Abstract
All retroviruses encode a Gag polyprotein containing an N-terminal matrix domain (MA) that anchors Gag to the plasma membrane and recruits envelope glycoproteins to virus assembly sites. Membrane binding by the Gag protein of HIV-1 and most other lentiviruses is dependent on N-terminal myristoylation of MA by host Nmyristoyltransferase enzymes (NMTs), which recognize a six-residue “myristoylation signal” with consensus sequence: M1GXXX[ST]. For unknown reasons, the feline immunodeficiency virus (FIV), which infects both domestic and wild cats, encodes a non-consensus myristoylation sequence not utilized by its host or by other mammals (most commonly: M1GNGQG). To explore the evolutionary basis for this sequence, we compared the structure, dynamics, and myristoylation properties of native FIV MA with a mutant protein containing a consensus feline myristoylation motif (MANOS) and examined the impact of MA mutations on virus assembly and ability to support spreading infection. Unexpectedly, myristoylation efficiency of MANOS in E. coli by co-expressed mammalian NMT was reduced by ~70% compared to the wild-type protein. NMR studies revealed that residues of the N-terminal myristoylation signal are fully exposed and mobile in the native protein but partially sequestered in the MANOS chimera, suggesting that the unusual FIV sequence is conserved to promote exposure and efficient myristoylation of the MA N-terminus. In contrast, virus assembly studies indicate that the MANOS mutation does not affect virus assembly, but does prevent virus spread, in feline kidney cells. Our findings indicate that residues of the FIV myristoylation sequence play roles in replication beyond NMT recognition and Gag–membrane binding.
Keywords: feline immunodeficiency virus (FIV), matrix protein (MA), myristoylated (myr), unmyristoylated (myr(−)), nitric oxide synthase (NOS), nuclear magnetic resonance (NMR), N-myristoyltransferase (NMT)
Introduction
The feline immunodeficiency virus (FIV) is a lentivirus that infects about 8% of household cats in the United States and causes an immunodeficiency-like syndrome with similarities to AIDS [1]. Felines have immune systems that are similar to those of humans and are easily housed and maintained at a cost about ten times less than that of simians [2], and therefore serve as attractive models for development and evaluation of HIV therapeutics and vaccines [1, 3–8]. Felines served as the initial animal model used in FDA-approved HIV-1 integrase and reverse transcriptase (RT) inhibitor studies [9] and for in vivo assessment of anti-HIV-1 nucleoside analogs [10, 11]. The genomic organization and viral replication cycle of FIV are similar to those of HIV, and some studies of FIV biology have been shown to be translatable to HIV [9, 12–15]. The FIV genome, like that of HIV, encodes a structural polyprotein called Gag that directs virus assembly. Gag is targeted to the inner leaflet of the plasma membrane (PM) [16–18] by its N-terminally myristoylated matrix domain (MA), which recognizes lipid bilayers enriched in cholesterol and acidic phospholipids including phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] [18–25]. Phosphatidylinositol phosphates are members of a family of signaling molecules that normally help target cellular proteins to specific intracellular membranes [21], and PI(4,5)P2 is considered a major PM landmark and determinant for HIV and FIV PM targeting [18, 23]. HIV-1 MA also plays a critical role in recruiting the viral envelope glycoproteins (Env) to assembling virions. Point mutations in MA can inhibit Env incorporation, and this defect can be rescued by truncation of the cytoplasmic tail of Env [26]. These and other studies [26–33] are consistent with a mechanism in which MA trimerization and a well-defined higher-order MA lattice are important for both recruiting and accommodating the cytoplasmic tail of Env in the Gag lattice of assembling particles [33]. Studies also implicate MA in an early, post-entry function that has yet to be defined [34].
The membrane binding activity of HIV-1 MA is dependent on N-terminal myristoylation and requires a conserved patch of basic residues located on the surface of the MA domain [16, 35–38]. As for cellular proteins, MA myristoylation is catalyzed co-translationally [39, 40] by N-myristoyltransferase (NMT), a cellular enzyme that recognizes a stretch of residues at the N-terminus of the protein (the myristoylation signal) after removal of the initiator Met residue [41]. Mammals generally utilize the conserved sequence (G2-[ζ(uncharged)]-[ϕ]-[ϕ][ST]; G=glycine, S=serine, T=threonine, ζ=residues with polar side chains, and ϕ = residues with hydrophobic side chains) [41, 42], and HIV has evolved this same sequence, apparently to ensure efficient myristoylation. Interestingly, whereas feline proteins also contain the common mammalian myristoylation sequence, FIV evolved a distinct myristoylation sequence that is not associated with feline or other mammalian proteins (G2-N-[GEN][QS]-G; N = asparagine, Q = glutamine, E = glutamic acid) [23]. The FIV myristoylation signal appears to be less robust than the common mammalian signal that is coopted by other retroviruses, and efforts to myristoylate recombinant FIV MA in vitro using co-expressed yeast NMT were unsuccessful despite the fact that the yeast enzyme efficiently myristoylates HIV MA and other viral proteins [23, 43]. For this reason, in vitro studies of FIV MA myristoylation have thus far only been carried out with a mutated construct that contains an HIV-like (consensus mammalian) myristoylation signal that could be myristoylated by a yeast NMT in a commonly utilized co-expression vector [23]. The myristoylated FIV MA mutant protein exhibited structural properties and myristoyl switch behavior similar to those of the native HIV protein, suggesting that the MA domains of these viruses are functionally and mechanistically similar [23]. However, to date, no structural or biophysical studies have been reported for the native, myristoylated FIV MA protein, and it remains unclear why the virus evolved this unusual myristoylation signal.
We have developed a co-expression system for myristoylation of wild-type FIV MA protein using mammalian NMT that enabled structural, dynamical, and biophysical studies of native FIV MA containing the most commonly observed FIV MA myristoylation signal, G2-N-G-Q-G (66% of reported FIV sequences). Comparative studies were conducted with a chimeric MA construct that contains a consensus feline myristoylation motif utilized by the feline nitric oxide synthase (G4L/Q5K/G6S; MANOS). Virus production and spreading infection assays were also conducted with native and mutant virus constructs to assess the role of the noncanonical myristoylation signal on assembly and replication. Our in vitro studies revealed that the wild type MA and MANOS proteins adopt similar structures, but that MANOS is less efficiently myristoylated in E. coli by the mammalian NMT enzyme than the WT protein. These findings initially suggested that the native FIV myristoylation signal evolved to enhance interactions with the NMT enzyme and thereby promote virus assembly. However, virus assembly studies conducted in both human and feline cells indicate that the wild type- and MANOScontaining viruses are myristoylated and assemble with similar efficiencies. Despite the lack of an assembly defect, the MANOS chimeric virus was unable to replicate in a spreading infection assay in feline kidney cells. The implications of these findings for the non-canonical N-terminal residues of FIV MA in virus replication are presented.
Results
Construct design and preparation of samples
Previous X-ray crystallographic studies of the unmyristoylated (myr(−)) FIV MA protein revealed that the twenty C-terminal residues are largely disordered, except for a six-residue helical turn that contributes to a threeresidue crystal contact [43]. These studies also showed that a 14-residue C-terminal deletion did not affect the structure of the globular form of the protein [43]. NMR studies confirmed that C-terminal residues Asp 116 - Tyr 135 are disordered in the myristoylated Q5A/G6S mutant [23]. The present studies therefore focused on 16residue deletion constructs (hereafter MA), which exhibited fewer overlapping NMR signals and improved solubility under conditions of the NMR experiments (Fig. S1).
Efforts to prepare native FIV MA through use of the dual expression vector incorporating yeast NMT, as employed for preparation of HIV-1 and HIV-2 MA, did not lead to effective myristoylation [23, 44], likely due to the inability of yeast NMT to recognize the amino acid sequence downstream from the required N-terminal glycine [45]. Previous work by Towler et al. indicate similar yet distinct differences in myristoylation signal recognition of yeast and mammalian NMTs [46], thus efforts were initiated to design a co-expression system employing mammalian NMT to myristoylate FIV MA. Since the sequence of the feline NMT was unknown when this work was initiated, we prepared a dual-expression vector containing the FIV MA and human NMT genes for myristoylation in E. coli. FIV MA was expressed and purified as described previously [23], and myristoylation was verified by mass spectrometry and NMR techniques (≥ 95% myristoylation using endogenous myristoyl CoA). As this work was being completed, the feline NMT sequence was reported (National Center for Biotechnology Information Database; Reference Sequence: XP_003997047.3). A BLAST analysis revealed that the feline and human NMT proteins share 99% sequence identity [47], with only three amino acid differences present within the 392-residue catalytic domain of the enzyme. All three substitutions map to surface sites on the human NMT X-ray crystal structure that are removed from the catalytic center [48], including the peptide and myristoyl-CoA binding sites (Fig. S2). This observation, and the fact that humans and felines utilize the same NMT recognition signals, suggest that the human NMT should be an appropriate substitute for the feline enzyme.
