Abstract
The essential SecA motor ATPase acts in concert with the SecYEG translocon to secrete proteins into the periplasmic space of Escherichia coli. In aqueous solutions, SecA exists largely as dimers, but the oligomeric state on membranes is less certain. Crystallographic studies have suggested several possible solution dimeric states, but its oligomeric state when bound to membranes directly or indirectly via the translocon is controversial. We have shown using disulfide crosslinking that the principal solution dimer, corresponding to a crystallographic dimer (PDB 1M6N), binds only weakly to large unilamellar vesicles (LUV) formed from E. coli lipids. We report here that other soluble crosslinked crystallographic dimers also bind weakly, if at all, to LUV. Furthermore, using a simple glutaraldehyde crosslinking scheme, we show that SecA is always monomeric when bound to LUV formed from E. coli lipids.
Keywords: protein secretion, large unilamellar vesicles (LUV), protein partitioning, disulfide crosslinking, glutaraldehyde crosslinking
1. INTRODUCTION
The translocation of proteins across cell membranes is a conserved and essential process in all living cells that is carried out by the general secretory (Sec) system. In Escherichia coli, secretion of proteins across the inner membrane into the periplasm is performed primarily by the soluble SecA ATPase motor protein that forms a secretion-competent complex with the SecYEG channel (reviewed in [1]). In vivo, SecA appears to be distributed equally between the cytosol and the inner membrane [2] where negatively charged lipids are required for efficient protein translocation [3, 4]. In vitro, SecA interacts strongly with lipid monolayers [5] and bilayers containing negatively charged lipids [6–10]. While it is generally accepted that SecA is a homodimer in solution [11–15], the oligomeric state in cell membranes has been controversial. In some reports, dimeric SecA appears to enhance protein secretion [12, 16–18], while in others monomeric SecA is sufficient for secretion [14, 19–21]. X-ray- [22, 23] and cryo-EM [24] structures show monomeric SecA bound to SecYEG, although high salt concentrations and detergents used during sample preparation might have affected the oligomeric state. For the cryo-EM structure, an engineered SecA-SecY-Preprotein complex expressed in E. coli [21] was used, leaving open the question exactly how SecA gains access to SecY in vivo. The oligomeric state of SecA during preprotein translocation is thus perplexing (For reviews, see [25–27]).
Because recent in vitro studies suggest that SecA gains access to the translocon via a membrane-associated intermediate [4, 28], it is crucial to examine the role of the lipid bilayer in determining the oligomeric state of SecA. A logical first step was taken by Lill et al. [6] who examined the properties of SecA in the presence of liposomes. They established that acidic lipids are sufficient for SecA binding. They further established that binding activated SecA’s ATPase activity, which was stimulated by preproteins. After it was later established that SecA is homodimeric in solution [11], Or et al. [14] provided evidence that dimeric SecA dissociates into monomers upon binding, consistent with a subsequent study showing that long-chain phospholipid analogs can cause dissociation of SecA dimers [29]. Or et al. used Förster resonance energy transfer (FRET) between coumarin- and fluorescein-labeled SecA molecules to determine the extent of dimerization in the presence or absence of liposomes formed from various phospholipids. Here, we reëxamine the effect of lipids on SecA dimerization using a more direct approach by asking if SecA can bind significantly as a dimer to large unilamellar vesicles (LUV) of different lipid compositions.
Six SecA x-ray crystal structures [30–34] have suggested several possible dimeric states in solution, but it is likely that there is a dynamic equilibrium between the different dimers and the monomer in solution [14, 26, 35]. Exactly which dimer, if any, represents the functional cytoplasmic dimer in vivo has been controversial, but the consensus is that the anti-parallel dimer structure determined for Bacillus subtilis SecA (PDB: 1M6N) represents the dominant dimer [16, 36–39]. We recently examined the partitioning of two disulfide-stabilized 1M6N dimers based and found that the dimer binds only weakly to large unilamellar vesicles (LUV) formed from E. coli lipids [10]. Given that multiple dimeric states in solution are likely, one can imagine that one of the other dimeric species determined by x-ray crystallography might bind strongly to LUV and shift the population toward a membrane-bound dimeric state. We report here that three other crystallographic dimers—3JV2 [40], 2FSF [34], and 1NL3 [31]—bind weakly, if at all, to LUV. Because this result does not rule out the possibility that some other dimeric SecA form of undetermined structure might bind strongly, we adopted a more general crosslinking approach. We used a short, seconds-long crosslinking procedure designed not to perturb the equilibrium distribution of SecA monomers and dimers between the aqueous and membrane phases. The results indicate that SecA binds only as monomers to LUVs formed from E. coli lipids.
2. Materials and Methods
2.1. Materials
All phospholipids were purchased form Avanti Polar Lipids (Alabaster, AL): E. coli total extract (catalog number 100500), 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC, 850457), 1-palmytoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE, 850757), 1-palmioyl-2-oleoyl-sn-glycero-3-phospho-(1’rac-glycerol) (POPG, 840457), and cardiolipin (841199).
2.2. Construction of the cysteine-mutants
Plasmids pT7-C4-secA (cysteine-less) and pT7-G11C+S661C-secA encoding for Escherichia coli SecA mutants were a generous gift from Donald Oliver [41]. Starting from the cysteine-less construction, single amino acid substitution mutagenesis was performed by PCR mutagenesis using the primers shown in Table 1. Mutations were confirmed via DNA sequencing.
Table 1 – Identification of the modified SecA residues.
