Abstract
Nanometer-sized features and molecular recognition properties make DNA a useful material for nanoscale construction, but degradation in biological fluids poses a considerable roadblock to biomedical applications of DNA nanotechnology. Here we report the remarkable biostability of a multi-stranded motif called paranemic crossover (PX) DNA. Compared to double stranded DNA, PX DNA has dramatically enhanced (sometimes >1000 fold) resistance to degradation by four different nucleases, bovine and human serum, and human urine. We trace the cause of PX’s biostability to DNA crossovers, showing a continuum of protection that scales with the number of crossovers. These results suggest that enhanced biostability can be engineered into DNA nanostructures by adopting PX-based architectures or by strategic crossover placement.
Graphical Abstract
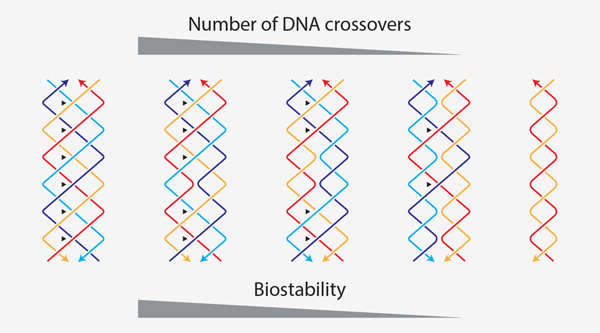
Introduction
Some daring purveyors of science fiction have imagined superior beings with more than two strands of DNA. In the 1997 movie The Fifth Element, for example, the heroine Leeloo is considered a perfect human-like being in part because of her 8-stranded helical DNA that is “tightly packed with infinite genetic knowledge”. While this is notably incorrect, there are instances both in biology and biotechnology where DNA structures can have more than two strands. Triplexes, for example can form a single helix using three strands,1 while guanine tetrads can form four-stranded DNA complexes.2
For DNA-based nanoscale assembly, synthetic DNA strands are designed and integrated together to form different motifs that serve as the building blocks for bottom-up construction.3 These (usually) multi-stranded structures typically contain double helical domains that are connected together by strand crossovers. A wide variety of structures have been made using bottom-up DNA construction, ranging from small objects and devices to larger, trigger-responsive “cages” that have emerging applications in drug delivery. To succeed as drug delivery vehicles, DNA objects must overcome a major challenge of surviving harsh in vivo environments such as blood.4 Strategies to improve the biostability of DNA structures include polymer5 and protein-based6–8 coating, viral capsid encapsulation,9 modified nucleotides10,11 and crosslinking.8,12 One possibility that has been largely overlooked is that the inherent design of these DNA nanostructures can be altered to change biostability.
Construction of DNA nanostructures is based on robust starting units. One example is the double crossover (DX) motif that has two adjacent double helices connected by 2 crossover points.13 Design rules and construction parameters established on such DNA motifs are applied to other strategies and hierarchical assemblies (for example, the multi-crossover DNA origami).14 Another (less common) DNA motif is paranemic crossover (PX) DNA, a four-stranded DNA structure that consists of two adjacent and connected double helical DNA domains (Figure 1a).15,16 The motif is formed by creating crossovers between strands of the same polarity at every possible point between two side-by-side helices.16 Each duplex domain of PX DNA contains alternating major (wide) groove (denoted by W) or a minor (narrow) groove separation (denoted by N) flanking the central dyad axis of the structure, with one helical repeat containing a mixture of four half turns. Previous studies have reported PX DNA with different major/minor groove separations (W:N), with the most stable complexes containing 6, 7 or 8 nucleotides in the major groove and 5 nucleotides in the minor groove (PX 6:5, 7:5 and 8:5 respectively).15,16
Figure 1. Design and validation of paranemic crossover (PX) DNA.
