Abstract
The metalloprotease ADAM17 (a disintegrin and metalloprotease 17) regulates EGF-receptor and TNFα signaling, thereby protecting the skin and intestinal barrier but also contributing to autoimmunity. ADAM17 can be rapidly activated by many stimuli through its transmembrane domain (TMD), with the seven membrane-spanning inactive Rhomboids (iRhom) 1 and 2 implicated as candidate regulatory partners. However, several alternative models of ADAM17 regulation exist that don’t involve the iRhoms, such as regulation through disulfide bond exchange or through interaction with charged phospholipids. Here, we report that a non-activatable mutant of ADAM17 with the TMD of betacellulin can be rescued by restoring residues from the ADAM17 TMD, but only in Adam17−/− cells, which contain iRhoms, not in iRhom1/2−/− cells. We also provide the first evidence that the extracellular juxtamembrane domains (JMDs) of ADAM17 and iRhom2 regulate the stimulation and substrate selectivity of ADAM17. Interestingly, a point mutation in the ADAM17 JMD identified in a patient with Tetralogy of Fallot, a serious heart valve defect, affects the substrate selectivity of ADAM17 towards HB-EGF, a crucial regulator of heart valve development in mice. These findings provide new insights into the regulation of ADAM17 through an essential interaction with the TMD1 and JMD1 of iRhom2.
Keywords: ADAM17 (a disintegrin and metalloprotease 17), iRhom1/2 (inactive Rhomboid like protein 1/2), Rhbdf1/2 (rhomboid family member 1/2), TNFα (Tumor necrosis factor α), EGFR (epidermal growth factor receptor), TGFα (transforming growth factor α)
Introduction
The membrane-anchored metalloprotease ADAM17 (a disintegrin and metalloprotease 17) has emerged as a key regulator of the TNFα and EGF-receptor signaling pathways due to its essential role in the processing of TNFα and of several ligands of the EGFR (1–5). ADAM17-dependent signaling through the EGFR pathway is important for mouse development (3, 5–7), protection of the skin and intestinal barrier during adulthood (8–10) and it can contribute to the pathogenesis and progression of cancers (11, 12). ADAM17 also liberates soluble TNFα from immune cells such as macrophages and thus has important roles in TNFα-dependent pathologies such as sepsis and rheumatoid arthritis (1–3, 7, 13). Moreover, ADAM17 has numerous other substrates (14), including the IL-6R, which also has well-established roles in supporting inflammation[dummy] (15–18). ADAM17 can be rapidly and post-translationally activated by a variety of signaling pathways, such as tyrosine kinases and G-protein coupled receptors (GPCRs) (5, 19–23). The physiological relevance of the regulation of ADAM17 by a GPCR is well established for hair follicle development (24) and the rapid response of ADAM17 to many different stimuli is thought to be important for its ability to protect the skin and intestinal barriers upon injury and to maintain their function under physiological conditions.
Elucidating the precise mechanisms which control the function of ADAM17 is critical to understanding the posttranslational regulation of its proteolytic activity. The current understanding of the regulation of ADAM17 is multifaceted and has given rise to a variety of different models regarding the underlying mechanism. Studies from our lab support a model in which the activation of ADAM17 by various physiological stimuli is a rapid and reversible process that requires its transmembrane domain (TMD) (22) and several studies have concluded that its cytoplasmic domain is dispensable for this process (22, 25–27). The discovery of the seven-membrane spanning inactive rhomboid proteins, iRhom1 and iRhom2 (iR1 and iR2) as crucial regulators of the maturation and function of ADAM17 uncovered attractive candidate molecules that could control ADAM17 through an interaction with its TMD (28–32). Additional evidence for an interaction between ADAM17 and iR2 via their TMDs was provided by a point mutation in the first TMD (TMD1) of iR2, the I387F sinecure mutation, which blocks shedding of iR2-selective substrates such as murine Kit-Ligand 2 (KL2) or the EGFR-ligand epiregulin (EREG) in mouse embryonic fibroblasts and TNFα release in myeloid cells (30, 31, 33). In addition, point mutations in predicted iR2-interacting sites in the TMD of ADAM17 abolished shedding of the ADAM17/iR2-selective substrates KL2 and EREG without affecting the ADAM17/iR1/2-dependent shedding of TGFα (33). Recently, cytoplasmic binding partners of iR2 were discovered that regulate the function of the ADAM17/iR2 complex (34–37). The phosphorylation of iR2 is thought to activate ADAM17 on the cell surface, providing a plausible explanation for why the cytoplasmic domain of ADAM17 is dispensable for its rapid activation (34, 36).
Although there is now considerable evidence in support of a model in which iR2 and the related iR1 are key regulators of ADAM17, several alternative mechanisms for the activation of ADAM17 have been proposed, which are summarized in recent reviews (14, 38, 39). Cytoplasmic phosphorylation of ADAM17 has been suggested to regulate its activation (14, 40, 41), as has rapid transport to the cell surface (41). Other studies have concluded that ADAM17 is regulated by a reversible interaction with TIMP3 (42) or integrins (39, 43). In addition, the extracellular domain of ADAM17 has been suggested to control the stimulation of ADAM17 (reviewed in (39)) either by mediating a reversible interaction with the cell membrane (44, 45) or as a target of disulfide isomerase activity (39, 46, 47), although the disulfide isomerase(s) that are essential for ADAM17 activity remain to be identified (48). Finally, it has been suggested that the catalytic domain of ADAM17 is constitutively active and that stimulated substrate release is accomplished by exposure of negatively charged phosphatidylserine residues that pull the catalytic domain to the cell membrane (49, 50).
The goal of this study was to test our hypothesis that the iRhoms are key regulators of ADAM17 by exploring the role of the extracellular juxtamembrane domains (JMDs) and TMDs of iR2 and ADAM17 in modulating ADAM17-dependent protein ectodomain shedding. We analyzed how the ADAM17 TMD affects its activity in cells lacking ADAM17 versus cells lacking both iR1 and 2. Moreover, we used a constitutively active mutant of ADAM17 with the TMD of betacellulin that cannot be stimulated in gain-of-function experiments (22) to determine which amino acid residues in the ADAM17 TMD are sufficient for restoring stimulated activity. Since the TMDs of ADAM17 and iR2 are required for iR2/ADAM17-dependent shedding, this raised the possibility that their interaction extends to the extracellular JMDs. We therefore asked whether the JMDs of iR2 and A17 also contribute to the regulation of ADAM17. Finally, we tested whether a point mutation in the JMD of ADAM17 (L659P) that was recently identified in a patient suffering from severe defects in heart valve development (Tetralogy of Fallot) (51), affects the regulation of ADAM17 by iR2. Our findings provide the first evidence that disruptions in either the JMD of ADAM17 or iR2 interfere with the function of ADAM17. These results lend strong support to a model in which the TMD and JMD of ADAM17 and iR2 are crucial for the regulation and substrate selectivity of the iR2/ADAM17 complex and are discussed in the context of other proposed models of the regulation of ADAM17.
Materials and Methods
Cell lines and reagents
All reagents were purchased from Sigma Aldrich (St. Louis, MO, USA) except indicated otherwise. The following antibodies were used: Rat anti-HA monoclonal (clone 3F10) (Roche, Mannheim, Germany); mouse anti-T7 tag monoclonal (Novagen, Darmstadt, Germany); rabbit α-Tubulin polyclonal (Cell Signaling Technology, Danvers, MA, USA). Rabbit anti-ADAM17 polyclonal antibodies have been described previously (52). The antibodies against iRhom2 were generated by injecting a gst-fusion protein with the cytoplasmic domain of mouse iRhom2 (amino acid residues 1–376) into rabbits (ProSci Inc., Poway, CA, USA). Anti-rat (Sigma Aldrich, St. Louis, MO, USA), anti-rabbit and anti-mouse HRP-tagged antibodies (Promega, Madison, WI, USA) were used as secondary antibodies. The isolation and culture of mouse embryonic fibroblasts (mEFs) from Adam17−/−, iR1/2−/−, WT, and Adam10/17−/− mice has been described previously (21, 25, 31, 32). mEFs were cultured in DMEM with antibiotics and 10% fetal calf serum.
