Abstract
Offline gains in motor performance after initial motor learning likely depend on sleep, but the molecular mechanisms by which this occurs are understudied. Regulation of mRNA translation via p70 S6 kinase 1 (S6K1) signaling represents one potential mechanism, as protein synthesis is thought to be increased during sleep compared to wake and is necessary for several forms of long-term memory. Using phosphorylation of ribosomal protein S6 (RpS6) as a readout of S6K1 activity, we demonstrate that a period of 10 h of acute sleep disruption impairs both S6K1 signaling and offline gains in motor performance on the rotarod in adult wild type C57/Bl6 mice. Rotarod motor learning results in increased abundance of RpS6 in the striatum, and inhibition of S6K1 either indirectly with rapamycin or directly with PF-4708671 diminished the offline improvement in motor performance without affecting the initial acquisition of rotarod motor learning when sleep is normal. In sum, S6K1 activity is required for sleep-dependent offline gains in motor performance and is inhibited following acute sleep disruption, while motor learning increases the abundance of striatal RpS6. Thus, S6K1 signaling represents a plausible mechanism mediating the beneficial effects of sleep on motor performance.
Keywords: rotarod, mTORC1, ribosomal protein S6, rapamycin, PF-4708671, protein synthesis, mRNA translation
Statement of Significance.
The molecular mechanisms by which sleep can augment particular types of learning and memory remains poorly understood. The current study identifies signaling through S6 kinase 1, a key regulator of mRNA translation, as a putative mechanism by which sleep-dependent offline gains in motor learning may occur in a mouse model. A richer understanding of such molecular mechanisms can help to identify targets capable of enhancing sleep-dependent motor learning and recovery following clinical injuries such as stroke and traumatic brain injury.
Introduction
Offline gains in motor performance have been observed in several motor learning tasks in human subjects including the finger tapping motor sequence task (MST) [1, 2] and mirror drawing task [3]. Such offline gains appear to be influenced by sleep, as the degree of offline improvement was shown to be greater across sleep than across equivalent periods of wake, regardless of whether the period of sleep occurred across the subjective day or night [1]. The benefit of sleep was greater as the complexity of the motor learning task increased [4, 5], and sleep disruption from obstructive sleep apnea (OSA) reduced the degree of benefit from sleep on MST learning [6].
The molecular mechanisms by which sleep imparts such benefits on motor learning are poorly understood. Signaling pathways that regulate new protein synthesis represent intriguing targets as several lines of evidence point toward a significant contribution of new rounds of mRNA translation in mediating rodent motor learning. At the same time, sleep is thought to promote protein synthesis, whereas sleep disruption impairs protein synthesis.
Injection of the protein synthesis inhibitor anisomycin into rat primary motor cortex significantly attenuated skilled reach performance, with no such impairment when anisomycin was injected into parietal cortex or cerebellum [7]. A smaller but significant effect on skilled reach performance was appreciated when anisomycin was injected into dorsal striatum [8]. Anisomycin similarly degraded cortical motor maps resulting from skilled reach training and reduced motor cortical synapse density [9]. Inhibition of protein synthesis with systemic administration of cyclohexamide [10], intraventricular administration of anisomycin [11], or systemic administration of rapamycin [12] impaired rotarod learning.
Despite this generalized role of protein synthesis in motor learning, the precise molecular mechanisms regulating translation remain unknown. Signaling through the mammalian target of rapamycin complex 1 (mTORC1) is a leading possibility as it has been shown to be crucial for other forms of rodent learning and memory [13]. mTORC1, formed by the protein kinase mammalian target of rapamycin (mTOR) and its adaptor protein Raptor, triggers cap-dependent translation initiation through phosphorylation of eukaryotic initiation factor (eIF) 4E-binding proteins (4E-BPs) and S6K1 when activated [14, 15]. Phosphorylated 4E-BP2 releases the translation factor eIF4E, thereby permitting eIF4F (eIF4E + eIF4G + eIF4A) to form and initiation to proceed [16]. mTORC1 also impacts translation by phosphorylating S6K1, which then phosphorylates downstream targets such as eIF4B to promote initiation [17–19], eukaryotic elongation factor 2 kinase (eEF2K) to promote elongation [20], and ribosomal protein S6 [21].
With regard to sleep, radiolabeled amino acid uptake in rat cortex occurred in a state-dependent fashion, with larger increases during slow wave sleep in comparison to wake or rapid eye movement (REM) sleep [22]. A similar finding correlating protein synthesis with deep sleep was observed in non-human primates [23]. Studies comparing transcript expression in sleep versus wake found an upregulation of mRNA for genes associated with mRNA translation, including eukaryotic elongation factor 2 (eEF2) and eukaryotic initiation factor 4A2 (eIF4A2) in sleep [24].
Sleep deprivation, on the other hand, acts to impair signaling pathways that promote protein synthesis. Analysis of mouse hippocampus after sleep deprivation showed a decrease in transcripts related to mRNA translation along with decreased total and phosphorylated levels of mTOR [25]. In the cortex, sleep deprivation resulted in increased phosphorylation of the ER-stress kinase PERK and its downstream target eukaryotic initiation factor 2α (eIF2α), which are both markers of decreased mRNA translation [26]. Finally, rapamycin infused into the visual cortex of cats blocked formation of ocular dominance column plasticity during subsequent sleep. This subsequent sleep resulted in increased phosphorylation of 4E-BP, which was abrogated if sleep was instead deprived [27].
These observations raise the possibility that mTORC1 signaling is required for motor learning and that sleep disruption following motor learning impairs this signaling pathway. The current study focuses on mTORC1 signaling, specifically through the S6K1 signaling pathway. We demonstrate that acute sleep disruption impairs motor learning, that motor learning itself results in increased striatal RpS6 levels without a change in the phosphorylated fraction, and that pharmacological attenuation of S6K1 signaling preceding motor learning impairs the expected offline change.
