Abstract
In bacteria, the rates of transcription elongation and translation elongation are coordinated, changing together in response to growth conditions. It has been proposed that this is due to physical coupling of RNA polymerase and the lead ribosome on nascent mRNA, an interaction important for preventing premature transcription termination by Rho factor. Recent studies challenge this view and provide evidence that coordination is indirect, mediated in E. coli by the alarmone (p)ppGpp. Here, we discuss these new findings and how they shape our understanding of the functional relationship between RNA polymerase and the ribosome as well as the basis of transcriptional polarity.
Keywords: transcription, translation, ppGpp, Rho, premature transcription termination
Graphical abstract
Transcription and translation take place together in the cytoplasm in Bacteria and Archaea [1]. It has been known for decades that translating ribosomes can functionally impact RNA polymerase (RNAP), exemplified by the phenomena of transcriptional polarity and transcription attenuation. In the 1960s, investigators noticed that nonsense or frameshift mutations in the lacZ gene abolished synthesis of LacY and LacA, encoded downstream in the same operon [2–4]. This resulted from an absence of downstream mRNA, due to premature transcription termination (PTT) by Rho [5]. This phenomenon, known as transcriptional polarity, was later reported for the trp operon [6], the ilv operon [7], and the his operon [8], and hence seemed to be general. In the 1970s, it was found that ribosomes can influence RNAP in another way, a phenomenon termed transcription attenuation. In the trp operon, the leader region contains a small open reading frame with tandem tryptophan codons upstream of a transcription terminator. Stalling of the ribosome at these codons due to low levels of Trp-tRNATrp promotes formation of an anti-terminator structure and continuation of transcription of the entire operon [9–13]. Attenuation is a well-known mechanism to regulate the biosynthesis of amino acids and other metabolites in bacteria [14–17].
Vogel and Jensen (1994) first showed that rates of polypeptide and mRNA chain elongation are normally correlated [18]. Using the lacZ gene, they measured the appearance of full-length mRNA and protein products upon isopropyl-β-d-thiogalactopyranoside (IPTG) induction, allowing calculation of chain elongation rates for both transcription and translation. Cells growing rapidly exhibited the highest rates of chain elongation, while cells growing at progressively slower rates showed reductions in transcription and translation elongation rates, always of equivalent magnitude. Such coordination of chain elongation rates implies an important functional purpose.
A model of RNAP-ribosome coupling
In 2010, Nudler and coworkers confirmed that transcription and translation of lacZ are tightly coordinated under different growth conditions. They also provided compelling evidence that slowing the ribosome, by using a mutation in rpsL (encoding ribosomal protein S12) or the antibiotic chloramphenicol (Cm, which inhibits peptidyl transfer), also slows RNAP to the same degree. The authors proposed that by “pushing” the RNAP forward and preventing it from backtracking, the lead ribosome can control the rate of transcription [19]. In other words, the two macromolecules work like coupled train locomotives, each being able to influence the other [20, 21]. Concurrent work by Gottesman and coworkers showed that the transcription factor NusG can interact with both RNAP and ribosomal protein S10 (also known as NusE) [22]. It was envisaged that NusG acts as the molecular coupler, physically linking RNAP to the lead ribosome [23–25].
A number of groups have since explored the idea of physical coupling between RNAP and the ribosome, using cryo-electron microscopy (cryo-EM) and biochemical approaches. In 2017, the structure of an RNAP-ribosome complex formed in an in vitro transcription-translation system, termed the “expressome”, was solved by cryo-EM [26]. In this structure, the mRNA exit tunnel of RNAP docks right onto the mRNA entry tunnel of the 30S subunit of the ribosome, resulting in seamless protection of the mRNA. In independent work, the structure of RNAP bound to the small subunit (30S) of the ribosome was solved by cryo-EM [27]. In this structure, the mRNA exit region of RNAP is near the mRNA exit tunnel of the 30S subunit, a conformation at odds with the “expressome” structure and difficult to rationalize functionally. Recent biochemical studies showed that RNAP core enzyme is capable of interacting with the 30S subunit, the 50S subunit, and the 70S ribosome, all with a similar affinity [28]. These structural and biochemical data are puzzling and lend little congruent support for the coupling model.
