Abstract
Nearly 7% of the world’s population lives with a hemoglobin variant. Hemoglobins S, C, and E are the most common and significant hemoglobin variants worldwide. Sickle cell disease, caused by hemoglobin S, is highly prevalent in sub-Saharan Africa and in tribal populations of Central India. Hemoglobin C is common in West Africa, and hemoglobin E is common in Southeast Asia. Screening for significant hemoglobin disorders is not currently feasible in many low-income countries with the high disease burden. Lack of early diagnosis leads to preventable high morbidity and mortality in children born with hemoglobin variants in low-resource settings. Here, we describe HemeChip, the first miniaturized, paper-based, microchip electrophoresis platform for identifying the most common hemoglobin variants easily and affordably at the point-of-care in low-resource settings. HemeChip test works with a drop of blood. HemeChip system guides the user step-by-step through the test procedure with animated on-screen instructions. Hemoglobin identification and quantification is automatically performed, and hemoglobin types and percentages are displayed in an easily understandable, objective way. We show the feasibility and high accuracy of HemeChip via testing 768 subjects by clinical sites in the United States, Central India, sub-Saharan Africa, and Southeast Asia. Validation studies include hemoglobin E testing in Bangkok, Thailand, and hemoglobin S testing in Chhattisgarh, India, and in Kano, Nigeria, where the sickle cell disease burden is the highest in the world. Tests were performed by local users, including healthcare workers and clinical laboratory personnel. Study design, methods, and results are presented according to the Standards for Reporting Diagnostic Accuracy (STARD). HemeChip correctly identified all subjects with hemoglobin S, C, and E variants with 100% sensitivity, and displayed an overall diagnostic accuracy of 98.4% in comparison to reference standard methods. HemeChip is a versatile, mass-producible microchip electrophoresis platform that addresses a major unmet need of decentralized hemoglobin analysis in resource-limited settings.
Keywords: global health, hemoglobinopathies, sickle cell anemia, point-of-care diagnostics
Graphical Abstract

(Illustration Credit: Grace Gongaware, Cleveland Institute of Art)
INTRODUCTION
Hemoglobin disorders are among the world’s most common monogenic diseases. Nearly 7% of the world’s population carry hemoglobin gene variants, with the most prevalent hemoglobinopathies or structural hemoglobin variants being the recessive β-globin gene mutations, βS or S, βC or C, and βE or E [1–3]. Hemoglobin S is highly prevalent in sub-Saharan Africa [4] and in tribal populations of Central India [5]. Hemoglobin C is common in West Africa [6], and hemoglobin E is common in Southeast Asia [7]. Hemoglobin S results from a single point mutation in the 6th codon on the β-globin gene replacing the normal amino acid glutamine with the hydrophobic amino acid valine [8–10]. Sickle cell disease (SCD) causes the highest morbidity and mortality among hemoglobin disorders [11]. SCD arises when these mutations are inherited homozygously (Hb SS or SCD-SS) or paired with another β-globin gene mutation, such as hemoglobin C (Hb SC or SCD-SC) or β-thalassemia (compound heterozygous, Hb Sβthal+/0). In SCD, abnormal polymerization of deoxygenated hemoglobin S makes red blood cells (RBCs) stiff, changes membrane properties, alters shape, and triggers deleterious activation of inflammatory and endothelial cells [12–14]. Sickled RBCs are non-deformable and adhesive in the microcirculation [15–20], particularly in parts of the body where the oxygen tension is relatively low, such as the kidney or spleen [13, 17, 21, 22]. Abnormally shaped RBCs result in microvascular occlusion and a vicious cycle of enhanced sickling, hemoglobin desaturation, and further vascular occlusion [23–25]. In childhood, recurrent splenic infarction from sickled RBCs increases the risk for life-threatening infections [10, 26–29]. In addition, young children with SCD are at risk of life-threatening cerebral vasculopathy [29]. Afflicted patients who survive to adulthood can suffer both acute and chronic painful crises as well as cumulative organ damage and early mortality [29, 30]. Infections, stroke, and numerous other SCD-related complications can be mitigated by newborn/neonatal screening and comprehensive medical care [29, 31, 32]. Individuals who inherit one copy of hemoglobin S and one copy of the normal hemoglobin A have sickle cell trait (Hb AS or SCD Trait). These people are healthy carriers, but have a 25% chance of transmitting SCD to their offspring. Individuals who carry one copy of hemoglobin C or hemoglobin βthal+/0 and one copy of hemoglobin A may likewise transmit hemoglobin C disease or thalassemia [33]. Approximately 70% of individuals with SCD have homozygous Hb SS and over 25% have compound heterozygous Hb SC or Hb Sβthal+/0 [8].
Hemoglobin E is another common structural hemoglobin variant, but it is unique in that it also decreases expression of the β-globin gene [7, 34]. Individuals with hemoglobin E trait (Hb AE or Hb E Trait) are asymptomatic, and homozygous hemoglobin E (Hb EE or Hb E Disease) causes a mild microcytic anemia. However, hemoglobin E in combination with βthal (Hb Eβthal+/0) causes thalassemia of varying severity [35]. Hemoglobin SE disease is similar to Hb Sβthal in terms of clinical manifestations and treatment, though uncommon due to the geographical separation of the S and E variants [35, 36]. With increasing migration and inter-racial marriages, combinations of these hemoglobin diseases are being encountered all around the world [35, 36]. Regardless of their geographic origin, all of these common hemoglobin variants need to be screened for, so that individuals with disease can be diagnosed early and managed timely. Newborn/neonatal screening, currently available in resource-rich countries, is needed for optimal management of hemoglobinopathies worldwide [37–39]. For example, in high-income countries, which have less than 1% of the global disease burden, over 90% of babies born with SCD survive into adulthood due to established national screening programs and comprehensive care [29]. In contrast, in low-income countries, due to lack of nationwide screening and comprehensive care programs, up to 80% of babies born with SCD are undiagnosed and less than half of them survive beyond 5 years of age [29]. More than 300,000 babies are born each year with SCD, with 2–3 per 100 births in some high-burden countries with expected under-5 mortality of 50–90%. It is projected that by 2050, about 400,000 babies will be born with SCD annually worldwide [29, 40]. Screening for hemoglobin disorders is not currently feasible in many low-income countries with the high disease burden [29]. For example, in Nigeria, 150,000 babies are born with the disease every year [41]. An estimated 50–90% of these babies die before age 5, in part because they are not diagnosed and hence not treated [15, 42–45].
Hemoglobinopathy screening after birth is mandated by all 50 states in the United States and in the District of Columbia, as well as in many other high-resource states, including The United Kingdom and France [45–49]. Newborn screening programs in the United States and other high-resource countries typically involve collection and shipping of blood samples to centralized laboratories [46, 50, 51]. While decentralized blood sample collection and centralized testing works in high-resourced settings, this approach is not practical in resource-limited regions due to the logistical and infrastructural limitations. Hemoglobinopathy screening studies conducted in low-resource settings, using centralized laboratories, have reported that up to 50% of test results never reach to the patients due to infrastructural challenges [52–54].
In resource-rich countries, standard clinical laboratory tests (high-performance liquid chromatography (HPLC) and hemoglobin electrophoresis) are typically used in the diagnosis of hemoglobin disorders [33]. Additionally, genetic testing, usually polymerase chain reaction-based, can be used to precisely identify globin gene mutations [55]. However, these advanced laboratory techniques require trained personnel and state-of-the-art facilities, which are lacking or in short supply in countries where the prevalence of hemoglobin disorders is the highest [29]. Furthermore, these tests are costly in terms of time, labor, and resources [15, 56]. For example, it may take days to weeks to receive the test results from centralized clinical laboratories [56], and when screening is conducted in remote areas, locating those who test positive may be difficult or impossible [15, 44, 56]. Therefore, there is a need for affordable, portable, easy-to-use, accurate point-of-care tests to facilitate decentralized hemoglobin testing in resource-constrained countries [20, 29]. The realities of resource-limited environments demand a fundamentally different approach to diagnosis: one that is affordable, portable, and easily administered by entry-level healthcare workers in local health service settings or in rural areas at the point-of-need [15, 44, 56]. Importantly, test results must be available while the patient is still present so that the test result can be given to the patient or legal guardian(s) immediately, and the treatment and education can begin without losing the patient to follow-up. The WHO estimates that more than 70% of SCD related deaths are preventable with early diagnosis and widely available cost-efficient interventions (e.g., pneumococcal vaccinations, daily penicillin, hydroxyurea) [27–29, 45, 57–62]. For example, hydroxyurea use has been shown to reduce the vaso-occlusive crises, stroke, infections, malaria, transfusions, and death in children with SCD, warranting the need for wider access to this treatment [29, 59–62]. Hydroxyurea treatment has been shown to be feasible, safe, and effective in children with SCD living in India [63] and in sub-Saharan Africa [29, 59, 64, 65].
In this manuscript, we describe the design, development, manufacturing, and clinical testing of a novel point-of-care hemoglobin test, HemeChip. HemeChipis a paper-based, microchip electrophoresis technology that helps with diagnosis of hemoglobin disorders in resource-limited settings. HemeChip system is composed of a single-use, disposable cartridge and a portable, affordable reader. HemeChip cartridge can be mass-produced at low cost. HemeChip reader houses the cartridge and it is used to separate, image, and track hemoglobin variants in real-time during electrophoresis. The fundamental principle behind the HemeChip technology is hemoglobin electrophoresis, in which different variants can be separated based on charge differences, when subjected to an electric field in the presence of a carrier substrate [66, 67]. The HemeChip test works with a standard finger-prick or a heel-prick blood sample, or with a venous blood sample. HemeChip test is completed in less than ten minutes, and can be run at the point-of-care, hence, the results would be available during a patient’s visit. Furthermore, the compact design of the HemeChip system allows portability for decentralized testing and use at the point-of-need, which eliminates blood sample transfer to central laboratories.
MATERIALS AND METHODS
Fully integrated microchip electrophoresis cartridge allows high-volume manufacturing
The HemeChip cellulose acetate paper-based microchip electrophoresis system (Fig. 1) facilities, for the first time, real-time tracking and quantitative analysis of hemoglobin electrophoresis process (Video S1). The HemeChip cartridge is composed of two injection molded plastic parts made of Optix® CA-41 Polymethyl Methacrylate Acrylic. This single-use, cartridge-based design was transformed from a proof-of-concept laboratory prototype to a version that supports low-cost mass-production via injecting molding (Fig. S1) as described in Supplementary Materials. The top and bottom parts (Fig. S2) were manufactured with a 1+1 injection mold. Cartridge design embodies specific geometrical features and a precisely designed energy director (Fig. S2) for rapid ultrasonic welding after assembly. Cellulose acetate paper was chosen because of its stability over environmental conditions [68]. Cartridge layout, plastic material selection, and injection molding process were engineered to achieve structural integrity, uniform optical clarity, and high light transmission (up to 80%) in the visible spectrum (Fig. S3). The injection molded HemeChip cartridge embodies a pair of round corrosion-resistant [69], biomedical grade stainless steel 316 electrodes [70]. HemeChip electrodes provide oxidative resistance, stability against electrochemical reactions during operation, and reliability of electrical connection with the power source (Fig. 1A). The combination of high stability cellulose acetate paper [68], injection molded Polymethyl Methacrylate Acrylic plastic, and corrosion-resistant biomedical grade stainless-steel electrodes [69, 70] result in a shelf life of at least two years. The cartridge also houses a pair of buffer pools that are in direct contact with the electrode top surface and the cellulose acetate paper strip (Fig. 1B&C). One corner of the HemeChip cartridge is chamfered to facilitate correct orientation during use (Fig. 1A&B).
Fig. 1. HemeChip: paper-based microchip electrophoresis for hemoglobin testing.
