Abstract
Cell cycle alterations are among the principle hallmarks of cancer. Consequently, the study of cell cycle regulators has emerged as an important topic in cancer research, particularly in relation to environmental exposure. Particulate matter and coal dust around coal mines have the potential to induce cell cycle alterations. Therefore, in the present study, we performed chemical analyses to identify the main compounds present in two mineral coal samples from Colombian mines and performed systems chemo-biology analysis to elucidate the interactions between these chemical compounds and proteins associated with the cell cycle. Our results highlight the role of oxidative stress generated by the exposure to the residues of coal extraction, such as major inorganic oxides (MIOs), inorganic elements (IEs) and polycyclic aromatic hydrocarbons (PAH) on DNA damage and alterations in the progression of the cell cycle (blockage and/or delay), as well as structural dysfunction in several proteins. In particular, IEs such as Cr, Ni, and S and PAHs such as benzo[a]pyrene may have influential roles in the regulation of the cell cycle through DNA damage and oxidative stress. In this process, cyclins, cyclin-dependent kinases, zinc finger proteins such as TP53, and protein kinases may play a central role.
Keywords: Coal, Colombia, cell cycle, systems chemo-biology
Introduction
One of the largest open-pit coal mines in the world is located in northern Colombia Huertas et al. (2012a). According to the 2015 BP Statistical Energy Survey, Colombia aimed to increase its coal production by 35% to 115,000 tons per year by 2015 from 85,000 tons in 2011. Open-pit mines were forcast to account for almost 50% of this increase (BP, 2014). According to Chaulya (2004) and Huertas et al. (2012b), activities associated with coal extraction during surface coal mining release major air pollutants into the atmosphere as particulate matter (PM) and coal dust. These activities include: topsoil removal, drilling, blasting, overburden loading and unloading, coal transport over unpaved roads and wind erosion of exposed surfaces. In addition to the coal itself, PM and coal dust around coal mines can also contain O, N, H, trace species, and several inorganic minerals. The trace species may include SiO2, Cu, Al, Ni, Cd, B, Sb, Fe, Pb, and Zn (Huertas et al., 2012a). In mining, excessive occupational exposure to metals is considered to be the leading cause of metal-related cancers (Gloscow, 2007). Additionally, in open-cast coal mines, coal is stored at elevated ambient temperatures, where combustion may lead to the emission of polycyclic aromatic hydrocarbons (PAHs) (Liu et al., 2008), most of which exhibit mutagenic and carcinogenic properties (Celik et al., 2007).
There is a growing body of evidence that links long-term exposure to coal mining residues with increased risks of cardiovascular mortality (Pope and Dockery, 2006; Brook et al., 2010), premature mortality (Callén et al., 2009) and cancer (Pope 3rd et al., 2002, 2011). However, the mechanisms underlying the development of these adverse effects are poorly understood. In vitro toxicological studies have found that exposure to PM induces cell damage including genotoxicity (de Kok et al., 2005; Billet et al., 2008), cell death (Hsiao et al., 2000; Alfaro-Moreno et al., 2002), cell cycle alterations (Poma et al., 2006) and the stimulation of pro-inflammatory cytokine production (Schins and Borm, 1999). Some of the mechanisms proposed for these effects include the occurrence of oxidative damage through the production of reactive oxygen species (ROS) (Valko et al., 2006); the release of growth factors, such as TGF-β (Borm, 1997; Sambandam et al., 2015), and reduced proliferation associated with cell cycle arrest in response to genotoxic stresses and structural dysfunction of proteins (Kocbach et al., 2008; Gualtieri et al., 2011). Furthermore, a recent study (Espitia-Pérez et al., 2018) revealed a highly significant correlation between PM2.5 levels around the coal mining areas of northern Colombia and incidences of mitotic arrest, centromere damage, kinetochore malfunction and disruption of the mitotic spindle in local populations.
It has been shown that oxidative stress can override the spindle checkpoint (D’Angiolella et al., 2007), inducing microtubule depolymerization (Parker et al., 2014) and alterations in the spindle structure (Choi et al., 2007). This observation supports prior results showing that the organic components of PM2.5, particularly PAHs, have deleterious effects on the cell cycle and cause DNA damage (Longhin et al., 2013). DNA-integrity checkpoints G1/S and G2/M and metaphase–anaphase (M/A) transitions are particularly implicated in cell cycle delay (Branzei and Foiani, 2008).
Considering that one of the main characteristics of cancer is cell cycle alterations (Otto and Sicinski, 2017). The study of cell cycle regulators, particularly in terms of exposure to environmental stresors, has emerged as a pertinent avenue of research in cancer studies (Puente et al., 2014). Populations are rarely exposed to single air pollutants; therefore, experimental investigations which have focused on single-pollutant effects do not accurately assess real-world exposure risks. Consequently, a multi-pollutant perspective should be the focus of air quality management, rather than adhering to a single-pollutant viewpoint (Huang et al., 2012). Furthermore, although several recent studies have investigated the combined toxicity of complex mixtures of chemicals (Labranche et al., 2012), detailed investigations into synergistic toxicity and the possible mechanisms involved in biological responses to complex exposures remain scarce (Ku et al., 2017). Therefore, in the present study, we performed a chemical analysis of mineral coals from two different Colombian mines to identify the main compounds present. We then performed systems chemo-biology analyses to reveal the interactions between these compounds and proteins associated with the cell cycle, elucidating their underlying regulatory mechanisms.
Material and Methods
Coal sample collection
To construct a chemo-biology interactome network for the proteins associated with the cell cycle and the major chemical constituents present in the coal samples, we chemically characterized bituminous and sub-bituminous coal samples, each collected from a different open-pit mine in Colombia. The samples were collected from coalfaces at the ‘El Cerrejón’ (La Guajira, Colombia) and ‘Guacamaya’ (Puerto Libertador, Córdoba, Colombia) coal mines in December 2013 (Figure S1 (218.8KB, pdf) ). Six random points at each mine were sampled; samples were then prepared as a homogeneous pool. Coals from El Cerrejón are typically bituminous with a volatile content of 37.4% and an ash content of 6.8% (dry basis) (Feng et al., 2003). Coals from Guacamaya are sub-bituminous with a high S content (2.30% total S with 1.06% as pyritic, 1.10% as organic and 0.14% from sulfates) and high volatile content (Prada et al., 2016). While detailed chemical characterizations of El Cerrejón coal have been reported elsewhere (Nathan et al., 1999), other Colombian coals, such as those obtained from the Guacamaya mine, have not been sufficiently characterized.
Analytical methods
Chemical analysis of the coal samples included identification of the major inorganic oxides (MIOs) in the coal ashes, inorganic element (IE) determination and quantification of PAHs, described in detail below.
Analysis of MIOs in coal ashes
A fraction of bituminous and sub-bituminous coal samples were incinerated separately at 815°C. The resulting ashes were processed according to the methods described by Norrish and Hutton (1969). Finally, the detection of MIOs was performed using X-ray fluorescence spectrometry (XRF) in a Philips PW2400 spectrometer system equipped with SuperQ software.
IE measurements by particle-induced X-ray emission (PIXE) assay
The elemental composition of each coal sample was measured by the conventional in vacuo PIXE assay, as described by Johansson et al. (1995). Individual portions of each coal sample were homogenized using a mortar, pressed into pellets, and then placed in the reaction chamber (at ∼ 10-5 mbar), in a 3-MV Tandetron accelerator equipped with an energy resolution of ∼ 155 eV to 5.9 keV for obtaining the spectra. The spectra were analyzed using GUPIXWIN software (Campbell et al., 2010), and expressed in parts per million. Each sample was evaluated three times in independent replicates to obtain the mean and standard deviation.
Measurement and quantification of PAHs
The PAH contents of the coal samples were quantified using the HPLC-UV/Vis method, according to Sun et al. (1998) and Cavalcante et al. (2008). Briefly, 5 g of each coal sample was dried at 30 °C for 24 h (in duplicate) for later extraction. The extraction was performed by ultrasonication in 5 mL acetone/hexane (1:1, v/v) for 15 min. The filtrate was concentrated on a rotary evaporator and then further under a stream of nitrogen gas to ∼2 mL. A clean glass column was used for adsorption chromatography. The concentrated extracts were fractionated using a 20 × 1.5-cm column containing pre-cleaned silica gel (20 h at 110 °C). The column was first eluted with 20 mL hexane/dichloromethane (9:1, v/v), then with 30 mL hexane/dichloromethane (4:1, v/v) and finally with 10 mL dichloromethane/methanol (9:1, v/v). The eluted volumes were reduced to 1 mL, and finally, each extract was injected into a HPLC-UV system. The chromatographic conditions were as follows: 5 μm Kromasil C18 reverse-phase column (250 × 4.6 mm); injection volume: 20 μL; mobile phase (A): acetonitrile; mobile phase (B): MilliQ water; gradient method: 0 min (1:1), 10 min (7:3), 20 min (8:2), 25 min (8:2), 28 min (1:1), 30 min (1:1) and λ = 254 nm. Analytical curves were created using external standardization for quantification. In our study, we detected 11 PAHs in the samples. The PAHs detected and their limits of detection were: naphthalene (1.7976 g L−1), acenaphthylene (0.0041 g L−1), phenanthrene (0.1758 g L−1), anthracene (0.0339 g L−1), fluoranthene (0.3787 g L−1), benzo[a]anthracene (0.3411 g L−1), benzo[b]fluoranthene (0.0691 g L−1), dibenzo[a,h]anthracene (1.1110 g L−1), benzo[k]fluoranthene (2.2221 g L−1), indene[1,2,3-cd]pyrene (3.5788 g L−1) and benzo[g,h,i]perylene (0.0005 g L−1). All chromatographic measurements were performed in duplicate at ambient temperature.
Interactome data mining and design of the chemo-biology network
To design the interactome network among the main chemical substances present in the coal samples and their potential interactions with Homo sapiens proteins involved in the cell cycle, we used the STITCH search engine version 5.0 [http://stitch.embl.de/] and STRING 10.0 [http://http://string-db.org/newstring_cgi/show_input_page.pl/] (Snel et al., 2000; Jensen et al., 2008). A total of 36 chemical elements were detected in the chemical analysis of both coal samples using the XRF, PIXE, and HPLC/UV/Vis methods, and these were used for the exploration of networks within the STITCH metasearch engine. While STITCH allows visualization of the physical interactions between chemical elements and proteins, the STRING metasearch engine generates protein-protein interactions (PPIs) (Feltes et al., 2013). Each chemical–protein interaction (CPI) and PPI has a confidence level between 0 and 1.0 (where 1.0 indicates the highest confidence). Parameters used by the STITCH and STRING metasearch engines were as follows: all predictive methods were enabled except text mining; interactions: 50; degree of confidence: 0.7 and network depth: 1. The results were combined and analyzed using Cytoscape 3.4.0 (Shannon et al., 2003) and the search engine GeneCards (Rebhan et al., 1997; Safran et al., 2010) using the default parameters.
The chemical elements not involved in interactions according to STITCH were excluded. Then, using Cytoscape 3.4.0., we created the interactome that fused the small CPI and PPI networks (not shown individually) that were generated by STITCH and STRING, respectively.
Centrality analysis
To evaluate the node degree, betweenness, and to identify the ‘central’ nodes (chemical compounds/proteins) in the interactome, a centrality analysis of the interactome was performed using CentiScaPe 2.1 in Cytoscape (Scardoni et al., 2009).
Modular analysis of the major CPI-PPI network
In the interactome or CPI-PPI network, we analyzed clusters or highly connected regions that are indicative of functional protein complexes. These regions were identified using the Molecular Complex Detection application (MCODE) (Bader and Hogue, 2003; Scott, 2017). The MCODE application is included within the Cytoscape program and was used with the following parameters: loops; grade limit: 2; cluster expansion by a neighbor shell allowed; removal of a single connected node from the clusters; cut-off node density: 0.1; node score limit: 0.2; score: 2 and maximum network depth: 100.
Gene ontology (GO) analysis
The genetic ontology analysis was performed using the Biological Networks GO tool (BiNGO 3.0.3) (Maere et al., 2005), which is an application installed in Cytoscape. The clusters obtained with MCODE were analyzed to determine the main bioprocesses associated with each cluster. The degree of functional enrichment was evaluated quantitatively using the hypergeometric distribution by group and category (p-value). The false discovery rate algorithm (Benjamini and Hochberg, 1995) was used to correct for multiple tests, as implemented in BINGO with a significance of p <0.05.
