Abstract
More than 20 genetic variants associated with late-onset Alzheimer’s disease (LOAD) have been identified by genome-wide association studies (GWAS). However, these variants are tag markers for nearby genetic variants in linkage disequilibrium (LD) and may not be themselves functional. Moreover, most of the significant variants identified by the International Genomics of Alzheimer’s Project (IGAP) meta-analysis are in non-protein-coding regions, implicating gene regulatory mechanisms as underlying the association signal. There are several previously identified coding variant associations with AD, notably the strong APOE signal, but all the novel signals identified in the IGAP GWAS were noncoding.
We set out to characterize the causal variants, regulatory mechanisms, tissue contexts, and target genes underlying noncoding LOAD-associated genetic signals. To do so, we applied our INFERNO method to the IGAP GWAS data, annotating all LD-expanded potentially causal variants at each locus with tissue-specific regulatory activity. Bayesian co-localization analysis of GWAS summary statistics and eQTL data was performed to identify tissue-specific target genes.
INFERNO identified enhancer dysregulation in all 19 tag regions analyzed, significant enrichments of enhancer overlaps in the immune-related blood category, and co-localized eQTL signals overlapping enhancers from the matching tissue class in ten regions (ABCA7, BIN1, CASS4, CD2AP, CD33, CELF1, CLU, EPHA1, FERMT2, ZCWPW1). In several cases, we identified dysregulation of long noncoding RNA (lncRNA) transcripts and applied the lncRNA target identification algorithm from INFERNO to characterize their downstream biological effects. We also validated the allele-specific effects of several variants on enhancer function using luciferase expression assays.
Integrating functional genomics with GWAS signals yielded insights into the regulatory mechanisms, tissue contexts, genes, and biological processes affected by noncoding genetic variation associated with LOAD risk.
Keywords: Alzheimer’s Disease, Bioinformatics, Genetics, Genomics, Long noncoding RNA
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia in the United States [1], but no effective therapies for treatment or prevention exist. Late-onset Alzheimer’s disease (LOAD), defined by age-at-onset after 60 years, is the most common form of AD. Heritability estimates for LOAD stand at 60–80%, implicating genetics as an important factor in disease development [2]. While the APOE locus shows the strongest association [3], LOAD is complex and polygenic [4], and genome-wide association studies (GWAS) have successfully associated over 20 other genetic variants with LOAD [5,6]. Recent studies have implicated a number of different biological processes in LOAD susceptibility such as microglial-mediated innate immunity [7–9].
The majority of top GWAS variants reside in noncoding regions of the genome outside of protein-coding sequences [10]. Any variant in linkage disequilibrium (LD) with a top GWAS variant could be responsible for the association signal, and GWAS data alone lacks the granularity to identify these causal variants. In addition, noncoding variants presumably affect gene regulatory elements, and the affected target genes are often not the closest ones [11]. Thus, functional annotation is needed to reveal the causal variants, regulatory mechanisms, tissue context, and target genes underlying GWAS signals.
Enhancers, which modulate the expression of a target gene independently of orientation and distance, are one of the most common regulatory elements in the noncoding genome [12–14]. Several consortia have generated large-scale functional genomics datasets to characterize regulatory activity in the noncoding genome across different tissue contexts [15–19]. Previous studies used these data to identify noncoding genetic variants with regulatory potential for Alzheimer’s disease [20–22], diabetes [23,24] and schizophrenia [25], but such studies often assume that the relevant tissue context is known a priori.
We hypothesize that noncoding LOAD GWAS signals modulate disease risk by perturbing genomic elements that regulate genes involved in pathogenesis. To explore this hypothesis, we applied our bioinformatics pipeline, INFERNO (INFERring the molecular mechanisms of NOncoding genetic variants) [26] to LOAD GWAS data from the International Genomics of Alzheimer’s Project (IGAP) [6]. INFERNO characterizes noncoding GWAS signals by integrating information across diverse functional genomics data sources to identify causal noncoding variants and the regulatory mechanisms, tissue contexts, and target genes they affect (Figure 1a). INFERNO identified several putatively causal genetic variants in ten GWAS regions and uncovered strong functional evidence of their effects on immune- and brain-related regulatory mechanisms. Using luciferase reporter assays, we validated the enhancer activity and allelic differences of causal variants in three regions prioritized by relevant tissue context, strength of annotation support, and prior literature.
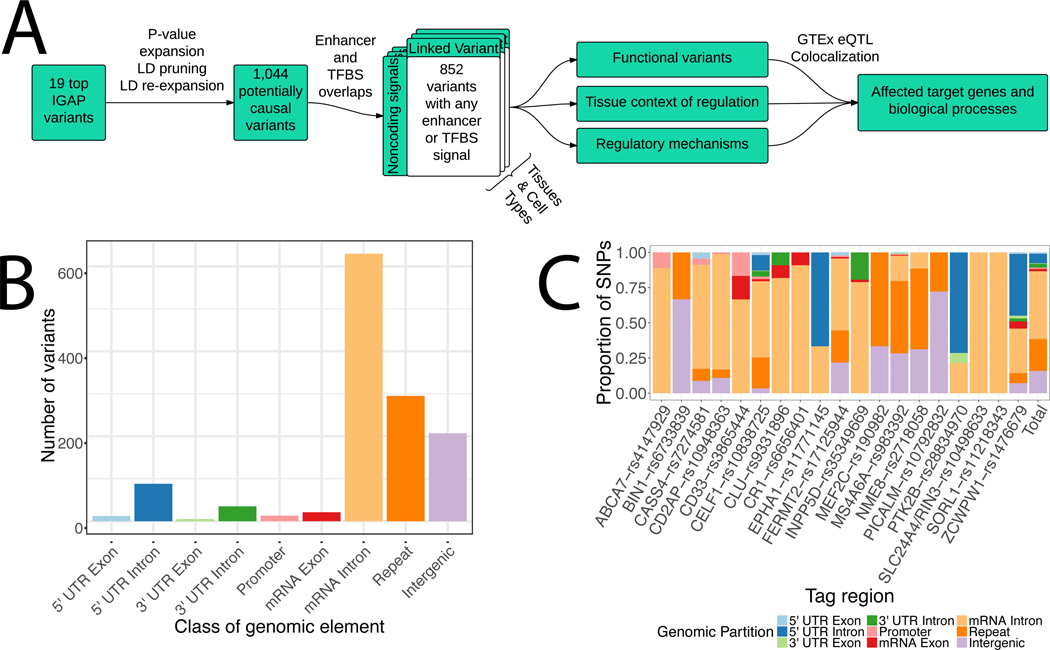
Figure 1: LD expansion and functional annotation of top IGAP hits.
a) Flowchart of analysis approach. b) Genomic localization of all variants in P value- and LD-expanded set. c) Genomic partition proportions split by tag regions.
Materials and Methods
INFERNO analysis of IGAP top hits
INFERNO (details of the algorithm and data sources are described in [26]) was used to analyze 19 top variants from Phase 1 of the IGAP study, excluding the locus near DSG2 (tagged by rs8093731) which did not replicate in Phase 2 and the HLA-DRB5 locus (rs9271192) which is difficult to analyze due to the dense LD structure in the major histocompatibility (MHC) region caused by population-specific selective pressure [27]. INFERNO was run using P value expansion within one order of magnitude and 500 kilobases (kb) of each tagging variant, and the European population from the 1,000 Genomes Project [28] Phase 1 Release v3 (2010/11/23) was used for LD pruning and expansion. All rsID annotations are from dbSNP build 137. For both pruning and expansion, a threshold of r2 >= 0.7 was used to define LD blocks. All downstream analyses including lncRNA correlation and pathway analysis were performed as defined in [26]: correlation values of each lncRNA with every other expressed gene in the transcriptome were calculated using GTEx RNA-sequencing data and the lncRNA-gene pairs meeting thresholds of 0.5 on the absolute value of both the Spearman and Pearson correlations were considered as targets. WebGestalt was used for all pathway analyses [29–31]. Supplementary Note 1 highlights a few possible ways to prioritize the causal variants for further mechanistic studies.
Luciferase validation
From patient DNA with each allele of interest, insert sequences including the enhancers (800–1,739 base pairs (bp) in length, Supplementary Table 1) overlapping each prioritized variant were PCR-amplified using the Phusion DNA polymerase (New England Biolabs) and primers (Integrated DNA Technologies) with a HindIII overhang (Supplementary Figure 1). The HindIII high fidelity enzyme (New England Biolabs) was used to digest the PCR product along with the vector plasmid containing a minimal promoter and the luciferase reporter gene, pGL4.23[luc2/minP] (Promega). Enhancer constructs and plasmids were then ligated using the T4 DNA Ligase (New England Biolabs). These ligated vectors were used to transform One Shot TOP10 Competent cells (Invitrogen), and colonies containing the full enhancer sequences along with either allele of the prioritized variants were identified. These clones were amplified using the EndoFree Plasmid Maxi Kit (Qiagen) to generate high levels of DNA suitable for mammalian cell line transfection. Two different vectors were generated this way for each prioritized variant: one with the minor allele of the prioritized variant and one with the major allele. Additionally, we generated vectors containing a minimal promoter with no enhancer inserted and another negative control vector with a minimal promoter and a ~1kb random genomic heterochromatin insert. 300ng of each vector was mixed with one-tenth the amount of a Renilla expressing vector, allowing us to normalize Luciferase expression for transfection efficiency. This mixture was transfected into separate aliquots of K562 (ATCC) cells using the Lonza Nucleofector Device with Kit V. A mock sample was run through the same transfection procedure with no DNA to account for background luminescence. The Promega Dual-Glo system was used to measure Luciferase and Renilla expression after 24 hours of incubation for the cells to express the luciferase vector. Background-subtracted Luciferase luminescence levels were divided by the corresponding background-subtracted Renilla luminescence, and all ratios were normalized to the average of the minimal promoter condition for quantitative analysis. A total of n = 5 biological replicate experiments were carried out, each including 4 technical replicates per condition. Statistical analysis was performed using a linear mixed model treating experimental days as random effects and alleles as fixed effect using the lmerTest package [32] in R v3.4.4 [33]. P values for the comparisons between conditions were obtained by analysis of variance (ANOVA) using Satterthwaite’s approximation for degrees of freedom.
eGWAS data processing and sampling analysis
For the Zou et al 2012 eGWAS, eQTL data tables using HapMap2-imputed genotypes were downloaded from the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site at the University of Pennsylvania [34]. Custom awk scripts were used to investigate INFERNO-prioritized variants, matching by rsID and/or by hg18 genome position.
Results
Expansion and annotation of IGAP loci
To identify genetic variants with regulatory potential for LOAD, we used INFERNO to analyze the 19 genome-wide significant loci from Phase I of IGAP (Figure 1a, Table 1) [6]. The region tagged by each top variant is referred to by the name of the nearest gene by convention, although these genes are not necessarily causal for the association signals. For each top variant, we identified all variants within 500kb that had a P value within an order of magnitude and the same minor allele effect direction (i.e. variants whose minor alleles are associated with increased AD risk if the top variant is a risk variant, or decreased AD risk if the top variant is protective), identifying a total of 496 variants (Table 1). These variants come from various haplotypes that may underlie the observed association signal at each locus, so to define a comprehensive set of potentially causal variants, we pruned the p-value expanded sets by LD into 52 independent variants tagging each potentially causal haplotype. Then, we re-expanded these by LD, yielding 1,044 unique potentially causal variants (Table 1) used for all subsequent analyses. These variants were primarily in introns and intergenic regions, with only 17 in mRNA exons (Figure 1b-c).