NMR studies of FIV MA and myr(−)MA
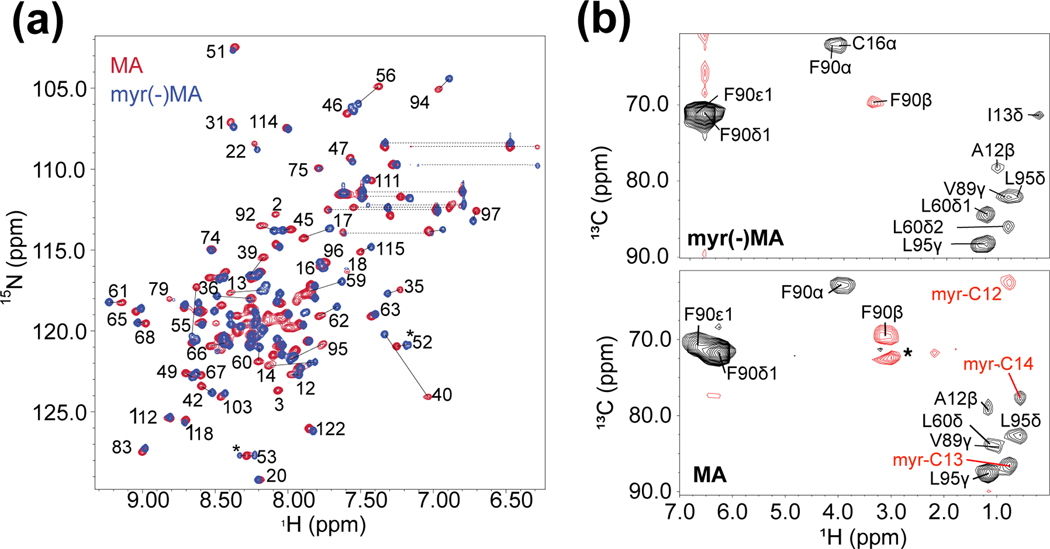
2D-[1H-15N] heteronuclear single quantum coherence (HSQC) spectra obtained for MA and myr(−)MA exhibited good spectral quality and were readily assigned using standard 1H,15N,13C triple resonance NMR methods [49–56] (Fig. 1a). Spectra for both proteins were similar except for minor chemical shift variations for Ser 17, Phe 35 - Arg 40, Gln 59 – Leu 62, Phe 90, Val 92, and Leu 95 (Fig. 1a). The 1H and 15N chemical shifts were generally insensitive to variations in protein concentration (50 μM to ~ 1 mM) and sample pH (MA: 6.0–8.0; myr(−)MA 5.0–8.0) (Fig. S3), consistent with earlier findings for HIV-2 MA and a mutant (Q5A/G6S) FIV MA [23, 44]. These findings contrast with those obtained for HIV-1 MA, in which a subset of signals shifted progressively toward the chemical shifts of myr(−)MA as the protein concentration was elevated or the pH was decreased [57]. The changes in the HIV-1 MA NMR spectra reflect a rapid equilibrium between myristoylexposed and -sequestered states, with the myristoyl-exposed form being favored at elevated protein concentrations and/or lower pH conditions. We also observed significant precipitation of MA within a few hours at concentrations over 300 μM and/or pH of 7 or lower, sufficient time to allow for HSQC analysis but inadequate for triple resonance and NOE-based NMR experiments. These finding are consistent with observations of reduced solubility of HIV-1 MA under these conditions due to low pH-induced myristate exposure [57]. Although we were unable to detect signals consistent with a myristoyl-exposed FIV MA conformer by NMR as detected for HIV-1 MA, our findings suggest that FIV MA may also transiently expose its myristoyl moiety, thereby reducing protein solubility at NMR concentrations (> 50 μM) and lower pH (at or below 7.3).
Figure 1.
Myristoyl sequestration does not significantly influence NMR spectra with the exception of residues interacting with the myristoyl group. (a) Superposed [1H-15N] HSQC spectra of MA (red) and myr(−)MA (blue) (both 300 μM; pH 8.0) obtained at 25 °C demonstrate the overall structural similarity between the two proteins aside from a subset of signals. Significant shifts are observed for amino-terminal residues and core residues that are near the hydrophobic cleft for myristoyl moiety sequestration. Assignments are shown for signals in lesscrowded regions of the overlaid spectra. Dashed lines represent signals that correspond to Asn and Gln sidechains. Asterisks denote signals observed as the result of deamidation of myr(−)MA; (b) Representative 4D CCNOE spectra of the side chain of Phe 90 in FIV myr(−)MA (top) and FIV MA (bottom) showing that their environments are similar with the exception of myristoyl group signals detected in FIV MA. Asterisks represent artifacts observed in the spectra.
Solubility of FIV myr(−)MA constructs was pH independent; however, several backbone amide signals, including those corresponding to the N-terminus, were absent at pH 6.0 or greater. Backbone assignments of native myr(−)MA were carried out at pH 5.0. The native myr(−)MA 1H-15N HSQC, 15N-edited 1H-1H NOESY and HNCACB spectra exhibited two sets of backbone amide signals associated with residues Asn 3 through Gly 6. Signal doubling was not detected for the remaining residues of the protein. These findings indicate that the Nterminal residues either exist as two distinct conformers, or that a portion of the sample has undergone modification. The NMR chemical shifts and NOE patterns indicate that the N-terminal residues of both species are disordered. Notably, the HNCACB spectrum exhibited intensity-inverted signals for the Gly 4 to Asn 3 correlations (Fig. S4). This pattern is consistent with an iso-aspartate peptidyl linkage resulting from deamidation of Asn 3 [58]. Mass spectrometric analyses confirmed the presence of a succinimide species, together with an unmodified protein species, in samples incubated at pH 8 and 5. Non-enzymatic deamidation of Asn and Gln residues is an irreversible post-translational modification that occurs both in vitro and in vivo [59–63]. Although we were unable to examine the NMR spectrum of the myristoylated protein at lower pH due to poor protein solubility, mass spectral studies indicated that the myristoylated MA protein does not undergo deamidation. Deamidation is influenced by several factors, including 3D structure, pH, temperature, ionic strength, buffer ions [60, 61, 64], and samples of FIV myr(−)MA prepared and purified at higher pH exhibited greater percentages of the deamidated species. Studies with peptides indicate that the identity of neighboring residues contributes significantly to deamidation frequency [59, 60], with Asn(i)-Gly(i+1) exhibiting the greatest degree of deamidation [62, 65]. Thus, the Gly 2-Asn 3-Gly 4-Gln 5-Gly 6 sequence of the myr(−)MA N-terminus and its apparent lack of structure make it a prime candidate for deamidation. The finding that the myristoylated protein does not undergo deamidation, despite having the same peptide sequence, is likely due to conformational restraints imposed by the myristoyl group. Because deamidation occurs slowly at physiological pH (significantly slower than the rate of virus assembly), we believe the process is unlikely to be important for viral replication and carried out expression as well as purification at low pH to delay production of the deamidated species for analysis of myr(−)MA.
Structure of FIV MA
Interproton distance information was obtained from 2D 1H-1H NOE (in 99% D2O), 3D 15N-edited NOE, and 4D 15N,13C- and 13C,13C-edited NOE [66–69] NMR spectra collected for 15N- and 15N,13C-labeled MA and myr(−)MA protein samples (50 mM sodium phosphate, pH 8.0, 10 mM DTT). The amides of residues Gly 4 and Gly 6 and Gly 2 – Trp 9 were undetected for both MA and myr(−)MA under these conditions due to rapid exchange with water protons. Unlike the myristoylated protein, myr(−)MA is soluble under conditions of lower pH (50 mM sodium phosphate, pH 5.0, 10 mM DTT), which enabled detection of amide signals for the N-terminal residues. For the myristoylated protein, unique 1H-13C myristoyl moiety correlation signals were readily identified by comparison of 2D 1H-13C HMQC NMR spectra obtained for proteins prepared using endogenous 13C-myristoylCoA in 13C-enriched minimal media and media supplementation with 12C-myristoyl-CoA (Fig. S5). 2D 1H-1H NOESY and 4D 13C,13C-edited NOE spectra exhibited NOEs between the degenerate carbons 4–10 of the myristoyl group and side chains of Trp 9, Ile 53, and Glu 55, and between the terminal methyl group of the myristoyl moiety and the side chains of Phe 35, Ile 39, Leu 60, Leu 87, and Phe 90.
