Abstract
Recently, a novel coronavirus initially designated 2019-nCoV but now termed SARS-CoV-2 has emerged and raised global concerns due to its virulence. SARS-CoV-2 is the etiological agent of “coronavirus disease 2019”, abbreviated to COVID-19, which despite only being identified at the very end of 2019, has now been classified as a pandemic by the World Health Organization (WHO). At this time, no specific prophylactic or postexposure therapy for COVID-19 are currently available. Viral entry is the first step in the SARS-CoV-2 lifecycle and is mediated by the trimeric spike protein. Being the first stage in infection, entry of SARS-CoV-2 into host cells is an extremely attractive therapeutic intervention point. Within this review, we highlight therapeutic intervention strategies for anti-SARS-CoV, MERS-CoV, and other coronaviruses and speculate upon future directions for SARS-CoV-2 entry inhibitor designs.
Graphical Abstract
INTRODUCTION
Coronaviruses (CoVs) are enveloped positive-stranded RNA viruses. They belong to the order of Nidovirales and are classified into four genera: α, β, γ, and δ.1 Coronaviruses are animal viruses with circulating reservoirs in mammals and birds. For most coronaviruses, the lifecycle can be dissected into four steps, including viral entry, replication, assembly, and release.2
Until last year, six strains of coronaviruses have been identified that are pathogenic to humans. Among them are CoV-NL63, CoV-OC43, CoV-HKU1, and CoV-229E that could cause mild respiratory tract diseases.3 However, two of the β-CoVs, the severe acute respiratory syndrome coronavirus (SARS-CoV), and the Middle East respiratory syndrome coronavirus (MERS-CoV) have caused severe epidemics in the past.4,5 In April 2003, SARS-CoV was responsible for 8098 infections, with a fatality rate of ~10% by the end of September 2003.6 MERS-CoV emerged from its zoonotic reservoir in 2012 and infected 2494 people with a fatality rate of ~34% by the end of 2019.7 Both outbreaks having such high fatality rates, highlight the need for surveillance of coronavirus emergence. While efforts for the development of antivirals against SARS-CoV or MERS-CoV are still in process, a new coronavirus (SARS-CoV-2) has emerged from an epicenter located in Wuhan, China, in December 2019.8 SARS-CoV-2 is highly contagious and has quickly spread in and beyond China. As of May 28, 2020, there have been more than 5 596 550 diagnosed cases around the world, with 353 373 confirmed deaths (Figure 1).9 The United States of America and Brazil reporting the majority of the confirmed cases in the Americas, with 1 658 896 and 391 222 cases, respectively.
Figure 1.
Countries with reported SARS-CoV-2 infections.10 Countries with reported infections in blue and countries/areas with no reported infections in yellow (North Korea, Turkmenistan, and Western Sahara).
Recently the genome of SARS-CoV-2 was determined, which revealed 80% identity with that of some SARS-CoV strains (GZ02, BJ01, Tor2, SZ3, PC4-227) and interestingly 96% identity to the bat coronavirus BatcoV RaTG13.11 The receptor-binding spike (S) protein is highly divergent from other CoVs and displays nucleotide sequence identities of 75% or less to all other previously described SARS-CoVs. However, again, the new SARS-CoV-2 S protein shares 93.1% identity to the RaTG13 S protein.11
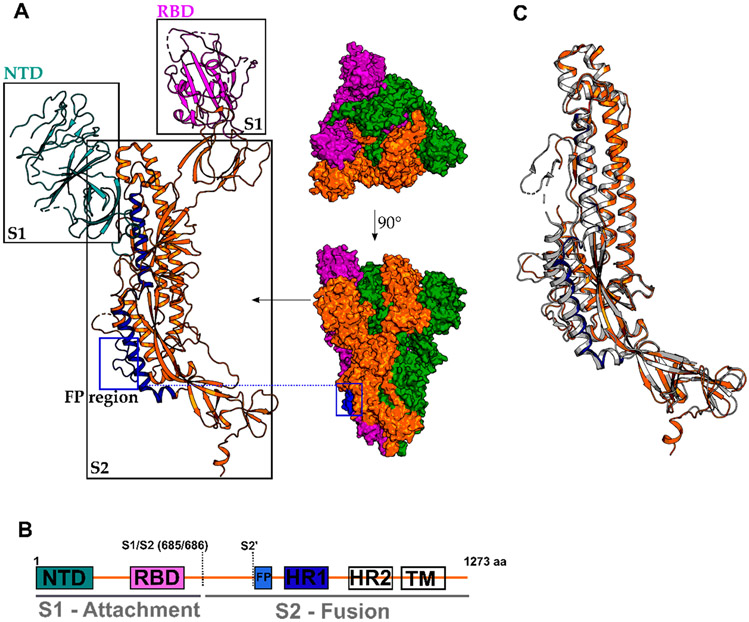
The glycoprotein or S protein is responsible for receptor recognition and viral entry into host cells. The spike protein can be divided into two domains; S1 is responsible for angiotensin-converting enzyme II(ACE2) recognition, the recently identified host cell receptor, and S2 mediates membrane fusion (Figure 2).12 Structural alignment of SARS-CoV-2 S protein with SARS-CoV S protein shows that both S proteins are similarly with a root-mean-square deviation (RMSD) of 3.8 Å over 959 Cα atoms, while the S2 domain, responsible for membrane fusion, display the most substantial similarities with an RMSD of 2.0 Å (Figure 2C).
Figure 2.
Structure of the SARS-CoV-2 Spike (S) protein in its prefusion conformation (PDB 6VSB). (A) Cryo-EM structure of the trimeric and monomeric S protein and (B) domain architecture with colored domains and not resolved/missing regions in white. NTD, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide region; HR1/2, heptad repeat 1/2; TM, transmembrane domain S1/S2; S2′: protease cleavage sites. (C) Structural alignment of SARS-CoV-2 S (in orange and HR1 in dark-blue, PDB 6VSB) and SARS-CoV S (in gray, PDB 6CRZ).
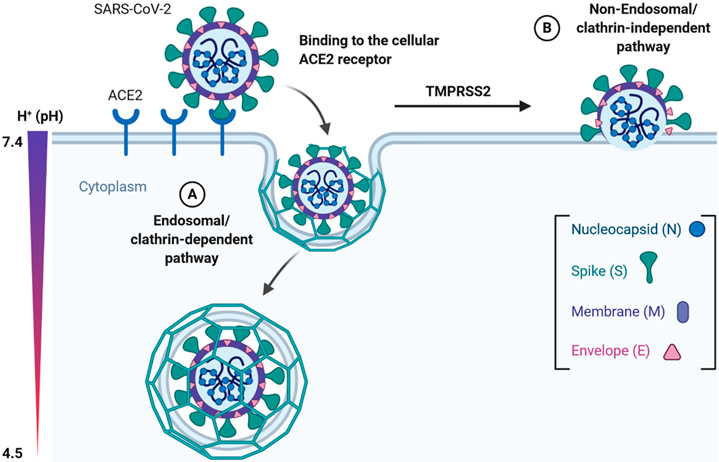
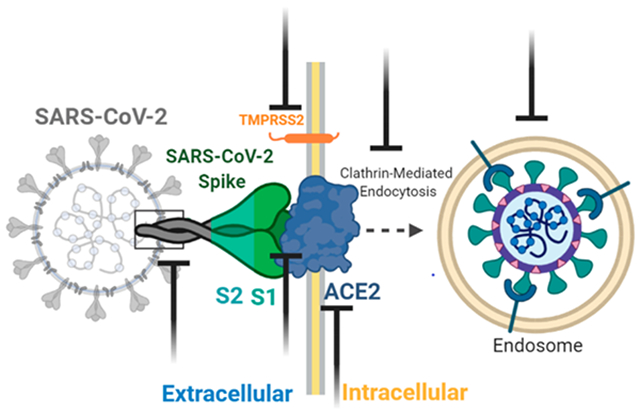
Engagement of the host cell receptor ACE2 is important for viral entry; however, subsequent entry steps can vary and are cell-type specific. SARS-CoV can enter the host cell via both clathrin (endosomal) and nonclathrin pathways (nonendosomal); however, both pathways are dependent upon ACE2 binding.13,14 The clathrin-mediated pathway includes the S protein binding to ACE2 and subsequent dynamin/clathrin-mediated internalization of endosomal vesicles that maturate to late endosomes. Within the late endosomes and lysosomes, acidification of the internalized endosomes and H+-dependent activation of the cellular cathepsin L proteinase takes place that cleaves and activates the S protein, therefore initiating viral fusion with the endosomal/lysosomal membrane (Figure 3). In the case of SARS-CoV, cell culture studies revealed that the entry process is delayed with a lag phase of around 30 min, suggesting substantial maturation requirements.15 In accordance with findings that mouse hepatitis coronavirus (MHV) and feline coronavirus (FCV) infections of HeLa cells are also heavily dependent on endosomal maturation, the clathrin-dependent entry and endosomal maturation are key to entry across Coronaviridae.16 For SARS-CoV-2, a recent study also confirms that virus can use host cell receptor CD147 to gain entry into the host cells besides ACE2.17
Figure 3.
Entry model of SARS-CoV-2 into the host cell. Binding of the S1 domain within the spike (S) protein to the cellular ACE2 receptor triggers conformational changes in the S2 domain that results in internalization and subsequent membrane fusion ((A) endosomal/clathrin-dependent pathway). The endosomal pathway is facilitated by a low pH and the pH-dependent cysteine protease cathepsin L. Alternatively, SARS-CoV-2 can enter the cell via the nonendosomal/clathrin-independent pathway (B). During this route, ACE2 recognition by the SARS-CoV-2 S protein (comparable to route A) is followed by additional activation/cleavage of the S protein into S1 and S2 domains by cell membrane-associated serine proteases such as TMPRSS2 and TMPRSS11D. The figure was prepared with https://biorender.com/.
In addition to the endosome-mediated entry pathway, host proteases also play critical roles in the nonendosomal entry of coronaviruses.5 Host proteases such as the transmembrane protease serine 2 (TMPRSS2) and TMPRSS11D can cleave the S protein at the S1/S2 cleavage site (Figure 2) to prime and activate the S protein for membrane fusion during the nonendosomal pathway.18 A recent study also confirms that TMPRSS2 expressing VeroE6 cells are highly susceptible to SARS-CoV-2 infection, highlighting the importance of TMPRSS2 in the replication cycle.19
MERS-CoV can also be activated by furin (serine endoprotease) to initiate the nonclathrin mediated membrane fusion event.20 Interestingly, in the new SARS-CoV-2 S protein, additional amino acid insertions at the S1/S2 cleavage site results in an “RRAR” furin recognition site absent in SARS-CoV S protein.21 This polybasic insertion sequence has possible implications for the SARS-CoV-2 replication cycle and its increased pathogenicity. Indeed, polybasic furin sites have been observed in hemagglutinin (HA) proteins of highly virulent avian and human influenza viruses, and similar furin-like processing events are also observed for other RNA viruses such as Ebola virus and Marburg virus, human immune deficiency virus (HIV), and flaviviruses.22
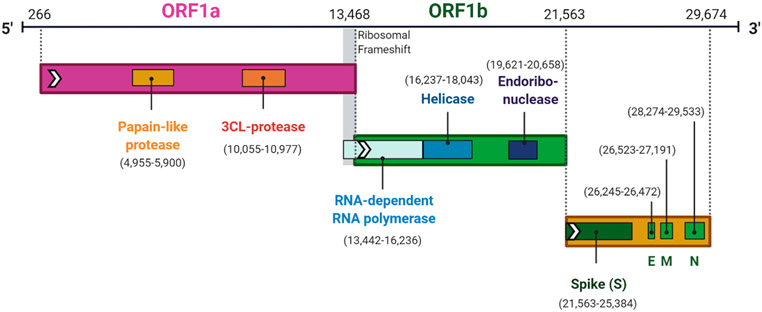
To activate the S protein for membrane fusion with the cellular membrane, structural rearrangements within the S2 domain are required. Two heptad repeats, HR1 (dark blue in Figure 2) and HR2 can interact to form a six-helix bundle (6-HB), a common postfusion structure shared by all type I viral glycoproteins, to bring viral and cellular membranes in close proximity. Additionally, the S2 domain contains a membrane interacting domain or fusion peptide that is exposed upon specific triggers such as receptor binding or low endosomal pH. To date, three membrane interacting regions with host-membrane destabilizing effects have been identified in the SARS-CoV S protein: two conserved sequences across coronaviridae, with residues 798–81523 and residues 864–886,24 both C-terminal positioned at the second cleavage site in the S protein termed S2′ at Arg 797 and a less conserved third region with membrane disordering properties residues 770–788.25 Once in the host cell, the viral particle uncoats and is ready for transcription and translation.26 The first ORF codes for approximately 67% of the genome and is separated into open reading frames (ORF) 1a and 1b (Figure 4). ORF1a and ORF1b are translated into polyproteins pp1a (4382 amino acids) and pp1ab (7073 amino acids) that are processed by 3-C-like protease (3Clpro) and papain-like protease (Plpro). The processing of these polyproteins produces a variety of nonstructural proteins (NSPs), including RNA-dependent RNA polymerase (RdRp) and helicase, to catalyze viral genome replication and protein synthesis.27 The remaining ORFs in the SARS-CoV-2 genome code for accessory and structural proteins. Following further assembly, the mature virions are transported to the cell surface in vesicles and released by exocytosis.28 Any protein involved in the replication process could be a potential target for the development of antiviral agents.
Figure 4.
Genome organization of SARS-CoV-2. Genome organization of the SARS-CoV-2 and location the central genes within the genome (numbers in brackets).29 The figure was prepared with https://biorender.com/.