There are other caveats to the physical coupling model. First, it has been shown that the elongation rate of ribosomal RNA (rRNA) transcription also varies as a function of growth rate, in a similar manner as mRNA transcription [18]. By necessity, rRNA synthesis rate itself cannot depend on translating ribosomes. Second, S10 is known to play moonlighting roles off the ribosome [29, 30], for example as part of the λN and rrn antitermination complexes, which also include NusA, NusB and NusG [31–33]. Whether the role of S10 in transcription processivity involves the ribosome or another S10-containing complex is difficult to address and remains unclear.
Evidence that coordination of transcription and translation is indirect
Chen and Fredrick (2018) tested the idea that RNAP is normally coupled to the lead ribosome on the nascent mRNA chain [34]. They reasoned that if this is true, the ratio of proteins templated by polycistronic mRNA and produced by the lead ribosome should match that of transcription (1:1), even when the ratio of protein products from multiple-round translation differs (e.g., 3:1) (Fig. 1A–B). They identified several operons in which genes 1 and 2 were differentially translated, based on published ribosome profiling datasets, and engineered strains, each containing lacZ translationally fused to a given gene at its native chromosomal locus. Encoded within the lacZ reporter was an efficient hammerhead ribozyme (with or without active-site mutations), allowing direct comparison of relative yields of co-transcriptional (‘single’-round) versus multiple-round translation (Fig. 1C–D). The data revealed that the ratio of protein products came no closer to unity (1:1) when rounds of translation was substantially reduced via hammerhead cleavage, arguing against the strict-coupling model. In fact, only in one case when transcription elongation was artificially slowed (using a mutation in RNAP) did the ratio of protein products approach 1:1. Based on the collective data, it was deduced that there exists no general mechanism to ensure coupling, and any coupling is stochastic [34].
