Abstract
Purpose
Protein kinase C α (PRKCA) is involved in multiple functions and has been implicated in heart failure risks and treatment outcomes. This study aims to identify regulatory variants affecting PRKCA expression in human heart, and evaluate attributable risk of heart disease.
Methods
mRNA expression quantitative trait loci (eQTLs) were extracted from the Genotype and Tissue Expression Project (GTEx). Allelic mRNA ratios were measured in 51 human heart tissues to identify cis-acting regulatory variants. Potential regulatory regions were tested with luciferase reporter gene assays and further evaluated in GTEx and genome-wide association studies.
Results
Located in a region with robust enhancer activity in luciferase reporter assays, rs9909004 (T > C, minor allele frequency =0.47) resides in a haplotype displaying strong eQTLs for PRKCA in heart (p = 1.2 × 10−23). The minor C allele is associated with both decreased PRKCA mRNA expression and decreased risk of phenotypes characteristic of heart failure in GWAS analyses (QT interval p = 3.0 × 10−14). While rs9909004 is the likely regulatory variant, other variants in high linkage disequilibrium cannot be excluded. Distinct regulatory variants appear to affect expression in other tissues.
Conclusions
The haplotype carrying rs9909004 influences PRKCA expression in the heart and is associated with traits linked to heart failure, potentially affecting therapy of heart failure.
Keywords: association, gene expression, genetic variant, heart failure, polymorphism, proteinkinaseCα(PRKCA)
INTRODUCTION
Protein kinase C α (PRKCA [MIM 176960]) is a member of the protein kinase C family of serine-threonine-specific protein kinases (1), which is involved in regulation of cell proliferation, apoptosis, differentiation, migration, cardiac hypertrophy, and inflammation (2, 3). Expressed in all human tissues, PRKCA spans over a 500 kb genomic region. Genome-wide association studies (GWAS) reveal that PRKCA variants are associated with cardiovascular disease traits (4, 5), schizophrenia (6), neural basis of episodic remembering (7), body mass index and asthma (8), multiple sclerosis (9), regulating bone architecture and osteoblast activity (10), diabetes (11), and pain (12). Further studies have reported associations with hypertension and response to hydrochlorothiazide (5, 13). As the majority of significant SNPs reside outside the protein coding regions, it is likely that multiple variants regulate PRKCA expression in a tissue-specific fashion.
For many PRKCA SNPs implicated in diseases and phenotypic traits, the functional relevance remains uncertain, most being surrogate markers rather than causative variants. Among significant GWAS hits, two reported functional variants modulating PRKCA gene expression and alternative splicing have been identified in a multiple sclerosis cohort study (14), including a GCC microsatellite VNTR located in the 5′ regulatory region of PRKCA. The 9–12 repeats are associated with increased PRKCA expression and protection against multiple sclerosis, and 3–6 repeats with lower expression and increased risk. Another in/del polymorphism (rs35476409/rs61762387, with 1 or 2 GGTG tandem repeats) is also associated with multiple sclerosis, with the 2-repeat allele (exon 3* + 5nt) as the risk factor.
It remains unclear how many distinct causative variants exist in PRKCA. Since PRKCA plays an important physiological role, resolving the mechanistic basis of PRKCA-association with diseases and traits is critical. Regulatory variants can be identified as expression quantitative trait loci (eQTLs) in the Genotype-Tissue Expression (GTEx) project, providing mRNA expression profiles in multiple human tissues along with genome-wide SNP data (15).
The most significant eQTLs in GTEx are found in heart tissue, suggesting cardiac-selective effects. Indeed, PRKCA is a key regulator of cardiac contractility and Ca2+ homeostasis in myocytes in mice, with PRKCA overexpression reducing cardiac contractility and PRKCA deficiency preventing heart failure (16). As PRKCA is a key downstream signaling molecule of the adrenergic beta-1 receptor, regulatory PRKCA variants active in the heart have the potential to affect treatment outcomes with beta-blockers in heart failure. The responsible genetic variant remains to be ascertained, since the best scoring eQTLs are part of a long haplotype with multiple SNPs in high linkage disequilibrium (LD) with each other. Moreover, we wanted to determine whether this eQTL haplotype is selective for heart tissues or active in other tissues as well.
Beyond eQTL analysis, regulatory variants can be detected by measuring allelic mRNA expression in the target tissue, using marker SNPs located in the transcribed region. In carriers heterozygous for a marker SNP, a significant deviation of the allelic mRNA expression ratios from that of allelic gDNA ratios, also referred to as allelic expression imbalance (AEI), reveals the presence of regulatory variants (17). In previous studies, we have used AEI to identify a series of regulatory variants in pharmacologically and medically relevant genes (18–23).
In this study, to identify the regulatory genetic PRKCA variants in heart, we determined allelic PRKCA mRNA ratios in human left ventricular heart tissues, and we compared the results with AEI ratios from kidney, liver, and blood. This approach led us to identify a regulatory variant/haplotype modulating PRKCA expression in human heart, followed by testing its association with cardiovascular traits in available clinical studies.
MATERIALS AND METHODS
Databases
Genome-wide significant SNPs assigned to PRKCA were collected from the GWAS Catalog (https://www.ebi.ac.uk/gwas/) and GRASP (http://grasp.nhlbi.nih.gov/Search.aspx). PRKCA eQTLs were acquired from the Genotype and Tissue Expression Project (GTEx Analysis Release V6p, dbGaP Accession phs000424.v6.p1). Chromatin marks and transcription factors (TF) binding site predictions were obtained from HaploReg v4.1. Linkage disequilibrium (LD) data were obtained from the 1000 Genome Project.
