Abstract
The pathogenesis of Salmonella Typhimurium depends on the bacterium’s ability to survive and replicate within host cells. The formation and maintenance of a unique membrane-bound compartment, termed the Salmonella-containing vacuole (SCV), is essential for S. Typhimurium pathogenesis. SCV-bound S. Typhimurium induces formation of filamentous tubules that radiate outwards from the SCV, termed Salmonella-induced filaments (SIFs). SIF formation is concomitant with the onset of replication within host epithelial cells. SIF biogenesis, formation and maintenance of the SCV, and the intracellular positioning of the SCV within the host cell requires translocation of bacterial proteins (effectors) into the host cell. Effectors secreted by the type III secretion system encoded on Salmonella pathogenicity island 2 (T3SS2) function to interfere with host cellular processes and promote both intracellular survival and replication of S. Typhimurium. Seven T3SS2-secreted effectors, SifA, SopD2, PipB2, SteA, SseJ, SseF, and SseG have previously been implicated to play complementary, redundant, and/or antagonistic roles with respect to SIF biogenesis, intracellular positioning of the SCV, and SCV membrane dynamics modulation during infection. We undertook a systematic study to delineate the contribution of each effector to these processes by (i) deleting all seven of these effectors in a single S. Typhimurium strain; and (ii) deleting combinations of multiple effectors based on putative effector function. Using this deletion mutant library, we show that each of SIF biogenesis, intracellular SCV localization, intramacrophage replication, colonization, and virulence depends on the activities of multiple effectors. Together, our data demonstrates the complex interplay between these seven effectors and highlights the necessity to study T3SS2-secreted effectors as groups, rather than studies of individual effectors.
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a Gram-negative foodborne pathogen that commonly causes non-typhoidal salmonellosis (gastroenteritis) in humans. The success of Salmonella as an intracellular pathogen is largely due to its ability to evade host defense mechanisms by invading and residing within intestinal epithelial cells and phagocytic cells of the host [1,2]. Upon invasion of the host cell, S. Typhimurium resides within a unique compartment called the Salmonella-containing vacuole (SCV). Formation and maintenance of the SCV during infection is critical to survival within phagocytes and plays an important role in promoting S. Typhimurium replication in non-phagocytic cells [3].
The SCV, while a unique compartment distinct from host organelles, initially exhibits features similar to maturing endosomes. Upon invasion of the host cell, the nascent SCV (early SCV) carries the same membrane markers that characterize early endosomes such as EEA1 and transferrin receptor [4]. Subsequent SCV maturation occurs through a series of controlled selective sequential interactions with the host’s endocytic pathway [4,5]. During maturation, the SCV—like late endosomes—accumulates lysosomal glycoproteins such as lysosomal associated membrane proteins (LAMP) 1 and 2 within the SCV membrane and the SCV lumen acidifies. However, unlike late endosomes, the SCV does not fully mature into a lysosome owing to manipulation of the host cell by intravacuolar S. Typhimurium [6–9].
S. Typhimurium possesses two distinct type III secretion systems (T3SSs) encoded on Salmonella pathogenicity islands 1 and 2 (T3SS1 and T3SS2 respectively). These two T3SSs, along with additional virulence factors, allow S. Typhimurium to invade, survive, and replicate within host cells [2]. Whereas the T3SS1-secreted effectors are primarily associated with facilitating invasion of non-phagocytic cells and initial formation of the early SCV, the T3SS2-secreted effectors generally function to promote replication within both phagocytic and non-phagocytic cells [10–12]. The T3SS2-secreted effectors exert a wide variety of functions during infection including, but not limited to, maintaining the SCV membrane, regulating intracellular SCV positioning, and forming the membranous filament-like extensions that radiate outwards from the SCV, termed Salmonella-induced filaments (SIFs) [13]. SIF biogenesis begins at 4–6 hours post-infection, concomitant with the onset of intercellular bacterial replication in human epithelial cells [14–16]. SIFs are thought to play a number of important roles during infection including nutrient acquisition from the host and cell-to-cell transfer [17–20].
A subset of seven T3SS2-secreted effectors have been shown to play a role in SIF biogenesis, intracellular positioning of the SCV, and in controlling SCV membrane dynamics. These effectors of interest include: SifA, SseF, SseG, SteA, PipB2, SopD2, and SseJ [10,11,21–23]. The single deletion mutant of each of these seven effectors results in attenuation of virulence in the mouse model of systemic infection [13,22,24–27] and all but PipB2 contribute to survival in mouse macrophages [10]. These data highlight the importance of these effectors in both in vitro and in vivo infection models. The precise function of the seven effectors of interest is known for some but unclear for others, and their contribution to formation of the intracellular replication niche remains ambiguous. Each of SifA, SseF, SseG, SteA, PipB2, SopD2, and SseJ contribute to at least one, if not several of the following roles during infection: SIF biogenesis, precise intracellular positioning of the SCV, SCV membrane stability, SCV membrane modification, microtubule recruitment, and/or regulation of microtubule motor activity at the SCV membrane. The effectors’ overlapping roles during infection make it difficult to determine precise effector function when studying a single effector at a time. Increasing evidence suggests that T3SS2-secreted effectors cooperate to facilitate the interaction of S. Typhimurium with host cell machinery, leading to events such as SIF biogenesis and SCV movement [28–31].
In this study, we systematically constructed a S. Typhimurium SL1344-based strain that lacks all seven of our effectors of interest, as well as multiple effector deletion combinations. We show that LAMP1+-tubule (SIF) extension is not exclusively driven by SifA, but rather, likely requires the activity of other effectors. We also demonstrate that LAMP1+-tubule extension, intracellular positioning, intramacrophage replication, and replication in vivo all require the action of multiple effectors. One effector alone does not solely mediate a single process.
Results
Construction of multi-effector deletion mutants
Through an extensive literature search we identified seven effectors of interest implicated in SIF biogenesis, SCV membrane maintenance, and intracellular SCV localization. These effectors include SseF, SseG, SteA, PipB2, SopD2, SseJ, and SifA (summarized in [32]). In order to address the redundancy and coordination of these effectors, we constructed a series of effector-deletion mutants (see Table 1) in the wild type S. Typhimurium SL1344 genetic background. We generated multiple-effector deletion mutants of the seven effectors of interest in a stepwise manner, using a suicide vector-based approach and homologous recombination [33], to generate a strain lacking all seven effectors, as well as specific combinations of effectors (see Table 1). The ΔsseFG strain does not express the two T3SS2-secreted effectors SseF or SseG encoded by the genes sseF and sseG, respectively, which are a part of the sseABCDEFG operon [34]. Within epithelial cells the replication, SCV localization, and appearance and frequency of SIFs in the ΔsseF single-effector deletion mutant very closely resembles both the ΔsseG single-effector deletion mutant and the ΔsseFG double deletion mutant, likely owing to the functional link between the two effectors [35,36]. We therefore consider the double-deletion mutant, ΔsseFG, effectively as a single-effector deletion mutant. Deleting one, or multiple coding regions for T3SS2-secreted effectors does not significantly impair the fitness of the effector deletion strains in LB broth (S1 Fig).
Table 1. Bacterial strains used in the study.
| Escherichia coli Strains | ||||
| Strain Designation | Relevant Characteristics/Genotype | Source/Reference | ||
| MC1061λpir | hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 gal1 galK rpsL thiλpir | [37] | ||
| MFDpir | MG1655 RP4-2-TC::[ΔMu1::aac(3)IV-ΔaphA-Δnic35-ΔMu2::zeo] ΔdapA::(erm-pir) ΔrecA | [38] | ||
| DH10B | F- araDJ39 Δ(ara, leu)7697 ΔlacX74 galU galK rpsL deoR ɸ80dlacZΔM15 endAI nupG recAl mcrA Δ(mrr hsdRMS mcrBC) | [39] | ||
| Salmonella Typhimurium Strains | ||||
| Strain Designation | Relevant Characteristics/Genotype | Source/Reference | ||
| SL1344 | Wild type stain, hisG | [40] | ||
| Single-effector deletion mutants | ΔsteA | SL1344ΔsteA | This study | |
| ΔpipB2 | SL1344ΔpipB2 | This study | ||
| ΔsopD2 | SL1344ΔsopD2 | This study | ||
| ΔsseJ | SL1344ΔsseJ | This study | ||
| ΔsifA | SL1344ΔsifA | This study | ||
| ΔssaR | SL1344ΔssaR | [41] | ||
| ΔsseFG | SL1344ΔsseFΔsseG | This study | ||
| Multi-effector deletion mutants | Sequential-effector deletion mutants | ΔsseFGΔsteA | SL1344ΔsseFΔsseGΔsteA | This study |
| ΔsseFGΔsteAΔpipB2 | SL1344ΔsseFΔsseGΔsteAΔpipB2 | This study | ||
| ΔsseFGΔsteAΔpipB2ΔsopD2 | SL1344ΔsseFΔsseGΔsteAΔpipB2ΔsopD2 | This study | ||
| ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJ | SL1344ΔsseFΔsseGΔsteAΔpipB2ΔsopD2ΔsseJ | This study | ||
| ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA | SL1344ΔsseFΔsseGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA | This study | ||
| ΔsseFGΔsseJ | SL1344ΔsseFΔsseGΔsseJ | This study | ||
| ΔsseFGΔsopD2 | SL1344ΔsseFΔsseGΔsopD2 | This study | ||
| ΔsifAΔsseJ | SL1344 sifAΔsseJ | This study | ||
| ΔsifAΔsopD2 | SL1344ΔsifAΔsopD2 | This study | ||
| ΔsifAΔsseJΔsteA | SL1344ΔsifAΔsseJΔsteA | This study | ||
| ΔsifAΔsseJΔsopD2 | SL1344ΔsifAΔsseJΔsopD2 | This study | ||
Formation of LAMP1-positive tubules is dependent on multiple effectors
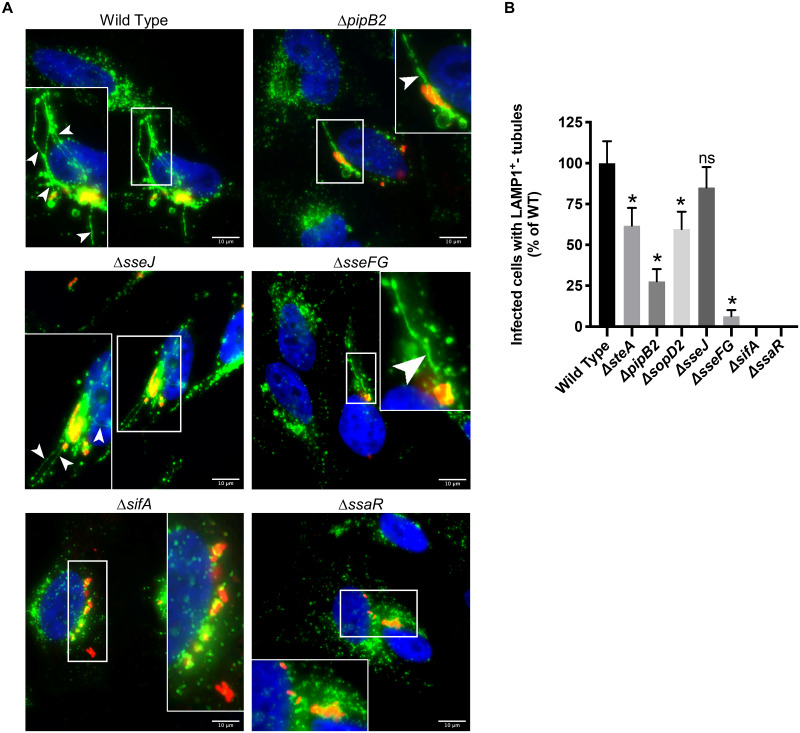
We analyzed the contribution of each of the effectors of interest to the formation of LAMP1-positive (LAMP1+) tubules that radiate outwards from the SCV. SIFs, the first of the Salmonella-induced tubules to be described [15,42,43], are identified by the presence of the host membrane protein LAMP1 within their membranes [23,44]. HeLa human epithelial cells were infected with the various effector deletion mutants and evaluated for the frequency of SIF formation in infected cells. Infected HeLa cells were fixed at 8 hours post-infection and immunolabeled with an anti-LAMP1 antibody to label LAMP1-positive compartments (SCVs) and tubules (SIFs) and anti-Salmonella antibody to label intracellular S. Typhimurium. Labeled cells were analyzed by indirect immunofluorescence microscopy. SIFs are, by definition, LAMP1+-tubules comprised of an inner and outer membrane that extend outwards from the SCV [43]. As we are unable to evaluate whether the SIFs observed are single- or double-membraned using this methodology, we will hereafter refer to them as LAMP1+-tubules.
