Abstract
Respiratory syncytial virus (RSV) is the most common cause of lower respiratory tract infection and hospitalization in infants. It is well-established that both CD4+ and CD8+ T cells are critical for mediating viral clearance but also contribute to the induction of immunopathology following RSV infection. C57BL/6 mice are often utilized to study T cell responses following RSV infection given the wide variety of genetically modified animals available. To date, few RSV-derived CD4+ and CD8+ T cell epitopes have been identified in C57BL/6 mice. Utilizing an overlapping peptide library spanning the entire RSV proteome, intracellular cytokine staining for IFN-γ was performed to identify novel CD4+ and CD8+ T cell epitopes in C57BL/6 mice. We identified two novel CD4+ T cell epitopes and three novel CD8+ T cell epitopes located within multiple RSV proteins. Additionally, we characterized the newly described T cell epitopes by determining their TCR Vβ expression profiles and MHC-restriction. Overall, the novel RSV-derived CD4+ and CD8+ T cell epitopes identified in C57BL/6 mice will aid in future studies of RSV-specific T cell responses.
Introduction
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infection in infants and young children (1). RSV represents a substantial healthcare burden worldwide, causing approximately three-four million hospitalizations and 200,000 deaths annually (1). However, despite the immense burden of RSV-associated disease, there remains no licensed RSV vaccine. The peak of disease following RSV infection coincides with the onset of the host immune response, including the expansion of RSV-specific T cells (2). Studies in mouse models of RSV infection have clearly established that both CD4+ and CD8+ T cells contribute to RSV-associated disease, despite their vital role in mediating viral clearance (3). T cells have also been associated with enhanced viral control and exacerbated disease following RSV infection in humans (4–7). Therefore, both CD4+ and CD8+ T cells play a critical role in determining the severity of disease during RSV infection in both mice and humans.
C57BL/6 mice are susceptible to RSV infection and are often utilized as an animal model for RSV. Given the wide variety of genetically modified mice that are available on the H-2b genetic background, C57BL/6 mice are frequently used for studying RSV-specific T cells. Several T cell epitopes have been defined in C57BL/6 mice using prediction algorithms and peptide library screening techniques. Three RSV-derived CD4+ T cell epitopes in the RSV matrix (M), attachment glycoprotein (G), and M2–1 proteins (G168–185, M209–223, and M2–126–39) have been previously described (8–10). CD8+ T cell epitopes against RSV in C57BL/6 mice have also been identified, including the immunodominant epitope within the RSV M protein, M187–195 (11). Several additional subdominant CD8+ T cell epitopes have also been described within the RSV nucleoprotein (N), fusion glycoprotein (F), and G proteins (N57–64, N360–368, F250–258, F433–442, and G177–188). Therefore, known CD4+ or CD8+ T cell epitopes in C57BL/6 mice are located within the RSV M, G, N, F, and M2–1 proteins. However, the remaining RSV proteins lack defined RSV-derived T cell epitopes in C57BL/6 mice.
In this study, we utilized a peptide library spanning the entire RSV proteome to identify novel RSV-derived CD4+ and CD8+ T cell epitopes in C57BL/6 mice. We discovered two novel CD4+ T cell epitopes and three novel CD8+ T cell epitopes against RSV in C57BL/6 mice. Additionally, we defined the TCR Vβ profiles and the MHC-restriction of the newly identified T cell epitopes. Overall, the discovery of these novel CD4+ and CD8+ T cell epitopes will provide valuable tools for the study of RSV-specific T cell responses in C57BL/6 mice.
Materials and Methods
Mice and infection
Female C57BL/6 mice between 6–8 weeks of age were purchased from National Cancer Institute (Frederick, MD). The A2 strain of RSV (RSV-A2) was a gift from Dr. Barney Graham (National Institutes of Health, Bethesda, MD) and was propagated on HEp-2 cells (ATCC). Mice were infected intranasally with 1.6–2.5 × 106 PFU RSV-A2. The A/Puerto Rico/8/34 strain of influenza (IAV) was a gift from Dr. Kevin Legge (University of Iowa). Mice were infected intranasally with a 0.1 LD50 dose of IAV. All experimental procedures utilizing mice were approved by the University of Iowa Animal Care and Use Committee. The experiments were performed under strict accordance to the Office of Laboratory Animal Welfare guidelines and the PHS Policy on Humane Care and Use of Laboratory Animals.
Peptides and epitope mapping
A peptide library spanning the entire RSV-A2 proteome consisting of 889 peptides (15-mer overlapping every 10 amino acids) was utilized to screen for novel T cell epitopes (Mimotopes Pty. Ltd., Roseville, MN). For initial screening, the software program Deconvolute This (Version 2.0; courtesy of Dr. Mario Roederer, Vaccine Research Center, NIAID, NIH) was used to generate a set of peptide pools, such that each library peptide was represented in two unique pools of 10 peptides each (12). Deconvolute This software was utilized to identify potential peptides contributing to a positive response, which was defined as being ≥ 3-SD over the average of no peptide controls in both unique pools in both of two independent experiments. The identified potential epitopes were screened individually, and peptides eliciting a response ≥ 3-SD above the average of no peptide controls in both of two independent experiments were considered confirmed epitopes. CD8+ T cell minimal core epitopes were predicted using the immune epitope database (IEDB) analysis resource consensus tool (13), which combines predictions from artificial neural networks, also known as NetMHC (14–16), stabilized matrix method (17), and comblib (18). 9-mer peptides of the predicted CD8+ T cell minimal core epitopes were synthesized for epitope confirmation (Biosynthesis, Lewisville, TX).
Ex vivo peptide stimulation
Lungs were harvested from RSV-infected mice on day 8 post-infection (p.i.), and single-cell suspensions were generated as previously described (19, 20). Cells were stimulated with either peptide pools or individual peptides at 1 μM per peptide for 5 hrs at 37°C in the presence of 10 μg/ml brefeldin A (BFA; Sigma-Aldrich) in 10% FCS-supplemented RPMI. In certain experiments, the peptide concentrations were titrated from 10 μM to 0.000001 μM per well. No peptide controls were incubated with 10 μg/ml BFA in 10% FCS-supplemented RPMI in the absence of peptides for 5 hrs at 37°C.
MHC class I restriction assay
Parent P815 cells not expressing H-2b MHC class I molecules and P815 cells stably expressing H-2Db or H-2Kb MHC class I molecules were a gift from Dr. John Harty (University of Iowa). P815 cell lines were pulsed with 1 μM peptide at 37°C for 1 hour and washed twice. CD8+ T cells were isolated from the lungs using anti-CD8a microbeads (Miltenyi Biotec, Auburn, CA) and pre-sorted via positive selection on a MidiMACS separator (Miltenyi Biotec). Positively selected cells were stained extracellularly with a monoclonal antibody specific to CD8 (clone 53–6.7) and purified populations of CD8+ T cells were sorted using a FACSAria (BD Biosciences). Sort-purified CD8+ T cells were stimulated with either peptide-pulsed P815 cells or unpulsed P815 cells at a 2:1 effector-to-stimulator ratio for 5 hrs at 37°C in the presence of 10 μg/ml BFA in 10% FCS-supplemented RPMI.
Flow cytometry analysis
Following stimulation, cells were stained extracellularly with monoclonal antibodies specific to CD90.2 (BioLegend; clone 53–2.1), CD4 (clone RM4–5), CD8 (clone 53–6.7), and CD11a (clone M17/4) in FACS buffer (PBS, 2% FCS, 0.02% sodium azide) for 30 mins at 4°C. For TCR Vβ usage analysis, cells were stained with one antibody from a 15 antibody TCR Vβ staining panel (BD Biosciences). Background staining was detected using fluorescence minus one controls and subtracted from the frequency of Vβ+ T cells. After extracellular staining, cells were fixed using 1-step Fix/Lyse Solution for 10 mins at room temperature (eBioscience). Cells were stained intracellularly for IFN-γ (clone XMG1.2) in FACS buffer containing 0.5% saponin (Sigma-Aldrich) for 30 mins at 4°C. All antibodies were purchased from eBioscience unless otherwise indicated. Samples were run on a BD LSRFortessa and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). Cell types were gated as follows: CD4+ T cells (CD90.2+CD4+) and CD8+ T cells (CD90.2+CD8+).
Statistical analysis
All statistical analyses were performed using Prism software (GraphPad Software, San Diego, CA). Data were evaluated using Student’s t test for two groups or two-way ANOVA with Tukey’s post-test for more than two groups as indicated in the figure legends to determine if there was a statistical significance of at least α = 0.05.
Results
Identification of candidate CD4+ T cell epitopes
An 889 15-mer peptide library, overlapping every 10 amino acids, spanning the entire RSV proteome was utilized to identify novel RSV T cell epitopes in C57BL/6 mice (21). To increase assay efficiency and reduce the number of animals required, library peptides were divided into peptide pools using Deconvolute This software for initial screening (12). Pools containing 10 peptides were created such that each library peptide was included within two unique pools for 179 total pools. Lung cells from day 8 RSV-infected C57BL/6 mice were stimulated with the peptide pools, and CD4+ T cell reactivity was measured by intracellular cytokine staining (ICS) for IFN-γ. A positive pool was defined as having results ≥ 3-SD above the average of no peptide controls.