Expression vectors
In order to generate expression constructs for the murine ADAM17 mutants used in this study, the transmembrane and cytoplasmic domains of A17-BTC, A17-cytoBTC, A17-tmBTC and A17-BTC1–3 (see Figure 2A and Supplementary Figure S1A for details) were generated by GENEWIZ (South Plainsfield, NJ, USA) and sub-cloned into a pcDNA3.1(+) murine ADAM17 plasmid that had been prepared by enzymatic digestion at the BsiWI/NotI restriction site. The A17-E>A, A17, A17-BTC and A17-CD62L plasmids were described previously (22). Site-directed mutagenesis was performed with the QuikChange® Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). All oligonucleotide primer sequences used for site-directed mutagenesis can be found in Table S1. The iRhom deletion mutants were generated by PCR and cloned into pcDNA3.1(+) (see Table S2 for details). All mutant constructs were sequenced to ensure that the desired mutations were present and to rule out other mutations in the coding sequence.
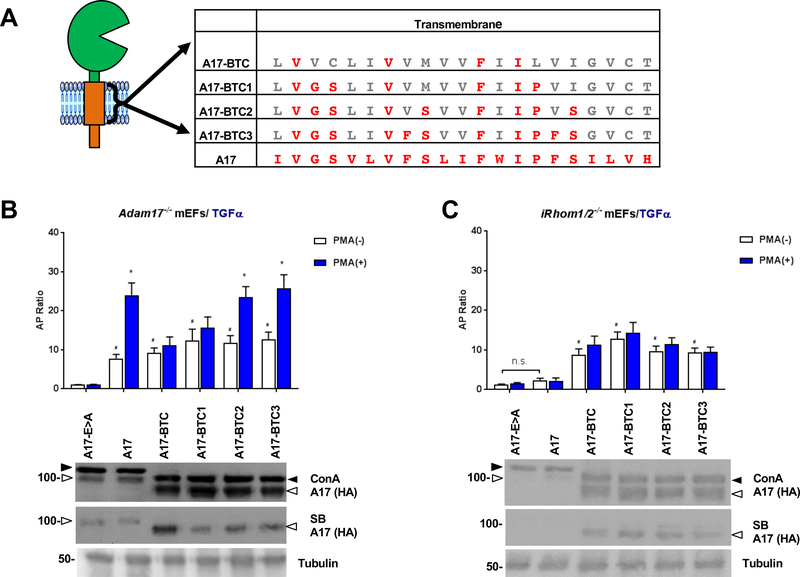
Figure 2. Step-by-step conversion of amino acid residues of the TMD of A17-BTC to those in the TMD of A17 defines residues that are sufficient to support stimulated ADAM17-dependent shedding.
(A) The TMD of A17-BTC is shown with amino acid residues that are conserved with the A17-TMD highlighted in red. A17-BTC1 – 3 represent A17-BTC mutants that are progressively turned into A17 through point mutations, as indicated in red letters. (B, C) Ectodomain shedding assays were performed in Adam17−/− mEFs (B) and iR1/2−/− mEFs (C) following cotransfection of AP-tagged TGFα and A17-E>A, A17 or A17-BTC or A17-BTC1 – 3. Expression of ADAM17 and the mutant forms of ADAM17 was detected by Western Blot against the C-terminal HA-tag for Adam17−/− mEFs (B, top panel of Western blot) and for iR1/2−/− mEFs (C, top panel), whereas the presence of these proteins at the cell surface was detected by cell surface biotinylation (B, C, middle panels), with α-tubulin serving as a loading control (B, C, lower panel). Results are shown as mean ± SEM (n=3). # denotes the comparison of constitutive activity between a sample and the mutant E>A where P≤0.05. * denotes P≤0.05 between the PMA (−) and PMA (+) condition for a given sample. Western blots are representative of at least three independent experiments.
Table 1-.
Site-directed mutagenesis primers
| Mutations | Forward Primer | Reverse Primer |
|---|---|---|
| BTC, M44S | TGCCTGATCGTGGTGTCCGTGGTGTTCATCATC | GATGATGAACACCACGGACACCACGATCAGGCA |
| BTC, I52S | TTCATCATCCTGGTGTCCGGCGTGTGCACCTGC | GCAGGTGCACACGCCGGACACCAGGATGATGAA |
| BTC1, M44S | CTGATCGTGGTGTCCGTGGTGTTCATC | GATGAACACCACGGACACCACGATCAG |
| BTC1, I52S | ATCATCCCCGTGTCCGGCGTGTGCACC | GGTGCACACGCCGGACACGGGGATGAT |
| iR2-JMD1–1 V406E, Q409D | TGTTCTTCAGCACCAGATCGGTGGTCTCGTGCTGGGCAAAGCC | GGCTTTGCCCAGCACGAGACCACCGATCTGGTGCTGAAGAACA |
| iR2-JMD1–2 A403S, L410S | GTGGGCTTTTCCCAGCACGAGACCACCGATTCCGTGCTGAAG | CTTCAGCACGGAATCGGTGGTCTCGTGCTGGGAAAAGCCCAC |
| iR2-JMD1–3 A403S, V406E, Q409D, L410S | GTGGGCTTTTCCCAGCACGTTACCACCCAGTCCGTGCTGAAG | CTTCAGCACGGACTGGGTGGTAACGTGCTGGGAAAAGCCCAC |
| ADAM17 L659P | GATTTCATTGACCAGCCTAGCATCAACACTTTTG | CAAAAGTGTTGATGCTAGGCTGGTCAATGAAATC |
| ADAM17 6A.1 (first mutagenesis step) | GATTTCATTGACCAGCTGGCCGCCGCCACTTTTGGGAAGTTTCTG | CAGAAACTTCCCAAAAGTGGCGGCGGCCAGCTGGTCAATGAAATC |
| ADAM17 6A.2 (second mutagenesis step) | CAGAAACTTCCCAAAAGTGGCGGCGGCCAGCTGGTCAATGAAATC | GTTATCTGCCAGAAACTTGGCGGCGGCGGCGGCGGCCAGCTGGTC |
Table 2-.
Primer sequence used for juxtamembrane domain deletion of iR2
| Mutations | Forward Primer | Reverse Primer |
|---|---|---|
| iR2ΔJM1 | GGCATCGCACCTGTGGGCTTTGCCCAGAGAGGCGTGTATGAGAGCGTGAAGTAC | GTACTTCACGCTCTCATACACGCCTCTCTGGGCAAAGCCCACAGGTGCGATGCC |
| iR2ΔJM2 | TTAGACAAGGTGTGTGGGCTCCTGCCTTACCGGATCTGGCTGTCTTTATTCCTG | CAGGAATAAAGACAGCCAGATCCGGTAAGGCAGGAGCCCACACACCTTGTCTAA |
Western Blot Analysis
All wild type and mutant mEFs used here were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and the indicated plasmids. The day after transfection, cells were washed with PBS and lysed in 1 ml lysis buffer (PBS, 1% Triton X-100, protease inhibitor cocktail (2μg/ml Leupeptin; 0.4 mM Benzamidine; 10μg/ml Soybean Trypsin Inhibitor; 0.5mM Iodoacetamide), and 10mM 1,10-phenanthroline). Concanavalin A beads were used for selective enrichment of glycoproteins (29). Comparable amounts of protein from a single well of a 6-well plate (9.5cm2 growth area) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% nonfat milk for one hour at room temperature. Primary antibodies were added overnight at 40C, then the membranes were washed three times in PBS, 0.05% Tween-20, probed with anti-HRP secondary antibody and bound antibodies were detected using Luminata (Millipore/Sigma, Burlington, MA, USA) and a Chemidoc image analyzer (BioRad, Hercules, CA, USA).
Cell Surface Biotinylation
Adam17−/− and iR1/2−/− mEFs were grown to 80% confluency in 6-well plates and transfected with C-terminally HA-tagged wild type ADAM17, inactive ADAM17 (E>A), or ADAM17 BTC mutants (21, 25). A day after transfection, the cells were pre-incubated with Dulbecco’s Phosphate Buffered Saline (DPBS, with Ca++/Mg++, Corning, Corning, NY, USA) for 30 minutes at 40C, and then the cells were incubated with 1mg/ml of NHS-LC-Biotin for 30 minutes at 40C, followed by 100mM of Glycine in Dulbecco’s Phosphate Buffered Saline (DPBS) to quench the reaction. Cells were lysed as described above and cell lysates were incubated with NeutraAvidin Agarose resin (Thermo Scientific, Rockford, IL, USA) overnight at 40C. Isolated biotinylated proteins were detected by Western blot, as described above.
