The work presented here solved a long-standing paradox of the negative effects of certain missense alleles of envZ, which codes for kinase of the EnvZ/OmpR two-component system, on the expression of ferric uptake genes. The data revealed that the constitutive envZ alleles activate the Feo- and OmpC-mediated ferrous uptake pathway to flood the cytoplasm with accessible ferrous iron. This activates the ferric uptake regulator, Fur, which inhibits ferric uptake system but cannot inhibit the feo operon due to the positive effect of activated EnvZ/OmpR. The data also revealed the importance of porins in iron homeostasis.
KEYWORDS: iron homeostasis, two-component signal transduction systems, porins, ferric transport, ferrous transport, EnvZ/OmpR
ABSTRACT
Escherichia coli secretes high-affinity Fe3+ chelators to solubilize and transport chelated Fe3+ via specific outer membrane receptors. In microaerobic and anaerobic growth environments, where the reduced Fe2+ form is predominant, ferrous transport systems fulfill the bacterial need for iron. Expression of genes coding for iron metabolism is controlled by Fur, which when bound to Fe2+ acts as a repressor. Work carried out here shows that the constitutively activated EnvZ/OmpR two-component system, which normally controls expression of the ompC and ompF porin genes, dramatically increases the intracellular pool of accessible iron, as determined by whole-cell electron paramagnetic resonance spectroscopy, by inducing the OmpC/FeoB-mediated ferrous transport pathway. Elevated levels of intracellular iron in turn activated Fur, which inhibited the ferric transport pathway but not the ferrous transport pathway. The data show that the positive effect of constitutively activated EnvZ/OmpR on feoB expression is sufficient to overcome the negative effect of activated Fur on feoB. In a tonB mutant, which lacks functional ferric transport systems, deletion of ompR severely impairs growth on rich medium not supplemented with iron, while the simultaneous deletion of ompC and ompF is not viable. These data, together with the observation of derepression of the Fur regulon in an OmpC mutant, show that the porins play an important role in iron homeostasis. The work presented here also resolves a long-standing paradoxical observation of the effect of certain mutant envZ alleles on iron regulon.
INTRODUCTION
Iron, used as a redox center by many enzymes, is an essential trace metal required by almost all living organisms. The intracellular level of free catalytically active iron is typically kept low due to its toxic effects. Free ferrous iron reacts with hydrogen peroxide, a natural by-product of aerobic respiration, to generate highly toxic hydroxyl radicals (OH⋅) via the Fenton reaction (1). Due to this potentially damaging property of iron, there exists an intricate balance between iron transport, utilization, and storage. Most bacteria possess mechanisms to import iron in its oxidized ferric state (Fe3+), reduced ferrous state (Fe2+), or both (for reviews, see references 2 and 3). The solubility of these two iron forms differs drastically at neutral pH: ferric iron has extremely low solubility at 10−18 M, whereas ferrous iron is readily soluble at 10−1 M. To take up ferric iron, bacteria have developed high-affinity ferric iron chelators called siderophores to capture, solubilize, and deliver insoluble iron into the cell (4). Unlike the Fe3+ transport system, which requires a number of proteins involved in siderophore synthesis and Fe3+ siderophore acquisition, the Fe2+ transport system appears to consist of mainly one protein, FeoB (5). The FeoB protein is synthesized from the feoABC operon, whose expression is activated by Fnr, an anaerobic transcriptional regulator (5). FeoB is a highly conserved, 773-residue inner membrane protein that contains several GTP-binding motifs (6–8). In the absence of FeoB or FeoA, Fe2+ uptake is either virtually abolished (ΔfeoB) or mildly reduced (ΔfeoA) (5). The function of FeoC, which is present only in members of the Enterobacteriaceae family, is unknown (7, 8). FeoB and its homologs are required for full virulence in many bacteria, including Escherichia coli (9), Salmonella enterica serovar Typhimurium (10, 11), and Helicobacter pylori (12).
Fur (ferric uptake regulator) in E. coli and its orthologs in many Gram-negative and Gram-positive bacteria are the master regulator of genes encoding both ferric and ferrous iron acquisition functions, as well as siderophore synthesis and uptake (13, 14). Cells lacking Fur experience iron overload that causes oxidative damage and mutagenesis (15). Fur-regulated genes contain one or more Fur-binding sites around the −35 and −10 regions of the promoter, often referred to as the Fur boxes (16, 17). Fur uses Fe2+ as a cofactor: when the level of available Fe2+ increases in the cell, it binds to Fur and enhances its affinity for DNA by almost 1,000-fold (2). The active Fur-Fe2+ complex then binds to a Fur box and represses transcription of the iron acquisition gene. RyhB is a small regulatory RNA whose transcription is also repressed by Fur-Fe2+ (18). Consequently, when Fur is active, the levels of RyhB are low, resulting in stabilization and translation of over a dozen mRNAs encoding nonessential iron utilization proteins, including those that store iron (Bfr), detoxify superoxide (SodB), and catalyze steps of the tricarboxylic acid cycle (AcnA and SdhCDAB) (19). Thus, excess Fe2+ activates Fur to halt further iron uptake and at the same time, promotes the utilization of Fe2+, and inversely, low intracellular iron level induces iron uptake and utilization (20). Recent genome-wide analyses revealed a more comprehensive profile of Fur and RyhB regulons (21, 22).
Whereas Fur and RyhB are the principal determinants of iron homeostasis in E. coli, evidence exists supporting the involvement of some two-component signal transduction systems (TCS) in iron homeostasis. EnvZ and OmpR are the archetypal TCS in E. coli, where EnvZ serves as a sensor kinase and OmpR as a response regulator (23). They respond to medium osmolarity and influence the expression of OmpC and OmpF, the two major porins that facilitate the diffusion of small hydrophilic solutes (∼600 Da) across the outer membrane (24). OmpC is preferentially expressed in high osmolarity, whereas OmpF expression is favored in low osmolarity (25). Microarray data from an ΔompR ΔenvZ background showed a significant increase in the expression of a number of Fur-regulated genes, particularly those involved in enterobactin siderophore synthesis and transport (26). Over 3 decades ago, Lundrigan and Earhart (27) reported that in a perA (envZ) mutant background, the levels of three iron-regulated proteins were significantly reduced. These authors suggested that this could be due to a posttranscriptional defect. Later, it was speculated that this inhibition could be due to the indirect effects of envZ/ompR, leading to alterations in the structure and diffusion properties of the outer membrane (28). While characterizing revertants of an E. coli mutant defective in outer membrane biogenesis, we discovered several pleiotropic envZ alleles conferring an OmpC+ OmpF− LamB− phenotype (29). These alleles were hypothesized to biochemically lock EnvZ into a conformation that causes increased OmpR phosphorylation. This activated EnvZ/OmpR state is thought to enable OmpR to bind to promoters with weak OmpR-binding affinities. One such pleiotropic envZ allele, envZR397L, was characterized in detail (29). The preliminary whole genomic microarray analysis of the envZR397L mutant carried out in our laboratory found that the largest group of genes (>50) affected by the activated EnvZR397L/OmpR+ background belonged to the Fur regulon (30; unpublished data).
In this study, we show that EnvZR397L exerts its effect on the Fur regulon in part by increasing the accessible intracellular pool of iron via the OmpC-FeoB-mediated Fe2+ transport pathway. This, in turn, activates Fur and downregulates the Fe3+ transport pathway. Our analyses also revealed the critical roles of EnvZ/OmpR and porins in iron homeostasis in the ΔtonB background where high-affinity iron transport systems are nonfunctional.
RESULTS
Effects of envZR397L on the ferric transport system.
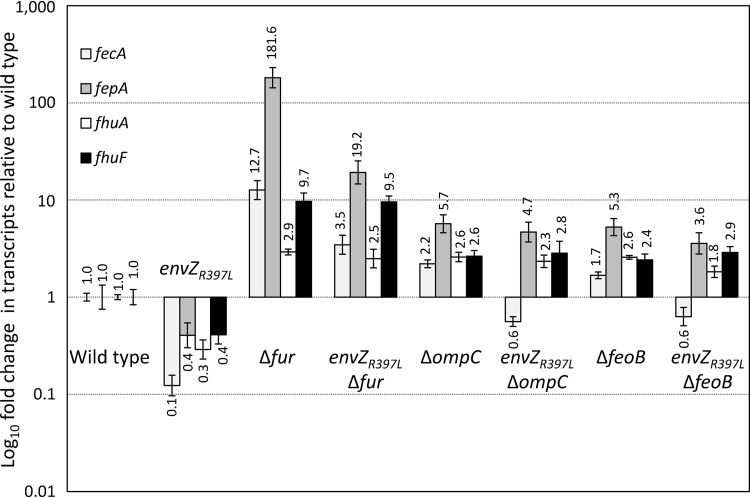
We first set out to investigate the effects of envZR397L on the Fur regulon. RNA isolated from mid-log-phase grown cells was converted to cDNA, and the levels of various transcripts were analyzed by real-time quantitative PCR (RT-qPCR). The data in Fig. 1 show relative transcript levels of four Fur-regulated genes: fecA, fepA, fhuA, and fhuF. In the envZR397L background, their transcript levels went down 10 (fecA)-, 3 (fhuA)-, and 2.5 (fepA and fhuF)-fold relative to the wild-type (EnvZ+) strain. As expected, in a Δfur background their expression was derepressed, resulting in a dramatic increase in their transcripts (Fig. 1). In that background, the presence of envZR397L was still able to reduce fecA and fepA transcript levels 3.6- and 9.5-fold, respectively, but not that of fhuA and fhuF, which experienced a <20% reduction (Fig. 1). Using the chromosomally integrated fepA::lacZ and fhuA::lacZ gene fusion constructs, we were able to recapitulate the key RNA data shown in Fig. 1 (see Fig. S1 in the supplemental material). This indicated that EnvZR397L/OmpR or factors under the activated TCS control could also downregulate fecA and fepA transcription in the absence of Fur. In contrast, the negative effect of envZR397L on fhuA and fhuF expression requires Fur. Moreover, the repressive effect of envZR397L on fecA and fepA in the fur+ background was found to be independent of OmrA and OmrB (Fig. S2), the two EnvZ/OmpR-dependent small regulatory RNAs whose overexpression from plasmids was previously shown to downregulate fecA, fepA, and other Fur-regulated genes (31). It is worth mentioning that the envZR397L allele has been previously shown to increase omr::lacZ expression almost 10-fold (29).
FIG 1.