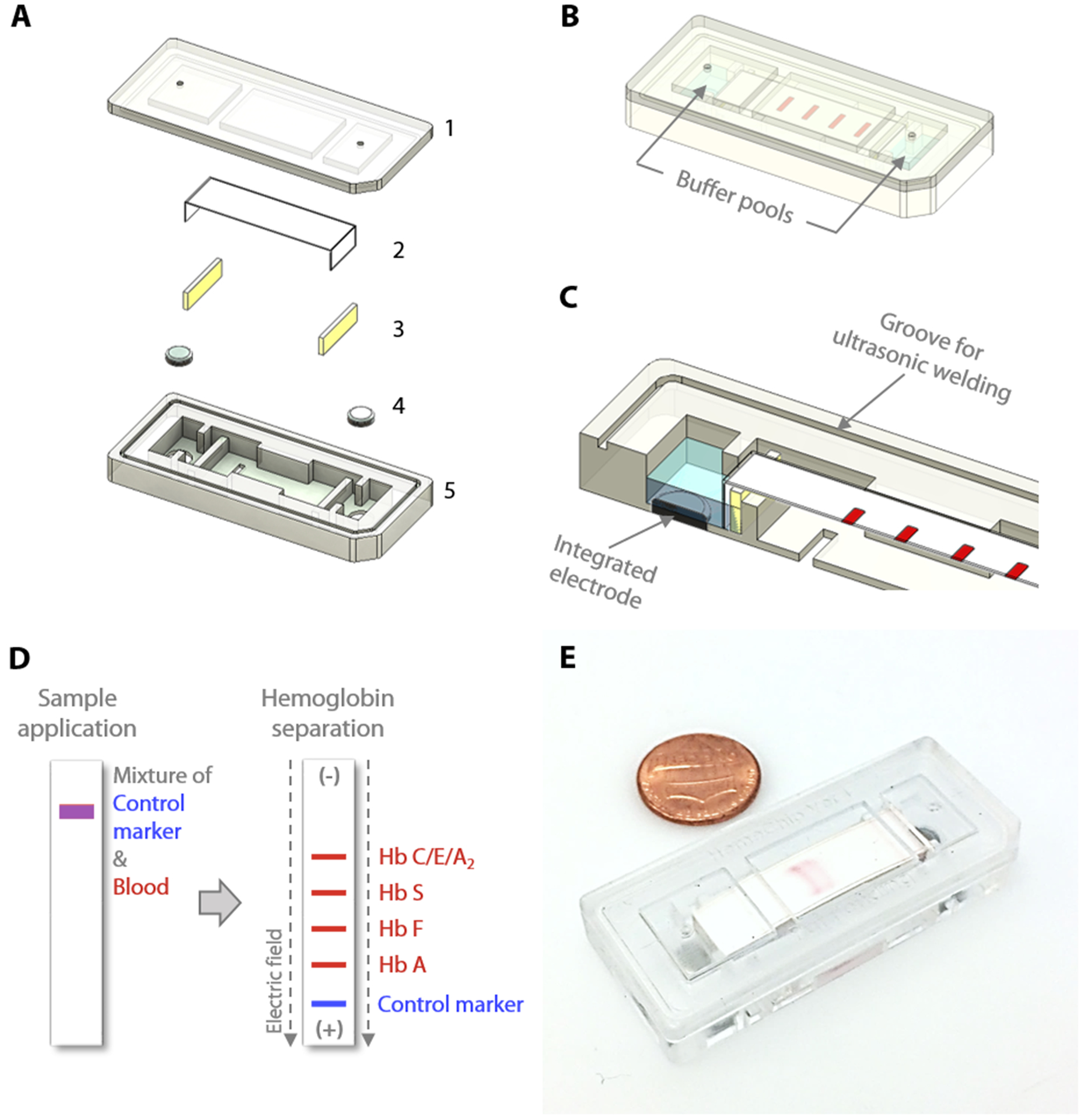
HemeChip is a miniaturized, fully integrated cartridge-based microchip electrophoresis system that can be mass-produced. (A) Top (1) and bottom (5) plastic parts are manufactured via injection molding. Cartridge contains a single strip of cellulose acetate paper (2), a pair of blotting pads (3), and integrated stainless-steel electrodes (4). (B) HemeChip cartridge design is compact, fully integrated, and self-contained, including liquid compartments (buffer pools). One corner of the HemeChip cartridge is chamfered to restrict orientation and facilitate correct placement during use. (C) A partial section view of the internal components of HemeChip, showing the cross-section of an electrode and the cellulose acetate paper. (D) A schematic representation of the separation of hemoglobin variants in HemeChip: normal hemoglobin (Hb A), fetal hemoglobin (Hb F), sickle hemoglobin (Hb S), and hemoglobins C/E/A2 that co-migrate. A blue control marker (xylene cyanol) is pre-mixed with blood before sample application into the cartridge. (E) A fully assembled injection molded HemeChip is shown after a completed test with hemoglobin S and C bands that are visible.
Design transformation of HemeChip from laboratory prototype to a mass-producible cartridge
The HemeChip cartridge was previously fabricated by a lamination approach composed of five distinct plastic layers and did not incorporate integrated electrodes (Fig. S1A&B). In the injected molded design, the number of plastic parts was reduced to two, a top part, and a bottom part, which can be ultrasonically welded to encompass the internal components and integrated stainless-steel electrodes (Fig. S1C&D). This reduction in the number of injection-molded parts reduced the final assembly time and effort. The current design employs integrated stainless-steel electrodes (Fig. S1C&D). The choice for electrode material was constrained by the cost and the corrosion resistance of the electrode material. The use of noble metals and/or their alloys, which are the preferred materials for specialized applications [71–74], would have increased the cost of the chips significantly. In addition, noble metals can corrode when subjected to electrochemical processes [74–77]. The 316 stainless steel, which we employed, is known for its high corrosion resistance [70]. It is inexpensive and is available in a variety of stock shapes that are easily machined or processed into the required dimensions. Although the design is sophisticated in view of all the delicate interior features, this adaptation provided more control over the design of the complex interior features, which were unattainable with the lamination-based fabrication approach (Fig. S1A). These interior features include custom designed buffer pools, buffer basin ribs, metal electrode enclosures, alignment and positioning features for the cellulose acetate paper strips, sample and buffer ports, fiducial markers, and product artwork (Fig. 2A&B). Another improvement to the HemeChip design was the inclusion of the in-chip blotting mechanism (Fig. S1C). The blotting mechanism is designed to handle any excess TBE buffer that may be applied to the cellulose acetate paper. A notable addition to the transformed design is the energy director and ultrasonic welding groove for the purpose of ultrasonic welding to fully seal the HemeChip cartridge top and bottom pieces (Fig. S2A&B).
Fig. 2. Overview of HemeChip operation and hemoglobin variant separation with a control marker.
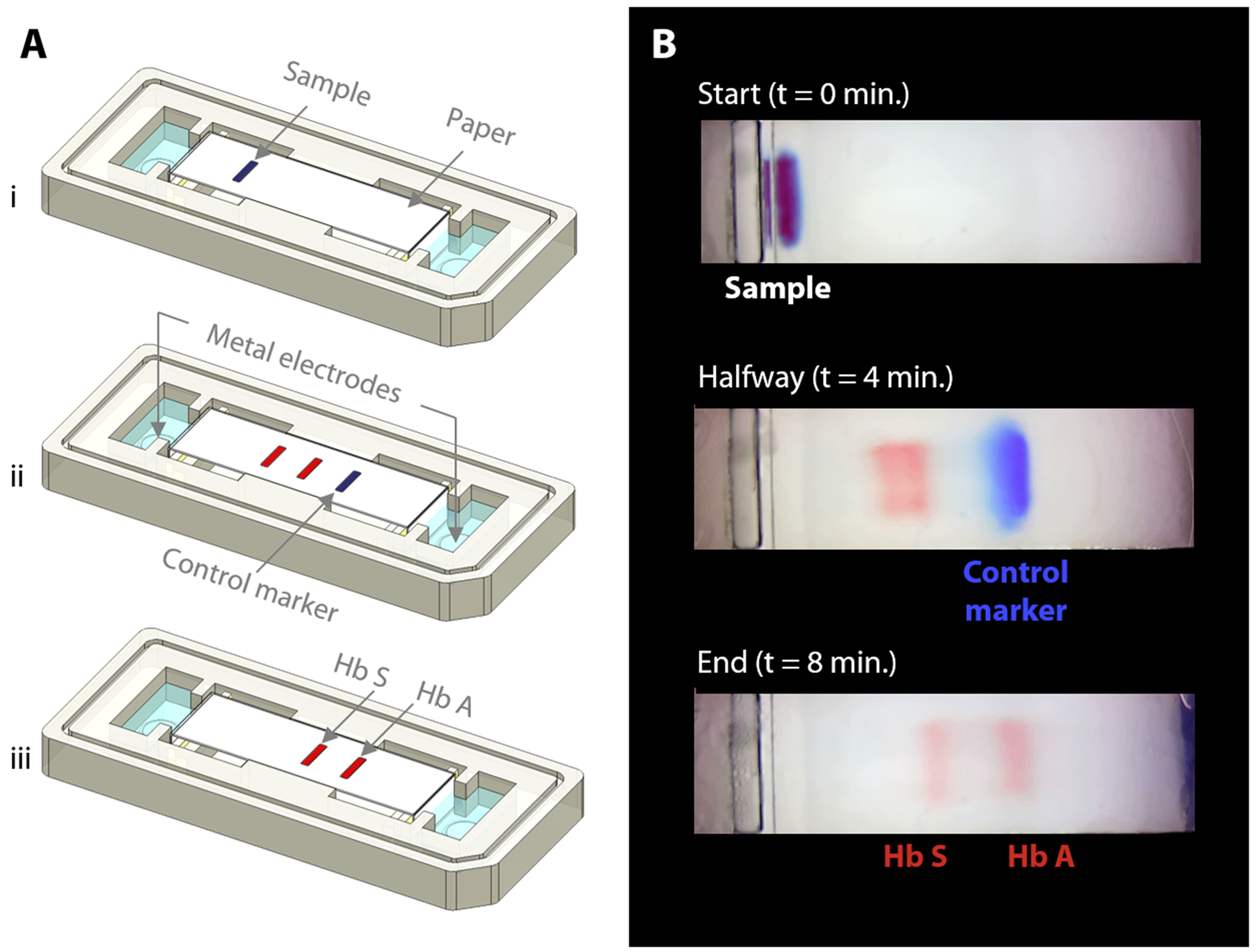
(A) Schematic illustration of hemoglobin separation and the blue control marker (xylene cyanol) migration in HemeChip. (i) Beginning of the test is shown with blood sample and marker mixture applied. (ii) Hemoglobin bands and the control marker start migrating. Migration of the blue control marker is used image processing algorithm to confirm that the test is running as expected. (iii) Fully separated hemoglobin bands appear at the end of the test. Control marker migrates all the way to the end and leaves the field of view, at which time, hemoglobin bands are imaged and analyzed in their final positions. (B) Time lapse images captured during a HemeChip test demonstrate the separation of hemoglobin A and S bands, and the migration of the control marker.
HemeChip portable reader design
One advantage of point-of-care technologies is that they can be used in remote locations where use of existing technologies is not feasible [78–80]. HemeChip has been designed as a battery-powered, portable test platform to enable hemoglobin testing in remote locations (Fig. 3). HemeChip reader consists of a rechargeable battery power supply, a data acquisition system, and an imaging and image analysis unit (Fig. S4). The portable HemeChip Reader, once fully charged, allows a minimum of ten hours of testing, which corresponds to at least 48 tests per charge. The reader is equipped with a rechargeable 12V lithium ion battery with a capacity of 11,000 mAh, yielding 132-Watt hours. Each test consumes about 2-Watt hours. The reader enables automated interpretation of test results, local and remote test data storage, and includes geolocation (Global Positioning System). The user is guided through the test process, and the test result is shown on the reader’s display and the reader stores the results. The reader can link via Bluetooth or Wi-Fi to a cell phone or PC to transfer the information for review or retention. The reader is equipped to send a PDF version of the results page to a wireless network printer. The data acquisition system is a national instruments data acquisition board (USB-6001) controlled by a custom-built LabView program that records the voltage and current values for the duration of the test. The imaging system consists of an ELP video camera (ELP-USB500W02M) that captures real-time video and the run-time images (Fig. S4). The imaging system includes 85° wide angle 5 mega pixel HD USB camera and the camera optics are modified with a 3.6 mm, F1.3 mega pixel, CCTV Board Lens. The imaging system and the developed software are calibrated to correct for the fish-eye effect (due to the wide-angle lens) and to normalize the nonlinear pixels-to-distance relation. The power supply, imaging system and data acquisition system are assembled inside a rugged Pelican 1400 case (Fig. 3A). An embedded tablet computer runs the custom-built software to control the Reader as well as to acquire, process, and analyze the run-time data. The chamber door houses the backlight used in the imaging system and a magnetic door sensor to ensure that the high voltage supply turns on only when the door is closed (Fig. 3B). An array of white light LEDs is used as the light source for the imaging system. The HemeChip cartridges are imaged using a transmissive light mode.
Fig. 3. Portable reader for HemeChip.
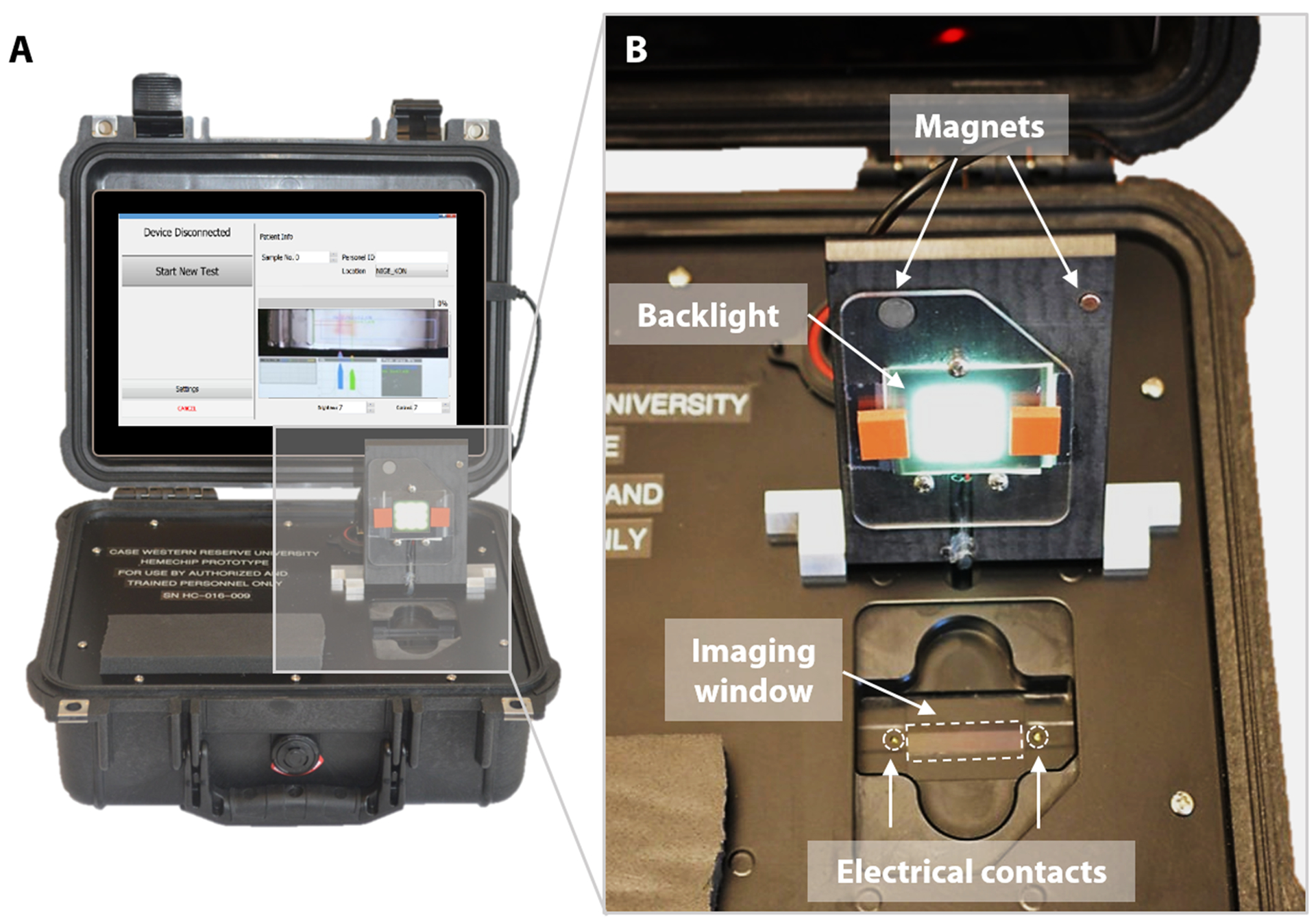
(A) The portable HemeChip prototype readers were built for research use only in a robust Pelican Protector Case for clinical studies worldwide. The reader includes a rechargeable battery, a touch-screen tablet computer, and an integrated imaging system. The reader guides the user through the test procedure via on-screen instructions, allows real- time imaging, automated data analysis, result storage, and wireless transmission of test results. (B) The cartridge chamber in the reader houses electrical contacts, an imaging window, and a backlight for imaging in transmission mode. The chamber door is equipped with magnetic contacts as a safety feature for sensing the door status as open or closed.
