low dose), since this isolate of U. parvum serovar 14 caused the highest rate of preterm birth as well as a significant rate of neonatal mortality (Fig. 2). The gestational length of dams intra-amniotically inoculated with U. parvum serovar 14 and treated with CLR was significantly longer than those treated with vehicle control, resulting in a reduced rate of preterm birth (Fig. 7b). Moreover, treatment with CLR significantly reduced neonatal mortality in dams intra-amniotically inoculated with U. parvum serovar 14 (Fig. 7c). These results demonstrate that the adverse pregnancy and neonatal consequences caused by intra-amniotic infection with U. parvum serovar 14 can be ameliorated using clarithromycin, a recently recommended antibiotic.
FIG 7.
Clarithromycin treatment after intra-amniotic inoculation with U. parvum serovar 14. (a) Experimental design. Dams were intra-amniotically inoculated with U. parvum isolate 4 (1.0 × 104 cells/25 μl) and subcutaneously treated with clarithromycin (CLR; 75 mg/kg) or vehicle control (DMSO) at 6, 18, 30, 54, and 78 h post-inoculation and monitored until delivery (n = 6 each). (b) Kaplan-Meier survival curves showing the gestational lengths at delivery. The P value was determined by a Gehan-Breslow-Wilcoxon test. (c) Neonatal mortality. The P value was determined by an unpaired t test. *, P < 0.05.
DISCUSSION
The present study provides evidence of a causal link between clinically isolated Ureaplasma species and adverse pregnancy and neonatal outcomes. First, taxonomic characterization of four Ureaplasma isolates from women with intra-amniotic infection revealed that the most common Ureaplasma species in this clinical condition is U. parvum. However, no differences in known genetic virulence factors were found among clinically obtained Ureaplasma isolates. Therefore, the bacterial virulence cannot be the sole factor accounting for disease severity among the four included patients, suggesting that host-pathogen interactions may dictate the pregnancy outcome. Next, using animal models, we showed that intra-amniotic inoculation with clinically obtained Ureaplasma isolates induces preterm birth and neonatal mortality to variable degrees. Further, we demonstrated that U. parvum serovar 14 induces a severe intra-amniotic inflammatory response in the absence of a maternal systemic inflammatory response. Importantly, U. parvum serovar 14 invaded the fetal lung, intestine, and spleen, inducing fetal inflammatory response syndrome. This bacterium was also found in the fetal membranes, placenta, and decidua as well as the reproductive tissues, inducing severe inflammatory responses primarily in the fetal membranes. In vitro studies showed the relevance of our in vivo findings by demonstrating that inoculation with U. parvum serovar 14 triggers proinflammatory responses in human amniocytes. Lastly, we demonstrated the efficacy of clarithromycin, a recently recommended yet not widely utilized antibiotic, in abrogating the adverse pregnancy and neonatal outcomes induced by Ureaplasma species.
Initially, we identified taxonomic differences among the four clinically obtained Ureaplasma isolates, indicating that there is diversity in the Ureaplasma species that invade the amniotic cavity. However, the broad genomic analysis performed here did not allow us to identify differences in known virulence factors among Ureaplasma isolates obtained from women with intra-amniotic infection. Possible explanations for why the difference in these genomic factors is unable to distinguish the virulence of Ureaplasma species in the context of preterm delivery include the following: (i) unknown strain-specific virulence factors exist and affect the pregnancy outcomes; (ii) the inflammation induced by Ureaplasma species not only depends on this bacterium but also on host factors such as genetic predisposition (36); or (iii) intra-amniotic infection with Ureaplasma species slightly alters the timing of birth in all humans/animals in a manner that was imperceptible in the present study. In addition, the number of Ureaplasma isolates included in this study may have limited our capacity to detect subtle differences between strains. A deeper whole-genome analysis is required to identify novel strain-specific virulence factors that may dictate pregnancy disease. In the present study, we focused on the in vivo effects of Ureaplasma species invading the amniotic cavity.
Intra-amniotic inoculation with clinically isolated Ureaplasma species caused preterm birth and neonatal mortality that varied between isolates. Importantly, U. parvum serovar 14 obtained from a woman who delivered preterm caused the highest rates of preterm birth and neonatal mortality, indicating that this specific isolate displayed an intrinsically greater degree of virulence than other isolates. To our knowledge, this is the first demonstration that clinically isolated U. parvum serovar 14 has been shown to induce preterm birth in an animal model. In our study, we also found that intra-amniotic inoculation with U. parvum serovar 3 induced a low rate of preterm birth but high rates of neonatal mortality. This finding contrasts with a previous report showing that intra-amniotic inoculation with U. parvum serovar 3 via mini-laparotomy did not induce preterm birth (21). The variability in the rates of preterm birth and neonatal mortality caused by the different Ureaplasma isolates indicates that individual host susceptibility to intra-amniotic infection contributes to the severity of adverse pregnancy outcomes.
In humans, Ureaplasma-associated intra-amniotic infection causes a local inflammatory response in the absence of maternal systemic inflammation, preventing noninvasive diagnosis of this clinical condition (37). This finding is consistent with the present study, in which Ureaplasma inoculation neither induced a maternal inflammatory response nor were Ureaplasma strains detected in systemic maternal organs such as the brain, lung, liver, and lymphatic organs. On the other hand, intra-amniotic inoculation with Ureaplasma isolates induced high rates of neonatal mortality, which coincided with intra-amniotic and fetal inflammatory responses. Importantly, intra-amniotic inoculation with Ureaplasma species induced a surge of proinflammatory cytokines (e.g., IL-6 and CXCL10), which are conventionally measured for the diagnosis of intra-amniotic inflammation (28, 38). However, this intra-amniotic cytokine storm alone is insufficient to cause fetal disease; therefore, we also investigated the microbial burden and the inflammatory response in the fetus. Consistent with human research in the fetal inflammatory response syndrome (a clinical condition resulting from fetal exposure to microbial invasion of the amniotic cavity [39, 40]), the fetuses of dams intra-amniotically inoculated with U. parvum displayed severe inflammatory responses primarily in the lung, spleen, and liver. The most affected fetal organ was the lung, which also contained the highest microbial burden. These observations are supported by previous reports showing that fetal exposure to Ureaplasma species induces an increased expression of proinflammatory cytokines in the fetal lung of mice (21) as well as an influx of immune cells in that of rhesus macaques (19). Fetal lung inflammation characterized by immune cell influx and elevated cytokine expression is also observed after chronic Ureaplasma infection in the amniotic cavity in ovine models (41). Moreover, clinical studies have described that prenatal exposure to Ureaplasma species is associated with chronic lung disease (42) or bronchopulmonary dysplasia (43, 44). The association of Ureaplasma species with fetal lung complications observed in multiple animal models and in humans may be explained by the fact that the fetal lung is directly exposed to bacteria in the amniotic cavity, as the fetus can aspirate amniotic fluid (45). In the present study, we also found that Ureaplasma species were detected in the fetal intestine, which is in tandem with a clinical demonstration showing that fetuses exposed to these bacteria can develop necrotizing enterocolitis in the neonatal period (46). Therefore, it is likely that the intense inflammatory processes induced by Ureaplasma species in mice can subsequently lead to organ damage, as observed in humans (47), which may explain the high rate of neonatal mortality.
U. parvum was not only detected in the fetal organs but also in the gestational tissues and at the maternal-fetal interface (i.e., the decidua). These observations coincide with clinical studies showing the presence of bacteria in the placenta (32), chorioamniotic membranes (31), and choriodecidua (48) in women with intra-amniotic infection. The microbial invasion of these tissues is strongly associated with a proinflammatory milieu (49), as observed in this study. However, we found that, among the fetal and maternal tissues, U. parvum invasion was greatest in the fetal membranes and coincided with a severe inflammatory response, which is consistent with recent findings showing that vaginal inoculation with U. parvum results in an ascending infection that reaches the fetal membranes, inducing local inflammation (50). Interestingly, the immune response induced by inoculation with U. parvum involves the activation of the NLRP3 inflammasome, an important pathway for preterm parturition in the context of intra-amniotic infection (51–53). Of note, our in vitro studies showed that the incubation of primary amniocytes with U. parvum also induced an inflammatory response in the absence of the upregulation of the NLRP3 transcript, suggesting that the choriodecidua is central in the intra-amniotic activation of the inflammasome in humans.
