Abstract
Background
The new coronavirus (SARS-CoV-2), which has been responsible for the recent coronavirus disease 2019 (COVID-19) pandemic, uses the cell receptor angiotensin converting enzyme-2 (ACE2) for entry and the serine protease TMPRSS2 for spike (S) protein priming. Meanwhile, the presence of B0AT1 (SLC6A19) may prevent TMPRSS2 from accessing the cutting position on ACE2. Identifying the expression of these cell receptor-related genes of SARS-CoV-2 is critical for understanding the pathogenesis of SARS-CoV-2 in various tissues, especially in the kidney.
Methods
The single-cell transcription datasets of the human cell landscape (HCL) and other relevant single-cell transcription databases were used to analyze the expression of ACE2, TMPRSS2, and SLC6A19 in various organs and tissues, but mainly in the kidney.
Results
ACE2 was significantly expressed in the S1, S2, and S3 segments of proximal tubule (PT) cells. TMPRSS2 was widely expressed in several renal tubule populations extending from the PT cells to the collection system cell type, of which intercalated cells and the distal convoluted tubule cells showed more significant expression than PT cells. Unexpectedly, although expressed on various renal tubule populations, SLC6A19 was mainly enriched in PT cells, in line with ACE2 expression. Although alveolar-type (AT) 2 cells of the lung and intestinal epithelial cells expressed ACE2 as well as PT cells, AT 2 cells significantly expressed TMPRSS2 but not SLC6A19, while all 3 genes were significantly expressed in intestinal epithelial cells.
Conclusions
ACE2 was widely expressed in specific cell subgroups of various human tissues, especially in intestinal epithelial cells, kidney PT cells, and also AT 2 cells of the lung. These 3 types of cells demonstrated significant differences in the distribution of the cell receptor-related genes of SARS-CoV-2, which may indicate the diversity of cell surface structures and differences in the affinity between SARS-CoV-2 and cells.
Keywords: SARS-CoV-2, Single-cell transcriptome sequencing, Proximal tubule cells, Angiotensin-converting enzyme-2
Introduction
The rapid national and international spread of the disease caused by SARS-CoV-2 (COVID-19) has posed a global health threat and has infected millions of people [1]. Although SARS-CoV-2 is closely related to SARS-CoV in gene sequence (about 79% identity), it has no evolutionary relationship with SARS-CoV and belongs to the subgenus sarbecovirus of the genus betacoronavirus [2, 3]. The spike (S) protein of SARS-CoV-2, composed of the amino N-terminal receptor-binding S1 and carboxyl C-terminal membrane fusion S2 subunits, mediates virus entry into target cells [4, 5]. During viral infection, the S protein is cleaved into S1 and S2 subunits, and the S1 subunit contains the receptor-binding domain, which directly binds to the peptidase domain of ACE2 [4, 6]. When the receptor-binding domain binds to the ACE2, another cleavage site on S2 is exposed and cleaved by host proteases, which is critical for viral infection [7, 8, 9]. Hoffmann et al. [10] have documented that host cell entry of SARS-CoV-2 depends on ACE2 and can be blocked by the cellular type II transmembrane serine protease TMPRSS2. The arginine and lysine residues in amino acids 697–716 of ACE2 are essential for cleavage by TMPRSS2, and ACE2 processing is necessary for augmentation of SARS-S-driven entry by these proteases [8]. The cryo-electron-microscopic structure of full-length human ACE2 has been resolved in the presence of the amino acid transporter B0AT1 (also known as SLC6A19), and it is found that residues 697–716 form the 3rd and 4th helices in the neck domain [11]. The ACE2-B0AT1 exists as a dimer of heterodimers, and each B0AT1 interacts with the neck in the adjacent ACE2 [11]. This structure suggests that the presence of B0AT1 (SLC6A19) may prevent TMPRSS2 from accessing the cutting position on ACE2. Hence, according to the present studies, SARS-CoV-2 may infect target cells through synaptic effects of viral spike protein, ACE2, and TMPRSS2 while inhibited by B0AT1.
Cell subgroups expressing ACE2 may be SARS-CoV-2-targeted cells, and the co-expression of TMPRSS2 indicates that they may have a direct risk of infection. Previous studies have found that ACE2 is strongly expressed in the renal tubules [12], and recent single-cell transcriptome analyses have revealed that proximal tubule (PT) cells significantly express ACE2 [13]. Yet, the incidence of acute kidney injury (AKI) in COVID-19 patients varies in various studies [14, 15, 16, 17, 18]. Meanwhile, there is still debate about the presence of viruses in the urine [19, 20, 21]. SARS-CoV-2 entry into target cells is an elegantly regulated multistep processes, of which binding to ACE2 is only the first. Identifying the expression of cell receptor-related genes of SARS-CoV-2 in the kidney is critical for understanding the pathogenesis of SARS-CoV-2. In this study, we analyze the expression of genes related to SARS-CoV-2 entry into target cells through single-cell transcriptome databases in order to objectively reflect the relationship between kidney cells and SARS-CoV-2.
Concise Methods
Analysis of ACE2 Expression in Human Organs and Cell Types
We queried the NCBI website to obtain ACE2 bulk expression data among human organs. We used 3 different databases (https://www.ncbi.nlm.nih.gov/gene/?term = 59272): human protein atlas RNA-seq normal tissue, RNA sequencing of total RNA from 20 human tissues, and the Illumina BodyMap 2 transcriptome (online suppl. Fig. 1a–c; for all online suppl. material, see www.karger.com/doi/10.1159/000508162).
