Abstract
Checkpoint kinase 1 (Chk1) is a key element in the DNA-damage response pathway that is required for maintaining genomic stability. To study the potential role of Chk1 in mammary tumorigenesis, we disrupted it using a Cre/loxP system. We showed that although Chk1 heterozygosity caused abnormal development of the mammary gland, it was not sufficient to induce tumorigenesis. Simultaneous deletion of one copy of p53 failed to rescue the developmental defects; however, it synergistically induced mammary tumor formation in Chk1+/−; MMTV-Cre animals with a median time to tumor latency of about 10 months. Chk1 deficiency caused a preponderance of abnormalities, including prolongation, multipolarity, misalignment, mitotic catastrophe and loss of spindle checkpoint, that are accompanied by reduced expression of several cell cycle regulators, including Mad2. On the other hand, we also showed that Chk1 deficiency inhibited mammary tumor formation in mice carrying a homozygous deletion of p53, uncovering a complex relationship between Chk1 and p53. Furthermore, inhibition of Chk1 with a specific inhibitor, SB-218078, or acute deletion of Chk1 using small hairpin RNA killed mammary tumor cells effectively. These data show that Chk1 is critical for maintaining genome integrity and serves as a double-edged sword for cancer: although its inhibition kills cancer cells, it also triggers tumorigenesis when favorable mutations are accumulated for cell growth.
Keywords: Chk1, mitotic catastrophe, genome integrity, mammary cancer, SB-218078
Introduction
The majority of cancers are characterized by abnormalities of the cell cycle (Kastan and Bartek, 2004; Deng, 2006). Most commonly, G1/S checkpoint abrogation figures prominently in these abnormalities, primarily because of mutations in the broadly important tumor suppressor p53, which enforces the checkpoint through p21 (Harper et al., 1993; el-Deiry et al., 1993; Brugarolas et al., 1995; Deng et al., 1995). Mitotic catastrophe, however, can be avoided if these cells retain a functional G2/M checkpoint. Loss of this checkpoint can send cancer cells down an apoptotic pathway, which explains why abrogation of the G2/M checkpoint is an interest target for antitumor therapy, particularly when combined with traditional DNA-damaging agents. An especially attractive aspect of this approach is that cells with mutated p53 may be especially vulnerable to such treatment, whereas normal cells with functional p53 and an intact G1/S checkpoint should be relatively unaffected. If this is the case, drugs that abrogate the G2/M checkpoint represent a new class of anticancer agents that target p53-null tumors, which may otherwise be resistant to chemotherapy and radiation (Lowe et al., 1994).
The regulation of cell cycle progression involves overlapping pathways, but it has become clear that the checkpoint kinase 1 (Chk1) is essential in regulating progression in multiple phases of the cell cycle, including the G1/S and G2/M transition, as well as cytokinesis (Liu et al., 2000; Zhao et al., 2002; Xiao et al., 2003; Peddibhotla et al., 2009). It has been shown that Chk1 mediates signals from the upstream DNA-damage response kinases ATR and ATM, and acts principally by phosphorylating Cdc25A at inhibitory sites, blocking cellular entry into mitosis (Bartek and Lukas, 2003; Zhang et al., 2008). Chk1 also has a role in delaying the onset of anaphase in mitotic cells when there is DNA damage (Collura et al., 2005). It has been shown that the hereditary breast cancer-associated gene-1 (BRCA1), which has a vital role in genome surveillance and represses breast cancer formation, is also essential in activating Chk1 in response to DNA damage (Miki et al., 1994; Yarden et al., 2002; Deng and Wang, 2003).
Studying functions of Chk1 in tumorigenesis and progression is made difficult by the fact that Chk1−/− embryos die at E6.5 (Liu et al., 2000; Takai et al., 2000). Using a Cre-LoxP-mediated approach, however, indicated that haploid loss or complete loss of Chk1 in the mouse mammary gland failed to cause tumor formation, and instead resulted in extensive underdevelopment (Lam et al., 2004). It is noted that Chk1 heterozygous mammary epithelial cells exhibited three distinct haploinsufficient phenotypes, including inappropriate S-phase entry, accumulation of DNA damage during replication, and premature mitotic entry. It was therefore proposed that Chk1 had multiple functions critical to tumor suppression, and that Chk1 haploinsufficient phenotypes could cause mammary tumorigenesis. However, experimental evidence of tumor formation in mammary epithelial-specific Chk1 mutant mice has not yet been provided (Lam et al., 2004).
To investigate whether long-term Chk1 deficiency could induce tumorigenesis and how Chk1 deficiency affects cells differently depending on their p53 status, we created an animal model carrying conditional knockout of both Chk1 and p53 in the mammary gland. Our analysis revealed that Chk1 is critical for viability of both normal mammary epithelial cells and cancer cells, providing a molecular basis for the observed dual function of Chk1 in tumor inhibition and promotion under different conditions.
Results
Targeted disruption of Chk1 and p53 in mouse mammary epithelium
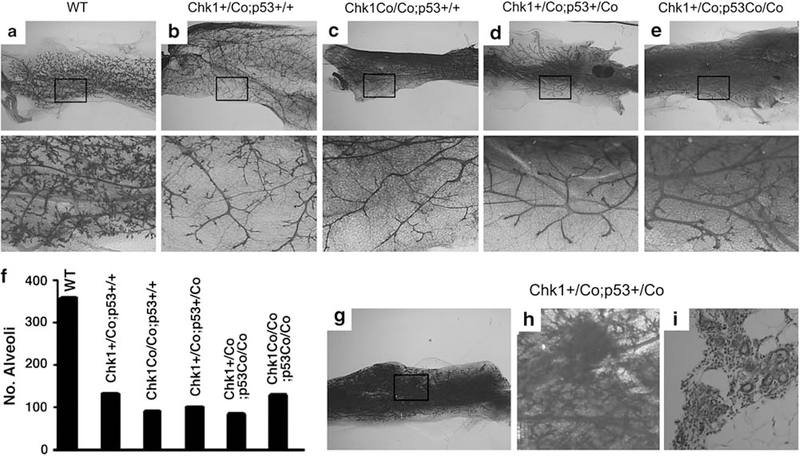
Chk1 was disrupted in mammary tissue by crossing a mouse strain that carries a loxP-flanked exon 2 of Chk1 (Liu et al., 2000) with MMTV-Cre transgenic mice (Wagner et al., 1997). Analysis of the mammary glands of Chk+/Co;MMTV-Cre and Chk1Co/Co;MMTV-Cre mice at 2 months to 12 months of age detected abnormal mammary development (Figures 1a–c). The mutant glands appeared less dense than controls, and this abnormality was more impressive when the alveoli were visualized in high power images. Our previous studies indicated that mammary defects caused by Brca1 mutation could be rescued by p53 deficiency (Xu et al., 2001; Li et al., 2007). As Chk1 activity is regulated at least in part by Brca1 (Yarden and Brody, 2001), we were interested in investigating whether the developmental defect associated with Chk1 mutation could be rescued by simultaneous deletion of p53. Therefore, the Chk1Co/+;MMTV-Cre mice were further crossed with p53Co/+ mice (Jonkers et al., 2001) to generate mice with various genotypes. Analysis of these mutant mice indicated that mammary tissues of Chk1+/Co;p53+/Co;MMTV-Cre, Chk1+/Co;p53Co/Co;MMTV-Cre and Chk1Co/Co;p53Co/Co;MMTV-Cre mice were also underdeveloped (Figures 1d, e, and data not shown). Quantitative measurement of mammary branches and alveoli of mutant and control mice within arbitrary areas of the same size revealed that haploid loss or complete loss of Chk1 significantly impaired mammary branch morphogenesis and alverolization, and that these defects could not be rescued by haploid or complete loss of p53 (Figure 1f).