Comet assay
The alkaline comet assay was carried out according to Singh et al. (1988) and Tice et al. (2000) with several modifications for a high-throughput comet assay version, which allows the processing of multiple samples (Tice et al., 2000). The high-throughput “96-mini gel format” is an 8x12 multi-array on GelBond® film (Lonza, Rockland Inc. ME, USA) (McNamee, 2000) described by Kiskinis et al. (2002). Briefly, 6 × 104 V79 cells per well were seeded in 12-well cell culture plates and incubated for 24 h; plates were subsequently treated with a 0.15 mg/mL coal dilution from either El Cerrejón or Guacamaya for 24 h. The negative control was incubated with DMEM medium (FBS free), and the positive control was treated with 150 μM H2O2 for 3 h. For semi-automated scoring, stained cells were analyzed using an Olympus BX51 fluorescence microscope (Olympus, Japan) and examined at 40X magnification under a green filter (540 nm). We analyzed 100 randomly selected nuclei, 50 from each of the two replicate slides (Gutzkow et al., 2013). % tail DNA was scored using the Comet Assay IV software (Perceptive Instruments, Haverhill, UK). The alkaline comet assay using the lesion-specific enzyme Formamidopyrimidine DNA glycosylase (FPG) (New England Biolabs, MA, USA) was used to detect oxidized purines (Collins, 2009). The protocol was used as previously described with minor modifications for the high-throughput comet assay (Kushwaha, 2011). FPG recognizes oxidized purines, specifically 8-oxo-guanine (Kushwaha, 2011). All experiments were performed in triplicate.
The normality of the data was evaluated using the Kolmogorov–Smirnov test, while the Student's t-test was used to compare results of the comet assay with and without the FPG enzyme. P ≤ 0.05 was considered statistically significant. All analyses were performed using the Graphpad PRISM statistical software (Graphpad Inc., San Diego, CA).
Results
Chemical characterization, interactome data mining and design of the chemo-biology network
The chemical characterizations of the El Cerrejón and Guacamaya coal samples are shown in Tables S1 (51.8KB, pdf) –S3 (54.1KB, pdf) . Chemical analysis by XRF revealed a similar oxide composition for each coal ash (Table S1 (51.8KB, pdf) ). A total of 10 different oxides were identified. As expected, samples from El Cerrejón showed a bulk chemical composition containing several metal oxides in the order SiO2 > Al2O3 > Fe2O3 > K2O > MgO. Ashes from the sub-bituminous coal samples from Guacamaya showed higher concentrations of CaO, MgO, and SO3 and lower concentrations of SiO2 and Al2O3 than those reported in similar studies on bituminous and sub-bituminous coals (Blissett and Rowson, 2012).
As shown in Table S2 (58.8KB, pdf) , 15 IEs were identified by PIXE. Typically, bituminous samples from El Cerrejón showed higher concentrations of Si, Al, S, and Fe than those from the sub-bituminous samples of Guacamaya. Conversely, relatively high concentrations of Na, Ca, and Mg were present in the Guacamaya samples. Sr was detected only in the Guacamaya samples. Finally, concentration data for the 11 PAHs identified by HPLC/UV/Vis are shown in Table S3 (54.1KB, pdf) . For both samples, the most abundant PAHs detected were naphthalene, phenanthrene, anthracene, fluoranthene and benzo[a]anthracene. In general, however, higher concentrations of all PAHs were found in the El Cerrejón samples.
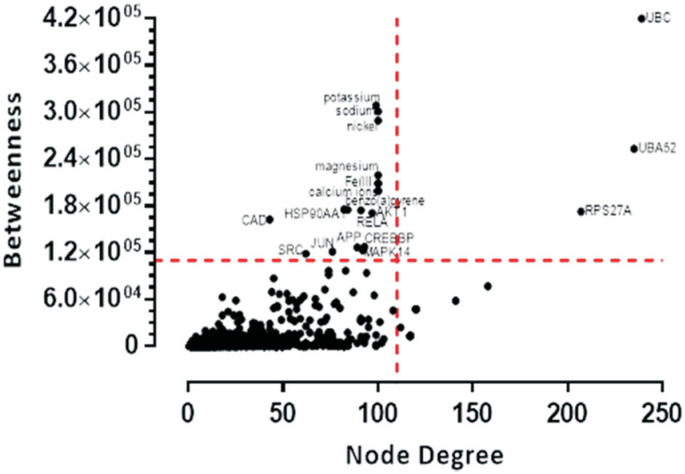
Chemical characterization of the bituminous and sub-bituminous coal samples revealed no significant differences in their chemical compositions. 36 compounds (i.e., 10 MIOs detected in coal ash, 15 IEs, and 11 PAHs) were used to construct the chemo-biology interactome. Once unconnected compounds were excluded, the remaining 24 protein-interacting compounds were used to generate 48 small CPI-PPI networks using the STRING and STITCH metasearch engines (Table 1). All the small networks were combined, resulting in a large CPI-PPI network with 2,057 nodes and 24,957 edges (Figure S2 (312.8KB, pdf) ). This large CPI-PPI network was then analyzed using CentiScaPe 2.1 to identify the nodes (proteins) occupying central positions in the network architecture. In this context, nodes known as hub-bottlenecks (HBs) are the most important and combine hub (high degree) and bottleneck (high betweenness) characteristics according to Azevedo and Moreira-Filho (2015). Through centrality analysis, we observed three HB nodes (UBC, UBA52, and RPS27A) and 15 bottlenecks (HSP90AA1, CAD, SRC, JUN, MAPK14, APP, CREBBP, AKT1, K, Na, Ni, Mg, Fe, benzo[a]pyrene and Cr) (Figure 1 and Table S4 (66.8KB, pdf) ).
Table 1. Chemical constituents of coal samples found in the major CPI-PPI network.
| Compound | Classification | Chemical classification |
|---|---|---|
| Acenaphthene | Organic | Polycyclic Aromatic Hydrocarbon |
| Anthracene | Organic | Polycyclic Aromatic Hydrocarbon |
| Benzo(a)pyrene | Organic | Polycyclic Aromatic Hydrocarbon |
| Benzo(b)fluoranthene | Organic | Polycyclic Aromatic Hydrocarbon |
| Fluoranthene | Organic | Polycyclic Aromatic Hydrocarbon |
| Naphthalene | Organic | Polycyclic Aromatic Hydrocarbon |
| Phenanthrene | Organic | Polycyclic Aromatic Hydrocarbon |
| SiO2 | Inorganic | Oxide of silicon |
| TiO2 | Inorganic | Oxide of titanium |
| Fe2O3 | Inorganic | Oxide of iron |
| Al | Inorganic | Metal |
| Ca | Inorganic | Alkaline earth metal |
| Cl | Inorganic | Halogen |
| Cr | Inorganic | Transition metal |
| Fe | Inorganic | Transition metal |
| K | Inorganic | Alkali metal |
| Mg | Inorganic | Alkaline earth metal |
| Mn | Inorganic | Transition metal |
| Na | Inorganic | Alkali metal |
| Ni | Inorganic | Transition metal |
| S | Inorganic | Non-metal |
| Sr | Inorganic | Alkaline earth metal |
| Ti | Inorganic | Transition metal |
| Zn | Inorganic | Transition metal |
Figure 1. Scatter plot of degree and betweenness values for all nodes. Hubs (high degree), bottlenecks (high betweenness), and nodes with high relative values in both parameters are identified.
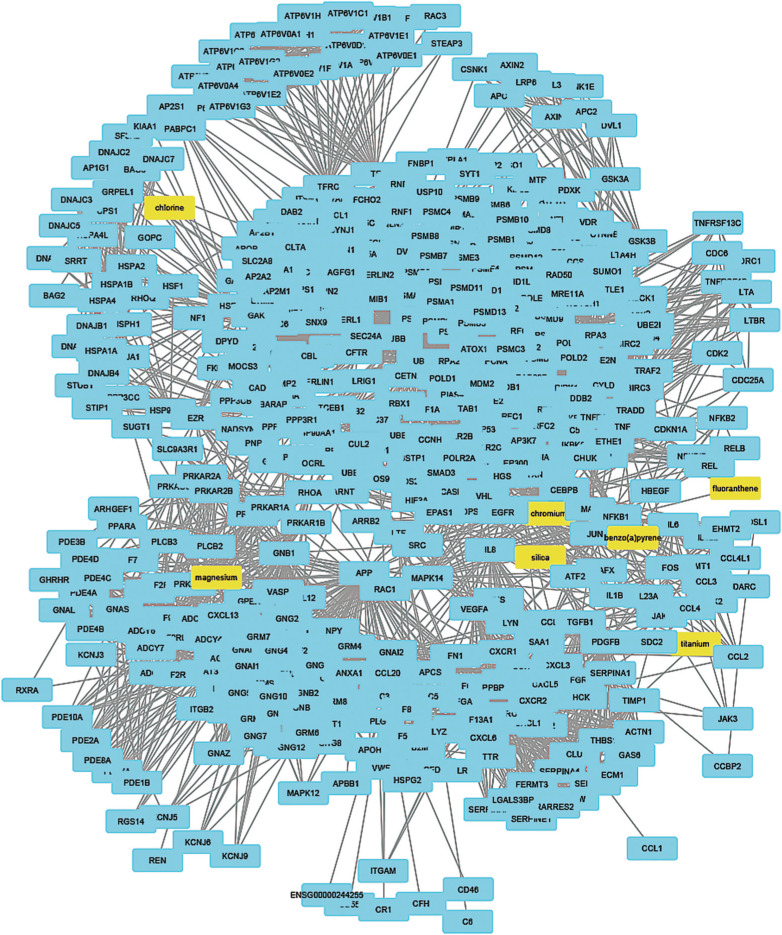
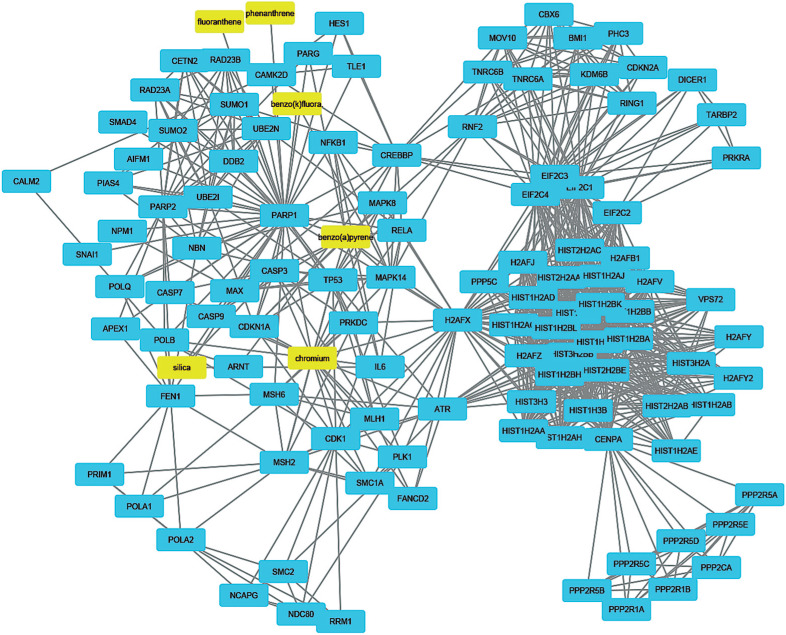
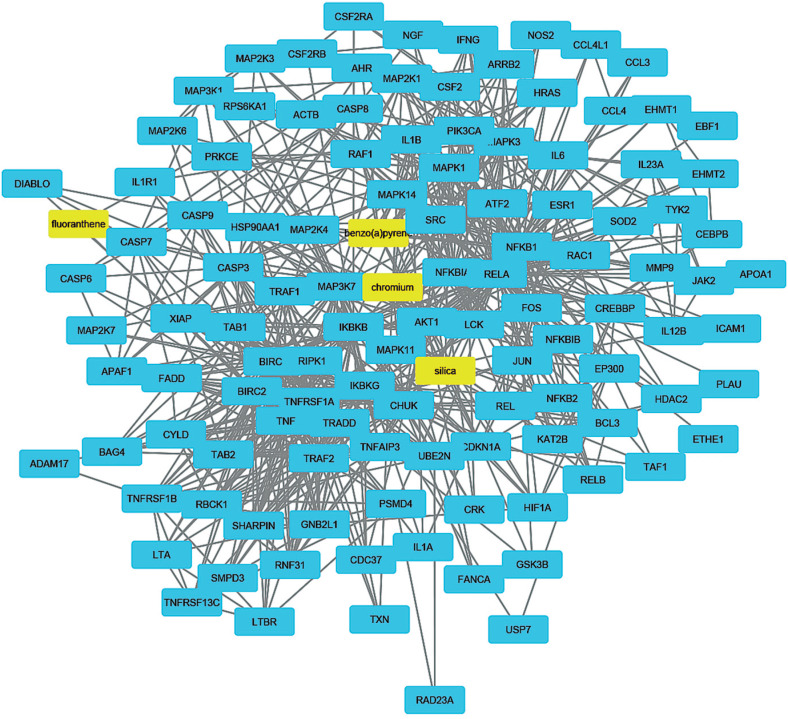
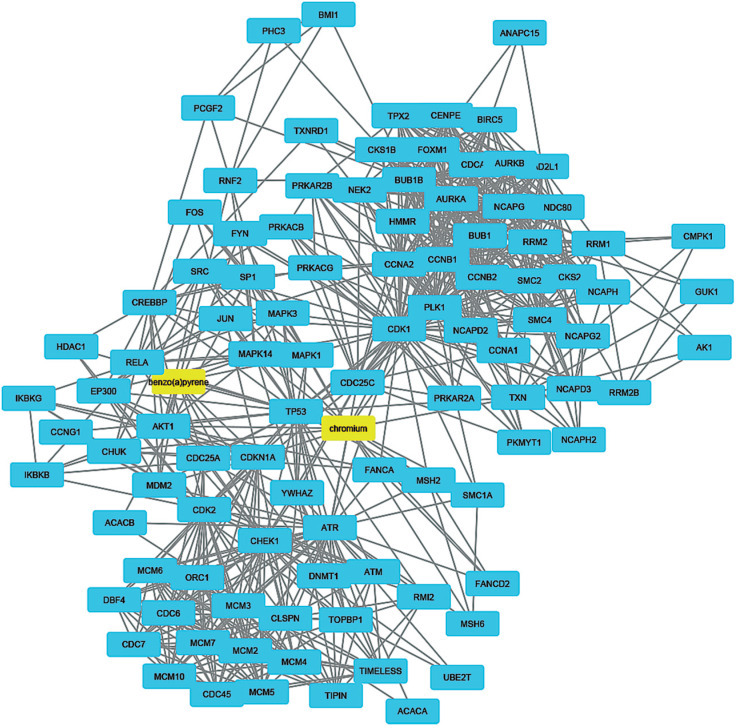
To understand how coal chemical constituents interact with cell cycle processes, we identified the modules in the main CPI-PPI network using the MCODE program. From these analyses, we obtained eight significant modules related to cell cycle processes (Figures 2 - 9). Clusters 6 (Figure 2), 11(Figure 3), 13 (Figure 4), and 14 (Figure 5) are associated with MIOs, IEs and PAHs; clusters 9 (Figure 6) and 12 (Figure 7) appear to be associated with IEs and PAHs; finally clusters 2 (Figure 8) and 4 (Figure 9) are associated with IEs only. The analysis revealed 15 common proteins associated with different cell cycle processes.