It is worth mentioning that intra-amniotic inoculation with U. parvum also resulted in the detection of this bacterium in the uterus and cervix. There are two plausible explanations for this finding: (i) the intra-amniotic infection progressed beyond the amniotic cavity into the reproductive tissues, and/or (ii) inoculation with U. parvum, or the bacterium itself, induced rupture of the fetal membranes, thus facilitating the spread of infection. Regardless, the presence of U. parvum in the uterus and cervix resulted in the upregulation of several inflammatory mediators that, in turn, led to the activation of the common pathway of parturition (5).
Here, we are the first to demonstrate that clarithromycin abrogates Ureaplasma-induced preterm birth and neonatal mortality. Notably, recent studies have shown that clarithromycin can be used together with ceftriaxone and metronidazole to treat preterm birth in women with intra-amniotic infection (22–24). Clarithromycin is a macrolide antibiotic, which acts against Mollicutes (including Ureaplasma species) by inhibiting bacterial protein synthesis (54). In addition to the bacteriostatic function against Ureaplasma species, this antibiotic exhibits immunomodulatory effects by suppressing the NF-κB pathway (55), which was shown in this and other studies to be activated by Ureaplasma species (56). Together, these results provide mechanistic evidence that the recent recommendations to use clarithromycin in the treatment of women with intra-amniotic infection (22–24) are substantiated in clinical practice.
In summary, the present study provides a taxonomic characterization of Ureaplasma species found in women with intra-amniotic infection. In addition, mechanistic experimentation shows that U. parvum induces adverse pregnancy and neonatal outcomes by causing a severe inflammatory response in the amniotic cavity, the fetus, the gestational and reproductive tissues, and the maternal-fetal interface. Importantly, our data provide solid evidence demonstrating that treatment with clarithromycin, an antibiotic that is not yet widely utilized, must be implemented in the management of women with intra-amniotic infection, given that Ureaplasma species are the most commonly detected microbes in such patients (Fig. 8). Collectively, our investigations provide insights into the maternal-fetal immunobiology of intra-amniotic infection, the most established link to prematurity and neonatal morbidity and mortality worldwide.
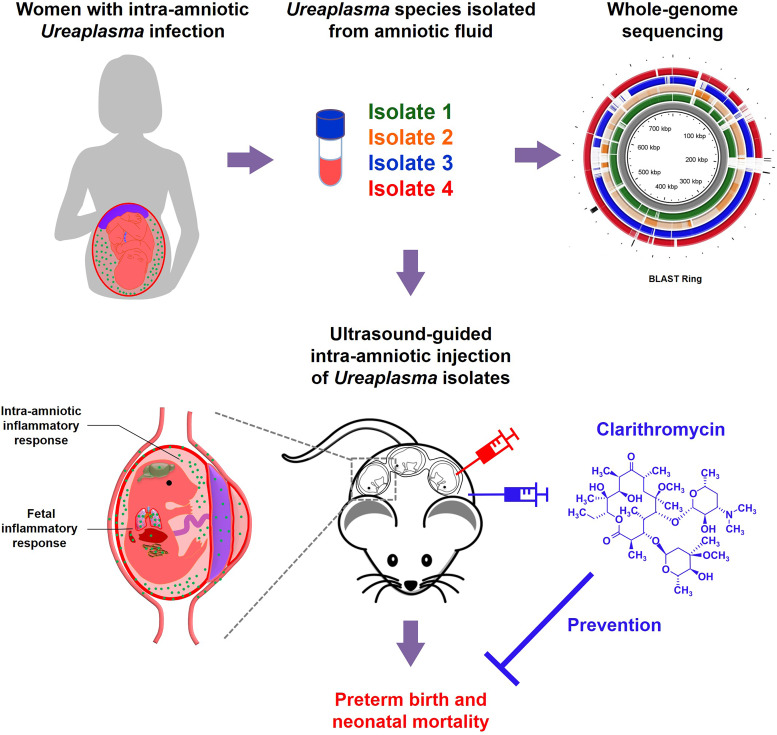
FIG 8.
Graphical abstract. Ureaplasma species were isolated from amniotic fluid of four women with intra-amniotic Ureaplasma infection. Whole-genome sequencing of each Ureaplasma isolate was performed (BLAST Ring shown in Fig. 1), and the isolates were taxonomically characterized, indicating that U. parvum was the most common genital mycoplasma in the amniotic cavity. The ultrasound-guided intra-amniotic injection of Ureaplasma species into pregnant mice resulted in adverse pregnancy and neonatal outcomes driven by severe intra-amniotic and fetal inflammatory responses, as well as the invasion of Ureaplasma species into specific tissues. Importantly, treatment with clarithromycin successfully prevented Ureaplasma-induced adverse pregnancy and neonatal outcomes.
MATERIALS AND METHODS
Ethics statement. (i) Human subjects and clinical specimens.
Human amniotic fluid samples and chorioamniotic membranes were obtained at the Perinatology Research Branch, an intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services, Wayne State University (Detroit, MI), and the Detroit Medical Center (Detroit, MI). The collection and use of human materials for research purposes were approved by the Institutional Review Boards of the National Institute of Child Health and Human Development and Wayne State University. All participating women provided written informed consent prior to sample collection.
(ii) Mice.
Animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Wayne State University (protocols A 07-03-15 and 18-03-0584). We adhered to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Amniotic fluid sample collection.
Amniotic fluid was retrieved by transabdominal amniocentesis under antiseptic conditions using ultrasound monitoring from women who were suspected to have intra-amniotic inflammation/infection (see clinical definitions below). All samples were collected to assess the presence of intra-amniotic infection (see clinical definitions below). The clinical and demographic characteristics of the study population are shown in Table S1 in the supplemental material. Amniotic fluid samples were transported to the clinical laboratory in a capped sterile syringe to perform research cultures for Ureaplasma species. In addition, an aliquot of amniotic fluid was transported to the clinical laboratory for culture of aerobic/anaerobic bacteria and genital mycoplasmas, as previously described (1, 7). The clinical and research tests also included the determination of an amniotic fluid white blood cell count, Gram stain examination, glucose concentration, and IL-6 concentration, as previously performed (57).
Clinical definitions and placental histopathologic examination.
The presence of viable bacteria in the amniotic cavity was evaluated by amniotic fluid culture as previously described and included culture for genital mycoplasmas. Intra-amniotic inflammation was defined as an amniotic fluid IL-6 concentration of ≥2.6 ng/ml (28). Microbial-associated intra-amniotic inflammation, or intra-amniotic infection, was defined as the presence of cultivable bacteria together with intra-amniotic inflammation (57). Histopathological examination of the placenta was performed by perinatal pathologists blinded to clinical diagnoses and obstetrical outcomes according to standardized Perinatology Research Branch protocols (58). Acute inflammatory lesions of the placenta (maternal inflammatory response and fetal inflammatory response) were diagnosed according to established criteria, including staging and grading (58). Preterm delivery was defined as delivery <37 weeks of gestation whereas term delivery occurred after 37 weeks of gestation.
Isolation of Ureaplasma species from amniotic fluid.
Amniotic fluid samples were inoculated in SP4 broth with urea (catalog no. R87; Hardy Diagnostics, Santa Maria, CA) to selectively culture Ureaplasma species. A change in the pH of the media resulted in a color change from yellow to red, which visually indicated the growth of bacteria. The color-changed media (n = 4) was mixed with 50% glycerol (Teknova, Hollister, CA; with a final concentration of glycerol of 15 to 25%), aliquoted, and stored at –80°C (original stocks, isolates 1 to 4). These isolates were used for in vivo and in vitro experiments. The clinical characteristics of these four patients are shown in Table S1.
Isolation of DNA from Ureaplasma isolates.
Amniotic fluid Ureaplasma isolates were recovered from original stocks (isolates 1 to 4) in SP4 broth with urea. Bacterial cells were pelleted by centrifugation at 10,000 × g at 4°C for 20 min. DNA was extracted from the pellets of bacterial cells using the DNeasy PowerSoil kit (Qiagen, Hilden, Germany) with minor modifications to the manufacturer’s protocol. Specifically, the pellets were resuspended in 200 μl of the bead solution, and the suspension was returned to the bead tube. Bacterial cells were lysed by mechanical disruption for 30 s using a bead beater (Mini-Beadbeater 16; Biospec Products, Bartlesville, OK). After centrifugation, the supernatants were transferred to new tubes, 100 μl of solution C2 and 100 μl of solution C3 were added, and the tubes were incubated at 4°C for 5 min. Finally, 60 μl of solution C6 was placed on the filter columns and incubated for 5 min before centrifuging for 30 s to elute the extracted DNA. Purified DNA was stored at –20°C.
