Abstract
Defining the interplay between genetic events and microenvironmental contexts necessary to initiate tumorigenesis in normal cells is a central endeavor in cancer biology. We found that RTK/Ras oncogenes reprogram normal, freshly explanted primary mouse and human cells into tumor precursors, in a process requiring increased force transmission between oncogene-expressing cells and their surrounding extracellular matrix (ECM). Microenvironments approximating the normal softness of healthy tissues, or blunting cellular mechanotransduction, prevent oncogene-mediated cell reprogramming and tumor emergence. However, RTK/Ras oncogenes empower a disproportional cellular response to the mechanical properties of the cell's environment, such that when cells experience even subtle supraphysiological ECM rigidity they are converted into tumor-initiating cells. These regulations rely on YAP/TAZ mechanotransduction, and YAP/TAZ target genes account for a large fraction of the transcriptional responses downstream of oncogenic signaling. This work lays the groundwork for exploiting oncogenic mechanosignaling as vulnerability at the onset of tumorigenesis, including tumor prevention strategies.
The minimal compendium of genetic/epigenetic changes sufficient to turn a normal cell into a tumorigenic one remains a central unanswered question in cancer biology. Normal tissues have been recently shown to carry clones of healthy cells bearing a host of oncogenic alterations1–3. Thus, oncogenes appear insufficient to drive tumor initiation in living tissues, consistent with the view that cancer is not just a genetic disease, but, rather, the product of a wrecked-havoc communication between the cell and its microenvironment.
In this vein, the mechanical signals that the cell receives from its surroundings are emerging as overarching determinants of its behavior4,5, and are known to foster tumor progression6. However, what remains unexplored is the role of altered cell mechanics in the pivotal preceding step - i.e. reprogramming of normal cells into cells endowed with tumorigenic properties. Here we set to determine whether and how common genetic drivers of tumorigenesis can reprogram normal cells into tumor precursor cells (hereafter: oncogenic reprogramming) by establishing an interplay between the physical attributes of cells and their microenvironment at the beginning of tumorigenesis. We focused on the RTK/Ras cascade, because of its widespread relevance for human cancer7.
Results
Mechanical signaling is required for transformation of primary cells
As first paradigm, we focused on mammary gland tumorigenesis. Mammary tumors emerge from luminal cells acquiring self-renewing potential and ability to seed tumors when transplanted into an immunocompromised host8. RTK/Ras signaling is a potent driver of mammary tumorigenesis due to overexpression of growth factor receptors, such as HER2, EGFR or FGFR. We first tested whether freshly explanted luminal differentiated cells (LD) from healthy mouse and human mammary glands, normally void of any proliferative potential9,10, can be reprogrammed into cells able to proliferate and self-renew by expression of a constitutive active form of HER2 (HER2-CA). FACS purified LD cells (see methods) were plated on collagen-coated dishes and transduced with lentiviral vectors encoding for either EGFP or for the activated version of HER2 (see schemes in Extended Data Fig. 1a, c). Remarkably, oncogenic signaling conferred to mouse and human LD cells the ability to form self-renewing colonies (Fig. 1a-d), that developed into solid organoids entirely composed of K8+ luminal cells, a hallmark of human HER2+ breast cancer (Fig. 1e).
Fig. 1. Abnormal substrate rigidity is required for oncogenes to reprogram normal mammary cells into tumorigenic ones.
a,b, Representative images and quantifications of murine LD (mLD)-derived colonies. Murine basal cells (mBasal)-derived colonies are shown for comparison. Scale bars, 170 μm. ****p-value=9.9x10-5. See also Extended Data Fig. 1b and 1e.
c, d, Representative images (c) and quantifications (d) of colonies formed by human LD (hLD) cells. Colonies formed by human mammary basal cells (hBasal) are shown for comparison. Scale bars, 170 μm. ****p-value=2.0x10-5. See also Extended Data Fig. 1d, f.
e, Representative bright field and immunofluorescence images (n= 3 independent experiments) of organoids formed by mLD cells expressing HER2-CA. Organoids formed by mBasal cells are shown for comparison. K14 and K8 serve as markers of basal and luminal cell identity, respectively. Scale bars, 400 μm, left and 17 μm, right.
f-h, Representative images and quantifications of solid colonies formed by mLD (f), hLD (g) or murine LP (mLP) (h) cells, cultured on 40 kPa or on 0.5 kPa hydrogels, and then seeded in clonogenic medium. Scale bars, 170 μm. **** p-value=5.2x10-12 (f),1.69x10-11 (g) and 1.68x10-12 (h). See also Extended Data Fig. 1g, h.
i, Representative pictures (n=5) of the in vivo outgrowths generated from mLD cells. T1/2, C3H10T1/2 fibroblasts. See Extended Data Fig. 1m, demonstrating increased fibrillar collagen organization by LOX-L2.
j, Representative images (n=4) of subcutaneous tumors and lung-disseminated GFP-traced tumor cells derived from indicated hLD cells. Scale bars, 80 μm for primary tumor and 25 μm for lung.
k, Quantifications of the volume of subcutaneous outgrowths generated from control or the indicated reprogrammed hLDs injected within hydrogels of the indicated stiffness. Data are mean +/- s.d. of n=4 independent experiments.
a,b,d, f-h, Data are mean + s.d. of n=3 biologically independent samples.
P-values were determined by unpaired two-sided Student’s t-test (a, d) or one-way ANOVA with Sidak’s multiple comparisons test (f-h).
To test the relevance of ECM rigidity in HER2-driven oncogenic reprogramming, we repeated the above experiment, but this time plating primary cells on soft adhesive hydrogels of 0.5 kPa, phenocopying the compliance of the normal mammary gland11. Remarkably, oncogenic activation was unable to induce reprogramming of cells experiencing these physiological levels of substrate stiffness (Fig. 1f, g), as these cells invariably remained as differentiated single cells. Similar experiments were carried out using FACS-purified luminal progenitors (LP). Upon plating in colony medium, LP cells formed cyst-like outgrowths that, as previously reported, lacked self-renewal potential10. As control, when HER2-expressing LD or LP cells were plated on the same hydrogels just tuned at a higher rigidity (40 kPa), they were converted into proliferating cells that formed self-renewing solid colonies (Fig. 1f-h) and organoids composed of K8+ cells (Extended Data Fig. 1i), similarly to what observed above on tissue culture plastics (3 GPa). Thus, an appropriate mechanical environment is required for oncogenic reprogramming of primary cells.
Next, we aimed to establish whether mechanical inputs are also instrumental in vivo at reprogramming oncogene-expressing cells into cells endowed with tumorigenic properties. We tested this idea first in mouse and then in human primary luminal cells, using independent means to manipulate cell mechanics in vivo. First, HER2-expressing mouse LD cells were transplanted in the cleared mammary fat pad of immunocompromised mice, together with C3H10T1/2 fibroblasts expressing LOX-L2, a secreted enzyme well known to increase ECM stiffness by promoting collagen crosslinking12. Remarkably, we found that this combination was sufficient to induce tumor formation (ductal carcinoma in situ, DCIS) (Fig. 1i and Extended Data Fig. 1j, k). Co-transplantation with control (empty-vector-expressing) C3H10T1/2 fibroblasts was instead insufficient to induce any tumor formation, as co-transplanted HER2-LD cells generated only short, monostratified luminal duct primordia (Fig. 1i and Extended Data Fig. 1j, k), similarly to HER2-LD cells alone. Similar conclusions could be inferred from ex vivo experiments involving mammary organoid cultures expressing HER2 and treated with LOX-L2 (Extended Data Fig. 1l).
As independent procedure to demonstrate the relevance of mechanical signaling for oncogenic cell reprogramming, we co-expressed HER2-CA with Itgb1V737N, a self-clustering integrinβ1 mutant (Itgb1V737N), that mimics, cell-autonomously, the effect of a stiff ECM at adhesion sites13. This combination also induced DCIS formation, whereas LD cells expressing Itgb1V737N alone formed no outgrowth (Fig. 1i and Extended Data Fig. 1j, k, n).
Third, we used HER2-expressing and GFP-labeled human primary luminal cells and injected these into recipient mice embedded in either soft (0.5 kPa) or stiff (9 kPa) hyaluronic acid (HA)-based hydrogels. Palpable tumors could be detected after 7 weeks, only from HER2-CA-expressing cells embedded within the stiff HA (Fig. 1j, k). By histological examination, these neoplastic lesions were characterized by poor cellularity, massive ECM deposition and aggressiveness, with cells radiating out of the tumor center through collective migration, or as stretched tumor cells aligned to stromal fibers (Fig. 1j and Extended Data Fig. 1o). Intriguingly, immunostaining revealed the presence of individual human cells homed in the mouse lung, yet in an ostensibly quiescent state (Fig. 1j). These results indicate that raised mechanical signaling from the microenvironment is crucial for oncogenes to initiate tumorigenesis, while the normal softness of healthy tissues averts oncogene function.
Oncogenes empower disproportional mechanical responsiveness
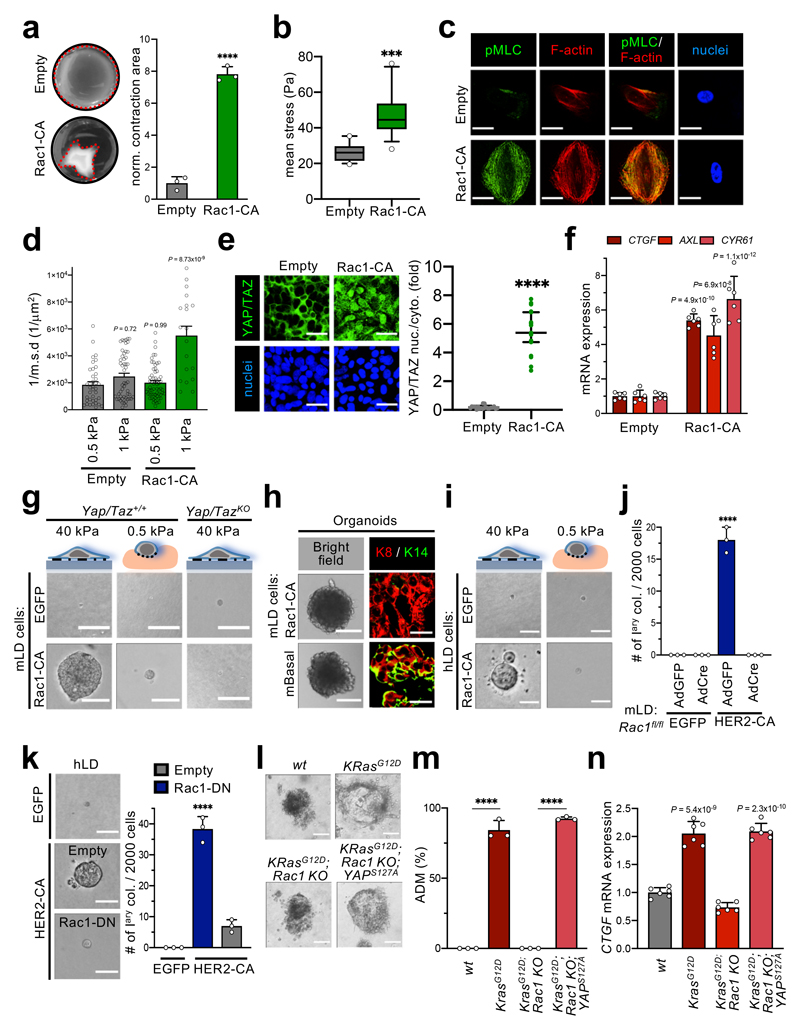
Our findings indicate that a dual requirement needs to be met for a normal cell to undergo oncogenic reprogramming: a cell-autonomous event, i.e. oncogenic activation, and a non-cell-autonomous one, i.e. increased stiffness of their environment. Are these parallel inputs or two sides of the same coin? The ability of cells to respond to mechanical signals is intimately connected to the cell's ability to pull against the ECM, as determined by the tensional and architectural organization of the cytoskeleton5,14,15. We thus assessed the traction forces developed by non-tumorigenic immortalized mammary cells (MCF10A) induced to express KRasG12V or HER2-CA. We first measured the ability of these cells to contract a collagen gel in which they were embedded. We found that oncogenes greatly empower cell contractility (Fig. 2a). In line, as assessed by traction force microscopy (TFM), cells expressing KRasG12V or HER2-CA apply significantly stronger traction stresses when compared to parental (empty vector-transduced) cells (Fig. 2b). Consistently, transformed cells display increased contractile actomyosin bundles as visualized by increased Phospho-Myosin-light chain (pMLC) (Fig. 2c and Extended Data Fig. 2b). Increased intracellular contractility also directly impacts formation and maturation of focal adhesions (FA)16, and, in line, HER2- or KRas-expressing cells displayed increased FA formation and maturation (Extended Data Fig. 2c, d). YAP/TAZ are two highly related transcriptional regulators serving as well-established sensors of mechanotransduction4,15. We found that cell-autonomous changes in cell mechanics induced by oncogenes lead to massive activation of YAP/TAZ, as visualized by their nuclear accumulation (Fig. 2d), YAP/TAZ binding to their DNA-binding platform TEAD (Extended Data Fig. 2e), and induction of YAP/TAZ-dependent transcriptional activity (Extended Data Fig. 2f, g). These data suggest that activated oncogenes raise cellular mechanoperception.