Determination of fecA, fepA fhuA, and fhuF expression in different genetic backgrounds by real-time quantitative PCR (RT-qPCR). RNA was isolated from bacterial cultures grown to mid-log phase. Relative quantification of transcripts was performed using the 2–ΔΔCT method, with ftsL and purC serving as the reference genes. The relative fold changes in gene expression and error bars representing standard deviations are shown. The bacterial strains used included RAM1292 (wild type), RAM1541 (envZR397L), RAM2697 (Δfur), RAM2698 (envZR397L Δfur), RAM2699 (ΔompC), RAM2700 (envZR397L ΔompC), RAM2701 (ΔfeoB), and RAM2702 (envZR397L ΔfeoB).
Determination of fepA, fhuA, and feo expression using chromosomally integrated lacZ fusions. β-Galactosidase activities (shown in Miller units) were measured from at three independent biological replicates in various genetic backgrounds as shown. Error bars represent standard deviations. Download FIG S1, TIF file, 0.6 MB (638.9KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effects of OmrA and OmrB deletion on fecA and fepA in wild-type and EnvZR397L backgrounds. Determination of fecA and fepA expression was carried out using RT-qPCR. Data were obtained from two independent biological samples and two technical replicates. Error bars represent standard deviations. Download FIG S2, TIF file, 0.2 MB (172KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of OmpC is activated constitutively in the envZR397L background, while that of OmpF and LamB is severely inhibited (29). To determine whether OmpC is somehow involved in the envZR397L-mediated downregulation of fecA, fepA, fhuA, and fhuF, we examined their transcript levels in the ΔompC and ΔompC envZR397L backgrounds. Remarkably, without OmpC, envZR397L was unable to exert any significantly negative effect on fepA, fhuA, and fhuF expression, while the effect on fecA diminished from 10-fold in the presence of OmpC to <2-fold without OmpC (Fig. 1). Interestingly, the levels of all four transcripts went up in ΔompC cells (Fig. 1). We theorize that without OmpC, diffusion of Fe2+ into the cell is decreased and the less active Fur fails to fully repress fecA, fepA, fhuA, and fhuF expression.
If the intake of Fe2+ by OmpC porin increases active Fur-Fe2+ levels, then the absence of FeoB, the Fe2+-specific iron transporter, should also interfere with this activation and abrogate the Fur-mediated effects of envZR397L on fecA, fepA, fhuA, and fhuF. Indeed, just like in the ΔompC background, fecA, fepA, fhuA, and fhuF transcript levels went up in the ΔfeoB background, and envZR397L could either no longer impose a significant negative effect (fepA, fhuA, and fhuF) or the effect was significantly reduced (fecA).
Effects of envZR397L on the ferrous transport system.
The data presented in Fig. 1 show the involvement of the FeoB ferrous iron transporter and OmpC porin in envZR397L-mediated downregulation of the ferric iron transport system. While ompC expression increases in the envZR397L background (29), the status of the feo operon in this background is unknown. The feo operon is under the negative control of Fur (5). Consequently, if higher Fur-Fe2+ activity is present in the envZR397L background, as we have suggested above, then the expression of the feo operon, like that of fecA, fepA, fhuA, and fhuF, should also be inhibited. This, however, will be incongruent with our data showing envZR397L’s dependence on feoB for its effects. We therefore hypothesized that feo expression, like that of ompC, is activated by envZR397L to such a degree that it more than compensated for the feo downregulation by increased Fur-Fe2+ activity.
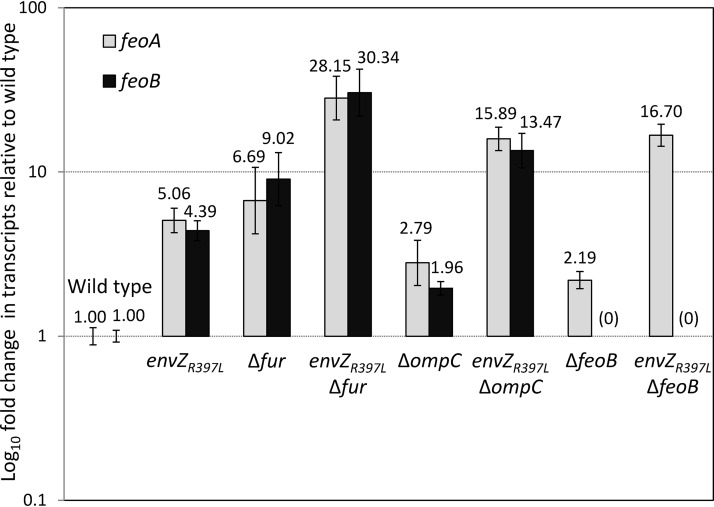
To test these possibilities, we analyzed feoA and feoB transcript levels in different genetic backgrounds by RT-qPCR (Fig. 2). Note that feoABC are part of a contiguous operon and therefore likely expressed from a polycistronic message. Consequently, feoA and feoB transcript analysis probes their respective coding regions in a polycistronic message. In the EnvZR397L background, feoA and feoB transcript levels went up dramatically over those in the envZ+ control strain. As expected, their levels also went up in the Δfur background. Interestingly, in the envZR397L Δfur background feoA and feoB transcript levels increased well above those in the individual mutation backgrounds, indicating that envZR397L and Δfur act independently and synergistically to enhance feo expression. Again, these observations were recapitulated using the chromosomally integrated feo::lacZ fusion (Fig. S1). These data support our hypothesis that envZR397L activates feo expression in a fashion that counteracts repression by higher levels of Fur-Fe2+.
FIG 2.
Determination of the relative gene expression of feoA and feoB by RT-qPCR. RNA was isolated from bacterial cultures grown to mid-log phase. Relative quantification of transcripts in various genetic backgrounds was performed using the 2–ΔΔCT method, with ftsL and purC serving as reference genes. The relative fold changes in gene expression and error bars representing standard deviations are shown. The bacterial strains used included RAM1292 (wild type), RAM1541 (envZR397L), RAM2697 (Δfur), RAM2698 (envZR397L Δfur), RAM2699 (ΔompC), RAM2700 (envZR397L ΔompC), RAM2701 (ΔfeoB), and RAM2702 (envZR397L ΔfeoB).
We then examined the effects of envZR397L on feoA and feoB transcript levels in the absence of OmpC or FeoB. Without OmpC or FeoB, a modest 2-fold increase in feoA and feoB (ΔompC) or feoA (ΔfeoB) transcripts was observed (Fig. 2). We interpret this to reflect a modest relief in the Fur-mediated repression of the feo operon, since we have already implicated OmpC and FeoB in the ferrous iron transport and increase in Fur-Fe2+ levels (Fig. 1). The presence of envZR397L in the ΔompC or ΔfeoB background led to an increase in feoA and feoB, or feoA transcripts, respectively, in a synergistic fashion, which is likely due to the simultaneous activation of feo expression by envZR397L and a modest decrease in the Fur-mediated repression of feo from the absence of OmpC and FeoB. These data showed that envZR397L inhibits ferric transport pathway but activates ferrous transport pathway.
Intracellular iron levels in the envZR497L mutant.
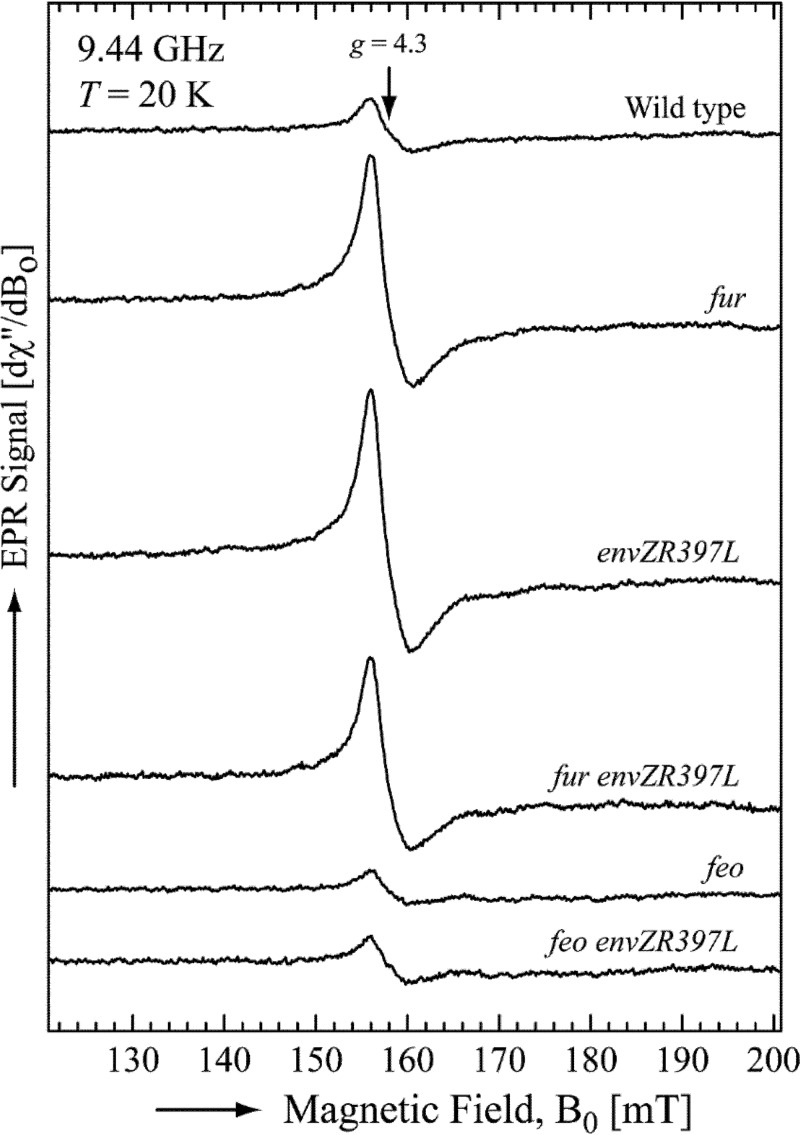
The OmpC/Feo-mediated increase in Fur-Fe2+ activity in the envZR397L background implies that the cytoplasm of the envZR397L mutant contains higher levels of accessible iron than that in the cytoplasm of the EnvZ+ cell. To test this directly, we measured the intracellular pool of accessible iron by whole-cell electron paramagnetic resonance (EPR) spectroscopy, a method established in the Imlay laboratory (32). The data presented in Fig. 3 show that the wild-type (EnvZ+) strain had 32 μM of accessible intracellular iron. Expectedly, this level rose 4-fold to 120 μM in the Δfur mutant. Remarkably, the level of accessible iron in the envZR397L was also very high (135 μM) and remained high in the Δfur envZR397L double mutant (105 μM), thus supporting the notion that a higher pool of accessible iron in the envZR397L background is responsible for the higher levels of active Fur-Fe2+.
FIG 3.