Figure 1.
Haploid or complete loss of Chk1 impaired mammary gland development independent of p53. (a–e) Whole-mount imaging of mammary glands from 12-month-old parous WT (a), Chk1+/Co;p53+/+;MMTV-Cre (b), Chk1Co/Co;p53+/+;MMTV-Cre (c), Chk1+/Co;p53+/Co;MMTV-Cre (d) and Chk1+/Co;p53Co/Co;MMTV-Cre (e) mice. Boxed areas are enlarged and placed underneath. (f) Quantitative measurement of mammary branches and alveoli of mutant and control mice within arbitrary areas of equal size, which were counted under high power magnification. (g–i) Whole-mount imaging of a mammary gland from a 12-month-old parous Chk1+/Co;p53+/Co;MMTV-Cre mouse showing increased branching morphogenesis and hyperplastic foci (g). The boxed area is enlarged (h) and sectioned (i).
Mammary hyperplasia and tumor formation in Chk1 and p53 double-mutant mice
Despite the observation that impaired p53 function did not rescue the growth defects, hyperplastic and noninvasive focal lesions were detected in the mammary glands of some Chk1+/Co;p53+/Co;MMTV-Cre mice starting at 12 months of age (Figures 1g–i). In Chk1+/Co;p53Co/Co;MMTV-Cre glands, the formation of multifocal hyperplasia was more uniform than that in Chk1+/Co;p53+/Co;MMTV-Cre glands, suggesting that deletion of the remaining wild-type p53 allele had a stronger impact in inducing this lesion. Deletion of the remaining wild-type Chk1 allele, however, had a negative impact, as no hyperplastic foci were observed in the mammary glands of Chk1Co/Co;p53Co/Co;MMTV-Cre mice of similar age.
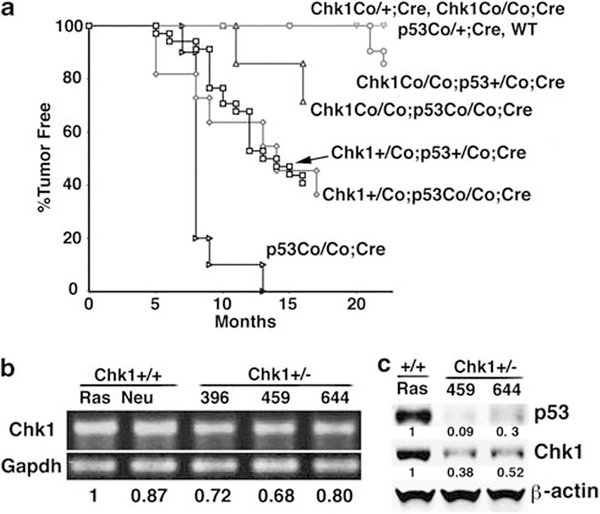
To study the potential impact of mutations of Chk1 and p53 on mammary neoplasia, we followed cohorts of animals that had gone through multiple cycles of pregnancy for tumor formation. Twenty out of 33 (60%) Chk1+/Co;p53+/Co;MMTV-Cre mice developed tumors, with a median time to tumor latency of approximately 10.5 months, whereas none of the control mice (p53+/Co;MMTV-Cre and Chk1+/Co;MMTV-Cre, Chk1Co/Co;MMTV-Cre, or MMTV-Cre mice, n412 of each) developed tumors during the same period of time (Figure 2a). These data suggest that haploid loss of Chk1 and p53 synergistically induces mammary tumor formation.
Figure 2.
Mammary tumor formation in mice carrying mammary-specific deletion of Chk1 and p53. (a) Kaplan–Meier survival curve of mice with different genotypes as indicated. (b) Transcriptional levels of Chk1 in three cell lines derived from three mammary tumors of Chk1+/Co;p53+/Co;MMTV-Cre mice (396 459, 644), and two cell lines derived from Chk1 wild-type MMTV-ras (Ras) and MMTV-neu (Neu) mice, as revealed by reverse transcription–PCR. (c) Protein levels of Chk1 and p53 in Chk1+/− (644 and 459) and Ras cell lines as revealed by western blot analysis. The intensity of bands was measured by Quantity One Software (Bio-Rad, Hercules, CA, USA) and normalized using intensity of Gapdh or β-actin. The quantified numbers are shown at the bottom of the pictures.
We further showed that 7 out of 11 Chk1+/Co;p53Co/Co;MMTV-Cre mice (64%) developed mammary tumors with a median time to tumor latency of 9 months. Thus, heterozygous loss of p53 allowed tumor formation in Chk1+/Co mice, whereas the loss of the remaining wild-type p53 allele slightly increased tumorigenesis (Figure 2a). Next, we studied the effect of homozygous loss of Chk1 on tumorigenesis. We showed that 3 out of 21 Chk1Co/Co;p53+/Co;MMTV-Cre mice (14%) developed mammary tumors with a median time to tumor latency of 21 months. Two out of seven Chk1Co/Co;p53Co/Co;MMTV-Cre mice (29%) developed mammary tumors with a median time to tumor latency of 14 months (Figure 2a). This observation indicates that although Chk1 heterozygosity promotes tumorigenesis, loss of both copies of Chk1 attenuates the synergistic effect in tumor formation. This observation also suggests that a certain level of Chk1 function is needed to support tumor formation. Consistent with this, our study indicated that mammary tumors derived from the Chk1+/Co;p53+/Co;MMTV-Cre mice and cell lines isolated from these tumors still expressed Chk1, although at a lower level compared with Chk1 wild-type controls (Figures 2b and c), whereas all tumors analyzed from these mice lost p53 protein (Figure 2c, and data not shown). These data suggest that Chk1 behaves in a haploinsufficient manner for mammary tumor formation in a p53-deficient background.
