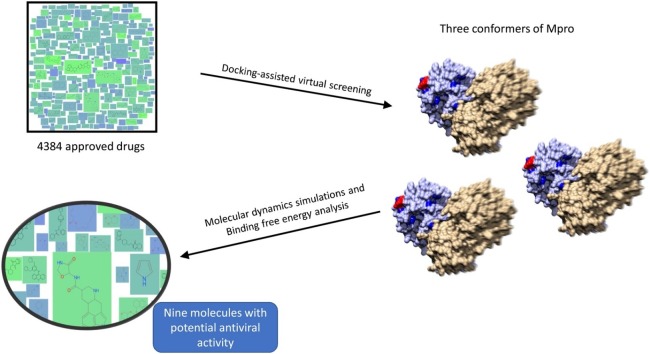
Graphical abstract
Keywords: Mpro, SARS-CoV-2, COVID-19, Docking, Molecular dynamics simulation
Highlights
-
•
The main protease of SARS-CoV-2 has a highly flexible active site.
-
•
Drug repurposing is a treatment opportunity in the current COVID-19 pandemic.
-
•
Nine drugs merit testing as inhibitors of the SARS-CoV-2 main protease.
Abstract
The global emergency caused by COVID-19 makes the discovery of drugs capable of inhibiting SARS-CoV-2 a priority, to reduce the mortality and morbidity of this disease. Repurposing approved drugs can provide therapeutic alternatives that promise rapid and ample coverage because they have a documented safety record, as well as infrastructure for large-scale production. The main protease of SARS-CoV-2 (Mpro) is an excellent therapeutic target because it is critical for viral replication; however, Mpro has a highly flexible active site that must be considered when performing computer-assisted drug discovery. In this work, potential inhibitors of the main protease (Mpro) of SARS-Cov-2 were identified through a docking-assisted virtual screening procedure. A total of 4384 drugs, all approved for human use, were screened against three conformers of Mpro. The ligands were further studied through molecular dynamics simulations and binding free energy analysis. A total of nine currently approved molecules are proposed as potential inhibitors of SARS-CoV-2. These molecules can be further tested to speed the development of therapeutics against COVID-19.
1. Introduction
In December 2019, a cluster of atypical pneumonia cases was reported in Wuhan, China (Wu et al., 2020; Zhu et al., 2020). Coronavirus disease (COVID-19) rapidly spread to other countries and by March 11th, the disease was declared a pandemic by the World Health Organization (WHO) (Cucinotta and Vanelli, 2020). According to the WHO, to date (Situation Report 83, April 15th, 2020) there are 1914916 COVID-19 cases and more than 120,000 COVID-19-associated deaths worldwide (World Health Organization, 2019). The causal agent of COVID-19 is a SARS-related coronavirus named SARS-CoV-2, which was likely passed to humans in live-animal markets (Jalava, 2020). There is strong evidence that SARS-CoV-2 originated in bats but was likely transmitted to humans through an intermediate animal, probably the pangolin (Lam et al., 2020).
SARS-CoV-2 is a betacoronavirus closely related to MERS-CoV and SARS-CoV, which have also caused outbreaks with pandemic potential (Chen et al., 2020a). These outbreaks have affected the global economy, causing high economic losses (Tripp and Tompkins, 2019). The SARS-CoV-2 genome is a single-stranded positive-sense RNA of about 30 kb in length and contains at least six open reading frames (ORFs) that code for a minimum of 16 non-structural proteins and 4 structural proteins (Chen et al., 2020a; Lai et al., 2020). ORF 1a/b is translated into a large protein that undergoes extensive proteolytic processing to yield the replicase complex, which mediates viral transcription and replication (Bartlam et al., 2005; Chen et al., 2020a). The protease responsible for the proteolytic processing is the main protease (Mpro) or 3C-like protease (3CLpro), which is matured by auto-cleavage into the dimeric active conformation (Xia and Kang, 2011). Given the relevance for the viral replication cycle, Mpro has been proposed as a target in the development of inhibitors against coronaviruses (Bartlam et al., 2005; Yang et al., 2006).
Drug repurposing, also known as drug repositioning, is the use of an active pharmaceutical ingredient to treat a novel medical condition different from the original intended condition and has arisen mainly by serendipity when beneficial off-target or secondary effects are noticed (Pacios et al., 2020; Pushpakom et al., 2019). The use of currently approved drugs to treat different diseases has the advantage of assuring medical safety because the drugs have already been tested in animal models and undergone clinical trials. Additionally, the infrastructure to manufacture at large-scale is already in place (Cha et al., 2018; Pushpakom et al., 2019). Drug repurposing is also a strategy that has been used to discover novel antibiotics or antiviral drugs (Dyall et al., 2018; Pacios et al., 2020). In the case of SARS-CoV-2, many drugs with repurposing potential are already being tested (Li and De Clercq, 2020). The attractiveness of repurposing has led to the evaluation of at least 35,000 drugs for more than one medical condition (Baker et al., 2018).
Another advantage of drug repurposing is a quick approval in emergencies such as the current COVID-19 pandemic. Taking this into consideration, we performed in silico evaluation of a set of approved drugs as potential inhibitors of Mpro from SARS-CoV-2; our findings show that several molecules warrant further analysis as treatment options against COVID-19.