Sample Preparation
Fifty-one left ventricle heart tissues, twenty kidney tissues and twenty liver tissues were obtained from the Cooperative Human Network Midwestern and Western Division. Heart tissues were obtained from patients undergoing heart transplantation. Of these, 41 are Caucasians, and 10 are African Americans. Kidney and liver tissues were from autopsies and biopsies, respectively. EB-transformed B lymphocytes were obtained from the Coriell Institute Cell Repositories. Ohio State University Institutional Review Board (IRB) approved the tissue studies. DNA and RNA were isolated from tissues as described (17, 24, 25). cDNA was synthesized with SuperScript III (Invitrogen, Carlsbad, CA, USA) using gene-specific primers and oligo-dT.
Microsatellite and SNP Genotyping
The 15 × GCC microsatellite VNTR (chr17:64298451–64298495 in hg19 coordinate) and rs35476409/rs61762387 (1 or 2 GGTG tandem repeats) (14) were amplified using JumpStart ™ Taq DNA Polymerase (Sigma-Aldrich, St. Louis, MO, USA) using a 5′-labeled forward fluorescent primer (6-FAM), and the labeled products separated on an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). Data were analyzed with GeneMapper software v3.0 (Applied Biosystems, Foster City, CA, USA). SNPs rs2227857 G > A, rs2228945 A > G, rs9910355 C > A, rs9303504 G > C, rs12601850 G > A, rs17633437 A > G, rs9909004 C > T and rs7210446 G > A were genotyped with a SNaPshot primer extension assay (Life Technologies, Carlsbad, CA, USA), using extension primers designed to anneal immediately adjacent to the SNP (26). Genomic DNA and cDNA containing the target SNP were amplified by PCR and the products subjected to the SNaPshot procedure (Applied Biosystems, Foster City, CA, USA) (17). Then the extended primers were separated on an ABI3730 DNA analyzer and genotypes determined for each SNP. Primer sequences and PCR conditions are listed in Supplemental Table S1.
Allelic mRNA Expression Analysis
The SNaPshot primer extension assay described above was also used to measure allelic PRKCA mRNA expression ratios (17). rs2227857 G > A located in exon 8 and rs2228945 A > G in exon 16 were used as marker SNPs. Peak area ratios were measured for both genomic DNA and mRNA (cDNA). For heterozygous carriers of a marker SNP, the genomic DNA allele ratios are expected to be 1; in the samples analyzed we did not find a case of gDNA copy number variants detectable by significant ratio deviations from unity (mean of all gDNA allelic ratios). Therefore, cDNA ratios were normalized to the average of genomic DNA ratios. Each cDNA sample was analyzed in triplicate, and an allelic mRNA ratio of <0.77 or >1.3 was considered as significant AEI (approximate 3x SD from 1.0). Under the assumption of a single causative variant, all samples heterozygous for the causative SNP must show AEI, while homozygous samples should not.
Cell Culture and Luciferase Reporter Gene Assays
Cells were cultured at 37°C in humidified air at 5% CO2 in DMEM/F12 (HEK-293) containing 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Luciferase assay vectors were prepared using pGL3-Basic plasmid (Promega, Madison, WI, USA). The Infusion Cloning System (Clontech, Mountain View, CA, USA) was used to insert the fragment into the cloning site of pGL3-Basic. For each candidate SNP, the region around the SNP of ~500 bps was amplified by PCR. The fragments containing either the major or the minor allele were inserted at the KpnI or XhoI multiple cloning sites of pGL3-Basic (see Supplemental Table S2 for primers and cloning sites). Then the constructs were transformed into the competent cells (Clontech, Mountain View, CA, USA) and plated on LB agar with carbenicillin. Individual clones were screened and plasmids isolated using the Zyppy MiniPrep Kit (Zymo Research, Irvine, CA, USA). The sequence of plasmids with correct insertion was confirmed by Sanger sequencing. Positive clones were digested with KpnI or XhoI and the insert was again subcloned into pGL3-Basic vectors. The clones with either forward or reverse insert orientation were identified using restriction enzyme digestion and gel electrophoresis, and plasmids isolated using the ZymoPURE MidiPrep Kit (Zymo Research, Irvine, CA, USA). HEK293 cells were incubated in 24-well plates for 24h and each well of cells were transfected with 1 μg target construct and 0.2 μg renilla luciferase control vector using lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) in 50 μL OptiMem Media (Thermo Fisher Scientific, Waltham, MA, USA). Cell culture media containing ampicillin replace the OptiMem Media 6 h after transfection. The Dual-Glo Luciferase Assay System (Promega, Madison,WI, USA) was used to measure the firefly and renilla luciferase signals on a Packard Fusion Plate Reader (PerkinElmer Life and Analytical Sciences, Shelton, CT, USA). The pGL3-Basic empty vectors without any insert were also transfected into cells as controls. All transfections were carried out in triplicate. For each well, luciferase activity was normalized to renilla fluorescence, and the ratio was further normalized to the ratio of pGL3-Basic empty vectors.
Statistical Analyses
SNP & Variation Suite (Golden Helix, Bozeman, MT, USA) was used to calculate D’ and R2 values between SNPs using the Expectation Maximization (EM) algorithm. Chi-squared test and statistical analyses were performed using the GraphPad Prism software package (version 3, GraphPad Software, Inc., La Jolla, CA) and R version 3.2.3.
RESULTS
GWAS SNPs Assigned to PRKCA
The PRKCA SNPs with genome-wide significance (p < 5 × 10−8) are listed in Table I, plus candidate SNPs with p values < 10−6. The low linkage disequilibrium (LD) between significant single nucleotide polymorphisms (SNPs) in these various traits (Table I) indicates that distinct functional variants affect different traits. In two study cohorts of individuals of European ancestry, two SNPs in complete LD, rs9892651 and rs9912468, are significantly associated with QT interval and QRS interval respectively (4, 27), while rs9912468 is also associated with QT interval but with a higher p value (3.7 × 10−6) (Table I, #13 study (27)). In addition, PRKCA SNPs are associated with other diseases or traits such as transmission distortion, LDL cholesterol level and height in women (Table I). Taking rs9892651 as reference SNP, minor allele frequency and LD with the other SNPs vary with phenotypes independent of GWAS p values (Table I), suggesting distinct causative genetic variants for different traits.