As mentioned previously, Zhang et al. determined the full-length genome sequence of SARS-CoV-2 and revealed that the virus was very similar (89.1% nucleotide similarity) to a group of SARS-like coronaviruses.30 Simultaneously, Shi et al. found that SARS-CoV-2 shares 96% sequence identity at a whole-genome level to a bat coronavirus, and importantly, they confirmed that SARS-CoV-2 utilizes the same cell entry receptor, ACE2, as SARS-CoV.11 Recently, the cryo-EM structure of full-length human ACE2 bound to the RBD of the SARS-CoV-2 was solved, providing an important structural foundation for intervention strategies.31 Conservation analysis also revealed that the RdRp and the 3CLpro are highly conserved between SARS-CoV-2 and SARS-CoV.32 Therefore, it is widely accepted that SARS-CoV-2 would behave similarly to SARS-CoV with regards to viral entry and replication.
Being the first step in the infection process, the entry of pathogenic viruses into susceptible cells is an extremely attractive intervention point. As with other well-known viruses, such as HIV-1 and Ebola, viral entry of coronaviruses is a complex multiple-step process with numerous interactions and processing points that, in theory, could be targeted.33 In this review, we summarize case studies and highlight efforts in designing entry inhibitors against SARS-CoV, MERS-CoV, and other coronaviruses that can provide important information to combat the current SARS-CoV-2 outbreak.
HOST CELL ACE2 RECEPTOR RECOGNITION BY THE SARS-COV-2 SPIKE (S) AS A PROMISING ANTIVIRAL TARGET
Binding of the SARS-CoV-2 spike (S) protein to the cellular ACE2 receptor represents the first encounter (in both the endosomal and nonendosomal pathway) in the viral replication cycle and provides prophylactic intervention opportunities.34 SARS-CoV-2 spike (S) recognizes with its RBD the cellular ACE2 receptor with high affinity (Kd = 14.7 nM)12 as judged by surface plasmon resonance (SPR) interaction analysis, and intervention at the RBD-ACE2 interface can potentially disrupt infection efficiency.
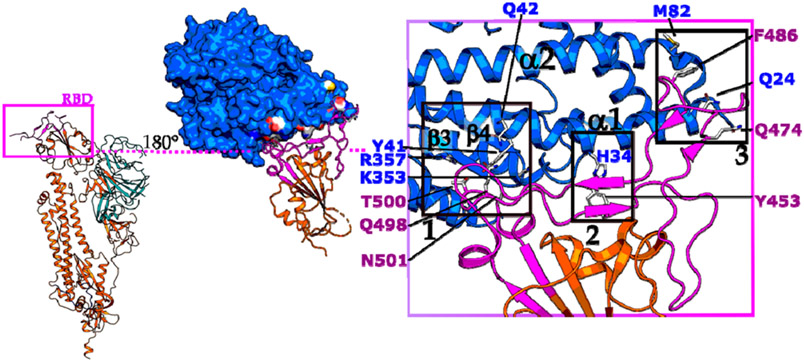
Recently the cryo-EM and crystal structures of SARS-CoV-2’s RBD in complex with ACE2 were solved and provide important structural guidance for inhibitor design (Figure 5).31 The interface can be divided into three contact sides, mainly polar in nature, and is similar to the SARS-CoV-ACE2 complex.35,36 In this structure, an extended loop of the RBD contacts an arch-like helix α1 of the proteolytic domain (PD) of ACE2 via an N- (cluster 1), central (cluster 2), and C-terminal (cluster 3) portion (Figure 5 purple box). Additionally, helix α2 and loop 3–4 (connecting β3 and β4) of ACE2 provide limited contacts. At the N terminus of α1 (cluster 1), Gln498, Thr500, and Asn501 of the RBD interact via hydrogen bonds with Tyr41, Gln42, Lys353, and Arg357 from ACE2. The middle portion (cluster 2) of the RBD loop contacts via Tyr453, the ACE2 PD at residue His34. At the C terminus of α1 (cluster 3), Gln474 of RBD contacts Gln24 of ACE2, and Phe486 of RBD interacts with Met82 of ACE2 through van der Waals interactions (Figure 5).
Figure 5.
SARS-CoV-2-RBD and ACE2 interface. ACE2 (in blue) is contacting via its proteolytic domain (PD) with helix α1 the extended loop region (in purple) of SARS-CoV-2 RBD, mainly via polar interactions. In addition, helix α2 and the loop 3–4 connecting β3 and β4 are also contributing to the interface. SARS-CoV-2 S protein monomer was obtained from PDB 6VSB and RBD-ACE2 complex from PDB 6VW1. Boxes 1, 2, and 3 highlight polar clusters 1, 2, and 3, respectively.
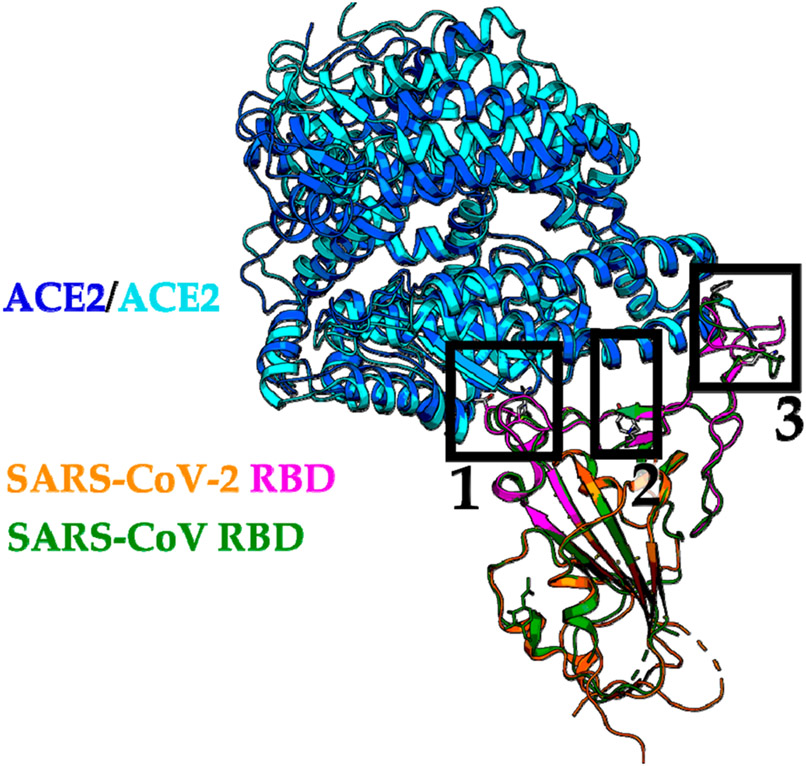
The structures of the RBDs from the SARS-CoV-2-ACE2 complex and the SARS-CoV-ACE2 complex are quite similar, with an RMSD of 0.68 Å over 139 Cα atoms (Figure 6).31 A comparison of both structures, however, also highlights some deviations at all three clusters summarized in Table 1. These deviations need to be considered carefully during the inhibitor design process.
Figure 6.
Structural alignment of the RBD-ACE2 interface from SARS-CoV-2 and SARS-CoV. The SARS-CoV-2 RBD-ACE complex (PDB 6VW1) with ACE2 in blue and RBD in purple/orange are superimposed to the SARS-CoV RBD-ACE complex (PDB 2AJF) with ACE2 in cyan and RBD in green. N-Terminal, central, and C-terminal clusters are highlighted in black boxes with 1, 2, and 3, respectively.
Table 1.
Amino Acid Alterations between SARS-CoV-2 and SARS-CoV RBD-ACE2 Interface
| SARS-COV RBD | SARS-COV-2 RBD (with corresponding altered residue) |
|
|---|---|---|
| cluster 1 (N-terminus) | Arg426, Tyr484, Thr487 | Asn439, Gln498, Asn501 |
| cluster 2 (central) | Val404, Tyr442, Leu443, Phe460, Asn479 | Lys417, Leu455, Phe456, Tyr473, Gln493 |
| cluster 3 (C-terminus) | Leu472 | Phe486 |
Numbering corresponds to the individual RBD–ACE2 complex. For a more detailed insight, we refer to ref 30.
TARGETING THE RBD
Peptide Analogues, Monoclonal Antibodies, and Protein Chimeras as RBD Inhibitors.
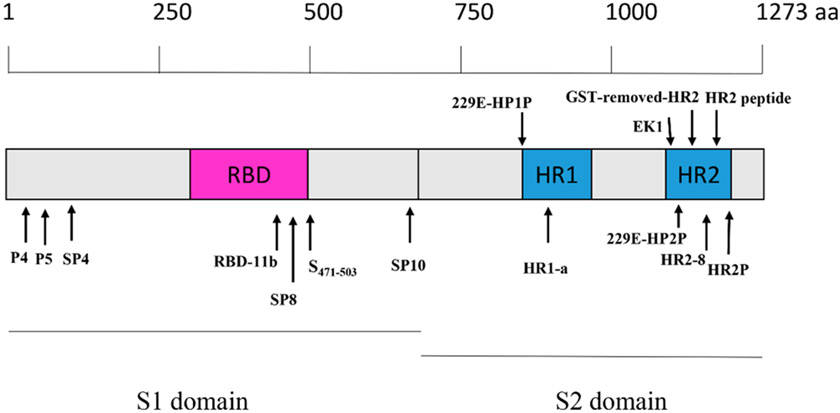
Both SARS-CoV and SARS-CoV-2 use ACE2 to gain entry into the host cells. As such, this critical interaction can be blocked to stop viral entry.19 This strategy was first demonstrated by Hsiang et al. Using a biotinylated enzyme-linked immunosorbent assay (ELISA), Hsiang et al. reported the disruption of the SARS-CoV S protein-ACE2 interaction by small peptides. From a total of 14 designed peptides, peptides SP-4, SP-8, and SP-10 (Figure 7 and Table 2) significantly blocked the interaction of the SARS-CoV S protein with ACE2 with IC50 values of 4.30, 6.99, and 1.88 nM, respectively. Additional immunofluorescence assay (IFA) studies with S-protein-pseudotyped retroviruses, revealed a novel mechanism of infection inhibition of Vero E6 cells by SP-10.37 Structural investigation of the RBD-ACE2 complex by Michael et al. revealed crucial charged residues between positions 22 and 57 for SARS-CoV viral entry. This structural information resulted in the design of two longer peptides P4 and P5 with IC50 values of around 50 and 6 μM, respectively. Glycine linkage of peptide P4 (residue 22–47) with an ACE2 derived peptide (residue 351–357) further improved antiviral activity against a SARS-CoV pseudovirus with an IC50 of 100 nM and no cytotoxicity up to 200 μM.38 In light of the successful inhibition of SARS-CoV with this linked peptide, a similar strategy could potentially be effective against the new SARS-CoV-2. The recently solved cryoEM structure of SARS-CoV-2 in complex with the human ACE2 receptor can provide a structural rationale for the peptide design.31
Figure 7.
Location of synthetic peptides derived from the S1 and S2 domain of the spike protein.
Table 2.
