Abstract
The study of the genetic structure of different countries within Europe has provided significant insights into their demographic history and population structure. Although France occupies a particular location at the western part of Europe and at the crossroads of migration routes, few population genetic studies have been conducted so far with genome-wide data. In this study, we analyzed SNP-chip genetic data from 2184 individuals born in France who were enrolled in two independent population cohorts. Using FineSTRUCTURE, six different genetic clusters of individuals were found that were very consistent between the two cohorts. These clusters correspond closely to geographic, historical, and linguistic divisions of France, and contain different proportions of ancestry from Stone and Bronze Age populations. By modeling the relationship between genetics and geography using EEMS, we were able to detect gene flow barriers that are similar across the two cohorts and correspond to major rivers and mountain ranges. Estimations of effective population sizes also revealed very similar patterns in both cohorts with a rapid increase of effective population sizes over the last 150 generations similar to other European countries. A marked bottleneck is also consistently seen in the two datasets starting in the 14th century when the Black Death raged in Europe. In conclusion, by performing the first exhaustive study of the genetic structure of France, we fill a gap in genetic studies of Europe that will be useful to medical geneticists, historians, and archeologists.
Subject terms: Population genetics, Genetics
Introduction
Gallia est omnis divisa in partes tres [Commentarii de Bello Gallico [1] was one of the earliest demographic description of antique France (known as Gaul). These three parts were Aquitania, in the south-west, with Garonne and the Pyrenees mountains as borders; Belgia in the north-west, following the Seine as Southern border; and finally, what we know as Celtic Gaul, that spanned from the Atlantic Ocean to the Rhine River and Alps. A fourth part of the present-day French territory, already part of Romanized territories at this time, was Gallia Transalpina, a strip of land from Italy to Iberia, with the Alps and Cevennes mountains as the northern border.
The area that constitutes today the modern France was subject to successive population migrations: Western Hunter-Gatherers (15 kya), Neolithic farmers (7 kya), and later Steppe Eneolithic Age populations [2, 3], Celtic expansion, integration in the Roman empire, Barbarian migrations, whose demographical importance remains to be assessed. France’s position at the western part of Europe has made it not only the final goal of a large number of, potentially massive, migrations but also a place of transit either to the North (British Isles) or to the South of Europe (Iberian Peninsula) and North Africa, as well as an important crossroad for trade and exchanges [4].
Before France became a single political entity, its territory was divided into various kingdoms and later provinces, which often displayed a fiercely independent spirit towards the central power. Pre-Roman Gaul was divided into politically independent territories. After the fall of the Roman Empire, the modern French territory was divided into Barbarian Germanic kingdoms (Franks, Visigoths, and Burgunds). After a short period of reunification and extension into the Carolingian Empire (VIIth century), the weakening of the central power led to the reduction of Occidental France at its western part and the rise of local warlords gaining high levels of independence within the Kingdom itself. The feudal period created provinces that were effectively independent, although nominally linked through the oath of allegiance to the King of France (Fig. S1) [4] .
Through the centuries, in spite of reverses such as the Hundred Years War, the French Kings managed to slowly integrate the Eastern lands as well as Brittany, enforcing central power until the French Revolution. However, each province maintained political, cultural, and linguistic differences, which could have left imprints in the genetic structure of modern French populations [4, 5].
Geographically, modern France is a continental country surrounded by natural borders: the Atlantic Ocean to the West, the Channel to the North, mountains (Pyrenees and Alps) closing the south/west and east/south-east borders, as well as the Mediterranean Sea to the south (Fig. S2). The eastern side has the Rhine as a natural border along 166 km while the Northeastern borders show no notable obstacle and exhibits a continuum with Belgium and Germany (Fig. S2). This complex history and varied geography are expected to have shaped the genetic make-up of the current French population and left some footprints in its genetic structure.