User interface and automated image analysis
We developed a user-friendly user interface (UI) that guides the user through the test, performs quality checks, and analyzes the results. This UI has the following functionalities (Fig. S5): (i) perform hemoglobin separation in HemeChip cartridge by controlling the Reader electronic circuit, (ii) collect runtime data for quality checks, (iii) guide the user through the test procedure using step-by-step instructions (Fig. S6, Video S2), and (iii) analyze post-run data analysis from the collected real-time data and images. The UI performs the test control and run-time data collection simultaneously and automatically without any interference or assistance from the user. The UI is designed to generate the data files with specific file name and format based on the information provided before the test, which contains the patient, sample and test identifier. HemeChip tests are monitored real-time with a custom developed application. The real-time tracking of hemoglobin bands (Fig. 4, Video S1) and automated image analysis provide necessary information for the identification and quantification of the test results. The application is configured to automatically detect the sample application point and the separated hemoglobin bands. Relative pixel intensities along the paper are used to identify the peaks corresponding to each type of hemoglobin band. The area under each peak is calculated to obtain the relative hemoglobin percentages. The area under each peak is outlined using the valley-to-valley method commonly used in gas chromatography [81]. The HemeChip image analysis algorithm was developed to find the position and intensity of the single hemoglobin band (for homogeneous hemoglobin types) or multiple hemoglobin bands (for heterogeneous hemoglobin types) that appear on the cellulose acetate paper strip inside the HemeChip after a test has been performed. The captured RGB image is analyzed to determine how far the hemoglobin band(s) propagated and the relative percentage of each band based on pixel intensity data.
Fig. 4. Real-time imaging, image analysis, and tracking of control marker and hemoglobin bands in HemeChip.
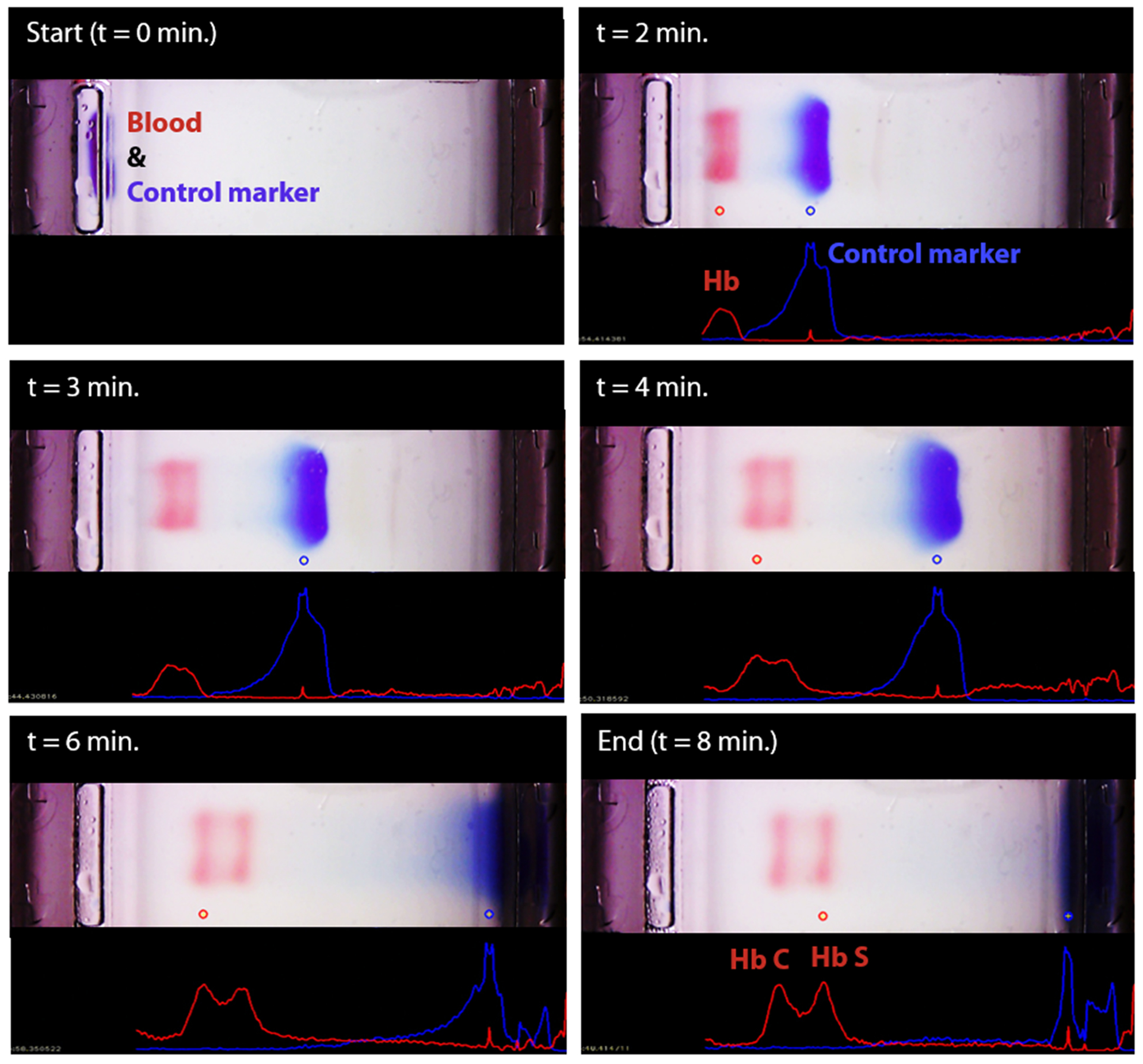
The time-lapse images show the real-time tracking of the blue control marker and hemoglobin bands. Hemoglobin bands, and the control marker are imaged, automatically recognized, and identified. Movement of the control marker is tracked and used to confirm that the test is running as expected. In this example, hemoglobin types S and C were identified by the algorithm.
Clinical study design and participants
Clinical study design, study participants, sample size calculation, and details on test methods are described according to the Standards for Reporting Diagnostic Accuracy (STARD) 2015 guidelines [82]. Institutional Review Board (IRB) approved study protocols included the following common objectives: to validate HemeChip technology as a point-of-care platform for hemoglobin testing, to compare the screening results obtained from HemeChip with that obtained from laboratory electrophoresis and/or HPLC as the standard reference methods, to determine the diagnostic accuracy of the test including sensitivity and specificity, and to determine the feasibility of using HemeChip as a point-of-care testing platform in low and middle income countries. We obtained approvals from Institutional Review Boards at University Hospitals Cleveland Medical Center (UHCMC IRB# 04-17-15), University of Nebraska Medical Center (UNMC IRB# 754–17-CB), Amino Kano Teaching Hospital in Kano, Nigeria (AKTH/MAC/SUB/12A/P-3/VI/2102), the Kano State Ministry of Health in Nigeria (MOH/Off/797/T.I/377). The study was registered in United States Library of Medicine’s ClinicalTrials.gov (Identifier: NCT03948516). The study protocol in India was approved by the institutional ethical committee of ICMR (Indian Council of Medical Research) National Institute of Research in Tribal Health, Jabalpur, Madhya Pradesh, India (Letter vide no. NIRTH/IEC/1153/2017). The study protocol in Thailand was approved by the Siriraj Institutional Review Board (SiRB Protocol no. 906/2561(EC1)). Informed consents were obtained from all participants of these clinical studies.
Clinical sample size estimation was based on published methods by incorporating disease prevalence into sample size calculation [83] with 95% confidence interval and with estimated sensitivity and specificity of 98% (Table S3). We estimated the SCD-SS (homozygous S) disease prevalence in the study population in Nigeria at 3% [84], which resulted in a minimum sample size of 251. SCD-SS prevalence was estimated at 2.7% in Central India [85], which resulted in a minimum sample size of 279 in India. In Bangkok, Thailand, since homozygous Hb E disease prevalence is low (1.8%), we utilized a combined homozygous and heterozygous Hb E (with or without α-thalassemia) prevalence estimated at 8.7% [86], which resulted in a minimum sample size of 87.
315 children (6 weeks to 5 years of age) were tested in Kano, Nigeria. Study participants were enrolled as part of the existing Community-Acquired Pneumonia and Invasive Bacteremia Disease study at three government hospitals, Amino Kano Teaching Hospital, Murtala Muhammad Specialist Hospital, and Hasiya Bayero Pediatric Hospital. Inclusion criteria for the Nigeria study included: 6 weeks to 5 years of age, fever or hypothermia and one of the following conditions: prostration, excessive crying, poor feeding, altered consciousness, convulsion, difficulty breathing, profuse vomiting, diarrhea, and a provision of signed and dated informed consent by the parent or guardian. Exclusion criteria for the Nigeria study included, parent or child choosing to opt out of the study after initial consent, and blood transfusion within 3 months of the study enrollment. In Cleveland, Ohio, 32 adult participants were recruited to the study as part of an ongoing clinical trial (ClinicalTrials.gov Identifier: NCT02824471). The main purpose of the clinical study in Cleveland was to test SCD-SC blood samples, which are rare in Nigeria and India.
124 subjects (7 weeks to 63 years old) were tested in Bangkok, Thailand by local laboratory personnel. Surplus blood samples were obtained from patients undergoing clinical laboratory testing at Siriraj Hospital in Bangkok. Hb AA, Hb AE, and Hb EE (with or without alpha-thalassemia) samples were identified by laboratory personnel using a combination of complete blood count, low-pressure liquid chromatography (LPLC) or HPLC, and clinical history of hemoglobin disorders. Inclusion criteria for the Thailand study included: any age or gender, residual blood sample available with a known date of collection within 1 month, and predicted hemoglobin types of Hb AA, Hb AE, or Hb EE. Exclusion criteria for the Thailand study included: unknown date of sample collection, blood sample older than 1 month at the time of use, any findings on complete blood count, LPLC or HPLC, or clinical history to suggest the presence of a beta-globin variant other than Hb E, such as a high quantitative Hb A2 in the range suggestive of beta-thalassemia.
298 subjects (8 months to 65 years old) were tested at a referral testing facility of ICMR-National Institute of Research in Tribal Health located at Late Baliram Kashayap Memorial Medical College, Jagdalpur, Chhattisgarh, India. Inclusion criteria included: subjects of 6 weeks or older, with or without the signs and symptoms of pallor, jaundice, abdominal pain, joint pain and provision of a signed informed consent (either by the subject or by the parent as appropriate if the subject is a minor). Subjects who withdrew their consent after enrolling were excluded from the study.
Blood sample acquisition and testing
In all clinical studies, blood samples were collected as part of the standard clinical care and only surplus de-identified blood samples were utilized for testing. The whole blood samples were tested with both HemeChip and the reference standard HPLC (VARIANT™ II, Bio-Rad Laboratories, Inc., Hercules, California) in Cleveland, Ohio, Nigeria, and Thailand. In India, blood samples were tested with both HemeChip and the standard laboratory-based reference standard cellulose acetate electrophoresis, followed by HPLC testing for discordant results. HPLC and cellulose acetate electrophoresis procedures were performed according to manufacturer’s guidelines. HPLC was considered to be the reference standard for our comparisons. In any disagreement between HemeChip, cellulose acetate electrophoresis and HPLC, HPLC result was considered to be correct. HPLC %Hb results were normalized to a total of 100% for the cases in which the summation of reported Hb% values were less than 100%. In India, hemoglobin percentage values were reported for only Hb S, A2, F by the standard reference laboratory, hence the balance percentage was assumed to be Hb A. The HemeChip reader guides the user step-by-step through the test procedure (Fig. S6) with animated on-screen instructions to minimize user errors. Local users were trained to use a custom-designed micro-applicator (Fig. S7), which is included in a kit (Fig. S8) with graphical instructions for use (Fig. S9). Hemoglobin identification and quantification is automatically performed with a custom software on the reader and results are reported to the user in a clear and objective way (Fig. S10–S15).