To validate our experimental approach, we determined whether the previously defined CD4+ T cell epitopes in C57BL/6 mice, M209–223 and M2–126–39, and pools containing their corresponding peptides within the peptide library elicited a positive response (9). Both M209–223 and M2–126–39 CD4+ T cell epitopes stimulated responses exceeding the 3-SD threshold (Fig. 1A). Importantly, all pools containing their corresponding library peptides, M211–225 (pools 65 and 144) and M2–126–40 (pools 36 and 150), also produced positive responses above the 3-SD threshold (Fig. 1). These results indicate that the peptide pooling approach is sufficient to identify potential CD4+ T cell epitopes. Stimulation with the 179 peptide pools resulted in a total of 17 positive pools exceeding the 3-SD threshold for positivity (Fig. 1B). Individual peptides potentially contributing to a positive response were detected using the Deconvolute This software and defined as peptides contained in two positive pools in both of two independent experiments. A total of 16 individual peptides within the pools were identified as candidate CD4+ T cell epitopes for further analysis.
Figure 1. RSV peptide library screen for candidate CD4+ T cell epitopes.
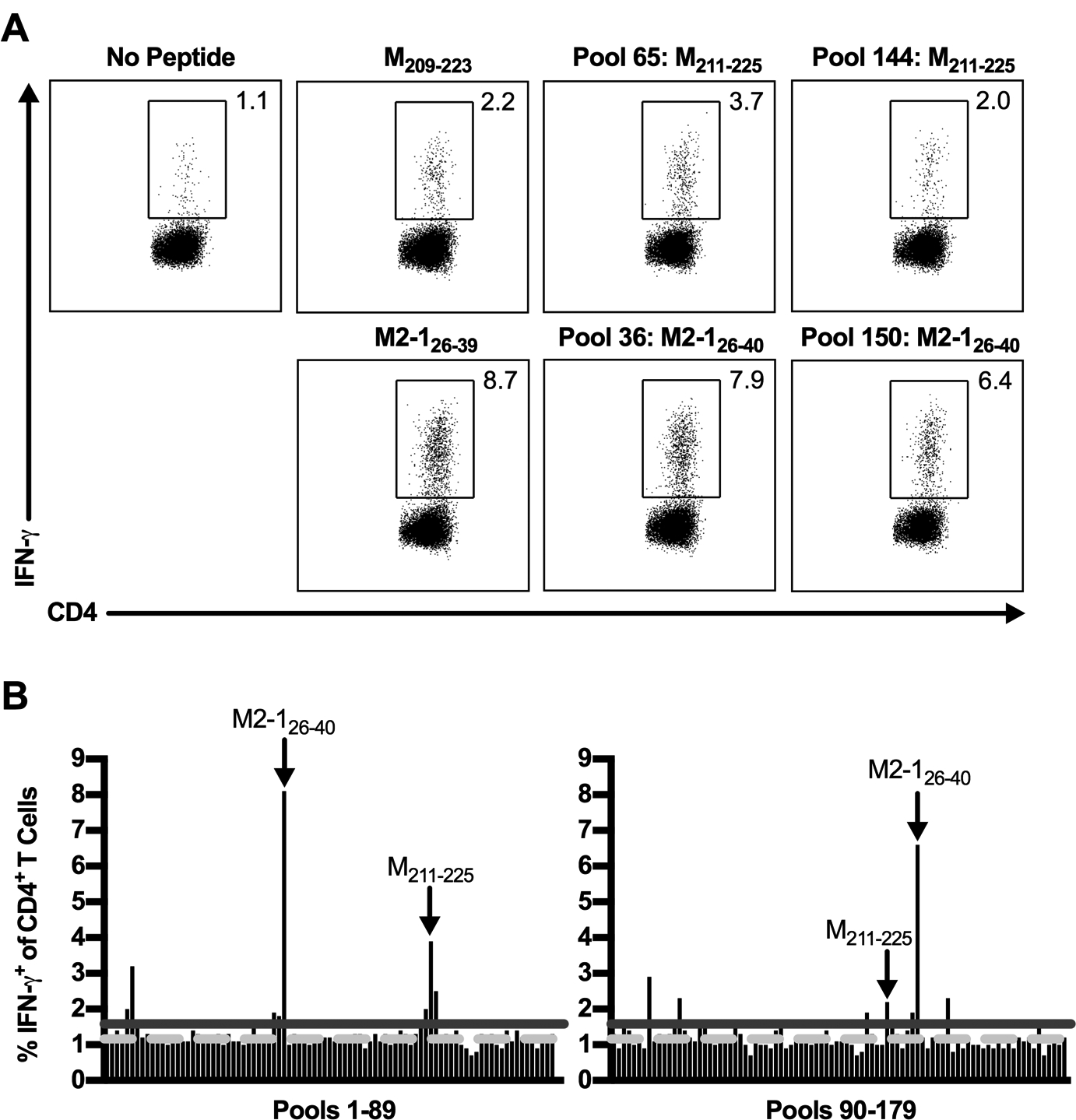
C57BL/6 mice were infected with RSV, and lungs were harvested on day 8 p.i. Lung cells were either incubated with BFA alone or stimulated with a pool containing 10 individual RSV-derived peptides from a peptide library spanning the entire RSV proteome. (A) Dot plots show the frequency of IFN-γ+ CD4+ T cells elicited following peptide stimulation with previously defined CD4+ T cell epitopes, M209–223 and M2–126–39, or pools containing their corresponding peptides in the peptide library, M211–225 and M2–126–40. (B) The frequency of IFN-γ+ CD4+ T cells induced by peptide stimulation with all peptide pools. The dashed light gray line represents the average of no peptide controls (y=1.16), and the solid dark gray line represents the 3-SD threshold for positivity (y=1.58). Arrows indicate pools containing the previously defined M209–223 and M2–126–39 CD4+ T cell epitopes. Data are representative of 1 of 2 experiments with 24 mice pooled per experiment.
Confirmation of novel CD4+ T cell epitopes
To confirm the CD4+ T cell epitope candidates, lung cells from RSV-infected C57BL/6 mice were stimulated with each candidate peptide individually and assessed for IFN-γ production by ICS. A total of four candidate peptides elicited a frequency of IFN-γ+ CD4+ T cells that exceeded the 3-SD threshold for positivity (Fig. 2A). Two of the identified CD4+ T cell epitopes, M211–225 and M2–126–40, contain the previously defined epitopes M209–223 and M2–126–39. Two novel RSV CD4+ T cell epitopes in C57BL/6 mice were identified: M166–180 and N346–360. Importantly, CD4+ T cells harvested from either naive or IAV-infected C57BL/6 mice did not produce IFN-γ above background levels following stimulation with M166–180 and N346–360 peptides (Fig. 2B, 2C). These results confirm that M166–180 and N346–360 are true RSV-derived CD4+ T cell epitopes, as cross-recognition by non-RSV-specific CD4+ T cells did not occur. Both M166–180 and N346–360 epitopes are H2-I-Ab MHC class II-restricted, as C57BL/6 mice lack expression of the mature H2-I-Eb MHC class II molecule (22).
Figure 2. Identification of novel RSV CD4+ T cell epitopes.