Determination of the intracellular free iron concentration. The averages of five ferric-chelate EPR scans per strain are shown. All scans were normalized to the final culture OD600 used in the measurements. The EPR parameters were as follows: microwave power, 10 mW; microwave frequency, 9.44 GHz; center field, 160 mT; sweep width, 80 mT; modulation amplitude, 1.25 mT; and modulation frequency, 100 kHz. The free intracellular iron concentrations, calculated as described in Materials and Methods, were as follows: wild type, 32 μM; Δfur, 120 μM; envZR397L, 135 μM; Δfur envZR397L, 105 μM; ΔfeoB, 20 μM; and ΔfeoB envZR397L, 29 μM. The bacterial strains used included RAM1292 (wild type), RAM1541 (envZR397L), RAM2697 (Δfur), RAM2698 (envZR397L Δfur), RAM2701 (ΔfeoB), and RAM2702 (envZR397L ΔfeoB).
Next, we tested whether the FeoB-mediated ferrous transport pathway is responsible for the elevated level of accessible iron in the envZR397L mutant. The accessible iron level in the ΔfeoB mutant was 20 μM or 35% less than the parental feoB+ strain (Fig. 3), explaining the observed deregulation of the Fur regulon in the ΔfeoB mutant (Fig. 1 and 2). Strikingly, without feoB, envZR397L failed to increase intracellular iron levels (Fig. 3), thus confirming the involvement of the FeoB-mediated ferrous transport in elevating the intracellular pool of iron, which, in turn, would increase Fur-Fe2+ levels and repress expression of fecA, fepA, fhuA, and fhuF. As described below, EnvZ/OmpR play a more direct role in activating feo expression to overcome the Fur-mediated downregulation.
Effects of envZR397L on fepA and feo requires phosphorylated OmpR.
Previously, it was shown that the pleiotropic effects of the mutant envZ allele, envZ473 with its V241G substitution, is mediated through OmpR (33). In that study, the authors did not analyze the iron regulon. Here, we sought to test whether the effect of envZR397L on iron regulon requires functional OmpR. We used a missense allele of ompR with a D55Y substitution, which confers a null phenotype with respect to ompC and ompF expression, presumably due to the inability of the mutant OmpR to be phosphorylated. The conserved D55 residue of OmpR is the site of phosphorylation (34). The ompRD55Y allele was isolated in a fepA::lacZ envZR397L background among Lac+ revertants (R. Misra, unpublished data). Using a linked chloramphenicol resistance (Cmr) marker, we transduced the ompRD55Y envZR397L mutations into a feo::lacZ background so that the effects of the mutant ompR and envZ alleles on feo expression can be determined. It is worth noting that although ompR/envZ are highly linked to the feo operon, we were able to construct the above strain since feo::lacZ is marked by the kanamycin resistance (Kmr) gene and the mutant ompR/envZ alleles produce a distinct porin phenotype.
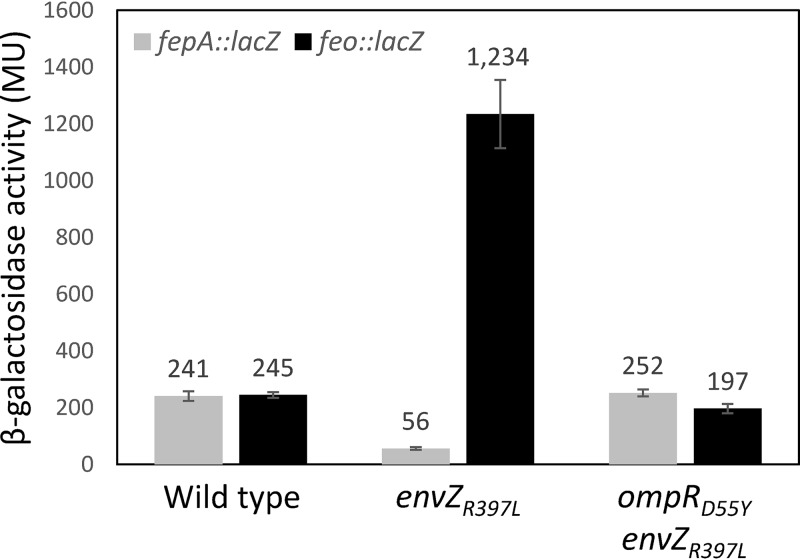
Data presented in Fig. 4 show that envZR397L reduced fepA::lacZ expression ∼4-fold, whereas ompRD55Y abolished this effect of envZR397L. Likewise, the presence of envZR397L elevated feo::lacZ expression 5-fold and again ompRD55Y abolished this increase in feo expression. Curiously, feo::lacZ expression in the ompRD55Y envZR397L background was slightly lower than that seen in the wild-type background, suggesting a role for functional OmpR in the expression of the feo operon. Together, these data show unambiguously that the negative and positive effects of envZR397L on fepA and feo, respectively, require functional OmpR.
FIG 4.
Determination of fepA::lacZ and feo::lacZ activities in various genetic backgrounds. The β-galactosidase activities were measured from two independent overnight grown cultures. Error bars represent standard deviations. The bacterial strains used included RAM2920 (ompR+ envZ+ fepA::lacZ), RAM2921 (ompR+ envZR397L fepA::lacZ), RAM2922 (ompRD55Y envZR397L fepA::lacZ), RAM2923 (ompR+ envZ+ feo::lacZ), RAM2924 (ompR+ envZR397L feo::lacZ), and RAM2925 (ompRD55Y envZR397L feo::lacZ).
Direct regulation of feoABC operon by EnvZ/OmpR.
The data in Fig. 2 and 4 showed a dramatic increase in the feo transcript/transcription levels in the envZR397L/ompR+ background. This could be due to the direct regulation of feo by OmpR or an effect of an OmpR-controlled factor on the feo promoter or feo transcript. We took cues from an earlier publication that showed overexpression of RstA, the response regulator of the RstB/RstA TCS, upregulated feoB expression and repressed the Fur regulon in Salmonella Typhimurium (35). Electrophoretic mobility shift assays (EMSAs) showed direct binding of RstA to the feo promoter sequence (35). Moreover, these authors identified the “RstA motif” (TACA-N6-TACA) upstream of the S. Typhimurium feoA gene of the feo operon (35). Although OmpR recognition sequences are quite degenerate (36, 37), one of the motifs–GTTACANNNN–resembles that of RstA (Fig. 5A). Indeed, both RstA and OmpR regulate some of the same genes by binding to overlapping promoter sequences (38). Our initial assessment detected two potential sequences (−294)-TTATCAtttcaTTAACA-(–278) and (−165)-CCAACAttcgCACACA-(–150) upstream of the feoA ATG codon that might contain both RstA and OmpR binding motifs (Fig. 5A).
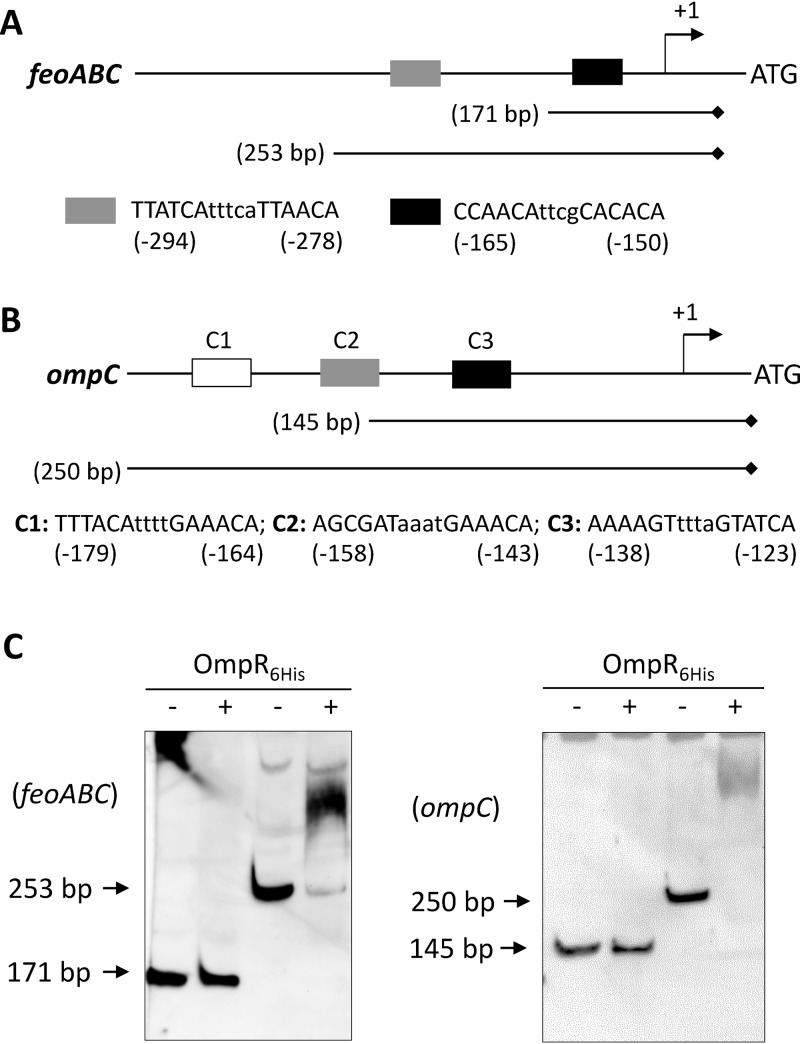
FIG 5.
In vitro binding of purified OmpR6His to the feoABC and ompC promoter regions. DNA binding was examined by EMSA using biotin-labeled DNA fragments of various lengths generated by PCR. (A) Diagram showing the regulatory region of the feoABC operon (not drawn to scale). Gray and black boxes represent possible OmpR binding sequences. Nucleotide numberings are relative to the feoA start codon. The relative positions and lengths of the two DNA fragments used in EMSA are shown. Diamond marks the biotin-labeled end of the DNA probe. (B) Diagram showing the regulatory region of the ompC gene (not drawn to scale). Three boxes represent the known OmpR binding sites; the DNA sequences of all three OmpR binding sites—C1, C2, and C3—are shown. Nucleotide numberings are relative the ompC start codon. The relative positions and lengths of the two DNA fragments used in the EMSA are shown. A diamond indicates the biotin-labeled end of the DNA probe. (C) Polyacrylamide gels showing EMSA results. Plus and minus signs denote the presence and absence, respectively, of OmpR in the reaction mixture prior to gel electrophoresis. Gels were electroblotted, and DNA bands were detected by treating membranes with stabilized streptavidin-HRP conjugate, followed by luminol/enhancer and stable peroxide. Arrows point to positions of unshifted DNA fragments.