Clinical information and reference test results were not available to the performers of the test or the study team at the time of testing. Similarly, HemeChip test results were not available to the performers of the standard reference tests. In Nigeria, HemeChip tests were performed on eHealth Africa campus in Kano, Nigeria, by local healthcare workers using blood samples collected at the nearby Hasiya Bayero Pediatric Hospital, Murtala Muhammad Specialist Hospital, and Aminu Kano Teaching Hospital. Clinical standard (HPLC) testing was done independently by the International Foundation Against Infectious Disease in Nigeria (IFAIN, Abuja, Nigeria) for the blood samples obtained in Kano. In Cleveland, Ohio, HemeChip tests were performed at Case Biomanufacturing and Microfabrication laboratory by the research team members. HPLC testing was done independently by the University Hospitals Cleveland Medical Center Clinical Laboratories (Cleveland, Ohio) for the blood samples obtained in Cleveland. In Thailand, HemeChip tests were performed at Siriraj Thalassemia Center by local laboratory personnel. Reference standard testing (HPLC) was independently performed at ATGenes Co. Ltd. (Bangkok, Thailand). In India, HemeChip tests and standard reference cellulose acetate electrophoresis tests were performed by local laboratory personnel at the testing facility of ICMR-National Institute of Research in Tribal Health located at Late Baliram Kashyap Memorial Medical College located at Jagdalpur, Chhattisgarh, and HPLC testing was performed at the central laboratory of ICMR-National Institute of Research in Tribal Health, Jabalpur, Madhya Pradesh, India.
Statistical Methods
Hemoglobin band separation data was analyzed statistically using one-way Analysis of Variance (ANOVA) test with p-value set at 0.001 for statistically significant difference (Fig. 6A–F). Tukey post-hoc test was used for multiple comparisons, where applicable. Hemoglobin percent quantification agreement between HemeChip and the reference standard method (HPLC) was assessed by Bland-Altman analysis for heterozygous samples tested (Fig. 6G–I). For homozygous samples tested, Bland-Altman analysis was not performed, since for such samples, both HemeChip and the reference standard method display only one Hb%, which is greater than 90%. The Bland-Altman method was used to determine the repeatability of HemeChip quantification using residual analysis by comparison to the standard method. The coefficient of repeatability was set as 1.96 times the standard deviations of the differences between the two measurements. Diagnostic accuracy of HemeChip test in comparison to HPLC was determined as the percent ratio of correct tests results, and summation of correct tests results and incorrect test results for a hemoglobin variant category (Table 2). Sensitivity was determined as the ratio of true positive results divided by the summation of true positive results and false negative results. Specificity was determined as the ratio of true negative results divided by the summation of true negative results and false positive results (Table 3). Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were analyzed and reported for binary pairs of SCD-SS vs. others, SCD-SC vs. others, SCD Trait vs. others, Hb E Disease vs. others, Hb E Trait vs. others (Table 3).
Fig. 6. HemeChip hemoglobin band separation and quantification.
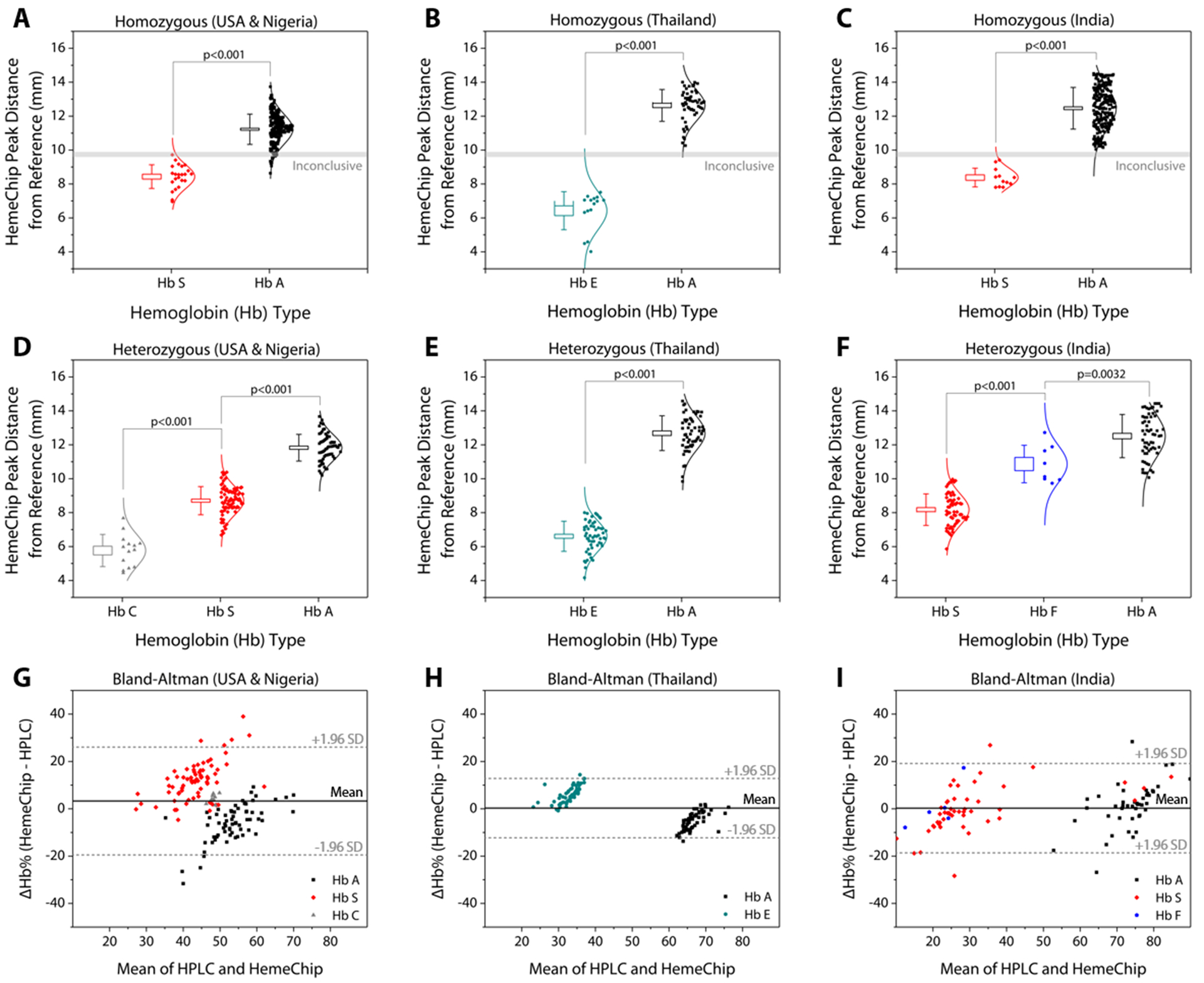
The top row are data distribution plots that show the hemoglobin band separation for homozygous subjects tested in: (A) the USA (9 HbSS) and Nigeria (13 Hb SS, 212 Hb AA), (B) Thailand (15 Hb EE, 50 Hb AA), and (C) India (11 Hb SS, 219 Hb AA). The middle row are data distribution plots for heterozygous subjects tested in: (D) the USA (7 Hb AS, 14 Hb SC) and Nigeria (60 Hb AS), (E) Thailand (57 Hb AE), and (F) India (60 Hb AS, 7 Hb SF, 1 Hb AF). The horizontal lines between hemoglobin positions represent statistically significant differences based on one-way Analysis of Variance (ANOVA) with Tukey post-hoc test for multiple comparisons, where applicable. In whisker plots, brackets represent standard deviation of the mean, and boxes represent standard error of the mean. Lower row are Bland-Altman analysis plots comparing HemeChip with reference standard method (HPLC) for hemoglobin percentage quantification for the heterozygous subjects tested in the USA and in Nigeria (G), Thailand (H), and India (I). In Bland-Altman plots, solid line indicates the mean difference and dashed lines indicate 95% limits of agreement defined as mean difference ± 1.96 times standard deviation (SD). HemeChip reports hemoglobin C/E/A2. Test location was used to differentiate co-migrating hemoglobins C and E.
Table 2:
HemeChip diagnostic accuracy in comparison to reference standard method.
| Category | Hemoglobin type | Correct | Incorrect | Accuracy |
|---|---|---|---|---|
| SCD-SS | Hb SS | 33 | 0 | 100% |
| SCD-SC | Hb SC | 14 | 0 | 100% |
| SCD Trait | Hb AS | 129 | 0 | 100% |
| Hb E Disease | Hb EE | 15 | 0 | 100% |
| Hb E Trait | Hb AE | 57 | 0 | 100% |
| Normal (no abnormal Hb) | Hb AA | 472 | 9* | 98.1% |
| SCD-Sβ+, SCD-Sβ0 | Hb SAA2, Hb SFA2 | 0 | 3† | - |
| All categories | - | 720 | 12 | 98.4% |
Nine subjects with normal hemoglobin (Hb AA) were identified as SCD by HemeChip.
Three subjects with compound heterozygous Sβ-thalassemia+/0 were identified as SCD by HemeChip.
HemeChip reports hemoglobin C/E/A2. Test location was used to differentiate co-migrating hemoglobins C and E.
Table 3:
HemeChip diagnostic sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) in comparison to reference standard method.
| SCD-SS vs. others | SCD-SC vs. others | SCD Trait vs. others | Hb E Disease vs. others | Hb E Trait vs. others | |
|---|---|---|---|---|---|
| True positive, TP | 33 | 14 | 129 | 15 | 57 |
| True negative, TN | 687 | 706 | 591 | 705 | 663 |
| False Positive, FP | 9 | 0 | 0 | 0 | 0 |
| False negative, FN | 0 | 0 | 0 | 0 | 0 |
| Sensitivity, TP/(TP + FN) | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Specificity, TN/(TN + FP) | 98.7% | 100.0% | 100.0% | 100.0% | 100.0% |
| PPV, TP/(TP + FP) | 78.6% | 100.0% | 100.0% | 100.0% | 100.0% |
| NPV, TN/(TN + FN) | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
HemeChip reports hemoglobin C/E/A2. Test location was used to differentiate co-migrating hemoglobins C and E.
RESULTS
HemeChip separates, images, and tracks hemoglobin variants real-time during electrophoresis
The fundamental principle behind the HemeChip technology is hemoglobin electrophoresis, in which different hemoglobin variants can be separated based on electric charge differences when subjected to an electric field in the presence of a carrier substrate [66, 67]. The HemeChip test works with a standard finger-prick or a heel-prick blood sample that is collected according to the World Health Organization (WHO) guidelines for drawing blood [87], which typically yields about 25 μL per drop [88]. Test also works with venous blood samples. For ease of handling, 20 μL of blood is collected and diluted with 40 μL of lysing solution. Next, less than 1 μL (0.56 μL ± 0.17 μL, N=5) of lysed blood sample is transferred into the cartridge for electrophoresis (Fig. 1D). Actual blood volume utilized per test is approximately 0.2 μL. The HemeChip separates hemoglobins A, F, S, C/E/A2 on cellulose acetate paper that is subjected to an electric field (Fig. 1D). Tris/Borate/EDTA (TBE) buffer is used to provide the necessary ions for electrical conductivity at a pH of 8.4 in the cellulose acetate paper [66, 67]. The pH induced net negative charges of the hemoglobins cause them to travel from the negative to the positive electrode when placed in an electric field (Fig. 1D, Video S1). Differences in hemoglobin mobilities due to their negative electrical charge allow hemoglobin separation to occur (Fig. 1D) with visible results at the end of a HemeChip test (Fig. 1E). Among the four major hemoglobin types that we tested, hemoglobin C/E/A2 co-migrate and they are the slowest. Hemoglobin A is the fastest moving hemoglobin in alkaline solution. A unique feature of HemeChip is that it utilizes a blue control marker (xylene cyanol) that is pre-mixed with the blood sample before application into the cartridge (Fig. 1D&2A). Towards the end of the test, the blue control marker reaches to the end of the cellulose acetate paper strip (Fig. 1D) and disappears into the buffer pool, leaving behind the separated hemoglobin variants (Fig. 2B). At this point, the hemoglobin bands are automatically identified by a custom software based on their respective locations on the cellulose acetate paper strip relative to the sample application point (Fig. 3). Hemoglobins in the blood sample form visible bands at the end of the test due to the natural bright red color of the protein (Fig. 1E&2B). This feature of naturally red, visible hemoglobin, combined with optically clear HemeChip cartridge (Fig. S3) in transmission imaging mode (Fig. 2B) within the reader’s imaging chamber (Fig. 3A&B), negates the need for picrosirius red staining, which is typically utilized in benchtop cellulose acetate hemoglobin electrophoresis [66, 67].