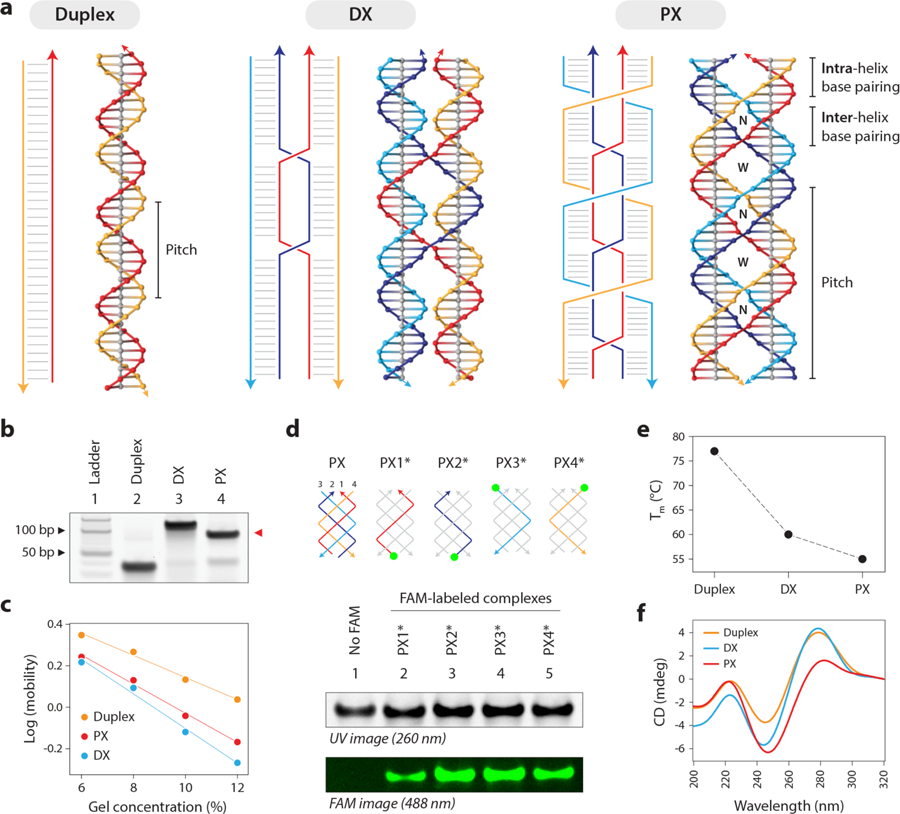
(a) Schematic and molecular models of a B-DNA duplex, a double crossover (DX) motif and paranemic crossover (PX) DNA. (b) Non-denaturing PAGE showing formation of structures as predominant products in each lane, with PX migrating slightly faster than its DX counterpart. (c) Ferguson plot showing gel mobility characteristics of the control and PX structures as a function of gel concentration. (d) Validating incorporation of all four strands in the PX by making four structures each with a single FAM-labeled strand. Gel image under UV is shown for reference, with a control PX lane omitting FAM labels. (e) Melting temperatures determined from UV melting experiment show a decrease in thermal stability from duplex to DX to PX. (f) Circular dichroism spectra of the tested structures show that characteristics of PX is similar to those previously reported.
In DNA nanotechnology, PX DNA has been used to construct objects such as an octahedron17 and a triangle,18 as well as one- and two-dimensional arrays.19,20 PX DNA has also been a component of nanomechanical devices21 that are used in molecular assembly lines22 and DNA-based computation.23 In biology, PX DNA is studied for its involvement in double stranded DNA homology recognition due to its ability to relax supercoiled DNA.24 Recent studies sought out proteins in the cell that can structure-specifically bind to PX DNA, so as to elucidate its biological function.25,26 These studies found that DNA polymerase I (Pol I) and T7 endonuclease I can bind to PX DNA, supporting the notion of its biological relevance. Despite the interest in PX DNA in nanotechnology and biological contexts, the biostability of the structure has not yet been explored.
In this work, we discovered that PX DNA has a remarkable ability to resist nuclease degradation compared to normal DNA duplexes. We show that this increased resistance is varied in magnitude but persistent across multiple enzymes and biological fluids. We find that PX DNA appears almost indestructible to a few nucleases, and can withstand 3 biological fluids for 24 hours with no evidence of damage. Exploring the cause of this enhanced stability, we find a clear dependence on the number of crossovers, with each additional crossover conferring additional protection. The results suggest that enhanced biostability can be designed into DNA nanostructures.
Results
Design and characterization of PX DNA and control structures
In this study, we used a PX 6:5 molecule (6 nucleotides in the major groove and 5 nucleotides in the minor groove),16,25 where the helical repeat of each strand is 22 nucleotides, making the pitch of the PX DNA roughly twice of that of B-DNA (with a net twist half of that of B-DNA) (Figure 1a, right). We made two control structures: one DNA duplex with the sequence matching half of the PX motif (Figure 1a, left) and a double crossover (DX) motif with sequence similar to the PX but has only two crossover points (Figure 1a, middle). Figure 1 illustrates the crossover points in the strand diagrams for each structure and their respective molecular models (sequences are shown in Figure S1). We annealed the motifs in Tris-acetate-EDTA-Mg2+ (TAE) buffer and checked their formation using non-denaturing polyacrylamide gel electrophoresis (PAGE) (Figure 1b and Figure S2). We analyzed the electrophoretic mobility of the different motifs as a function of gel concentration using a Ferguson plot (Figure 1c and Figure S3), where the slope provides an estimate of the retardation (frictional) coefficient of the structures. The plot shows that the slope of the PX molecule is comparable to that of the DX, and distinct from a regular double stranded DNA, a trend consistent with previous results.16 To further confirm the formation of the four-stranded PX, we used fluorescein (FAM)-labeled strands and annealed four different PX complexes each containing one of the FAM-labeled strands (green circles in Figure 1d, labeled strand is indicated by *). The presence of a fluorescent band in each of these four lanes on a non-denaturing PAGE indicated that all four strands are present in the complex (lanes 2–5) compared to a non-labeled complex (lane 1) (gel image in Figure 1d and Figure S4). We then analyzed the thermal melting profiles of the structures and found that the thermal stability decreases from the duplex (77 °C) to the DX (60 °C) and PX (55 °C) as the number of crossovers increases (Figure 1e and Figure S5). Circular dichroism profiles of the duplex, DX and PX were also consistent with previous reports available for these structures (Figure 1f).27
Nuclease resistance of PX DNA and control structures
For the first test of PX DNA biostability, we chose DNase I, one of the most widely used endonucleases in molecular biology that nonspecifically cleaves both strands of double stranded DNA.28 DNase I performs optimally at the physiological temperature of 37 °C, so we first confirmed that our DNA structures were stable at this temperature for at least 24 hours (Figure S6). We probed enzymatic degradation by incubating the PX DNA or relevant controls with DNase I enzyme for different times at 37 °C and quantifying the reduction of the band representing the structure on PAGE gels (Figure 2a and Figure S7 & S8). With 0.1 units of DNase I, the duplex and DX structures were over 90% degraded within a few minutes, while PX DNA had less than 5% degradation even after 1 hour. At higher concentrations of DNase I the digestion of all products accelerated but maintained similar trends (Figure S7 & S9).