Figure 2. Cluster analysis of the major CPI-PPI network showing the association of cluster 6 with MIOs, IEs, and PAHs (yellow). The cluster is composed of 487 nodes and 5,545 edges, with Ci = 22,725. The associated constituents are SiO2, Ti, Mg, Cr, Cl, fluoranthene, and benzo[a]pyrene.
Figure 9. Cluster analysis of the major CPI-PPI network showing association of cluster 4 with IEs (yellow). It is composed of 208 nodes and 3,134 edges, with Ci = 2,999, associated with Cr and S.
Figure 3. Cluster analysis of the major CPI-PPI network showing the association of cluster 11 with MIOs, IEs, and PAHs (yellow). It is composed of 117 nodes and 867 edges, with Ci = 14,695. The associated constituents are SiO2, Cr, benzo[b]fluoranthene, fluoranthene, phenanthrene and benzo[a]pyrene.
Figure 4. Cluster analysis of the major CPI-PPI network showing the association of cluster 13 with MIOs, IEs, and PAHs (yellow). It is composed of 118 nodes and 732 edges, with Ci = 12,303. The associated constituents include SiO2, Cr, fluoranthene, and benzo[a]pyrene.
Figure 5. Cluster analysis of the major CPI-PPI network showing the association of cluster 14 with MIOs, IEs, and PAHs (yellow). It is composed of 432 nodes and 2,520 edges, with Ci = 1,164. The associated constituents are S, Mn, Mg, Fe, Fe2O3 and benzo[a]pyrene.
Figure 6. Cluster analysis of the major CPI-PPI network showing the association of cluster 9 with IEs and PAHs (yellow). It is composed of 249 nodes and 2,180 edges, with Ci = 17,44. The associated compounds are S, Cr, Ti, and benzo[a]pyrene.
Figure 7. Cluster analysis of the major CPI-PPI network showing the association of cluster 12 with IEs and PAHs (yellow). It is composed of 102 nodes and 741 edges, with Ci = 14,388. The associated compounds are Cr and benzo[a]pyrene.
Figure 8. Cluster analysis of the major CPI-PPI network showing association of cluster 2 with IEs (yellow). It is composed of 250 nodes and 5,976 edges, with Ci = 47,618, associated with Mg.
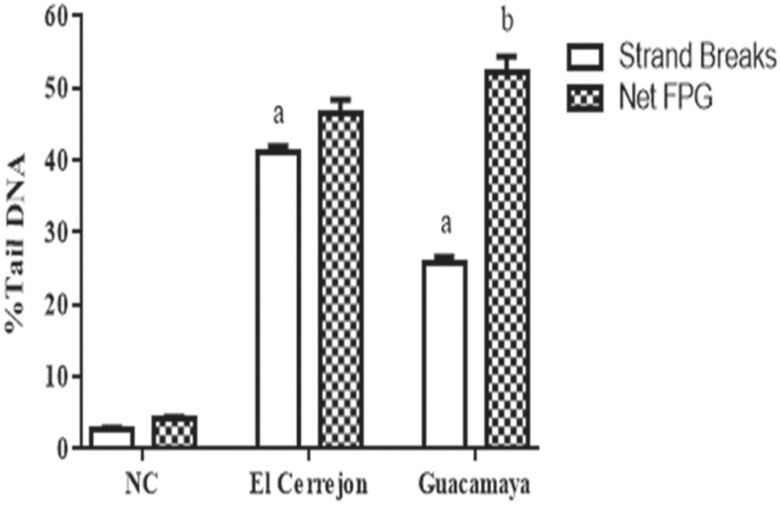
The DNA damage induced by El Cerrejón and Guacamaya coal was determined by the modified alkaline high-throughput version of the comet assay and evaluated by the % tail DNA. The results of the comet assay showed statistically significant differences in relation to the negative control (NC) without enzyme (P <0.05) and the % DNA tail increase. Additionally, the results of the modified comet assay showed a statistically significant difference when compared with the same sample group (P < 0.05) (Figure 10).
Figure 10. Percent tail DNA in the alkaline comet assay (strand breaks in white) and oxidized purines (grid) in a modified comet assay with FPG in V79 cells under 24 h exposure with El Cerrejon and Gucamaya coal. a) Statistically significant differences in relation negative control (NC) without enzyme P <0.05. b) Statistically significant differences in relation to the same sample group with an enzyme. The results are shown as the mean ± SEM.
Discussion
Ubiquitin (UBC) and two ubiquitin-coding genes (UBA52 and RPS27A) demonstrated the highest node degree and betweenness values, thus representing highly central proteins inside the network (Feltes et al., 2013). UBC is a small 76-amino acid protein that is involved in several different pathways within the cell, including the clearing of damaged/misfolded proteins during proteotoxic stress (Bianchi et al., 2015). UBC genes are upregulated in response oxidative stress (Lee and Ryu, 2017), thereby increasing cellular UBC above threshold levels and conferring resistance to oxidative damage.
Systemic effects of MIOs, IEs and PAHs in the cell cycle and DNA damage
Tables 2–4 show the results of the GO analysis for each cluster and the cell cycle process categories. The main biological processes linked to clusters 6, 11, 13 and 14 included the following: (i) cell cycle process, (ii) mitotic cell cycle, (iii) cell cycle, (iv) cell cycle checkpoint, (v) regulation of cell cycle and (vi) cell cycle arrest (Table 2). Interestingly, DNA repair bioprocesses were found in this module only in co-occurrence with MIOs, IEs and PAHs. The particular combination of these compounds is associated with increased DNA damage in cell systems in vitro (Leon-Mejia et al., 2016) and human populations in coal mining environments (Leon-Mejia et al., 2011). The primary mechanism proposed for these effects involves oxidative damage through the production of ROS (Valko et al., 2006). In this regard, within the same module, proteins regulated by oxidative stress inside the cell were identified as bottlenecks (AKT, APP, JUN and CREBBP). While AKT has been reported to be regulated by oxidative stress for cell survival (Wang et al., 2000), several studies have indicated that oxidative stress participates in events that enhance amyloidogenic APP processing in neurons (Lin and Beal, 2006; Mouton-Liger et al., 2012) and in events that affect cerebrovascular endothelial APP processing (Muche et al., 2017). ROS-facilitated protein phosphorylation can also lead to kinase-mediated activation of transcription factors, such as the JUN group (Nathan and Cunningham-Bussel, 2013), affecting cell cycle progression by their ability to regulate the expression and function of cell cycle regulators such as cyclins (Schreiber et al., 1999; Chiba et al., 2017), and apoptosis (Meixner et al., 2010). Together with JUN, CREBBP is also involved in cell division and cell proliferation, and it is upregulated by the oxidative stress response in retinoblastoma cells (Meixner et al., 2010).
Table 2. Major cell cycle bioprocesses in clusters 6, 11, 13 and 14 associated with MIOs, IEs and PAHs.
| GO ID | p-value | corr p-value | k* | n# | Description | Genes in test set |
|---|---|---|---|---|---|---|
| 22402 | 1,64E-22 | 9,74E-21 | 77 | 582 | cell cycle process | APP|CDKN1A|CETN2|CLTC|UBE2D1|PSMD8|PSMD9|PPP3CA|PSMD7|PSMD4|PSMD2|PSMD3|PSMD1|AKT1|IL12B|NBN|POLE|APC2|H2AFX|CDC25A|DNM2|PSMA5|PSMA6|DNAJC2|PSMA3|ADAM17|PSMA4|PSMA1|PSME3|PSME1|PSME2|TP53|PSMD10|PSMD12|PSMD11|RGS14|PSMD13|CUL2|THBS1|EGFR|PSMB10|PSMB6|PSMB7|PSMB4|C6|PSMB5|PSMB2|PSMB3|POLD1|PSMB1|CLTCL1|APBB1|UBE2I|TGFB1|SMAD3|VDR|RPA1|MRE11A|CDC6|HSPA2|PSMB8|MAPK12|PSMB9|PPP5C|RAD50|PSMC6|PSMC3|APC|IL8|PSMC4|PSMC1|PSMC2|CDK2|MDM2|CTNNB1|CALR|SUGT1 |
| 278 | 9,54E-21 | 5,08E-19 | 60 | 380 | mitotic cell cycle | APP|CDKN1A|CETN2|CLTC|UBE2D1|PSMD8|PSMD9|PPP3CA|PSMD7|PSMD4|PSMD2|PSMD3|PSMD1|AKT1|POLE|APC2|CDC25A|DNM2|PSMA5|PSMA6|DNAJC2|PSMA3|ADAM17|PSMA4|PSMA1|PSME3|PSME1|PSME2|PSMD10|PSMD12|PSMD11|RGS14|PSMD13|CUL2|EGFR|PSMB10|PSMB6|PSMB7|PSMB4|C6|PSMB5|PSMB2|PSMB3|POLD1|PSMB1|CLTCL1|UBE2I|CDC6|PSMB8|PSMB9|PPP5C|PSMC6|PSMC3|APC|PSMC4|PSMC1|PSMC2|CDK2|MDM2|SUGT1 |
| 7049 | 3,12E-18 | 1,43E-16 | 84 | 794 | cell cycle | APP|CDKN1A|STEAP3|CCNH|CETN2|CLTC|UBE2D1|PSMD8|PSMD9|PPP3CA|PSMD7|PSMD4|PSMD2|PSMD3|PSMD1|AKT1|IL12B|EP300|NBN|POLE|APC2|ANXA1|H2AFX|CDC25A|DNM2|PSMA5|GAK|PSMA6|DNAJC2|PSMA3|ADAM17|PSMA4|PSMA1|PSME3|PSME1|PSME2|TP53|PSMD10|PSMD12|PSMD11|RGS14|PSMD13|CUL2|THBS1|EGFR|PSMB10|PSMB6|PSMB7|PSMB4|C6|PSMB5|PSMB2|PSMB3|POLD1|PSMB1|CLTCL1|APBB1|UBE2I|TGFB1|SMAD3|VDR|RPA1|MRE11A|CDC6|HSPA2|PSMB8|MAPK12|PSMB9|CYLD|PPP5C|RAD50|PSMC6|PSMC3|APC|IL8|PSMC4|PSMC1|PSMC2|CDK2|MDM2|CTNNB1|REN|CALR|SUGT1 |
| 22402 | 4,62E-06 | 1,46E-04 | 22 | 582 | cell cycle process | CDKN1A|NPM1|UBE2I|CDKN2A|CETN2|PLK1|H2AFX|NCAPG|SMC1A|MLH1|CENPA|NDC80|SMC2|MSH6|PPP2CA|POLA1|PPP5C|MSH2|FANCD2|CDK1|NBN|TP53 |
| 75 | 5,61E-06 | 1,73E-04 | 11 | 107 | cell cycle checkpoint | CDKN1A|MSH2|CDKN2A|PLK1|H2AFX|CDK1|NBN|PPP2R5C|SMC1A|TP53|ATR |
| 6281 | 7,00E-12 | 4,71E-10 | 22 | 298 | DNA repair | POLQ|FEN1|PARP1|PRKDC|PARP2|H2AFX|RAD23A|SMC1A|MLH1|RAD23B|DDB2|MSH6|POLB|POLA1|MSH2|SUMO1|FANCD2|APEX1|UBE2N|NBN|TP53|ATR |
| 51726 | 1,37E-05 | 1,78E-04 | 19 | 446 | regulation of cell cycle | MAP2K1|JUN|CREBBP|CDKN1A|HDAC2|NGF|TNF|CYLD|KAT2B|IL1A|ADAM17|IFNG|CDC37|CASP3|IL1B|IL12B|AKT1|HRAS|MAP2K6 |
| 51726 | 5,25E-11 | 1,98E-09 | 48 | 447 | regulation of cell cycle | CDS1|CDKN1A|HDAC2|TRRAP|HDAC1|CITED2|CUL1|ILK|FOXO4|ETS1|EGFR|SOX2|CCND3|CCND1|CDH1|AKT1|IL12B|SFN|PRKACA|BTRC|JUNB|HRAS|MEN1|APC2|TCF7L2|JUN|CREBBP|MAP2K1|TIPIN|SMAD3|CDKN2A|GSS|INSR|PTPN11|CDC25C|CDC25A|SMARCA4|FOSL1|KAT2B|COPS5|APC|PKIA|MDM2|TIMELESS|ATM|TCF4|TCF3|TP53 |
| 22402 | 9,37E-06 | 1,83E-04 | 46 | 583 | cell cycle process | CAMK2B|CDKN1A|NCAPG2|CUL1|UBE2D1|ILK|FOXO4|EGFR|SOX2|PPP2CA|PPP3CA|CCND1|CDH1|RUVBL1|ABL1|AKT1|IL12B|BTRC|HRAS|MEN1|SKP1|APC2|TCF7L2|MAP2K1|TIPIN|UBE2I|SMAD3|CSNK1A1|CDKN2A|GSS|CDC25C|CDC25A|KAT2B|PPP5C|APC|MDM2|TIMELESS|CTNNB1|NCAPD2|MDM4|ATM|TCF4|NCAPD3|TCF3|TP53|TAF1 |
| 7049 | 2,41E-05 | 4,31E-04 | 55 | 795 | cell cycle | CDKN1A|STEAP3|NCAPG2|UBE2D1|ILK|CDC73|SOX2|PPP3CA|CCND1|CDH1|RUVBL1|AKT1|IL12B|EP300|BTRC|HRAS|MEN1|SKP1|APC2|MAP2K1|TIPIN|ANXA1|DUSP1|FBXW11|CDC25C|CDC25A|KAT2B|TIMELESS|TP53|CDS1|CAMK2B|CUL1|FOXO4|EGFR|RNF2|PPP2CA|ABL1|TCF7L2|UBE2I|SMAD3|CSNK1A1|CDKN2A|GSS|PPP1CA|PPP5C|APC|MDM2|CTNNB1|NCAPD2|MDM4|ATM|TCF4|NCAPD3|TCF3|TAF1 |
| 7050 | 2,63E-05 | 4,64E-04 | 18 | 109 | cell cycle arrest | TCF7L2|CDKN1A|MAP2K1|SMAD3|CDKN2A|GSS|CUL1|ILK|FOXO4|SOX2|KAT2B|APC|IL12B|ATM|TCF4|TP53|HRAS|MEN1 |
total number of nodes in the gene ontology (GO) annotation;
number of nodes related to a given GO in the network.