Fig. 1.
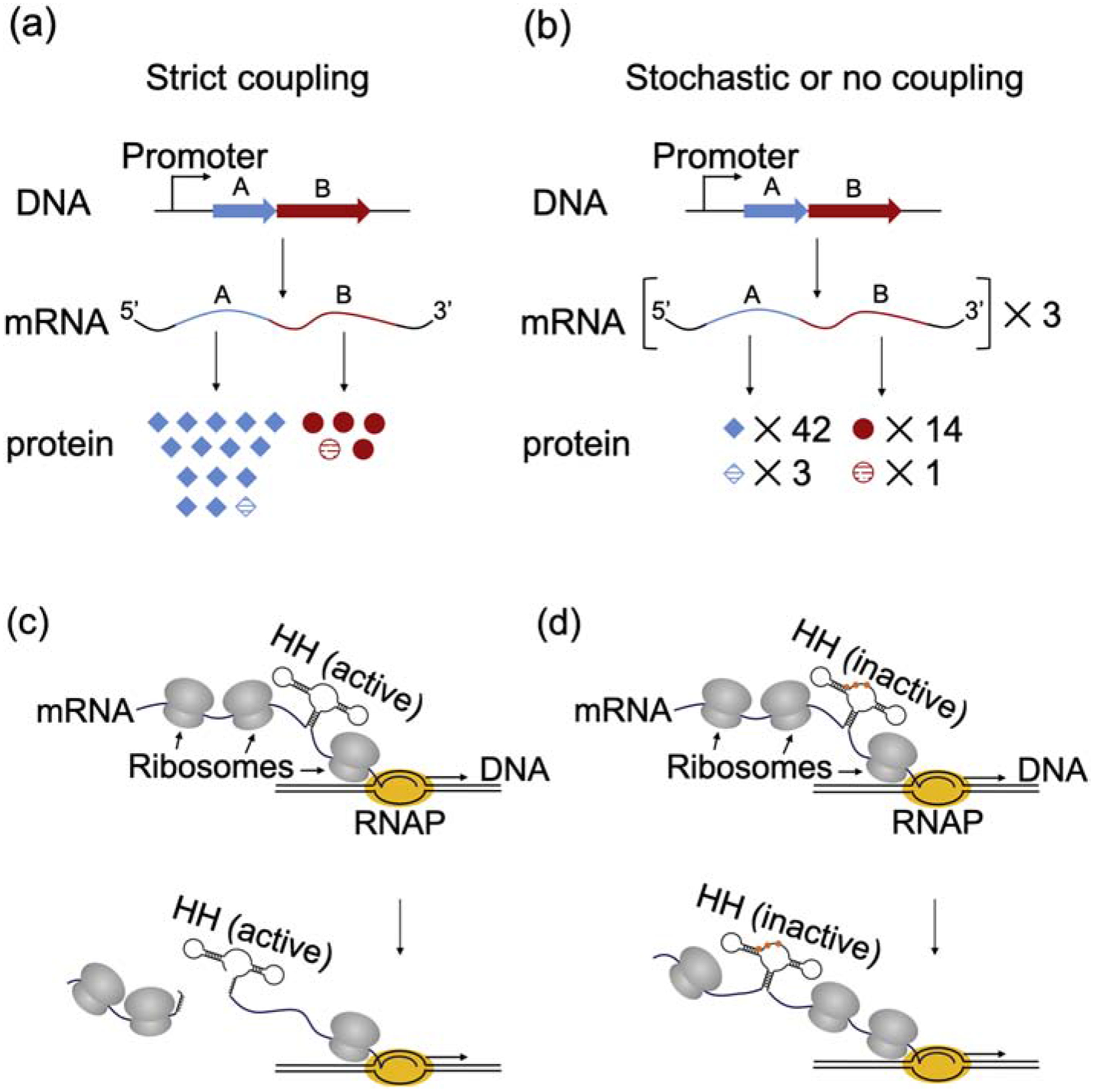
Rationale and approach of Chen and Fredrick (2018). (A-B) Hypothetical protein products of strict versus stochastic coupling. (A) A scenario in which RNAP and the lead ribosome are strictly coupled. Genes A and B are co-transcribed, resulting in stoichiometric levels of A and B mRNA (1:1). The first round of translation by the RNAP-coupled ribosome generates equivalent amounts of proteins A and B (1:1, striped symbols), whereas multiple-round translation yields different amounts of total protein (3:1, all symbols). (B) A scenario in which transcription and translation are uncoupled or stochastically coupled. In this case, RNAP has effectively no impact on the lead ribosome, and hence the ratio of protein products made during transcription (3:1, striped symbols) matches that of multiple-round translation (3:1, all symbols). (C-D) A hammerhead ribozyme enables direct comparison of ‘single’- (co-transcriptional) versus multiple-round translation. (C) The active hammerhead (HH) quickly self-cleaves after being made, so only a ribosome tailgating RNAP will produce full-length LacZ. (D) Three point mutations (colored dots) inactivate the hammerhead without altering the encoded polypeptide, enabling multiple-round translation to be measured.
Further evidence that functional interplay between RNAP and the ribosome is stochastic came from another study by Shi and coworkers [35]. They generated a series of constructs in which the distance between an intrinsic terminator and the stop codon was incrementally varied. A gradual and smooth increase in termination efficiency was observed as this distance increased, data which were best fit to a stochastic interaction model. They also showed that a terminator embedded within the 3’ portion of a gene is repressed more by translation than one located in the 5’ portion of the gene. Presumably the ribosome has more time to catch up to RNAP and prevent transcription termination in the former case. These data suggest that RNAP and the lead ribosome move independently and interact stochastically.