Table I.
Top Scoring Genome-wide Significant and Candidate SNPs Assigned to PRKCA, Associated with Traits/Diseases in GWAS
| # | Disease/Trait | rs# | MAF | R2/D’a | Context | P value | Population | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | QT interval | rs9892651 | 0.47 | 1/1 | intron 2 | 3.0 × 10 −14 | European | (4) |
| 2 | QRS interval | rs9912468 | 0.47 | 1/1 | intron 2 | 1.1 × 10 −08 | European | (27) |
| 3 | Transmission distortion | rs41461845 | 0.004 | 0/0 | intron 2 | 7.6 × 10 −36 | European | (28) |
| 4 | Maternal transmission distortion | rs41461845 | 0.004 | 0/0 | intron 2 | 1.4 × 10 −32 | European | (28) |
| 5 | Height (females) | rs3889237 | 0.04 | 0/0 | intron 14 | 3.0 × 10 −08 | African American | (29) |
| 6 | BP Responses to hydrochlorothiazide | rs16960228 | 0.06 | 0.008/0.356 | intron 16 | 3.3 × 10 −08 | European | (5) |
| 7 | Blood pressure measurement | rs11867410 | 0.03 | 0.006/0.41 | upstream | 3.0 × 10 −07 | Chinese-Han | (13) |
| 8 | β2 Glycoprotein I levels | rs10048158 | 0.46 | 0.68/0.85 | upstream | 1.0 × 10 −06 | European | (30) |
| 9 | Longevity in long-living individuals | rs6504441 | 0.34 | 0.001/0.04 | intron 3 | 1.1 × 10 −06 | Italian | (31) |
| 10 | Heschl’s gyrus morphology | rs4791051 | 0.48 | 0.008/0.116 | intron 3 | 2.0 × 10 −06 | European | (32) |
| 11 | Coronary artery calcification | rs1165l708 | 0.34 | 0.002/0.057 | intron 3 | 3.0 × 10 −06 | African-American | (33) |
| 12 | Left ventricular wall thickness | rs9896894 | 0.34 | 0.027/0.227 | intron 3 | 3.2 × 10 −06 | European | (34) |
| 13 | QT interval | rs9912468 | 0.47 | 1/1 | intron 2 | 3.7 × 10 −06 | European | (27) |
| 14 | Post-traumatic stress disorder | rs7207499 | 0.28 | 0.011/0.277 | intron 2 | 4.6 × 10 −06 | African-American | (35) |
| 15 | Height (females) | rs4254365 | 0.04 | 0/0 | intron 16 | 4.7 × 10 −06 | African-American | (29) |
MAF minor alele frequency
The LD (R2/D’) between rs9892651 and each other SNP was calculated from the 1000genome database
eQTLs of PRKCA in Human Heart Left Ventricle and Thyroid
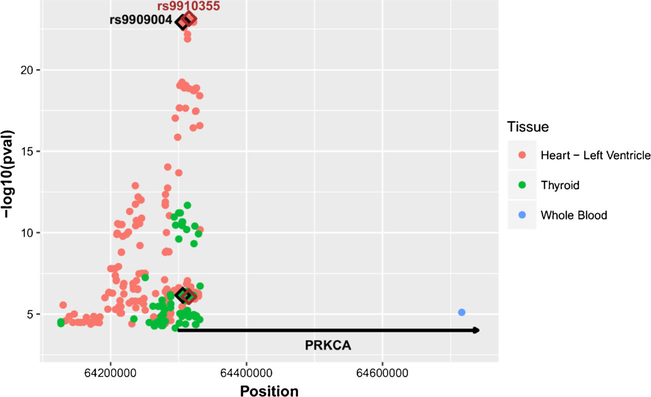
The GTEx Portal lists 183 eQTLs in human heart left ventricle, 88 eQTLs in human thyroid and 1 eQTL in whole blood with p values < 10−5. Shown in Fig. 1, PRKCA heart eQTLs are distinct from thyroid eQTLs and whole blood eQTLs, indicating the presence of selective eQTLs in heart. Several SNPs have similarly high eQTL scores (low p value) in heart tissues, the top scoring SNP (9 × 10−23) being rs9910355 (Fig. 1) located in intron 2 in a haplotype block with high LD between a number of eQTL SNPs (Table II). The remaining eQTLs with higher p values have progressively lesser LD with the top scoring SNPs, indicating that the genetic influence on mRNA expression in the heart is mainly determined by a single regulatory element. Any of the top scoring SNPs in high LD (in Caucasians) could be the causative SNPs, while we cannot exclude the possibility that more than one regulatory SNP resides on the same haplotype. In Africans, fewer SNPs have near perfect LD, indicating the possibility to rule out some variants with use of AEI analysis, a more precise indicator of a regulatory variant in each individual subject.
Fig. 1.
Manhattan plot of eQTLs (p = <10−5) for PRKCA mRNA expression in human heart left ventricle, thyroid and whole blood. The Y-axis shows −log10 (p value) and X-axis shows genomic position (chr17 with hg19 coordinates). The red dots represent the eQTLs in heart, the green dots represent the eQTLs in thyroid, and the blue dot represents the eQTL in whole blood. eQTLs of interest are highlighted with a diamond: rs9909004 (black), and rs9910355 (brown). The position of PRKCA is represented by the horizontal line.
Table II.