Cells infected with the wild type strain and single-deletion mutants (Fig 1) exhibit LAMP1+-tubule formation consistent in both morphology and frequency to previous reports [14,15,28,36,43,45,46]. We observed “bulky” LAMP1+-tubules extending outwards from the SCV of ΔpipB2 infected cells, consistent with previous reports [46] (Fig 1A). All single-effector deletion mutant strains, with the exception of ΔsseJ, had significantly fewer LAMP1+-tubule-positive infected cells relative to wild type, while the ΔsifA and ΔssaR strains failed to form LAMP1+-tubules.
Fig 1. LAMP1+-tubule extension from single deletion mutants.
(A) Comparison of frequency of LAMP1+-tubule formation of WT and isogenic single-effector deletion mutants in HeLa cells after 8 hours of infection. Cells were immunostained for Salmonella (red) and LAMP1 (green), and the nucleus was stained with DAPI (blue). Representative images of select strains are shown. The white boxes indicate zoomed-in region in inset. Arrowheads indicate LAMP1+-tubules. Scale Bar = 10 μm. (B) Quantification of LAMP1+-tubule frequency in HeLa cells infected with the single deletion mutants for 8 hours. The average frequency of infected cells with LAMP1+-tubules relative to wild type infected cells ± standard error of the mean for three separate experiments is shown (n = 3). At least 100 infected cells per strain were blindly analyzed in each experiment. An asterisk indicates a significant difference between the indicated mutant strain LAMP1+-tubule frequency and the corresponding WT LAMP1+-tubule frequency (p < 0.02) as determined by Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison post-test.
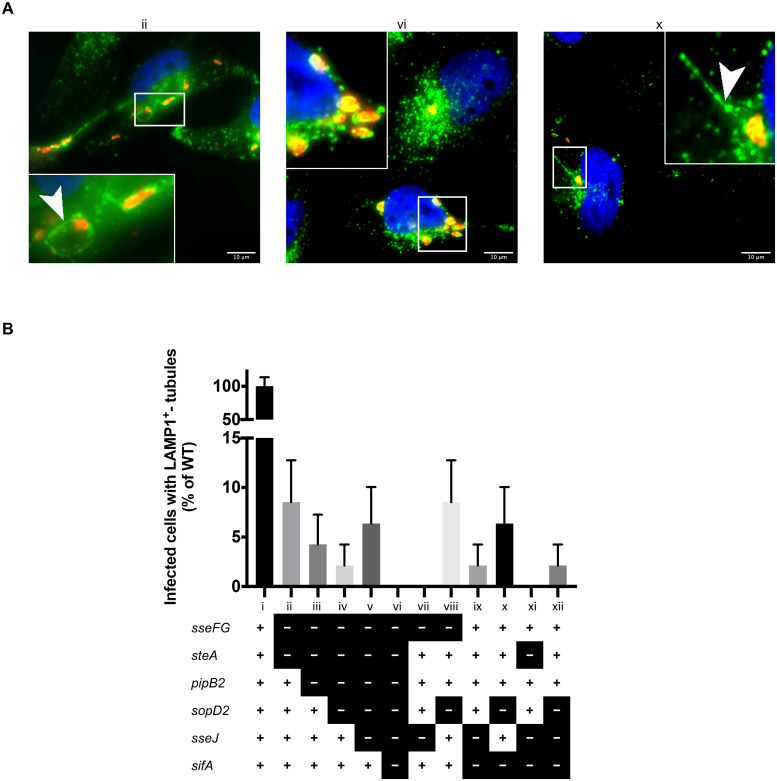
Cells infected with the multiple-effector deletion mutant strains (Table 1) exhibit a dramatic decrease in the frequency of LAMP1+-tubule formation relative to both the wild type strain (Fig 2B) and the corresponding single-effector deletion mutants (Fig 1B). The sequential-effector deletion mutants (Fig 2B, strains ii-vi)—a subset of the multiple-effector deletion mutants—were found to have LAMP1+-tubules extending outwards from intracellular Salmonella in 2–8% of infected cells relative to wild type infected cells (Fig 2, strain i). The frequency of LAMP1+-tubule-positive infected cells was not statistically different between the sequential-effector deletion mutants. The sequential deletion of effectors does not dramatically reduce LAMP1+-tubule frequency (i.e. ΔsseFGΔsteA vs. ΔsseFGΔsteAΔpipB2 vs. ΔsseFGΔsteAΔpipB2ΔsopD2). The sequential-effector deletion mutants ΔsseFGΔsteA, ΔsseFGΔsteAΔpipB2, ΔsseFGΔsteAΔpipB2ΔsopD2, and ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJ (Fig 2, strains ii-v) can all induce formation LAMP1+-tubules and only the sequential-effector deletion mutant with all seven effectors deleted (ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA) fails to induce LAMP1+-tubules. This is consistent with previous evidence suggesting that SifA plays a major role in inducing LAMP1+-tubules [47] as all the sequential-effector deletion mutants are able to induce LAMP1+-tubule formation except for the strain with the sifA deletion.
Fig 2. LAMP1+-tubule extension from the SCV results from the actions of several effectors.
(A) Comparison of frequency of LAMP1+-tubule formation of WT and isogenic multiple-effector deletion mutants in HeLa cells after 8 hours of infection. Cells were fixed at 8 hours post-infection, immunostained, and analyzed as described in the legend of Fig 1. Representative images of select strains are shown. Strain designation (ii, vi, and x) corresponds to strains described in the legend of (B). (B) Quantification of LAMP1+-tubule frequency in HeLa cells infected with the multiple-effector deletion mutants for 8 hours. LAMP1+-tubule frequency was quantified and analyzed as described in the legend of Fig 1. Strain legend: “+” = gene present, “-” = gene deleted. A “+” for all genes indicates wild type (strain i). Results analyzed by a Kruskal-Wallis test with Dunn’s correction for multiple comparisons. All LAMP1+-tubule frequencies in the multiple-deletion mutant strains were significantly different from wild type, however there was no significance between the multiple-deletion mutants themselves.
Infection of HeLa cells with the remainder of the multiple-effector deletion mutants with different combinations of deleted effectors reveals that the mechanism of LAMP1+-tubule extension is indeed very intricate. The strain ΔsseFGΔsseJ (Fig 2, strain vii) is unable to form LAMP1+-tubules even though this strain has SifA. This contrasts with the above results from the sequential-effector deletion mutants suggesting that mutant strains can form LAMP1+-tubules so long as sifA was not deleted. Unlike ΔsseFGΔsseJ, the strain ΔsseFGΔsopD2 (Fig 2, strain viii) was able to form LAMP1+-tubules. This may suggest an interaction or coordinated roles between SseJ and SseF/G that facilitates LAMP1+-tubule extension. For example, SseF/G may require the activity of SseJ in order to induce LAMP1+-tubulation which could explain the presence of LAMP1+-tubules in ΔsseFGΔsopD2 infected cells but not ΔsseFGΔsseJ infected cells. Further studies are required to delve deeper into the reasons behind the formation of LAMP1+-tubules in some multiple-effector deletion strains and not in others.
The ability to form SIFs (LAMP1+-tubules) has long been thought to be heavily dependent on SifA as ectopic expression of SifA in HeLa cells induces LAMP1+-tubule formation [24,47]. We observed LAMP1+-tubules radiating outwards from the SCV in cells infected with both ΔsifAΔsseJ and ΔsifAΔsopD2 double deletion mutants (Fig 2, strains ix and x, respectively), albeit at a very low frequency. Both strains lack the gene encoding sifA, yet they retain the ability to form LAMP1+-tubules. Intriguingly, we did not observe any LAMP1+ tubules in the ΔsifAΔsseJΔsopD2 triple-effector deletion mutant (Fig 2, strain xi). This may indicate a required sequential or coordinated actions of SifA, SopD2, and SseJ, in order to extend LAMP1+-tubules.
Intracellular localization of S. Typhimurium is modulated by multiple effectors
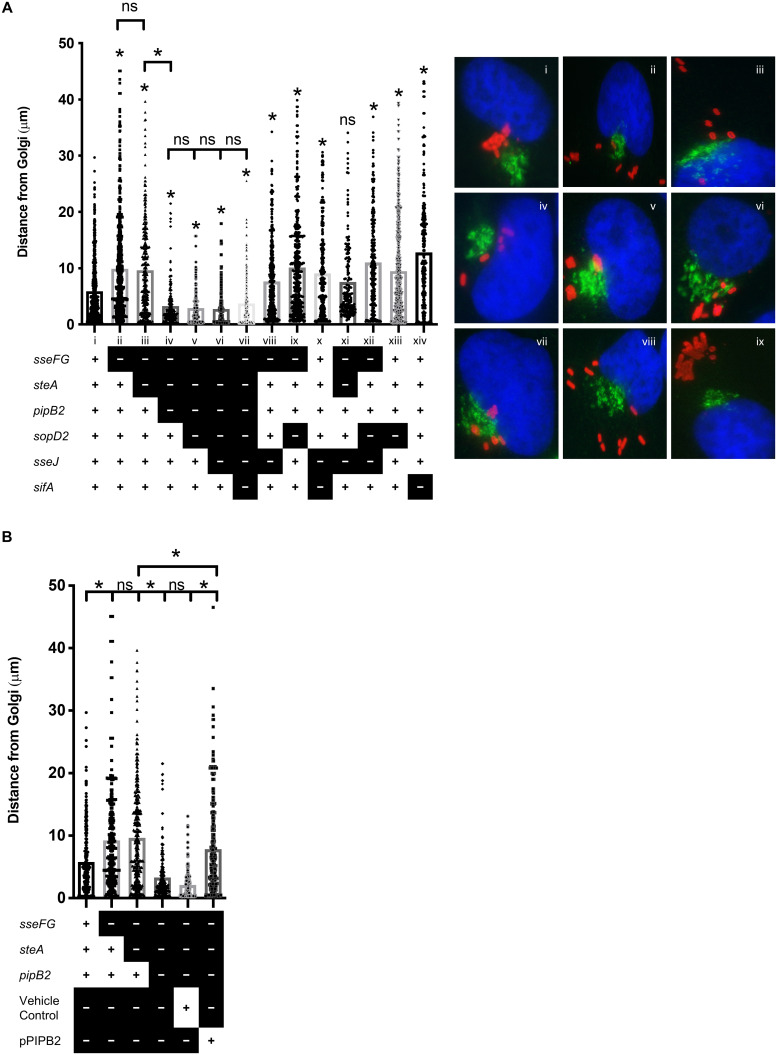
Intracellular S. Typhimurium typically forms microcolonies near the microtubule-organizing center and Golgi-complex several hours post-infection in infected epithelial cells [35,48–50]. The T3SS2-secreted effectors SseF, SseG, SifA, PipB2, and SteA have been individually implicated in SCV localization during infection [35,48–53]. We used our library of multiple-effector deletion mutants to examine the effect of multiple effector deletions on intracellular localization. We quantified the distribution of S. Typhimurium relative to the Golgi complex by measuring the distance between intracellular S. Typhimurium and the Golgi complex 8 hours after infection in HeLa cells immunostained for S. Typhimurium and Golgin-97 (Fig 3).