Amino Acid Sequences of Peptide Inhibitors
| peptide | amino acid sequence (from N- to C-terminus) |
|---|---|
| SP-4 | GFLYVYKGYQPI |
| SP-8 | FYTTTGIGYQPY |
| SP-10 | STSQKSIVAYTM |
| EEQAKTFLDKFNHEAEDLFYQSS | |
| P-5 | EEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEE |
| S471–503 | ALNCYWPLNDYGFYTTTGIGYQPYRVVVLSFEL |
| RBD-11B | YKYRYL |
| DX600 | GDYSHCSPLRYYPWWKCTYPDPEGGG |
| HR2-8 | ELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIK |
| HR1-A | YENQKQIANQFNKAISQIQESLTTTSTA |
| GST-REMOVED-HR2 | DVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYI |
| HR2 | ISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL |
| HR2P | SLTQINTTLLDLTYEMLSLQQVVKALNESYIDLKEL |
| HR2P-M2 | SLTQINTTLLDLEYEMKKLEEVVKKLEESYIDLKEL |
| EK1 | SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL |
| 229E-HR1P | AASFNKAMTNIVDAFTGVNDAITQTSQALQTVATALNKIQDVVNQQGNSLNHLTSQ |
| 229E-HR2P | VVEQYNQTILNLTSEISTLENKSAELNYTVQKLTQTLIDNINSTLVDLKWL |
Monoclonal antibodies (mAb) have potential applications for diagnosis, prophylaxis, and treatment of established and evolving viral infections.39-41 Prabhakar et al. isolated specific antibodies from B cells in XenoMouse immunized with SARS-CoV. Further investigation revealed that several Abs directly react with the RDB domain, and a combination of two Abs (4D4 and 3C7) displayed near-complete neutralization efficiency as compared to a single Ab application.42 Two additional potent monoclonal antibodies, mAb201 and mAb68, could be isolated from transgenic mice immunized with the soluble ectodomain of SARS-CoV S protein.43 This mAb could bind SARS-CoV S protein directly with affinities of 34 nM (mAb 201) and 83 nM (mAb 68) as judged by SPR analysis. Mice that received 40 mg/kg of mAb 201 or mAb 68 before SARS-CoV infection showed complete protection from reinfection of lung tissues.43,44 Cross-reactivity of mAbs is highly desirable, and Dimitrov et al. identified the human mAb m396 that binds SARS-CoV with high affinity (Kd = 20 nM).45 Mice that received 200 μg of m396 were nearly completely protected from infection by Urbani and GD03 virus strains.46 M396 did compete with the SARS-CoV receptor, ACE2, for binding to the RBD, suggesting that m396 inhibits SARS-CoV-ACE2 binding as the predominant mechanism of action.45 However, SARS-COV-2 showed some complexities for RBD directed antibodies. For instance, Wrapp et al. tested cross-reactivity of three antibodies, including S230, m396, and 80R, against SARS-COV-2 RBD. Despite the partly high degree of structural homology between the SARS-COV-2 and SARS-COV, no binding to the SARS-COV-2 RBD was detected for any of the three antibodies at the concentration of 1 μM. It can be concluded that SARS-COV antibodies will not necessarily be cross-reactive for SARS-COV-2.12
In a different approach, Hu et al. generated a novel chimeric recombinant protein recently by connecting the extracellular domain of human ACE2 to the Fc region of human immunoglobulin IgG1. These chimeric constructs displayed high-affinity for the SARS-CoV-2 and SARS-CoV RBD binding and potently neutralized SARS-CoV and SARS-CoV-2 in vitro, with IC50 values between 0.8 and 0.1 μM, respectively. These recombinant chimeras also showed cross-reactivity and could have, therefore, useful applications for diagnosis, prophylaxis, and treatment of SARS-CoV-2.47
Using the VelocImmune platform, Pascal et al. generated several human, noncompeting monoclonal antibodies that target MERS-CoV S protein and block viral entry into host cells. Among them, two antibodies, REGN3051 and REGN3048, can significantly inhibit MERS-CoV pseudoparticles, with IC50 values of 460 and 180 pM, respectively.48 In addition, REGN3051 and REGN3048 showed a good performance in a novel transgenic mouse model, which was developed by replacing the mouse DPP4 coding sequence with that encoding human DPP4. Results suggested that both REGN3051 and REGN3048 were able to potently reduce MERS-CoV specific RNA levels in the lungs at a 200 μg per mouse dose compared with the isotype control antibody. At the 20 μg dose, REGN3051 was more effective at decreasing MERS-CoV RNA levels compared with REGN3048 at the same dose.48 Recently, in the common marmoset model of MERS-CoV infection, de Wit et al. tested the prophylactic and therapeutic efficacy of REGN3051 and REGN3048. Data demonstrated that their protection might be more effective in a prophylactic treatment process rather than treatment of MERS-CoV.49 In the latest attempt, Chen et al. identified SARS-CoV-2 RBD specific antibodies from samples of 26 recovered COVID-19 patients using an RBD-specific ELISA binding study. Among them, 311mab-31B5 and 311mab-32D4 effectively neutralized pseudovirus entry, with IC50 values of 0.0338 and 0.0698 μM, respectively.50 Recently, in an ELISA based (cross)reactivity assay, assessing antibody-containing supernatants of a collection of 51 SARS-S hybridoma’s derived from immunized transgenic H2L2 mice that encode chimeric immunoglobulins, Wang et al. identified a chimeric mAb 47D11 that targets RBD. 47D11 exhibited cross-neutralizing activity of SARS-CoV-S protein and SARS-CoV-2-S protein pseudotyped VSV infection with IC50 values of 0.19 and 0.57 μM, respectively.51
Brouwer et al. used cross-sectional blood samples from three PCR-confirmed SARS-CoV-2-infected individuals to screen for binders to a soluble prefusion-stabilized S protein of SARS-CoV-2 using an ELISA-based approach. All three blood samples did bind to the prefusion-stabilized S protein and prompted subsequent sorting of SARS-CoV-2 S protein-specific B cells for mAb isolation. Nineteen Nabs could be identified that target a diverse range of antigenic sites on the S protein and showed remarkable picomolar inhibiting activities with the two most potent IC50 values of 0.010 and 0.007 μg/mL (COVA1-18 and COVA2-15, respectively) against live SARS-CoV-2 virus.52
Large antibody libraries are crucial in response to rapidly emerging pathogens. Using eight large phage-displayed VH, scFv, and Fab libraries and panning against the RBD of the SARS-CoV-2, Li et al. identified an exceptional potent (Kd to RBD of 160 pM as judged by biolayer interferometry) mAb IgG1 ab1 that competes with ACE2 in vitro and protected transgenic mice expressing hACE2 from high-titer intranasal SARS-CoV-2 challenge.53 In two different assays using replication-competent SARS-CoV-2 in a microneutralization-based assay, 100% neutralization at <400 nM, and in a luciferase reporter gene assay, an IC50 of 200 nM was reported. Moreover, transgenic mice expressing human ACE2 administrated with 0.3 mg of Ig1 ab1 prior intranasal infection with SARS-CoV-2 did not show any detectable replication-competent virus, demonstrating the preventive effect of IgG1 ab1.53
Small Molecules Targeting the RBD.
Besides peptides, mAb, and protein chimeras, small molecules are still the preferred modality for a drug. This is due to improved pharmacokinetics, stability, and dosage logistics compared to proteins or peptides.54,55 In addition, small molecules have advantages compared to peptides/proteins regarding dissemination logistics in remote areas and the high expenses of peptide/protein production.54,55
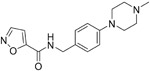
To identify small molecule entry inhibitors against the SARS-CoV S protein, Sarafianos et al. screened a chemical library composed of 3000 compounds according to Lipinski’s rule of five56 and identified an oxazole-carboxamide derivative, SSAA09E2 (1, Table 3), that blocks the binding of the RBD of SARS-CoV S protein and ACE2 with an IC50 value of 3.1 μM and CC50 value of >100 μM. Further investigation confirmed that 1 does not alter ACE2 expression but most likely blocks directly ACE2 recognition by interfering with the RBD.57
Table 3.
Small Molecules Targeting RBD and ACE2
| Name | Chemical structure | IC50 | Targets | Ref |
|---|---|---|---|---|
| SSAA09E2 (1) |
|
3.1 μM | RBD | 57 |
| K22 (2) |
|
0.7 μM | RBD | 58 |
| NAAE (3) |
|
57 μM | ACE2 | 66 |
| Chloroquine (4) |
|
1.13-4.4 μM | ACE2 | 67,68 |
| Hydroxychloroquine (5) |
|
0.72 μM | ACE2 | 70,71 |
| GW280264X (6) |
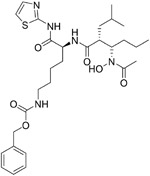
|
1 nM | ACE2 | 76 |
| TAPI-0 (7) |
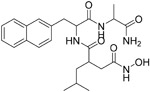
|
100 nM | ACE2 | 75 |
| TAPI-2 (8) |
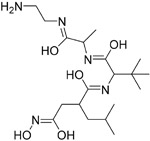
|
200 nM | ACE2 | 75 |
| MLN-4760 (9) |
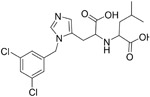
|
440 pM | ACE2 | 77 |
| HTCC (10) |
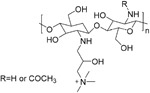
|
~50 nM | ACE2 | 78 |
| HM-HTCC (11) |
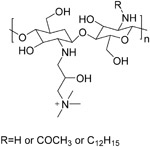
|
~230nM | ACE2 | 78 |
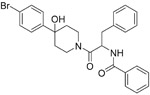
Lundin et al. screened a library of 16 671 diverse compounds and found a small molecule inhibitor, K22 (2), which was able to inhibit HCoV-229E with an IC50 value of 0.7 μM and CC50 value of 110 μM. Studies for mechanism showed that K22 targeted a very early step in the HCoV-229E life cycle and may interact with viral particles, thus inactivating their binding.58
TARGETING THE CELLULAR RECEPTOR
Peptide Analogues as ACE2 Inhibitors.
Human angiotensin-converting enzyme (ACE) is a highly glycosylated type I integral membrane protein and has been identified as a fundamental regulator of the renin–angiotensin system (RAS) in humans and is an important target in regulation of blood pressure homeostasis. ACE2 is a human homologue of ACE.59 It contains a single zinc-binding catalytic domain, which is 42% similar to the human ACE active region.60 ACE2 can catalyze the cleavage of angiotensin I into angiotensin 1-9, and angiotensin II into the vasodilator angiotensin 1-7 and its organ- and cell-specific expression also suggests a role in the regulation of cardiovascular and renal function and fertility.60 ACE2 is a functional receptor to the SARS-CoV during viral entry, and recent research demonstrated that SARS-CoV-2 also utilizes ACE2 for infection.61 However, ACE2 cannot be inhibited by ACE inhibitors, so there is an urgent need to develop specific ACE2 inhibitors that would prevent infection by both SARS-CoV and SARS-CoV-2.
One of the first efforts to target the ACE2 receptors was documented by Liu et al. Using a novel epitope assembling assay, Liu et al. identified linear B-cell immuno-cross-reactive epitopes of SARS-CoV S protein by synthesizing 22 longer peptides. Five of these peptides showed serologically highly cross-reactivity in all tested SARS patients sera. Among them, peptide S471-503 could significantly block the binding of RBD to ACE2. S471-503, derived from the S1 fragment (Figure 7 and Table 2) could target ACE2, and showed antiviral activity against SARS-CoV infection in vitro, with an EC50 value of 41.6 μM, providing an important basis to explore the antiviral potential of S471-503 against SARS-CoV-2.62
Another peptide derived from the RBD, RBD-11b, located in S1 of the SARS-CoV S protein, is crucial for binding to the host cells ACE2 receptor62 (Figure 7 and Table 2). Given the vital role of this motif, Meyer et al. confirmed the binding to ACE2 of a synthesized peptide mimicking this region (438YKYRYL443) with a Kd of around 46 μM. Moreover, RBD-11b displays no toxicity, as judged by an MTT (3-(4,5)-dimethylthiahiazo-(-z-y1)-3,5-di-phenytetrazoliumbromide) cell proliferation assay, on VeroE6 cells. In addition, RBD-11b showed antiviral activity to HCoV-NL63 at a peptide concentration of 7 mM in CaCo2 cells, which also used ACE2 as a functional receptor.63
Constrained peptides are receiving more attention in the drug development field, combining the best attributes of antibodies and small molecules. Linear peptides are often highly flexible and unstructured in solution, only forming structures upon target binding. This can sometimes reduce the affinity of such peptides for their target by an entropic penalty mechanism. However, stabilization methods such as cyclization or hydrocarbon stapling can increase the physicochemical characteristics and drug-like properties while negating the entropic penalty of binding and having a positive impact on affinity.64
Using a constrained peptide library displayed on filamentous phages, Ladner et al. identified several peptides inhibiting ACE2 function with the most potent being DX600 (Table 2). DX600, an N-terminal acetylated and C terminal amidated peptide, was a potent ACE2 peptide inhibitor with an IC50 value of 10 nM and a Ki value of 2.8 nM. DX600 did not inhibit ACE activity and thus is specific to ACE2. In addition, DX600 was chemically stable and not hydrolyzable by ACE2.65 Although it is not clear whether DX600 can inhibit coronavirus, as an effective ACE2 inhibitor, anticoronavirus tests should be conducted in the future.
Small Molecule as ACE2 Inhibitors.
As discussed previously, peptide and constrained peptide inhibitors have inherent caveats concerning their use as drugs.64 Therefore, screening for small molecule inhibitors, guided by information gleaned from the previous studies is the next logical step. A virtual screen targeting the ACE2 catalytic site with around 140 000 compounds combined with a molecular docking approach led to the identification of NAAE (N-(2-amino-ethyl)-1 aziridine-ethanamine) (3, Table 3). 3 showed a dose-dependent inhibition of ACE2 catalytic activity with an IC50 value of 57 μM and a Ki of 459 μM. Despite its micromolar potency in inhibiting a SARS-CoV pseudotyped virus, cytotoxicity data is not available to date.66
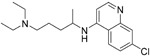
Chloroquine (4) currently has applications for malaria and amoebiasis treatment. Interestingly, Nichol et al. showed that chloroquine could also block the interaction of RBD of SARS-CoV to ACE2 under cell culture conditions with an ED50 value of 4.4 μM.67 Recently, Wang et al. found that 4 blocked SARS-CoV-2 virus infection, with an IC50 value of 1.13 μM and a CC50 > 100 μM in Vero E6 cells.68 Chloroquine possibly increases endosomal pH required for virus/cell fusion as well as impairs with the terminal glycosylation of the cellular ACE2 receptor, thereby reducing the affinity of SARS-CoV/SARS-CoV-2 to ACE2. Besides its antiviral activity, chloroquine may synergistically enhance its antiviral effect with immune-modulating activity in vivo.68 At present, chloroquine is carried out in clinical research in China for the treatment of SARS-CoV-2 (ChiCTR2000029609).69 Hydroxychloroquine (5) is an analogue of chloroquine, which shares the same mechanism of action as chloroquine but displays a more tolerable safety profile.70 Yao et al. showed that 5 had an IC50 value of 0.72 μM after a 48 h incubation time. In physiological-based pharmacokinetic (PBPK) models, Yao et al. found 5 exhibited better in vitro anti-SARS-CoV-2 activity than 4.71 Recent studies suggest that 4 and 5 could cause ventricular arrhythmias,72 QT prolongation,72,73 retinopathy,74 and other cardiac-related toxicity, which may pose a particular risk to critically ill patients. Although both show antiviral activity, safety, and effectiveness, they require further clinical research.
Turner et al. identified that the SARS-CoV receptor, ACE2, undergoes proteolytic shedding, releasing an enzymatically active ectodomain during viral entry.75 Further research identified that a disintegrin and metalloproteinase (ADAM17) is responsible for shedding regulation of ACE2. Inhibiting ADAM activity with the ADAM-specific inhibitor GW280264X (6) reduced shedding of ACE2 at 1 nM against SARS-CoV.76 Another enzyme involved in ACE2 shedding is TACE (TNF-α converting enzyme, a member of the ADAM family). Two TACE inhibitors, TAPI-0 (7) and TAPI-2 (8), reduced ACE2 shedding against SARS-CoV, with IC50 values of 100 and 200 nM, respectively.75
Perhaps the most promising small molecule described to date is the very potent ACE2 inhibitor MLN-4760 (9). 9 can inhibit the catalytic activity of ACE2 with an IC50 of around 440 pM.77 The crystal structure of the apo and 9 bound ACE2 complex revealed a significant subdomain movement of the N-terminal and C-terminal subdomains of ACE2 upon 9 binding. This movement is important to position critical residues to stabilize the bound inhibitor. Its high potency makes 9 a very attractive candidate for SARS-CoV-2 interference; however, no antiviral coronavirus data is available at this time.