EMSA was carried out to test whether OmpR can bind directly to the feo promoter region. The coding region of ompR was cloned into an expression vector, pET24d(+). To aid in protein purification, six consecutive histidine codons were included at the 3′ end of the gene during cloning, and the protein was purified to near homogeneity by metal affinity chromatography (Fig. S3). The purified protein was used directly without in vitro phosphorylation. Using biotinylated primers, two DNA templates of the feo regulatory region, encompassing the predicted OmpR binding motifs, were amplified by PCR (Fig. 5A). As a positive control for OmpR binding, two ompC DNA fragments were also included for EMSA (Fig. 5B). No DNA gel shift occurred with the smaller feoB DNA fragment containing one of the predicted OmpR binding motifs (Fig. 5C). However, the larger feo promoter fragment, containing the upstream predicted OmpR binding motif, displayed shifts after incubation with purified OmpR6His (Fig. 5C). Consistent with these in vitro data, we found that overexpression of OmpR6His from a pBAD24 replicon increased feo::lacZ expression 2-fold (from 140 ± 8 Miller units in the pBAD24 vector containing strain to 296 ± 25 Miller units in the strain containing pBAD24-ompR6His). OmpR bound to the ompC promoter fragment containing the high-affinity OmpR-binding motif C1 (39), but not with the one containing the partial C2 and the entire C3 motif (Fig. 5C). Incidentally, only the ompC fragment, containing all three OmpR motifs, expressed the promoterless lacZ gene in an OmpR-dependent manner (Fig. S4), thus corroborating the EMSA data. Together, these data indicated that OmpR positively regulates feo expression by directly binding to the feo promoter region.
SDS-PAGE analysis of fractions containing OmpR6His obtained after nickel affinity chromatography. Protein samples were visualized after Coomassie brilliant blue staining. Fractions shown in the box were pooled and used for DNA binding studies. The position of OmpR6His is shown. Download FIG S3, TIF file, 0.9 MB (926.2KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Determination of ompC::lacZ activities of constructs carrying two different lengths of the ompC promoter regions in front of a promoterless lacZ gene. ompC145::lacZ and ompC250::lacZ contain 145 and 250 bp, respectively, of the ompC promoter region, including ATG. The β-galactosidase activities in ompR+ or ΔompR backgrounds were measured from two independent cultures grown to late log phase. Error bars represent standard deviations. Download FIG S4, TIF file, 0.8 MB (805.8KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Role of porins in iron homeostasis.
The data in Fig. 1 and 4 revealed a possible mechanism by which a pleiotropic allele of envZ downregulates the ferric transport systems by employing the OmpC/FeoB-mediated ferrous transport pathway. While these data implicated EnvZ/OmpR and OmpC in iron transport, the use of a pleiotropic envZ allele may have created an unnatural genetic environment in which EnvZ/OmpR and porins become involved in iron homeostasis. To eliminate this possibility, we determined the roles EnvZ/OmpR and porins in iron transport using the null alleles of ompR and the porin genes. Before testing their roles, we disabled the high-affinity ferric transport system, since porins likely mediate iron transport by simple diffusion of ferrous or small iron-chelated compounds, and this passive activity of porins will likely be masked by the high-affinity iron transport system. In E. coli, the high-affinity iron transport principally involves a ferric chelator, enterobactin, and TonB that interacts with the outer membrane iron receptors for the release of chelator-Fe3+ complexes bound to the receptor. Consequently, we disabled the ferric iron transport by deleting aroB, tonB or both. The aroB gene encodes 3-dehydroquinate synthase, which is required for the second step of the chorismate pathway in the synthesis of enterobactin, aromatic amino acids, and other important compounds (40).
We first determined the iron dependency of wild-type, ΔaroB, ΔtonB, and ΔaroB ΔtonB strains by growing them on Lysogeny broth agar (LBA), LBA supplemented with 40 μM FeCl3 and LBA containing 200 μM 2,2′-dipyridyl (DP), a synthetic iron chelator (Fig. 6). Bacterial growth in the absence of aroB was unaffected on LBA+FeCl3 or LBA (Fig. 6A and B). However, significant growth impairment of the ΔaroB strain occurred on LBA+DP plates (Fig. 6C), reflecting the loss of a major, enterobactin-mediated iron transport system. In contrast to ΔaroB, the deletion of tonB impaired bacterial growth even on LBA (Fig. 6B), which contains around 6 μM iron, and completely prevented growth on LBA+DP medium (Fig. 6C). The ΔtonB strain grew like WT on LBA+FeCl3, showing that the growth impairment of this strain on LBA was due to low accessibility to iron. Interestingly, growth of the ΔaroB ΔtonB double mutant improved slightly on LBA compared to the ΔtonB strain (Fig. 6B) but ceased again on LBA+DP (Fig. 6C). An improvement in growth of the double mutant compared to the ΔtonB strain on LBA may be due to the absence of extracellular enterobactin-Fe3+ complexes, which, when allowed to accumulate outside the ΔtonB cells, would sequester iron from the medium and further exacerbate growth defects (41). Because of the greater growth dependence of the ΔtonB and ΔtonB ΔaroB strains on external iron sources than the ΔaroB strain, we selected the former two genetic backgrounds to examine the effects of EnvZ/OmpR and porins in iron transport. It is worth noting that we did not determine bacterial growth rates by monitoring growth of liquid cultures because the ΔtonB strain frequently reverts without supplemented iron, and these faster-growing revertants take over the population to artificially display better-than-expected growth.
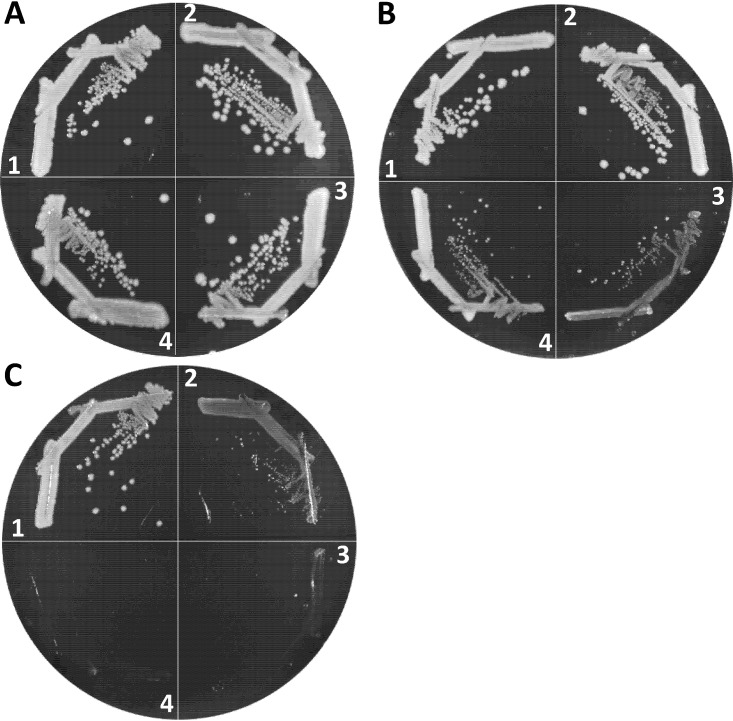
FIG 6.
Effects of ΔtonB and ΔaroB mutations on bacterial growth under iron-replete and iron-depleted conditions. Bacterial growth on LBA plus 40 μM FeCl3 (A), LBA (B), and LBA plus 200 μM 2,2′-dipyridyl (C) was recorded after petri plates were incubated at 37°C for 24 h. Bacterial strains used: 1, RAM1292 (wild type); 2, RAM2553 (ΔaroB); 3, RAM2572 (ΔtonB); and 4, RAM2574 (ΔaroB ΔtonB).
We employed two different null ompR alleles, ompR101 and ΔompR::Kmr, both of which produce the OmpC– OmpF– phenotype. The ompR101 allele was transduced into a ΔtonB background using a linked tetracycline resistance (Tcr) marker, malPQ::Tn10, while ΔompR::Kmr was transduced directly using the Kmr gene that replaced the deleted ompR gene. Although both ompR alleles could be transduced in the ΔtonB strain when transductants were selected on LBA+FeCl3 containing appropriate antibiotics, the resulting null ompR ΔtonB transductants grew poorly compared to the ompR+ ΔtonB strain (Fig. 7A, sectors 4 and 5). In contrast, ompR101 and ΔompR::Kmr severely compromised growth of the ΔtonB strain on LBA not supplemented with FeCl3 (Fig. 7B, sectors 4 and 5). Similar to the ΔtonB ompR101 strain, we were able to construct the ΔaroB ΔtonB ompR101 strain on LBA+FeCl3 medium, where it grew poorly (Fig. 7A, sector 8) but not as poorly as on LBA without FeCl3, where the strain failed to form single colonies (Fig. 7B, sector 8). These observations pointed to a critical role for the EnvZ/OmpR TCS in iron transport in the absence of the high-affinity iron transport system.
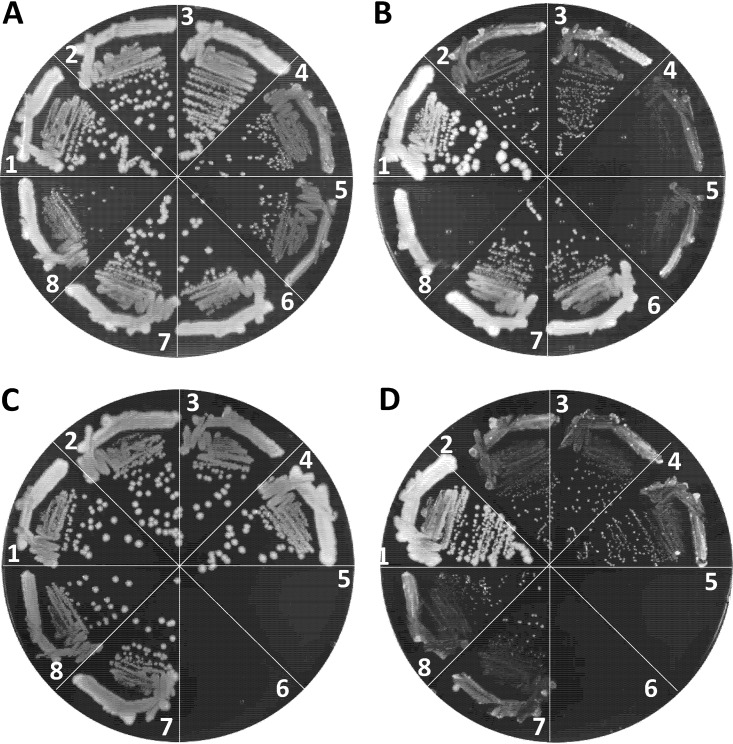
FIG 7.