Forward primers used for recombinant PCR to introduce amino acid substitutions into E. coli SecA.
| Structure | Mutation(s) | Primer |
|---|---|---|
| 3JV2 | G765C | 5’-CTTCGAGAAATGCGTCATGCTGC-3’ |
| 2FSF | A521C | 5’-TGCTCGGTGGTAGCTGGCAGTGTGAAGTTGCCGCG-3’ |
| 1NL3 | G605C | 5’-GAGTATCCTGCATGATGCG-3’ |
| G788C | 5’-TCTGCGTCAGTGTATCCACCTG-3’ | |
2.3. SecA Protein production
E. coli WT-SecA, G11C+S661C-SecA (1M6N), G765C-SecA (3JV2), A521C-SecA (2FSF) and G605C+G788C-SecA (1NL3) were obtained from E. coli BL21 cells carrying the corresponding secA gene with a C-terminal His6-tag under the control of the T5 promotor. Cells were grown in LB medium at 37°C with constant shaking. Log-phase cultures (OD 0.8) were stimulated with IPTG (0.5 mM) for 2 hours at 30°C. Cells were then harvested by centrifugation at 4,000 rpm for 15 minutes and the resulting pellet was stored at −20°C until needed.
2.4. SecA Protein purification
All protein purification steps and centrifugations were performed at 4°C. Bacterial pellets (from 400 mL culture) were solubilized in 48 mL of Buffer A (50 mM Hepes-NaOH pH 7.4, 10 mM imidazole, and 50 mM KCl) for 10 minutes at room temperature. Protease inhibitor cocktail (Roche) was added before the cell suspension was passed through a French Pressure Cell (SLM-Amico) at 10,000 lb/in2. The suspension was then centrifuged at 13,000 g for 15 minutes in a Fisherbrand Accuspin 17R centrifuge to pellet the membrane fraction. The supernatant was loaded onto a Talon-Resin (1.5×5 cm) previously equilibrated with 25 mL of Buffer A. His-tagged SecA protein was then eluted with Buffer B (50 mM Hepes-NaOH pH 7.4, 500 mM imidazole, 50 mM KCl, and 1 mM GSH). Two-mL fractions were collected, and the protein profile analyzed using SDS-PAGE. Fractions containing 100 kDa SecA were then pooled, concentrated, and loaded onto a Superdex 200 increase 10/300 GL (GElifescience, Reference 28990944) equilibrated in 50 mM Hepes-NaOH pH 7.4, 1 mM GSH, 1 mM MgCl2, and 50 mM KCl). SecA was then eluted at a flow rate of 0.5 mL/minute and protein elution monitored by optical absorbance at 280 nm. 500-μL fractions were collected and the protein profile analyzed using SDS-PAGE. Fractions containing the SecA protein were then pooled and the protein concentration was estimated using the BioRad Assay with BSA as a reference and a molecular weight of 102 kDa.
2.5. Liposome preparation
Phospholipids dissolved in chloroform were dried under a stream of nitrogen and further dried under vacuum overnight. Lipids were then suspended in 25 mM Hepes-NaOH pH 7.4 and vortexed 15 minutes. Large unilamellar vesicles (LUVs) were prepared by extrusion through a polycarbonate membrane (100 nm pore diameter). Lipid concentrations were determined according to the procedure of Bartlett [42].
2.6. Crosslinking of cysteine-mutants using Glutathione
Proteins (G11C+S661C, G765C, A521C, and G605C+G788C) were purified in the presence of 1 mM reduced glutathione (GSH). The formation of disulfide bridge was induced by reducing the GSH concentration to 0.1 mM and introducing 5 mM oxidized glutathione (GSSG) using a PD-10 desalting column, before running the samples on a non-reducing denaturing gel.
2.7. Crosslinking of SecA in solution using glutaraldehyde
SecA (1 μM) in 50 mM Hepes-KOH pH 8.0, 100 mM KGlu, 2 mM MgCl2 and 0.5 mM DTT were incubated for 30 minutes at 37°C. Crosslinking was initiated by adding glutaraldehyde (GA) at a final concentration of 0.15%. The reaction was allowed for 15 seconds and then stopped by adding an excess of Tris-HCl pH 7.0 (final concentration of 100 mM) before running the samples on a denaturing gel.
2.8. Titration of GA-crosslinked SecA monitored by tryptophan fluorescence
GA-crosslinked sample was loaded onto a desalting PD-10 column previously equilibrated in 50 mM Hepes-NaOH pH 7.4, 100 mM KGlu, 1 mM EDTA, 2 mM MgCl2, and 0.5 mM DTT. Lipid vesicles made from E. coli lipids were then added to SecA (final dimeric concentration of 1 μM) and binding was then allowed for at least 30 minutes. Fluorescence spectra were recorded at 37°C using a SLM 8000c spectrophotometer (Urbana, FL) using an excitation wavelength of 295 nm. Excitation slits were not wider than 8 nm; emission slits were 4 nm. Unless otherwise indicated, the emission polarizer was oriented at 0° relative to the vertical and the excitation polarizer at 90°. Spectra were measured using a 10 × 2 mm quartz cuvette. Spectra were collected in the region of 310–400 nm in increments of 1 nm. Ten or more spectra were averaged to achieve an adequate signal-to-noise ratio. Fluorescence intensities, excited at 295 nm (1 nm slit) were recorded between 310 and 400 nm (4 nm slit). The same conditions were used for recording scans in buffer alone, which were subtracted from the appropriate peptide spectra. Fluorescence signals were corrected for light scattering as previously described [43]. Three or more data sets were collected for all experiments.
2.8. Crosslinking of SecA by glutaraldehyde after partitioning into LUVs
SecA (1 μM) in 50 mM Hepes-KOH pH 8.0, 100 mM KGlu, 2 mM MgCl2 and 0.5 mM DTT was incubated for 30 minutes at 37°C in the presence of lipid vesicles (made from POPC, POPC:POPG (0.7:0.3), or POPC:CL (0.7:0.3) at different concentrations to allow the binding of the protein to membranes. Crosslinking was then initiated by adding glutaraldehyde (GA) at a final concentration of 0.15% for 15 seconds and then stopped by adding an excess of Tris-HCl pH 7.0 (final concentration of 100 mM) before running on a denaturing gel.