Methods
Animals
Adult C57BL/6 male mice (2–6 months of age) were kept on a 12 h/12 h light/dark schedule with lights on at 9:00 am (zeitgeber time (ZT) 0). Mice were group housed (4–5 mice per cage), and food and water were available ad libitum, including during periods of sleep disruption. Mice undergoing surgery were transiently housed singly. All experiments were approved by the Institution of Animal Care and Use Committee of both New York University and Mount Sinai School of Medicine and were carried out in accordance with all National Institutes of Health guidelines.
Surgery
Surgical and implantation procedures were performed as previously described [28]. Mice were anesthetized with inhaled isoflurane and placed in a stereotaxic apparatus (David Kopf). After exposing the skull, five electrodes were positioned. Two subdural electrodes (2.5 mm diameter screws with tapered tips, Pinnacle Technologies), symmetrically placed over left and right primary motor cortices (1.5 mm anterior to Bregma, ±2.0 mm lateral to the midline) served as electroencephalogram (EEG) electrodes. Two epidural screw electrodes were placed above the cerebellum to serve as reference and ground. A bipolar, twisted stainless steel electrode (California Fine Wire Co.) inserted into the nuchal muscles served as an electromyogram (EMG) site. After implantation, a six-pin connector (Millmax) was centered over the skull with dental cement (Dentsply) and the animal was placed in its home cage on top of a heating pad set to 37°C (Harvard Apparatus) until fully ambulatory. All animals were supplied with subcutaneous hydration and pain control (buprenorphine) following surgery.
Sleep recordings and sleep disruption
EEG/EMG data acquisition
Mice were housed individually for at least a week after surgery in the room where EEG was conducted so that they would acclimate to the recording environment. A 24 h tethered session served as acclimation prior to sleep recordings. Recordings were performed in a cylindrical chamber with an approximately 12 inch base coupled with a multichannel commutator (Pinnacle Technologies) to allow freedom of movement with access to food pellets and water over the 24 h recording session.
Signals were acquired at 1000 Hz sampling rate and bandpass filtered from 0.5 to 100 Hz (Pinnacle). Simultaneous video was recorded continuously at 10 frames per second (synchronized with the EEG record) during both light and dark periods using an infrared LED camera with Sirenia Video Acquisition software (Pinnacle).
EEG/EMG data analysis
Data analysis was performed using MATLAB (MathWorks) with the Statistics/Signal Processing toolboxes and the FieldTrip toolbox [29].
Sleep/wake analysis
Sleep/wake scoring was performed as previously described [28]. Video-EEG/EMG was analyzed continuously to characterize behavioral states in 1 s epochs. Behavioral states (wakefulness, non-rapid eye movement (NREM) and REM sleep) were classified in an epoch-free approach based on these criteria:
-
•
Time-varying ratio of theta over delta power (θ, 5–10 Hz; δ, 1–4 Hz) using both the right and left primary motor cortex lead.
-
•
Presence of slow waves (delta power, 1–4 Hz) defined as segments with greater than 1 zscore analyzed from both the right and left primary motor cortex lead across the entire recording.
Movement, detected by EMG and confirmed by simultaneous manual review of video.
REM sleep was defined by a high ratio of theta/delta power (ratio >2.5), and little or no movement of the body (based on EMG <1 zscore). In addition, a criterion for REM sleep was that the prior behavioral state was NREM sleep (which is the normal pattern for sleep in rodents). REM sleep epochs separated by less than 3 s were merged because these were periods when small twitches or slight posture changes appeared to interrupt an otherwise continuous period of REM sleep. If movement was less than 1 zscore, but other criteria for REM were not met, the behavioral state was classified as NREM sleep or quiet wakefulness. NREM was discriminated from quiet wakefulness based on power in the delta band and presence of putative spindles. Thus, NREM sleep showed a lower ratio of theta/delta power (<2.5) than quiet wakefulness. Sleep episodes were confirmed manually by reviewing video and finding that mice had a curled position. Periods with relatively low delta power (<1 zscore) and minimal movement (<1 zscore) were designated as quiet wakefulness. Periods with movement for greater than 3 s were classified as active wakefulness and included exploration/walking, grooming, sniffing, consummatory behavior (eating/drinking), and arousals from sleep (both spontaneous and induced by the sleep disruption chamber). All spectral thresholds were verified manually for each recording. Continuity of behavioral states was assessed using a resampling cumulative distribution approach [30, 31].
Mechanical sleep disruption
Animals were placed in a custom designed chamber with an approximately 12 inch diameter slowly rotating round floor in which wires forming an “X” hung about 1 cm above the floor. During sleep, a somatosensory stimulus created by the motionless mouse meeting the wire occurred automatically once every 10 s. Food and water were available ad libitum. Sleep disruption occurred during the light phase between ZT2 and ZT12.
Rotarod
Mice were habituated to the room containing the rotarod for 30 min (ZT0 to ZT0.5) before initiating behavioral training. Mice were placed on an accelerating rotarod (Ugo Basile) in which the rod accelerated from 4 to 40 RPM over 300 s. Mice completed 10 consecutive rotarod trials with an inter-trial interval of 3 min. Each trial terminated when the mouse fell off the rod, when the animal clung to the rod for a full 360 degree rotation, or when the maximum time of 300 s was reached. All rotarod training took place between ZT0.5 and ZT2.
Drug preparation and administration
Rapamycin (LC Laboratories, Woburn, MA) was dissolved in a vehicle solution of 1% dimethylsulfoxide (DMSO), 5% Tween-80 and 1× phosphate buffered saline (PBS) and injected at a dose of 40 mg/kg. PF-4708671 (Tocris, Bristol, England, UK) was dissolved in a vehicle solution of 17% DMSO, 10% Tween-80 and 1× PBS and injected at a dose of 50 mg/kg. All drugs were injected intraperitoneally at volumes of 5–10 mL/kg relative to body weight. Control mice received equivalent volumes of vehicle solution.