Taxonomic identification of Ureaplasma isolates.
The 16S rRNA gene of each Ureaplasma isolate was amplified using the 27F/1492R primer set (5′-AGAGTTTGATCMTGGCTCAG-3′ / 5′-TACCTTGTTACGACTT-3′) and then bidirectionally Sanger sequenced by Genewiz (South Plainfield, NJ) using the 515F/806R primer set (5′-GTGYCAGCMGCCGCGGTAA-3′ / 5′-GGACTACNVGGGTWTCTAAT-3′), which targets the V4 hypervariable region of the 16S rRNA gene. Forward and reverse reads were trimmed using Chromatogram Explorer Lite (Heracle BioSoft S.R.L, Arges, Romania) with default settings; the reads were then assembled using the CAP (contig assembly program) of BioEdit software (v7.0.5.3), also with default settings. The taxonomic identities of individual bacterial isolates were determined using the Basic Local Alignment Search Tool (BLAST) (59). If a matching bacterial strain in the nucleotide collection (nr/nt) database had more than a single copy of the 16S rRNA gene, the percent nucleotide similarity values between the isolate and the strain were reported for the best matching sequence.
Genome sequencing, assembly, and analysis.
Amniotic fluid Ureaplasma isolates were recovered from original stocks in SP4 broth with urea. Bacterial cells were pelleted by centrifugation at 3,000 × g at room temperature for 30 min. The cells were washed with Tris-EDTA (TE) buffer (Teknova), resuspended in TE buffer, stored at –80°C, and sent to CosmosID (Rockville, MD) for DNA isolation, genomic sequencing, contig assembly, SNP analysis, phylogenetic analysis, antibiotic resistance analysis, and virulence factor analysis. Raw sequence reads were trimmed through the BBduk (BBMap package) package tool (60) using minlen = 25, and contig sequences were assembled in SPAdes (61) using 77, 99, and 127 k-mer sizes. Coding sequences were then annotated using the Prokka pipeline (62). For identification of antibiotic resistance genes and virulence factor genes, contigs were compared against CosmosID’s in-house databases. Antibiotic resistance and virulence factor genes were considered to be present if their sequences matched with those in an assembled genome at a >90% identity with 60% coverage. Phylograms were constructed from the alignment of the SNPs extracted for each isolate using Harvest Suite (including Parsnp and Gingr) (63).
Secondarily, contigs were assembled from raw sequence reads using CLC Genomics Workbench version 12 (Qiagen, Valencia, CA). Contig assembly settings included mapping reads back to the assembled contigs, updating contigs, automatic word and bubble sizes, and limiting the minimum contig length to 200 bp. Genome scaffolds were then generated by mapping contigs to a reference genome, either ATCC 27815 (CP000942) for U. parvum or ATCC 33699 (CP001184) for U. urealyticum. Multiple genome alignment was then performed using the BLAST Ring Image Generator (BRIG) (64) to compare whole-genome sequence similarities.
Mice.
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred in the animal care facility at the C. S. Mott Center for Human Growth and Development at Wayne State University (Detroit, MI). All mice were kept under a circadian cycle (light:dark, 12:12 h). Females, 8 to 12 weeks old, were bred with males of proven fertility. Female mice were checked daily between 8:00 a.m. and 9:00 a.m. for the appearance of a vaginal plug, which indicated 0.5 days post coitum (dpc). Females were then housed separately from the males, their weights were monitored daily, and a gain of two or more grams by 12.5 dpc confirmed pregnancy.
Preparation of Ureaplasma isolates for intra-amniotic inoculation.
The original stocks of the four clinical Ureaplasma isolates (isolates 1 to 4) were inoculated in SP4 broth and cultured at 37°C for 18 to 24 h. The culture medium was then mixed with the same volume of 50% glycerol (final concentration of glycerol, 25%), aliquoted, and stored at –80°C until use (working stocks). Aliquots of the working stocks were inoculated in SP4 broth and cultured at 37°C for 4 to 12 h to reach the exponential phase based on the growth rates of the individual working stocks. After incubation, the culture medium was collected and centrifuged at 3,000 × g for 30 min at room temperature. The resulting Ureaplasma pellet was resuspended with 1 ml of SP4 broth and kept on ice until intra-amniotic inoculation (inoculation suspension). The Ureaplasma cell count in the inoculation suspension was measured using flow cytometry by following a modified version of a previously described method for counting Mycoplasma (65) with a bacterial counting kit (catalog no. L34856; Live/Dead BacLight bacterial viability and counting kit for flow cytometry; Invitrogen by Thermo Fisher Scientific, Carlsbad, CA). Briefly, a fraction of the inoculation suspension (50 μl) was taken, diluted 10× with 1× PBS, and then stained with SYTO9 (final concentration, 10 μM) for 15 min at room temperature in the dark. Stained bacteria were centrifuged at 3,000 × g for 30 min at room temperature, resuspended with 500 μl of 1× PBS, and acquired using the BD LSR II flow cytometer (BD Bioscience, San Jose, CA) and BD FACSDiva 6.0 software (BD Bioscience). The absolute number of cells was determined using counting beads included in the kit. The Ureaplasma suspension was diluted with SP4 broth based on the obtained count. As the final yield of Ureaplasma cells varied in each inoculation suspension, those with final concentrations of Ureaplasma from ≥1 × 103 to ≤1 × 104 cells/25 μl were used for intra-amniotic inoculation of pregnant mice within a single study group (referred to as “low dose”), and those from >1 × 104 to ≤1 × 105 cells/25 μl were used for intra-amniotic inoculation of pregnant mice assigned to a second group (referred to as “high dose”). These doses were chosen based on concentrations of Ureaplasma species recovered from amniotic fluid of women with intra-amniotic infection. PBS and SP4 broth were used as the sham control and vehicle control, respectively. Since the intra-amniotic injection of SP4 broth alone caused increased concentrations of specific cytokines/chemokines in amniotic fluid (see Fig. S2) and upregulation of inflammatory genes in primary human amniocytes (Fig. S8), all experiments with Ureaplasma were compared to SP4 broth as the vehicle control treatment.
Animal model of Ureaplasma-induced intra-amniotic infection.
Dams were anesthetized at 16.5 dpc by inhalation of 2% isoflurane (Aerrane; Baxter Healthcare Corporation, Deerfield, IL) and 1 to 2 liters/min of oxygen in an induction chamber. This gestational age (16.5 dpc) was chosen for its equivalency to the third trimester of human pregnancy, when preterm birth most frequently occurs (6, 66). Anesthesia was maintained with a mixture of 1.5 to 2% isoflurane and 1.5 to 2 liters/min of oxygen. Mice were positioned on a heating pad and stabilized with adhesive tape. Fur removal from the abdomen and thorax was achieved by applying Nair cream (Church & Dwight Co., Inc., Ewing, NJ) to those areas. Body temperature was maintained in the range of 37 ± 1°C and detected with a rectal probe (VisualSonics, Toronto, Ontario, Canada), and respiratory and heart rates were monitored by electrodes embedded in the heating pad. An ultrasound probe was fixed and mobilized with a mechanical holder, and the transducer was slowly moved toward the abdomen. Ultrasound-guided intra-amniotic inoculation with Ureaplasma (low dose or high dose) was performed in each amniotic sac using a 30G needle (BD PrecisionGlide needle; Becton Dickinson, Franklin Lakes, NJ). Controls were injected with 25 μl of SP4 broth or sterile 1× PBS. The syringe was stabilized by a mechanical holder (VisualSonics). After the ultrasound examination, the mice were placed under a heat lamp for recovery (defined as when the mouse resumed normal activities such as walking and responding), which typically occurred 10 to 20 min after removal from anesthesia. After recovery, mice were video-monitored to observe pregnancy outcomes.
Video monitoring of pregnancy outcomes.
Pregnancy parameters, including the rates of preterm birth and pup mortality, were recorded via video camera (Sony Corporation, Tokyo, Japan). Preterm birth was defined as delivery occurring before 18.5 dpc, and its rate was represented by the percentage of females delivering preterm among the total number of mice injected. The rate of pup mortality for each litter was defined as the proportion of delivered pups found dead among the total litter size.