An interesting finding came from analyzing p53Co/Co;MMTV-Cre mice, which were initially set up as a control for tumorigenesis in Chk1+/Co;p53Co/Co;MMTV-Cre mice and Chk1Co/Co;p53Co/Co;MMTV-Cre mice. Among a studying population of 10 p53Co/Co;MMTV-Cre mice, 4 developed lymphoma at 7–8 months of age, 5 developed mammary tumor at 8 months and the remaining 1 developed a mammary tumor at 13 months (Figure 2a). The higher tumor incidence of p53Co/Co;MMTV-Cre mice than Chk1+/Co;p53Co/Co;MMTV-Cre mice and Chk1Co/Co;p53Co/Co;MMTV-Cre mice suggests that impaired Chk1 function inhibits tumorigenesis induced by p53 deficiency. Thus, the relationship between Chk1 and p53 in tumorigenesis is complex with the following features: (1) Chk1+/Co, Chk1Co/Co or p53+/Co mutation alone in the mammary gland does not cause mammary tumor formation; (2) Chk1+/Co;MMTV-Cre or Chk1Co/Co;MMTV-Cre together with p53+/Co mutation synergistically induces mammary tumorigenesis; and (3) homozygous loss of p53 causes mammary tumor formation at a relatively high frequency, which is reduced by heterozygous loss of Chk1 (Chk1+/Co;p53Co/Co;MMTV-Cre), and is further inhibited by homozygous loss of Chk1 (Chk1+/Co;p53Co/Co;MMTV-Cre).
Mammary tumors exhibited diverse histopathological features
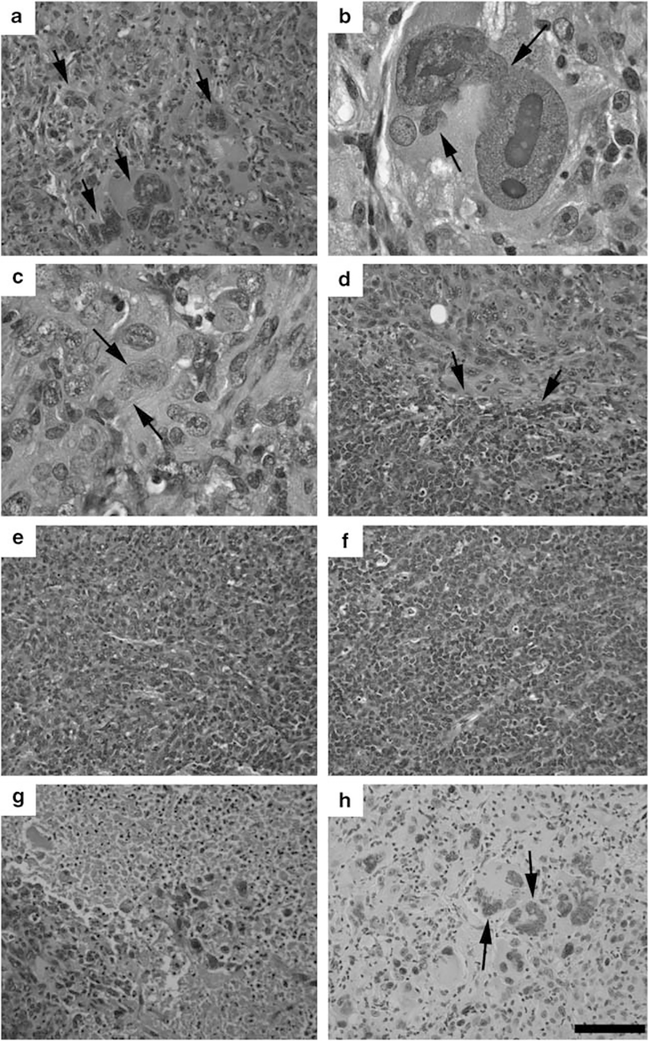
Mammary tumors developed in the Chk1+/Co;p53+/Co;MMTV-Cre mice were analyzed for their histopathological features. The majority of tumors were adenocarcinomas, but were highly diverse in histopathology. A characteristic feature of the tumors was the presence of giant cells that had nuclear diameters many times larger than those of surrounding cells (Figures 3a and b). The nucleus of the giant cells was usually misshapen, containing smaller projections seeming to be in a process of separation (Figure 3b). Most tumor cells exhibited nuclear polymorphism (Figure 3c). Sometimes, a distinct histological boundary was formed within tumors, where cells with a distinct nuclear histology were adjacent (Figure 3d, arrows). The diverse histological appearance suggests that the impaired function of Chk1 and p53 might lead to random genetic alterations that initiated the tumorigenesis. A majority of tumors exhibited extensive angiogenesis (Figure 3e), although small patches of tumor cells of uniform size and with less angiogenesis were also observed (Figure 3f). We had also analyzed three tumors of varying sizes (0.5, 1 and 2 cm) that were developed in a single mouse. The data indicated that all these tumors contained many necrotic cells, with a higher number of necrotic cells in larger tumors (Figure 3g), possibly due to an increasing difficulty in getting nutrition supply for these cells. We further showed that only about 1% of cancer cells were 5-bromodeoxyuridine labeled, suggesting that these tumors were not highly proliferative. It is noted that all giant cells were 5-bromodeoxyuridine negative (Figure 3h), and PCNA negative and phospho-p27 positive, suggesting that they were in G1 phase (Supplementary Figure 1).
Figure 3.
Histologic diversity of mammary tumors found in Chk1+/Co;p53+/Co;MMTV-Cre mice as revealed in hematoxylin and eosinstained sections. (a) An adenocarcinoma-containing cell with a giant nucleus. (b) An enlarged image of the nucleus of a giant cell. (c) Nuclear polymorphism (arrows). (d) Distinct histological boundaries (arrows). (e, f) Most tumors exhibited extensive angiogenesis (e), although small patches of tumor cells of uniform size, and less angiogenesis were also observed (f). (g) Areas of necrotic cells. More necrotic areas can be found in tumors with larger sizes. (h) 5-Bromodeoxyuridine (BrdU) labeling showing that all giant cells are BrdU negative (arrows).
It is also noted that tumors derived from Chk1Co/Co;p53Co/Co;MMTV-Cre mice exhibited similar diverse histopathological features similar to those derived from Chk1+/Co;p53+/Co;MMTV-Crie mice (Supplementary Figures 2a–c). In contrast, tumors derived from p53Co/Co;MMTV-Cre mice displayed uniform histological features and were highly proliferative (Supplementary Figure 2d), which is consistent with the view that loss of Chk1 suppressed p53-null status-induced mammary tumorigenesis.