2. Methods
2.1. SARS-CoV-2 genome sequences retrieval and homology modeling of Mpro
A total of 111 SARS-CoV-2 genome sequences were retrieved from the GISAID platform (Shu and McCauley, 2017) and aligned with Clustal Omega through the UGENE platform (Okonechnikov et al., 2012; Sievers and Higgins, 2014). For homology modeling, the BetaCoV/Wuhan/WIV02/2019 genome was analyzed with VGAS (Zhang et al., 2019) to predict the Open Reading Frame (ORF) corresponding to ORF1a, which contains the Mpro sequence. This sequence was used to predict the structure of Mpro in its biologically active conformation (dimer) by using Modeller (Ho et al., 2015; Webb and Sali, 2016); the following structures were used as templates (PDB ids): 2AMD, 1WOF, 2AMQ, 2D2D, 3E91, and 3EA7 (Yang et al., 2005). A total of 20 models were generated and the DOPE (Discrete Optimized Protein Energy) score was used to select the best structural model. Global and local structural quality was evaluated with QMEAN, which is a scoring function that measures the global and local quality of protein models, estimating the degree of structural ‘nativeness’. QMEAN uses a linear combination of structural descriptors that include long-range interactions, torsion angles, and solvation potential. Scores calculated form the structural descriptors are transformed into Z-scores to compare them with high-resolution crystal structures. QMEAN is available in the SWISS-MODEL server (Benkert et al., 2011; Waterhouse et al., 2018). Sequence conservation analysis was done with Chimera (Pettersen et al., 2004).
2.2. Molecular dynamics simulation of Mpro
The predicted structural model was submitted to the CHARMM-GUI server to prepare the system (Brooks et al., 2009; Jo et al., 2014, 2008; Lee et al., 2016). The Solution Builder module was used to prepare the protein inside a water cube (TIP3P model) and potassium chloride (KCl) was used to neutralize the system charge and to adjust the salt concentration to 0.15 M. The CHARMM36m force field was used and input files for GROMACS were generated and downloaded (Huang et al., 2017). The molecular dynamics simulation was performed with GROMACS (Abraham et al., 2019, 2015) in three stages: first, a minimization stage (steepest descent) consisting of 5000 steps was performed to eliminate major atomic clashes in the system. Then, an equilibration stage was performed in which protein movement was constrained to allow the solvent and ions to contact the protein. Harmonic force constants of 400 kJ mol−1 nm-2 for protein backbone and 40 kJ mol−1 nm-2 for sidechains were used, with a total equilibration time of 250 ps and a time step of 1 fs at 310 K. Lastly, the production stage was performed without position restrains for a total of 100 ns with a timestep of 2 fs at 310 K. The resulting trajectory was visualized and analyzed with VMD (Humphrey et al., 1996) and GROMACS. Conformers from the resulting trajectory were clustered with the GROMOS method (Daura et al., 1999), which generates non-overlapping clusters. Briefly, the method starts with the RMSD calculation between all pairs of conformers in the trajectory. The first cluster is formed when the biggest set of conformers sharing less than the cut-off is detected. These conformers are eliminated from the pool of clusters. The process is repeated for remaining conformers. A cut-off of 0.18 nm was used to generate approximately 20 clusters from the 1000 conformers resulting from the molecular dynamics simulation.
2.3. Docking-mediated virtual screening
The strategy to discover molecules that potentially inhibit Mpro involved molecular docking of each molecule into the active site of Mpro. iDock (Li et al., 2012) was used as the molecular docking engine and the procedure was first validated by redocking ligand 9NI to the SARS-Mpro structure (PDB id: 2AMD). iDock is a molecular docking program that was developed with a focus on virtual screening, being capable of processing a massive number of ligands in a relatively short time. It is similar to AutoDock Vina (a fast and accurate evolution of AutoDock) with regard to the scoring function and to the optimization algorithm, with emphasis on a faster execution. The SARS-CoV-2 Mpro structure and two of its main conformers, extracted from the molecular dynamics simulation trajectory file, were processed with AutoDockTools (Morris et al., 2009). The small-molecule database consisted of 4384 molecules and was downloaded from ZINC15 (Sterling and Irwin, 2015). These molecules corresponded to the “world” subset; according to ZINC15, these molecules have been approved for human use in major jurisdictions, including the U.S. Food and Drug Administration. The database was processed with the prepare_ligand4.py script included in AutoDockTools to obtain the input files needed for virtual screening. Box coordinates for virtual screening were center (x, y, z) = 93.3, 10.5, 2.0 and size (x, y, z) = 22.3, 22.1, 24.2. The candidate molecules were selected according to the docking score predicted by iDock; the top 10 molecules from each conformer were further analyzed to select the potential inhibitors of Mpro.