Table 4. Major cell cycle bioprocesses in clusters 2 and 4 associated with IEs.
| GO ID | p-value | corr p- | k* | n# | Description | Genes in test set |
|---|---|---|---|---|---|---|
| 278 | 6,09E-38 | 2,63E-36 | 60 | 380 | mitotic cell cycle | CLTC|BUB1B|PSMD8|CDC20|PSMD9|PSMD7|CDC23|PSMD2|PSMD3|CDC27|PSMD1|AKT1|ANAPC7|RPS6|DNM2|PSMA5|PSMA6|PSMA3|ADAM17|PSMA4|PSMA1|PSME3|PSME1|RPL24|PSME2|ANAPC4|ANAPC5|ANAPC1|ANAPC2|PSMD10|ANAPC13|PSMD12|PSMD11|CUL5|PSMD13|CUL2|ANAPC10|PSMB10|ANAPC11|PSMB6|PSMB7|PSMB4|CCNB1|FZR1|PSMB5|PSMB2|PSMB3|PSMB1|CLTCL1|PSMB8|PSMB9|MAD2L2|PSMC6|PSMC3|PSMC4|PSMC1|CDC16|PSMC2|MDM2|MAD2L1 |
| 22402 | 4,93E-32 | 1,84E-30 | 65 | 582 | cell cycle process | CLTC|BUB1B|PSMD8|CDC20|PSMD9|PSMD7|CDC23|PSMD2|PSMD3|CDC27|PSMD1|AKT1|ANAPC7|RPS6|DNM2|RAD51B|PSMA5|PSMA6|PSMA3|ADAM17|RAD51C|PSMA4|PSMA1|PSME3|PSME1|RPL24|PSME2|ANAPC4|ANAPC5|TP53|ANAPC1|ANAPC2|PSMD10|ANAPC13|PSMD12|PSMD11|CUL5|PSMD13|CUL2|ANAPC10|PSMB10|ANAPC11|PSMB6|PSMB7|PSMB4|CCNB1|FZR1|PSMB5|PSMB2|PSMB3|PSMB1|CLTCL1|BARD1|PSMB8|PSMB9|MAD2L2|PSMC6|RAD51|PSMC3|PSMC4|PSMC1|CDC16|PSMC2|MDM2|MAD2L1 |
| 7049 | 1,12E-25 | 3,45E-24 | 67 | 794 | cell cycle | CLTC|BUB1B|BRCA1|PSMD8|CDC20|PSMD9|PSMD7|CDC23|PSMD2|PSMD3|CDC27|PSMD1|AKT1|ANAPC7|RPS6|DNM2|RAD51B|PSMA5|GAK|PSMA6|PSMA3|ADAM17|RAD51C|PSMA4|PSMA1|PSME3|PSME1|RPL24|PSME2|ANAPC4|ANAPC5|TP53|ANAPC1|ANAPC2|PSMD10|ANAPC13|PSMD12|PSMD11|CUL5|PSMD13|CUL2|ANAPC10|PSMB10|ANAPC11|PSMB6|PSMB7|PSMB4|CCNB1|FZR1|PSMB5|PSMB2|PSMB3|PSMB1|CLTCL1|BARD1|PSMB8|PSMB9|MAD2L2|PSMC6|RAD51|PSMC3|PSMC4|PSMC1|CDC16|PSMC2|MDM2|MAD2L1 |
| 22403 | 3,84E-07 | 8,30E-06 | 30 | 435 | cell cycle phase | ANAPC13|CUL5|CLTC|CUL2|BUB1B|ANAPC10|ANAPC11|CDC20|CCNB1|FZR1|CDC23|CDC27|CLTCL1|AKT1|ANAPC7|RPS6|DNM2|RAD51B|MAD2L2|ADAM17|RAD51C|RAD51|CDC16|MDM2|RPL24|ANAPC4|ANAPC5|ANAPC1|MAD2L1|ANAPC2 |
| 87 | 5,19E-06 | 1,03E-04 | 21 | 239 | M phase of mitotic cell cycle | ANAPC13|ANAPC7|CLTC|RPS6|BUB1B|ANAPC10|ANAPC11|CDC20|MAD2L2|CCNB1|FZR1|CDC23|CDC27|CDC16|CLTCL1|RPL24|ANAPC4|ANAPC5|ANAPC1|MAD2L1|ANAPC2 |
| 7049 | 5,73E-35 | 4,37E-33 | 71 | 794 | cell cycle | CCNK|CDKN1A|CCNT2|CCNT1|MCM7|CCNH|CETN2|BUB1B|BRCA1|CKS1B|CDC20|CDC23|EXO1|CHEK1|CDC27|EP300|NBN|POLK|POLE|TIPIN|LIG1|ANAPC7|LIG4|LIG3|RAD51B|MSH6|CCNA2|CCNA1|RAD51C|DBF4|MSH2|CKS2|TIMELESS|MCM3|ANAPC4|ANAPC5|MCM6|TP53|ANAPC1|ANAPC2|MCM2|ANAPC13|BLM|ANAPC10|ANAPC11|CCNB2|CCNB1|CDC45|POLD1|CLSPN|BARD1|UBE2I|UBE2B|RPA1|MRE11A|CDC7|CDC6|MAD2L2|POLA1|RAD50|CDK6|RAD51|CDC16|CDK2|MDM2|CDK1|ATM|MNAT1|ATR|MAD2L1|TAF1 |
| 22403 | 2,64E-31 | 1,79E-29 | 53 | 435 | cell cycle phase | CCNK|CDKN1A|CETN2|BUB1B|CDC20|CDC23|EXO1|CHEK1|CDC27|NBN|POLK|POLE|TIPIN|ANAPC7|LIG3|RAD51B|MSH6|CCNA2|CCNA1|RAD51C|DBF4|CKS2|TIMELESS|ANAPC4|ANAPC5|ANAPC1|ANAPC2|ANAPC13|BLM|ANAPC10|ANAPC11|CCNB2|CCNB1|POLD1|UBE2I|UBE2B|RPA1|MRE11A|CDC7|CDC6|MAD2L2|POLA1|RAD50|CDK6|RAD51|CDC16|CDK2|MDM2|CDK1|ATM|MNAT1|MAD2L1|TAF1 |
| 22402 | 7,25E-28 | 4,15E-26 | 56 | 582 | cell cycle process | CCNK|CDKN1A|CETN2|BUB1B|CDC20|CDC23|EXO1|CHEK1|CDC27|NBN|POLK|POLE|TIPIN|ANAPC7|LIG3|RAD51B|MSH6|CCNA2|CCNA1|RAD51C|DBF4|MSH2|CKS2|TIMELESS|ANAPC4|ANAPC5|TP53|ANAPC1|ANAPC2|ANAPC13|BLM|ANAPC10|ANAPC11|CCNB2|CCNB1|POLD1|BARD1|UBE2I|UBE2B|RPA1|MRE11A|CDC7|CDC6|MAD2L2|POLA1|RAD50|CDK6|RAD51|CDC16|CDK2|MDM2|CDK1|ATM|MNAT1|MAD2L1|TAF1 |
| 51726 | 1,75E-27 | 9,46E-26 | 50 | 446 | regulation of cell cycle | CCNK|CDKN1A|CCNT2|CCNT1|BUB1B|BRCA1|CKS1B|CCND3|CDC23|CASP3|CHEK1|NBN|TIPIN|DDB1|CCNA2|MSH2|CKS2|TIMELESS|TP53|ANAPC2|BLM|ANAPC10|BRIP1|CCNB1|CDC45|RBBP8|CLSPN|BARD1|JUN|CREBBP|UBE2B|MRE11A|CDC7|GTF2H1|CDC6|MAD2L2|CDK7|COPS5|CDK6|ERCC3|CDC16|FAM175A|CDK2|ERCC2|MDM2|CDK1|ATM|MNAT1|ATR|MAD2L1 |
| 75 | 5,24E-20 | 2,13E-18 | 25 | 107 | cell cycle checkpoint | CDKN1A|BLM|BUB1B|BRCA1|BRIP1|CCNB1|CDC45|CHEK1|RBBP8|NBN|CLSPN|TIPIN|CDC6|DDB1|MAD2L2|CCNA2|MSH2|ERCC3|FAM175A|ERCC2|CDK1|ATM|TP53|ATR|MAD2L1 |
| 278 | 4,71E-19 | 1,88E-17 | 39 | 380 | mitotic cell cycle | ANAPC13|CCNK|CDKN1A|BLM|CETN2|BUB1B|ANAPC10|ANAPC11|CDC20|CCNB2|CCNB1|CDC23|POLD1|CDC27|POLK|POLE|TIPIN|UBE2I|ANAPC7|CDC7|CDC6|MAD2L2|CCNA2|POLA1|CCNA1|CDK6|DBF4|CDC16|CDK2|TIMELESS|MDM2|CDK1|ANAPC4|ANAPC5|MNAT1|ANAPC1|MAD2L1|ANAPC2|TAF1 |
| 87 | 3,12E-14 | 1,00E-12 | 28 | 239 | M phase of mitotic cell cycle | ANAPC13|CCNK|CETN2|BUB1B|ANAPC10|ANAPC11|CDC20|CCNB2|CCNB1|CDC23|CDC27|POLK|TIPIN|UBE2I|ANAPC7|CDC6|MAD2L2|CCNA2|CCNA1|CDC16|CDK2|TIMELESS|CDK1|ANAPC4|ANAPC5|ANAPC1|MAD2L1|ANAPC2 |
| 51329 | 2,90E-11 | 6,99E-10 | 18 | 102 | interphase of mitotic cell cycle | CDKN1A|BLM|CDC7|CDC6|ANAPC10|POLA1|CCNB1|CDC23|CDK6|DBF4|POLD1|CDK2|MDM2|ANAPC4|ANAPC5|MNAT1|POLE|TAF1 |
| 51327 | 1,02E-07 | 1,44E-06 | 15 | 102 | M phase of meiotic cell cycle | UBE2B|RPA1|MRE11A|LIG3|RAD51B|MSH6|CCNA1|RAD50|RAD51C|RAD51|EXO1|CHEK1|CKS2|ATM|NBN |
| 51321 | 1,18E-07 | 1,65E-06 | 15 | 103 | meiotic cell cycle | UBE2B|RPA1|MRE11A|LIG3|RAD51B|MSH6|CCNA1|RAD50|RAD51C|RAD51|EXO1|CHEK1|CKS2|ATM|NBN |
| 10564 | 8,19E-06 | 9,75E-05 | 15 | 138 | regulation of cell cycle process | TIPIN|CREBBP|CDKN1A|UBE2B|MRE11A|CDC7|BRCA1|ANAPC10|MAD2L2|CCNB1|CDC23|CDC16|TIMELESS|MDM2|ATM |
| 7093 | 2,08E-05 | 2,38E-04 | 10 | 52 | mitotic cell cycle checkpoint | MAD2L2|CCNA2|CDKN1A|CCNB1|CDK1|BUB1B|ATM|NBN|TP53|MAD2L1 |
| 7346 | 2,63E-05 | 2,95E-04 | 16 | 174 | regulation of mitotic cell cycle | CDKN1A|BUB1B|CDC6|ANAPC10|MAD2L2|CCNA2|CCNB1|CDC23|CDC16|CDK2|MDM2|CDK1|ATM|NBN|TP53|MAD2L1 |
| 86 | 9,00E-05 | 9,82E-04 | 7 | 21 | G2/M transition of mitotic cell cycle | CDKN1A|CCNB1|CDK2|ANAPC4|ANAPC5|ANAPC10|TAF1 |
total number of nodes in the gene ontology (GO) annotation;
number of nodes related to a given GO in the netwo
Oxidative stress can generate alterations in the progression of the cell cycle (blockage and/or delay), as well as structural dysfunction in several proteins. DNA-integrity checkpoints G1/S and G2/M, and M/A transitions determine cell cycle delays (Rieder, 2011) depending on the cyclin-dependent kinase (Cdk)/cyclin system, such as Cdk1/cyclin B1, which drives the progression from G2 to the mitotic phase (Pearce and Humphrey, 2001). The protein kinases ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) promote DNA damage response and stimulate the checkpoint protein kinases Chk1/2, that can influence cell cycle arrest. CDK1 and other important proteins related to cell cycle checkpoints (e.g. CDC25C and CDC25A), and DNA damage, were found to be the critical proteins inside this cluster. Oxidative stress often induces cell cycle arrest (Klein and Ackerman, 2003; Pyo et al., 2013), in part through the degradation of the CDC25C protein through a Chk1 protein kinase-dependent pathway (Savitsky and Finkel, 2002).