Tissue sampling from dams intra-amniotically inoculated with Ureaplasma isolates.
Dams were intra-amniotically inoculated with 1 × 104 cells/sac of Ureaplasma isolate 3 or Ureaplasma isolate 4, or injected with 25 μl of sterile 1× PBS or SP4 broth at 16.5 dpc, as described above. Mice were euthanized at 17.5 dpc (16 h post-inoculation), and maternal blood was collected by cardiac puncture and placed into a 1.5-ml Safe-Lock Eppendorf tube (Fisher Scientific, Hanover Park, IL). Serum (n = 6 each) was separated from the maternal peripheral blood and stored at −20°C until analysis. Animal dissection to obtain the maternal brain, maternal lung, maternal liver, maternal spleen, maternal para-aortic lymph node (PALN), decidua, uterus, cervix, fetal membranes, placenta, fetal brain, fetal lung, fetal thymus, fetal spleen, fetal liver, and fetal intestine (n = 6 each) was performed. Tissues were snap-frozen in liquid nitrogen and stored at –80°C until analysis or placed in RNAlater stabilization solution (Invitrogen/Thermo Fisher Scientific) according to the manufacturer’s instructions. Amniotic fluid was also collected from each amniotic sac with a 26G needle and placed into a 0.5-ml Safe-Lock Eppendorf tube (Fisher Scientific). Amniotic fluid samples were centrifuged at 1,300 × g for 10 min at 4°C, and the supernatants were separated and stored at −20°C until analysis.
Determination of cytokine concentrations in amniotic fluid and maternal serum.
Amniotic fluid and maternal serum samples were assessed for cytokine/chemokine concentrations. A ProcartaPlex mouse cytokine and chemokine panel 1A 36-plex (Invitrogen/Thermo Fisher Scientific) was used to measure the concentrations of IFNα, IFNγ, IL-12p70, IL-1β, IL-2, TNFα, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-18, IL-17A, IL-22, IL-23, IL-27, IL-9, IL-15/IL-15R, IL-13, IL-4, IL-5, IL-6, IL-10, Eotaxin (CCL11), IL-28, IL-3, LIF, IL-1α, IL-31, GRO-α (CXCL1), MIP-1α (CCL3), IP-10 (CXCL10), MCP-1 (CCL2), MCP-3 (CCL7), MIP-1β (CCL4), MIP-2 (CXCL2), RANTES (CCL5), G-CSF, M-CSF, and ENA-78 (CXCL5) in the serum and amniotic fluid samples, according to the manufacturer’s instructions. Plates were read by using the Luminex FLEXMAP 3D (Luminex, Austin, TX), and analyte concentrations were calculated with MILLIPLEX Analyst v5.1 Flex software (Vigene Tech, Carlisle, MA). The sensitivities of the assays were as follows: 3.03 pg/ml (IFNα), 0.09 pg/ml (IFNγ), 0.21 pg/ml (IL-12p70), 0.14 pg/ml (IL-1β), 0.10 pg/ml (IL-2), 0.39 pg/ml (TNFα), 0.19 pg/ml (GM-CSF), 9.95 pg/ml (IL-18), 0.08 pg/ml (IL-17A), 0.24 pg/ml (IL-22), 2.21 pg/ml (IL-23), 0.34 pg/ml (IL-27), 0.28 pg/ml (IL-9), 0.42 pg/ml (IL-15/IL-15R), 0.16 pg/ml (IL-13), 0.03 pg/ml (IL-4), 0.32 pg/ml (IL-5), 0.21 pg/ml (IL-6), 0.69 pg/ml (IL-10), 0.01 pg/ml (Eotaxin/CCL11), 20.31 pg/ml (IL-28), 0.11 pg/ml (IL-3), 0.28 pg/ml (LIF), 0.32 pg/ml (IL-1α), 0.45 pg/ml (IL-31), 0.05 pg/ml (GRO-α/CXCL1), 0.13 pg/ml (MIP-1α/CCL3), 0.26 pg/ml (IP-10/CXCL10), 3.43 pg/ml (MCP-1/CCL2), 0.15 pg/ml (MCP-3/CCL7), 1.16 pg/ml (MIP-1β/CCL4), 0.37 pg/ml (MIP-2/CXCL2), 0.35 pg/ml (RANTES/CCL5), 0.19 pg/ml (G-CSF), 0.02 pg/ml (M-CSF), and 5.67 pg/ml (ENA-78/CXCL5). Interassay and intra-assay coefficients of variation were less than 10%.
Bacterial burden determination of U. parvum in maternal and fetal tissues and at the maternal-fetal interface by real-time quantitative PCR.
Total DNA was isolated from the frozen tissues of the maternal brain, maternal lung, maternal liver, maternal spleen, maternal PALN, decidua, uterus, cervix, fetal membranes, placenta, fetal brain, fetal lung, fetal thymus, fetal spleen, fetal liver, and fetal intestine by using DNeasy blood and tissue kits (Qiagen), according to the manufacturer’s instructions. DNA concentrations were assessed with a Qubit dsDNA HS assay kit and a Qubit 3 fluorometer (Invitrogen/Thermo Fisher Scientific) according to the manufacturer’s instructions.
U. parvum DNA abundances in the mouse tissues were measured by qPCR using taxon-specific primers (5′-CCAGGTAAATTAGTACCAGG-3′ and 5′-CCTGATGGAATATCGAAACG-3′) targeting the ureB gene, as previously described (67). Each 20-μl reaction contained 0.6 μM concentrations of each primer, 10.0 μl of 2× QuantiTect SYBR green PCR kit (Qiagen), and 3.0 μl of purified DNA diluted to a maximum concentration of 50.0 ng/μl. Cycling conditions for the assay were as follows: 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 25 s. All qPCRs and data collection were carried out in 96-well plates with an ABI 7500 Fast real-time PCR system using software v2.3 (Applied Biosystems, Alameda, CA). Duplicate reactions were carried out for all samples, and all samples were run across a total of 12 batches.
DNA derived from the clinical U. parvum isolate (isolate 1) was quantified using a Qubit 3.0 fluorometer with a Qubit dsDNA HS assay kit (Invitrogen/Thermo Fisher Scientific) and subsequently used for the generation of standard curves. It was estimated that the U. parvum isolate genome has a mass of 4.78 × 105 kDa and contains a single copy of the ureB gene. A standard curve based on triplicate 10-fold serial dilutions ranging from 1.70 × 106 to 1.70 × 101 ureB gene copies was included in each qPCR run. The standard curves were used to evaluate the performance of the qPCR assay by estimation of its efficiency based on the slope of regression lines, according to a previous study (68). Raw amplification data were normalized to the ROX passive reference dye and analyzed using an automatic baseline setting and a threshold setting of 0.05. Quantification cycle (Cq) values for samples were calculated based on the mean number of cycles required for normalized fluorescence to exponentially increase.
After plotting a regression of log(ureB gene copy number) and Cq values for standard curves included in each qPCR run, the ureB gene copy number in mouse samples was calculated using the equation Xo = EAMP(b–Cq), where EAMP is the exponential amplification value for the qPCR assay, calculated as EAMP = 10(−1/m), and b and m are the intercept and slope of the regression (69).
RNA isolation, cDNA synthesis, and reverse transcription-quantitative PCR analysis of murine tissues.
Total RNA was isolated from the decidua, uterus, cervix, fetal membranes, placenta, fetal brain, fetal lung, fetal thymus, fetal spleen, fetal liver, and fetal intestine using QIAshredders, RNase-free DNase sets, and RNeasy Mini kits (all from Qiagen) according to the manufacturer’s instructions. RNA concentrations and integrity were evaluated with the Bioanalyzer 2100 (Agilent Technologies, Wilmington, DE). Complementary (c)DNA was synthesized by using SuperScript III first-strand synthesis supermix (Invitrogen/Thermo Fisher Scientific). Gene expression profiling was performed on the BioMark system for high-throughput RT-qPCR (Fluidigm, San Francisco, CA) with the TaqMan gene expression assays (Applied Biosystems/Life Technologies Corporation, Foster City, CA) listed in Table S2.
Mouse and human primers. Download Table S2, DOCX file, 0.02 MB (21.2KB, docx) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primary cell culture of human amnion epithelial cells.