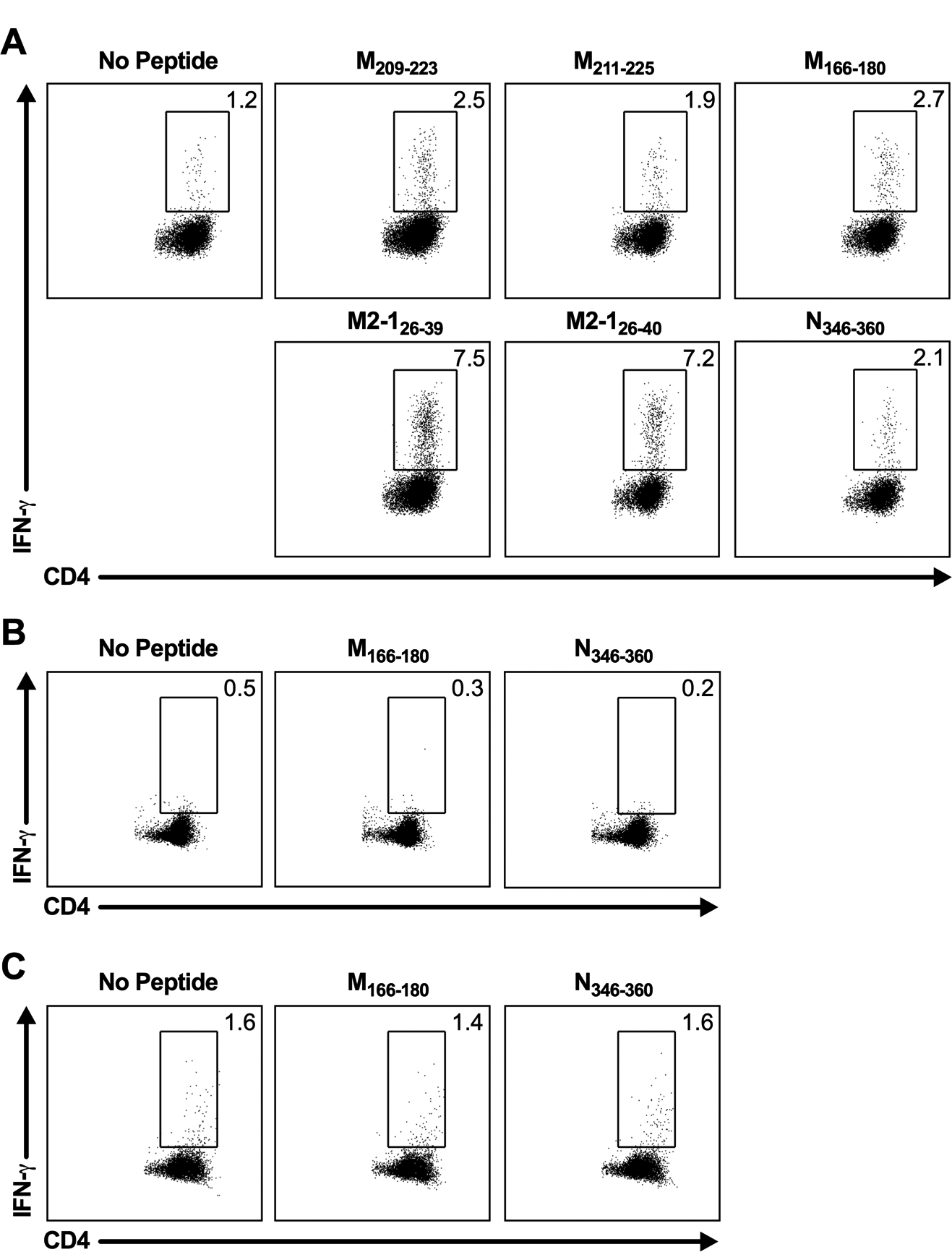
(A) C57BL/6 mice were infected with RSV, and lungs were harvested on day 8 p.i. Lung cells were either incubated with BFA alone or stimulated with individual candidate CD4+ T cell epitopes. Dot plots show the frequency of IFN-γ+ CD4+ T cells elicited by the previously defined CD4+ T cell epitopes, M209–223 and M2–126–39, and the individual peptides that are ≥ the 3-SD threshold for positivity, M166–180 and N346–360. (B/C) Lung cells from (B) naive C57BL/6 mice or (C) day 8 IAV-infected C57BL/6 mice were either incubated with BFA alone or stimulated with M166–180 or N346–360 peptides and stained intracellularly for IFN-γ. Data are representative of 1 of 2 experiments with (A) 30 mice pooled per experiment or (B/C) 4 mice per group per experiment.
Because we have previously found that CD4+ T cells that recognize the RSV I-Ed-restricted T cell epitope G183–195 predominately express TCR Vβ14, we evaluated the CD4+ T cell repertoire of the novel CD4+ T cell epitopes by staining with a panel of TCR Vβ-specific antibodies (23, 24). As expected, lung CD4+ T cells from naive C57BL/6 mice exhibit a polyclonal TCR repertoire (Fig. 3A). Vβ8.1/8.2 is the TCR Vβ chain most highly expressed by naive CD4+ T cells. Vβ4, Vβ6, and Vβ14 are represented at comparable levels with smaller contributions from the other Vβ chains. Similar to naive CD4+ T cells, M166–180-specific CD4+ T cells exhibit a polyclonal TCR repertoire in which Vβ8.1/8.2 is most highly represented, followed by smaller frequencies of Vβ3-, Vβ6-, and Vβ8.3-expressing cells (Fig. 3B). Vβ8.1/8.2 is also highly represented in the N346–360-specific CD4+ T cell TCR repertoire, followed closely by a population of cells expressing Vβ6 (Fig. 3C). These results are similar to the TCR repertoires of previously defined epitopes M209–223 and M2–126–39, which are also highly represented by Vβ8.1/8.2-expressing cells (25).
Figure 3. TCR Vβ usage of novel CD4+ T cell epitopes.
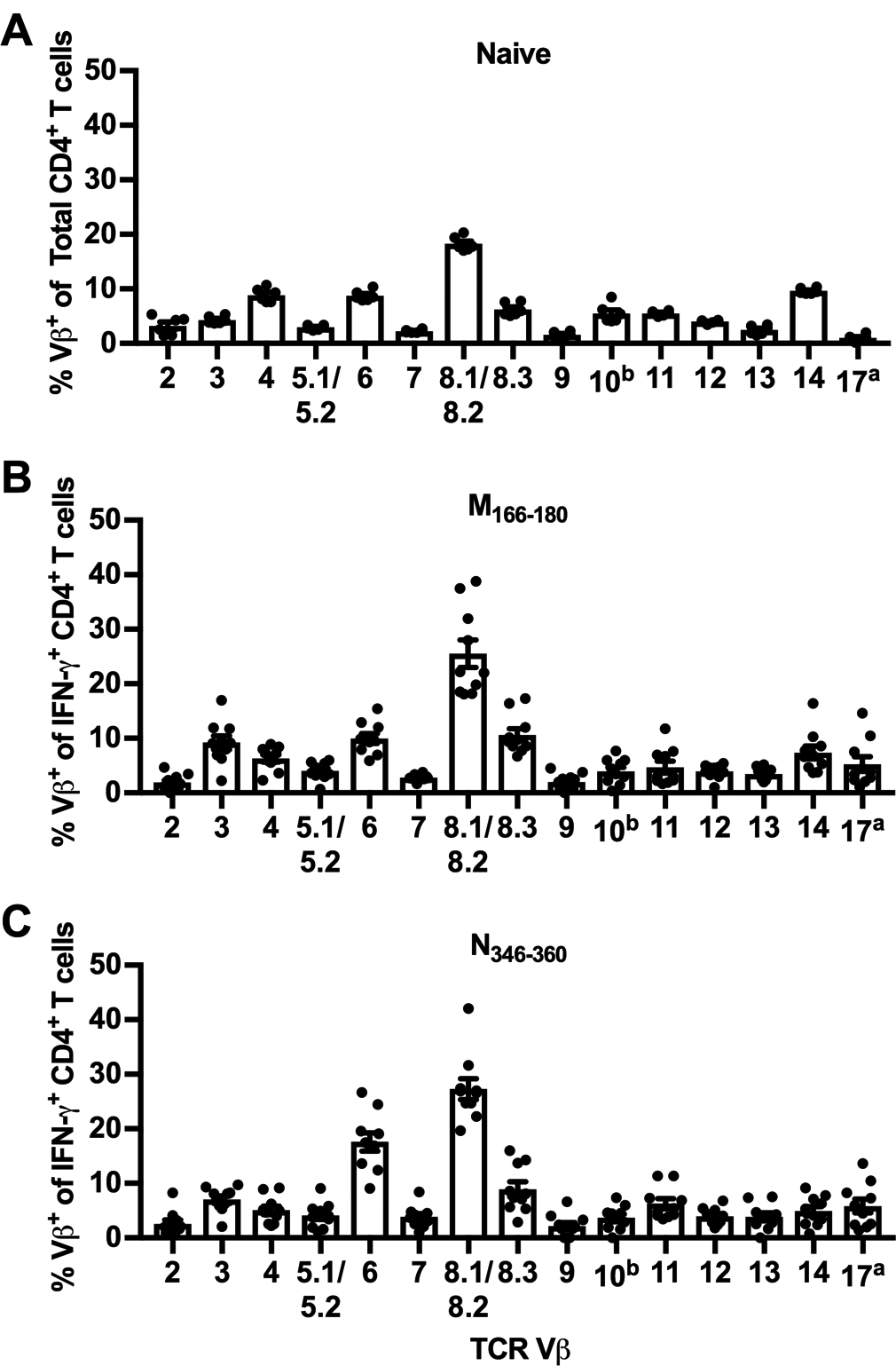
(A) TCR Vβ usage of CD4+ T cells in the lungs of naive C57BL/6 mice. (B/C) C57BL/6 mice were infected with RSV, and lungs were harvested on day 8 p.i. Lung cells were stimulated with M166–180 or N346–360 peptides, stained extracellularly with a panel of TCR Vβ-specific antibodies, and stained intracellularly for IFN-γ. Lung cells were gated on IFN-γ+ CD4+ T cells and graphs show the TCR Vβ usage of (B) M166–180- and (C) N346–360-specific CD4+ T cells. Data are presented as mean ± SEM of 2–3 independent experiments. Each dot represents an individual mouse (n=6–10).