Fig. 2. Ras/RTK oncogenes change the mechanical and material properties of cells.
a, Representative images (n=3) and quantifications of collagen contraction assays performed with MCF10A cells. Data are mean + s.d. of n= 3 biologically independent samples. (Empty is empty vector). See controls in Extended Data Fig. 2a.
b, Traction force microscopy measurements of mean stresses exerted by the indicated MCF10A cells on 3 kPa substrates. Data are box-and-whiskers plots (whiskers: 10th to 90th percentile; box: from 25th to 75th percentile; line within the box: median) of n>12 independent samples. ** p-value=0.0034, * p-value=0.049.
c, Representative immunofluorescence images for pMLC of the indicated MCF10A cells. Scale bars, 24 μm. Images are representative of n=3 independent experiments. Quantifications are in Extended Data Fig. 2b.
d, Representative immunofluorescence images and quantifications of the subcellular localization of YAP/TAZ in post-confluent MCF10A cells. Scale bars, 16 μm. Data are mean +/- s.d. of n>17 independent samples. **** p-value=10-15. YAP/TAZ are downstream of oncogene-induced contractility (Extended Data Fig. 2h).
e, Particle-tracking microrheology of the indicated MCF10A cells seeded on 0.5 kPa and 1 kPa hydrogels. Data are mean + s.d. of n>35 biologically independent samples.
f, Representative immunofluorescence images and quantifications of YAP/TAZ localization in single MCF10A cells on 0.5 and 1 kPa hydrogels. Scale bars, 8 μm. Data are mean +/- s.d. of n>17 independent samples. See also Extended Data Fig. 2i, k.
g, qRT-PCRs assessing the expression levels of the YAP/TAZ endogenous target CTGF in MCF10A cells seeded on 0.5 or on 1 kPa hydrogels. Data are mean + s.d. of n= 3 biologically independent samples.
P-values were determined by unpaired two-sided t-test (a, b) or one-way ANOVA with Sidak’s (d-g) multiple comparisons test.
We then sought to determine whether oncogenes change the cell's mechanical properties, and the dependency of this response on the substrate material properties. For this, we used multiple particle-tracking microrheology, a procedure that probes the mechanical attributes of the F-actin cytoskeleton providing a global picture of the stiffness of an individual cell17. Control and oncogene-expressing cells remained equally soft when challenged at levels of ECM rigidity similar to those experienced by mammary cells within the healthy mammary gland (0.5 kPa) (Fig. 2e). However, only oncogene-expressing cells were able to stiffen when challenged at rigidity values (1 kPa) starting to approach those of mammary tumors11, even if these correspond to an ostensibly modest change in substrate's rheology when compared to normal mammary tissue (Fig. 2e). Thus, oncogenes should enable epithelial cells to biologically respond to mechanical stimuli that are otherwise sub-threshold for non-oncogene-expressing cells. Consistently, oncogenes specifically empower the ability to engage YAP/TAZ activation already at 1 kPa, yet not at 0.5 kPa (Fig. 2f, g and Extended Data Fig. 2 j).
Thus, cell contractility and microrheology assays support the view that oncogenic signaling disproportionally enhances cellular mechanotransduction in response to the physical inputs received from the environment. Oncogene activity alone is insufficient to reprogram normal cells in absence of sufficiently matching, and mechanically empowering, extracellular rigidity. Cutting extracellular stiffness down to the very compliant values of natural tissues inevitably drops cell rigidity down to levels becoming impossible to compensate by RTK/Ras oncogenes, and this averts cell reprogramming toward a tumorigenic fate, as visualized in vivo (Fig. 1).
The above findings predict that even a relatively modest increase in substrate rigidity of the microenvironment in which oncogene-expressing cells are embedded should be sufficient to ignite oncogenic mechano-signaling and empower oncogene-mediated cell reprogramming. To test this hypothesis, and expand the generality of our conclusions, we focused on pancreatic tumorigenesis. Human pancreatic neoplasias are characterized by activating mutations in KRas at exceedingly high frequencies (>90%)18, and this is recapitulated in mouse models, where Ras mutation is indeed a potent driver for pancreatic tumorigenesis19. That said, even in these cases, tumor emergence is extraordinary rare if one considers that most cells remain normal and healthy throughout the mouse lifespan in spite of continuous expression of mutant KRas19. Thus, additional events appear ostensibly required for initiation of tumorigenesis in pancreata expressing oncogenic Ras. The cell of origin of pancreatic cancer is the exocrine acinar cell that, after Ras activation, undergoes a change of fate known as acinar-to-ductal metaplasia (ADM), accompanied by reactivation of cell proliferation20. This step can be recapitulated in ex vivo cultures by activating KRasG12D in freshly explanted acini, that, after seeding in 3D collagen I ECM, convert to self-expanding ductal organoids (see scheme of Extended Data Fig. 3a).
To test whether KRasG12D-mediated cell reprogramming is blunted at physiological stiffness levels but promoted by quantal increases in substrate mechanics, we embedded KRas expressing acini in distinct synthetic HA-based matrices offering comparable adhesivity but tuned at different rigidities15,21 (see scheme in Extended Data Fig. 3b). Remarkably, KRas was insufficient to induce ADM at the lower stiffness (0.1 kPa); however, KRas-expressing acini started to reprogram at intermediate rigidity levels (0.2 kPa) and wholesale converted into ADM at higher rigidities (>0.5 kPa) (Fig. 3a). By gene expression, KRas-expressing acini experiencing stiffer ECM lose expression of the differentiation marker amylase and acquire the expression of ductal/progenitor markers, such as K19 and Sox9, whereas KRas-expressing acini embedded in soft ECM remain fully differentiated, indistinguishable from control (Fig. 3b). As readout of Ras-induced mechanosignaling in pancreatic acini we monitored YAP/TAZ nuclear localization and found that oncogenic Ras induced YAP/TAZ activation at permissive ECM rigidity and not in compliant conditions (Fig. 3c and Extended Data Fig. 3c). We conclude from these experiments that oncogenic signaling must be matched by sufficient ECM rigidity to promote the early step of pancreatic tumorigenesis.
Fig. 3. Oncogenes empower a disproportional cellular response to ECM mechanical properties to drive pancreatic tumorigenesis.
a, Representative bright field images and quantifications of R26-rtTAM2; KRasG12D pancreatic acini embedded in 3D hydrogels. Images are representative of n=3 biologically independent experiments. Scale bars, 200 μm. Data are mean + s.d.
b, qRT-PCRs for the differentiated acinar cells marker Amylase (Amy) and the ductal/progenitor markers K19 and Sox9 in pancreatic acini, treated as in (a). Data are presented as mean + s.d. of n=3 independent samples. ****p-value=2.39x10-6. Data are normalized to 18-S rRNA.
c, Representative YAP/TAZ immunofluorescence pictures (n=3) of pancreatic acini expressing KRasG12D and embedded in 0.1 kPa or 0.5 kPa hydrogels, as in (a). Scale bars, 50 μm.
d, Representative bright field and YAP/TAZ immunofluorescence pictures of the indicated pancreatic acini treated with Defactinib or cytochalasin-D (Cyto.D). Images are representative of n=3 biologically independent experiments. Scale bars, 400 μm in top panels and 50 μm in bottom panels.
e, Quantifications of the percentage of ADM events in samples treated as in (d). Data are presented as mean + s.d. of n=3 independent samples. ****p-value=10-15. See controls in Extended Data Fig. 3f.
f, g, Representative (n=3 independent samples) histological stainings, YAP/TAZ immunohistochemical and pMLC immunofluorescence images of the indicated pancreata treated with the LOX-inhibitor BAPN (f, see scheme of Extended Data Fig. 3g) or with the ROCK inhibitor Fasudil (g). Genotypes analyzed were: Ptf1aCreERTM(wt), Ptf1aCreERTM;KRas+/LSL-G12D(KRasG12D), Ptf1aCreERTM;KRas+/LSL-G12D;p53+/LSL-R172H(KRasG12D; p53R172H). Scale bars, 100 μm for histological and immunohistochemical stainings, 19 μm for immunofluorescence. See also Extended Data Fig. 3f-l.
P-values were calculated by Sidak’s (a,b,e) multiple comparisons tests.
Our model predicts that experimentally lowering cell mechanics by interfering with its tensional state should prevent oncogenic cell reprogramming. Indeed, KRasG12D-expressing acini treated with inhibitors of focal adhesions (FAK) or using low (vital) dose of the F-actin inhibitor Cytochalasin-D (Cyto.D)4, remained fully differentiated without any sign of ADM (Fig. 3d, e and Extended Data Fig. 3d); this correlates with failure to activate YAP/TAZ (Fig. 3d and Extended Data Fig. 3e).
In light of the above results, we asked whether tumor initiation in vivo could be prevented by blunting the mechanical interplay between oncogene-expressing cells and their substrate. We did so by lowering either ECM or intracellular mechanics. For this, we took advantage of mice conditionally expressing oncogenic KRasG12D and mutant p53R172H in adult pancreatic acini (Ptf1a-CreERTM; KRasLSL-G12D; p53LSL-R172H). After tamoxifen administration, these mice developed neoplasias (PanINs) at high frequency (Fig. 3f, and Extended Data Fig. 3h). Of note, pancreatic cancer is known for being a very rigid tumor, and indeed KRas/mutant-p53 tumors were surrounded by a collagen-rich fibrous ECM, and displayed elevated pMLC2 staining since the earliest stage of tumorigenesis (PanIN1; Fig. 3f, and Extended Data Fig. 3i, j). Yet, the role of altered cell mechanics in the early step of tumor emergence remains unclear. To lower ECM mechanics in vivo, mice expressing oncogenic Ras and mutant-p53 in their acinar cells were treated with BAPN, an inhibitor of the LOX family of collagen cross-linking enzymes (see scheme in Extended Data Fig. 3g). Strikingly, we found that no neoplastic lesions whatsoever (i.e., neither PanINs nor PDACs) could develop in BAPN-treated pancreata (Fig. 3f and Extended Data Fig. 3h), with pancreatic parenchyma remaining ostensibly normal over the several (11) weeks during which we monitored these mice. BAPN treatment inhibited deposition of collagen fibers, normalized the cells' tensional state (Extended Data Fig. 3i, j), and prevented YAP/TAZ activation (Fig. 3f).
Complementarily, to blunt intracellular mechanics, we treated Ras-expressing Ptf1a-CreERTM; KRasLSL-G12D adult mice with Fasudil, a FDA-approved drug that inhibits ROCK, a main orchestrator of actomyosin contractility (see scheme of Extended Data Fig. 3g). Remarkably, similarly to BAPN-mediated tumor prevention, Fasudil administration also abolished any emergence of neoplasia (Fig. 3g and Extended Data Fig. 3k). Oncogenic Ras induced massive YAP/TAZ nuclear accumulation that was blunted by Fasudil treatment, correlating with normalization of cellular tension as visualized by pMLC (Fig. 3g and Extended Data Fig. 3l).
Oncogenic mechanosignaling mediated by YAP/TAZ
We next asked by what means changes in cell mechanical and material properties induced by oncogenes could convert normal cells into tumor-precursor cells. Clearly, this had to ultimately involve changes in the cell's transcriptome, and YAP/TAZ mechanotransduction represented the most likely culprits as mediator of these responses4. Indeed, YAP/TAZ are essential mediators of tumor emergence in the mammary gland and pancreas22,23, a finding also replicated in our hands for this study (Extended Data Fig. 4a, b). To test whether YAP/TAZ act as nuclear effectors of oncogenic mechanosignaling for primary cell reprogramming we carried out YAP/TAZ knockout in explanted mammary LD or pancreatic acinar cells expressing oncogenes and challenged at ECM rigidities permissive for transformation. For this, we used primary cells explanted from mice bearing YAP/TAZ floxed alleles (Yapfl/fl;Tazfl/fl). Remarkably, we found that YAP/TAZ are genetically required to sustain HER2-induced reprogramming of primary mouse LD cells, and to sustain KRas induced pancreatic reprogramming in organoids (Fig. 4a, b and Extended Data Fig. 4c). Crucially, by using cells explanted from doxycycline-inducible tetO-YAPS127A transgenic mice, we also found that YAP overexpression could single-handedly rescue oncogene-induced pancreatic cell reprogramming in mechanical conditions otherwise prohibitory for oncogene function (Fig. 4c, d and Extended Data Fig. 4e, g).