Top Scoring eQTLs in Human Heart Left Ventricle
| SNPs | Location | Minor Allele | Major Allele | MAFa | P value# |
|---|---|---|---|---|---|
| rs9910355 | intron 2 | A | C | 0.47 | 7.2 × 10−24 |
| rs9303504 | intron 2 | G | C | 0.47 | 8.2 × 10−24 |
| rs9909004 | intron 2 | C | T | 0.47 | 1.2 × 10−23 |
| rs12601850 | intron 2 | A | G | 0.42 | 8.5 × 10−20 |
| rs17633437 | intron 1 | G | A | 0.42 | 9.2 × 10−20 |
| rs7210446 | intron 2 | G | A | 0.47 | 1.2 × 10−23 |
| rs9910577 | intron 2 | T | C | 0.47 | 7.7 × 10−24 |
| rs9890911 | intron 2 | T | C | 0.47 | 8.2 × 10−24 |
| rs9912468 | intron 2 | G | C | 0.47 | 8.2 × 10−24 |
| rs11867573 | intron 2 | A | G | 0.47 | 8.2 × 10−24 |
| rs8071250 | intron 2 | C | T | 0.47 | 8.3 × 10−24 |
| rs989265l | intron 2 | C | T | 0.47 | 1.2 × 10−23 |
| rs11079650 | intron 2 | A | C | 0.47 | 1.2 × 10−23 |
| rs4577128 | intron 2 | T | C | 0.47 | 1.3 × 10−23 |
| rs9893075 | intron 2 | C | T | 0.47 | 1.3 × 10−23 |
| rs11658550 | intron 2 | C | T | 0.47 | 1.3 × 10−23 |
| rs11658630 | intron 2 | G | A | 0.47 | 1.3 × 10−23 |
| rs129406l 0 | intron 2 | A | G | 0.47 | 1.3 × 10−23 |
| rs4335805 | intron 2 | T | G | 0.47 | 1.7 × 10−23 |
| rs4328478 | intron 2 | C | T | 0.47 | 2.7 × 10−23 |
| rs7406054 | intron 2 | G | A | 0.47 | 6.6 × 10−23 |
| rs7406066 | intron 2 | G | A | 0.47 | 1.3 × 10−22 |
| rs35183571 | intron 2 | C | T | 0.42 | 5.7 × 10−20 |
| rs12944131 | intron 2 | A | C | 0.42 | 8.5 × 10−20 |
Minor allele frequency data from CEU population in the 1000genome database
P value is from GTEx Analysis Release V6p
Allelic PRKCA mRNA Ratios and AEI Analysis
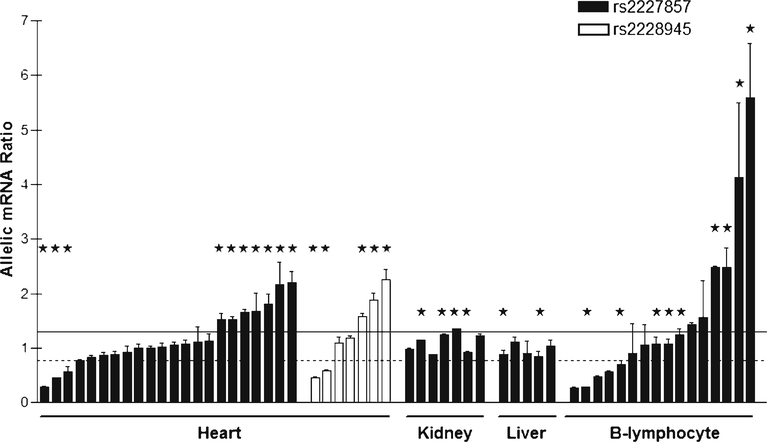
To identify causative SNPs, we measured AEI in human heart tissues, followed by genotyping to scan for causative variants (18–23). Two common SNPs located in the PRKCA coding region (rs2227857 in exon 8 and rs2228945 in exon 16) were used as marker SNPs to measure allelic mRNA ratios in human heart, kidney, liver, and B-lymphocytes. The two marker SNPs are not in LD with each other, with heterozygous carriers not overlapping between the two markers. With marker rs2227857, 10 of 22 heterozygous heart samples displayed AEI, with AEI ratios in the range of 1.5–3.5 fold (Fig. 2). With rs2228945, 5 of 7 heart samples showed AEI with ratios of 1.6–2.3 (Fig. 2). Significant AEI ratios (major/minor allele) were both below and above 1. These results indicate the presence of one or more frequent regulatory variants, not in LD with the marker SNPs, of substantial effect on mRNA expression. We also performed allelic PRKCA mRNA analyses in human kidney, liver, and EB-virus transformed B-lymphocytes. Absence of allelic ratios deviating significantly from unity in kidney and liver (Fig. 2) is consistent with absence of significant PRKCA eQTLs in kidney and liver in the GTEx portal database. B-lymphocytes display frequent AEI (Fig. 2), consistent with the presence of regulatory variants in blood (14). These results indicate that regulatory variants of PRKCA are tissue specific.
Fig. 2.
Allelic mRNA expression ratios of PRKCA in heart and other tissues. Ratios represent major over minor alleles for two marker SNPs located in exon 8 (rs2227857) and exon 16 (rs2228945). Tissues heterozygous for rs9909004 are indicated with *. The horizontal dotted line represents the lower (0.77) and the solid line the upper threshold (1.3) for declaring the presence of AEI (defined as ~3× SD of mean allelic mRNA ratios). Each bar is the mean allelic ratio (± SD) per tissue.
