Abstract
Introduction
Many bacteria are responsible for infections in humans and plants, being found in vegetables, water, and medical devices. Most bacterial detection methods are time-consuming and take days to give the result. Aptamers are a promising alternative for a quick and reliable measurement technique to detect bacteria present in food products. Selected aptamers are DNA or RNA oligonucleotides that can bind with bacteria or other molecules with affinity and specificity for the target cells by the SELEX or cell-SELEX technique. This method is based on some rounds to remove the non-ligand oligonucleotides, leaving the aptamers specific to bind to the selected bacteria. Compared with conventional methodologies, the detection approach using aptamers is a rapid, low-cost form of analysis.
Objective
This review summarizes obtention methods and applications of aptamers in the food industry and biotechnology. Besides, different techniques with aptamers are presented, which enable more effective target detection.
Conclusion
Applications of aptamers as biosensors, or the association of aptamers with nanomaterials, may be employed in analyses by colorimetric, fluorescence, or electrical devices. Additionally, more efficient ways of sample preparation are presented, which can support food safety to provide human health, with a low-cost method for contaminant detection.
|
Key points • Aptamers are promising for detecting contaminants outbreaks. • Studies are needed to identify aptamers for different targets. |
Keywords: Biosensor, DNA Aptamer, Cell-SELEX, Bacteria
Introduction
The growth of epidemiologic diseases has been increasing due to food production and consumption trends, such as production processes, globalization of consumer goods, and high demand for raw or undercooked foods (Cheung and Kam 2012). Gram-positive and gram-negative bacteria are responsible for infectious diseases even at low concentrations (Soundy and Day 2017). Many methods may be applied to identify these bacteria, such as conventional bacterial culture, immunofluorescence, and polymerase chain reaction (PCR). However, these techniques are often time-consuming, without quantitative response, and often they are high-cost analyses (Lavu et al. 2016). In the last decades, rapid and efficient methods of detection are being developed to decrease food contamination and improve human health (Amaya-González et al. 2013).
Alternative methods to the detection of microorganisms are the use of single strands of DNA or RNA, called aptamers, which are capable to bind to non-nucleic acid molecules. The technique known as systematic evolution of ligands by exponential enrichment (SELEX) combines different steps such as incubation of oligonucleotides library, separation, amplification by polymerase chain reaction (PCR), and purification (Liu and Zhang 2015). This approach has been used since the 1990s when Tuerk and Gold (1990) discovered an RNA aptamer capable to bind to the T4 DNA polymerase, and Ellington and Szostak (1990) studied the bind of RNA aptamer to organic dyes. Over the years many publications were published on the use of aptamers in different ways, eventually conjugated with metallic, oxide or polymeric nanoparticles, and carbon nanotubes (Liu and Zhang 2015).
This review aims to address the techniques for obtaining aptamers, and their use particularly for contaminants detection in food products. The structure and properties of aptamers, as well as nanomaterials commonly used to be conjugated with aptamers, are presented. Moreover, recent applications and market perspectives are also introduced.
Aptamers structure and properties
Aptamers are single-stranded DNA or RNA molecules (oligonucleotides) with 50 to 100 nucleotides bases, 1 to 2 nm in size, and 7.5 to 32 kDa, with high affinity and specificity to bind to a target molecule (Ellington and Szostak 1990; Stoltenburg et al. 2005). According to Radom et al. (2013), DNA aptamers are molecules more stable than RNA; however, no significant differences were found between their specificities and binding capacities.
Aptamers can assume stable three-dimensional configurations in the aqueous phase, such as lops, triplexes, pseudoknots, G-quadruplexes, and staples (Bing et al. 2017). Due to the ability to change their shape, they can bind to different targets such as amino acids, vitamins, nucleotides, proteins, pesticides, drugs, bacteria, and inorganic or organic compounds. Moreover, they can be used as enzyme inhibitors to recognize proteins and nucleic acids, as well as inhibitors of toxins and hormones, applied in the detection of molecules in complex mixtures, purification, and biosensors (Liu and Zhang 2015; Peterson et al. 2015).
The affinity with the target molecule is associated with the dissociation constant that ranges from picomoles per liter to nanomoles per liter, being calculated through thermodynamic stability (Takenaka et al. 2017). Besides, aptamers may be stored and transported at room temperature due to the stability in the environmental conditions (Missailidis and Perkins 2007). Also, modifications in the aptamers can make them more resistant, stable, and improve the targeting ability, as shown by Ni et al. (2017). According to the authors, aptamers for the therapeutic area are susceptible to nuclease degradation and can be excreted by renal filtration even before they bind to the target. Thus, to increase the resistance to nuclease degradation, aptamers can be modified in the 3′ end with inverted thymidine or conjugated with biotin or biotin-streptavidin. Another modification is at 5′ position, which consists of the addition of cholesterol to increase the aptamer stability in plasma (Ni et al. 2017).
In the same way, it is possible to combine aptamers with nanostructures, such as gold nanoparticles or carbon nanotubes (Radom et al. 2013). They can be labeled with compounds such as dye or biotin. This process is widely used as systematic evolution of ligands by exponential enrichment (SELEX) to control the binding properties (Missailidis and Perkins 2007; Radom et al. 2013).
Systematic evolution of ligands by exponential enrichment
SELEX is a technique used to obtain aptamers through the in vitro selection of oligonucleotides over many rounds (Radom et al. 2013). In the 1990s, the first reports using the SELEX technique were published with RNA aptamers bounded to T4 DNA polymerase with high affinity (Tuerk and Gold 1990). In addition, Ellington and Szostak (1990) studied in vitro selection of organic dyes-binding RNA, and Robertson and Joyce (1990) selected the tetrahymena ribozyme and through amplifications obtained an RNA sequence able to cleave a specific DNA sequence.
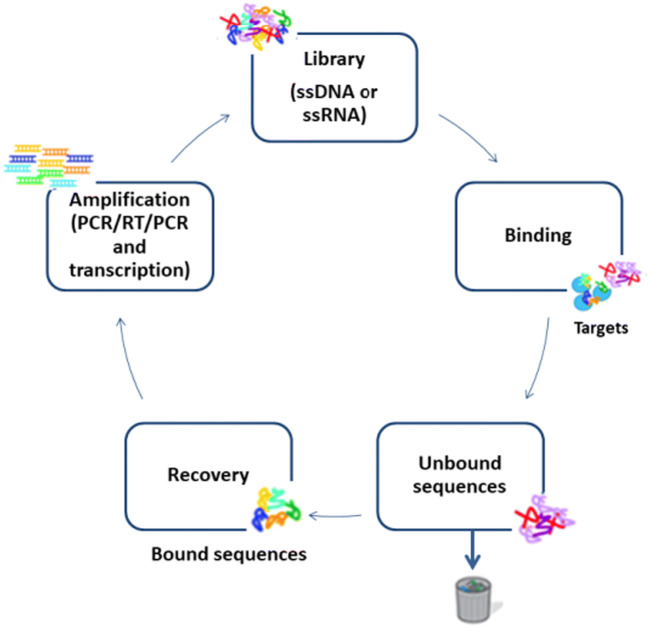
This technique allows the selection of specific oligonucleotides that can be chemically synthesized, without the need to be produced or selected in a living organism (Stoltenburg et al. 2007). As shown in Fig. 1, SELEX first step is to obtain a library of random oligonucleotides, which can be single strands of DNA or RNA, with 1014 to 1015 different sequences. This library, also called a bank, has a fixed region and a random region of 20 to 80 nucleotides, which allows the molecules to form different structures as already mentioned, ready to bind the desired molecules (Stoltenburg et al. 2005). The fixed region, which is the library extremities, allows the amplification (Stoltenburg et al. 2005).
Fig. 1.