Table 1:
IGAP top hits expansion counts and annotation overlaps
| Tag variant | Nearest Gene | # P value expanded | # LD pruned | # LD re-expanded | # FANTOM5 overlaps | # Roadmap ChromHMM enhancer overlaps | # HOMER TFBS overlaps |
|---|---|---|---|---|---|---|---|
| rs4147929:A>G | ABCA7 | 7 | 2 | 9 | 0 | 8 | 6 |
| rs35349669:C>T | BIN1 | 2 | 1 | 3 | 0 | 3 | 1 |
| rs7274581:T>C | CASS4 | 20 | 2 | 23 | 4 | 19 | 10 |
| rs10948363:A>G | CD2AP | 80 | 5 | 83 | 0 | 45 | 30 |
| rs3865444:C>A | CD33 | 3 | 1 | 6 | 0 | 3 | 4 |
| rs10838725:T>C | CELF1 | 92 | 7 | 264 | 13 | 147 | 110 |
| rs28834970:T>C | CLU | 10 | 1 | 11 | 0 | 11 | 7 |
| rs6656401:A>G | CR1 | 20 | 1 | 22 | 0 | 12 | 9 |
| rs11771145:G>A | EPHA1 | 9 | 3 | 9 | 2 | 16 | 2 |
| rs17125944:T>C | FERMT2 | 32 | 7 | 92 | 2 | 79 | 43 |
| rs6733839:C>T | INPP5D | 65 | 2 | 114 | 0 | 66 | 54 |
| rs190982:G>A | MEF2C | 2 | 1 | 3 | 0 | 2 | 1 |
| rs983392:A>G | MS4A6A | 80 | 3 | 173 | 5 | 91 | 75 |
| rs1476679:C>T | TXNDC3 / NME8 | 44 | 5 | 96 | 4 | 44 | 47 |
| rs10792832:A>G | PICALM | 2 | 1 | 18 | 2 | 16 | 11 |
| rs9331896:C>T | PTK2B | 7 | 2 | 14 | 3 | 14 | 8 |
| rs10498633:G>T | SLC24A4 | 5 | 3 | 5 | 4 | 4 | 4 |
| rs11218343:T>C | SORL1 | 1 | 1 | 1 | 0 | 1 | 1 |
| rs2718058:A>G | ZCWPW1 | 15 | 4 | 98 | 0 | 78 | 28 |
| Total | - | 496 | 52 | 1,044 | 39 | 659 | 451 |
Next, we sought to compute the number of overlaps with different functional genomics data, before statistically quantifying the enrichments of variants overlapping FANTOM5 enhancers, Roadmap enhancers, or both within INFERNO tissue categories (Figure 2b). We overlapped these variants with enhancers defined by bidirectional enhancer RNA (eRNA) transcription in 112 tissues and cell types from the FANTOM5 consortium [17] and by the ChromHMM epigenomic state-based method [35] in 127 tissues and cell types from the Roadmap Epigenomics Project [15,36,37]. This identified 39 variants overlapping FANTOM5 enhancers in 9 tag regions (Table 1). The FANTOM5 data source with the most enhancer-overlapping variants was monocyte cells, with 25 overlapping variants, whereas the brain harbored only 6 variants (Supplementary Figure 2a). For the Roadmap data, variants were overlapped with a total of 15 ChromHMM states including 3 types of enhancer states (enhancers, genic enhancers, and bivalent enhancers). 659 variants representing all 19 tag regions were found to overlap a ChromHMM-defined enhancer state in at least one tissue (Table 1). Like the FANTOM5 results, primary monocytes from peripheral blood had the most overlapping variants (149 unique variants, Supplementary Figure 2), but 146 unique variants overlapped enhancers in at least one of the brain-related Roadmap datasets.
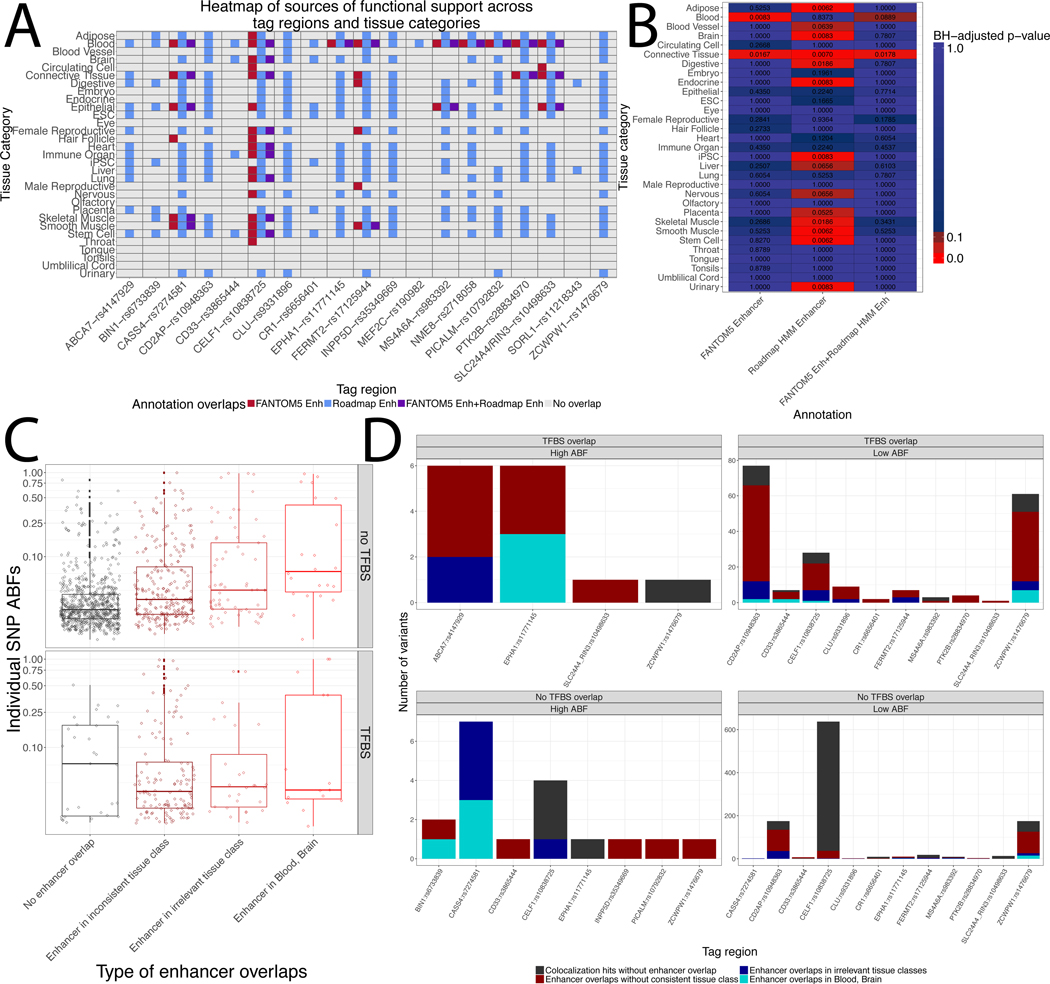
Figure 2: Integrative analysis of annotations for IGAP top hits.
a) Integrative tissue context analysis of enhancer overlaps from FANTOM5 and Roadmap datasets. b) Results of LD-collapsed bootstrapping for enhancer annotation overlap enrichments c) Distributions of variant probability of underlying highly colocalized signals stratified by annotation overlap. d) Barplots of numbers of variant – eQTL comparisons across tag regions stratified by motif overlap, enhancer support, and concordant support in a relevant tissue class.
We also used INFERNO to identify variants overlapping tissue-specific small noncoding RNA (sncRNA) loci from our DASHR database [38,39] as well as those disrupting microRNA (miRNA) seed sites within the 3’ untranslated region (UTR) of miRNA targets predicted by TargetScan [40] since studies have implicated sncRNA dysregulation in LOAD pathogenesis [41]. The DASHR overlap analysis found only one sncRNA overlap, where the variant rs4543938 in the CELF1 region overlapped the piRNA piR-56133. Analysis of miRNA seed disruption found that 46 variants across 5 tag regions (CELF1, FERMT2, INPP5D, MS4A6A, and ZCWPW1) overlapped binding sites for 40 miRNAs in 11 target genes (Supplementary Table 2).
We also used INFERNO to find variants affecting transcription factor binding sites (TFBSs) as identified by HOMER [42]. This identified 451 variants representing all 19 tag regions that either increased or decreased TFBS strength (measured by the change in the positional weight matrix, ΔPWM) for 191 unique transcription factors (Supplementary Figure 3). The majority of these overlaps had negative ΔPWM values, reflecting TFBSs disruptions.
Integrative analysis of enhancer enrichment patterns
Using INFERNO’s tissue categorization approach [26] that groups each functional genomics dataset into one of 32 high-level tissue categories, we identified 36 variants from nine tag regions (the CASS4, CELF1, EPHA1, FERMT2, MS4A6A, NME8, PICALM, PTK2B, and SLC24A4/RIN3 regions) that overlapped concordant FANTOM5 and Roadmap ChromHMM enhancers in a tissue category (Figure 2a). All of these regions harbored at least one variant with concordant support in the blood category, supporting the hypothesis of immune mechanisms underlying LOAD genetic signals [7–9]. The CELF1 region was the only one to harbor variants with concordant overlaps of brain enhancers, supporting the approach of performing functional annotation in all available tissues rather than focusing on a single predetermined tissue context. However, we point out that this region contains concordant enhancer overlaps in 9 tissue contexts (Figure 2a), so this regulatory mechanism may exert effects in several tissues.
Many variants overlapped FANTOM5 enhancers in the blood category, which included all the immune-related cell lines such as monocytes and macrophages in addition to whole blood (Supplementary Figure 4a). All 22 of the tissue categories sampled by the Roadmap Epigenomics consortium contained ChromHMM-defined enhancer-related states that harbored at least one variant in the expanded set (Supplementary Figure 4b). Again, many variants overlapped Roadmap enhancers in the blood category.
INFERNO includes a method to statistically quantify the enrichments of variants overlapping FANTOM5 enhancers, Roadmap enhancers, or both in each tissue category. This revealed significant enrichments of variants overlapping both FANTOM5 and concordant enhancers in the blood category and a significant enrichment of Roadmap enhancers in the brain category (Figure 2b) as well as in 13 other tissue contexts including connective tissue, which contains fibroblasts, nervous, and stem cell. Some of these tissue categories had even stronger enrichments than brain or blood, supporting our hypothesis generating approach of performing functional annotation and enrichment analysis in a wide range of tissue contexts. One possible scenario explaining these results is that AD-associated genetic variation could also affect regulatory mechanisms that are active in earlier life and in non-brain regions.