2.4. Molecular dynamics simulations of Mpro-ligand complexes
The 30 complexes were inspected visually with Pymol (https://pymol.org) and Discovery Studio Visualizer (https://www.3dsbiovia.com); those molecules that made contact with the catalytic residues of Mpro (His41 and Cys145) were subjected to molecular dynamics simulations. The input files were prepared with CHARMM-GUI as mentioned above, with simulation times of 20 ns. The resulting trajectories were visualized and analyzed with VMD and GROMACS. The stability of the ligands in the active site of Mpro was estimated by calculating the RMSD (Root-Mean-Square Deviation) and the binding energy (ΔG) was calculated with the CaFE plugin (Liu and Hou, 2016) in VMD. The CaFE plugin consists of a set of Tcl scripts that allow the calculation of binding free energies through the MM/PBSA and LIE methods and is powered by VMD, NAMD (Phillips et al., 2005) and APBS (Jurrus et al., 2018) programs. Binding free calculations were performed with the MM/PBSA method. Briefly, receptor and ligand conformations are extracted from the trajectories, then three energetic components are calculated: The gas-phase energy difference between the complex, the receptor, and the ligand is obtained by calling NAMD. Then the polar solvation free energy is calculated with APBS. Subsequently, the difference of solvent accessible surface area (SASA) is measured and the nonpolar solvation free energy is estimated by its approximate linear relation with SASA. Finally, the binding free energy is summed and averaged throughout the selected conformations.
3. Results
3.1. Homology modeling of Mpro and molecular dynamics simulations
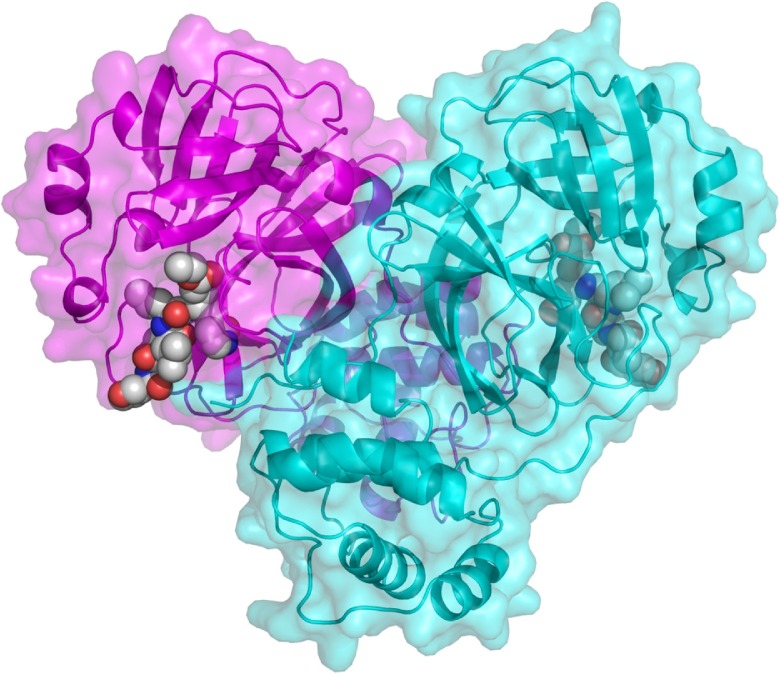
There are several experimental structures reported for the SARS-CoV-1 Mpro, which vary in amino acid sequence, resolution, and presence of additional molecules, such as inhibitors or solvents. Consequently, we decided to include structures 2AMD, 1WOF, 2AMQ, 2D2D, 3E91, and 3EA7 to model the structure of the SARS-CoV-2 main protease. Sequence identity of SARS-CoV-2 Mpro with SARS-CoV-1 sequences ranged from 94.4 % (sequence corresponding to PDB code 2AMQ) to 95.1 % (PDB codes 2AMD and 1WOF). The resulting model is in the classical heart-shaped conformation (Fig. 1 ), with the binding site near the dimer interface. The selected model had an RMSD=0.266 Å when compared with template 2AMD.
Fig. 1.
Modeled structure of the SARS-CoV-2 Mpro dimeric protease. One monomer is shown in magenta and the other in cyan. Inhibitor 9IN, from structure 2AMD, is shown as spheres. Image generated with Pymol.
The final Mpro model was analyzed with the SWISS-MODEL server, resulting in a global QMEAN score of -0.41 with only minor deviations in the N- and C-termini (Fig. S1). These values indicate a good structural model, suitable for use in the following analyses. To observe the amino acid conservation of Mpro, amino acid sequences from 111 reported isolates were aligned and the degree of conservation was mapped into the Mpro structure. The most variable position had a 97.7 % conservation, which indicates that the active site of Mpro is highly conserved among the analyzed isolates (Fig. S2).
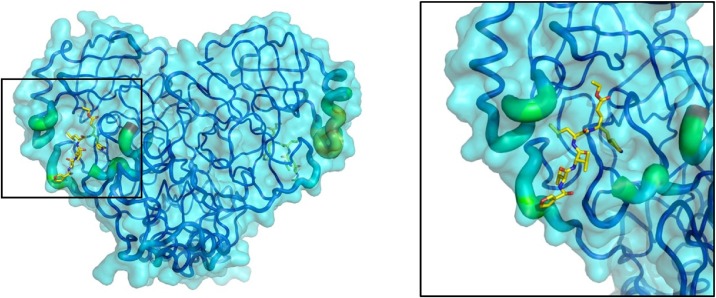
Next, solvent-explicit molecular dynamics simulations were performed on Mpro; the resulting trajectory showed that the protein has a highly flexible active site as the amino acids surrounding the binding site had high RMSF (Root-Mean-Square Fluctuation) values. The regions surrounding the binding site were the most mobile during the simulation (Figs. 2 and S3). This flexibility makes drug discovery a challenge because a ligand that could bind to the closed site could not bind to the open state and vice versa; therefore, we proceeded to analyze the MD trajectory, and from 19 clusters that encompass the trajectory, the two most representative conformers were selected (Fig. S4). Thus, the virtual screening was done with the starting structure, as well as two conformers: Mpro_0 (initial structure), Mpro_412 (conformer 412, which is representative of cluster 1), and Mpro_837 (conformer 837, which is representative of cluster 2).