2.9. Partition coefficients and free energies of transfer from water to bilayer
We measured partitioning by titrating a peptide or protein solution of fixed concentration with lipid vesicles and calculating the fraction fp of the total amount of peptide or protein partitioned into the lipid vesicles as a function of lipid concentration [L] (see [10, 44]). The partition coefficient Kx from the aqueous phase to the bilayer is given by
| (1) |
where [P]bil, [P]w, [L], and [W] are the concentration of peptide in the bilayer phase, concentration peptide in the aqueous phase, concentration of lipid, and concentration of water, respectively. Given that [P]total = [P]bil + [P]w, one can show that
| (2) |
Note in this equation, which assumes a fixed amount of protein in the aqueous phase during titration, that Kx is constant and the only variable is the lipid concentration. Thus, as [L] increases through titration, fP increases. For high values of Kx, nearly all of the peptide can be bound at sufficiently high lipid concentrations. But regardless of [L], Kx remains constant.
The fraction of SecA bound to lipid vesicles using the relative optical intensities of Coomassie-blue stained SDS-PAGE gels were calculated from
| (3) |
where [M] is the scanned optical intensity of the monomer band on the gel and [D] is the intensity the dimer band.
The free energy of transfer of a peptide from water to bilayer is given by
| (4) |
3. RESULTS AND DISCUSSION
The principal cytoplasmic salt of E. coli is potassium glutamate (KGlu) [45–48], which we have shown thermostabilizes SecA in solution and when partitioned into LUV formed from E. coli lipids [49]. Therefore, all experiments reported below were carried in 100 mM KGlu solutions.
3.1. SecA does not bind to LUVs as one of the crystallographic dimers
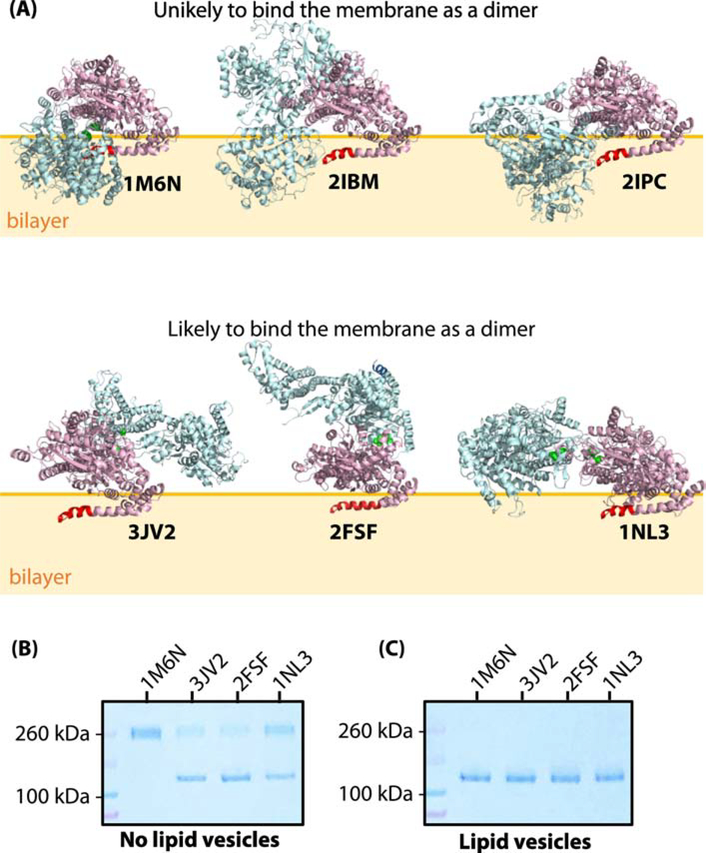
SecA binds principally to negatively charged bilayers via its first fifteen or so N-terminal residues as an amphiphilic helix [10, 50]. We examined the structures of the six crystallographic dimers to determine which ones might remain dimeric while bound to membranes via their N-termini. To be a candidate for dimeric binding, we required that the N-terminus have access the bilayer without burial of significant portions of the rest of the dimer in the bilayer. It appeared to us that the 1M6N, 2IBM, and 2IPC dimers could not bind to a bilayer by means of the N-terminus without the soluble polar domains of the homodimer being buried deep within the bilayer (Figure 1A), suggesting that those crystallographic dimers were unlikely to be able to bind to lipid bilayer without a major rearrangement of the protomers. We recently demonstrated that the 1M6N dimer, stabilized by disulfide bridges [41] binds only weakly, if at all, to liposomes made from E. coli lipids [10], consistent with our reasoning. Therefore, we focused our attention on the binding of the three remaining dimers, 3JV2, 2FSF, and 1NL3 (Figure 1A). We engineered Cys residues into their dimer interfaces to stabilize their dimeric structures under oxidizing conditions (Figure S1).
Figure 1.
SecA does not bind as one of the crystallographic dimers to large unilamellar vesicles (LUV) formed from Escherichia coli lipids. (A) Three-dimensional structures of six reported crystallographic dimers. The N-terminus of SecA, which is necessary for membrane binding [10, 50], is highlighted in red in one of the protomers. The top panel corresponds to the dimer species that are unlikely to bind to the membrane without major steric clashes with the membrane. In support of this hypothesis, we note that an earlier study we showed that the 1M6N dimer cannot bind significantly to E. coli LUV [10]. The bottom panel shows the dimer species that potentially could exist on the lipid bilayer without steric clashes with the bilayer. (B) Coomassie-blue stained SDS-PAGE denaturing gel of cysteine-mutants of SecA (1 μM) in the absence of LUVs under oxidizing conditions. Note the presence of both monomers and dimers for 3JV2, 2FSF, and 1NL3 but not 1M6N. (C) Coomassie-blue stained SDS-PAGE gel of cysteine-mutants of SecA (1 μM) in the presence of LUVs (E. coli lipids, 6 mM). Partitioning of SecA was allowed for 30 minutes prior to introducing the oxidizer. In this case, no dimers are observed.