Co-localization analysis with GTEx eQTLs
To identify target genes affected by dysregulated enhancers, INFERNO uses expression quantitative trait loci (eQTLs) – variants whose alleles are correlated with differing levels of a target gene – from the Genotype-Tissue Expression (GTEx) project [16] across 44 tissues. Of the 1,044 potentially causal variants, 750 were significant eQTLs in at least one tissue (corrected q-value ≤ 0.05 on empirical p-values from Matrix eQTL [43]). However, due to dense LD structures in many of our significant regions, this direct overlap approach may yield false positive variants in LD with the truly causal eQTL variant. To address this issue, INFERNO incorporates a Bayesian statistical model (COLOC [44]) to identify co-localized GWAS/eQTL signals with shared causal variants, quantified as the posterior probability P(H4). The COLOC method also computes the probability of any individual variant being the shared causal variant, quantified as their Approximate Bayes Factors (ABFs).
We applied COLOC to compare GWAS signals across all 19 tag regions with tissue-specific eQTL signals for 884 unique genes in 44 tissues (median number of genes within each region = 34) for 25,435 tests of GWAS – tissue-specific eQTL co-localization (Supplementary Figure 5a, Methods). We identified 153 co-localized GWAS/eQTL signals (P(H4) >= 0.5 representing strong support for a shared causal signal [26]) representing 16 tag regions, 37 tissues and 71 target genes (Supplementary Figure 5b). These co-localization signals were found in 16 tissue categories, with the majority of signals in the brain category. For 32 of these 153 GWAS/eQTL signals, COLOC identified individual variants with ABF >= 0.5, but in the majority of cases COLOC was not able to prioritize a single causal variant. This is likely caused by dense LD structures where GWAS and eQTL signals are dispersed across all variants in the LD block (Supplementary Figure 5c). Thus, for each co-localized GWAS/eQTL signal we sampled the highest ABF variants until their sum was 0.5 or greater (Supplementary Figure 5d, [26]). Across the 153 co-localized signals, this yielded 1,291 unique variant–tissue–target gene relationships accounted for by 286 unique variants, 182 of which were in the LD-expanded set.
Comparison of enhancer overlaps with eQTL co-localization signals
We next used the INFERNO tissue categorizations to stratify variants in the ABF-expanded sets by whether they affected a TFBS, overlapped any enhancer, and whether the enhancer came from the concordant tissue category matching the eQTL (Figure 2c). To identify the variants with the strongest INFERNO support, we began by filtering the ABF-expanded variant sets to include only those variants overlapping concordant enhancers, and took two complementary approaches for further variant prioritization: requiring that the variant also overlapped a TFBS (TFBS prioritization) and/or requiring that the variant was the only one in the ABF-expanded set (e.g. ABF ≥ 0.5; ABF prioritization). TFBS prioritization identified 20 unique variants involved in 43 unique variant–tissue–gene sets across 8 tag regions (Figure 2d, top row) including 15 in the brain or blood categories. ABF prioritization identified 6 unique variants involved in 14 variant–tissue–gene sets across 5 tag regions (Figure 2d, left column), including 2 variants which also had TFBS overlaps. Together, these two approaches identified potentially causal variants in 10 tag regions (Table 2, Supplementary Tables 3-4). We further prioritized four of these signals for experimental validation based on prior literature, strength of annotation support, and relevant tissue contexts: EPHA1, CD33, BIN1, and CD2AP.
Table 2.
Summary of colocalization results in 4 top prioritized tag regions.
| Tag Region | Top affected mechanism(s) and evidence | Direction of effect | Experimental validation performed? |
|---|---|---|---|
| EPHA1 | Whole blood eQTL for EPHA1-AS1 supported by high ABF and increased CEBP TFBS variant rs11765305 | Protective haplotype has strong increase in EPHA1-AS1 expression | Yes, enhancer activity and significant allelic difference |
| CD33 | Whole blood eQTL for CD33 with high ABF tag variant rs3865444 | Protective tag variant decreases CD33 expression | Yes, strong enhancer activity and significant allelic difference |
| BIN1 | Lymphocyte eQTLs for BIN1, high ABF variant rs4663105 | Risk haplotype decreases BIN1 expression | Yes, moderate enhancer activity with significant allelic difference |
| CD2AP | Cerebellar eQTL for RP11–385F7.1 and fibroblast eQTL for CD2AP with TF disrupting variant rs9367279 | Risk haplotype with moderate, inconsistent effects on lncRNA expression in brain, decrease in CD2AP expression | Yes, no K562 enhancer activity or allelic difference |
EPHA1 region functional variant upregulates lncRNA affecting the JAK2 signaling axis
The strongest signal by both annotation and ABF evidence was in the EPHA1 region, where the variant rs11765305:C>G (chr7.hg19:g.143111112C>G, R2 = 1 with tag variant rs11771145) had an ABF of 0.999 underlying an eQTL for the EPHA1-AS1 antisense long non-coding RNA (lncRNA) in whole blood (P(H4) = 0.516). This variant also colocalized with whole blood eQTLs for the TAS2R60 taste receptor gene (P(H4) = 0.516, ABF = 1.00) and the TAS2R62P taste receptor gene (P(H4) = 0.537, ABF = 0.714) (Supplementary Table 3). rs11765305 overlapped FANTOM5 and Roadmap enhancers in the blood category, including white blood cells in the myeloid lineage such as monocytes and macrophages (Figure 3a), and creates a stronger binding site for CEBPB (ΔPWM score = 1.53), an enhancer-binding transcription factor that is associated with immune-related gene regulation [45]. This increase in TF binding is consistent with the positive effect of the rs11765305 minor allele on EPHA1-AS1 expression observed in GTEx (β = 1.25, where a β greater than 1 reflects an increase in gene expression).
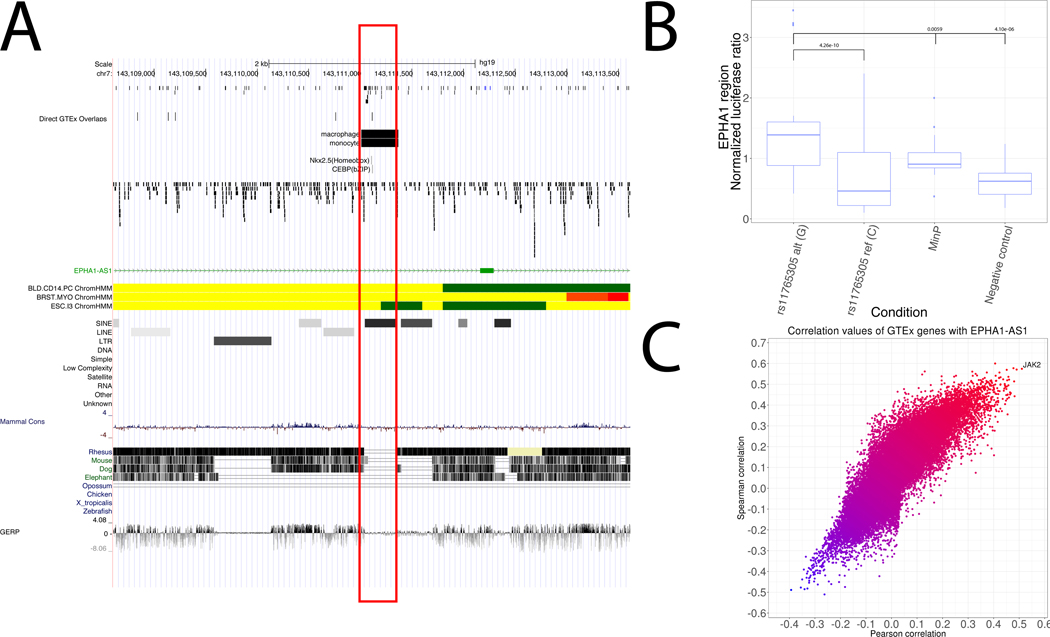
Figure 3: Functional variant in EPHA1 region upregulates EPHA1-AS1 lncRNA which regulates the JAK2 signaling axis.
a) Genome browser view of the region around rs11765305 (in red box) including relevant FANTOM5 and Roadmap enhancer annotations. b) Luciferase assay results for rs11765305 in K562 cells. Luciferase expression is normalized against Renilla expression in the same well. Negative control is randomly sampled heterochromatin insert. A linear mixed model was applied to 5 biological replicates per condition, each with 4 technical replicates per experimental day. c) Scatterplot of Pearson and Spearman correlations between expression of EPHA1-AS1 and all other genes in the genome across all GTEx tissues.
To compare enhancer activity between the major and minor alleles of rs11765305, we performed luciferase assays in K562 leukemia cells, which are from the same myeloid cell lineage as monocytes. Although the major allele had no significant luciferase expression compared to controls, the minor allele had significantly higher expression compared to both controls and the major allele (Figure 3b). These results confirm the predicted monocyte enhancer activity in this region and are consistent with the mechanism that the minor allele of rs11765305 creates a stronger CEBPB TFBS, increasing the activity of an enhancer regulating EPHA1-AS1, TAS2R60, and TAS2R62P.
We next set out to computationally identify the downstream effects of EPHA1-AS1, as lncRNAs can modulate gene expression through recruitment of regulatory proteins or binding to target transcripts [46]. INFERNO uses GTEx RNA-seq data to identify genes whose expression is correlated with that of a lncRNA using a threshold of 0.5 on both Pearson and Spearman correlations across 44 tissues [26]. Note that correlation between the expression of genes does not imply causal regulation. We introduced this computational approach as part of INFERNO for identifying gene hubs for functional enrichment analyses. Experimental approaches (such as knocking down the lncRNAs in the relevant tissues/cell types) could provide mechanistic validation to identify the downstream effects of the identified lncRNAs in followup studies.
For EPHA1-AS1, this correlation approach yielded one gene, JAK2 (Pearson r2 = 0.517, Spearman r2 = 0.582) (Figure 3c). JAK2 is part of the JAK2/STAT3 signaling axis, whose disturbance by amyloid-β leads to memory impairment [47]. The tag variant in this region is protective and rs11765305 has the same effect direction, so INFERNO prioritized a mechanism whereby the protective minor allele of rs11765305 increases EPHA1-AS1 expression which in turn increases the activity of the JAK2/STAT3 signaling axis, implying that JAK2/STAT3 activation may protect against LOAD.
To investigate these signals in an eQTL dataset specific to AD, we analyzed the Mayo eGWAS dataset comparing AD patients (n=202) to subjects with non-AD neurodegenerative diseases (n=197) [48] (Methods). This study used expression microarrays and genotyping to perform eQTL mapping in cerebellum and temporal cortex on AD cases, non-AD cases, and the combined patient population. In this study, rs11765305 was not genotyped or imputed, so we investigated the tag variant rs11771145 as a proxy which is in perfect LD. This variant had two significant eQTL signals in the cerebellum combined condition (q-value = 0.02254) and the temporal cortex non-AD patient condition (q-value = 0.02215), both for the gene ZYX. One limitation of the expression microarray approach is that the expression of long noncoding RNAs such as EPHA1-AS1 were not quantified, so this dataset cannot recapitulate these INFERNO results. However, ZYX was the most strongly correlated gene with EPHA1-AS1 by Spearman correlation (r = 0.6095), suggesting that these eQTL signals may reflect other downstream effects of EPHA1-AS1. ZYX encodes the zyxin protein which is part of the actin cytoskeleton, can be induced by amyloid-β, and may interact with SIRT1, an important protein in the aging process [49,50]. The eGWAS data could not recapitulate the JAK2 signal because it is on a different chromosome and the eGWAS analysis only tested cis-eQTLs within 200kb of any gene of interest.