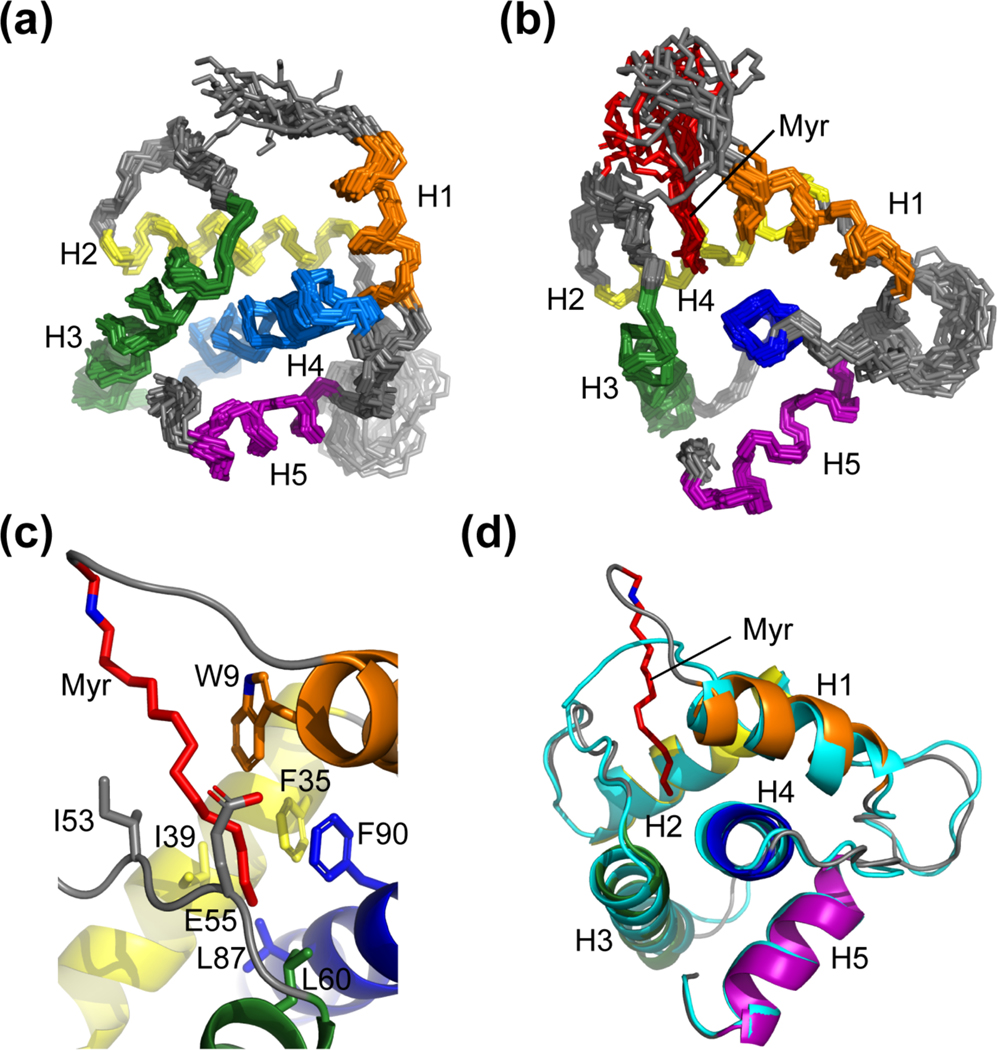
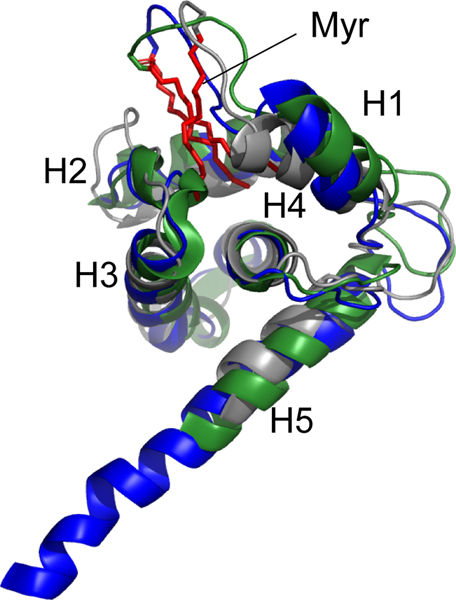
NMR structures of MA and myr(−)MA were determined using interproton distance restraints derived from the above experiments. Ensembles of 20 structures with the lowest target functions (MA: 0.08 ± 0.01 Å2; myr()MA: 0.12 ± 0.026 Å2) exhibited good convergence, with best-fit superpositions of backbone heavy atoms affording pairwise RMS deviations of 1.13 ± 0.27 Å and 1.10 ± 0.34 Å for MA and myr(−)MA, respectively (Table 1 and Figs. 2a–b). Both proteins adopt structures similar to that observed in the X-ray crystal structure of FIV myr(−)MA and the NMR structure of the myristoylated Q5A/G6S MA mutant [23, 43], in which four α-helices (helix-I, Arg 7-Asn 18; helix-II, Glu 32-Thr 46; helix-III, Leu 57-Phe 74; and helix-V, Ala 103-Met 113) pack around a central α-helix (helix-IV, Ser 77-Leu 96,) (Figs. 2c–d). This architecture is similar to that observed for other retroviral MA proteins (Fig. 3) [23, 38, 44, 70–81].
Table 1.
Structural restraint and refinement statistics for FIV MA and related constructs
| MA | myr(−)MA | myr(−)MANOS | |
|---|---|---|---|
| NMR-derived restraints | |||
| Intraresidue | 295 | 308 | 274 |
| Sequential (|i-j| = 1) | 304 | 307 | 277 |
| Medium/long range (|i-j| > 1) | 353 | 359 | 322 |
| Hydrogen bonds | 306 | 306 | 306 |
| Protein-myristate | 10 | N/A | N/A |
| Total restraints | 1,268 | 1,280 | 1,179 |
| Average restraints per residue | 10.7 | 10.8 | 10.0 |
| Residual Restraint Violations | |||
| CYANA target function (Å2) | |||
| Mean (SD) | 0.075 (0.013) | 0.12 (0.026) | 0.14 (0.059) |
| Maximum violations (Å2) | |||
| Upper limits | 0.0076 (0.0008) | 0.0093 (0.0015) | 0.0071 (0.0024) |
| Lower limits | 0.0053 (0.0005) | 0.0003 (0.0002) | 0.0003 (0.0001) |
| Van der Waals | 1.3 (0.2) | 1.8 (0.3) | 1.9 (0.5) |
| Structure Convergence | |||
| Pairwise rms deviations [(Å2); Mean (SD)] | |||
| Backbone heavy atoms | 1.13 (0.27) | 1.10 (0.34) | 1.27 (0.25) |
| All heavy atoms | 1.83 (0.32) | 1.84 (0.45) | 2.08 (0.43) |
| Ramachandran Analyses | |||
| Most favored regions (%) | 88.8 | 92.2 | 91.4 |
| Additional allowed regions (%) | 8.6 | 7.8 | 8.6 |
| Generously allowed regions (%) | 2.6 | N/A | N/A |
Figure 2.
The NMR structures of the myristoylated and unmyristoylated FIV MA. (a) Overlay of the 20 refined structures calculated for FIV myr(−)MA and (b) FIV MA. Both (a) and (b) are superpositioned based on backbone heavy atoms of the following residues: Arg 7 – Asn 18 (orange), Glu 32 – Thr 46 (yellow), Leu 57 – Phe 74 (green), Ser 77 – Leu 96 (blue), Ala 103 – Met 113 (purple). The orientation of helices and the position of the myristoyl moiety (red) are demonstrated; (c) Residues that compose the hydrophobic pocket to support myristoyl group sequestration; and (d) Ribbon diagram of myr(−)MA (cyan) fitted to the structure of MA, demonstrating that structural differences between the two proteins as observed in the HSQC are localized to the regions that comprise the hydrophobic pocket, where helix I of MA is drawn toward the myristoyl group
Figure 3.
Superposed ribbon diagrams of FIV MA (grey), HIV-1 MA (blue, PDB 2H3I), and HIV-2 MA (green, PDB 2K4H) showing the structural similarity and location of the myristoyl groups (red).
Mutant MANOS protein with canonical myristoylation signal is inefficiently myristoylated in E. coli
We previously showed that felines evolved the same consensus myristoylation signals utilized by other mammals [23, 42]. To understand why FIV did not evolve the same mammalian myristoylation signal, we mutated the native FIV MA/mammalian NMT expression vector such that the N-terminal MA residues that matched those of the feline endothelial nitric oxide synthase, which contains a consensus feline (mammalian) myristoylation sequence (MANOS; M1-G2-N3-L4-K5-S6). Myristoylation efficiency was determined by measurement of relative myr(−)MA backbone amide signal intensities in MA 1H-15N HSQC spectra (Fig. 1a). Whereas wild-type MA produced under these conditions was efficiently myristoylated (~96 %, Table 2), the MANOS chimera containing the consensus mammalian myristoylation signal exhibited significantly lower levels of myristoylation (~ 31 %, Table 2). These findings indicate that the canonical mammalian myristoylation signal is attenuated in the context of the FIV MA protein and suggest that that FIVs may have evolved their rare myristoylation signal to enhance myristoylation efficiency.
Table 2.
Quantitative assessment of FIV MA myristoylation efficiency by mammalian NMT
| Protein | Myristoylation signal | FIV MA myristoylation efficiency (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Average1 | Range | |||||||
| FIV MA | M | G | N | G | Q | G | 96.16 | 86.17 – 99.93 |
| FIV MANOS | M | G | N | L | K | S | 30.60 | 17.31 – 37.73 |
Average and range of the relative amount of myristoylated versus non-myristoylated proteins (%) from a total of three experiments (see text for details).