The study of the genetic structure of human populations is of major interest in many different fields. It is informative about the demographic history of populations and how they have formed and expanded in the past, and the consequences of these movements on the distribution of traits. Genetic differences between populations can identify genetic variants likely to play a major role on different phenotypes, including disease [6]. This explains the growing interest in human population studies that aim to describe genetic diversity, now facilitated by the rich genetic information available over the entire genome. In the last decades, several studies were performed using genome-wide SNP data collected for genome-wide association studies. These studies have first shown that there exist allele frequency differences at all geographic scales and that these differences increase with geographic distances. The first studies showed differences between individuals of different continental origins [7–9] and then, as more data were collected and marker density increased, these differences were found within continents, including Europe [10, 11]. Several studies at the scale of a single country have shown that differences also exist within countries. This was for instance observed in Sweden, where Humphreys et al. [12] reported strong differences between the far northern and the remaining counties, partly explained by remote Finnish or Norwegian ancestry. More recent studies have shown structure in the Netherlands [13], Ireland [14], UK [15], or Iberian peninsula [16]. Previous studies of population stratification in France have examined only Western France (mainly Pays de la Loire and Brittany) and detected a strong correlation between genetics and geography [17]. However, no study so far has investigated the fine-scale population structure of France in its entirety using unbiased samples from individuals with ancestries all over the country.
In this paper, we applied haplotype-based methods that have been shown to provide higher resolution than allele-based approaches [15] to investigate the pattern of fine-scale population stratification in France. To assess the genetic structure of the French population and make inference about demographic history, we used two independent cohorts with genome-wide genotype data, 3C and SU.VI.MAX with more than 2000 individuals whose birthplace covered continental France.
Material and methods
Data from SU.VI.MAX and 3C studies
Genetic data were obtained from two French studies, SU.VI.MAX [18] and the Three-Cities study [19] (3C) with the idea to compare whether, analyzing them independently, concordant results could be obtained. One major drawback of genetic inferences obtained on population samples is the fact that they can strongly depend on how individuals were sampled and which genetic markers were used. Here, by using data from two studies that sampled individuals using different criteria and genotyped them on different SNP arrays, we should be able to draw more robust inferences.
For every individual, information on places of birth was available, either the exact location (3C study) or the “département” (SU.VI.MAX). Départements are the smallest administrative subdivisions of France. There are a total of 101 French départements and 94 of them are located in continental France. These units were created in 1789, during the French Revolution, partly based on historical counties.
3C Study: The Three-City Study was designed to study the relationship between vascular disease and dementia in 9294 persons aged 65 years and over. Individuals were recruited from three French cities: Bordeaux (south-west), Dijon (north-east), and Montpellier (south-east). For more details on the study, see http://www.three-city-study.com/the-three-city-study.php. Analyses were performed on individuals who were free of dementia or cognitive impairment by the time their blood sample was taken and who were previously genotyped [20]. Although individuals were recruited only in three French cities, we had access to their places of birth that covered all geographic regions of France. The geographical locations of individuals were defined according to the latitude and longitude of their place of birth, declared at enrollment. Individuals with missing place of birth or born outside continental France were excluded. A total of 4659 individuals were included in the present study. We assumed that these places of birth were a good surrogate for the geographical origin of individuals and that sampled individuals were good representatives of the genetic diversity in their region of origin. This might not be completely true especially in the 3C where individuals born outside the three cities where the recruitment was performed are in fact individuals who have moved away from their region of origin and might in fact be different from the people who stayed there (see for example [21]).
SU.VI.MAX: The study was initiated in 1994 with the aim of collecting information on food consumption and health status of French people. Healthy volunteers were recruited through a national multimedia campaign. They were invited to visit a medical exam mobile medical unit or a preventive health center where blood samples were collected. In the present study, we only considered a subset of 1416 volunteers for whom genome-wide SNP-chip data was available and who were born in any of the 94 continental French départements. The geographic coordinates of each département were approximated based on the coordinates of the corresponding main city. Compared with the 3C study, the SU.VI.MAX study has a more geographically uniform coverage. However, we cannot exclude that the included volunteers are not fully representative of the rest of the population on sociological parameters, especially as urban populations are over-represented. The impact it could have on genetic structure inferences is difficult to measure. It highlights the interest of comparing results obtained using two different sampling schemes as proposed here with 3C and SU.VI.MAX.