Fig. 4. YAP/TAZ are the nuclear effectors downstream of the changes in the cell's mechanical and material properties induced by oncogenes.
a, Quantifications of the mammary colonies formed by mLD cells derived from Yapfl/fl;Tazfl/fl mice, infected with AdCre or AdGFP, and transduced with lentiviral constructs expressing EGFP or HER2-CA. Data are mean + s.d. of n= 3 biologically independent experiments. ****p-value=3,66x10-10, one-way ANOVA with Sidak’s multiple comparisons test.
b, Representative bright field and Ki67 immunofluorescence pictures of KRas+/LSL-G12D (KRasG12D) or KRas+/LSL-G12D;Yapfl/fl ;Tazfl/fl (KRasG12D;Yap/Taz KO) pancreatic acini. Images are representative of 6 biologically independent experiments. Scale bars, 50 μm. See controls in Extended Data Fig. 4d.
c, Representative bright field images of R26-rtTAM2; KRasG12D (KRasG12D) or R26-rtTAM2; KRasG12D; tetOYapS127A (KRasG12D; YAPS127A) pancreatic acini embedded in 3D hydrogels of increasing stiffness. Images are representative of n=3 biologically independent experiments. Panels show the same set of controls related to Fig. 3a. Scale bars, 200 μm.
d, Representative bright field images of wild-type, KRasG12D, or KRasG12D;YAPS127A pancreatic acini treated with Defactinib or Dasatinib. Images are representative of at n=3 biologically independent experiments. Panels show the same set of controls related to Fig. 3d. Scale bars, 400 μm. For c and d, see controls of YAP expression in Extended Data Fig. 4h, f.
e, Hierarchical clustering of gene expression profiles from RNA-seq data of control (Empty vector) MCF10A cells (lane 2), HER2-CA expressing cells plated on stiff substrates and transfected with Control siRNA (siCo, lane 1) or YAP/TAZ siRNAs (siY/T, lane 4), or HER2-CA cells cultured on 0.5 kPa hydrogels (lane 3). The heatmap shows row-wise standardized expression of genes significantly upregulated (fold change ≥1.33, Benjamini-Hochberg FDR ≤5%) in MCF10A cells by HER2-CA in standard culture conditions. Genes are ordered according to decreasing average expression in HER2-CA expressing cells plated on stiff substrates (lane 1).
Finally, we aimed to determine, unbiasedly and at the genome-wide level, to what extent YAP/TAZ-dependent transcription serves as mediator of oncogene-induced mechanosignaling. For this, we compared by RNA-seq the transcriptome of HER2-expressing immortalized mammary cells, either control or YAP/TAZ depleted, experiencing two extreme rigidities: stiff, fibronectin-coated tissue culture dishes (a paradigm of standard tissue culture conditions) vs. fibronectin-coated soft substrate (0.5 kPa) (Fig. 4e). By hierarchical clustering of HER2-induced genes, the transcriptome of HER2-expressing cells either experiencing a soft ECM or depleted of YAP/TAZ closely matched each other (lane 3 and 4); in turn, the latters display a gene-expression pattern resembling control (non-HER2-expressing) cells (lane 2). Overall, a staggering amount of the genes differentially induced by HER2 in standard tissue culture conditions, but not in non-permissive mechanical conditions (0.5 kPa), are YAP/TAZ-regulated (68%, n=1348). Of note, more than 40% (47% or 41% depending on the considered anti-YAP/TAZ siRNA pair) of the entire list of HER2-induced genes (n=3183) are YAP/TAZ-dependent, all in all highlighting the relevance of these factors and cell mechanics for RTK/Ras oncogene function (See Supplementary Table 1 and Extended Data Fig. 4i for main biological processes regulated by RTK/Ras-YAP/TAZ oncogenic mechanosignaling).
Oncogenic mechanosignaling mediated by Rac1
Next, we aimed to determine how activated oncogenes intercept YAP/TAZ mechanotransduction. Mechanical activation of YAP/TAZ has been shown to occur through Hippo-dependent and -independent mechanisms4. We first excluded the involvement of the Hippo pathway, as the inhibitory phosphorylation of YAP by the LATS kinases remained unchanged in transformed cells (Extended Data Fig. 5a); then, direct measurement of Phospho-LATS, identifying the active pool of these kinases, showed that LATS activity remains constitutive, unaltered by oncogenes (Extended Data Fig. 5b). Thus, YAP/TAZ activation by oncogenic mechano-signaling occurs independently from Hippo pathway regulation.
Downstream of RTK/Ras signaling, the Rho-family of small GTPases serves as pivotal regulators of cytoskeletal biology24. A body of work in vitro and in vivo suggested Rac1 and Rac1 GEFs as the most promising effector of Ras-induced transformation, yet through poorly understood mechanisms25,26. We thus hypothesized that, at least in part, oncogenes could modify the cell's material properties and mechanoresponsiveness by in fact hijacking Rac1, and, in so doing, inducing oncogenic reprogramming. Several lines of evidence support this prediction. First, we discovered that a constitutive active version of Rac1 (CA, i.e., GTP-bound form) phenocopied the effects of HER2-CA or oncogenic KRas, leading to massive raise in cellular contractility, cell stiffening and YAP/TAZ activation (Fig. 5a-f and Extended Data Fig. 5c-e). Of note, Rac1-induced cell contractility was blunted by ROCK inhibitors (Fasudil and Y27632), highlighting a previously underappreciated positive connection between Rac1 and ROCK for induction of contractility in epithelial cells (Extended Data Fig. 5f).
Fig. 5. Oncogenes modify the cell's mechanical properties through Rac1 activation.
a, Rac1-induced MCF10A collagen contraction assays. Data are mean + s.d. of n= 3 independent samples. ****p-value=4,56x10-5.
b, Traction force microscopy measurements of MCF10A cells on 3 kPa substrates. Control (Empty) data are the same of Fig. 2b. Data are box-and-whiskers plots (whiskers: 10th to 90th percentile; box: from 25th to 75th percentile; line within the box: median) of n>12 independent samples. ***p-value=2,80x10-5.
c, Representative pMLC immunofluorescence images (n=3) of MCF10A cells. Scale bars, 40 μm.
d, Particle-tracking microrheology of MCF10A cells seeded on 0.5 kPa and 1 kPa hydrogels. Control (Empty) data are the same of Fig. 2e. Data are mean + s.d. of n>19 independent samples.
e, YAP/TAZ localization. Data are mean +/- s.d. of n>19. Scale bars, 38 μm. **** p-value=10-15.
f, qRT-PCRs of YAP/TAZ targets in MCF10A cells seeded on 1 kPa hydrogels. Data are mean + s.d. of n=6 independent samples. Data are normalized to GAPDH.
g, Representative images (n=3) of Yap+/+;Taz+/+ or YapKO;TazKO mLD cells cultured on 40 kPa or on 0.5 kPa hydrogels, and then seeded in clonogenic medium. Scale bars, 170 μm.
h, Representative bright field and immunofluorescence images (n= 3) of Rac1-CA-expressing mLD organoids and mBasal organoids, as in Fig. 1e. Scale bars, 400 μm, left 17 μm, right.
i, Representative images (n=3) of hLD cells and outgrowths on 40 kPa or on 0.5 kPa hydrogels. Scale bars, 170 μm.
j, Quantifications of the colonies formed by Rac1fl/fl mLD cells. Data are mean + s.d. of n= 3 independent experiments. ****p-value=1.14x10-7.
k, Representative images (n=3) and quantifications of colonies formed by hLD cells expressing Rac1-DN. Scale bars, 170 μm. Data are mean + s.d. of n=3 independent samples.****p-value=1.84x10-5.
l, m, Representative bright field images (n=3) (l) and quantifications (m) of ADM events of pancreatic acini of the indicated genotypes (see methods). Scale bars, 70 μm. Data are mean + s.d. of n= 3 independent experiments. ****p-value=5.25x10-9.
n, qRT-PCRs of YAP/TAZ endogenous target Ctgf, in pancreatic acini treated as in l. Data are presented as mean + s.d. of n=6 independent samples. Data are normalized to 18-S rRNA. See also Extended Data Fig. 5k, l.
P-values were calculated by unpaired two-sided t-test (a,b,e) and by one-way ANOVA with Sidak’s multiple comparisons test (d,f,j,k,m,n).
We next tested the role of Rac1 for oncogene-induced mechanosignaling and cell reprogramming by gain- and loss-of-function assays. By gain of function, Rac1-CA reprograms freshly explanted and FACS purified mouse and human LD cells into cells able to form colonies and self-renewing organoids (Fig. 5g-i and Extended Data Fig. 5g-i) closely reminiscent those induced by HER2 expression. Cell reprogramming occurred only when cells were seeded on rigid substrates (plastic or rigid hydrogels) whereas Rac1-CA was inconsequential in LD cells experiencing a hydrogel of physiological compliance (Fig. 5g, i and Extended Data Fig. 5g, i). Importantly, the reprogramming ability of Rac1 activation was completely dependent on YAP/TAZ (Fig. 5g and Extended Data Fig. 5g), all in all indicating, along the above results, that Rac1 acts upstream of YAP.
To validate the role of Rac1 by loss-of-function, we inactivated Rac1 in HER2-CA transduced primary cells using independent experimental set-ups, that is ex vivo AdCre genetic ablation of Rac1 in LD cells from Rac1fl/fl mice, or expression of a widely-used Dominant-Negative(DN)-Rac1 mutant (Rac1N17) in human LD cells. As shown in Fig. 5j and 5k, loss-of-Rac1 activity by either procedure abolished the effects of HER2. Furthermore, Rac1 is required downstream of oncogenes to promote YAP/TAZ activity (Extended Data Fig. 5j). Extending the functional requirement of Rac1 for oncogene-induced mechanosignaling to pancreatic cells, we employed two strategies, one based on pharmacological inhibition of the Rac-specific GEFs Tiam1/Trio (NSC23766, Extended Data Fig. 5m), and the other making use of conditional Rac1fl/fl alleles (Rac1 KO, Fig. 5l-n) and consistently found that Rac1 was required for ADM downstream of KRas. Crucially, by using cells explanted from doxycycline-inducible tetO-YAPS127A transgenic mice, we also found that YAP overexpression could single-handedly rescue oncogene-induced pancreatic cell reprogramming in cells genetically depleted of Rac1, or treated with Rac1-GEF inhibitors (Fig. 5l-n and Extended Data Fig. 5j, k, m). Collectively, this suggests that YAP/TAZ are the nuclear effectors of the RTK/Ras-Rac1 cascade.
Discussion
Here we report that RTK/Ras signaling, main drivers of tumorigenesis, must regulate the cell's mechanical properties as an integral element of their ability to reprogram normal cells into tumor precursor cells. This process leads to the build-up of cytoskeletal tension and cell stiffening, with ensuing activation of YAP/TAZ, whose target genes account for an unexpectedly large fraction of the transcriptional responses downstream of oncogenic signaling. Here we further highlight the role of Rac1 as mediator of oncogenic mechanosignaling, sufficient and required to empower in primary epithelial cells an enhanced responsiveness to changes in the rigidity of the surrounding ECM. Our findings offer mechanistic support to previous observations connecting YAP activity to Ras-induced tumorigenesis23,27–29. Future studies are warranted to explore the contribution of more complex mechanical properties of living tissues and natural ECM, including viscoelastic behaviors30,31 (see Supplementary Discussion).
The interplay here highlighted between a minimal set of genetic and environmental changes required to initiate tumor emergence holds a number of implications for tumor biology. To start, the data suggest that the high compliance (i.e. softness) of normal epithelial organs represents an overarching tumor suppressor system at the tissue level. In an extreme interpretation, one may envision that even prototypic oncogenic lesions may not be truly causative, but just permissive of later tumor-initiating events in the form of altered mechanical properties of the ECM. At the same time, we show that tumorigenic cells can be generated de novo from normal primary cells as soon as these cells express an activated oncogene and are concomitantly allowed to experience supra-physiological mechanical inputs. These findings offer a window of opportunity for tumor prevention. It is tempting to speculate that ECM alterations typical of ageing, inflammation, tissue damage, fibrosis, diabetes and smoke4 may be associated with an increased rate of tumor emergence due to increased mechanical activation of epithelial cells.
Of note, the mechanical framework here proposed for tumor initiation may incorporate oncogenic lesions other than those directly involving RTK/Ras signaling. For example, Src, hyperactive in several malignancies, is another oncogene upstream of YAP/TAZ33. Another possible example is mutation of p53 whose oncogenic effects have been recently causally linked to the activation of Rho/Rac GTP-ases, as such controlling cell mechanics and YAP/TAZ activity32,33.