In 2019, Hwa and coworkers revisited the question of transcription-translation coordination in a comprehensive way and showed that translation is not required to maintain the speed of transcription elongation [36]. Using lacZ, they quantified the elongation speed of transcription and translation under five different growth conditions and found the rates to be tightly correlated. When cells were treated with sublethal concentrations of Cm, the elongation rate of neither translation nor transcription was reduced [36, 37], in contrast to the earlier report by Nudler and coworkers [19]. While the basis of this discrepancy remains unclear, the more recent data have higher time resolution and higher signal-to-noise [36, 37]. Importantly, when Hwa and coworkers challenged cells with fusidic acid (FA), an antibiotic that targets ribosome-bound EFG, translation elongation speed was clearly reduced whereas transcription elongation speed was unaffected [36]. The authors also showed that a nonsense mutation in lacZ has no effect on transcription elongation, but strongly reduced the level of full-length mRNA. In other words, eliminating ribosome traffic reduces transcription processivity without altering elongation kinetics.
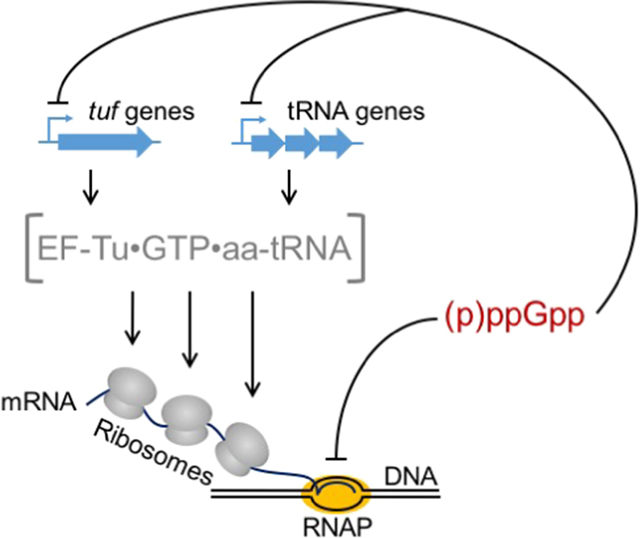
Guanosine penta/tetra-phosphate (p)ppGpp is a key molecule for regulation of cell growth in E. coli [38]. In starved cells, (p)ppGpp accumulates, binds to RNAP, and inhibits initiation of rRNA transcription [39]. Several studies suggest that (p)ppGpp also slows RNAP elongation and plays a role in coordinating transcription and translation [40–44]. Hwa and coworkers progressively increased the concentration of (p)ppGpp in the cell and observed corresponding decreases in transcription elongation speed [36]. Because (p)ppGpp levels also control production of EFTu•GTP•aa-tRNA ternary complexes and translation elongation speed depends on ternary complex concentration [37, 45, 46], it was suggested that (p)ppGpp functions as the global coordinator, modulating transcription and translation speeds simultaneously [36].
Transcriptional polarity results from artificial disruption of transcription-translation coordination
Hwa and coworkers also described transcriptional polarity in quantitative terms [36]. As mentioned above, introduction of a nonsense mutation in lacZ reduced its transcript levels markedly. A graded reduction of mRNA as a function of gene position was observed, consistent with premature transcription termination (PTT). This was largely eliminated by bicyclomycin (a Rho inhibitor), demonstrating that the PTT observed was mediated by Rho. Similarly, translation inhibitors FA and Cm both caused obvious PTT, even though Cm did not affect measured translation elongation rates. In these cases, PTT was less pronounced than in the case of the nonsense mutation. The authors suggest that in contrast to FA, which slows translocation of all ribosomes, Cm gains access to only a subset of ribosomes but fully stalls their progress. Such distinct modes of inhibition can explain the in vivo effects of the two antibiotics [36]. Importantly, PTT was only observed under situations when translation is artificially inhibited, for example by nonsense mutation or antibiotic treatment. No PTT was seen when nutrient limitation is used to slow translation [36].