Effects of ompR and porin gene mutations on the growth of ΔtonB or ΔtonB ΔaroB mutants. Bacterial growth was monitored on LBA plus 40 μM FeCl3 (A and C) and LBA (B and D) after incubation of petri plates at 37°C for 24 h. Relevant genotypes of strains used in panels A and B: 1, RAM1292 (wild type); 2, RAM2572 (ΔtonB); 3, RAM2765 (ΔtonB malPQ::Tn10); 4, RAM2766 (ΔtonB malPQ::Tn10 ompR101); 5, RAM2767 (ΔtonB ΔompR::Kmr); 6, RAM2574 (ΔtonB ΔaroB::Kmr); 7, RAM2771 (ΔtonB ΔaroB::Kmr malPQ::Tn10); and 8, RAM2772 (ΔtonB ΔaroB::Kmr malPQ::Tn10 ompR101). Relevant genotypes of strains used in panels C and D: 1, RAM1292 (wild type); 2, RAM2572 (ΔtonB); 3, RAM2769 (ΔtonB ΔompC::Cmr); 4, RAM2768 (ΔtonB ΔompF::Kmr); 5 and 6, no bacteria; 7, RAM2792 (ΔtonB ΔompC::Cmr ΔompF::Kmr/pompC); and 8, RAM2790 (ΔtonB ΔompF::Kmr ΔompC::Cmr/pompF). pompF and pompC are pTrc99A plasmid clones expressing ompF and ompC, respectively. The expression of these plasmid-coded genes did not require induction by an inducer.
Although the porin genes are the main targets of the EnvZ/OmpR regulatory system, transcription of other genes is also affected either directly or indirectly in the ompR-null mutant (26). Therefore, to establish unambiguously the importance of porins in iron transport, we attempted to delete the porin genes in a background devoid of the high-affinity transport system. In the ΔtonB background, the deletion of ompC or ompF individually did not significantly influence growth on LBA (Fig. 7C and D, compare sectors 3 and 4 to sector 2). Strikingly, however, we failed to delete ompC and ompF simultaneously, via P1 transduction of ΔompF::Kmr and ΔompC::Cmr alleles, in the ΔtonB background even when transductants were selected on LBA+FeCl3 plates carrying appropriate antibiotics. In contrast, when the ΔtonB ΔompC or ΔtonB ΔompF double mutant was first complemented with a plasmid expressing one of the porin genes, the uncomplemented porin gene from the chromosome could be readily deleted by P1 transduction. The plasmid-complemented triple mutants displayed growth behavior similar to the uncomplemented ΔtonB ΔompC and ΔtonB ΔompF double mutants on LBA+FeCl3 or LBA (Fig. 7C and D, compare sectors 3 and 4 with sectors 7 and 8). It is worth noting that the ΔtonB ΔompC and ΔtonB ΔompF strains were not defective in P1 transduction, since drug resistant markers not associated with the porin genes or their regulators could be transduced readily into these strains. Moreover, unlike the ΔtonB strain, in the wild-type and ΔaroB backgrounds the ompC and ompF genes could be deleted simultaneously without causing iron dependency or a significant growth defect (Fig. S5). These data indicated for the first time that the OmpC and OmpF porins play a critical role in iron intake when the high-affinity iron transport system is blocked.
Effects of ΔompC and ΔompF gene mutations on the growth of wild-type and ΔaroB strains. Bacterial growth was monitored on an LBA plate incubated at 37°C for 24 h. The relevant genotypes are shown. Download FIG S5, TIF file, 0.5 MB (560.6KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Although the EnvZ/OmpR TCS is classically associated with the regulation of the OmpC and OmpF porins in response to medium osmolarity (23), recent transcriptomics and chromatin immunoprecipitation analyses showed that it is a global regulatory system (26, 37, 42). Indeed, missense alleles of envZ, called perA and tpo, isolated over 3 decades ago, were shown to also influence nonporin regulons, including pho and mal (43–45). A separate study revealed that perA lowered the expression of three iron-regulated proteins without an apparent reduction in the rate of enterobactin secretion (27). This led these authors to suggest that the effect of the perA (envZ) allele on the expression of iron-regulated proteins is most likely posttranscriptional (27).
In the present study, we sought to resolve the mechanism by which the activated EnvZ/OmpR TCS reduces expression of genes involved in iron homeostasis and determine the role of porins in iron acquisition. We used the envZR397L allele, which is phenotypically similar to the pleotropic perA and tpo alleles of envZ, i.e., in the envZR397L background, OmpC levels go up, while those of OmpF and LamB go down dramatically (29). The RT-qPCR (this work) and the whole-genome microarray data (30; unpublished data) showed that in the presence of envZR397L the transcript levels of several Fur-controlled genes, including fecA, fepA, fhuA, and fhuF, decreased significantly. In the case of fhuA and fhuF, the effects of envZR397L required Fur, while expression of fecA and fepA was still reduced by envZR397L in the absence of Fur. These observations indicated the involvement of at least two different mechanisms by which envZR397L affected iron regulon. In support of the Fur-dependent mechanism, the whole-cell EPR data confirmed the presence of significantly elevated levels of accessible iron in the envZR397L strain. Several observations supported the hypothesis that in the envZR397L mutant, FeoB and OmpC are responsible for increased intracellular Fur-Fe2+ level (Fig. 8). First, unlike the expression of genes involved in ferric iron transport or metabolism, expression of the feoAB genes involved in ferrous iron transport went up dramatically in the envZR397L background. This increase in the expression of the ferrous iron transport system had an adverse effect on the ferric iron transport system, since the absence of FeoB, the ferrous permease, abolishes or significantly reduces the negative effects of envZR397L on ferric transport/metabolic genes. Second, like FeoB, the absence of OmpC (envZR397L already severely represses ompF expression [29]) largely negated the inhibitory effects of envZR397L on ferric transport/metabolic genes. Because the single deletion of feoB or ompC and the simultaneous deletion of feoB and ompC reversed the effects of envZR397L on fecA, fepA, fhuA, and fhuF to the same extent, it indicated that FeoB and OmpC must act in the same pathway to transport ferrous iron into the cell and elevate Fur-Fe2+ levels. Third, the absence of FeoB or OmpC in an EnvZ+ background caused derepression of six Fur-controlled genes, indicating that the ferrous iron transport pathway is active under our experimental conditions and that envZR397L enhances this pathway to achieve its inhibitory effects on the ferric transport system. Lastly, we provided direct evidence of excessive iron inside the envZR397L mutant by whole-cell EPR spectroscopy measurements, which showed that, as in the Δfur mutant, the level of accessible iron in the envZ mutant rose 4-fold over that present in the parental strain. Moreover, this increase in the intracellular free pool of iron in the envZR397L mutant was dependent on FeoB. From these observations, we conclude that the upregulation of the OmpC-FeoB ferrous iron transport pathway by envZR397L elevates the intracellular Fur-Fe2+ level, which, in turn, represses the expression of iron-regulated genes (Fig. 8). These effects of envZR397L required functional OmpR since the presence of ompRD55Y, which confers a null phenotype, neutralized all envZR397L phenotypes.
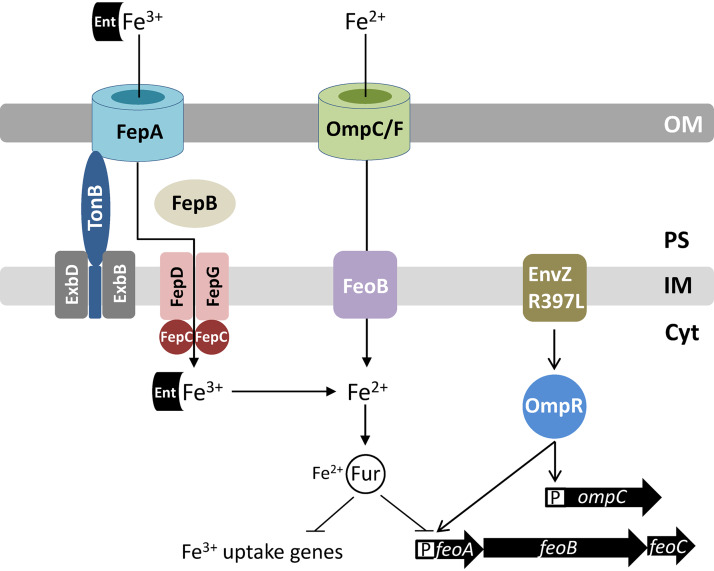
FIG 8.
Diagram showing regulation of the ferric (Fe3+) and ferrous (Fe2+) uptake systems in E. coli. Fur-Fe2+ is the master regulator of transcription of genes involved in iron metabolism. Under aerobic growth conditions, where Fe3+ is the major source of iron, E. coli secretes enterobactin (Ent) in the medium to chelate Fe3+. The Fe3+-chelate complex is transported back into the cell through the outer membrane receptor protein, FepA. The TonB-ExbB-ExbD complex of the inner membrane facilitates FepA channel opening. In the periplasm, FepB interacts with the Fe3+-chelate and delivers it to the FepDGC complex for transport into the cytoplasm. Under microaerobic or anaerobic growth conditions, Fe2+ is the main source of iron. It is brought into the cell via porins OmpC and OmpF and FeoB. The EnvZ/OmpR two-component system, classically known for regulating the expression of the ompC and ompF porin genes, also induces feo expression when hyperactivated due to a specific mutation in envZ (envZR397L). This positive effect of EnvZR397L/OmpR on feo expression can overcome the negative effect of Fur-Fe2+ on feo expression, thus tipping the balance in favor of ferrous over ferric transport. Porins and EnvZ/OmpR play a crucial role in iron acquisition in a TonB-deficient background that lacks functional ferric transport systems. Abbreviations: OM, outer membrane; PS, periplasm; IM, inner membrane; Cyt, cytoplasm; P, promoter.
Whereas envZR397L-mediated reduction in fhuA and fhuF transcript levels required Fur, the effects of envZR397L on fepA and fecA transcripts did not. This suggested the existence of another regulatory mechanism responsible for the envZR397L-mediated downregulation of fepA and fecA that did not involve Fur. Previous studies showed that the plasmid-mediated overexpression of OmrA and OmrB small RNAs, whose expression is under the EnvZ/OmpR control, can downregulate fepA and fecA transcript levels (31, 46). We have shown that envZR397L increases OmrA expression almost 10-fold (29). This increase in OmrA expression could contribute to the downregulation of fepA and fecA. However, the fact that deleting ompC or feoB in a Fur+ background abolishes envZR397L-mediated downregulation of the ferric iron transport genes suggests that envZR397L-mediated increase in OmrA and OmrB levels contributes little, if any, to fepA and fecA repression. Consistent with this notion, we found that deletions of ΔomrA and ΔomrB failed to reverse the negative effect of envZR397L on fepA or fecA (Fig. S2). We conclude that a mechanism independent of Fur and OmrA and OmrB must also exist for the envZR397L-mediated downregulation of fepA and fecA. A direct role of EnvZ/OmpR has not been ruled out.