Mammary tumors exhibited marked aneuploidy and chromosomal aberrations
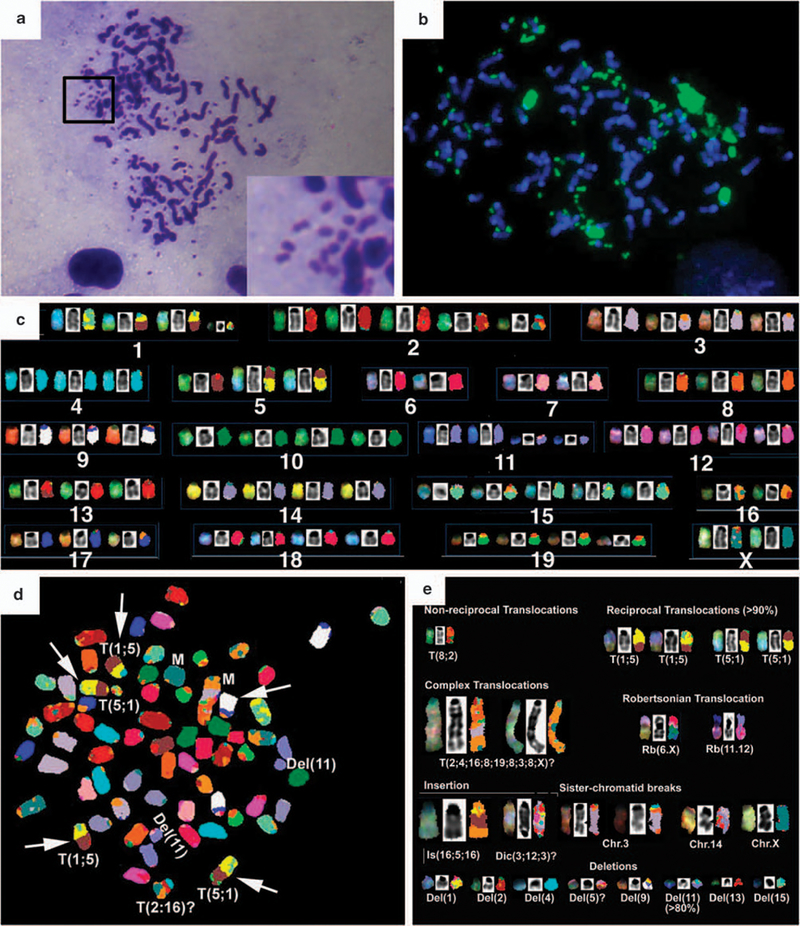
The nuclear polymorphism of these tumors prompted us to investigate the status of chromosomes derived from tumor cells. Chromosome spreads of primary tumor cells from Chk1+/Co;p53+/Co;MMTV-Cre mice cultured overnight showed that ~68% cells were aneuploid (Figure 4a). In nearly 40% of the spreads, distinct double minute chromosomes were detected that mapped to chromosome 9, as confirmed by fluorescence in situ hybridization (Figure 4b). To further characterize chromosomal aberrations, we performed spectral karyotyping on metaphase spreads derived from early passages of two primary tumors.
Figure 4.
Spectral karyotyping (SKY) analysis of chromosomes prepared from primary mammary tumors developed in Chk1+/Co;p53+/Co;MMTV-Cre mice and a cancer cell line, 644. (a) Metaphases of cells showing polypoidy and double minute chromosomes. (b) Fluorescence in situ hybridization on metaphase spreads from D644, showing double minutes that map to chromosome 9 (green) using a whole-chromosome probe for chromosome 9. (c) SKY classification of chromosomes. (d) A representative SKY image showing chromosomal aberrations as indicated. (e) A summary of all chromosomal aberrations from D644.
Twelve spreads were analyzed from the tumor, D644. These cells showed a wide variation in ploidy levels (68–86 chromosomes), with near tetraploid to hypertetraploid cells (Figure 4c). A variety of structural chromosomal aberrations were identified by spectral karyotyping analysis (Figure 4c). It is noted that a reciprocal translocation of T(1;5)(5;1) was detected clonally (>90%). In single spreads, chromosomal aberrations such as non-reciprocal and Robertsonian translocations were also identified that were non-clonal. Complex translocations occurred because of fusions from parts of different chromosomes (Chr2, 3, 4, 8, 16, 19 and X). These aberrant fusions resulted in elongated chromosomes and were seen clonally in two metaphase spreads. A striking structural aberration was a sister chromatid break independently detected on Chr3, Chr14 and ChrX. A variety of chromosomal deletions were also found, notable among which was a deletion of Chr11 in >80% of the spreads analyzed. The second tumor, D643, contained an average number of chromosomes (~80, near tetraploid) and also structural aberrations, including chromosome translocations, insertions, deletions and fusions (Supplementary Figure 3).
Chk1 haploinsufficiency resulted in mitotic catastrophe
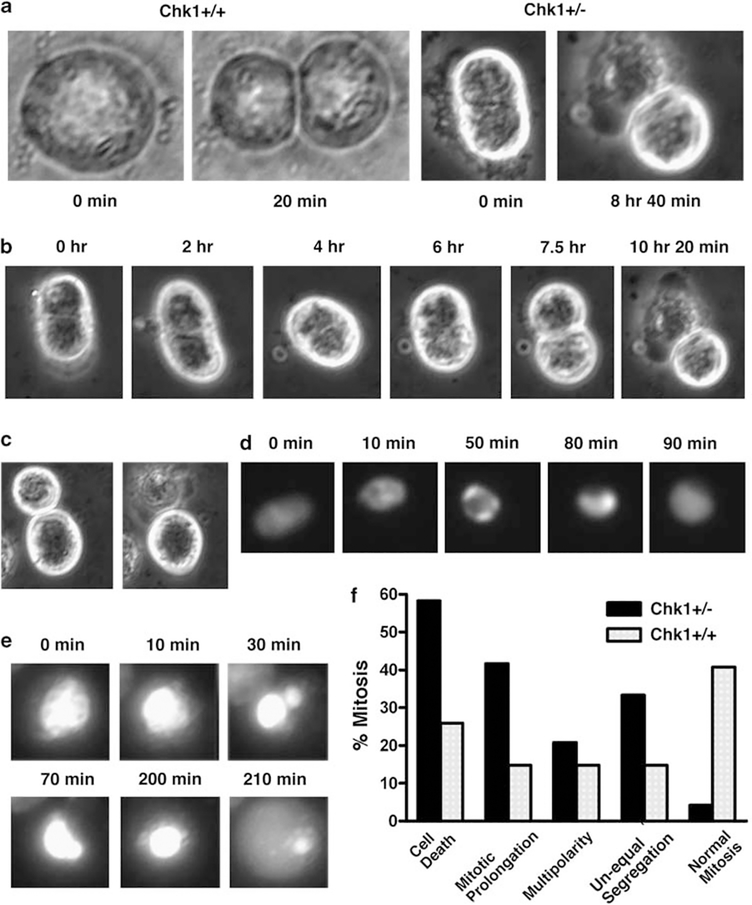
To further investigate the source of this chromosome damage, as well as the origin of the polynuclear features in Chk1-associated tumors, we examined mitoses in three cell lines (369, 459 and 644) derived from Chk1+/Co;p53+/Co;MMTV-Cre tumors using time-lapse photography. Although mitosis of most Chk1+/+control tumor cells (MMTV-neu, a cell line that was derived from a mammary tumor of a MMTV-neu,p53+/− mouse; or MMTV-ras, a cell line that was derived from a mammary tumor of a MMTV-ras,p53+/+ mouse) (Brodie et al., 2001b) was completed within 60 min, mitosis in many Chk1 mutant cells could last more than 8 h, which often led to cell death (Figure 5a). Many mutant cells encountered difficulties in cytokinesis. This cell (Figure 5b) already had a cleavage furrow at the time we noticed it. The cell became slightly elongated with a clearer cleavage furrow at 2 h. However, the cleavage was aborted and the cell maintained single at 4 h. At 6 h, the cell began a new round of cell division and formed a new cleavage furrow at 7.5 h. This time the cleavage was effective, but it resulted in the death of one daughter cell (Figure 5b). We also found that many divisions yielded daughter cells of different sizes, and that the smaller cells always died shortly after the division (Figure 5c).