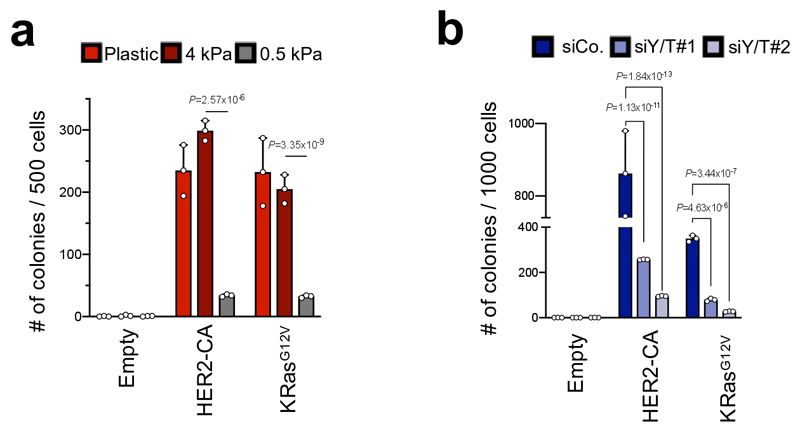
In hindsight, the crucial role of mechanical signals, here highlighted for primary cells, might be also a general trait of cell transformation of immortalized cell lines, classically visualized by induction of clonogenic potential and bypass of contact inhibition. We notice that these assays have been carried out on tissue culture surfaces (glass or plastic)34, whose rigidities (gigapascals) exceeds many folds the physiological stiffness of real tissues15, as such raising questions on to what extent the mechanical signal intrinsic to tissue culture conditions may have contributed to the discovery of many oncogenes. Intriguingly, when we used immortalized MCF10A cells and monitored their transformation by transduced oncogenes, we found that overexpression of HER2 or KRas was inconsequential in cells experiencing adhesion to a physiologically soft hydrogel (0.5 kPa) (Extended Data Figure 6a, b). This at least suggests a link between conventional transformation assays and oncogenic reprogramming of primary cells.
We further note that our findings help to solve a conundrum in cancer biology, as YAP/TAZ are broadly activated in most, if not all, solid malignancies, although never directly targeted by activating mutations35, possibly because unleashing full YAP/TAZ activity may be unattainable by sole mutational events36–39. Finding that YAP/TAZ are activated by oncogenic mechanosignaling initiated by activation of common drivers provides a solution to this conundrum.
Finally, our data may inform on potential routes to exploit oncogenic mechanosignaling as vulnerability at the onset of tumorigenesis, including tumor prevention strategies akin to those used by normal tissues to prevent cancer emergence.
Methods
Plasmids and reagents
pBABEpuro (#1764), pBABE-Puro-KRas*G12V (#46746), pCDNA3-HER2-CA (HER2 V659E) (#16259), FUdeltaGW-rtTA (#19780), FUW-tetO-MCS (#84008), pRRLSIN.cPPT.PGK-GFP.WPRE (#12252), pRK5 beta1 (#16042), pRK5myc RAC1 L61 (Rac1-CA) and pRK5myc Rac1-DN (RAC1 T17N) (#15904) were purchased from Addgene. FUW-tetO-EGFP (#84041) was previously reported10, phRG-TK-Renilla was purchased from Promega. CSII-CMV-MCS-IRES-puro was subcloned from CSII-CMV-MCS-IRES-bsd. pBABEpuro-HER2-CA, FUW-tetO-HER2-CA and CSII-CMV-MCS-IRES-HER2-CA were subcloned from pCDNA3-HER2-CA, into pBABEpuro, FUW-tetO-MCS and CSII-CMV-MCS-IRES-puro, respectively. pCS2-KRas*G12V was subcloned from pBABE-KRas*G12V into pCS2+40. CSII-CMV-MCS-IRES-puro-Itgb1V737N was cloned by PCR mutagenesis from pRK5 beta1 into CSII-CMV-MCS-IRES-puro. CSII-CMV-MCS-IRES-puro-LOXL2 was subcloned from pCMV6-myc-ddk-LOXL2 (Origene #RC200455) into CSII-CMV-MCS-IRES-puro. pBABEpuro-RAC1-CA and FUW-tetO-Rac1-CA were subcloned from pRK5myc RAC1-CA into pBABEpuro and FUW-tetO-MCS, respectively. FUW-tetO-Rac1-DN was subcloned from pRK5myc RAC1-DN into FUW-tetO-MCS. 8xGTIIC–LUX (a synthetic luciferase reporter of YAP/TAZ-dependent transcription) was previously described38.
FBS, HS, L-Glutamine, Pen/Strep (10,000 U/mL), EMEM, DMEM/F12, DMEM, Waymouth’s, AdvancedDMEM/F12 media, Trypsin-EDTA 0,05%, Prolong-DAPI, Dispase, ITS-X, BPE supplement were from Life Technologies. Acrylamide, Bis-acrilamide, APS, TEMED, Formaldehyde, Cytochalasin D, (Trimethoxysilyl)propyl methacrylate (TMSPM), Hydrochortisone, Cholera Toxin, Heparin Sodium Salt, Collagenase type I, Hyaluronidase, Forskolin, SBTI, Dexamethasone, NSC23766, Dasatinib, BAPN, Tamoxifen, Doxycycline hyclate, Rat Tail Collagen I (coating), Phoshatase Inhibitor Cocktail 2, DL-Dithiothreitohl, PolyFreeze (OCT) were from Sigma Aldrich. Tripure Isolation Reagent was from Roche. Matrigel Growth Factor Reduced Basement Membrane Matrix, Phenol Red-Free was from Corning. Rat Tail Collagen I for 3D culture was from Cultrex. Fibronectin was from Santa Cruz. Recombinant human Noggin, human EGF, human bFGF, murine EGF were from Peprotech. Glycosil, Gelin and Extralink were from ESI Bio. Recombinant human R-spondin-1 was from Sino Biological. Lipofectamine RNAi-MAX, Alexa Fluor 568 Phalloidin, B27 were from Thermo Fisher Scientific. Fasudil and Y27632 were from Tocris Bioscience. TransIT-LT1 was from Mirus Bio. Cre- and GFP-expressing adenoviruses were from University of Iowa, Gene Transfer Vector Core.
Mice
CD-1 mice (IMSR Cat# CRL:22), BALB/c mice (IMSR Cat# CRL:28) and NOD-SCID mice (IMSR Cat# CRL:394) were purchased from Charles River. Transgenic lines used in the experiments were gently provided by: Duojia Pan (Yap1fl/fl); Fernando Camargo (tetO-YAPS127A); Guillermina Lozano (p53+/LSLR172H); Tyler Jacks (KRas+/LSLG12D), Jens Siveke (Rac1fl/fl). Tazfl/fl and double Yapfl/fl; Tazfl/fl conditional knock-out mice were previously described41. Ptf1a-CreERTM (IMSR Cat# JAX:019378), and R26-rtTAM2mice (IMSR Cat# JAX:006965) were purchased from The Jackson Laboratory. Animals were genotyped with standard procedures42 and with the recommended set of primers. Animal experiments were performed adhering to our institutional guidelines as approved by OPBA and authorized by the Ministry of Health.
Cell culture
MCF10A cells were a gift from F. Miller (Karmanos) and were cultured in DMEM/F12 (Gibco) with 5% horse serum, glutamine and antibiotics, freshly supplemented with insulin (Sigma-Aldrich), EGF (Peprotech), hydrocortisone (Sigma-Aldrich) and cholera toxin (Sigma-Aldrich). HEK293 cells were from ATCC and were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS), glutamine and antibiotics. HEK293 and MCF10A cells were authenticated by DSMZ/Eurofins Genomics. KRasG12V, HER2-CA and Rac1-CA– expressing MCF10A cells were obtained by transduction with retroviral constructs (pBABEpuro-KRas*G12V, pBABEpuro-HER2-CA V659E and pBABEpuro-RAC1-CA); control cells were transduced with empty retroviral constructs (pBABEpuro). C3H10T1/2 fibroblasts were from ATCC and were cultured in EMEM, 10% FBS, Glutamine and antibiotics. All cell lines tested negative for mycoplasma contamination. To exclude any toxic effect associated with drug treatments performed throughout, maximal dose (Dasatinib 100 nM; Defactinib 5 μM; Fasudil 10 μM; CytochalasinD 200 nM; BAPN 500 μM; Y27632 50 μM) was administered to MCF10A cells for 24 hours and viability was assessed with CellTiter-Glo Luminescent Cell Viability assay (Promega), according to manufacturer instructions; treatment with high dose of puromycin (20 ng/μl) was used as comparison.
Human mammary tissue
Discard tissue was collected from anonymized healthy women undergoing reduction mastoplasty surgery with informed consent according to our institutional guidelines and by the Azienda Ospedaliera di Padova Ethics Committee (CESC).
2D mechanical challenging
For mechanical challenging of MCF10A, cells were plated on standard fibronectin-coated tissue culture dishes or on fibronectin-coated hydrogels of the indicated stiffness (kPa) (produced as described previously38. Hydrogel formulations in Acrylamide (AA), Bis-acrylamide (BA), Tetramethylethylenediamine (TEMED) and ammonium persulfate (APS) were as follows: 4% w/V AA, 0.03% w/V BA, 1:1000 V/V TEMED, 0.1 % w/V APS (0.5 kPa); 4 % w/V AA, 0.06% w/V BA, 1:1000 V/V TEMED, 0.1 % w/V APS (1 kPa); 4 % w/V AA, 0.1% w/V BA, 1:1000 V/V TEMED, 0.1 % w/V APS (2 kPa); 5 % w/V AA, 0.15% w/V BA, 1:1000 V/V TEMED, 0.2 % w/V APS (4 kPa); 8 % w/V AA, 0.48% w/V BA, 1:1000 V/V TEMED, 0.2 % w/V APS (40 kPa). Cells (4x104/cm2) were seeded in a drop of complete culture medium on top of fibronectin-coated hydrogels; after attachment, the hydrogel containing wells were filled with the appropriate culture medium. Cells were collected for immunofluorescence, or RNA extraction after 24 h or were trypsinized and recovered after 48 hours for soft agar assays. For mechanical challenging of primary murine and human mammary luminal cells, cells were plated on collagen I-coated tissue culture dishes or on collagen I-coated hydrogels of the indicated stiffness in a drop of complete culture medium (see below for details).
Rheological measurements of PAA were performed to verify that hydrogel compositions fitted a purely elastic behavior with negligible contribution of the loss modulus (Extended Data Fig. 7). Rheology was carried out using rotating parallel plates at room temperature (Dynamic Stress Rheometer DSR, Rheometrics Inc., New Jersey, USA). The hydrogels (24mm diameter, 1mm height) were bonded between two coverslips and measured after swelling overnight. Storage (G’) and loss (G”) moduli were calculated as a function of the frequency (from 0.01 to 1.259 Hz) in a constant strain mode. Young’s modulus E was calculated from the elastic shear modulus, E=3G, on the base of rubber elasticity theory.
Preparation of synthetic hyaluronan and gelatin-based 3D hydrogels
3D mechanically defined hydrogels were designed as previously reported43, with minor modifications. Briefly, under sterile conditions thiolate hyaluronic acid (Glycosil, ESI Bio), thiolate gelatin (Gelin, ESI Bio) and PEGDA (Extralink, ESI Bio) were dissolved in degassed water according to the manufacturer’s directions. For the preparation of hydrogels of defined stiffness for subcutaneous injections, stock concentrations of 10 mg/ml Glycosil, 10 mg/ml Gelin, and 5 mg/ml Extralink and concentrated stocks of 2x Glycosil and 5x Extralink were prepared. Solutions were prepared at 1:5 ratios of Extralink: (Glycosil + Gelin) for soft (0.5 kPa) hydrogels and 5xExtralink: (2xGlycosil + Gelin) for stiff (9 kPa) hydrogels. For the preparation of hydrogels with a defined stiffness gradient for pancreatic acinar cultures, we prepared stock concentrations of 10 mg/ml Glycosil and 10 mg/ml Gelin and 3 different stock concentrations of Extralink: 4mg/mL, 3mg/mL and 2 mg/mL, for stiff (0.5 kPa), medium (0.2 kPa) and soft (0.1 kPa) hydrogels, respectively. Solutions were prepared at 1:5 ratios of Extralink: (Glycosil + Gelin).