Illustration of DNA aptamers selection by SELEX. Adapted from Liu and Zhang (2015)
The binding step occurs by the incubation of the bank of ligands (aptamer candidates) and the target molecule in a buffer solution. After this, the bounded oligonucleotides are separated from non-bounded by physical separation and PCR amplification is carried out on the recovered bounded oligonucleotides, thus the first SELEX cycle started (Radom et al. 2013). Different techniques can be used in the separation process, such as centrifugation (Soundy and Day 2017; Ramlal et al. 2018); capillary electrophoresis (Tang et al. 2006); ultrafiltration with nitrocellulose filter (Joshi et al. 2009); flow cytometry (Davis et al. 1996); and affinity chromatography using agarose, sepharose, magnetic beads and microwell plate, or sol-gel channels to immobilize the target (Mckeague et al. 2009; Bae et al. 2013; Kim et al. 2014).
The separation of aptamer candidates from the target molecule can be carried out by heat treatment, competing for ligand elution, urea addition, EDTA, or SDS techniques (Weiss et al. 1997; Bianchini et al. 2001; Theis et al. 2004; Stoltenburg et al. 2005). According to Ellington and Szostak (1990), the affinity chromatography technique is used when it is desired to obtain aptamers for small targets.
For the second SELEX cycle, the amplified product (double-stranded DNA or RNA) should be converted by PCR to a single-stranded. According to Stoltenburg et al. (2005), if the library is DNA, the most used methods are biotin/streptavidin-added electrophoresis for strand distinction (desired and unwanted), size-difference primers where the unwanted strand is modified, modified primers at tip 3′ (addition of a ribose), or hexamethylene glycol spacer primers. For the last two methods, electrophoresis is used to check the size difference and separate the unwanted tape. If the double strand is RNA, dsDNA is transcribed into RNA with T7 RNA polymerase. After the simple tapes are incubated again with the target, one more SELEX cycle occurs (Stoltenburg et al. 2005).
The cycles are performed until a binder with high affinity and specificity is obtained (Fig. 2), and in this case, 5 to 20 cycles can be performed (Tuerk and Gold 1990; Missailidis and Perkins 2007; Liu and Zhang 2015). This number depends on some factors such as the selected library, selection conditions of oligonucleotides, the concentration of incubated target, as well as the concentration of aptamer candidates (Radom et al. 2013). Once obtained, the high affinity and specificity aptamers are cloned into bacterial vectors, usually, Escherichia coli, sequenced and characterized (Tuerk and Gold 1990; Stoltenburg et al. 2005; Missailidis and Perkins 2007).
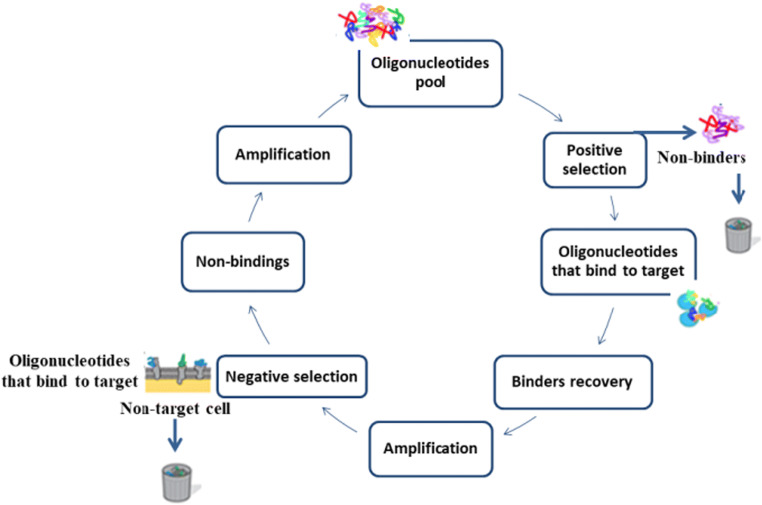
Fig. 2.
cell-SELEX steps to obtain a DNA aptamer. Adapted from Ye et al. (2012)
Several aptamers have already been selected for a range of targets as inorganic ions (Na+) (Zhou et al. 2016), dopamine, and other organic compounds such as amino acids like L-tryptophan (Idili et al. 2019), proteins (Deng et al. 2014), antibiotics (Smart et al. 2020), and microorganisms (bacteria, viruses, and fungi) (Soundy and Day 2017; Smart et al. 2020). When the aptamers are obtained by whole cells, the technique is called cell-SELEX (Radom et al. 2013).
Contrariwise to the SELEX approach, which uses purified targets, the cell-SELEX technique allows the use of whole cells where no knowledge of target conformation or protein purification is required, and whole cells remain in their natural state throughout the selection process. This technique is used because in some cases, when the target is purified, the native configuration can be lost, and the target is masked. So, the candidate aptamers may not bind since the natural structure of the targets is not recognized (Ye et al. 2012). The cell-SELEX cycle follows the same structure as the SELEX technique, but with the addition of negative selection. This approach uses different cells that are non-target for reducing the number of aptamers, which bind with non-specific cells, thus increasing aptamer specificity (Fig. 2). In the negative selection, Ye et al. (2012) described that non-binding aptamers are discarded and those targeting cell-binding are eluted and amplified by PCR. On the other hand, to the negative control, non-target cells are incubated with amplified library and non-binding cells are separated and amplified by PCR and so on until high affinity and specificity aptamers are obtained (Ye et al. 2012). In the cell-SELEX, the candidate aptamers can bind to the three-dimensional configuration of the target (Ye et al. 2012).
Cell-SELEX is reported, in the literature, to select aptamers, i.e., Salmonella enterica serovar Typhimurium (Duan et al. 2013; Lavu et al. 2016), Pseudomonas aeruginosa (Soundy and Day 2017), Neisseria meningitidis (Mirzakhani et al. 2018), Escherichia coli O157:H7 (Amraee et al. 2017), Streptococcus pyogenes (Hamula et al. 2011), Staphylococcus aureus (Ramlal et al. 2018), Haemophilus influenzae (Bitaraf et al. 2016), Trypanosoma cruzi (Ulrich et al. 2002), tumor liver cells (Mi et al. 2009), and mouse stem cells (Guo et al. 2007).
In this way, the chosen method depends on the target. Many works employed the cell-SELEX technique to obtain aptamers for bacteria detection, providing high affinity and selectivity. On the other hand, the SELEX technique can be used to identify bacteria and other compounds such as organic and inorganic molecules, viruses, and tumors.
Aptamers conjugation
Aptamers can have diverse applications, from basic research in medicine, pharmaceuticals, diagnostics, therapy, and drug development to pathogen detection, which encompasses the medical field and the food industry (Tuerk and Gold 1990; Lavu et al. 2016). In therapy, aptamers act as inhibitors of targets, as nucleolin inhibition (Radom et al. 2013), while for food safety, aptamers are used to detect contaminants (Amaya-González et al. 2013).
To improve the application range, aptamers may be conjugated to nanostructures, which assist in the identification of the target compounds. Common conjugates for aptamers are metal or silica nanoparticles, hydrogels, and even carbon nanomaterials, due to their biocompatibility, controllable chemical and physical properties, and stability (Liu and Zhang 2015; Yang et al. 2015).
Among the conjugation applications, one can be the aptamer conjugation for colorimetric detection. This type of detection is the most attractive and widely used since the target is detected through visual observation with the aid of colored reagent without the use of analytical instruments as a spectrophotometer. For this kind of application, gold, magnetic, or cerium oxide nanoparticles, carbon nanotubes, graphene oxide, or even polymers may be conjugated to the aptamers (Sharma et al. 2015). These nanostructured supports have been commonly synthesized and applied (Almeida et al. 2017; Valério et al. 2017; Chiaradia et al. 2018; Hoelscher et al. 2018; Maass et al. 2019).