Fig 3. Multiple effectors drive SCV movement away from the Golgi complex.
HeLa cells were infected with the indicated S.
Typhimurium strains for 8 hours, fixed, and immunostained for
Salmonella (red) and Golgin-97 (green), and the
nucleus was stained with DAPI (blue). (A) Quantification of
S. Typhimurium position relative to the Golgi. The
distances from the center of individual bacteria to the nearest edge of
the Golgi complex was measured in infected cells. Strain legend: “+” =
gene present, “-” = gene deleted. All data points are shown to
accurately indicate the spread of the data. The averages for three
separate experiments are shown (n = 3). An asterisk
indicates a significant difference (p < 0.003)
between the indicated mutant strain and the corresponding WT strain or
other strain if indicated by  as determined by a Kruskal-Wallis one-way ANOVA
with Dunn’s multiple comparison post-test. ns = not significant.
(B) Select representative images used to enumerate
distances in (A). (C) SL1344 strain
ΔsseFGΔsteAΔpipB2
was complemented with a low-copy plasmid expressing a functional copy of
PipB2. HeLa cells were infected, fixed, stained, and analyzed as
described in (A).
as determined by a Kruskal-Wallis one-way ANOVA
with Dunn’s multiple comparison post-test. ns = not significant.
(B) Select representative images used to enumerate
distances in (A). (C) SL1344 strain
ΔsseFGΔsteAΔpipB2
was complemented with a low-copy plasmid expressing a functional copy of
PipB2. HeLa cells were infected, fixed, stained, and analyzed as
described in (A).
Consistent with previous reports, the deletion of ΔsseFG alters SCV localization such that ΔsseFG mutants are scattered throughout the host cell cytoplasm, rather than remaining in close proximity to the Golgi apparatus like wild type S. Typhimurium (Fig 3A, strains ii and i, respectively) [21,35,48,49,51,54,55]. The additional deletion of steA (resulting in the ΔsseFGΔsteA triple-effector deletion mutant, strain iii in Fig 3A) does not significantly alter S. Typhimurium positioning relative to the Golgi as compared to the ΔsseFG double deletion mutant, which is consistent with the findings of Domingues et al., (2014). Further deletion of PipB2 (resulting in the ΔsseFGΔsteAΔpipB2 quadruple-effector deletion mutant, strain iv in Fig 3) results in a strain that remains closer to the Golgi than wild type S. Typhimurium. ΔpipB2 and ΔsteA single-effector deletion mutants have previously been shown to be positioned close to the nucleus at 8–14 hours-post infection and a ΔsseFΔpipB2 double deletion mutant is found scattered throughout the cytosol [19,53]. Our results indicate potential interplay between SteA and PipB2 to promote movement away from the Golgi as both effectors must be deleted in the ΔsseFG background to maintain close apposition to the Golgi. Subsequent sequential deletions of sopD2, sseJ, and sifA (Fig 3A, strains v, vi, and vii, respectively) do not impact intracellular localization of these S. Typhimurium mutant strains as they all reside very close to the Golgi. This is unexpected as the single-effector deletion mutants ΔsseFG, ΔsopD2, and ΔsifA all have a scattered distribution SCVs throughout the host cell (Fig 3A, strains ii, xiii, and xiv, respectively). In fact, strains ΔsseFGΔsteAΔpipB2ΔsopD2, ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJ, ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA are positioned closer to the Golgi than even wild type S. Typhimurium. The PipB2-dependent SCV scattering was complemented when a plasmid expressing PipB2 was introduced in the ΔsseFGΔsteAΔpipB2 mutant strain (Fig 3B). All the remainder of the multiple-effector deletion strains in Fig 3, apart from ΔsseFGΔsteAΔsseJ, have altered SCV positioning relative to wild type. Our results indicate that these effectors are involved in keeping intracellular S. Typhimurium close to the Golgi and that it seems likely that there is interplay between PipB2 and SteA which plays a critical role in SCV localization during infection.
Multiple-effector deletion mutants of S. Typhimurium do not replicate in macrophages
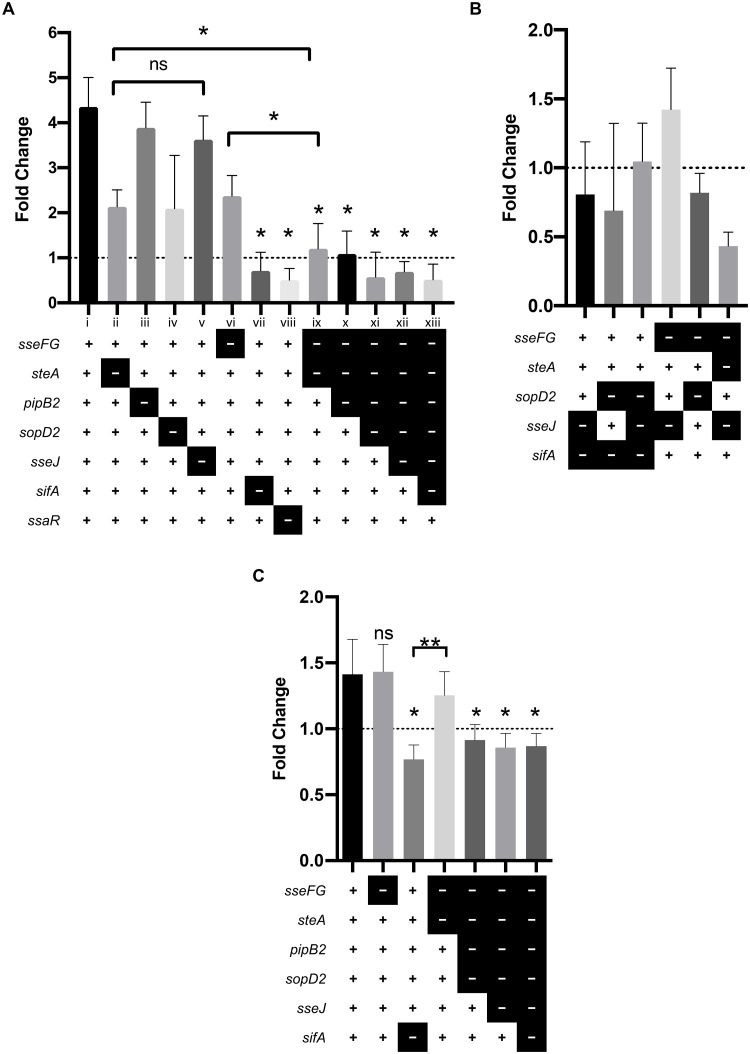
The effectors SseJ, SopD2, SifA, PipB2, and SteA modulate SCV membrane dynamics to promote intracellular replication [32]. In macrophages, S. Typhimurium that escape the SCV and enter the host cell cytoplasm are eliminated by host defenses [56]. Therefore, maintaining the SCV membrane is critical to replication in macrophages. Previous studies have demonstrated that multiple T3SS2-secreted effectors, including the seven effectors in this study, contribute to replication within mouse macrophages [25,57]; specifically, the single-effector deletion mutants sifA, sseJ, sopD2, and sseFG exhibit decreased replication relative to wild type S. Typhimurium [57]. Given that several effectors are implicated in promoting replication in macrophages, we wanted to evaluate if our effectors of interest act independently, sequentially, or cooperatively to promote intramacrophage replication.
RAW 264.7 mouse macrophages were infected with our library of deletion mutants and CFUs enumerated at 2 hours and 24 hours post-infection. Most single-effector deletion mutants replicated in RAW 264.7 cells as indicated by a fold change greater than 1, while the T3SS2-secretion negative control ΔssaR mutant and the ΔsifA mutant did not replicate which is consistent with previous reports (Fig 4A) [24,58]. Conversely, the sequential deletion mutants (strains ix-xiii in Fig 4A) were unable to replicate as were the multiple-effector deletion mutants shown in Fig 4B.
Fig 4. Multiple effectors are required for replication in macrophages.
Strain legend: “+” = gene present, “-” = gene deleted. A “+” for all
genes indicates wild type. (A) Replication of
S. Typhimurium single- and sequential- deletion
mutants in RAW 264.7 macrophages. RAW 264.7 cells were infected with the
indicated strains at a MOI of 10. Fold change was determined by dividing
CFU counts at 24 hours post-infection by CFU counts at 2 hours
post-infection. The average fold change ± standard deviation for three
experiments is shown (n = 3). An asterisk indicates a
significant difference (p < 0.02) between the
indicated mutant strain and WT or other strain if indicated by
 as determined by a
Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison post-test.
ns = not significant. (B) Replication of
S. Typhimurium multiple-effector deletion mutants in
RAW 264.7 macrophages. Experiment was performed and analyzed as
described in (A). (C) Replication of S.
Typhimurium strains in THP-1 monocytes. THP-1 monocytes were infected
with select S. Typhimurium strains and results analyzed
as described in (A). Representative results from one experiment is
shown. An asterisk indicates a significant difference
(p < 0.03) between the indicated mutant strain
and WT or other strain if indicated by
as determined by a
Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison post-test.
ns = not significant. (B) Replication of
S. Typhimurium multiple-effector deletion mutants in
RAW 264.7 macrophages. Experiment was performed and analyzed as
described in (A). (C) Replication of S.
Typhimurium strains in THP-1 monocytes. THP-1 monocytes were infected
with select S. Typhimurium strains and results analyzed
as described in (A). Representative results from one experiment is
shown. An asterisk indicates a significant difference
(p < 0.03) between the indicated mutant strain
and WT or other strain if indicated by  as determined by a Kruskal-Wallis one-way ANOVA
with Dunn’s multiple comparison post-test. ns = not significant.
as determined by a Kruskal-Wallis one-way ANOVA
with Dunn’s multiple comparison post-test. ns = not significant.