Western blotting
Mice were killed by cervical dislocation immediately following rotarod motor learning, brains were microdissected for striatum and cerebellum, and tissue was snap-frozen on dry ice. The tissue was homogenized and analyzed using standard Western blotting procedures as previously described [32]. Primary antibodies for Western blotting included phospho-RpS6 (240/244) (1:1000) (cat # 2215), total RpS6 (1:1000) (cat # 2217) (Cell Signaling Technology, Danvers, MA), and actin (1:5000) (cat # A2228) (Sigma Aldrich, St. Louis, MO).
Immunohistochemistry
Mouse brains were prepared for immunohistochemistry as described previously with minor modifications [33]. Free-floating sections were cut at 40 µm in the coronal plane with a vibratome, and primary and secondary antibodies were applied in 12-well plates containing 2 mL volume. Following washes, sections were mounted on Plus slides and staining revealed with 3,3′-Diaminobenzidine (DAB). The number of immunoreactive striatal cell soma within a 0.05 mm2 area was tabulated within four separate striatal regions, and the average value per animal per condition was recorded.
Statistical analysis
Data were analyzed using SigmaPlot version 11.0 and Matlab (R 2018b). Data normality testing was done with the Shapiro–Wilk test. Comparisons between sleep physiology variables were performed using paired t-tests for normally distributed data and mean values ± the standard error of the mean are reported. Sleep continuity was assessed as duration of sleep runs, defined as the duration of consecutive epochs of sleep scored as non-REM or REM sleep, terminated by one or more epochs scored as another stage, including wake. Kolmogorov–Smirnov tests were used to compare disrupted versus ad libitum sleep conditions on the survival curves.
For the rotarod motor learning behavior, the primary metric of interest was offline change in performance between day 1 and day 2. We assessed this first by examining the median latency to fall during the first 3 trials of day 2 (F3D2) compared to the median latency to fall during the last 3 trials of day 1 (L3D1) within each condition using Wilcoxon signed rank paired non-parametric tests given that the data were not normally distributed. Mann–Whitney rank sum unpaired non-parametric tests were used to compare the baseline L3D1 between conditions. As a secondary measure of offline change, we calculated the ratio of F3D2/L3D1 for each individual mouse and compared this value across conditions using the Mann–Whitney rank-sum test.
Western blotting and immunohistochemical data were compared within brain region across conditions using unpaired Wilcoxon signed rank non-parametric and parametric t-tests, respectively.
Results
Automated acute sleep disruption reduces and fragments sleep
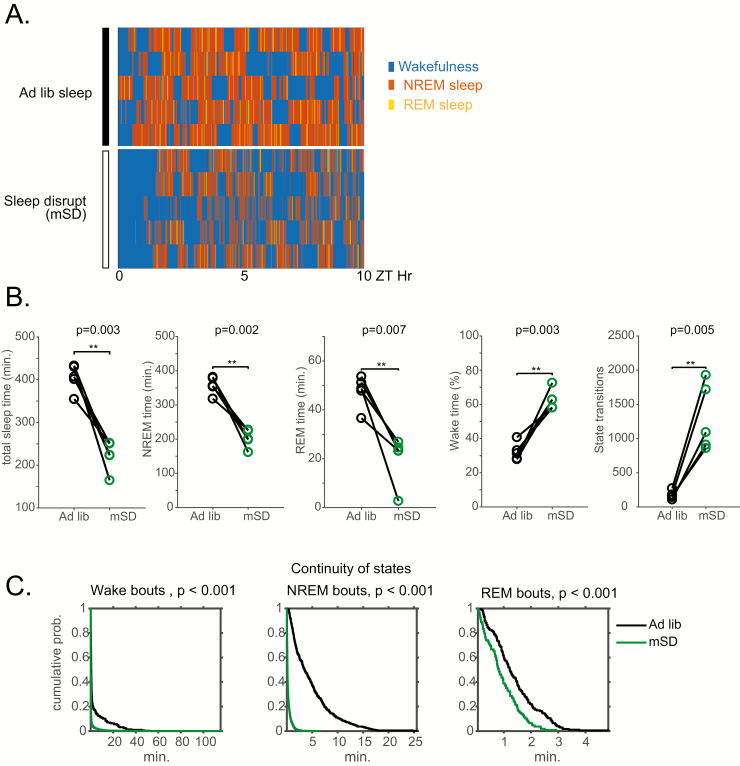
In order to test the effect of mechanical sleep disruption (mSD) on motor learning and S6K1 signaling, mice were placed in an automated sleep disruption chamber in which an intermittent somatosensory stimulus was delivered every 10 s for 10 h between ZT2 and ZT12. In comparison to a period of baseline sleep without the intermittent somatosensory stimulus between ZT2 and ZT12, animals placed in the same chamber experiencing sleep disruption (n = 5) displayed significant decreases in non-REM sleep (202 ± 12 min for mSD vs 358 ± 11 min for ad libitum sleep, paired t-test, p = 0.002) and REM sleep (20 ± 4 min for mSD vs 48 ± 3 min for ad libitum sleep, paired t-test, p = 0.007) and significant increases in sleep fragmentation evidenced by increased state transitions (1301 ± 219 transitions for mSD vs 174 ± 27 transitions for ad libitum sleep, paired t-test, p = 0.005). The increase in sleep fragmentation was also reflected in significant leftward shift in the cumulative duration probability distribution of both non-REM and REM sleep during mSD, indicating that sleep occurred in smaller bouts than during baseline ad libitum sleep. Effects of sleep disruption are summarized in Figure 1.
Figure 1.
Effects of 10 h of automated sleep disruption on sleep physiology. (A) Hypnograms between ZT0 and ZT10 for 5 mice during baseline undisrupted sleep (top) and during 10 h of sleep disruption (mSD) (bottom). (B) Mice undergoing 10 h of sleep disruption show significant decreases in total sleep time, NREM duration, and REM duration, and significant increases in percent wake time and number of state transitions. (C) Survival curves (cumulative probability distributions) of wake, NREM, and REM runs in the normal baseline sleep and sleep disrupted conditions demonstrate significant fragmentation of all vigilance states. ** p < 0.01.