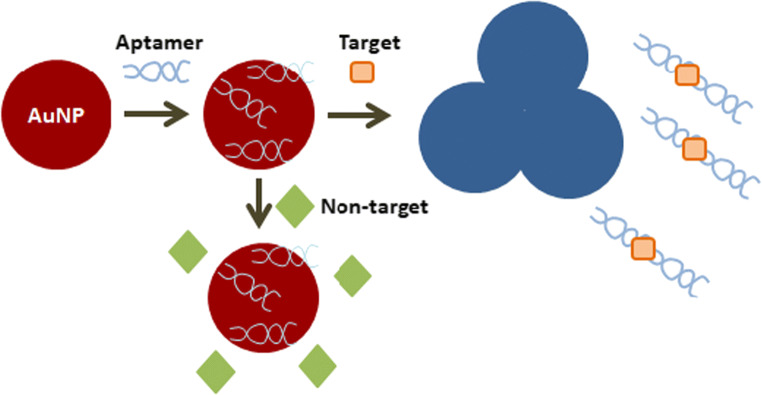
Gold nanoparticles are widely used because they decrease the distance between dispersed particles and increase the size after aggregation resulting in red to blue colors, as shown in Fig. 3 (Sharma et al. 2015). Some authors reported the use of silver ions to improve detection sensitivity. In this case, silver ions adhere to the surface of AuNPs reducing silver atoms by electrons released from the reducing agent around the gold nanoparticles. Thus, the nucleation reaction increases the gold nanoparticles size changing the color making possible a visible identification (Liu et al. 2014).
Fig. 3.
Aptamer conjugation with gold nanoparticles. Note: Adapted from Sharma et al. (2015)
Fluorescence is the emission of light from an excited molecule, a dye or even a nanomaterial that then returns to its initial state (Sharma et al. 2015). Both colorimetric and fluorescence assays are widely used in aptamer studies, as they present high sensitivity, high efficiency, and easy operation. Those techniques require the use of fluorophore and chromophore dyes for measurable signal emission (Sharma et al. 2015).
According to Sharma et al. (2015), besides dyes, some nanomaterials provide fluorescence emission, which are economically viable but have a time consuming laborious process that can affect the selectivity of the aptamer to the target binding. Assays without fluorescent markers consist of the use of DNA intercalators, base site binding dyes, and metallic nanomaterials with the fluorescence emission.
Organic dyes such as FAM (fluorescein amidite) are commonly bounded to oligonucleotides but have some limitations such as broad emission range, low photostability, low absorption, and photodegradation (Li et al. 2008). To improve these limitations, inorganic fluorescents like quantum dots can be used, since their optical characteristics depend on the size (1 to 10 nm). They have a broad absorption spectrum, narrow emission spectrum, and long life fluorescence. However, their drawbacks are the high-cost synthesis and toxicity associated (Sharma et al. 2015). An example of quantum dots is cadmium selenide, which has high luminescence and good quantum yield (Xu et al. 2015) as already reported to detect the presence of E. coli O157: H7 by Xu et al. (2015).
Yang et al. (2011) reported the use of carbon nanoparticles for fluorescence testing, highlighting material advantages such as low cost, high quantum productivity, simple preparation, low toxicity, good biocompatibility, good aqueous solubility, and superior photoluminescence properties. In addition to fluorescent dyes, nucleic acid dyes may be used (SYBR green I, AccuBlue, and PicoGreen). The drawbacks of the nucleic acid dyes are the low fluorescence intensity and the requirement to interleaving with the aptamer DNA sequence (Duan et al. 2014a).
The chemiluminescence technique also uses optical detection and is mainly applied in food safety since it produces energy through chemical reaction without the need for excitation source as in the fluorescence technique (Sharma et al. 2015). Chemiluminescence signals can be increased using AuNPs as catalysts enhancing biocompatibility and stability (Yang et al. 2011). DNA aptamers that are rich in guanine (G) can react with 3,4,5-trimethoxylphenylglyoxal (TMPG), which forms an energy-rich compound that emits light or transfers energy to some aptamer-coupled dye, such as 6-FAM (6-carboxyfluorescein) with green light emission and can be employed in target detection studies (Kwon et al. 2015). Thus, aptamers can be conjugated to different nanoparticles to increase the selectivity to the target. Besides increasing the selectivity, these nanomaterials allow visual detection.
Aptamers applications
Several aptamers have been developed for different applications, as reported by Chan et al. (2008) that employed PEG-conjugated aptamers RB006 against coagulation. Wu et al. (2008) developed a PO RO10–60 aptamer to stimulate the immune response against pathogens by delaying symptoms and allowing the use of antibiotics. There are also electrochemical sensors, which are aptamers that act in real-time detection of cocaine in fetal bovine serum (Swensen et al. 2009).
Aptamer-based nanostructures have also been widely used in medicine, biology, and nanoelectronics due to the high stability, as shown in Table 1. The AS1411 aptamer conjugated with gold nanoclusters was tested in mice cancer cells and demonstrated to be a good radiosensitizer (Ghahremani et al. 2018). Through cell-SELEX, the JHIT2 aptamer was selected and labeled with FAM and iodine-131 to detect human hepatoma cell line HepG2 by a fluorescent signal (Zhang et al. 2020). All these studies show that the aptamers can assist in the rapid detection of different targets, shortening the time to start treatments that are important for human health.
Table 1.
Examples of aptamers application to different targets in different areas
| Target | Method | Sample | Reference |
|---|---|---|---|
| Streptococcus pyogenes | Cell-SELEX | Cooked chicken | (Huang et al. 2018) |
| Salmonella typhimurium | Cell-SELEX | Pasteurized milk | (Duan et al. 2014b) |
| Salmonella typhimurium and Vibrio parahemolyticus | Cell-SELEX | Frozen shrimp, chicken breasts | (Duan et al. 2014a) |
| Salmonella | – | Pork | (Ma et al. 2014) |
| Escherichia coli | Cell-SELEX | Milk and tap water and pond | (Kim et al. 2013; Jin et al. 2017) |
| Staphylococcus aureus | – | Fresh fish | (Jia et al. 2014) |
| Staphylococcus aureus | Cell-SELEX | Pork meat | (Hao et al. 2017) |
| Staphylococcus aureus | Cell-SELEX | Milk | (Yuan et al. 2014) |
| Listeria monocytogenes | SELEX | Liced beef, chicken, turkey | (Ohk et al. 2010) |
| Campylobacter jejuni | SELEX | Live cell | (Bruno et al. 2009) |
| Lactobacillus acidophilus | Cell-SELEX | Oxidized PSi Fabry-Pérot thin films | (Urmann et al. 2016) |
| Francisella tularensis | SELEX | Bacterial antigen | (Vivekananda and Kiel 2006) |
| Mycobacterium tuberculosis | SELEX | Live cell | (Chen et al. 2012) |
| Vibrio parahemolyticus | Cell-SELEX | Live cell | (Duan et al. 2012) |
| Shigella sonnei | Cell-SELEX | Live cell | (Song et al. 2017) |
| C. jejuni | Cell-SELEX | Live cell | (Dwivedi et al. 2010) |
| Vaccinia virus | SELEX | Vaccinia intacto | (Labib et al. 2012) |
| herpes simplex virus | SELEX | Gd protein of HSV-1 | (Gopinath et al. 2012) |
| Hepatitis C and hepatitis B virus | SELEX | Hepatitis C virus | (Kumar et al. 1997) |
| Human immunodeficiency virus | In vitro selection | Human immunodeficiency virus type-1 | (Boiziau et al. 1999) |
| Influenza virus | SELEX | Hemagglutinin protein of human influenza virus B | (Gopinath et al. 2005) |
| Severe Acute Respiratory Syndrome (SARS) coronavirus | SELEX | Live cell | (Jang et al. 2008) |
| Trypanosoma spp. | SELEX | Plasma of T. cruzi infected mice | (Nagarkatti et al. 2014) |
| Leishmania spp. | SELEX | Live cell | (Guerra-Pérez et al. 2015) |
| Plasmodium spp. | – | P. falciparum para-sites | (Cheung et al. 2018) |
| Cryptosporidium parvum | SELEX | Fresh fruits | (Iqbal et al. 2015) |
| Entamoeba histolytica | SELEX | Live cell | (Ospina-Villa et al. 2015) |
| MCF-7 breast cancer cells | – | Target cancer cells | (Wang et al. 2015) |
| Leukemia CCRF-CEM cells | – | Human leukemia CCRF-CEM cells | (Ye et al. 2015) |
| Metastatic tumor tissues | Cell-SELEX | Colon cancer cell SW620 | (Li et al. 2015) |
| Ochratoxin A (OTA) | SELEX | Immobilized OTA | (Cruz-Aguado and Penner 2008) |
| Bacterial endotoxins | SELEX | Lipopolysaccharide | (Kim et al. 2012) |
| Copper | – | Lake samples | (Chen et al. 2011) |
| Arsenic | – | Aqueous solution | (Oroval et al. 2017) |
| Acetamiprid | – | Wastewater and tomatoes | (Fan et al. 2013) |
| Herbicides | SELEX | Atrazine | (Sinha et al. 2010) |
| Milk allergen | SELEX | β-LG variants A and B | (Eissa and Zourob 2017) |
| Bisphenol A | – | Aqueous solution | (Chen et al. 2017) |
| Beta1-adrenoreceptor autoantibodies | – | Serum of patients | (Wallukat et al. 2016) |
| Lung cancer | SELEX | Cells | (Bates et al. 2009) |
| Colorectal cancer | – | Camptothecin loaded-pegylated dendrimer | (Alibolandi et al. 2017) |
| Breast cancer | – | Breast cancer tissues | (Wang et al. 2017) |
Lavu et al. (2016) studied the use of gold nanoparticles with aptamers for the detection of Salmonella enterica. The aptamer SAL 26 was conjugated with gold nanoparticles at room temperature and in the presence of NaCl keeping the solution red, and when in the presence of Salmonella enterica (102 to 106 CFU/mL), the solution turned blue after 30 min. According to the authors, the color change is associated with the formation of a tertiary structure with the target cell that has no affinity for gold nanoparticles, resulting in salt-induced aggregation.
