Abstract
We report on a microsystem that couples high-throughput bacterial immunomagnetic capture to contact-free cell lysis using an alternating current magnetic field (AMF) to enable downstream molecular characterization of bacterial nucleic acids. Traditional methods for cell lysis rely on either dilutive chemical methods, expensive biological reagents, or imprecise physical methods. We present a microchip with a magnetic polymer substrate (Mag-Polymer microchip), which enables highly controlled, on-chip heating of biological targets following exposure to an AMF. First, we present a theoretical framework for the quantitation of power generation for single-domain magnetic nanoparticles embedded in a polymer matrix. Next, we demonstrate successful bacterial DNA recovery by coupling (1) high-throughput, sensitive microfluidic immunomagnetic capture of bacteria to (2) on-chip, contact-free bacterial lysis using an AMF. The bacterial capture efficiency exceeded 76% at 50 ml/h at cell loads as low as ∼10 CFU/ml, and intact DNA was successfully recovered at starting bacterial concentrations as low as ∼1000 CFU/ml. Using the presented methodology, cell lysis becomes non-dilutive, temperature is precisely controlled, and potential contamination risks are eliminated. This workflow and substrate modification could be easily integrated in a range of micro-scale diagnostic systems for infectious disease.
I. INTRODUCTION
Microfluidic platforms have emerged as a popular alternative to traditional macro-scale diagnostic methods.1–5 Microfluidic systems enable extremely precise fluid control and manipulation and have demonstrated their ability to isolate and detect rare cells from both environmental and biological samples by harnessing a variety of physical and chemical separation methods.6–10 The ability to rapidly isolate and specifically detect bacterial pathogens has applications in infectious disease, biosecurity, and food and water quality monitoring.11–13 Integrated micro-scale systems could aid in shortening diagnostic timelines due their demonstrated efficacy as high-throughput, sensitive, and specific biomarker isolation and detection platforms.
Numerous pathogen characterization methods rely on access to intracellular proteins and nucleic acids, requiring cell lysis following pathogen isolation.14–17 Traditional methods for cell lysis rely on either dilutive chemical methods (e.g., detergents), expensive biological reagents (e.g., lysozyme), or imprecise physical methods.18,19 There is a significant need for a rapid, precise, and reagent-free bacterial lysis method that can be easily integrated with upstream microfluidic enrichment processes. This need for highly controlled, non-dilutive cell lysis becomes especially relevant when targeting the isolation and analysis of rare cells, which is relevant to a range of clinical scenarios including the diagnosis of bloodstream infections and prosthetic joint infections.20–22
Here, we utilize microfluidic immunomagnetic separation methods to rapidly and specifically capture and concentrate bacteria of interest on the surface of our microchip.23–25 The microchip substrate is composed of a unique three-layer magnetic polymer (Mag-Polymer), which consists of single-domain magnetic nanoparticles mixed into a polydimethylsiloxane (PDMS) matrix. Mechanisms of heat generation from magnetic nanoparticles have been comprehensively studied.26–29 Most often, these studies are performed within the context cancer therapy. Specifically, these studies investigate the use of in vivo localized magnetic nanoparticles coupled to an external alternating magnetic field for the hyperthermia of cancerous cells and/or tissues. Significant effort has been dedicated to optimizing therapeutic effects through rational design of magnetic nanoparticle characteristics such as size, geometry, and composition in an effort to limit field intensity requirements.26,30–32 For cancer cell hyperthermia, target temperatures range from approximately 40 to 45 °C33; however, in this work, we aim to reach significantly higher temperatures (i.e., 80–110 °C) to enable the highly controlled, on-chip, thermal lysis of bacteria.18,34
The presented methodology couples microfluidic bacterial enrichment with contact-free lysis using an AC magnetic field (AMF). Following exposure to an AMF, bacteria are thermally lysed, enabling additional on-chip and/or downstream nucleic acid amplification and analysis (Fig. 1). First, we present a theoretical framework for the optimization of Mag-Polymer microchip heating as a function of magnetic field strength, field frequency, and magnetic nanoparticle characteristics. Next, we demonstrate an optimized microfluidic bacterial immunomagnetic enrichment system, which enables high-throughput sample processing, while achieving extremely low limits-of-detection. Finally, we provide an experimental characterization of microchip heating and demonstrate successful recovery of double-stranded bacterial DNA for downstream molecular characterization.