As stated above, unlike the ferric transport genes, expression of the ferrous transport genes feoAB, which are also under the control of Fur, went up in the envZR397L background. At first glance, this appears inconsistent with the notion that an increase in the Fur-Fe2+ level by envZR397L should also decrease feoAB expression. Our data suggest that envZR397L overcomes the repressive effect of Fur-Fe2+ on feoAB by activating their expression. Moreover, because feoAB expression in the envZR397L background increases dramatically without Fur, it shows that Fur-Fe2+ does repress feoAB in the envZR397L background, but the positive effect of envZR397L on feoAB expression overwhelms the negative effect of Fur on these genes. The EMSA data showed that purified OmpR binds to the feo regulatory region containing a putative OmpR binding site, indicating that OmpR directly activates feoAB expression. Interestingly, the whole-genome microarray data from an ΔompR ΔenvZ (porin-minus) strain showed that feoB transcript levels decreased 2-fold, whereas those of fepA and fecA increased 2-fold (26). These observations are consistent with our proposal that the EnvZ/OmpR TCS directly stimulates the FeoB-OmpC pathway to increase intracellular Fe2+ levels and thus active Fur-Fe2+ complexes, which then downregulate the expression of the ferric iron transport genes.
The proposed role of EnvZ/OmpR in iron homeostasis is similar to that suggested for RstA in S. Typhimurium (35). The authors found that overexpression of RstA increased feoAB expression and repressed fhuA and fhuF expression. A RstA binding site was identified in the feo promoter and the EMSA data confirmed that RstA bound there (35). The RstA binding motif “TACAtntngtTACA” resembles that of OmpR’s “GTTACAnnnnGTTACA” and not surprisingly, both proteins regulate overlapping genes by binding to the similar sequences (38). Our EMSA data showed that OmpR binds to the feo promoter region. Specifically, it binds to a DNA fragment containing the sequence “ttATCAtttcattAACA” located 278 bp upstream of the start codon of feoA. The OmpR binding studies carried out here involved the purified protein not modified by in vitro phosphorylation. Therefore, it is possible that stronger binding and/or additional binding sites may be discovered with phosphorylated OmpR. It is worth noting that in previous EMSAs, unphosphorylated RstA from S. Typhimurium and E. coli was shown to bind to their target promoter sequences (35, 47). Further work will be required to identify the exact binding sequences and to determine whether OmpR and RstA bind to the same, overlapping, or distinct regulatory sequences of the feo operon.
Our work also revealed for the first time the essential role of OmpC and OmpF porins in iron acquisition when the TonB-dependent ferric transport pathways are inoperative. The absence of OmpC or OmpF produced no growth defects in the ΔtonB background on LBA supplemented with iron, but the construction of a triple-knockout mutant (ΔtonB ΔompC ΔompF) required the expression of at least one of the porin genes from a plasmid replicon. Interestingly, unlike the porin-devoid triple-knockout mutant, we were able to construct ΔtonB ΔompR and ΔtonB ΔaroB ΔompR mutants, albeit only on LBA supplemented with iron. In the ΔompR background, ompC and ompF porin expression is extremely low but presumably not zero, which is the case in the ΔtonB ΔompC ΔompF mutant. We think that this extremely low porin expression permits the construction of the ΔtonB ΔompR strain, which can form single, albeit very small colonies, but only on LBA supplemented with iron. Whereas the diffusion of ferrous iron across the outer membrane occurs via OmpC or OmpF channels, at least three proteins—FeoB, MntH, and ZupT—can transport ferrous iron across the inner membrane of E. coli cells (48). Consistent with this, a ΔtonB ΔfeoB double mutant is viable and grows like the ΔtonB mutant (data not shown).
Although the essential role of porins in iron acquisition becomes apparent without the TonB-dependent, high-affinity ferric iron transport systems, their derepression without OmpC or FeoB indicate that the porin-mediated iron transport is active even in the presence of the TonB-dependent high-affinity iron transport systems. The importance of the porin-FeoB pathway for bacterial growth should further increase as E. coli cells enter microaerobic or anaerobic environments where the ferrous species predominates. The involvement of porin and FeoB in iron-dependent growth and/or virulence has been reported for several bacteria, including E. coli (9), S. Typhimurium (10; 11), Helicobacter pylori (12), Vibrio cholerae (49), and Mycobacterium smegmatis (50). Interestingly, M. smegmatis porins increase ferric citrate uptake (50). Similarly, a study reported liganded iron uptake via the OprF porin in Pseudomonas aeruginosa (51). There are no definitive reports in E. coli showing the involvement of porins in liganded iron transport, even for ferric citrate, whose size is below the diffusion limits of the porins (52). Regardless of these ambiguities, published reports and the work carried out here highlight the importance of the porin/FeoB-mediated iron transport pathways in iron homeostasis.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
Escherichia coli K-12 strains used in this study were constructed from MC4100 (53) and are listed in Table 1. Lysogeny broth (LB) was prepared using LB broth EZMix powder (Lennox). LB agar (LBA) medium contained LB plus 1.5% agar (Becton Dickinson). ONPG (2-ortho-nitrophenyl-β-d-galactopyranoside) was purchased from Acros. Diethylenetriaminepentaacetic acid (DTPA) and desferrioxamine were obtained from Sigma-Aldrich. All other chemicals were of analytical grade. The growth medium was supplemented with ampicillin (50 μg/ml), chloramphenicol (12.5 μg/ml), kanamycin (25 μg/ml), or tetracycline (10 μg/ml) when necessary. To induce plasmid-borne gene expression, 0.2% l-arabinose or 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the medium.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| RAM1292 | MC4100 (Δ[argF-lac]169 λ– e14– flhD5301 Δ[fruK-yeiR]725 relA1 rpsL150 (Strr) rbsR22 Δ[fimB-fimE]632 deoC1) Δara714 | 61 |
| RAM1541 | RAM1292 envZR397L | 29 |
| RAM2697 | RAM1292 Δfur::scar | This study |
| RAM2698 | RAM1292 envZR397L Δfur::scar | This study |
| RAM2699 | RAM1292 ΔompC::scar | This study |
| RAM2700 | RAM1292 envZR397L ΔompC::scar | This study |
| RAM2701 | RAM1292 ΔfeoB::scar | This study |
| RAM2702 | RAM1292 envZR397L ΔfeoB::scar | This study |
| RAM2703 | RAM1292 envZR397L ΔfeoB::scar ΔompC::scar | This study |
| RAM2704 | RAM1292 ΔompR::scar | This study |
| RAM2705 | RAM1292 ΔompR::scar pBAD24-ompR6His | This study |
| RAM2707 | RAM1292 ΔompR::scar pBAD24 | This study |
| RAM2708 | RAM1292 pBAD24 | This study |
| RAM2709 | RAM1292 pBAD24-ompR6His | This study |
| RAM2711 | RAM1292 ΔfeoA::lacZ-Kmr | This study |
| RAM2712 | RAM1292 ΔfeoA::lacZ-Kmr envZR397L | This study |
| RAM2713 | RAM1292 ΔfeoA::lacZ-Kmr Δfur::scar | This study |
| RAM2714 | RAM1292 ΔfeoA::lacZ-Kmr Δfur::scar envZR397L | This study |
| RAM2715 | BL21(DE3) pET24d-ompR6His | This study |
| RAM2469 | RAM1292 ΔfepA::lacZ-Kmr | This study |
| RAM2470 | RAM1292 ΔfepA::lacZ-Kmr envZR397L | This study |
| RAM2471 | RAM1292 ΔfepA::lacZ-Kmr Δfur | This study |
| RAM2472 | RAM1292 ΔfepA::lacZ-Kmr Δfur envZR397L | This study |
| RAM2473 | RAM2469 ΔompC::Cmr | This study |
| RAM2474 | RAM2469 ΔfeoB::Cmr | This study |
| RAM2475 | RAM2470 ΔompC::Cmr | This study |
| RAM2476 | RAM2470 ΔfeoB::Cmr | This study |
| RAM2477 | RAM2469 ΔompF::Cmr | This study |
| RAM2478 | RAM2470 ΔompF::Cmr | This study |
| RAM2505 | RAM1292 ΔtonB::Kmr | This study |
| RAM2553 | RAM1292 ΔaroB::Kmr | This study |
| RAM2572 | RAM2505 ΔtonB::scar (via pCP20) | This study |
| RAM2574 | RAM2572 ΔaroB::Kmr | This study |
| RAM2623 | RAM2553 ΔaroB::scar | This study |
| RAM2625 | RAM2574 ΔaroB::scar (via pCP20) | This study |
| RAM2765 | RAM2572 malPQ::Tn10-ompR+ | This study |
| RAM2766 | RAM2572 malPQ::Tn10-ompR101 | This study |
| RAM2767 | RAM2572 ΔompR::Kmr | This study |
| RAM2768 | RAM2572 ΔompF::Kmr | This study |
| RAM2769 | RAM2572 ΔompC::Cmr | This study |
| RAM2771 | RAM2574 malPQ::Tn10-ompR+ | This study |
| RAM2772 | RAM2574 malPQ::Tn10-ompR101 | This study |
| RAM2790 | RAM2768 (pTrc99A-ompF) | This study |
| RAM2792 | RAM2769 (pTrc99A-ompC) | This study |
| RAM2795 | RAM2790 ΔompC::Cmr | This study |
| RAM2796 | RAM2792 ΔompF::Kmr | This study |
| RAM2920 | RAM2469 Cmr linked to envZ+ | This study |
| RAM2921 | RAM2470 Cmr linked to envZR397L | This study |
| RAM2922 | RAM2799 Cmr linked to ompRD55Y envZR397L | This study |
| RAM2924 | RAM2711 Cmr linked to envZ+ | This study |
| RAM2925 | RAM2711 Cmr linked to envZR397L | This study |
| RAM2926 | RAM2711 Cmr linked to ompRD55Y envZR397L | This study |
| RAM2928 | RAM2711 pBAD24 (Apr) | This study |
| RAM2932 | RAM2711 pBAD24-ompR6His | This study |
Genetic and DNA methods.