Cell cycle arrest associated with complex mixtures of PAHs, metals, and other organic compounds upon exposure to coal mining residues has been observed in vitro (Tucker and Ong, 1985) and in vivo (Espitia-Perez et al., 2018). More recently, exposure to benzo[a]pyrene (also present in the cluster) has been reported to induce cell cycle arrest and apoptosis in human choriocarcinoma cancer cells through the generation of ROS (Kim et al., 2017).
Systemic effects of IEs and PAHs in the cell cycle
As shown in Table 3, the GO analysis of clusters 9 and 12 revealed 14 main process annotations associated with the cell cycle and particularly Cr and benzo[a]pyrene. The main biological processes found in these clusters included the following: i) regulation of mitotic cell cycle, ii) cell cycle checkpoint and iii) the interphase of mitotic cell cycle. Several reports have demonstrated that more-than-additive mortality is common for IE/PAH mixtures. The PAH toxicity in individual aspects suggests that they modify the accumulation of IEs and improve element-derived reactive ROS. Redox-active elements (e.g., Cu and Ni) are also capable of enhancing the redox cycling of PAHs (Gauthier et al., 2015). Several reports have implicated IEs as modifiers of P450 function and regulation, which implies that such elements could alter P450-mediated PAH mutagenicity and carcinogenicity (Peng et al., 2015). Cr is typically used in coal mining processes (Pandey et al., 2014) and is particularly associated with the fine fractions of PM (Kothai et al., 2009). The genotoxic effects of Cr are predominantly the formation of oxidative adducts and apurinic/apyrimidinic lesions, eventually resulting in DNA breakage (Vasylkiv et al., 2010). Additionally, Cr(VI) has been shown to be aneugenic, as revealed by both chromosome assays and centromere-positive micronuclei assays (Wise and Wise, 2010). However, the combined toxicity of Cr and benzo[a]pyrene has rarely been studied.
Table 3. Major cell cycle bioprocesses in clusters 9 and 12 associated with IEs and PAHs.
| GO ID | p-value | corr p- | k* | n# | Description | Genes in test set |
|---|---|---|---|---|---|---|
| 22402 | 2,71E-34 | 3,52E-31 | 67 | 582 | cell cycle process | UBE2D1|BUB1B|CDC20|PPP3CA|CDC23|EXO1|CHEK1|CDC27|IL12B|AKT1|NEK2|NBN|HRAS|TIPIN|ANAPC7|H2AFX|CDC25C|RAD51B|MSH6|CCNA2|CCNA1|RAD51C|MSH2|IFNG|CKS2|TIMELESS|ANAPC4|BIRC5|ANAPC5|TP53|ANAPC1|ANAPC2|ANAPC13|BLM|CUL5|CUL2|NCAPG|CDCA8|PKMYT1|CENPA|THBS1|ANAPC10|EGFR|AURKB|ANAPC11|AURKA|CCNB2|CCNB1|FZR1|BUB1|BARD1|UBE2I|TGFB1|PLK1|MRE11A|CDC6|MLH1|NDC80|TPX2|CENPE|RAD50|RAD51|CDC16|CDK2|CDK1|ATM|MAD2L1 |
| 7049 | 3,93E-33 | 3,40E-30 | 75 | 794 | cell cycle | UBE2D1|BUB1B|FOXM1|CKS1B|CDC20|PPP3CA|CDC23|EXO1|CHEK1|CDC27|IL12B|AKT1|EP300|NEK2|NBN|HRAS|TIPIN|ANAPC7|H2AFX|CDC25C|RAD51B|MSH6|CCNA2|CCNA1|RAD51C|MSH2|IFNG|CKS2|TIMELESS|ANAPC4|BIRC5|ANAPC5|TP53|ANAPC1|ANAPC2|ANAPC13|BLM|CUL5|CUL2|NCAPG|CDCA8|PKMYT1|CENPA|THBS1|ANAPC10|EGFR|AURKB|ANAPC11|AURKA|CCNB2|CCNB1|FZR1|CDC45|MAPK1|CLSPN|BUB1|MAPK3|BARD1|UBE2I|TGFB1|PLK1|MRE11A|CDC6|MLH1|NDC80|TPX2|CENPE|RAD50|RAD51|CDC16|CDK2|CDK1|ATM|ATR|MAD2L1 |
| 22403 | 2,04E-31 | 1,06E-28 | 57 | 435 | cell cycle phase | BUB1B|CDC20|PPP3CA|CDC23|EXO1|CHEK1|CDC27|AKT1|NEK2|NBN|TIPIN|ANAPC7|H2AFX|CDC25C|RAD51B|MSH6|CCNA2|CCNA1|RAD51C|CKS2|TIMELESS|ANAPC4|BIRC5|ANAPC5|ANAPC1|ANAPC2|ANAPC13|BLM|CUL5|CUL2|NCAPG|CDCA8|PKMYT1|ANAPC10|EGFR|AURKB|ANAPC11|AURKA|CCNB2|CCNB1|FZR1|BUB1|UBE2I|PLK1|MRE11A|CDC6|MLH1|NDC80|TPX2|CENPE|RAD50|RAD51|CDC16|CDK2|CDK1|ATM|MAD2L1 |
| 51726 | 1,03E-27 | 3,84E-25 | 54 | 446 | regulation of cell cycle | BUB1B|FOXM1|CKS1B|CDC23|CHEK1|IL12B|AKT1|NEK2|NBN|PRKACA|HRAS|TIPIN|H2AFX|CDC25C|CCNA2|MSH2|IFNG|CDC37|CKS2|TIMELESS|BIRC5|TP53|ANAPC2|BLM|CUL5|CUL2|PKMYT1|THBS1|ANAPC10|EGFR|BRIP1|CCNB1|FZR1|CDC45|RBBP8|CLSPN|BUB1|BARD1|JUN|CREBBP|TGFB1|PLK1|MRE11A|CDC6|TPX2|CENPE|COPS5|CDC16|FAM175A|CDK2|CDK1|ATM|ATR|MAD2L1 |
| 278 | 6,00E-23 | 8,21E-21 | 46 | 380 | mitotic cell cycle | ANAPC13|BLM|CUL5|CUL2|NCAPG|CDCA8|UBE2D1|BUB1B|PKMYT1|CENPA|ANAPC10|EGFR|AURKB|ANAPC11|AURKA|CDC20|PPP3CA|CCNB2|CCNB1|FZR1|CDC23|CDC27|AKT1|NEK2|BUB1|TIPIN|UBE2I|ANAPC7|PLK1|CDC6|CDC25C|NDC80|CCNA2|TPX2|CENPE|CCNA1|CDC16|CDK2|TIMELESS|CDK1|ANAPC4|BIRC5|ANAPC5|ANAPC1|MAD2L1|ANAPC2 |
| 87 | 1,10E-22 | 1,36E-20 | 38 | 239 | M phase of mitotic cell cycle | ANAPC13|NCAPG|CDCA8|BUB1B|PKMYT1|ANAPC10|AURKB|ANAPC11|AURKA|CDC20|CCNB2|CCNB1|FZR1|CDC23|CDC27|NEK2|BUB1|TIPIN|UBE2I|ANAPC7|PLK1|CDC6|CDC25C|NDC80|CCNA2|TPX2|CENPE|CCNA1|CDC16|CDK2|TIMELESS|CDK1|ANAPC4|BIRC5|ANAPC5|ANAPC1|MAD2L1|ANAPC2 |
| 75 | 8,32E-21 | 6,55E-19 | 27 | 107 | cell cycle checkpoint | BLM|BUB1B|BRIP1|CCNB1|FZR1|CDC45|CHEK1|RBBP8|NBN|CLSPN|HRAS|BUB1|TIPIN|TGFB1|PLK1|H2AFX|CDC6|CCNA2|CENPE|MSH2|FAM175A|CDK1|BIRC5|ATM|TP53|ATR|MAD2L1 |
| 7346 | 1,07E-11 | 3,66E-10 | 24 | 174 | regulation of mitotic cell cycle | TGFB1|PLK1|BUB1B|CDC6|CDC25C|PKMYT1|ANAPC10|EGFR|CCNA2|TPX2|CENPE|CCNB1|CDC23|CDC16|CDK2|CDK1|BIRC5|ATM|NEK2|NBN|TP53|HRAS|BUB1|MAD2L1 |
| 10564 | 1,19E-08 | 2,53E-07 | 19 | 138 | regulation of cell cycle process | TIPIN|CREBBP|TGFB1|MRE11A|CDC25C|PKMYT1|FOXM1|ANAPC10|TPX2|CENPE|CCNB1|FZR1|CDC23|CDC16|TIMELESS|BIRC5|ATM|NEK2|BUB1 |
| 7093 | 3,57E-07 | 5,87E-06 | 12 | 52 | mitotic cell cycle checkpoint | CCNA2|CENPE|CCNB1|TGFB1|CDK1|BUB1B|ATM|NBN|TP53|HRAS|BUB1|MAD2L1 |
| 51327 | 1,15E-06 | 1,75E-05 | 15 | 102 | M phase of meiotic cell cycle | H2AFX|MRE11A|MLH1|RAD51B|MSH6|CCNA1|RAD50|RAD51C|RAD51|EXO1|CHEK1|CKS2|ATM|NEK2|NBN |
| 51329 | 1,15E-06 | 1,75E-05 | 15 | 102 | interphase of mitotic cell cycle | BLM|CUL5|CUL2|CDC6|CDC25C|ANAPC10|EGFR|PPP3CA|CCNB1|CDC23|CDK2|AKT1|ANAPC4|BIRC5|ANAPC5 |
| 51321 | 1,33E-06 | 1,99E-05 | 15 | 103 | meiotic cell cycle | H2AFX|MRE11A|MLH1|RAD51B|MSH6|CCNA1|RAD50|RAD51C|RAD51|EXO1|CHEK1|CKS2|ATM|NEK2|NBN |
| 7049 | 4,53E-45 | 6,63E-42 | 59 | 794 | cell cycle | CDKN1A|MCM7|NCAPG2|BUB1B|FOXM1|SMC4|SMC2|CKS1B|CHEK1|EP300|AKT1|NEK2|TIPIN|CDC25C|SMC1A|CDC25A|MSH6|CCNA2|CCNA1|DBF4|MSH2|FANCD2|TIMELESS|MCM3|CKS2|BIRC5|MCM6|TP53|MCM2|NCAPG|CDCA8|PKMYT1|NCAPH|RNF2|AURKB|AURKA|CCNB2|CCNB1|CDC45|MAPK1|CLSPN|BUB1|MAPK3|PLK1|FANCA|CDC7|CDC6|NDC80|TPX2|CENPE|CDK2|CCNG1|MDM2|CDK1|ATM|NCAPD2|NCAPD3|ATR|MAD2L1 |
| 22403 | 7,21E-37 | 5,27E-34 | 44 | 435 | cell cycle phase | CDKN1A|NCAPG2|BUB1B|NCAPG|CDCA8|PKMYT1|SMC4|NCAPH|AURKB|SMC2|AURKA|CCNB2|CCNB1|CHEK1|AKT1|NEK2|BUB1|TIPIN|PLK1|FANCA|CDC7|CDC6|CDC25C|SMC1A|CDC25A|NDC80|MSH6|CCNA2|TPX2|CCNA1|CENPE|DBF4|FANCD2|CDK2|CCNG1|TIMELESS|MDM2|CDK1|CKS2|BIRC5|ATM|NCAPD2|NCAPD3|MAD2L1 |
| 22402 | 5,73E-34 | 2,79E-31 | 46 | 582 | cell cycle process | CDKN1A|NCAPG2|BUB1B|NCAPG|CDCA8|PKMYT1|SMC4|NCAPH|AURKB|SMC2|AURKA|CCNB2|CCNB1|CHEK1|AKT1|NEK2|BUB1|TIPIN|PLK1|FANCA|CDC7|CDC6|CDC25C|SMC1A|CDC25A|NDC80|MSH6|CCNA2|TPX2|CCNA1|CENPE|DBF4|MSH2|FANCD2|CDK2|CCNG1|TIMELESS|MDM2|CDK1|CKS2|BIRC5|ATM|NCAPD2|NCAPD3|TP53|MAD2L1 |
| 278 | 4,13E-32 | 1,21E-29 | 39 | 380 | mitotic cell cycle | CDKN1A|NCAPG2|BUB1B|NCAPG|CDCA8|PKMYT1|SMC4|NCAPH|RNF2|AURKB|SMC2|AURKA|CCNB2|CCNB1|AKT1|NEK2|BUB1|TIPIN|PLK1|CDC7|CDC6|CDC25C|SMC1A|CDC25A|NDC80|CCNA2|TPX2|CCNA1|CENPE|DBF4|CDK2|CCNG1|TIMELESS|MDM2|CDK1|BIRC5|NCAPD2|NCAPD3|MAD2L1 |
| 87 | 1,23E-30 | 3,01E-28 | 33 | 239 | M phase of mitotic cell cycle | NCAPG2|BUB1B|NCAPG|CDCA8|PKMYT1|SMC4|NCAPH|AURKB|SMC2|AURKA|CCNB2|CCNB1|NEK2|BUB1|TIPIN|PLK1|CDC6|CDC25C|SMC1A|CDC25A|NDC80|CCNA2|TPX2|CCNA1|CENPE|CDK2|CCNG1|TIMELESS|CDK1|BIRC5|NCAPD2|NCAPD3|MAD2L1 |