Milewska et al. synthesized several polymer-based compounds showing prominent anticoronaviral activity. Among them, a cationically modified chitosan derivative, N-(2-hydroxypropyl)-3-trimethylammonium chitosan chloride (HTCC, 10), and hydrophobically modified HTCC (HM-HTCC, 11) were found that could inhibit HCoV-NL63 replication. For both tested polymers, their IC50 values were relatively low in LLC-MK2 cells, amounting to ~50 nM for 10 and ~230 nM for 11. CC50 values were ~0.8 and ~1 μM for 10 and 11, respectively.78 Recent research showed that 10 and 11 blocked the interaction of HCoV-NL63 with its ACE2 receptor and thus interfered with the process of viral entry.79
Despite the availability of many compounds with inhibitory effects on ACE2, the corresponding ADMET data in a preclinical model is not available. Regardless, direct inhibition of ACE2 is probably not a viable therapeutic modality, however. This is due to its important normal physiological roles, in addition to its lung injury protective role in acute respiratory distress syndrome from a variety of causes, including SARS-CoV infection.80,81 As such, directly inhibiting ACE2 as an antiviral strategy appears to be physiologically unsound, and virally targetted blockers of its interaction with the SARS-CoV/SARS-CoV-2 S protein hold greater promise.
INTERFERENCE WITH MEMBRANE FUSION OF THE SPIKE PROTEIN
Membrane fusion is a crucial step in the MERS/SARS infection cycle in both described pathways (see section 1). Within the endosomal/clathrin-dependent route, internalized viral particles need to fuse with the endosomal membrane to escape the endosomal/lysosomal environment. This is achieved via a conformational change of the S protein (S2 domain) within the acidic milieu followed by membrane fusion activation by the host protease cathepsin L. Membrane fusion is also essential during the nonendosomal/clathrin-independent route to fuse with cellular membranes facilitated by host protease cleavage of the S protein by cell membrane-associated proteases such as TMPRSS2.19 In conclusion, the S2 domain of the SARS-CoV S protein and host proteases such as cathepsin L and TMPRSS2 are very attractive therapeutic targets.82,83 Therefore, we highlight in the following section a few examples of peptide analogues, mAbs, and small molecules that target the S2 domain or inhibit directly host proteases that are crucial for the S protein processing and fusion event.
INHIBITORS TARGETING THE S2 DOMAIN
Peptide Analogues and Monoclonal Antibodies Targeting the S2 Domain.
The heptad repeat (HR) regions in the S2 domain are crucial for the viral membrane fusion event.84,85 HR1 and HR2 can interact with each other to form a 6-HB to bring viral and cellular membranes close (for exact location, see Figure 2). On the basis of this requirement, Bosch et al. obtained peptides corresponding to region HR2 within the HR. HR2-8 displayed in an infection inhibition assay with pseudotyped SARS-CoV S protein in Vero cells an EC50 value of 17 μM (Figure 7 and Table 2).84 Moreover, HR2-8 demonstrated concentration-dependent inhibition of HCoV-NL63 infection with an IC50 value of 0.5 μM and a CC50 value of 20 μM.86 On the basis of these initial results, further development of the HR2-8 peptide is necessary to develop a more potent human coronaviruse (HCoV) peptide inhibitor. Similarly, Ngai et al. obtained three HR derived peptides, including HR1-a, GST-removed-HR2, and HR2 peptide, with remarkable inhibitory activity against SARS-CoV (Figure 7 and Table 2). Virus entry inhibition studies suggested that HR1-A, derived from the HR1 region, had an EC50 value of 1.61 μM. GST-removed-HR2 peptide and HR2 peptide, derived from the HR2 region, had EC50 values of 2.15 and 0.34 μM, respectively.87 HR2P, spanning residues 1251–1286 in HR2 domains, could effectively inhibit MERS-CoV infection and S protein-mediated membrane fusion (Figure 7 and Table 2). This study indicates that HR2P could specifically inhibit MERS-CoV in Vero cells, with an IC50 value of ~0.6 μM and a CC50 value of >1000 μM. HR2P also demonstrated high selectivity, as indicated by its high selectivity index (SI > 1667). Importantly, the introduction of Arg, Lys, or Glu residues into the HR2P peptide increased stability, solubility, and anti-MERS-CoV activity.88 To improve the stability, solubility, and antiviral activity of HR2P, Channappanavar et al. designed and synthesized an HR2P analogue named HR2P-M2. HR2P-M2 strongly blocked S protein-mediated cell–cell fusion in a dose-dependent manner at IC50 values of 0.55 μM in vitro. In vivo, HR2P-M2 intranasal administration to Ad5/hDPP4 transgenic mice protected them from MERS-CoV infection and reduced the lung viral titers by more than 1000-fold. Moreover, combination treatment with IFN-β was demonstrated to enhance the protective effect.89
The development of a drug with broad-spectrum HCoV inhibitory activity is increasingly becoming an attractive approach. Xia et al. found that the EK1 peptide showed pan-CoV fusion inhibitory activity against multiple HCoVs (Figure 7 and Table 2).90 Further investigation revealed that EK1 directly reacts with the HR1 region and can competitively inhibit viral 6-HB formation. The pseudovirus assay suggested that the antiviral activity of EK1 against HCoV-OC43, HCoV-NL63, and HCoV-229E infection with IC50 values of 1.81, 6.02, and 3.35 μM, respectively. In vitro cytotoxicity assay determined that EK1 is not cytotoxic at concentrations up to 1 mM. Mice that received 5 mg/kg of EK1 were nearly completely protected from infection by HCoV-OC43 and 200 μg of EK1 against MERS-CoV infection. Recently, this team found that EK1 could also potentially inhibit SARS-CoV-2 with an IC50 value of 2.38 μM in pseudovirus assay and an IC50 value of 0.19 μM in fusion inhibitory assay.91 To improve the inhibitory activity of EK1 against SARS-CoV-2, they conjugate the cholesterol molecule to the EK1 peptide and found that a new peptide, EK1C4, exhibited highly potent inhibitory activity inhibit SARS-CoV-2 S-mediated membrane fusion and pseudovirus infection with IC50 values of 1.3 and 15.8 nM, The CC50 of EK1C4 was 5 μM, and the selectivity index was >136. In the OC43-infected mouse model, mice that received 0.5 mg/kg of EK1C4 were nearly completely protected from infection by HCoV-OC43. These data suggested that EK1C4 could be used for inhibition and treatment of infection by currently circulating SARS-CoV-2.92
MERS-5HB, a polypeptide derived from the HR1 and HR2 region, was synthesized by Gong et al., and affinity analysis demonstrated a low Kd value of 0.24 nM, and an IC50 value of 1 μM against MERS-CoV and CC50 > 50 μM. HR derived peptides is a highly promising strategy for viral fusion inhibition. Successful HR peptides have been used in the past to block entry of other virus families such as the HIV with the gp41 derived peptide Fuzeon (T20), the only approved fusion inhibitor for HIV-1 treatment to date.93 Therefore, HR derived peptides highlight a promising strategy for inhibitor development combating the new SARS-CoV-2.
Xia et al. reported that two peptide-based membrane fusion inhibitors, 229E-HR1P and 229E-HR2P (Figure 7 and Table 2), targeting the HCoV-229E S protein HR1 and HR2 domains, could competitively inhibit the viral autologous 6-HB formation and inhibit HCoV-229E S protein-mediated virus-cell membrane fusion with IC50 values of 5.7 and 0.3 μM, respectively. Moreover, neither 229E-HR1P nor 229E-HR2P had significant cytotoxicity to Huh-7 and A549 cells at concentrations up to 1000 μM. In addition, 229E-HR2P potentially inhibited pseudotyped and live HCoV-229E infection with IC50 values of 0.5 and 1.7 μM, respectively.94
The S2 domain is the most conserved motif between the SARS-CoV and the new SARS-CoV-2 S protein.92 It represents an ideal immunogen for the generation of a novel or repurposing SARS-CoV S2 domain targeting mAbs with cross-reactive potential.95 Sasazuki et al., for example, could successfully isolate the human mAb 5H10 from immunized Kunming (KM) mice. 5H10 displayed an anti-SARS-CoV neutralizing activity of around 5 μg/mL. Cell fusion assays indicate that 5H10 can inhibit viral fusion and entry rather than viral attachment to the surface of host cells or cleavage of the S protein. Consequently, the S protein of SARS-CoV might be the direct target of 5H10; however, further studies are required to confirm this hypothesis.96 Tan et al. identified mAb 1A9 (IC50 value between 25 and 50 μg/mL), an anti-SARS-CoV S2 domain mAb, that binds to a conserved loop region between the HR1 and HR2 domains of the S2 domain.97
Tsunetsugu-Yokota et al. found that antibody SKOT20 can inhibit SARS-CoV with an EC50 value of 5 μg/mL in Vero E6 cells SARS-CoV. Mutational studies indicate that SKOT20 restrict conformational changes within the S2 domain, essential for viral entry.98 Taken together, the here presented peptide and mAb candidates targeting the S2 domain derived from previous SARS-CoV studies could potentially help develop effective vaccines to combat SARS-CoV-2.
Small Molecules as S2 Inhibitors.
Although broadly neutralizing antibodies (bnAbs) targeting the S2 domain of SARS-CoV S protein have been studied in the past, Abs are generally not suitable for oral delivery, limiting their potential application.99,100 Small molecules mimicking bnAbs as fusion inhibitors have also been described for other viruses such as influenza virus or HIV-1 and represent a promising strategy.99,100 Xu et al. developed a two-step screening method to identify inhibitors that potentially block SARS-CoV entry into the host cells.99 On the basis of this approach, they identified two small molecules, TGG (12, Table 4) and luteolin (13), that can bind avidly to the SARS-CoV S2 protein and inhibit viral entry of SARS-CoV into Vero E6 cells with IC50 values of 4.5 and 10.6 μM, respectively. Cytotoxicity assay showed that the CC50 of 12 and 13 were 1.08 and 0.155 mM, respectively. Therefore, the selectivity index of 12 and 13 were 240.0 and 14.62, respectively. Further acute toxicity suggested that the 50% lethal doses of 12 and 13 were ~456 and 232.2 mg/kg, respectively. These indicated that these small molecules could be used at relatively high concentrations in mice.98 Quercetin (14), an analogue of 13, also showed antiviral activity against SARS-CoV, with an IC50 value of 83.4 μM and a CC50 value of 3.32 mM.101
Table 4.
Small Molecules Targeting S2 Domain and Proteolytic Processing
| Name | Chemical structure | IC50 | Targets | Ref |
|---|---|---|---|---|
| TGG (12) |
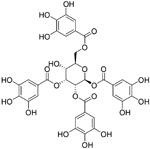
|
4.5 μM | S2 domain | 99 |
| luteolin (13) |
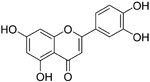
|
10.6 μM | S2 domain | 99 |
| Quercetin (14) |
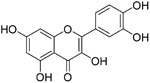
|
83.4 μM | S2 domain | 101 |
| ADS-J1 (15) |
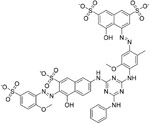
|
0.6-3.89 μM | S2 domain | 102 |
| Arbidol (16) |
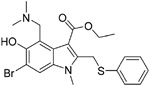
|
4.11 μM | S2 domain | 104 |
| MDL28170 (17) |
|
2.5 nM | cathepsin L | 83 |
| CID 16725315 (18) |
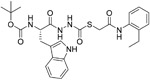
|
56 nM | cathepsin L | 114 |
| CID23631927 (19) |
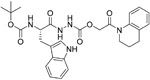
|
6.9 nM | cathepsin L | 114 |
| SSAA09E1 (20) |
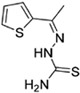
|
5.33 μM | cathepsin L | 57 |
| K11777 (21) |
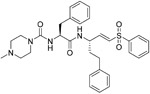
|
0.68-46.12 nM | cathepsin L | 115 |
| Camostat (22) |
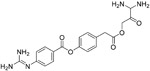
|
10 μM | TMPRSS2 | 116,19 |
| dec-RVKR-CMK (23) |
|
75 μM | furin | 20 |
| MI-1851 (24) |
|
- | furin | 117 |
ADS-J1 (15), a potential viral entry inhibitor, was reported by Ngai et al. The IC50 of 15 was 3.89 μM. Molecular docking analysis suggests that 15 can bind into a deep pocket of the SARS-CoV S protein HR region and block the SARS-CoV entry into host cells.43 Recently, Zhao et al. demonstrated that 15 could also inhibit MERS-CoV infection in a pseudovirus-based inhibition assay, with an IC50 value of 0.6 μM, a CC50 value of 26.9 μM, and providing a selectivity index of almost 45.102
Arbidol (16), a broad-spectrum drug, has been licensed for decades in Russia and China against influenza by binding to the HA protein to block the viruses–cell fusion.103 Recently, Wang et al. identified that 16 efficiently inhibited SARS-CoV-2 virus infection in vitro with an IC50 value of 4.11 μM, a CC50 value of 31.79 μM, and an SI of 7.73.104 Vankadari compared protein sequence analysis and found that a small region of the S2 domain (aa947–aa1027) of the SARS-CoV-2 spike glycoprotein resembles that of the influenza virus H3N2 HA. So the mechanism of 16 was to target the SARS-CoV-2 spike glycoprotein and blocked its trimerization, which may inhibit host cell adhesion and hijacking.105 In January 2020, in Wuhan, China, a clinical pilot trial conducted with 36 patients with SARS-CoV-2 virus infection received 400 mg 16 three times a day for 9 days; 31 untreated SARS-CoV-2 patients served as a control group. In this trial, patients with 16 showed a tendency to decrease viral load as determined by RT-PCR and reduced mortality (0% vs 16%), as compared to the control group.106
The HR regions of SARS-CoV and SARS-CoV-2 S protein share a high degree of conservation, and the described small molecules as fusion inhibitors can have potential applications in inhibiting SARS-CoV-2 fusion. Indeed, targeting virus surface protein is a promising antiviral strategy, whether inhibiting RBD or S2 domain.