Figure 5.
Mitotic catastrophe and abortive chromosome segregation revealed by time-lapse experiment. (a) Images showing abnormal (from a Chk1+/− cell) and normal (from an MMTV-neu cell that is wild type for Chk1) mitoses. (b) Time-lapse images showing abnormal mitosis of one Chk1+/− cell. (c) Asymmetric division of Chk1+/− cells produces cells of different size and causes the subsequent death of a smaller cell. (d, e) Abnormal mitosis observed in cells carrying a stably expressed histone H2B-GFP expression vector, allowing their DNA to be visualized. (f) Summary of mitotic abnormalities in Chk1+/− mutant and Neu cells.
It is conceivable that smaller cells might obtain less amount of DNA, and could not therefore maintain their normal cell cycle progression. To investigate this, we transfected mutant cells with histone H2B-GFP expression vector so that their chromosomes could be monitored during cell division. We found that many cells entered mitosis, but they were arrested at metaphase and then directly returned to interphase (Figure 5d). In some cases, cells underwent unequal segregation of their DNA. Such segregations were usually not successful and resulted in mitotic abortion (Figure 5e). As summarized in Figure 5f, the time-lapse study revealed a range of mitotic catastrophe in Chk1 mutant cells. These include: (1) cell death during mitosis; (2) mitotic prolongation, defined as delayed cytokinesis beyond 1 h; (3) multipolarity, defined as the presence of more than two poles of cytokinesis; and (4) asymmetric cleavage furrow, leading to the formation of daughter cells of different size. About 95% of mitotic figures exhibited one or more of these abnormalities. We have also examined mitotic figures of tumor cells derived from MMTV-neu mice under the same condition and found that over 40% of cells were normal (Figure 5f). Although the above abnormalities could be detected in the MMTV-neu cells, they were much milder than those observed in Chk1 mutant cells.
To investigate whether the mitotic catastrophe phenotypes could also be recapitulated in the cells that carry acute knockdown of Chk1, we performed small hairpin RNA knockdown of Chk1 in both MMTV-neu and MMTV-ras cells. Our analysis indicated that suppression of Chk1 in both cell lines significantly reduced their proliferation (Supplementary Figure 4). Time lapse revealed similar mitotic catastrophe in these cells (Supplementary Figure 5 and Figure 6) compared with the Chk1+/− cells shown earlier (Figure 5). Altogether, these data revealed an essential role of Chk1 in maintaining genome integrity.
Figure 6.
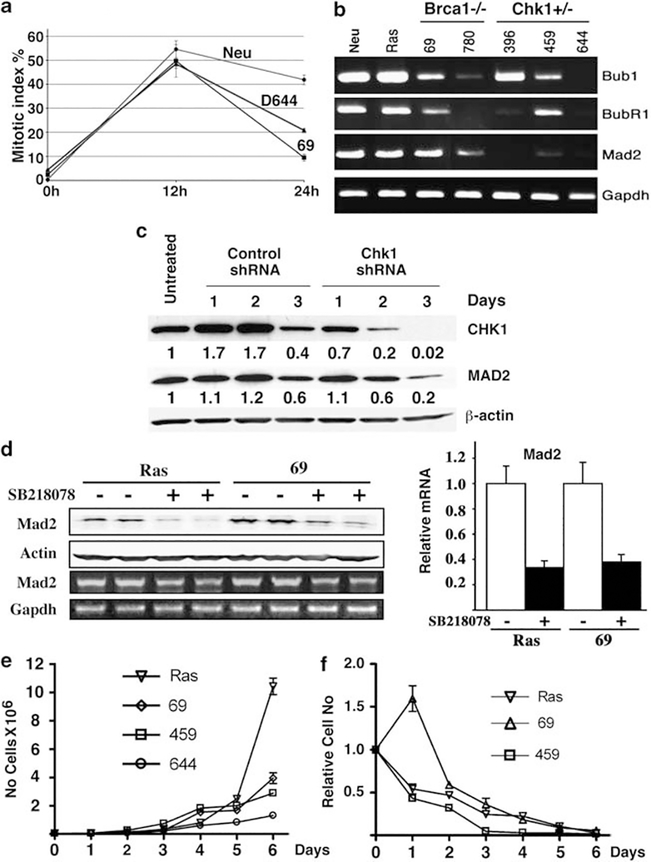
Haploid loss of Chk1 impaired by the spindle checkpoint is partially mediated by Mad2. (a) Mitotic index of cells at 12 and 24 h after treatment with nocodazole. (b) Expression of several genes involved in the spindle checkpoint pathway as revealed by reverse transcription (RT)–PCR. (c) Small hairpin RNA-mediated knockdown of Chk1 in MMTV-neu cells decreases Mad2 as revealed by western blot analysis (left), which is quantified on the basis of untransfected cells (right). (d) Treatment of Chk1 wild-type cells with SB-218078 results in reduced levels of Mad2 as revealed by western blot analysis (left) and real-time RT–PCR (right). A quantitative measurement is provided (right). (e) Growth curve of cell lines without receiving SB-218078 (Cat. no. 559402, Calbiochem, La Jolla, CA, USA). (f) Relative response of cell lines to SB-218078 (1 μg/ml) treatment compared with untreated cells.