Ectodomain Shedding Assays
The wt and mutant mEFs were grown to 80% confluency in 12-well plates and co-transfected with alkaline phosphatase (AP)- tagged transforming growth factor α (TGFα) (4) or Kit Ligand 2 (KL2) (53) together with ADAM17 or iR2 mutant constructs. The day after transfection, cells were washed with Optimem for 30 minutes. Then fresh Optimem was added with or without 25ng/ml of Phorbol 12-myristate 13-acetate (PMA) for one hour to stimulate shedding. The alkaline phosphatase substrate, 4-nitrophenyl phosphate, was added to the supernatant and cell lysate and the absorbance at A405nm was measured to assess the AP activity (54). Measurements were conducted in triplicates and the ratio between the AP activity in the supernatant and the AP-activity in the cell lysate plus supernatant was calculated. Each experiment was performed at least three times with a total of three wells per experiment.
Statistical Analysis
All data are expressed as mean ± S.E.M with representative data from at least three independent experiments. Statistical analyses were performed using an unpaired, two-tailed student’s t-test. Multiple comparisons between experimental groups were analyzed by two-way ANOVA in conjunction with Fisher’s LSD test. P values of ≤ 0.05 were considered statistically significant.
Results
The ADAM17 TMD has different effects on protein ectodomain shedding in the presence or absence of iR1 and 2
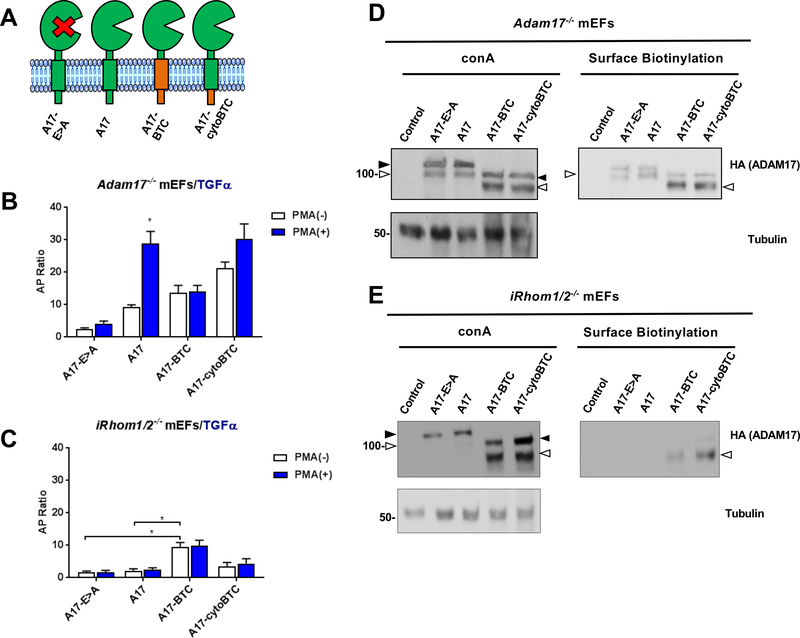
To explore the potential contribution of the iRhoms to the regulation of ADAM17 by its TMD, we tested the function of wild type (wt) and mutant forms of ADAM17 in the presence or absence of the iRhoms by performing rescue experiments in Adam17−/− or iR1/2−/− mouse embryonic fibroblasts (mEFs). Constitutive and stimulated shedding of the ADAM17-substrate TGFα in Adam17−/− mEFs can be rescued by wt ADAM17, but not by the inactive ADAM17E>A mutant (Fig. 1A, B, see also (22)), whereas both wt and E>A mutant ADAM17 were not active in iR1/2−/− double knockout mEFs, in which endogenous ADAM17 exists only in its inactive pro-form (32) (Figure 1C). However, a mutant ADAM17 with the TMD and cytoplasmic domain of betacellulin (A17-BTC), which had previously been shown to restore constitutive, but not stimulated TGFα shedding in Adam17−/− mEFs ((22), also shown in Fig. 1B) also restored constitutive, but not stimulated TGFα shedding in iR1/2−/− mEFs (Fig. 1C). When only the cytoplasmic domain, but not the TMD of ADAM17 was replaced with that of BTC (A17-cytoBTC), this increased constitutive TGFα shedding compared to wt ADAM17 in Adam17−/− mEFs, which could be further enhanced by PMA stimulation (Fig. 1B). Stimulated TGFα shedding from iR1/2−/− mEFs transfected with A17-cytoBTC was not significantly increased compared to the inactive A17E>A control (Fig. 1C, see Supp. Fig. 1 for sequence information). A construct with the TMD of BTC and the cytoplasmic domain of A17 (A17-tmBTC) restored constitutive TGFα shedding in A17−/− mEFs, but only had slightly increased activity compared to A17E>A or wt A17 in iR1/2−/− mEFs (Suppl. Fig. 2A-C). When a mutant ADAM17 with the TMD and cytoplasmic domain of a different membrane protein, CD62L (A17-CD62L), was introduced into iR1/2−/− mEFs, it also rescued constitutive, but not stimulated TGFα shedding, just like A17-BTC (Suppl. Fig. 3). A Western blot analysis confirmed similar expression of the pro-form of ADAM17E>A and wt ADAM17 in Adam17−/− as well as in iR1/2−/− mEFs (Fig. 1D, E). The mature form of the overexpressed wt ADAM17 and the E>A mutant could be detected by Western blot and cell surface biotinylation in Adam17−/− mEFs, but not in iR1/2−/− mEFs (Fig. 1D, E), consistent with the properties of endogenous ADAM17 in iR1/2−/− mEFs (32). The three mutants containing the TMD or cytoplasmic domain of BTC or both (A17-BTC, A17-cytoBTC, A17-tmBTC) had relatively higher levels of mature ADAM17 when compared to ADAM17E>A (A17E>A) or wild type ADAM17 (A17) in Adam17−/− and iR1/2−/− mEFs (Fig. 1D,E, Suppl. Fig. 2D,E). Moreover, whereas A17E>A and A17 could be biotinylated on the cell surface when expressed in Adam17−/− mEFs (Fig. 1D, Suppl. Fig. 2D), only the ADAM17/BTC chimera (A17-BTC, A17-cytoBTC, A17-tmBTC) could be biotinylated on the cell surface of iR1/2−/− mEFs (Fig. 1E, Suppl. Fig. 2E). These results suggest that ADAM17 contains a retention motif that prevents its transport to the cell surface in the absence of the iRhoms, which can be disrupted by replacing the TMD or cytotail or both domains with the equivalent domains of BTC.
Figure 1. Role of the ADAM17 transmembrane domain in regulating protein ectodomain shedding in the presence or absence of iRhoms 1 and 2.
(A) Schematic representation of the catalytically inactive ADAM17-E>A mutant (A17-E>A) and wild type ADAM17 (A17), as well as ADAM17 mutants containing the transmembrane domain (TMD) and the cytoplasmic domain (cyto) of betacellulin (A17-BTC), or the TMD of ADAM17 and the cytoplasmic domain of BTC (A17-cytoBTC). (B, C) Shedding experiments in which Adam17−/− mEFs (B) or iR1/2−/− mEFs (C) were co-transfected with alkaline phosphatase (AP) -tagged transforming growth factor α (TGFα) and the ADAM17 constructs shown in (A). Constitutive and PMA-stimulated shedding (treatment with 25ng/ml PMA) were measured after 1 hour. Results are shown as mean ± SEM (n=3), * denotes P≤0.05 between the PMA (−) and PMA (+) condition for a given sample. (D, E) Western blot analysis and cell surface biotinylation of the overexpressed ADAM17 and ADAM17 mutants in Adam17−/− mEFs and iR1/2−/− mEFs. Concanavalin A beads were used for selective enrichment of glycoproteins, which were detected with antibodies against the C-terminal HA-tag. α-Tubulin served as a loading control. The ADAM17 pro-form is indicated by a black arrowhead and the mature form by a white arrowhead. Please note that overexpressed pro-ADAM17 can sometimes be detected on the cell surface of transfected Adam17−/− mEFs, as in panel D, which is not the case for endogenous ADAM17 in wild type mEFs or in transfected iR1/2−/− mEFs. These Western blots are representative of at least three independent experiments.