| 51726 | 1,02E-26 | 1,36E-24 | 37 | 446 | regulation of cell cycle | CDKN1A|HDAC1|BUB1B|PKMYT1|FOXM1|CKS1B|CCNB1|CDC45|CHEK1|AKT1|NEK2|CLSPN|BUB1|TIPIN|JUN|CREBBP|PLK1|CDC7|CDC6|CDC25C|SMC1A|CDC25A|CCNA2|TPX2|CENPE|MSH2|CDK2|CCNG1|TIMELESS|MDM2|CDK1|CKS2|BIRC5|ATM|TP53|ATR|MAD2L1 |
| 75 | 2,29E-21 | 2,57E-19 | 21 | 107 | cell cycle checkpoint | TIPIN|CDKN1A|PLK1|BUB1B|CDC6|SMC1A|CCNA2|CENPE|CCNB1|CDC45|MSH2|CHEK1|CCNG1|CDK1|BIRC5|ATM|CLSPN|TP53|BUB1|ATR|MAD2L1 |
| 7346 | 2,52E-15 | 2,30E-13 | 20 | 174 | regulation of mitotic cell cycle | CDKN1A|PLK1|BUB1B|CDC6|CDC25C|PKMYT1|CCNA2|TPX2|CENPE|CCNB1|CDK2|CCNG1|MDM2|CDK1|BIRC5|NEK2|ATM|TP53|BUB1|MAD2L1 |
| 10564 | 4,70E-13 | 3,44E-11 | 17 | 138 | regulation of cell cycle process | TIPIN|CREBBP|CDKN1A|CDC7|CDC25C|PKMYT1|FOXM1|SMC1A|TPX2|CENPE|CCNB1|TIMELESS|MDM2|BIRC5|NEK2|ATM|BUB1 |
| 7093 | 4,22E-10 | 1,81E-08 | 11 | 52 | mitotic cell cycle checkpoint | CCNA2|CENPE|CCNB1|CDKN1A|CCNG1|CDK1|BUB1B|ATM|TP53|BUB1|MAD2L1 |
| 51329 | 9,39E-07 | 2,41E-05 | 11 | 102 | interphase of mitotic cell cycle | CCNB1|CDKN1A|DBF4|CDK2|MDM2|AKT1|BIRC5|CDC7|CDC6|CDC25C|CDC25A |
total number of nodes in the gene ontology (GO) annotation;
number of nodes related to a given GO in the network.
Interestingly, in vitro cell cycle analysis has demonstrated that mixtures of benzo[a]pyrene and metals reduce the cell population in the G1 phase and increase cell arrest or accumulation in the G2/M phase (Muthusamy et al., 2018). Once more, the mechanisms suggested include oxidative stress (Fischer et al., 2005), DNA repair alteration (Tran et al., 2002), and suppressor protein TP53 inhibition (Chiang and Tsou, 2009). Particularly, in vitro exposure to a combination of benzo[a]pyrene with As, Cr and Pb increases the ROS-mediated oxidative stress in HepG2 cells (Muthusamy et al., 2018). In this regard, within the same module, proteins regulated by oxidative stress and DNA damage inside the cell were also identified as bottlenecks (AKT1, JUN, and CREBBP) together with benzo[a]pyrene. Other trace species found in our IE analysis, such as SiO2, have also been found to cause DNA damage, oxidative stress, cell cycle arrest at the G2/M checkpoint and apoptosis synergistically in co-exposure with benzo[a]pyrene (Asweto et al., 2017).
Systemic effects of IEs in the cell cycle
This cluster (composed of clusters 2 and 4) addresses a particular area of interest in relation to whether metal ions and IEs interfere with other cellular responses to DNA damage, such as cell cycle progression and control. In clusters 2 and 4, AKT1, JUN and CREBBP and the TP53, CCNB1, CCNA2, CDK6, CDK2, CDK1, ATM, ATR, and CDK7 proteins were found to be bottlenecks together with Cr and S. The biological processes linked to this and its respective proteins are presented in Table 4.
Among all the chemical species present in coal mining environments, IEs, in particular, are capable of causing the most oxidative damage through the generation of ROS (Valko et al., 2006). IEs can enter the body through inhalation or consumption of contaminated meals and then accumulate in the bloodstream (Schweinsberg and Von Karsa, 1990). These elements are deposited in tissues by various mechanisms (Bridges and Zalups, 2005) and may cause DNA damage. In this cluster, together with proteins regulated by oxidative stress and DNA damage, we also found proteins such as cyclins and cyclin-dependent kinases that have been reported to be down-regulated in response to ROS and are implicated in the induction of cell cycle arrest as one of the immediate defense mechanisms against genotoxic damage from oxidative stress (Burch and Heintz, 2005). Particularly, CCNB1 seems to be depleted in response to oxidative stress, causing the regulation of G2/M transit via the Chk1-Cdc2 DNA damage checkpoint pathway (Pyo et al., 2013). Conversely, because altered cell cycle progression and/or cell cycle control and DNA repair inhibition have been observed under low, non-cytotoxic concentrations of metal compounds, some authors have suggested that inhibition could also be a result of the ability of metal ions to modify zinc finger proteins involved in cell cycle control and DNA repair (Hartwig et al., 2002). Interestingly, some authors have reported the suppression of TP53-mediated cell cycle arrest in human breast cancer cells MCF7, as a response to DNA damage caused by Cd(II) (Méplan et al., 1999). Other IEs involved in the modification of zinc finger proteins include Ni and Co (Hartwig and Schwerdtle, 2002). However, no similar implications have been reported for Cr and S.
As discussed in the previous section, Cr(VI) has been demonstrated to be consistently mutagenic in bacterial and mammalian model systems, and its carcinogenic activity is thought to be due to the induction of DNA damage generated by reactive intermediates, eventually resulting in DNA breakage (Vasylkiv et al., 2010). Free radicals from SO2 metabolism, such as SO3 ._, SO4 ._, SO5 ._ may also induce DNA strand breaks (Meng et al., 2005), and recent studies have confirmed that SO2 derivatives (bisulfite and sulfite) cause mitotic delay in cultured human blood lymphocytes in a dose-dependent manner (Uren et al., 2014).
Effect of El Cerrejón and Guacamaya coal exposure on alkaline and FPG high-throughput Comet assay
The results of the alkaline comet assay showed the presence of primary lesions (% DNA tail increase) in V79 cells exposed to ECCS (bituminous coal from El Cerrejón mine) and LGCS (sub-bituminous coal from La Guacamaya mine) for 24 h. Additionally, the results of the modified comet assay show that the cultures exposed to ECCS maintain the same levels of % tail DNA, wheras the cultures exposed to LGCS showed an increase in % tail DNA, when compared to the no-enzyme groups. These results could indicate oxidative damage. Previous studies on coal and its products demonstrated resulting DNA damage and oxidative stress induced by the presence of IE and PAH (Valko et al., 2006; da Silva, 2016). Such results may also be due to compounds identified in the current study, in which we report various levels of inorganic elements (heavy metals) in the bituminous coal from ECCS and sub-bituminous coal from LGCS and high levels of chromium in the coal from LGCS. It was known that some IEs (heavy metals) could generate oxidative damage by generating ROS (Valko et al., 2006). Multiple cellular processes including cell cycle checkpoint activation and DNA repair are typically initiated in response to such DNA damage (Dasika et al., 1999; Lima et al., 2016).
Conclusions
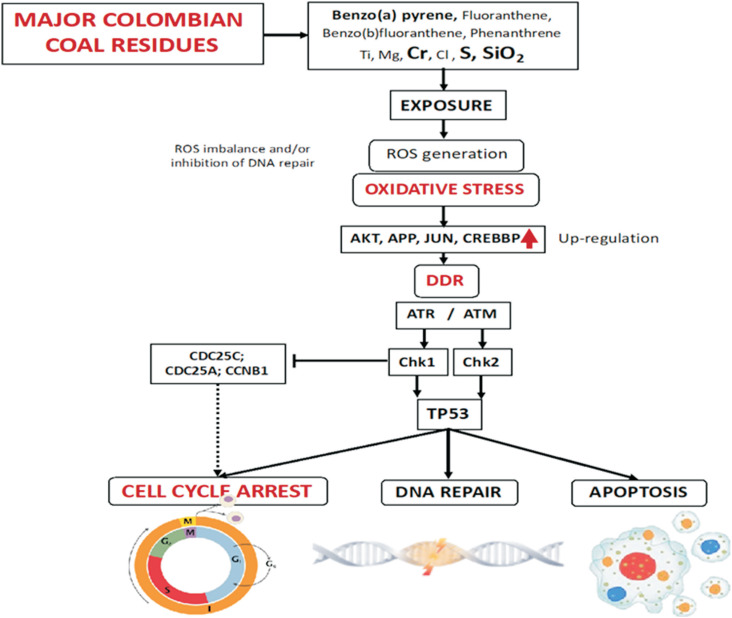
Using a systems chemo-biology approach, we examined how some of the major chemical constituents of coal dust and PM derived from coal mining activities interact with specific biological processes relation to the cell cycle. The main proteins and compounds present in the network were taken into account to construct a molecular model characterizing the effects of major coal residues on the cell cycle (Figure 11). The analysis performed in the present study suggests that coal residue MIOs (SiO2), IEs (Ti, Mg, Cr, Cl and S) and PAHs (benzo[a]pyrene, fluoranthene, benzo[b]fluoranthene and phenanthrene) can generate ROS. The resultant oxidative stress can induce cell cycle arrest through the upregulation of proteins such as AKT, APP, JUN and CREBBP, leading to DNA damage response activation by ATM/ATR and Chk1/Chk2 or by CDC25C or CCNB1 degradation. The model also suggested that protein p53 could be activated by Chk1/Chk2 and induce cell cycle arrest, senescence or apoptosis.