The human cell landscape (HCL) single-cell transcriptome sequencing datasets were clustered into cell types using the annotation provided online [22]. The database is maintained by a single laboratory through the microwell-seq experimental method that essentially covers most tissues and organs of the human body [23]. After normalization and scaling the expression data, we further compared the mean expression of ACE2 among the HCL cell types and showed the top 50 cell types (those with highest ACE2 expression) in the t-distributed stochastic neighbor embedding plot according to the organs and cell types (Fig. 1a).
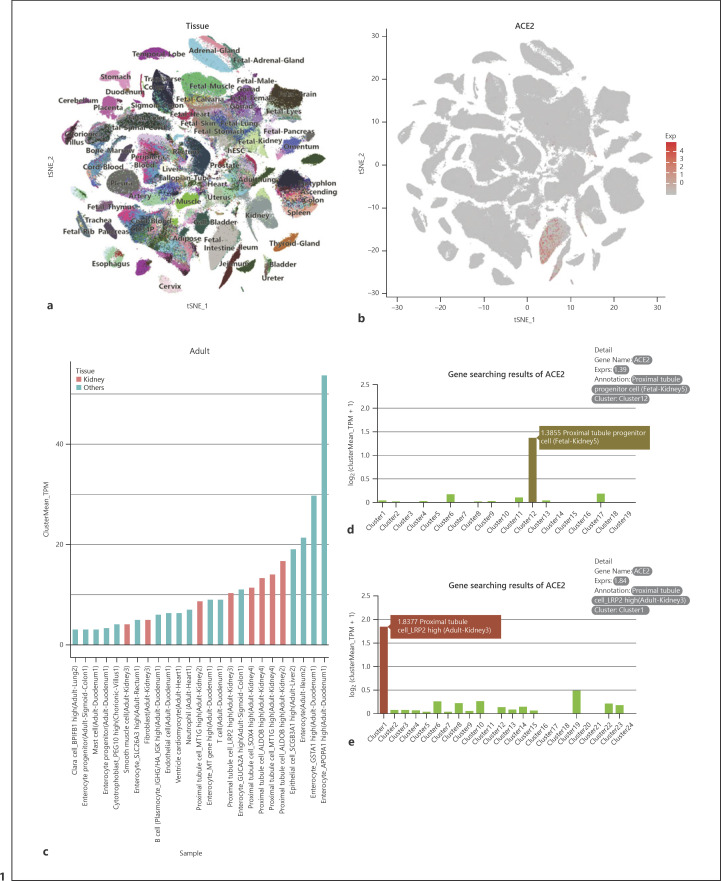
Fig. 1.
ACE2 expression in human cell landscape (HCL). a Two-dimensional t-distributed stochastic neighbor embedding plot displaying major cell clusters in HCL. b Scatter plots showing clusters of cells with ACE2 expression. c Histograms of ACE2 expression in adult tissues in HCL. d, e Histograms represent a subgroup of kidney cells that significantly express ACE2 in “adult kidney−3” (d) and in “fetal kidney-5” (e) in HCL, respectively (reproduced from http://bis.zju.edu.cn/HCL/index.html).
Gene Expression Analysis in Kidney Single-Cell Transcriptome Sequencing Data Sets
Using the GSE140989 database with the 10X genomic single-cell RNA sequencing (scRNA-seq) protocol, combined processing of 24 samples, including 16 tumor nephrectomy, 5 surveillance, and 3 preperfusion biopsies, yielded 22,268 cells (4,690 from surveillance biopsies, 16,491 from tumor nephrectomies, and 2,834 cells from preperfusion biopsies) [24]. The GSE121862 database is a combination of data from different experiments and institutions, and studies have suggested that single-nuclear transcriptome sequencing technology is more suitable in the kidney than other methods of gene expression analysis [25]. The average sequencing depth of each nucleus is 1,082 genes, and 17,659 nuclear transcripts are divided into 30 different cell clusters [26]. The dataset GSE131685 contains a 10X genomic single-cell transcriptome sequencing dataset of 23,366 high-quality human kidney cells from 3 Chinese donors, including 20,308 PT cells [27].
In the standard single-cell transcriptome sequencing analysis pipeline, we first filtered cells with fewer than 200 genes, and genes with fewer than 3 cells as a quality control step [28]. After normalizing and scaling the data, we selected highly variable genes using Seurat to find highly variable gene functions with default parameters [29]. For dataset GSE121862, we directly used the cluster annotation provided in the reference article and visualized the result in a UMAP embedding space using 20 principal components in the principal component analysis (PCA) and UMAP function of Seurat. We performed Harmony for batch correction of the GSE131685 dataset and visualized the result in a UMAP embedding space using 20 principal components in PCA with a resolution 0.25 to find the cluster function. For the dataset GSE140989, we used 20 principal components and 1.2 resolution in the FindClusters function in order to obtain the similar cluster in the reference article. The detailed cluster markers and annotations of datasets GSE131685 and GSE140989 can be accessed in online supplementary Table 2. Finally, we visualized the expression level of the ACE2 gene in the embedding space and compared the ACE2 gene expression among the kidney cell types.
Differential Gene Expression Analysis in PT Cells
In order to further analyze the genetic and functional characteristics of ACE2-expressing cells, we divided kidney cells into ACE2+ and ACE2– cells according to the expression level of ACE2. The differential expression test was performed using the Seurat FindMarkers function with Wilcoxon rank sum test and further visualized in a volcano plot. Genes with log average fold change larger than 1 were defined as up-regulation genes and those smaller than −1 as down-regulation genes. The GO enrichment analysis of the top 100 different expression genes was performed using Metascape, an online gene annotation tool [30]. Detailed run scripts are included in the supplementary materials and also available from https://github.com/JiaqiLiZju/ACE2-in-kidney.