Each SecA cysteine mutant was expressed and purified under reducing conditions (Materials and Methods). An oxidizer (oxidized glutathione, GSSG) was added in excess to allow the formation of the disulfide bridges in the absence or presence of E. coli LUV. The protein was then applied to non-reducing denaturing SDS-PAGE gels, electrophoresed, and stained with Coomassie-blue. In the absence of lipid vesicles (Figure 1B), the addition of the oxidizer resulted in the formation of SecA dimers, confirming the formation of the specific disulfides bridges. Interestingly, the 1M6N-like dimer was observed exclusively as a dimer, while other cysteine-mutants were only partially cross-linked so that both dimers and monomers were visible on the gels. This is in agreement with the 1M6N-like dimer being the dominant dimer in solution, but in equilibrium with other dimers [14, 26, 35]. When the oxidizer was added after SecA was allowed to bind to LUVs made form E. coli lipids, only monomers were observed (Figure 1C), suggesting that dimeric SecA dissociated into monomers upon binding. It thus appears that SecA cannot bind to vesicles as one of the tested crystallographic dimers (1M6N, 3JV2, 1NL3, and 2FSF).
As a negative control, we created a mutant lacking the Cys788 residue of the Cys788-Cys605 disulfide-forming pair (1NL3 structure Figure S1). This single-Cys mutant did not result in the formation of dimers, thus confirming the specificity of our protocol (see Figure S2A). To eliminate the possibility that the vesicles might have interfered with disulfide bridge formation, we used a mutant containing two cysteines (Cys268-Cys597) forming an intra-monomer disulfide bridge [51]. No differences were observed in the formation of this intra-monomer disulfide bridge in the absence or presence of lipid vesicles under the same protocol (Figure S2B). We conclude that lipid vesicles do not interfere with disulfide bridge formation.
3.2. Gluteraldehyde-crosslinked SecA does not apparently partition into LUV
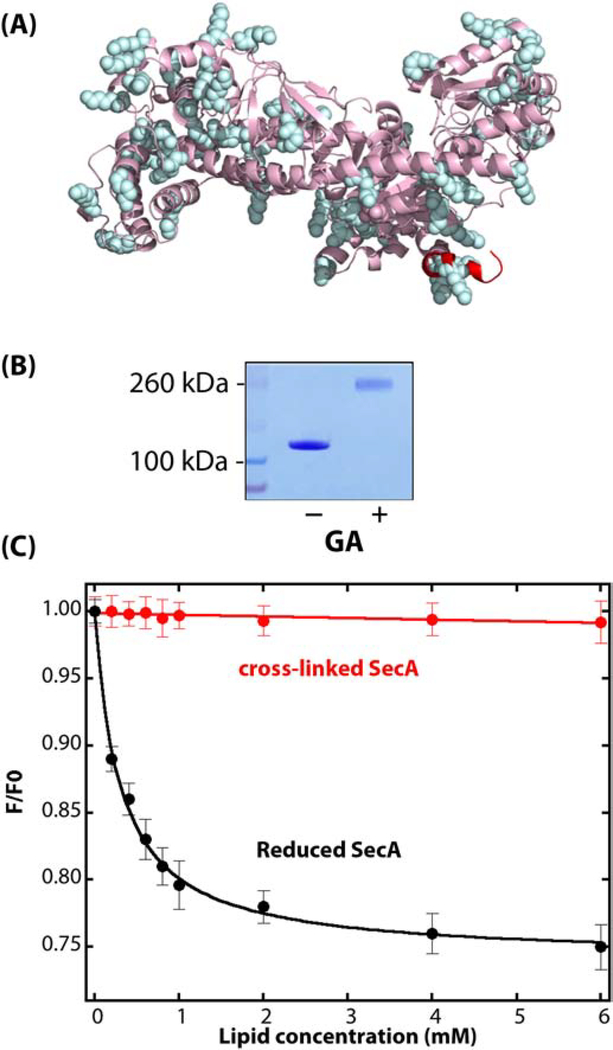
The cysteine-crosslinking measurements suggested that none of the disulfide-stabilized crystallographic dimers tested associate with LUVs. However, crosslinking can be quite sensitive to the quaternary structure of the protein and even a slight reorganization of the protomers might prevent dimerization, resulting in the observation of monomers on denaturing gels. In order to seek possible dimer partitioning into the membrane, we used glutaraldehyde (GA) as a non-specific crosslinking reagent, which has been previously used with SecA to demonstrate that the protein exists as a homodimer in solution [11, 29]. Because Lys residues carrying the tertiary amine groups that are crosslinked via GA are distributed reasonably uniformly over the surface of SecA (Figure 2A), we reasoned that if there were a dimer that could bind to the membrane, we should be able to capture it on LUVs using glutaraldehyde fixation. Figure 2B shows that SecA dimers in solution are readily produced by short exposure to GA (15 seconds, before halting the reaction with the addition of excess of Tris-HCl pH 7.0); no tetramers or higher order multimers were observed on the gel under our conditions (see Figure S4). However, the porosity of the gels used probably did not allow higher order multimers to be observed if they existed. Because of the non-specific nature of the method, the crosslinked product is probably a mixture of various dimers that were in equilibrium in solution. Can these dimers partition significantly into LUV formed from E. coli lipids? To answer that question, we measured the partitioning of GA-crosslinked SecA dimers into LUVs using the Trp fluorescence titration protocol that we used earlier to determine the partitioning of SecA into LUV [10] (Materials and Methods). In those experiments, the ratio of SecA Trp fluorescence in the absence (F0) and presence of LUV (F) was measured as a function of the concentration of lipid. Because of small conformational changes in SecA, F decreases upon partitioning. Consequently, the ratio F/F0 reports on the extent of partitioning of SecA into the LUV. For untreated SecA (Figure 2C, black curve), F decreases with increasing lipid concentration due to conformational changes in bound SecA that increase aqueous exposure of Trp residues (data from Roussel and White [10]).
Figure 2.