Ribosome traffic and its role in transcription processivity
It has been suggested that physical coupling between RNAP and the lead ribosome normally prevents PTT [47–49]. But, as detailed above, a growing body of evidence argues against any stable linkage between RNAP and the lead ribosome. And yet, ribosome traffic can clearly influence RNAP processivity. How can these observations be explained? Fig. 2 depicts the fate of a paused transcription elongation complex under three different conditions. When ribosome traffic on a natural mRNA is high (Fig. 2A), due to high translation initiation rate, there is a large probability that RNAP will escape the pause, either spontaneously or in a ribosome-assisted manner. Ribosome traffic will occlude Rho, even when Rho-utilization (rut) sites are present, and the lead ribosome may push RNAP forward (or inhibit backtracking). When translation of the same mRNA is artificially inhibited, for example due to introduction of a nonsense mutation (Fig. 2B), Rho will have open access to the rut-containing nascent chain and PTT will likely occur. Importantly, this differs from the case of a natural mRNA which normally exhibits light ribosome traffic, due to low translation initiation rate (Fig. 2C). Even though light ribosome traffic may be sufficient to largely inhibit Rho [50, 51], other determinants are likely at play. These mRNAs may have idiosyncratically evolved to become Rho-resistant, for example via increased structure and the lack of rut sites. Consistent with this possibility, coding regions of polycistronic mRNAs fold independently, and the degree of structure in these ORF-centric domains is inversely correlated with translation efficiency [52].
Fig. 2.
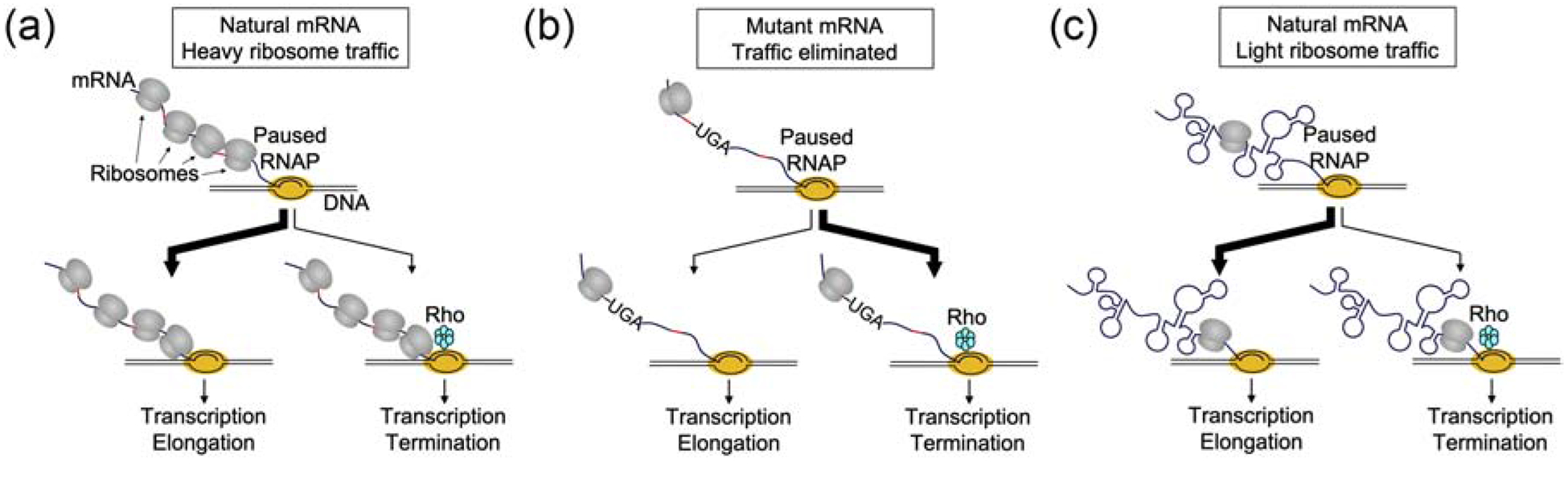
Premature transcription termination (PTT) depends on various factors. (A) When RNAP pauses during transcription of a natural mRNA that is highly translated, ribosomes occlude Rho from nascent chain rut sites (red) and may facilitate pause escape by “nudging” RNAP forward. Hence, the chances of Rho-dependent PTT are low. (B) When the same gene depicted in panel A contains a nonsense mutation, ribosome traffic is eliminated. This gives Rho access to rut sites (red) and PTT becomes favorable. (C) When RNAP pauses during transcription of a natural mRNA that is normally translated with a low initiation rate, other mechanisms must be employed to prevent Rho-dependent PTT. These likely include RNA structures and omission of rut sites.