Sequential replacement of BTC-TMD residues with sequences from the ADAM17 TMD restores iRhom1/2-dependent stimulated ADAM17 activity
To identify sequences in the ADAM17 TMD that are critical for its activation, we used gain-of-function experiments, in which we performed stepwise replacements of residues in the A17-BTC TMD with ADAM17 TMD residues. The mutants possess progressively more similarity to wt A17 and were named A17-BTC1, 2, and 3 (Fig. 2A). We found that A17-BTC1 behaved similarly to A17-BTC in that it restored constitutive, but not PMA-stimulated TGFα shedding in Adam17−/− mEFs (Fig. 2B). Remarkably, introduction of two serine residues into A17-BTC1 to yield A17-BTC2 (S44, S52) restored PMA-stimulated shedding of TGFα, as did A17-BTC3 (Fig. 2B). However, introduction of individual serine mutations (S44 or S52) into A17-BTC or A17-BTC1 was not sufficient to restore PMA stimulated TGFα shedding (Suppl. Fig. 4). In iR1/2−/− mEFs, all A17-BTC mutants supported constitutive, but not PMA-stimulated TGFα shedding, whereas A17E>A and A17 were inactive (Fig. 2C). The mature forms of A17-BTC and A17-BTC1 – 3 could be detected by cell surface biotinylation in Adam17−/− and iR1/2−/− mEFs, whereas mature A17 or A17E>A was only detected on the cell surface of Adam17−/− mEFs, but not iR1/2−/− mEFs (Fig. 2B, C, lower panels, see also Fig. 1).
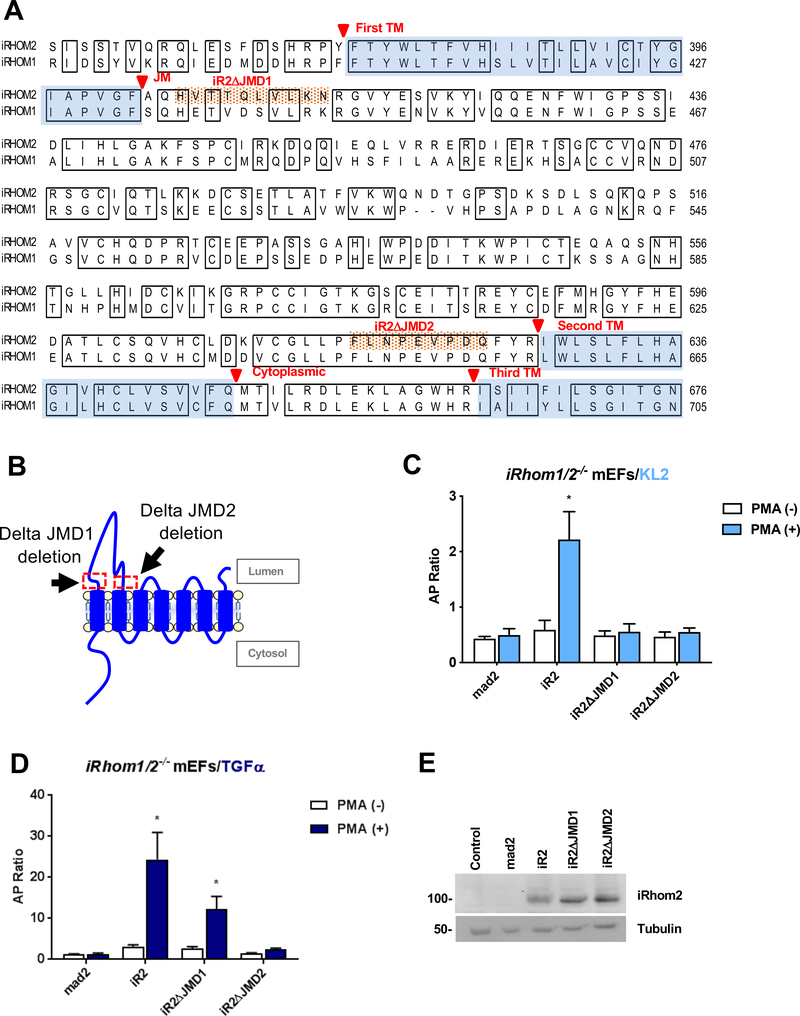
The extracellular juxtamembrane domains next to iR2 TMD1 and TMD2 affect ADAM17-dependent stimulated shedding
Given the essential role of the ADAM17 TMD and the TMD1 of iR2 in stimulated iR2/ADAM17-dependent shedding (Fig. 1, see also (33), we asked whether the extracellular juxtamembrane domains (JMD) next to TMD1 and TMD2 iR2 also contribute to this process. Sequence alignment of the extracellular JMD adjacent to the iR2 TMD1 (referred to as JMD1) showed that only five out of the 12 residues were conserved in iR1 and 2, whereas the last 18 amino acid residues in the JMD2 preceding TMD2 were completely conserved (Fig. 3A). To test whether these short juxtamembrane regions affect the function of iR2, we generated two deletion mutants, iR2ΔJMD1 and iR2ΔJMD2 (Fig. 3A, B). In rescue experiments, we found that both mutants abolished constitutive and stimulated shedding of the iR2-selective substrate Kit-Ligand2 (KL2) in iR1/2−/− mEFs (Fig. 3C). Interestingly, iR2ΔJMD1 supported stimulated shedding of the iR1/2-substrate TGFα, albeit at reduced levels compared to wt iR2, whereas iR2ΔJMD2 did not (Fig. 3D). A Western blot analysis confirmed that both mutants were expressed at similar levels as wt iR2 in the iR1/2 −/− mEFs (Fig. 3E). These findings provide the first evidence that the iR2 JMD1 has a role in the stimulated substrate selective activity of ADAM17, whereas the highly conserved JMD2 is required for the stimulated shedding of both the iR2-selective KL2 and the iR1/2 substrate TGFα by ADAM17.
Figure 3. Characterization of the role of the iR2 extracellular JMD 1 or 2 on stimulated ADAM17-dependent shedding.
(A) Amino acid sequence alignment of partial sequences of iR1 and iR2, starting with a part of the N-terminal cytoplasmic domain, followed by the TMD1, the extracellular loop domain, and TMD2 and a part of TMD3. Identical sequences are boxed and transmembrane domains are shaded in light blue. The extracellular juxtamembrane sequences that are deleted in iR2ΔJMD1 and iR2ΔJMD2 are highlighted with orange dots. (B) Schematic indicating the position of the deleted regions in the iR2 juxtamembrane domain in iR2ΔJMD1 and iR2ΔJMD2. (C, D) Ectodomain shedding assays in iR1/2−/− mEFs after co-transfection of the iR2-dependent substrate AP-tagged Kit-ligand 2 (KL2, C) or AP-tagged TGFα (D) along with iR2, iR2ΔJMD1 and iR2ΔJMD2. Results are shown as mean ± SEM (n=3). *P<0.05. (E) Western blot analysis confirmed similar expression of iR2, iR2ΔJMD1 and iR2ΔJMD2 in iR1/2−/− mEFs. Western blots are representative of at least three independent experiments.