PROTEOLYTIC PROCESSING INHIBITORS
Antibiotics that Target the Cathepsin L Proteinase.
During clathrin-dependent viral entry, the host cellular cathepsin L protease plays a key role in infection efficiency by activation of the S protein into a fusogenic state to escape the late endosomes, and cathepsin L (lysosomal endopeptidase) cleavage is believed to expose a hydrophobic fusion peptide essential to initiate membrane fusion.107 In light of its vital role in the SARS CoV infection cycle, cathepsin L is a desirable target to interfere with virus–cell entry.83
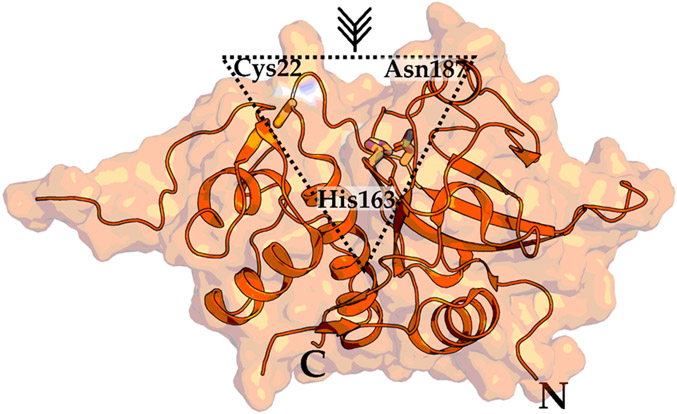
Cathepsin L consists of a pro- and a mature-domain. In a low pH milieu, the pro-domain is autocatalytically cleaved to obtain the papain-like folded mature-domain consisting of an N-terminal helical domain and a C-terminal β-sheet domain (Figure 8).108 A well conserved Cys-His-Asn triad in the active site is crucial for substrate binding and catalysis. In light of its importance in the SARS-CoV-2 replication cycle, cathepsin L is a highly desirable target that will be described in the following section.109
Figure 8.
Crystal structure of the mature-domain of cathepsin L. The catalytic triad (Cys22, His163, Asn187) essential for proteolytic activity are highlighted in a dashed triangle. N and C represent N- and C-terminus, respectively. PDB 5I4H.
Teicoplanin is a glycopeptide antibiotic, with applications in the treatment of serious infections caused by Gram-positive bacteria such as Streptococcus and Staphylococcus aureus.110 Interestingly, teicoplanin was shown to block the entry of SARS, MERS, and Ebola virus by specifically inhibiting the cathepsin L activity.111 More recently, Zhang et al. showed that teicoplanin could also block the entry of the new SARS-CoV-2 pseudoviruses with an IC50 value of 1.66 μM. As a routinely used clinical antibiotic, teicoplanin could be potentially used immediately to combat the current SARS-CoV-2 outbreak.112
Small Molecules as Cathepsin L Inhibitors.
Human cathepsin L plays numerous critical roles in diverse cellular settings associated with human diseases.113 Previous studies also highlighted the feasibility of targeting this cysteine endopeptidase with small molecules with implications for possible intervention strategies of SARS-CoV-2 infection.113
A high-throughput screen (HTS) of a 1000-compound library that resulted in the identification of MDL28170 (17, Table 4) by Bates et al., and in an antiviral activity assay, 17 specifically inhibited cathepsin L-mediated substrate cleavage and blocked SARS-CoV viral entry, with an IC50 value of 2.5 nM and EC50 value in the range of 100 nM. However, despite its potent inhibitory activity, no cytotoxicity data for 17 is currently available.83
Two small molecules, CID 16725315 (18) and CID 23631927 (19), were reported by Diamond et al. as viral entry inhibitors of the SARS-CoV. In a cathepsin L inhibition assay, 19 could block cathepsin L with an IC50 value of 6.9 nM, while 18 showed slightly weaker potency with an IC50 value of 56 nM. Interestingly, besides inhibiting SARS-CoV, compound 19 (EC50 value of 273 nM) showed some inhibition activity for Ebola virus infection (EC50 value of 193 nM) of human embryonic kidney 293T cells. Importantly, 19 did not show any sign of toxicity to human aortic endothelial cells at 100 μM. This data offers a new promising point for the treatment of SARS and Ebola virus infections.114 Recently, in a cell-based assay screen of ~14000 compounds, SSAA09E1 (20) was identified that could specifically bind to the cathepsin L proteinase and interference SARS-S protein during viral entry, with an IC50 value of 5.33 μM. In a pseudotype-based assay in 293T cells, the EC50 value of 20 was around 6.4 μM, and no cytotoxicity was detected below 100 μM.57
Using SARS-CoV entry assays, Zhou et al. screened 2100 cysteine protease inhibitors with confirmed activity to inhibit human cathepsins. Among them, K11777 (21) demonstrated the most robust activity. Results demonstrated that 21 blocked SARS-CoV pseudovirus entry at an IC50 value of 0.68 nM while no toxicity was observed, CC50 value >10 μM. Interestingly, for other coronaviruses, 21 showed broad-spectrum antiviral activity with IC50 values of 1.48, 6.78, and 46.12 nM against HCoV-229E, HCoV-NL63, and MERS-CoV, respectively.115
Inhibitors of Cell Membrane-Associated TMPRSS2.
Either the endosomal cysteine proteases cathepsin L or the cell membrane-associated serine protease TMPRSS2 can facilitate SARS-CoV virus entry into host cells by cleavage of the viral S protein.19 This cleavage exposes fusion-competent motifs known as fusion peptides, and importantly, for SARS-CoV, the interference of both proteases is required for efficient inhibition of virus replication.19 Matsuyama et al. identified Camostat (22, Table 4), a commercially available serine protease inhibitor that can efficiently prevent SARS-CoV infections at 10 μM by inhibiting TMPRSS2 activity. However, even at high concentrations (100 μM) of 22, the inhibition of viral entry via SARS S protein-mediated cell fusion never exceeded 65% (inhibition efficiency), indicating that despite the inhibition of TMPRSS2, 35% of virus entry takes place via the endosomal cathepsin pathway. Therefore, they examined the activity of pseudotyped viruses when treated with a combination of (23,25)trans-epoxysuccinyl-l-leucylamindo-3-methylbutane ethyl ester (EST, a cathepsin inhibitor) and 22. The results suggested that simultaneous treatment with EST and 22 remarkably blocked infection (>95%).116 Similarly, Pohlmann et al. reported that 22 could prevent the viral entry of SARS-CoV-2. Importantly, full inhibition efficiency was attained when treated with both 22 and E-64d (a cathepsin inhibitor). Both studies indicate that SARS-CoV and SARS-CoV-2 enter cells via a similar mechanism, showing the potential of 22 as a promising candidate for further development as a SARS-CoV-2 treatment.19
Inhibitors of the Furin Cleavage Site in the Coronavirus Spike Proteins.
Elevated levels of furin expression were able to facilitate MERS-CoV pseudovirion infection, and viral entry could be reduced by furin siRNA silencing.20 Decanoyl-RVKR-chloromethylketone (23, dec-RVKR-CMK), a furin inhibitor, was shown to block MERS-CoV S protein-mediated entry as well as virus infection, with an IC50 value of 75 μM in HEK-293T cells. Furthermore, when cathepsin inhibitor camostat was used in combination with 23, a significant inhibition in infectivity was characterized compared to camostat alone.20 Recently, Bestle et al., showed that the potent peptidomimetic inhibitor MI-1851 (24) could prevent proteolytic processing of the S protein from SARS-CoV-2 by endogenous furin in HEK293 cells. However, no antiviral data is available for 24 yet.117 The peculiar furin-like cleavage site (S1/S2-site in Figure 2) in SARS-CoV-2 that is absent in the SARS-CoV and other SARS-like CoVs indicates that furin inhibitors could play a significant role in blocking the viral entry process.117,118
HOST FACTOR INHIBITORS
SARS-CoV-2 cell entry also relies on host cell factors. Therefore, these host cell factors can play an essential role as targets for SARS-CoV-2 inhibition.119 Chlorpromazine (25 Table 5) is an antipsychotic drug developed for the treatment of schizophrenia. It has also been reported to inhibit the infection of hepatic C virus (HCV),120 mouse hepatitis virus (MHV-2), 27 and alphavirus.121 Recently, Liu et al. demonstrated that 25 could inhibit the clathrin-mediated endocytosis of MERS-CoV cell entry, with an IC50 of 7.24 μM and CC50 > 40 μM.122 Additionally, fluphenazine (26) and promethazine (27) showed a similar inhibitory effect against MERS-CoV, with IC50 values of 3.23 and 7.48 μM, respectively.122
Table 5.
Small Molecules Targeting Host Factors and Unknown Targets
| Name | Chemical structure | IC50 | Targets | Ref |
|---|---|---|---|---|
| Chlorpromazine (25) |
|
7.24 μM | clathrin-mediated endocytosis | 122 |
| fluphenazine (26) |
|
3.23 μM | clathrin-mediated endocytosis | 122 |
| promethazine (27) |
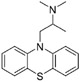
|
7.48 μM | clathrin-mediated endocytosis | 122 |
| imatinib (28) |
|
10 μM | Abl2 | 123, 124 |
| dasatinib (29) |
|
2.1-5.4 μM | Abl | 125 |
| saracatinib (30) |
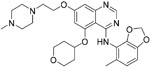
|
2.4-5.1 μM | SFK signaling pathways | 126 |
| emodin (31) |
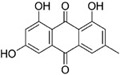
|
200 μM | - | 131 |
| SSAA09E3 (32) |
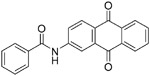
|
9.7 μM | - | 57 |
| VE607 (33) |
|
1.6 μM | - | 132 |
Machamer et al. found imatinib (28), an Abelson kinase signaling pathway inhibitor that could inhibit Abelson tyrosine–protein kinase 2 (Abl2) to block MERS-CoV virion fusion with endosomal membranes with an IC50 value of 10 μM. 28 showed no cytotoxic effects in Vero cells at 100 μM.123,124 Another Abl inhibitor, dasatinib (29), was active against both MERS-CoV and SARS-CoV, with IC50 values of 5.4 and 2.1 μM, respectively.125
On the basis of an HTS assay using cytopathic-effect (CPE), Shin et al. identified saracatinib (30), a potent inhibitor of the Src-family of tyrosine kinases (SFKs), that can block the early process of the MERS-CoV life cycle, possibly through inhibition of the SFK signaling pathways. 30 exhibited prominent antiviral activity with an IC50 value of 2.9 μM and a CC50 value of >50 μM, resulting in an SI value of approximately >17. Moreover, 30 showed a broad-antiviral activity against hCoV-229E and hCoV-OC43, with an IC50 value of 2.4 and 5.1 μM, respectively.126
PHENOTYPIC SCREENING FOR NEW ENTRY INHIBITORS
Phenotypic screening methods are usually used to identify first-in-class drugs without knowing the actual target and mechanism of action of the drug, while target-based screening identifies best-in-class drugs.127-129 Although the phenotypic screening approach often is limited in terms of capacity compared to in silico target-based screening, it can have advantages in identifying cell-active compounds providing information on drug solubility or cell uptake.127-129
Many drugs, especially natural products, have an unknown mechanism of action but were shown to inhibit coronavirus entry.130 Hsiang et al. screened a library of 121 Chinese herbs using a biotinylated enzyme-linked immunosorbent assay to search for active compounds that could potentially inhibit SARS-CoV S protein binding to ACE2. Further studies identified emodin (31, Table 5), the active component from Polygonum multiflorum and Rheum officinale, could block the interaction of SARS-CoV S protein to ACE2, with an IC50 value of 200 μM in an S protein-pseudotyped retrovirus assay using Vero E6 cells. However, the mechanism of action of 31 still needs to be determined.131 Sarafianos et al. found that SSAA09E3 (32), a benzamide derivative, could prevent SARS-CoV virus–cell membrane fusion in pseudotyped-based and antiviral-based assays, with an IC50 value of 9.7 μM, but a CC50 value of 20 μM indicates additional unknown cellular targets.57
Out of an HTS, VE607 (33) was identified using a phenotype-based screen from a 50 240 structurally diverse small-molecule compound library. Pseudotype virus entry assay suggested VE607 can specifically inhibit SARS-CoV virus entry into cells with an EC50 value of 3 μM and inhibited SARS-CoV plaque formation with an IC50 of 1.6 μM.132 A similar HTS approach was employed by Zhang et al. for screening a compound library consisting of 727 structurally diverse small molecules. eighty-four compounds were identified with significant anticoronavirus potential. Further studies revealed that 51 compounds inhibited virus entry, while 19 others interfered with viral replication.133 Natural products should, however, be considered with caution due to their unknown mechanism of action and possible toxic side effects.