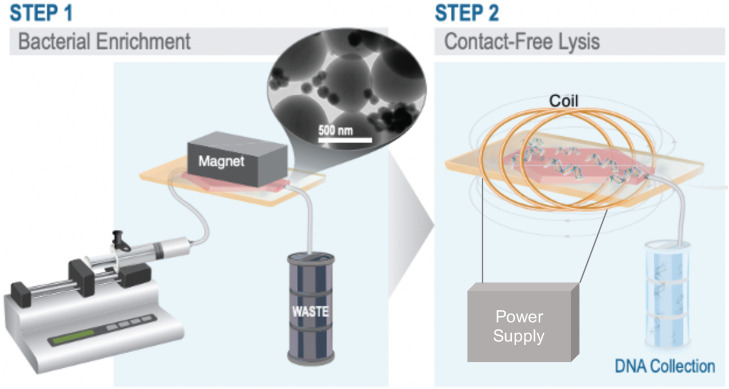
FIG. 1.
Overview schematic of bacterial enrichment and contact-free lysis driven by an AC magnetic field. Step 1. The syringe pump pushes sample through hexagonal microchannel. The external magnet retains bacteria bound to functionalized magnetic nanoparticles within the microchannel, while waste products are collected as the output. TEM image of S. aureus bound (∼0.5 μm) bound to magnetic nanoparticles (∼150 nm) (top right). Step 2. Overview schematic of contact-free cell lysis. External magnet is removed, microchip is placed in coil, and microchip is exposed to an AMF. Bacteria are thermally lysed, enabling downstream nucleic acid collection and analysis.
II. MATERIALS AND METHODS
A. Bacterial strains and culture conditions
Staphylococcus aureus (ATCC #27660) was pre-cultured overnight in 5 ml tryptic soy broth (TSB) (37 °C, 250 rpm shaking) (Becton Dickenson, Franklin Lakes, NJ). The pre-culture was inoculated 1:1000 into 25 ml fresh TSB in a 250 ml Erlenmeyer flask and cultured for 12 h under identical conditions (37 °C, 250 rpm shaking). The sample was centrifuged (12 100×g, 4 °C 10 min), and the supernatant was aspirated. Bacteria were resuspended in fresh TSB and 50% glycerol (1:1), aliquoted, and stored at −20 °C until use.
B. Functionalization of magnetic nanoparticles
150 nm streptavidin coated magnetic nanoparticles (SV0150, Ocean Nanotech, San Diego, CA) were functionalized with biotinylated anti-S. aureus polyclonal antibody (PA1–73174, ThermoFisher Scientific, Waltham, MA). First, magnetic nanoparticles (MNPs) were washed three times with PBS. Next, approximately 20 μg of IgG was added to 1 mg of suspended MNPs. The mixture was incubated for 30 min at room temperature with gentle rotation. Finally, conjugated MNPs were washed four times with 0.1% bovine serum albumin (BSA) in PBS and adjusted to a final concentration of 1 mg/ml. Functionalized MNPs were stored at 4 °C until use.
C. Mag-Polymer microchip fabrication
To fabricate the magnetic polymer, 30 nm iron oxide (Fe3O4) nanoparticles (Nanostructured & Amorphous Materials Inc., Katy, TX) were mixed with Sylgard-184 polydimethylsiloxane (PDMS) (Dow Corning, Midland, MI) to create a 35% (w/w) mixture. The curing agent was added to the mixture at a ratio of 1:5 (w/w). The mixture was manually stirred and degassed for 60 min. The mixture was then spin coated onto a glass slide at 600 rpm for 30 s and baked at 150 °C for 10 min. This step was repeated a total of three times to create three polymer layers, having a total thickness equal to ∼200 μm. The three-layer polymer structure was selected after experimentation with varying layering and weight density structures.