Genotype Association With AEI in Heart
We applied the AEI ratios in heart to scan the PRKCA gene locus for causative variants. Among the 29 samples with AEI data, 21 samples are from Caucasians and 8 from African Americans. To identify likely causative SNPs that account for AEI status in the heart, we used the top eQTL SNP (rs9910355) to search for SNPs that are in high LD with rs9910355 and also show histone marks indicative of possible regulatory function (Supplemental Table S3). Using HaploReg v4.1, six SNPs were selected (Table III) and genotyped in all heart samples (n = 51). In addition, two proposed regulatory variant active in blood previously reported (14), the 15 × GCC microsatellite and rs35476409/rs61762387 (1 or 2 GGTG tandem repeats), were also genotyped in all samples (Table III). The minor allele frequency (MAF) and p values reflecting association with incidents of AEI are listed in Table III, while LD between the variants is summarized in Table IV. Of the eight SNPs genotyped, only rs9909004 and rs7210446 completely match the AEI status in all samples (Fig. 2), and both have the lowest p value for association with AEI (5.4 × 10−7, Table III), the minor allele associating with reduced PRKCA expression. SNP rs12601850 was ruled out because it was homozygous in a tissue showing AEI (1.9-fold ratio). Similarly, rs9910355 and rs9303504, in high LD (R2 = 0.98, D’ = 0.99) with rs9909004, are less likely causative candidates because one sample with AEI (1.6-fold) is homozygous for both variants. We repeated the AEI analysis for this sample three times, confirming a finding of AEI (data not shown). SNPs rs17633437, the 15 × GCC microsatellite, and rs35476409/rs61762387 do not account for AEI status independently, nor in combination with other SNPs (Table III), indicating these two variants, reported to be regulatory in blood (14), are not active in heart tissue. In contrast, none of the SNPs in Table III can account for the AEI in B-lymphocytes (Fig. 2), indicating tissue selective regulatory variant in heart. Since genome-wide genotyping data are available for the B-lymphocytes used in this study, we obtained all SNPs within PRKCA locus and 50kb up- and down-stream of PRKCA and tested association with AEI status. This approach failed to identify any SNPs that can fully account for AEI in B-lymphocytes, suggesting the presence of multiple regulatory variants or other regulatory mechanisms in B-lymphocytes.
Table III.
Association of Genotyped Variants with AEI Status (Measured with Two Marker SNPs) in Heart Tissues
| SNP | Location | Minor allele | MAF | Chi-squared-test p |
|---|---|---|---|---|
| rs9910355 | intron 2 | A | 0.49 | 3.2 × 10 −6 |
| rs9303504 | intron 2 | G | 0.49 | 3.2 × 10 −6 |
| rs9909004 | intron 2 | C | 0.49 | 5.4 × 10 −7 |
| rs12601850 | intron 2 | A | 0.46 | 6.5 × 10 −5 |
| rs17633437 | intron 1 | G | 0.41 | 0.016 |
| rs7210446 | intron 2 | G | 0.49 | 5.4 × 10 −7 |
| rs35476409/rs61762387 | intron 3 | 1 repeat (GGTG) | 0.33 | 0.82 |
| 15 × GCC | promoter | (GCC)7 | 0.21 | 0.093 |
Table IV.
LD Values (R2/D’) for Genotyped SNPs
| rs17633437 | rs12601850 | rs9909004 | rs7210446 | rs9910355 | rs9303504 | rs2227857 | rs2228945 | |
|---|---|---|---|---|---|---|---|---|
| rs17633437 | 1/1 | . | . | . | . | . | . | . |
| rs12601850 | 1/1 | 1/1 | . | . | . | . | . | . |
| rs9909004 | 0.89/0.99 | 0.89/0.99 | 1/1 | . | . | . | . | . |
| rs7210446 | 0.89/0.99 | 0.89/0.99 | 1/1 | 1/1 | . | . | . | . |
| rs9910355 | 0.90/0.99 | 0.90/0.99 | 0.98/0.99 | 0.98/0.99 | 1/1 | . | . | . |
| rs9303504 | 0.90/1.0 | 0.90/1.0 | 0.98/0.99 | 0.98/0.99 | 1/1 | 1/1 | . | . |
| rs2227857 | 0.01/0.12 | 0.01/0.12 | 0.009/0.12 | 0.009/0.12 | 0.01/0.12 | 0.009/0.11 | 1/1 | . |
| rs2228945 | 0.004/0.24 | 0.004/0.24 | 0.004/0.23 | 0.004/0.23 | 0.006/0.28 | 0.006/0.28 | 0.0002/0.06 | 1/1 |
Chromatin State and Histone Marks for Candidate SNPs in Heart and Other Tissues
To determine whether the top scoring heart eQTL SNPs have a potential functional role, we searched for nearby chromatin marks in HaploReg v4.1. In heart left ventricle, rs9910355, rs9303504, rs7210446, and rs9909004 locate in a genomic region marked with H3K4me1, enriched in enhancers (Supplemental Table S3). Most of the other SNPs do not locate in genomic regions with the core 15 state, H3K4me1 and H3K29ac. Combined with the AEI results, rs9909004 and rs7210446 appear to be the most likely regulatory variants in the heart. Contributions from other variants in the same haplotypes cannot be excluded.