Murine mammary luminal cells isolation and culture
Primary mammary cells were isolated from the mammary glands of 8- to 12-week-old virgin CD-1 mice (experiments of Fig. 1a, b, d, f, h and Extended Data Fig. 1b, e, g, i), BALB/c (experiments of Fig. 1i and Extended Data Fig. 1j-n), Yapfl/fl; Tazfl/fl mice (experiments of Fig. 4a and Fig. 5g) or Rac1fl/fl (experiments of Fig. 5j), as previously described44. Briefly, mammary glands were minced and digested before sorting. To separate various subpopulations, cells were stained for 30 min at 4°C with antibodies against CD49f (BD Biosciences Cat# 551129), CD29 (BioLegend Cat# 102222), CD61 (BD Biosciences Cat# 553347), EpCAM (BioLegend Cat# 118208) and lineage markers (BD Biosciences Cat# 558074) in DMEM/F12. The stained cells were then resuspended in PBS/BSA 0,1% and sorted on a BD FACS Aria sorter (BD Biosciences) into luminal differentiated (LD) cells, luminal progenitor (LP) cells and basal cells. The identity of the sorted populations was confirmed by qRT-PCR for lineage-specific markers (Extended Data Fig. 1b). After FACS purification, cells were seeded in collagen-I coated 24-well plates or PAAm hydrogels and cultured in Mouse Mammary epithelial Growth Medium (MMGM: DMEM/F12 supplemented with glutamine, antibiotics, 10 ng/ml murine EGF, 10 ng/ml murine bFGF, and 4 μg/ml heparin with 2% FBS). Cells were infected with lentiviral vectors (virus suspension was mixed 1:1 with medium). For primary mammary cell reprogramming, LD or LP cells were transduced for 48 hr with FUW-tetO-HER2-CA or FUW-tetO-Rac1-CA in combination with rtTA-encoding lentiviruses (FudeltaGW-rtTA). As a (negative) control, LD cells were transduced with EGFP-expressing vector (FUW-tetO-EGFP) in combination with rtTA-encoding lentiviruses. For the experiments depicted in Fig. 4a and Fig. 5g, j, LD cells were transduced for 24 hr with lentiviral vectors as above and subsequently transduced for 24 hr with either Ad-Cre or Ad-GFP, as control. For the experiments depicted in Extended Data Fig. 1n, LD cells were transduced for 48 hr with lentiviral vectors encoding for HER2-CA (CSII-CMV-MCS-IRES-HER2V659E), with a self-clustering Integrinβ1 mutant13 (CSII-CMV-MCS-IRES-puro-Itgb1V737N), or with empty vector as control (CSII-CMV-MCS-IRES-puro). After infection, adherent cells were washed and treated with 2 μg/ml doxycycline for 7 days in Mouse Mammary epithelial Growth Medium for activating tetracycline-inducible gene expression. After one week mammary cells were detached with trypsin and seeded at a density of 2000 cells/well in 24-well ultralow attachment plates (Corning) in mammary clonogenic suspension medium (DMEM/F12 containing glutamine, antibiotics, 5% Matrigel, 5% FBS, 10 ng/ml murine EGF, 20 ng/ml murine bFGF, and 4 μg/ml heparin) containing doxycycline (2 μg/ml). Primary colonies were counted 14 days after seeding. To show the self-renewal capacity of primary colonies, these were recovered from the clonogenic medium by collecting and incubating cell suspensions with an excess volume of ice cold HBSS in order to solubilize Matrigel. After 1 hour, colonies were rinsed 3 times in cold HBSS by centrifugation at 1000 rpm for 5 min and incubated in trypsin 0.05% for 30 min to obtain a single cell suspension. Cells were counted and re-seeded at 2000 cells/well in 24-well ultralow attachment plates in mammary clonogenic suspension medium. For mammary organoid formation, primary colonies were recovered from clonogenic medium in cold HBSS and transferred in 100% Matrigel. After Matrigel formed a gel at 37°C, organoid medium was added (Advanced DMEM/F12 supplemented with HEPES, GlutaMax, antibiotics, B27 1X, 50 ng/ml murine EGF, 20 ng/ml murine bFGF, 4 μg/ml heparin, 100 ng/ml Noggin and 1 μg/ml R-Spondin1) containing doxycycline (2 μg/ml). 2 weeks after seeding, organoids were removed from Matrigel, trypsin-dissociated and transferred to fresh Matrigel. Passages were performed in a 1:4-1:8 split ratio every 2 weeks. For analysis, organoids were recovered from Matrigel and embedded in OCT medium (PolyFreeze, Sigma Aldrich) to obtain frozen sections for immunofluorescence.
Cleared mammary fat pad transplantation of murine mammary cells
Mammary fat pad transplantations were performed as previously described10, with minor modifications. Briefly, HER2-CA expressing mammary organoids (derived from LD cells) were transduced by spinfection with lentiviral vectors encoding for a self-clustering Integrinβ1 mutant13 (CSII-CMV-MCS-IRES-puro-Itgb1V737N), or with empty vector as control (CSII-CMV-MCS-IRES-puro), all in combination (1:1 ratio) with EGFP-encoding lentiviruses (pRRLSIN.cPPT.PGK-GFP.WPRE) for cell tracing. After infection, organoids were transferred in 100% Matrigel and cultured in organoid medium containing doxycycline (2 μg/ml) for 7 days. HER2-CA/Integrinβ1V737N expressing organoids were resuspended in 50 μl PBS/10% Matrigel; HER2-CA/empty vector expressing organoids were resuspended in 50 μl PBS/10% Matrigel either alone or in combination with C3H10T1/2 murine fibroblast transduced with lentiviruses encoding the collagen crosslinking enzyme LoxL212 (CSII-CMV-MCS-IRES-puro-LOXL2), or with empty vector (CSII-CMV-MCS-IRES-puro) as control. Cellular suspensions were injected into the inguinal mammary fat pads of NOD-SCID mice (Charles River), which had been cleared of endogenous mammary epithelium at 3 weeks of age. Animals were then administered doxycycline in the drinking water for 10-12 weeks. Transplanted mammary fat pads were examined by GFP immunofluorescence on sections from paraffin-embedded biopsies.
Human mammary luminal cells isolation and culture
Single cell suspensions of primary human mammary cells were generated as previously described9, with minor modifications. Briefly, the ductal tree was mechanically minced and enzymatically digested in tissue dissociation medium (Advanced DMEM-F12 supplemented with HEPES, Glutamax 1,5%, 600 U/ml collagenase and 200 U/ml hyaluronidase at 37°C over-night). Cells were spun down 3 minutes at 700 rpm and the pellet was further dissociated in 0.25% Trypsin-EDTA for 5 minutes followed by addition of 5 mg/ml dispase and 1μg/mL DNaseI for further 10 minutes. Digestion was stopped in Advanced DMEM 10%FBS and cells were filtered through a 40 μm strainer, to remove residual tissue fragments and cell aggregates. Single-cell suspensions of primary mammary cells were stained with CD31 (BioLegend Cat# 303119), CD45 (BD Biosciences Cat# 557833), CD49f (BD Biosciences Cat# 555736), EpCAM (BD Biosciences Cat# 347197) in DMEM for 30 minutes at 4ºC. After excluding CD31+/CD45+ (Lin+) cells, mammary cells were sorted into four populations: Luminal differentiated cells (LD): CD49f-/EpCAM+; Luminal progenitor cells (LP): CD49f+/ EpCAM+; Basal cells: CD49f+/EpCAM−; and Stromal cells: CD49f- /EpCAM−, using a FACS Aria III (BD Biosciences). The identity of the separated populations was validated by gene expression analysis (see Extended Data Fig. 1d). Freshly isolated primary LD cells were seeded in Human Mammary epithelial Growth Medium (HMGM: Advanced DMEM/F12 supplemented with HEPES, Glutamax, 0.5% FBS, 4ul/mL BPE, 10ng/mL hEGF, 10 mM Y27632, 10mM Forskolin and antibiotics). After FACS purification, cells were seeded in collagen-I coated 24-well plates or PAAm hydrogels and transduced with lentiviral vectors (virus suspension was mixed 1:1 with medium). For primary mammary cell reprogramming, LD cells were transduced for 48 hr with FU-tetO-HER2-CA or FUW-tetO-Rac1-CA in combination with rtTA-encoding lentiviruses (FudeltaGW-rtTA). As a (negative) control, LD cells were transduced with EGFP-expressing vector (FUW-tetO-EGFP) in combination with rtTA-encoding lentiviruses. For the experiments depicted in Fig. 5k, LD cells were transduced for 48 hr with lentiviral vectors encoding for HER2-CA either alone or together with lentiviral vectors encoding for Rac1-DN (FUW-tetO-Rac1-DN), in combination with rtTA-encoding lentiviruses.
After infection, adherent cells were washed and treated with 2 μg/ml doxycycline for 7 days in Mammary epithelial Growth Medium for activating tetracycline-inducible gene expression. After one week, mammary cells were detached with trypsin and seeded at a density of 2000 cells/well in 24-well ultralow attachment plates (Corning) in mammary clonogenic suspension medium (Advanced DMEM/F12 containing HEPES, Glutamax, antibiotics, 5% Matrigel, 2% FBS, 10 ng/ml human EGF, 4 μL/mL BPE, 10mM Forskolin and 2 μg/ml doxycycline). Primary colonies were counted 14 days after seeding. To show the self-renewal capacity of primary colonies, these were recovered from the clonogenic medium by collecting and incubating cell suspensions with an excess volume of ice cold HBSS in order to solubilize Matrigel. After 1 hour, colonies were rinsed 3 times in cold HBSS by centrifugation at 1000 rpm for 5 min and incubated in trypsin 0.05% for 30 min to obtain a single cell suspension. Cells were counted and re-seeded at 2000 cells/well in 24-well ultralow attachment plates in mammary clonogenic suspension medium.
Subcutaneous injection of primary human mammary cells
Human luminal cells isolated and cultured as above were transduced with lentiviral vectors encoding for HER2-CA (CSII-CMV-MCS-IRES-HER2-CA) or with empty vector (CSII-CMV-MCS-IRES-puro) as a (negative) control. All cells were co-transduced with EGFP encoding lentiviruses (pRRLSIN.cPPT.PGK-GFP.WPRE) for in vivo cell tracing. After one week, mammary cells were trypsinized and embedded in stiff (9 kPa) or soft (0.5 kPa) synthetic hyaluronan and gelatin-based 3D hydrogels (106 cells/100μL). Cell suspensions were subcutaneously injected prior to hydrogel gelification into the flank on immunocompromised 4-6 weeks old NOD/SCID female mice (100μL for each injection). Tumor masses were removed and fixed in 4% PFA for subsequent analyses 7 weeks after the injection.
3-D collagen contraction assays
Collagen contraction assays were performed as previously described45, with minor modifications. Briefly, collagen I solution (1.5 mg/mL) was prepared by neutralizing the pH of acid-solubilized rat tail collagen I (Cultrex) with NaOH and PBS buffer. MCF10A cells (4x105 cells/well) were embedded in 300 μl/well of neutralized collagen in 24-well ultra-low attachment plates, the collagen hydrogel was polymerized at 37°C for 40 minutes and culture medium was added on top. For experiments in Extended Data Fig. 2a conditioned medium (CM) was added to collagen gels. CMs were obtained from post-confluent culture of MCF10A cells transduced with Empty-vector or retroviral vectors encoding for HER2-CA or KrasG12V. For experiments in Extended Data Fig. 5f Rac1-CA expressing cells or control cells (empty vector-transduced) were treated for 24h before embedding in collagen with the following drugs: Fasudil 10μM (ROCK inhibitor, Tocris Biosciences), Defactinib 5μM (FAK inhibitor, Selleckchem), Dasatinib 100 nM (Src inhibitor, Sigma Aldrich), Y27632 50μM (ROCK inhibitor, Tocris Biosciences), or with DMSO as control. For experiments in Extended Data Fig. 2h, MCF10A cells were transfected with siRNAs 24h before embedding in collagen. After overnight incubation, the extent of contraction was assessed by subtracting the area of the gel from well area using ImageJ software. Experiments were performed in triplicate at least twice.
Traction force microscopy
Polyacrylamide substrates were prepared as described in46 by using a combination of acrylamide (4% wt/vol) and methylene-bis-acrylamide (0.1% wt/vol). Carboxyl-modified fluorescent polystyrene particles (0.50 μm diameter, Life Technologies) were suspended in the polyacrylamide solutions in a volume ratio of 1:100. The substrates were functionalized with fibronectin at a final concentration of 50 μg/ml. Cells were seeded on polyacrylamide substrates and allowed to adhere for 24 h. Images of cells and fluorescent substrates-embedded beads were acquired by using a wide field fluorescent microscope (Olympus Cell-R) equipped with a UPLSAPO 20× objective (NA = 0.75). Then, cells were detached by trypsinization to capture images of undeformed substrates. Images of fluorescent particles with and without cells were combined into a stack and aligned using the ImageJ plugin “align slices in the stack”. Displacement fields were obtained by a custom-written particle image velocimetry (PIV) ImageJ plugin (http://imagej.net/PIV_analyser). The plugin performs an iterative algorithm, which recursively calculated the displacement in a smaller region (interrogation window) using information from the previous computations to filter out wrong displacement vectors. The interrogation window size was progressively reduced (128, 64 and 32 pixels with 0.181 μm/px) to obtain a better PIV resolution. From the displacement field, Fourier transform traction cytometry (FTTC) was then used to estimate traction stress by using the FTTC ImageJ plugin. Average stresses were calculated on a ROI of 174.46x149 μm2 around each cell.
Multiple particle tracking microrheology
Multiple particle tracking microrheology was performed as previously described47, with minor modifications. Carboxyl-modified fluorescent polystyrene particles (0.50 mm diameter, Polyscience, Inc.) were introduced into adherent cells by using a ballistic gun (Bio-Rad, Hercules, CA). Helium gas at 2000 psi was used to force a macro-carrier disk coated with particles to crash into a stopping screen. The force of collision was transferred to the particles, causing their dissociation from the macrocarrier and the bombardment of cells. Once bombarded, cells were extensively washed with phosphate-buffered saline (PBS) and left to recover 24 hours in culture medium. After recovery, cells were seeded (2x104/cm2) on 1 kPa or on 0.5 kPa PAAm hydrogels, formulated as above and covalently attached to 3-(Trimethoxysilyl)propyl methacrylate (TMSPM, Sigma-Aldrich)-treated fluorodish in a 13 μL drop covered with a 2.54 cm2 Kapton disk (DuPont), in order to obtain a maximal hydrogel height of 120 μm. After 24 hours, the motion of intra-cellular fluorescent beads was recorded for a total of 6 s at 100 frames per second (yielding a total of 600 frames per video) using a digital camera (Hamamatsu, ORCA- Flash 2.8) attached to a PC and Cam control video capture software mounted on an inverted fluorescence microscope (Olympus IX81) equipped with a fluorescent mercury lamp (Olympus U-LH100L- 3). A water immersion objective (60x objective with NA = 1.35) at 1.6x magnification was used for particle tracking. Videos were kept short (the total duration was 6 seconds) to avoid photo-bleaching of particles. The total number of analyzed particles was at least 200 from more than 19 cells and regions for each sample. Particle tracking microrheology allows the monitoring of local viscoelastic properties of living cells with a high spatio-temporal resolution, collecting and analyzing the Brownian motions of particles embedded in the cytoplasm. Once the nanoparticle trajectories had been obtained, mean squared displacements (MSDs) at 0.5 seconds were calculated from Eq.
where angular brackets mean time average, is the time scale and t the elapsed time.