De Girolamo et al. (2011) developed a DNA aptamer to detect OTA (Ochratoxin A) mycotoxin produced by Aspergillus ochraceus and Penicillium verrucosum, found in wheat. They showed a system able to detect OTA in a range from 0.4 to 500 ng. Chen et al. (2015) reported the direct detection of FB1 (fumonisin B1) in maize samples by using gold nanoparticles conjugated to modified aptamers (5′-SH-(CH2)6-AGCAGCACAGAGGTCAGATGCGATCTGGATATTATTTTTGATACCCCTTTGGGGAGACATCCTATGCGTGCTACCGTGAA-3′). The authors reported an accrued detection after 40 min at room temperature for FB1 concentrations above to 2 pM. Another mycotoxin that is toxic to humans is zearalenone (ZEN). It is found in cereal crops and produced by Fusarium graminearum (Luo et al. 2020). To detect ZEN, the mycotoxin was extracted of cereal crops and different solutions were prepared and analyzed by the aptamer conjugated with zinc oxide-nitrogen doped graphene quantum dots (ZnO-NGQDs), which was capable to detect 3.3 × 10−14 g.mL−1 (Luo et al. 2020).
Heavy metals present in milk and dairy products, fish, eggs, oils, and seeds can be also detected by aptamers. Hazardous metals, such as arsenic and mercury, can affect human health by interfering with the central nervous system and endocrine system. Thereby, colorimetric detection by DNA aptamers has been reported by Li et al. (2009), who used aptamer (5′-TTTTTTTTTT-3′) conjugated with AuNPs (13 nm) incubated with mercury (1 × 10−4 mol.L−1) at room temperature. After the addition of 50 μL of 0.5 M NaCl, it was observed that the solution turned to blue confirming the presence of metal. Wu et al. (2014) investigated DNA aptamer conjugated with gold nanoparticles to cadmium and reported high-affinity detection for an aqueous solution containing cadmium at lower concentration (4.6 nM).
The green malachite fungicide is widely used in aquaculture and can contaminate fish and their eggs, posing a risk to those who consume them (Stead et al. 2010). In 2010, it was reported the first malachite green (MG) detection by RNA aptamer (5’-GGAUCCCGACUGGCGAGAGCCAGGUAACGAAUGGAUCC-3′) in fish skin samples, and the developed approach was able to quickly confirm the contamination after 15–20 min at 2 μg.kg−1 of salmon tissue (Stead et al. 2010).
Many types of pesticides are used to prevent contamination by bacteria, fungi, and viruses, and the detection methods should be efficient even at low pesticide concentrations. However, detection by liquid and gas chromatography are expensive and time-consuming (Fan et al. 2013). To reduce the costs and analysis time, new technologies are employed, such as the use of biosensors. Fan et al. (2013) developed a conjugated aptamer with a gold nanoparticle that generates a signal of impedance to identify acetamiprid with a detection limit of 1 nM in wastewater and tomatoes and the process takes up to 3 h.
For herbicides, widely used in the cultivation of corn and oilseed rape, with the consequent environment and human life issue, different DNA aptamers with affinity to atrazine were studied by Williams et al. (2014). The double-stranded DNA aptamer for fluorescence detection was drawn to detect fipronil insecticide in river water samples and showed high sensitivity (Hong et al. 2018).
Moreover, the C07 aptamer was developed to detect Sudan dye III in chili sauce. From the study, it was reported a fast and accurate binding to the target, and according to the authors, 100 nM of aptamer was enough to detect 400 ng of Sudan dye III (Wang et al. 2018). Besides, organic molecules, such as bisphenol A (BPA), present in some food products are harmful to the human endocrine system, and in 2010, the US and Canadian governments banned their use (Mckeague et al. 2009). In this sense, Lee et al. (2011) studied the detection of BPA by aptamers conjugated with carbon nanotube as a biosensor and showed a detection limit at low concentrations (10 fM).
In this context, Smart et al. (2020) reported several promising biosensors for agribusiness. In some cases, aptamers were conjugated to nanoparticles forming carbon electrodes for detection of pesticides, toxins, antibiotics, microorganisms, vitamins, fructose, and lactate. In addition, Yan et al. (2020) showed different photoelectrochemical and electrochemiluminescent apta-sensors capable to detect food contaminants and pollutants.
Conclusions and market perspectives
There are several studies in the literature related to the development of aptamers for different targets. They are widely studied in the therapeutic area, to identify cancer cells, bacterial contamination, and viruses. Moreover, aptamers are being developed for food safety since there is a high diversity of pathogens in food products from different origins (animal, vegetable, processed), as well as contamination by packaging or transportation.
Despite being basic, inexpensive and selective, most existing aptamers are still not currently used in the industries and agribusiness routine. Through the aptamer applications, a quick analysis system can be launched as a biosensor, bringing advantages to the market as visual detection, low-cost compared with conventional techniques, as well as delivering robustness and selectivity.
Chemiluminescence detection is one of the most studied technics since it does not require equipment for signal detection. However, for many targets, studies are needed to ensure that sensitivity and specificity are enhanced using nanoparticles. Further investigation should be directed to sample preparation methods.
Many of the aptamers are not able to recognize samples in the raw phase and need to be prepared in aqueous solutions. To reduce the gap between lab-scale and industrial large-scale applications, advances in fast and efficient detection for food safety are increasing, but some aspects still need to be improved, such as sample preparation, concentration, and the presence of contaminants from raw materials.
Due to the difficulties related to food safety, the companies are looking for cheaper and faster alternatives. Whereas the world population is expected to reach 8.5 billion people by 2030 (United Nations 2019), the food industry faces problems with changes in food production and supply, increased imports, changes in the environment that lead to contamination, development of outbreaks, or pests on crops in different locations. Herewith, the market is turning to faster and more effective sensors to prevent detect contamination. In this context, aptamers come to the market as an alternative (Liu and Zhang 2015).