Functional validation of blood regulation of CD33
In the CD33 region, COLOC identified co-localized GWAS/eQTLs for CD33 itself in whole blood (P(H4) = 0.955) and for AC018755.1 (P(H4) = 0.683) in brain hypothalamus. In both cases, rs12459419:C>T (chr19.hg19:g.51728477C>T) was prioritized by concordant enhancer and motif overlap. However, the tag variant rs3865444:C>A (chr19.hg19:g.51727962C>A) had a higher ABF in both cases (0.491 and 0.489, respectively) and is in perfect LD (R2 = 1) with rs12459419. rs3865444 overlaps Roadmap enhancers in 6 cell lines including primary monocytes and primary T regulatory cells from peripheral blood. In contrast, rs12459419 only overlapped Roadmap enhancers from 3 cell lines including primary T regulatory cells from peripheral blood and fetal brain but lacked the monocyte enhancer overlap (Supplementary Table 3). Additionally, rs3865444 has been extensively studied, with previous work showing that the protective minor allele (A) decreases the levels of CD33 protein [51], decreases CD33 mRNA expression consistent with the direction of the GTEx eQTL effect (β = 0.352) [52], and reduces cell surface expression of CD33 in monocytes [53].
Based on the prior literature, the strong ABF signal, perfect LD with rs12459419, and the monocyte enhancer overlap, we analyzed rs3865444 in our luciferase assays. This found significant increases for the major allele and significant decreases for the minor allele relative to the controls, as well as a striking decrease in enhancer activity of the minor allele relative to the major allele (Figure 4a). This was consistent with prior reports and the GTEx eQTL direction for this variant (β = 0.352). We also investigated rs3865444 in the eGWAS dataset but found no significant eQTL signals for that variant or any of the 5 other variants in the LD-expanded set in the CD33 region. This is consistent with the putative monocyte mechanism affected in this region, which we would not expect to detect in bulk brain tissue samples as they only include a small fraction of monocytes [54].
Figure 4: Luciferase and lncRNA analysis in the BIN1, CD33, and CD2AP regions.
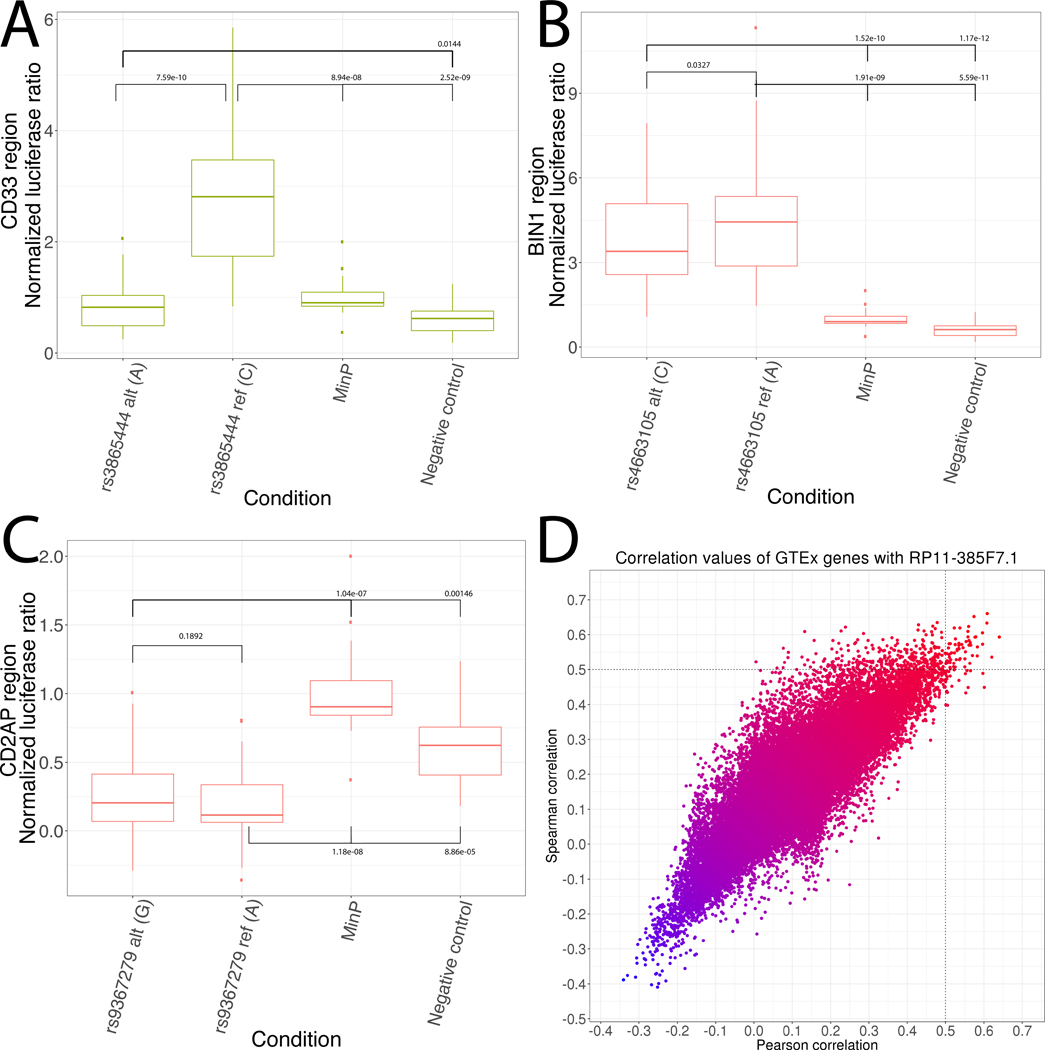
a) Luciferase validation in the CD33 region. b) Luciferase validation in the BIN1 region. c) Luciferase validation in the CD2AP region. For all luciferase analyses, a linear mixed model was applied to 5 biological replicates per condition, each with 4 technical replicates per experimental day. d) Scatterplot of Pearson and Spearman correlations between expression of RP11–385F7.1 (CD2AP region) and all other genes in the genome across all GTEx tissues.
Functional validation of lymphocyte regulation of BIN1
In the BIN1 region, INFERNO identified a co-localized GWAS/eQTL for BIN1 in EBV-transformed lymphocytes (P(H4) = 0.652) with the variant rs4663105:A>C (chr2.hg19:g.127891427A>C) prioritized by ABF (ABF = 0.777). This variant overlaps Roadmap enhancers in primary monocyte cells and placenta but does not overlap any TFBSs and is in strong LD with the tag variant rs6733839 (R2 = 0.8968). rs4663105 has been previously associated with LOAD risk, and an insertion in that region was associated with increased BIN1 expression [55]. This previous study found no difference in luciferase activity between the two alleles of rs4663105 in SKNSH-SY5Y and HEK cells. However, their construct only spanned 60bp around the variant, whereas the monocyte enhancer is 800bp. Therefore, we cloned the full Roadmap enhancer region (Supplementary Table 1) for luciferase assays in K562 cells, which are more relevant to the functional annotations in this region. This found significantly increased enhancer activity for both alleles of rs4663105 relative to the control vectors, and a slight but significant decrease of the minor allele relative to the major allele (p = 0.0328, Figure 4b), consistent with the direction of the GTEx eQTL (β = 0.496). We investigated these signals in the Mayo eGWAS datasets, but none of the variants in the BIN1 region were genotyped or imputed in any samples.
CD2AP region variants modulate lncRNA with widespread brain regulatory effects
Finally, in the CD2AP region, INFERNO prioritized several co-localized signals including RP11–385F7.1 in brain cerebellar hemisphere and cerebellum (P(H4) = 0.904 and 0.923, respectively) and an eQTL for CD2AP in fibroblasts (P(H4) = 0.801). TFBS prioritization implicated rs9367279:A>G (chr6.hg19:g.47448336A>G), which overlaps Roadmap enhancers in 33 tissues/cell lines from 13 tissue categories and disrupts a CArG-box binding site (ΔPWM = −1.38) for the MADS-box family of transcription factors, which includes the enhancer-related factors SRF and MEF2A [56,57]. CD2AP encodes a scaffolding molecule that regulates the actin cytoskeleton and is involved in endocytic processes [58], while RP11–385F7.1 is a lncRNA near the promoter for the CD2AP gene.
We performed luciferase assays, but both alleles of rs9367279 had significantly decreased enhancer activity relative to the controls, and there was no strong difference between the two alleles, suggesting that this enhancer may not be active in K562 cells (Figure 4c, p = 0.1892). RP11–385F7.1 was strongly correlated with 64 transcripts (Figure 4d, Supplementary Table 5), and we performed pathway analysis of these targets using the WebGestalt tool [29,30] to interpret the increased number of targets relative to the single target of EPHA1-AS1, but found no enrichments after controlling for false discovery rate. The gene with the strongest Pearson correlation was PPP1R16A (Pearson r2 = 0.641, Spearman r2 = 0.593) and the gene with the strongest Spearman correlation was COQ4 (Pearson r2 = 0.608, Spearman r2 = 0.660). PPP1R16A, also known as MYPT3, directs the protein phosphatase PP1c to its targets and is involved in actin binding and G-protein coupled receptor pathways [59]. COQ4 is part of the coenzyme Q biosynthesis pathway, an antioxidant that may modify LOAD-associated oxidative damage [60]. The eQTL effects of rs9367279 on RP11–385F7.1 are weak and inconsistent between the two brain regions (β = 0.969 in cerebellar hemisphere and 1.194 in cerebellum), suggesting that rs9367279 contributes to fine-scale regulation of RP11–385F7.1 in brain, although it has a relatively strong repressive effect on CD2AP in fibroblasts (β = 0.505). We also investigated this variant in the Mayo eGWAS data, but it was only tested for association with CD2AP and not any lncRNAs, and it was not a significant eQTL for CD2AP in any condition.
Discussion
Our application of INFERNO to LOAD GWAS data prioritized perturbations of tissue-specific regulatory mechanisms in 10 IGAP tag regions (Table 2, Supplementary Table 4). In the EPHA1, CD2AP, CELF1, and CASS4 regions, the target genes of the co-localized GWAS/eQTL signals included lncRNAs, so identifying affected enhancers and target genes may be only the first step towards understanding genetic effects on regulatory networks contributing to disease pathogenesis. The tissue classification approach implemented in INFERNO also enabled the unbiased investigation of the relevant tissue contexts affected by each genetic signal. Limiting our analysis to only brain datasets would have missed the blood-category signals that we detected. These immunity-related signals are in line with other recent work highlighting neuroinflammation as a crucial component of LOAD pathogenesis and etiology [7–9]. Our validation experiments used K562 as an immune-related cell line to test that the enhancers predicted by FANTOM5 and Roadmap were indeed active and that there was differential activity between the alleles of our prioritized variants. This cell line does not model specific aspects of AD pathology, but these experiments were aimed at validating the enhancer mechanisms prioritized by INFERNO, and followup studies will elucidate the effects of these regulatory mechanisms in AD-specific contexts.