Gene Expression Analysis in Lung and Small-Intestine Single-Cell Transcriptome Sequencing Datasets
The dataset GSE125970 contains a single-cell transcriptome sequencing dataset of 14,537 epithelial cells from adult human ileum, colon, and rectum [31]. Fresh resected human lung tissue was obtained from a transplant donor and profiled using single cell RNA sequencing technology (GSM3489185) [32]. We used Seurat V3.0 to discriminate different cell types. The data were first normalized using the LogNormalize method. For the dataset GSE125970, the cluster annotation provided in the reference article is applied, and cell scatter plots were obtained using UMAP. For the dataset GSM3489185, seven clusters were identified using the 12 canonical correlation vectors with the resolution parameter set to 0.1. Marker genes for the 7 clusters were defined using the ROC test in the Seurat package, which allowed for cell types to be assigned to each cluster, with the intent to be similar to the reference article.
Differential Gene Expression Analysis in Enterocytes and Alveolar Type 2 Cells
In addition to further analyze the genetic and functional characteristics of ACE2-expressing enterocytes and alveolar type (AT) 2 cells, we divided cells into ACE2+ and ACE2– cells according to the expression level of ACE2. The differential expression analysis was performed by the function FindMarkers in Seurat using a Wilcoxon rank sum test on all genes in ACE2+ and ACE2– cells detected within a cluster. Differentially expressed genes with a fold change more than 0.5 (log scale) or less than 0.5 were considered to be marker genes. Gene expression of selected markers was visualized in a volcano plot using R package. The GO enrichment analysis of differential expression genes was performed using Metascape, an online gene annotation tool [30]. Detailed run scripts are included in the supplementary materials (also available from https://github.com/JiaqiLiZju/ACE2-in-kidney).
Results
ACE2 Expression in Human Tissue
To truly reflect the expression level of ACE2 in various tissues and organs, we used the recent HCL database to analyze the global expression of ACE2 in human tissue. According to this database, we found that ACE2 is mainly enriched in the small intestine (Fig. 1a, b). In addition, ACE2 expression is also present in kidney cell subgroups (Fig. 1a, b). This result is consistent with the NCBI transcriptome data (online suppl. Fig. 1) and the human protein atlas. We analyzed the top 50 subgroups of cells in the HCL in terms of ACE2 expression level and divided them into 2 groups: adult and embryo