We expected that sequential and successive deletion of effectors would have a cumulatively negative impact on intramacrophage replication. As such, we expected a strain with three effectors deleted would experience decreased replication as compared to a strain with only two effectors deleted. However, while deletion of a single effector is permissive of intramacrophage replication, deletion of two or more effectors results in an inability to replicate within the macrophage. For example, the double-effector deletion strain ΔsseFG (considered a single-deletion mutant, Fig 4A strain vi) and the single-effector deletion strain ΔsteA (Fig 4A, strain ii) are able to replicate within RAW 264.7 cells (fold change > 1), however the triple-effector deletion mutant ΔsseFGΔsteA (Fig 4A, strain ix) is unable to replicate (fold change < 1). We found a similar effect with select multiple-effector deletion mutants in human THP1 monocytes (Fig 4C). The inability of the sequential- and multiple-effector deletion mutants to replicate within macrophages was not driven by the specific effectors deleted, but rather by the number of effectors deleted. Simply put, intramacrophage replication does not occur when two or more of the effectors examined in this study are deleted. We can therefore conclude that intramacrophage replication is driven by the actions of multiple effectors.
Multiple effector deletion mutants of S. Typhimurium have impaired virulence in a mouse model of infection
We have demonstrated that these seven effectors (SseF, SseG, SteA, PipB2, SopD2, SseJ, and SifA) are all required to establish an intracellular replicative niche within both epithelial cells and macrophages. While these models provide insight into the cellular events within host cells, they do not provide any information regarding virulence. We therefore investigated the contribution of these seven effectors to virulence in an in vivo infection model. We chose to use a low-dose streptomycin pre-treatment murine model of gastroenteritis as it more closely models S. Typhimurium infections in humans and produces consistent results in our hands. In this model, pre-treatment of C57BL/6 mice with low-dose of streptomycin induces susceptibility to gastroenteritis upon infection with wild type S. Typhimurium [59].
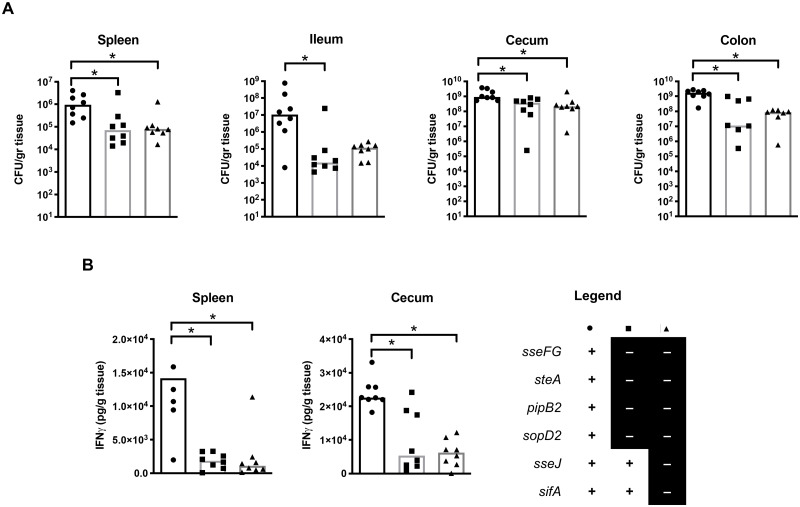
Streptomycin-treated mice were infected with either wild type SL1344, ΔsseFGΔsteAΔpipB2ΔsopD2, or ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA multiple-effector deletion strains. These two multiple-effector deletion strains were selected as they both exhibit reduced frequency of LAMP1+-tubulation and an inability to replicate in macrophages. S. Typhimurium colonization in intestinal and systemic sites were determined at three days post-infection by CFU counts from the spleen, cecum, ileum, and colon. Both multiple-effector deletion strains colonized the spleen, cecum, and colon significantly less than the wild type strain (Fig 5A). Both multiple-effector deletion strains also colonized the ileum to a lesser extent than the wild type strain, though the ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA was not statistically significantly different from wild type (Fig 5A). There was no significant difference in colonization at the four sites between ΔsseFGΔsteAΔpipB2ΔsopD2 and ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA. This suggests that successful colonization, at both intestinal and systemic sites, requires the action of multiple effectors.
Fig 5. Gastroenteritis model of S. Typhimurium infection.
Mice were treated with streptomycin for two days prior to oral infection with select S. Typhimurium strains to induce gastroenteritis as per Sekirov et al., 2008. Strain legend found on bottom right of figure: “+” = gene present, “-” = gene deleted. A “+” for all genes indicates wild type. A single asterisk indicates a significant difference between the indicated strains (p < 0.03) as determined by a Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison post-test. (A) Bacterial counts were recovered from systemic and intestinal organs of mice three days post-infection. Counts given represent colony forming units per gram of tissue. The median CFU/g for three separate experiments is shown with individual data points visible to accurately represent the spread of the data. (B) IFNγ-levels as determined by ELISA. The median amount of IFNγ (pg of IFNγ/g of tissue) is shown with individual data points visible to accurately represent the spread of the data.
One of the hallmarks of non-typhoidal salmonellosis infection is acute intestinal inflammation [60]. Therefore, elevated levels of gastrointestinal inflammation—reflected by the levels of IFNγ—indicate S. Typhimurium infection within the intestinal epithelium. We found significantly decreased IFNγ in the spleen and cecum of mice infected with the multiple-effector deletion strains ΔsseFGΔsteAΔpipB2ΔsopD2 and ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA as compared to mice infected with wild type strain (Fig 5B). The additional deletion of sifA and sseJ to the ΔsseFGΔsteAΔpipB2ΔsopD2 strain does not further decrease colonization or inflammation, mirroring our results in the macrophage infection models (both RAW 264.7 and THP-1 cells). This suggests that the multiple-effector deletion strains did not elicit as strong of an inflammatory response as the wild type strain. The combination of decreased colonization and decreased inflammation induced by the multiple-effector deletion strains suggests that multiple effectors are required for both colonization and virulence of Salmonella within a gastroenteritis model of infection.
Discussion
The importance of T3SS2-secreted effectors during infection is widely recognized, but the precise biochemical activity and function of many of these effectors is poorly understood. Previous attempts to identify the functions and targets of T3SS2-secreted effectors often involve studying effectors separately [22,55,61,62]; however increasing evidence suggests that the effectors have overlapping yet distinct roles during infections. As infections involve dynamic and complex processes, the effect of one effector may require prior action by another effector, or their activities may be linked. It is therefore necessary to study effector activities in the presence or absence of other related effectors to discover the precise function of each effector.
SIF biogenesis is a complex and dynamic process involving the action of several effectors as shown by multiple studies [16,43,45]. Here, we demonstrate the complexity of LAMP1+-tubule extension (i.e. SIF biogenesis) by infecting HeLa cells with a library of single-effector and multiple-effector deletion strains. Most single-effector deletion mutant strains are capable of forming LAMP1+-tubules at a frequency of at least 25% relative to wild type, whereas all 11 multiple-effector deletion strains fail to induce LAMP1+-tubules at a frequency greater than 10% relative to wild type. The fact that single-effector deletion mutants form more LAMP1+-tubules as compared to the multiple-effector deletion mutants implies that more than one effector is required to extend LAMP1+-tubules and that at least two or more of the effectors are working in conjunction with one another. Furthermore, the sequential deletion of effectors does not decrease the frequency of LAMP1+-tubule formation in a step wise manner indicating unequal contribution by each effector to this process. The severe and non-cumulative defects in LAMP1+-tubule formation observed in the multiple-effector deletion mutants strongly suggests that extensive LAMP1+-tubule extension requires multiple effectors mediating the process.
Previous studies show that multiple T3SS2-secreted effectors are required to mediate LAMP1+-tubule extension [31,35]. While one study indicates that ectopically expressed sifA in HeLa cells is sufficient to induce LAMP1+-tubulation, others studies report significantly higher frequencies of LAMP1+-tubulation when sifA is co-expressed with either sopD2 or sseJ [27,31]. SifA and SseJ cooperate through interactions with the host kinesin-binding protein SKIP and RhoA family GTPases to induce LAMP1+-tubulation [31]. We observed similar frequencies of LAMP1+-tubulation in a strain with functional SifA and SseJ (ΔsseFGΔsteAΔpipB2ΔsopD2) and a strain with only functional SifA (ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJ), while LAMP1+-tubules were not observed in the strain that lacks all seven effectors of interest (ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA). From this we can conclude that SifA is sufficient to induce LAMP1+-tubule formation on its own. However, the frequency of LAMP1+-tubulation was not significantly higher when both SifA and SseJ are functional than with SifA alone, suggesting that the previously described cooperative actions between SifA and SseJ, resulting in the increased LAMP1+-tubulation, possibly involves other S. Typhimurium effectors or other host proteins to increase the frequency of LAMP1+-tubules. The discrepancy between our results and others, regarding the cooperation between SifA and SseJ, may be explained by previous studies observing increased LAMP1+-tubule frequencies with ectopically expressed SifA and SseJ [30,31] whereas our study is in the context of a S. Typhimurium infection. Our results strongly suggest that multiple T3SS2-secreted effectors are required to facilitate efficient formation and extension of LAMP1+-tubules.
Previous studies have demonstrated that neither the ΔsifA single-effector deletion mutant nor the ΔsifAΔsseJ double-effector deletion mutant form SIFs. However, the ΔsifA mutant escapes the SCV while the ΔsifAΔsseJ mutant remains in the SCV. [15,24,30,63,64]. While the N-terminal domain of SifA is required for interactions with the host protein SKIP (PLEKHM2) and PLEKHM1 [48,62,65–67], the C-terminal domain promotes LAMP1 recruitment to Salmonella-induced tubules via interactions with the GTPase Arl8b [68]. Studies have demonstrated that Arl8b controls membrane fusion events with late endocytic compartments and is associated with LAMP1 accumulation in SIFs [68,69]. In our study, the ΔsifA deletion mutant was unable to extend LAMP1+-tubules, yet we observed LAMP1+-tubules in HeLa cells infected with ΔsifAΔsseJ and ΔsifAΔsopD2 strains, suggesting that LAMP1 recruitment, and subsequent LAMP1+-tubule extension, can occur via a SifA-independent mechanism. Further work is required to determine if LAMP1 recruitment in these strains is mediated by an additional effector interacting with Arl8b, or if an alternative Arl8b-independent mechanism is at play. The fact that the additional deletion of sseJ or sopD2 in the ΔsifA background restores the ability to extend LAMP1+-tubules indicates potential antagonistic action between SseJ or SopD2 and other effectors that mediate SIF biogenesis.
The low frequency of LAMP1+-tubule extension observed in multiple-effector deletion mutants lacking ΔsseFG may be directly related to the role of SseF and SseG during infection. Both SseF and SseG, while not required for the formation of single membrane SIFs, are required for the conversion of single-membraned SIFs (also known as pseudo-SIFs) to double-membrane SIFs [43]. The inability to convert from single- to double-membraned SIFs may explain the thinner appearance of SIFs in cells infected ΔsseF/G strains as compared to wild type infected cells [36,46]. The methods used in our study may fail to detect these thinner pseudo-SIFs, and may therefore account for low frequency of LAMP1+-tubules in the ΔsseFG mutant strains.