In our primary INFERNO analysis, we considered GTEx eQTLs that come from healthy, non-diseased tissues, rather than eQTLs from AD patients. We believe that this is a desirable approach for our goal of characterizing the molecular mechanisms underlying AD GWAS signals, which measure genetic susceptibility conferred by inherited (i.e. germline) genetic variation. These susceptibility signals reflect the life-time risk of developing AD, which is present from conception and conferred well before the neurodegenerative disease process begins. The molecular mechanisms underlying these associations might exert their effects beginning in the normal state and continuing throughout the life-time. On the other hand, AD eQTL data measures genetic variants associated with AD-associated differential gene expression which could be either up- or down-stream of the actual pathologic processes and susceptibility mechanisms. Also, the onset of AD results in neurodegeneration and the loss of specific cell types which may drive expression changes, but bulk tissue datasets may not account for these proportional changes. Thus, we used GTEx data to generate hypotheses about regulatory mechanisms underlying genetic susceptibility and then incorporated the eGWAS dataset to explore if these mechanisms were recapitulated in the disease state. Consistent with the majority of our signals reflecting immune-related processes and the lack of lncRNA profiling by microarray, the majority of our signals were not detected in the eGWAS dataset, supporting our approach of using the GTEx data to generate hypotheses.
INFERNO did not identify regulatory mechanisms in all 19 of the IGAP regions, and this may be driven by several aspects of this analysis. We are limited by the sample sizes and sets of tissues that were assayed by the FANTOM5, Roadmap, and GTEx consortia and the number of datasets that went into each tissue category, with some categories being much more sparsely sampled than others [26], especially the brain tissues which have some of the smallest GTEx sample sizes [16]. Another issue with this bulk tissue-based approach is that tissue samples are a mixture of a large variety of different cell types such as neurons, astrocytes, and microglia in brain [61], and so the eQTL and enhancer signals that INFERNO analyzes may be mainly driven by the most prevalent cell type in each tissue, potentially missing signals involving rarer cell types. Emerging single cell technologies aimed at analyzing homogeneous cell populations will help to address this gap, but we are not currently aware of any data sources measuring enhancer or eQTL signals in single brain cells.
Our application of INFERNO to IGAP GWAS data for late-onset Alzheimer’s Disease yielded insights into the regulatory mechanisms affected by noncoding LOAD-associated genetic variants. Experimental validation supported our computationally predicted regulatory effects, suggesting that our approach is able to prioritize truly causal regulatory mechanisms at GWAS loci for post-GWAS experiments. Incorporating more functional genomics data as it is generated in concert with more refined validation experiments using a broader range of cell types and molecular techniques will yield insights into a range of phenotypes.
Supplementary Material
Supplementary Note 1: Guidelines to prioritize the causal variants for further mechanistic studies
Supplementary Table 1: Luciferase enhancer regions (Excel file)
Supplementary Table 2: microRNA seed sequence disruptions by INFERNO-expanded variants (Excel file).
Supplementary Table 3: Full colocalization annotation results. Top signals in each region are highlighted. (Excel file)
Supplementary Table 4: Summary of colocalization results in 6 non-prioritized regions. (Excel file).
Supplementary Table 5. Highly correlated genes with RP11-385F7.1. (Excel table)
Flowchart of luciferase-based enhancer validation assays.
Number of enhancer overlaps across individual tissues from FANTOM5 and Roadmap. a) Number of variants overlapping eRNA-defined enhancers across 112 FANTOM5 tissue and cell type facets. b) Number of variants overlapping each type of ChromHMM-defined enhancer state across 127 Roadmap tissues and cell types.
HOMER motif overlap ΔPWM distributions
Number of enhancer overlaps across tissue categories sampled in FANTOM5 and Roadmap. a) Number of variants overlapping eRNA-defined enhancers in each tag region across the 28 tissue categories that include FANTOM5 samples. b) Number of variants overlapping any of the three ChromHMM-defined enhancer states in each tag region across the 22 tissue categories that include Roadmap samples.
Colocalization analysis of GTEx eQTLs with IGAP GWAS signals. a) Distributions of the 5 colocalization hypotheses across all tissues and tag regions. b) Histograms of P(H4) for highly colocalized (P(H4) >= 0.5) signals across tag regions. c) Histograms of the approximate Bayes factor values of the most supported variants across tag regions. d) Histograms of the number of variants required to cumulatively account for 50% of the individual variant probability (ABFs) for each colocalization signal across tag regions.
Acknowledgements
This work was supported by the National Institutes of Health [grant numbers U01-AG032984, UF1-AG047133, U54-AG052427, U24-AG041689, R01-GM099962, P30-AG010124, RF1-AG055477, U54-NS100693, and T32-AG00255]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We gratefully acknowledge Adam Naj, Ian Mellis, and Eddie Lee for providing writing help and feedback on these results. We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer’s Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
Footnotes
Conflict of Interest
The authors have no conflict of interest to report.
References
- 1.Association A (2015) 2015 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 11, 332–384. [DOI] [PubMed] [Google Scholar]
- 2.Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer J a, Posner SF, Viitanen M, Winblad B, Ahlbom A (1997) Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. Journals Gerontol. Ser. A, Biol. Sci. Med. Sci. 52, M117–M125. [DOI] [PubMed] [Google Scholar]
- 3.Corder EH, Saunders a M, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses a D, Haines JL, Pericak-Vance M a (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science (80-. ). 261, 921–923. [DOI] [PubMed] [Google Scholar]
- 4.Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, Morgan K, Passmore P, Holmes C, Powell J, Brayne C, Gill M, Mead S, Goate A, Cruchaga C, Lambert JC, Van Duijn C, Maier W, Ramirez A, Holmans P, Jones L, Hardy J, Seshadri S, Schellenberg GD, Amouyel P, Williams J (2015) Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 138, 3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider J a, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha G a, Jin L-W, Johnson N, Karlawish J, Karydas A, Kaye J a, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller C a, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary R a, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer Ma, Smith CD, Sonnen Ja, Spina S, Stern Ra, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters H V, Vonsattel JP, Weintraub S, Welsh-Bohmer K a, Williamson J, Woltjer RL, Cantwell LB, Dombroski B a, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett D a, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull W a, Foroud TM, Haines JL, Mayeux R, Pericak-Vance Ma, Farrer L a, Schellenberg GD (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert JC, Ibrahim-Verbaas C a, Harold D, Naj a C, Sims R, Bellenguez C, DeStafano a L, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells T a, Jones N, Smith a V, Chouraki V, Thomas C, Ikram M a, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston J a, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate a M, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers a J, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Bennett D a, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull W a, Hannequin D, Powell JF, Nalls M a, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans P a, Lathrop M, Pericak-Vance M a, Launer LJ, Farrer L a, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heneka MT, Golenbock DT, Latz E (2015) Innate immunity in Alzheimer’s disease. Nat. Rev. Immunol. 16, 229–236. [DOI] [PubMed] [Google Scholar]
- 8.Heppner FL, Ransohoff RM, Becher B (2015) Immune attack: the role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 16, 358–372. [DOI] [PubMed] [Google Scholar]
- 9.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, Martin ER, Grenier-Boley B, Heilmann-Heimbach S, Chouraki V, Kuzma AB, Sleegers K, Vronskaya M, Ruiz A, Graham RR, Olaso R, Hoffmann P, Grove ML, Vardarajan BN, Hiltunen M, Nöthen MM, White CC, Hamilton-Nelson KL, Epelbaum J, Maier W, Choi S-H, Beecham GW, Dulary C, Herms S, Smith A V, Funk CC, Derbois C, Forstner AJ, Ahmad S, Li H, Bacq D, Harold D, Satizabal CL, Valladares O, Squassina A, Thomas R, Brody JA, Qu L, Sánchez-Juan P, Morgan T, Wolters FJ, Zhao Y, Garcia FS, Denning N, Fornage M, Malamon J, Naranjo MCD, Majounie E, Mosley TH, Dombroski B, Wallon D, Lupton MK, Dupuis J, Whitehead P, Fratiglioni L, Medway C, Jian X, Mukherjee S, Keller L, Brown K, Lin H, Cantwell LB, Panza F, McGuinness B, Moreno-Grau S, Burgess JD, Solfrizzi V, Proitsi P, Adams HH, Allen M, Seripa D, Pastor P, Cupples LA, Price ND, Hannequin D, Frank-García A, Levy D, Chakrabarty P, Caffarra P, Giegling I, Beiser AS, Giedraitis V, Hampel H, Garcia ME, Wang X, Lannfelt L, Mecocci P, Eiriksdottir G, Crane PK, Pasquier F, Boccardi V, Henández I, Barber RC, Scherer M, Tarraga L, Adams PM, Leber M, Chen Y, Albert MS, Riedel-Heller S, Emilsson V, Beekly D, Braae A, Schmidt R, Blacker D, Masullo C, Schmidt H, Doody RS, Spalletta G, Jr WTL, Fairchild TJ, Bossù P, Lopez OL, Frosch MP, Sacchinelli E, Ghetti B, Yang Q, Huebinger RM, Jessen F, Li S, Kamboh MI, Morris J, Sotolongo-Grau O, Katz MJ, Corcoran C, Dunstan M, Braddel A, Thomas C, Meggy A, Marshall R, Gerrish A, Chapman J, Aguilar M, Taylor S, Hill M, Fairén MD, Hodges A, Vellas B, Soininen H, Kloszewska I, Daniilidou M, Uphill J, Patel Y, Hughes JT, Lord J, Turton J, Hartmann AM, Cecchetti R, Fenoglio