Fig. 2.
Flexibility of Mpro as determined from molecular dynamics simulations. The image on the left shows Mpro rendered as a transparent surface and the backbone as “putty”. Regions with high RMSF values are shown as thicker; colors range from blue (most stable) to red (most flexible). The highlighted area is zoomed in on the right. Ligand IN9 is displayed as “sticks” and is shown to identify the active site.
3.2. Virtual screening of approved drugs
Virtual screening was performed with the Mpro conformers and the 4384-molecule bank. The top 10 binders for each conformer were selected for further analysis (Table 1 ). Docking score values ranged from −9.17 kcal/mol (dihydroergotamine) to −10.25 kcal/mol (bisoctrizole). Noticeably, there are several drug metabolites (glucuronides) and drugs used in chemotherapy (sorafenib beta-d-glucoronide, N-Trifluoroacetyladriamycin, amrubicin, daunorubicin, idarubicin, midostaurin, maraviroc, and celsentri). Ligands that contacted catalytic amino acids His41 or Cys145 were the most promising candidates and thus were selected for further molecular dynamics simulations.
Table 1.
Top 10 molecules with the best docking score.
| Ligand Name | ZINC ID | iDock score (kcal/mol) |
|---|---|---|
| Mpro_0 | ||
| Bisoctrizole | ZINC000011677911 | −10.25 |
| Ironophore | ZINC000017545546 | −10.08 |
| Sorafenib Beta-d-Glucuronidea | ZINC000077313075 | −10.08 |
| Oxazepam Glucuronide | ZINC000031460595 | −10.04 |
| Ergoloidb | ZINC000003995616 | −9.95 |
| Eltrombopag | ZINC000011679756 | −9.85 |
| Telmisartan | ZINC000001530886 | −9.74 |
| Bromocriptine | ZINC000053683151 | −9.73 |
| Dutasteride | ZINC000003932831 | −9.72 |
| Ergotamine | ZINC000052955754 | −9.67 |
| Mpro_412 | ||
| N-Trifluoroacetyladriamycinc | ZINC000095618916 | −9.96 |
| Amrubicin | ZINC000003780800 | −9.77 |
| Daunorubicin | ZINC000003917708 | −9.75 |
| 5a Reductase inhibitor | ZINC000014880001 | −9.43 |
| Carindacillin | ZINC000003871889 | −9.42 |
| Simeprevir | ZINC000253632968 | −9.41 |
| Idarubicin | ZINC000003920266 | −9.38 |
| Midostaurin | ZINC000100013130 | −9.38 |
| Lorazepam Glucuronide | ZINC000031290884 | −9.33 |
| Meclocycline | ZINC000084402690 | −9.30 |
| Mpro_837 | ||
| Maraviroc | ZINC000101160855 | −9.38 |
| Glycyrrhizinate Dipotassium | ZINC000096015174 | −9.33 |
| Teniposide | ZINC000004099009 | −9.32 |
| Indomethacin Glucuronide | ZINC000084386263 | −9.32 |
| Celsentri | ZINC000003817234 | −9.31 |
| Lorazepam Glucuronide | ZINC000031290884 | −9.26 |
| Estrone Glucuronide | ZINC000004099004 | −9.23 |
| 5a Reductase inhibitor | ZINC000014880001 | −9.22 |
| Ketotifen-N-Glucuronide | ZINC000095618575 | −9.20 |
| Dihydroergotamine | ZINC000003978005 | −9.17 |
Glucuronides are products of metabolization mainly by the liver.
Names in bold correspond to molecules that contacted either catalytic amino acid (His41 or Cys145).
A metabolite of Valrubicin.