Reporter Gene Assays for Enhancer Activity Using Luciferase Expression
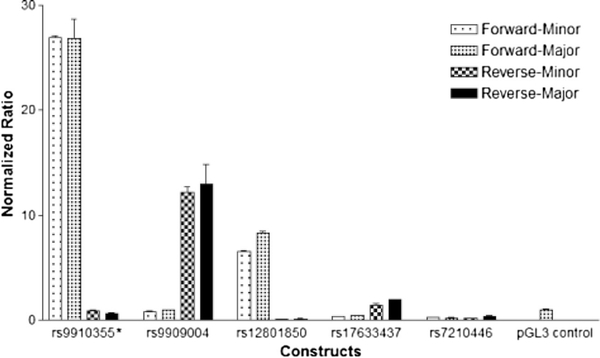
To determine whether the regions surrounding six highly linked PRKCA heart eQTL SNPs, we used a luciferase reporter gene assay in HEK-293 cells. The plasmid constructs containing either forward or reverse inserts for each the major allele or minor allele were transfected into the HEK-293 cells and luciferase activity measured. Shown in Fig. 3, constructs containing the reverse insert carrying rs9909004 increased expression 12–13 fold over pGL3-Basic vector, whereas the rs7210446 region insert did not when inserted in either direction. The inserts of the rs9910355 and rs12801850 region also yielded robust increases, but only in the forward orientation, whereas the other three SNP regions caused little or no increases. The results demonstrate that enhancer orientation is important to consider in reporter gene assays. However, none of the minor alleles resulted in detectable differences in luciferase activity. These results indicate that rs9909004 and two additional eQTL SNPs reside in putative enhancer regions whereas no allele effects are detectable in HEK293 cells (derived from the kidney where AEI was not detectable). Attempts failed to transfect the reporter gene plasmid into IMR90 cells that show similar a chromatin state as reported for heart tissue (Supplemental Table S3), possibly expressing needed transcription factors, and no further attempts were made in consideration of the tissue specific nature of the regulatory genetic effect.
Fig. 3.
Relative luciferase activity of twenty PRKCA luciferase reporter gene constructs in HEK293 cells. Luciferase activity was normalized to renilla luciferase activity in each sample, and adjusted to the activity of the respective pGL3-Basic vectors [Construct (Firefly/Renilla)/pGL3 (Firefly/Renilla)]. *rs9910355 and rs9303504 are in close proximity so that the region inserted into the plasmid contained both SNPs. The vector constructs with inserts in the forward or reverse orientation are listed.
DISCUSSION
PRKCA is expressed throughout the body and has been associated with multiple traits and diseases, including the regulation of the cardiac contractility. In patients with dilated cardiomyopathy, PRKCA is overexpressed (36). Mice lacking PRKCA have enhanced cardiac ventricular performance whereas transgenic overexpression of PRKCA impairs cardiac performance (16, 37), effects mimicked by pharmacological PRKCA inhibition in PRKCA wild-type mice (37). These results strongly support a deleterious effect of elevated levels of PRKCA in the heart and predict a protective effect by genetic variants that reduce PRKCA heart expression. Strong PRKCA eQTLs in human heart tissues – with distinct patterns or undetectable in other tissues – support the presence of heart-specific regulatory variants responsible for association with cardiac phenotypes. LD patterns among these eQTL SNPs indicate that a single variant or haplotype accounts for cardiac PRKCA eQTLs. AEI analysis and bioinformatics characterization have identified rs9909004 as a likely causative variant, consistent with its clinical association with deleterious heart contractility measures (Table I), the minor C allele being associated with reduced PRKCA mRNA expression and therefore conveying a protective role. Presence of strong eQTLs in thyroid tissues, one reported eQTL in blood of moderate strength, and AEI in blood lymphocytes in our study demonstrate the influence of regulatory PRKCA variants in other tissues, but the eQTL and AEI patterns point to distinct PRKCA variants, associated with distinct clinical phenotypes.
AEI status can be established with high confidence in a single individual. Only rs9909004 and rs7210446, in complete LD (R2 = 1, D’ = 1) with each other, fully account for AEI status, implicating both as top candidates for a causative variant. However, only rs9909004 resides in a region with strong enhancer activity in a reporter gene assay (Fig. 3). It is noteworthy that the insertion direction of tested genomic regions had substantial impact on enhancer activity in the reporter gene assays; testing both insertion directions revealed strong enhancer activity of the rs9909004 region, located in intron 2, only in the reverse direction. Matching an additional criterion of a regulatory variant, the rs9909004 C allele is predicted to decrease occupancy of certain transcription factors (TFs) compared to the T allele (p = 0.005) (38), while TF binding changes are less pronounced for rs7210446, rs12601850, and rs17633437. Further studies are needed to identify the relevant transcription factor(s). Taken together, rs9909004 is the likely functional variant regulating the PRKCA gene expression in the heart.
Two GWAS report significant associations of QT interval and QRS interval with rs9892651 and rs9912468 in a European population (Table I) (4, 27). The rs9892651 T and rs9912468 C alleles are associated with prolonged QT-intervals, increasing the risk of ventricular conduction defects. Both variants are in complete LD (R2 = 1, D’ = 1) with rs9909004 in Europeans. However, neither variant is in proximity of any chromatin marks in heart tissue, leaving rs9909004 as the strongest candidate regulatory variant. Increased expression of PRKCA in heart associated with the rs9909004 T allele can reduce phosphorylation of phospholamban (PLB), the sarcoplasmic reticulum Ca2+ ATPase-2 (SERCA-2) pump inhibitory protein (16). The dephosphorylated PLB interacts with SERCA-2 and inhibits its activity, slowing transport of Ca2+ back to the sarcoplasmic reticulum of myocytes, and thereby prolonging the QT interval. Thus, heart failure patients with different PRKCA expression levels may require different treatment strategies. A main therapeutic strategy, beta-blocker therapy has been shown to vary with genetic variants in the beta-1 adrenergic receptor (ADRB1); therefore, we propose that rs9909004 can also modulate beta-blocker therapy, which may be more effective when PRKCA activity is high. Clinical trials should be done to test the combined effect of these genetic variants on the outcome of betablocker therapy (39, 40).
According to GTEx Analysis Release V6p, the population consists of 84.3% White, 13.7% African-American and 1% Asian. With rs9909004 showing highly significant eQTL p values across these population, the causative variant regulating PRKCA expression in the heart is present across population. All heart tissue donors in our study had been diagnosed with heart failure, whereas only a small portion of GTEx autopsy tissue donors had been diagnosed with heart failure, indicating the regulatory variant effect is not contingent on disease status.