Reduced exposure and flexibility of the myr(−)MANOS myristoylation signal
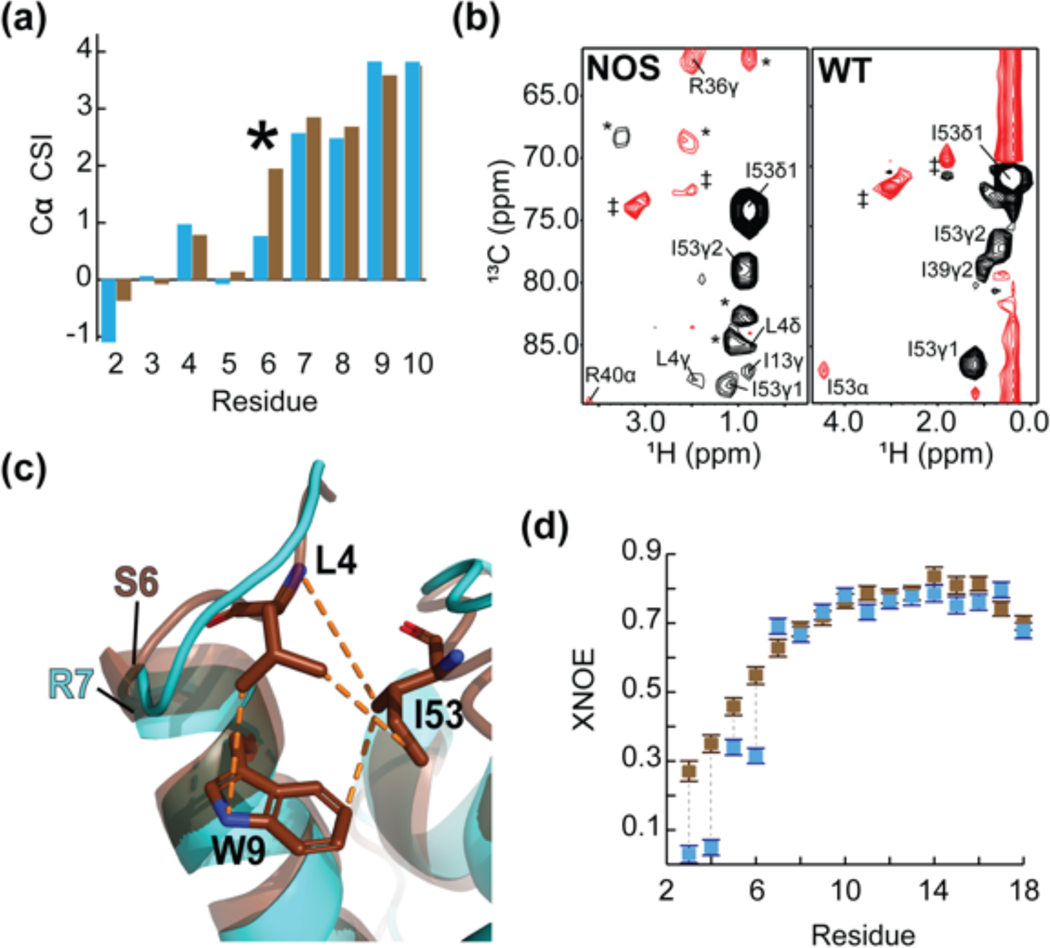
To help understand the variations in observed myristoylation efficiency, NMR and structural studies were conducted with myr(−)MANOS using the same methods and conditions described above for myr(−)MA. Except for a small subset of signals for residues located primarily near the sites of mutation, the 1H-15N HSQC spectrum of myr(−)MANOS closely resembles that of myr(−)MA (Fig. S6. Twenty NOE-derived structures of myr(−)MANOS with mean target functions (0.14 ± 0.059 Å2) exhibited good convergence (pairwise RMS deviations = 1.27 ± 0.25 Å for backbone heavy atoms of residues Gly 2 – Pro 119) (Table 1). The overall structure of myr(−)MANOS closely resembles that of myr(−)MA (Fig. 4). However, several small, but apparently important, differences were observed. First, helix I of myr(−)MANOS (Ser 6 – Asn 18) is one residue longer than that of myr(−)MA (Arg 7 through Asn 18), based on the NOEs and chemical shift indices (Fig. 4a). Second, the side chain of residue Leu 4 exhibited long-range NOEs to core residues in myr(−)MANOS, whereas no long-range NOEs were observed for the side chain nuclei of any residue preceding Arg 7 of myr(−)MA (Fig. 4b). Notably, the NOE data indicate that the side chain of Leu 4 in myr(−)MANOS packs into the hydrophobic cleft that becomes occupied by the myristoyl moiety in MA (Fig. 4c).
Figure 4.
The consensus feline protein myristoylation signal causes sequestration of FIV MA’s N-terminus. (a) A portion of the Cα CSI indicating that the N-termini of wild-type myr(−)MA and myr(−)MANOS are very similar. Consecutive positive deviations from random coil Cα chemical shift values are suggestive of α-helical secondary structure. In myr(−)MA (cyan), Arg 7 leads helix I as opposed to Ser 6 in myr(−)MANOS (brown); (b) Representative 4D CCNOE spectra of the side chain of Ile 53 in myr(−)MANOS and myr(−)MA demonstrate that the Leu 4 side chain of myr(−)MANOS is sequestered and is close to Ile 53 in comparison to the unstructured Gly 4 in the wildtype myr(−)MA. (* = breakthrough peaks from adjacent planes; ‡ = noise); (c) Ribbon diagrams of myr(−)MA (cyan), and myr(−)MANOS (brown) showing the high degree of structural similarity between the two proteins despite differences in the leading residue of helix I and burial of Leu 4 in myr(−)MANOS; and (d) A portion of the XNOE data of myr(−)MA (cyan) and myr(−)MANOS (brown) showing that the amino-terminus (residues Asn 3 – Gly 6) of myr(−)MA is more mobile than that of myr(−)MANOS whereas first helices of both proteins (residues Arg 7- Asn 18) exhibit similar flexibility.
The above findings suggest that differences in myristoylation efficiency in E. coli may be due to differences in accessibility of the myristoylation signal residues by the NMT enzyme. To further test this hypothesis, backbone 1H-15N heteronuclear NOE (XNOE) data were obtained for myr(−)MA and myr(−)MANOS. These data provide information on high-frequency backbone motions and internal mobility, with XNOE values below ~ 0.8 indicative of high rotational flexibility. Although XNOE values for most residues of myr(−)MA and myr(−)MANOS were similar (Fig. S7), values associated with residues Asn 3 through Gly 6 of myr(−)MA were significantly smaller than those for myr(−)MANOS (Fig. 4d), indicating that the N-terminal residues of myr(−)MA are more dynamic than those of myr(−)MANOS.
MANOS mutations impair FIV replication but not particle production
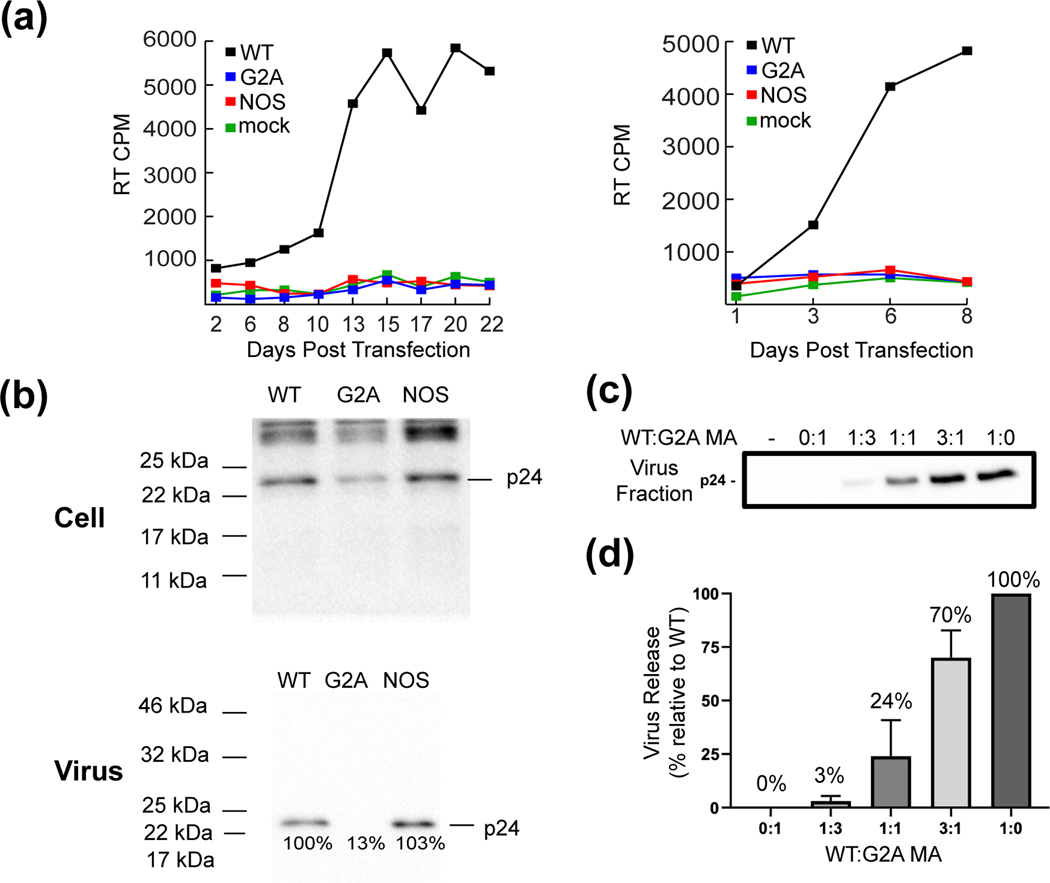
The conservation of a non-canonical myristoylation signal in FIV MA suggests that a canonical mammalian myristoylation signal would be detrimental to viral replication. To test this hypothesis, we examined the ability of FIV molecular clones containing native and mutant myristoylation signals to support viral replication in transfected Crandall feline kidney (CrFK) cells [6]. Studies were conducted with three molecular clones encoding the wild-type MA sequence (positive control), or bearing a myristoylation-deficient mutation that blocks virus assembly (G2A) (negative control), or the MANOS mutation. Replication was assessed by measuring the RT activity in the extracellular supernatant over time. Whereas the molecular clone containing the WT MA sequence replicated efficiently, no replication was detected for the G2A- or NOS-containing clones, even after 22 days posttransfection (Fig. 5a). These data indicate that the NOS myristoylation sequence abrogates virus replication.