Primary pancreatic acinar cells isolation and culture
Primary pancreatic acini were isolated and cultured as previously described44 from the pancreas of 6- to 9-week-old mice. Explants were seeded in neutralized rat tail collagen type I (Cultrex)/acinar culture medium (1:1), overlaid with acinar culture medium (Waymouth’s medium supplemented with 0.1% FBS, 0.1% BSA, 0.2 mg/ml SBTI, 1x ITS-X, 50 μg/ml BPE, 1μg/ml dexamethasone, and antibiotics) once collagen formed a gel. ADM events were assessed 5 days after seeding. To activate KRas overexpression and to concomitantly delete Yap/Taz or Rac1 alleles, acinar cells derived from KRas+/LSL-G12D; Yapfl/fl; Tazfl/fl, or from KRas+/LSL-G12D; Rac1fl/fl mice were infected with Ad-Cre for 3 hours at 37°C before seeding, or with Ad-GFP as (negative) control. To induce YAPS127A expression, acinar cells derived from R26-rtTAM2; tetO-YAPS127A mice were cultured in the presence of doxycycline 2 μg/ml.
For the experiment described in Fig. 3d, e, Fig. 4d, Extended Data Fig. 4g, h and Extended Data Fig. 5m acinar culture medium was supplemented either with the FAK inhibitor Defactinib (Selleckchem) 10μM, with the Src inhibitor Dasatinib (Sigma Aldrich) 100 nM, with CytochalasinD (Sigma Aldrich) 200 nM, with the Rac1-GEF inhibitor NSC23766 100μM (Sigma Aldrich) or with DMSO as control.
For the experiment described in Fig. 3a-c Fig. 4c, pancreatic acini were embedded in synthetic hyaluronan and gelatin-based 3D hydrogels and overlaid with acinar culture medium (Waymouth’s medium supplemented with 0.1% FBS, 0.1% BSA, 0.2 mg/ml SBTI, 1x ITS-X, 50 μg/ml BPE, 1μg/ml dexamethasone, and antibiotics) once the hydrogels gellified.
RNA-seq
Cells were harvested by the RNeasy Mini Kit (Qiagen) for total RNA extraction and contaminant DNA was removed by the RNase-Free DNase Set (Qiagen).
RNA-seq libraries for deep-sequencing were prepared with the Illumina TruSeq Standard Total RNA with Ribo-Zero GOLD kit, and sequencing was performed with Illumina HiSeq2500. About 40M reads/sample were obtained. Raw reads were aligned using STAR48 version 2.5.3a to build version hg19 of the human genome. Counts for UCSC annotated genes were calculated from the aligned reads using featureCounts function of the Rsubread R package49. Normalization and differential analysis were carried out using edgeR package50 and R (version 3.3.1). Raw counts were normalized to obtain Counts Per Million mapped reads (CPM) and Reads Per Kilobase per Million mapped reads (RPKM). Only genes with a CPM greater than 1 in at least 1 sample were retained for differential analysis. Genes were considered significantly regulated when fold changes were equal or greater than 1.33 for upregulated genes or equal or lower than 0.75 for downregulated genes, with a Benjamini-Hochberg FDR less than or equal to 5%. Hierarchical clustering was carried out using the Hierarchical Clustering of the MultiExperiment Viewer (MeV 4.8; http://mev.tm4.org) package with Euclidean distance metric and average linkage clustering, using the row-wise standardized RPKM of genes with a RPKM greater than 1 in siCo.-transfected HER2CA-MCF10A cells plated on stiff substrates that are significantly upregulated compared to control MCF10A cells on stiff substrates. Gene Ontology (GO) analyses were performed using Enrichr51. The list of GO terms significantly enriched (p-value ≤ 0.05) in HER2-induced genes whose expression is dependent on YAP/TAZ and substrate stiffness is presented in Supplementary Table 1.
KRas-driven pancreatic tumorigenesis in vivo
For induction of recombination in pancreatic acini of adult mice, Ptf1a-CreERTM (control), or mice of the indicated genotypes received three intra-peritoneal injections of 1 mg each of tamoxifen (Sigma Aldrich) dissolved in corn oil (10 mg/ml) at 4 weeks of age and were sacrificed after 6 months. For pharmacologic inhibition of collagen crosslinking, mice were administered with 3mg/mL of the lysyl oxidase inhibitor β-aminopropionitrile (BAPN, Sigma-Aldrich) in drinking water from 5 weeks of age until sacrifice. For pharmacologic inhibition of intracellular acto-myosin contractility, mice received each week four intra-peritoneal injections of 10 mg/kg each of the ROCK inhibitor Fasudil (Tocris Bioscience) dissolved in water, from 5 weeks of age until sacrifice. All experimental groups treated with BAPN or Fasudil were sacrificed at 16 weeks of age and pancreata were collected for analyses.
Luciferase reporter assays
Luciferase assays were performed in HEK293 cells transfected with the TEAD reporter (8xGTIIC–LUX). The luciferase reporter (25 ng/cm2) was transfected together with phRG-TK-Renilla (25 ng/cm2) to normalize for transfection efficiency. Expression construct doses were the following: HER2-CA 62.5 ng/cm2, KRasG12V 2.5 ng/cm2, Rac1-DN 37.5 ng/cm2. DNA content transfected in all samples was kept uniform by adding a pBluescript plasmid up to 250 ng/cm2. Cells were plated at 40% confluence (day 0), transfected with the indicated siRNAs (day1) and the following day (day 2) transfected with DNA. Cells were harvested after 48 h of treatment (day 3). Firefly luciferase activity was measured with an Infinite F200PRO plate reader (Tecan). Data are presented as firefly/Renilla luciferase activity.
RNA interference
siRNA transfections were done with Lipofectamine RNAi-MAX (Thermo Fisher Scientific) in antibiotics-free medium according to the manufacturer’s instructions. Sequences of siRNAs are provided in Supplementary Table 2.
Quantitative real-time PCR (qRT-PCR)
MCF10A cells were collected using the RNeasy Mini Kit (Qiagen) for total RNA extraction, and contaminant DNA was removed by DNase treatment. Total RNA from pancreatic or mammary primary cells was extracted using TriZOL reagent (ThermoFisher). qRT-PCR analyses were carried out on reverse-transcribed cDNAs with QuantStudio5 (applied Biosystems, ThermoFisher Scientific) and analysed with QuantStudio Design & Analysis software (version 1.4.3). Expression levels are always normalized to GAPDH, or to 18-S rRNA for experiments with primary pancreatic cells. PCR oligonucleotide sequences are listed Supplementary Table 3.
Western blots
Immunoblots were carried out as previously described52. Primary antibodies used were: anti-YAP/TAZ (Santa Cruz Biotechnology, sc-101199), anti-P-YAP (Cell Signaling Technology, 4911), anti-LATS (Cell Signaling Technology, 3477), anti-p-LATS (Cell Signaling Technology, 8654) and anti-GAPDH (Millipore, MAB374). Uncropped immunoblots are presented in Supplementary Figure 1.
Immunofluorescence
Immunofluorescence on PFA-fixed cells and on PFA-fixed paraffin-embedded tissue slices was performed as previously described10. Primary antibodies were: YAP/TAZ (Santa Cruz Biotechnology Cat# sc-101199), Ki67 (Spring Bioscience Cat# M3062), phospho-MLC (Cell Signaling Technology Cat# 3671, for staining on fixed cells or Cat# 3675, for staining on paraffin sections), FAK (Millipore Cat# 06-543), phospho-FAK (BD Biosciences Cat# 611722), Vinculin (Sigma-Aldrich Cat# V9264), Paxillin (Abcam Cat# ab32084), K8 (Abcam Cat# ab14053), K14 (Abcam Cat# ab7800), GFP (Abcam Cat# ab13970). F-actin was stained with Alexa Fluor 568 Phalloidin (Thermo Fisher). Secondary antibodies (1:200) were from Molecular Probes. Samples were counterstained with ProLong-DAPI (Molecular Probes, Life Technologies) to label cell nuclei. Confocal images were obtained with a Leica TCS SP5 equipped with a CCD camera and analyzed using Volocity software (PerkinElmer, version 5.5.1). For immunofluorescence on mammary organoids, outgrowths freshly recovered from Matrigel were embedded in OCT tissue-freezing medium (PolyFreeze, Sigma Aldrich) and frozen on dry ice. 8 μm cryostat sections were cut at –20 °C. Sections were mounted on glass slides and dried for at least 30 min. The sections were then fixed with 4% formaldehyde for 10 min. After washing with PBS the sections were processed as previously described10. For immunofluorescence on pancreatic primary cells, pancreatic cells were fixed overnight in PBS 4%PFA at 4°C, permeabilized with two washes in PBS 0.5% NP40 for 20 minutes at 4°C, followed by one wash in PBS 0.3% Triton X-100 for 20 minutes at room temperature. After two washes in PBS 0.1% Triton X-100 (PBST) for 15 minutes at room temperature, cells were blocked with two washes in PBST 10% Goat Serum (GS) for 1 hour at room temperature, and incubated overnight with primary antibodies. The following day, cells were washed twice in PBST 2% GS for 15 minutes at 4°C, and five more times in PBST 2% GS for 1 hour at 4°C. Secondary antibodies were incubated overnight. The third day, cells were washed five times in PBST for 15 minutes, incubated 20 min with DAPI solution and mounted in glycerol. For immunofluorescence on mammary, lung tissue and subcutaneous tumor masses, biopsies were fixed with PFA, paraffin-embedded and cut in 10 μm-thick sections. Sections were re-hydrated and antigen retrieval was performed by incubation in citrate buffer (Sodium Citrate Dihydrate 0.01 M, Tween 0,05% pH 6) at 95°C for 20 minutes. Slides were then permeabilized (10 min at RT with PBS 0.3% Triton X-100) and processed as described above. Cytoskeletal features (average number of FAs and pMLC staining / cell) and YAP/TAZ nuclear vs. cytoplasmic ratios were quantified by using NIH ImageJ analysis software with the same threshold among each stain. Immunohistochemical staining experiments were performed on PFA-fixed, paraffin-embedded tissue sections as previously described52. For immunohistochemistry antibodies used were: anti-YAP (Proteintech Group Cat# 13584-1-AP) and anti-TAZ (Sigma-Aldrich Cat# HPA007415).
Picrosirius red collagen staining
Collagen visualization on PFA fixed, paraffin embedded tissue sections was carried out by Picrosirius red staining. Briefly, sections were dewaxed with following incubation steps: 10 minutes in xylene 100%, 10 minutes in xylene:ethanol 1:1 V/V, 10 minutes in ethanol 100%, 5 minutes in ethanol:distilled H2O 1:1 V/V. Nuclei were stained in Weigert’s iron hematoxylin for (Sigma, HT1079) according to manufacturer’s instructions; sections were washed 10 minutes in running tap water and collagen was stained in Picrosirius red solution (0,5 g of Direct Red 80 (Sigma, 365548) into 500 ml of Picric Acid Solution 1,3% (Sigma, P6744) for one hour. Following collagen staining, sections were washed twice in acidified water (glacial acetic acid 1:200 V/V) and dehydrated in ethanol 100%, cleared in xylene and mounted with Eukitt Quick-hardening mounting medium (Sigma, 03989). Bright field images and collagen birefringence53 images were collected with a Leica 5000B microscope under parallel and orthogonal polarized light, respectively, and quantified using ImageJ software; signal threshold was maintained equal for all images across all conditions.
In situ proximity ligation assay (PLA)
In situ PLAs were performed with Duolink In Situ Detection Reagents (Sigma-Aldrich). MCF10A cells were plated on fibronectin-coated glass chamber slides. After 24 h, after cells reached confluency, cells were fixed in 4% PFA for 10 min at room temperature and subjected to PLA, following the manufacturer’s instructions. Primary antibodies used in the PLA were: anti-TEF1 (BD Biosciences, 610923) and anti-YAP (Proteintech, 13584-1-AP). Images were acquired with a Leica TCS SP5 confocal microscope equipped with a CCD camera and analyzed using Volocity software (PerkinElmer, version 5.5.1).
Lenti- and retrovirus preparation
Lentiviral particles were prepared as previously described52. Briefly, HEK293T (ATCC) cells were transiently transfected with lentiviral vectors (10 μg per 60-cm2 dish) together with packaging vectors pMD2-VSVG (2.5 μg) and pPAX2 (7.5 μg) using TransIT-LT1 (Mirus Bio) according to the manufacturer’s instructions.