Immediately after collection, the amnion membrane was manually peeled from the underlying chorion layer of the chorioamniotic membranes and then dissected into small pieces. The amnion fragments were rinsed in 0.05% (wt/vol) trypsin/EDTA (Life Technologies) and incubated with 15 ml of fresh trypsin-EDTA at 37°C with gentle shaking for 10 min. The trypsin digestion supernatant was discarded, and the amnion fragments were placed into fresh trypsin-EDTA solution and amnion epithelial cells (AECs) were obtained by digesting at 37°C for 40 min with gentle shaking. The total digestion/incubation process was repeated twice. Fetal bovine serum (FBS; Thermo Fisher Scientific) was added to the supernatant to stop digestion between each incubation period. Finally, digested tissues were filtered through a 100-μm cell strainer (Fisher Scientific, Durham, NC). The resulting cell suspensions were centrifuged at 300 × g for 10 min, and the cells were cultured in Dulbecco modified Eagle medium (Life Technologies) containing 10% FBS and 100 U/ml penicillin and streptomycin (Thermo Fisher Scientific) at 37°C with 5% CO2. The absence of mycoplasma contamination in AEC cultures was confirmed by qPCR using the MycoSensor QPCR assay kit (catalog no. 302107; Agilent Technologies, Santa Clara, CA). Immunofluorescence staining was performed with cells at passage 1. Cells at passage 2 were used for incubation with Ureaplasma isolates.
Immunofluorescence and confocal microscopy.
Amnion epithelial cells were cultured in four-well Lab-Tek chamber slides (Nunc, Rochester, NY) at 1 × 104 cells per well. Next, cells were fixed using 4% paraformaldehyde (Electron Microscopy Sciences Hatfield, PA) for 20 min at room temperature, rinsed with 1× PBS, and permeabilized using 0.25% Triton X-100 (EMD Millipore, Billerica, MA) for 5 min at room temperature. After 30 min of incubation with an Image-iT Fx signal enhancer (Invitrogen by Thermo Fisher Scientific), nonspecific antibody interactions were blocked using antibody diluent/block (catalog no. ARD1001EA; Perkin-Elmer, Boston, MA) for 30 min at room temperature. The cells were then stained with a mouse anti-human EpCAM (catalog no. MA5-12436; Invitrogen) antibody at room temperature for 1 h. Mouse IgG (Invitrogen/Thermo Fisher Scientific) was used as a negative control. After staining, the cells were washed with 1× PBS containing 0.1% Tween 20 (PBST; Sigma-Aldrich, Saint Louis, MO). After a wash step, secondary goat anti-mouse IgG–Alexa Fluor 594 (catalog no. A11032; Life Technologies) and Alexa Fluor 488 phalloidin (catalog no. A12379; Life Technologies) were added, and the cells were incubated for 1 h at room temperature in the dark. Finally, the cells were washed with PBST and mounted using ProLong Diamond antifade mountant with DAPI (4′,6′-diamidino-2-phenylindole; Life Technologies). Immunofluorescence was visualized using a Zeiss LSM 780 laser scanning confocal microscope (Carl Zeiss Microscopy, Jena, Germany) at the Microscopy, Imaging, and Cytometry Resources Core at the Wayne State University School of Medicine (http://micr.med.wayne.edu/).
Determination of inflammatory genes in primary amnion epithelial cells.
Amnion epithelial cells were plated into six-well tissue culture plates (Corning, Inc., Corning, NY) at 1.5 × 105 cells/well and cultured at 37°C with 5% CO2. The cells were then incubated with Ureaplasma isolate 3 or isolate 4 (1.3 × 105 cells/well with serum-free Opti-MEM [Life Technologies]), sterile 1× PBS with serum-free Opti-MEM, or SP4 broth with serum-free Opti-MEM as controls. After 24 h of treatment, total RNA was isolated from cells by using the RNeasy Mini kits (Qiagen) according to the manufacturer’s instructions. RNA concentrations were assessed with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized using a high-capacity cDNA reverse transcription kit with RNase inhibitor (Thermo Fisher Scientific, Waltham, MA), according to the manufacturer’s instructions. cDNA was amplified using TaqMan PreAmp master mix on an ABI 7500 Fast real-time PCR system. For all genes besides NLRP7, mRNA expression was determined by qPCR utilizing a BioMark high-throughput qRT-PCR system (Fluidigm) with the TaqMan gene expression assays (Applied Biosystems) listed in Table S2. For NLRP7, mRNA expression was determined by qPCR using an ABI 7500 Fast real-time PCR system with the TaqMan gene expression assays.
Clarithromycin treatment of mice intra-amniotically inoculated with Ureaplasma parvum.
Dams were intra-amniotically inoculated with 1 × 104 cells/25 μl of U. parvum (isolate 4) in each amniotic sac under ultrasound guidance at 16.5 dpc and then monitored via video camera until delivery in order to obtain the gestational age and rate of preterm birth. The dams were injected subcutaneously with 75 mg/kg of clarithromycin (CLR; Sigma-Aldrich) or its vehicle control (dimethyl sulfoxide [DMSO]) at 6, 18, 30, 54, and 78 h after intra-amniotic inoculation with U. parvum. Clarithromycin (reconstituted with DMSO) or DMSO (vehicle control) was diluted with sterile 5% dextrose water. The dose of CLR was determined based on a previous study (70) showing that a 75 mg/kg/day dose of CLR in mice was equivalent to human oral CLR dosages (ranging from 250 mg twice daily to 500 mg twice daily in adults [71]). The latter dose has been demonstrated to eradicate intra-amniotic infection or inflammation in a subset of women with preterm labor and intact membranes (23), cervical insufficiency (24), or preterm prelabor rupture of the membranes (22) when it is administered together with other antibiotics (ceftriaxone and metronidazole).
Statistical analysis.
Statistical analyses were performed using Prism v8 (GraphPad, San Diego, CA), the R package (https://www.r-project.org/), or Subio Platform v1.23.5551 (Subio, Inc., Kagoshima, Japan). To determine rates of preterm birth, a Fisher's exact test was used. Kaplan-Meier survival curves were used to plot and compare the gestational length data (a Gehan-Breslow-Wilcoxon test). The unpaired t test was performed for the statistical analysis of neonatal mortality. For multiplex assays, the concentrations of the cytokines/chemokines were log transformed and statistically tested by one-way analysis of variance (ANOVA), followed by a Tukey’s test to determine differences between groups. The statistical difference of the ureB gene copies between isolate 3 and isolate 4 was tested by the Mann-Whitney U test. For qPCR analysis in mouse tissues and AECs, negative delta threshold cycle (–ΔCT) values were determined using multiple reference genes (Gusb, Hsp90ab1, Gapdh, and Actb for mouse tissues or RPLP0, GAPDH, and ACTB for AECs) averaged within each sample to determine gene expression levels. In mouse tissues, the –ΔCT values were normalized by calculating the Z-score of each gene, and then heatmaps were created, which represent the mean of the Z-score of –ΔCT and the hierarchical clustering using uncentered correlation. Relative fold changes for mRNA expression were calculated using the 2–ΔΔCT method (72) and as shown as the log2(fold change). One-way ANOVA, followed by a Tukey’s test, was performed to assess the statistical significance of group comparisons for mRNA expression. A P value of <0.05 was considered statistically significant.
Data availability.
Most relevant data are present within the manuscript and in the supplemental material.
Mouse raw data may be requested from the corresponding author. Ureaplasma genome sequence files have been deposited to the NCBI Sequence Read Archive under the BioProject accession number PRJNA615648 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA615648).
ACKNOWLEDGMENTS
We gratefully acknowledge Marcia Arenas-Hernandez and George Schwenkel for their assistance with Ureaplasma cultures, Rona Wang and Hong Meng for their assistance with DNA isolation and PCR assays, and Bogdan Done for his support in statistical analyses. We thank the physicians and nurses from the Center for Advanced Obstetrical Care and Research and the Intrapartum Unit for their help in collecting human samples. We also thank the staff members of the PRB Clinical Laboratory and the PRB Histology/Pathology Unit for the processing and examination of the pathological sections.