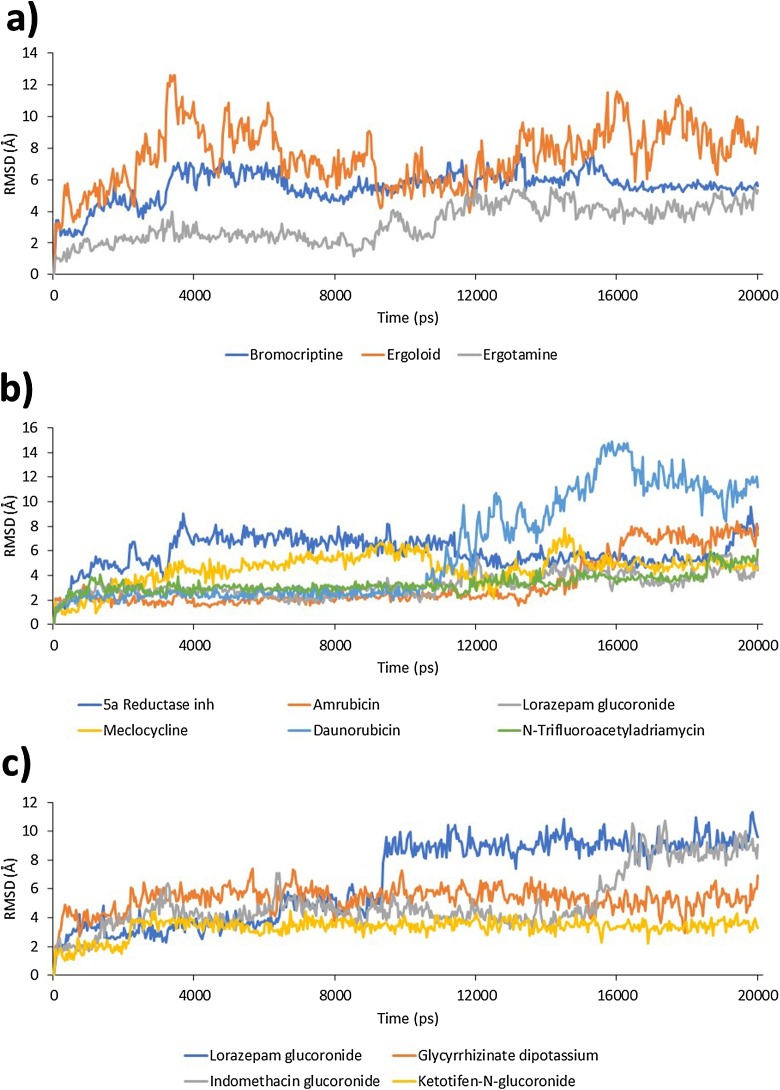
MD simulations for the selected Mpro-ligand complexes were carried out and the relative mobility of the ligand within the active site of Mpro was monitored through evaluation of ligand RMSD (Fig. 3 ). In the case of Mpro_0, the most mobile ligand was ergoloid, with an average RMSD of 7.6 Å, and the most stable ligand was ergotamine, with an average RMSD of 3.6 Å. In the case of Mpro_412, the most mobile ligand was daunorubicin with an average RMSD of 7.2 Å, and the most stable ligand was lorazepam glucoronide, with an average RMSD of 3.4 Å. Lastly, for Mpro_837 the most mobile ligand was lorazepam glucoronide with an average RMSD of 7.8 Å, and the most stable ligand was ketotifen-N-glucoronide, with an average RMSD of 3.5 Å.
Fig. 3.
Ligand RMSD within the binding site of Mpro. a) Ligands complexed with Mpro_0; b) ligands complexed with Mpro_412; and c) ligands complexed with Mpro_837. Vertical axis units are Å and horizontal axis units are picoseconds.
Binding free energy was calculated for each complex (the initial 5 ns were not considered for calculation) and the results are indicated in Table 2 . It is noteworthy that complexes with conformer Mpro_0 and Mpro_412 had the lowest binding free energy values. After considering all the bioinformatics analyses, we posit that ligands daunorubicin, ergotamine, bromocriptine, meclocycline, amrubicin, ergoloid, ketotifen-N-glucoronide, N-trifluoroacetyladriamycin, 5a reductase inhibitor are good candidates as Mpro inhibitors and therefore warrant further evaluation as potential treatment against SARS-CoV-2 infection.
Table 2.
Binding free energies of Mpro-ligand complexes.
| Conformer | ZINC code | Drug | ΔG (kcal/mol) |
|---|---|---|---|
| Mpro_412 | ZINC000003917708 | Daunorubicin | −138.8 |
| Mpro_0 | ZINC000052955754 | Ergotamine | −119.2 |
| Mpro_412 | ZINC000003780800 | Amrubicin | −117.5 |
| Mpro_0 | ZINC000053683151 | Bromocriptine | −116.7 |
| Mpro_412 | ZINC000084402690 | Meclocycline | −115.1 |
| Mpro_0 | ZINC000003995616 | Ergoloid | −109.1 |
| Mpro_837 | ZINC000095618575 | Ketotifen-N-Glucuronide | −89.0 |
| Mpro_412 | ZINC000095618916 | N-Trifluoroacetyladriamycin | −46.2 |
| Mpro_412 | ZINC000014880001 | 5α Reductase Inhibitor | −31.0 |
| Mpro_412 | ZINC000031290884 | Lorazepam Glucuronide | −1.1 |
| Mpro_837 | ZINC000031290884 | Lorazepam Glucuronide | −0.5 |
| Mpro_837 | ZINC000084386263 | Indomethacin Glucuronide | 11.9 |
| Mpro_837 | ZINC000096015174 | Glycyrrhizinate Dipotassium | 36.1 |
4. Discussion
A race against time is currently underway to develop safe and effective antivirals against SARS-CoV-2. Drug repurposing is a straightforward strategy when time is a critical factor, such as in this COVID-19 pandemic (Phadke and Saunik, 2020; Pushpakom et al., 2019). Indeed, the U.S. Food and Drug Administration has encouraged the use of the App CURE ID for health care professionals to report novel uses of existing medicines for the treatment of infectious diseases (https://cure.ncats.io/), and there are several studies, some in clinical phases, that involve the repurposing of approved drugs (Pacios et al., 2020). On the other hand, bioinformatic studies are useful in detecting drugs that can be used in in vitro studies or clinical trials as treatment options against COVID-19. In this work, a set of nine currently approved drugs was identified as having the potential to inhibit the SARS-CoV-2 Mpro and therefore could be used as a COVID-19 treatment, either alone or in combination with other drugs.