Figure 2. Exceptional nuclease resistance of PX DNA.
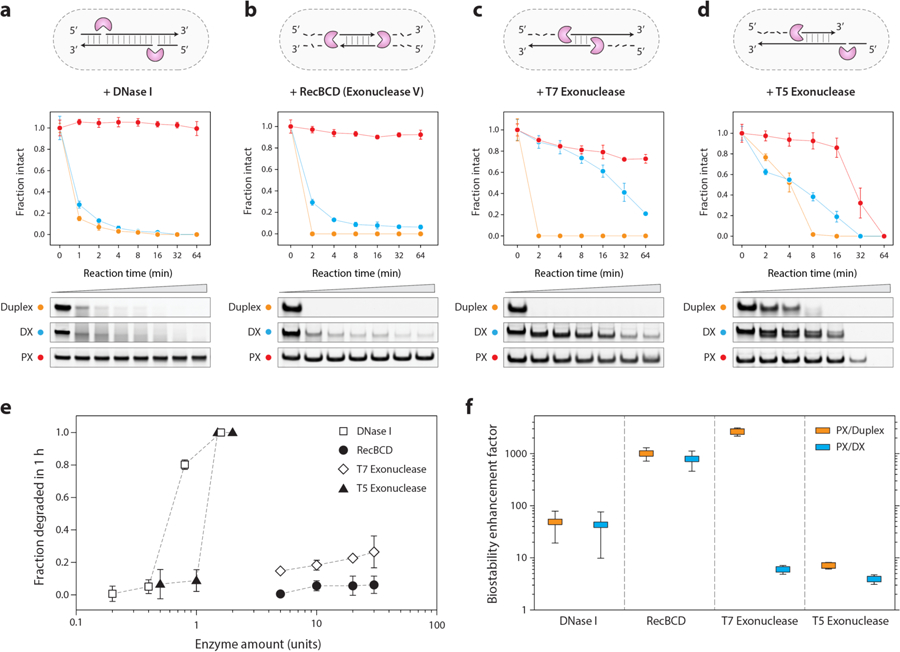
(a) Degradation of DNA motifs treated with 0.1 unit of DNase I enzyme. (b, c, d) Degradation of structures treated with RecBCD (exonuclease V), T7 exonuclease and T5 exonuclease, respectively. The known activity of the tested enzymes on duplex DNA is shown as a cartoon in the corresponding figure panels. (e) Activity of different nucleases on PX DNA. (f) Biostability enhancement factor (fold-increase) of PX DNA as compared to duplex and DX. Error bars are standard deviations calculated from experiments performed in triplicates.
Based on these results, we next tested whether this nuclease resistance of PX holds for nucleases other than DNase I. Since different nucleases can have different activities, mechanisms, substrates, and polarities of digestion, we chose a representative cross section of common nucleases that act on double stranded DNA. We explored three exonucleases in detail, two with 5’ to 3’ directionality (T7 and T5), and one with bidirectionality (Rec BCD). For all three enzymes, we found that PX was the most stable (Figure 2b–d and Figure S10). We then assessed PX degradation by screening different concentrations of these enzymes. We found that different nucleases had vastly different activity on PX DNA. In a one-hour assay, DNase I and T5 exonuclease fully digested the PX DNA with ~1 unit enzyme while RecBCD and T7 exonuclease were unable to fully digest the PX even with the highest possible enzyme amounts (30 units) (Figure 2e and Figure S11).
To estimate the biostability enhancement of PX, we identified an optimal concentration for each enzyme to quantify the decay kinetics of PX, DX, and duplex (Figure S12 & S13). We then calculated the fold change in degradation kinetics for PX relative to DX and duplex (Figure 2f). The enhancement factor of PX/duplex ranges from a low of ~7-fold for T5 exonuclease to a high of ~2800-fold in T7 exonuclease, with an enhancement of T7 Exo > RecBCD > DNase I > T5 Exo. For PX/DX, the trend was RecBCD > DNase I > T7 exonuclease > T5 exonuclease, with lower enhancement values in general. The data suggests that the enhanced biostability of PX is a somewhat general phenomenon, but that certain enzymes struggle more than others with the digestion.
PX DNA does not interfere with fundamental biological processes
The exceptional biostability of PX DNA suggests that the motif may be useful for biological applications of DNA nanotechnology. Considering this important aspect, we asked whether PX DNA interferes with any biological or cellular processes. To address this, we performed cellular viability assay (MTT assay) and cellular differentiation assay (myoblast differentiation assay). We first tested cellular viability in the presence of PX DNA using MTT assays in mouse (C2C12 myoblast) and human (HeLa) cell lines. We incubated the cells either with different concentrations of PX DNA or 1X TAE (control; PX DNA was assembled in 1X TAE) and performed MTT assay (Figure S14). We did not observe any significant changes in cellular viability compared to control cells after 24, 48 and 72 hours of incubation with 100 nM PX DNA (Figure 3a).
Figure 3. PX DNA does not affect cellular viability or influence cellular differentiation.