Non-specific cross-linking of WT-SecA using glutaraldehyde (GA). (A) Side view of monomeric Escherichia coli SecA (PDB: 2FSF) with the membrane-partitioning N-terminus colored in red and lysine residues that are potential glutaraldehyde cross-linking sites highlighted in cyan. (B) Coomassie-blue stained SDS-PAGE gel of SecA protein (1 μM) in solution in the absence or presence of 0.15% GA. SecA in solution in the absence of vesicles was exposed to GA for 15 secs before halting the reaction with Tris-HCl. (C) GA-crosslinked SecA dimers do not partition significantly into E. coli LUV. The black curve shows the partitioning of untreated SecA (data from [49]). Titration of GA-crosslinked SecA (1 μM; 0.15% GA for 15 seconds) with LUV monitored by the change in intrinsic fluorescence is shown by the red curve. F0 is the fluorescence intensity at 340 nm in the absence of lipids, and F is the intensity in the presence of vesicles.
We repeated those experiments under the same conditions as the black curve except that we used GA-crosslinked dimers. The red curve shows that for crosslinked SecA, F changes little with increasing lipid concentration, suggesting that GA-crosslinked SecA does not apparently partition into LUVs. However, because of internal (Lys-Lys) crosslinking, it could be that SecA is stabilized in a way that prevents changes in the aqueous exposure of Trp that is necessary for monitoring partitioning. To determine the oligomeric state of membrane-bound SecA, we instead applied GA-crosslinking after SecA was allowed to partition into the LUV.
3.3. Short-time GA crosslinking after SecA LUV partitioning reveals no evidence of dimer formation
To determine the oligomeric state of membrane bound SecA, we introduced GA after partitioning into LUVs. However, because POPE—the principal uncharged component of the E. coli lipids—contains a tertiary amine group that could be targeted by GA, we replaced POPE with POPC to form POPC:POPG:CL (0.7:0.2:0.1) LUV, which is sufficient to allow the binding of SecA [10] to vesicles and to stimulate its ATPase activity [6]. If SecA partitions into LUV only as monomers, we hypothesized that only monomers would be observed by crosslinking after partitioning into LUV.
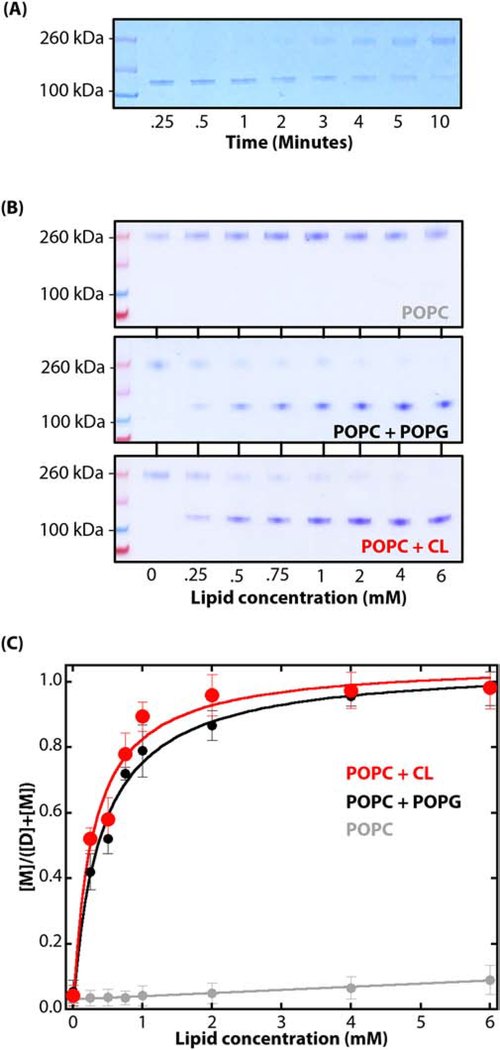
However, under equilibrium partitioning conditions there will always be some SecA in solution available for crosslinking regardless of the dimeric status of the SecA bound to the LUV membranes. We established earlier by means of Trp fluorescence that in solutions containing 4 μM SecA, more than 98% of the SecA partitions into E. coli LUV at a lipid concentration of 6 mM. But a small fraction of SecA must remain in solution under equilibrium conditions [10]. This means that if we exposed SecA to GA long enough in the presence of LUV, all of the protein would eventually be converted to dimers, crosslinked, and be found solely in the aqueous phase, because the crosslinked dimer in solution does not apparently bind to LUV (Figure 2C). To test this hypothesis, we added GA to 6 mM LUV suspensions (POPC:POPG:CL) in the presence of 1 μM SecA and then crosslinked SecA for systematically longer times. The results presented in Figure 3A reveal a shift from monomers at short crosslinking times (< 2 min.) to mostly dimers at long times. This result supports the hypothesis that SecA binds to LUV only as monomers and that prolonged crosslinking shifts the monomer:dimer equilibrium toward non-binding crosslinked dimers. The membrane-bound monomer observed must be internally crosslinked, because its thermal stability is significantly increased after the GA treatment (Figure S4B). This is additional evidence that GA-crosslinking can occur on the surface of the LUVs.
Figure 3.