Offline gains in rotarod motor learning are minimized by 10 h of acute sleep disruption
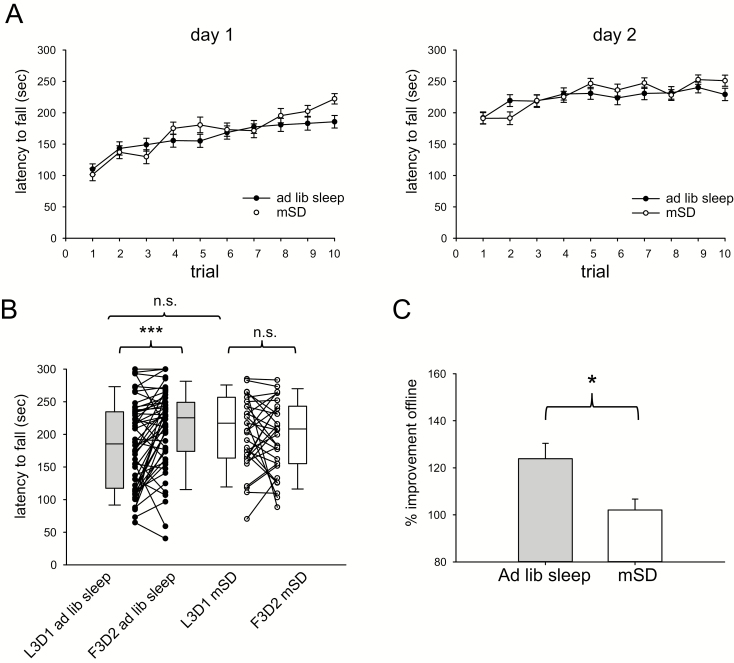
Mice were placed on a rotarod accelerating from 4 to 40 RPM over 300 s for 10 consecutive trials with an inter-trial interval of 3 min. This occurred on 2 consecutive days between the hours of ZT0 and ZT2. Following completion of training on the first day, animals either slept normally (ad libitum sleep) or experienced 10 h of mSD from ZT2 to ZT12 and then returned to their home cages. Offline changes in motor performance were evaluated by comparing the average of the last 3 trials on day 1 (L3D1) to the average of the first 3 trials on day 2 (F3D2). By this metric, mice allowed to sleep ad libitum showed significant improvements (185.3 s (inter-quartile range (IQR) 117–235 s) L3D1 vs 225.3 s (IQR 174–249 s) F3D2, n = 52, p < 0.001, Wilcoxon signed rank test) whereas mice experiencing sleep disruption did not show such improvements (217 s (IQR 164–257 s) L3D1 vs 208.3 s (IQR 155–243 s) F3D2, n = 43, p = 0.90, Wilcoxon signed rank test). Additionally, the individual change for each mouse was expressed as F3D2/L3D1. By this metric, normally sleeping mice displayed significantly greater gain in offline performance than mice experiencing mSD (123.8% ± 7% for ad lib sleep vs 102.1% ± 5% for mSD, p = 0.01). There were no significant differences in mean day 1 performance (p = 0.44) or L3D1 performance (p = 0.08) between mice assigned to the ad libitum sleep and mSD groups. Rotarod performance data as a function of sleep condition is summarized in Figure 2.
Figure 2.
Offline gains in rotarod motor learning are minimized by 10 h of acute sleep disruption. (A) Trial-by-trial rotarod performance on day 1 and day 2 for mice experiencing ad libitum sleep (n = 52) or 10 h acute sleep disruption (mSD) (n = 43). (B) Mice experiencing ad libitum sleep demonstrate significant improvement in the first 3 trials of day 2 (F3D2) compared to the last 3 trials of day 1 (L3D1) whereas mice experiencing 10 h acute sleep disruption do not. There was no significant difference in L3D1 performance between conditions. (C) Offline improvement in motor performance expressed as F3D2/L3D1 was significantly higher in mice experiencing ad libitum sleep vs. acute sleep disruption. *** p < 0.001. * p < 0.05. n.s. = not significant.
Changes in RpS6 but not its relative phosphorylation following rotarod motor learning
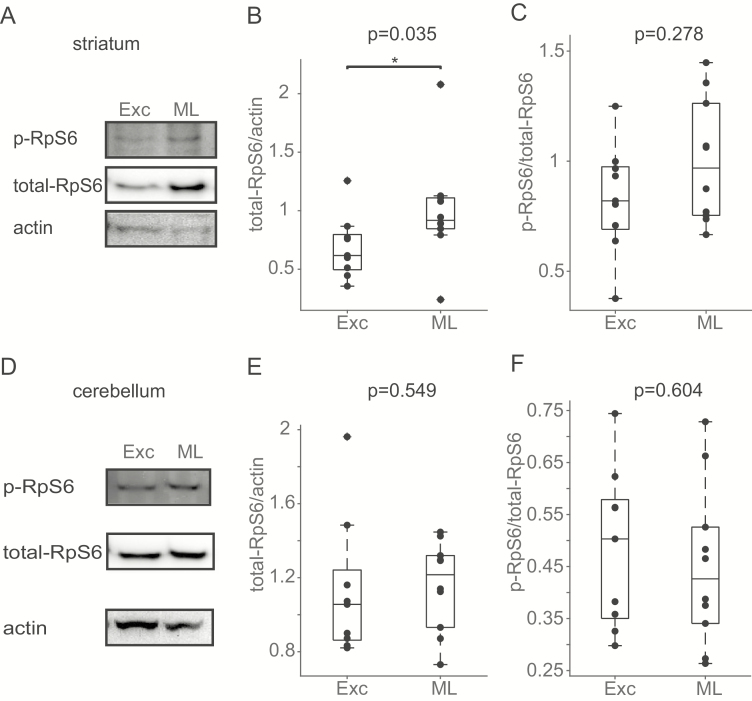
To assess changes to S6K signaling following motor learning, adult male C57BL/6 mice were trained on 10 consecutive trials of rotarod per the same paradigm described in Figure 2 and then immediately sacrificed. Brains were microdissected for bilateral striatum and cerebellum with the abundance of total RpS6 and phospho-RpS6 for each region measured by Western blotting. Control mice underwent 10 consecutive trials of rotarod at a constant 4 RPM across each 5 min trial as an exercise control. Total RpS6 levels (Wilcoxon rank sum p = 0.035, Figure 3B) but not phospho-RpS6 (Wilcoxon rank sum p = 0.278, Figure 3C) were higher in the striatum immediately after motor learning compared to exercise. The cerebellum did not exhibit such differences for both total-RpS6 (Wilcoxon rank sum p = 0.549, Figure 3E) or phosphor-RpS6 (Wilcoxon rank sum p = 0.604, Figure 3F).