Figure 5.
Cell-based assays demonstrate that the MANOS mutant is defective for multi-round virus replication but exhibits WT levels of particle assembly and release. (a) CrFK cells were transfected with MAWT, MANOS, and MAG2A-containing clones. Virus replication was monitored by measuring RT activity in the supernatant. Two independent experiments are shown. (b) CrFK cells were transfected with MAWT, MANOS, and MAG2A-containing clones. Cell- and virion-associated p24(CA) levels were evaluated by western blotting. Similar results were obtained in multiple independent experiments. (c) Titration assays performed via co-transfection of CrFK cells with MAWT and MAG2A-containing clones at the indicated molar DNA ratios. Levels of virion-associated p24(CA) were evaluated by western blotting; blots were quantified in (d) from 4 independent experiments. Error bars indicate SDs.
To test whether the defect in replication exhibited by the MANOS mutant is due to impaired virus assembly, we measured virus release efficiency with the MAWT, MANOS, and MAG2A-containing clones. Particle production was assessed by western blot analysis of p24gag levels in the supernatant 48 h post-transfection. As expected, the G2A substitution blocked particle production, whereas the MANOS chimera supported wild-type levels of particle assembly and release (Fig. 5b). Similar results were obtained in the human 293T cell line (Fig. S8.). To evaluate the possibility that FIV assembly and release might tolerate incomplete Gag myristoylation, we performed assembly assays using cells co-transfected with different ratios of the MAG2A and MAWT molecular clones. The results of this analysis demonstrated that virus release was linearly dependent on the MAWT/MAG2A ratio (Fig. 5c–d); for example, at a 1:1 MAWT/MAG2A ratio, virus release efficiency was 24% that of WT alone, and at a 3:1 ratio it was 70% that of the WT level. These results indicate that FIV assembly and release are highly sensitive to reductions in the efficiency of MA myristoylation. Because the MAWT and MANOS clones promoted virus assembly and release with similar efficiencies, these findings suggest that the MANOS mutant is myristoylated with an efficiency similar to that of the wild-type, despite the prior observation of decreased myristoylation efficiency of MANOS in E. coli (see above).
Discussion
All retroviruses except FIV that require N-terminal Gag myristoylation evolved myristoylation signals similar or identical to those of their hosts, as might be expected since both the viral and host proteins are substrates for the same host NMT enzymes. The FIV myristoylation signal, M-G-N-G-Q-G, is unusual in that the conserved Ser/Thr residue at position 6 is substituted by Gly, and this substitution is conserved among all reported sequences of FIV isolated from domesticated felines [23], despite the fact that myristoylated feline proteins generally contain the consensus mammalian Ser or Thr at this position [41, 42, 46]. The Ser to Gly substitution might be expected to negatively influence substrate binding, and hence myristoylation efficiency, due to loss of intermolecular hydrogen bonding with the Ser (or Thr) side chain. Indeed, in vitro kinetic studies with short peptides indicate that yeast and murine NMTs have similar substrate preferences, both exhibiting optimal binding and catalytic properties with a Ser residue at position 6 [46, 82]. Recent X-ray crystallographic studies showed that octapeptides with different sequences bind with essentially identical extended backbone geometries to the substrate-binding pocket of the Homo sapiens NMT [48]. In these peptide-NMT structures, the backbone and side chain atoms of Ser 6 participate in extensive intermolecular hydrogen bonding [48, 83, 84]. Despite these preferences for a Ser at position 6, we observed significant myristoylation (~ 96%) of the wild-type MA protein in E. coli using our MA/NMT dual expression system. Unexpectedly, the MANOS chimera containing a consensus mammalian myristoylation signal was inefficiently myristoylated (~ 31%) in our E. coli dual MA/NMT expression system. These findings indicate that factors other than simply the NMT recognition sequence can influence myristoylation of FIV MA, at least using our dual expression system in E. coli.
The finding that wild-type MA containing a non-canonical myristoylation signal was efficiently myristoylated in E. coli by a mammalian NMT but the MANOS construct containing a canonical myristoylation signal was not prompted us to conduct NMR studies to determine if the differences in myristoylation efficiencies might be due to differences in protein structure or dynamics. The overall 3D structural architectures determined for the wild-type FIV MA, myr(−)MA, and myr(−)MANOS proteins were similar to each other and to X-ray and NMR structures reported previously for myr(−)MA [43] and a MA mutant that could be myristoylated by the yeast NMT [23]. In both the wild-type and NOS-containing myristoylated MA proteins, the myristoyl group is partially sequestered within a hydrophobic pocket and makes contacts with the side chains of residues W9, F35, I39, I53, E55, L87, L60, and F90. Based on comparisons with the myr(−)MA proteins, it appears that myristate exposure, which would be required for membrane binding, is accompanied by a small shift in the position of helix I. These structural changes are similar to those observed for myristoylated and unmyristoylated forms of the HIV-1 MA protein and consistent with MA myristoyl switch mechanisms proposed for membrane binding by HIV Gag [38, 57].
Although the structures of the myristoylated MA and MANOS proteins are similar, small but significant structural differences were observed for the N-terminal residues of the unmyristoylated myr(−)MA and myr()MANOS proteins that may explain the differences in myristoylation efficiencies in E. coli. Whereas Gly 6 is disordered and does not appear to make long-range contacts in the wild-type protein, Ser 6 is folded within the Nterminal end of helix I of myr(−)MANOS. Chemical shift index analyses indicate that Ser 6 does not adopt the commonly observed N-terminal capping structure [85] but instead exhibits a standard α-helical backbone geometry. In addition, the side chain of residue Leu 4 of myr(−)MANOS packs within the hydrophobic pocket that would normally be occupied by the myristoyl group. Consistent with these findings, 1H-15N heteronuclear NOE data indicate that the distal residues of the myr(−)MANOS myristoylation signal are more dynamically restricted than those of myr(−)MA. Interestingly, the N-terminal residues of the structurally characterized HIV and SIV MA proteins are fully exposed and disordered, despite the fact that these proteins contain a canonical mammalian myristoylation sequence [44, 57, 79]. Although the structure of the endothelial NOS myristoylation sequence is unknown [86, 87], it is likely that its N-terminal residues are also unstructured since known myristoylated mammalian proteins recoverin [88], neurocalcin [89], myristoylated alanine-rich C kinase substrate (MARCKS) [90], and fibroblast growth factor receptor substrate 2 [91] all feature unstructured N-termini. Studies suggest that fewer than 100 amino acid residues are translated before myristoylation occurs [39], and it is thus possible that co-translational folding of the 113-residue globular portion of the FIV MA domain occurs at a rate sufficient to inhibit co-translational N-myristoylation in E. coli.
Although the above studies support a model in which the non-canonical myristoylation signal evolved to promote myristoylation and virus assembly, the virology experiments paint a different picture. The clone containing the canonical (NOS) myristoylation signal was unable to establish a spreading infection, consistent with a critical requirement for the non-canonical, wild-type FIV myristoylation signal. However, virus release experiments revealed that clones containing MAWT and MANOS promoted assembly and release with similar efficiencies. Co-transfection experiments conducted with differing ratios of MAWT and MAG2A-containing clones revealed an inverse relationship between MAG2A/MAWT ratio and particle assembly/release, suggesting that, as expected, assembly and release of FIV Gag is highly sensitive to defects in Gag myristoylation. These experiments collectively indicate that the MAWT- and MANOS-containing Gag proteins are myristoylated with similar efficiencies in transfected feline CrFK cells and that the replication defect observed for the MANOS construct is unrelated to membrane binding by Gag or particle production.
We speculate that the differences in myristoylation efficiencies observed in the E. coli and CrFK assays could be due to potential differences in the NMT isoforms used in E. coli versus CrFK cells. Higher eukaryotes often contain two NMTs, Types I and II, both of which exhibit a high degree of conservation across species [92]. Divergence between Types I and II NMTs exists mainly at their N-termini and well-removed from the catalytic site. These variations do not appear to influence catalytic activity, and it has been suggested that the divergence plays a role in sub-cellular localization [93]. Thus, the isoforms utilized in feline CrFK cells (and human 293T cells) could be targeted to sub-cellular sites that are better optimized for co-translational myristoylation of FIV Gag.