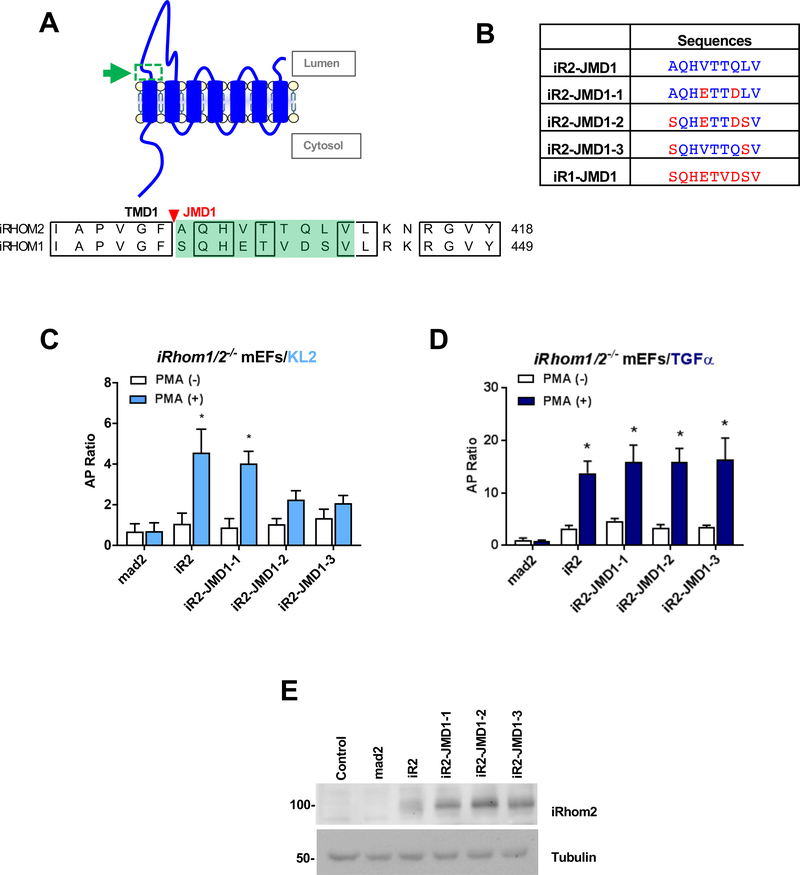
To follow up on the substrate selective effect of deleting the JMD1 in iR2, we attempted to identify amino acid residues in the mouse iR2 JMD1 that control the substrate selectivity of ADAM17. For this purpose, we generated three mutants, in which specific iR2 JMD1 residues were replaced with the corresponding sequences from the JMD1 of mouse iR1. We named these mutants iR2-JMD1–1, −2, and −3 (Fig. 4A, B). In iR1/2−/− mEFs, iR2-JMD1–1 rescued stimulated KL2 shedding as well as wt iR2, whereas iR2-JMD1–2 and iR2-JMD1–3 did not (Fig. 4C). In contrast, the stimulated shedding of TGFα was comparable between iR2 and all three mutants (Fig. 4D), which were expressed at similar levels (Fig. 4E). These results provide the first evidence that specific residues in the iR2 JMD1 have a role in regulating the substrate selectivity of ADAM17, presumably by interacting with the corresponding JMD in ADAM17.
Figure 4. Contribution of the extracellular JMD1 of iR2 to the substrate selectivity of ADAM17.
(A) Schematic of the domain organization of iR2 with a green arrow and box highlighting JMD1 (top panel). Amino acid sequence alignment of the JMD1 domain (shaded in green) of iR1 and iR2 with identical regions boxed (lower panel). (B) iR2 mutants (iR2-JMD1–1 to 1–3) were generated by replacing specific juxtamembrane amino acid residues with the corresponding iR1 residues. (C, D) Rescue experiments were conducted in iR1/2−/− mEFs to test the ability of iR2-JMD1–1 to 1–3 to restore stimulated ADAM17-dependent shedding of KL2 (iR2 selective) versus TGFα (iR1/2 selective). Results are shown as mean ± SEM (n=3). *P<0.05. (E) Western blot control for the expression levels of iR2-JMD1–1 to 1–3.
The extracellular JMD of ADAM17 contributes to its substrate selectivity
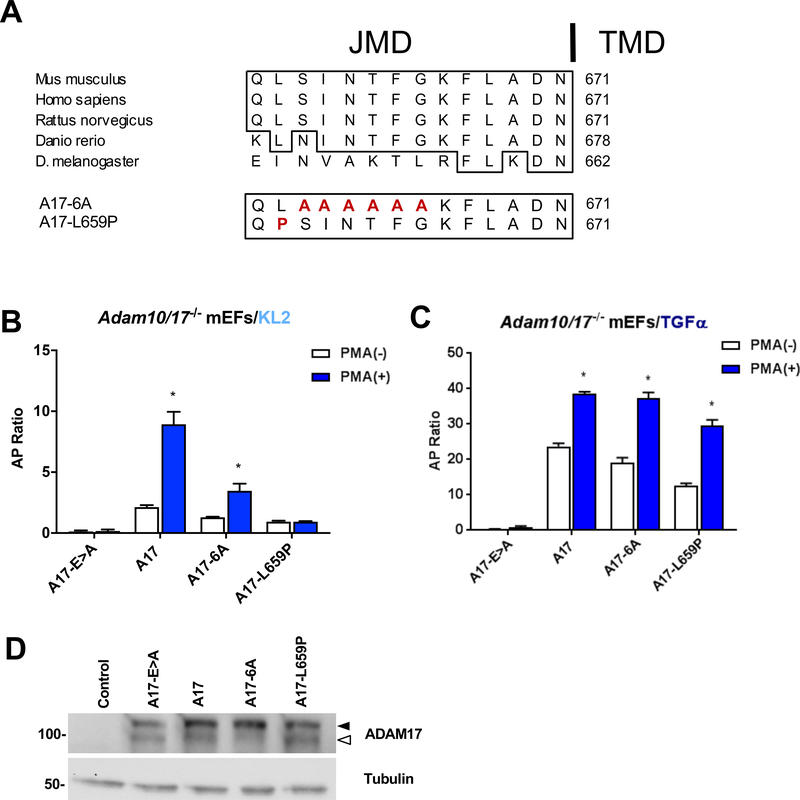
The juxtamembrane domain of ADAM17 is highly conserved in vertebrates (e.g. mice, humans, rats, zebrafish), but not in D. melanogaster, which use an active Rhomboid protease instead of iRhom/ADAM17 to activate the EGFR (55) (Fig. 5A). Given the role of the iR2-JMD1 and TMD1 in regulating the substrate selectivity of ADAM17 (see Figs. 3, 4 and (33)), we hypothesized that the extracellular JMD of ADAM17 acts as a counterpart for the iR2-JMD1 and therefore also contributes to the substrate selective activity of ADAM17. To test this hypothesis, we generated a mutant ADAM17, in which six amino acid residues in the extracellular JMD domain were changed to alanines (Fig. 5A, mutant A17–6A). In rescue experiments in A10/17−/− mEFs, iR2-selective KL2 shedding supported by the A17–6A mutant was significantly lower compared to wt A17 (Fig. 5B), whereas A17–6A rescued constitutive and stimulated shedding of the iR1/2 substrate TGFα to a similar degree as wt A17 (Fig. 5C). We also tested a missense variant in the JMD of A17 (A17-L659P) that was recently identified in a patient suffering from Tetralogy of Fallot (TOF), a severe congenital heart defect (51). Rescue experiments with the corresponding mouse A17-L659P mutant showed significantly increased TGFα shedding following stimulation with PMA, albeit slightly reduced compared to the wt control (Fig. 5C). However, the A17-L659P mutant only supported very low levels of constitutive KL2 shedding, which could not be further stimulated (Fig. 5B). A Western blot analysis confirmed comparable expression levels of the mutant and wt forms of ADAM17 (Fig. 5D). These results provided the first evidence that the membrane proximal JMD of ADAM17 has a role in determining its selectivity towards an iR2-dependent substrate (KL2), but not an iR1-dependent substrate (TGFα, (31)), suggesting that it is more important for the interaction with iR2 than with iR1.
Figure 5. The extracellular JMD of ADAM17 has a role in the substrate selectivity of stimulated ADAM17.
(A) Amino acid sequence alignment of the ADAM17 extracellular JMD in four vertebrates and D. melanogaster, which uses active Rhomboids to activate the EGFR (identical sequences boxed). A series of six conserved JMD residues found in vertebrates but not D. melanogaster was mutagenized to alanine (ADAM17–6A). In addition, a point mutation found in a human patient suffering from Tetralogy of Fallot, a congenital heart defect, is indicated (L659P). (B, C) Rescue experiments were conducted in Adam17−/− mEFs to assess the ability of the ADAM17 JMD mutants to support stimulated ADAM17-dependent shedding of KL2 (B) versus TGFα (C). Results are shown as mean ± SEM (n=3). * denotes P≤0.05 between the PMA (−) and PMA (+) condition for a given sample. (D) Comparable expression of ADAM17 and the ADAM17 JMD mutants was confirmed by Western Blot analysis, which showed slightly less mature A17–6A compared to the other mutants, despite comparable activity on TGFα (C). The pro-form of ADAM17 is marked by a black arrowhead and the mature form by a white arrowhead. Western blots are representative of at least three independent experiments.
Discussion
The main goal of this study was to explore the mechanism underlying the rapid and posttranslational stimulation of ADAM17. The results presented here are the first to uncover a crucial role of the extracellular JMDs of ADAM17 and iR2 in regulating the function of ADAM17, with implications for how a point mutation in the JMD of ADAM17 could contribute to a severe developmental abnormality of the heart. The results presented here support the concept that the iRhoms are crucial for the regulation of ADAM17.
