Abstract
Metabolic syndrome is a complex disorder that is comprised of several other complex disorders, including obesity, hypertension, dyslipidemia, and diabetes. There are several rat models that encompass component features of MetS. Some models are inbred strains selected for one or more traits underlying MetS; others are population models with genetic risk for MetS traits, are induced by environmental stressors such as diet, are spontaneous monogenic mutant models, or are congenic strains derived from a combination of these models. Together they can be studied to identify the genetic and physiological underpinnings of MetS to identify candidate genes or mechanisms for study in human MetS subjects.
1. Introduction
The Metabolic Syndrome, sometimes referred to as Syndrome X or MetS, is a coincident occurrence of disorders that substantially increase the risk for mortality from heart disease, stroke, and renal failure. Syndrome X was described by Reaven in 1988 as the cooccurrence of insulin resistance, hyperglycemia, hyperinsulinemia, hyperlipidemia and hypertension [1]. While the specific clinical criteria have been debated over the past three decades, the internationally recognized clinical diagnosis for Metabolic Syndrome is three or more of the following: elevated waist circumference (a measure of abdominal obesity); elevated triglycerides; reduced high-density lipoprotein (HDL) cholesterol; elevated blood pressure; elevated fasting glucose [2]. Globally, the prevalence of MetS is estimated to be 25% [3] and its prevalence in children and young adults is estimated at 6.5% [4]. While these numbers vary by population and by criteria used to define MetS, the dramatic increase over the past two to three decades is a major health concern.
While MetS is a syndrome in name, it is actually a complex disorder that is comprised of several other complex disorders, including obesity, hypertension, and diabetes. MetS is caused by both heritable and environmental influences [5-7]. Each defining feature of MetS also has a genetic component, with strong influences by environmental stimuli [6,5,8]. To effectively treat hypertension in MetS, it is important to determine what genes and mechanisms underlie MetS and its individual components, a goal that is substantially benefitted by physiological and genetic studies in animal models of MetS and/or its defining traits.
There are several rat models that encompass component features of MetS. Some models are inbred strains selected for one or more traits underlying MetS; others are population models with genetic risk for MetS traits, are induced by environmental stressors such as diet, are spontaneous monogenic mutant models, or are congenic strains derived from a combination of these models. Together they can be studied to identify the genetic and physiological underpinnings of MetS to identify candidate genes or mechanisms for study in human MetS subjects.
Some rat strains have several MetS traits, allowing for identification of putative pleiotropic genes while some have only a small subset, avoiding the confounding effects of other MetS traits. A comprehensive list of rat strains was annotated as obesity/metabolic syndrome models in the Rat Genome Database Obesity Portal [9]. Table 1 lists examples of different strains with two or more MetS traits, along with their defining features of MetS. Because waist circumference is not readily translatable to rats, obesity was included as a defining feature. Because of species differences in lipid profiles between human and rats [10-12], increased total cholesterol was included with low HDL as a defining feature of MetS for this review. Below are some examples of the different rat models of MetS.
Table 1:
Selected Rat Models of Metabolic Syndrome. TC = Total cholesterol; HDL = high density lipoprotein cholesterol; LDL = low density lipoprotein cholesterol. Strain details are provided in the text.
| Traits Defining Metabolic Syndrome | |||||||
|---|---|---|---|---|---|---|---|
| Strain | Disease Type | Strain Type | Hypertension | Obesity/ ↑Adiposity |
Hyperglycemia/ Glucose intolerance/ Insulin resistance |
Elevated triglycerides |
Reduced HDL/ Increased TC |
| LH | Spontaneous | Inbred selection | √ | √ | √ | √ | √ |
| WOKW | Spontaneous | Inbred selection | √ | √ | √ | √ | √ |
| OLETF | Spontaneous | Inbred selection | √ | √ | √ | √ | √ |
| SHR | Spontaneous | Inbred selection | √ | √ | √ | √ | |
| HTG | Spontaneous | Inbred selection | √ | √ | √ | ||
| PD/Cub | Spontaneous | Inbred selection | √ | √ | √ | ||
| GK | Spontaneous | Inbred selection | √ | √ | |||
| SDT | Spontaneous | Inbred selection | √ | √ | |||
| CRDH | Diet induced | Inbred | √ | √ | |||
| Wistar | Diet induced | Outbred | √ | √ | √ | √ | √ |
| SD | Diet induced | Outbred | √ | √ | √ | √ | √ |
| Obese Koletsky | Monogenic | Inbred | √ | √ | √ | √ | √ |
| Obese Zucker | Monogenic | Inbred | √ | √ | √ | √ | |
| ZDF | Monogenic | Inbred | √ | √ | |||
2. Inbred selection models of MetS
Selective breeding followed by inbreeding has been used to generate numerous polygenic models of human behaviors or conditions in rats ranging from learning and alcohol preference to hypertension and diabetes [13-16]. Selective breeding involves identifying and breeding animals with a specific trait over several generations, thereby ‘fixing’ the genetic factors contributing to the trait. Subsequent inbreeding while maintaining the trait results in a genetically predisposed inbred strain that can be used in physiological and genetic studies. There are several inbred selection rat models that have all or a subset of characteristic features of MetS.
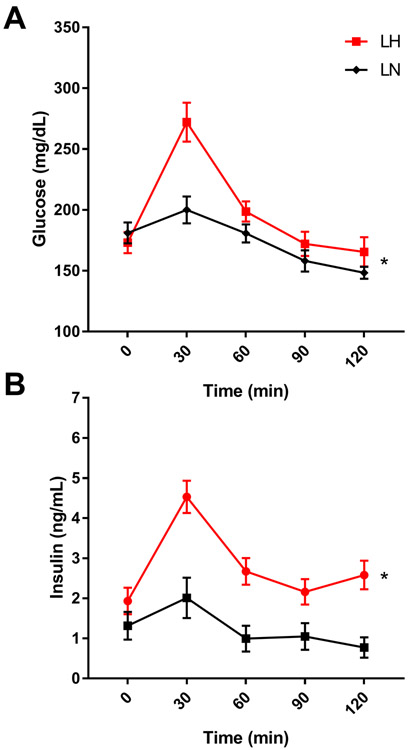
The Lyon Hypertensive (LH/Mav; RGDID:10021) rat model was selectively bred for spontaneous hypertension from a small founder population of six SD rats [17]. Selection for normotensive (LN/Mav; RGDID:10022) and hypotensive (LL/Mav; RGDID:1581645) rats were done in parallel from the same founders. After as few as three generations of selection, LH rats were indeed spontaneously hypertensive, LN rats were normotensive, and LL were hypotensive. However, upon further inbreeding, the LL rats lost the hypotensive phenotype but remained normotensive, providing a second control strain for the LH strain [18,19]. LH rats are both spontaneously hypertensive and salt sensitive [20]. Further study determined that, in addition to hypertension, LH rats have increased body weight, serum lipids including triglycerides and cholesterol, and altered insulin/glucose ratios [21,22] compared to the LN control strain. While initial studies indicate the LH rat is not diabetic or insulin resistant [23], we demonstrate male LH/MRrrcAek (RGDID:10755352) rats have altered glucose tolerance (Fig 1A) and are hyperinsulinemic (Fig 1B) compared to the control male LN (LN/MRrrcAek; RGDID:10755354) rats when fed a low fat diet (D12450H, Research Diets, Inc.). Overall, the LH strain is a comprehensive MetS model for both physiological and genetic studies. Comprehensive genetic studies in the LH rat determined that MetS is polygenic, with both independent and pleiotropic genetic effects on MetS traits, and combined fine-mapping and systems genetic studies have identified multiple loci and candidate genes [24,22,25-28].