D. Sample preparation, processing, and quantification
To prepare a sample, S. aureus was diluted in PBS to the desired concentration and volume (1 ml) and combined with functionalized MNPs. Samples were incubated for 1 h at room temperature with gentle rotation. Samples were pushed through the microchip at flow rates ranging from 5 ml/h to 50 ml/h with a syringe pump (Harvard Apparatus PHD Ultra, Holliston, MA). Flow rate optimization experiments were conducted at a bacterial load on the order of 103 CFU/ml and were combined with 25 μg functionalized MNPs per sample. Magnetic nanoparticle mass optimization experiments were conducted at bacterial load on the order of 103 CFU/ml at a flow rate of 10 ml/h. System sensitivity experiments were conducted at a flow rate of 50 ml/h and were combined with 100 μg functionalized MNPs per sample. Bacteria were quantified using traditional plate counting methods on TSB agar plates. Capture efficiency was calculated by comparing the number of viable bacteria in the input sample to the number of viable bacteria in the output sample. Paired control samples, containing viable bacteria and without magnetic nanoparticles, were processed on the system to quantify potential bacterial loss and/or death within the microsystem. System sterilization was performed by pushing 5 ml of 70% ethanol at 0.5 ml/h, followed by 10 ml of PBS at 1 ml/h and ∼2 ml of air to clear the microsystem prior to sample processing.
E. AC magnetic field, DNA quantification, and cell viability
The AMF induction coil used in these experiments was a single-turn solenoid coil which was custom built by the Hoopes lab at Dartmouth College.35 It is powered by a 25-kW generator (Radyne, Milwaukee, WI) and cooled by a 3-ton ethylene glycol cooling system (Tek-Temp Instruments, Croydon, PA). The field was tuned to 165 kHz. The microchip surface temperature was measured using a thermal camera (Model SC325, FLIR Systems, Wilsonville, OR). Following a 60 s exposure to the AMF (200 Oe, 30 s; 500 Oe, 30 s), DNA was quantified using a Qubit 3.0 Fluorometer dsDNA High Sensitivity Assay Kit (ThermoFisher Scientific, Waltham, MA). Cell viability was determined using the plate counting method via 10 μl drop plates.
III. RESULTS and DISCUSSION
A. Theoretical framework for magnetic polymer heating
Previous work by Tong et al. identified that magnetic iron oxide particles ranging from 30 to 40 nm have a specific absorption rate (SAR) approaching the theoretical limit when exposed to a clinically relevant alternating magnetic field (AMF).28 Thus, ∼30 nm iron oxide nanoparticles were selected as the magnetic component of our polymeric material. These magnetic nanoparticles were spiked into polydimethylsiloxane (PDMS); this magnetic polymer was spin-coated onto a glass substrate to create a three-layer polymer structure for microchip heating [Figs. 2(a) and 2(b)]. The design of the heating layers was guided by an attempt to maximize heating efficiency, while limiting multi-layer microfabrication requirements. The three-layer polymer structure was selected as the optimal outcome. First, we sought to maximize the weight density of the iron oxide in the polymer matrix, while still achieving reliable and repeatable polymer cross-linking. Next, we sequentially added polymer layers until target temperatures were achieved. Importantly, we wanted to preserve ease-of-fabrication and repeatability by employing a spin-coating fabrication methodology.
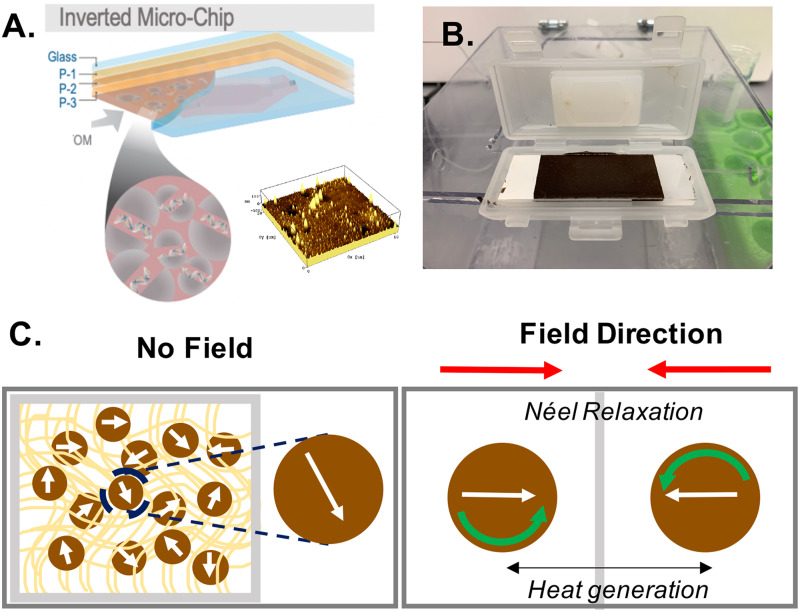
FIG. 2.