tissues (Fig. 1c). In adult tissues, ACE2 is mainly expressed in enterocytes of the duodenum, ileum, and colon, epithelial cells of the liver, and PT cells of the kidney, and it is also expressed in Clara cells of the lung (Fig. 1c, d). Moreover, in fetal tissues, ACE2 is significantly expressed in enterocytes, enterocyte progenitor cells, smooth muscle cells, proliferating cells, fibroblasts, and goblet cells of the fetal intestine, hepatocyte-like cells of the fetal adrenal gland, and PT progenitor cells of the fetal kidney (Fig. 1e).
Expression of ACE2 in the Kidney
Single-cell transcription data from 3 adult kidneys and 3 fetal kidneys from the HCL database indicate that ACE2 is mainly concentrated in PT cells of adult kidneys (Fig. 1d; online suppl. Table 1) and in PT progenitor cells of fetal kidneys (Fig. 1e; online suppl. Table 1). The basic information of the 6 patients is shown in online supplementary Table 1.
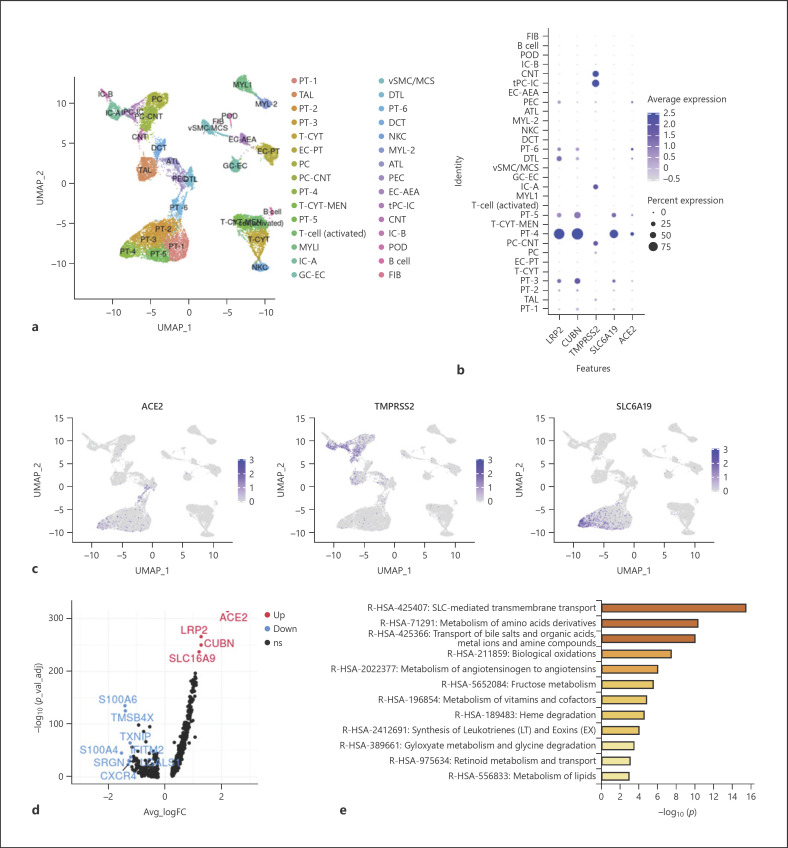
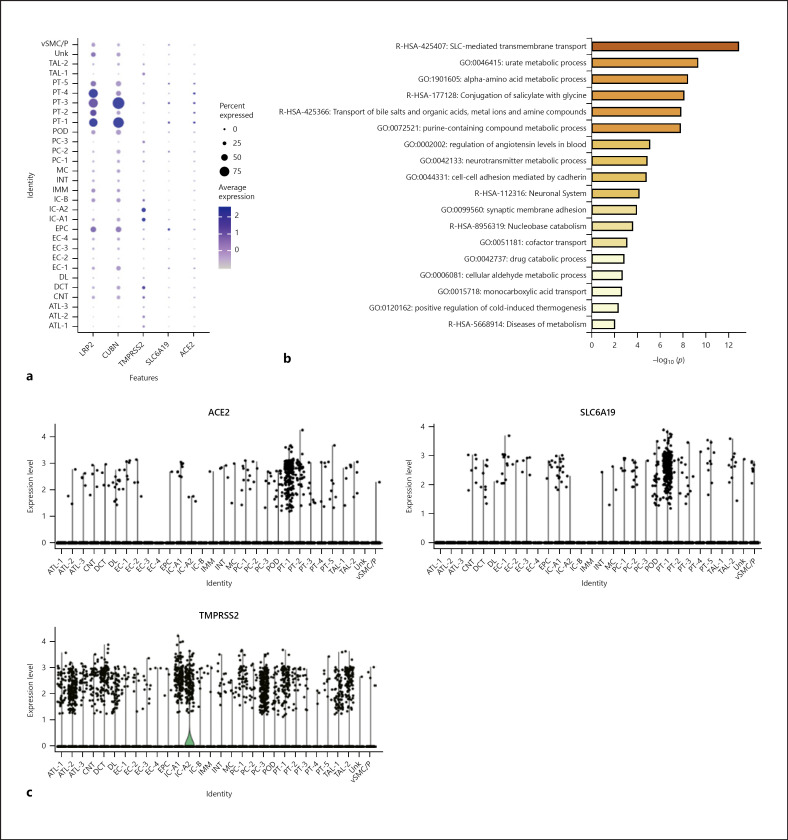
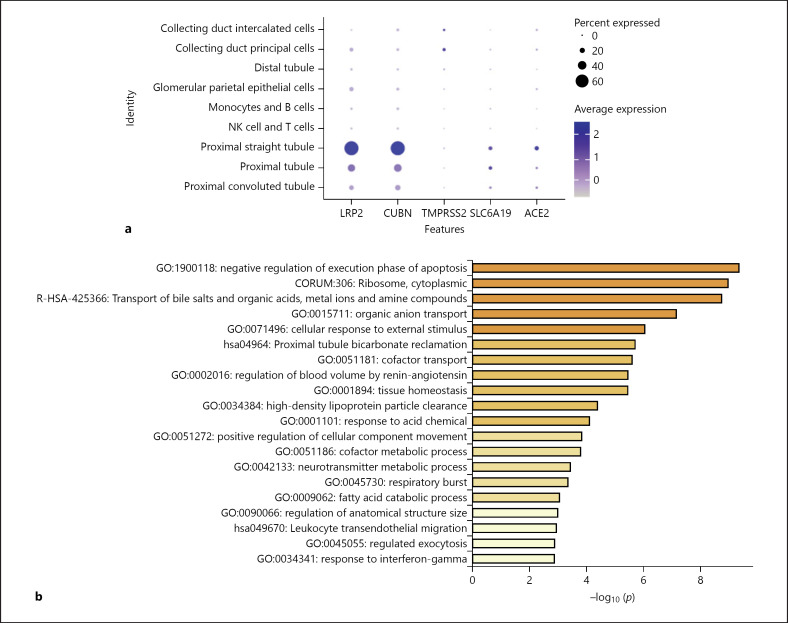
Several studies have provided high-quality renal single-cell transcription databases that allow us to analyze RNA information from individual cells. In the latest human kidney single-cell transcriptome database (scRNA-seq) from the Broad Institute and the University of Michigan [24], we found significant expression of ACE2 in PT cells, glomerular parietal epithelial cells, and descending thin limb cells (Fig. 2b, c; online suppl. Fig. 2). ACE2 was significantly expressed in all 6 PT cell clusters, and most significantly in PT-4 (Fig. 2b; online suppl. Fig. 2). Through the single-nuclear transcription database (snRNA-seq) from human adult kidneys [26], we analyzed the expression of ACE2 in 30 subgroups of kidney cells (online suppl. Fig. 3a). We found that variable kidney cells express ACE2, including podocytes and epithelial cells, but PT cells had the highest expression level (Fig. 3a, c). Furthermore, ACE2 was mainly expressed in PT-1, PT-2, and PT-5 clusters (Fig. 3c), which correspond to S1, S2, and S3 PT segments, respectively [26]. Meanwhile, we analyzed the kidney single-cell transcription database (Asian scRNA-seq) from Asian populations (online suppl. Fig. 4a) [27]. The results showed that convoluted PT (S1 and S2 segments) and straight PT (S3 segment) highly expressed ACE2 (Fig. 4a; online suppl. Fig. 4c, d). From the above results, all the single-cell transcriptome analyses of adult kidneys from different races and different methods demonstrated that ACE2 is expressed in the S1, S2, and S3 segments of PT cells.
Fig. 2.