Standard bacterial genetic methods, including P1 transduction and plasmid transformation, were carried out as described by Silhavy et al. (54). To clone the ompR and rstA genes into pBAD24 (55) and pET24d(+) (Novagen), DNA corresponding to their open reading frames (ORFs) were amplified by PCR using primers that carried appropriate restriction enzyme sites for cloning. The reverse primers used for cloning into pBAD24 additionally contained nucleotides encoding six consecutive histidine codons. (Primer sequences are available upon request.) Deletion of the fepA, feoA, feoB, and feoAB genes from their chromosomal locations and subsequent scaring of the antibiotic-resistant marker at the deletion sites were done using the λ-Red-mediated gene recombination method (56). Deletions were confirmed by PCR and DNA sequence analyses. In some instances, promoterless lacZY genes were recombined at the deletion scar site by the method of Ellermeier et al. (57).
RNA isolation, real-time quantitative PCR, and microarray analyses.
Total RNA was extracted from 5 ml cells grown to log phase (OD600 ∼0.6) at 37°C using TRIzol Max bacterial RNA isolation kit (Invitrogen). RNA was further purified using the RNeasy kit (Qiagen), and the quality of RNA was assessed by using an Agilent 2100 bioanalyzer (Agilent Technologies). The purified RNA was then converted to either single-stranded cDNA for use in RT-qPCR or double-stranded cDNA for use in DNA microarray analysis.
For RT-qPCR, single-stranded cDNA was synthesized from 10 μg of RNA using 100 pM random hexamer primer (Integrated DNA Technologies) and M-MuLV reverse transcriptase (New England Biolabs). After reverse transcription, cDNA was treated with 5 U of RNase H (New England Biolabs) for 20 min at 37°C, followed by purification with a QIAquick PCR purification kit (Qiagen). To quantify the RNA transcripts, 300 nM concentrations of primer specific to the gene of interest and 20 ng of cDNA was added to SYBR green PCR master mix (Applied Biosystems) in a 20-μl reaction. Primers were designed according to the manufacturer’s protocol included with the SYBR green PCR master mix and RT-qPCR reagents. Critical threshold (CT) values were determined by using an ABI Prism 7900HT sequence detection system (Applied Biosystems). The relative quantification of target transcripts was calculated according to the 2–ΔΔCT method (58) using ftsL and purC as the endogenous control genes. Briefly, changes in CT value (ΔCT) for the gene of interest were calculated by subtracting that gene’s average CT from the average CT for the endogenous control gene. The ΔCT for the mutant was then subtracted from the wild-type strain’s ΔCT value to give the ΔΔCT value. Each PCR was performed in triplicate and fold changes in transcript levels, along with the standard deviations, were calculated from at least two experiments (n ≥ 2).
For microarray analysis, an Invitrogen superscript double-stranded cDNA synthesis kit was used to generate double-stranded cDNA according to the manufacturer’s instructions. Single-stranded cDNA was synthesized from 10 μg of RNA using a 100 pM concentration of random hexamer primer (Integrated DNA Technologies) and Superscript II reverse transcriptase. Second strand synthesis was performed according to the manufacturer’s instructions, and the reaction was stopped with 0.5 M EDTA. RNA was then digested using RNase A (25 μg/ml final concentration), followed by treatment with phenol-chloroform and precipitation with ethanol. Double-stranded cDNA was further purified with a QIAquick PCR purification kit (Qiagen) and quality tested by using an Agilent bioanalyzer. Cy3 fluorescently labeled cDNA was used to probe array slides printed with 4,254 E. coli ORFs. Array slides contained 8 probes per gene (in duplicate) corresponding to roughly 72,000 probes per sample. Sample labeling with Cy3 fluorescent dye, hybridization to the 4-plex array (0771112 E. coli K-12 EXP X4, catalog number A6697-00-01), washing, and one-color scanning were performed by Roche Nimblegen in accordance with their standard protocol. Analysis of gene expression profiles was performed using ArrayStar 2.0 software (DNAStar) with a focus on genes with a ≥2-fold change in gene expression. P values were generated with the Student t test, and false positives were minimized using false discovery rate analysis (59).
Enzymatic assays.
A β-galactosidase assay was performed according to the Miller method (60). Assays were carried out with at least two independent cultures. The β-galactosidase activity was expressed as Miller units (60). In some instances, kinetic analysis of enzyme activity was carried out using a VersaMax (Molecular Dynamics) microtiter plate reader in quadruplicate, and the activity was measured as the rate of ONPG cleavage divided by the cell density in each well.
Electron paramagnetic resonance spectroscopy.
Free iron concentration in whole cells was determined by EPR spectroscopic analysis (32) with some modifications. Briefly, overnight grown bacterial cultures were diluted 1:100 in 200 ml of LB and grown shaking at 37°C until the optical density at 600 nm (OD600) reached 0.8. Cells were pelleted by centrifugation in a GSA rotor (Sorvall) for 10 min at 6,000 × g. Pellets were resuspended in 10 ml of LB containing 10 mM DTPA (to chelate extracellular iron) and 20 mM desferrioxamine (to chelate intracellular free or accessible ferric iron) and incubated with shaking for 37°C for 15 min. Cells were pelleted as described above and washed twice with 5 ml of ice-cold 20 mM Tris-HCl (pH 7.4). The final cell pellet was resuspended in 0.3 ml of ice-cold 20 mM Tris-HCl (pH 7.4) containing 30% glycerol. A 250-μl aliquot of this cell suspension was placed in a quartz EPR tube (length, 250 mm; external diameter, 4 mm [Wilmad-Labglass]). Tubes were frozen in loosely packed dry ice and then transferred to –80°C until the EPR analysis. The remaining cells were diluted 103-fold to determine the OD600. Iron standards were prepared from a freshly prepared 10 mM FeCl3⋅6H2O stock in a buffer containing 20 mM Tris-HCl (pH 7.4) and 1 mM desferrioxamine. Theoretical concentrations of iron standards were 100, 50, 25, 10, 5, and 0 μM. The actual iron concentrations were determined by measuring the OD420 of each standard and using the formula following: molar concentration = A420/ε, where [Fe]ε is 2.865 mM−1 Cm−1. A 250-μl aliquot of each standard was placed in separate EPR tubes that were then frozen. The standard curve was generated by plotting EPR signals against actual iron concentrations (Fig. S6). The free iron concentration for each strain was determined from the EPR data and the standard curve. The intracellular free iron concentration was then deduced by integrating the intracellular volume of the cell (1 ml of 1.0 OD600 cells has an intracellular volume of 0.00052 ml; Jim Imlay, unpublished data) and using the following formula: intracellular free iron concentration = [Fe] from standard curve/cell paste OD600 × 0.00052 ml.
Standard curve of ferric chloride solutions of known concentrations. The actual concentrations of ferric chloride solutions were determined by measuring the absorbance at 420 nm and then utilizing the following formula: molar concentration = A420/ε, where [Fe]ε is 2.865 mM−1 Cm−1. These values were plotted against the values obtained for the same solutions by EPR spectroscopy. We used peak-to-peak EPR measurements instead of the double integration values due to a greater confidence of the former at lower ferric chloride concentrations. The standard curve was used to measure iron concentrations in the whole cell. Download FIG S6, TIF file, 0.5 MB (504.3KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
EPR measurements were carried out at the EPR Facility at Arizona State University. Continuous wave EPR spectra were recorded using an ELEXSYS E580 CW X-band spectrometer (Bruker, Rheinstetten, Germany) equipped with a model 900 EPL liquid helium cryostat (Oxford Instruments, Oxfordshire, UK). For all measurements, the magnetic field modulation frequency was 100 kHz, the amplitude was 1.25 mT, the microwave power was 10 mW, the microwave frequency was 9.44 GHz, the sweep time was 42 s, and the temperature was 20 K.
OmpR purification.
OmpR was purified from BL21(DE3) cultures carrying a pET24-ompR6His plasmid. Overnight cultures, grown without IPTG, were diluted 1:100 in 1 liter of LB, grown with vigorous shaking for 90 min, and then supplemented with IPTG and grown for another 2 h. The cells were pelleted, washed with 10 mM Tris-HCl (pH 7.5), resuspended in lysis buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 μg/ml lysozyme), and incubated on ice for 30 min. MgCl2 (10 mM final), phenylmethylsulfonyl fluoride (2 mM final), and DNase I (40 μg/ml final) were then added to the cell suspension. Cells were lysed by passage through a French pressure cell three times, and the lysate was centrifuged at low speed to remove unlysed cells. Envelopes were removed from the lysate by ultracentrifugation at 105,000 × g for an hour at 4°C. Supernatant was filtered through a 0.45-μm syringe filter, and the filtrate was subjected to nickel affinity column chromatography using buffers for protein binding (20 mM sodium phosphate [pH 7.4], 20 mM imidazole, and 50 mM NaCl), washing (20 mM sodium phosphate [pH 7.4], 50 mM imidazole, and 300 mM NaCl), and elution (20 mM sodium phosphate [pH 7.4], 300 mM imidazole, and 300 mM NaCl). Samples from eluted fractions were analyzed by SDS-PAGE, and protein bands were visualized after Coomassie blue staining (Fig. S3). Fractions representing OmpR6His peaks were pooled and dialyzed against a buffer containing 20 mM sodium phosphate (pH 7.4) and 300 mM NaCl. Purified proteins were stored at 4°C in the dialysis buffer supplemented with glycerol (5% final concentration), EDTA (0.1 mM, final concentration), and dithiothreitol (0.1 mM, final concentration).
Electrophoretic mobility gel shift assays.
EMSAs were carried out using a LightShift chemiluminescent EMSA kit (Thermo Scientific). ompC and feoABC promoter fragments were generated by PCR using primers specific to the region of interest, with one of the primers biotinylated. Biotin-labeled DNA probes (20 fmol), purified OmpR6His (100 pmol), and other relevant reagents provided with the kit were incubated for 20 min at room temperature, and the reaction was stopped by adding 5× loading buffer. The mixture was analyzed by 5% acrylamide gel electroblotted onto polyvinylidene difluoride Immobilon-P membrane (Millipore) using a Mini Trans-Blot cell (Bio-Rad). After transfer, DNA was cross-linked to the membrane using Hoefer UV Crosslinker and incubated with stabilized streptavidin-HRP conjugate for an hour. DNA was detected by a molecular imager ChemiDoc XRS system (Bio-Rad) after the membrane was incubated for 5 min with freshly mixed luminol/enhancer and stable peroxide solutions.
ACKNOWLEDGMENTS
This study was supported in part by grants from the National Institutes of Health (GM048167 and AI117150, both now completed) and the School of Life Sciences, Arizona State University (ASU).
We thank Ananya Sen and Yidan Zhou in Jim Imlay’s lab for training R.M. in EPR analysis. The initial EPR work at the University of Illinois Urbana-Champaign facility was supported by GM49640 to Jim Inlay. The final EPR analysis was conducted at the ASU EPR facility. We thank Marco Flores for assistance in EPR analysis at ASU and in constructing Fig. 3.