The very different behavior of the ADAM17 TMD mutants with and without iRhoms strongly supports a model in which iR1 and 2 are critical for the activation of ADAM17 (Fig. 6A). The constitutive activity of the A17-BTC and ADAM17-CD62L mutants even in the absence of the iRhoms demonstrate that the iRhoms are not required for the extracellular domain of ADAM17 to fold into a functional and constitutively active protease. This interpretation is consistent with a previous study, in which the inactive pro-ADAM17 isolated from bone marrow-derived macrophages lacking iRhoms could be activated by addition of the pro-protein convertase furin (28). Interestingly, replacing only the cytoplasmic domain of ADAM17 with that of BTC while retaining the ADAM17 TMD allowed similar levels of transport to the cell surface as seen for A17-BTC in iR1/2−/− mEFs. Nevertheless, the A17-cytoBTC had much less activity than A17-BTC in iR1/2−/− mEFs, suggesting that the A17 TMD prevents the constitutive activity of mature ADAM17 if there are no iRhoms to interact with.
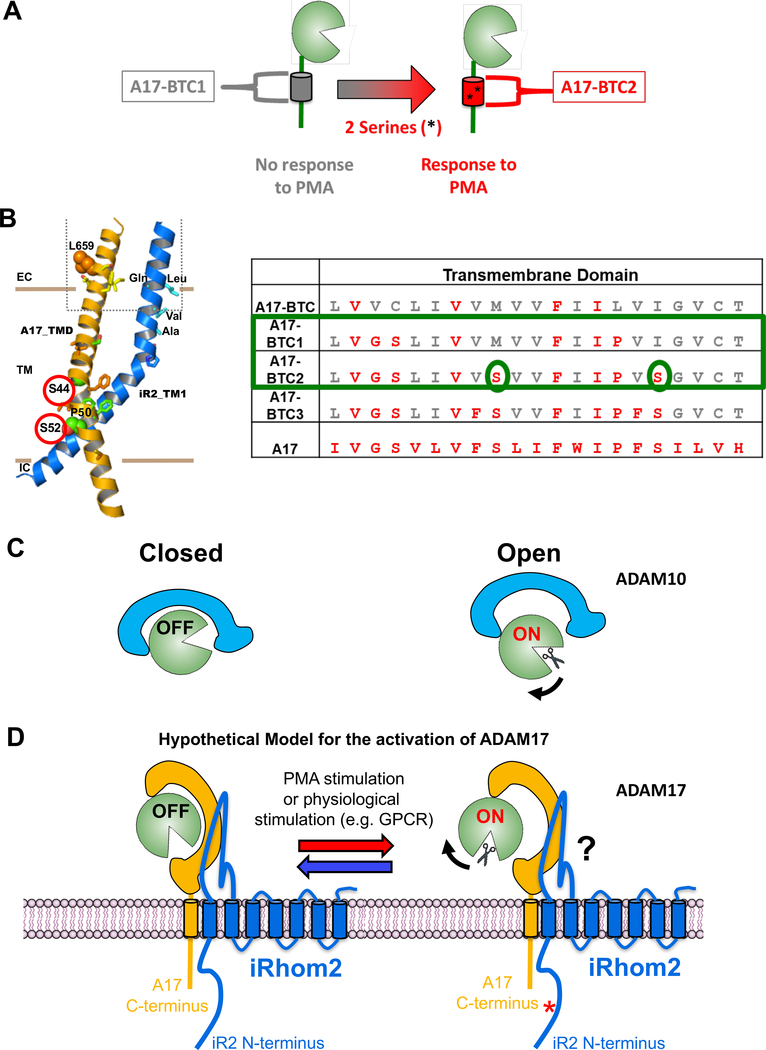
Figure 6. Diagrams indicating the predicted interactions between iRhom2 and ADAM17, the location of TMD serine residues that are required for ADAM17 activity and of a hypothetical model of the possible regulation of ADAM17 by iRhom2.
(A) Diagram of the role of Serine S44 and S52 in restoring PMA-stimulated activity to ADAM17-BTC1. (B) A Molecular Dynamics modeling of the predicted interaction between the ADAM17 TMD and the iRhom2 TMD1 (33), with S44 and S52 highlighted by red circles. The amino acid sequences of the TMD of A17-BTC, A17-BTC1 – 3 is shown for comparison, with S44 and S52 highlighted by green ovals. (C,D) Model of the possible allosteric regulation of the extracellular domain of ADAM17, based on the recent crystallographic identification of an “on” and “off” position in the highly related ADAM10 (C, adapted from (56)). We hypothesize that the interaction between the TMD/JMD of ADAM17 and the TMD1/JMD1 of iRhom2 provides a fulcrum that allows the complex to reversibly toggle between two allosteric conformations, one “on” and the second “off” (D). This switch could be activated by phosphorylation of the cytoplasmic tail of iRhom2 (34). Further studies will be necessary to understand how this switch is controlled and how it could exert allosteric control over the activation of ADAM17.
Gain-of-function experiments, in which residues in the TMD of A17-BTC were sequentially replaced with those of ADAM17 further supported the concept of a functionally relevant interaction between the ADAM17 TMD and the iRhoms. The most informative mutants were A17-BTC1 and A17-BTC2, which differ in 2 serine residues that restored stimulated shedding in A17-BTC2, but only in the presence of iR1/2, not in their absence (see model in Fig. 6B). Each of the two serine residues have also been implicated in the shedding of iR2 selective substrates in loss of function experiments (33). In a computational model of the interaction between the TMD of ADAM17 and TMD1 of iR2, these two serine residues flank a proline that introduces a kink into the TMD of ADAM17 (see model in Fig. 6B). They could therefore conceivably help ADAM17 to dock or clamp onto the TMD1 of iR2 and perhaps also iR1. This notion is supported by our observation that single mutations, in which only one serine was introduced into BTC1, were not sufficient to restore stimulated A17 activity. These results argue against models that invoke regulation of ADAM17 by its extracellular domain, for example through disulfide isomerase-mediated exchange of disulfide bonds or through an interaction between charged residues in the extracellular domain with charged phospholipids in the plasma membrane (39, 45–47, 49, 50), at least without an involvement of the iRhoms, since the TMD mutations described here that affect the stimulation of ADAM17 leave putative regulatory sequences in the extracellular domain intact.
To further explore how iR2 regulates ADAM17, we tested whether the extracellular extension that protrudes from the TMD1, the first juxtamembrane domain of iR2 (JMD1), is involved in regulating iR2/A17-dependent shedding. A short deletion within the JMD1 in iR2 abolished stimulated shedding of the iR2-dependent KL2, whereas stimulated shedding of TGFα, which can be supported by iR1 and iR2 was only reduced, but not abolished. Point mutations in the iR2 JMD1 recapitulated the effect of deleting the JMD1, arguing against structural rearrangements in the large extracellular loop domain as cause of the substrate selective effects. On the other hand, JMD2, which is at the C-terminus of the iR2 loop domain adjacent to TMD2 was important for iR2 and iR1/2 selective shedding, suggesting that JMD2 serves an important function in the synthesis, maturation or functional regulation of ADAM17.
The substrate selective effects of mutations in JMD1 of iR2 raised questions about the contribution of the corresponding JMD of A17 to substrate selective shedding. We found that the two ADAM17 JMD mutants tested here reduced the shedding of iR2-selective substrate KL2 more strongly than that of the iR1/2 substrate TGFα, suggesting that the A17 JMD interacts differently with iR2 compared to iR1. The L659P point mutation the ADAM17 JMD was initially identified in a patient suffering from TOF, a serious developmental defect in the outflow tract of the heart (51). A17 L659P affects the shedding of HB-EGF (51), an iR2-selective substrate that is crucial for proper heart valve morphogenesis in mice (6, 31). The substrate selective effects of the ADAM17 JMD mutations resembled the effect of deleting iR2 on the function of wt ADAM17. Further studies will be necessary to understand the underlying mechanism of the dominant effect of this mutation.