Overview of device substrate and heating mechanism. (a) Mag-Polymer microchip. Substrate modification consists of three identical spin coated polymer layers (P-1–P-3). Magnetic nanoparticles mixed within the polymer (PDMS) enable thermal lysis of bacteria, making molecules of interest available (i.e., DNA) for analysis (left). Atomic force microscopy image (AFM) displaying topography of magnetic polymer surface (right). (b) Image of magnetic polymer-coated microchip in microfluidic cartridge. (c) Schematic of heating mechanism for magnetic nanoparticles embedded in polymer matrix (left). Néel relaxation—the rapid change in magnetic moment in opposition to the nanoparticle's crystalline structure—drives heat generation (right).
Below, we present a theoretical framework for the quantitation of power generation from single-domain magnetic nanoparticles confined in a polymer matrix when exposed to an AMF.
Power dissipation () from magnetic nanoparticles following exposure to an AC magnetic field can be modeled using the Rosensweig equation,27,28
| (1) |
where is the permeability constant of free space (4π × 10−7 N/A2), is the magnetic susceptibility of the particles, H is the magnetic field strength, f is the magnetic field frequency, and τ is the effective relaxation time. When exposed to an alternating magnetic field, magnetic nanoparticles produce heat via three main mechanisms: hysteresis, Brownian motion, and Néel relaxation. Given the paramagnetic properties of the single-domain iron oxide nanoparticles (<30 nm) and their confinement in a polymer matrix, we can assume that magnetization reversal and heat generation is primarily limited to Néel relaxation (spin relaxation), which is dictated by the anisotropy energy of the nanoparticles [Fig. 2(c)].26 Therefore, τ can be defined as
| (2) |
where is the attempt time/period; , the anisotropy energy, is the product of the magnetocrystalline anisotropy () and particle volume (); , the thermal energy, is the product of Boltzmann constant () and absolute temperature ().29,30 By combining Eqs. (1) and (2), we can define power generation from a paramagnetic nanoparticle embedded in a polymer as follows:
| (3) |
Aside from thoughtful and intentional particle selection, power dissipation from the magnetic polymer can be increased by increasing the strength of the magnetic field (), and by optimizing the field frequency () such that f is equal to τ−1.29
B. Microfluidic immunomagnetic bacterial enrichment
Bacterial samples were processed through a hexagonal-shaped microchannel (30 × 20 mm2) exposed to an optimized external magnetic field. The specifications of this platform have been previously reported for the isolation of circulating tumor cells (CTCs).36,37 In this work, the microchip glass substrate was modified with a three-layer, spin-coated magnetic polymer to enable contact-free heating of the microchip immediately following cell capture.
Magnetic nanoparticles for cell capture were functionalized with an anti-S. aureus polyclonal antibody to selectively bind target bacteria [Fig. 3(a)]. The microfluidic bacterial capture system was optimized to maximize system sensitivity and sample throughput. First, bacterial capture efficiency was evaluated as a function of sample flow rate. Bacterial samples were continuously flowed through the microchannel at flow rates ranging from 5 ml/h to 50 ml/h, but no significant difference in capture efficiency was observed [Fig. 3(b)]. This finding suggests that our microfluidic immunomagnetic capture system is robust to high flow rates, enabling rapid sample processing and target biomarker enrichment. Next, bacterial capture efficiency was evaluated as a function of magnetic nanoparticle mass. We observed that bacterial capture efficiency significantly increased with increasing magnetic nanoparticle mass [Fig. 3(c)]. These initial experiments allowed for the implementation of optimized assay parameters to asses system limit-of-detection.