This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), and, in part, with Federal funds from NICHD/NIH/DHHS under contract HHSN275201300006C (Funder ID https://doi.org.proxy.lib.wayne.edu/10.13039/100009633). R.R. has contributed to this work as part of his official duties as an employee of the U.S. Federal Government. This research was also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal and Child Health. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Motomura K, Romero R, Xu Y, Theis KR, Galaz J, Winters AD, Slutsky R, Garcia-Flores V, Zou C, Levenson D, Para R, Ahmad MM, Miller D, Hsu C-D, Gomez-Lopez N. 2020. Intra-amniotic infection with Ureaplasma parvum causes preterm birth and neonatal mortality that are prevented by treatment with clarithromycin. mBio 11:e00797-20. https://doi.org/10.1128/mBio.00797-20.
REFERENCES
- 1.Romero R, Miranda J, Kusanovic JP, Chaiworapongsa T, Chaemsaithong P, Martinez A, Gotsch F, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM. 2015. Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques. J Perinat Med 43:19–36. doi: 10.1515/jpm-2014-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SE, Romero R, Kim CJ, Shim SS, Yoon BH. 2006. Funisitis in term pregnancy is associated with microbial invasion of the amniotic cavity and intra-amniotic inflammation. J Matern Fetal Neonatal Med 19:693–697. doi: 10.1080/14767050600927353. [DOI] [PubMed] [Google Scholar]
- 3.Berger A, Witt A, Haiden N, Kretzer V, Heinze G, Pollak A. 2004. Amniotic cavity cultures, blood cultures, and surface swabs in preterm infants: useful tools for the management of early-onset sepsis? J Perinat Med 32:446–452. doi: 10.1515/JPM.2004.145. [DOI] [PubMed] [Google Scholar]
- 4.Romero R, Mazor M, Wu YK, Sirtori M, Oyarzun E, Mitchell MD, Hobbins JC. 1988. Infection in the pathogenesis of preterm labor. Semin Perinatol 12:262–279. [PubMed] [Google Scholar]
- 5.Romero R, Dey SK, Fisher SJ. 2014. Preterm labor: one syndrome, many causes. Science 345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chawanpaiboon S, Vogel JP, Moller AB, Lumbiganon P, Petzold M, Hogan D, Landoulsi S, Jampathong N, Kongwattanakul K, Laopaiboon M, Lewis C, Rattanakanokchai S, Teng DN, Thinkhamrop J, Watananirun K, Zhang J, Zhou W, Gulmezoglu AM. 2019. Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob Health 7:e37–e46. doi: 10.1016/S2214-109X(18)30451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero R, Sirtori M, Oyarzun E, Avila C, Mazor M, Callahan R, Sabo V, Athanassiadis AP, Hobbins JC. 1989. Infection and labor. V. Prevalence, microbiology, and clinical significance of intraamniotic infection in women with preterm labor and intact membranes. Am J Obstet Gynecol 161:817–824. doi: 10.1016/0002-9378(89)90409-2. [DOI] [PubMed] [Google Scholar]
- 8.Yoon BH, Chang JW, Romero R. 1998. Isolation of Ureaplasma urealyticum from the amniotic cavity and adverse outcome in preterm labor. Obstet Gynecol 92:77–82. doi: 10.1016/S0029-7844(98)00122-7. [DOI] [PubMed] [Google Scholar]
- 9.DiGiulio DB, Romero R, Kusanovic JP, Gomez R, Kim CJ, Seok KS, Gotsch F, Mazaki-Tovi S, Vaisbuch E, Sanders K, Bik EM, Chaiworapongsa T, Oyarzun E, Relman DA. 2010. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol 64:38–57. doi: 10.1111/j.1600-0897.2010.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vornhagen J, Quach P, Boldenow E, Merillat S, Whidbey C, Ngo LY, Adams Waldorf KM, Rajagopal L. 2016. Bacterial hyaluronidase promotes ascending GBS infection and preterm birth. mBio 7:e00781-16. doi: 10.1128/mBio.00781-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doster RS, Sutton JA, Rogers LM, Aronoff DM, Gaddy JA. 2018. Streptococcus agalactiae induces placental macrophages to release extracellular traps loaded with tissue remodeling enzymes via an oxidative burst-dependent mechanism. mBio 9:e03084-18. doi: 10.1128/mBio.02084-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson JA, Stemke GW, Davis JW Jr, Harasawa R, Thirkell D, Kong F, Shepard MC, Ford DK. 2002. Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974) Robertson et al. 2001. Int J Syst Evol Microbiol 52:587–597. doi: 10.1099/00207713-52-2-587. [DOI] [PubMed] [Google Scholar]
- 13.Kim M, Kim G, Romero R, Shim SS, Kim EC, Yoon BH. 2003. Biovar diversity of Ureaplasma urealyticum in amniotic fluid: distribution, intrauterine inflammatory response and pregnancy outcomes. J Perinat Med 31:146–152. doi: 10.1515/JPM.2003.020. [DOI] [PubMed] [Google Scholar]
- 14.Kong F, James G, Ma Z, Gordon S, Bin W, Gilbert GL. 1999. Phylogenetic analysis of Ureaplasma urealyticum–support for the establishment of a new species, Ureaplasma parvum. Int J Syst Bacteriol 49(Pt 4):1879–1889. doi: 10.1099/00207713-49-4-1879. [DOI] [PubMed] [Google Scholar]
- 15.Oh KJ, Lee KA, Sohn YK, Park CW, Hong JS, Romero R, Yoon BH. 2010. Intraamniotic infection with genital mycoplasmas exhibits a more intense inflammatory response than intraamniotic infection with other microorganisms in patients with preterm premature rupture of membranes. Am J Obstet Gynecol 203:211.e1–211.e8. doi: 10.1016/j.ajog.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh KJ, Romero R, Park JY, Hong JS, Yoon BH. 2019. The earlier the gestational age, the greater the intensity of the intra-amniotic inflammatory response in women with preterm premature rupture of membranes and amniotic fluid infection by Ureaplasma species. J Perinat Med 47:516–527. doi: 10.1515/jpm-2019-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber S, Vial Y, Hohlfeld P, Witkin SS. 2003. Detection of Ureaplasma urealyticum in second-trimester amniotic fluid by polymerase chain reaction correlates with subsequent preterm labor and delivery. J Infect Dis 187:518–521. doi: 10.1086/368205. [DOI] [PubMed] [Google Scholar]
- 18.Gravett MG, Witkin SS, Haluska GJ, Edwards JL, Cook MJ, Novy MJ. 1994. An experimental model for intraamniotic infection and preterm labor in rhesus monkeys. Am J Obstet Gynecol 171:1660–1667. doi: 10.1016/0002-9378(94)90418-9. [DOI] [PubMed] [Google Scholar]
- 19.Novy MJ, Duffy L, Axthelm MK, Sadowsky DW, Witkin SS, Gravett MG, Cassell GH, Waites KB. 2009. Ureaplasma parvum or Mycoplasma hominis as sole pathogens cause chorioamnionitis, preterm delivery, and fetal pneumonia in rhesus macaques. Reprod Sci 16:56–70. doi: 10.1177/1933719108325508. [DOI] [PubMed] [Google Scholar]
- 20.Senthamaraikannan P, Presicce P, Rueda CM, Maneenil G, Schmidt AF, Miller LA, Waites KB, Jobe AH, Kallapur SG, Chougnet CA. 2016. Intra-amniotic Ureaplasma parvum-induced maternal and fetal inflammation and immune responses in rhesus macaques. J Infect Dis 214:1597–1604. doi: 10.1093/infdis/jiw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Normann E, Lacaze-Masmonteil T, Eaton F, Schwendimann L, Gressens P, Thébaud B. 2009. A novel mouse model of Ureaplasma-induced perinatal inflammation: effects on lung and brain injury. Pediatr Res 65:430–436. doi: 10.1203/PDR.0b013e31819984ce. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Romero R, Kim SM, Chaemsaithong P, Yoon BH. 2016. A new antibiotic regimen treats and prevents intra-amniotic inflammation/infection in patients with preterm PROM. J Matern Fetal Neonatal Med 29:2727–2737. doi: 10.3109/14767058.2015.1103729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon BH, Romero R, Park JY, Oh KJ, Lee J, Conde-Agudelo A, Hong JS. 2019. Antibiotic administration can eradicate intra-amniotic infection or intra-amniotic inflammation in a subset of patients with preterm labor and intact membranes. Am J Obstet Gynecol 211:142.e1–142.e22. doi: 10.1016/j.ajog.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh KJ, Romero R, Park JY, Lee J, Conde-Agudelo A, Hong JS, Yoon BH. 2019. Evidence that antibiotic administration is effective in the treatment of a subset of patients with intra-amniotic infection/inflammation presenting with cervical insufficiency. Am J Obstet Gynecol 211:140.e1–140.e18. doi: 10.1016/j.ajog.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glass JI, Lefkowitz EJ, Glass JS, Heiner CR, Chen EY, Cassell GH. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757–762. doi: 10.1038/35037619. [DOI] [PubMed] [Google Scholar]