Figure 1:
Male LH/MavRrrcAek rats are glucose intolerant and hyperinsulimemic compared to LN/MavRrrcAek rats. Rats were fed a 10% fat diet (Research Diets D12450H) from wean. At 12 weeks of age Intraperitoneal Glucose Tolerance Tests (IPGTT) were performed on animals fasted 6 hours (+/− 0.5 hr) then given 2g glucose/kg body wt IP. Blood glucose and insulin levels were measured at 0, 30, 60, 90 and 120 minutes following glucose challenge. * P < 0.05, two-way ANOVA.
The WOKW (Wistar Ottawa Karlsburg W (RT1u); RGDID:67935) and OLETF (Otsuka Long-Evans Tokushima Fatty; RGDID:61014) rat strains are also spontaneous genetic models of MetS with obesity, hypertension, hypertriglyceridemia, abnormal plasma cholesterol levels, glucose intolerance, and hyperinsulinemia [29-34]. Through genetic mapping and fine-mapping in congenic strains, several QTL and candidate genes for MetS traits have been identified in WOKW [35-37] and OLETF rats [34,38-40,33]. Both strains have spontaneous risk for all major features of MetS, although the progression can be associated with age and sex. Interestingly, the OLETF was found to have a spontaneous mutation in the CCK1 receptor that contributes to its hyperphagia and obesity [41].
The Spontaneously Hypertensive Rat (SHR; RGDID:61000) is arguably the most comprehensively studied model of genetic hypertension with over 19,000 publications in PubMed (query SHR AND rat). Like the LH rat, SHR rats are also genetically predisposed to several features of MetS including hypertension and dyslipidemia [42-47] although they are not generally considered obese. The breeding history of the SHR is complex, and over 40 substrains of SHR are annotated in the Rat Genome Database. The stroke-prone SHRSP strains are well recognized SHR substrains derived from SHR progenitors with spontaneous stroke and then inbred. There are over 20 substrains of SHRSP. Gene identification using traditional positional cloning, comprehensive systems genetics, and gene targeting have been an active area of study in the SHR rat [48,45,49]. Dysregulation of several genes involved in MetS related traits have been identified in the SHR rat through genetic and genomic approaches (e.g. CD36, Ogn, Srebpf, Folr1), and validated in transgenic and gene targeted models [50,51,49,52].
There are several additional inbred selection strains having a subset of MetS traits. For instance, the Prague hereditary hypertriglyceridemic (HTG; RGD ID:1302795) strain is a non-obese model with high plasma triglycerides, hypertension and insulin resistance [53-55]. This model was selectively bred for high plasma triglycerides on a high-sucrose diet, suggesting possible pleiotropic genetic effects on regulation of lipids, blood pressure, and glucose, although genetic mapping studies failed to identify a single locus involved in all phenotypes [56,53]. The Polydactylous rat strain (PD/Cub; RGDID:728161) has a spontaneous mutation causing polydactyly-luxate syndrome, but also shows features of MetS, including increased adiposity, insulin resistance and hypertriglyceridemia [57]. A comprehensive genetic study that included dietary and pharmacologic stressors in an intercross between PD/Cub and BN/Cub identified several loci for MetS related traits, some with independent effects and others showing epistatic interactions [58].
Two non-obese, spontaneously diabetic rat strains are the Spontaneously Diabetic Torii (SDT/Jcl; RGDID: 631219) and Goto-Kakizaki (GK/Jcl; RGDID: 13506737) strains. The SDT strain is a Type 2 diabetes model that also shows hypertriglyceridemia severe diabetic retinopathy, nephropathy, and neuropathy [59,60]. Several loci for hyperglycemia and glucose intolerance and diabetes have been identified in SDT rats [61,62], and genetically induced islet inflammation is thought to play a role in developing diabetes [61]. Glucose intolerance and Type 2 diabetes in the GK rat have been extensively studied, and further studies revealed the GK strain also shows increased basal blood pressure compared to Wistar controls and salt sensitivity [63]. Several genetic loci have been identified that regulate not only the GK phenotypes but also its transcriptome and metabolome [64,65]. Furthermore, the specific genetic contributions evolve over the lifetime of the animal and the disease progression [66,67]. There are over 10 different substrains of GK rat annotated in the Rat Genome Database or reviewed in Portha and colleagues [66]; therefore, one must be cognizant of the substrain(s) being studied as there are substantive phenotype differences between them [66].
3. Diet Induced Models of MetS
Most of the models described above show spontaneous features of MetS. However, induced models also exist whereby MetS traits are only evident after an environmental stressor such as diet. For example, when the inbred Cohen Rosenthal Diabetic Hypertensive rats (CDRH; RGDID:68019) are fed a high-sucrose, copper-poor diet, they become hypertensive and diabetic but not obese [68]. This strain was derived by crossing Cohen Diabetic Rats (CDR; RGDID:1357178) with SHR rats, then selectively breeding for both high blood glucose and high blood pressure for several generations while being fed the copper-poor, high sucrose diet. This unique model has been used to study the effects of various anti-hypertensives on metabolic parameters in diabetic hypertension [69-71].
The Dahl Salt Sensitive rat (SS; RGDID:69369) is likely the most studied salt-induced hypertension model, with nearly 3000 publication (query Dahl AND rat). While SS rats are a well-established inbred model of salt sensitive hypertension, studies have also found SS rats display insulin resistance and dyslipidemia on normal and high salt diets ([72,73]; further reviewed in [74]). Furthermore, SS rats can develop hypertension, insulin resistance, and dyslipidemia when fed diets high in fat, fructose and/or sucrose [75-78]. Finally, dietary protein levels and even protein source influences the phenotypes of SS rats. A low-protein diet or replacing casein with wheat gluten as the dietary protein source lowered both blood pressure and body weight, potentially through modulation of the immune system [79,80].
Some outbred strains also show diet induced MetS. The two most prevalent models are Sprague Dawley (SD; RGDID:70508) and Wistar (WI; RGDID:13508588) rats fed high energy (fat and/or sugar) diets, sometimes called DIO rats [81-83]. The SD DIO rats were developed after the observation that SD rat populations show a bimodal body weight distribution when fed a high energy diet [84]. Levin and colleagues performed several generations of selective breeding in the obesity prone SD rats to develop the DIO and in the obesity resistant SD to develop the DR rats, with both lines being fed a diet high in fat and sucrose [81]. While they went through selection, there is not clear evidence that isogenic inbred strains were developed. Nevertheless, in addition to increased body weight and adiposity, most features of MetS have been observed in SD derived DIO rats including glucose intolerance, dyslipidemia and hypertension [85,81]. Another outbred strain identified to have a bimodal weight distribution on high fat diet is the Wistar strain. Studies of diet induced obesity using this model continue to be performed in commercially available outbred Wistar rats, stratified into diet induced or diet resistant categories after exposure to the obesogenic moderately high fat diet with or without high sugar [86-88]. Again, all features defining MetS have been observed in the Wistar DIO rats (reviewed in [89]). Of note, the Wistar DIO rats show evident sex differences [86].
Outbred rats such as SD and Wistar are often studied to recapitulate the heterogeneity of human populations. However, we and others have shown that phenotypes in outbred strains vary widely by source due to differing breeding histories of the strains; therefore, care must be taken when generalizing results from strains such as SD from differing vendors as they do not represent the same population sampling [90,91].