Identification of candidate CD8+ T cell epitopes
We used a similar strategy as outlined for the CD4+ T cells to identify candidate RSV CD8+ T cell epitopes in C57BL/6 mice. Similar to the CD4+ T cell epitope screen, positive pools were defined as those having CD8+ T cell IFN-γ responses ≥ 3-SD above the average of no peptide controls. To verify the approach for the identification of CD8+ T cell epitopes, we evaluated whether pools containing the peptides encompassing the previously defined immunodominant CD8+ T cell epitope in C57BL/6 mice, M187–195, elicited positive responses (11). As expected, M187–195 stimulated robust CD8+ T cell responses well above the 3-SD threshold (Fig. 4A). Similarly, all pools containing its corresponding library peptides, M181–195 (pools 65 and 116) and M186–200 (pools 5 and 108), exhibited positive IFN-γ responses above the 3-SD threshold (Fig. 4). Stimulation with the 179 peptide pools resulted in a total of 127 positive pools meeting the 3-SD threshold for positivity, much greater than the number of positive responses observed during the CD4+ T cell epitope screen (Fig. 4B). Following this initial screening, Deconvolute This software identified 510 individual peptides potentially contributing to the positive responses. To reduce the overall number of peptides requiring individual testing, a second peptide pool screen (204 total pools of 5 peptides each) for CD8+ T cell reactivity was performed yielding 155 positive pools (data not shown). Deconvolution of this response identified a total of 352 candidate CD8+ T cell epitopes for individual screening.
Figure 4. RSV peptide library screen for candidate CD8+ T cell epitopes.
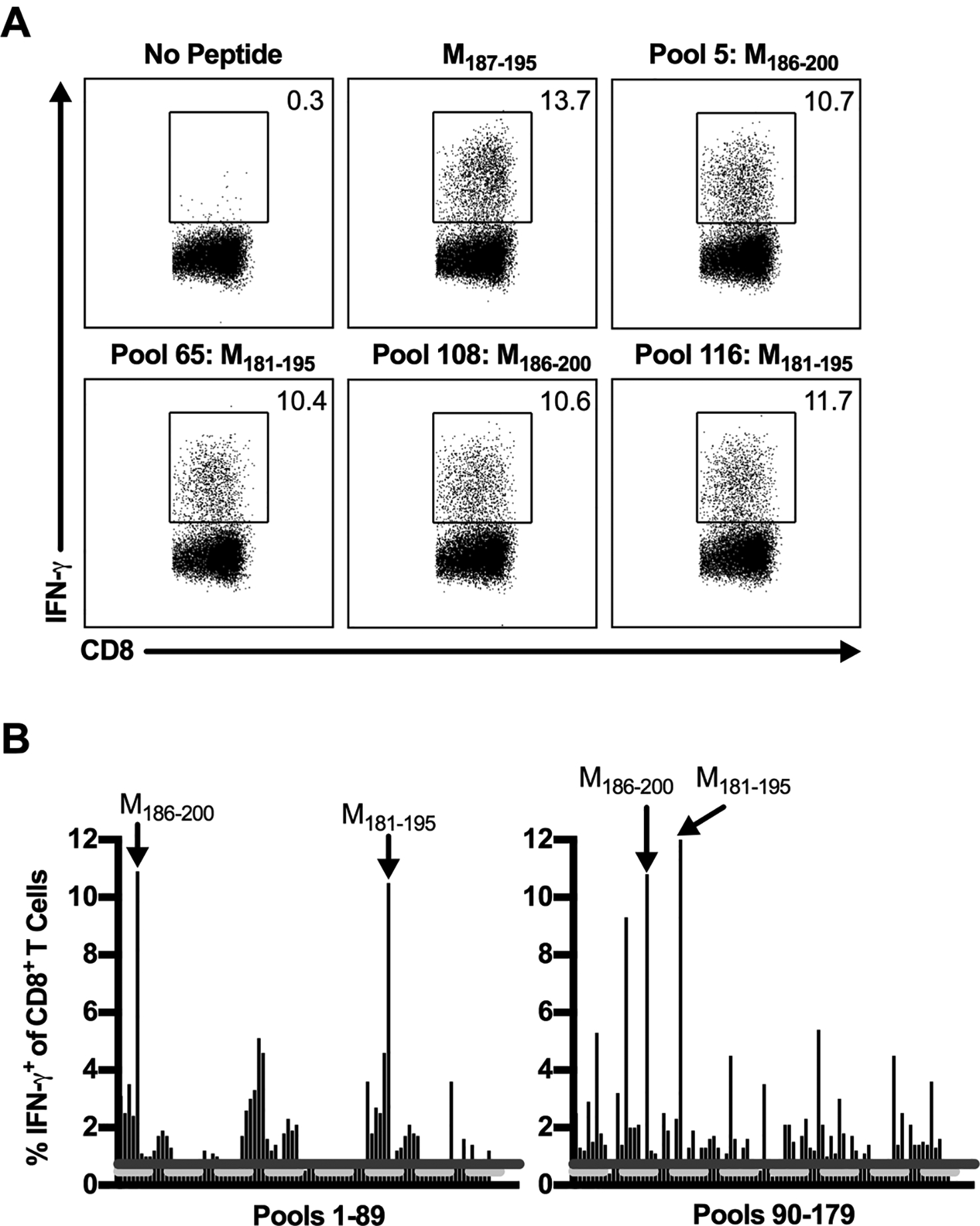
C57BL/6 mice were infected with RSV, and lungs were harvested on day 8 p.i. Lung cells were either incubated with BFA alone or stimulated with a pool containing 10 individual RSV-derived peptides from a peptide library spanning the entire RSV proteome. (A) Dot plots show the frequency of IFN-γ+ CD8+ T cells elicited following peptide stimulation with the previously defined CD8+ T cell epitope, M187–195, or pools containing its corresponding peptides in the peptide library, M181–195 and M186–200. (B) The frequency of IFN-γ+ CD8+ T cells induced by peptide stimulation with all peptide pools. The dashed light gray line represents the average of no peptide controls (y=0.47), and the solid dark gray line represents the 3-SD threshold for positivity (y=0.73). Arrows indicate pools containing the previously defined M187–195 CD8+ T cell epitope. Data are representative of 1 of 2 experiments with 24 mice pooled per experiment.
Confirmation and evaluation of novel CD8+ T cell epitopes
To confirm the CD8+ T cell epitope candidates, IFN-γ production following stimulation of lung cells with each of the 352 candidate peptides individually was evaluated by ICS. A total of 72 candidate peptides elicited a positive IFN-γ response above the 3-SD threshold in both of two individual experiments (Fig. 5A, Supplemental Table 1). Included within the positive candidate peptides are three peptides containing either the entire or partial peptide sequence of the previously defined CD8+ T cell epitope M187–195 (Fig. 5, Supplemental Table 1). In total, 69 candidate peptides were identified as potential novel CD8+ T cell epitopes, although the overall number of CD8+ T cell epitopes is likely fewer given the overlapping nature of the peptide library utilized (Supplementary Table 1).
Figure 5. Identification of novel RSV CD8+ T cell epitopes.