Having established that the NOS sequence does not lead to a myristoylation or assembly/release defect in CrFK or human cells, the replication defect associated with the canonical myristoylation signal must be due to disruption of a downstream function. One possibility is that the N-terminal residues of the wild-type protein contribute to MA-MA or MA-Env interactions that are required for the incorporation of Env into assembling particles. Virions that assemble and bud efficiently but lack the Env glycoprotein would be unable to infect new cells. Although similar phenotypes have been observed for HIV-1 MA mutants, the substitutions responsible for defects in Env incorporation involved residues known to be important for MA trimerization or higher-order oligomerization when bound to membranes (e.g., residues 62 and 74) or were located around a central aperture in the hexameric Gag lattice (e.g., residues 12, 30 and 34) [26–33]. In this regard, it is notable that myr(−)MA proteins of the lentiviruses HIV-1 and SIV contain similar trimer interfaces in high-resolution crystal structures [73, 74, 78, 79] whereas the equine infectious anemia virus (EIAV) and FIV myr(−)MA protein crystalize with dimeric intermolecular interfaces, despite adopting a similar overall fold [43]. The intermolecular interface in the FIV dimer involves unstructured residues of the C-terminal tail of one molecule and the globular portion of the adjacent molecule, and the relative orientations of the two molecules appear incompatible with formation of the immature Gag lattice. The intermolecular interfaces observed in all myr(−)MA X-ray crystal structures involve relatively small surface areas, and all retroviral myr(−)MA proteins examined in solution by NMR are predominantly monomeric, even at relatively high protein concentrations (~0.5 mM). It is thus conceivable that FIV MA may also adopt a trimeric structure when it associates with the membrane. Importantly, residues of the N-terminal myristoylation signal of FIV and HIV MA are well removed from sites that promote trimerization of HIV MA. Although it remains possible that the non-canonical myristoylation signal evolved to promote a MA oligomerization structure that supports Env incorporation, the interactions and models would have to deviate from those established for HIV.
A third possibility is that the non-canonical FIV MA myristoylation signal evolved to support functions after the virus enters a new host cell. In this regard, studies have implicated HIV-1 MA in an undefined but critical post-entry activity [34]. Proposals that MA plays a role in nuclear localization of the viral capsid [94–96] have largely been refuted [97–99], but other potential functions warrant examination. Studies to test these hypotheses are underway.
Materials and Methods
Plasmid cloning to prepare FIV MA constructs
The previously described FIV MA/yeast NMT co-expression vector was used as a template to PCR amplify the FIV MA gene and polyhistidine affinity tag for construction of FIV MA plasmids using the 5ʹ-primer CGCGCGCCATGGGCAATGGACAGGGGCGAGATTGGA (NcoI site underlined) and the 3ʹ-primer CGCGCGGGATCCCTCGAGTTAATGATGATGATGATG (BamHI and stop codon underlined). The product from PCR amplification was digested using NcoI and BamHI and ligated by T4 DNA ligase into a pET Duet dualexpression plasmid (Novagen, Madison, WI, USA) for preparation of FIV MA or the single-expression vector pET-19b for synthesis of FIV myr(−)MA. A pUC57 cloning vector containing the Escherichia coli codon optimized gene for human NMT type 1 (hNMT, accession number NP_066565) was purchased from Genewiz, Inc. (South Plainfield, NJ, USA), PCR amplified using the 5ʹ-primer CGCGCGCATATGGCCGACGAGAGTGAAACCGCAGTT (NdeI site underlined) and the 3ʹ-primer GCGCGCGGCCCGCCTAGGTTATTGTAAAACCAGACCCACTTTTTC (AvrII site and stop codon underlined), digested, and ligated into the pET Duet vector in order to myristoylate native FIV MA.
To improve FIV MA solubility, site-directed mutagenesis was performed on the FIV MA gene such that the 3ʹ nucleotides which code for amino acid residues 120–135 were removed using QuikChange XL Kit according to the manufacturer’s instructions (Stratagene, La Jolla, CA, USA). The pET-Duet-1 and pET-19b vectors containing the truncated MA gene were subjected to site directed mutagenesis for preparation of the MANOS variant. All constructs were sequenced through Genewiz, Inc. (South Plainfield, NJ, USA). Sample purity and myristoylation efficiency were established by mass spectrometry, gel electrophoresis, and NMR.
Protein expression
The respective vectors for the expression of FIV MA constructs were transformed into BL21 DE3(RIL) E. coli cells (Agilent Technologies). A starter culture of 200 mL of Lysogeny Broth medium (LB) was inoculated to prepare natural abundance samples and grown at 37 °C overnight. The starter culture was added to 2 L LB and grown at 37 °C until the OD600 reached ~ 0.6–0.7 at which point protein expression was induced with isopropyl-β-D-1-thiogalactoside (IPTG, Sigma Aldrich) which was added to a final concentration of 1 mM. The cells shook for 4 hours at 37 °C before centrifugation to harvest.
For isotopically enriched samples, 7 mL of Luria Broth medium were inoculated and grown at 37 °C overnight. Next, the starter culture was used to inoculate 200 mL of M9 minimal medium containing 15N-NH4Cl and/or 13C-glucose (Cambridge Isotope, Tewksbury, MA, USA) as the sole nitrogen and carbon source for 24 hours then transferred to 2 L of M9 isotopically enriched minimal medium. The cells were incubated at 37 °C until the OD600 reached between 0.6–0.7 before induction with 1 mM IPTG (Sigma Aldrich). Cells expressing the 15N and/or 13C labeled samples were incubated at 30 °C for 16–18 hours before harvesting by centrifugation. For samples that required incorporation of exogenous, natural abundance myristate, 50 mg/L myristoyl coenzyme A was added once the OD600 reached ~ 0.2–0.3.
The cell pellets were resuspended in 50 mM sodium phosphate pH 8.0, 250 mM NaCl, 5 mM TCEP and lysed using a microfluidizer (Microfluidics, Westwood, MA, USA). The lysate was clarified by centrifugation and proteins were purified by cobalt affinity chromatography (GE Healthcare, Issaquah, WA, USA). The eluted proteins were dialyzed against 50 mM sodium phosphate pH 8.0 with 10 mM DTT and further purified by cation exchange column chromatography (GE Life Science, Piscataway, NJ, USA) using an increasing gradient of NaCl. The purified protein samples were dialyzed into 50 mM sodium phosphate pH 8.0 with 10 mM DTT and analyzed by electrospray ionization mass spectrometry in order to confirm the molecular weights prior to NMR analysis [myr(−)MA Mmeas = 13,686.5 ± 1.7 Da, Mcalc = 13,689 Da; MA Mmeas = 14,172.5 ± 0.6 Da, Mcalc = 14,173 Da, ~ 95% myristoylated] (David King, UC Berkeley, or Molecular Characterization and Analysis Complex, UMBC).
NMR spectroscopy
Samples for NMR experiments on MA, myr(−)MA, and myr(−)MANOS were prepared in 50 mM sodium phosphate, 10 mM DTT, 5 mM sodium chloride, pH 8.0. A subset of myr(−)MA and myr(−)MANOS data were acquired using 50 mM sodium phosphate, 10 mM DTT, 5 mM sodium chloride, pH 5.0. All structural NMR data were collected using ~ 2.5 mg protein. NMR spectroscopy data were acquired at 25°C on Bruker 600 MHz spectrometers equipped with a cryogenic probe or a 500 MHz Bruker DMX spectrometer equipped with a room temperature probe, processed using NMRPipe/nmrDraw or NMRFx [100, 101], and analyzed with NMRViewJ [102, 103]. A combination of two-, three-, and four-dimensional NOESY data were collected for combinations of natural abundance, 15N-, 13C-, and 15N,13C-labeled protein samples. Standard triple resonance approaches were employed to assign the protein backbone signals. Myristoyl moiety chemical shifts were assigned by comparison of 1H-13C HMQC spectra of 13C-enriched FIV MA with an unlabeled myristoyl moiety to 13C-labeled FIV MA and 13C-labeled myristate. NOEs between the myristoyl group and MA were assigned using 2D 1H-1H NOESY and 4D NOESY datasets.
NMR myristoylation efficiency measurement
The 1H-15N HSQC spectra of MA show a strong, well-resolved signal for Arg 40 and a weak signal corresponding to Arg 40 of myr(−)MA. The intensities of FIV MA and myr(−)MA spectra were normalized according to the amide signal of Phe 74, and the relative intensity of the myr(−)MA Arg 40 signal was measured in order to determine the myristoylation efficiency of FIV MA samples.
Structure calculations
CYANA was used to calculate structures in torsion angle space [104]. Upper interproton distance bounds of 2.7, 3.3 and 5.0 Å (with appropriate corrections for pseudoatoms) were employed for NOE cross-peaks of strong, medium, and weak intensity, respectively. PyMOL was used to prepare structure figures [105].
Replication Kinetics
Transfection of CrFK cells was carried out in a 12-well plate using the FIV Orf2rep clone [106]. Cells were transfected using lipofectamine 2000 reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. Cells were split 1:3 every 2–3 days and supernatants were collected to monitor spreading replication via RT activity using methods described for HIV-1 [107].
Virus Release Assay
CrFK cells were plated in a 12-well plate and transfected according to the lipofectamine 2000 protocol (Invitrogen, Waltham, MA, USA). Cells were transfected with wild-type, G2A, and NOS clones. A titration was performed by transfecting different ratios of wild-type (WT):G2A MA (untransfected, G2A alone, 1WT:3G2A, 1WT:1G2A, 3WT:1G2A, and WT alone). Cell and virus lysate were collected 48-hour post-transfection. Virus release was analyzed via western blot analysis using mouse anti-FIV p24gag (clone PAK3–2C1). Virus release efficiency was calculated as the amount of virion-associated Gag as a fraction of total (cell plus virion) Gag, normalized to samples containing WT only.
Database Depositions
Atomic coordinates and NMR chemical shifts/structure refinement parameters have been deposited in the RSCB and BMRB databases, respectively: FIV MA (6WA3, 30738); myr(−)MA (6WA4, 30739); and myr()MANOS (6WA5, 30740).