4. Population Models of MetS
Another unique source of outbred rats was first developed by Hansen and colleagues at the National Institutes of Health (NIH), called N/NIH heterogeneous stock (HS) (RGDID:728185; [92]). HS rats are descendants of founders from eight inbred strains - ACI/N, BN/SsN, BUF/N, F344/N, M520/N, MR/N, WKY/N and WN/N – that represent high genetic diversity and span the phylogenetic tree of the laboratory rat [93]. Crossbreeding between these eight founder strains was performed to minimize inbreeding and thus maximizing genetic diversity in the heterogeneous stock, making them a strong population model which is also amenable to genetic studies. While no intentional phenotypic selection was performed in the development of N/NIH, they have been shown to segregate many traits [94-96]. An advantage of the HS rats is that they segregate many traits without intentional selection. They are thus a strong model for human population studies of exposures, for example to drugs and chemicals. The conditions most studied in HS rats include those related to psychiatric disorders and substance abuse, but also include symptoms of MetS. Solberg Woods and colleagues have performed extensive phenotyping and genotyping of HS rats, followed by genome wide association studies to identify loci and candidate genes for adiposity and glucose homeostasis in HS rats [97,98].
In the mid-1990s Drs. Koch and Britton selectively bred the N/NIH rats for low and high capacity exercise endurance, respectively [99]. Different from many selection models, the Low Capacity Runners (LCR; RGDID:2314396) and High Capacity Runners (HCR; RGDID:2314397) rats have been intentionally maintained as outbred stock. These rats have been extensively studied at the physiological level and studies determined the LCR rats are a robust MetS model [100,101]. Genetic studies suggest high heritability of several metabolic traits and the potential of QTL mapping in crosses between LCR and HCR [102].
5. Genetic Models of MetS
Models of human disease often arise through spontaneous or targeted mutation of a gene that is sufficient to cause a phenotype. The most prominent monogenic rat models of MetS arose through two independent spontaneous mutations in the gene encoding the leptin receptor (Lepr). The Obese Zucker rat (ZUC-Leprfa; RGDID:629464) was described in the early 1960s [103] as a spontaneously obese rat in an outbred colony that was later found to be due to a homozygous missense mutation (Gln269Pro) in Lepr [104], called fatty or fa that resulted in constitutive receptor activity [105]. Several other rat strains were derived from the Obese Zucker rat that included other MetS traits. The Zucker Diabetic Fatty Rat (ZDF-Leprfa/Drt; RGDID:12859287) originated from the original Zucker colony by selecting breeding rats that were both obese and diabetic [106]. Inbred MetS strains with the fa mutation include the obese diabetic KZF (KZ-Leprfa/Tky; RGDID:1302693) and models where the fa allele was bred onto hypertensive strains such as the SS (SS.ZUC-Leprfa−/−/Slc; RGDID:13432148) and SHRSP (SHRSP.ZUC-(D5Rat4-D5Rat36)/IzmDmcr; RGDID: 2300018) and show all defining MetS features [107,108].
The cp or corpulent allele of Lepr was identified in the Koletsky or Obese SHR rat strain (SHROB/KolGmiCrl-Leprcp; RGDID: 2311049) which arose as a spontaneous Lepr mutation in a cross between a hypertensive SHR female and a SD male [109]. These rats were obese, hypertensive and dyslipidemic but not diabetic [110]. The cp mutation was found to be a nonsense mutation, causing truncation at amino acid position 763 [111]. The cp mutation has also been bred into the Diabetes Resistant BioBreeding (BBDR/Rhw; RGDID: 10003) rat to develop an obese, hypertensive, dyslipidemic, diabetic model (BBDR.LA-(D5Rat98-D5Rat233)/Rhw; RGDID: 6893530), where the phenotype was found to be sex-dependent [112].
While the leptin receptor mutant rats were the only monogenic model of MetS for decades, transgenic rat models were developed to study MetS traits, for example transgenic expression of Cd36 and Srebf1 in the SHR (RGDIDs: 2302148-2302151; RGDIDs: 2300216, 2313693) [113,114] and SS rats overexpressing human CETP (RGDID: 2290429) [115]. Furthermore, the successful culture of rat ES lines [116,117] and other applications of targeted mutagenesis (e.g. using Zinc Finger Nucleases, TALENS, or CRISPR-Cas9) is opening up new opportunities to study pathobiological mechanisms in single gene rat MetS models [118-121], including melanocortin receptor (Mc3r and Mc4r) and ghrelin receptor (Ghsr) mutations in outbred Wistar rats and Angiopoietin-like protein 8 (Angptl8) in inbred F344 rats [122-124].
6. Notes
Herein we described numerous rat models that display key features of the human metabolic syndrome. The models range from single gene mutations, to inbred selection models with polygenic inheritance, to population models that perhaps best mimic the heterogeneity of human MetS. Each model has its own benefits and limitations. Some models have all defining traits of MetS and could be useful for identifying gene pleiotropy or common underlying pathophysiology linking seemingly independent traits. However, identifying which mechanism(s) are causal may be confounded by the presence of other traits. The population models may better represent the general human clinical condition and can map traits to smaller genomic intervals than is possible with other approaches such as F2 intercrosses. A disadvantage of these models is that once a locus is identified, identifying the causal variant(s) can be confounded by a mixed genomic background. Single gene models can facilitate thorough mechanistic understanding of the resulting phenotype. However, single gene MetS disorders are extremely rare in humans and do not represent the highly complex genetic, epigenetic, and environmental contributions to MetS. As such, one must carefully consider which type of model answers the research question at hand. Regardless, the rat is a rich resource for polygenic models such as the metabolic syndrome.