Expression of genes associated with SARS-CoV-2 entry in single-cell transcriptome sequencing database. a UMAP visualization displaying major cell clusters (22,268 single cells). b Dot plot of LRP2 and CUBN (marker genes of PT cells), ACE2, TMPRSS2, and SLC6A19; dot size represents the fraction of cells expressing a cell type, and color intensity binned count-based expression level amongst expressing cells. c UMAP projection, points colored by detection of ACE2 (left), TMPRSS2 (middle), and SLC6A19 (right). Blue, RNA positive; gray, RNA negative. d Volcano plot displaying differential expression of genes between ACE2+ and ACE2– PT cells. Red, up-regulated genes; blue, down-regulated genes. e Bar plot presenting significantly enriched GO terms obtained from GO enrichment analysis performed with differential expression of genes between ACE2+ and ACE2– PT cells.
Fig. 3.
Expression of genes associated with SARS-CoV-2 entry in single-nuclear transcriptome. a Dot plot of LRP2 and CUBN (marker genes of PT cells), ACE2, TMPRSS2, and SLC6A19; dot size represents the fraction of cells expressing each cell type, and color intensity binned count-based expression level amongst expressing cells. b Bar plot presenting significantly enriched GO terms obtained from GO enrichment analysis performed with differential expression of genes between ACE2+ and ACE2– PT cells. c Violin plot showing ACE2, SLC6A19, and TMPRSS2 expression distribution among different cell clusters.
Fig. 4.
Expression of genes associated with SARS-CoV-2 entry in a single-cell transcriptome sequencing database of Asians. a Dot plot of LRP2 and CUBN (marker genes of PT cells), ACE2, TMPRSS2, and SLC6A19; dot size represents the fraction of cells expressing each cell type, and color intensity binned count-based expression level amongst expressing cells. b Bar plot presenting significantly enriched GO terms obtained from GO enrichment analysis performed with differential expression of genes between ACE2+ and ACE2– PT cells.
Expression of Cell Receptor-Related Genes of SARS-CoV-2 in the Kidney
In addition to dissecting the expression of ACE2, identifying the expression of TMPRSS2 and SLC6A19 may also contribute to understanding the relationship between SARS-CoV-2 and kidney cells. TMPRSS2 is widely expressed in several renal tubule populations extending from the PT cells to the collection system cell type. The expression of TMPRSS2 is enriched in transitioning principal cells (PC)/intercalated cells (IC), intercalated α cells (IC-A), connecting tubule (CNT), and cortical CNT-principal cells (PC-CNT) in scRNA-seq of the human kidney (Fig. 2b, c; online suppl. Fig. 2). In snRNA-seq of the human kidney, PC, IC, and the distal convoluted tubule cells express TMPRSS2 more significantly than PT cells (Fig. 3a, c; online suppl. Fig. 3b). The expression of TMPRSS2 in the Asian scRNA-seq is also similar, mainly concentrated in IC and PC (Fig. 4a; online suppl. Fig. 4c, d). Unexpectedly, single-cell transcription database analysis results consistently suggest that despite the expression of various renal tubules, SLC6A19 is mainly enriched in PT cells, in line with ACE2 expression (Fig. 2b, 3a, 4a). Moreover, epithelial cells express SLC6A19 clearly in snRNA-seq of the human kidney (Fig. 3a).
Differential Genes and Functions of ACE2+ PT Cells
To further analyze the genetic and functional characteristics of ACE2-expressing PT cells, we artificially divided PT cells into PT cells with and without ACE2 expression (ACE2+ PT cells and ACE2– PT cells, respectively) instead of 2 different subgroups of cells obtained by cluster analysis. In scRNA-seq of the human kidney, compared with ACE2– PT cells, up-regulated genes in ACE2+ PT cells were LRP2, CUBN, and SLC16A9 (Fig. 2d). In general, these genes, considered to be marker genes of PT cells, are related to cell transmembrane transport function. In the snRNA-seq of the human kidney and the Asian scRNA-seq, there were nonsignificant differences in gene expression of the 2 cell types (online suppl. Fig. 3c, 4b). From different single-cell transcription databases, GO analysis of differential genes between the 2 cell types were enriched in SLC-mediated transmembrane transport, amino acid metabolism, and transport of bile salts and organic acids, metal ions, and amine compounds (Fig. 2e, 3b, 4b). The above results indicate that the transmembrane transport function of ACE2+ PT cells is more obvious.