Supplementary Material
Highlights.
First structure of myristoylated FIV MA containing the native (non-canonical) myristoylation signal.
Mutant with canonical feline myristoylation signal is inefficiently myristoylated in E. coli.
Canonical signal is partially sequestered and less dynamic than that of the wild-type MA protein.
Canonical signal supports virus assembly and budding but not spreading replication.
Myristoylation signal residues implicated in non-assembly replication functions.
Acknowledgements
This work was supported by NIH grant 8R01 AI50498 (to MFS). JB was supported by an NIH grant graduate training at the chemistry biology interface (T32GM066706), LB was supported by an NSF pre-doctoral grant DGE-1144243, and LB and JB were supported by an NIH grant that promotes doctoral diversity (IMSD R25 GM-055036). CN was supported by an NSF REU grant 1460653 (to UMBC). We thank the Howard Hughes Medical Institute (HHMI) staff at University of Maryland Baltimore County (UMBC), Dr. Joshua Wilhide and Ian Shaffer (UMBC), and Dr. David King (University of California, Berkeley) for technical support. Research in the Freed laboratory is supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH.
References
- [1].Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–3. [DOI] [PubMed] [Google Scholar]
- [2].Dias AS, Bester MJ, Britz RF, Apostolides Z. Animal models used for the evaluation of antiretroviral therapies. Curr HIV Res. 2006;4:431–46. [DOI] [PubMed] [Google Scholar]
- [3].Bendinelli M, Pistello M, Lombardi S, Poli A, Garzelli C, Matteucci D, et al. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin Microbiol Rev. 1995;8:87–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Elder JH, Lin YC, Fink E, Grant CK. Feline immunodeficiency virus (FIV) as a model for study of lentivirus infections: parallels with HIV. Curr HIV Res. 2010;8:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Luttge BG, Freed EO. FIV Gag: virus assembly and host-cell interactions. Veterinary immunology and immunopathology. 2010;134:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Luttge BG, Shehu-Xhilaga M, Demirov DG, Adamson CS, Soheilian F, Nagashima K, et al. Molecular characterization of Feline Immunodeficiency Virus budding J Virol. 2007;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burkhard M, Dean G. Transmission and Immunopathogenesis of FIV in Cats as a Model for HIV. Current HIV Research. 2003;1:15–29. [DOI] [PubMed] [Google Scholar]
- [8].Pedersen NC, Yamamoto JK, Ishida T, Hansen H. Feline immunodeficiency virus infection. Veterinary immunology and immunopathology. 1989;21:111–29. [DOI] [PubMed] [Google Scholar]
- [9].Hatziioannou T, Evans DT. Animal models for HIV/AIDS research. Nat Rev Microbiol. 2012;10:852–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Egberink H, Borst M, Niphuis H, Balzarini J, Neu H, Schellekens H, et al. Suppression of feline immunodeficiency virus infection in vivo by 9-(2-phosphonomethoxyethyl)adenine. Proc Natl Acad Sci U S A. 1990;87:3087–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hartmann K, Donath A, Beer B, Egberink H, Horzinek M, Lutz H, et al. Use of two virustatica (AZT, PMEA) in the treatment of FIV and of FeLV seropositive cats with clinical symptoms. Veterinary immunology and immunopathology. 1992;35:167–75. [DOI] [PubMed] [Google Scholar]
- [12].Yamamoto JK, Sanou MP, Abbott JR, Coleman JK. Feline immunodeficiency virus model for designing HIV/AIDS vaccines. Curr HIV Res. 2010;8:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wongsrikeao P, Saenz D, Rinkoski T, Otoi T, Poeschla E. Antiviral restriction factor transgenesis in the domestic cat. Nature methods. 2011;8:853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kenyon JC, Lever A. The molecular biology of feline immunodeficiency virus (FIV). Viruses. 2011;3:2192–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Elder JH, Sundstrom M, de Rozieres S, de Parseval A, Grant CK, Lin Y-C. Molecular mechanisms of FIV infection. Veterinary immunology and immunopathology. 2008;123:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Manrique ML, Celma CC, Gonzalez SA, Affranchino JL. Mutational analysis of the feline immunodeficiency virus matrix protein. Virus research. 2001;76:103–13. [DOI] [PubMed] [Google Scholar]
- [17].Cerfoglio JCA, Gonzalez SA, Affranchino JL. Structural elements in the Gag polyprotein of feline immunodeficiency virus involved in Gag self-association and assembly. J Gen Virol. 2014;95:2050–9. [DOI] [PubMed] [Google Scholar]
- [18].Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zimmerman C, Klein KC, Kiser PK, Singh AR, Bonnie FL, Riba SC, et al. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. [DOI] [PubMed] [Google Scholar]
- [21].Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. [DOI] [PubMed] [Google Scholar]
- [22].Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci U S A. 2006;103:2641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brown LA, Cox C, Baptiste J, Summers H, Button R, Bahlow K, et al. NMR structure of the myristylated feline immunodeficiency virus matrix protein. Viruses. 2015;7:2210–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dick RA, Goh SL, Feigenson GW, Vogt VM. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci U S A. 2012;109:18761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Alfadhli A, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol. 2009;83:12196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Freed OE, Martin AM. Virion Incorporation of Envelope Glycoproteins with Long but Not Short Cytoplasmic Tails Is Blocked by Specific, Single Amino Acid Substitutions in the Human Immunodeficiency Virus Type 1 Matrix. Journal of Virology. 1995;69:1984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Freed EO, Martin AM. Domains of the Human Immonodeficiency Virus Type 1 Matrix and gp41 Cytoplasmic Tail Required for Envelope Incorporation into Virions. Journal of Virology. 1996;70:341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ono A, Huang M, Freed EO. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol. 1997;71:4409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kiernan RE, Ono A, Freed EO. Reversion of a Human Immunodeficiency Virus Type 1 Matrix Mutation Affecting Gag Membrane Binding, Endogenous Reverse Transriptase Activity, and Virus Infectivity. Journal of Virology. 1999;73:4728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Murakami T, Freed E. Genetic Evidence for an Interaction between Human Immunodeficiency Virus Type 1 Matrix and α-Helix 2 of the gp41 Cytoplasmic Tail. J Virol. 2000;74:3548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tedbury PR, Ablan SD, Freed EO. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog. 2013;9:e1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tedbury PR, Novikova M, Ablan SD, Freed EO. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc Natl Acad Sci U S A. 2016;113:E182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tedbury PR, Novikova M, Alfadhli A, Hikichi Y, Kagiampakis I, KewalRamani VN, et al. HIV-1 Matrix Trimerization-Impaired Mutants Are Rescued by Matrix Substitutions That Enhance Envelope Glycoprotein Incorporation. J Virol. 2019;94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kiernan RE, Ono A, Englund G, Freed EO. Role of Matrix in an Early Postentry step in the Human Immunodeficiency Virus Type 1 Life Cycle. Journal of Virology. 1998;72:4116–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lindwasser OW, Resh MD. Myristoylation as a target for inhibiting HIV assembly: Unsaturated fatty acids block viral budding. Proc Natl Acad Sci USA. 2002;99:13037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Freed EO, Englund G, Martin AM. Role of the Basic Domain of Human Immondeficiency Virus Type 1 Matrix in Macrophage Infection. Journal of Virology. 1995;69:3949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ono A, Freed EO. Binding of Human Immunodeficiency Virus Type 1 gag to membrane: Role of the matrix amino terminus. J Virol. 1999;73:4136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mercredi PY, Bucca N, Loeliger B, Gaines CR, Mehta M, Bhargava P, et al. Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein. J Mol Biol. 2016;428:1637–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Deichaite I, Casson LP, Ling HP, Resh MD. In vitro synthesis of pp60v-src: Myristylation in a cellfree system. Mol Cell Biol. 1988;8:4295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wilcox C, Hu J-S, Olson EN. Acylation of Proteins with Myristic Acid Occurs Cotranslationally. Science. 1987;238:1275–8. [DOI] [PubMed] [Google Scholar]
- [41].Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochimica et biophysica acta. 1999;1451:1–16. [DOI] [PubMed] [Google Scholar]
- [42].Resh MD. Covalent lipid modifications of proteins. Current biology : CB. 2013;23:R431–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Serriere J, Robert X, Perez M, Gouet P, Guillon C. Biophysical characterization and crystal structure of the Feline Immunodeficiency Virus p15 matrix protein. Retrovirology. 2013;10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, et al. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382:434–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Towler DA, Eubanks SR, Towery DS, Adams SP, Glaser L. Amino-terminal Processing of Proteins by N-Myristoylation. J Biol Chem. 1987;262:1030–6. [PubMed] [Google Scholar]
- [46].Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, et al. Myristyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J Biol Chem. 1988;263:1784–90. [PubMed] [Google Scholar]
- [47].Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Castrec B, Dian C, Ciccone S, Ebert CL, Bienvenut WV, Le Caer JP, et al. Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern. Nature chemical biology. 2018. [DOI] [PubMed] [Google Scholar]