HemeChip automatically identifies hemoglobin variants and determines their relative percentages
The blue control marker mobility on the cellulose acetate paper strip is tracked real-time by the image processing and decision algorithm (Fig. S5). The mobility of the control marker is then used to confirm that the test is running as expected (Fig. 4, Video S1). Briefly, the algorithm searches and finds the sample application mark (Fig. 4, t=0 min.), and the intensity curve is generated for the red hemoglobin bands and the blue control marker. The intensity curve is evaluated to detect and track the movement of the peaks throughout the process (Fig. 4). The peaks are identified based on their final locations at the end of the test (Fig. 4, t=8 min.). The distance between each peak and the application point and the relative percentage of the areas under the peaks are evaluated to determine whether they fit into the categories pre-defined in the software for hemoglobin C/E/A2, hemoglobin S, hemoglobin F, and hemoglobin A. The amount of hemoglobin is presented as a percentage that is relative to that of the other hemoglobin bands in the sample. In the case of a single hemoglobin variant detected, such as Hb AA (Fig. 5A) or Hb SS (Fig. 5B), the HemeChip software reports a percentage value greater of >90%, which agrees with the results reported by the reference standard method (HPLC). If there is more than one peak identified, then the areas under each of the peaks are calculated and the relative percentages are reported, for example in the cases of Hb AS (Fig. 5C), Hb SC (Fig. 5D), Hb SS blood containing fetal hemoglobin (Hb F) (Fig. 5E), and Hb AE (Fig. 5F). Hemoglobin A and hemoglobin S separation is significantly greater (p<0.001) than hemoglobin F and hemoglobin S separation (Fig. S16), which can be used to distinguish between Hb SF and Hb AS.
Fig. 5. Identification of hemoglobin types and quantification of hemoglobin percentages by HemeChip.
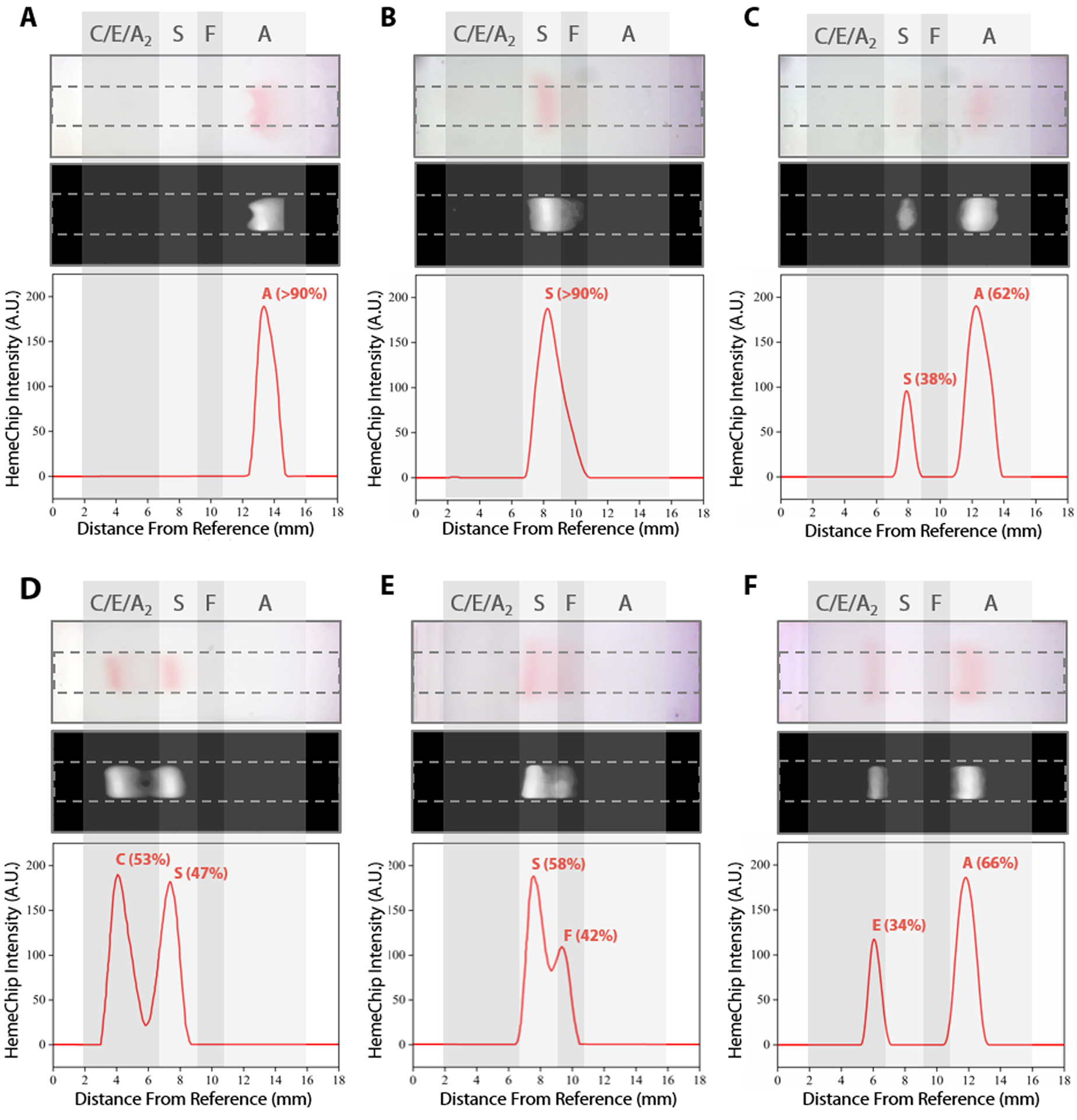
Typical HemeChip results are presented for the main hemoglobin variants tested. For each subset, the top image represents the raw image captured by the HemeChip reader at the end of the test. The middle black and white image is generated by the image analysis software algorithm to produce the HemeChip intensity curves (bottom image) for peak identification and Hb % quantification. Hb % values displayed were determined by HemeChip, which showed agreement with the reference standard method (HPLC). Gray regions indicate the approximate zones in which A, F, S, and C/E/A2 bands appear at the end of the test. (A) Normal (no abnormal Hb) with Hb AA (>90%). (B) SCD-SS with Hb SS (>90%). (C) SCD Trait with Hb AS (A: 62%, S: 38%). (D) SCD-SC with Hb SC (S: 47%, C: 53%). (E) SCD-SS with Hb SF (S: 58%, F: 42%). (F) Hb E Trait with Hb AE (A: 66%, E: 34%).
HemeChip accurately identifies hemoglobin bands and quantifies hemoglobin variants in blood
Band positions for hemoglobin C/EA2, hemoglobin S, hemoglobin F, and hemoglobin A were reported for homozygous and heterozygous subjects (Fig. 6). The band separation between each hemoglobin type was significantly different (Fig. 6A–F). For homozygous subjects tested in Nigeria (Fig. 6A), one Hb SS and eleven Hb AA test results were in the ‘Inconclusive’ zone (9.59 mm to 9.89 mm) between A and S bands (Table 1). Nine Hb AA subjects tested appeared below the ‘Inconclusive’ zone, resulting in false-positives or incorrect identification as Hb SS (Table 2). No test results were in the inconclusive zone for the subjects tested in Thailand (Fig. 6B) or in India (Fig. 6C). Heterozygous subjects tested include Hb AS and Hb SC in Nigeria and the USA (Fig. 6D), Hb AE in Thailand (Fig. 6E), and Hb AS, Hb SF, and Hb AF in India (Fig. 6F). In heterozygous subjects tested, appearance of two bands in different locations (Fig. 5C–F) negates the need for an ‘Inconclusive’ zone. Bland-Altman analysis showed an agreement between quantitative HemeChip results and HPLC results for the hemoglobin percent measurements (Fig. 6G–I). For subjects tested in the USA and in Nigeria, Bland-Altman analysis showed a mean bias of +3.27% for HemeChip in comparison to HPLC with a systematic over-estimation of %Hb S at higher mean values and under-estimation of %HbA in lower mean values (Fig. 6G). For subjects tested in Thailand, Bland-Altman analysis showed a mean bias of +0.27% for HemeChip compared to HPLC (Fig. 6H). For subjects tested in India, Bland-Altman analysis showed a mean bias of +0.25% for HemeChip in comparison to HPLC (Fig. 6I).
Table 1:
Overview of clinical studies with HemeChip.
| Total | Valid tests | Uninterpretable tests | Inconclusive tests | |
|---|---|---|---|---|
| Subjects and tests | 768 | 732 | 24 | 12 |
| Percentage | 100% | 95.3% | 3.1% | 1.6% |
768 subjects were tested in clinical studies with HemeChip and the reference standard method.
315 subjects (6 weeks to 5 years old) were tested in Kano, Nigeria by local users.
124 subjects (7 weeks to 63 years old) were tested in Bangkok, Thailand by local users.
298 subjects (8 months to 65 years old) were tested in Jagdalpur, Chhattisgarh, India by local users.
31 subjects (20 to 59 years old) were tested in Cleveland, Ohio, United States, by the research team.
Clinical testing of HemeChip in the United States, Nigeria, Thailand, and India
We report the clinical study results and HemeChip diagnostic accuracy according to the Standards for Reporting Diagnostic Accuracy (STARD) 2015 guidelines [82]. A detailed description of the study design, participants, test methods, and analyses are available in the Methods section. In clinical validation studies, we tested a total of 768 subjects with both HemeChip and reference standard methods, in the United States, Nigeria, Thailand, and India (Table 1). We defined ‘Valid’, ‘Uninterpretable’, and ‘Inconclusive’ tests according to published recommendations in literature [89] and STARD guidelines [82]. A ‘Valid’ test was defined as a test that performed as expected according to objective standards, such as the normal movement of the blue control marker (Fig. 4). Examples of ‘Valid’ test results are presented in Supplementary Information (Figs. S10–13). An ‘Uninterpretable’ test was defined as a test that failed to produce any results and that did not meet the minimum set of objective criteria which constitute a valid or adequate test. Reasons for ‘Uninterpretable’ tests include the poor migration of the blue control marker, electrical connectivity issues, or faulty cartridges. An example ‘Uninterpretable’ test is shown in Supplementary Information (Fig. S14). An ‘Inconclusive’ test was defined as a test that performed adequately according to an objective set of standards, such as the satisfactory movement of the blue control marker and appearance of hemoglobin bands, but the test algorithm could not make an identification of hemoglobin types present. Reasons for ‘Inconclusive’ tests include appearance of a band or bands at or close to the borderline region between two adjacent detection windows (Fig. 6A). An example ‘Inconclusive’ test is shown in Supplementary Information (Fig. S15). An ‘Uninterpretable’ or ‘Inconclusive’ test can be recognized by the image analysis algorithm during or at the end of the test. In addition to the ‘Valid’, ‘Uninterpretable’, and ‘Inconclusive’ tests reported here, a number of tests were not included in the data analysis due to the following reasons: missing reference standard results, sample labeling errors, and the tests performed for user training or algorithm development. Among the total 768 test results reported here, 732 tests were ‘Valid’ (95.3%), 24 tests were ‘Uninterpretable’ (3.1%), and 12 were ‘Inconclusive’ (1.6%) (Table 1). An ‘Uninterpretable’ or ‘Inconclusive’ test does not result in a diagnostic decision [89]. As such, when an ‘Uninterpretable’ or ‘Inconclusive’ test is encountered, the test can be repeated or other methods of testing can be utilized, which are common practices in diagnostics [89], as well as in hemoglobin testing [66, 67]. Therefore, based on recommended practices in the literature [89] and the STARD guidelines [82], the uninterpretable and inconclusive test results were reported separately in this study (Table 1).
Results of clinical studies with HemeChip indicate high diagnostic accuracy in hemoglobin testing
In clinical studies, HemeChip test results included the following (Table 2): SCD-SS (Hb SS), SCD-SC (Hb SC), SCD Trait (Hb AS), Hb E Disease (Hb EE), Hb E Trait (Hb AE), and Normal (no abnormal Hb, or Hb AA). HemeChip identified SCD-SS, SCD-SC, SCD Trait, Hb E Disease, and Hb E Trait with 100% accuracy (Table 2). Nine subjects with no abnormal Hb (Hb AA) were identified as SCD-SS. Three subjects with compound heterozygous Sβ-thalassemias (2 subjects with SCD-Sβ+, 1 subject with SCD-Sβ0) were identified as SCD by HemeChip (Table 2). Sensitivity and negative predictive value (NPV) were 100% for all hemoglobin types tested (Table 3). Specificity was 98.7% for SCD-SS vs. others, and 100% for all other types (Table 3). Positive predictive value was 78.6% for SCD-SS vs. others, and 100% for all other types (Table 3). For studies reporting diagnostic accuracy, one of the recommendations in the published guidelines is that, inconclusive results are excluded from binary statistics (i.e., sensitivity and specificity analyses), but an additional summary statistic is reported that accounts for them [89]. Based on these recommendations, if the twelve inconclusive results are considered as incorrect, then the overall diagnostic accuracy of HemeChip would be 96.8%. When the inconclusive results are reported separately according to the published guidelines [82, 89], the overall diagnostic accuracy of HemeChip is 98.4% in identifying SCD-SS, SCD-SC, SCD Trait, Hb E Disease, Hb E Trait, and Normal (Table 2).
DISCUSSION
HemeChip combines the benefits of standard hemoglobin electrophoresis with the benefits of a point-of-care test. HemeChip technology has a number of fundamental differences and unique features when compared to standard hemoglobin electrophoresis techniques. For example, HemeChip replaces the benchtop laboratory setup [66, 67] with a portable reader, and replaces the hemoglobin controls [66, 67] with a blue control marker (Fig. 4). HemeChip provides hemoglobin type identification and quantitative results of relative hemoglobin percentages (Fig. 5). The overall simplicity of the HemeChip enables any user to quickly and accurately screen for SCD and other hemoglobin disorders. The user is guided through the step-by-step process with on-screen animated instructions, which minimizes errors (Fig. S6, Video S2). HemeChip does not require a dedicated lab environment and battery operation allows use in remote places lacking electrical power. These critical features distinguish HemeChip from currently available laboratory methods (Table S1) and other emerging point-of-care technologies for hemoglobin testing (Table S2).