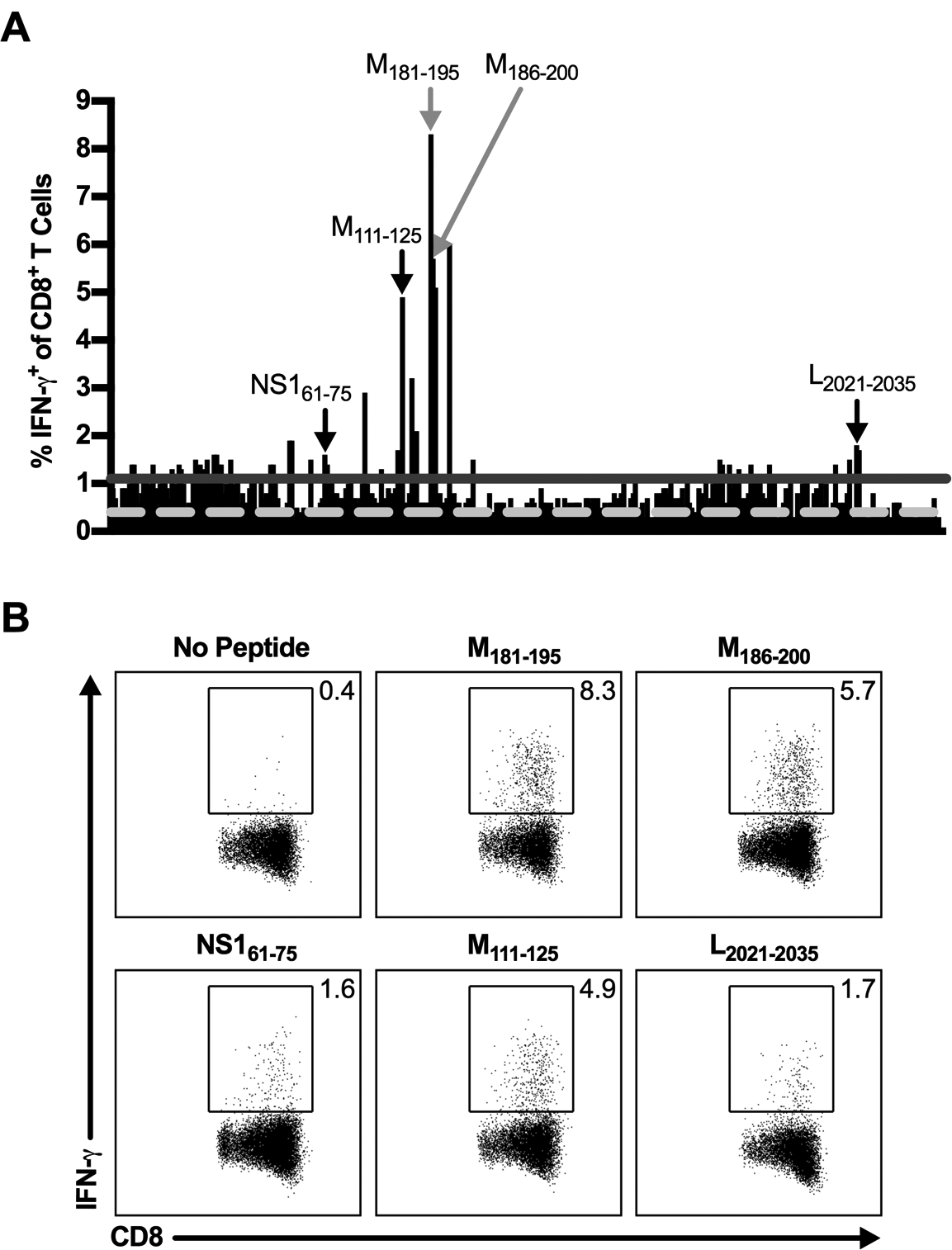
C57BL/6 mice were infected with RSV, and lungs were harvested on day 8 p.i. Lung cells were either incubated with BFA alone or stimulated with individual candidate CD8+ T cell epitopes. (A) The frequency of IFN-γ+ CD8+ T cells induced by peptide stimulation with individual candidate CD8+ T cell epitopes. The dashed light gray line represents the average of no peptide controls (y=0.40), and the solid dark gray line represents the 3-SD threshold for positivity (y=1.10). Gray arrows indicate library peptides containing the previously defined M187–195 CD8+ T cell epitope. Black arrows indicate select CD8+ T cell epitopes utilized for further study. (B) Dot plots show the frequency of IFN-γ+ CD8+ T cells elicited by select individual peptides that are ≥ the 3-SD threshold for positivity. Data are representative of 1 of 2 experiments with 30 mice pooled per experiment.
Of the 69 potential novel CD8+ T cell epitopes identified, we selected three CD8+ T cell epitopes to confirm and utilize for further study: NS161–75, M111–125, and L2021–2035 (Fig. 5). We selected NS161–75 and L2021–2035 because no H-2b-restricted CD8+ T cell epitopes are currently known within the RSV NS1 and L proteins. M111–125 was chosen to identify a potential subdominant CD8+ T cell epitope within the RSV M protein to evaluate in addition to the immunodominant CD8+ T cell epitope, M187–195. To confirm these novel CD8+ T cell epitopes, we first utilized cell lines expressing H-2Db and H-2Kb MHC class I molecules to determine their MHC class I-restriction. Sort-purified CD8+ T cells isolated from the lungs of RSV-infected C57BL/6 mice were stimulated with peptide-pulsed parent P815 cells or P815 cells expressing either H-2Db or H-2Kb and assessed for IFN-γ production by ICS. As previously described, the M187–195 CD8+ T cell epitope is H-2Db MHC class I-restricted (Fig. 6A) (11). Similarly, the three newly identified epitopes NS161–75, M111–125, and L2021–2035 are all also H-2Db MHC class I-restricted (Fig. 6A). Importantly, CD8+ T cells did not produce IFN-γ following stimulation with P815 cells that were not pulsed with peptides, confirming that they are responding specifically to the RSV-derived peptides bound to the H-2Db MHC class I molecule (Fig. 6A). After determining the MHC restriction of the newly identified CD8+ T cell epitopes, we predicted their minimal core CD8+ T cell epitope sequence using the IEDB analysis resource (13). The IEDB resource identified the minimal 9-mer sequences with the highest percentile rank of NS161–75, M111–125, and L2021–2035 epitopes to be NS165–73 (ICPNNNIVV), M112–120 (IKACSLTCL), and L2021–2029 (NVVQNAKLI), respectively. Peptides for the three core minimal epitopes were synthesized and used to stimulate lung cells for confirmation. NS165–73, M112–120, and L2021–2029 minimal core epitopes all elicited strong IFN-γ production by CD8+ T cells (Fig. 6B). Additionally, the vast majority of IFN-γ+ CD8+ T cells express high levels of the integrin CD11a after peptide stimulation, indicating that they are antigen-experienced cells (Fig. 6B) (26). To confirm that the 9-mer minimal core epitope sequences were recognized at enhanced or similar levels as the 15-mer library peptides, the peptide concentration used for stimulation was titrated from 101 to 10−6 μM (Fig. 6C). NS165–73 and L2021–2029 stimulation exhibited increased IFN-γ+ CD8+ T cells compared to stimulation with their 15-mer peptide library counterparts down to 10−4 μM, indicating that they are strongly recognized (Fig. 6C). In contrast, M112–120 stimulation resulted in similar levels of IFN-γ+ CD8+ T cells as stimulation with its 15-mer peptide library counterpart at all peptide concentrations (Fig. 6C). Three additional peptides within the M111–125 15-mer library peptide sequence were identified by the IEDB resource as having medium-high percentile rankings, M113–121, M115–123, and M117–125, but did not elicit IFN-γ production following stimulation at any peptide concentration (data not shown). Therefore, given the consistent and robust results obtained with M112–120, it is highly likely that M112–120 is the confirmed minimal CD8+ T cell epitope within the M111–125 sequence. Similar to the novel CD4+ T cell epitopes, stimulation with NS165–73, M112–120, and L2021–2029 did not induce IFN-γ production above background levels by CD8+ T cells harvested from either naive or IAV-infected C57BL/6 mice, confirming that they are bona-fide RSV-derived CD8+ T cell epitopes (Fig. 6D, 6E).
Figure 6. Confirmation of novel RSV CD8+ T cell epitopes.