Dimeric SecA dissociates into monomers upon binding to LUV. (A) In the presence of LUV, there is a progressive shift from monomers to dimers with crosslinking time. After binding to 6 mM LUV (POPC:POPG:CL=0.7:0.2:0.1) for 30 minutes at 37°C, GA was introduced (0.15%) and allowed to crosslink for the periods shown before quenching the reaction. We conclude that as the cross-linking time increases, there is a shift from membrane-bound monomers to unbound dimers in solution. (B) Coomassie-blue stained SDS-PAGE denaturing gels showing the results of titrating SecA (1 μM) with lipid vesicles made of POPC (grey), POPC:POPG (0.7:0.3, black), or POPC:CL (0.7:0.3, red). In each case, partitioning was allowed for 30 minutes at 37°C before introducing GA (0.15%) for 15 seconds before stopping the reaction by the addition of an excess of 100 mM Tris-HCl pH 7.0). Note the progressive shift from dimers to monomers with increases in lipid concentration. (C) The changes in relative intensities of the bands on the gel as the lipid concentration is increased allows one to compute the fraction of SecA partitioned into the LUV at a given lipid concentration. The fraction partitioned fP is given by [M]/([M] + [D]) where [M] is given by the intensity of the monomer band and [D] is the intensity of dimer band. The color code is the same as panel B. The water-to-bilayer free energies of transfer ΔGwb for LUV formed from POPC:POPG and POPC:CL as −7.0 ± 0.3 and −7.1 ± 0.2 kcal.mol−1, respectively. These values agree well with the value of ΔGwb determined by fluorescence measurements of SecA partitioning into LUV formed from E. coli lipids (−7.4 ± 0.1 kcal mol−1) [10]
3.4. SecA partitioning into LUV can be measured accurately using short-time glutaraldehyde crosslinking
It occurred to us that we might be able to measure the partitioning of SecA into to LUV quantitatively using short-time GA crosslinking as a measure of partitioning. For this purpose, we used LUV formed from POPC, POPC:POPG (0.7:0.3), and POPC:CL (0.7:0.3). We followed our earlier approach for measuring partitioning by titrating SecA solutions (1 μM) with LUV concentrations ranging from 0 to 6 mM. At the end of each 30-minute equilibration period following the LUV addition, cross-linking was initiated by the addition of GA (0.15%) for 15 seconds before halting the reaction with the addition of excess of Tris-HCl pH 7.0 (100 mM) to the sample and loading onto a SDS:PAGE gel. Figures 3B and S5 show Coomassie-blue stained denaturing gels for the three lipid compositions. For POPC alone, only SecA dimers were seen, consistent with the lack of binding of SecA to POPC LUVs [10], and also consistent with GA crosslinking of dimers in solution. For the POPC:POPG and POPC:CL mixtures, the binding of SecA is apparent from the decreasing dimer and increasing monomer intensities with increasing lipid concentration. As a measure of SecA partitioning, we measured the relative intensities of the monomer and dimer bands and assumed the band intensities were proportional to the concentrations of monomer [M] and dimer [D]. With these assumptions, we computed the fraction fP of bound SecA using fP = [M]/([M] + [D]). Figure 3C reveals binding curves very similar to those we determined using Trp fluorescence [10] (see Figure 2). Using our earlier approach for measuring partition coefficients and partitioning free energies [10, 44] (see Materials and Methods), we determined water-to-bilayer free energies of transfer ΔGwb for LUV formed from POPC:POPG and POPC:CL as −7.0 ± 0.3 and −7.1 ± 0.2 kcal.mol−1, respectively. The value of ΔGwb determined by fluorescence measurements of SecA partitioning into LUV formed from E. coli lipids was −7.4 ± 0.1 kcal mol−1 [10]. These results generally validate our crosslinking approach and the hypothesis that SecA partitions only as monomers into LUV formed from E. coli lipids.
Benach et al. [29] observed in their study of phospholipid-induced monomerization that signal peptides induced oligomerization of SecA based upon GA crosslinking. Keeping in mind that SecA ATPase activity is observed when bound to lipid vesicles [6], we examined the effects of prePhoA signal peptide, ADP, and the non-hydrolyzable ATP analog AMP-PNP on dimerization. We found no effect of these compounds on the dimerization of SecA in the presence of E. coli LUV (Figure S6). A possible explanation is the crosslinking time. Benach et al. used GA crosslinking times of 3 minutes at 37° C or 5 minutes at 20° C. These relatively long crosslinking likely caused a significant non-equilibrium shift toward the dimeric state (Figure 3A).
4.0. Conclusions
We conclude from these in vitro results that dimeric SecA in solution dissociates into monomers upon binding to LUV membranes. Our results are in agreement with in vitro observations [14, 19, 20, 29] and with the idea that lipids might activate SecA for high-affinity binding to SecYEG [28]. Our results seem to be at odds with in vivo measurements showing that SecA functions as a dimer [16, 41]. A possible explanation is that LUV formed from E. coli lipids do not reflect accurately the complex properties of the E. coli inner membranes in vivo that may include an “integral form” of SecA [52–54], which is presumably a SecA:SecY complex. Linda Randall and her colleagues [55, 56] have shown that coassembly of SecYEG and SecA into E. coli lipid vesicles support protein secretion much more effectively than when SecA is added separately to LUV containing only SecYEG. Further, they found that the transport effectiveness of their co-assembled SecYEG∙SecA proteoliposome system was similar to that of inverted native urea-washed E. coli membrane vesicles [57]. These results point to the possibility that the dimerization state on intact E. coli membranes might be affected by interactions with the SecYEG translocon, the SecB chaperone, and preproteins [55], as previously proposed [18, 58]. Our results demonstrate that the lipid bilayer itself induces the dissociation of dimeric SecA into monomers. If SecA associated with intact E. coli membranes is dimeric in vivo, it will be essential to determinate which partner stabilizes dimeric SecA.
Supplementary Material
Highlights.
SecA is dimeric in solution, but does it bind to lipid vesicles as a dimer?
Known crystallographic SecA dimers do not bind to E. coli-lipid vesicles
A short-term glutaraldehyde crosslinking protocol revealed no binding of SecA dimers
SecA-bilayer interactions cause dissociation of SecA dimers into monomers
Acknowledgements
This work was supported by grant GM-74637 from National Institute of General Medical Sciences. We thank Dr. Eric Lindner for invaluable discussions and Dr. Gargi Dasgupta for excellent technical support.