While the reporter gene assays performed in HEK293 cells derived from human embryonic kidney cells did reveal strong enhancer activity of the rs9909004 region, it failed to reveal a distinct effects of the rs9909004 alleles on mRNA expression. Although reporter gene analysis is considered the gold standard for promoter and enhancer activity, it fails to recapilutate genetic influence observed in vivo, possibly caused by temporal and tissue specific expression of TFs and TF networks, or changes in chromatin states/epigenetic modifications that are not reflected in reporter gene assays as we have reported for VKORC1 (20). Serving as an eQTL in the heart, rs9909004 is not an eQTLs in kidneys nor in other tissues available in GTEx (over 70 tissues). Therefore, failure to detect allelic effects is likely caused by the tissue specific nature of the cardiac eQTLs, requiring tissue specific TFs or TF networks for the regulatory genetic effect to be detectable. In addition, we cannot exclude the possibility that other variants in high LD with rs9909004 are regulatory or contribute to the effect of rs9909004 when combined. As several SNPs are in high LD with rs9909004 in most populations, these SNPs can serve as markers for clinical associations, specifically for cardiac phenotypes.
In summary, our results support the conclusion that rs9909004 affects PRKCA expression in the heart, consistent with a significant association with heart disease via affecting cardiac contractility. We cannot exclude the possibility that other SNPs in high LD, for example rs7210446, could be responsible or contribute to the regulatory effect of rs9909004. On the basis of these results and the strong rational for PRKCA’s role in cardiac functions, rs990904 should be assessed as a potential biomarker predictive of PRKCA activity in the heart and risk of heart disease and personalized treatment of heart diseases.
Supplementary Material
ACKNOWLEDGMENTS AND DISCLOSURES
We thank Rosanna Asselta for giving us the primer sequences and PCR condition of rs35476409/rs61762387 and 15 × GCC microsatellite. This study was supported by National Institutes of Health Pharmacogenetics Research Network grant U01 GM092655 (WS), U01-GM074492 (JAJ), the National Institute of Health grant R01HL126969 (DW), and partially by a grant from the National Center for Research Resources (UL1RR025755). We also acknowledge support from the Ohio Supercomputer Center (grant #PAS0885). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Center for Research Resources. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health. Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\SAIC-Frederick, Inc. (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by a supplement to University of Miami grants DA006227 & DA033684 and to contract N01MH000028. Statistical Methods development grants were made to the University of Geneva (MH090941 & MH101814), the University of Chicago (MH090951, MH090937, MH101820, MH101825), the University of North Carolina - Chapel Hill (MH090936 & MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University St Louis (MH101810), and the University of Pennsylvania (MH101822). The data used for the analyses described in this manuscript were obtained from GTEx Analysis Release V6p, dbGaP Accession phs000424.v6.p1. Disclosures None.
ABBREVIATIONS
- ADRB1
beta-1 adrenergic receptor
- AEI
Allelic expression imbalance
- eQTL
Expression quantitative trait loci
- GTEx
Genotype-Tissue Expression
- GWAS
Genome-wide association studies
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- PLB
Phospholamban
- PRKCA
Protein kinase C α subunit
- SERCA-2
Sarcoplasmic reticulum Ca2+ ATPase-2
- SNPs
Single nucleotide polymorphisms
- TF
Transcription factors
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11095-017-2102-x) contains supplementary material, which is available to authorized users.
REFERENCES
- 1.Coussens L, Parker PJ, Rhee L, Yang-Feng TL, Chen E, Waterfield MD, et al. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science. 1986;233(4766):859–66. [DOI] [PubMed] [Google Scholar]
- 2.Dempsey EC, Newton AC, Mochly-Rosen D, Fields AP, Reyland ME, Insel PA, et al. Protein kinase C isozymes and the regulation of diverse cell responses. Am J Physiol Lung Cell Mol Physiol. 2000;279(3):L429–38. [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Usuwanthim K, Munawara U, Quach A, Gorgani NN, Abbott CA, et al. Protein kinase cα regulates the expression of complement receptor Ig in human monocyte-derived macrophages. J Immunol. 2015;194(6):2855–61. [DOI] [PubMed] [Google Scholar]
- 4.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46(8):826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner ST, Boerwinkle E, O’Connell JR, Bailey KR, Gong Y, Chapman AB, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62(2):391–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll LS, Williams NM, Moskvina V, Russell E, Norton N, Williams HJ, et al. Evidence for rare and common genetic risk variants for schizophrenia at protein kinase C, alpha. Mol Psychiatry. 2010;15(11):1101–11. [DOI] [PubMed] [Google Scholar]
- 7.MacLeod CA, Donaldson DI. PRKCA polymorphism changes the neural basis of episodic remembering in healthy individuals. PLoS One. 2014;9(5), e98018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy A, Tantisira KG, Soto-Quirós ME, Avila L, Klanderman BJ, Lake S, et al. PRKCA: a positional candidate gene for body mass index and asthma. Am J Hum Genet. 2009;85(1):87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saarela J, Kallio SP, Chen D, Montpetit A, Jokiaho A, Choi E, et al. PRKCA and multiple sclerosis: association in two independent populations. PLoS Genet. 2006;2(3), e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galea GL, Meakin LB, Williams CM, Hulin-Curtis SL, Lanyon LE, Poole AW, et al. Protein kinase Cα (PKCα) regulates bone architecture and osteoblast activity. J Biol Chem. 2014;289(37): 25509–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quack I, Woznowski M, Potthoff SA, Palmer R, Königshausen E, Sivritas S, et al. PKC alpha mediates beta-arrestin2-dependent nephrin endocytosis in hyperglycemia. J Biol Chem. 2011;286(15):12959–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopach O, Viatchenko-Karpinski V, Atianjoh FE, Belan P, Tao YX, Voitenko N. PKCα is required for inflammation-induced trafficking of extrasynaptic AMPA receptors in tonically firing lamina II dorsal horn neurons during the maintenance of persistent inflammatory pain. J Pain. 2013;14(2):182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He J, Kelly TN, Zhao Q, Li H, Huang J, Wang L, et al. Genomewide association study identifies 8 novel loci associated with blood pressure responses to interventions in Han Chinese. Circ Cardiovasc Genet. 2013;6(6):598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paraboschi EM, Rimoldi V, Soldà G, Tabaglio T, Dall’Osso C, Saba E, et al. Functional variations modulating PRKCA expression and alternative splicing predispose to multiple sclerosis. Hum Mol Genet. 2014;23(25):6746–61. [DOI] [PubMed] [Google Scholar]