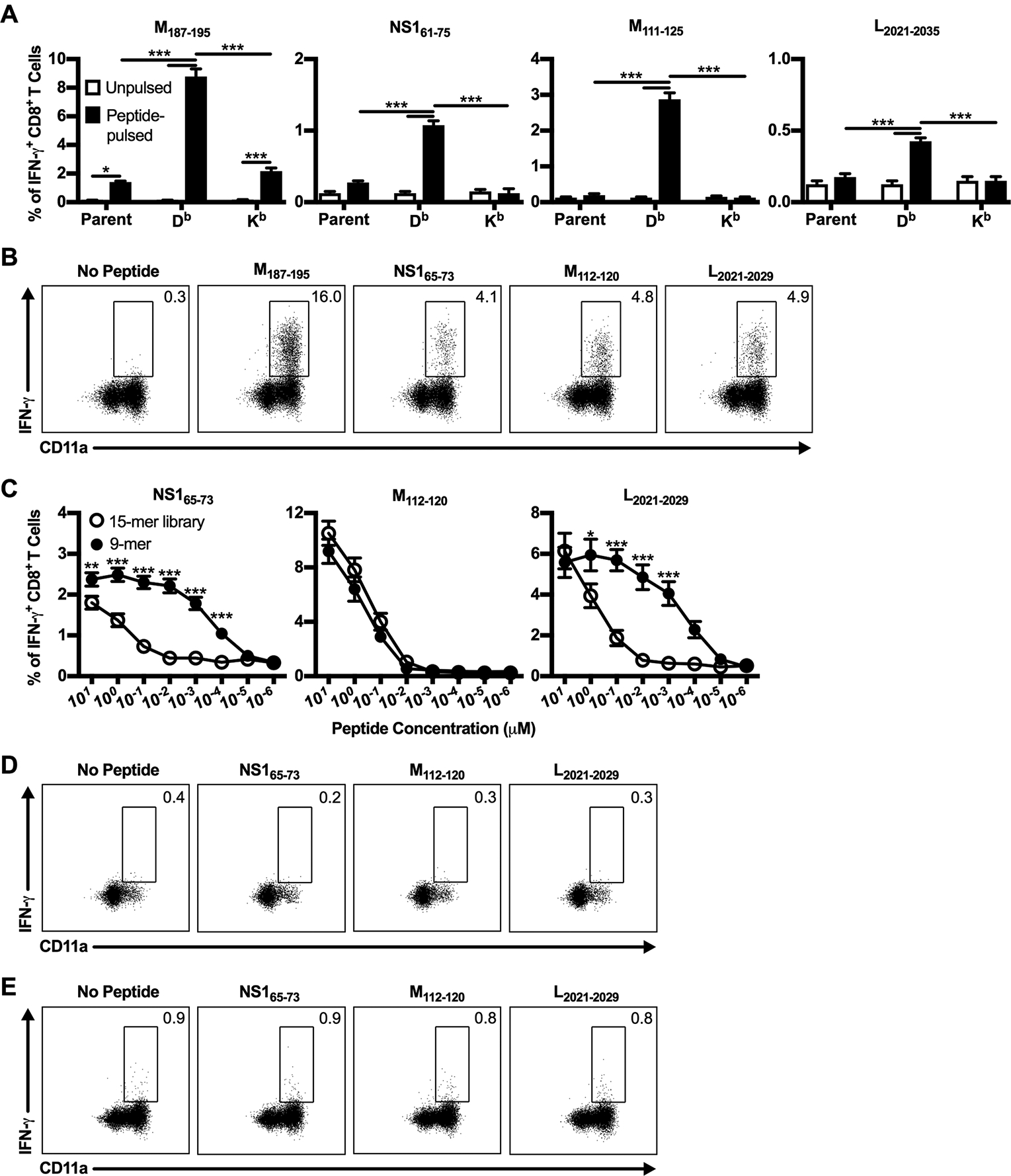
(A-C) C57BL/6 mice were infected with RSV, and lungs were harvested on day 8 p.i. (A) Sort-purified CD8+ T cells were stimulated with either unpulsed (open bars) or peptide-pulsed (closed bars) MHC class I-expressing P815 cell lines at a 2:1 effector-to-stimulator ratio and stained intracellularly for IFN-γ. Parent indicates untransfected P815 cells, and Db and Kb indicate P815 cells transfected with H-2Db and H-2Kb MHC class I molecules, respectively. Data are presented as mean ± SEM of 1 of 2 independent experiments (n=4). Groups were compared using two-way ANOVA with Tukey’s post-test. (B) Lung cells were either incubated with BFA alone or stimulated with CD8+ T cell minimal core epitope 9-mer peptides at a 1 μM concentration and stained intracellularly for IFN-γ. Dot plots are previously gated on CD8+ T cells and show the frequency of IFN-γ+ CD8+ T cells that are CD11ahi. Data are representative of 1 of 4 experiments (n=4–6). (C) Lung cells were stimulated with either 15-mer library peptides (open circles) or CD8+ T cell minimal core epitope 9-mer peptides (closed circles) at the indicated peptide concentrations and stained intracellularly for IFN-γ. Data are presented as mean ± SEM of 2 independent experiments (n=8). Groups were compared using Student’s t test. (D/E) Lung cells from (D) naive C57BL/6 mice or (E) day 8 IAV-infected C57BL/6 mice were either incubated with BFA alone or stimulated with CD8+ T cell minimal core epitope 9-mer peptides at a 1 μM concentration and stained intracellularly for IFN-γ. Data are representative of 1 of 2 experiments (n=4). * p<0.05, ** p<0.01, *** p<0.001.
We also determined the epitope-specific CD8+ T cell repertoire of the novel CD8+ T cell epitopes by staining with a panel of TCR Vβ-specific antibodies. Similar to naive CD4+ T cells, CD8+ T cells from the lungs of naive C57BL/6 mice exhibit a polyclonal TCR repertoire in which Vβ8.1/8.2 is most highly expressed, followed closely by Vβ5.1/5.2 (Fig. 7A). Vβ6, Vβ7, Vβ8.3, Vβ10, Vβ11, and Vβ13 are also well-represented in naive CD8+ T cells within the lung. NS165–73-specific CD8+ T cells also express a polyclonal TCR repertoire, but the expression of individual Vβ chains are highly variable between mice, unlike in naive CD8+ T cells (Fig. 7B). Both M112–120 and L2021–2029 exhibit highly skewed CD8+ T cell repertoires that are dominated by Vβ9 with smaller contributions made by Vβ6 and Vβ8.1/8.2 for M112–120 and Vβ13 for L2021–2029 (Fig. 7C–D). The oligoclonal TCR repertoires of M112–120 and L2021–2029 are similar to that of the previously described CD8+ T cell epitope M187–195, which also predominately expresses Vβ9 following RSV infection (27).
Figure 7. TCR Vβ usage of novel CD8+ T cell epitopes.
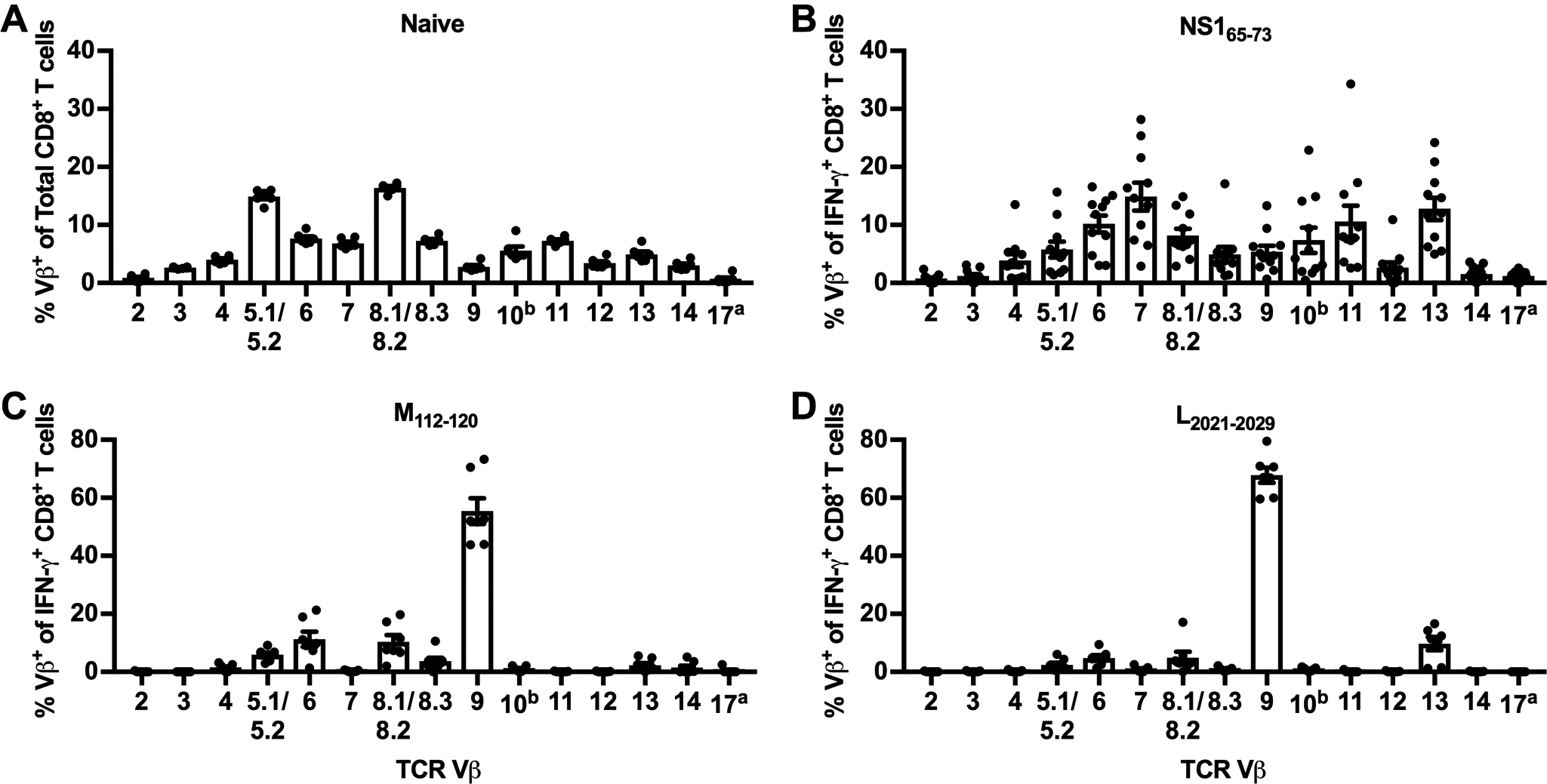
(A) TCR Vβ usage of CD8+ T cells in the lungs of naive C57BL/6 mice. (B/C) C57BL/6 mice were infected with RSV, and lungs were harvested on day 8 p.i. Lung cells were stimulated with NS165–73, M112–120, or L2021–2029 peptides, stained extracellularly with a panel of TCR Vβ-specific antibodies, and stained intracellularly for IFN-γ. Lung cells were gated on IFN-γ+ CD8+ T cells and graphs show the TCR Vβ usage of (B) NS165–73-, (C) M112–120-, and (D) L2021–2029-specific IFN-γ+ CD8+ T cells. Data are presented as mean ± SEM of 2–3 independent experiments. Each dot represents an individual mouse (n=6–11).