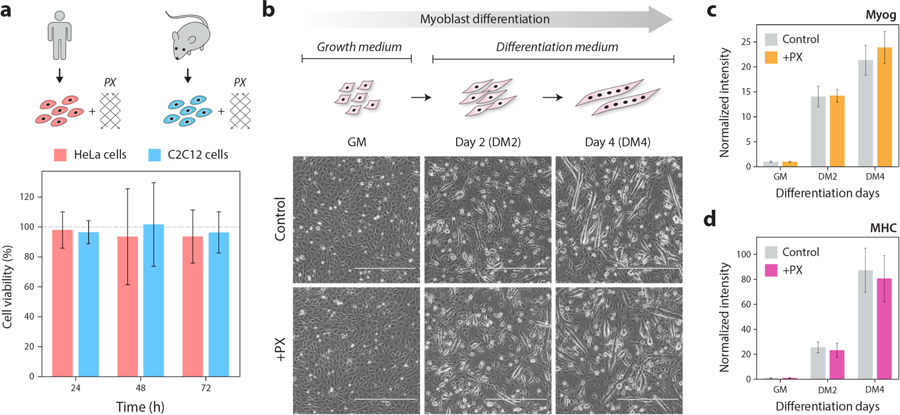
(a) Cell viability from MTT assay when HeLa cells and C2C12 cells were incubated with 100 nM PX DNA for 24–72 hours. Bars represent % of cell viability as compared to the control cells without PX (indicated by grey dashed line). (b) Incubation with PX DNA does not affect differentiation of myoblast cells (C2C12 cell line). Images show undifferentiated (GM), early (DM2) and late (DM4) differentiated cells. Scale bars are 400 μm. (c, d) qRT-PCR results of myogenic differentiation markers Myog and MHC both of which are upregulated in DM2 and DM4 cells in both the control set and cells incubated with PX DNA. The Myog and MHC values are normalized to GM values (as 1) to indicate fold change over time. Values presented are mean ± standard deviation calculated from biological triplicates.
Next, we examined whether PX DNA affects cellular differentiation using C2C12 myoblast differentiation assays. These cells proliferate in the presence of high serum (Growth Medium; GM) and differentiate in low serum (Differentiation Medium; DM). We incubated PX DNA or 1X TAE while culturing the cells either in GM or in DM. We first imaged and later harvested these undifferentiated (GM) and early and late stages of differentiated cells (DM2 and DM4 respectively) for measuring myogenic markers. As shown in the micrographs, PX DNA did not interfere with the normal process of cellular differentiation (Figure 3b). It is apparent from this experiment that PX DNA also did not change the kinetics of differentiation as compared to control (Figure 3b). We further confirmed these findings by quantitating an early myogenic marker, Myogenin (Myog) and a late myogenic marker, Myosin Heavy Chain (MHC) by qRT-PCR analysis. PX DNA did not change the levels of Myog or MHC transcript level (Figure 3c–d). These findings suggest that PX DNA does not affect basic biological or cellular properties and can be used for biological applications in the future.
Removal of crossovers in PX DNA reduces nuclease resistance
Considering the unusual nuclease resistance of PX DNA, we hypothesized that the nuclease activity may be obstructed by the frequent crossovers in the structure. To test this hypothesis, we constructed variations of the PX DNA motif that have sequentially fewer crossover points, called juxtaposed crossover (JX) DNA motifs (Figure 4a). The JX motifs are denoted with numbers signifying the missing crossovers compared to PX structure (e.g. JX2 denotes two missing crossovers) (Figure 4b and Figure S15). Based on the PX, we designed strands to make JX motifs JX1, JX2 and JX3. Following similar protocols for formation of PX, we formed all three structures (Figure 4c and Figure S16). To our knowledge, the JX1 and JX2 structures have been previously demonstrated in the lab,21,29 while JX3 has been simulated by molecular dynamics. Simulations predicted these structures to be stable to varying degrees, with a stability trend of PX > JX1 > JX2 > JX3.30,31 When testing at 37 °C, we found that the JX1 and JX2 structures remained stable but not the JX3, and so we omitted this structure from further experiments (Figure 4d and Figure S17). To test the degradation of the JX1 and JX2 structures, we incubated them with 0.1 unit DNase I for different time periods (Figure 5a and Figure S18). The results, combined with those showing PX, DX, and duplex, clearly demonstrate that there is a hierarchy of nuclease resistance that follows the trend PX > JX1 > JX2 > DX > Duplex. These results support our hypothesis that the nuclease resistance is crossover-dependent.
Figure 4. Construction of PX DNA analogs (called JX) with fewer crossovers.