Why are transcription and translation elongation rates coordinated?
The fact that transcription elongation is at least as fast as translation elongation makes sense. This prevents excessive queuing of ribosomes, which would effectively limit translational control. But, why does transcription elongation speed match rather than exceed translation elongation speed? We suspect that, fundamentally, this is driven by the economics of cell growth. In coordinating chain elongation rates, the cell makes a given mRNA no faster than it can be translated—in other words, with optimal efficiency. Synthesizing even part of an mRNA beforeit can be used provides no obvious benefit, only potential costs. Regulatory mechanisms, such as Rho-dependent PTT and transcriptional attenuation, certainly rely on the coordination between RNAP and the ribosome. But such regulatory mechanisms may have evolved later to sense transcription-translation coordination, which was already in place. Notably, almost all studiesthat have looked at transcription-translation coordination have used the model organism E. coli. Comparative studies with other bacteria may provide new mechanistic insight and evolutionary perspective on this important aspect of gene expression.
Highlights.
Rates of transcription elongation and translation elongation normally match one another, and the basis of this coordination has remained unclear.
Recent evidence suggests that RNA polymerase and the lead ribosome move independently, and coordination is mediated by the alarmone (p)ppGpp.
Transcription is normally processive; premature transcription termination by Rho occurs when ribosome traffic is eliminated via nonsense mutation or antibiotic treatment.
Acknowledgements
We thank M. Ibba, I. Artsimovitch, and T. Hwa for helpful feedback on the manuscript. Work in the Fredrick laboratory is supported by grants from the National Institutes of Health (GM072528) and the National Science Foundation (MCB-1614990).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gowrishankar J, Harinarayanan R, Why is transcription coupled to translation in bacteria?, Mol. Microbiol 54 (2004) 598–603. [DOI] [PubMed] [Google Scholar]
- [2].Jacob F, Monod J, Genetic regulatory mechanisms in the synthesis of proteins, J. Mol. Biol 3 (1961) 318–356. [DOI] [PubMed] [Google Scholar]
- [3].Franklin NC, Luria SE, Transduction by bacteriophage P-1 and the properties of the lac genetic region in E. coli and S. dysenteriae, Virology 15 (1961) 299–311. [DOI] [PubMed] [Google Scholar]
- [4].Newton WA, Beckwith JR, Zipser D, Brenner S, Nonsense mutants and polarity in the lac operon of Escherichia coli, J. Mol. Biol 14 (1965) 290–296. [DOI] [PubMed] [Google Scholar]
- [5].Adhya S, Gottesman M, Control of transcription termination, Annu. Rev. Biochem 47 (1978) 967–996. [DOI] [PubMed] [Google Scholar]
- [6].Yanofsky C, Ito J, Nonsense codons and polarity in the tryptophan operon, J. Mol. Biol 21 (1966) 313–334. [DOI] [PubMed] [Google Scholar]
- [7].Wek RC, Sameshima JH, Hatfield GW, Rho-dependent transcriptional polarity in the ilvGMEDA operon of wild-type Escherichia coli K12, J. Biol. Chem 262 (1987) 15256–15261. [PubMed] [Google Scholar]
- [8].Martin RG, Silbert DF, Smith WE, Whitfield HJ, Polarity in the histidine operon, J. Mol. Biol 21 (1966) 357–369. [DOI] [PubMed] [Google Scholar]
- [9].Jackson EN, Yanofsky C, Thr region between the operator and first structural gene of the tryptophan operon of Escherichia coli may have a regulatory function, J. Mol. Biol 76 (1973) 89–101. [DOI] [PubMed] [Google Scholar]
- [10].Lee F, Squires CL, Squires C, Yanofsky C, Termination of transcription in vitro in the Escherichia coli tryptophan operon leader region, J. Mol. Biol 103 (1976) 383–393. [DOI] [PubMed] [Google Scholar]