Figure 11. Molecular model illustrating how major coal residues potentially affect cell cycle progression: Exposure to major coal residues, such as benzo[a]pyrene, fluoranthene, benzo[b]fluoranthene, phenanthrene, Ti, Mg, Cr, Cl, S, and SiO2, can generate ROS via several pathways (e.g., Fenton-like reactions). The ROS imbalance and/or inhibition of the DNA repair process can lead to oxidative stress and the upregulation of several proteins associated with the oxidative response (AKT, APP, JUN and CREBBP) which are also involved in the control of the cell cycle. DNA and protein damage caused by the oxidative damage triggers DNA damage response mechanisms (DDR), including the protein kinase cascades ATM-Chk2/ATR-Chk1, which may result in cell cycle arrest. Oxidative stress can also induce cell cycle arrest through the degradation of CDC25C via the Chk1 protein kinase-dependent pathway. ATR phosphorylates and activates Chk1, which in turn, phosphorylates and inhibits Cdc25 phosphatases. Cdc25 inhibition ends up causing cell cycle arrest. Cdc25A phosphorylation by Chk1 triggers its degradation in a ubiquitin/proteasome-dependent manner. Both kinases phosphorylate TP53. In response to DNA damage, the activation of TP53 activates the expression of numerous genes involved in cell cycle arrest, DNA repair, apoptosis, and many other processes.
Acknowledgments
This work was supported by a grant from Conselho Nacional para o Desenvolvimento Científico e Tecnológico-CNPq, Brazil; Universal Grant Number 454288/2014-0.
Supplementary Material:
The following online material is available for this article:
Coal sample collection sites in Colombia.
Main CPI-PPI network generated by the Cytoscape 3.4.0 program.
Major inorganic oxide components in coal ashes (%wt) as identified by XRF.
IEs concentrations in coal samples as revealed by the PIXE assay (mean ± standard deviation).
Polycyclic aromatic hydrocarbon concentrations per sample (mean ± standard deviation) as revealed by HPLC/UV/Vis.
Proteins involved in hub-bottlenecks (HBs) and their function.
Footnotes
Associate Editor: Regina C. Mingroni-Netto
REFERENCES
- Alfaro-Moreno E, Martínez L, García-Cuellar C, Bonner JC, Murray JC, Rosas I, Rosales SP, Osornio-Vargas AR. Biologic effects induced in vitro by PM10 from three different zones of Mexico City. Environ Health Persp. 2002;110:715–720. doi: 10.1289/ehp.02110715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asweto CO, Wu J, Hu H, Feng L, Yang X, Duan J, Sun Z. Combined Effect of Silica Nanoparticles and Benzo[a]pyrene on Cell Cycle Arrest Induction and Apoptosis in Human Umbilical Vein Endothelial Cells. Int J Environ Res Public Health. 2017;14:289–289. doi: 10.3390/ijerph14030289. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Azevedo H, Moreira-Filho CA. Topological robustness analysis of protein interaction networks reveals key targets for overcoming chemotherapy resistance in glioma. Sci Rep-UK. 2015;5:16830–16830. doi: 10.1038/srep16830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2–2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bianchi M, Giacomini E, Crinelli R, Radici L, Carloni E, Magnani M. Dynamic transcription of ubiquitin genes under basal and stressful conditions and new insights into the multiple UBC transcript variants. Gene. 2015;573:100–109. doi: 10.1016/j.gene.2015.07.030. [DOI] [PubMed] [Google Scholar]
- Billet S, Abbas I, Le Goff J, Verdin A, Andre V, Lafargue PE, Hachimi A, Cazier F, Sichel F, Shirali P, Garcon G. Genotoxic potential of Polycyclic Aromatic Hydrocarbons-coated onto airborne Particulate Matter (PM 2.5) in human lung epithelial A549 cells. Cancer Lett. 2008;270:144–155. doi: 10.1016/j.canlet.2008.04.044. [DOI] [PubMed] [Google Scholar]
- Blissett RS, Rowson NA. A review of the multi-component utilisation of coal fly ash. Fuel. 2012;97:1–23. [Google Scholar]
- Borm PJ. Toxicity and occupational health hazards of coal fly ash (CFA). A review of data and comparison to coal mine dust. Ann Occup Hyg. 1997;41:659–676. doi: 10.1016/S0003-4878(97)00026-4. [DOI] [PubMed] [Google Scholar]
- British Petroleum . BP Statistical Review of World Energy. British Petroleum; London: 2014. [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Bio. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Bridges CC, Zalups RK. Molecular and ionic mimicry and the transport of toxic metals. Toxicol Appl Pharmacol. 2005;204:274–308. doi: 10.1016/j.taap.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA, 3rd, Brook JR, Bhatnagar A, Diez-Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, et al. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation. 2010;121:2331–2378. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- Burch PM, Heintz NH. Redox regulation of cell-cycle re-entry: cyclin D1 as a primary target for the mitogenic effects of reactive oxygen and nitrogen species. Antioxid Redox Signal. 2005;7:741–751. doi: 10.1089/ars.2005.7.741. [DOI] [PubMed] [Google Scholar]
- Callen MS, de la Cruz MT, Lopez JM, Navarro MV, Mastral AM. Comparison of receptor models for source apportionment of the PM10 in Zaragoza (Spain) Chemosphere. 2009;76:1120–1129. doi: 10.1016/j.chemosphere.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Campbell JL, Boyd NI, Grassi N, Bonnick P, Maxwell JA. The Guelph PIXE software package IV. Nucl Instrum Meth B. 2010;268:3356–3363. [Google Scholar]
- Cavalcante RM, de Lima DM, Correia LM, Nascimento RF, Silveira ER, Freire GS, Viana RB. Técnicas de extrações e procedimentos de clean-up para a determinação de hidrocarbonetos policílicos aromáticos (HPA) em sedimentos da costa do Ceará. Quím Nova. 2008;31:1371–1377. [Google Scholar]
- Celik M, Donbak L, Unal F, Yuzbasioglu D, Aksoy H, Yilmaz S. Cytogenetic damage in workers from a coal-fired power plant. Mutat Res. 2007;627:158–163. doi: 10.1016/j.mrgentox.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Chaulya S. Assessment and management of air quality for an opencast coal mining area. J Environ Manage. 2004;70:1–14. doi: 10.1016/j.jenvman.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Chiang HC, Tsou TC. Arsenite enhances the benzo[a]pyrene diol epoxide (BPDE)-induced mutagenesis with no marked effect on repair of BPDE-DNA adducts in human lung cells. Toxicol In Vitro. 2009;23:897–905. doi: 10.1016/j.tiv.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Chiba S, Okayasu K, Tsuchiya K, Tamaoka M, Miyazaki Y, Inase N, Sumi Y. The C-jun N-terminal kinase signaling pathway regulates cyclin D1 and cell cycle progression in airway smooth muscle cell proliferation. Int J Clin Exp Med. 2017;10:2252–2262. [Google Scholar]
- Choi WJ, Banerjee J, Falcone T, Bena J, Agarwal A, Sharma RK. Oxidative stress and tumor necrosis factor-alpha-induced alterations in metaphase II mouse oocyte spindle structure. Fertil Steril. 2007;88:1220–1231. doi: 10.1016/j.fertnstert.2007.02.067. [DOI] [PubMed] [Google Scholar]
- Collins AR. Investigating oxidative DNA damage and its repair using the comet assay. Mut Res. 2009;681:24–32. doi: 10.1016/j.mrrev.2007.10.002. [DOI] [PubMed] [Google Scholar]
- D’Angiolella V, Santarpia C, Grieco D. Oxidative stress overrides the spindle checkpoint. Cell Cycle. 2007;6:576–579. doi: 10.4161/cc.6.5.3934. [DOI] [PubMed] [Google Scholar]
- da Silva J. DNA damage induced by occupational and environmental exposure to miscellaneous chemicals. Mutat Res. 2016;770:170–182. doi: 10.1016/j.mrrev.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Dasika GK, Lin SCJ, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncog. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- de Kok TM, Hogervorst JG, Briede JJ, van Herwijnen MH, Maas LM, Moonen EJ, Driece HA, Kleinjans JC. Genotoxicity and physicochemical characteristics of traffic-related ambient particulate matter. Environ Mol Mutagen. 2005;46:71–80. doi: 10.1002/em.20133. [DOI] [PubMed] [Google Scholar]
- Espitia-Pérez L, da Silva J, Espitia-Pérez P, Brango H, Salcedo-Arteaga S, Hoyos-Giraldo LS, de Souza CT, Dias JF, Agudelo-Castañeda D, Valdés Toscano A, et al. Cytogenetic instability in populations with residential proximity to open-pit coal mine in Northern Colombia in relation to PM10 and PM2.5 levels. Ecotox and Environ Saf. 2018;148:453–466. doi: 10.1016/j.ecoenv.2017.10.044. [DOI] [PubMed] [Google Scholar]
- Feltes BC, de Faria Poloni J, Notari DL, Bonatto D. Toxicological Effects of the Different Substances in Tobacco Smoke on Human Embryonic Development by a Systems Chemo-Biology Approach. PLoS ONE. 2013;8:e61743. doi: 10.1371/journal.pone.0061743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Jensen A, Bhatia SK, Dam-Johansen K. Activation energy distribution of thermal annealing of a bituminous coal. Energ Fuel. 2003;17:399–404. [Google Scholar]
- Fischer JM, Robbins SB, Al-Zoughool M, Kannamkumarath SS, Stringer SL, Larson JS, Caruso JA, Talaska G, Stambrook PJ, Stringer JR. Co-mutagenic activity of arsenic and benzo[a]pyrene in mouse skin. Mutat Res. 2005;588:35–46. doi: 10.1016/j.mrgentox.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Gauthier PT, Norwood WP, Prepas EE, Pyle GG. Metal–polycyclic aromatic hydrocarbon mixture toxicity in Hyalella azteca. 2. Metal accumulation and oxidative stress as interactive co-toxic mechanisms. Environ Sci Technol. 2015;49:11780–11788. doi: 10.1021/acs.est.5b03233. [DOI] [PubMed] [Google Scholar]
- Gloscow EJ. New Research on Genomic Instability. Nova Science Publishers; Nova York: 2007. [Google Scholar]
- Gualtieri M, Øvrevik J, Mollerup S, Asare N, Longhin E, Dahlman HJ, Camatini M, Holme JA. Airborne urban particles (Milan winter-PM 2.5) cause mitotic arrest and cell death: effects on DNA, mitochondria, AhR binding and spindle organization. Mutat Res. 2011;713:18–31. doi: 10.1016/j.mrfmmm.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Gutzkow KB, Langleite TM, Meier S, Graupner A, Collins AR, Brunborg G. High-throughput comet assay using 96 minigels. Mutagen. 2013;28:333–340. doi: 10.1093/mutage/get012. [DOI] [PubMed] [Google Scholar]
- Hartwig A, Asmuss M, Ehleben I, Herzer U, Kostelac D, Pelzer A, Schwerdtle T, Bürkle A. Interference by toxic metal ions with DNA repair processes and cell cycle control: molecular mechanisms. Environ Health Perspect 110 Suppl. 2002;5:797–799. doi: 10.1289/ehp.02110s5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A, Schwerdtle T. Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol Lett. 2002;127:47–54. doi: 10.1016/s0378-4274(01)00482-9. [DOI] [PubMed] [Google Scholar]
- Hsiao WL, Mo ZY, Fang M, Shi XM, Wang F. Cytotoxicity of PM(2.5) and PM(2.5—10) ambient air pollutants assessed by the MTT and the Comet assays. Mutat Res. 2000;471:45–55. doi: 10.1016/s1383-5718(00)00116-9. [DOI] [PubMed] [Google Scholar]
- Huang YC, Rappold AG, Graff DW, Ghio AJ, Devlin RB. Synergistic effects of exposure to concentrated ambient fine pollution particles and nitrogen dioxide in humans. Inhal Toxicol. 2012;24:790–797. doi: 10.3109/08958378.2012.718809. [DOI] [PubMed] [Google Scholar]
- Huertas JI, Huertas ME, Izquierdo S, González ED. Air quality impact assessment of multiple open pit coal mines in northern Colombia. J Environ Manage. 2012a;93:121–129. doi: 10.1016/j.jenvman.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Huertas JI, Huertas ME, Solis DA. Characterization of airborne particles in an open pit mining region. Sci Total Environ. 2012b;423:39–46. doi: 10.1016/j.scitotenv.2012.01.065. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson SAE, Campbell JL, Malmqvist KG. Particle-Induced X-Ray Emission Spectrometry (PIXE) Wiley-Blackwell; Hoboken: 1995. [Google Scholar]
- Kim SM, Lee HM, Hwang KA, Choi KC. Benzo(a)pyrene induced cell cycle arrest and apoptosis in human choriocarcinoma cancer cells through reactive oxygen species-induced endoplasmic reticulum-stress pathway. Food Chem Toxicol. 2017;107:339–348. doi: 10.1016/j.fct.2017.06.048. [DOI] [PubMed] [Google Scholar]