According to their advantages, aptamers can supply the agribusiness as well as food industrialization needs, being increasingly used worldwide to speed up the food safety analysis, to avoid products recall and unnecessary business expenses around the world (Amaya-González et al. 2013; Aptamer Group 2016).
Authors’ contributions
All authors wrote, read, assisted in the article correction, and approved the manuscript.
Funding information
We thank the Foundation for Research and Innovation of the State of Santa Catarina (FAPESC) and the Program CAPES-PRINT (project numbers 88887.310560/2018-00 and 88887.310727/2018-00), for financially supporting this research.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alibolandi M, Taghdisi SM, Ramezani P, Hosseini Shamili F, Farzad SA, Abnous K, Ramezani M. Smart AS1411-aptamer conjugated pegylated PAMAM dendrimer for the superior delivery of camptothecin to colon adenocarcinoma in vitro and in vivo. Int J Pharm. 2017;519:352–364. doi: 10.1016/j.ijpharm.2017.01.044. [DOI] [PubMed] [Google Scholar]
- Almeida ÉS, de Oliveira D, Hotza D. Characterization of silver nanoparticles produced by biosynthesis mediated by Fusarium oxysporum under different processing conditions. Bioprocess Biosyst Eng. 2017;40:1291–1303. doi: 10.1007/s00449-017-1788-9. [DOI] [PubMed] [Google Scholar]
- Amaya-González S, de-los- Santos-Alvarez N, Miranda-Ordieres AJ, Lobo-Castañón MJ. Aptamer-based analysis: a promising alternative for food safety control. Sensors (Basel) 2013;13:16292–16311. doi: 10.3390/s131216292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amraee M, Oloomi M, Yavari A, Bouzari S. DNA aptamer identification and characterization for E. coli O157 detection using cell based SELEX method. Anal Biochem. 2017;536:36–44. doi: 10.1016/j.ab.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Aptamer Group (2016) Aptamers in Agri-Tech and Food Safety
- Bae H, Ren S, Kang J, Kim M, Jiang Y, Jin MM, Min IM, Kim S. Sol-gel SELEX circumventing chemical conjugation of low molecular weight metabolites discovers Aptamers selective to xanthine. Nucleic Acid Ther. 2013;23:443–449. doi: 10.1089/nat.2013.0437. [DOI] [PubMed] [Google Scholar]
- Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 AS a novel treatment for cancer. Exp Mol Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini M, Radrizzani M, Brocardo MG, Reyes GB, Gonzalez Solveyra C, Santa-Coloma TA. Specific oligobodies against ERK-2 that recognize both the native and the denatured state of the protein. J Immunol Methods. 2001;252:191–197. doi: 10.1016/s0022-1759(01)00350-7. [DOI] [PubMed] [Google Scholar]
- Bing T, Zheng W, Zhang X, Shen L, Liu X, Wang F, Cui J, Cao Z, Shangguan D. Triplex-quadruplex structural scaffold: a new binding structure of aptamer. Sci Rep. 2017;7:15467. doi: 10.1038/s41598-017-15797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitaraf FS, Rasooli I, Mousavi Gargari SL. DNA aptamers for the detection of Haemophilus influenzae type b by cell SELEX. Eur J Clin Microbiol Infect Dis. 2016;35:503–510. doi: 10.1007/s10096-015-2567-7. [DOI] [PubMed] [Google Scholar]
- Boiziau C, Dausse E, Yurchenko L, Toulme JJ. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J Biol Chem. 1999;274:12730–12737. doi: 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- Bruno JG, Phillips T, Carrillo MP, Crowell R. Plastic-adherent DNA aptamer-magnetic bead and quantum dot sandwich assay for campylobacter detection. J Fluoresc. 2009;19:427–435. doi: 10.1007/s10895-008-0429-8. [DOI] [PubMed] [Google Scholar]
- Chan MY, Cohen MG, Dyke CK, Myles SK, Aberle LG, Lin M, Walder J, Steinhubl SR, Gilchrist IC, Kleiman NS, Vorchheimer DA, Chronos N, Melloni C, Alexander JH, Harrington RA, Tonkens RM, Becker RC, Rusconi CP. Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation. 2008;117:2865–2874. doi: 10.1161/CIRCULATIONAHA.107.745687. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li L, Mu X, Zhao H, Guo L. Electrochemical aptasensor for detection of copper based on a reagentless signal-on architecture and amplification by gold nanoparticles. Talanta. 2011;85:730–735. doi: 10.1016/j.talanta.2011.04.056. [DOI] [PubMed] [Google Scholar]
- Chen F, Zhang X, Zhou J, Liu S, Liu J. Aptamer inhibits Mycobacterium tuberculosis (H37Rv) invasion of macrophage. Mol Biol Rep. 2012;39:2157–2162. doi: 10.1007/s11033-011-0963-3. [DOI] [PubMed] [Google Scholar]
- Chen X, Huang Y, Ma X, Jia F, Guo X, Wang Z (2015) Impedimetric aptamer-based determination of the mold toxin fumonisin B1. Microchim Acta 182:1709–1714. 10.1007/s00604-015-1492-x
- Chen M, Chen J, Ding L, Xu Z, Wen L, Wang L, Cheng Y (2017) Study of the detection of bisphenol A based on a nano-sized metal–organic framework crystal and an aptamer. Anal Methods:906–909. 10.1039/c6ay03151j
- Cheung P-Y, Kam KM. Salmonella in food surveillance: PCR, immunoassays, and other rapid detection and quantification methods. Food Res Int. 2012;45:802–808. doi: 10.1016/j.foodres.2011.12.001. [DOI] [Google Scholar]
- Cheung Y-W, Dirkzwager RM, Wong W-C, Cardoso J, D’Arc Neves Costa J, Tanner JA. Aptamer-mediated Plasmodium-specific diagnosis of malaria. Biochimie. 2018;145:131–136. doi: 10.1016/j.biochi.2017.10.017. [DOI] [PubMed] [Google Scholar]
- Chiaradia V, Polloni AE, de Oliveira D, de Oliveira JV, Araújo PHH, Sayer C. Polyester nanoparticles from macrolactones via miniemulsion enzymatic ring-opening polymerization. Colloid Polym Sci. 2018;296:861–869. doi: 10.1007/s00396-018-4306-y. [DOI] [Google Scholar]
- Cruz-Aguado JA, Penner G. Determination of Ochratoxin A with a DNA Aptamer. J Agric Food Chem. 2008;56:10456–10461. doi: 10.1021/jf801957h. [DOI] [PubMed] [Google Scholar]
- Davis KA, Abrams B, Lin Y, Jayasena SD. Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids Res. 1996;24:702–706. doi: 10.1093/nar/24.4.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Girolamo A, McKeague M, Miller JD, DeRosa MC, Visconti A. Determination of ochratoxin A in wheat after clean-up through a DNA aptamer-based solid phase extraction column. Food Chem. 2011;127:1378–1384. doi: 10.1016/j.foodchem.2011.01.107. [DOI] [PubMed] [Google Scholar]
- Deng B, Lin Y, Wang C, Li F, Wang Z, Zhang H, Li X-F, Le XC. Aptamer binding assays for proteins: the thrombin example—a review. Anal Chim Acta. 2014;837:1–15. doi: 10.1016/j.aca.2014.04.055. [DOI] [PubMed] [Google Scholar]
- Duan N, Wu S, Chen X, Huang Y, Wang Z. Selection and identification of a DNA aptamer targeted to Vibrio parahemolyticus. J Agric Food Chem. 2012;60:4034–4038. doi: 10.1021/jf300395z. [DOI] [PubMed] [Google Scholar]
- Duan N, Wu S, Chen X, Huang Y, Xia Y, Ma X, Wang Z. Selection and characterization of Aptamers against Salmonella typhimurium using whole-bacterium systemic evolution of ligands by exponential enrichment (SELEX) J Agric Food Chem. 2013;61:3229–3234. doi: 10.1021/jf400767d. [DOI] [PubMed] [Google Scholar]