- 26.Momynaliev K, Klubin A, Chelysheva V, Selezneva O, Akopian T, Govorun V. 2007. Comparative genome analysis of Ureaplasma parvum clinical isolates. Res Microbiol 158:371–378. doi: 10.1016/j.resmic.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Yoder PR, Gibbs RS, Blanco JD, Castaneda YS, St Clair PJ. 1983. A prospective, controlled study of maternal and perinatal outcome after intra-amniotic infection at term. Am J Obstet Gynecol 145:695–701. doi: 10.1016/0002-9378(83)90575-6. [DOI] [PubMed] [Google Scholar]
- 28.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, Jun JK. 2001. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol 185:1130–1136. doi: 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 29.Romero R, Espinoza J, Goncalves LF, Kusanovic JP, Friel L, Hassan S. 2007. The role of inflammation and infection in preterm birth. Semin Reprod Med 25:21–39. doi: 10.1055/s-2006-956773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero R, Grivel JC, Tarca AL, Chaemsaithong P, Xu Z, Fitzgerald W, Hassan SS, Chaiworapongsa T, Margolis L. 2015. Evidence of perturbations of the cytokine network in preterm labor. Am J Obstet Gynecol 213:836.e1–836.e18. doi: 10.1016/j.ajog.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MJ, Romero R, Gervasi MT, Kim JS, Yoo W, Lee DC, Mittal P, Erez O, Kusanovic JP, Hassan SS, Kim CJ. 2009. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab Invest 89:924–936. doi: 10.1038/labinvest.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikhney J, von Schoning D, Steding I, Schulze J, Petrich A, Hiergeist A, Reischl U, Moter A, Thomas A. 2017. Is Ureaplasma spp. the leading causative agent of acute chorioamnionitis in women with preterm birth? Clin Microbiol Infect 23:e1–e7. doi: 10.1016/j.cmi.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Swanson KV, Deng M, Ting JP. 2019. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19:477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Glanville B. 2014. Activation and regulation of the innate immune system in response to Ureaplasma infection. PhD thesis Cardiff University, Cardiff, United Kingdom. [Google Scholar]
- 35.Renaudin H, Bebear C. 1990. Comparative in vitro activity of azithromycin, clarithromycin, erythromycin and lomefloxacin against Mycoplasma pneumoniae, Mycoplasma hominis and Ureaplasma urealyticum. Eur J Clin Microbiol Infect Dis 9:838–841. doi: 10.1007/BF01967388. [DOI] [PubMed] [Google Scholar]
- 36.Strauss JF III, Romero R, Gomez-Lopez N, Haymond-Thornburg H, Modi BP, Teves ME, Pearson LN, York TP, Schenkein HA. 2018. Spontaneous preterm birth: advances toward the discovery of genetic predisposition. Am J Obstet Gynecol 218:294–314.e2. doi: 10.1016/j.ajog.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cobo T, Kacerovsky M, Jacobsson B. 2018. Noninvasive sampling of the intrauterine environment in women with preterm labor and intact membranes. Fetal Diagn Ther 43:241–249. doi: 10.1159/000480232. [DOI] [PubMed] [Google Scholar]
- 38.Romero R, Chaemsaithong P, Chaiyasit N, Docheva N, Dong Z, Kim CJ, Kim YM, Kim JS, Qureshi F, Jacques SM, Yoon BH, Chaiworapongsa T, Yeo L, Hassan SS, Erez O, Korzeniewski SJ. 2017. CXCL10 and IL-6: markers of two different forms of intra-amniotic inflammation in preterm labor. Am J Reprod Immunol 78:e12685. doi: 10.1111/aji.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romero R, Gomez R, Ghezzi F, Yoon BH, Mazor M, Edwin SS, Berry SM. 1998. A fetal systemic inflammatory response is followed by the spontaneous onset of preterm parturition. Am J Obstet Gynecol 179:186–193. doi: 10.1016/S0002-9378(98)70271-6. [DOI] [PubMed] [Google Scholar]
- 40.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. 1998. The fetal inflammatory response syndrome. Am J Obstet Gynecol 179:194–202. doi: 10.1016/S0002-9378(98)70272-8. [DOI] [PubMed] [Google Scholar]
- 41.Moss TJ, Knox CL, Kallapur SG, Nitsos I, Theodoropoulos C, Newnham JP, Ikegami M, Jobe AH. 2008. Experimental amniotic fluid infection in sheep: effects of Ureaplasma parvum serovars 3 and 6 on preterm or term fetal sheep. Am J Obstet Gynecol 198:122.e1–122.e8. doi: 10.1016/j.ajog.2007.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yada Y, Honma Y, Koike Y, Takahashi N, Momoi MY. 2010. Association of development of chronic lung disease of newborns with neonatal colonization of Ureaplasma and cord blood interleukin-8 level. Pediatr Int 52:718–722. doi: 10.1111/j.1442-200X.2010.03095.x. [DOI] [PubMed] [Google Scholar]
- 43.Inatomi T, Oue S, Ogihara T, Hira S, Hasegawa M, Yamaoka S, Yasui M, Tamai H. 2012. Antenatal exposure to Ureaplasma species exacerbates bronchopulmonary dysplasia synergistically with subsequent prolonged mechanical ventilation in preterm infants. Pediatr Res 71:267–273. doi: 10.1038/pr.2011.47. [DOI] [PubMed] [Google Scholar]
- 44.Glaser K, Gradzka-Luczewska A, Szymankiewicz-Breborowicz M, Kawczynska-Leda N, Henrich B, Waaga-Gasser AM, Speer CP. 2019. Perinatal Ureaplasma exposure is associated with increased risk of late onset sepsis and imbalanced inflammation in preterm infants and may add to lung injury. Front Cell Infect Microbiol 9:68. doi: 10.3389/fcimb.2019.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero R, Mazor M. 1988. Infection and preterm labor. Clin Obstet Gynecol 31:553–584. doi: 10.1097/00003081-198809000-00006. [DOI] [PubMed] [Google Scholar]
- 46.Okogbule-Wonodi AC, Gross GW, Sun CC, Agthe AG, Xiao L, Waites KB, Viscardi RM. 2011. Necrotizing enterocolitis is associated with Ureaplasma colonization in preterm infants. Pediatr Res 69:442–447. doi: 10.1203/PDR.0b013e3182111827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silwedel C, Speer CP, Glaser K. 2017. Ureaplasma-associated prenatal, perinatal, and neonatal morbidities. Expert Rev Clin Immunol 13:1073–1087. doi: 10.1080/1744666X.2017.1381559. [DOI] [PubMed] [Google Scholar]
- 48.Steel JH, Malatos S, Kennea N, Edwards AD, Miles L, Duggan P, Reynolds PR, Feldman RG, Sullivan MH. 2005. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr Res 57:404–411. doi: 10.1203/01.PDR.0000153869.96337.90. [DOI] [PubMed] [Google Scholar]
- 49.Romero R, Chaemsaithong P, Docheva N, Korzeniewski SJ, Kusanovic JP, Yoon BH, Kim JS, Chaiyasit N, Ahmed AI, Qureshi F, Jacques SM, Kim CJ, Hassan SS, Chaiworapongsa T, Yeo L, Kim YM. 2016. Clinical chorioamnionitis at term VI: acute chorioamnionitis and funisitis according to the presence or absence of microorganisms and inflammation in the amniotic cavity. J Perinat Med 44:33–51 doi: 10.1515/jpm-2015-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pavlidis I, Spiller OB, Sammut Demarco G, MacPherson H, Howie SEM, Norman JE, Stock SJ. 2020. Cervical epithelial damage promotes Ureaplasma parvum ascending infection, intrauterine inflammation and preterm birth induction in mice. Nat Commun 11:199. doi: 10.1038/s41467-019-14089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomez-Lopez N, Romero R, Xu Y, Plazyo O, Unkel R, Leng Y, Than NG, Chaiworapongsa T, Panaitescu B, Dong Z, Tarca AL, Abrahams VM, Yeo L, Hassan SS. 2017. A role for the inflammasome in spontaneous preterm labor with acute histologic chorioamnionitis. Reprod Sci 24:1382–1401. doi: 10.1177/1933719116687656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faro J, Romero R, Schwenkel G, Garcia-Flores V, Arenas-Hernandez M, Leng Y, Xu Y, Miller D, Hassan SS, Gomez-Lopez N. 2019. Intra-amniotic inflammation induces preterm birth by activating the NLRP3 inflammasomedagger. Biol Reprod 100:1290–1305. doi: 10.1093/biolre/ioy261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez-Lopez N, Motomura K, Miller D, Garcia-Flores V, Galaz J, Romero R. 2019. Inflammasomes: their role in normal and complicated pregnancies. J Immunol 203:2757–2769. doi: 10.4049/jimmunol.1900901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardy DJ, Guay DR, Jones RN. 1992. Clarithromycin, a unique macrolide: a pharmacokinetic, microbiological, and clinical overview. Diagn Microbiol Infect Dis 15:39–53. doi: 10.1016/0732-8893(92)90055-x. [DOI] [PubMed] [Google Scholar]