- [11].Lee F, Yanofsky C, Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination, Proc. Natl. Acad. Sci. U. S. A 74 (1977) 4365–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Oxender DL, Zurawski G, Yanofsky C, Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region, Proc. Natl. Acad. Sci. U. S. A 76 (1979) 5524–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yanofsky C, Transcription attenuation: once viewed as a novel regulatory strategy, J. Bacteriol 182 (2000) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Johnston HM, Barnes WM, Chumley FG, Bossi L, Roth JR, Model for regulation of the histidine operon of Salmonella, Proc. Natl. Acad. Sci. U. S. A 77 (1980) 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gavini N, Pulakat L, Role of translation of the pheA leader peptide coding region in attenuation regulation of the Escherichia coli pheA gene, J. Bacteriol 173 (1991) 4904–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Roland KL, Liu CG, Turnbough CL, Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12, Proc. Natl. Acad. Sci. U. S. A 85 (1988) 7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Donahue JP, Turnbough CL, Nucleotide-specific transcriptional pausing in the pyrBI leader region of Escherichia coli K-12, J. Biol. Chem 269 (1994) 18185–18191. [PubMed] [Google Scholar]
- [18].Vogel U, Jensen KF, The RNA chain elongation rate in Escherichia coli depends on the growth rate, J. Bacteriol 176 (1994) 2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Proshkin S, Rahmouni AR, Mironov A, Nudler E, Cooperation between translating ribosomes and RNA polymerase in transcription elongation, Science 328 (2010) 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Roberts JW, Molecular biology. Syntheses that stay together, Science 328 (2010) 436–437. [DOI] [PubMed] [Google Scholar]
- [21].Klaholz BP, The Ribosome Holds the RNA Polymerase on Track in Bacteria, Trends Biochem. Sci 42 (2017) 686–689. [DOI] [PubMed] [Google Scholar]
- [22].Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, et al. , A NusE:NusG complex links transcription and translation, Science 328 (2010) 501–504. [DOI] [PubMed] [Google Scholar]
- [23].Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, et al. , An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor, Cell 150 (2012) 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Saxena S, Myka KK, Washburn R, Costantino N, Court DL, Gottesman ME, Escherichia coli transcription factor NusG binds to 70S ribosomes, Mol. Microbiol(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Strauß M, Vitiello C, Schweimer K, Gottesman M, Rösch P, Knauer SH, Transcription is regulated by NusA:NusG interaction, Nucleic Acids Res. 44 (2016) 5971–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P, Architecture of a transcribing-translating expressome, Science 356 (2017) 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Demo G, Rasouly A, Vasilyev N, Svetlov V, Loveland AB, Diaz-Avalos R, et al. , Structure of RNA polymerase bound to ribosomal 30S subunit, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fan H, Conn AB, Williams PB, Diggs S, Hahm J, Gamper HB, et al. , Transcription-translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits, Nucleic Acids Res. 45 (2017) 11043–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Weisberg RA, Transcription by moonlight: structural basis of an extraribosomal activity of ribosomal protein S10, Mol. Cell 32 (2008) 747–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chen SS, Sperling E, Silverman JM, Davis JH, Williamson JR, Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry, Mol. Biosyst 8 (2012) 3325–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, et al. , Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex, Mol. Cell 32 (2008) 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vogel U, Jensen KF, NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA, J. Biol. Chem 272 (1997) 12265–12271. [DOI] [PubMed] [Google Scholar]