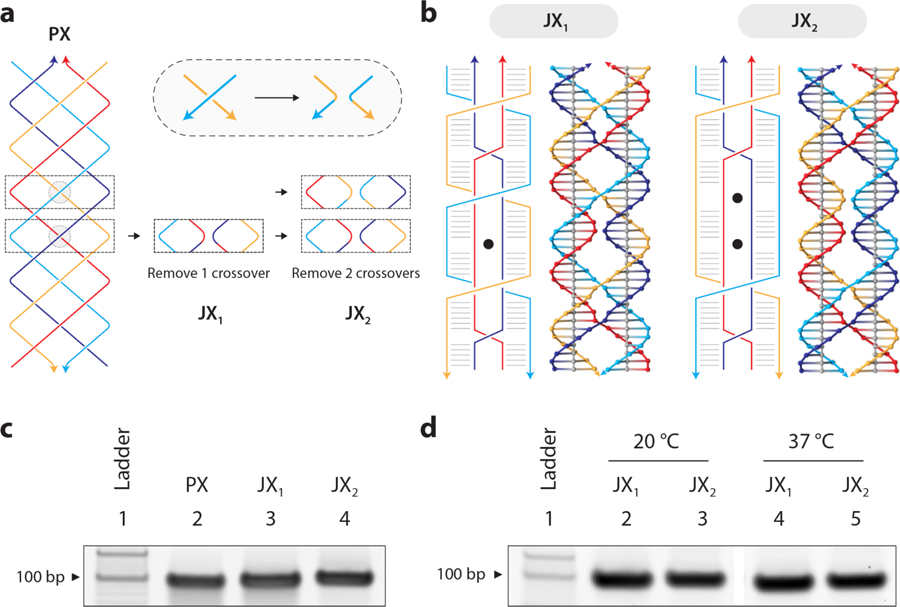
(a) Deriving structures with lesser number of crossovers from PX DNA. (b) Schematic and molecular models of JX1 and JX2 that lack 1 and 2 crossovers respectively (shown as black dots in the structural diagram). (c) Non-denaturing PAGE showing formation of JX1 and JX2 structures. (d) Stability of JX1 and JX2 motifs at 37 °C.
Figure 5. Crossover-dependent biostability.
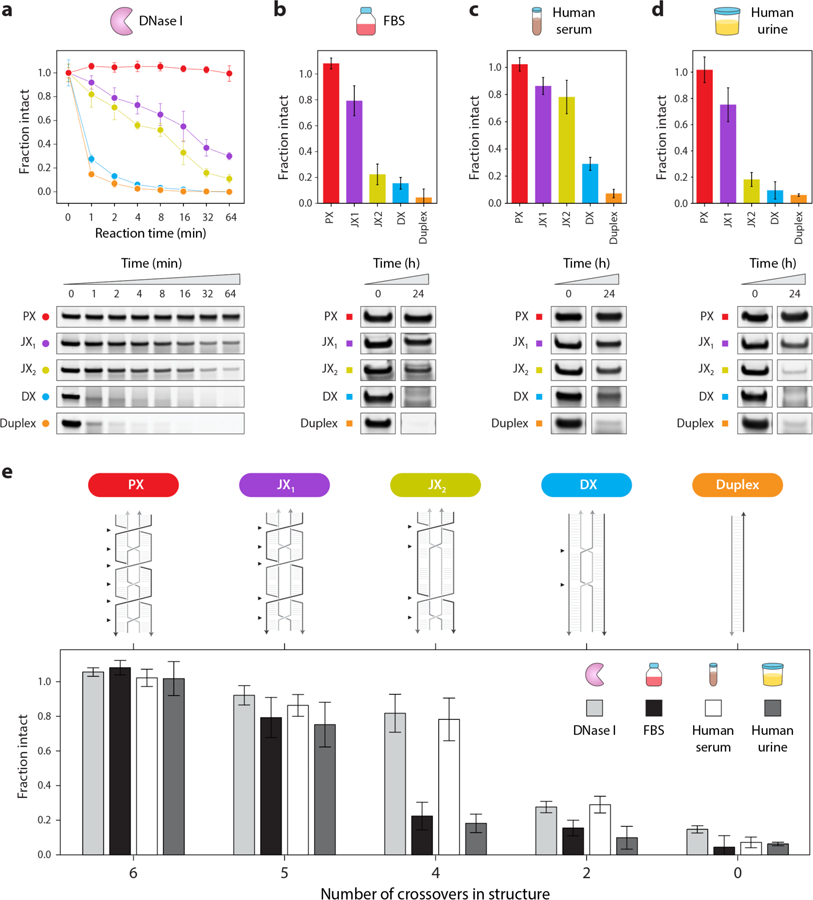
(a) Degradation of PX DNA and control structures when treated with 0.1 unit of DNase I enzyme. (b, c, d) Non-denaturing gels and degradation plots of control structures (duplex and DX) and PX, JX1, and JX2 incubated for 24 hours at 37 °C in 10% fetal bovine serum (FBS), human serum, and human urine respectively. (e) Comparative analysis of the tested structures (PX, JX1, JX2, DX and duplex) in DNase I enzyme (at 1 min reaction time), 10% FBS (at 24 hours), 10% human serum (at 24 hours) and 10% human urine (at 24 hours). Positions of crossovers in each structure is indicated by black arrows. Results show an upward trend in nuclease resistance and biostability with increasing number of crossovers in the structure (PX > JX1 > JX2 > DX > duplex). Error bars are standard deviations calculated from experiments performed in triplicates.