References:
- 1.Reaven GM (1988) Banting lecture 1988. Role of insulin resistance in human disease. Diabetes 37 (12):1595–1607 [DOI] [PubMed] [Google Scholar]
- 2.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC Jr., International Diabetes Federation Task Force on E, Prevention, Hational Heart L, Blood I, American Heart A, World Heart F, International Atherosclerosis S, International Association for the Study of O (2009) Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 120 (16):1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644 [DOI] [PubMed] [Google Scholar]
- 3.Saklayen MG (2018) The Global Epidemic of the Metabolic Syndrome. Current hypertension reports 20 (2):12. doi: 10.1007/s11906-018-0812-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nolan PB, Carrick-Ranson G, Stinear JW, Reading SA, Dalleck LC (2017) Prevalence of metabolic syndrome and metabolic syndrome components in young adults: A pooled analysis. Prev Med Rep 7:211–215. doi: 10.1016/j.pmedr.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin HF, Boden-Albala B, Juo SH, Park N, Rundek T, Sacco RL (2005) Heritabilities of the metabolic syndrome and its components in the Northern Manhattan Family Study. Diabetologia 48 (10):2006–2012. doi: 10.1007/s00125-005-1892-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henneman P, Aulchenko YS, Frants RR, van Dijk KW, Oostra BA, van Duijn CM (2008) Prevalence and heritability of the metabolic syndrome and its individual components in a Dutch isolate: the Erasmus Rucphen Family study. J Med Genet 45 (9):572–577. doi: 10.1136/jmg.2008.058388 [DOI] [PubMed] [Google Scholar]
- 7.Khan RJ, Gebreab SY, Sims M, Riestra P, Xu R, Davis SK (2015) Prevalence, associated factors and heritabilities of metabolic syndrome and its individual components in African Americans: the Jackson Heart Study. BMJ Open 5 (10):e008675. doi: 10.1136/bmjopen-2015-008675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dongen J, Willemsen G, Chen WM, de Geus EJ, Boomsma DI (2013) Heritability of metabolic syndrome traits in a large population-based sample. Journal of lipid research 54 (10):2914–2923. doi: 10.1194/jlr.P041673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayman GT, Laulederkind SJ, Smith JR, Wang SJ, Petri V, Nigam R, Tutaj M, De Pons J, Dwinell MR, Shimoyama M (2016) The Disease Portals, disease-gene annotation and the RGD disease ontology at the Rat Genome Database. Database (Oxford) 2016. doi: 10.1093/database/baw034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oschry Y, Eisenberg S (1982) Rat plasma lipoproteins: re-evaluation of a lipoprotein system in an animal devoid of cholesteryl ester transfer activity. Journal of lipid research 23 (8):1099–1106 [PubMed] [Google Scholar]
- 11.Bergen WG, Mersmann HJ (2005) Comparative aspects of lipid metabolism: impact on contemporary research and use of animal models. J Nutr 135 (11):2499–2502. doi: 10.1093/jn/135.11.2499 [DOI] [PubMed] [Google Scholar]
- 12.Yin W, Carballo-Jane E, McLaren DG, Mendoza VH, Gagen K, Geoghagen NS, McNamara LA, Gorski JN, Eiermann GJ, Petrov A, Wolff M, Tong X, Wilsie LC, Akiyama TE, Chen J, Thankappan A, Xue J, Ping X, Andrews G, Wickham LA, Gai CL, Trinh T, Kulick AA, Donnelly MJ, Voronin GO, Rosa R, Cumiskey AM, Bekkari K, Mitnaul LJ, Puig O, Chen F, Raubertas R, Wong PH, Hansen BC, Koblan KS, Roddy TP, Hubbard BK, Strack AM (2012) Plasma lipid profiling across species for the identification of optimal animal models of human dyslipidemia. Journal of lipid research 53 (1):51–65. doi: 10.1194/jlr.M019927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tryon RC (1940) Genetic differences in maze-learning ability in rats. In: Thirty-ninth ye3arbook of the National Society for the Study of Education. Intelligence: Its nature and nurture, Part 1. Comparative an critical exposition. Public School Publishing Co, Bloomington, pp 111–119 [Google Scholar]
- 14.Eriksson K (1968) Genetic selection for voluntary alcohol consumption in the albino rat. Science (New York, NY 159 (3816):739–741. doi: 10.1126/science.159.3816.739 [DOI] [PubMed] [Google Scholar]
- 15.Rapp JP (2000) Genetic analysis of inherited hypertension in the rat. Physiol Rev 80 (1):135–172 [DOI] [PubMed] [Google Scholar]
- 16.Goto Y, Kakizaki M, Masaki N (1976) Production of spontaneous diabetic rats by repetition of selective breeding. Tohoku J Exp Med 119 (1):85–90 [DOI] [PubMed] [Google Scholar]
- 17.Dupont J, Dupont JC, Froment A, Milon H, Vincent M (1973) Selection of three strains of rats with spontaneously different levels of blood pressure. Biomedicine 19 (1):36–41 [PubMed] [Google Scholar]
- 18.Su DF, Cerutti C, Barres C, Vincent M, Sassard J (1986) Blood pressure and baroreflex sensitivity in conscious hypertensive rats of Lyon strain. Am J Physiol 251 (6 Pt 2):H1111–1117 [DOI] [PubMed] [Google Scholar]
- 19.Sassard J, Vincent M, Orea V, Privat P, Bataillard A (1997) Genetics of blood pressure and associated phenotypes in the Lyon rat. Clin Exp Hypertens 19 (5-6):567–575 [DOI] [PubMed] [Google Scholar]
- 20.Florin M, Lo M, Liu KL, Sassard J (2001) Salt sensitivity in genetically hypertensive rats of the Lyon strain. Kidney Int 59 (5):1865–1872 [DOI] [PubMed] [Google Scholar]
- 21.Sassolas A, Vincent M, Benzoni D, Sassard J (1981) Plasma lipids in genetically hypertensive rats of the Lyon strain. J Cardiovasc Pharmacol 3 (5):1008–1014 [DOI] [PubMed] [Google Scholar]
- 22.Vincent M, Boussairi EH, Cartier R, Lo M, Sassolas A, Cerutti C, Barres C, Gustin MP, Cuisinaud G, Samani NJ, et al. (1993) High blood pressure and metabolic disorders are associated in the Lyon hypertensive rat. Journal of hypertension 11 (11):1179–1185 [PubMed] [Google Scholar]
- 23.Boulanger M, Duhault J, Broux O, Bataillard A, Sassard J (1997) Lack of insulin resistance in the Lyon hypertensive rat. Fundam Clin Pharmacol 11 (6):546–549 [DOI] [PubMed] [Google Scholar]
- 24.Vincent M, Cartier R, Privat P, Benzoni D, Samani NJ, Sassard J (1996) Major cardiovascular risk factors in Lyon hypertensive rats. A correlation analysis in a segregating population. Journal of hypertension 14 (4):469–474 [PubMed] [Google Scholar]
- 25.Dubay C, Vincent M, Samani NJ, Hilbert P, Kaiser MA, Beressi J-P, Kotelevtsev Y, Beckmann JS, Soubrier F, Sassard J, Lathrop GM (1993) Genetic determinants of diastolic and pulse pressure map to different loci in Lyon hypertensive rats. Nature genetics 3 (4):354–357 [DOI] [PubMed] [Google Scholar]