Retroviral particles were prepared as previously described52. Briefly, GP2-293 (Takara) were transiently transfected with retroviral vectors (15 μg per 60-cm2 dish) together with pMD2-Env (5 μg per 60-cm2 dish) using TransIT-LT1. Infections were carried out as previously described52.
Agar colony formation assay
For experiments in Extended Data Fig. 6a, MCF10A cells (4 x 104/cm2) were seeded on 2D hydrogels of the indicated stiffness, or on fibronectin-coated cell culture plastic dishes, and then embedded in agar 48 hours later. For experiments in Extended Data Fig. 6b, MCF10A cells (4 x 104/cm2) were transfected with the indicated siRNAs, and embedded in agar after 48 hours. Effective YAP/TAZ downregulation was assessed by immunoblotting. Clonogenic assays were performed as previously described22; briefly, MCF10A cells (0,5 - 1 x 103) were resuspended in complete growth medium with 0.3% agarose (Invitrogen) and were layered onto 0.6% agar beds in six-well plates. Complete medium was added on top of cells and was replaced with fresh medium twice a week for 3 weeks.
Statistics
The number of biological and technical replicates and the number of animals are indicated in figure legends, main text and methods section. All tested animals were included. Animal ages and sex are specified in the text and methods section. Sample size was not predetermined. Randomization was not applicable for our experiments with cell lines. Student’s t-test and ANOVA analyses were performed, as indicated in the captions of the figures and supplementary figures, with GraphPad Prism 8.0.2 for Mac software.
Extended Data
Extended Data Fig. 1. Abnormal substrate rigidity is required for oncogenes to reprogram normal cells into tumorigenic ones.
a, Schematic representation of the FACS purification strategy and of the experiments performed with mLD cells. LP: luminal progenitor cells.
b, qRT-PCR analyses for the indicated basal and luminal mammary cell markers in mBasal, mLP, and mLD cells obtained by FACS, as in Extended Data Fig. 1a. Data are normalized to Gapdh and are referred to Basal cells levels for basal genes, and to LD cells levels for all the other luminal markers (each set to 1). Results are representative of n=3 independent experiments (each using mammary glands from n=20 mice) performed with similar results.
c, Schematic representation of the FACS purification strategy and of the experiments performed with human primary mammary luminal cells.
d, qRT-PCR analyses for the indicated basal and luminal mammary cell markers in human Basal, LP, and LD cells obtained by FACS, as in Extended Data Fig. 1c. Data are normalized to GAPDH and are referred to Basal cells levels for basal genes, and to LD cells levels for all the other luminal markers (each set to 1). Results are representative of n=3 independent experiments performed with similar results.
e-h, Lower magnification images of experiments shown in Fig. 1a (e), in Fig. 1c (f), in Fig. 1f (g) and in Fig. 1g (h). Scale bars, 600 μm.
i, Representative bright field and immunofluorescence images (n=3 independent experiments) of organoids formed by mLP cells expressing HER2-CA. K14 serves as marker of basal cell identity and K8 serves as marker of luminal cell identity. Scale bars, 400 μm, left and 17 μm, right.
j, Quantifications of the frequency of different outgrowths emerging from fat pad injection of the same samples of Fig. 1i. Data are representative of n>5 independent samples.
k, Representative GFP-immunofluorescence pictures (n>5 independent samples) of outgrowths depicted in Fig. 1i, showing that outgrowths emerge form injected mLD cells. Scale bar, 50 μm.
l, Representative immunofluorescence images (n=3 independent experiments) of ex-vivo 3D outgrowths formed by mLD cells expressing HER2-CA, embedded in a collagen-based ECM and treated with conditioned media from Empty-vector (CM: Empty) or LOXL2-transduced C3H10T1/2 fibroblasts (CM: LOXL2). While HER2-expressing cells formed round spheroids/organoids in absence of LOXL2, those embedded in LOXL2-modified collagen formed highly proliferating and invasive outgrowths. This effect relied on LOXL2 enzymatic activity as it was abolished by concomitant treatment with LOX-inhibitor BAPN. Scale bars, 260 μm.
m, Bright field and birefringence Picrosirius Red images of the same samples shown in Fig. 1i, showing LOX-L2 mediated increased fibrillar collagen deposition. Scale bars, 100 μm. Quantifications of total fibrillar collagen by birefringence signal in the same sections are presented as mean + s.d. of n=4 independent samples. ** p-value=7.13x10-3.
n, Representative images and quantifications of colonies formed by mLD cells transduced with the indicated constructs. The expression of the sole integrinβ1 mutant is inconsequential for colony formation by mLD cells. Scale bars, 170 μm. Images and data are representative of n=4 independent experiments. Data are mean + s.d. **** p-value=4.03x10-6.
o, Low magnification of the representative histological staining of the subcutaneous outgrowth shown in Fig. 1j (corresponding to the frame).
P-values were calculated by one-way ANOVA with Sidak’s multiple comparisons test.
Extended Data Fig. 2. Ras/RTK oncogenes change the mechanical and material properties of cells.
a, Representative images and quantifications of collagen contraction assays performed with control MCF10A cells transduced with empty vectors and embedded in 3-D collagen hydrogels with conditioned media obtained from control MCF10A cells (CM: Empty) or cells expressing KrasG12V (CM: KrasG12V) or HER2-CA (CM: HER2-CA). Compared with Fig. 2a, this shows that oncogene-mediated cell contractility does not rely on secreted factors. Images and data are representative of n=3 independent samples. Data are mean + s.d.
b, Quantification of the colocalization of the cytoskeletal stainings for pMLC and F-actin shown in Fig. 2c. Data are as mean + s.d. of n=3 independent samples.
c, Representative immunofluorescence images and quantifications of FA formation by colocalization of focal adhesion kinase (FAK) and Vinculin in MCF10A cells transduced with HER2-CA or KRasG12V. Scale bars, 24 μm. Images and data are representative of n=3 independent experiments. Data are mean + s.d.
d, Representative immunofluorescence images and quantifications FA maturation by colocalization of phospho-focal adhesion kinase (pFAK) and Paxillin in MCF10A cells transduced with HER2-CA or KRasG12V. Scale bars, 24 μm. Images and data are representative of n=3 independent experiments. Data are mean + s.d.
e, Representative pictures (n=3 independent experiments) and quantifications of proximity ligation assay (PLA) detecting the nuclear interaction between endogenous YAP and endogenous TEAD37 in control (Empty vector) MCF10A cells or in cells overexpressing KrasG12V or HER2-CA. Scale bars, 9 μm. Data are mean + s.d. of n>200 cells. **** p-value=10-15.
f, Luciferase assay in post-confluent HEK293 cells transfected with a synthetic reporter for TEAD-dependent transcription (8xGTIIC-Lux) and with the indicated siRNAs and control (Empty) or oncogene-expressing vectors. Data are mean + s.d. of n= 3 independent samples. ****p-value=10-15.
g, qRT-PCRs of the YAP/TAZ endogenous targets CTGF and AXL, in MCF10A cells transduced with the indicated oncogene-expressing vectors. Data are mean + s.d. of n= 3 independent samples.
h, Collagen contraction assays performed with MCF10A cells transduced with the indicated vectors, transfected with control or YAP/TAZ targeted siRNAs, showing that YAP/TAZ are downstream of cell contractility induced by oncogenes. Data are mean + s.d. of n= 3 independent samples. Immunoblots are shown for validation of effective YAP/TAZ depletion. GAPDH serves as loading control.
i, Representative immunofluorescence images and quantifications of YAP/TAZ localization in MCF10A cells transduced with empty or oncogene-expressing vectors, and seeded on 40 kPa hydrogels. Scale bars, 8 μm. Data are mean +/- s.d. of n>17 independent samples.
j, qRT-PCRs of the YAP/TAZ endogenous target CYR61 in MCF10A cells transduced with the indicated vectors and seeded on 0.5 or on 1 kPa hydrogels, as in Fig. 2g. Data are mean + s.d. of n= 3 independent samples.
k, Representative immunofluorescence images of F-actin of the same MCF10A cells shown in Fig. 2f to visualize cell shape. F-actin was stained with fluorescently-labeled phalloidin and nuclei were counterstained with DAPI. Scale bars, 16 μm.
P-values were calculated by unpaired two-sided Student’s t-test (a), one-way ANOVA with Sidak’s multiple comparisons test (b-f) or two-way ANOVA with Sidak’s multiple comparisons test (g,h, j).
Extended Data Fig. 3. Oncogenes empower a disproportional cellular response to ECM mechanical properties to drive pancreatic tumorigenesis.
a, Schematic representation of the experimental strategy employed to study pancreatic ADM ex vivo. See also methods section for details.
b, Schematic representation of the chemistry of designer hydrogels employed in the experiments of Fig. 3a-c, Fig. 4c, Extended Data Fig. 3c and Extended Data Fig. 4e, f (see Methods). SH, sulfhydryl group; PEGDA, Poly(ethylene glycol) diacrylate. Hydrogel stiffness was raised by increasing the PEGDA crosslinker concentration, without changing Gelatin and Hyaluronic Acid content.
c, Quantification of nuclear YAP/TAZ levels in pancreatic acini shown in Fig. 3c Data mean + s.d. of n=4 independent samples. ****p-value=4.03x10-7.
d, qRT-PCRs assessing the expression levels of the acinar marker Amylase (Amy) and of the ductal markers K19 in pancreatic acini of the indicated genotypes, showing that KRas-expressing acini treated with FA or F-actin inhibitors (as in Fig. 3d) remain fully differentiated recapitulating the effects of a soft ECM. Data are mean + s.d. of n= 3 independent experiments. ns, p-value=0.99. Data are normalized to 18-s rRNA.
e, Quantification of nuclear YAP/TAZ levels in pancreatic acini shown in Fig. 3d, showing that oncogenic KRas promoted YAP/TAZ activation in a manner opposed by FA and F-actin inhibitors. Data are mean + s.d. of n>4 independent samples. **** p-value=10-15.
f, Dot-plot depicting cell viability assays comparing MCF10A cells treated with the indicated mechano-inhibitory drugs (at maximal doses, see methods for details) with untreated cells. Drug treatments employed throughout the study do not affect cell viability, at difference with Puromycin (Puro.) treatment, shown as positive control for reduced cell viability. Data are mean +/- s.d. of n=5 independent samples.
g, Schematic representation of the experimental strategy employed to oppose either ECM stiffness (by BAPN treatment) or intracellular contractility (by Fasudil treatment) at the onset of pancreatic tumorigenesis in vivo.
h, Quantifications of tumor areas in the samples depicted in Fig. 3f. Data are mean +/- s.d. of n=3 independent samples.
i, Quantifications of pMLC stainings shown in Fig. 3f, showing that BAPN treatment normalized the tensional state of oncogene-expressing pancreatic cells. Data are mean + s.d. of n=3 independent samples. ***p-value=6.6x10-4.
j, Representative bright field and birefringence images of Picrosirius red staining of the same samples shown in Fig. 3f, showing that BAPN treatment inhibited deposition and fibrillar organization of collagen. Quantifications of total fibrillar collagen obtained from birefringence are presented as mean + s.d. of n=3 independent samples. ****p-value=1.80x10-5.
k, Quantifications of tumor areas in the samples depicted in Fig. 3g. Data are mean +/- s.d.
l, Quantifications of pMLC stainings shown in Fig. 3g. Data are mean + s.d. of n=3 independent samples. ****p-value=8.88x10-5.
P-values were calculated by unpaired two-sided Student’s t-test (c) and by one-way ANOVA with Sidak’s multiple comparisons test (d-f,g,i,l).
Extended Data Fig. 4. YAP/TAZ are the nuclear effectors downstream of the changes in the cell's mechanical and material properties induced by oncogenes.
a, Representative H&E stainings and quantifications of tumor areas showing that initiation of pancreatic tumorigenesis form acinar cells is dependent on YAP/TAZ. Development of neoplastic lesions was assessed 6 months after tamoxifen administration to mice of the following genotypes: Ptf1aCreERTM(wt), Ptf1aCreERTM;KRas+/LSL-G12D(KRasG12D), Ptf1aCreERTM;KRas+/LSLG12D;Yapfl/fl;Tazfl/fl (KRasG12D;Y/T KO), Ptf1aCreERTM;KRas+/LSLG12D;p53+/LSLR172H (KRasG12D;p53R172H), Ptf1aCreERTM;KRas+/LSLG12D;p53+/LSL-R172H;Yapfl/fl;Tazfl/fl(KRasG12D;p53R172H;Y/T KO).