CONCLUSIONS AND PROSPECTS
The recent SARS-CoV-2 outbreak, with its high fatality rate, has raised global concerns and was declared as a global pandemic by the WHO. The number of infections continues to rise, and numerous research groups around the globe have prioritized the identification and development of new COVID-19 treatments. Still, there are no effective treatments to date. Viral entry is the first step in the viral life cycle and represents an attractive intervention point by blocking the coreceptor interaction or the virus–cell membrane fusion event. SARS-CoV-2 and other coronaviruses have similar infection mechanisms. This is especially true for SARS-CoV and CoV-NL63, which share the same human ACE2 receptor crucial for viral entry. Therefore, already developed inhibitors against known hCoVs could potentially be used to combat SARS-CoV-2. These efforts identified a large number of inhibitors, including peptides, antibodies, small-molecule compounds, and natural products with anticoronavirus activity. Although many inhibitors demonstrated efficacy in inhibiting coronavirus virus infection, no specific prophylactic or postexposure therapy is currently available for HCoVs. One of the main reasons causing this is that most of the potenial agents were not adequately evaluated for in vitro and in vivo studies. Most drugs are in the preclinical stage and stopped in animal models due to poor bioavailability, safety, and pharmacokinetics so that few entered human trials. In light of the urgency of the current outbreak, repositioning of already approved drugs is increasingly becoming a promising approach, especially with toxicity and safety data in hand.
The most effective measure to prevent viral diseases is vaccination. Coronavirus vaccine development mainly focused on S protein, and some of them reported can inhibit SARS,134-136 and MERS.137 Although vaccination strategies were developed in the context of previous epidemics, no vaccine for SARS-CoV-2 infections is yet available. Since the recent SARS-CoV-2 outbreak, research groups around the world are now stepping up to develop vaccines targeting SARS-CoV-2, and vaccine research routes include nucleic acid vaccines, viral vector vaccines, inactivated vaccines, and recombinant protein vaccines. Typical vaccine development is time, resource, and financially consuming, although this pandemic has created initiatives that hope to speed the development of a SARS-CoV-2 vaccine. Even the most optimiztic views regarding an effective SARS-CoV-2 vaccine being created are at least one year away. Even after creation, other hurdles for the SARS-CoV-2 include global implementation and distribution, and different strategies for containing this contagion should be explored simultaneously as the vaccine efforts.
In addition to small-molecule inhibitors, monoclonal antibodies, and vaccine development, convalescent sera from SARS-CoV-2 survivors (convalescent-phase sera) is an additional option for COVID-19 treatment. Passive immunization was well established for viral infection prophylaxis.138 By meta-analysis of studies about the 1918 influenza, H1N1 influenza epidemic demonstrated that early treatment of convalescent blood products decreased the risk ratio caused by pneumonia from 37% to 16%.139 Nevertheless, the appropriate titer of the convalescent-phase sera antibody remains to be determined, which was required for therapeutic efficacy to inhibit SARS-CoV-2. Research carried out with MERS-CoV suggested that sera from patients recovering from infections did not contain sufficient antibody titers for therapeutic use.140
Recent initiatives such as the governmental (USA) Operation Warp Speed (OWS) to support the development, manufacturing, and distribution of COVID-19 vaccines, therapeutics, and diagnostics or the Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) public–private partnership coordinated by the National Institutes of Health (NIH) are crucial milestones in a coordinated effort to accelerate and prioritize the development of the most promising vaccines and treatments. Initiatives like these that bridge government, academia, and industry should also be continued past the current COVID-19 crisis so that we can respond to future novel outbreaks rapidly and adequately.
ACKNOWLEDGMENTS
Financial support from the National Natural Science Foundation of China (NSFC nos. 81973181, 81773574), Key Project of NSFC for International Cooperation (no. 81420108027), Key Research and Development Project of Shandong Province (nos. 2017CXGC1401, 2019JZZY021011, 2020SFXGFY08), and NIH grant R01AI150491 (Cocklin, PI) are gratefully acknowledged. A.D. is partially funded by NS089435 (Cocklin, Co–I [Nonnemacher, PI]). We thank Megan Meuser for comments that greatly improved the manuscript.
ABBREVIATIONS USED
- ACE2
angiotensin-converting enzyme II
- ADAM
A disintegrin and metalloproteinase
- bnAbs
broadly neutralizing antibodies
- COVID-19
coronavirus disease 2019
- CPE
cytopathic effect
- ELISA
enzyme-linked immunosorbent assay
- FP
fusion peptide
- FCV
feline coronavirus
- HR
heptad repeat
- HA
hemagglutinin
- HIV
human immune deficiency virus
- HCoV
human coronaviruse
- HTS
high-throughput screen
- HCV
hepatic C virus
- IFA
immunofluorescence assay
- KM
immunized Kunming
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MHV
mouse hepatitis coronavirus
- NTD
N-terminal domain
- PD
proteolytic domain
- PBPK
physiologically based pharmacokinetic
- PCR
polymerase chain reaction
- RMSD
root-mean-square deviation
- RBD
receptor-binding domain
- RdRp
RNA-dependent RNA polymerase
- RAS
renin-angiotensin system
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SPR
surface plasmon resonance
- SFKs
Src-family of tyrosine kinases
- TM
transmembrane domain
- TMPRSS2
transmembrane protease serine 2
- WHO
World Health Organization
- 3CLpro
3-chymotrypsin-like protease
- 6-HB
six-helix bundle
Biographies
Siyu Xiu received his bachelor’s degree in pharmacy from Shandong First Medical University, China, in 2018. Currently, he is a master graduate student in medicinal chemistry at the School of Pharmaceutical Sciences, Shandong University, working under the supervision of Professor Xinyong Liu and Associate Professor Peng Zhan. His research interests focus on rational design, synthesis, and biological evaluation of novel potent inhibitors of influenza virus hemagglutinin.
Alexej Dick is currently a postdoctoral fellow at Drexel University, Philadelphia, PA, USA. He received his Ph.D. in 2016 at the Max Delbrück Center (MDC) for Molecular Medicine and Free University of Berlin, Germany, in Biochemistry/Structural Biology, working in the Laboratory of Prof. Dr. Oliver Daumke on the Myxovirus resistant protein family (MxA and MxB) and Orthomyxoviruses. After his Ph.D., he joined the group of Dr. Irwin Chaiken at Drexel Univerity, Philadelphia, PA, USA, to study cyclic peptides triazoles, an anti-HIV molecule class interfering with the virus entry/fusion. In 2019, he joined the group of Dr. Simon Cocklin and is currently working in infectious disease-related (HIV-1, Ebola virus, SARS-CoV-2) computationally aided drug design supported by biochemical, virological, molecular biological, and structural methodologies.
Han Ju was born in Weihai, China, in 1993. He completed his B.S. degree from Shenyang Pharmaceutical University in 2015 and received his Master’s degree from Shandong University in 2018. He is working in the School of Pharmaceutical Sciences of Shandong University as a Ph.D. candidate, supervised by Professor Xin-Yong Liu. His current research focuses on the discovery of novel anti-influenza drugs.
Sako Mirzaie is a visiting Professor in the Leslie Dan Faculty of Pharmacy at the University of Toronto. He is an Assistant Professor at Azad University of Sanandaj. Sako received his Ph.D. in Biochemistry from the science and research branch, Azad University of Tehran. His research interests lie in the area of protein engineering, drug design, and discovery. Sako has collaborated actively with researchers in several other disciplines of biological science. He has more than 40 papers in high-ranked international journals.
Fatemeh Abdi is a Postdoctoral fellowship in the University Health Network at the University of Toronto. She earned her Ph.D. in Biochemistry in 2018 from North Tehran Branch, Azad University. Then, Fatemeh joined the Azad University of QAZVIN as a lecturer. Her research focused on drug design, discovery, and computational biology. She is a consultant for chemical and pharmaceutical companies in Iran. She also was a QC manager of Toli Pers Co.
Simon Cocklin obtained his B.Sc. degree in Genetics from Newcastle University, UK, in 1997. He then earned his Ph.D. in Molecular Genetics from Newcastle University in 2001. He is currently an Associate Professor in the Department of Biochemistry and Molecular Biology at Drexel University College of Medicine. He has over 22 years of experience with surface plasmon resonance (SPR) interaction analysis and 18 years’ experience in the HIV-1 field. His research interests center around the discovery of novel antiviral inhibitors primarily to HIV-1 and targeting the Gag and Env structural proteins. His interests have since expanded to include inhibitor discovery against Ebola and SARS-CoV-2 viruses. His research spans multiple disciplines, including computational, biochemical, virological, molecular biological, and structural methodologies.
Peng Zhan obtained his B.S. degree from Shandong University, China, in 2005. Then he earned his M.S. degree and Ph.D. in medicinal chemistry from Shandong University in 2008 and 2010, respectively. He subsequently joined the research group of Professor Xinyong Liu as a Lecturer (2010–2012). From 2012 to 2014, he worked as a Postdoctoral Fellow funded by JSPS (Japan Society for the Promotion of Science) in the Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Japan. He is currently an associate professor in the Institute of Medicinal Chemistry, Shandong University. His research interests involve the discovery of novel antiviral, anticancer, and neurodegenerative diseases-related agents based on rational drug design and combinatorial chemistry approaches.
Xinyong Liu received his B.S. and M.S. degrees from the School of Pharmaceutical Sciences, Shandong University, in 1984 and 1991, respectively. From 1997 to 1999, he worked at the Instituto de Quimica Medica (CSIC) in Spain as a senior visiting scholar. He obtained his Ph.D. from Shandong University in 2004. He is currently a full Professor of the Institute of Medicinal Chemistry, Shandong University. His research interests include rational drug design, synthesis, and antiviral evaluation of a variety of molecules that interact with specific enzymes and receptors in the viral life cycle. Other ongoing programs include studies of the molecular modification and structure–activity relationships of some natural products used to treat cardiovascular diseases, and drug delivery research using PEGylated small-molecular agents.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Siyu Xiu, Department of Medicinal Chemistry, School of Pharmaceutical Sciences, Shandong University, Jinan 250012, China.
Alexej Dick, Department of Biochemistry & Molecular Biology, Drexel University College of Medicine, Philadelphia, Pennsylvania 19102, United States.
Han Ju, Department of Medicinal Chemistry, School of Pharmaceutical Sciences, Shandong University, Jinan 250012, China.
Sako Mirzaie, Department of Biochemistry, Sanandaj Branch, Islamic Azad University, Sanandaj 6616935391, Iran.
Fatemeh Abdi, Department of Cellular and Molecular Biology, Islamic Azad University, Tehran 1651153311, Iran.
Simon Cocklin, Department of Biochemistry & Molecular Biology, Drexel University College of Medicine, Philadelphia, Pennsylvania 19102, United States.
Peng Zhan, Department of Medicinal Chemistry, School of Pharmaceutical Sciences, Shandong University, Jinan 250012, China.
Xinyong Liu, Department of Medicinal Chemistry, School of Pharmaceutical Sciences, Shandong University, Jinan 250012, China.