Haploid loss of Chk1 impaired the spindle checkpoint
It was shown previously that Chk1 deficiency in yeast and chicken DT40 cells caused defective spindle checkpoint (Collura et al., 2005; Zachos et al., 2007). As Chk1+/− cells exhibited mitotic catastrophe, we suspected that the spindle checkpoint might be impaired in these cells. To examine this, we treated Chk1+/− and control cells with nocodazole, which depolymerizes microtubules and activates the spindle checkpoint. We found that although Chk1+/+ cells (MMTV-neu) were able to maintain a relatively high mitotic index 48 h after treatment (~40%) (Figure 6a), the mitotic index of Chk1+/− and Brca1−/− cells declined to ~20%, which is closer to Brca1 mutant cells (~10%) that are known to be defective in the spindle checkpoint (Wang et al., 2004). These data suggest that the haploid loss of Chk1 impairs the spindle checkpoint, highlighting an essential role for Chk1 in this process.
Multiple factors have a role in the spindle checkpoint (Deng, 2006). We found that Chk1 mutant cells had reduced levels of Bub1, BubR1 and Mad2, with the latter reduced in all three Chk1+/− cell lines examined (Figure 6b). This observation suggests that Mad2 has an important role in mediating Chk1 function in the spindle checkpoint. To investigate whether reduced Chk1 activity is a direct cause for the reduced levels of Mad2, we performed small hairpin RNA-mediated knockdown of Chk1 in MMTV-neu cells. Our data indicated that as Chk1 levels gradually reduced after lentiviral-mediated infection of small hairpin RNA specific for Chk1, Mad2 expression also decreased (Figure 6c). Next, we treated these with SB-218078, a Chk1-specific inhibitor (Jackson et al., 2000), and found that inhibition of Chk1 resulted in the downregulation of Mad2 (Figure 6d). Thus, it is conceivable that reduced expression of Mad2 triggered by Chk1 haplo-insufficiency cannot hold cells in the metaphase, thereby leading to premature entry into anaphase and mitotic abnormalities.
Inhibition of Chk1 with SB-218078 effectively killed mammary tumor cells
Corresponding to the defective spindle checkpoint and high rate of mitotic catastrophe, Chk1+/− mutant cancer cells grew much slower than did the cancer cell lines driven by MMTV-ras and MMTV-neu (Figure 6e, and data not shown). The reduced growth of the Chk1+/− mutant cancer cells was even more pronounced than the Brca1−/− cancer cells (Figure 6e), which also suffer mitotic catastrophe but to a lesser extent (Xu et al., 1999; Wang et al., 2004). Moreover, our earlier data indicated that Chk1 heterozygosity or Chk1 homozygosity (complete loss of Chk1) inhibited tumorigenesis in p53Co/Co;MMTV-Cre mice (Figure 2a). This observation prompted us to investigate the effect of inhibition of Chk1 using a Chk1 inhibitor, SB-218078, on these tumor cell lines. We found that this drug, which is a more specific inhibitor for Chk1 than some other Chk1 inhibitors (Jackson et al., 2000), effectively killed three mammary tumor cell lines, MMTV-ras, Brca1−/− (69) and Chk1+/− (459) (Figure 6f). It is noted that Chk1+/− cells were most sensitive to SB-218078 treatment, perhaps due to their lower levels of Chk1, compared with Ras and Brca1−/− cells. We had also compared the behavior of the SB-218078-treated cells and the Chk1-small hairpin RNA-treated cells during mitosis using time lapse, and found that these cells similarly exhibited high levels of mitotic catastrophe under both conditions (Supplementary Figures 7 and 8). These data suggest that inhibition of Chk1 with a specific inhibitor is effective to block growth of a variety of tumor cells.
Discussion
Previous investigations indicated that Chk1 deficiency induced a profound growth defect and cell cycle abnormalities (Liu et al., 2000; Takai et al., 2000; Lam et al., 2004; Peddibhotla et al., 2009). However, whether Chk1 loss can cause tumorigenesis is unknown. Using conditional Chk1 mutant mice, we found that either complete or heterozygous loss of Chk1 in the mammary gland causes profound growth defect, similar to early Chk1−/− embryos that cannot be overcome by p53 deficiency. These data suggest that Chk1 is critical for the viability of mammary epithelial cells. However, we found that the loss of one or both p53 alleles in the Chk1+/Co and Chk1Co/Co mammary gland was eventually able to cause hyperplastic foci formation and resulted in tumor formation after a long latency. This indicates that some Chk1+/− and Chk1−/− cells are able to overcome the growth defect, perhaps after accumulating multiple changes.
Why does the absence of p53 not rescue the developmental defects, but allows tumor formation associated with Chk1 deficiency? Previous investigations showed that Chk1 is an essential kinase involved in the regulation of G1/S and G2/M cell cycle checkpoints and in cytokinesis (Liu et al., 2000; Zhao et al., 2002; Xiao et al., 2003; Peddibhotla et al., 2009). Reduced Chk1 function in Pten-deficient cells leads to the accumulation of double-stranded DNA breaks and genetic instability, particularly at fragile sites (Puc and Parsons, 2005; Puc et al., 2005; Durkin et al., 2006). It was also shown that Chk1 has a regulatory role in the spindle checkpoint in chicken DT40 cells (Zachos et al., 2007) and in U2OS cells (Carrassa et al., 2009). Here, our data provide in vivo evidence that Chk1 is essential for the spindle checkpoint in mouse mammary epithelial cells and mammary cancer cells. Thus, Chk1 deficiency yields profound abnormalities, as evidenced by mitotic catastrophe, which is much more severe than that caused by Brca1 mutation (Shen et al., 1998; Xu et al., 1999; Wang et al., 2004). This may account for the reason why absence of p53 could not rescue the lethality caused by Chk1 deficiency, although the absence of p53 is sufficient to suppress the lethality and growth defects associated with Brca1 mutation (Xu et al., 2001; Li et al., 2007). However, the abnormalities in multiple cell cycle checkpoints and cytokinesis in Chk1 mutant cells could eventually result in accumulation of genetic alterations that may cooperate with p53 deficiency to induce tumor formation.
As evidence for this view, we found numerous chromosome structural aberrations in mammary cancer cells. We also showed that a majority of tumor cells contained 80 or more chromosomes, suggesting that tetraploidization may serve as a major mechanism that contributes to tumorigenesis, that is, increase gene dose to overcome profound growth defects associated with impaired Chk1 function. Chromosome tetraploidization frequently occurs before aneuploidy during tumorigenesis (Margolis, 2005). It also occurs in mammary cancers developed in transgenic mice with overexpression of oncogenes, such as Aurora-A (Wang et al., 2006). We suspect that the failure of cytokinesis in the Chk1 mutant cells could be, in part, responsible for this phenotype.