- 26.Bilusic M, Bataillard A, Tschannen MR, Gao L, Barreto NE, Vincent M, Wang T, Jacob HJ, Sassard J, Kwitek AE (2004) Mapping the genetic determinants of hypertension, metabolic diseases, and related phenotypes in the lyon hypertensive rat. Hypertension 44 (5):695–701. doi: 10.1161/01.HYP.0000144542.57306.5e [DOI] [PubMed] [Google Scholar]
- 27.Ma MCJ, Pettus JM, Jakoubek JA, Traxler MG, Clark KC, Mennie AK, Kwitek AE (2017) Contribution of independent and pleiotropic genetic effects in the metabolic syndrome in a hypertensive rat. PloS one 12 (8):e0182650. doi: 10.1371/journal.pone.0182650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Ma MC, Mennie AK, Pettus JM, Xu Y, Lin L, Traxler MG, Jakoubek J, Atanur SS, Aitman TJ, Xing Y, Kwitek AE (2015) Systems biology with high-throughput sequencing reveals genetic mechanisms underlying the metabolic syndrome in the Lyon hypertensive rat. Circ Cardiovasc Genet 8 (2):316–326. doi: 10.1161/CIRCGENETICS.114.000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kloting N, Bluher M, Kloting I (2006) The polygenetically inherited metabolic syndrome of WOKW rats is associated with insulin resistance and altered gene expression in adipose tissue. Diabetes/metabolism research and reviews 22 (2):146–154. doi: 10.1002/dmrr.582 [DOI] [PubMed] [Google Scholar]
- 30.van den Brandt J, Kovacs P, Kloting I (2002) Metabolic syndrome and aging in Wistar Ottawa Karlsburg W rats. Int J Obes Relat Metab Disord 26 (4):573–576 [DOI] [PubMed] [Google Scholar]
- 31.van den Brandt J, Kovacs P, Kloting I (2000) Features of the metabolic syndrome in the spontaneously hypertriglyceridemic Wistar Ottawa Karlsburg W (RT1u Haplotype) rat. Metabolism 49 (9):1140–1144. doi: 10.1053/meta.2000.8610 [DOI] [PubMed] [Google Scholar]
- 32.Kovacs P, Voigt B, Berg S, Vogt L, Kloting I (1997) WOK.1W rats. A potential animal model of the insulin resistance syndrome. Ann N Y Acad Sci 827:94–99 [DOI] [PubMed] [Google Scholar]
- 33.Yagi K, Kim S, Wanibuchi H, Yamashita T, Yamamura Y, Iwao H (1997) Characteristics of diabetes, blood pressure, and cardiac and renal complications in Otsuka Long-Evans Tokushima Fatty rats. Hypertension 29 (3):728–735 [DOI] [PubMed] [Google Scholar]
- 34.Kawano K, Hirashima T, Mori S, Saitoh Y, Kurosumi M, Natori T (1992) Spontaneous long-term hyperglycemic rat with diabetic complications. Otsuka Long-Evans Tokushima Fatty (OLETF) strain. Diabetes 41 (11):1422–1428 [DOI] [PubMed] [Google Scholar]
- 35.Weingarten A, Turchetti L, Krohn K, Kloting I, Kern M, Kovacs P, Stumvoll M, Bluher M, Kloting N (2016) Novel genes on rat chromosome 10 are linked to body fat mass, preadipocyte number and adipocyte size. Int J Obes (Lond) 40 (12):1832–1840. doi: 10.1038/ijo.2016.127 [DOI] [PubMed] [Google Scholar]
- 36.Baguhl R, Wilke B, Kloting N, Kloting I (2009) Genes on rat chromosomes 3, 5, 10, and 16 are linked with facets of metabolic syndrome. Obesity (Silver Spring) 17 (6):1215–1219. doi: 10.1038/oby.2008.658 [DOI] [PubMed] [Google Scholar]
- 37.Kloting N, Wilke B, Kloting I (2007) Triplet repeat in the Repin1 3'-untranslated region on rat chromosome 4 correlates with facets of the metabolic syndrome. Diabetes/metabolism research and reviews 23 (5):406–410. doi: 10.1002/dmrr.713 [DOI] [PubMed] [Google Scholar]
- 38.Kose H, Moralejo DH, Ogino T, Mizuno A, Yamada T, Matsumoto K (2002) Examination of OLETF-derived non-insulin-dependent diabetes mellitus QTL by construction of a series of congenic rats. Mamm Genome 13 (10):558–562. doi: 10.1007/s00335-002-2199-y [DOI] [PubMed] [Google Scholar]
- 39.Muramatsu Y, Yamada T, Taniguchi Y, Ogino T, Kose H, Matsumoto K, Sasaki Y (2005) Pnlip encoding pancreatic lipase is possible candidate for obesity QTL in the OLETF rat. Biochemical and biophysical research communications 331 (4):1270–1276. doi: 10.1016/j.bbrc.2005.04.040 [DOI] [PubMed] [Google Scholar]
- 40.Watanabe TK, Suzuki M, Yamasaki Y, Okuno S, Hishigaki H, Ono T, Oga K, Mizoguchi-Miyakita A, Tsuji A, Kanemoto N, Wakitani S, Takagi T, Nakamura Y, Tanigami A (2005) Mutated G-protein-coupled receptor GPR10 is responsible for the hyperphagia/dyslipidaemia/obesity locus of Dmo1 in the OLETF rat. Clin Exp Pharmacol Physiol 32 (5-6):355–366. doi: 10.1111/j.1440-1681.2005.04196.x [DOI] [PubMed] [Google Scholar]
- 41.Moran TH (2008) Unraveling the obesity of OLETF rats. Physiol Behav 94 (1):71–78. doi: 10.1016/j.physbeh.2007.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swales JD (1994) Textbook of hypertension. Blackwell Scientific Publications, Oxford; Boston [Google Scholar]
- 43.Wd Jong (1984) Experimental and genetic models of hypertension. Volume 4 Volume 4. [Google Scholar]
- 44.Coan PM, Hummel O, Garcia Diaz A, Barrier M, Alfazema N, Norsworthy PJ, Pravenec M, Petretto E, Hubner N, Aitman TJ (2017) Genetic, physiological and comparative genomic studies of hypertension and insulin resistance in the spontaneously hypertensive rat. Dis Model Mech 10 (3):297–306. doi: 10.1242/dmm.026716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pravenec M, Kren V, Landa V, Mlejnek P, Musilova A, Silhavy J, Simakova M, Zidek V (2014) Recent progress in the genetics of spontaneously hypertensive rats. Physiol Res 63 Suppl 1:S1–8 [DOI] [PubMed] [Google Scholar]
- 46.Tofovic SP, Jackson EK (2003) Rat models of the metabolic syndrome. Methods Mol Med 86:29–46. doi: 10.1385/1-59259-392-5:29 [DOI] [PubMed] [Google Scholar]
- 47.Aitman TJ, Gotoda T, Evans AL, Imrie H, Heath KE, Trembling PM, Truman H, Wallace CA, Rahman A, Dore C, Flint J, Kren V, Zidek V, Kurtz TW, Pravenec M, Scott J (1997) Quantitative trait loci for cellular defects in glucose and fatty acid metabolism in hypertensive rats. Nature genetics 16 (2):197–201. doi: 10.1038/ng0697-197 [DOI] [PubMed] [Google Scholar]
- 48.Doris PA (2017) Genetics of hypertension: an assessment of progress in the spontaneously hypertensive rat. Physiological genomics 49 (11):601–617. doi: 10.1152/physiolgenomics.00065.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pravenec M, Kurtz TW (2010) Recent advances in genetics of the spontaneously hypertensive rat. Current hypertension reports 12 (1):5–9. doi: 10.1007/s11906-009-0083-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J (1999) Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nature genetics 21 (1):76–83 [DOI] [PubMed] [Google Scholar]
- 51.Pravenec M, Kozich V, Krijt J, Sokolova J, Zidek V, Landa V, Mlejnek P, Silhavy J, Simakova M, Skop V, Trnovska J, Kazdova L, Kajiya T, Wang J, Kurtz TW (2016) Genetic Variation in Renal Expression of Folate Receptor 1 (Folr1) Gene Predisposes Spontaneously Hypertensive Rats to Metabolic Syndrome. Hypertension 67 (2):335–341. doi: 10.1161/HYPERTENSIONAHA.115.06158 [DOI] [PubMed] [Google Scholar]