REFERENCES
- (1).Pillaiyar T; Meenakshisundaram S; Manickam M Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today 2020, 25, 668–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Weiss SR; Navas-Martin S Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev 2005, 69, 635–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Tong TR Therapies for coronaviruses. Part I of II – viral entry inhibitors. Expert Opin. Ther. Pat 2009, 19, 357–367. [DOI] [PubMed] [Google Scholar]
- (4).Dyall J; Gross R; Kindrachuk J; Johnson RF; Olinger GG; Hensley LE; Frieman MB; Jahrling PB Middle east respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs 2017, 77, 1935–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Millet JK; Whittaker GR Host cell proteases: ritical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Skowronski DM; Astell C; Brunham RC; Low DE; Petric M; Roper RL; Talbot PJ; Tam T; Babiuk L Severe acute respiratory syndrome (SARS): a year in review. Annu. Rev. Med 2005, 56, 357–381. [DOI] [PubMed] [Google Scholar]
- (7).Middle East Respiratory Syndrome Coronavirus (MERS-CoV); World Health Organization, 2020; https://www.who.int/emergencies/mers-cov/en/ (accessed 2020-05-29). [Google Scholar]
- (8).Ling R; Dai Y; Huang B; Huang W; Yu J; Lu X; Jiang Y In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides 2020, 130, 170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Coronavirus Disease (COVID-2019) Situation Reports; World Health Organization, 2020; https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed 2020-05-29). [Google Scholar]
- (10).Coronavirus Disease 2019 (COVID-19); Centers for Disease Control and Prevention, 2020; https://www.cdc.gov/coronavirus/2019-ncov/locations-confirmed-cases.html#map (accessed 2020-05-27 2020). [Google Scholar]
- (11).Zhou P; Yang XL; Wang XG; Hu B; Zhang L; Zhang W; Si HR; Zhu Y; Li B; Huang CL; Chen HD; Chen J; Luo Y; Guo H; Jiang RD; Liu MQ; Chen Y; Shen XR; Wang X; Zheng XS; Zhao K; Chen QJ; Deng F; Liu LL; Yan B; Zhan FX; Wang YY; Xiao GF; Shi ZL A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wrapp D; Wang N; Corbett KS; Goldsmith JA; Hsieh CL; Abiona O; Graham BS; McLellan JS Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Inoue Y; Tanaka N; Tanaka Y; Inoue S; Morita K; Zhuang M; Hattori T; Sugamura K Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol 2007, 81, 8722–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang H; Yang P; Liu K; Guo F; Zhang Y; Zhang G; Jiang C SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Mingo RM; Simmons JA; Shoemaker CJ; Nelson EA; Schornberg KL; D’Souza RS; Casanova JE; White JM Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J. Virol 2015, 89, 2931–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Burkard C; Verheije MH; Wicht O; van Kasteren SI; van Kuppeveld FJ; Haagmans BL; Pelkmans L; Rottier PJ; Bosch BJ; de Haan CA Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014, 10, e1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wang K; Chen W; Zhou Y; Lian J; Zhang Z; Du P; Gong L; Zhang L; Cui H; Geng J; Wang B; Sun X; Wang C; Yang X; Lin P; Deng Y; Wei D; Yang X; Zhu Y; Zhang K; Zheng Z; Miao J; Guo T; Shi Y; Zhang J; Fu L; Wang Q; Bian H; Zhu P; Chen Z SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv. 2020, 988345, . [Google Scholar]
- (18).Shirato K; Kawase M; Matsuyama S Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol 2013, 87, 12552–12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hoffmann M; Kleine-Weber H; Schroeder S; Krüger N; Herrler T; Erichsen S; Schiergens TS; Herrler G; Wu NH; Nitsche A; Muller MA; Drosten C; Pohlmann S SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Millet JK; Whittaker GR Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 15214–15219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Coutard B; Valle C; de Lamballerie X; Canard B; Seidah NG; Decroly E The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020, 176, 104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chen J; Lee KH; Steinhauer DA; Stevens DJ; Skehel JJ; Wiley DC Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 1998, 95, 409–417. [DOI] [PubMed] [Google Scholar]
- (23).Madu IG; Roth SL; Belouzard S; Whittaker GR Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol 2009, 83, 7411–7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Petit CM; Melancon JM; Chouljenko VN; Colgrove R; Farzan M; Knipe DM; Kousoulas KG Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology 2005, 341, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sainz B Jr.; Rausch JM; Gallaher WR; Garry RF; Wimley WC Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J. Virol 2005, 79, 7195–7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Prajapat M; Sarma P; Shekhar N; Avti P; Sinha S; Kaur H; Kumar S; Bhattacharyya A; Kumar H; Bansal S; Medhi B Drug targets for corona virus: a systematic review. Indian J. Pharmacol 2020, 52, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pu Y; Zhang X Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of eps15. J. Virol 2008, 82, 8112–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Graham RL; Donaldson EF; Baric RS A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol 2013, 11, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Severe Acute Respiratory Syndrome Coronavirus 2 Isolate Wuhan-Hu-1, Complete Genome; National Center for Biotechnology Information, 2020; https://go.usa.gov/xwGE3 (accessed 2020-06-12). [Google Scholar]
- (30).Wu F; Zhao S; Yu B; Chen YM; Wang W; Song ZG; Hu Y; Tao ZW; Tian JH; Pei YY; Yuan ML; Zhang YL; Dai FH; Liu Y; Wang QM; Zheng JJ; Xu L; Holmes EC; Zhang YZ A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yan R; Zhang Y; Li Y; Xia L; Guo Y; Zhou Q Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Morse JS; Lalonde T; Xu S; Liu WR Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem 2020, 21, 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gu J; Korteweg C Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol 2007, 170, 1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ortega JT; Serrano ML; Pujol FH; Rangel HR Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020, 19, 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Song W; Gui M; Wang X; Xiang Y Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018, 14, e1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Li F; Li W; Farzan M; Harrison SC Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [DOI] [PubMed] [Google Scholar]
- (37).Ho TY; Wu SL; Chen JC; Wei YC; Cheng SE; Chang YH; Liu HJ; Hsiang CY Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2006, 69, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Han DP; Penn-Nicholson A; Cho MW Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology 2006, 350, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Dibo M; Battocchio EC; dos Santos Souza LM; da Silva MDV; Banin-Hirata BK; Sapla MMM; Marinello P; Rocha SPD; Faccin-Galhardi LC Antibody therapy for the control of viral diseases: an update. Curr. Pharm. Biotechnol 2019, 20, 1108–1121. [DOI] [PubMed] [Google Scholar]
- (40).Meulen J.t. Monoclonal antibodies for prophylaxis and therapy of infectious diseases. Expert Opin. Emerging Drugs 2007, 12, 525–540. [DOI] [PubMed] [Google Scholar]
- (41).Xiao X; Dimitrov DS Monoclonal antibodies against viruses and bacteria: a survey of patents. Recent Pat. Anti-Infect. Drug Discovery 2007, 2, 171–177. [DOI] [PubMed] [Google Scholar]
- (42).Coughlin MM; Babcook J; Prabhakar BS Human monoclonal antibodies to SARS-coronavirus inhibit infection by different mechanisms. Virology 2009, 394, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Greenough TC; Babcock GJ; Roberts A; Hernandez HJ; Thomas WD Jr.; Coccia JA; Graziano RF; Srinivasan M; Lowy I; Finberg RW; Subbarao K; Vogel L; Somasundaran M; Luzuriaga K; Sullivan JL; Ambrosino DM Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis 2005, 191, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Coughlin M; Lou G; Martinez O; Masterman SK; Olsen OA; Moksa AA; Farzan M; Babcook JS; Prabhakar BS Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse. Virology 2007, 361, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Prabakaran P; Gan J; Feng Y; Zhu Z; Choudhry V; Xiao X; Ji X; Dimitrov DS Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem 2006, 281, 15829–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhu Z; Chakraborti S; He Y; Roberts A; Sheahan T; Xiao X; Hensley LE; Prabakaran P; Rockx B; Sidorov IA; Corti D; Vogel L; Feng Y; Kim JO; Wang LF; Baric R; Lanzavecchia A; Curtis KM; Nabel GJ; Subbarao K; Jiang S; Dimitrov DS Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lei CH; Fu WY; Qian KW; Li T; Zhang S; Ding M; Hu S Potent neutralization of 2019 novel coronavirus by recombinant ACE2-Ig. BioRxiv. 2020, 929976, DOI: . [Google Scholar]
- (48).Pascal KE; Coleman CM; Mujica AO; Kamat V; Badithe A; Fairhurst J; Hunt C; Strein J; Berrebi A; Sisk JM; Matthews KL; Babb R; Chen G; Lai KMV; Huang TT; Olson W; Yancopoulos GD; Stahl N; Frieman MB; Kyratsous CA Pre- and postexposure efficacy of fully human antibodies against spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).de Wit E; Feldmann F; Okumura A; Horne E; Haddock E; Saturday G; Scott D; Erlandson KJ; Stahl N; Lipsich L; Kyratsous CA; Feldmann H Prophylactic and yherapeutic efficacy of mAb treatment against MERS-CoV in common marmosets. Antiviral Res. 2018, 156, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chen X; Li R; Pan Z; Qian C; Yang Y; You R; Zhao J; Liu P; Gao L; Li Z; Huang Q; Xu L; Tang J; Tian Q; Yao W; Hu L; Yan X; Zhou X; Wu Y; Deng K; Zhang Z; Qian Z; Chen Y; Ye L Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol 2020, 17, 647–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wang C; Li W; Drabek D; Okba NMA; van Haperen R; Osterhaus ADME; van Kuppeveld FJM; Haagmans BL; Grosveld F; Bosch BJ A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun 2020, 11, 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Brouwer PJM; Caniels TG; van der Straten K; Snitselaar JL; Aldon Y; Bangaru S; Torres JT; Okba NMA; Claireaux M; Kerster G; Bentlage AEH; van Haaren MM; Guerra D; Burger JA; Schermer EE; Verheul KD; van der Velde N; van der Kooi A; van Schooten J; van Breemen MJ; Bijl TPL; Sliepen K; Aartse A; Derking R; Bontjer I; Kootstra NA; Wiersinga J; Vidarsson G; Haagmans BL; Ward AB; de Bree GJ; Sanders RW; van Gils MJ Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, eabc5902, DOI: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Li W; Drelich A; Martinez DR; Gralinski LE; Chen C; Sun Z; Schäfer A; Leist SR; Liu X; Zhelev DV; Zhang L; Peterson EL; Conard A; Mellors JW; Tseng CT; Baric RS; Dimitrov DS Rapid selection of a human monoclonal antibody that potently neutralizes SARS-CoV-2 in two animal models. BioRxiv 2020, 093088, DOI: . [Google Scholar]
- (54).Gurevich EV; Gurevich VV Therapeutic potential of small molecules and engineered proteins. Handb. Exp. Pharmacol 2014, 219, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Ngo HX; Garneau-Tsodikova S What are the drugs of the future? MedChemComm 2018, 9, 757–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Lipinski CA; Lombardo F; Dominy BW; Feeney PJ Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46, 3–26. [DOI] [PubMed] [Google Scholar]
- (57).Adedeji AO; Severson W; Jonsson C; Singh K; Weiss SR; Sarafianos SG Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol 2013, 87, 8017–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Lundin A; Dijkman R; Bergström T; Kann N; Adamiak B; Hannoun C; Kindler E; Jónsdóttir HR; Muth D; Kint J; Forlenza M; Müller MA; Drosten C; Thiel V; Trybala E Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the Middle East respiratory syndrome virus. PLoS Pathog. 2014, 10, e1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Turner AJ Angiotensin-converting enzyme 2: cardioprotective player in the renin-angiotensin system? Hypertension 2008, 52, 816–817. [DOI] [PubMed] [Google Scholar]
- (60).Donoghue M; Hsieh F; Baronas E; Godbout K; Gosselin M; Stagliano N; Donovan M; Woolf B; Robison K; Jeyaseelan R; Breitbart BE; Acton S A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res 2000, 87, e1–e9. [DOI] [PubMed] [Google Scholar]
- (61).Zhang H; Penninger JM; Li Y; Zhong N; Slutsky AS Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Hu H; Li L; Kao RY; Kou B; Wang Z; Zhang L; Zhang H; Hao Z; Tsui WH; Ni A; Cui L; Fan B; Guo F; Rao S; Jiang C; Li Q; Sun M; He W; Liu G Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J. Comb. Chem 2005, 7, 648–656. [DOI] [PubMed] [Google Scholar]
- (63).Struck AW; Axmann M; Pfefferle S; Drosten C; Meyer B A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antiviral Res. 2012, 94, 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Robertson NS; Spring DR Using peptidomimetics and constrained peptides as valuable tools for inhibiting protein–protein interactions. Molecules 2018, 23, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Huang L; Sexton DJ; Skogerson K; Devlin M; Smith R; Sanyal I; Parry T; Kent R; Enright J; Wu QL; Conley G; DeOliveira D; Morganelli L; Ducar M; Wescott CR; Ladner RC Novel peptide inhibitors of angiotensin-converting enzyme 2. J. Biol. Chem 2003, 278, 15532–15540. [DOI] [PubMed] [Google Scholar]
- (66).Huentelman MJ; Zubcevic J; Hernandez Prada JA; Xiao X; Dimitrov DS; Raizada MK; Ostrov DA Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension 2004, 44, 903–906. [DOI] [PubMed] [Google Scholar]
- (67).Vincent MJ; Bergeron E; Benjannet S; Erickson BR; Rollin PE; Ksiazek TG; Seidah NG; Nichol ST Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J 2005, 2, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wang M; Cao R; Zhang L; Yang X; Liu J; Xu M; Shi Z; Hu Z; Zhong W; Xiao G Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in Vitro. Cell Res. 2020, 30, 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Gao J; Tian Z; Yang X Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. BioSci. Trends 2020, 14, 72–73. [DOI] [PubMed] [Google Scholar]
- (70).Rainsford KD; Parke AL; Clifford-Rashotte M; Kean WE Therapy and pharmacological properties of hydroxychloroquine and chloroquine in treatment of systemic lupus erythematosus, rheumatoid arthritis and related diseases. Inflammopharmacology 2015, 23, 231–269. [DOI] [PubMed] [Google Scholar]
- (71).Yao X; Ye F; Zhang M; Cui C; Huang B; Niu P; Liu X; Zhao L; Dong E; Song C; Zhan S; Lu R; Li H; Tan W; Liu D In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis 2020, ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Chen C; Wang F; Lin C Chronic hydroxychloroquine use associated with qt prolongation and refractory ventricular arrhythmia. Clin. Toxicol 2006, 44, 173–175. [DOI] [PubMed] [Google Scholar]
- (73).Stas P; Faes D; Noyens P Conduction disorder and qt prolongation secondary to long-term treatment with chloroquine. Int. J. Cardiol 2008, 127, e80–e82. [DOI] [PubMed] [Google Scholar]
- (74).Yaylali SA; Sadigov F; Erbil H; Ekinci A; Akcakaya AA Chloroquine and hydroxychloroquine retinopathy-related risk factors in a turkish cohort. Int. Ophthalmol 2013, 33, 627–634. [DOI] [PubMed] [Google Scholar]