Our data revealed that Chk1 mutant tumor cells lost the spindle checkpoint that is accompanied by reduced expression levels of several members in two evolutionarily conserved protein families: Bub1, BubR1 and, most pronounced, Mad2. It is known that Mad2 binds to unattached kinetochores and inhibits anaphase-promoting complex together with BubR1 (Sudakin et al., 2001; Tang et al., 2001). Mad2 deficiency results in premature anaphase onset, chromosome missegregation and apoptosis, leading to early lethality at embryonic day 5, whereas Mad2 haploinsufficiency causes lung tumors after a long latency (Dobles et al., 2000; Michel et al., 2001). We have also shown previously that Mad2 has an essential role in mediating functions of Brca1 in the spindle checkpoint (Wang et al., 2004). On the basis of these findings, we postulated that the reduced levels of Mad2 might be responsible, at least in part, for the spindle checkpoint defect observed here. Consistently, our data revealed that inhibition of Chk1 reduces Mad2 expression and triggers cell death, although it remains unclear how Chk1 affects Mad2 expression. This is an interesting issue that will be investigated further in future studies.
Although our study reveals that absence of p53 enhances tumorigenesis in Chk1 mutant cells, it also indicates that loss of Chk1 inhibits p53 deficiency-associated tumor formation. Several lines of evidence indicate that Chk1 is critical for viability of cancer cells, which may provide a clue to understand this complex pattern of Chk1 and p53 interaction. First, in all mammary tumor formation in Chk1+/Co;p53+/Co;MMTV-Cre mice, there was no loss of heterozygosity for Chk1, although p53-heterozygous mice did lose their remaining copy of p53 wild-type allele in all tumors examined. Second, tumor incidence of p53Co/Co;MMTV-Cre mice is much higher than that of Chk1+/Co;p53Co/Co;MMTV-Cre and Chk1Co/Co;p53Co/Co;MMTV-Cre mice. Finally, we found that acute knockdown of Chk1 in both Neu and Ras cells, which were derived from mammary cancers of MMTV-neu and MMTV-ras transgenic mice (Muller et al., 1988; Brodie et al., 2001a), resulted in profound growth defects and mitotic catastrophe. These observations prompted us to hypothesize that complete loss of Chk1 is harmful to cancer cell growth because of profound genetic instability and growth defects. This would also explain why treatment with a pharmaceutical antagonist to Chk1 effectively kills cancer cells, perhaps due to the fact that the treated cells do not have sufficient time to undergo profound genome alterations to overcome the growth defect triggered by Chk1 inhibition.
In summary, using the Cre/loxP system to mutate both Chk1 and p53 in mammary tissue, we showed that Chk1 acts as a haploinsufficient tumor suppressor and cooperates with p53 to inhibit mammary tumor formation. We also provide in vivo evidence that Chk1 has an essential role in the spindle checkpoint to maintain genome integrity. As Chk1 is critical for viability of cancer cells, we have tested the effect of SB-218078 on cell growth in vitro, and our data indicate that treatment of SB-218078 can efficiently inhibit growth of multiple tumor cells. Chk1 inhibition is being pursued as a promising target for the treatment of cancer (Blagden and de Bono, 2005). The oldest of such drugs, UCN-01, has been in development for the longest period of time and is now in Phase II clinical trials (Hotte et al., 2006). However, phase I trials revealed certain disadvantages of this drug, including dose-limiting toxicities (Kortmansky et al., 2005) and extremely long half-life due to binding to a1-acid glycoprotein (Fuse et al., 1998). Thus, our data suggest that SB-218078, which is a more specific inhibitor for Chk1 than UCN01 (Jackson et al., 2000), may also be tested in the therapeutic treatment of breast cancers.
Materials and methods
Mice and mating
The Chk1Co/+ (Liu et al., 2000), p53Co/+ (Jonkers et al., 2001) and MMTV-Cre (Wagner et al., 1997) mice were crossed to generate mice of different genotypes, including Chk1+/Co;MMTV-Cre, Chk1Co/Co;MMTV-Cre, Chk1+/Co;p53+/Co;MMTV-Cre, Chk1+/Co;p53Co/Co;MMTV-Cre, Chk1Co/Co;p53+/Co;MMTV-Cre, p53+/Co;MMTV-Cre, p53Co/Co;MMTV-Cre and Chk1Co/Co;p53Co/Co;MMTV-Cre. Genotyping of these mice was performed as described (Wagner et al., 1997; Liu et al., 2000; Jonkers et al., 2001). The female mice were kept with males for continuous mating and the number of pregnancies was recorded. When killed, one of the fourth glands was used for whole-mount preparation and the others used for DNA, RNA and/or histological analysis (Deng and Xu, 2004). The protocol for animal studies was approved by the ‘Animal Care and Use Committee’ of the National Institute of Diabetes and Digestive and Kidney Diseases.
RNA isolation and reverse transcription–PCR
Total RNA was isolated from cells or tissues with STAT-60 following the manufacturer’s protocol (TEL-TEST, Friendswood, TX, USA). Complementary DNA was synthesized with Cells-to-cDNAII (Ambion, Austin, TX, USA). Primer sequences are as follows:
Gapdh: forward 5′-ACAGCCGCATCTTCTTGTGC-3′, reverse 5′-CACTTTGCCACTGCAAATGG-3′; Chk1: forward 5′-TTTGGGAGAAGGTGCCTATG-3′, reverse 5′-TTC TGGACAGTCTATGGCCC-3′.
Cell culture, chromosome spread and spectral karyotyping analysis
Brca1 mutant cell lines 69 and 780, and MMTV-ras and MMTV-neu were as described (Brodie et al., 2001b). Establishment of cell lines 399, 459 and 644, from primary mammary tumors in Chk1+/Co;p53+/Co;MMTV-Cre mice, and chromosomal spread were as described (Deng and Xu, 2004). Spectral karyotyping was performed as described (Padilla-Nash et al., 2006). Fluorescence in situ hybridization was performed on metaphase spreads by hybridization with whole chromosome paints against target chromosomes.
Whole-mount staining of mammary glands, histology, immunohistochemical staining and western blotting
Whole-mount staining of mammary glands was carried out as described (Deng and Xu, 2004). For histology, tissues were fixed in 10% formalin, blocked in paraffin, sectioned, stained with hematoxylin and eosin, and examined by light microscopy. Detection of primary antibodies was performed using the Zymed Histomouse SP Kit (Zymed, South San Francisco, CA, USA) according to the manufacturer’s instructions. Western analysis was performed using standard procedures. Antibodies for Chk1 and p53 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibody for 5-bromodeoxyuridine was purchased from Covance (Princeton, NJ, USA).