Figure 3.
Total and phosphorylated RpS6 protein levels following motor learning. (A) Example Western blotting bands from striatum for phosphorylated and total RpS6 and actin following motor learning (ML) or the exercise control (Exc) condition. Western blotting immunoreactivity to total RpS6 was greater in striatum after motor learning compared to exercise control (B) without a significant increase in the ratio of phosphorylated RpS6 at site Ser240/244 (C). (D) Example Western blotting bands from cerebellum for phosphorylated and total RpS6 and actin following motor learning (ML) or the exercise control (Exc) condition. No differences in Western blotting immunoreactivity to total RpS6 (E) or phosphorylated RpS6 (F) were observed in the cerebellum following motor learning compared to the exercise control. N = 10 mice per condition. * p < 0.05.
Reduction in RpS6 phosphorylation following 10 h of acute sleep disruption
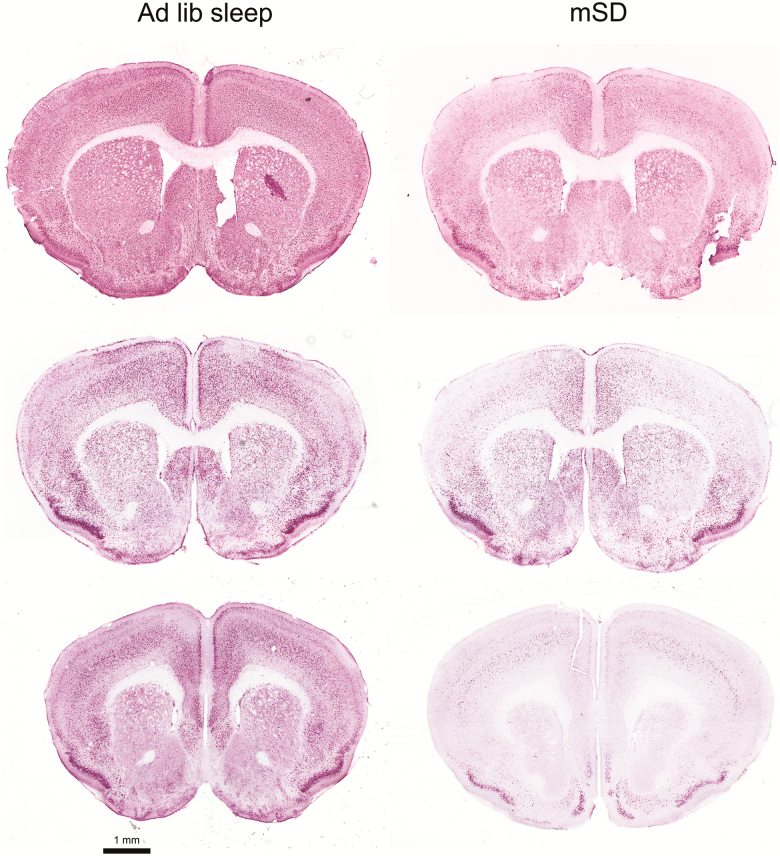
In order to examine the isolated effect of 10 h of acute mSD on phosphorylation of RpS6, naïve animals experienced mSD between ZT2 and ZT12 and were subsequently sacrificed with brains prepared for immunohistochemistry. Immunoreactivity to phosphorylated RpS6 (Ser 240/244) was reduced across several brain regions (Figure 4), with mice experiencing mSD showing reduced numbers of immunoreactive striatal cell soma (17.4 ± 1.0 cells per unit area following ad libitum sleep vs 7.8 ± 3.1 cells per unit area following mSD, p = 0.042).
Figure 4.
Ten (10) h of acute sleep disruption results in decreased phosphorylation of RpS6. Anterior coronal brain sections showing immunohistochemical immunoreactivity to phosphorylated RpS6 (Ser240/244) from ad libitum sleeping (left column) and sleep disrupted (mSD) mice (right column).