Data are presented as mean +/- s.d. of n=3 independent samples. These experiments are similar to what reported in Ref. 23, although in a different experimental setting, as we tested Yap/Taz requirement in absence of experimentally induced pancreatitis.
b, Immunoblot validating effective Yap and Taz in vivo knock out in pancreata shown in a. GAPDH serves as loading control. Results are representative of n=3 independent experiments.
c, Quantifications of the percentage of ADM events in pancreatic acini depicted in Fig. 4b. Data are presented as mean + s.d. of n=5 independent samples.****p-value=10-15.
d, qRT-PCRs validating effective Yap and Taz ex vivo knock out in pancreatic acini shown in Fig. 4b. Data are presented as mean + s.d. of n=3 independent samples. Data are normalized to 18-s rRNA.
e, Quantifications of the percentage of ADM events in pancreatic acini depicted in Fig. 4c. Data are presented as mean + s.d. of n=3 independent samples.
f, qRT-PCRs validating YAPS127A overexpression in pancreatic acini shown in Fig. 4c. Data are mean + s.d. of n=3 independent samples. Data are normalized to 18-s rRNA.
g, Quantifications of the percentage of ADM events in pancreatic acini depicted in Fig. 4d. Data are mean + s.d. of n=3 independent samples. ****p-value=10-15.
h, qRT-PCRs validating YAPS127A overexpression in pancreatic acini shown in Fig. 4d. Data are mean + s.d. of n=3 independent samples. Data are normalized to 18-s rRNA.
i, Pie chart of the main categories of GO entries associated to the HER2-induced genes that are dependent on YAP/TAZ and substrate stiffness, derived from the RNA-seq of Fig. 4e. Gene Ontology (GO) analysis was performed on the list of HER2-induced genes whose expression is dependent on YAP/TAZ and substrate stiffness (Supplementary Table 1). Of these genes, about 30% are linked to processes related to Cell Cycle progression, whereas others were classified as genes involved in Cell Fate Specification (10%), Signal Transduction (4%), RNA processing (22%) and Transcription (24%).
P-values were calculated by unpaired two-sided Student’s t-test (d), one-way ANOVA with Sidak’s multiple comparisons test (c,e,f,h) and two-way ANOVA with Sidak’s multiple comparisons test (g).
Extended Data Fig. 5. Oncogenes modify the cell's mechanical properties through Rac1 activation.
a, Western blots showing that the levels of YAP phosphorylation on the key LATS target residue S127 do not decrease after HER2-CA or KRasG12V expression in MCF10A cells. GAPDH serves as loading control. Phosphorylated vs. total YAP signal ratio (Empty: 1,19; HER2-CA: 1,24; KRasG12V: 0,93) was quantified by ImageJ analysis software. Results are representative of n=3 independent experiments, performed with similar results.
b, Western blots showing that HER2-CA or KRasG12V expression in MCF10A cells does not affect the levels of the active pool of LATS kinases, as assessed by monitoring LATS1-activating phosphorylation on its key residue Thr1079. GAPDH serves as loading control. Phosphorylated vs. total LATS signal ratio (Empty: 1,28; HER2-CA: 1,12; KRasG12V: 1,11) was quantified by ImageJ analysis software. Results are representative of n=3 independent experiments, performed with similar results.
c, Quantification of the colocalization of the cytoskeletal stainings for pMLC and F-actin shown in Fig. 5c. Data are as mean + s.d. of n=3 independent samples. ****p-value=2.42x10-6.
d, Representative immunofluorescence images and quantifications showing that MCF10A cells transduced with Rac1-CA display increased formation of focal adhesion as visualized and quantified by colocalization of focal adhesion kinase (FAK) and Vinculin. Scale bars, 24 μm. Images and data are representative of n=3 independent experiments. Data are mean + s.d. ****p-value=6.27x10-4.
e, Representative immunofluorescence images and quantifications showing that MCF10A cells transduced with HER2-CA or KRasG12V display increased maturation of focal adhesions as visualized and quantified by colocalization of phospho-focal adhesion kinase (pFAK) and Paxillin. Scale bars, 24 μm. Images and data are representative of n=3 independent experiments. Data are mean + s.d. *** p-value=2.41x10-4.
f, Representative images and quantifications of collagen contraction assays performed with MCF10A cells transduced with Rac1-CA and treated with the indicated drugs. MCF10A cells transduced with empty vector are shown as negative control. Data are mean + s.d. of n>3 independent samples. ****p-value=4.51x10-6.
g, Quantifications of the primary mammary colonies formed from Yap+/+;Taz+/+ or YapKO;TazKO mLD cells, treated as in Fig. 5g. Data are mean + s.d. of n= 3 independent experiments.
h, Quantifications of the secondary and tertiary colonies derived from primary colonies formed by Rac1-CA-transduced mLD cells. Data are mean + s.d. of n= 3 independent experiments.
i, Quantifications of colonies formed by hLD cells treated as in Fig. 5i. Data are mean + s.d. of n= 3 independent experiments. ****p-value=1.18x10-6.
j, Luciferase assay in post-confluent HEK293 cells transfected with 8xGTIIC-Lux, and with empty vector or the indicated oncogene expressing vectors, alone or in combination with vectors coding for Rac1-DN. Data are mean + s.d. of n= 3 independent samples.
k, qRT-PCRs of the ductal marker K19 in the pancreatic acini depicted in Fig 5l. Data are mean + s.d. of n=3 independent samples.
l, qRT-PCRs showing effective YAPS127A overexpression and Rac1 depletion in pancreatic acini depicted in Fig. 5l. Data are presented as mean + s.d. of n=3 independent samples.
m, Representative bright field images and quantifications of ADM events of pancreatic acini of the indicated genotypes treated with the Rac-specific GEFs Tiam1/Trio inhibitor NSC23766 (100 μM), or left untreated (Control). Images and data are representative of n=3 independent experiments. Scale bars, 70 μm. Data are mean + s.d. **** p-value=3.2x10-5.
P-values were calculated by one-way ANOVA with Sidak’s multiple comparisons test (c-g,i,l,m) and two-way ANOVA with Sidak’s multiple comparisons test (j).
Extended Data Fig. 6. Transformation of immortalized cells requires sufficient substrate rigidity and YAP/TAZ activity.
a, Quantifications of colonies formed by control or oncogene-expressing MCF10A cells, cultured at the three indicated stiffnesses before embedding in agar. Data are mean + s.d. of n=3 independent samples. Controls showing the requirement of YAP/TAZ in similar cell transformation assays are shown in b.
b, Quantifications of colonies formed by control or oncogene-expressing MCF10A cells, transfected with the indicated siRNAs before embedding in agar. YAP/TAZ depletion blunts oncogene-induced cell transformation. Data are mean + s.d. of n=3 independent samples.
P-values were calculated by two-way ANOVA with Sidak’s multiple comparisons test.
Extended Data Fig. 7. Rheological measurements validating the purely elastic behavior of PAA hydrogels used in the study.
a, Rheological measurements showing Storage (G’) and loss (G”) moduli of PAA hydrogels used in the study as function of the frequency (from 0.01 to 1.259 Hz) in a constant strain mode. Young’s modulus E=3G, is indicated above each panel. Results are representative of n=3 independent experiments, performed with similar results.
Supplementary Material
Acknowledgements
We thank V. Guzzardo for histology; C. Frasson for FACS; M. Ventre and P. Netti for rheology, E. Armato Smaniotto dai Roveri for human bioptic sample procurement, D.J. Pan, F. Camargo, G. Lozano, T. Jacks, and J. Siveke for gifts of mice. The research leading to these results has received funding from the following agencies: the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (DENOVOSTEM grant agreement No 670126) to S.P.; Fondazione AIRC under 5 per Mille 2019 – ID. 22759 program to S.P.; the Italian Ministry of Education, University and Research (MIUR), MIUR-FARE (No R16SXW55W4) to S.P., and PRIN 2017 grants to T.P. (No 2017L8FWY8_004) and S.P. (No 2017HWTP2K_001); Fondazione CARIPARO, under “CARIPARO Starting Grant” (No C94I19001680001) to T.P. and under “CARIPARO Ricerca Scientifica di Eccellenza 2018” grant (No 52008 - 2019.0356) to S.P.; and from University of Padua PRID grant to S.P. and L.A. (No CPDA135844).
Footnotes
Author Contributions
T.P. and A.C. performed all in vitro and in vivo experiments. A.R. helped with mammary gland transplantation. T.P and D.D.B., performed human mammary gland experiments, S.G., G.Br. and A.G., hydrogel preparation, L.A. mouse genetics, M.F., M.C. and S.B. performed bioinformatic analysis, V.P. and S.F. performed microrheology and traction force microscopy, M.Fa., histology and histopathological evaluations, V.V., F.B., M.Fa., A.P.D.T, for human samples, T.P., M.C. and S.P. conceived the initial hypothesis and experimental design, and planned, discussed and organized the work. T.P., A.C., M.C. and S.P. wrote the manuscript.
Competing Financial Interests
The authors declare no competing interests.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors on reasonable request. All relevant data used to generate Figures 1-5 and Extended Data Figures 1-7 panels are included in the paper as Source Data. RNA-seq data from this study have been deposited in the GEO database under accession number GSE128037.
Materials & Correspondence
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Stefano Piccolo (piccolo@bio.unipd.it).
References
- 1.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama A, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312–317. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 3.Lee-Six H, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574:532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 4.Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northey JJ, Przybyla L, Weaver VM. Tissue Force Programs Cell Fate and Tumor Aggression. Cancer Discov. 2017;7:1224–1237. doi: 10.1158/2159-8290.CD-16-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Vega F, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337 e310. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 9.Linnemann JR, et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development. 2015;142:3239–3251. doi: 10.1242/dev.123554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panciera T, et al. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell stem cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acerbi I, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb) 2015;7:1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Gauthier NC, Masters TA, Sheetz MP. Mechanical feedback between membrane tension and dynamics. Trends Cell Biol. 2012;22:527–535. doi: 10.1016/j.tcb.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Brusatin G, Panciera T, Gandin A, Citron A, Piccolo S. Biomaterials and engineered microenvironments to control YAP/TAZ-dependent cell behaviour. Nat Mater. 2018;17:1063–1075. doi: 10.1038/s41563-018-0180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng Y, Kole TP, Wirtz D. Micromechanical mapping of live cells by multiple-particle-tracking microrheology. Biophys J. 2002;83:3162–3176. doi: 10.1016/S0006-3495(02)75319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Balmain A, Counter CM. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer. 2018;18:767–777. doi: 10.1038/s41568-018-0076-6. [DOI] [PubMed] [Google Scholar]
- 19.Guerra C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp JL, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burdick JA, Prestwich GD. Hyaluronic acid hydrogels for biomedical applications. Adv Mater. 2011;23:H41–56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanconato F, et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med. 2018;24:1599–1610. doi: 10.1038/s41591-018-0158-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber R, et al. YAP1 and TAZ Control Pancreatic Cancer Initiation in Mice by Direct Up-regulation of JAK-STAT3 Signaling. Gastroenterology. 2016;151:526–539. doi: 10.1053/j.gastro.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scita G, et al. Signaling from Ras to Rac and beyond: not just a matter of GEFs. EMBO J. 2000;19:2393–2398. doi: 10.1093/emboj/19.11.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazanietz MG, Caloca MJ. The Rac GTPase in Cancer: From Old Concepts to New Paradigms. Cancer Res. 2017;77:5445–5451. doi: 10.1158/0008-5472.CAN-17-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heid I, et al. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 2011;141:719–730. doi: 10.1053/j.gastro.2011.04.043. 730 e711-717. [DOI] [PubMed] [Google Scholar]
- 27.Kapoor A, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caliari SR, Burdick JA. A practical guide to hydrogels for cell culture. Nat Methods. 2016;13:405–414. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon SH, et al. p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression. Cell. 2019;176:564–580 e519. doi: 10.1016/j.cell.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 34.Shih C, Shilo BZ, Goldfarb MP, Dannenberg A, Weinberg RA. Passage of phenotypes of chemically transformed cells via transfection of DNA and chromatin. Proc Natl Acad Sci U S A. 1979;76:5714–5718. doi: 10.1073/pnas.76.11.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 37.Chang L, et al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 2018;563:265–269. doi: 10.1038/s41586-018-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 39.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 40.Azzolin L, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 41.Azzolin L, et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Morsut L, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 43.Meng Z, et al. RAP2 mediates mechanoresponses of the Hippo pathway. Nature. 2018;560:655–660. doi: 10.1038/s41586-018-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panciera T, et al. De Novo Generation of Somatic Stem Cells by YAP/TAZ. J Vis Exp. 2018 doi: 10.3791/57462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanz-Moreno V, et al. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell. 2011;20:229–245. doi: 10.1016/j.ccr.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 46.Panzetta V, et al. ECM Mechano-Sensing Regulates Cytoskeleton Assembly and Receptor-Mediated Endocytosis of Nanoparticles. Acs Biomater Sci Eng. 2017;3:1586–1594. doi: 10.1021/acsbiomaterials.7b00018. [DOI] [PubMed] [Google Scholar]
- 47.Panzetta V, et al. Mechanical phenotyping of cells and extracellular matrix as grade and stage markers of lung tumor tissues. Acta Biomater. 2017;57:334–341. doi: 10.1016/j.actbio.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 53.Rittie L. Method for Picrosirius Red-Polarization Detection of Collagen Fibers in Tissue Sections. Methods Mol Biol. 2017;1627:395–407. doi: 10.1007/978-1-4939-7113-8_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.