Footnotes
This article is a direct contribution from Rajeev Misra, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Renato Morona, University of Adelaide, and Stanley Maloy, San Diego State University.
Citation Gerken H, Vuong P, Soparkar K, Misra R. 2020. Roles of the EnvZ/OmpR two-component system and porins in iron acquisition in Escherichia coli. mBio 11:e01192-20. https://doi.org/10.1128/mBio.01192-20.
REFERENCES
- 1.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 3.Grass G. 2006. Iron transport in Escherichia coli: all has not been said and done. Biometals 19:159–172. doi: 10.1007/s10534-005-4341-2. [DOI] [PubMed] [Google Scholar]
- 4.Pollack JR, Neilands JB. 1970. Enterobactin, an iron transport compound. Biochem Biophys Res Commun 38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 5.Kammler M, Schon C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol 175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hantke K. 2003. Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol 11:192–195. doi: 10.1016/s0966-842x(03)00100-8. [DOI] [PubMed] [Google Scholar]
- 7.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo-transport of ferrous iron into bacteria. Biometals 19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 8.Lau CKY, Krewulak KD, Vogel HJ. 2016. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev 40:273–298. doi: 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- 9.Stojiljkovic I, Cobeljic M, Hantke K. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol Lett 108:111–115. doi: 10.1111/j.1574-6968.1993.tb06082.x. [DOI] [PubMed] [Google Scholar]
- 10.Tsolis RM, Baumler AJ, Heffron F, Stojiljkovic I. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella Typhimurium in the mouse. Infect Immun 64:4549–4556. doi: 10.1128/IAI.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer E, Bergevin I, Malo D, Gros P, Cellier MF. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect Immun 70:6032–6042. doi: 10.1128/iai.70.11.6032-6042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velayudhan J, Hughes NJ, McColm AA, Bagshaw J, Clayton CL, Andrews SC, Kelly DJ. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol Microbiol 37:274–286. doi: 10.1046/j.1365-2958.2000.01987.x. [DOI] [PubMed] [Google Scholar]
- 13.Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 14.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 15.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol 177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lorenzo V, Wee S, Herrero M, Neilands JB. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol 169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. 2007. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res 35:6762–6777. doi: 10.1093/nar/gkm631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masse E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masse E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masse E, Arguin M. 2005. Ironing out the problem: new mechanisms of iron homeostasis. Trends Biochem Sci 30:462–468. doi: 10.1016/j.tibs.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Seo SW, Kim D, Latif H, O’Brien EJ, Szubin R, Palsson BO. 2014. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 5:4910. doi: 10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beauchene NA, Myers KS, Chung D, Park DM, Weisnicht AM, Keleş S, Kiley PJ. 2015. Impact of anaerobiosis on the expression of the iron-responsive Fur and RyhB regulons. mBio 6:e01947–e01915. doi: 10.1128/mBio.01947-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall MN, Silhavy TJ. 1981. Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol 151:1–15. doi: 10.1016/0022-2836(81)90218-7. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alphen WV, Lugtenberg B. 1977. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J Bacteriol 131:623–630. doi: 10.1128/JB.131.2.623-630.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 27.Lundrigan M, Earhart CF. 1981. Reduction in three iron-regulated outer membrane proteins and protein a by the Escherichia coli K-12 perA mutation. J Bacteriol 146:804–807. doi: 10.1128/JB.146.2.804-807.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earhart CF. 2004. Iron uptake via the enterobactin system, p 133–146. In Crosa JH, Mey AR, Payne SM (ed), Iron transport in bacteria. ASM Press, Washington, DC. [Google Scholar]
- 29.Gerken H, Charlson ES, Cicirelli EM, Kenney LJ, Misra R. 2009. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol Microbiol 72:1408–1422. doi: 10.1111/j.1365-2958.2009.06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerken HG. 2009. Novel aspects of bacterial envelope response pathways. PhD dissertation, publication 3391808 Arizona State University, Tempe, AZ. [Google Scholar]
- 31.Guillier M, Gottesman S. 2006. Remodeling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 32.Woodmansee AN, Imlay JA. 2002. Quantitation of intracellular free iron by electron paramagnetic resonance spectroscopy. Methods Enzymol 349:3–9. doi: 10.1016/s0076-6879(02)49316-0. [DOI] [PubMed] [Google Scholar]
- 33.Slauch JM, Garrett S, Jackson DE, Silhavy TJ. 1988. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol 170:439–441. doi: 10.1128/jb.170.1.439-441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delgado J, Forst S, Harlocker S, Inouye M. 1993. Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol Microbiol 10:1037–1047. doi: 10.1111/j.1365-2958.1993.tb00974.x. [DOI] [PubMed] [Google Scholar]
- 35.Jeon J, Kim H, Yun J, Ryu S, Groisman EA, Shin D. 2008. RstA-promoted expression of the ferrous iron transporter FeoB under iron-replete conditions enhances Fur activity in Salmonella enterica. J Bacteriol 190:7326–7334. doi: 10.1128/JB.00903-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuong P, Misra R. 2011. Guide to genome-wide bacterial transcription factor binding site prediction using OmpR as model, p 41–56. In Xia X. (ed), Selected works in bioinformatics. InTech, London, United Kingdom. [Google Scholar]
- 37.Quinn HJ, Cameron ADS, Dorman CJ. 2014. Bacterial regulon evolution: distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet 10:e1004125. doi: 10.1371/journal.pgen.1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. 2010. Regulation of Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 39.Head CG, Tardy A, Kenney LJ. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol 281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 40.Bender SL, Mehdi S, Knowles JR. 1989. Dehydroquinate synthase: the role of divalent metal cations and of nicotinamide adenine dinucleotide in catalysis. Biochem 28:7555–7560. doi: 10.1021/bi00445a009. [DOI] [PubMed] [Google Scholar]
- 41.Qiu N, Misra R. 2019. Overcoming iron deficiency of an Escherichia coli tonB mutant by increasing outer membrane permeability. J Bacteriol 201:e00340-19. doi: 10.1128/JB.00340-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins TT, Davies MR, Klemm EJ, Rowley G, Wileman T, James K, Keane T, Maskell D, Hinton JCD, Dougan G, Kingsley RA. 2013. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol Microbiol 87:526–538. doi: 10.1111/mmi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanner BL, Sarthy A, Beckwith J. 1979. Escherichia coli pleiotropic mutant that reduces amounts of several periplasmic and outer membrane proteins. J Bacteriol 140:229–239. doi: 10.1128/JB.140.1.229-239.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wandersman C, Moreno F, Schwartz M. 1980. Pleiotropic mutations rendering Escherichia coli K-12 resistant to bacteriophage TP1. J Bacteriol 143:1374–1383. doi: 10.1128/JB.143.3.1374-1383.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Case CC, Bukau B, Granett S, Villarejo MR, Boos W. 1986. Contrasting mechanisms of envZ control of mal and pho regulon genes in Escherichia coli. J Bacteriol 166:706–712. doi: 10.1128/jb.166.3.706-712.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guillier M, Gottesman S. 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogasawara H, Hasegawa A, Kanda E, Miki T, Yamamoto K, Ishihama A. 2007. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J Bacteriol 189:4791–4799. doi: 10.1128/JB.00319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. 2005. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J Bacteriol 187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol 188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones CM, Niederweis M. 2010. Role of orin in iron uptake in Mycobacterium smegmatis. J Bacteriol 192:6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer J-M. 1992. Exogenous siderophore-mediated iron uptake in Pseudomonas aeruginosa: possible involvement of porin in iron translocation. J Gen Microbiol 138:951–958. doi: 10.1099/00221287-138-5-951. [DOI] [PubMed] [Google Scholar]
- 52.Wagegg W, Braun V. 1981. Ferric citrate transport in Escherichia coli requires outer membrane protein receptor protein FecA. J Bacteriol 145:156–163. doi: 10.1128/JB.145.1.156-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casadaban MJ. 1976. Transposition and fusion of the lac genes to select promoters in Escherichia coli using bacteriophage Lambda and Mu. J Mol Biol 104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 54.Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 55.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ellermeier CD, Janakiraman A, Slauch J. 2002. Construction of targeted single copy lac fusions using λ Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Benjamin Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 60.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, p 71–74. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 61.Werner J, Misra R. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol 57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of fepA, fhuA, and feo expression using chromosomally integrated lacZ fusions. β-Galactosidase activities (shown in Miller units) were measured from at three independent biological replicates in various genetic backgrounds as shown. Error bars represent standard deviations. Download FIG S1, TIF file, 0.6 MB (638.9KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effects of OmrA and OmrB deletion on fecA and fepA in wild-type and EnvZR397L backgrounds. Determination of fecA and fepA expression was carried out using RT-qPCR. Data were obtained from two independent biological samples and two technical replicates. Error bars represent standard deviations. Download FIG S2, TIF file, 0.2 MB (172KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SDS-PAGE analysis of fractions containing OmpR6His obtained after nickel affinity chromatography. Protein samples were visualized after Coomassie brilliant blue staining. Fractions shown in the box were pooled and used for DNA binding studies. The position of OmpR6His is shown. Download FIG S3, TIF file, 0.9 MB (926.2KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Determination of ompC::lacZ activities of constructs carrying two different lengths of the ompC promoter regions in front of a promoterless lacZ gene. ompC145::lacZ and ompC250::lacZ contain 145 and 250 bp, respectively, of the ompC promoter region, including ATG. The β-galactosidase activities in ompR+ or ΔompR backgrounds were measured from two independent cultures grown to late log phase. Error bars represent standard deviations. Download FIG S4, TIF file, 0.8 MB (805.8KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effects of ΔompC and ΔompF gene mutations on the growth of wild-type and ΔaroB strains. Bacterial growth was monitored on an LBA plate incubated at 37°C for 24 h. The relevant genotypes are shown. Download FIG S5, TIF file, 0.5 MB (560.6KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Standard curve of ferric chloride solutions of known concentrations. The actual concentrations of ferric chloride solutions were determined by measuring the absorbance at 420 nm and then utilizing the following formula: molar concentration = A420/ε, where [Fe]ε is 2.865 mM−1 Cm−1. These values were plotted against the values obtained for the same solutions by EPR spectroscopy. We used peak-to-peak EPR measurements instead of the double integration values due to a greater confidence of the former at lower ferric chloride concentrations. The standard curve was used to measure iron concentrations in the whole cell. Download FIG S6, TIF file, 0.5 MB (504.3KB, tif) .
Copyright © 2020 Gerken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.