Clinical studies with HemeChip in the US, sub-Saharan Africa, Central India, and Southeast Asia demonstrated 100% sensitivity and specificity for SCD-SC, SCD Trait, Hb E Disease, and Hb E Trait (Table 3); and 100% sensitivity and 98.7% specificity for SCD-SS. The 100% sensitivity achieved in these clinical studies suggests that no individuals with SCD-SS, SCD-SC, SCD Trait, Hb E Disease, and Hb E Trait would be missed by HemeChip hemoglobin testing. Common practice in hemoglobin testing is that all positive test results (e.g., SCD-SS) are confirmed with a secondary method prior to final diagnostic decision and treatment initiation [90]. Therefore, all SCD-SS positive tests would likely result in a secondary confirmatory test that should rule out any false positives.
Affordability has been one of the primary objectives of HemeChip design. The projected sale price per test, including all consumables, distribution, and support cost, is estimated at $2.00 USD. A rugged, portable battery-powered reader can be priced as low as $700 USD, and designed to last at least 5 years or 10,000 tests (Fig. S17). The portable reader would add about 7 cents to the $2.00 USD test price. The projected HemeChip end user price of $2.07 USD per test (including the reader cost), thus clearly offers substantial cost savings over current practice and it is significantly lower than the estimated price threshold in the aforementioned study [91]. Deploying a robust, portable, battery operated platform as part of a point-of-care test has several advantages over alternatives (Table S2). For example, the reader includes a touchscreen display that guides and helps the user with the test procedure, reduces the training time and minimizes user errors. The portable reader also allows digital data entry (Fig. S17), including patient demographics. The reader also enables secure, encrypted data storage and wireless transmission to the cloud, which would enable tracking and follow-up of patients using electronic records and mobile networks [29]. The reader displays the results on the screen in a clear and objective way (Figs. S10–S15), removing user interpretation errors (Figs. S18–S32). In comparison, point-of-care lateral-flow assays lack a portable reader for objective analysis or electronic record keeping (Table S2). Therefore, these methods rely on the user to visually interpret the results and manually record the clinical data, which is prone to errors. In fact, in clinical research databases, user misinterpretation and data entry errors have been reported to range between 2.3% to 26.9% [92].
Hemoglobin electrophoresis techniques share a common limitation. Some hemoglobin types appear in the same electrophoretic window, as they exhibit the same or similar electrophoretic mobility in a given condition. For example, in capillary zone electrophoresis, it can be challenging to quantify hemoglobin A2 in the presence of hemoglobin C, due to partial overlap between the two zones for these two hemoglobin types [93, 94], which may be improved using curve fitting methods. Hemoglobin G and hemoglobin D are difficult to separate because they have identical migration in gel electrophoresis, in capillary electrophoresis, and they have overlapping elution times in HPLC. This sharing of detection window or peak overlapping is also a challenge for the reference standard method, HPLC as well as its alternatives [93, 95, 96]. For example, in HPLC, hemoglobin S elutes in the S window with 28 other hemoglobin variants, including 10 other β-chain variants [97]. A similar issue occurs in the A2 window for HPLC, where hemoglobin E and 18 other hemoglobin variants elude in the same window, and 13 of these 18 variants are β-chain variants [97]. hemoglobins C, E, and A2, co-migrate in paper based hemoglobin electrophoresis [66, 67]. Hemoglobins C, A2, and E are all detectable, but it is not possible to differentiate them [66, 67]. Therefore, instead of reporting Hb C or E or A2 individually, HemeChip reports hemoglobin C/E/A2. Since HemeChip provides quantitative results, the relative percentages of these hemoglobins can be used to make a distinction (Figs. S18–S32). For example, hemoglobin A2 is typically found at lower percentages (3–10%) in most subjects [67, 98, 99], whereas hemoglobin C is typically much higher (greater than 40% for Hb SC, greater than 90% for Hb CC) [100]. When hemoglobin C and hemoglobin A2 are both present in the same blood sample, the HemeChip shows the hemoglobin C percentage plus hemoglobin A2 percentage [101]. Clinical decision-making is not affected by this overlap, for Hb SC, Hb AC, or Hb CC results, but it is a potential limitation for detecting Hb Sβ-thalassemia and Hb Eβ-thalassemia. The algorithm has not yet been trained to identify β-thal variants, however, it categorizes them as SCD. In our clinical studies, HemeChip identified three Hb Sβ-thal subjects as SCD (Table 2). Some hemoglobin variants display distinct geographical prevalence and distribution. For example, hemoglobin C is highly prevalent in West Africa [102], while hemoglobin E is the most prevalent in the Mediterranean region, Southeast Asia, and in the Indian subcontinent [7, 34–36, 43]. Test location, the ethnicity of the subject, the hemoglobin percentage (Figs. S18–S32), and clinical history can be used to help differentiate between co-migrating hemoglobin types.
Fetal hemoglobin expression represents 50–95% of total hemoglobin expression in newborn babies and declines significantly over the course of the first 6 months of life [103, 104]. Hemoglobin F at high levels might mask the hemoglobin variants in newborns [105]. Screening for hemoglobin variants can be integrated into established, highly effective immunization programs in low income countries [29]. Babies are typically vaccinated around 6–8 weeks of age [29, 40], and by this age, hemoglobin F levels decrease significantly as hemoglobin F is replaced by adult hemoglobins, which would be easier to detect. Non-β chain hemoglobinopathies, such as α-thalassemia, are also major world-wide health problems in children, especially in Asia. We are eager to extend our technology to this population in future studies. This technology is expected to detect additional hemoglobins, such as hemoglobin Bart’s or hemoglobin H, which are elevated in alpha thalassemia.
In summary, HemeChip enables, for the first time, cost-effective identification of hemoglobin variants at the point-of-need. The HemeChip reader guides the user step-by-step through the test procedure with animated on-screen instructions to minimize user errors. Hb identification and quantification is automatically performed, and Hb types and percentages are displayed in an easily understandable, objective, and quantitative way. HemeChip is a versatile, mass-producible microchip electrophoresis platform technology that may address unmet needs in biology and medicine, when rapid, decentralized hemoglobin or protein analysis is needed.
Supplementary Material
Fig. S1. Transforming HemeChip design for injection molded mass production.
Fig. S2. Injection moldable HemeChip design with bottom and top plastic parts.
Fig. S3. Injection molding of HemeChip and effect of the mold finish on optical quality.
Fig. S4. Electronic circuit design of the HemeChip Reader.
Fig. S5. Software algorithm for the HemeChip Reader user interface.
Fig. S6. Screen views of the step-by-step instructions provided by the HemeChip user interface.
Fig. S7. Micro-applicator design and precise application of blood samples into HemeChip.
Fig. S8. HemeChip test kit components.
Fig. S9. Blood collection and HemeChip test protocol steps.
Fig. S10. HemeChip test result screen for no abnormal hemoglobin (Hb AA).
Fig. S11. HemeChip test result screen for SCD Trait (Hb AS).
Fig. S12. HemeChip test result screen for SCD-SS (Hb SS).
Fig. S13. HemeChip test result screen for Hb EE (Hb E Disease).
Fig. S14. HemeChip test result screen for an ‘Uninterpretable’ or invalid test.
Fig. S15. HemeChip test result screen for an ‘Inconclusive’ test.
Fig. S16. Hemoglobin F separation and identification in HemeChip.
Fig. S17. New design of portable, affordable HemeChip reader: Gazelle™.
Fig. S18. Gazelle™ Hb Variant test result screen for Hb AA (normal).
Fig. S19. Gazelle™ Hb Variant test result screen for Hb AA (normal) for a <6 months old infant.
Fig. S20. Gazelle™ Hb Variant test result screen for Hb AS trait.
Fig. S21. Gazelle™ Hb Variant test result screen for Hb AS trait for a <6 months old infant.
Fig. S22. Gazelle™ Hb Variant test result screen for Hb AC or Hb AE trait.
Fig. S23. Gazelle™ Hb Variant test result screen for Hb AC or Hb AE trait for a <6 months old infant.
Fig. S24. Gazelle™ Hb Variant test result screen for Hb SS (SCD).
Fig. S25. Gazelle™ Hb Variant test result screen for Hb SS (SCD) for a <6 months old infant.
Fig. S26. Gazelle™ Hb Variant test result screen for Hb SC or SE.
Fig. S27. Gazelle™ Hb Variant test result screen for Hb SC or Hb SE for a <6 months old infant.
Fig. S28. Gazelle™ Hb Variant test result screen for Hb CC or EE.
Fig. S29. Gazelle™ Hb Variant test result screen for Hb CC or Hb EE for a <6 months old infant.
Fig. S30. Gazelle™ Hb Variant test result screen for Hb Sβ+.
Fig. S31. Gazelle™ Hb Variant test result screen for Hb Sβ+ for <6 months old infant.
Fig. S32. Gazelle™ Hb Variant test result screen for an unusual hemoglobin pattern.
Table S1. Comparison of HemeChip with standard laboratory methods for hemoglobin testing
Table S2. Comparison of HemeChip with emerging point-of-care technologies for hemoglobin testing
Table S3. Sample size calculation for clinical studies
Video S1. Real-time tracking of hemoglobin bands in HemeChip during electrophoresis process
Video S2. Step-by-step HemeChip test procedure and analysis of results
PUBLIC HEALTH SIGNIFICANCE AND FUTURE PERSPECTIVE.
Despite SCD’s historical recognition of being the first discovered molecular disease, the technological advances for this disease have been slow [106]. The promise of the development of accurate, low-cost, point-of-care diagnostics, especially for low-income countries, has come closer than ever before to being met with new technologies, such as HemeChip. HemeChip and its diagnostic promise will address the global health burden, help close the gap of information in our current state of science and healthcare delivery around hemoglobin diseases, and build space for further innovation and advancement for SCD and other hemoglobinopathies. Through multi-lateral partnerships with local governments, pharmaceutical companies, the technological industry, and university systems, the translation, implementation, and dissemination of affordable screening and diagnostic technologies throughout countries with limited resources and infrastructure has the potential to change the paradigm of SCD, hemoglobin disorders, and global health.
ACKNOWLEDGEMENTS
The development of this technology has been supported by the Clinical and Translational Science Collaborative of Cleveland, UL1TR002548 from the National Center for Advancing Translational Sciences component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Other funding sources include Case-Coulter Translational Research Partnership Program, NIH Center for Accelerated Innovation at Cleveland Clinic funded by U54HL119810, Ohio Third Frontier Technology Validation and Start-up Fund, Consortia for Improving Medicine with Innovation & Technology, National Institutes of Health Fogarty International Center (R21TW010610), National Heart Lung and Blood Institute Small Business Innovation Research Program (R44HL140739), National Heart Lung and Blood Institute R01HL133574, and National Science Foundation CAREER Award #1552782. Scientific illustration was made by Grace Gongaware and videos were made by Courtney Fleming, supported by a National Science Foundation CAREER Award #1552782 and Glennan Fellowship from University Center for Innovation in Teaching and Education. Clinical studies in Thailand were supported by the National Institutes of Health, Fogarty International Center Global Health Research Fellowship (2D43TW009337-07) and the Cooley’s Anemia Foundation Research Fellowship awarded to Julia Z. Xu. This article’s contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
COMPETING INTERESTS
MNH, AF, RA, YA, RU, CMP, JAL, and UAG are inventors of intellectual property (patent applications: PCT/US2015/042907 and US20170227495A1) licensed by Hemex Health Inc. for commercialization. These inventors and Case Western Reserve University have financial interest in Hemex Health Inc., including licensed intellectual property, stock ownership, and consulting. PT is an employee of Hemex Health. Competing interests of Case Western Reserve University employees are overseen and managed by the Conflict of Interests Committee according to a Conflict of Interest Management Plan.
DATA AND MATERIALS AVAILABILITY
All reasonable requests for materials and data will be fulfilled by the corresponding author of this publication.
We present a versatile, mass-producible, paper-based microchip electrophoresis platform that enables rapid, affordable, decentralized hemoglobin testing at the point-of-care.