Expression of Cell Receptor-Related Genes of SARS-CoV-2 in the Lung and the Small Intestine
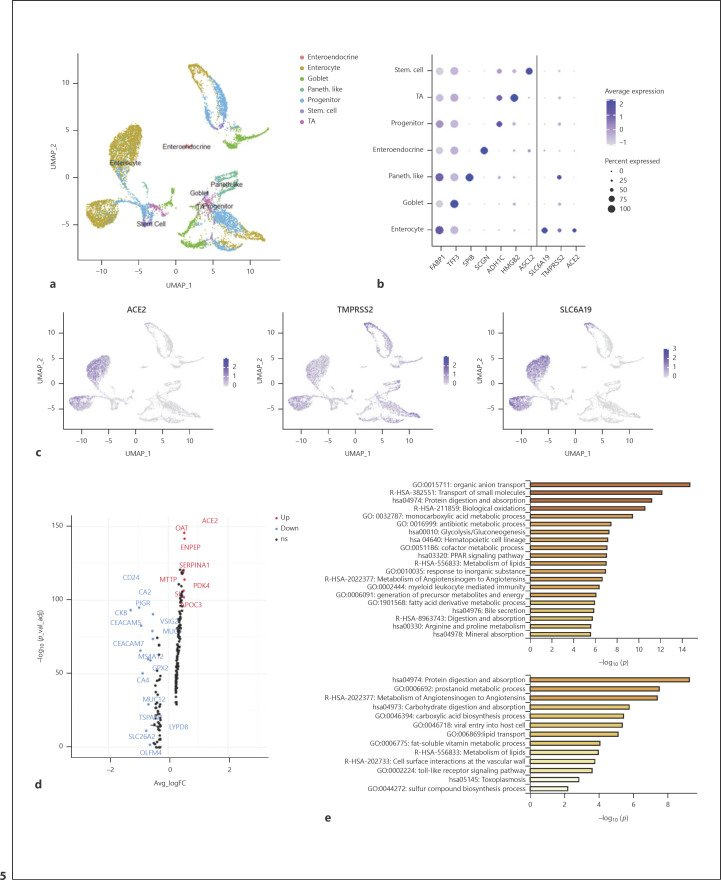
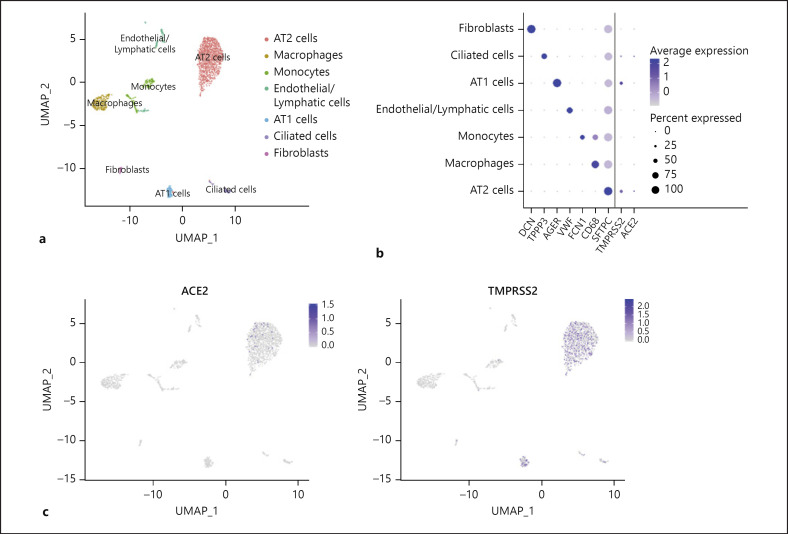
To compare cell receptor-related genes of SARS-CoV-2 in different tissues, high-quality single-cell transcriptome databases of the lung and small intestine were used for further analysis. Unsupervised cluster analysis of 14,537 cells from the human ileum, colon, and rectum yielded 7 cell subgroups (Fig. 5a). Consistent with the results of HCL database analysis, enterocytes express ACE2 significantly (Fig. 5b, c; online suppl. Fig. 5a). Unlike PT cells, enterocytes simultaneously express TMPRSS2 and SLC6A19 significantly (Fig. 5b, c; online suppl. Fig. 5b, 5c). The expression of ACE2 and SLC6A19 is mainly concentrated in enterocytes, while TMPRSS2 is widely expressed in various cell subgroups. In the lung, although there are only a small number of AT 2 cells and increased ACE2 expression of ciliated cells, the number of cells expressing TMPRSS2 in these cell subgroups is greater and more significant (Fig. 6b, c; online suppl. Fig. 6a, 6b). Surprisingly, no expression of SLC6A19 was found in all cell subpopulations. Considering that single-cell preparation and capture methods may cause errors in the number of rare cells, we did not compare the ratio of ACE2+/TMPRSS2+ cells, which is the highest-risk cell for SARS-CoV-2 in each tissue.
Fig. 5.
Expression of genes associated with SARS-CoV-2 entry in the small intestine. a UMAP visualization displaying major cell clusters (14,537 single cells). b Dot plot of marker genes for each cell type and ACE2, TMPRSS2, and SLC6A19; dot size represents the fraction of cells expressing each cell type, and color intensity binned count-based expression level amongst expressing cells. c UMAP projection: points colored by detection of ACE2 (left), TMPRSS2 (middle), and SLC6A19 (right). Blue, RNA positive; gray, RNA negative. d Volcano plot displaying differential expression genes between ACE2+ and ACE2– enterocytes. e Bar plot presenting significantly enriched GO terms obtained from GO enrichment analysis performed with differential expression genes between ACE2+ enterocytes and all other enterocytes (Up), ACE2+/TMPRSS2+ enterocytes, and all other enterocytes (Down).
Fig. 6.
Expression of genes associated with SARS-CoV-2 entry in the lung. a UMAP visualization displaying major cell clusters. b Dot plot of 3 marker genes for each cell type and ACE2, TMPRSS2, and SLC6A19; dot size represents the fraction of cells expressing each cell type, and color intensity binned count-based expression level amongst expressing cells. c UMAP projection: points colored by detection of ACE2 (left) and TMPRSS2 (right). Blue, RNA positive; gray, RNA negative.
Functional Analysis of Putative Target Cells for SARS-CoV-2
To identify enriched pathways within the putative target cells of SARS-CoV-2, we computed differentially expressed genes between ACE2+ and ACE2– enterocytes and AT2 cells, respectively. Compared with ACE2– enterocytes, up-regulated genes in ACE2+ enterocytes were OAT, ENPEP, SERPINA1, MTTP, and so on (Fig. 5d). GO analysis of differential genes between the 2 types of enterocytes (ACE2+ enterocytes and ACE2– enterocytes) were enriched in transmembrane transport and protein digestion and absorption (Fig. 5e). Furthermore, GO enrichment analysis was performed with differentially expressed genes between ACE2+/TMPRSS2+ enterocytes and all other enterocytes. Surprisingly, we discovered that there is a process for virus entry into the host (Fig. 5e). Because the number of ACE2– AT2 cells is much larger than ACE2+ AT2 cells, we did not find significant up- or down-regulated genes (logFC value of up-regulated gene SCGB3A1 >0.5 and logFC value of down-regulated gene SCGB3A2 >0.5) (online suppl. Fig. 6c). GO analyses of differential genes between the 2 types of AT2 cells were enriched in the PID ERBB1 downstream pathway, binding, and uptake of ligands by scavenger receptors, and regulation of bone resorption (online suppl. Fig. 6d).