Supplementary Material
Acknowledgements
We gratefully acknowledge members of Deng laboratory for a critical reading of the article. This work was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, USA.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Bartek J, Lukas J. (2003). Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421–429. [DOI] [PubMed] [Google Scholar]
- Blagden S, de Bono J. (2005). Drugging cell cycle kinases in cancer therapy. Curr Drug Targets 6: 325–335. [DOI] [PubMed] [Google Scholar]
- Brodie SG, Xu X, Li C, Kuo A, Leder P, Deng CX. (2001a). Inactivation of p53 tumor suppressor gene acts synergistically with c-neu oncogene in salivary gland tumorigenesis. Oncogene 20: 1445–1454. [DOI] [PubMed] [Google Scholar]
- Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX. (2001b). Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene 20: 7514–7523. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. (1995). Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377: 552–557. [DOI] [PubMed] [Google Scholar]
- Carrassa L, Sanchez Y, Erba E, Damia G. (2009). U2OS cells lacking Chk1 undergo aberrant mitosis and fail to activate the spindle checkpoint. J Cell Mol Med 13: 1565–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collura A, Blaisonneau J, Baldacci G, Francesconi S. (2005). The fission yeast Crb2/Chk1 pathway coordinates the DNA damage and spindle checkpoint in response to replication stress induced by topoisomerase I inhibitor. Mol Cell Biol 25: 7889–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. (1995). Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82: 675–684. [DOI] [PubMed] [Google Scholar]
- Deng CX. (2006). BRCA1: cell cycle checkpoint, genetic instability, DNA damage response, and cancer evolution. Nucleic Acids Res 34: 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX, Wang RH. (2003). Roles of BRCA1 in DNA damage repair: a link between development and cancer. Hum Mol Genet 12: R113–R123. [DOI] [PubMed] [Google Scholar]
- Deng CX, Xu X. (2004). Generation and analysis of Brca1 conditional knockout mice. Methods Mol Biol 280: 185–200. [DOI] [PubMed] [Google Scholar]
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. (2000). Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101: 635–645. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Arlt MF, Howlett NG, Glover TW. (2006). Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene 25: 4381–4388. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. (1993). WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825. [DOI] [PubMed] [Google Scholar]
- Fuse E, Tanii H, Kurata N, Kobayashi H, Shimada Y, Tamura T et al. (1998). Unpredicted clinical pharmacology of UCN-01 caused by specific binding to human alpha1-acid glycoprotein. Cancer Res 58: 3248–3253. [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816. [DOI] [PubMed] [Google Scholar]
- Hotte SJ, Oza A, Winquist EW, Moore M, Chen EX, Brown S et al. (2006). Phase I trial of UCN-01 in combination with topotecan in patients with advanced solid cancers: a Princess Margaret Hospital Phase II Consortium study. Ann Oncol 17: 334–340. [DOI] [PubMed] [Google Scholar]
- Jackson JR, Gilmartin A, Imburgia C, Winkler JD, Marshall LA, Roshak A. (2000). An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res 60: 566–572. [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. (2001). Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. (2004). Cell-cycle checkpoints and cancer. Nature 432: 316–323. [DOI] [PubMed] [Google Scholar]
- Kortmansky J, Shah MA, Kaubisch A, Weyerbacher A, Yi S, Tong W et al. (2005). Phase I trial of the cyclin-dependent kinase inhibitor and protein kinase C inhibitor 7-hydroxystaurosporine in combination with Fluorouracil in patients with advanced solid tumors. J Clin Oncol 23: 1875–1884. [DOI] [PubMed] [Google Scholar]
- Lam MH, Liu Q, Elledge SJ, Rosen JM. (2004). Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell 6: 45–59. [DOI] [PubMed] [Google Scholar]
- Li W, Xiao C, Vonderhaar BK, Deng CX. (2007). A role of estrogen/ERalpha signaling in BRCA1-associated tissue-specific tumor formation. Oncogene 26: 7204–7212. [DOI] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K et al. (2000). Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev 14: 1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE et al. (1994). p53 status and the efficacy of cancer therapy in vivo. Science 266: 807–810. [DOI] [PubMed] [Google Scholar]
- Margolis RL. (2005). Tetraploidy and tumor development. Cancer Cell 8: 353–354. [DOI] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W et al. (2001). MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409: 355–359. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S et al. (1994). A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71. [DOI] [PubMed] [Google Scholar]
- Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. (1988). Singlestep induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54: 105–115. [DOI] [PubMed] [Google Scholar]
- Padilla-Nash HM, Barenboim-Stapleton L, Difilippantonio MJ, Ried T. (2006). Spectral karyotyping analysis of human and mouse chromosomes. Nat Protoc 1: 3129–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. (2009). The DNA-damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci USA 106: 5159–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L et al. (2005). Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- Puc J, Parsons R. (2005). PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle 4: 927–929. [DOI] [PubMed] [Google Scholar]
- Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L et al. (1998). A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene 17: 3115–3124. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. (2001). Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T et al. (2000). Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev 14: 1439–1447. [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. (2001). Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 1: 227–237. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L et al. (1997). Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res 25: 4323–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Yu H, Deng CX. (2004). A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci USA 101: 17108–17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T et al. (2006). Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene 25: 7148–7158. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Chen Z, Gunasekera AH, Sowin TJ, Rosenberg SH, Fesik S et al. (2003). Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J Biol Chem 278: 21767–21773. [DOI] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA et al. (2001). Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet 28: 266–271. [DOI] [PubMed] [Google Scholar]
- Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW et al. (1999). Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell 3: 389–395. [DOI] [PubMed] [Google Scholar]
- Yarden RI, Brody LC. (2001). Identification of proteins that interact with BRCA1 by Far-Western library screening. J Cell Biochem 83: 521–531. [DOI] [PubMed] [Google Scholar]
- Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. (2002). BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat Genet 30: 285–289. [DOI] [PubMed] [Google Scholar]
- Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC et al. (2007). Chk1 is required for spindle checkpoint function. Dev Cell 12: 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Poh A, Fanous AA, Eastman A. (2008). DNA damage-induced S phase arrest in human breast cancer depends on Chk1, but G2 arrest can occur independently of Chk1, Chk2 or MAPKAPK2. Cell Cycle 7: 1668–1677. [DOI] [PubMed] [Google Scholar]
- Zhao H, Watkins JL, Piwnica-Worms H. (2002). Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci USA 99: 14795–14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.