In addition to the NS165–73, M112–120, and L2021–2029 CD8+ T cell epitopes identified, we attempted to confirm novel CD8+ T cell epitopes within eight additional RSV library peptides that elicited positive IFN-γ responses in the individual candidate peptide screen (Fig. 5A). Four 15-mer library peptides within the RSV M protein were evaluated for potential CD8+ T cell epitopes, three of which exhibited large IFN-γ production (>3%) in the candidate screen (Fig. 5A). Additionally, one 15-mer peptide within each of the RSV NS1, NS2, P, and L proteins were also tested for CD8+ T cell epitopes. However, we were not able to confirm RSV-derived CD8+ T cell epitopes within these other library peptides, as stimulation with predicted 9-mer amino acid sequences resulted in low IFN-γ production similar to background levels (data not shown). Alternatively, stimulation with multiple predicted 9-mer peptide sequences elicited IFN-γ production by CD8+ T cells above background levels. However, the responses observed were either significantly reduced compared to stimulation with the 15-mer library peptides or titration of the peptide concentration resulted in an immediate decline in IFN-γ production that was similar to background levels (data not shown). These results may help explain the large number of positive candidate peptides identified in the individual CD8+ T cell epitope screen (Fig. 5A). Therefore, while additional novel CD8+ T cell epitopes are likely present within the library peptides that were positive in the individual screen, only NS165–73, M112–120, and L2021–2029 have been confirmed to date.
Discussion
In this study, we utilized an overlapping peptide library spanning the entire RSV proteome to identify novel RSV-derived CD4+ and CD8+ T cell epitopes in C57BL/6 mice. The initial screening was performed using a peptide pooling approach, as has been described in the screening of T cell epitopes for a variety of other viruses (28–32). The peptide pooling approach was validated by confirming that pools containing library peptides that correspond to previously defined CD4+ and CD8+ T cell epitopes exhibited IFN-γ production above the 3-SD threshold for positivity. Library peptides containing the most widely used CD4+ T cell epitopes M209–223 and M2–126–39 and the immunodominant CD8+ T cell epitope M187–195 elicited positive responses, validating this experimental approach. In contrast, pools containing library peptides that represented the amino acid sequences of less widely utilized T cell epitopes against RSV in C57BL/6 mice did not produce positive responses, including the CD4+ T cell epitope G168–185 and the CD8+ T cell epitopes N57–64, N360–368, F250–258, F433–442, and G177–188. The differential ability for defined CD4+ and CD8+ T cell epitopes to induce positive IFN-γ responses within peptide pools may be due to a variety of factors. Given that there are multiple peptides in each pool, it is possible that peptides not corresponding to RSV T cell epitopes bind with a high affinity to the MHC molecule, preventing other peptides within the pool from attaching to the MHC molecule’s peptide binding groove. Alternatively, the 15-mer peptides utilized may not be the optimal size for eliciting a T cell response in vitro. We have previously shown that the use of 17-mer peptides induced an increased frequency of responding CD4+ T cells from RSV-infected BALB/c mice than 15-mer peptides (21). Therefore, it is likely that 17-mer peptides may also induce IFN-γ production from C57BL/6 CD4+ T cells more optimally, which may explain why the G168–185 CD4+ T cell epitope was negative in our initial screening. Additionally, the 15-mer peptides may not always be cleaved in the correct place to produce the proper amino acid sequence corresponding to a CD8+ T cell epitope that is typically 8–10 amino acids in length. Thus, the peptide library approach we have used here is likely an underestimate of the number of total T cell epitopes.
By screening individual library peptides, we identified two novel RSV-derived CD4+ T cell epitopes in C57BL/6 mice: M166–180 and N346–360. M166–180 provides an additional CD4+ T cell epitope within the RSV M protein that is of similar size to the previously defined M209–223 CD4+ T cell epitope. N346–360 represents the first CD4+ T cell epitope in C57BL/6 mice within the RSV N protein. Additionally, we confirmed three novel CD8+ T cell epitopes against RSV in C57BL/6 mice: NS165–73, M112–120, and L2021–2029. NS165–73 is the first RSV-derived T cell epitope to be described within the RSV NS1 protein in any mouse model. NS1 and NS2 are the first and most abundant viral gene products synthesized following RSV infection (33). Given the abundance of NS1 and NS2 proteins within a host cell, it is interesting that there is only a single defined murine T cell epitope within NS1 and none within NS2. Similarly, L2021–2029 is the first murine CD8+ T cell epitope identified within the RSV L protein. Lastly, M112–120 provides an additional subdominant CD8+ T cell epitope in C57BL/6 mice within the RSV M protein to study alongside the immunodominant CD8+ T cell epitope, M187–195.
In addition to confirming the novel CD4+ and CD8+ T cell epitopes, we characterized their TCR Vβ expression profiles. Lung CD4+ T cells from naive C57BL/6 mice express a polyclonal TCR Vβ repertoire in which Vβ8.1/8.2 is the most highly expressed. Similar to the naive CD4+ T cell repertoire, the previously defined CD4+ T cell epitopes M209–223 and M2–126–39 exhibit dominant responses by cells expressing Vβ8.1/8.2 (25). The novel CD4+ T cell epitopes M166–180 and N346–360 also exhibit polyclonal TCR Vβ repertoires in which Vβ8.1/8.2 is the most highly represented. Overall, RSV infection results in the expansion of CD4+ T cells of multiple specificities that predominately express TCR Vβ8.1/8.2. This is in contrast to the Vβ repertoire of the I-Ed-restricted T cell epitope G183–195 in BALB/c mice, which predominately express TCR Vβ14 (23, 24). The naive CD8+ T cell Vβ repertoire in the lung of C57BL/6 mice is also polyclonal with Vβ8.1/8.2 and Vβ5.1/5.2 expressed with the highest frequency. This is a similar pattern to what was previously observed on splenic naive CD8+ T cells in C57BL/6 mice (27). In contrast to the C57BL/6 CD4+ T cell epitopes which highly express Vβ8.1/8.2, Vβ9 is most highly represented on CD8+ T cells specific to the previously described CD8+ T cell epitope M187–195 (27). Similarly, the TCR Vβ usage of M112–120 and L2021–2029 CD8+ T cell epitopes is dominated by Vβ9 with very small frequencies of epitope-specific CD8+ T cells expressing other TCR Vβ chains. In contrast, NS165–73-specific CD8+ T cells exhibit a polyclonal Vβ repertoire that is highly variable between mice. It is particularly interesting that the Vβ repertoire for NS165–73-specific CD8+ T cells is highly variable, in contrast to other RSV-derived CD8+ T cell epitopes which do not exhibit TCR Vβ repertoire variability between individual mice (Figure 7) (27, 34). It is possible that the precursor frequency of NS165–73-specific CD8+ T cells may not be identical between individual C57BL/6 mice. Alternatively, the proliferation capacity of certain NS165–73-specific CD8+ T clones may be different between individual C57BL/6 mice. The mechanism underlying the variability of the NS165–73-specific CD8+ T cell Vβ repertoire is an interesting topic for future study. Overall, the TCR Vβ repertoire of RSV-specific T cells tends to skew towards Vβ8.1/8.2 for CD4+ T cells and Vβ9 for CD8+ T cells, with the exception of the CD8+ T cell epitope NS165–73.
In summary, we have identified two novel RSV-derived CD4+ T cell epitopes, M166–180 and N346–360, and three novel RSV-derived CD8+ T cell epitopes, NS165–73, M112–120, and L2021–2029, in C57BL/6 mice. We have also characterized the TCR Vβ expression profile as well as the MHC-restriction of each new T cell epitope described. In addition to utilizing these epitopes to perform ex vivo peptide stimulation, the characterization of their TCR Vβ expression profiles and MHC-restriction provides information that could aid in the generation of transgenic T cell lines and peptide-MHC tetramers to probe antigen-specific responses following RSV infection. While these novel T cell epitopes will be useful in future studies of C57BL/6 mice, they will also be important for the study of T cells in the CB6F1 model, a cross between C57BL/6 and BALB/c mice, which is also frequently used as a murine model for RSV infection (27, 34–40). Overall, the discovery of these novel T cell epitopes in C57BL/6 mice will provide valuable tools for further understanding RSV-specific T cell responses.
Supplementary Material
Acknowledgments
We thank Stacey Hartwig for excellent technical assistance and Justin Fishbaugh, Heath Vignes, and Michael Shey of the University of Iowa Flow Cytometry Facility for assistance with cell sorting.