Multiple studies have established that precise intracellular SCV positioning plays a key role during infection [48,49,54,70,71]. Mutant S. Typhimurium strains that fail to cluster near the Golgi at 8 hours-post infection in epithelial cells have lower frequencies of LAMP1+-tubule extension and impaired intracellular replication [35,49,72]. The sseF and/or sseG deletion mutants are found scattered throughout the host cell’s cytoplasm which could be a consequence of dysregulated microtubule motors [21]. Alternatively, the scattered phenotype caused by sseF/G deletion could also result from the absence of SseF and SseG mediated tethering to the Golgi-associated protein ACBD3 [55]. SifA and PipB2 also play a role in SCV localization during infection. T3SS2-secreted effector SifA inserts into the SCV membrane where it binds to the C-terminal PH domain of SKIP (PLEKHM2) [48,73]. Meanwhile, PipB2 tethers auto-inhibited kinesin-1 to the SCV membrane [22] in a process involving the small GTPase Arl8b [69,74]. Kinesin-1 is then activated by binding to the SifA-SKIP complex [65]. Deletion of pipB2 prevents centrifugal displacement of SCVs at later timepoints in infection [19]. SteA may play a role in SCV positioning as it is thought to activate kinesin-1 or inhibit dynein [53]. While we have many pieces of the puzzle, the exact mechanisms controlling SCV localization during infection remains unclear.
A previous study investigated the role of up to three effectors on the intracellular localization of S. Typhimurium during infection. The authors found that intracellular wild type, ΔsteA, and ΔpipB2 single-effector deletion mutants tend to reside close to the Golgi-apparatus at up to 14 hours post-infection, whereas any multiple deletion mutant that also had sseF and/or sseG deleted were more likely to be scattered throughout the host cell cytosol [19,53]. We found that the additional deletion of pipB2 in the ΔsseFGΔsteA background (resulting in ΔsseFGΔsteA ΔpipB2) restores SCV localization close to the Golgi-apparatus. Subsequent deletion of additional effectors in the ΔsseFGΔsteAΔpipB2 background did not alter this close apposition of the SCV to the Golgi. Conversely, SCVs in mutant strains with functional PipB2 and SteA tend to be scattered throughout the host cell cytosol. These results suggest that PipB2 may be the first effector in a series of events involving SteA that leads to outwards centrifugal movement of SCV at 8 hours post-infection (and later time points as well) and without the initial action of PipB2, the SCV remains in close proximity to the Golgi. The intracellular positioning of Salmonella must therefore rely on a delicate balance of effector actions to precisely regulate SCV positioning during infection. We can therefore conclude that multiple effectors are required to regulate positioning of the SCV during infection.
The question remains as to what the link is, if any, between SCV localization, LAMP1+-tubule extension, and intracellular replication? Does effector deletion alter SCV localization, which directly impairs LAMP1+-tubule extension, thereby limiting intracellular replication? Or does effector deletion itself impair LAMP1+-tubule extension, resulting in decreased intracellular replication and altered SCV localization is merely a coincidental phenotype? An example that brings causality into question is the effect of deletion of sseF and sseG (ΔsseFG). We, and others, have shown that deletion of sseFG results in altered SCV localization and decreased LAMP1+-tubule formation in HeLa cells, as well as reduced intracellular replication. Other studies have found that ΔsseFG strains also have altered SIF morphology [36,43]. SIFs (LAMP1+-tubules) are necessary for supplying intravacuolar S. Typhimurium with nutrients [20]. Intravacuolar S. Typhimurium forms SIFs by converting the host’s endosomal system into SIFs to siphon nutrients from the host [18]. So then, does the replication defect of the ΔsseFG double mutant result from altered SCV localization, or does the altered SIF morphology itself limit nutrient acquisition from the host causing impaired intracellular replication? Alternatively, does altered SIF morphology result directly from altered SCV localization, impacting interactions with the host’s endosomal system, and thus limiting nutrient acquisition, resulting in impaired intravacuolar replication? Further studies are necessary to elucidate the cause and effect of these processes. Intracellular localization, SIF formation, and replication are likely very intertwined and deletion of one effector critical to one of these processes could dramatically impact the others.
Multiple effectors are required to promote intracellular replication in macrophages. Most single-effector deletion mutants replicate within both RAW 264.7 macrophages and THP-1 monocytes, whereas multiple-effector deletion mutants do not. The precise reason for impaired replication is unclear, however a potential explanation is that deletion of multiple effectors alters interactions with the host’s endocytic pathway which would then alter or limit SIF formation and thereby decrease nutrient acquisition from the host [18,20,43,75]. The fact that all multiple-effector deletion mutants have impaired intramacrophage replication regardless of the effectors deleted, suggests that all effectors are required in combination with each other to successfully replicate within host cells.
The importance of these SPI-2 effectors during infection was reinforced by examining colonization and inflammatory response in an in vivo infection model. It has been reported that SifA is not required to induce inflammation in the colon of mice [76]; however another group showed that strains lacking SifA in addition to other T3SS2-decreted effectors (SseF, SseJ, SteA, and SpvB) have dramatically reduced inflammation during infection suggesting that intestinal inflammation requires the cooperative effects of at least these five effectors [77]. In line with these findings, both multiple-effector deletion strains in our study ΔsseFGΔsteAΔpipB2ΔsopD2 and ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA exhibited decreased virulence in a low-dose streptomycin pre-treatment mouse model of gastroenteritis. Both multiple-effector deletion strains are impaired to a similar degree with respect to colonization and inflammation in mice as compared to the mice infected with wild type S. Typhimurium. This means that SifA and SseJ are insufficient to mount a successful infection without the other five effectors of interest: SseF, SseG, PipB2, SteA, and SopD2. The impaired colonization observed in the two multiple-effector deletion mutants used in the mouse model of infection means that deletion of these effectors either impacts the ability of these strains to invade the host cells, the ability to replicate within host cells, or the ability to evade the host’s immune system. The decreased inflammation in mice infected with either multiple-effector deletion strain also suggests that these strains may not invade host cells as efficiently as wild type strains, or they were easily eliminated from the host before the end point of the experiment. Similar to the conclusions of Matsuda et al. (2019), we can surmise that the seven effectors of interest in this study are required to mount a successful infection in a mouse model of gastroenteritis.
We have shown that the processes of SIF biogenesis, intracellular localization, replication in macrophages, and colonization and inflammation in a mouse model, all require multiple effectors present and working together to successfully mount an infection. Our study highlights the fact that not one single effector of our seven of interest, is solely responsible for mediating complex infection phenotypes. It seems likely that several effectors act on the same process, either in conjunction with one another, or in a sequential manner. If these effectors work sequentially, then deletion of an effector that works early within the pathway will have dramatic results. As an example: deletion of PipB2 helps S. Typhimurium mutants remain very close to the Golgi during infection regardless of the presence or absence of other effectors, indicating that PipB2 likely acts early within the pathway. Similarly, these effectors could interact within much larger complexes, and deletion of a key effector could render the entire complex ineffective. If we want to elucidate the exact mechanisms underpinning Salmonella’s intracellular replicative niche, we must study the role of each effector in the context of other effectors, rather than deleting single effectors, or transfecting a single effector into tissue culture cells and examining their effect. Further studies are required to examine how these effectors interact with each other, or on similar host processes.
Materials and methods
Ethics statement
All animal experiments were performed according to protocol number A13-0265 approved by the University of British Columbia’s Animal Care Committee and in direct accordance with the Canadian Council of Animal Care (CACC) guidelines. Mice were euthanized at 3 days post-infection.
Bacterial strains and culture conditions
Bacterial strains used in this work are described in Table 1. All strains were routinely grown in Luria-Bertani (LB) medium at 37°C with shaking. For growth of the E. coli MFDpir strain, media was supplemented with DL-2,6-Diaminopimelic acid (DAP) at a final concentration of 0.3 mM when appropriate. Antibiotics were used at the following concentration when required: streptomycin 50 μg/mL, chloramphenicol 30 μg/mL.
Plasmid construction
Plasmids constructed and used in the study are listed in Table 2; primers used are described in Table 3. All plasmids were constructed using the Gibson Assembly method of cloning [78]. Complementation vectors and gene deletion vectors were routinely maintained in E. coli DH10B and MC1061 λpir respectively.
Table 2. Plasmids used in this study.
| Plasmid Designation | Relevant Characteristics/Genotype | Source/Reference |
|---|---|---|
| pRE112 | cat sacB oriVRGKγoriTRP4 CmR | [33] |
| pACYC184 | oriP15A, TetR, CmR | [79] |
| pRE112-ΔsseFΔsseG | Upstream region of sseF and downstream region of sseG from S. Typhimurium SL1344 in pRE112 | This study |
| pRE112-ΔsteA | Upstream and downstream regions of steA region from S. Typhimurium in pRE112 | This study |
| pRE112-ΔpipB2 | Upstream and downstream regions of pipB2 region from S. Typhimurium in pRE112 | This study |
| pRE112-ΔsopD2 | Upstream and downstream regions of sopD2 region from S. Typhimurium in pRE112 | This study |
| pRE112-ΔsseJ | Upstream and downstream regions of sseJ region from S. Typhimurium in pRE112 | This study |
| pPIPB2 | pipB2 under the control of its native promoter in pACYC184 | This study |
CmR = Chloramphenicol resistance, TetR = Tetracycline resistance
Table 3. Primers used in this study.
| Forward oligonucleotide | Reverse Oligonucleotide | |
|---|---|---|
| sseF 5’ flanking | GAACTGCATGAATTCCCGGGCTGGACAGTTTTATCCGCCG | CGGTATATACCTGAAAACGATTACATATTTCGTTCTGTTATTTAAGCAATAAG |
| sseG 3’ flanking | GAAATATGTAATCGTTTTCAGGTATATACCGG | CAAGCTTCTTCTAGAGGTACCGAAATAACAGACGCAGCGCC |
| steA 5’ flanking | GAACTGCATGAATTCCCGGGCCATCGCTTTGTGATACCCC | CATATCCTACTCCTTCAAATTTTGCTC |
| steA 3’ flanking | CAAAATTTGAAGGAGTAGGATATGTAAAAAGCGTTTATGTTTAGCC | CAAGCTTCTTCTAGAGGTACCCGGGATGAGACAGAATGACC |
| pipB2 5’ flanking | GAACTGCATGAATTCCCGGGGCTGCATCGTCATACTACGG | CATATATTTTCTCCCAGAGACAGCAAC |
| pipB2 3’ flanking | GTCTCTGGGAGAAAATATATGTAGCCCTTTTTGACGTAAATCTG | CCAAGCTTCTTCTAGAGGTACCCCTGGTAATATTTATCAGGCG |
| sopD2 5’ flanking | GTGAACTGCATGAATTCCCGGGGGGGTTTATGGACACATTCC | CTTTTTACATAATAACTCCCTTGATTATTTACCG |
| sopD2 3’ flanking | GTAAATAATCAAGGGAGTTATTATGTAAAAAGTCATTAAAAAGGCC | CAAGCTTCTTCTAGAGGTACCGTTCTGACCATTACTTCTAACG |
| sseJ 5’ flanking | CAAGCTTCTTCTAGAGGTACCCCCACTCCCCACGCTATTATG | CATAGTGTCCTCCTTACTTTATTAAACACG |
| sseJ 3’ flanking | CGTGTTTAATAAAGTAAGGAGGACACTATGTAAAGTTCCATCGGCTGCGG | ATGAATTCCCGGGAGAGCTCCCTGGCAACGGTTAAGGTGG |
| sifA 5’ flanking | CAAGCTTCTTCTAGAGGTACCCACCCCGAGCGCCGTTATTATC | CGTCTGATTTTACATATTAATCTCACTTATACTGGAG |
| sifA 3’ flanking | GAGATTAATATGTAAAATCAGACGACGCTTTCTCAGACG | ATGAATTCCCGGGAGAGCTCGACCGTGACGACCACAAACG |
| pRE112 plasmid backbone | GGTACCTCTAGAAGAAGCTTGGGA | CCCGGGAATTCATGCAGTTCAC |
| pACYC184 plasmid backbone | GCGGCCGCTCGATACCCATACG | CCCGAGATGCGCCGCGTGC |
| pipB2 complementation | GCACGCGGCGCATCTCGGGGAGTTGCAGGAAGGCGGCAAGC | GTATGGGTATCGAGCGGCCGCAATATTTTCACTATAAAATTCGTTAAAGAGTG |
The pRE112 plasmid backbone used for all gene deletion constructs was produced using the pRE112 backbone primer set to amplify linear pRE112 from the KpnI to SacI unique restriction sites (final size of 5749 bp). pRE112 plasmid backbone was subsequently digested with DpnI (NEB) to remove any remaining circular template DNA. To generate complete, unmarked deletions, the upstream homologous region of target genes up to an including the start codon, and the downstream homologous region of the target gene starting with the stop codon were amplified by PCR from the chromosomal DNA of wild type SL1344.