REFERENCES
- 1.Modell B and Darlison M, Global epidemiology of haemoglobin disorders and derived service indicators. Bulletin of the World Health Organization, 2008. 86: p. 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamison DT, Bank W, and Project DCP, Disease Control Priorities in Developing Countries. 2006: Oxford University Press. [PubMed] [Google Scholar]
- 3.Weatherall DJ and Clegg JB, Inherited haemoglobin disorders: an increasing global health problem. Bull World Health Organ, 2001. 79(8): p. 704–12. [PMC free article] [PubMed] [Google Scholar]
- 4.Williams TN, Sickle Cell Disease in Sub-Saharan Africa. Hematol Oncol Clin North Am, 2016. 30(2): p. 343–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colah R, Mukherjee M, and Ghosh K, Sickle cell disease in India. Curr Opin Hematol, 2014. 21(3): p. 215–23. [DOI] [PubMed] [Google Scholar]
- 6.Piel FB, Howes RE, Patil AP, Nyangiri OA, Gething PW, Bhatt S, Williams TN, Weatherall DJ, and Hay SI, The distribution of haemoglobin C and its prevalence in newborns in Africa. Scientific reports, 2013. 3: p. 1671–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fucharoen S and Weatherall DJ, The hemoglobin E thalassemias. Cold Spring Harbor perspectives in medicine. 2(8): p. a011734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rees DC, Williams TN, and Gladwin MT, Sickle-cell disease. Lancet, 2010. 376(9757): p. 2018–31. [DOI] [PubMed] [Google Scholar]
- 9.Bunn HF, Pathogenesis and treatment of sickle cell disease. N Engl J Med, 1997. 337(11): p. 762–9. [DOI] [PubMed] [Google Scholar]
- 10.Stuart MJ and Nagel RL, Sickle-cell disease. Lancet, 2004. 364(9442): p. 1343–60. [DOI] [PubMed] [Google Scholar]
- 11.Makani J, Ofori-Acquah SF, Nnodu O, Wonkam A, and Ohene-Frempong K, Sickle cell disease: new opportunities and challenges in Africa. ScientificWorldJournal, 2013. 2013: p. 193252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brittenham GM, Schechter AN, and Noguchi CT, Hemoglobin S polymerization: primary determinant of the hemolytic and clinical severity of the sickling syndromes. Blood, 1985. 65(1): p. 183–9. [PubMed] [Google Scholar]
- 13.Dong C, Chadwick RS, and Schechter AN, Influence of sickle hemoglobin polymerization and membrane properties on deformability of sickle erythrocytes in the microcirculation. Biophysical Journal, 1992. 63(3): p. 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manwani D and Frenette PS, Vaso-occlusion in sickle cell disease: pathophysiology and novel targeted therapies. Blood, 2013. 122(24): p. 3892–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alapan Y, Matsuyama Y, Little JA, and Gurkan UA, Dynamic deformability of sickle red blood cells in microphysiological flow. Technology, 2016. 4(2): p. 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alapan Y, Little JA, and Gurkan UA, Heterogeneous red blood cell adhesion and deformability in sickle cell disease. Sci Rep, 2014. 4: p. 7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M, Alapan Y, Adhikari A, Little JA, and Gurkan UA, Hypoxia-enhanced adhesion of red blood cells in microscale flow. Microcirculation, 2017. 24(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barabino GA, Platt MO, and Kaul DK, Sickle cell biomechanics. Annu Rev Biomed Eng, 2010. 12: p. 345–67. [DOI] [PubMed] [Google Scholar]
- 19.Alapan Y, Kim C, Adhikari A, Gray KE, Gurkan-Cavusoglu E, Little JA, and Gurkan UA, Sickle cell disease biochip: a functional red blood cell adhesion assay for monitoring sickle cell disease. Transl Res, 2016. 173: p. 74–91 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alapan Y, Fraiwan A, Kucukal E, Hasan MN, Ung R, Kim M, Odame I, Little JA, and Gurkan UA, Emerging point-of-care technologies for sickle cell disease screening and monitoring. Expert Review of Medical Devices, 2016. 13(12): p. 1073–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebbel RP, Beyond hemoglobin polymerization: the red blood cell membrane and sickle disease pathophysiology. Blood, 1991. 77(2): p. 214–37. [PubMed] [Google Scholar]
- 22.Schaer DJ, Buehler PW, Alayash AI, Belcher JD, and Vercellotti GM, Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood, 2013. 121(8): p. 1276–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verduzco LA and Nathan DG, Sickle cell disease and stroke. Blood, 2009. 114(25): p. 5117–25. [DOI] [PubMed] [Google Scholar]
- 24.Caboot JB and Allen JL, Hypoxemia in Sickle Cell Disease: Significance And Management. Paediatric Respiratory Reviews, 2014. 15(1): p. 17–23. [DOI] [PubMed] [Google Scholar]
- 25.Hargrave DR, Wade A, Evans JPM, Hewes DKM, and Kirkham FJ, Nocturnal oxygen saturation and painful sickle cell crises in children. Blood, 2003. 101(3): p. 846. [DOI] [PubMed] [Google Scholar]
- 26.Steinberg MH, Management of Sickle Cell Disease. New England Journal of Medicine, 1999. 340(13): p. 1021–1030. [DOI] [PubMed] [Google Scholar]
- 27.Booth C, Inusa B, and Obaro SK, Infection in sickle cell disease: a review. Int J Infect Dis, 2010. 14(1): p. e2–e12. [DOI] [PubMed] [Google Scholar]
- 28.Obaro SK and Iroh Tam PY, Preventing Infections in Sickle Cell Disease: The Unfinished Business. Pediatr Blood Cancer, 2016. 63(5): p. 781–5. [DOI] [PubMed] [Google Scholar]
- 29.Mburu J and Odame I, Sickle cell disease: Reducing the global disease burden. International Journal of Laboratory Hematology, 2019. 41(S1): p. 82–88. [DOI] [PubMed] [Google Scholar]
- 30.Kapoor S, Little JA, and Pecker LH, Advances in the Treatment of Sickle Cell Disease. Mayo Clin Proc, 2018. 93(12): p. 1810–1824. [DOI] [PubMed] [Google Scholar]
- 31.Vichinsky E, Hurst D, Earles A, Kleman K, and Lubin B, Newborn screening for sickle cell disease: effect on mortality. Pediatrics, 1988. 81(6): p. 749–55. [PubMed] [Google Scholar]
- 32.Frempong T and Pearson HA, Newborn screening coupled with comprehensive follow-up reduced early mortality of sickle cell disease in Connecticut. Conn Med, 2007. 71(1): p. 9–12. [PubMed] [Google Scholar]
- 33.Nagel RL, Fabry ME, and Steinberg MH, The paradox of hemoglobin SC disease. Blood Rev, 2003. 17(3): p. 167–78. [DOI] [PubMed] [Google Scholar]
- 34.Warghade S, Britto J, Haryan R, Dalvi T, Bendre R, Chheda P, Matkar S, Salunkhe Y, Chanekar M, and Shah N, Prevalence of hemoglobin variants and hemoglobinopathies using cation-exchange high-performance liquid chromatography in central reference laboratory of India: A report of 65779 cases. J Lab Physicians, 2018. 10(1): p. 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masiello D, Heeney MM, Adewoye AH, Eung SH, Luo HY, Steinberg MH, and Chui DH, Hemoglobin SE disease: a concise review. Am J Hematol, 2007. 82(7): p. 643–9. [DOI] [PubMed] [Google Scholar]
- 36.Xu JZ, Riolueang S, Glomglao W, Tachavanich K, Suksangpleng T, Ekwattanakit S, and Viprakasit V, The origin of sickle cell disease in Thailand. Int J Lab Hematol, 2019. 41(1): p. e13–e16. [DOI] [PubMed] [Google Scholar]
- 37.Wastnedge E, Waters D, Patel S, Morrison K, Goh MY, Adeloye D, and Rudan I, The global burden of sickle cell disease in children under five years of age: a systematic review and meta-analysis. Journal of Global Health, 2018. 8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams TN, Sickle Cell Disease in Sub-Saharan Africa. Hematology-Oncology Clinics of North America, 2016. 30(2): p. 343-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piel FB, The Present and Future Global Burden of the Inherited Disorders of Hemoglobin. Hematology-Oncology Clinics of North America, 2016. 30(2): p. 327-+. [DOI] [PubMed] [Google Scholar]
- 40.Aygun B and Odame I, A global perspective on sickle cell disease. Pediatr Blood Cancer, 2012. 59(2): p. 386–90. [DOI] [PubMed] [Google Scholar]
- 41.Sickle Cell Anemia, Report by the Secreteriat, 2006: World Health Organization Fifty-Ninth World Health Assembly. p. A59/9. [Google Scholar]
- 42.Grosse SD, Odame I, Atrash HK, Amendah DD, Piel FB, and Williams TN, Sickle cell disease in Africa: a neglected cause of early childhood mortality. Am J Prev Med, 2011. 41(6 Suppl 4): p. S398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colah R, Gorakshakar A, and Nadkarni A, Global burden, distribution and prevention of beta-thalassemias and hemoglobin E disorders. Expert Rev Hematol, 2010. 3(1): p. 103–17. [DOI] [PubMed] [Google Scholar]
- 44.Odame I, Perspective: We need a global solution. Nature, 2014. 515: p. S10. [DOI] [PubMed] [Google Scholar]
- 45.Piel FB, Hay SI, Gupta S, Weatherall DJ, and Williams TN, Global burden of sickle cell anaemia in children under five, 2010–2050: modelling based on demographics, excess mortality, and interventions. PLoS Med, 2013. 10(7): p. e1001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benson JM and Therrell BL, History and Current Status of Newborn Screening for Hemoglobinopathies. Seminars in Perinatology, 2010. 34(2): p. 134–144. [DOI] [PubMed] [Google Scholar]
- 47.Bain BJ, Neonatal/newborn haemoglobinopathy screening in Europe and Africa. Journal of Clinical Pathology, 2009. 62(1): p. 53. [DOI] [PubMed] [Google Scholar]
- 48.Streetly A, Latinovic R, Hall K, and Henthorn J, Implementation of universal newborn bloodspot screening for sickle cell disease and other clinically significant haemoglobinopathies in England: screening results for 2005–7. Journal of Clinical Pathology, 2009. 62(1): p. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bardakdjian-Michau J, Bahuau M, Hurtrel D, Godart C, Riou J, Mathis M, Goossens M, Badens C, Ducrocq R, Elion J, and Perini JM, Neonatal screening for sickle cell disease in France. J Clin Pathol, 2009. 62(1): p. 31–3. [DOI] [PubMed] [Google Scholar]
- 50.Therrell BL, Padilla CD, Loeber JG, Kneisser I, Saadallah A, Borrajo GJC, and Adams J, Current status of newborn screening worldwide: 2015. Seminars in Perinatology, 2015. 39(3): p. 171–187. [DOI] [PubMed] [Google Scholar]
- 51.Streetly A, Sisodia R, Dick M, Latinovic R, Hounsell K, and Dormandy E, Evaluation of newborn sickle cell screening programme in England: 2010–2016. Archives of Disease in Childhood, 2018. 103(7): p. 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wanjiku CM, Njuguna F, Asirwa FC, Njuguna C, Roberson C, and Greist A, Validation and Feasibility of a Point of Care Screening Test for Sickle Cell Disease in a Resource Constrained Setting — a New Frontier. Blood, 2018. 132(Suppl 1): p. 2229. [Google Scholar]
- 53.Tluway F and Makani J, Sickle cell disease in Africa: an overview of the integrated approach to health, research, education and advocacy in Tanzania, 2004–2016. Br J Haematol, 2017. 177(6): p. 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGann PT, Ferris MG, Macosso P, de Oliveira V, Ramamurthy U, Luis AR, Bernardino L, and Ware RE, A Prospective Pilot Newborn Screening and Treatment Program for Sickle Cell Anemia in the Republic of Angola. Blood, 2012. 120(21): p. 480. [Google Scholar]
- 55.Clark BE and Thein SL, Molecular diagnosis of haemoglobin disorders. Clinical & Laboratory Haematology, 2004. 26(3): p. 159–176. [DOI] [PubMed] [Google Scholar]
- 56.McGann PT and Hoppe C, The pressing need for point-of-care diagnostics for sickle cell disease: A review of current and future technologies. Blood Cells, Molecules, and Diseases, 2017. 67: p. 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makani J, Cox SE, Soka D, Komba AN, Oruo J, Mwamtemi H, Magesa P, Rwezaula S, Meda E, Mgaya J, Lowe B, Muturi D, Roberts DJ, Williams TN, Pallangyo K, Kitundu J, Fegan G, Kirkham FJ, Marsh K, and Newton CR, Mortality in sickle cell anemia in Africa: a prospective cohort study in Tanzania. PLoS One, 2011. 6(2): p. e14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Obaro S, Preventable deaths in sickle-cell anaemia in African children. Lancet, 2010. 375(9713): p. 460; author reply 461. [DOI] [PubMed] [Google Scholar]
- 59.Tshilolo L, Tomlinson G, Williams TN, Santos B, Olupot-Olupot P, Lane A, Aygun B, Stuber SE, Latham TS, McGann PT, and Ware RE, Hydroxyurea for Children with Sickle Cell Anemia in Sub-Saharan Africa. N Engl J Med, 2019. 380(2): p. 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voskaridou E, Christoulas D, Bilalis A, Plata E, Varvagiannis K, Stamatopoulos G, Sinopoulou K, Balassopoulou A, Loukopoulos D, and Terpos E, The effect of prolonged administration of hydroxyurea on morbidity and mortality in adult patients with sickle cell syndromes: results of a 17-year, single-center trial (LaSHS). Blood, 2010. 115(12): p. 2354–63. [DOI] [PubMed] [Google Scholar]