- Duan N, Wu S, Ma X, Xia Y, Wang Z. A universal fluorescent aptasensor based on AccuBlue dye for the detection of pathogenic bacteria. Anal Biochem. 2014;454:1–6. doi: 10.1016/j.ab.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Duan YF, Ning Y, Song Y, Deng L. Fluorescent aptasensor for the determination of Salmonella typhimurium based on a graphene oxide platform. Microchim Acta. 2014;181:647–653. doi: 10.1007/s00604-014-1170-4. [DOI] [Google Scholar]
- Dwivedi HP, Smiley RD, Jaykus L-A. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl Microbiol Biotechnol. 2010;87:2323–2334. doi: 10.1007/s00253-010-2728-7. [DOI] [PubMed] [Google Scholar]
- Eissa S, Zourob M. In vitro selection of DNA aptamers targeting β-lactoglobulin and their integration in graphene-based biosensor for the detection of milk allergen. Biosens Bioelectron. 2017;91:169–174. doi: 10.1016/j.bios.2016.12.020. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fan L, Zhao G, Shi H, Liu M, Li Z. A highly selective electrochemical impedance spectroscopy-based aptasensor for sensitive detection of acetamiprid. Biosens Bioelectron. 2013;43:12–18. doi: 10.1016/j.bios.2012.11.033. [DOI] [PubMed] [Google Scholar]
- Ghahremani F, Kefayat A, Shahbazi-Gahrouei D, Motaghi H, Mehrgardi MA, Haghjooy-Javanmard S. AS1411 aptamer-targeted gold nanoclusters effect on the enhancement of radiation therapy efficacy in breast tumor-bearing mice. Nanomedicine. 2018;13:2563–2578. doi: 10.2217/nnm-2018-0180. [DOI] [PubMed] [Google Scholar]
- Gopinath SCB, Kawasaki K, Kumar PKR. Selection of RNA-aptamer against human influenza B virus. Nucleic Acids Symp Ser (Oxf) 2005;49:85–86. doi: 10.1093/nass/49.1.85. [DOI] [PubMed] [Google Scholar]
- Gopinath SCB, Hayashi K, Kumar PKR. Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J Virol. 2012;86:6732–6744. doi: 10.1128/JVI.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Pérez N, Ramos E, García-Hernández M, Pinto C, Soto M, Martín ME, González VM. Molecular and functional characterization of ssDNA aptamers that specifically bind Leishmania infantum PABP. PLoS One. 2015;10:e0140048. doi: 10.1371/journal.pone.0140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K-T, Schafer R, Paul A, Ziemer G, Wendel HP. Aptamer-based strategies for stem cell research. Mini-Rev Med Chem. 2007;7:701–705. doi: 10.2174/138955707781024481. [DOI] [PubMed] [Google Scholar]
- Hamula CLA, Zhang H, Li F, Wang Z, Chris Le X, Li XF. Selection and analytical applications of aptamers binding microbial pathogens. TrAC - Trends Anal Chem. 2011;30:1587–1597. doi: 10.1016/j.trac.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Gu H, Duan N, Wu S, Ma X, Xia Y, Tao Z, Wang Z. An enhanced chemiluminescence resonance energy transfer aptasensor based on rolling circle amplification and WS2 nanosheet for Staphylococcus aureus detection. Anal Chim Acta. 2017;959:83–90. doi: 10.1016/j.aca.2016.12.045. [DOI] [PubMed] [Google Scholar]
- Hoelscher F, Machado TO, de Oliveira D, Hermes de Araújo PH, Sayer C. Enzymatically catalyzed degradation of poly (thioether-ester) nanoparticles. Polym Degrad Stab. 2018;156:211–217. doi: 10.1016/j.polymdegradstab.2018.09.007. [DOI] [Google Scholar]
- Hong S-T, Cheon H, Lee M-J. Social conditions of village democracy in South Korea. Dev Sociol. 2018;47:85–117. doi: 10.21588/dns/2018.47.1.004. [DOI] [Google Scholar]
- Huang Y, Wang X, Duan N, Xia Y, Wang Z, Che Z, Wang L, Yang X, Chen X. Selection and characterization, application of a DNA aptamer targeted to Streptococcus pyogenes in cooked chicken. Anal Biochem. 2018;551:37–42. doi: 10.1016/j.ab.2018.04.015. [DOI] [PubMed] [Google Scholar]
- Idili A, Gerson J, Parolo C, Kippin T, Plaxco KW. An electrochemical aptamer-based sensor for the rapid and convenient measurement of l-tryptophan. Anal Bioanal Chem. 2019;411:4629–4635. doi: 10.1007/s00216-019-01645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A, Labib M, Muharemagic D, Sattar S, Dixon BR, Berezovski MV. Detection of Cryptosporidium parvum oocysts on fresh produce using DNA Aptamers. PLoS One. 2015;10:e0137455. doi: 10.1371/journal.pone.0137455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KJ, Lee N-R, Yeo W-S, Jeong Y-J, Kim D-E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem Biophys Res Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Duan N, Wu S, Ma X, Xia Y, Wang Z, Wei X. Impedimetric aptasensor for Staphylococcus aureus based on nanocomposite prepared from reduced graphene oxide and gold nanoparticles. Microchim Acta. 2014;181:967–974. doi: 10.1007/s00604-014-1195-8. [DOI] [Google Scholar]
- Jin B, Wang S, Lin M, Jin Y, Zhang S, Cui X, Gong Y, Li A, Xu F, Lu TJ. Upconversion nanoparticles based FRET aptasensor for rapid and ultrasensitive bacteria detection. Biosens Bioelectron. 2017;90:525–533. doi: 10.1016/j.bios.2016.10.029. [DOI] [PubMed] [Google Scholar]
- Joshi R, Janagama H, Dwivedi HP, Senthil Kumar TMA, Jaykus LA, Schefers J, Sreevatsan S. Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol Cell Probes. 2009;23:20–28. doi: 10.1016/j.mcp.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Kim S-E, Su W, Cho M, Lee Y, Choe W-S. Harnessing aptamers for electrochemical detection of endotoxin. Anal Biochem. 2012;424:12–20. doi: 10.1016/j.ab.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Kim YS, Song MY, Jurng J, Kim BC. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell-systematic evolution of ligands by exponential enrichment approach. Anal Biochem. 2013;436:22–28. doi: 10.1016/j.ab.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Kim C-H, Lee L-P, Min J-R, Lim M-W, Jeong S-H. An indirect competitive assay-based aptasensor for detection of oxytetracycline in milk. Biosens Bioelectron. 2014;51:426–430. doi: 10.1016/j.bios.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Kumar PK, Machida K, Urvil PT, Kakiuchi N, Vishnuvardhan D, Shimotohno K, Taira K, Nishikawa S. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology. 1997;237:270–282. doi: 10.1006/viro.1997.8773. [DOI] [PubMed] [Google Scholar]
- Kwon M, Park Y, Lee JH. Guanine chemiluminescent biosensor capable of rapidly sensing mercury in a sample. RSC Adv. 2015;5:94629–94634. doi: 10.1039/C5RA17407D. [DOI] [Google Scholar]
- Labib M, Zamay AS, Muharemagic D, Chechik AV, Bell JC, Berezovski MV. Aptamer-based viability impedimetric sensor for viruses. Anal Chem. 2012;84:1813–1816. doi: 10.1021/ac203412m. [DOI] [PubMed] [Google Scholar]
- Lavu PSR, Mondal B, Ramlal S, Murali HS, Batra HV. Selection and characterization of aptamers using a modified whole cell bacterium SELEX for the detection of Salmonella enterica Serovar Typhimurium. ACS Comb Sci. 2016;18:292–301. doi: 10.1021/acscombsci.5b00123. [DOI] [PubMed] [Google Scholar]