Some of the data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine / Holden Comprehensive Cancer Center core research facility at the University of Iowa. The facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, Iowa City Veteran’s Administration Medical Center, and the National Center for Research Resources of the National Institutes of Health under Award Number 1 S10 OD016199-01A1. This work was supported by the Department of Microbiology and Immunology at the University of Iowa (to SMV) and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R01AI124093 (to SMV) and T32AI007485 (to MES). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used in this article:
- RSV
respiratory syncytial virus
- IAV
influenza
- IEDB
immune epitope database
- p.i.
post-infection
- BFA
brefeldin A
- ICS
intracellular cytokine staining
References
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, and Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375: 1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Openshaw PJ, and Tregoning JS. 2005. Immune responses and disease enhancement during respiratory syncytial virus infection. Clin Microbiol Rev 18: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham BS, Bunton LA, Wright PF, and Karzon DT. 1991. Role of T lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. J Clin Invest 88: 1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Saleeby CM, Suzich J, Conley ME, and DeVincenzo JP. 2004. Quantitative effects of palivizumab and donor-derived T cells on chronic respiratory syncytial virus infection, lung disease, and fusion glycoprotein amino acid sequences in a patient before and after bone marrow transplantation. Clin Infect Dis 39: e17–20. [DOI] [PubMed] [Google Scholar]
- 5.Fishaut M, Tubergen D, and McIntosh K. 1980. Cellular response to respiratory viruses with particular reference to children with disorders of cell-mediated immunity. J Pediatr 96: 179–186. [DOI] [PubMed] [Google Scholar]
- 6.Hall CB, Powell KR, MacDonald NE, Gala CL, Menegus ME, Suffin SC, and Cohen HJ. 1986. Respiratory syncytial viral infection in children with compromised immune function. N Engl J Med 315: 77–81. [DOI] [PubMed] [Google Scholar]
- 7.Jozwik A, Habibi MS, Paras A, Zhu J, Guvenel A, Dhariwal J, Almond M, Wong EH, Sykes A, Maybeno M, Del Rosario J, Trujillo-Torralbo MB, Mallia P, Sidney J, Peters B, Kon OM, Sette A, Johnston SL, Openshaw PJ, and Chiu C. 2015. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat Commun 6: 10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock GE, Tebbey PW, Scheuer CA, Pryharski KS, Heers KM, and LaPierre NA. 2003. Immune responses to the nonglycosylated ectodomain of respiratory syncytial virus attachment glycoprotein mediate pulmonary eosinophilia in inbred strains of mice with different MHC haplotypes. J Med Virol 70: 301–308. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Ruckwardt TJ, Chen M, Johnson TR, and Graham BS. 2009. Characterization of respiratory syncytial virus M- and M2-specific CD4 T cells in a murine model. J Virol 83: 4934–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srikiatkhachorn A, Chang W, and Braciale TJ. 1999. Induction of Th-1 and Th-2 responses by respiratory syncytial virus attachment glycoprotein is epitope and major histocompatibility complex independent. J Virol 73: 6590–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutigliano JA, Rock MT, Johnson AK, Crowe JE Jr., and Graham BS. 2005. Identification of an H-2D(b)-restricted CD8+ cytotoxic T lymphocyte epitope in the matrix protein of respiratory syncytial virus. Virology 337: 335–343. [DOI] [PubMed] [Google Scholar]
- 12.Roederer M, and Koup RA. 2003. Optimized determination of T cell epitope responses. J Immunol Methods 274: 221–228. [DOI] [PubMed] [Google Scholar]
- 13.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, and Peters B. 2015. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43: D405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreatta M, and Nielsen M. 2016. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32: 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, and Nielsen M. 2008. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36: W509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, and Lund O. 2003. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters B, and Sette A. 2005. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics 6: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, and Peters B. 2008. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulton RB, Olson MR, and Varga SM. 2008. Regulation of cytokine production by virus-specific CD8 T cells in the lungs. J Virol 82: 7799–7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss KA, Christiaansen AF, Fulton RB, Meyerholz DK, and Varga SM. 2011. Multiple CD4+ T cell subsets produce immunomodulatory IL-10 during respiratory syncytial virus infection. J Immunol 187: 3145–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDermott DS, Knudson CJ, and Varga SM. 2014. Determining the breadth of the respiratory syncytial virus-specific T cell response. J Virol 88: 3135–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PP, Murphy DB, and McDevitt HO. 1981. Variable synthesis and expression of E alpha and Ae (E beta) Ia polypeptide chains in mice of different H-2 haplotypes. Immunogenetics 12: 321–337. [DOI] [PubMed] [Google Scholar]
- 23.Varga SM, Wissinger EL, and Braciale TJ. 2000. The attachment (G) glycoprotein of respiratory syncytial virus contains a single immunodominant epitope that elicits both Th1 and Th2 CD4+ T cell responses. J Immunol 165: 6487–6495. [DOI] [PubMed] [Google Scholar]
- 24.Varga SM, Wang X, Welsh RM, and Braciale TJ. 2001. Immunopathology in RSV infection is mediated by a discrete oligoclonal subset of antigen-specific CD4(+) T cells. Immunity 15: 637–646. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Cao S, Peppers G, Kim SH, and Graham BS. 2014. Clonotype-specific avidity influences the dynamics and hierarchy of virus-specific regulatory and effector CD4(+) T-cell responses. Eur J Immunol 44: 1058–1068. [DOI] [PubMed] [Google Scholar]
- 26.Rai D, Pham NL, Harty JT, and Badovinac VP. 2009. Tracking the total CD8 T cell response to infection reveals substantial discordance in magnitude and kinetics between inbred and outbred hosts. J Immunol 183: 7672–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Billam P, Bonaparte KL, Liu J, Ruckwardt TJ, Chen M, Ryder AB, Wang R, Dash P, Thomas PG, and Graham BS. 2011. T Cell receptor clonotype influences epitope hierarchy in the CD8+ T cell response to respiratory syncytial virus infection. J Biol Chem 286: 4829–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy RB, and Poland GA. 2010. The identification of HLA class II-restricted T cell epitopes to vaccinia virus membrane proteins. Virology 408: 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee LY, Ha do LA, Simmons C, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend AR, Askonas BA, Rowland-Jones S, and Dong T. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest 118: 3478–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leligdowicz A, Yindom LM, Onyango C, Sarge-Njie R, Alabi A, Cotten M, Vincent T, da Costa C, Aaby P, Jaye A, Dong T, McMichael A, Whittle H, and Rowland-Jones S. 2007. Robust Gag-specific T cell responses characterize viremia control in HIV-2 infection. J Clin Invest 117: 3067–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Precopio ML, Butterfield TR, Casazza JP, Little SJ, Richman DD, Koup RA, and Roederer M. 2008. Optimizing peptide matrices for identifying T-cell antigens. Cytometry A 73: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richmond M, McKinnon LR, Kiazyk SA, Wachihi C, Kimani M, Kimani J, Plummer FA, and Ball TB. 2011. Epitope mapping of HIV-specific CD8+ T cell responses by multiple immunological readouts reveals distinct specificities defined by function. J Virol 85: 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins PL, and Wertz GW. 1983. cDNA cloning and transcriptional mapping of nine polyadenylylated RNAs encoded by the genome of human respiratory syncytial virus. Proc Natl Acad Sci U S A 80: 3208–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruckwardt TJ, Malloy AM, Gostick E, Price DA, Dash P, McClaren JL, Thomas PG, and Graham BS. 2011. Neonatal CD8 T-cell hierarchy is distinct from adults and is influenced by intrinsic T cell properties in respiratory syncytial virus infected mice. PLoS Pathog 7: e1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Haddad EK, Marceau J, Morabito KM, Rao SS, Filali-Mouhim A, Sekaly RP, and Graham BS. 2016. A Numerically Subdominant CD8 T Cell Response to Matrix Protein of Respiratory Syncytial Virus Controls Infection with Limited Immunopathology. PLoS Pathog 12: e1005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malloy AM, Ruckwardt TJ, Morabito KM, Lau-Kilby AW, and Graham BS. 2017. Pulmonary Dendritic Cell Subsets Shape the Respiratory Syncytial Virus-Specific CD8+ T Cell Immunodominance Hierarchy in Neonates. J Immunol 198: 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morabito KM, Erez N, Graham BS, and Ruckwardt TJ. 2016. Phenotype and Hierarchy of Two Transgenic T Cell Lines Targeting the Respiratory Syncytial Virus KdM282–90 Epitope Is Transfer Dose-Dependent. PLoS One 11: e0146781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruckwardt TJ, Bonaparte KL, Nason MC, and Graham BS. 2009. Regulatory T cells promote early influx of CD8+ T cells in the lungs of respiratory syncytial virus-infected mice and diminish immunodominance disparities. J Virol 83: 3019–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruckwardt TJ, Luongo C, Malloy AM, Liu J, Chen M, Collins PL, and Graham BS. 2010. Responses against a subdominant CD8+ T cell epitope protect against immunopathology caused by a dominant epitope. J Immunol 185: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruckwardt TJ, Malloy AM, Morabito KM, and Graham BS. 2014. Quantitative and qualitative deficits in neonatal lung-migratory dendritic cells impact the generation of the CD8+ T cell response. PLoS Pathog 10: e1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