- Kiskinis E, Suter W, Hartmann A. High throughput comet assay using 96-well plates. Mutagen. 2002;17:37–43. doi: 10.1093/mutage/17.1.37. [DOI] [PubMed] [Google Scholar]
- Klein JA, Ackerman SL. Oxidative stress, cell cycle, and neurodegeneration. J Clin Invest. 2003;111:785–793. doi: 10.1172/JCI18182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocbach A, Herseth JI, Låg M, Refsnes M, Schwarze PE. Particles from wood smoke and traffic induce differential pro-inflammatory response patterns in co-cultures. Toxicol Appl Pharmacol. 2008;232:317–326. doi: 10.1016/j.taap.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Kothai P, Prathibha P, Saradhi I, Pandit G, Puranik V. Characterization of atmospheric particulate matter using pixe technique. Int J Environ Sci Eng. 2009;3:39–42. [Google Scholar]
- Ku T, Chen M, Li B, Yun Y, Li G, Sang N. Synergistic effects of particulate matter (PM 2.5) and sulfur dioxide (SO 2) on neurodegeneration via the microRNA-mediated regulation of tau phosphorylation. Toxicol Res (Camb) 2017;6:7–16. doi: 10.1039/c6tx00314a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha S, Vikram A, Trivedi PP, Jena GB. Alkaline, Endo III and FPG modified comet assay as biomarkers for the detection of oxidative DNA damage in rats with experimentally induced diabetes. Mutat Res. 2011;726:242–250. doi: 10.1016/j.mrgentox.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Labranche N, El Khattabi C, Dewachter L, Dreyfuss C, Fontaine J, Van De Borne P, Berkenboom G, Pochet S. Vascular oxidative stress induced by diesel exhaust microparticles: synergism with hypertension. J Cardiovasc Pharmacol. 2012;60:530–537. doi: 10.1097/FJC.0b013e318270f1a8. [DOI] [PubMed] [Google Scholar]
- Lee D, Ryu KY. Effect of cellular ubiquitin levels on the regulation of oxidative stress response and proteasome function via Nrf1. Biochem Biophys Res Commun. 2017;485:234–240. doi: 10.1016/j.bbrc.2017.02.105. [DOI] [PubMed] [Google Scholar]
- Leon-Mejia G, Espitia-Perez L, Hoyos-Giraldo LS, da Silva J, Hartmann A, Henriques JA, Quintana M. Assessment of DNA damage in coal open-cast mining workers using the cytokinesis-blocked micronucleus test and the comet assay. Sci Total Environ. 2011;409:686–691. doi: 10.1016/j.scitotenv.2010.10.049. [DOI] [PubMed] [Google Scholar]
- Leon-Mejia G, Silva LF, Civeira MS, Oliveira ML, Machado M, Villela IV, Hartmann A, Premoli S, Correa DS, da Silva J, et al. Cytotoxicity and genotoxicity induced by coal and coal fly ash particles samples in V79 cells. Environ Sci Pollut Res Int. 2016;23:24019–24031. doi: 10.1007/s11356-016-7623-z. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lima M, Bouzid H, Soares DG, Selle F, Morel C, Galmarini CM, Henriques JAP, Larsen AK, Escargueil AE. Dual inhibition of ATR and ATM potentiates the activity of trabectedin and lurbinectedin by perturbing the DNA damage response and homologous recombination repair. Oncotarget. 2016;7:25885–25901. doi: 10.18632/oncotarget.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Niu Z, Van Niekerk D, Xue J, Zheng L. Polycyclic aromatic hydrocarbons (PAHs) from coal combustion: emissions, analysis, and toxicology. Rev Environ Contam Toxicol. 2008;192:1–28. doi: 10.1007/978-0-387-71724-1_1. [DOI] [PubMed] [Google Scholar]
- Longhin E, Holme JA, Gutzkow KB, Arlt VM, Kucab JE, Camatini M, Gualtieri M. Cell cycle alterations induced by urban PM2.5 in bronchial epithelial cells: characterization of the process and possible mechanisms involved. Part Fibre Toxicol. 2013;10:63–63. doi: 10.1186/1743-8977-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee J. Comet assay: rapid processing of multiple samples. Mutat Res Toxicol Environ Mutagen. 2000;466:63–69. doi: 10.1016/s1383-5718(00)00004-8. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Meixner A, Karreth F, Kenner L, Penninger JM, Wagner EF. Jun and JunD-dependent functions in cell proliferation and stress response. Cell Death Differ. 2010;17:1409–1419. doi: 10.1038/cdd.2010.22. [DOI] [PubMed] [Google Scholar]
- Meng Z, Qin G, Zhang B. DNA damage in mice treated with sulfur dioxide by inhalation. Environ Mol Mutagen. 2005;46:150–155. doi: 10.1002/em.20142. [DOI] [PubMed] [Google Scholar]
- Meplan C, Mann K, Hainaut P. Cadmium induces conformational modifications of wild-type p53 and suppresses p53 response to DNA damage in cultured cells. J Biol Chem. 1999;274:31663–31670. doi: 10.1074/jbc.274.44.31663. [DOI] [PubMed] [Google Scholar]
- Mouton-Liger F, Paquet C, Dumurgier J, Bouras C, Pradier L, Gray F, Hugon J. Oxidative stress increases BACE1 protein levels through activation of the PKR-eIF2alpha pathway. Biochim Biophys Acta. 2012;1822:885–896. doi: 10.1016/j.bbadis.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Muche A, Arendt T, Schliebs R. Oxidative stress affects processing of amyloid precursor protein in vascular endothelial cells. PloS One. 2017;12:e0178127. doi: 10.1371/journal.pone.0178127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy S, Peng C, Ng JC. Genotoxicity evaluation of multi-component mixtures of polyaromatic hydrocarbons (PAHs), arsenic, cadmium, and lead using flow cytometry based micronucleus test in HepG2 cells. Mutat Res Genet Toxicol Environ Mutagen. 2018;827:9–18. doi: 10.1016/j.mrgentox.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Nathan C, Cunningham-Bussel A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat Rev Immunol. 2013;13:349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan Y, Dvorachek M, Pelly I, Mimran U. Characterization of coal fly ash from Israel. Fuel. 1999;78:205–213. [Google Scholar]
- Norrish K, Hutton JT. An accurate X-ray spectrographic method for the analysis of a wide range of geological samples. Geochim Cosmochim Ac. 1969;33:431–453. [Google Scholar]
- Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey B, Agrawal M, Singh S. Assessment of air pollution around coal mining area: Emphasizing on spatial distributions, seasonal variations and heavy metals, using cluster and principal component analysis. Atmospheric Pollution Research. 2014;5:79–86. [Google Scholar]
- Parker AL, Kavallaris M, McCarroll JA. Microtubules and their role in cellular stress in cancer. Front Oncol. 2014;4:153–153. doi: 10.3389/fonc.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce AK, Humphrey TC. Integrating stress-response and cell-cycle checkpoint pathways. Trends Cell Biol. 2001;11:426–433. doi: 10.1016/s0962-8924(01)02119-5. [DOI] [PubMed] [Google Scholar]
- Peng C, Muthusamy S, Xia Q, Lal V, Denison MS, Ng JC. Micronucleus formation by single and mixed heavy metals/loids and PAH compounds in HepG2 cells. Mutagen. 2015;30:593–602. doi: 10.1093/mutage/gev021. [DOI] [PubMed] [Google Scholar]
- Poma A, Limongi T, Pisani C, Granato V, Picozzi P. Genotoxicity induced by fine urban air particulate matter in the macrophages cell line RAW 264.7. Toxicol In Vitro. 2006;20:1023–1029. doi: 10.1016/j.tiv.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Pope CA, 3rd, Burnett RT, Thun MJ, Calle EE, Krewski D, Ito K, Thurston GD. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. Jama. 2002;287:1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, 3rd, Burnett RT, Turner MC, Cohen A, Krewski D, Jerrett M, Gapstur SM, Thun MJ. Lung cancer and cardiovascular disease mortality associated with ambient air pollution and cigarette smoke: shape of the exposure-response relationships. Environ Health Perspect. 2011;119:1616–1621. doi: 10.1289/ehp.1103639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, Dockery DW. Health effects of fine particulate air pollution: lines that connect. J Air Waste Manag Assoc. 2006;56:709–742. doi: 10.1080/10473289.2006.10464485. [DOI] [PubMed] [Google Scholar]
- Prada-Fonseca M, Caicedo-Pineda G, Márquez-Godoy M. Comparación del potencial oxidativo de Acidithiobacillus ferrooxidans, en un proceso de biodesulfurización de carbón. Rev Colomb Biotecnol. 2016;18:57–64. [Google Scholar]
- Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, Grinsfelder D, Rothermel BA, Chen R, Garcia JA, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo CW, Choi JH, Oh SM, Choi SY. Oxidative stress-induced cyclin D1 depletion and its role in cell cycle processing. Biochim Biophys Acta. 2013;1830:5316–5325. doi: 10.1016/j.bbagen.2013.07.030. [DOI] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163–163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- Rieder CL. Mitosis in vertebrates: the G2/M and M/A transitions and their associated checkpoints. Chromosome Res. 2011;19:291–306. doi: 10.1007/s10577-010-9178-z. [DOI] [PubMed] [Google Scholar]
- Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010:baq020–baq020. doi: 10.1093/database/baq020. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam B, Devasena T, Islam VI, Prakhya BM. Characterization of coal fly ash nanoparticles and their induced in vitro cellular toxicity and oxidative DNA damage in different cell lines. Indian J Exp Biol. 2015;53:585–593. [PubMed] [Google Scholar]
- Savitsky PA, Finkel T. Redox regulation of Cdc25C. Int J Biol Chem. 2002;277:20535–20540. doi: 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schins RP, Borm PJ. Mechanisms and mediators in coal dust induced toxicity: a review. Ann Occup Hyg. 1999;43:7–33. doi: 10.1016/s0003-4878(98)00069-6. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg F, von Karsa L. Heavy metal concentrations in humans. Comp Biochem Physiol C. 1990;95:117–123. doi: 10.1016/0742-8413(90)90092-n. [DOI] [PubMed] [Google Scholar]
- Scott J. Social Network Analysis. SAGE Publications; Nova York: 2017. [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice R, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Littlejohn D, Gibson MD. Ultrasonication extraction and solid phase extraction clean-up for determination of US EPA 16 priority pollutant polycyclic aromatic hydrocarbons in soils by reversed-phase liquid chromatography with ultraviolet absorption detection. Anal Chim Acta. 1998;364:1–11. [Google Scholar]
- Tice R, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single-cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Env Mol Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Tran HP, Prakash AS, Barnard R, Chiswell B, Ng JC. Arsenic inhibits the repair of DNA damage induced by benzo(a)pyrene. Toxicol Lett. 2002;133:59–67. doi: 10.1016/s0378-4274(02)00088-7. [DOI] [PubMed] [Google Scholar]
- Tucker JD, Ong TM. Induction of sister chromatid exchanges by coal dust and tobacco snuff extracts in human peripheral lymphocytes. Environ Mutagen. 1985;7:313–324. doi: 10.1002/em.2860070308. [DOI] [PubMed] [Google Scholar]
- Uren N, Yuksel S, Onal Y. Genotoxic effects of sulfur dioxide in human lymphocytes. Toxicol Ind Health. 2014;30:311–315. doi: 10.1177/0748233712457441. [DOI] [PubMed] [Google Scholar]
- Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Vasylkiv OY, Kubrak OI, Storey KB, Lushchak VI. Cytotoxicity of chromium ions may be connected with induction of oxidative stress. Chemosphere. 2010;80:1044–1049. doi: 10.1016/j.chemosphere.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Wang X, McCullough KD, Franke TF, Holbrook NJ. Epidermal growth factor receptor-dependent Akt activation by oxidative stress enhances cell survival. J Biol Chem. 2000;275:14624–14631. doi: 10.1074/jbc.275.19.14624. [DOI] [PubMed] [Google Scholar]
- Wise SS, Wise JP. Aneuploidy as an early mechanistic event in metal carcinogenesis. Biochem Soc Trans. 2010;38:1650–1654. doi: 10.1042/BST0381650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coal sample collection sites in Colombia.
Main CPI-PPI network generated by the Cytoscape 3.4.0 program.
Major inorganic oxide components in coal ashes (%wt) as identified by XRF.
IEs concentrations in coal samples as revealed by the PIXE assay (mean ± standard deviation).
Polycyclic aromatic hydrocarbon concentrations per sample (mean ± standard deviation) as revealed by HPLC/UV/Vis.
Proteins involved in hub-bottlenecks (HBs) and their function.