- Lee J, Jo M, Kim TH, Ahn J-Y, Lee D, Kim S, Hong S. Aptamer sandwich-based carbon nanotube sensors for single-carbon-atomic-resolution detection ofnon-polar small molecular species. Lab Chip. 2011;11:52–56. doi: 10.1039/C0LC00259C. [DOI] [PubMed] [Google Scholar]
- Li Y, Guo L, Zhang Z, Tang J, Xie J. Recent advances of aptamer sensors. Sci China Ser B Chem. 2008;51:193–204. doi: 10.1007/s11426-008-0001-z. [DOI] [Google Scholar]
- Li L, Li B, Qi Y, Jin Y. Label-free aptamer-based colorimetric detection of mercury ions in aqueous media using unmodified gold nanoparticles as colorimetric probe. Anal Bioanal Chem. 2009;393:2051–2057. doi: 10.1007/s00216-009-2640-0. [DOI] [PubMed] [Google Scholar]
- Li X, An Y, Jin J, Zhu Z, Hao L, Liu L, Shi Y, Fan D, Ji T, Yang CJ. Evolution of DNA aptamers through in vitro metastatic-cell-based systematic evolution of ligands by exponential enrichment for metastatic cancer recognition and imaging. Anal Chem. 2015;87:4941–4948. doi: 10.1021/acs.analchem.5b00637. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang X. Aptamer-based technology for food analysis. Appl Biochem Biotechnol. 2015;175:603–624. doi: 10.1007/s12010-014-1289-0. [DOI] [PubMed] [Google Scholar]
- Liu R, Zhang Y, Zhang S, Qiu W, Gao Y. Silver enhancement of gold nanoparticles for biosensing: from qualitative to quantitative. Appl Spectrosc Rev. 2014;49:121–138. doi: 10.1080/05704928.2013.807817. [DOI] [Google Scholar]
- Luo L, Liu X, Ma S, Li L, You T. Quantification of zearalenone in mildewing cereal crops using an innovative photoelectrochemical aptamer sensing strategy based on ZnO-NGQDs composites. Food Chem. 2020;322:126778. doi: 10.1016/j.foodchem.2020.126778. [DOI] [PubMed] [Google Scholar]
- Ma X, Jiang Y, Jia F, Yu Y, Chen J, Wang Z. An aptamer-based electrochemical biosensor for the detection of Salmonella. J Microbiol Methods. 2014;98:94–98. doi: 10.1016/j.mimet.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Maass D, Valério A, Lourenço LA, de Oliveira D, Hotza D. Biosynthesis of iron oxide nanoparticles from mineral coal tailings in a stirred tank reactor. Hydrometallurgy. 2019;184:199–205. doi: 10.1016/j.hydromet.2019.01.010. [DOI] [Google Scholar]
- Mckeague M, Giamberardino A, Derosa MC (2009) Advances in aptamer-based biosensors for food safety. In: Environmental Biosensors. IntechOpen
- Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol. 2009;6:22. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzakhani K, Gargari SLM, Rasooli I, Rasoulinejad S. Development of a DNA aptamer for screening Neisseria meningitidis serogroup b by cell SELEX. Iran Biomed J. 2018;22:193–201. doi: 10.22034/ibj.22.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missailidis S, Perkins A. Update: Aptamers as novel radiopharmaceuticals: their applications and future prospects in diagnosis and therapy. Cancer Biother Radiopharm. 2007;22:453–468. doi: 10.1089/cbr.2007.357. [DOI] [PubMed] [Google Scholar]
- Nagarkatti R, de Araujo FF, Gupta C, Debrabant A. Aptamer based, non-PCR, non-serological detection of Chagas disease biomarkers in Trypanosoma cruzi infected mice. PLoS Negl Trop Dis. 2014;8:e2650. doi: 10.1371/journal.pntd.0002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni S, Yao H, Wang L, Lu J, Jiang F, Lu A, Zhang G (2017) Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int J Mol Sci 18. 10.3390/ijms18081683 [DOI] [PMC free article] [PubMed]
- Ohk SH, Koo OK, Sen T, Yamamoto CM, Bhunia AK. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J Appl Microbiol. 2010;109:808–817. doi: 10.1111/j.1365-2672.2010.04709.x. [DOI] [PubMed] [Google Scholar]
- Oroval M, Coll C, Bernardos A, Marcos MD, Martínez-Máñez R, Shchukin DG, Sancenón F. Selective fluorogenic sensing of as(III) using aptamer-capped nanomaterials. ACS Appl Mater Interfaces. 2017;9:11332–11336. doi: 10.1021/acsami.6b15164. [DOI] [PubMed] [Google Scholar]
- Ospina-Villa JD, Dufour A, Weber C, Ramirez-Moreno E, Zamorano-Carrillo A, Guillen N, Lopez-Camarillo C, Marchat LA, Zheng CY, Pestilli F, Rokem A, Wang KK, Fan DD, Liu Y, Wang E, Ye X, Shi H, He X, Wang KK, He D, Yan L, Xu F, Lei Y, Tang J, Yu Y, Li X, An Y, Jin J, Zhu Z, Hao L, Liu L, Shi Y, Fan DD, Ji T, Yang CJ, Wu X, Zhao Z, Bai H, Fu T, Yang CJ, Hu X, Liu Q, Champanhac C, Teng I-T, Ye M, Tan W, Cruz-Aguado JA, Penner G, Kim S-E, Su W, Cho M, Lee Y, Choe W-S, Chen Z, Li L, Mu X, Zhao H, Guo L, Oroval M, Coll C, Bernardos A, Marcos MD, Martínez-Máñez R, Shchukin DG, Sancenón F, Fan L, Zhao G, Shi H, Liu M, Li Z. Determination of ochratoxin A with a DNA aptamer. ACS Appl Mater Interfaces. 2015;87:11332–11336. doi: 10.1016/j.bios.2012.11.033. [DOI] [Google Scholar]
- Peterson AM, Jahnke FM, Heemstra JM. Modulating the substrate selectivity of DNA aptamers using surfactants. Langmuir. 2015;31:11769–11773. doi: 10.1021/acs.langmuir.5b02818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom F, Jurek PM, Mazurek MP, Otlewski J, Jeleń F. Aptamers: molecules of great potential. Biotechnol Adv. 2013;31:1260–1274. doi: 10.1016/j.biotechadv.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Ramlal S, Mondal B, Lavu PS, Bhavanashri N, Kingston J. Capture and detection of Staphylococcus aureus with dual labeled aptamers to cell surface components. Int J Food Microbiol. 2018;265:74–83. doi: 10.1016/j.ijfoodmicro.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- Sharma R, Ragavan KV, Thakur MS, Raghavarao KSMS. Recent advances in nanoparticle based aptasensors for food contaminants. Biosens Bioelectron. 2015;74:612–627. doi: 10.1016/j.bios.2015.07.017. [DOI] [PubMed] [Google Scholar]
- Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Smart A, Crew A, Pemberton R, Hughes G, Doran O, Hart JP. Screen-printed carbon based biosensors and their applications in agri-food safety. TrAC Trends Anal Chem. 2020;127:115898. doi: 10.1016/j.trac.2020.115898. [DOI] [Google Scholar]
- Song M-S, Sekhon SS, Shin W-R, Kim HC, Min J, Ahn J-Y, Kim Y-H. Detecting and Discriminating Shigella sonnei Using an Aptamer-Based Fluorescent Biosensor Platform. Molecules. 2017;22:22. doi: 10.3390/molecules22050825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundy J, Day D. Selection of DNA aptamers specific for live Pseudomonas aeruginosa. PLoS One. 2017;12:1–11. doi: 10.1371/journal.pone.0185385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead SL, Ashwin H, Johnston BH, Dallas A, Kazakov SA, Tarbin JA, Sharman M, Kay J, Keely BJ. An RNA-Aptamer-based assay for the detection and analysis of malachite green and leucomalachite green residues in fish tissue. Anal Chem. 2010;82:2652–2660. doi: 10.1021/ac902226v. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R, Reinemann C, Strehlitz B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal Bioanal Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R, Reinemann C, Strehlitz B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical Aptamer-based sensor. J Am Chem Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Okumura Y, Amino T, Miyachi Y, Ogino C, Kondo A. DNA-duplex linker for AFM-SELEX of DNA aptamer against human serum albumin. Bioorg Med Chem Lett. 2017;27:954–957. doi: 10.1016/j.bmcl.2016.12.080. [DOI] [PubMed] [Google Scholar]