Some of the most promising strategies involve the use of chloroquine alone or combined with azithromycin, as well as remdesivir (Gautret et al., 2020; Wang et al., 2020a). However, the efficacy of these therapies is as yet unknown. Regarding Mpro, currently, there is at least one clinical trial involving the use of protease inhibitors as a treatment for COVID-19 (clinical trial NCT04303299) and there are several reports of potential inhibitors of SARS-CoV-2 Mpro (Chen et al., 2020b; Khan et al., 2020; Li and De Clercq, 2020; Ton et al., 2020; Zhang et al., 2020). These reports use computational approaches to identify molecules with Mpro inhibitory potential; however, our work is the only one that considers the flexibility of the active site, which allowed us to propose a greater diversity of potential Mpro inhibitors. Mpro is an excellent target for computer-assisted drug discovery because it is critical in the early stages of viral replication (Chen et al., 2020a); however, we found that the substrate-binding site is highly flexible, therefore virtual-assisted drug design approaches should take flexibility into account. When this work started, there were no structures available for SARS-CoV-2 Mpro; however, two experimental structures have been reported to date (PDB id: 6LU7 and 6W63) (Jin et al., 2020). A rapid analysis showed that the b-factors of 6LU7 agree with our results (Supplementary Fig. S5), therefore the homology model constructed in this work is valid for our purposes.
MD simulations had a duration of 20 ns, and the first 5 ns were discarded for binding-free energy analysis to allow the system to equilibrate. There is no consensus about the duration of a molecular dynamics simulation, but it has been found that longer simulations do not necessarily improve binding free energy calculations (Hou et al., 2011). We chose 20 ns as a result of a literature search, in which simulation times ranged from 15 ns (one protein-ligand complex) to 200 ns (several protein-ligand complexes) (Kumar et al., 2019a; Mittal et al., 2020). To define the duration of a simulation run, the complexity of the phenomena under study and the size of the system should be considered, also depending directly on the available computer power. Overall, studies involving one complex normally used longer simulation times (Kumar et al., 2019b; Wang et al., 2020b) and studies involving several complexes used shorter simulation times (Ge et al., 2013; Razzaghi-Asl et al., 2018).
Binding free energies ranged from −138.8 kcal/mol to 36. 1 kcal/mol. At first sight, these values are noticeably higher than expected, considering that the experimental ΔG value for the avidin-biotin complex is −20.4 kcal/mol. The reason for this discrepancy is the solute dielectric variable used for our calculations; in fact, the use of different solute dielectric values can significatively shift the binding free energy calculation to a more negative or positive value (Li et al., 2018). In the case of the avidin-biotin complex, a solute dielectric variable of 1.0 agrees well with experimental values since calculations made with the CaFE plugin indicate a ΔG = −20.8 kcal/mol when a solute dielectric = 1.0 was used; this value is close to the experimental binding free energy (-20.4 kcal/mol) (Green, 1975). The choice of the solute dielectric value depends on the charges present at the binding interface and increases as more charges are present. Usually, values of 1.0, 2.0 and 4.0 are used depending on the nature of the binding site (Hou et al., 2011). More often, values ranging from 2.0 to 4.0 are used when studying binding sites with polar or charged amino acids are present. For proteases similar to Mpro, the solute dielectric value that correlates best with experimental results is 4.0 (Sun et al., 2014).
Three of the proposed drugs are used in chemotherapy (daunorubicin, amrubicin, and the valrubicin metabolite N-trifluoroacetyladriamycin) (Piska et al., 2017). These drugs as well as meclocycline, a highly toxic antibiotic, should be treated carefully because of side effects in the organism. Two of the drugs identified in this study, indomethacin and glycyrrhizinate, have also been tested in vitro and proposed as potential therapies against coronavirus (Amici et al., 2006; Hoever et al., 2005). However, the binding free energy analysis suggests that Mpro might not be the primary target of these molecules.
Despite the increasing number of publications related to virtual screening focused on Mpro (Jin et al., 2020; Ton et al., 2020; Wang, 2020), there is little agreement between them on potential candidates identified. Of all the reported drugs found in this work, only valrubicin also was identified in another study as a potential inhibitor of Mpro (Wang, 2020). This may be explained by the diversity of the selected databases and the algorithms used in docking and virtual screening. However, if the original purpose of the drug is considered, similarities arise between our findings and those of Jin et al. (2020) where drugs used in chemotherapy and antihistamines potentially inhibit Mpro. Given the emergency we are currently facing, this diversity of results may turn out to be beneficial because more options are available to repurpose these drugs as COVID-19 treatment options. Again, the drugs mentioned in this article should not be used as treatment against COVID-19 unless they have been tested in proper clinical trials.
5. Conclusions
The main protease of SARS-CoV-2 has a very flexible active site, an aspect that must be considered in rational drug design. Our approach involved the use of three conformers of Mpro and led to the identification of nine molecules that warrant in vivo testing or even in clinical trials so they can be repurposed as treatment of COVID-19.