- [49].Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amideproton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. Journal of Magnetic Resonance. 1993;Series B 101:201–5. [Google Scholar]
- [50].Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–8. [Google Scholar]
- [51].Grzesiek S, Bax A. The Importance of Not Saturating H2O in Protein NMR. Application to Sensitivity Enhancement and NOE Measurement. J Am Chem Soc. 1993;115:12593–4. [Google Scholar]
- [52].Muhandiram DR, Kay L. J Magn Reson Ser B. 1994;103:203. [Google Scholar]
- [53].Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. Journal of Magnetic Resonance. 1992;96:432–40. [Google Scholar]
- [54].Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. Journal of Magnetic Resonance. 1992;99:201–7. [Google Scholar]
- [55].Kay LE, Xu GY, Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. Journal of Magnetic Resonance, Series A. 1994;109:129–33. [Google Scholar]
- [56].Clubb RT, Thanabal V, Wagner G. A constant-time three-dimensional triple-resonance pulse scheme to correlate intraresidue 1HN, 15N, and 13C’ chemical shifts in 15N-13C-labeled proteins. J Magn Reson. 1992;97:213–7. [Google Scholar]
- [57].Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci USA. 2004;101:517–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].O’Connell NE, Lelli K, Mann RS, Palmer AG. Asparagine deamidation reduces DNA-binding affinity of the Drosophila melanogaster Scr homeodomain. FEBS Lett. 2015;589:3237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Robinson AB, McKerrow JH, Cary P. Controlled deamidation of peptides and proteins: An experimental hazard and a possible biological timer. Proc Natl Acad Sci U S A. 1970;66:753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Robinson NE, Robinson AB. Molecular Clocks. Proc Natl Acad Sci U S A. 2001;98:944–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Robinson NE, Robinson AB. Prediction of protein deamidation rates from primary and threedimensional structure. Proc Natl Acad Sci U S A. 2001;98:4367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Robinson NE, Robinson ZW, Robinson BR, Robinson AL, Robinson JA, Robinson ML, et al. Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J Pept Res. 2004;63:426–36. [DOI] [PubMed] [Google Scholar]
- [63].Flatmark T On the heterogeneity of beef heart cytochrome c. 3: A kinetic study of the nonenzymatic deamidation of the main subfractions (Cy I-Cy III). Acta Chemistry Scand A. 1966;20:1487–96. [DOI] [PubMed] [Google Scholar]
- [64].Tyler-Cross R, Schirch V. Effects of amino acid sequence, buffers, and ionic strength on the rate and mechanism of deamidation of asparagine residues in small peptides. J Biol Chem. 1991;266:22549–56. [PubMed] [Google Scholar]
- [65].Hao P, Ren Y, Alpert AJ, Sze SK. Detection, evaluation and minimization of nonenzymatic deamidation in proteomic sample preparation. Mol Cell Proteomics. 2011;10:O111.009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kay LE, Clore GM, Bax A, Gronenborn AM. Four-dimensional heteronuclear triple-resonance NMR spectroscopy of interleukin-1 beta in solution. Science. 1990;249:411–4. [DOI] [PubMed] [Google Scholar]
- [67].Kay LE, Ikura M, Bax A. Proton-proton correlation via carbon-carbon couplings: a threedimensional NMR approach for the assignment of aliphatic resonances in proteins labeled with carbon13. J Am Chem Soc. 1990;112:888–9. [Google Scholar]
- [68].Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, et al. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1b. Biochemistry. 1989;28:6150–6. [DOI] [PubMed] [Google Scholar]
- [69].Vuister GW, Clore GM, Gronenborn AM, Powers R, Garrett DS, Tschudin R, et al. Increased Resolution and Improved Spectral Quality in Four-Dimensional 13C/13C-Separated HMQC-NOESYHMQC Spectra Using Pulsed Field Gradients. Journal of Magnetic Resonance, Series B. 1993;101:210.3. [Google Scholar]
- [70].Christensen AM, Massiah MA, Turner BG, Sundquist WI, Summers MF. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J Mol Biol. 1996;264:1117–31. [DOI] [PubMed] [Google Scholar]
- [71].Conte MR, Klikova M, Hunter E, Ruml T, Matthews S. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. The EMBO journal. 1997;16:5819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Conte MR, Matthews S. Retroviral Matrix Proteins: A Structural Perspective. Virology. 1998;246:191–8. [DOI] [PubMed] [Google Scholar]
- [73].Hatanaka H, Iourin O, Rao Z, Fry E, Kingsman A, Stuart DI. Structure of the Equine Infectious Anemia Virus Matrix protein. J Virol. 2002;76:1876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal Structures of the Trimeric HIV-1 Matrix Protein: Implications for Membrane Association. Proceedings of the National Academy of Sciences USA. 1996;93:3099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. Three dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. [DOI] [PubMed] [Google Scholar]
- [76].Massiah MA, Worthylake D, Christensen AM, Sundquist WI, Hill CP, Summers MF. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: Evidence for conformational changes during viral assembly. Protein Sci. 1996;5:2391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Matthews S, Barlow P, Boyd J, Barton G, Russell R, Mills H, et al. Structural similarity between the p17 matrix protein of HIV-1 and interferon-g. Nature (London). 1994;370:666–8. [DOI] [PubMed] [Google Scholar]
- [78].Matthews S, Barlow P, Clark N, Kingsman S, Kingsman A, Campbell I. Refined solution structure of p17, the HIV matrix protein. Biochem Soc Trans. 1995;23:725–8. [DOI] [PubMed] [Google Scholar]
- [79].Rao Z, Belyaev AS, Fry E, Roy P, Jones IM, Stuart DI. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–7. [DOI] [PubMed] [Google Scholar]
- [80].Riffel N, Harlos K, Lourin O, Rao Z, Kingsman A, Stuart D, et al. Atomic Resolution Structure of Moloney Leukemia virus Matrix protein and Its Relationship to Other Retroviral Matrix Proteins. Structure. 2002;10:1627–36. [DOI] [PubMed] [Google Scholar]
- [81].Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, et al. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol. 2007;366:574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, et al. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc Natl Acad Sci U S A. 1987;84:2708–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–4. [DOI] [PubMed] [Google Scholar]
- [84].Farazi TA, Waksman G, Gordon JI. Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis. Biochemistry. 2001;40:6335–43. [DOI] [PubMed] [Google Scholar]
- [85].Gronenborn AM, Clore GM. Identification of N-terminal helix capping boxes by means of 13C chemical shifts. Journal of biomolecular NMR. 1994;4:455–8. [DOI] [PubMed] [Google Scholar]
- [86].Rosenfeld RJ, Garcin ED, Panda K, Andersson G, Åberg A, Wallace AV, et al. Conformational changes in nitric oxide synthases induced by chlorozoxazone and nitroindazoles: Crystallographic and computational analyses of inhibitor potency. Biochemistry. 2002;41. [DOI] [PubMed] [Google Scholar]
- [87].Li H, Jamal J, Plaza C, Pineda SH, Chreifi G, Jing Q, et al. Structures of human constituitive nitric oxide synthases. Acta Crystallog D Biol Crystallogr. 2014;70:2667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ames JB, Levay K, Wingard JN, Lusin JD, Slepak VZ. Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J Biol Chem. 2006;281:37237–45. [DOI] [PubMed] [Google Scholar]
- [89].Vijay-Kumar S, Kumar VD. Crystal structure of recombinant bovine neurocalcin. Nature. 1999;6:80–8. [DOI] [PubMed] [Google Scholar]
- [90].Yamauchi E, Nakatsu T, Matsubara M, Kato H, Taniguchi H. Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca2+-calmodulin. Nature. 2003;10:226–31. [DOI] [PubMed] [Google Scholar]
- [91].Zeng L, Kuti M, Mujtaba S, Zhou M-M. Structural insights into FRS2α PTB domain recognition by neurotrophin receptor TrkB. Proteins: Struct, Funct, Bioinf. 2014;82:1534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Giang DK, Cravatt BF. A second mammalian N-myristoyltransferase. J Biol Chem. 1998;273:6595–8. [DOI] [PubMed] [Google Scholar]
- [93].Glover CJ, Hartman KD, Felsted RL. Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J Biol Chem. 1997;272:28680–9. [DOI] [PubMed] [Google Scholar]
- [94].Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubei A, Spitz L, et al. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–88. [DOI] [PubMed] [Google Scholar]
- [96].Gallay P, Swingler S, Song J, Bishman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–76. [DOI] [PubMed] [Google Scholar]
- [97].Freed EO, Martin MA. HIV-1 Infection of Non-Dividing Cells. Nature. 1994;369:107–8. [DOI] [PubMed] [Google Scholar]
- [98].Fouchier RAM, Meyer BE, Simon JHM, Fischer U, Malim MH. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Freed EO, Englund G, Maldarelli F, Martin MA. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell. 1997;88:171–3. [DOI] [PubMed] [Google Scholar]
- [100].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. [DOI] [PubMed] [Google Scholar]
- [101].Norris M, Fetler B, Marchant J, Johnson BA. NMRFx Processor: a cross-platform NMR data processing program. Journal of biomolecular NMR. 2016:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. [DOI] [PubMed] [Google Scholar]
- [103].Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J Biomol NMR. 1994;4:603–14. [DOI] [PubMed] [Google Scholar]
- [104].Güntert P Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–78. [DOI] [PubMed] [Google Scholar]
- [105].DeLano WL. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- [106]. Waters AK, De Parseval AP, Lerner DL, Neil JC, Thompson FJ, Elder JH. Influence of ORF2 on Host Cell Tropism of Feline Immunodeficiency Virus. Virology. 1996;215:10–6. [DOI] [PubMed] [Google Scholar]
- [107].Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, et al. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.