ABBREVIATIONS
- KGlu
Potassium glutamate
- LUV
large unilamellar vesicle
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmytoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
1-palmioyl-2-oleoyl-sn-glycero-3-phospho-(1’rac-glycerol)
- CL
cardiolipin
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- GA
glutaraldehyde
Footnotes
Conflicts of interest
The authors declare no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Crane JM, Randall LL, The Sec system: Protein export in Escherichia coli, EcoSalplus, (2017) 10.1128/ecosalplus.ESP-0002-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cabelli RJ, Dolan KM, Qian L, Oliver DB, Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli, J. Biol. Chem, 266 (1991) 24420–24427. [PubMed] [Google Scholar]
- [3].de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B, Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes, Nature, 334 (1988) 173–175. [DOI] [PubMed] [Google Scholar]
- [4].Prabudiansyah I, Kusters I, Caforio A, Driessen AJM, Characterization of the annular lipid shell of the Sec translocon, Biochim. Biophys. Acta, 1848 (2015) 2050–2056. [DOI] [PubMed] [Google Scholar]
- [5].Breukink E, Demel RA, de Korte-Kool G, de Kruijff B, SecA insertion into phospholipids is stimulated by negatively charged lipids and inhibited by ATP: a monolayer study, Biochemistry, 31 (1992) 1119–1124. [DOI] [PubMed] [Google Scholar]
- [6].Lill R, Dowhan W, Wickner W, The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins, Cell, 60 (1990) 271–280. [DOI] [PubMed] [Google Scholar]
- [7].Ulbrandt ND, London E, Oliver DB, Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding, J. Biol. Chem, 267 (1992) 15184–15192. [PubMed] [Google Scholar]
- [8].Keller RCA, Snel MME, de Kruijff B, Marsh D, SecA restricts, in a nucleotide-dependent manner, acyl chain mobility up to the center of a phospholipid bilayer, FEBS Lett, 358 (1995) 251–254. [DOI] [PubMed] [Google Scholar]
- [9].Ahn T, Kim J-S, Lee B-C, Yun C-H, Effects of lipids on the interaction of SecA with model membranes, Arch. Biochem. Biophys, 395 (2001) 14–20. [DOI] [PubMed] [Google Scholar]
- [10].Roussel G, White SH, Binding of SecA ATPase monomers and dimers to lipid vesicles, Biochim. Biophys. Acta, 1862 (2020) 183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Akita M, Shinkai A, S.-i. Matsuyama, S. Mizushima, SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer, Biochem. Biophys. Res. Commun, 174 (1991) 211–216. [DOI] [PubMed] [Google Scholar]
- [12].Driessen AJM, SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer, Biochemistry, 32 (1993) 13190–13197. [DOI] [PubMed] [Google Scholar]
- [13].Nishiyama KI, Suzuki T, Tokuda H, Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation, Cell, 85 (1996) 71–81. [DOI] [PubMed] [Google Scholar]
- [14].Or E, Navon A, Rapoport T, Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane, EMBO J, 21 (2002) 4470–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Woodbury RL, Hardy SJS, Randall LL, Complex behavior in solution of homodimeric SecA, Protein Sci, 11 (2002) 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jilaveanu LB, Zito CR, Oliver D, Dimeric SecA is essential for protein translocation, Proc. Natl. Acad. Sci. U.S.A, 102 (2005) 7511–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kusters I, van den Bogaart G, Kedrov A, Krasnikov V, Fulyani F, Poolman B, Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level, Structure, 19 (2011) 430–439. [DOI] [PubMed] [Google Scholar]
- [18].Gouridis G, Karamanou S, Sardis MF, Schärer MA, Capitani G, Economou A, Quaternary dynamics of the SecA motor drive translocase catalysis, Mol. Cell, 52 (2013) 655–666. [DOI] [PubMed] [Google Scholar]
- [19].Duong F, binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase, EMBO J, 22 (2003) 4375–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Or E, Boyd D, Gon S, Beckwith J, Rapoport T, The bacterial ATPase SecA functions as a monomer in protein translocation, J. Biol. Chem, 280 (2005) 9097–9105. [DOI] [PubMed] [Google Scholar]
- [21].Sugano Y, Furukawa A, Nureki O, Tanaka Y, Tsukazaki T, SecY-SecA fusion protein retains the ability to mediate protein transport, PLoS One, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zimmer J, Nam Y, Rapoport TA, Structure of a complex of the ATPase SecA and the protein-translocation channel, Nature, 455 (2008) 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li L, Park E, Ling J, Ingram J, Ploegh H, Rapoport TA, Crystal structure of a substrate-engaged SecY protein-translocation channel, Nature, 531 (2016) 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ma C, Wu X, Sun D, Park E, Catipovic MA, Rapoport TA, Gao N, Li L, Structure of the substrate-engaged SecA-SecY protein translocation machine, Nature Communications, 10 (2019) 2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rusch SL, Kendall DA, Oligomeric states of SecA and SecYEG core components of the bacterial Sec translocon, Biochim. Biophys. Acta, 1768 (2007) 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sardis MF, Economou A, SecA: a tale of two promoters, Mol. Microbiol, 76 (2010) 1070–1081. [DOI] [PubMed] [Google Scholar]
- [27].Chatzi KE, Sardis MF, Economou A, Karamanou S, SecA-mediated targeting and translocation of secretory proteins, Biochim. Biophys. Acta, 1843 (2014) 1466–1474. [DOI] [PubMed] [Google Scholar]
- [28].Koch S, de Wit JG, Vos L, Birkner JP, Gordiichuk P, Herrmann A, van Oijen AM, Driessen AJM, Lipids activate SecA for high affinity binding to the SecYEG complex, J. Biol. Chem, 291 (2016) 22534–22543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Benach J, Chou Y-T, Fak JJ, Itkin A, Nicolae DD, Smith PC, Wittrock G, Floyd DL, Golsaz CM, Gierasch LM, Hunt JF, Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA, J. Biol. Chem, 278 (2003) 3628–3638. [DOI] [PubMed] [Google Scholar]
- [30].Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J, Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA, Science, 297 (2002) 2018–2026. [DOI] [PubMed] [Google Scholar]