- (75).Lambert DW; Yarski M; Warner FJ; Thornhill P; Parkin ET; Smith AI; Hooper NM; Turner AJ Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J. Biol. Chem 2005, 280, 30113–30119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Haga S; Nagata N; Okamura T; Yamamoto N; Sata T; Yamamoto N; Sasazuki T; Ishizaka Y TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. Antiviral Res. 2010, 85, 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Towler P; Staker B; Prasad SG; Menon S; Tang J; Parsons T; Ryan D; Fisher M; Williams D; Dales NA; Patane MA; Pantoliano MW ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem 2004, 279, 17996–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Milewska A; Ciejka J; Kaminski K; Karewicz A; Bielska D; Zeglen S; Karolak W; Nowakowska M; Potempa J; Bosch BJ; Pyrc K; Szczubialka K Novel polymeric inhibitors of HCoV-NL63. Antiviral Res. 2013, 97, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Milewska A; Kaminski K; Ciejka J; Kosowicz K; Zeglen S; Wojarski J; Nowakowska M; Szczubialka K; Pyrc K HTCC: broad range inhibitor of coronavirus entry. PLoS One 2016, 11, e0156552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Imai Y; Kuba K; Rao S; Huan Y; Guo F; Guan B; Yang P; Sarao R; Wada T; Leong-Poi H; Crackower MA; Fukamizu A; Hui CC; Hein L; Uhlig S; Slutsky AS; Jiang C; Penninger JM Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Imai Y; Kuba K; Penninger JM The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp. Physiol 2008, 93, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Kilianski A; Baker SC Cell-based antiviral screening against coronaviruses: developing virus-specific and broad-spectrum inhibitors. Antiviral Res. 2014, 101, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Simmons G; Gosalia DN; Rennekamp AJ; Reeves JD; Diamond SL; Bates P Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 11876–11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Bosch BJ; Martina BEE; van der Zee R; Lepault J; Haijema BJ; Versluis C; Heck AJR; de Groot R; Osterhaus AEME; Rottier PJM Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Deng Y; Liu J; Zheng Q; Yong W; Lu M Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure 2006, 14, 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Pyrc K; Bosch BJ; Berkhout B; Jebbink MF; Dijkman R; Rottier P; van der Hoek L Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother 2006, 50, 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Chu LH; Chan SH; Tsai SN; Wang Y; Cheng CH; Wong KB; Waye MM; Ngai SM Fusion core structure of the severe acute respiratory syndrome coronavirus (SARS-CoV): in search of potent SARS-CoV entry inhibitors. J. Cell. Biochem 2008, 104, 2335–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Lu L; Liu Q; Zhu Y; Chan KH; Qin L; Li Y; Wang Q; Chan JF; Du L; Yu F; Ma C; Ye S; Yuen KY; Zhang R; Jiang S Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun 2014, 5, 3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Channappanavar R; Lu L; Xia S; Du L; Meyerholz DK; Perlman S; Jiang S Protective effect of intranasal regimens containing peptidic middle east respiratory syndrome coronavirus fusion inhibitor against mers-cov infection. J. Infect. Dis 2015, 212, 1894–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Xia S; Yan L; Xu W; Agrawal AS; Algaissi A; Tseng CK; Wang Q; Du L; Tan W; Wilson IA; Jiang S; Yang B; Lu L A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv 2019, 5, eaav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Xia S; Zhu Y; Liu M; Lan Q; Xu W; Wu Y; Ying T; Liu S; Shi Z; Jiang S; Lu L Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol 2020, DOI: DOI: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Xia S; Liu M; Wang C; Xu W; Lan Q; Feng S; Qi F; Bao LL; Du LY; Liu SW; Qin C; Sun F; Shi ZL; Zhu Y; Jiang SB; Lu L Inhibition of SARS-CoV-2 infection (previously 2019-nCoV) by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Sun Y; Zhang H; Shi J; Zhang Z; Gong R Identification of a novel inhibitor against Middle East respiratory syndrome coronavirus. Viruses 2017, 9, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Xia S; Xu W; Wang Q; Wang C; Hua C; Li W; Lu L; Jiang S Peptide-based membrane fusion inhibitors targeting hcov-229e spike protein HR1 and HR2 domains. Int. J. Mol. Sci 2018, 19, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Ahmed SF; Quadeer AA; McKay MR Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Miyoshi-Akiyama T; Ishida I; Fukushi M; Yamaguchi K; Matsuoka Y; Ishihara T; Tsukahara M; Hatakeyama S; Itoh N; Morisawa A; Yoshinaka Y; Yamamoto N; Lianfeng Z; Chuan Q; Kirikae T; Sasazuki T Fully human monoclonal antibody directed to proteolytic cleavage site in severe acute respiratory syndrome (SARS) coronavirus S protein neutralizes the virus in a rhesus macaque SARS model. J. Infect. Dis 2011, 203, 1574–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Ng OW; Keng CT; Leung CS; Peiris JS; Poon LL; Tan YJ Substitution at aspartic acid 1128 in the SARS coronavirus spike glycoprotein mediates escape from a S2 domain-targeting neutralizing monoclonal antibody. PLoS One 2014, 9, e102415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Mitsuki YY; Ohnishi K; Takagi H; Oshima M; Yamamoto T; Mizukoshi F; Terahara K; Kobayashi K; Yamamoto N; Yamaoka S; Tsunetsugu-Yokota Y A single amino acid substitution in the S1 and S2 Spike protein domains determines the neutralization escape phenotype of SARS-CoV. Microbes Infect. 2008, 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).van Dongen MJP; Kadam RU; Juraszek J; Lawson E; Brandenburg B; Schmitz F; Schepens WBG; Stoops B; van Diepen HA; Jongeneelen M; Tang C; Vermond J; van Eijgen-Obregoso Real A; Blokland S; Garg D; Yu W; Goutier W; Lanckacker E; Klap JM; Peeters DCG; Wu J; Buyck C; Jonckers THM; Roymans D; Roevens P; Vogels R; Koudstaal W; Friesen RHE; Raboisson P; Dhanak D; Goudsmit J; Wilson IA A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science 2019, 363, eaar6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Frey G; Rits-Volloch S; Zhang XQ; Schooley RT; Chen B; Harrison SC Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Yi L; Li Z; Yuan K; Qu X; Chen J; Wang G; Zhang H; Luo H; Zhu L; Jiang P; Chen L; Shen Y; Luo M; Zuo G; Hu J; Duan D; Nie Y; Shi X; Wang W; Han Y; Li T; Liu Y; Ding M; Deng H; Xu X Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol 2004, 78, 11334–11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Zhao G; Du L; Ma C; Li Y; Li L; Poon VK; Wang L; Yu F; Zheng BJ; Jiang S; Zhou Y A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol. J 2013, 10, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Kadam RU; Wilson IA Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Wang X; Cao R; Zhang H; Liu J; Xu M; Hu H; Li Y; Zhao L; Li W; Sun X; Yang X; Shi Z; Deng F; Hu Z; Zhong W; Wang M The anti-influenza virus drug, Arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery 2020, 6, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Vankadari N Arbidol: A Potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents 2020, 105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Wang Z; Yang B; Li Q; Wen L; Zhang R Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis 2020, ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Bosch BJ; Bartelink W; Rottier PJ Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol 2008, 82, 8887–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Verma S; Dixit R; Pandey KC Cysteine Proteases: modes of activation and future prospects as pharmacological targets. Front. Pharmacol 2016, 7, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Sosnowski P; Turk D Caught in the act: the crystal structure of cleaved cathepsin L bound to the active site of cathepsin L. FEBS Lett. 2016, 590, 1253–1261. [DOI] [PubMed] [Google Scholar]
- (110).Glupczynski Y; Lagast H; Van Der Auwera P; Thys J; Crokaert F; Yourassowsky E; Meunier-Carpentier F; Klastersky J; Kains JP; Serruys-Schoutens E Clinical evaluation of teicoplanin for therapy of severe infections caused by gram-positive bacteria. Antimicrob. Agents Chemother 1986, 29, 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Zhou N; Pan T; Zhang J; Li Q; Zhang X; Bai C; Huang F; Peng T; Zhang J; Liu C; Tao L; Zhang H Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biol. Chem 2016, 291, 9218–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Zhang JS; Ma XC; Yu F; Liu J; Zou F; Pan T; Zhang H Teicoplanin potently blocks the cell entry of 2019-nCoV. BioRxiv. 2020, 935387 DOI: . [Google Scholar]
- (113).Dana D; Pathak SK A review of small molecule inhibitors and functional probes of human cathepsin L. Molecules 2020, 25, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Shah PP; Wang T; Kaletsky RL; Myers MC; Purvis JD ; Jing H; Huryn DM; Greenbaum DC; Smith AB; Bates P; Diamond SL A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol 2010, 78, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Zhou Y; Vedantham P; Lu K; Agudelo J; Carrion R Jr; Nunneley JW; Barnard D; Pohlmann S; McKerrow JH; Renslo AR; Simmons G Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015, 116, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Kawase M; Shirato K; van der Hoek L; Taguchi F; Matsuyama S Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol 2012, 86, 6537–6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Bestle D; Heindl MR; Limburg H; van TVL; Pilgram O; Moulton H; Stein DA; Hardes K; Eickmann M; Dolnik O; Rohde C; Becker S; Klenk HD; Garten W; Steinmetzer T; Böttcher-Friebertshäuser E TMPRSS2 and furm are both essential for proteolytic activation and spread of SARS-CoV-2 in human airway epithelial cells and provide promising drug targets. BioRxiv 2020, 042085 DOI: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Hoffmann M; Kleine-Weber H; Pöhlmann S A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 2020, 78, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Cherian SS; Agrawal M; Basu A; Abraham P; Gangakhedkar RR; Bhargava B Perspectives for repurposing drugs for the coronavirus disease 2019. Indian J. Med. Res 2020, 151, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Blanchard E; Belouzard S; Goueslain L; Wakita T; Dubuisson J; Wychowski C; Rouille Y Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol 2006, 80, 6964–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Pohjala L; Utt A; Varjak M; Lulla A; Merits A; Ahola T; Tammela P Inhibitors of alphavirus entry and replication identified with a stable chikungunya replicon cell line and virus-based assays. PLoS One 2011, 6, e28923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Liu Q; Xia S; Sun Z; Wang Q; Du L; Lu L; Jiang S Testing of Middle East respiratory syndrome coronavirus replication inhibitors for the ability to block viral entry. Antimicrob. Agents Chemother 2015, 59, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Sisk JM; Frieman MB; Machamer CE Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J. Gen. Virol 2018, 99, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Coleman CM; Sisk JM; Mingo RM; Nelson EA; White JM; Frieman MB Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol 2016, 90, 8924–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Dyall J; Coleman CM; Hart BJ; Venkataraman T; Holbrook MR; Kindrachuk J; Johnson RF; Olinger GG; Jahrling PB; Laidlaw M; Johansen LM; Lear-Rooney CM; Glass PJ; Hensley LE; Frieman MB Repurposing of clinically developed drugs for treatment of middle east respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother 2014, 58, 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Shin JS; Jung E; Kim M; Baric RS; Go YY Saracatinib inhibits middle east respiratory syndrome-coronavirus replication in vitro. Viruses 2018, 10, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Swinney DC Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharmacol. Ther 2013, 93, 299–301. [DOI] [PubMed] [Google Scholar]
- (128).Wu G; Zhao T; Kang D; Zhang J; Song Y; Namasivayam V; Kongsted J; Pannecouque C; De Clercq E; Poongavanam V; Liu X; Zhan P Overview of recent strategic advances in medicinal chemistry. J. Med. Chem 2019, 62, 9375–9414. [DOI] [PubMed] [Google Scholar]
- (129).Du J; Guo J; Kang D; Li Z; Wang G; Wu J; Zhang Z; Fang H; Hou X; Huang Z; Li G; Lu X; Liu X; Ouyang L; Rao Z; Zhan P; Zhang X; Zhang Y; New techniques and strategies in drug discovery. Chin. Chem. Lett 2020. DOI: DOI: 10.1016/j.cclet.2020.03.028. [DOI] [Google Scholar]
- (130).Islam MT; Sarkar C; El-Kersh DM; Jamaddar S; Uddin SJ; Shilpi JA; Mubarak MS Natural products and their derivatives against coronavirus: a review of the non-clinical and preclinical data. Phytother. Res 2020. DOI: DOI: 10.1002/ptr.6700. [DOI] [PubMed] [Google Scholar]
- (131).Ho TY; Wu SL; Chen JC; Li CC; Hsiang CY Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007, 74, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Kao RY; Tsui WH; Lee TS; Tanner JA; Watt RM; Huang JD; Hu L; Chen G; Chen Z; Zhang L; He T; Chan KH; Tse H; To AP; Ng LW; Wong BC; Tsoi HW; Yang D; Ho DD; Yuen KY Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol 2004, 11, 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Cao J; Forrest JC; Zhang X A screen of the NIH clinical collection small molecule library identifies potential anti-coronavirus drugs. Antiviral Res. 2015, 114, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Bisht H; Roberts A; Vogel L; Bukreyev A; Collins PL; Murphy BR; Subbarao K; Moss B Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 6641–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Chen Z; Zhang L; Qin C; Ba L; Yi CE; Zhang F; Wei Q; He T; Yu W; Yu J; Gao H; Tu X; Gettie A; Farzan M; Yuen KY; Ho DD Recombinant modified vaccinia virus ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol 2005, 79, 2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Woo PCY; Lau SKP; Tsoi HW; Chen Z; Wong BHL; Zhang L; Chan JKH; Wong LP; He W; Ma C; Chan KH; Ho DD; Yuen KY SARS coronavirus spike polypeptide dna vaccine priming with recombinant spike polypeptide from escherichia coli as booster induces high titer of neutralizing antibody against SARS coronavirus. Vaccine 2005, 23, 4959–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Zhou Y; Jiang S; Du L Prospects for a MERS-CoV spike vaccine. Expert Rev. Vaccines 2018, 17, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Raab CP Passive immunization. Prim Care. 2011, 38, 681–691. [DOI] [PubMed] [Google Scholar]
- (139).Luke TC; Kilbane EM; Jackson JL; Hoffman SL Meta-analysis: convalescent blood products for spanish influenza pneumonia: a future h5n1 treatment? Ann. Intern. Med 2006, 145, 599–609. [DOI] [PubMed] [Google Scholar]
- (140).Arabi Y; Balkhy H; Hajeer AH; Bouchama A; Hayden FE; Al-Omari A; Al-Hameed FM; Taha Y; Shindo N; Whitehead J; Merson L; AlJohani S; Al-Khairy K; Carson G; Luke TC; Hensley L; Al-Dawood A; Al-Qahtani S; Modjarrad K; Sadat M; Rohde G; Leport C; Fowler R Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: a study protocol. SpringerPlus 2015, 4, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]