Pharmacological inhibition of mTORC1 or S6K1 signaling impairs offline gains in rotarod motor learning
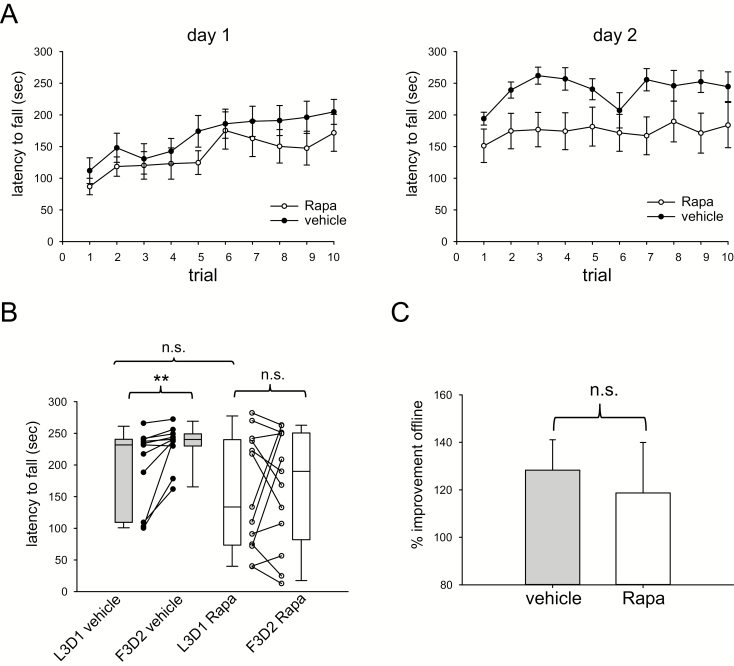
Having established that total RpS6 protein levels increase in the striatum following motor learning, we investigated the necessity of mTORC1 and S6K1 signaling in mediating the offline changes in motor learning. Mice were treated systemically with either the mTORC1 inhibitor rapamycin (40 mg/kg) or with the S6K1 inhibitor PF-4708671 (50 mg/kg) 1 h before the onset of rotarod trials. Biochemical effects of these drugs in reducing RpS6 phosphorylation are shown in Supplementary Figure S1. Mice treated with vehicle and allowed to sleep ad libitum showed significant offline improvements (231.7 s (IQR 109–231 s) L3D1 vs 240.3 s (IQR 230–249 s) F3D2, n = 11, p = 0.002, Wilcoxon rank-sum test) whereas mice receiving rapamycin sleeping ad libitum did not show such improvements (133.7 s (IQR 73–240 s) L3D1 vs 190 s (IQR 82–251 s) F3D2, n = 13, p = 0.84, Wilcoxon rank-sum test). However, the individual gain for each mouse expressed as F3D2/L3D1 was not different between the two groups (128.3% ± 13% for vehicle vs 118.6% ± 21% for rapamycin, p = 0.71, t-test). Although performance appeared to plateau earlier on day 1 in rapamycin treated mice, there were not differences in mean day 1 performance (p = 0.23) or L3D1 performance (p = 0.35) between the two groups. The effect of rapamycin on offline change in motor learning is shown in Figure 5.
Figure 5.
Inhibition of mTORC signaling with rapamycin prior to motor learning impairs offline gains in motor performance. (A) Trial-by-trial rotarod performance on day 1 and day 2 for mice treated with rapamycin (Rapa, n = 13) or vehicle control (vehicle, n = 11). (B) Mice treated with vehicle demonstrate significant improvement in the first 3 trials of day 2 (F3D2) compared to the last 3 trials of day 1 (L3D1) whereas mice treated with rapamycin do not. There was no significant difference in L3D1 performance between treatments. (C) Offline improvement in motor performance expressed as F3D2/L3D1 was not significantly different between the two treatments. ** p < 0.01. n.s. = not significant.
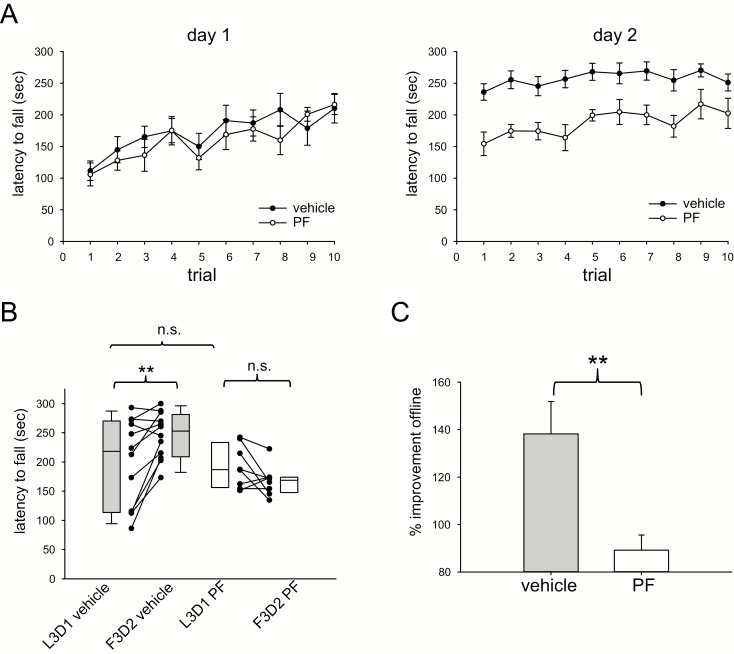
In comparison, in experiments in which treatment with vehicle was compared to treatment with the S6K1 inhibitor PF-4708671, mice treated with vehicle and allowed to sleep ad libitum showed significant offline improvements (218.2 s (IQR 113–270 s) L3D1 vs 252.8 s (IQR 209–281 s) F3D2, n = 12, p = 0.009, Wilcoxon rank-sum test) whereas mice receiving PF-4708671 sleeping ad libitum did not show such improvements (186.8 s (IQR 156–233 s) L3D1 vs 168.5 s (IQR 148–174 s) F3D2, n = 8, p = 0.16, Wilcoxon rank-sum test). In contrast to the results with rapamycin, the individual gain for each mouse expressed as F3D2/L3D1 was significantly lower in PF-4708671 treated mice compared to vehicle treated mice (138.2% ± 14% for vehicle vs 89.2% ± 6% for PF-4708671, p = 0.01, t-test). There were not differences in mean day 1 performance (p = 0.56) or L3D1 performance (p = 0.67) between the PF-4708671 and vehicle treated groups. The effect of PF-4708671 on offline change in motor learning is shown in Figure 6.
Figure 6.
Inhibition of S6 kinase signaling with PF-4708671 prior to motor learning impairs offline gains in motor performance. (A) Trial-by-trial rotarod performance on day 1 and day 2 for mice treated with PF-4708671 (PF, n = 8) or vehicle control (vehicle, n = 12). (B) Mice treated with vehicle demonstrate significant improvement in the first 3 trials of day 2 (F3D2) compared to the last 3 trials of day 1 (L3D1) whereas mice treated with PF-4708671 do not. There was no significant difference in L3D1 performance between treatments. (C) Offline improvement in motor performance expressed as F3D2/L3D1 was significantly higher in mice treated with vehicle vs. PF-4708671. ** p < 0.01. n.s. = not significant.