The results presented here support a model in which the TMD of ADAM17 has a key role in regulating the function of vertebrate ADAM17 by interacting with the iRhoms. We propose that the interaction of ADAM17 with the iRhoms helps to posttranslationally regulate the activity of the ADAM17 extracellular domain. The crystal structure of the related ADAM10 suggested that its catalytic activity can be controlled by an extracellular C-terminal regulatory domain (56) (light blue domain in model in Fig. 6C). It is tempting to speculate that the C-terminal part of the extracellular domain of ADAM17 has a similar regulatory function, allowing ADAM17 to be switched on and off and then back on again (22) (dark blue domain in model in Fig. 6D). Perhaps the interaction between ADAM17 and the iRhoms regulates the activation of ADAM17 in response to cytoplasmic phosphorylation of the iRhoms, as has been shown for iR2 (34, 36), although the details of this putative molecular switch remain to be elucidated. Moreover, the extracellular ADAM17 JMD and the JMD1 of iR2 are important for iR2-dependent shedding, suggesting that the interaction between ADAM17 and iR2 extends from the TMD to the membrane proximal JMD domains. Since the ADAM17 JMD mutations selectively affect iR2-dependent KL2 shedding without abolishing the iR1/2-dependent TGFα shedding, this suggests that the JMD of ADAM17 and JMD1 of iR2 are involved in ensuring the proper alignment or presentation of ADAM17 or of its iR2-dependent substrates or both, but not the activation of ADAM17. Given the proposed role of iR2/ADAM17-dependent release of TNFα and of ligands of the EGFR in autoimmune diseases such as Rheumatoid Arthritis and in cancer, these findings could help identify novel inhibitors of dysregulated and pathogenic iR2/ADAM17 signaling pathways.
Supplementary Material
Acknowledgements
We thank Mr. Daniel Li for excellent technical assistance. This study was supported by NIH GM64750 to CPB.
Nonstandard Abbreviations
- ADAM17
A disintegrin and metalloprotease 17
- BTC
Betacellulin
- CD62L
L-Selectin
- EGFR
Epidermal growth factor receptor
- EREG
Epiregulin
- GPCR
G-protein coupled receptor
- HB-EGF
Heparin-binding epidermal growth factor like growth factor
- IL-6R
Interleukin-6 receptor
- iR1 and iR2
inactive Rhomboids 1 and 2
- JMD
Juxtamembrane domain
- KL2
Kit ligand 2
- mEFs
mouse embryonic fibroblasts
- TMD
Transmembrane domain
- TGFα
Transforming growth factor α
- TNFα
Tumor necrosis factor α
Footnotes
Declaration of Interests
Drs. McIlwain, Mak, Maretzky and Blobel hold a patent on a method of identifying agents for combination with inhibitors of iRhoms. Dr. Blobel and the Hospital for Special Surgery have identified iRhom inhibitors and have co-founded the start-up company SciRhom in Munich to commercialize these inhibitors.
References
- 1.Black R, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, and Cerretti DP (1997) A metalloprotease disintegrin that releases tumour-necrosis factor-a from cells. Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 2.Moss ML, Jin S-LC, Milla ME, Burkhart W, Cartner HL, Chen W-J, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Lessnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su J-L, Warner J, Willard D, and Becherer JD (1997) Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-a. Nature 385, 733–736 [DOI] [PubMed] [Google Scholar]
- 3.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russel WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, and Black RA (1998) An essential role for ectodomain shedding in mammalian development. Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 4.Sahin U, Weskamp G, Zhou HM, Higashiyama S, Peschon JJ, Hartmann D, Saftig P, and Blobel CP (2004) Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR-ligands. J. Cell Biol. 164, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blobel CP (2005) ADAMs: key players in EGFR-signaling, development and disease. Nat. Rev. Mol. Cell. Bio. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 6.Jackson LF, Qiu TH, Sunnarborg SW, Chang A, Zhang C, Patterson C, and Lee DC (2003) Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. Embo J. 22, 2704–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horiuchi K, Kimura T, Miyamoto T, Takaishi H, Okada Y, Toyama Y, and Blobel CP (2007) Cutting Edge: TNF-{alpha}-Converting Enzyme (TACE/ADAM17) Inactivation in Mouse Myeloid Cells Prevents Lethality from Endotoxin Shock. J Immunol 179, 2686–2689 [DOI] [PubMed] [Google Scholar]
- 8.Chalaris A, Adam N, Sina C, Rosenstiel P, Lehmann-Koch J, Schirmacher P, Hartmann D, Cichy J, Gavrilova O, Schreiber S, Jostock T, Matthews V, Hasler R, Becker C, Neurath MF, Reiss K, Saftig P, Scheller J, and Rose-John S (2010) Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J Exp Med 207, 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaydon DC, Biancheri P, Di WL, Plagnol V, Cabral RM, Brooke MA, van Heel DA, Ruschendorf F, Toynbee M, Walne A, O’Toole EA, Martin JE, Lindley K, Vulliamy T, Abrams DJ, MacDonald TT, Harper JI, and Kelsell DP (2011) Inflammatory skin and bowel disease linked to ADAM17 deletion. N Engl J Med 365, 1502–1508 [DOI] [PubMed] [Google Scholar]
- 10.Franzke CW, Cobzaru C, Triantafyllopoulou A, Loffek S, Horiuchi K, Threadgill DW, Kurz T, van Rooijen N, Bruckner-Tuderman L, and Blobel CP (2012) Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand-dependent terminal keratinocyte differentiation. J Exp Med 209, 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy G (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer 12, 929–941 [DOI] [PubMed] [Google Scholar]
- 12.Ardito CM, Gruner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, Delgiorno KE, Carpenter ES, Halbrook CJ, Hall JC, Pal D, Briel T, Herner A, Trajkovic-Arsic M, Sipos B, Liou GY, Storz P, Murray NR, Threadgill DW, Sibilia M, Washington MK, Wilson CL, Schmid RM, Raines EW, Crawford HC, and Siveke JT (2012) EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 22, 304–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Issuree PD, Maretzky T, McIlwain DR, Monette S, Qing X, Lang PA, Swendeman SL, Park-Min KH, Binder N, Kalliolias GD, Yarilina A, Horiuchi K, Ivashkiv LB, Mak TW, Salmon JE, and Blobel CP (2013) iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J Clin Invest 123, 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zunke F, and Rose-John S (2017) The shedding protease ADAM17: Physiology and pathophysiology. Biochim Biophys Acta Mol Cell Res 1864, 2059–2070 [DOI] [PubMed] [Google Scholar]
- 15.Scheller J, Chalaris A, Garbers C, and Rose-John S (2011) ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol 32, 380–387 [DOI] [PubMed] [Google Scholar]
- 16.Scheller J, Garbers C, and Rose-John S (2014) Interleukin-6: from basic biology to selective blockade of pro-inflammatory activities. Semin Immunol 26, 2–12 [DOI] [PubMed] [Google Scholar]
- 17.Weber S, and Saftig P (2012) Ectodomain shedding and ADAMs in development. Development 139, 3693–3709 [DOI] [PubMed] [Google Scholar]
- 18.Garbers C, Heink S, Korn T, and Rose-John S (2018) Interleukin-6: designing specific therapeutics for a complex cytokine. Nat Rev Drug Discov 17, 395–412 [DOI] [PubMed] [Google Scholar]
- 19.Gschwind A, Hart S, Fischer OM, and Ullrich A (2003) TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells. Embo J. 22, 2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maretzky T, Evers A, Zhou W, Swendeman SL, Wong PM, Rafii S, Reiss K, and Blobel CP (2011) Migration of growth factor-stimulated epithelial and endothelial cells depends on EGFR transactivation by ADAM17. Nat Commun 2, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Gall S, Bobe P, Reiss K, Horiuchi K, Niu X-D, Lundell D, Gibb D, Conrad D, Saftig P, and Blobel C (2009) ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as TGFα, L-Selectin and TNFα. Mol Biol Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Gall SM, Maretzky T, Issuree PDA, Niu X-D, Reiss K, Saftig P, Khokha R, Lundell D, and Blobel CP (2010) ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J. Cell Science 123, 3913–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue A, Ishiguro J, Kitamura H, Arima N, Okutani M, Shuto A, Higashiyama S, Ohwada T, Arai H, Makide K, and Aoki J (2012) TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods 9, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 24.Inoue A, Arima N, Ishiguro J, Prestwich GD, Arai H, and Aoki J (2011) LPA-producing enzyme PA-PLA(1)alpha regulates hair follicle development by modulating EGFR signalling. EMBO J 30, 4248–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, and Blobel C (2007) Substrate Selectivity of EGF-Receptor Ligand Sheddases and Their Regulation by Phorbol Esters and Calcium Influx. Mol. Biol. Cell. 18, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doedens JR, Mahimkar RM, and Black RA (2003) TACE/ADAM-17 enzymatic activity is increased in response to cellular stimulation. Biochem. Biophys. Res. Commun. 308, 331–338 [DOI] [PubMed] [Google Scholar]