Enhanced, crossover dependent biostability is observed for multiple biofluids
The ultimate goal for many practitioners of DNA nanotechnology is to provide solutions to biomedical problems such as drug delivery and biosensing. In such cases, DNA nanostructures may need to survive for hours, days, or weeks in complex biological fluids such as serum or urine. To see if the exceptional and crossover-dependent stability holds for biological fluids, we tested fetal bovine serum (FBS), human serum and human urine. We incubated each of the five DNA motifs in 10% solutions of these fluids for various time points up to 24 hours at 37 °C, and analyzed the treated samples on non-denaturing PAGE (Figure 5b–d and Figure S19–S20). Similar to the nuclease assays, quantified results showed that PX DNA was the most stable in all cases, with no discernable degradation even after 24 hours. We also tested the duplex, DX and PX DNA in different percentages of FBS and PX was more stable in all cases (Figure S21). The duplex and DX structures, on the other hand, were almost completely degraded in the same conditions while the JX structures were again intermediate. From the results of nuclease resistance and biofluids experiments, we did a comparative analysis of all the tested structures to find out the biostability trend as a function of the number of crossovers in the structure (Figure 5e). The overall stability of the structures in all these conditions was PX > JX1 > JX2 > DX > duplex (with 6, 5, 4, 2 and 0 crossover points respectively), reflecting the effect of crossovers on nuclease resistance and biostability.
Discussion
Here we have discovered that PX DNA has dramatically enhanced biostability compared to normal double stranded DNA that can sometimes exceed 1000-fold. This remarkable difference could have a number of plausible explanations that may merit further study. Enzymatic activity on DNA is known to depend on the helical twist of DNA molecules,32 with DNase I in particular dependent on groove width and flexibility of the duplex.33 Thus, the enhanced resistance of PX DNA to DNase I could be in part due to the difference in helical parameters compared to that of regular DNA duplexes. DNase I has also been previously shown to require a substrate of typically 6–8 base pairs,33,34 and digestion of branched junctions has shown that several nucleotides near the branch point are protected from DNase I cleavage.35 In PX DNA, crossovers occur every half turn and thus the available double helical region between consecutive crossovers is only 5 or 6 baes pairs (alternating half turns). These small regions may make DNase I binding difficult or impossible and slow the degradation. Our results on the JX motifs are consistent with these possible explanations, as removal of each crossover will both extend potential binding regions and relax the DNA in that region. This explanation might also hold true for the other exonucleases we tested, most of which require flexibility and unwinding of the DNA substrate for processing.36
DNA nanostructures have previously been shown to exhibit enhanced nuclease resistance compared to oligonucleotides or plasmid DNA,37 but stability of DNA nanostructures remains a major problem in the field.38 Previous studies have shown that DNA nanostructures incubated with 10% FBS degraded within a few hours.39–41 Some stabilization strategies have been proposed, including heat treatment of FBS,39 addition of actin protein to inhibit nuclease activity,39 or chemical modification of component DNA strands.41 However, these strategies are not without disadvantages. Heat treatment and addition of external proteins can affect the physiological environment and are probably not feasible in vivo, while chemically modified nucleic acids can sometimes be toxic or induce unwanted immune responses.
Our findings suggest that PX DNA based nanostructures should be inherently more biostable than typical DNA nanostructures. While direct quantitative comparison against previous studies is difficult, it is worth noting that our PX DNA showed no signs of degradation at 24 hours in 10% FBS, while DNA origami objects were largely destroyed under similar conditions.39 Since DNA nanostructures tend to be more nuclease resistant than their individual structural components, we predict that larger DNA objects constructed from PX motifs will be more enzyme resistant than duplex based nanostructures. A few DNA nanostructures including the recently developed single-stranded origami42 have utilized PX DNA, but the relative biostability of similar PX and non-PX nanostructures has not yet been investigated to see if our predictions hold true.
Beyond using PX DNA motifs to increase robustness for biological applications of DNA nanostructures, our discovery of crossover-dependent biostability suggests that biostability can be engineered into DNA nanostructures. For example, it could be possible to add protection to structures by strategic placement of crossovers at areas that are especially exposed to nucleases. Such a strategy could lead to DNA nanostructures with a “tunable” biostability dictated by bottom-up design principles. In drug delivery applications, this type of tailored biostability would facilitate timed degradation for fast or slow release of the encapsulated cargo. Further studies in DNA nanostructures containing different number or arrangement of crossovers are certainly needed to see if such speculation is fully grounded in reality. Coming full circle back to our introduction, the sci-fi writers of The Fifth Element were not always (or ever) grounded in reality, but they were inadvertently on to something with their idea of enhancement by more than two DNA strands. At least for the 4-stranded PX DNA structure, perhaps enhanced biostability is “the fifth element”.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the NIH through NIGMS under award R35GM124720 to K.H. and by the American Heart Association under award 17SDG33670339 to B.K.D. We thank Dr. Jibin Abraham Punnoose for discussions on the project, Johnsi Mathivanan for assistance with CD experiments, and Andrew Hayden for the pop-culture reference to multi-stranded DNA in The Fifth Element.
Footnotes
Competing interests
The authors declare no competing interests.
Supporting Information
Materials and methods, sequences used, additional experimental results and full gel images.