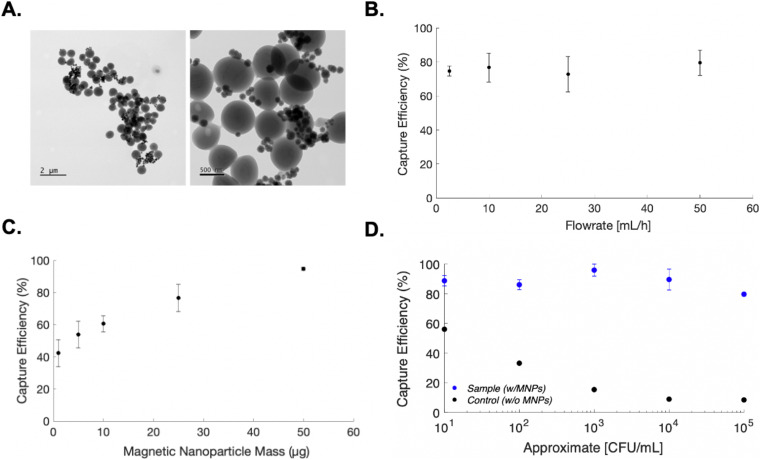
FIG. 3.
Microfluidic immunomagnetic bacterial capture. (a) Transmission Electron Microscopy (TEM) images of S. aureus bound to 150 nm magnetic nanoparticles. (b) Bacterial capture efficiency as a function of flow rate. (c) Bacterial capture efficiency as a function of magnetic nanoparticle mass. (d) Bacterial capture efficiency as a function of cell concentration. Control samples contained no functionalized magnetic particles and were evaluated to account for any potential bacterial loss and/or gain within the micro-system. All samples were evaluated in triplicate. Standard error of mean is reported.
Our optimized flow-through immunomagnetic capture platform demonstrated successful bacterial capture at high flow rates (50 ml/h), while still achieving low limits-of-detection (∼10 CFU/ml). By employing optimized assay conditions, Staphylococcus aureus capture efficiency ranged from 86.1% ± 3.34% to 95.93% ± 4.07% for 105 CFU/ml and 103 CFU/ml, respectively. Notably, at bacterial concentrations on the order or 101 CFU/ml, capture efficiency exceeded 80% for all samples evaluated, with a mean of 88.7% ± 3.49% [Fig. 3(d)]. The data presented suggest that our proposed immunomagnetic enrichment platform can rapidly concentrate bacteria at extremely low cell loads, which is relevant to a range of infectious disease diagnostic applications.5,38,39
C. Microchip heating and quantification of recovered DNA
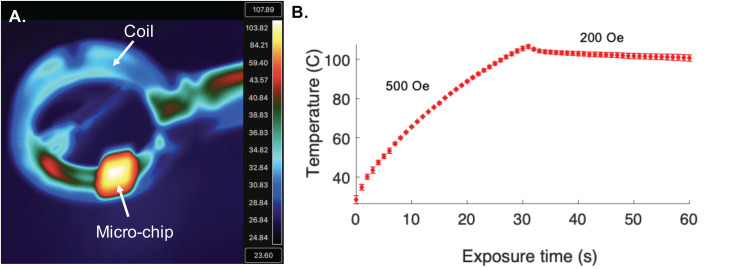
Following S. aureus capture, the microchip was exposed to an AMF for 60 s [Fig. 4(a)]. The field strength was optimized to result in a microchip temperature that maximized bacterial lysis, while preserving biological molecules of interest (i.e., dsDNA).34 For the first 30 s, the microchip was exposed to a field of approximately 500 Oe to rapidly achieve the target temperature (105.5 °C ± 0.92 °C). Once the target temperature was achieved, the field strength was lowered to approximately 200 Oe for an additional 30 s to maintain an exposure temperature ranging from 105.5 °C ± 0.92 °C to 100.6 °C ± 0.92 °C [Fig. 4(b)]. In addition to the heating profile evaluated here, the magnetic polymer substrate enables extremely precise heating at a range of biologically relevant temperature profiles. In comparison to other off-chip thermal lysis methodologies (e.g., heating block), the Mag-Polymer substrate modification allows for fine-tuning of the thermal gradient and localized heating on rationally patterned regions of the microchip surface. Additionally, thermal exposure is highly homogenous and extremely precise, as is indicated by the relatively small standard error observed in reported temperatures across multiple devices. This heating modality also moves toward the design of a fully integrated microsystem for nucleic acid recovery from biological samples.
FIG. 4.
Mag-Polymer microchip heating. (a) Representative thermal image of microchip in coil after 30 s exposure to AMF. (b) Temperature of the microchip as a function of time. Temperature data were collected using a thermal camera. Three unique devices were evaluated, and each device was tested in triplicate. Standard error of the mean is reported.