Discussion
In the current study, we demonstrated that offline gains in motor performance on rotarod occur when followed by a period of ad libitum sleep, and such gains are significantly attenuated when followed by a 10 h period of sleep disruption. This observation is consistent with prior observations showing that 7 h of sleep disruption following rotarod learning significantly attenuates offline gains in performance [34], but differs from work showing a benefit of sleep only on a more challenging complex wheel task, but not on standard rotarod protocols [5]. Factors contributing to such differences are most likely to include the age (and thus weight) of the mice, the genotype, the sex distribution, and precise parameters of rotarod acceleration. A benefit of sleep and detriment of acute sleep disruption has been observed in alternative motor learning tasks such as the mouse skilled reaching task [35].
Prior work has demonstrated that acute sleep disruption results in decreased mRNA translation [36, 37], likely through sleep disruption-associated decreases in mTOR at the transcript and protein level, as well as decreased phosphorylation of mTOR, which is thought to reflect reduced mTORC1 activity [25]. However, investigations into how acute sleep disruption impacts signaling downstream of mTORC1 have been limited. Mice experiencing acute sleep disruption via gentle handling for 5 h were shown to have decreased levels of hippocampal 4E-BP phosphorylation, but surprisingly, no significant changes in phosphorylation of either S6K1 or its downstream target RpS6 in the hippocampus at the Ser235/236 site [37]. It bears noting that multiple kinases have the capacity to phosphorylate the Ser235/236 site, whereas only S6 kinase is known to phosphorylate the Ser240/244 site [38, 39] Our current observations of decreased RpS6 phosphorylation may be related to longer duration of sleep disruption (10 h), method of sleep disruption (gentle handling vs. automated methods), or precise phosphorylation site as we examined the Ser240/244 site. Of note, 5 h of sleep disruption using an automated device (Pinnacle) resulted in significant reductions in S6K1 phosphorylation in hippocampus [40].
We found that motor learning increases striatal total RpS6 protein compared to an exercise control. While the fraction of phosphorylated RpS6 was not significantly changed following motor learning, the ability to maintain levels of RpS6 phosphorylation at the Ser240/244 site in the face of increased abundance of RpS6 proteins implicates required S6 kinase activity. Inhibition of mTORC1 signaling with rapamycin has been shown to impair offline gains in motor learning previously [12]. Our current findings support and extend this observation by demonstrating that inhibition of S6K1 alone is sufficient for the impairment in offline motor learning gains, as such offline gains in motor learning were minimized by inhibition of S6K1 with PF, leaving signaling through 4E-BP intact. PF did not impair the acquisition of motor learning on day 1, and, although rapamycin treated animals appear to trail off in trials 7–10 on day 1, there was no statistically significant difference in mean latency to fall between rapamycin and vehicle treated mice in the last L3D1. The mutual observations that sleep disruption impairs RpS6 phosphorylation (suggesting reduced S6K1 activity) and that blocking S6K1 activity diminishes the offline gains in motor learning suggest that reduced S6K1 activity is a plausible mechanism by which acute sleep disruption mediates its deleterious effects. If this mechanism is accurate, rescue of S6K1 activity during periods of sleep disruption would be expected to counteract the deleterious effect of sleep disruption on the offline gains in motor learning. Future experiments may take advantage of viral constructs expressing constitutively active S6K1 [41] to increase S6K1 activity during periods of mSD in a brain-region specific fashion, as one limitation of the current work is the lack of anatomical specificity associated with use of systemic pharmacological injections.
Cortical synaptogenesis [42] and dendritic spine formation [34, 43] are observed in the cortex following motor training, and the role of S6K1 in regulating de novo protein synthesis may be important for these processes. S6K1 has also been implicated in actin dynamics largely in non-neuronal cells [44], but there is evidence S6K1 can impact the neuronal cytoskeleton [45–47]. Alternatively, the AMPA receptor subunit GluR1 has been identified as a substrate for S6K1 at T840 [48], and S6K1-mediated phosphorylation at this site has been implicated in fear extinction learning [49].
In summary, we have identified that offline gains in rotarod motor learning are dependent on both sleep and S6K1 activity. Acute sleep disruption impinges on S6K1 signaling and may serve as a mechanistically important contributor to the neuronal plasticity involved in the offline gains in motor learning. Future work will be needed to confirm this role and identify both the downstream mechanisms and neural circuits utilizing such plasticity processes during sleep.
Supplementary Material
Acknowledgments
We thank Drs. Ricardo Osorio, Indu Ayappa, and David Rapoport for reading the manuscript and providing useful insights. This work was supported by the philanthropy of the James Kuhn Friends of Sleep Medicine, the Leon Levy Foundation Neuroscience Fellowship (A.W.V.), the American Sleep Medicine Foundation Bridge to Success Award (A.W.V.), a Friedman Brain Institute Scholars Award (A.W.V.), NINDS grants NS034007 and NS047384 (E.K.), and by NIA grants AG056682 and AG059179 (A.W.V.).
Conflict of interest statement. None declared.