References
- (1).Chandrasekaran AR; Rusling DA Triplex-Forming Oligonucleotides: A Third Strand for DNA Nanotechnology. Nucleic Acids Res 2018, 46 (3), 1021–1037. 10.1093/nar/gkx1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lipps HJ; Rhodes D G-Quadruplex Structures: In Vivo Evidence and Function. Trends Cell Biol 2009, 19 (8), 414–422. 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- (3).Seeman NC; Sleiman HF DNA Nanotechnology. Nat. Rev. Mater 2018, 3 (1), 17068 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- (4).Madhanagopal BR; Zhang S; Demirel E; Wady H; Chandrasekaran AR DNA Nanocarriers: Programmed to Deliver. Trends Biochem. Sci 2018, 43 (12), 997–1013. 10.1016/j.tibs.2018.09.010. [DOI] [PubMed] [Google Scholar]
- (5).Agarwal NP; Matthies M; Gür FN; Osada K; Schmidt TL Block Copolymer Micellization as a Protection Strategy for DNA Origami. Angew. Chem. Int. Ed 2017, 56 (20), 5460–5464. 10.1002/anie.201608873. [DOI] [PubMed] [Google Scholar]
- (6).Ponnuswamy N; Bastings MMC; Nathwani B; Ryu JH; Chou LYT; Vinther M; Li WA; Anastassacos FM; Mooney DJ; Shih WM Oligolysine-Based Coating Protects DNA Nanostructures from Low-Salt Denaturation and Nuclease Degradation. Nat. Commun 2017, 8 (1), 1–9. 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Auvinen H; Zhang H; Nonappa; Kopilow A; Niemelä EH; Nummelin S; Correia A; Santos HA; Linko V; Kostiainen MA Protein Coating of DNA Nanostructures for Enhanced Stability and Immunocompatibility. Adv. Healthc. Mater 2017, 6 (18), 1700692 10.1002/adhm.201700692. [DOI] [PubMed] [Google Scholar]
- (8).Anastassacos FM; Zhao Z; Zeng Y; Shih WM Glutaraldehyde Cross-Linking of Oligolysines Coating DNA Origami Greatly Reduces Susceptibility to Nuclease Degradation. J. Am. Chem. Soc 2020, 142 (7), 3311–3315. 10.1021/jacs.9b11698. [DOI] [PubMed] [Google Scholar]
- (9).Perrault SD; Shih WM Virus-Inspired Membrane Encapsulation of DNA Nanostructures To Achieve In Vivo Stability. ACS Nano 2014, 8 (5), 5132–5140. 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kim K-R; Kim HY; Lee Y-D; Ha JS; Kang JH; Jeong H; Bang D; Ko YT; Kim S; Lee H; Ahn D-R Self-Assembled Mirror DNA Nanostructures for Tumor-Specific Delivery of Anticancer Drugs. J. Controlled Release 2016, 243, 121–131. 10.1016/j.jconrel.2016.10.015. [DOI] [PubMed] [Google Scholar]
- (11).Liu Q; Liu G; Wang T; Fu J; Li R; Song L; Wang Z-G; Ding B; Chen F Enhanced Stability of DNA Nanostructures by Incorporation of Unnatural Base Pairs. ChemPhysChem 2017, 18 (21), 2977–2980. 10.1002/cphc.201700809. [DOI] [PubMed] [Google Scholar]
- (12).Cassinelli V; Oberleitner B; Sobotta J; Nickels P; Grossi G; Kempter S; Frischmuth T; Liedl T; Manetto A One-Step Formation of “Chain-Armor”-Stabilized DNA Nanostructures. Angew. Chem. Int. Ed 2015, 54 (27), 7795–7798. 10.1002/anie.201500561. [DOI] [PubMed] [Google Scholar]
- (13).Fu TJ; Seeman NC DNA Double-Crossover Molecules. Biochemistry 1993, 32 (13), 3211–3220. 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- (14).Rothemund PWK Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440 (7082), 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- (15).Wang X; Chandrasekaran AR; Shen Z; Ohayon YP; Wang T; Kizer ME; Sha R; Mao C; Yan H; Zhang X; Liao S; Ding B; Chakraborty B; Jonoska N; Niu D; Gu H; Chao J; Gao X; Li Y; Ciengshin T; Seeman NC Paranemic Crossover DNA: There and Back Again. Chem. Rev 2019, 119 (10), 6273–6289. 10.1021/acs.chemrev.8b00207. [DOI] [PubMed] [Google Scholar]
- (16).Shen Z; Yan H; Wang T; Seeman NC Paranemic Crossover DNA: A Generalized Holliday Structure with Applications in Nanotechnology. J. Am. Chem. Soc 2004, 126 (6), 1666–1674. 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Shih WM; Quispe JD; Joyce GFA 1.7-Kilobase Single-Stranded DNA That Folds into a Nanoscale Octahedron. Nature 2004, 427 (6975), 618 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- (18).Liu W; Wang X; Wang T; Sha R; Seeman NC PX DNA Triangle Oligomerized Using a Novel Three-Domain Motif. Nano Lett 2008, 8 (1), 317–322. 10.1021/nl072803r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ohayon YP; Sha R; Flint O; Liu W; Chakraborty B; Subramanian HKK; Zheng J; Chandrasekaran AR; Abdallah HO; Wang X; Zhang X; Seeman NC Covalent Linkage of One-Dimensional DNA Arrays Bonded by Paranemic Cohesion. ACS Nano 2015, 9 (10), 10304–10312. 10.1021/acsnano.5b04335. [DOI] [PubMed] [Google Scholar]
- (20).Shen W; Liu Q; Ding B; Shen Z; Zhu C; Mao C The Study of the Paranemic Crossover (PX) Motif in the Context of Self-Assembly of DNA 2D Crystals. Org. Biomol. Chem 2016, 14 (30), 7187–7190. 10.1039/C6OB01146B. [DOI] [PubMed] [Google Scholar]