C, Serpente M, Arcaro M, Caltagirone C, Orfei MD, Ciaramella A, Pichler S, Mayhaus M, Gu W, Lleó A, Fortea J, Blesa R, Barber IS, Brookes K, Cupidi C, Maletta RG, Carrell D, Sorbi S, Moebus S, Urbano M, Pilotto A, Kornhuber J, Bosco P, Todd S, Craig D, Johnston J, Gill M, Lawlor B, Lynch A, Fox NC, Hardy J, Albin RL, Apostolova LG, Arnold SE, Asthana S, Atwood CS, Baldwin CT, Barnes LL, Barral S, Beach TG, Becker JT, Bigio EH, Bird TD, Boeve BF, Bowen JD, Boxer A, Burke JR, Burns JM, Buxbaum JD, Cairns NJ, Cao C, Carlson CS, Carlsson CM, Carney RM, Carrasquillo MM, Carroll SL, Diaz CC, Chui HC, Clark DG, Cribbs DH, Crocco EA, DeCarli C, Dick M, Duara R, Evans DA, Faber KM, Fallon KB, Fardo DW, Farlow MR, Ferris S, Foroud TM, Galasko DR, Gearing M, Geschwind DH, Gilbert JR, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Honig LS, Huentelman MJ, Hulette CM, Hyman BT, Jarvik GP, Abner E, Jin L-W, Jun G, Karydas A, Kaye JA, Kim R, Kowall NW, Kramer JH, LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lunetta KL, Lyketsos CG, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Morris JC, Murrell JR, Myers AJ, O’Bryant S, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Perry W, Peskind E, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rogaeva E, Rosen HJ, Rosenberg RN, Sager MA, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters H V, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu C-E, Yu L, Garzia F, Golamaully F, Septier G, Engelborghs S, Vandenberghe R, De Deyn PP, Fernadez CM, Benito YA, Thonberg H, Forsell C, Lilius L, Kinhult-Stählbom A, Kilander L, Brundin R, Concari L, Helisalmi S, Koivisto AM, Haapasalo A, Dermecourt V, Fievet N, Hanon O, Dufouil C, Brice A, Ritchie K, Dubois B, Himali JJ, Keene CD, Tschanz J, Fitzpatrick AL, Kukull WA, Norton M, Aspelund T, Larson EB, Munger R, Rotter JI, Lipton RB, Bullido MJ, Hofman A, Montine TJ, Coto E, Boerwinkle E, Petersen RC, Alvarez V, Rivadeneira F, Reiman EM, Gallo M, O’Donnell CJ, Reisch JS, Bruni AC, Royall DR, Dichgans M, Sano M, Galimberti D, St George-Hyslop P, Scarpini E, Tsuang DW, Mancuso M, Bonuccelli U, Winslow AR, Daniele A, Wu C-K, Peters O, Nacmias B, Riemenschneider M, Heun R, Brayne C, Rubinsztein DC, Bras J, Guerreiro R, Al-Chalabi A, Shaw CE, Collinge J, Mann D, Tsolaki M, Clarimón J, Sussams R, Lovestone S, O’Donovan MC, Owen MJ, Behrens TW, Mead S, Goate AM, Uitterlinden AG, Holmes C, Cruchaga C, Ingelsson M, Bennett DA, Powell J, Golde TE, Graff C, De Jager PL, Morgan K, Ertekin-Taner N, Combarros O, Psaty BM, Passmore P, Younkin SG, Berr C, Gudnason V, Rujescu D, Dickson DW, Dartigues J-F, DeStefano AL, Ortega-Cubero S, Hakonarson H, Campion D, Boada M, Kauwe JK, Farrer LA, Van Broeckhoven C, Ikram MA, Jones L, Haines JL, Tzourio C, Launer LJ, Escott-Price V, Mayeux R, Deleuze J-F, Amin N, Holmans PA, Pericak-Vance MA, Amouyel P, van Duijn CM, Ramirez A, Wang L-S, Lambert J-C, Seshadri S, Williams J, Schellenberg GD (2017) Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Stamatoyannopoulos J a (2012) Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science (80-. ). 337, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corradin O, Scacheri PC (2014) Enhancer variants: evaluating functions in common disease. Genome Med. 6, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulger M, Groudine M (2010) Enhancers: The abundance and function of regulatory sequences beyond promoters. Dev. Biol. 339, 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittkopp PJ, Kalay G (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. [DOI] [PubMed] [Google Scholar]
- 14.Ong C-T, Corces VG (2011) Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium RE, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong N a, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer L a, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra M a, McManus MT, Sunyaev S, Thomson J a, Tlsty TD, Tsai L-H, Wang W, Waterland R a, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos J a, Wang T, Kellis M, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning A, Wang X, ClaussnitzerYaping Liu M, Coarfa C, Alan Harris R, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, David Hawkins R, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Scott Hansen R, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong N a, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Abdennur N, Adli M, Akerman M, Barrera L, Antosiewicz-Bourget J, Ballinger T, Barnes MJ, Bates D, Bell RJ a, Bennett D a, Bianco K, Bock C, Boyle P, Brinchmann J, Caballero-Campo P, Camahort R, Carrasco-Alfonso MJ, Charnecki T, Chen H, Chen Z, Cheng JB, Cho S, Chu A, Chung W-Y, Cowan C, Athena Deng Q, Deshpande V, Diegel M, Ding B, Durham T, Echipare L, Edsall L, Flowers D, Genbacev-Krtolica O, Gifford C, Gillespie S, Giste E, Glass I a, Gnirke A, Gormley M, Gu H, Gu J, Hafler D a, Hangauer MJ, Hariharan M, Hatan M, Haugen E, He Y, Heimfeld S, Herlofsen S, Hou Z, Humbert R, Issner R, Jackson AR, Jia H, Jiang P, Johnson AK, Kadlecek T, Kamoh B, Kapidzic M, Kent J, Kim A, Kleinewietfeld M, Klugman S, Krishnan J, Kuan S, Kutyavin T, Lee A-Y, Lee K, Li J, Li N, Li Y, Ligon KL, Lin S, Lin Y, Liu J, Liu Y, Luckey CJ, Ma YP, Maire C, Marson A, Mattick JS, Mayo M, McMaster M, Metsky H, Mikkelsen T, Miller D, Miri M, Mukame E, Nagarajan RP, Neri F, Nery J, Nguyen T, O’Geen H, Paithankar S, Papayannopoulou T, Pelizzola M, Plettner P, Propson NE, Raghuraman S, Raney BJ, Raubitschek A, Reynolds AP, Richards H, Riehle K, Rinaudo P, Robinson JF, Rockweiler NB, Rosen E, Rynes E, Schein J, Sears R, Sejnowski T, Shafer A, Shen L, Shoemaker R, Sigaroudinia M, Slukvin I, Stehling-Sun S, Stewart R, Subramanian SL, Suknuntha K, Swanson S, Tian S, Tilden H, Tsai L, Urich M, Vaughn I, Vierstra J, Vong S, Wagner U, Wang H, Wang T, Wang Y, Weiss A, Whitton H, Wildberg A, Witt H, Won K-J, Xie M, Xing X, Xu I, Xuan Z, Ye Z, Yen C, Yu P, Zhang X, Zhang X, Zhao J, Zhou Y, Zhu J, Zhu Y, Ziegler S, Beaudet AE, Boyer L a, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra M a, McManus MT, Sunyaev S, Thomson J a, Tlsty TD, Tsai L-H, Wang W, Waterland R a, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos J a, Wang T, Kellis M (2015) Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardlie KG, Deluca DS, Segre AV, Sullivan TJ, Young TR, Gelfand ET, Trowbridge CA, Maller JB, Tukiainen T, Lek M, Ward LD, Kheradpour P, Iriarte B, Meng Y, Palmer CD, Esko T, Winckler W, Hirschhorn JN, Kellis M, MacArthur DG, Getz G, Shabalin AA, Li G, Zhou Y-H, Nobel AB, Rusyn I, Wright FA, Lappalainen T, Ferreira PG, Ongen H, Rivas MA, Battle A, Mostafavi S, Monlong J, Sammeth M, Mele M, Reverter F, Goldmann JM, Koller D, Guigo R, McCarthy MI, Dermitzakis ET, Gamazon ER, Im HK, Konkashbaev A, Nicolae DL, Cox NJ, Flutre T, Wen X, Stephens M, Pritchard JK, Tu Z, Zhang B, Huang T, Long Q, Lin L, Yang J, Zhu J, Liu J, Brown A, Mestichelli B, Tidwell D, Lo E, Salvatore M, Shad S, Thomas JA, Lonsdale JT, Moser MT, Gillard BM, Karasik E, Ramsey K, Choi C, Foster BA, Syron J, Fleming J, Magazine H, Hasz R, Walters GD, Bridge JP, Miklos M, Sullivan S, Barker LK, Traino HM, Mosavel M, Siminoff LA, Valley DR, Rohrer DC, Jewell SD, Branton PA, Sobin LH, Barcus M, Qi L, McLean J, Hariharan P, Um KS, Wu S, Tabor D, Shive C, Smith AM, Buia SA, Undale AH, Robinson KL, Roche N, Valentino KM, Britton A, Burges R, Bradbury D, Hambright KW, Seleski J, Korzeniewski GE, Erickson K, Marcus Y, Tejada J, Taherian M, Lu C, Basile M, Mash DC, Volpi S, Struewing JP, Temple GF, Boyer J, Colantuoni D, Little R, Koester S, Carithers LJ, Moore HM, Guan P, Compton C, Sawyer SJ, Demchok JP, Vaught JB, Rabiner CA, Lockhart NC, Ardlie KG, Getz G, Wright FA, Kellis M, Volpi S, Dermitzakis ET (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science (80-. ). 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs a M, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume D a, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A (2014) An atlas of active enhancers across human cell types and tissues. Nature 507, 455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams B a., Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See L-H, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender M a., Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu Y-C, Rasmussen MD, Bansal MS, Kellis M, Keller C a., Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan C a., Rosenbloom KR, Lacerda de Sousa B, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, James Kent W, Ramalho Santos M, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh T a., Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Scott Hansen R, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang K-H, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel G a., Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou X-Q, Pazin MJ, Feingold E a., Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos J a., Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer M a., Ren B (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium TF, Pmi R, Dgt C, Forrest ARR, Kawaji H, Rehli M, Kenneth Baillie J, de Hoon MJL, Haberle V, Lassmann T, Kulakovskiy I V., Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jørgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva Y a., Plessy C, Vitezic M, Severin J, Semple C a., Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer J a. C, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake J a., Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Maxwell Burroughs A, Califano A, Cannistraci C V., Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis C a., Detmar M, Diehl AD, Dohi T, Drabløs F, Edge ASB, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov A V., Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furuno M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Peter Klinken S, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JFJ, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais D a., Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van Nimwegen E, Ninomiya N, Nishiyori H, Noma S, Nozaki T, Ogishima S, Ohkura N, Ohmiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov D a., Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JGD, Rackham OJL, Ramilowski J a., Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Satoh H, Savvi S, Saxena A, Schneider C, Schultes E a., Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, Shimoni Y, Shin JW, Simon C, Sugiyama D, Sugiyama T, Suzuki M, Suzuki N, Swoboda RK, ‘t Hoen P a. C, Tagami M, Takahashi N, Takai J, Tanaka H, Tatsukawa H, Tatum Z, Thompson M, Toyoda H, Toyoda T, Valen E, van de Wetering M, van den Berg LM, Verardo R, Vijayan D, Vorontsov IE, Wasserman WW, Watanabe S, Wells C a., Winteringham LN, Wolvetang E, Wood EJ, Yamaguchi Y, Yamamoto M, Yoneda M, Yonekura Y, Yoshida S, Zabierowski SE, Zhang PG, Zhao X, Zucchelli S, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume D a., Carninci P, Hayashizaki Y (2014) A promoter-level mammalian expression atlas. Nature 507, 462–470.24670764 [Google Scholar]