- 15.Consortium GTEx. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braz JC, Gregory K, Pathak A, Zhao W, Sahin B, Klevitsky R, et al. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat Med. 2004;10(3):248–54. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, Johnson AD, Papp AC, Kroetz DL, Sadée W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C > T affects mRNA stability. Pharmacogenet Genomics. 2005;15(10):693–704. [PubMed] [Google Scholar]
- 18.Johnson AD, Gong Y, Wang D, Langaee TY, Shin J, Cooper-Dehoff RM, et al. Promoter polymorphisms in ACE (angiotensin I-converting enzyme) associated with clinical outcomes in hypertension. Clin Pharmacol Ther. 2009;85(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith RM, Alachkar H, Papp AC, Wang D, Mash DC, Wang JC, et al. Nicotinic α5 receptor subunit mRNA expression is associated with distant 5′ upstream polymorphisms. Eur J Hum Genet. 2011;19(1):76–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112(4):1013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11(4):274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinsonneault JK, Han DD, Burdick KE, Kataki M, Bertolino A, Malhotra AK, et al. Dopamine transporter gene variant affecting expression in human brain is associated with bipolar disorder. Neuropsychopharmacology. 2011;36(8):1644–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrie ES, Weinshenker D, Verma A, Pendergrass SA, Lange LA, Ritchie MD, et al. Regulatory polymorphisms in human DBH affect peripheral gene expression and sympathetic activity. Circ Res. 2014;115(12):1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Papp AC, Binkley PF, Johnson JA, Sadée W. Highly variable mRNA expression and splicing of L-type voltage-dependent calcium channel alpha subunit 1C in human heart tissues. Pharmacogenet Genomics. 2006;16(10):735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Li CJ, Zhang YJ, Zheng L, Jiang HX, Si-Tu B. Simultaneous detection of CYP3A5 and MDR1 polymorphisms based on the SNaPshot assay. Clin Biochem. 2011;44(5–6):418–22. [DOI] [PubMed] [Google Scholar]
- 27.Sotoodehnia N, Isaacs A, de Bakker PI, Dörr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42(12):1068–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer WK, Arbeithuber B, Ober C, Ebner T, Tiemann-Boege I, Hudson RR, et al. Evaluating the evidence for transmission distortion in human pedigrees. Genetics. 2012;191(1):215–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carty CL, Johnson NA, Hutter CM, Reiner AP, Peters U, Tang H, et al. Genome-wide association study of body height in African Americans: the Women’s Health Initiative SNP Health Association Resource (SHARe). Hum Mol Genet. 2012;21(3): 711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Athanasiadis G, Sabater-Lleal M, Buil A, Souto JC, Borrell M, Lathrop M, et al. Genetic determinants of plasma β2 -glycoprotein I levels: a genome-wide association study in extended pedigrees from Spain. J Thromb Haemost. 2013;11(3):521–8. [DOI] [PubMed] [Google Scholar]
- 31.Malovini A, Illario M, Iaccarino G, Villa F, Ferrario A, Roncarati R, et al. Association study on long-living individuals from Southern Italy identifies rs10491334 in the CAMKIV gene that regulates survival proteins. Rejuvenation Res. 2011;14(3):283–91. [DOI] [PubMed] [Google Scholar]
- 32.Cai DC, Fonteijn H, Guadalupe T, Zwiers M, Wittfeld K, Teumer A, et al. A genome-wide search for quantitative trait loci affecting the cortical surface area and thickness of Heschl’s gyrus. Genes Brain Behav. 2014;13(7):675–85. [DOI] [PubMed] [Google Scholar]
- 33.Wojczynski MK, Li M, Bielak LF, Kerr KF, Reiner AP, Wong ND, et al. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med Genet. 2013;14:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302(2):168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74(9):656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowling N, Walsh RA, Song G, Estridge T, Sandusky GE, Fouts RL, et al. Increased protein kinase C activity and expression of Ca2 + -sensitive isoformsin the failing human heart. Circulation. 1999;99(3):384–91. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q, Chen X, Macdonnell SM, Kranias EG, Lorenz JN, Leitges M, et al. Protein kinase Cα, but not PKCβ or PKCγ, regulates contractility and heart failure susceptibility: implications for ruboxistaurin as a novel therapeutic approach. Circ Res. 2009;105(2):194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurano MT, Haugen E, Sandstrom R, Vierstra J, Shafer A, Kaul R, et al. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat Genet. 2015;47(12):1393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rau T, Düngen HD, Edelmann F, Waagstein F, Lainščak M, Dimković S, et al. Impact of the β1-adrenoceptor Arg389Gly polymorphism on heart-rate responses to bisoprolol and carvedilol in heart-failure patients. Clin Pharmacol Ther. 2012;92(1):21–8. [DOI] [PubMed] [Google Scholar]
- 40.Baudhuin LM, Miller WL, Train L, Bryant S, Hartman KA, Phelps M, et al. Relation of ADRB1, CYP2D6, and UGT1A1 polymorphisms with dose of, and response to, carvedilol or metoprolol therapy in patients with chronic heart failure. Am J Cardiol. 2010;106(3):402–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.