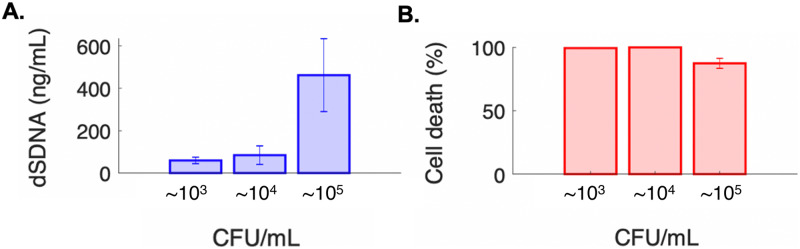
Following exposure to the AMF, the efficacy of our contact-free cell lysis platform was evaluated as a function of recovered dsDNA and cell death (Fig. 5). We demonstrate successful recovery of intact dsDNA for starting bacterial sample concentrations on the order of 103 CFU/ml (59.8 ng/ml ± 15.2 ng/ml). We hypothesize that these low detection limits are feasible as a direct result of our microfluidic enrichment step prior to cell lysis, which effectively localizes and concentrates bacterial nucleic acids. Specifically, the starting sample volume of 1 ml is effectively concentrated to an ∼5 μl sample on the surface of the microchip. Additionally, cell death was confirmed and ranged from 87.41% ± 3.95% to 99.98% ± 0.003% for bacterial sample concentrations on the order of 105 CFU/ml to 104 CFU/ml, respectively.
FIG. 5.
Recovered DNA and cell viability. (a) Total recovered DNA and (b) cell death as a function of cell load following 60 s exposure to AMF. All samples were evaluated in triplicate, with three unique devices used. Standard error of mean is reported.
IV. CONCLUSIONS
To the best of our knowledge, this is the first study to report on a contact-free lysis method coupled to a flow-through microfluidic cell capture platform. We describe a unique Mag-Polymer microchip design that enables controlled, contact-free, and non-dilutive cell lysis following exposure to an AMF. We also provide a theoretical framework for future work aimed at optimizing power dissipation from the material in an effort to limit required external power and equipment. We demonstrate extremely sensitive and high-throughput microfluidic immunomagnetic bacterial enrichment using a hexagonal microchannel and optimized external magnetic field. This enrichment platform has been previously demonstrated for the enrichment of circulating tumor cells (CTCs), but this study reports its first successful translation and application to bacterial enrichment.23,37 It is important to note that by performing bacterial enrichment prior to lysis, rare biomarker detection sensitivity is significantly increased.
Following experimental characterization of microchip heating, we demonstrate that this novel methodology is successful at lysing captured bacteria and recovering intact double-stranded DNA for downstream characterization of the captured pathogen. We think that this methodology is especially relevant to the micro-scale platforms that target the isolation and detection of rare biomarkers due to the ability to finely tune thermal exposure, and eliminate dilutive wash steps and/or chemical buffers. In addition to bacterial cell lysis, there are numerous other biological applications (i.e., PCR) that could utilize this Mag-Polymer microchip substrate modification and precise heating mechanism for on-chip molecular analysis and detection. Due to the relative simplicity of our magnetic polymer substrate fabrication method, we anticipate that this microchip substrate modification could be easily integrated in a range of micro-scale diagnostic systems for rapid, precise, and contact-free heating to enable comprehensive characterization of the disease-causing pathogens.
ACKNOWLEDGMENTS
The authors would like to acknowledge the support of the Ph.D. Innovation Program at the Thayer School of Engineering at Dartmouth College. Partial financial support was also from the National Institutes of Health (NIH) Director's Transformative Research Award (No. R01HL137157), National Science Foundation Award (No. ECCS1509369), and the startup fund from the Thayer School of Engineering at Dartmouth. The authors would also like to thank John H. Molinski, Congran Jin, and Lawrence Livermore National Laboratory.
Alison Burklund and John X. J. Zhang are both founders of nanopathdx LLC, a company working to develop in vitro diagnostic tests.
DATA AVAILABILITY
Additional data requests can be directed to the corresponding author.