Discussion
Due to the key role of ACE2 as a cellular surface receptor for SARS-CoV-2, and based on previous results that have shown that small-intestine [33], liver [34], and kidney [35] cells express ACE2, we analyzed the expression of ACE2 in various tissues and even individual cell subgroups. Using the latest HCL database to analyze the global expression of ACE2 is the first analysis of the distribution of ACE2 in whole tissues using a single database, effectively reducing the existence of false-positive results and making the results more credible [23, 36]. Compared with other tissues, our results showed that ACE2 is mainly expressed in enterocytes of the small intestine, epithelial cells of the liver, and PT cells of the kidney. Obviously, although ACE2 is expressed in the lung, the above tissue expression is more significant. This result indicates that the affinity of SARS-CoV-2 cannot be predicted only by the expression of ACE2.
The renin-angiotensin system has been classically conceived as a critical hormonal signaling pathway mainly responsible for blood pressure control and hydroelectrolyte balance. Previous studies have documented that in severe lung injury, the expression of ACE2 would be significantly down-regulated, and ACE2 blockage or genetic manipulation to delete Ace2 resulted in exacerbated lung injury [37, 38, 39, 40]. The recent research has confirmed that clinical-grade human recombinant soluble ACE2 can effectively inhibit SARS-CoV-2 infection [44]. The above results suggest that the role of ACE2 in COVID-19 patients is complicated and diverse [41]. SARS-CoV-2 entry into target cells is an elegantly regulated multistep process, of which binding to ACE2 is only the first. For the first time from the single-cell level analysis, our results demonstrate that cell receptor-related genes of SARS-CoV-2 are differentially expressed in cell subgroups of different tissues. AT 2 cells in the lung significantly express ACE2 and TMPRSS2, but not SLC6A19, and all 3 genes are significantly expressed in intestinal epithelial cells. Unlike other ACE2-expressing cells, PT cells in the kidney expressed SCL6A19 more significantly than TMPRSS2. These 3 types of cells have significant differences in the distribution of the cell receptor-related genes of SARS-CoV-2, which may indicate the diversity of the cell surface structure and the difference in the affinity between SARS-CoV-2 and cells.
In the literature, COVID-19 is characterized by symptoms of viral pneumonia, such as fever, cough, and lymphopenia [16, 17, 18, 42]. Except for causing pneumonia, COVID-19 may also damage other organs, such as the kidney [43]. A very recent study showed that SARS-CoV-2 can replicate in kidney organoids [44]. Diao et al. [45] found the nucleocapsid protein of SARS-CoV-2 virus accumulated in renal tubules, which indicates that SARS-CoV-2 directly infected the kidney. However, the incidence of AKI in COVID-19 patients is heterogeneous in various studies. Some data showed that almost 40% of hospitalized patients had proteinuria and hematuria [14, 15], while others suggested that the incidence of AKI is between 0.5 and 7% [16, 17, 18]. Among 116 hospitalized COVID-19-confirmed patients, all these patients did not meet the diagnostic criteria of AKI [46]. This result suggested that SARS-CoV-2 infection did not cause AKI or aggravate CKD in the COVID-19 patients. In a study by Ronco and Reis [47], the prevalence of direct kidney involvement in COVID-19 is low, and cytokine damage, organ cross talk, and systemic effects may be related to kidney involvement in COVID-19 patients. In addition, although it has been reported that SARS-CoV-2 can be found in the urine [19], more studies have not found its presence in urine [20, 21]. Our study is the first to present differences in the expression of cell receptor-related genes of SARS-CoV-2, rather than ACE2 alone, which provides somewhat more persuasive clues to explain kidney injury in COVID-19.
In this research, we computed differentially expressed genes between ACE2+ and ACE2– PT cells through 3 different single-cell transcription databases. However, considering that the different single-cell preparation and methods (scRNA-seq and snRNA-seq), the results of differential gene and GO analyses are different. Compared with ACE2– PT cells, the transmembrane transport function of ACE2+ PT cells is more active. Due to the small number of PT cells and AT2 cells co-expressed with ACE2 and TMPRSS2, we did not conduct further differential gene analysis. A comparison of ACE2+/TMPRSS2+ enterocytes with all other enterocytes revealed that there is a process for virus entry into the host. The results indicated that enterocytes co-expressing ACE2 and TMPRSS2 may have a direct risk of virus infection. With the promotion of single-cell transcriptome sequencing technology, we can obtain more information about rare cells, such as ACE2+/TMPRSS2+ cells in different tissues, so that we can have a more accurate understanding of such cells.
Next-generation sequencing and bioinformatics are changing the way we respond to outbreaks of infectious diseases [48]. The widespread use of sequencing technology has enabled humans to obtain viral gene sequences in the early stages of infectious disease outbreaks and to quickly predict virus types, transmission pathways, and mechanisms of infection through sequence analysis and structure prediction [49]. At the same time, the astonishing development of single-cell transcriptome sequencing technology makes it possible to predict the susceptibility not only of specific organs but also subgroups of cells to a virus.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported by the National Key Research and Development Program of China (2016YFC0901202).