- 27.Schwarz J, Broder C, Helmstetter A, Schmidt S, Yan I, Muller M, Schmidt-Arras D, Becker-Pauly C, Koch-Nolte F, Mittrucker HW, Rabe B, Rose-John S, and Chalaris A (2013) Short-term TNFalpha shedding is independent of cytoplasmic phosphorylation or furin cleavage of ADAM17. Biochim Biophys Acta 1833, 3355–3367 [DOI] [PubMed] [Google Scholar]
- 28.Adrain C, Zettl M, Christova Y, Taylor N, and Freeman M (2012) Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 335, 225–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, Lang KS, Haussinger D, Wakeham A, Itie-Youten A, Khokha R, Ohashi PS, Blobel CP, and Mak TW (2012) iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 335, 229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siggs OM, Xiao N, Wang Y, Shi H, Tomisato W, Li X, Xia Y, and Beutler B (2012) iRhom2 is required for the secretion of mouse TNFalpha. Blood 119, 5769–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maretzky T, McIlwain DR, Issuree PD, Li X, Malapeira J, Amin S, Lang PA, Mak TW, and Blobel CP (2013) iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc Natl Acad Sci U S A 110, 11433–11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Maretzky T, Weskamp G, Monette S, Qing X, Issuree PD, Crawford HC, McIlwain DR, Mak TW, Salmon JE, and Blobel CP (2015) iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. Proc Natl Acad Sci U S A 112, 6080–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Maretzky T, Perez-Aguilar JM, Monette S, Weskamp G, Le Gall S, Beutler B, Weinstein H, and Blobel CP (2017) Structural modeling defines transmembrane residues in ADAM17 that are crucial for Rhbdf2/ADAM17-dependent proteolysis. J Cell Sci 130, 868–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grieve AG, Xu H, Kunzel U, Bambrough P, Sieber B, and Freeman M (2017) Phosphorylation of iRhom2 at the plasma membrane controls mammalian TACE-dependent inflammatory and growth factor signalling. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunzel U, Grieve AG, Meng Y, Sieber B, Cowley SA, and Freeman M (2018) FRMD8 promotes inflammatory and growth factor signalling by stabilising the iRhom/ADAM17 sheddase complex. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavadas M, Oikonomidi I, Gaspar CJ, Burbridge E, Badenes M, Felix I, Bolado A, Hu T, Bileck A, Gerner C, Domingos PM, von Kriegsheim A, and Adrain C (2017) Phosphorylation of iRhom2 Controls Stimulated Proteolytic Shedding by the Metalloprotease ADAM17/TACE. Cell Rep 21, 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oikonomidi I, Burbridge E, Cavadas M, Sullivan G, Collis B, Naegele H, Clancy D, Brezinova J, Hu T, Bileck A, Gerner C, Bolado A, von Kriegsheim A, Martin SJ, Steinberg F, Strisovsky K, and Adrain C (2018) iTAP, a novel iRhom interactor, controls TNF secretion by policing the stability of iRhom/TACE. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambrecht BN, Vanderkerken M, and Hammad H (2018) The emerging role of ADAM metalloproteinases in immunity. Nat Rev Immunol 18, 745–758 [DOI] [PubMed] [Google Scholar]
- 39.Grotzinger J, Lorenzen I, and Dusterhoft S (2017) Molecular insights into the multilayered regulation of ADAM17: The role of the extracellular region. Biochim Biophys Acta Mol Cell Res 1864, 2088–2095 [DOI] [PubMed] [Google Scholar]
- 40.Xu P, and Derynck R (2010) Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell 37, 551–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soond SM, Everson B, Riches DW, and Murphy G (2005) ERK-mediated phosphorylation of Thr735 in TNFalpha-converting enzyme and its potential role in TACE protein trafficking. J Cell Sci 118, 2371–2380 [DOI] [PubMed] [Google Scholar]
- 42.Xu P, Liu J, Sakaki-Yumoto M, and Derynck R (2012) TACE activation by MAPK-mediated regulation of cell surface dimerization and TIMP3 association. Sci Signal 5, ra34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gooz P, Dang Y, Higashiyama S, Twal WO, Haycraft CJ, and Gooz M (2012) A disintegrin and metalloenzyme (ADAM) 17 activation is regulated by alpha5beta1 integrin in kidney mesangial cells. PLoS One 7, e33350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dusterhoft S, Hobel K, Oldefest M, Lokau J, Waetzig GH, Chalaris A, Garbers C, Scheller J, Rose-John S, Lorenzen I, and Grotzinger J (2014) A disintegrin and metalloprotease 17 dynamic interaction sequence, the sweet tooth for the human interleukin 6 receptor. J Biol Chem 289, 16336–16348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dusterhoft S, Michalek M, Kordowski F, Oldefest M, Sommer A, Roseler J, Reiss K, Grotzinger J, and Lorenzen I (2015) Extracellular Juxtamembrane Segment of ADAM17 Interacts with Membranes and Is Essential for Its Shedding Activity. Biochemistry 54, 5791–5801 [DOI] [PubMed] [Google Scholar]
- 46.Willems SH, Tape CJ, Stanley PL, Taylor NA, Mills IG, Neal DE, McCafferty J, and Murphy G (2010) Thiol isomerases negatively regulate the cellular shedding activity of ADAM17. Biochem. J. 428, 439–450 [DOI] [PubMed] [Google Scholar]
- 47.Dusterhoft S, Jung S, Hung CW, Tholey A, Sonnichsen FD, Grotzinger J, and Lorenzen I (2013) Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase. J Am Chem Soc 135, 5776–5781 [DOI] [PubMed] [Google Scholar]
- 48.Krossa S, Scheidig AJ, Grotzinger J, and Lorenzen I (2018) Redundancy of protein disulfide isomerases in the catalysis of the inactivating disulfide switch in A Disintegrin and Metalloprotease 17. Sci Rep 8, 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sommer A, Kordowski F, Buch J, Maretzky T, Evers A, Andra J, Dusterhoft S, Michalek M, Lorenzen I, Somasundaram P, Tholey A, Sonnichsen FD, Kunzelmann K, Heinbockel L, Nehls C, Gutsmann T, Grotzinger J, Bhakdi S, and Reiss K (2016) Phosphatidylserine exposure is required for ADAM17 sheddase function. Nat Commun 7, 11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reiss K, and Bhakdi S (2017) The plasma membrane: Penultimate regulator of ADAM sheddase function. Biochim Biophys Acta Mol Cell Res 1864, 2082–2087 [DOI] [PubMed] [Google Scholar]
- 51.Xie Y, Ma A, Wang B, Peng R, Jing Y, Wang D, Finnell RH, Qiao B, Wang Y, Wang H, and Zheng Y (2019) Rare mutations of ADAM17 from TOFs induce hypertrophy in human embryonic stem cell-derived cardiomyocytes via HB-EGF signaling. Clin Sci (Lond) 133, 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlöndorff J, Becherer JD, and Blobel CP (2000) Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE). Biochem. J. 347 Pt 1, 131–138 [PMC free article] [PubMed] [Google Scholar]
- 53.Kawaguchi N, Horiuchi K, Becherer JD, Toyama Y, Besmer P, and Blobel CP (2007) Different ADAMs have distinct influences on Kit ligand processing: phorbol-ester-stimulated ectodomain shedding of Kitl1 by ADAM17 is reduced by ADAM19. J Cell Sci 120, 943–952 [DOI] [PubMed] [Google Scholar]
- 54.Sahin U, and Blobel CP (2007) Ectodomain shedding of the EGF-receptor ligand epigen is mediated by ADAM17. FEBS Lett 581, 41–44 [DOI] [PubMed] [Google Scholar]
- 55.Urban S, Lee JR, and Freeman M (2001) Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182 [DOI] [PubMed] [Google Scholar]
- 56.Seegar TCM, Killingsworth LB, Saha N, Meyer PA, Patra D, Zimmerman B, Janes PW, Rubinstein E, Nikolov DB, Skiniotis G, Kruse AC, and Blacklow SC (2017) Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10. Cell 171, 1638–1648 e1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.