REFERENCES
- 1.Foudeh A. M. et al. , “Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics,” Lab Chip 12(18), 3249–3266 (2012). 10.1039/c2lc40630f [DOI] [PubMed] [Google Scholar]
- 2.Gossett D. R. et al. , “Label-free cell separation and sorting in microfluidic systems,” Anal. Bioanal. Chem. 397(8), 3249–3267 (2010). 10.1007/s00216-010-3721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kothari A., Morgan M., and Haake D. A., “Emerging technologies for rapid identification of bloodstream pathogens,” Clin. Infect. Dis. 59(2), 272–278 (2014). 10.1093/cid/ciu292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mairhofer J., Roppert K., and Ertl P., “Microfluidic systems for pathogen sensing: A review,” Sensors (Basel) 9(6), 4804–4823 (2009). 10.3390/s90604804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitt W. G. et al. , “Rapid separation of bacteria from blood-review and outlook,” Biotechnol. Prog. 32(4), 823–839 (2016). 10.1002/btpr.2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrera C. R., “Continuous concentration of bacteria in a microfluidic flow cell using electrokinetic techniques,” Electrophoresis. 22(2), 355–362 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Cho Y. K. et al. , “Bacteria concentration using a membrane type insulator-based dielectrophoresis in a plastic chip,” Electrophoresis 30(18), 3153–3159 (2009). 10.1002/elps.200900179 [DOI] [PubMed] [Google Scholar]
- 8.Jung J. H., Kim G. Y., and Seo T. S., “An integrated passive micromixer-magnetic separation-capillary electrophoresis microdevice for rapid and multiplex pathogen detection at the single-cell level,” Lab Chip 11(20), 3465–3470 (2011). 10.1039/c1lc20350a [DOI] [PubMed] [Google Scholar]
- 9.Mai J. et al. , “Rapid detection of trace bacteria in biofluids using porous monoliths in microchannels,” Biosens. Bioelectron. 54, 435–441 (2014). 10.1016/j.bios.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pivetal J. et al. , “Selective isolation of bacterial cells within a microfluidic device using magnetic probe-based cell fishing,” Sens. Actuators B 195, 581–589 (2014). 10.1016/j.snb.2014.01.004 [DOI] [Google Scholar]
- 11.Tay A. et al. , “Advances in microfluidics in combating infectious diseases,” Biotechnol. Adv. 34(4), 404–421 (2016). 10.1016/j.biotechadv.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bridle H., Miller B., and Desmulliez M. P. Y., “Application of microfluidics in waterborne pathogen monitoring: A review,” Water Res. 55, 256–271 (2014). 10.1016/j.watres.2014.01.061 [DOI] [PubMed] [Google Scholar]
- 13.Du G., Fang Q., and den Toonder J. M. J., “Microfluidics for cell-based high throughput screening platforms—A review,” Anal. Chim. Acta 903, 36–50 (2016). 10.1016/j.aca.2015.11.023 [DOI] [PubMed] [Google Scholar]
- 14.Váradi L. et al. , “Methods for the detection and identification of pathogenic bacteria: Past, present, and future,” Chem. Soc. Rev. 46(16), 4818–4832 (2017). 10.1039/C6CS00693K [DOI] [PubMed] [Google Scholar]
- 15.Liesenfeld O. et al. , “Molecular diagnosis of sepsis: New aspects and recent developments,” Eur. J. Microbiol. Immunol. 4(1), 1–25 (2014). 10.1556/EuJMI.4.2014.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun K. et al. , “Sepsis pathogen identification,” J. Lab. Autom. 20(5), 539–561 (2015). 10.1177/2211068214567345 [DOI] [PubMed] [Google Scholar]
- 17.Sinha M. et al. , “Emerging technologies for molecular diagnosis of sepsis,” Clin. Microbiol. Rev. 31(2), e00089–17 (2018). 10.1128/CMR.00089-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shehadul Islam M., Aryasomayajula A., and Selvaganapathy P. R., “A review on macroscale and microscale cell lysis methods,” Micromachines 8(3), 83 (2017). 10.3390/mi8030083 [DOI] [Google Scholar]
- 19.Kim J. et al. , “Microfluidic sample preparation: Cell lysis and nucleic acid purification,” Integr. Biol. 1(10), 574–586 (2009). 10.1039/b905844c [DOI] [PubMed] [Google Scholar]