Author Contributions
Q.L. Chen conceived of the project, interpreted the results of the analysis, made figures, and wrote the paper; J.Q. Li performed single-cell transcriptome sequencing analyses, made figures, and wrote the paper; Q.L. Chen and J.Q. Li contributed equally to this work; Z.D. Xiang discussed the draft paper and offered bioinformatic help; Yue Lang discussed the draft paper, and critically reviewed the manuscript; G.J. Guo and Zhi-Hong Liu conceived of and supervised the project, and reviewed the manuscript; all authors read and commented on the manuscript.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgment
We thank for sharing unpublished data (HCL), including single-cell transcriptome sequencing databases of various human tissues.
References
- 1.WHO Coronavirus disease (COVID-19) Situation dashboard. [cited 2020 April 4]. Available from: https://experience.arcgis.com/experience/685d0ace521648f8a5beeeee1b9125cd.
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Mar;579((7798)):270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 Feb;395((10224)):565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020 Apr;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013 Dec;87((23)):12552–61. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016 May;15((5)):327–47. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Kramer-Kuhl A, et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87((10)):5502–11. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88((2)):1293–307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85((2)):873–82. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020 Apr;181((2)):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar;367((6485)):1444–8. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lely AT, Hamming I, van Goor H, Navis GJ. Renal ACE2 expression in human kidney disease. J Pathol. 2004 Dec;204((5)):587–93. doi: 10.1002/path.1670. [DOI] [PubMed] [Google Scholar]
- 13.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020 Mar; doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Wu M, Yao J, Guo J, Liao X, Song S, et al. Caution on Kidney Dysfunctions of COVID-19 Patients. medRxiv. 2020;2020 02.08.20021212. [Google Scholar]
- 15.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, et al. Kidney impairment is associated with in-hospital death of COVID-19 patients. medRxiv. 2020;2020 doi: 10.1016/j.kint.2020.03.005. 02.18.20023242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020 Feb;323((11)):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb;395((10223)):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020 Apr;382((18)):1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. DOI: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. DOI: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. DOI: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Human Cell Landscape. Available from: http://bis.zju.edu.cn/HCL.
- 23.Han X, Zhou Z, Fei L, Sun H, Wang R, Chen Y, et al. Construction of a human cell landscape at single-cell level. Nature. 2020 doi: 10.1038/s41586-020-2157-4. DOI:10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- 24.Menon R, Otto EA, Hoover P, Eddy S, Mariani L, Godfrey B, et al. Nephrotic Syndrome Study Network (NEPTUNE) Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight. 2020 Mar;5((6)):133267. doi: 10.1172/jci.insight.133267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J Am Soc Nephrol. 2019 Jan;30((1)):23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lake BB, Chen S, Hoshi M, Plongthongkum N, Salamon D, Knoten A, et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun. 2019;10((1)):2832. doi: 10.1038/s41467-019-10861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao J, Yu Z, Chen Y, Bao M, Zou C, Zhang H, et al. Single-cell RNA sequencing of human kidney. Sci Data. 2020;7((1)):4. doi: 10.1038/s41597-019-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177((7)):1888–902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19((1)):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10((1)):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Song W, Wang J, Wang T, Xiong X, Qi Z, et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med. 2020;217((2)):pii–e20191130. doi: 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019 Jul;25((7)):1153–63. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012 Jul;487((7408)):477–81. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paizis G, Tikellis C, Cooper ME, Schembri JM, Lew RA, Smith AI, et al. Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut. 2005;54((12)):1790–6. doi: 10.1136/gut.2004.062398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warner FJ, Lew RA, Smith AI, Lambert DW, Hooper NM, Turner AJ. Angiotensin-converting enzyme 2 (ACE2), but not ACE, is preferentially localized to the apical surface of polarized kidney cells. J Biol Chem. 2005 Nov;280((47)):39353–62. doi: 10.1074/jbc.M508914200. [DOI] [PubMed] [Google Scholar]
- 36.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36((5)):421–7. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou Z, Yan Y, Shu Y, Gao R, Sun Y, Li X, et al. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat Commun. 2014 May;5((1)):3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye R, Liu Z. ACE2 exhibits protective effects against LPS-induced acute lung injury in mice by inhibiting the LPS-TLR4 pathway. Exp Mol Pathol. 2020 Apr;113:104350. doi: 10.1016/j.yexmp.2019.104350. [DOI] [PubMed] [Google Scholar]
- 39.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11((8)):875–9. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu H, Xie Z, Li T, Zhang S, Lai C, Zhu P, et al. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci Rep. 2016;6:19840. doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaduganathan M, Vardeny O, Michel T, McMurray JJ, Pfeffer MA, Solomon SD. Renin-Angiotensin-Aldosterone System Inhibitors in Patients with Covid-19. N Engl J Med. 2020 Apr;382((17)):1653–9. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 Mar;395((10229)):1054–62. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang T, Du Z, Zhu F, Cao Z, An Y, Gao Y, et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020 Mar;395((10228)):e52. doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. doi: 10.1016/j.cell.2020.04.004. DOI 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diao B, Wang C, Wang R, Feng Z, Tan Y, Wang H, et al. Human Kidney is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. medRxiv. 2020 doi: 10.1038/s41467-021-22781-1. 2020.03.04.20031120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Li X, Chen H, Yan S, Li D, Li Y, et al. Coronavirus Disease 19 Infection Does Not Result in Acute Kidney Injury: An Analysis of 116 Hospitalized Patients from Wuhan, China. Am J Nephrol. 2020 doi: 10.1159/000507471. DOI: 10.1159/000507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronco C, Reis T. Kidney involvement in COVID-19 and rationale for extracorporeal therapies. Nat Rev Nephrol. 2020 doi: 10.1038/s41581-020-0284-7. DOI:10.1038/s41581-020-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020 Feb;382((8)):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020 Mar;382((13)):1199–207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data