- 20.Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai L-H, Kellis M (2015) Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagliano SA, Pouget JG, Hardy J, Knight J, Barnes MR, Ryten M, Weale ME (2016) Genomics implicates adaptive and innate immunity in Alzheimer’s and Parkinson’s diseases. Ann. Clin. Transl. Neurol. 3, 924–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tansey KE, Cameron D, Hill MJ (2018) Genetic risk for Alzheimer’s disease is concentrated in specific macrophage and microglial transcriptional networks. Genome Med. 10, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Mägi R, Reschen ME, Mahajan A, Locke A, William Rayner N, Robertson N, Scott RA, Prokopenko I, Scott LJ, Green T, Sparso T, Thuillier D, Yengo L, Grallert H, Wahl S, Frånberg M, Strawbridge RJ, Kestler H, Chheda H, Eisele L, Gustafsson S, Steinthorsdottir V, Thorleifsson G, Qi L, Karssen LC, van Leeuwen EM, Willems SM, Li M, Chen H, Fuchsberger C, Kwan P, Ma C, Linderman M, Lu Y, Thomsen SK, Rundle JK, Beer NL, van de Bunt M, Chalisey A, Kang HM, Voight BF, Abecasis GR, Almgren P, Baldassarre D, Balkau B, Benediktsson R, Blüher M, Boeing H, Bonnycastle LL, Bottinger EP, Burtt NP, Carey J, Charpentier G, Chines PS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Doney ASF, Dorkhan M, Edkins S, Eriksson JG, Esko T, Eury E, Fadista J, Flannick J, Fontanillas P, Fox C, Franks PW, Gertow K, Gieger C, Gigante B, Gottesman O, Grant GB, Grarup N, Groves CJ, Hassinen M, Have CT, Herder C, Holmen OL, Hreidarsson AB, Humphries SE, Hunter DJ, Jackson AU, Jonsson A, Jørgensen ME, Jørgensen T, Kao W-HL, Kerrison ND, Kinnunen L, Klopp N, Kong A, Kovacs P, Kraft P, Kravic J, Langford C, Leander K, Liang L, Lichtner P, Lindgren CM, Lindholm E, Linneberg A, Liu C-T, Lobbens S, Luan J, Lyssenko V, Männistö S, McLeod O, Meyer J, Mihailov E, Mirza G, Mühleisen TW, Müller-Nurasyid M, Navarro C, Nöthen MM, Oskolkov NN, Owen KR, Palli D, Pechlivanis S, Peltonen L, Perry JRB, Platou CGP, Roden M, Ruderfer D, Rybin D, van der Schouw YT, Sennblad B, Sigurðsson G, Stančáková A, Steinbach G, Storm P, Strauch K, Stringham HM, Sun Q, Thorand B, Tikkanen E, Tonjes A, Trakalo J, Tremoli E, Tuomi T, Wennauer R, Wiltshire S, Wood AR, Zeggini E, Dunham I, Birney E, Pasquali L, Ferrer J, Loos RJF, Dupuis J, Florez JC, Boerwinkle E, Pankow JS, van Duijn C, Sijbrands E, Meigs JB, Hu FB, Thorsteinsdottir U, Stefansson K, Lakka TA, Rauramaa R, Stumvoll M, Pedersen NL, Lind L, Keinanen-Kiukaanniemi SM, Korpi-Hyövälti E, Saaristo TE, Saltevo J, Kuusisto J, Laakso M, Metspalu A, Erbel R, Jöcke K-H, Moebus S, Ripatti S, Salomaa V, Ingelsson E, Boehm BO, Bergman RN, Collins FS, Mohlke KL, Koistinen H, Tuomilehto J, Hveem K, Njølstad I, Deloukas P, Donnelly PJ, Frayling TM, Hattersley AT, de Faire U, Hamsten A, Illig T, Peters A, Cauchi S, Sladek R, Froguel P, Hansen T, Pedersen O, Morris AD, Palmer CNA, Kathiresan S, Melander O, Nilsson PM, Groop LC, Barroso I, Langenberg C, Wareham NJ, O’Callaghan CA, Gloyn AL, Altshuler D, Boehnke M, Teslovich TM, McCarthy MI, Morris AP (2015) Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 47,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur N a., Liu J, Svensson P-A, Hsu Y-H, Drucker DJ, Mellgren G, Hui C-C, Hauner H, Kellis M (2015) FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 150819140043007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, Stahl EA, Georgakopoulos A, Ruderfer DM, Charney A, Okada Y, Siminovitch KA, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Plenge RM, Raychaudhuri S, Fromer M, Purcell SM, Brennand KJ, Robakis NK, Schadt EE, Akbarian S, Sklar P (2014) A Role for Noncoding Variation in Schizophrenia. Cell Rep. 9, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amlie-Wolf A, Tang M, Mlynarski EE, Kuksa PP, Valladares O, Katanic Z, Tsuang D, Brown CD, Schellenberg GD, Wang L-S (2018) INFERNO: inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Res. 211599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evseeva I, Nicodemus KK, Bonilla C, Tonks S, Bodmer WF (2010) Linkage disequilibrium and age of HLA region SNPs in relation to classic HLA gene alleles within Europe. Eur. J. Hum. Genet. 18, 924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, Flicek P, Gabriel SB, Gibbs RA, Green ED, Hurles ME, Knoppers BM, Korbel JO, Lander ES, Lee C, Lehrach H, Mardis ER, Marth GT, McVean GA, Nickerson DA, Schmidt JP, Sherry ST, Wang J, Wilson RK, Gibbs RA, Boerwinkle E, Doddapaneni H, Han Y, Korchina V, Kovar C, Lee S, Muzny D, Reid JG, Zhu Y, Wang J, Chang Y, Feng Q, Fang X, Guo X, Jian M, Jiang H, Jin X, Lan T, Li G, Li J, Li Y, Liu S, Liu X, Lu Y, Ma X, Tang M, Wang B, Wang G, Wu H, Wu R, Xu X, Yin Y, Zhang D, Zhang W, Zhao J, Zhao M, Zheng X, Lander ES, Altshuler DM, Gabriel SB, Gupta N, Gharani N, Toji LH, Gerry NP, Resch AM, Flicek P, Barker J, Clarke L, Gil L, Hunt SE, Kelman G, Kulesha E, Leinonen R, McLaren WM, Radhakrishnan R, Roa A, Smirnov D, Smith RE, Streeter I, Thormann A, Toneva I, Vaughan B, Zheng-Bradley X, Bentley DR, Grocock R, Humphray S, James T, Kingsbury Z, Lehrach H, Sudbrak R, Albrecht MW, Amstislavskiy VS, Borodina TA, Lienhard M, Mertes F, Sultan M, Timmermann B, Yaspo M-L, Mardis ER, Wilson RK, Fulton L, Fulton R, Sherry ST, Ananiev V, Belaia Z, Beloslyudtsev D, Bouk N, Chen C, Church D, Cohen R, Cook C, Garner J, Hefferon T, Kimelman M, Liu C, Lopez J, Meric P, O’Sullivan C, Ostapchuk Y, Phan L, Ponomarov S, Schneider V, Shekhtman E, Sirotkin K, Slotta D, Zhang H, McVean GA, Durbin RM, Balasubramaniam S, Burton J, Danecek P, Keane TM, Kolb-Kokocinski A, McCarthy S, Stalker J, Quail M, Schmidt JP, Davies CJ, Gollub J, Webster T, Wong B, Zhan Y, Auton A, Campbell CL, Kong Y, Marcketta A, Gibbs RA, Yu F, Antunes L, Bainbridge M, Muzny D, Sabo A, Huang Z, Wang J, Coin LJM, Fang L, Guo X, Jin X, Li G, Li Q, Li Y, Li Z, Lin H, Liu B, Luo R, Shao H, Xie Y, Ye C, Yu C, Zhang F, Zheng H, Zhu H, Alkan C, Dal E, Kahveci F, Marth GT, Garrison EP, Kural D, Lee W-P, Fung Leong W, Stromberg M, Ward AN, Wu J, Zhang M, Daly MJ, DePristo MA, Handsaker RE, Altshuler DM, Banks E, Bhatia G, del Angel G, Gabriel SB, Genovese G, Gupta N, Li H, Kashin S, Lander ES, McCarroll SA, Nemesh JC, Poplin RE, Yoon SC, Lihm J, Makarov V, Clark AG, Gottipati S, Keinan A, Rodriguez-Flores JL, Korbel JO, Rausch T, Fritz MH, Stütz AM, Flicek P, Beal K, Clarke L, Datta A, Herrero J, McLaren WM, Ritchie GRS, Smith RE, Zerbino D, Zheng-Bradley X, Sabeti PC, Shlyakhter I, Schaffner SF, Vitti J, Cooper DN, Ball EV, Stenson PD, Bentley DR, Barnes B, Bauer M, Keira Cheetham R, Cox A, Eberle M, Humphray S, Kahn S, Murray L, Peden J, Shaw R, Kenny EE, Batzer MA, Konkel MK, Walker JA, MacArthur DG, Lek M, Sudbrak R, Amstislavskiy VS, Herwig R, Mardis ER, Ding L, Koboldt DC, Larson D, Ye K, Gravel S, Swaroop A, Chew E, Lappalainen T, Erlich Y, Gymrek M, Frederick Willems T, Simpson JT, Shriver MD, Rosenfeld JA, Bustamante CD, Montgomery SB, De La Vega FM, Byrnes JK, Carroll AW, DeGorter MK, Lacroute P, Maples BK, Martin AR, Moreno-Estrada A, Shringarpure SS, Zakharia F, Halperin E, Baran Y, Lee C, Cerveira E, Hwang J, Malhotra A, Plewczynski D, Radew K, Romanovitch M, Zhang C, Hyland FCL, Craig DW, Christoforides A, Homer N, Izatt T, Kurdoglu AA, Sinari SA, Squire K, Sherry ST, Xiao C, Sebat J, Antaki D, Gujral M, Noor A, Ye K, Burchard EG, Hernandez RD, Gignoux CR, Haussler D, Katzman SJ, James Kent W, Howie B, Ruiz-Linares A, Dermitzakis ET, Devine SE, Abecasis GR, Min Kang H, Kidd JM, Blackwell T, Caron S, Chen W, Emery S, Fritsche L, Fuchsberger C, Jun G, Li B, Lyons R, Scheller C, Sidore C, Song S, Sliwerska E, Taliun D, Tan A, Welch R, Kate Wing M, Zhan X, Awadalla P, Hodgkinson A, Li Y, Shi X, Quitadamo A, Lunter G, McVean GA, Marchini JL, Myers S, Churchhouse C, Delaneau O, Gupta-Hinch A, Kretzschmar W, Iqbal Z, Mathieson I, Menelaou A, Rimmer A, Xifara DK, Oleksyk TK, Fu Y, Liu X, Xiong M, Jorde L, Witherspoon D, Xing J, Eichler EE, Browning BL, Browning SR, Hormozdiari F, Sudmant PH, Khurana E, Durbin RM, Hurles ME, Tyler-Smith C, Albers CA, Ayub Q, Balasubramaniam S, Chen Y, Colonna V, Danecek P, Jostins L, Keane TM, McCarthy S, Walter K, Xue Y, Gerstein MB, Abyzov A, Balasubramanian S, Chen J, Clarke D, Fu Y, Harmanci AO, Jin M, Lee D, Liu J, Jasmine Mu X, Zhang J, Zhang Y, Li Y, Luo R, Zhu H, Alkan C, Dal E, Kahveci F, Marth GT, Garrison EP, Kural D, Lee W-P, Ward AN, Wu J, Zhang M, McCarroll SA, Handsaker RE, Altshuler DM, Banks E, del Angel G, Genovese G, Hartl C, Li H, Kashin S, Nemesh JC, Shakir K, Yoon SC, Lihm J, Makarov V, Degenhardt J, Korbel JO, Fritz MH, Meiers S, Raeder B, Rausch T, Stütz AM, Flicek P, Paolo Casale F, Clarke L, Smith RE, Stegle O, Zheng-Bradley X, Bentley DR, Barnes B, Keira Cheetham R, Eberle M, Humphray S, Kahn S, Murray L, Shaw R, Lameijer E-W, Batzer MA, Konkel MK, Walker JA, Ding L, Hall I, Ye K, Lacroute P, Lee C, Cerveira E, Malhotra A, Hwang J, Plewczynski D, Radew K, Romanovitch M, Zhang C, Craig DW, Homer N, Church D, Xiao C, Sebat J, Antaki D, Bafna V, Michaelson J, Ye K, Devine SE, Gardner EJ, Abecasis GR, Kidd JM, Mills RE, Dayama G, Emery S, Jun G, Shi X, Quitadamo A, Lunter G, McVean GA, Chen K, Fan X, Chong Z, Chen T, Witherspoon D, Xing J, Eichler EE, Chaisson MJ, Hormozdiari F, Huddleston J, Malig M, Nelson BJ, Sudmant PH, Parrish NF, Khurana E, Hurles ME, Blackburne B, Lindsay SJ, Ning Z, Walter K, Zhang Y, Gerstein