- [31].Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR Jr, Sacchettini JC, Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase, Proc. Natl. Acad. Sci. U.S.A, 100 (2003) 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vassylyev DG, Mori H, Vassylyev MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K, Crystal structure of the translocation TPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer, J. Mol. Biol, 364 (2006) 248–258. [DOI] [PubMed] [Google Scholar]
- [33].Zimmer J, Li W, Rapoport TA, A novel dimer interface and conformational changes revealed by an x-ray structure of B. subtilis SecA, J. Mol. Biol, 364 (2006) 259–265. [DOI] [PubMed] [Google Scholar]
- [34].Papanikolau Y, Papadovasilaki M, Ravelli RBG, McCarthy AA, Cusack S, Economou A, Petratos K, Structure of dimeric SecA, the Escherichia coli preprotein translocase motor, J. Mol. Biol, 366 (2007) 1545–1557. [DOI] [PubMed] [Google Scholar]
- [35].Yu D, Wowor AJ, Cole JL, Kendall DA, Defining the Escherichia coli SecA dimer interface residues through in Vivo site-specific photo-cross-linking, J. Bacteriol, 195 (2013) 2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wowor AJ, Yan Y, Auclair SM, Yu D, Zhang J, May ER, Gross ML, Kendell DA, Cole JL, Analysis of SecA dimerization in solution, Biochemistry, 53 (2014) 3248–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ding H, Hunt JF, Mukerji I, Oliver D, Bacillus subtilis SecA ATPase exists as an antiparallel dimer in solution, Biochemisty, 42 (2003) 8729–8738. [DOI] [PubMed] [Google Scholar]
- [38].Auclair SM, Oliver DB, Mukerji I, Defining the solution state dimer structure of Escherichia coli SecA using Förster resonance energy transfer, Biochemistry, 52 (2013) 2388–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen Y, Pan X, Tang Y, Quan S, Tai PC, Sui S-F, Full-length Escherichia coli SecA dimerizes in a closed conformation in solution as determined by cryo-electron microscopy, J. Biol. Chem, 283 (2008) 28783–28787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zimmer J, Rapoport TA, Conformational flexibility and peptide interaction of the translocation ATPase SecA, J. Mol. Biol, 394 (2009) 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jilaveanu LB, Oliver D, SecA dimer cross-linked at its subunit interface is functional for protein translocation, J. Bacteriol, 188 (2006) 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bartlett GR, Phosphorus assay in column chromatography, J. Biol. Chem, 234 (1959) 466–468. [PubMed] [Google Scholar]
- [43].Ladokhin AS, Jayasinghe S, White SH, How to measure and analyze tryptophan fluorescence in membranes properly, and why bother?, Anal. Biochem, 285 (2000) 235–245. [DOI] [PubMed] [Google Scholar]
- [44].White SH, Wimley WC, Ladokhin AS, Hristova K, Protein folding in membranes: Determining energetics of peptide-bilayer interactions, Methods Enzymol, 295 (1998) 62–87. [DOI] [PubMed] [Google Scholar]
- [45].Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz J, Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli, Nat. Chem. Biol, 5 (2009) 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sengupta R, Pantel A, Cheng X, Shkel I, Peran I, Stenzoski N, Raleigh DP, Record MT Jr., Positioning the intracellular salt potassium glutamate in the Hofmeister series by chemical unfolding studies of NTL9, Biochemistry, 55 (2016) 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cheng X, Guinn EJ, Buechel E, Wong R, Sengupta R, Shkel IA, Record MT Jr., The basis of protein stabilization by K glutamate: Unfavorable interactions with carbon, oxygen groups, Biophys. J, 111 (2016) 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Anumalla B, Prabhu NP, Glutamate induced thermal equilibirium intermediate and counteracting effect on chemical denaturation of proteins, J. Phys. Chem. B, 122 (2018) 1132–1144. [DOI] [PubMed] [Google Scholar]
- [49].Roussel G, Lindner E, White SH, Stabilization of SecA ATPase by the Primary cytoplasmic salt of Escherichia coli, Protein Sci, 28 (2019) 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Findik BT, Smith VF, Randall LL, Penetration into membrane of amino-terminal region of SecA when associated with SecYEG in active complexes, Protein Sci, 27 (2018) 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chatzi KE, Sardis MF, Tsirigotaki A, Koulaki M, Šoštarić N, Konijnenberg A, Sobott F, Kalodimas CG, Karamanou S, Economou A, Preprotein mature domains contain translocase targeting signals that are essential for secretion, J. Cell Biol, 216 (2017) 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chen X, Xu H, Tai PC, A significant fraction of functional SecA is permanently embedded in the membrane, J. Biol. Chem, 271 (1996) 29698–29706. [DOI] [PubMed] [Google Scholar]
- [53].Ramamurthy V, Oliver D, Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding, J. Biol. Chem, 272 (1997) 23239–23246. [DOI] [PubMed] [Google Scholar]
- [54].Chen X, Brown T, Tai PC, Identification and characterization of protease-resistant SecA fragments: SecA has two membrane-integral forms, J. Bacteriol, 180 (1998) 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mao C, Cheadle CE, Hardy SJS, Lilly AA, Suo Y, Gari RRS, King GM, Randall LL, Stoichiometry of SecYEG in the active translocase of Escherichia coli varies with precursor species, Proc. Natl. Acad. Sci. U.S.A, 110 (2013) 11815–11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bariya P, Randall LL, Coassembly of SecYEG and SecA fully restores the properties of the native translocon, J. Bacteriol, 201 (2019) e00493–00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Futai M, Orientation of membrane vesicles from Escherichia coli prepared by different methods, J. Membr. Biol, 15 (1974) 15–28. [DOI] [PubMed] [Google Scholar]
- [58].Gari RRS, Chattrakun K, Marsh BP, Mao C, Chada N, Randall LL, King GM, Direct visualization of the E. coli Sec translocase engaging precursor proteins in lipid bilayers, Science Advances, 5 (2019) eaav9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.