As sseF and sseG are in an operon [34], plasmid pRE112-ΔsseFΔsseG was generated by amplifying the upstream region of sseF and the downstream region of sseG using primer pairs sseF 5’ flanking and sseG 3’ flanking respectively. PCR products were ligated into the pRE112 vector using Gibson Assembly. Plasmids pRE112-ΔsteA, pRE112-ΔpipB2, pRE112-ΔsopD2, pRE112-ΔsseJ, and pRE112-ΔsifA using the respective 5’ flanking and 3’ flanking primers pairs shown in Table 3.
Generation of mutants by allelic exchange
Unmarked complete deletion mutants were generated as previously described [25]. Briefly, MFDpir strain transformed with the gene deletion plasmids were conjugated with different SL1344 based-strains. The unmarked SL1344 gene deletion were constructed by inserting the pRE112 plasmid constructs into the SL1344 chromosome. Post-conjugation single crossover mutants between pRE112 constructs and the SL1344 chromosome were selected on LB agar plates containing chloramphenicol. Sucrose counter-selection was performed as previously described [33] to select for the second crossover event, thus effectively deleting the gene of choice, leaving only the start and stop codons.
The unmarked SL1344 mutant strain ΔsseFG was constructed by inserting the homologous regions from the pRE112-ΔsseFΔsseG plasmid into the wild type chromosome. The unmarked SL1344 mutant strain ΔsseFGΔsteA was constructed by inserting the homologous regions from the pRE112-ΔsteA into the ΔsseFG mutant chromosome. The unmarked SL1344 mutant strain ΔsseFGΔsteAΔpipB2 was constructed by inserting the homologous regions from the pRE112-ΔpipB2 into the ΔsseFGΔsteA mutant chromosome. The unmarked SL1344 mutant strain ΔsseFGΔsteAΔpipB2ΔsopD2 was constructed by inserting the homologous regions from the pRE112-ΔsopD2 into the ΔsseFGΔsteAΔpipB2 mutant chromosome. The unmarked SL1344 mutant strain ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJ was constructed by inserting the homologous regions from the pRE112-ΔsseJ into the ΔsseFGΔsteAΔpipB2ΔsopD2 mutant chromosome. The unmarked SL1344 mutant strain ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJΔsifA was constructed by inserting the homologous regions from the pRE112-ΔsifA into the ΔsseFGΔsteAΔpipB2ΔsopD2ΔsseJ mutant chromosome. Homologous regions from plasmid pRE112-ΔsseJ was introduced into the chromosomes of SL1344 mutant strain ΔsseFG and ΔsifA, creating strains ΔsseFGΔsseJ and ΔsifAΔsseJ respectively. Homologous regions from the plasmid pRE112-ΔsopD2 was introduced into the chromosomes of SL1344 mutant strains ΔsseFG and ΔsifA, creating strains ΔsseFGΔsopD2 and ΔsifAΔsopD2 respectively. The strains ΔsifAΔsseJΔsteA and ΔsifAΔsseJΔsopD2 were generated by incorporating the homologous regions of the plasmids pRE112-ΔsteA and pRE112-ΔsopD2, respectively into the ΔsifAΔsseJ mutant chromosome. Successful gene deletions were verified by PCR and DNA sequencing.
Cell lines
HeLa (ATCC® CCL-2™), RAW 264.7 (ATCC® TIB-71™), and THP-1 (ATCC® TIB-202™) cells were directly obtained from ATCC. All cell lines were routinely maintained at 37°C in a 5% CO2 atmosphere. HeLa and RAW 264.7 cells were cultured in Dulbecco’s Modified Essential Medium (DMEM) (Hyclone) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Gibco), 1% (v/v) Glutamax (Gibco), and 1% (v/v) nonessential amino acids (Gibco). HeLa and RAW 264.7 cells were used up to passage 15. THP-1 cells were routinely maintained at a density of 2 x 105 to 1 x 106 cells/mL in Roswell Park Memorial Institute (RPMI) 1640 Medium (Gibco) supplemented with 10% (v/v) heat inactivated FBS, and 1% (v/v) nonessential amino acids. THP-1 cells were used up to passage 10.
HeLa cell infections
HeLa cells were seeded on 12 mm diameter glass coverslips in 24-well plates (Corning) at a density of 5 x 104 cells/well, 16–24 hours prior to infection. Overnight bacterial cultures were diluted 1:33 in LB without antibiotic and incubated for 3 hours at 37°C with shaking (late log-phase cultures). 1 mL of bacterial cultures were pelleted and resuspended in Dulbecco’s Phosphate-Buffered Saline (DPBS) (Hyclone), subsequently diluted in DMEM and added to the HeLa cells at a multiplicity of infection (MOI) of ≈ 100:1. The infection was allowed to proceed for 15 minutes at 37°C in 5% CO2. Non-internalized bacteria were removed by three washes in DPBS and cells incubated in growth media containing 100 μg/mL gentamicin until 2 hours post-infection, followed by growth media containing 10 μg/mL gentamicin for the remainder of the experiment. HeLa cells were infected for a total of 8 hours.
RAW 264.7 cell infections
RAW 264.7 cells were seeded in 24-well plates at a density of 1 x 105 cells/well 16–24 hours prior to infection. Overnight bacterial cultures (stationary phase) were pelleted and resuspended in DPBS, and subsequently opsonized in DPBS containing 10% normal mouse serum for 20 min at 37°C. Opsonized bacteria were diluted in DMEM and added to the monolayers at a MOI of ≈ 10:1, centrifuged at 170 g for 5 min at room temperature and incubated for 25 min at 37°C in 5% CO2. Non-internalized bacteria were removed by three washes in DPBS and cells incubated in growth media containing 100 μg/mL gentamicin until 2 hours post-infection, followed by growth media containing 10 μg/mL gentamicin for the remainder of the experiment. For enumeration of intravacuolar bacteria, macrophages were lysed in lysis buffer (1% Triton X-100, 0.1% SDS (Sigma) in DPBS) for 10 minutes and serial dilutions plated on LB agar containing 50 μg/mL streptomycin. CFU counts were taken at 2 hours, and 24 hours post-infection.
THP-1 cell infections
THP-1 cells were seeded in 24-well plates at a density of 2 x 105 cells/well in complete RPMI media supplemented with 100 nM phorbol myristate acetate (PMA) 16–24 hours prior to infection for differentiation. Overnight bacterial cultures (stationary phase) were pelleted and resuspended in DPBS, and subsequently diluted in RPMI and added to the THP-1 cells at a MOI of ≈ 50:1. Plates were centrifuged at 170 g for 5 min at room temperature and incubated for 25 min at 37°C in 5% CO2. Non-internalized bacteria were removed by three washes in DPBS and cells incubated in RPMI containing 100 μg/mL gentamicin until 2 hours post-infection, followed by RPMI containing 10 μg/mL gentamicin for the remainder of the experiment. CFU counts were taken at 2 hours, and 24 hours post-infection.
Antibodies
The goat polyclonal anti-Salmonella antibody CSA-1 (Kirkegaard and Perry Laboratories) was used at a dilution of 1:300; the mouse anti-LAMP1 antibody H4A3c developed by J.T. August and J. E. K. Hildreth, obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa (Department of Biological Sciences, Iowa, USA) was used at a dilution of 1:100. The mouse monoclonal anti-Golgin 97 antibody CDF4 (Molecular Probes) was used at a dilution of 1:100. Secondary antibodies were obtained from Thermo Fisher Scientific and used at a dilution of 1:500: Alexa 488-conjugated donkey anti-mouse, and Alexa 568-conjugated donkey anti-goat.
Immunofluorescence microscopy
Cell monolayers seeded on glass coverslips were fixed with 4% (vol/vol) paraformaldehyde in DPBS at room temperature for 10 minutes, and washed three times in DPBS. Excess paraformaldehyde was quenched in 50 mM ammonium chloride for 10 minutes at room temperature followed by two washes in DPBS.
For LAMP1+-tubules: Cells on coverslips were permeabilized in ice-cold acetone for 5 minutes at -20°C and then blocked in 1% Bovine Serum Albumin (BSA, wt/vol) (Sigma) in DPBS for 30 minutes at room temperature. Cells on coverslips were then incubated with primary antibodies diluted in 1% BSA in DPBS at room temperature for 1 hour followed by three washes in DPBS. Secondary antibodies diluted in 1% BSA in DPBS were added to the coverslips and incubated at room temperature for 1 hour and then washed once with DPBS. Cells were then incubated for 10 minutes at room temperature with DAPI (Invitrogen) in DPBS, followed by two DPBS washes. Cells were then washed in deionized water prior to mounting with ProLong Gold Antifade Mountant (Life Technologies) on glass slides. Microscopy was performed using Zeiss Axio Imager M2 (100x objective) and processed using Zeiss Zen Pro and ImageJ (NIH) softwares.
For Golgi-staining: Cells on coverslips were simultaneously permeabilized and blocked in 10% Normal Goat Serum (NGS) (Invitrogen) and 0.1% Triton X-100 in DPBS for 30 minutes at room temperature. Cells on coverslips were then incubated with primary antibodies diluted in 10% NGS and 0.1% Triton X-100 in DPBS and incubated at room temperature for 1 hour followed by three washes in DPBS. Secondary antibodies diluted in 10% NGS and 0.1% Triton X-100 in DPBS were then added to the coverslips and incubated at room temperature for 1 hour followed by three washes in DPBS. Coverslips were mounted on glass slides using ProLong® Gold Antifade Mountant with DAPI (Life Technologies) and incubated at room temperature for 24 h prior to sealing. Microscopy was performed using Olympus IX81 microscope (100x objective) and SlideBook 4.1.0 software. Distances were quantified using ImageJ software (NIH).