- 20.Yagupsky P. and Nolte F. S., “Quantitative aspects of septicemia,” Clin. Microbiol. Rev. 3(3), 269–279 (1990). 10.1128/CMR.3.3.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burklund A., Tadimety A., and Zhang J.X.J., “Micro-scale immunomagnetic bacterial enrichment coupled to nanoplasmonic sensing for rapid detection of pathogens in whole blood” in Conference Proceeding. MicroTAS 2019 (Chemical and Biological Microsystems Society, 2019). [Google Scholar]
- 22.Melendez D. P. et al. , “Evaluation of a genus- and group-specific rapid PCR assay panel on synovial fluid for diagnosis of prosthetic knee infection,” J. Clin. Microbiol. 54(1), 120 (2016). 10.1128/JCM.02302-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshino K. et al. , “Microchip-based immunomagnetic detection of circulating tumor cells,” Lab Chip 11, 3449–3457 (2011). 10.1039/c1lc20270g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper R. M. et al. , “A microdevice for rapid optical detection of magnetically captured rare blood pathogens,” Lab Chip 14(1), 182–188 (2014). 10.1039/C3LC50935D [DOI] [PubMed] [Google Scholar]
- 25.Burklund A. et al. , “Advances in diagnostic microfluidics,” Adv. Clin. Chem. 95(1), 1–72 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Pankhurst Q. A. et al. , “Applications of magnetic nanoparticles in biomedicine,” J. Phys. D Appl. Phys. 36(13), R167–R181 (2003). 10.1088/0022-3727/36/13/201 [DOI] [Google Scholar]
- 27.Rosensweig R. E., “Heating magnetic fluid with alternating magnetic field,” J. Magn. Magn. Mater. 252, 370–374 (2002). 10.1016/S0304-8853(02)00706-0 [DOI] [Google Scholar]
- 28.Tong S. et al. , “Size-Dependent heating of magnetic iron oxide nanoparticles,” ACS Nano 11(7), 6808–6816 (2017). 10.1021/acsnano.7b01762 [DOI] [PubMed] [Google Scholar]
- 29.Mohapatra J., Xing M., and Liu P. J., “Inductive thermal effect of ferrite magnetic nanoparticles,” Materials 12(19), 1–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis C. L. and Ivkov R., “Physics of heat generation using magnetic nanoparticles for hyperthermia,” Int. J. Hyperthermia 29(8), 715–729 (2013). 10.3109/02656736.2013.836758 [DOI] [PubMed] [Google Scholar]
- 31.Chang D. et al. , “Biologically targeted magnetic hyperthermia: Potential and limitations,” Front. Pharmacol. 9, 831–831 (2018). 10.3389/fphar.2018.00831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shubitidze F. et al. , “Magnetic nanoparticles with high specific absorption rate of electromagnetic energy at low field strength for hyperthermia therapy,” J. Appl. Phys. 117(9), 094302–094302 (2015). 10.1063/1.4907915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee D. K., Diagaradjane P., and Krishnan S., “Nanoparticle-mediated hyperthermia in cancer therapy,” Ther. Delivery 2(8), 1001–1014 (2011). 10.4155/tde.11.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Packard M. M. et al. , “Performance evaluation of fast microfluidic thermal lysis of bacteria for diagnostic sample preparation,” Diagnostics 3(1), 105–116 (2013). 10.3390/diagnostics3010105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoopes P. J. et al. , “Treatment of canine oral melanoma with nanotechnology-based immunotherapy and radiation,” Mol. Pharm. 15(9), 3717–3722 (2018). 10.1021/acs.molpharmaceut.8b00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y.-y. et al. , “Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system,” Biomed. Microdevices 15(4), 673–681 (2013). 10.1007/s10544-012-9718-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen W. et al. , “Combined immunomagnetic capture coupled with ultrasensitive plasmonic detection of circulating tumor cells in blood,” Biomed. Microdevices 20(4), 99 (2018). 10.1007/s10544-018-0333-1 [DOI] [PubMed] [Google Scholar]
- 38.Burklund A. and Zhang J. X. J., “Microfluidics-Based organism isolation from whole blood: An emerging tool for bloodstream infection diagnosis,” Ann. Biomed. Eng. 47(7), 1657–1674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y., Kim S., and Chae J., “Separating and detecting escherichia coli in a microfluidic channel for urinary tract infection applications,” J. Microelectromech. Syst. 20(4), 819–827 (2011). 10.1109/JMEMS.2011.2159095 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Additional data requests can be directed to the corresponding author.