- 55.Ichiyama T, Nishikawa M, Yoshitomi T, Hasegawa S, Matsubara T, Hayashi T, Furukawa S. 2001. Clarithromycin inhibits NF-κB activation in human peripheral blood mononuclear cells and pulmonary epithelial cells. Antimicrob Agents Chemother 45:44–47. doi: 10.1128/AAC.45.1.44-47.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimizu T, Kida Y, Kuwano K. 2008. Ureaplasma parvum lipoproteins, including MB antigen, activate NF-κB through TLR1, TLR2, and TLR6. Microbiology 154:1318–1325. doi: 10.1099/mic.0.2007/016212-0. [DOI] [PubMed] [Google Scholar]
- 57.Romero R, Miranda J, Chaiworapongsa T, Korzeniewski SJ, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Yeo L. 2014. Prevalence and clinical significance of sterile intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Reprod Immunol 72:458–474. doi: 10.1111/aji.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM. 2015. Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol 213:S29–S52. doi: 10.1016/j.ajog.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 60.Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner, abstr Abstracts 9th Annual Genomics of Energy and Environment Meeting, Walnut Creek, CA, March 17–20, 2014. [Google Scholar]
- 61.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 63.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Assuncao P, Diaz R, Comas J, de Galarreta CM, Gonzalez-Llamazares OR, Poveda JB. 2005. Evaluation of Mycoplasma hyopneumoniae growth by flow cytometry. J Appl Microbiol 98:1047–1054. doi: 10.1111/j.1365-2672.2005.02536.x. [DOI] [PubMed] [Google Scholar]
- 66.Blencowe H, Born Too Soon Preterm Birth Action Group, Cousens S, Chou D, Oestergaard M, Say L, Moller AB, Kinney M, Lawn J. 2013. Born too soon: the global epidemiology of 15 million preterm births. Reprod Health 10(Suppl 1):S2. doi: 10.1186/1742-4755-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishiumi F, Ogawa M, Nakura Y, Hamada Y, Nakayama M, Mitobe J, Hiraide A, Sakai N, Takeuchi M, Yoshimori T, Yanagihara I. 2017. Intracellular fate of Ureaplasma parvum entrapped by host cellular autophagy. Microbiologyopen 6:e00441. doi: 10.1002/mbo3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M. 2015. How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif 3:9–16. doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gallup J. 2011. qPCR inhibition and amplification of difficult templates, p 23–66. In Kennedy S, Oswald N (ed), PCR troubleshooting and optimization: the essential guide. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 70.Tagliabue C, Techasaensiri C, Torres JP, Katz K, Meek C, Kannan TR, Coalson JJ, Esposito S, Principi N, Leff R, Baseman JB, Hardy RD. 2011. Efficacy of increasing dosages of clarithromycin for treatment of experimental Mycoplasma pneumoniae pneumonia. J Antimicrob Chemother 66:2323–2329. doi: 10.1093/jac/dkr306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain R, Danziger LH. 2004. The macrolide antibiotics: a pharmacokinetic and pharmacodynamic overview. Curr Pharm Des 10:3045–3053. doi: 10.2174/1381612043383322. [DOI] [PubMed] [Google Scholar]
- 72.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and demographic characteristics of women from whose amniotic fluid Ureaplasma species were isolated. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pregnancy outcomes of dams injected with PBS or SP4 broth. (a) Rates of preterm birth of dams intra-amniotically injected with PBS or SP4 broth. The P values were determined by a Fisher's exact test. (b) Kaplan-Meier survival curve showing the gestational ages at delivery of dams intra-amniotically injected with PBS or SP4 broth. Statistical analysis was performed using a Gehan-Breslow-Wilcoxon test. (c) Mortality rates of neonates born to dams intra-amniotically injected with PBS or SP4 broth. Statistical analysis was performed using an unpaired t test. (d) Mortality rates of only those neonates born at term to dams intra-amniotically injected with PBS or SP4 broth. Statistical analysis was performed using an unpaired t test. The results of all statistical analyses were non-significant. Download FIG S1, TIF file, 1.3 MB (1.2MB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cytokine and chemokine concentrations in amniotic fluid from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. P values were determined by using unpaired t tests. *, P < 0.05; **, P < 0.01. Download FIG S2, TIF file, 0.1 MB (102.6KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cytokine and chemokine concentrations in maternal serum from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. Statistical analysis was performed by using unpaired t tests. The results of all statistical analyses were non-significant. Download FIG S3, TIF file, 0.1 MB (96.8KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial burden in the fetal, maternal, reproductive, and gestational tissues from dams intra-amniotically injected with PBS. (a) Copy numbers of the ureB gene in the fetal organs. (b) Copy numbers of the ureB gene in the maternal, reproductive, and gestational tissues. Download FIG S4, TIF file, 0.1 MB (53.3KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inflammatory gene expression in the fetal intestine from dams intra-amniotically inoculated with U. parvum or injected with SP4 broth or PBS. (a) Expression of Il1b in the fetal intestine from dams intra-amniotically inoculated with U. parvum serovar 14 isolate 3 or U. parvum serovar 14 isolate 4, or injected with SP4 broth (control). (b) Expression of Il1b in the fetal intestine from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. Statistical analysis was performed using one-way analysis of variance (ANOVA), followed by a Tukey’s test, to determine differences between groups (a), or an unpaired t test (b). **, P < 0.01; ***, P < 0.001. Download FIG S5, TIF file, 0.04 MB (45.7KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inflammatory gene expression in the fetal organs from dams intra-amniotically injected with PBS or SP4 broth. (a to c) Expression of specific inflammatory genes in the fetal lung (a), fetal spleen (b), and fetal liver (c) from dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. Statistical analysis was performed by using unpaired t tests. The results of all statistical analyses were non-significant. Download FIG S6, TIF file, 0.1 MB (149.4KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inflammatory gene expression in the reproductive and gestational tissues from dams intra-amniotically injected with PBS or SP4 broth. The expression of specific inflammatory genes in the fetal membranes, placenta, decidua, uterus, and cervix of dams intra-amniotically injected with PBS or SP4 broth. The data are shown as box-and-whisker plots where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum and maximum ranges. P values were determined by using unpaired t tests. *, P < 0.05. Download FIG S7, TIF file, 0.1 MB (118.2KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of specific inflammatory genes in primary human amniocytes incubated with U. parvum serovar 14, PBS, or SP4 broth. (a) Expression of NLRP3 in primary human amnion epithelial cells incubated with U. parvum isolate 3, isolate 4, or SP4 broth. (b) Expression of inflammatory genes in primary human amnion epithelial cells incubated with PBS or SP4 broth. The data are shown as medians with minimum and maximum ranges. P values were determined by one-way analysis of variance (ANOVA), followed by a Tukey’s test, to determine differences between groups (a), or unpaired t tests (b). **, P < 0.01; ***, P < 0.001. Download FIG S8, TIF file, 0.1 MB (130.2KB, tif) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mouse and human primers. Download Table S2, DOCX file, 0.02 MB (21.2KB, docx) .
Copyright © 2020 Motomura et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Most relevant data are present within the manuscript and in the supplemental material.
Mouse raw data may be requested from the corresponding author. Ureaplasma genome sequence files have been deposited to the NCBI Sequence Read Archive under the BioProject accession number PRJNA615648 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA615648).