Scoring of phenotypes by microscopy
To quantify the number of infected cells with LAMP1+-tubules, we surveyed cells infected with Salmonella and immunolabelled for Salmonella and LAMP1. Uninfected cells were discarded from consideration. Infected cells were then scored for presence or absence of LAMP1+-tubules radiating outwards from a labelled Salmonella. The number of tubules/Salmonella was not considered as we were only concerned with the presence or absence of LAMP1+-tubules per each bacterium. At least 100 infected cells were scored blind in each experiment, and each experiment was repeated at least three times.
To quantify the distance from the Golgi, we surveyed cells infected with Salmonella and immunolabelled for Salmonella and Golgin-97. Distanced from the Golgi was enumerated by measuring the distance from the center of individual Salmonella cells to the center of the Golgi (μM). At least 200 Salmonella-to-Golgi distances were measured blind in each experiment, and all experiments were repeated at least three times.
Murine gastroenteritis model
Specific pathogen free C57BL/6 female 6-week old mice were obtained from The Jackson Laboratory (Bar harbor, Maine, USA) and housed in the animal facility at the University of British Columbia. Mice were pre-treated with 450 mg/L of streptomycin in drinking water as previously described [59]. Mice were orally gavaged with 2.8 x 107 CFU/mouse from overnight cultures of wild type and mutant SL1344 strains suspended in 0.1 mL DPBS. Mice were euthanized three days post-infection by anesthesia with isoflurane followed by CO2 asphyxiation and tissues were aseptically harvested for further evaluation. Ceca, colons, ilea and spleens were collected in 1 mL of sterile DPBS and homogenized by a FastPrep Homogenizer (MP Biochemicals). CFU from each organ was enumerated by serial dilutions on LB agar plates containing 100 μg/mL of streptomycin.
ELISAs
Cecum and spleen homogenates were centrifuged twice for 10 minutes at 13,000 g, and the supernatants were collected, diluted 1:2 in DPBS, and stored at -20°C. Levels of interferon-γ (IFN-γ) were determined by enzyme-linked immunosorbent assays (ELISAs) using BD OptEIA Mouse IFN-γ ELISA set (BD Biosciences) according for the manufacturer’s instructions. IFN-γ levels were normalized to the weight of the organs.
Statistical analysis
Statistical analysis was performed using Prism8 (GraphPad). In vitro infections, S. Typhimurium colonization in mice, and cytokine levels were analyzed. All data was found to not adhere to a Gaussian distribution using the following normality tests: Shapiro-Wilk test, and the Kolmogorov-Smirnov normality test with Dallal-Wilkinson-Lilliefor P value test. Data sets were analyzed using Kruskal-Wallis one-way analysis of variance (ANOVA) with Dunn’s multiple comparison post-test.
Supporting information
Single- and multiple-effector deletion mutants do not have impaired growth in LB liquid culture. 3 mL cultures of each strains were grown for 16–20 hours in Luria-Bertani (LB) medium at 37°C with shaking. Cultures were diluted 1:1000 in fresh media in a volume of 200 μL in a 96-well plate. Cell density was determined by incubating the plate at 37°C in a BioTek plate reader that shook the plate for 5 minutes before each read, every 20 minutes. Absorbance was read at 600 nm. The change in OD600 (ΔOD600) was calculated by subtracting the OD600 at Time = 0 from the OD600 at each selected time point. (A) Growth of single-effector deletion mutants in LB. (B) Growth of sequential-effector deletion mutants in LB. (C) Growth of multiple-effector deletion mutants in LB.
(TIF)
(PZFX)
Acknowledgments
The authors would like to Wanyin Deng for many fruitful discussions and assistance with troubleshooting.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Research reported in this publication was supported by the Canadian Institutes of Health Research under the award number FDN-15993 (B.B.F). The funders had no role in study design, data collection, and analysis, decisions to publish, or preparation of the manuscript.
References
- 1.Eswarappa SM, Negi VD, Chakraborty S, Chandrasekhar Sagar BK, Chakravortty D. Division of the Salmonella-containing vacuole and depletion of acidic lysosomes in Salmonella-infected host cells are novel strategies of Salmonella enterica to avoid lysosomes. Infect Immun. 2010;78(1):68–79. 10.1128/IAI.00668-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008. January;6(1):53–66. 10.1038/nrmicro1788 [DOI] [PubMed] [Google Scholar]
- 3.Steele-Mortimer O, Brumell JH, Knodler LA, Méresse S, Lopez A, Finlay BB. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002;4(1):43–54. 10.1046/j.1462-5822.2002.00170.x [DOI] [PubMed] [Google Scholar]
- 4.Steele-Mortimer O, Meresse S, Gorvel JP, Toh BH, Finlay BB. Biogenesis of Salmonella Typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol. 1999;1(1):33–49. 10.1046/j.1462-5822.1999.00003.x [DOI] [PubMed] [Google Scholar]
- 5.Smith AC, Cirulis JT, Casanova JE, Scidmore MA, Brumell JH. Interaction of the Salmonella-containing vacuole with the endocytic recycling system. J Biol Chem. 2005;280(26):24634–41. 10.1074/jbc.M500358200 [DOI] [PubMed] [Google Scholar]
- 6.Buchmeier NA, Heffron F. Inhibition of macrophage phagosome-lysosome fusion by Salmonella Typhimurium. Infect Immun. 1991;59(7):2232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drecktrah D, Knodler LA, Howe D, Steele-Mortimer O. Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic. 2007;8(3):212–25. 10.1111/j.1600-0854.2006.00529.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garvis SG, Beuzón CR, Holden DW. A role for the PhoP/Q regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell Microbiol. 2001;3(11):731–44. 10.1046/j.1462-5822.2001.00153.x [DOI] [PubMed] [Google Scholar]
- 9.Hashim S, Mukherjee K, Basu SK, Chem JB, Mukhopadhyay A. Live Salmonella modulate expression of Rab proteins to persist in a specialized compartment and escape transport to lysosomes. J Biol Chem. 2000;275(21):16281–8. 10.1074/jbc.275.21.16281 [DOI] [PubMed] [Google Scholar]
- 10.Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 2012. May;158(5):1147–61. 10.1099/mic.0.058115-0 [DOI] [PubMed] [Google Scholar]
- 11.LaRock DL, Chaudhary A, Miller SI. Salmonellae interactions with host processes. Nat Rev Microbiol. 2015;13(4):191–205. 10.1038/nrmicro3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol. 2009. February;12(1):117–24. 10.1016/j.mib.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brumell JH, Goosney DL, Finlay BB. SifA, a type III secreted effector of Salmonella Typhimurium, directs Salmonella-induced filament (Sif) formation along microtubules. Traffic. 2002;3(6):407–15. 10.1034/j.1600-0854.2002.30604.x [DOI] [PubMed] [Google Scholar]
- 14.Birmingham CL, Jiang X, Ohlson MB, Miller SI, Brumell JH. Salmonella-induced filament formation is a dynamic phenotype induced by rapidly replicating Salmonella enterica serovar Typhimurium in epithelial cells. Infect Immun. 2005;73(2):1204–8. 10.1128/IAI.73.2.1204-1208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc Natl Acad Sci U S A. 1993;90(22):10544–8. 10.1073/pnas.90.22.10544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajashekar R, Liebl D, Seitz A, Hensel M. Dynamic remodeling of the endosomal system during formation of Salmonella-induced filaments by intracellular Salmonella enterica. Traffic. 2008;9(12):2100–16. 10.1111/j.1600-0854.2008.00821.x [DOI] [PubMed] [Google Scholar]
- 17.LaRock DL, Brzovic PS, Levin I, Blanc MP, Miller SI. A Salmonella Typhimurium-translocated glycerophospholipid:cholesterol acyltransferase promotes virulence by binding to the RhoA protein switch regions. J Biol Chem. 2012;287(35):29654–63. 10.1074/jbc.M112.363598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liss V, Swart AL, Kehl A, Hermanns N, Zhang Y, Chikkaballi D, et al. Salmonella enterica remodels the host cell endosomal system for efficient intravacuolar nutrition. Cell Host Microbe. 2017;21(3):390–402. 10.1016/j.chom.2017.02.005 [DOI] [PubMed] [Google Scholar]
- 19.Szeto J, Namolovan A, Osborne SE, Coombes BK, Brumell JH. Salmonella-containing vacuoles display centrifugal movement associated with cell-to-cell transfer in epithelial cells. Infect Immun. 2009;77(3):996–1007. 10.1128/IAI.01275-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popp J, Noster J, Busch K, Kehl A, zur Hellen G, Hensel M. Role of host cell-derived amino acids in nutrition of intracellular Salmonella enterica. Infect Immun. 2015;83(12):4466–75. 10.1128/IAI.00624-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsden AE, Holden DW, Mota LJ. Membrane dynamics and spatial distribution of Salmonella-containing vacuoles. Trends Microbiol. 2007;15(11):516–24. 10.1016/j.tim.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 22.Henry T, Couillault C, Rockenfeller P, Boucrot E, Dumont A, Schroeder N, et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc Natl Acad Sci U S A. 2006. September 5;103(36):13497–502. 10.1073/pnas.0605443103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder N, Mota LJ, Méresse S. Salmonella-induced tubular networks. Trends Microbiol. 2011;19(6):268–77. 10.1016/j.tim.2011.01.006 [DOI] [PubMed] [Google Scholar]
- 24.Beuzón CR, Méresse S, Unsworth KE, Ruíz-Albert J, Garvis S, Waterman SR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19(13):3235–49. 10.1093/emboj/19.13.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckner MMC, Croxen M, Arena ET, Finlay BB. A comprehensive study of the contribution of Salmonella enterica serovar Typhimurium SPI2 effectors to bacterial colonization, survival, and replication in typhoid fever, macrophage, and epithelial cell infection models. Virulence. 2011. May 1;2(3):208–16. 10.4161/viru.2.3.15894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman JA, Ohl ME, Miller SI. The Salmonella enterica serovar Typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect Immun. 2003;71(1):418–27. 10.1128/IAI.71.1.418-427.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Rossanese OW, Brown NF, Kujat-Choy S, Galán JE, Finlay BB, et al. The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Mol Microbiol. 2004;54(5):1186–98. 10.1111/j.1365-2958.2004.04344.x [DOI] [PubMed] [Google Scholar]
- 28.Knodler LA, Steele-Mortimer O. The Salmonella effector PipB2 affects late endosome/lysosome distribution to mediate Sif extension. Mol Biol Cell. 2005;16:4108–23. 10.1091/mbc.E05-04-0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao EA, Brittnacher M, Haraga A, Jeng RL, Welch MD, Miller SI. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol Microbiol. 2003;48(2):401–15. 10.1046/j.1365-2958.2003.t01-1-03456.x [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Albert J, Yu X-J, Beuzón CR, Blakey AN, Galyov EE, Holden DW. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol Microbiol. 2002;44(3):645–61. 10.1046/j.1365-2958.2002.02912.x [DOI] [PubMed] [Google Scholar]
- 31.Ohlson MB, Huang Z, Alto NM, Blanc M-P, Dixon JE, Chai J, et al. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008. November 13;4(5):434–46. 10.1016/j.chom.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jennings E, Thurston TLM, Holden DW. Salmonella SPI-2 type III secretion system effectors: Molecular mechanisms and physiological consequences. Cell Host Microbe. 2017;22(2):217–31. 10.1016/j.chom.2017.07.009 [DOI] [PubMed] [Google Scholar]