MB, Abyzov A, Chen J, Clarke D, Lam H, Jasmine Mu X, Sisu C, Zhang J, Zhang Y, Gibbs RA, Yu F, Bainbridge M, Challis D, Evani US, Kovar C, Lu J, Muzny D, Nagaswamy U, Reid JG, Sabo A, Yu J, Guo X, Li W, Li Y, Wu R, Marth GT, Garrison EP, Fung Leong W, Ward AN, del Angel G, DePristo MA, Gabriel SB, Gupta N, Hartl C, Poplin RE, Clark AG, Rodriguez-Flores JL, Flicek P, Clarke L, Smith RE, Zheng-Bradley X, MacArthur DG, Mardis ER, Fulton R, Koboldt DC, Gravel S, Bustamante CD, Craig DW, Christoforides A, Homer N, Izatt T, Sherry ST, Xiao C, Dermitzakis ET, Abecasis GR, Min Kang H, McVean GA, Gerstein MB, Balasubramanian S, Habegger L, Yu H, Flicek P, Clarke L, Cunningham F, Dunham I, Zerbino D, Zheng-Bradley X, Lage K, Berg Jespersen J, Horn H, Montgomery SB, DeGorter MK, Khurana E, Tyler-Smith C, Chen Y, Colonna V, Xue Y, Gerstein MB, Balasubramanian S, Fu Y, Kim D, Auton A, Marcketta A, Desalle R, Narechania A, Wilson Sayres MA, Garrison EP, Handsaker RE, Kashin S, McCarroll SA, Rodriguez-Flores JL, Flicek P, Clarke L, Zheng-Bradley X, Erlich Y, Gymrek M, Frederick Willems T, Bustamante CD, Mendez FL, David Poznik G, Underhill PA, Lee C, Cerveira E, Malhotra A, Romanovitch M, Zhang C, Abecasis GR, Coin L, Shao H, Mittelman D, Tyler-Smith C, Ayub Q, Banerjee R, Cerezo M, Chen Y, Fitzgerald TW, Louzada S, Massaia A, McCarthy S, Ritchie GR, Xue Y, Yang F, Gibbs RA, Kovar C, Kalra D, Hale W, Muzny D, Reid JG, Wang J, Dan X, Guo X, Li G, Li Y, Ye C, Zheng X, Altshuler DM, Flicek P, Clarke L, Zheng-Bradley X, Bentley DR, Cox A, Humphray S, Kahn S, Sudbrak R, Albrecht MW, Lienhard M, Larson D, Craig DW, Izatt T, Kurdoglu AA, Sherry ST, Xiao C, Haussler D, Abecasis GR, McVean GA, Durbin RM, Balasubramaniam S, Keane TM, McCarthy S, Stalker J, Chakravarti A, Knoppers BM, Abecasis GR, Barnes KC, Beiswanger C, Burchard EG, Bustamante CD, Cai H, Cao H, Durbin RM, Gerry NP, Gharani N, Gibbs RA, Gignoux CR, Gravel S, Henn B, Jones D, Jorde L, Kaye JS, Keinan A, Kent A, Kerasidou A, Li Y, Mathias R, McVean GA, Moreno-Estrada A, Ossorio PN, Parker M, Resch AM, Rotimi CN, Royal CD, Sandoval K, Su Y, Sudbrak R, Tian Z, Tishkoff S, Toji LH, Tyler-Smith C, Via M, Wang Y, Yang H, Yang L, Zhu J, Bodmer W, Bedoya G, Ruiz-Linares A, Cai Z, Gao Y, Chu J, Peltonen L, Garcia-Montero A, Orfao A, Dutil J, Martinez-Cruzado JC, Oleksyk TK, Barnes KC, Mathias RA, Hennis A, Watson H, McKenzie C, Qadri F, LaRocque R, Sabeti PC, Zhu J, Deng X, Sabeti PC, Asogun D, Folarin O, Happi C, Omoniwa O, Stremlau M, Tariyal R, Jallow M, Sisay Joof F, Corrah T, Rockett K, Kwiatkowski D, Kooner J, Tịnh Hiêǹ T, Dunstan SJ, Thuy Hang N, Fonnie R, Garry R, Kanneh L, Moses L, Sabeti PC, Schieffelin J, Grant DS, Gallo C, Poletti G, Saleheen D, Rasheed A, Brooks LD, Felsenfeld AL, McEwen JE, Vaydylevich Y, Green ED, Duncanson A, Dunn M, Schloss JA, Wang J, Yang H, Auton A, Brooks LD, Durbin RM, Garrison EP, Min Kang H, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Kirov S, Snoddy J (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Duncan D, Shi Z, Zhang B (2013) WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B (2017) WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuznetsova A, Brockhoff PB, Christensen RHB (2017) lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw 82,. [Google Scholar]
- 33.R Core Team (2014) R: A Language and Environment for Statistical Computing. [Google Scholar]
- 34.Kuzma A, Valladares O, Cweibel R, Greenfest-Allen E, Childress DM, Malamon J, Gangadharan P, Zhao Y, Qu L, Leung YY, Naj AC, Stoeckert CJ, Schellenberg GD, Wang L-S (2016) NIAGADS: The NIA Genetics of Alzheimer’s Disease Data Storage Site. Alzheimer’s Dement. 12, 1200–1203. [Google Scholar]
- 35.Ernst J, Kellis M (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leung YY, Kuksa PP, Amlie-Wolf A, Valladares O, Ungar LH, Kannan S, Gregory BD, Wang L-S (2016) DASHR: Database of Small human noncoding RNAs. Nucleic Acids Res. 44,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuksa PP, Amlie-Wolf A, Katanić Ž, Valladares O, Wang L-S, Leung YY (2018) DASHR 2.0: integrated database of human small non-coding RNA genes and mature products. Bioinformatics 0–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Femminella GD, Ferrara N, Rengo G (2015) The emerging role of microRNAs in Alzheimer’s disease. Front. Physiol. 6, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shabalin AA (2012) Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V (2014) Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 10,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulger M, Groudine M (2011) Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engreitz JM, Ollikainen N, Guttman M (2016) Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. [DOI] [PubMed] [Google Scholar]
- 47.Chiba T, Yamada M, Sasabe J, Terashita K, Shimoda M, Matsuoka M, Aiso S (2009) Amyloid-beta causes memory impairment by disturbing the JAK2/STAT3 axis in hippocampal neurons. Mol. Psychiatry 14, 206–222. [DOI] [PubMed] [Google Scholar]
- 48.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, Maharjan S, Nguyen T, Ma L, Malphrus KG, Palusak R, Lincoln S, Bisceglio G, Georgescu C, Kouri N, Kolbert CP, Jen J, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N (2012) Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet 8,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujita Y, Yamaguchi A, Hata K, Endo M, Yamaguchi N, Yamashita T (2009) Zyxin is a novel interacting partner for SIRT1. BMC Cell Biol. 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanni C, Necchi D, Pinto A, Buoso E, Buizza L, Memo M, Uberti D, Govoni S, Racchi M (2013) Zyxin is a novel target for beta-amyloid peptide: Characterization of its role in Alzheimer’s pathogenesis. J. Neurochem. 125, 790–799. [DOI] [PubMed] [Google Scholar]
- 51.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE (2013) Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, Estus S (2013) CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 33, 13320–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, The Alzheimer Disease Neuroimaging Initiative, Morris MC, Evans DA, Johnson K, Sperling RA, Schneider JA, Bennett DA, De Jager PL (2013) CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 16, 848–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lawson LJ, Perry VH, Dri P, Gordon S (1990) Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170. [DOI] [PubMed] [Google Scholar]
- 55.Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen K V, Geller F, Sottejeau Y, Harold D, Dourlen P, Grenier-Boley B, Kamatani Y, Delepine B, Demiautte F, Zelenika D, Zommer N, Hamdane M, Bellenguez C, Dartigues J-F, Hauw J-J, Letronne F, Ayral A-M, Sleegers K, Schellens A, Broeck L V, Engelborghs S, De Deyn PP, Vandenberghe R, O’Donovan M, Owen M, Epelbaum J, Mercken M, Karran E, Bantscheff M, Drewes G, Joberty G, Campion D, Octave J-N, Berr C, Lathrop M, Callaerts P, Mann D, Williams J, Buée L, Dewachter I, Van Broeckhoven C, Amouyel P, Moechars D, Dermaut B, Lambert J-C (2013) Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 18, 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM, Glass CK (2011) Serum Response Factor Utilizes Distinct Promoter-and Enhancer-Based Mechanisms To Regulate Cytoskeletal Gene Expression in Macrophages. Mol. Cell. Biol. 31, 861–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potthoff MJ, Olson EN (2007) MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140. [DOI] [PubMed] [Google Scholar]
- 58.Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, Corthals G, Cordwell S, Daly RJ (2003) A cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278, 21805–21813. [DOI] [PubMed] [Google Scholar]
- 59.Yong J, Tan I, Lim L, Leung T (2006) Phosphorylation of myosin phosphatase targeting subunit 3 (MYPT3) and regulation of protein phosphatase 1 by protein kinase A. J. Biol. Chem. 281, 31202–31211. [DOI] [PubMed] [Google Scholar]
- 60.Wadsworth TL, Bishop J a, Pappu AS, Woltjer RL, Quinn JF (2008) Evaluation of coenzyme Q as an antioxidant strategy for Alzheimer’s disease. J. Alzheimer’s Dis. 14, 225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Chen K, Sloan S a, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow S a, Zhang C, Daneman R, Maniatis T, Barres B a, Wu JQ (2014) An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci. 34, 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note 1: Guidelines to prioritize the causal variants for further mechanistic studies
Supplementary Table 1: Luciferase enhancer regions (Excel file)
Supplementary Table 2: microRNA seed sequence disruptions by INFERNO-expanded variants (Excel file).
Supplementary Table 3: Full colocalization annotation results. Top signals in each region are highlighted. (Excel file)
Supplementary Table 4: Summary of colocalization results in 6 non-prioritized regions. (Excel file).
Supplementary Table 5. Highly correlated genes with RP11-385F7.1. (Excel table)
Flowchart of luciferase-based enhancer validation assays.
Number of enhancer overlaps across individual tissues from FANTOM5 and Roadmap. a) Number of variants overlapping eRNA-defined enhancers across 112 FANTOM5 tissue and cell type facets. b) Number of variants overlapping each type of ChromHMM-defined enhancer state across 127 Roadmap tissues and cell types.
HOMER motif overlap ΔPWM distributions
Number of enhancer overlaps across tissue categories sampled in FANTOM5 and Roadmap. a) Number of variants overlapping eRNA-defined enhancers in each tag region across the 28 tissue categories that include FANTOM5 samples. b) Number of variants overlapping any of the three ChromHMM-defined enhancer states in each tag region across the 22 tissue categories that include Roadmap samples.
Colocalization analysis of GTEx eQTLs with IGAP GWAS signals. a) Distributions of the 5 colocalization hypotheses across all tissues and tag regions. b) Histograms of P(H4) for highly colocalized (P(H4) >= 0.5) signals across tag regions. c) Histograms of the approximate Bayes factor values of the most supported variants across tag regions. d) Histograms of the number of variants required to cumulatively account for 50% of the individual variant probability (ABFs) for each colocalization signal across tag regions.