- [33].Torres M, Balada JM, Zellars M, Squires C, Squires CL, In vivo effect of NusB and NusG on rRNA transcription antitermination, J. Bacteriol 186 (2004) 1304–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen M, Fredrick K, Measures of single- versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation, Proc. Natl. Acad. Sci. U. S. A 115 (2018) 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li R, Zhang Q, Li J, Shi H, Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the Rho-independent terminator, Nucleic Acids Res. 44 (2016) 2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu M, Mori M, Hwa T, Dai X, Disruption of transcription-translation coordination in Escherichia coli leads to premature transcriptional termination, Nat. Microbiol 4 (2019) 2347–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dai X, Zhu M, Warren M, Balakrishnan R, Patsalo V, Okano H, et al. , Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth, Nat. Microbiol 2 (2016) 16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Potrykus K, Cashel M, (p)ppGpp: still magical?, Annu. Rev. Microbiol 62 (2008) 35–51. [DOI] [PubMed] [Google Scholar]
- [39].Paul BJ, Ross W, Gaal T, Gourse RL, rRNA transcription in Escherichia coli, Annu. Rev. Genet 38 (2004) 749–770. [DOI] [PubMed] [Google Scholar]
- [40].Vogel U, Sørensen M, Pedersen S, Jensen KF, Kilstrup M, Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation, Mol. Microbiol 6 (1992) 2191–2200. [DOI] [PubMed] [Google Scholar]
- [41].Vogel U, Jensen KF, Effects of guanosine 3’,5’-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli, J. Biol. Chem 269 (1994) 16236–16241. [PubMed] [Google Scholar]
- [42].Kingston RE, Nierman WC, Chamberlin MJ, A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation, J. Biol. Chem 256 (1981) 2787–2797. [PubMed] [Google Scholar]
- [43].Furman R, Sevostyanova A, Artsimovitch I, Transcription initiation factor DksA has diverse effects on RNA chain elongation, Nucleic Acids Res. 40 (2012) 3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Iyer S, Le D, Park BR, Kim M, Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli, Nat. Microbiol 3 (2018) 741–748. [DOI] [PubMed] [Google Scholar]
- [45].Ryals J, Little R, Bremer H, Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate, J. Bacteriol 151 (1982) 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shibuya M, Kaziro Y, Studies on stringent control in a cell-free system. Regulation by guanosine-5’-diphosphate-3’-diphosphate of the synthesis of elongation factor Tu, J. Biochem 86 (1979) 403–411. [DOI] [PubMed] [Google Scholar]
- [47].McGary K, Nudler E, RNA polymerase and the ribosome: the close relationship, Curr. Opin. Microbiol 16 (2013) 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Burmann BM, Rösch P, The role of E. coli Nus-factors in transcription regulation and transcription:translation coupling: From structure to mechanism, Transcription 2 (2011) 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Svetlov V, Nudler E, Unfolding the bridge between transcription and translation, Cell 150 (2012) 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].de Smit MH, Verlaan PW, van Duin J, Pleij CW, Intracistronic transcriptional polarity enhances translational repression: a new role for Rho, Mol. Microbiol 69 (2008) 1278–1289. [DOI] [PubMed] [Google Scholar]
- [51].de Smit MH, Verlaan PW, van Duin J, Pleij CW, In vivo dynamics of intracistronic transcriptional polarity, J. Mol. Biol 385 (2009) 733–747. [DOI] [PubMed] [Google Scholar]
- [52].Burkhardt DH, Rouskin S, Zhang Y, Li GW, Weissman JS, Gross CA, Operonm RNAs are organized into ORF-centric structures that predict translation efficiency, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