- 52.Pravenec M, Zidek V, Landa V, Mlejnek P, Silhavy J, Simakova M, Trnovska J, Skop V, Markova I, Malinska H, Huttl M, Kazdova L, Bardova K, Tauchmannova K, Vrbacky M, Nuskova H, Mracek T, Kopecky J, Houstek J (2017) Mutant Wars2 gene in spontaneously hypertensive rats impairs brown adipose tissue function and predisposes to visceral obesity. Physiol Res 66 (6):917–924 [DOI] [PubMed] [Google Scholar]
- 53.Zicha J, Pechanova O, Cacanyiova S, Cebova M, Kristek F, Torok J, Simko F, Dobesova Z, Kunes J (2006) Hereditary hypertriglyceridemic rat: a suitable model of cardiovascular disease and metabolic syndrome? Physiol Res 55 Suppl 1:S49–63 [DOI] [PubMed] [Google Scholar]
- 54.Klimes I, Vrana A, Kunes J, Sebokova E, Dobesova Z, Stolba P, Zicha J (1995) Hereditary hypertriglyceridemic rat: a new animal model of metabolic alterations in hypertension. Blood Press 4 (3):137–142 [DOI] [PubMed] [Google Scholar]
- 55.Stolba P, Dobesova Z, Husek P, Opltova H, Zicha J, Vrana A, Kunes J (1992) The hypertriglyceridemic rat as a genetic model of hypertension and diabetes. Life Sci 51 (10):733–740 [DOI] [PubMed] [Google Scholar]
- 56.Ueno T, Tremblay J, Kunes J, Zicha J, Dobesova Z, Pausova Z, Deng AY, Sun YL, Jacob HJ, Hamet P (2004) Rat model of familial combined hyperlipidemia as a result of comparative mapping. Physiological genomics 17 (1):38–47 [DOI] [PubMed] [Google Scholar]
- 57.Sedova L, Kazdova L, Seda O, Krenova D, Kren V (2000) Rat inbred PD/cub strain as a model of dyslipidemia and insulin resistance. Folia Biol (Praha) 46 (3):99–106 [DOI] [PubMed] [Google Scholar]
- 58.Seda O, Liska F, Krenova D, Kazdova L, Sedova L, Zima T, Peng J, Pelinkova K, Tremblay J, Hamet P, Kren V (2005) Dynamic genetic architecture of metabolic syndrome attributes in the rat. Physiological genomics 21 (2):243–252. doi: 10.1152/physiolgenomics.00230.2004 [DOI] [PubMed] [Google Scholar]
- 59.Sasase T, Ohta T, Masuyama T, Yokoi N, Kakehashi A, Shinohara M (2013) The spontaneously diabetic torii rat: an animal model of nonobese type 2 diabetes with severe diabetic complications. J Diabetes Res 2013:976209. doi: 10.1155/2013/976209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shinohara M, Masuyama T, Shoda T, Takahashi T, Katsuda Y, Komeda K, Kuroki M, Kakehashi A, Kanazawa Y (2000) A new spontaneously diabetic non-obese Torii rat strain with severe ocular complications. Int J Exp Diabetes Res 1 (2):89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuse M, Yokoi N, Shinohara M, Masuyama T, Kitazawa R, Kitazawa S, Seino S (2008) Identification of a major locus for islet inflammation and fibrosis in the spontaneously diabetic Torii rat. Physiological genomics 35 (1):96–105. doi: 10.1152/physiolgenomics.90214.2008 [DOI] [PubMed] [Google Scholar]
- 62.Masuyama T, Fuse M, Yokoi N, Shinohara M, Tsujii H, Kanazawa M, Kanazawa Y, Komeda K, Taniguchi K (2003) Genetic analysis for diabetes in a new rat model of nonobese type 2 diabetes, Spontaneously Diabetic Torii rat. Biochemical and biophysical research communications 304 (1):196–206 [DOI] [PubMed] [Google Scholar]
- 63.Cheng ZJ, Vaskonen T, Tikkanen I, Nurminen K, Ruskoaho H, Vapaatalo H, Muller D, Park JK, Luft FC, Mervaala EM (2001) Endothelial dysfunction and salt-sensitive hypertension in spontaneously diabetic Goto-Kakizaki rats. Hypertension 37 (2 Pt 2):433–439 [DOI] [PubMed] [Google Scholar]
- 64.Kaisaki PJ, Otto GW, Argoud K, Collins SC, Wallis RH, Wilder SP, Yau ACY, Hue C, Calderari S, Bihoreau MT, Cazier JB, Mott R, Gauguier D (2016) Transcriptome Profiling in Rat Inbred Strains and Experimental Cross Reveals Discrepant Genetic Architecture of Genome-Wide Gene Expression. G3 (Bethesda) 6 (11):3671–3683. doi: 10.1534/g3.116.033274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dumas ME, Wilder SP, Bihoreau MT, Barton RH, Fearnside JF, Argoud K, D'Amato L, Wallis RH, Blancher C, Keun HC, Baunsgaard D, Scott J, Sidelmann UG, Nicholson JK, Gauguier D (2007) Direct quantitative trait locus mapping of mammalian metabolic phenotypes in diabetic and normoglycemic rat models. Nature genetics 39 (5):666–672. doi: 10.1038/ng2026 [DOI] [PubMed] [Google Scholar]
- 66.Portha B, Giroix MH, Tourrel-Cuzin C, Le-Stunff H, Movassat J (2012) The GK rat: a prototype for the study of non-overweight type 2 diabetes. Methods in molecular biology (Clifton, NJ 933:125–159. doi: 10.1007/978-1-62703-068-7_9 [DOI] [PubMed] [Google Scholar]
- 67.Nobrega MA, Woods LC, Fleming S, Jacob HJ (2009) Distinct genetic regulation of progression of diabetes and renal disease in the Goto-Kakizaki rat. Physiological genomics 39 (1):38–46. doi:90389.2008 [pii] 10.1152/physiolgenomics.90389.2008 [DOI] [PubMed] [Google Scholar]
- 68.Cohen AM, Rosenmann E, Rosenthal T (1993) The Cohen diabetic (non-insulin-dependent) hypertensive rat model. Description of the model and pathologic findings. Am J Hypertens 6 (12):989–995 [DOI] [PubMed] [Google Scholar]
- 69.Younis F, Leor J, Abassi Z, Landa N, Rath L, Hollander K, Naftali-Shani N, Rosenthal T (2018) Beneficial Effect of the SGLT2 Inhibitor Empagliflozin on Glucose Homeostasis and Cardiovascular Parameters in the Cohen Rosenthal Diabetic Hypertensive (CRDH) Rat. J Cardiovasc Pharmacol Ther 23 (4):358–371. doi: 10.1177/1074248418763808 [DOI] [PubMed] [Google Scholar]
- 70.Younis F, Stern N, Limor R, Oron Y, Zangen S, Rosenthal T (2010) Telmisartan ameliorates hyperglycemia and metabolic profile in nonobese Cohen-Rosenthal diabetic hypertensive rats via peroxisome proliferator activator receptor-gamma activation. Metabolism 59 (8):1200–1209. doi: 10.1016/j.metabol.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 71.Rosenthal T, Erlich Y, Rosenmann E, Cohen A (1997) Effects of enalapril, losartan, and verapamil on blood pressure and glucose metabolism in the Cohen-Rosenthal diabetic hypertensive rat. Hypertension 29 (6):1260–1264 [DOI] [PubMed] [Google Scholar]
- 72.Reaven GM, Twersky J, Chang H (1991) Abnormalities of carbohydrate and lipid metabolism in Dahl rats. Hypertension 18 (5):630–635 [DOI] [PubMed] [Google Scholar]
- 73.Somova L, Channa ML (1999) Glucose metabolism and insulin sensitivity in Dahl hypertensive rats. Methods Find Exp Clin Pharmacol 21 (6):421–425 [DOI] [PubMed] [Google Scholar]
- 74.Zicha J, Dobesova Z, Vokurkova M, Rauchova H, Hojna S, Kadlecova M, Behuliak M, Vaneckova I, Kunes J (2012) Age-dependent salt hypertension in Dahl rats: fifty years of research. Physiol Res 61 Suppl 1:S35–87 [DOI] [PubMed] [Google Scholar]