References
- 1. Fischer S, et al.. Sleep forms memory for finger skills. Proc Natl Acad Sci USA. 2002;99(18):11987–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walker MP, et al.. Sleep-dependent learning and memory consolidation. Neuron. 2004;44(1):121–133. [DOI] [PubMed] [Google Scholar]
- 3. Smith C, et al.. Impaired motor memory for a pursuit rotor task following Stage 2 sleep loss in college students. J Sleep Res. 1994;3(4):206–213. [DOI] [PubMed] [Google Scholar]
- 4. Kuriyama K, et al.. Sleep-dependent learning and motor-skill complexity. Learn Mem. 2004;11(6):705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagai H, et al. Sleep consolidates motor learning of complex movement sequences in mice. Sleep. 2017;40(2). doi: 10.1093/sleep/zsw059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Djonlagic I, et al.. Increased sleep fragmentation leads to impaired off-line consolidation of motor memories in humans. PLoS One. 2012;7(3):e34106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luft AR, et al.. Motor skill learning depends on protein synthesis in motor cortex after training. J Neurosci. 2004;24(29):6515–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wächter T, et al.. Motor skill learning depends on protein synthesis in the dorsal striatum after training. Exp Brain Res. 2010;200(3-4):319–323. [DOI] [PubMed] [Google Scholar]
- 9. Kleim JA, et al.. Functional organization of adult motor cortex is dependent upon continued protein synthesis. Neuron. 2003;40(1):167–176. [DOI] [PubMed] [Google Scholar]
- 10. Luft AR, et al.. Protein synthesis inhibition blocks consolidation of an acrobatic motor skill. Learn Mem. 2004;11(4):379–382. [DOI] [PubMed] [Google Scholar]
- 11. Peng JY, et al.. Protein synthesis is essential not only for consolidation but also for maintenance and post-retrieval reconsolidation of acrobatic motor skill in rats. Mol Brain. 2009;2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bergeron Y, et al.. mTOR signaling contributes to motor skill learning in mice. Front Mol Neurosci. 2014;7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costa-Mattioli M, et al.. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61(1):10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holz MK, et al.. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569–580. [DOI] [PubMed] [Google Scholar]
- 15. Richter JD, et al.. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23(1):1–11. [DOI] [PubMed] [Google Scholar]
- 16. Gingras AC, et al.. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15(7):807–826. [DOI] [PubMed] [Google Scholar]
- 17. Raught B, et al.. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23(8):1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuang E, et al.. Phosphorylation of eukaryotic translation initiation factor 4B (EIF4B) by open reading frame 45/p90 ribosomal S6 kinase (ORF45/RSK) signaling axis facilitates protein translation during Kaposi sarcoma-associated herpesvirus (KSHV) lytic replication. J Biol Chem. 2011;286(48):41171–41182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dennis MD, et al.. Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J Biol Chem. 2012;287(51):42890–42899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, et al.. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20(16):4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shahbazian D, et al.. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25(12):2781–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramm P, et al.. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol Behav. 1990;48(5):749–753. [DOI] [PubMed] [Google Scholar]
- 23. Nakanishi H, et al.. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur J Neurosci. 1997;9(2):271–279. [DOI] [PubMed] [Google Scholar]
- 24. Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41(1): 35–43. [DOI] [PubMed] [Google Scholar]
- 25. Vecsey CG, et al.. Genomic analysis of sleep deprivation reveals translational regulation in the hippocampus. Physiol Genomics. 2012;44(20):981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Naidoo N, et al.. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 2005;92(5):1150–1157. [DOI] [PubMed] [Google Scholar]
- 27. Seibt J, et al.. Protein synthesis during sleep consolidates cortical plasticity in vivo. Curr Biol. 2012;22(8):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kam K, et al.. Interictal spikes during sleep are an early defect in the Tg2576 mouse model of β-amyloid neuropathology. Sci Rep. 2016;6:20119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oostenveld R, et al.. FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput Intell Neurosci. 2011;2011:156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kishi A, et al.. Sleep-stage dynamics in patients with chronic fatigue syndrome with or without fibromyalgia. Sleep. 2011;34(11):1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varga AW, et al.. Apnea-induced rapid eye movement sleep disruption impairs human spatial navigational memory. J Neurosci. 2014;34(44):14571–14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bowling H, et al.. Altered steady state and activity-dependent de novo protein expression in fragile X syndrome. Nat Commun. 2019;10(1):1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Varga AW, et al.. Input-specific immunolocalization of differentially phosphorylated Kv4.2 in the mouse brain. Learn Mem. 2000;7(5):321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang G, et al.. Sleep promotes branch-specific formation of dendritic spines after learning. Science. 2014;344(6188):1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Varga AW, et al.. Effects of acute sleep deprivation on motor and reversal learning in mice. Neurobiol Learn Mem. 2014;114:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wisor JP. The sleep-deprived hippocampus: a loss in translation. Physiol Genomics. 2013;45(1):26–27. [DOI] [PubMed] [Google Scholar]
- 37. Tudor JC, et al.. Sleep deprivation impairs memory by attenuating mTORC1-dependent protein synthesis. Sci Signal. 2016;9(425):ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Biever A, et al.. Ribosomal protein S6 phosphorylation in the nervous system: from regulation to function. Front Mol Neurosci. 2015;8:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pende M, et al.. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24(8):3112–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frolinger T, et al.. Dietary polyphenols promote resilience against sleep deprivation-induced cognitive impairment by activating protein translation. FASEB J. 2018;32(10):5390–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dwyer JM, et al.. Ribosomal protein S6 kinase 1 signaling in prefrontal cortex controls depressive behavior. Proc Natl Acad Sci USA. 2015;112(19):6188–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kleim JA, et al.. Cortical synaptogenesis and motor map reorganization occur during late, but not early, phase of motor skill learning. J Neurosci. 2004;24(3):628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu T, et al.. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462(7275):915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ip CK, et al.. p70 S6 kinase and actin dynamics: a perspective. Spermatogenesis. 2012;2(1):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Franco-Villanueva A, et al.. Neuritic complexity of hippocampal neurons depends on WIP-mediated mTORC1 and Abl family kinases activities. Brain Behav. 2015;5(11):e00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun J, et al.. mTORC1-S6K1 inhibition or mTORC2 activation improves hippocampal synaptic plasticity and learning in Angelman syndrome mice. Cell Mol Life Sci. 2016;73(22):4303–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burnett PE, et al.. Neurabin is a synaptic protein linking p70 S6 kinase and the neuronal cytoskeleton. Proc Natl Acad Sci USA. 1998;95(14):8351–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Delgado JY, et al.. NMDA receptor activation dephosphorylates AMPA receptor glutamate receptor 1 subunits at threonine 840. J Neurosci. 2007;27(48):13210–13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huynh TN, et al.. Activation of a novel p70 S6 kinase 1-dependent intracellular cascade in the basolateral nucleus of the amygdala is required for the acquisition of extinction memory. Mol Psychiatry. 2018;23(6):1394–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.