- (21).Yan H; Zhang X; Shen Z; Seeman NC A Robust DNA Mechanical Device Controlled by Hybridization Topology. Nature 2002, 415 (6867), 62 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- (22).Gu H; Chao J; Xiao S-J; Seeman NC A Proximity-Based Programmable DNA Nanoscale Assembly Line. Nature 2010, 465 (7295), 202–205. 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chakraborty B; Jonoska N; Seeman NC A Programmable Transducer Self-Assembled from DNA. Chem. Sci 2011, 3 (1), 168–176. 10.1039/C1SC00523E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang X; Zhang X; Mao C; Seeman NC Double-Stranded DNA Homology Produces a Physical Signature. Proc. Natl. Acad. Sci 2010, 107 (28), 12547–12552. 10.1073/pnas.1000105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gao X; Gethers M; Han S; Goddard WA; Sha R; Cunningham RP; Seeman NC The PX Motif of DNA Binds Specifically to Escherichia Coli DNA Polymerase I. Biochemistry 2019, 58 (6), 575–581. 10.1021/acs.biochem.8b01148. [DOI] [PubMed] [Google Scholar]
- (26).Kizer M; Huntress ID; Walcott BD; Fraser K; Bystroff C; Wang X Complex between a Multicrossover DNA Nanostructure, PX-DNA, and T7 Endonuclease I. Biochemistry 2019, 58 (10), 1332–1342. 10.1021/acs.biochem.9b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Niu D; Jiang H; Sha R; Canary JW; Seeman NC The Unusual and Dynamic Character of PX-DNA. Nucleic Acids Res 2015, 43 (15), 7201–7206. 10.1093/nar/gkv739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Laskowski M 12 Deoxyribonuclease I In The Enzymes; Boyer PD, Ed.; Hydrolysis; Academic Press, 1971; Vol. 4, pp 289–311. 10.1016/S1874-6047(08)60372-8. [DOI] [Google Scholar]
- (29).Spink CH; Ding L; Yang Q; Sheardy RD; Seeman NC Thermodynamics of Forming a Parallel DNA Crossover. Biophys. J 2009, 97 (2), 528–538. 10.1016/j.bpj.2009.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Maiti PK; Pascal TA; Vaidehi N; Goddard WA The Stability of Seeman JX DNA Topoisomers of Paranemic Crossover (PX) Molecules as a Function of Crossover Number. Nucleic Acids Res 2004, 32 (20), 6047–6056. 10.1093/nar/gkh931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Santosh M; Maiti PK Structural Rigidity of Paranemic Crossover and Juxtapose DNA Nanostructures. Biophys. J 2011, 101 (6), 1393–1402. 10.1016/j.bpj.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lomonossoff GP; Butler PJG; Klug A Sequence-Dependent Variation in the Conformation of DNA. J. Mol. Biol 1981, 149 (4), 745–760. 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- (33).Weston SA; Lahm A; Suck D X-Ray Structure of the DNase I-d(GGTATACC)2 Complex at 2·3Å Resolution. J. Mol. Biol 1992, 226 (4), 1237–1256. 10.1016/0022-2836(92)91064-V. [DOI] [PubMed] [Google Scholar]
- (34).Suck D DNA Recognition by DNase I. J. Mol. Recognit 1994, 7 (2), 65–70. 10.1002/jmr.300070203. [DOI] [PubMed] [Google Scholar]
- (35).Lu M; Guo Q; Seeman NC; Kallenbach NR DNase I Cleavage of Branched DNA Molecules. J. Biol. Chem 1989, 264 (35), 20851–20854. [PubMed] [Google Scholar]
- (36).Ceska TA; Sayers JR Structure-Specific DNA Cleavage by 5′ Nucleases. Trends Biochem. Sci 1998, 23 (9), 331–336. 10.1016/S0968-0004(98)01259-6. [DOI] [PubMed] [Google Scholar]
- (37).Keum J-W; Bermudez H Enhanced Resistance of DNA nanostructures to Enzymatic Digestion. Chem. Commun 2009, No. 45, 7036–7038. 10.1039/B917661F. [DOI] [PubMed] [Google Scholar]
- (38).Stephanopoulos N Strategies for Stabilizing DNA Nanostructures to Biological Conditions. ChemBioChem 2019, 20 (17), 2191–2197. 10.1002/cbic.201900075. [DOI] [PubMed] [Google Scholar]
- (39).Hahn J; Wickham SFJ; Shih WM; Perrault SD Addressing the Instability of DNA Nanostructures in Tissue Culture. ACS Nano 2014, 8 (9), 8765–8775. 10.1021/nn503513p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Mei Q; Wei X; Su F; Liu Y; Youngbull C; Johnson R; Lindsay S; Yan H; Meldrum D Stability of DNA Origami Nanoarrays in Cell Lysate. Nano Lett 2011, 11 (4), 1477–1482. 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Conway JW; McLaughlin CK; Castor KJ; Sleiman H DNA Nanostructure Serum Stability: Greater than the Sum of Its Parts. Chem. Commun 2013, 49 (12), 1172–1174. 10.1039/C2CC37556G. [DOI] [PubMed] [Google Scholar]
- (42).Han D; Qi X; Myhrvold C; Wang B; Dai M; Jiang S; Bates M; Liu Y; An B; Zhang F; Yan H; Yin P Single-Stranded DNA and RNA Origami. Science 2017, 358 (6369), eaao2648 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