CRediT authorship contribution statement
Alicia Jiménez-Alberto: Conceptualization, Investigation, Writing - review & editing, Funding acquisition, Visualization. Rosa María Ribas-Aparicio: Conceptualization, Resources, Writing - review & editing, Funding acquisition, Supervision, Formal analysis. Gerardo Aparicio-Ozores: Conceptualization, Investigation, Writing - review & editing, Funding acquisition, Data curation. Juan A. Castelán-Vega: Methodology, Software, Validation, Formal analysis, Data curation, Writing - original draft, Visualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study received financial support from Instituto Politécnico Nacional (IPN) through SIP grants 20201445, 20201245, 20202136 y 20201438. JAC-V, AJ-A, GA-O, and RMR-A are recipients of Comisión de Operación y Fomento de Actividades Académicas (COFAA-IPN) grants, and JAC-V of the Programa de Estímulos al Desempeño de los Investigadores (EDI-IPN) grant. RMR-A and GA-O are recipients of Programa de Estímulo al Desempeño Docente (EDD) fellowships, all of them granted by Instituto Politécnico Nacional. Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.compbiolchem.2020.107325.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Abraham M.J., van der Spoel D., Lindahl E., Hess B., The Gromacs development team . 2019. GROMACS User Manual Version 2019.http://www.gromacs.org [Google Scholar]
- Amici C., Di Caro A., Ciucci A., Chiappa L., Castilletti C., Martella V., Decaro N., Buonavoglia C., Capobianchi M.R., Santoro M.G. Indomethacin has a potent antiviral activity against SARS coronavirus. Antivir. Ther. 2006;11:1021–1030. [PubMed] [Google Scholar]
- Baker N.C., Ekins S., Williams A.J., Tropsha A. A bibliometric review of drug repurposing. Drug Discov. Today. 2018;23:661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlam M., Yang H., Rao Z. Structural insights into SARS coronavirus proteins. Curr. Opin. Struct. Biol. 2005;15:664–672. doi: 10.1016/j.sbi.2005.10.004. Catalysis and regulation/Proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinforma. Oxf. Engl. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Brooks C.L., Mackerell A.D., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A.R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R.W., Post C.B., Pu J.Z., Schaefer M., Tidor B., Venable R.M., Woodcock H.L., Wu X., Yang W., York D.M., Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha Y., Erez T., Reynolds I.J., Kumar D., Ross J., Koytiger G., Kusko R., Zeskind B., Risso S., Kagan E., Papapetropoulos S., Grossman I., Laifenfeld D. Drug repurposing from the perspective of pharmaceutical companies. Br. J. Pharmacol. 2018;175:168–180. doi: 10.1111/bph.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Yiu C.-P.B., Wong K.-Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research. 2020;9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Bio-Medica Atenei Parm. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W.F., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. 1999;38:236–240. doi: 10.1002/(SICI)1521-3773(19990115)38:1/2<236::AID-ANIE236>3.0.CO;2-M. [DOI] [Google Scholar]
- Dyall J., Nelson E.A., DeWald L.E., Guha R., Hart B.J., Zhou H., Postnikova E., Logue J., Vargas W.M., Gross R., Michelotti J., Deiuliis N., Bennett R.S., Crozier I., Holbrook M.R., Morris P.J., Klumpp-Thomas C., McKnight C., Mierzwa T., Shinn P., Glass P.J., Johansen L.M., Jahrling P.B., Hensley L.E., Olinger G.G., Thomas C., White J.M. Identification of combinations of approved drugs with synergistic activity against ebola virus in cell cultures. J. Infect. Dis. 2018;218:S672–S678. doi: 10.1093/infdis/jiy304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ge H., Wang Y., Li C., Chen N., Xie Y., Xu M., He Y., Gu X., Wu R., Gu Q., Zeng L., Xu J. Molecular dynamics-based virtual screening: accelerating the drug discovery process by high-performance computing. J. Chem. Inf. Model. 2013;53:2757–2764. doi: 10.1021/ci400391s. [DOI] [PubMed] [Google Scholar]
- Green N.M. Avidin. In: Anfinsen C.B., Edsall J.T., Richards F.M., editors. Advances in Protein Chemistry. Academic Press; 1975. pp. 85–133. [DOI] [Google Scholar]
- Ho B.-L., Cheng S.-C., Shi L., Wang T.-Y., Ho K.-I., Chou C.-Y. Critical assessment of the important residues involved in the dimerization and catalysis of MERS coronavirus main protease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoever G., Baltina Lidia, Michaelis M., Kondratenko R., Baltina Lia, Tolstikov G.A., Doerr H.W., Cinatl J. Antiviral activity of glycyrrhizic acid derivatives against SARS−Coronavirus. J. Med. Chem. 2005;48:1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA Methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jalava K. First respiratory transmitted food borne outbreak? Int. J. Hyg. Environ. Health. 2020;226 doi: 10.1016/j.ijheh.2020.113490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang Xinglou, You T., Liu Xiaoce, Yang Xiuna, Bai F., Liu H., Liu Xiang, Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jo S., Cheng X., Islam S.M., Huang L., Rui H., Zhu A., Lee H.S., Qi Y., Han W., Vanommeslaeghe K., MacKerell A.D., Roux B., Im W. Chapter eight – CHARMM-GUI PDB manipulator for advanced modeling and simulations of proteins containing nonstandard residues. In: Karabencheva-Christova T., editor. Advances in Protein Chemistry and Structural Biology, Biomolecular Modelling and Simulations. Academic Press; 2014. pp. 235–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J., Liles K., Chun M., Li P., Gohara D.W., Dolinsky T., Konecny R., Koes D.R., Nielsen J.E., Head‐Gordon T., Geng W., Krasny R., Wei G.-W., Holst M.J., McCammon J.A., Baker N.A. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- Kumar A., Rathi E., Kini S.G. E-pharmacophore modelling, virtual screening, molecular dynamics simulations and in-silico ADME analysis for identification of potential E6 inhibitors against cervical cancer. J. Mol. Struct. 2019;1189:299–306. doi: 10.1016/j.molstruc.2019.04.023. [DOI] [Google Scholar]