- 61.Steinberg MH, McCarthy WF, Castro O, Ballas SK, Armstrong FD, Smith W, Ataga K, Swerdlow P, Kutlar A, DeCastro L, and Waclawiw MA, The risks and benefits of long-term use of hydroxyurea in sickle cell anemia: A 17.5 year follow-up. Am J Hematol, 2010. 85(6): p. 403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang WC, Ware RE, Miller ST, Iyer RV, Casella JF, Minniti CP, Rana S, Thornburg CD, Rogers ZR, Kalpatthi RV, Barredo JC, Brown RC, Sarnaik SA, Howard TH, Wynn LW, Kutlar A, Armstrong FD, Files BA, Goldsmith JC, Waclawiw MA, Huang X, and Thompson BW, Hydroxycarbamide in very young children with sickle-cell anaemia: a multicentre, randomised, controlled trial (BABY HUG). Lancet, 2011. 377(9778): p. 1663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deshpande SV, Bhatwadekar SS, Desai P, Bhavsar T, Patel A, Koranne A, Mehta A, and Khadse S, Hydroxyurea in Sickle Cell Disease: Our Experience in Western India. Indian J Hematol Blood Transfus, 2016. 32(2): p. 215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lagunju I, Brown BJ, Oyinlade AO, Asinobi A, Ibeh J, Esione A, and Sodeinde OO, Annual stroke incidence in Nigerian children with sickle cell disease and elevated TCD velocities treated with hydroxyurea. Pediatr Blood Cancer, 2019. 66(3): p. e27252. [DOI] [PubMed] [Google Scholar]
- 65.Opoka RO, Ndugwa CM, Latham TS, Lane A, Hume HA, Kasirye P, Hodges JS, Ware RE, and John CC, Novel use Of Hydroxyurea in an African Region with Malaria (NOHARM): a trial for children with sickle cell anemia. Blood, 2017. 130(24): p. 2585–2593. [DOI] [PubMed] [Google Scholar]
- 66.Bain BJ, Haemoglobinopathy Diagnosis. 2005: Wiley. [Google Scholar]
- 67.Bain BJ, Wild B, Stephens A, and Phelan L, Variant Haemoglobins: A Guide to Identification. 2011: Wiley. [Google Scholar]
- 68.Reilly JM, Rochester Institute of T, and Image Permanence I, IPI storage guide for acetate film : instructions for using the wheel, graphs, and table : basic strategy for film preservation. 1993, Rochester, NY: Image Permanence Institute, Rochester Institute of Technology. [Google Scholar]
- 69.Desanctis O, Gomez L, Pellegri N, Parodi C, Marajofsky A, and Duran A, Protective Glass Coatings on Metallic Substrates. Journal of Non-Crystalline Solids, 1990. 121(1–3): p. 338–343. [Google Scholar]
- 70.Hanawa T, 1 - Overview of metals and applications, in Metals for Biomedical Devices, Niinomi M, Editor. 2010, Woodhead Publishing; p. 3–24. [Google Scholar]
- 71.Guerra-Balcazar M, Morales-Acosta D, Castaneda F, Ledesma-Garcia J, and Arriaga LG, Synthesis of Au/C and Au/Pani for anode electrodes in glucose microfluidic fuel cell. Electrochemistry Communications, 2010. 12(6): p. 864–867. [Google Scholar]
- 72.Schrott W, Svoboda M, Slouka Z, and Snita D, Metal electrodes in plastic microfluidic systems. Microelectronic Engineering, 2009. 86(4–6): p. 1340–1342. [Google Scholar]
- 73.Qiang LL, Vaddiraju S, Rusling JF, and Papadimitrakopoulos F, Highly sensitive and reusable Pt-black microfluidic electrodes for long-term electrochemical sensing. Biosensors & Bioelectronics, 2010. 26(2): p. 682–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maharbiz MM, Holtz WJ, Howe RT, and Keasling JD, Microbioreactor arrays with parametric control for high-throughput experimentation. Biotechnol Bioeng, 2004. 85(4): p. 376–81. [DOI] [PubMed] [Google Scholar]
- 75.Fox MB, Esveld DC, Valero A, Luttge R, Mastwijk HC, Bartels PV, van den Berg A, and Boom RM, Electroporation of cells in microfluidic devices: a review. Analytical and Bioanalytical Chemistry, 2006. 385(3): p. 474–485. [DOI] [PubMed] [Google Scholar]
- 76.Lee SW and Tai YC, A micro cell lysis device. Sensors and Actuators a-Physical, 1999. 73(1–2): p. 74–79. [Google Scholar]
- 77.Fox M, Esveld E, Luttge R, and Boom R, A new pulsed electric field microreactor: comparison between the laboratory and microtechnology scale. Lab on a Chip, 2005. 5(9): p. 943–948. [DOI] [PubMed] [Google Scholar]
- 78.St John A and Price CP, Existing and Emerging Technologies for Point-of-Care Testing. Clin Biochem Rev, 2014. 35(3): p. 155–67. [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma S, Zapatero-Rodriguez J, Estrela P, and O’Kennedy R, Point-of-Care Diagnostics in Low Resource Settings: Present Status and Future Role of Microfluidics. Biosensors (Basel), 2015. 5(3): p. 577–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Biehl M and Velten T, Gaps and challenges of Point-of-Care technology. Ieee Sensors Journal, 2008. 8(5–6): p. 593–600. [Google Scholar]
- 81.Prichard E, - Practical Laboratory Skills Training Guides (Complete Set). p. - P001. [Google Scholar]
- 82.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Lijmer JG, Moher D, Rennie D, de Vet HCW, Kressel HY, Rifai N, Golub RM, Altman DG, Hooft L, Korevaar DA, and Cohen JF, STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ : British Medical Journal, 2015. 351: p. h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buderer NM, Statistical methodology: I. Incorporating the prevalence of disease into the sample size calculation for sensitivity and specificity. Acad Emerg Med, 1996. 3(9): p. 895–900. [DOI] [PubMed] [Google Scholar]
- 84.Adewoyin AS, Management of sickle cell disease: a review for physician education in Nigeria (sub-saharan Africa). Anemia, 2015. 2015: p. 791498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jain DL, Sarathi V, Upadhye D, Gulhane R, Nadkarni AH, Ghosh K, and Colah RB, Newborn screening shows a high incidence of sickle cell anemia in Central India. Hemoglobin, 2012. 36(4): p. 316–22. [DOI] [PubMed] [Google Scholar]
- 86.Nuinoon M, Kruachan K, Sengking W, Horpet D, and Sungyuan U, Thalassemia and hemoglobin e in southern thai blood donors. Advances in hematology, 2014. 2014: p. 932306–932306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.World Health Organization guidelines on drawing blood: best practices in phlebotomy, ed. W.H. Organization 2010: World Health Organization. [PubMed] [Google Scholar]
- 88.Peck HR, Timko DM, Landmark JD, and Stickle DF, A survey of apparent blood volumes and sample geometries among filter paper bloodspot samples submitted for lead screening. Clin Chim Acta, 2009. 400(1–2): p. 103–6. [DOI] [PubMed] [Google Scholar]
- 89.Shinkins B, Thompson M, Mallett S, and Perera R, Diagnostic accuracy studies: how to report and analyse inconclusive test results. BMJ : British Medical Journal, 2013. 346: p. f2778. [DOI] [PubMed] [Google Scholar]
- 90.Ryan K, Bain BJ, Worthington D, James J, Plews D, Mason A, Roper D, Rees DC, de la Salle B, and Streetly A, Significant haemoglobinopathies: guidelines for screening and diagnosis. Br J Haematol, 2010. 149(1): p. 35–49. [DOI] [PubMed] [Google Scholar]
- 91.Mvundura M, Kiyaga C, Metzler M, Kamya C, Lim JM, Maiteki-Sebuguzi C, and Coffey PS, Cost for sickle cell disease screening using isoelectric focusing with dried blood spot samples and estimation of price thresholds for a point-of-care test in Uganda. J Blood Med, 2019. 10: p. 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goldberg SI, Niemierko A, and Turchin A, Analysis of data errors in clinical research databases. AMIA Annu Symp Proc, 2008: p. 242–6. [PMC free article] [PubMed] [Google Scholar]
- 93.Keren DF, Hedstrom D, Gulbranson R, Ou CN, and Bak R, Comparison of Sebia Capillarys Capillary Electrophoresis With the Primus High-Pressure Liquid Chromatography in the Evaluation of Hemoglobinopathies. American Journal of Clinical Pathology, 2008. 130(5): p. 824–831. [DOI] [PubMed] [Google Scholar]
- 94.Borbely N, Phelan L, Szydlo R, and Bain B, Capillary zone electrophoresis for haemoglobinopathy diagnosis. J Clin Pathol, 2013. 66(1): p. 29–39. [DOI] [PubMed] [Google Scholar]
- 95.Nusrat M, Moiz B, Nasir A, and Rasool Hashmi M, An insight into the suspected HbA2’ cases detected by high performance liquid chromatography in Pakistan. BMC Research Notes, 2011. 4: p. 103–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sharma P and Das R, Cation-exchange high-performance liquid chromatography for variant hemoglobins and HbF/A2: What must hematopathologists know about methodology? World Journal of Methodology, 2016. 6(1): p. 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoyer JD and Scheidt RM, Identification of hemoglobin variants by HPLC. Clin Chem, 2005. 51(7): p. 1303–4; author reply 1305. [DOI] [PubMed] [Google Scholar]
- 98.Figueiredo MS, The importance of hemoglobin A2 determination. Rev Bras Hematol Hemoter, 2015. 37(5): p. 287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Giambona A, Passarello C, Renda D, and Maggio A, The significance of the hemoglobin A2 value in screening for hemoglobinopathies. Clinical biochemistry, 2009. 42(18): p. 1786–1796. [DOI] [PubMed] [Google Scholar]
- 100.George Tracy I., K.T.B., Perkins Sherrie L., Hemoglobin SC Disease, 2010. [Google Scholar]
- 101.Krauss JS, Drew PA, Jonah MH, Trinh M, Shell S, Black L, and Baisden CR, Densitometry and microchromatography compared for determination of the hemoglobin C and A2 proportions in hemoglobin C and hemoglobin SC disease and in hemoglobin C trait. Clin Chem, 1986. 32(5): p. 860–2. [PubMed] [Google Scholar]
- 102.Kreuels B, Kreuzberg C, Kobbe R, Ayim-Akonor M, Apiah-Thompson P, Thompson B, Ehmen C, Adjei S, Langefeld I, Adjei O, and May J, Differing effects of HbS and HbC traits on uncomplicated falciparum malaria, anemia, and child growth. Blood, 2010. 115(22): p. 4551–8. [DOI] [PubMed] [Google Scholar]
- 103.Thomas C and Lumb AB, Physiology of haemoglobin. Continuing Education in Anaesthesia, Critical Care & Pain, 2012. 12(5): p. 251–256. [Google Scholar]
- 104.Metaxotou-Mavromati AD, Antonopoulou HK, Laskari SS, Tsiarta HK, Ladis VA, and Kattamis CA, Developmental changes in hemoglobin F levels during the first two years of life in normal and heterozygous beta-thalassemia infants. Pediatrics, 1982. 69(6): p. 734–8. [PubMed] [Google Scholar]
- 105.Watson J, The significance of the paucity of sickle cells in newborn Negro infants. Am J Med Sci, 1948. 215(4): p. 419–23. [DOI] [PubMed] [Google Scholar]
- 106.Ware RE, Technological Advances in Sickle Cell Disease. Blood Cells Mol Dis, 2017. 67: p. 102–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Transforming HemeChip design for injection molded mass production.
Fig. S2. Injection moldable HemeChip design with bottom and top plastic parts.
Fig. S3. Injection molding of HemeChip and effect of the mold finish on optical quality.
Fig. S4. Electronic circuit design of the HemeChip Reader.
Fig. S5. Software algorithm for the HemeChip Reader user interface.
Fig. S6. Screen views of the step-by-step instructions provided by the HemeChip user interface.
Fig. S7. Micro-applicator design and precise application of blood samples into HemeChip.
Fig. S8. HemeChip test kit components.
Fig. S9. Blood collection and HemeChip test protocol steps.
Fig. S10. HemeChip test result screen for no abnormal hemoglobin (Hb AA).
Fig. S11. HemeChip test result screen for SCD Trait (Hb AS).
Fig. S12. HemeChip test result screen for SCD-SS (Hb SS).
Fig. S13. HemeChip test result screen for Hb EE (Hb E Disease).
Fig. S14. HemeChip test result screen for an ‘Uninterpretable’ or invalid test.
Fig. S15. HemeChip test result screen for an ‘Inconclusive’ test.
Fig. S16. Hemoglobin F separation and identification in HemeChip.
Fig. S17. New design of portable, affordable HemeChip reader: Gazelle™.
Fig. S18. Gazelle™ Hb Variant test result screen for Hb AA (normal).
Fig. S19. Gazelle™ Hb Variant test result screen for Hb AA (normal) for a <6 months old infant.
Fig. S20. Gazelle™ Hb Variant test result screen for Hb AS trait.
Fig. S21. Gazelle™ Hb Variant test result screen for Hb AS trait for a <6 months old infant.
Fig. S22. Gazelle™ Hb Variant test result screen for Hb AC or Hb AE trait.
Fig. S23. Gazelle™ Hb Variant test result screen for Hb AC or Hb AE trait for a <6 months old infant.
Fig. S24. Gazelle™ Hb Variant test result screen for Hb SS (SCD).
Fig. S25. Gazelle™ Hb Variant test result screen for Hb SS (SCD) for a <6 months old infant.
Fig. S26. Gazelle™ Hb Variant test result screen for Hb SC or SE.
Fig. S27. Gazelle™ Hb Variant test result screen for Hb SC or Hb SE for a <6 months old infant.
Fig. S28. Gazelle™ Hb Variant test result screen for Hb CC or EE.
Fig. S29. Gazelle™ Hb Variant test result screen for Hb CC or Hb EE for a <6 months old infant.
Fig. S30. Gazelle™ Hb Variant test result screen for Hb Sβ+.
Fig. S31. Gazelle™ Hb Variant test result screen for Hb Sβ+ for <6 months old infant.
Fig. S32. Gazelle™ Hb Variant test result screen for an unusual hemoglobin pattern.
Table S1. Comparison of HemeChip with standard laboratory methods for hemoglobin testing
Table S2. Comparison of HemeChip with emerging point-of-care technologies for hemoglobin testing
Table S3. Sample size calculation for clinical studies
Video S1. Real-time tracking of hemoglobin bands in HemeChip during electrophoresis process
Video S2. Step-by-step HemeChip test procedure and analysis of results