- Tang J, Xie J, Shao N, Yan Y. The DNA aptamers that specifically recognize ricin toxin are selected by two in vitro selection methods. Electrophoresis. 2006;27:1303–1311. doi: 10.1002/elps.200500489. [DOI] [PubMed] [Google Scholar]
- Theis MG, Knorre A, Kellersch B, Moelleken J, Wieland F, Kolanus W, Famulok M. Discriminatory aptamer reveals serum response element transcription regulated by cytohesin-2. Proc Natl Acad Sci U S A. 2004;101:11221–11226. doi: 10.1073/pnas.0402901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science (80- ) 1990;249:505 LP–505510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ulrich H, Magdesian MH, Alves MJM, Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J Biol Chem. 2002;277:20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- United Nations (2019) World Population Prospects 2019: Highlights. New York
- Urmann K, Arshavsky-Graham S, Walter JG, Scheper T, Segal E. Whole-cell detection of live lactobacillus acidophilus on aptamer-decorated porous silicon biosensors. Analyst. 2016;141:5432–5440. doi: 10.1039/c6an00810k. [DOI] [PubMed] [Google Scholar]
- Valério A, Feuser PE, Bubniak LDS, Santos-Silva MCD, Araújo PHH d, Sayer C. In vitro biocompatibility and macrophage uptake assays of poly(urea-urethane) nanoparticles obtained by miniemulsion polymerization. J Nanosci Nanotechnol. 2017;17:6. doi: 10.1166/jnn.2017.13434. [DOI] [Google Scholar]
- Vivekananda J, Kiel JL. Anti-Francisella tularensis DNA aptamers detect tularemia antigen from different subspecies by Aptamer-linked immobilized sorbent assay. Lab Investig. 2006;86:610–618. doi: 10.1038/labinvest.3700417. [DOI] [PubMed] [Google Scholar]
- Wallukat G, Muller J, Haberland A, Berg S, Schulz A, Freyse E-J, Vetter R, Salzsieder E, Kreutz R, Schimke I. Aptamer BC007 for neutralization of pathogenic autoantibodies directed against G-protein coupled receptors: a vision of future treatment of patients with cardiomyopathies and positivity for those autoantibodies. Atherosclerosis. 2016;244:44–47. doi: 10.1016/j.atherosclerosis.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Wang K, Fan D, Liu Y, Wang E. Highly sensitive and specific colorimetric detection of cancer cells via dual-aptamer target binding strategy. Biosens Bioelectron. 2015;73:1–6. doi: 10.1016/j.bios.2015.05.044. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen X, Tian B, Liu J, Yang L, Zeng L, Chen T, Hong A, Wang X. Nucleolin-targeted extracellular vesicles as a versatile platform for biologics delivery to breast cancer. Theranostics. 2017;7:1360–1372. doi: 10.7150/thno.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Qiao P, Jing L, Song Y, Zhang J, Chen Q, Han Q. Screening and application of a new aptamer for the rapid detection of Sudan dye III. Eur J Lipid Sci Technol. 2018;120:1700112. doi: 10.1002/ejlt.201700112. [DOI] [Google Scholar]
- Weiss S, Proske D, Neumann M, Groschup MH, Kretzschmar HA, Famulok M, Winnacker EL. RNA aptamers specifically interact with the prion protein PrP. J Virol. 1997;71:8790–8797. doi: 10.1128/JVI.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Crihfield CL, Gattu S, Holland LA, Sooter LJ (2014) In vitro selection of a single-stranded DNA molecular recognition element against atrazine. Int J Mol Sci 15:14332–14347. 10.3390/ijms150814332 [DOI] [PMC free article] [PubMed]
- Wu CCN, Sabet M, Hayashi T, Tawatao R, Fierer J, Carson DA, Guiney DG, Corr M. In vivo efficacy of a phosphodiester TLR-9 aptamer and its beneficial effect in a pulmonary anthrax infection model. Cell Immunol. 2008;251:78–85. doi: 10.1016/j.cellimm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhan S, Wang L, Zhou P. Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles. Analyst. 2014;139:1550–1561. doi: 10.1039/C3AN02117C. [DOI] [PubMed] [Google Scholar]
- Xu L, Callaway Z, Wang R, Wang H, Slavik M, Wang A, Li Y. A fluorescent aptasensor coupled with nanobeads-based immunomagnetic separator for simultaneous detection of four foodborne pathogenic bacteria. Trans ASABE (American Soc Agric Biol Eng) 2015;58:891–906. doi: 10.13031/trans.58.11089. [DOI] [Google Scholar]
- Yan S-R, Foroughi MM, Safaei M, Jahani S, Ebrahimpour N, Borhani F, Rezaei Zade Baravati N, Aramesh-Boroujeni Z, Foong LK. A review: recent advances in ultrasensitive and highly specific recognition aptasensors with various detection strategies. Int J Biol Macromol. 2020;155:184–207. doi: 10.1016/j.ijbiomac.2020.03.173. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhang X, Ye M, Jiang J, Yang R, Fu T, Chen Y, Wang K, Liu C, Tan W. Aptamer-conjugated nanomaterials and their applications. Adv Drug Deliv Rev. 2011;63:1361–1370. doi: 10.1016/j.addr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Han Q, Zhang Y, Wu J, Tang X, Dong C, Liu W. Determination of free tryptophan in serum with aptamer-comparison of two aptasensors. Talanta. 2015;131:672–677. doi: 10.1016/j.talanta.2014.08.023. [DOI] [PubMed] [Google Scholar]
- Ye M, Hu J, Peng M, Liu J, Liu J, Liu H, Zhao X, Tan W. Generating aptamers by cell-SELEX for applications in molecular medicine. Int J Mol Sci. 2012;13:3341–3353. doi: 10.3390/ijms13033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Shi H, He X, Wang K, He D, Yan L, Xu F, Lei Y, Tang J, Yu Y. Iodide-responsive cu–au nanoparticle-based colorimetric platform for ultrasensitive detection of target cancer cells. Anal Chem. 2015;87:7141–7147. doi: 10.1021/acs.analchem.5b00943. [DOI] [PubMed] [Google Scholar]
- Yuan J, Wu S, Duan N, Ma X, Xia Y, Chen J, Ding Z, Wang Z. A sensitive gold nanoparticle-based colorimetric aptasensor for Staphylococcus aureus. Talanta. 2014;127:163–168. doi: 10.1016/j.talanta.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Zhang G-X, Liu Y-L, Yang M, Huang W-S, Xu J-H. An aptamer-based, fluorescent and radionuclide dual-modality probe. Biochimie. 2020;171–172:55–62. doi: 10.1016/j.biochi.2020.02.007. [DOI] [PubMed] [Google Scholar]
- Zhou W, Ding J, Liu J. A highly specific sodium aptamer probed by 2-aminopurine for robust Na+ sensing. Nucleic Acids Res. 2016;44:10377–10385. doi: 10.1093/nar/gkw845. [DOI] [PMC free article] [PubMed] [Google Scholar]