- 75.Donnelly R, Ho H, Reaven GM (1995) Effects of low sodium diet and unilateral nephrectomy on the development of carbohydrate-induced hypertension. Blood Press 4 (3):164–169 [DOI] [PubMed] [Google Scholar]
- 76.Kotchen TA, Zhang HY, Covelli M, Blehschmidt N (1991) Insulin resistance and blood pressure in Dahl rats and in one-kidney, one-clip hypertensive rats. Am J Physiol 261 (6 Pt 1):E692–697 [DOI] [PubMed] [Google Scholar]
- 77.Nagae A, Fujita M, Kawarazaki H, Matsui H, Ando K, Fujita T (2009) Effect of high fat loading in Dahl salt-sensitive rats. Clin Exp Hypertens 31 (5):451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang HY, Reddy S, Kotchen TA (1999) A high sucrose, high linoleic acid diet potentiates hypertension in the Dahl salt sensitive rat. Am J Hypertens 12 (2 Pt 1):183–187 [DOI] [PubMed] [Google Scholar]
- 79.De Miguel C, Lund H, Mattson DL (2011) High dietary protein exacerbates hypertension and renal damage in Dahl SS rats by increasing infiltrating immune cells in the kidney. Hypertension 57 (2):269–274. doi: 10.1161/HYPERTENSIONAHA.110.154302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mattson DL, Meister CJ, Marcelle ML (2005) Dietary protein source determines the degree of hypertension and renal disease in the Dahl salt-sensitive rat. Hypertension 45 (4):736–741. doi: 10.1161/01.HYP.0000153318.74544.cc [DOI] [PubMed] [Google Scholar]
- 81.Levin BE, Dunn-Meynell AA, Balkan B, Keesey RE (1997) Selective breeding for diet-induced obesity and resistance in Sprague-Dawley rats. Am J Physiol 273 (2 Pt 2):R725–730. doi: 10.1152/ajpregu.1997.273.2.R725 [DOI] [PubMed] [Google Scholar]
- 82.Chang S, Graham B, Yakubu F, Lin D, Peters JC, Hill JO (1990) Metabolic differences between obesity-prone and obesity-resistant rats. Am J Physiol 259 (6 Pt 2):R1103–1110. doi: 10.1152/ajpregu.1990.259.6.R1103 [DOI] [PubMed] [Google Scholar]
- 83.Levin BE, Triscari J, Hogan S, Sullivan AC (1987) Resistance to diet-induced obesity: food intake, pancreatic sympathetic tone, and insulin. Am J Physiol 252 (3 Pt 2):R471–478. doi: 10.1152/ajpregu.1987.252.3.R471 [DOI] [PubMed] [Google Scholar]
- 84.Schemmel R, Mickelsen O, Gill JL (1970) Dietary obesity in rats: Body weight and body fat accretion in seven strains of rats. J Nutr 100 (9):1041–1048. doi: 10.1093/jn/100.9.1041 [DOI] [PubMed] [Google Scholar]
- 85.Dobrian AD, Davies MJ, Prewitt RL, Lauterio TJ (2000) Development of hypertension in a rat model of diet-induced obesity. Hypertension 35 (4):1009–1015 [DOI] [PubMed] [Google Scholar]
- 86.Giles ED, Jackman MR, MacLean PS (2016) Modeling Diet-Induced Obesity with Obesity-Prone Rats: Implications for Studies in Females. Front Nutr 3:50. doi: 10.3389/fnut.2016.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Panchal SK, Poudyal H, Iyer A, Nazer R, Alam A, Diwan V, Kauter K, Sernia C, Campbell F, Ward L, Gobe G, Fenning A, Brown L (2011) High-carbohydrate high-fat diet-induced metabolic syndrome and cardiovascular remodeling in rats. J Cardiovasc Pharmacol 57 (1):51–64. doi: 10.1097/FJC.0b013e3181feb90a [DOI] [PubMed] [Google Scholar]
- 88.Moreno-Fernandez S, Garces-Rimon M, Vera G, Astier J, Landrier JF, Miguel M (2018) High Fat/High Glucose Diet Induces Metabolic Syndrome in an Experimental Rat Model. Nutrients 10 (10). doi: 10.3390/nu10101502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wong SK, Chin KY, Suhaimi FH, Fairus A, Ima-Nirwana S (2016) Animal models of metabolic syndrome: a review. Nutr Metab (Lond) 13:65. doi: 10.1186/s12986-016-0123-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwitek AE, Jacob HJ, Baker JE, Dwinell MR, Forster HV, Greene AS, Kunert MP, Lombard JH, Mattson DL, Pritchard KA Jr., Roman RJ, Tonellato PJ, Cowley AW Jr. (2006) BN phenome: detailed characterization of the cardiovascular, renal, and pulmonary systems of the sequenced rat. Physiological genomics 25 (2):303–313. doi: 10.1152/physiolgenomics.00288.2005 [DOI] [PubMed] [Google Scholar]
- 91.Brower M, Grace M, Kotz CM, Koya V (2015) Comparative analysis of growth characteristics of Sprague Dawley rats obtained from different sources. Lab Anim Res 31 (4):166–173. doi: 10.5625/lar.2015.31.4.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hansen C, Spuhler K (1984) Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res 8 (5):477–479 [DOI] [PubMed] [Google Scholar]
- 93.Hermsen R, de Ligt J, Spee W, Blokzijl F, Schafer S, Adami E, Boymans S, Flink S, van Boxtel R, van der Weide RH, Aitman T, Hubner N, Simonis M, Tabakoff B, Guryev V, Cuppen E (2015) Genomic landscape of rat strain and substrain variation. BMC Genomics 16:357. doi: 10.1186/s12864-015-1594-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baud A, Guryev V, Hummel O, Johannesson M, Rat Genome S, Mapping C, Flint J (2014) Genomes and phenomes of a population of outbred rats and its progenitors. Sci Data 1:140011. doi: 10.1038/sdata.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Woods LC, Mott R (2017) Heterogeneous Stock Populations for Analysis of Complex Traits. Methods in molecular biology (Clifton, NJ 1488:31–44. doi: 10.1007/978-1-4939-6427-7_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alam I, Koller DL, Canete T, Blazquez G, Mont-Cardona C, Lopez-Aumatell R, Martinez-Membrives E, Diaz-Moran S, Tobena A, Fernandez-Teruel A, Stridh P, Diez M, Olsson T, Johannesson M, Baud A, Econs MJ, Foroud T (2015) Fine mapping of bone structure and strength QTLs in heterogeneous stock rat. Bone 81:417–426. doi: 10.1016/j.bone.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holl K, He H, Wedemeyer M, Clopton L, Wert S, Meckes JK, Cheng R, Kastner A, Palmer AA, Redei EE, Solberg Woods LC (2017) Heterogeneous stock rats: a model to study the genetics of despair-like behavior in adolescence. Genes Brain Behav. doi: 10.1111/gbb.12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keele GR, Prokop JW, He H, Holl K, Littrell J, Deal A, Francic S, Cui L, Gatti DM, Broman KW, Tschannen M, Tsaih SW, Zagloul M, Kim Y, Baur B, Fox J, Robinson M, Levy S, Flister MJ, Mott R, Valdar W, Solberg Woods LC (2017) Genetic Fine-Mapping and Identification of Candidate Genes and Variants for Adiposity Traits in Outbred Rats. Obesity (Silver Spring). doi: 10.1002/oby.22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koch LG, Britton SL (2001) Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiological genomics 5 (1):45–52. doi: 10.1152/physiolgenomics.2001.5.1.45 [DOI] [PubMed] [Google Scholar]