- 33.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998. January 30;207(2):149–57. 10.1016/S0378-1119(97)00619-7 [DOI] [PubMed] [Google Scholar]
- 34.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30(1):163–74. 10.1046/j.1365-2958.1998.01047.x [DOI] [PubMed] [Google Scholar]
- 35.Deiwick J, Salcedo SP, Boucrot E, Gilliland SM, Henry T, Petermann N, et al. The translocated Salmonella effector proteins SseF and SseG interact and are required to establish an intracellular replication niche. Infect Immun. 2006;74(12):6965–72. 10.1128/IAI.00648-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhle V, Hensel M. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell Microbiol. 2002;4(12):813–24. 10.1046/j.1462-5822.2002.00234.x [DOI] [PubMed] [Google Scholar]
- 37.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138(2):179–207. 10.1016/0022-2836(80)90283-1 [DOI] [PubMed] [Google Scholar]
- 38.Ferrieres L, Hemery G, Nham T, Guerout A-M, Mazel D, Beloin C, et al. Silent Mischief: Bacteriophage Mu Insertions Contaminate Products of Escherichia coli Random Mutagenesis Performed Using Suicidal Transposon Delivery Plasmids Mobilized by Broad-Host-Range RP4 Conjugative Machinery. J Bacteriol. 2010;192(24):6418–27. 10.1128/JB.00621-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant SGN, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A. 1990;87(12):4645–9. 10.1073/pnas.87.12.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoiseth SK, Stocker BAD. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. [Internet]. Vol. 291, Nature. 1981. p. 238–9. 10.1038/291238a0 [DOI] [PubMed] [Google Scholar]
- 41.Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun. 1999;67(11):5690–8. 10.1128/iai.67.11.5690-5698.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung KY, Finlay BB. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1991;88(24):11470–4. 10.1073/pnas.88.24.11470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krieger V, Liebl D, Zhang Y, Rajashekar R, Chlanda P, Giesker K, et al. Reorganization of the endosomal system in Salmonella-infected cells: the ultrastructure of Salmonella-induced tubular compartments. PLoS Pathog. 2014;10(9):e1004374 10.1371/journal.ppat.1004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder N, Henry T, de Chastellier C, Zhao W, Guilhon A-A, Gorvel J-P, et al. The virulence protein SopD2 regulates membrane dynamics of Salmonella-containing vacuoles. PLoS Pathog. 2010;6(7):e1001002 10.1371/journal.ppat.1001002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler L a., Schroer T a., et al. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic. 2008;9(12):2117–29. 10.1111/j.1600-0854.2008.00830.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajashekar R, Liebl D, Chikkaballi D, Liss V, Hensel M. Live cell imaging reveals novel functions of Salmonella enterica SPI2-T3SS effector proteins in remodeling of the host cell endosomal system. PLoS One. 2014;9(12):e115423 10.1371/journal.pone.0115423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brumell JH, Tang P, Mills SD, Finlay BB. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic. 2001;2(16):643–53. 10.1034/j.1600-0854.2001.20907.x [DOI] [PubMed] [Google Scholar]
- 48.Boucrot E, Henry T, Borg J-P, Gorvel J-P, Méresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Sci (New York, NY). 2005;308(5725):1174–8. 10.1126/science.1110225 [DOI] [PubMed] [Google Scholar]
- 49.Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003. October 1;22(19):5003–14. 10.1093/emboj/cdg517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsden AE, Mota LJ, Münter S, Shorte SL, Holden DW. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell Microbiol. 2007;9(10):2517–29. 10.1111/j.1462-5822.2007.00977.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abrahams GL, Müller P, Hensel M. Functional dissection of SseF, a type III effector protein involved in positioning the Salmonella-containing vacuole. Traffic. 2006. August;7(8):950–65. 10.1111/j.1600-0854.2006.00454.x [DOI] [PubMed] [Google Scholar]
- 52.Henry T, Gorvel JP, Méresse S. Molecular motors hijacking by intracellular pathogens. Cell Microbiol. 2006;8(1):23–32. 10.1111/j.1462-5822.2005.00649.x [DOI] [PubMed] [Google Scholar]
- 53.Domingues L, Holden DW, Mota LJ. The Salmonella effector SteA contributes to the control of membrane dynamics of Salmonella-containing vacuoles. Infect Immun. 2014;82(7):2923–34. 10.1128/IAI.01385-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrison RE, Brumell JH, Khandani A, Bucci C, Scott CC, Jiang X, et al. Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell. 2004;15(7):3146–54. 10.1091/mbc.E04-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X-J, Liu M, Holden DW. Salmonella effectors SseF and SseG interact with mammalian protein ACBD3 (GCP60) to anchor Salmonella-containing vacuoles at the Golgi network. MBio. 2016;7(4):e00474–16. 10.1128/mBio.00474-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beuzón CR, Salcedo SP, Holden DW. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology. 2002;148(9):2705–15. 10.1099/00221287-148-9-2705 [DOI] [PubMed] [Google Scholar]
- 57.Figueira R, Watson KG, Holden DW, Helaine S. Identification of Salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar Typhimurium: Implications for rational vaccine design. MBio. 2013;4(2):1–10. 10.1128/mBio.00065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 2001;3(2):75–84. 10.1046/j.1462-5822.2001.00087.x [DOI] [PubMed] [Google Scholar]
- 59.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76(10):4726–36. 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tükel Ç, Tsolis RM, et al. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17(11):498–506. 10.1016/j.tim.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Auweter SD, Bhavsar AP, de Hoog CL, Li Y, Chan YA, van der Heijden J, et al. Quantitative mass spectrometry catalogues Salmonella pathogenicity island-2 effectors and identifies their cognate host binding partners. J Biol Chem. 2011. July 8;286(27):24023–35. 10.1074/jbc.M111.224600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao W, Moest T, Zhao Y, Guilhon A-A, Buffat C, Gorvel J-P, et al. The Salmonella effector protein SifA plays a dual role in virulence. Sci Rep. 2015;5:12979 10.1038/srep12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stein MA, Leung KY, Zwick M, Garcia-del Portillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol. 1996;20(1):151–64. 10.1111/j.1365-2958.1996.tb02497.x [DOI] [PubMed] [Google Scholar]
- 64.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. SifA permits survival and replication of Salmonella Typhimurium in murine macrophages. Cell Microbiol. 2001;3(2):75–84. 10.1046/j.1462-5822.2001.00087.x [DOI] [PubMed] [Google Scholar]
- 65.Dumont A, Boucrot E, Drevensek S, Daire V, Gorvel JP, Poüs C, et al. SKIP, the host target of the Salmonella virulence factor SifA, promotes kinesin-1-dependent vacuolar membrane exchanges. Traffic. 2010;11(7):899–911. 10.1111/j.1600-0854.2010.01069.x [DOI] [PubMed] [Google Scholar]
- 66.McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, et al. PLEKHM1 Regulates Autophagosome-Lysosome Fusion through HOPS Complex and LC3/GABARAP Proteins. Mol Cell. 2015;57(1):39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 67.Diacovich L, Dumont A, Lafitte D, Soprano E, Guilhon A-A, Bignon C, et al. Interaction between the SifA virulence factor and its host target SKIP is essential for Salmonella pathogenesis. J Biol Chem. 2009. November 27;284(48):33151–60. 10.1074/jbc.M109.034975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moest T, Zhao W, Zhao Y, Schüssler JM, Yan W, Gorvel JP, et al. Contribution of bacterial effectors and host proteins to the composition and function of Salmonella-induced tubules. Cell Microbiol. 2018;20(12):1–13. 10.1111/cmi.12951 [DOI] [PubMed] [Google Scholar]
- 69.Kaniuk NA, Canadien V, Bagshaw RD, Bakowski M, Braun V, Landekic M, et al. Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell Microbiol. 2011;13(11):1812–23. 10.1111/j.1462-5822.2011.01663.x [DOI] [PubMed] [Google Scholar]
- 70.Marsman M, Jordens I, Kuijl C, Janssen L, Neefjes J. Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol Biol Cell. 2004;15(June):2954–64. 10.1091/mbc.e03-08-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wasylnka JA, Bakowski MA, Szeto J, Ohlson MB, Trimble WS, Miller SI, et al. Role for myosin II in regulating positioning of Salmonella-containing vacuoles and intracellular replication. Infect Immun. 2008;76(6):2722–35. 10.1128/IAI.00152-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuhle V, Abrahams GL, Hensel M. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic. 2006;7(6):716–30. 10.1111/j.1600-0854.2006.00422.x [DOI] [PubMed] [Google Scholar]
- 73.Reinicke AT, Hutchinson JL, Magee AI, Mastroeni P, Trowsdale J, Kelly AP. A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J Biol Chem. 2005;280(15):14620–7. 10.1074/jbc.M500076200 [DOI] [PubMed] [Google Scholar]
- 74.Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell. 2011. December 13;21(6):1171–8. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liss V, Hensel M. Take the tube: remodeling of the endosomal system by intracellular Salmonella enterica. Cell Microbiol. 2015;17(April):639–47. 10.1111/cmi.12441 [DOI] [PubMed] [Google Scholar]
- 76.Fierer J, Okamoto S, Banerjee A, Guineya DG. Diarrhea and colitis in mice require the Salmonella pathogenicity island 2-encoded secretion function but not SifA or Spv effectors. Infect Immun. 2012;80(10):3360–70. 10.1128/IAI.00404-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuda S, Haneda T, Saito H, Miki T, Okada N. Salmonella enterica effectors SifA, SpvB, SseF, SseJ, and SteA contribute to type III secretion system 1-independent inflammation in a streptomycin-pretreated mouse model of colitis. Infection and Immunity. 2019;e00872–18. 10.1128/IAI.00872-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison C a, Smith HO, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–5. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 79.Chang ACY, Cohen SN. Construction and characterization of amplifiable DNA cloning vectors derived from P15A cryptic miniplasmid. J Bacteriol. 1978;134(3):1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Single- and multiple-effector deletion mutants do not have impaired growth in LB liquid culture. 3 mL cultures of each strains were grown for 16–20 hours in Luria-Bertani (LB) medium at 37°C with shaking. Cultures were diluted 1:1000 in fresh media in a volume of 200 μL in a 96-well plate. Cell density was determined by incubating the plate at 37°C in a BioTek plate reader that shook the plate for 5 minutes before each read, every 20 minutes. Absorbance was read at 600 nm. The change in OD600 (ΔOD600) was calculated by subtracting the OD600 at Time = 0 from the OD600 at each selected time point. (A) Growth of single-effector deletion mutants in LB. (B) Growth of sequential-effector deletion mutants in LB. (C) Growth of multiple-effector deletion mutants in LB.
(TIF)
(PZFX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.