- Kumar N., Srivastava R., Prakash A., Lynn A.M. Structure-based virtual screening, molecular dynamics simulation and MM-PBSA toward identifying the inhibitors for two-component regulatory system protein NarL of Mycobacterium tuberculosis. J. Biomol. Struct. Dyn. 2019;0:1–15. doi: 10.1080/07391102.2019.1657499. [DOI] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S., Wei W., Cheung W.Y.-M., Li W.-J., Li L.-F., Leung G.M., Holmes E.C., Hu Y.-L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020:1–6. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., Jo S., Pande V.S., Case D.A., Brooks C.L., MacKerell A.D., Klauda J.B., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Li H., Leung K.-S., Wong M.-H. idock: A multithreaded virtual screening tool for flexible ligand docking. 2012 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB). Presented at the 2012 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB) 2012:77–84. doi: 10.1109/CIBCB.2012.6217214. [DOI] [Google Scholar]
- Li Y., Cong Y., Feng G., Zhong S., Zhang J.Z.H., Sun H., Duan L. The impact of interior dielectric constant and entropic change on HIV-1 complex binding free energy prediction. Struct. Dyn. 2018;5 doi: 10.1063/1.5058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hou T. CaFE: a tool for binding affinity prediction using end-point free energy methods. Bioinforma Oxf. Engl. 2016;32:2216–2218. doi: 10.1093/bioinformatics/btw215. [DOI] [PubMed] [Google Scholar]
- Mittal L., Kumari A., Srivastava M., Singh M., Asthana S. Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. J. Biomol. Struct. Dyn. 2020;0:1–19. doi: 10.1080/07391102.2020.1768151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M., UGENE team Unipro UGENE: a unified bioinformatics toolkit. Bioinforma. Oxf. Engl. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- Pacios O., Blasco L., Bleriot I., Fernandez-Garcia L., González Bardanca M., Ambroa A., López M., Bou G., Tomás M. Strategies to combat multidrug-resistant and persistent infectious diseases. Antibiot. Basel Switz. 2020;9 doi: 10.3390/antibiotics9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phadke M., Saunik S. COVID-19 treatment by repurposing drugs until the vaccine is in sight. Drug Dev. Res. 2020 doi: 10.1002/ddr.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piska K., Koczurkiewicz P., Bucki A., Wójcik-Pszczoła K., Kołaczkowski M., Pękala E. Metabolic carbonyl reduction of anthracyclines - role in cardiotoxicity and cancer resistance. Reducing enzymes as putative targets for novel cardioprotective and chemosensitizing agents. Invest. New Drugs. 2017;35:375–385. doi: 10.1007/s10637-017-0443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., Norris A., Sanseau P., Cavalla D., Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- Razzaghi-Asl N., Mirzayi S., Mahnam K., Sepehri S. Identification of COX-2 inhibitors via structure-based virtual screening and molecular dynamics simulation. J. Mol. Graph. Model. 2018;83:138–152. doi: 10.1016/j.jmgm.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. Clifton NJ. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Sterling T., Irwin J.J. ZINC 15 – ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Li Y., Tian S., Xu L., Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014;16:16719–16729. doi: 10.1039/C4CP01388C. [DOI] [PubMed] [Google Scholar]
- Ton A.-T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020 doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp R., Tompkins S. 1st ed. Springer; New York, NY: 2019. Roles of Host Gene and Non-coding RNA Expression in Virus Infection. 2018 edition. ed. [Google Scholar]
- Wang J. 2020. Fast Identification of Possible Drug Treatment of Coronavirus Disease -19 (COVID-19) Through Computational Drug Repurposing Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang Y., Gao Y., Niu X. Discovery of the novel inhibitor against New Delhi Metallo-β-Lactamase based on virtual screening and molecular modelling. Int. J. Mol. Sci. 2020;21:3567. doi: 10.3390/ijms21103567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2016;86 doi: 10.1002/cpps.20. 2.9.1-2.9.37. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2019. Novel Coronavirus (2019-nCoV) Situation Reports [WWW Document] URL https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (Accessed 4.1.20) [Google Scholar]
- Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;0 doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2:282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the Achilles’ heel of coronaviruses. Curr. Pharm. Des. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- Zhang K.-Y., Gao Y.-Z., Du M.-Z., Liu S., Dong C., Guo F.-B. Vgas: a viral genome annotation system. Front. Microbiol. 2019;10:184. doi: 10.3389/fmicb.2019.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Wei L., Niu P. The novel coronavirus outbreak in Wuhan, China. Glob. Health Res. Policy. 2020;5:6. doi: 10.1186/s41256-020-00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.