- 100.Koch LG, Britton SL, Wisloff U (2012) A rat model system to study complex disease risks, fitness, aging, and longevity. Trends Cardiovasc Med 22 (2):29–34. doi: 10.1016/j.tcm.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koch LG, Britton SL (2008) Development of animal models to test the fundamental basis of gene-environment interactions. Obesity (Silver Spring) 16 Suppl 3:S28–32. doi: 10.1038/oby.2008.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ren YY, Overmyer KA, Qi NR, Treutelaar MK, Heckenkamp L, Kalahar M, Koch LG, Britton SL, Burant CF, Li JZ (2013) Genetic analysis of a rat model of aerobic capacity and metabolic fitness. PloS one 8 (10):e77588. doi: 10.1371/journal.pone.0077588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zucker LM, Zucker TF (1961) FATTY, A NEW MUTATION IN THE RAT. Journal of Heredity 52 (6):275–278. doi: 10.1093/oxfordjournals.jhered.a107093 [DOI] [Google Scholar]
- 104.Takaya K, Ogawa Y, Isse N, Okazaki T, Satoh N, Masuzaki H, Mori K, Tamura N, Hosoda K, Nakao K (1996) Molecular cloning of rat leptin receptor isoform complementary DNAs--identification of a missense mutation in Zucker fatty (fa/fa) rats. Biochemical and biophysical research communications 225 (1):75–83. doi: 10.1006/bbrc.1996.1133 [DOI] [PubMed] [Google Scholar]
- 105.White DW, Wang DW, Chua SC Jr., Morgenstern JP, Leibel RL, Baumann H, Tartaglia LA (1997) Constitutive and impaired signaling of leptin receptors containing the Gln --> Pro extracellular domain fatty mutation. Proceedings of the National Academy of Sciences of the United States of America 94 (20):10657–10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peterson RG, Shaw WN, Neel M-A, Little LA, Eichberg J (1990) Zucker Diabetic Fatty Rat as a Model for Non-insulin-dependent Diabetes Mellitus. ILAR Journal 32 (3):16–19. doi: 10.1093/ilar.32.3.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hattori T, Murase T, Ohtake M, Inoue T, Tsukamoto H, Takatsu M, Kato Y, Hashimoto K, Murohara T, Nagata K (2011) Characterization of a new animal model of metabolic syndrome: the DahlS.Z-Lepr(fa)/Lepr(fa) rat. Nutr Diabetes 1:e1. doi: 10.1038/nutd.2010.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hiraoka-Yamamoto J, Nara Y, Yasui N, Onobayashi Y, Tsuchikura S, Ikeda K (2004) Establishment of a new animal model of metabolic syndrome: SHRSP fatty (fa/fa) rats. Clin Exp Pharmacol Physiol 31 (1-2):107–109 [DOI] [PubMed] [Google Scholar]
- 109.Koletsky S (1973) Obese spontaneously hypertensive rats--a model for study of atherosclerosis. Exp Mol Pathol 19 (1):53–60 [DOI] [PubMed] [Google Scholar]
- 110.Koletsky S (1975) Pathologic findings and laboratory data in a new strain of obese hypertensive rats. Am J Pathol 80 (1):129–142 [PMC free article] [PubMed] [Google Scholar]
- 111.Takaya K, Ogawa Y, Hiraoka J, Hosoda K, Yamori Y, Nakao K, Koletsky RJ (1996) Nonsense mutation of leptin receptor in the obese spontaneously hypertensive Koletsky rat. Nature genetics 14 (2):130–131. doi: 10.1038/ng1096-130 [DOI] [PubMed] [Google Scholar]
- 112.Moralejo DH, Hansen CT, Treuting P, Hessner MJ, Fuller JM, Van Yserloo B, Jensen R, Osborne W, Kwitek AE, Lernmark A (2010) Differential effects of leptin receptor mutation on male and female BBDR Gimap5-/Gimap5- spontaneously diabetic rats. Physiological genomics 41 (1):9–20. doi: 10.1152/physiolgenomics.00186.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qi NR, Wang J, Zidek V, Landa V, Mlejnek P, Kazdova L, Pravenec M, Kurtz TW (2005) A New Transgenic Rat Model of Hepatic Steatosis and the Metabolic Syndrome. Hypertension [DOI] [PubMed] [Google Scholar]
- 114.Pravenec M, Landa V, Zidek V, Musilova A, Kazdova L, Qi N, Wang J, St Lezin E, Kurtz TW (2003) Transgenic expression of CD36 in the spontaneously hypertensive rat is associated with amelioration of metabolic disturbances but has no effect on hypertension. Physiol Res 52 (6):681–688 [PubMed] [Google Scholar]
- 115.Herrera VL, Makrides SC, Xie HX, Adari H, Krauss RM, Ryan US, Ruiz-Opazo N (1999) Spontaneous combined hyperlipidemia, coronary heart disease and decreased survival in Dahl salt-sensitive hypertensive rats transgenic for human cholesteryl ester transfer protein. Nat Med 5 (12):1383–1389. doi: 10.1038/70956 [DOI] [PubMed] [Google Scholar]
- 116.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A (2008) Capture of authentic embryonic stem cells from rat blastocysts. Cell 135 (7):1287–1298 [DOI] [PubMed] [Google Scholar]
- 117.Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, Pera MF, Ying QL (2008) Germline competent embryonic stem cells derived from rat blastocysts. Cell 135 (7):1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ (2011) Knockout rats generated by embryo microinjection of TALENs. Nature biotechnology 29 (8):695–696. doi: 10.1038/nbt.1940 [DOI] [PubMed] [Google Scholar]
- 119.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Menoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R (2009) Knockout rats via embryo microinjection of zinc-finger nucleases. Science (New York, NY 325 (5939):433. doi:325/5939/433 [pii] 10.1126/science.1172447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li W, Teng F, Li T, Zhou Q (2013) Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nature biotechnology 31 (8):684–686. doi: 10.1038/nbt.2652 [DOI] [PubMed] [Google Scholar]
- 121.Meek S, Mashimo T, Burdon T (2017) From engineering to editing the rat genome. Mammalian Genome 28 (7):302–314. doi: 10.1007/s00335-017-9705-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Izumi R, Kusakabe T, Noguchi M, Iwakura H, Tanaka T, Miyazawa T, Aotani D, Hosoda K, Kangawa K, Nakao K (2018) CRISPR/Cas9-mediated Angptl8 knockout suppresses plasma triglyceride concentrations and adiposity in rats. Journal of lipid research 59 (9):1575–1585. doi: 10.1194/jlr.M082099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zallar LJ, Tunstall BJ, Richie CT, Zhang YJ, You ZB, Gardner EL, Heilig M, Pickel J, Koob GF, Vendruscolo LF, Harvey BK, Leggio L (2018) Development and initial characterization of a novel ghrelin receptor CRISPR/Cas9 knockout wistar rat model. Int J Obes (Lond). doi: 10.1038/s41366-018-0013-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.You P, Hu H, Chen Y, Zhao Y, Yang Y, Wang T, Xing R, Shao Y, Zhang W, Li D, Chen H, Liu M (2016) Effects of Melanocortin 3 and 4 Receptor Deficiency on Energy Homeostasis in Rats. Sci Rep 6:34938. doi: 10.1038/srep34938 [DOI] [PMC free article] [PubMed] [Google Scholar]