Quality control
Quality control of the genotypes was performed using the software PLINK version 1.9 [22, 23].
3C: raw genotype data were generated in the context of a previous study [20] on Illumina Human610-Quad BeadChip. Following the recommendations from Anderson et al. [24], individuals were removed if they had a call rate <99% or heterozygosity further than ±3 standard deviations (SD) from the mean. Cryptic relatedness was assessed by estimating pi_hat (the IBD test implemented in PLINK [23]) in each dataset after doing LD-based pruning. Individuals related to another individual from the sample with an IBD proportion of 0.1875 or above were removed (one individual was kept from each pair). As a final quality control to exclude outlier individuals from populations, we performed principal component analysis (PCA) using the smartpca software from the EIGENSOFT package version 6.0.1 [25] and removed outliers across the first ten eigenvectors. The default procedure was used for outlier removals with up to five iterative PCA runs and, at each run, removing of individuals with any of the top ten PCs departing from the mean by more than 6 SD. SNPs in strong linkage disequilibrium (LD) were pruned out with PLINK 1.9 (described in PCA section). Outlier individuals were removed prior to performing further analyses. Applying all these QC filters led to the removal of 226 individuals. To avoid redundant information from individuals born in the same place, when several individuals were born in the same location (same latitude and longitude of birth places), we randomly selected only one of these individuals. This was necessary to avoid oversampling in the three cities where recruitments took place. A total of 770 individuals covering the 94 continental French départements were included. All samples failing sample-level QC were removed prior to performing SNPs QC. Markers were removed if they had a genotype-missing rate >1%, a minor allele frequency <1%, or departed from Hardy–Weinberg proportion (P ≤ 10−7). After QC, there were 770 individuals and 490,217 autosomal SNPs.
SU.VI.MAX: Genotype data of the 2834 samples were available from previous studies using different SNP chips: 1978 with Illumina 300 k/300 k duo and 856 with Illumina 660 W. Individuals with an unknown birthplace or a birthplace outside of continental France were removed, 1416 samples were kept. Two individuals were removed because of a call rate <95%. IBD statistic, calculated in PLINK version 1.9, did not identify any related samples with a threshold of 0.1875. SNPs were removed if they had a genotype-missing rate >2%, a minor allele frequency <10 % or departed from Hardy–Weinberg proportion (P ≤ 10−5). After QC, there were 1414 individuals and 271,886 autosomal SNPs.
Population structure within France
ChromoPainter/FineSTRUCTURE analysis
For investigating fine-scale population structure, we used ChromoPainter version 2 and FineSTRUCTURE version 2.0.7 [26]. Data were phased with SHAPEIT v2.r790 [27] using the 1000 Genomes dataset as a reference panel [28]. In the 3C dataset, we removed 932 of these SNPs because of strand issues prior to phasing. Files were then converted to ChromoPainter format using the ‘impute2chromopainter2.pl’ script. ChromoPainter outputs from the different chromosomes were combined with chromocombine to generate a final coancestry matrix of chunck counts for FineSTRUCTURE. For the FineSTRUCTURE run we sampled values after successive series of 10,000 iterations for 1 million MCMC iterations following 10 million “burn-in” iterations. Starting from the MCMC sample with the highest posterior probability among all samples, FineSTRUCTURE performed 100,000 additional hill-climbing moves to reach its final inferred state (See Ref. [15] for details). The final tree was visualized in R with the help of FineSTRUCTURE and ‘dendextend’ libraries. We checked that the MCMC samples were independent of the algorithm’s initial position by visually comparing the results of two independent runs starting from different random seeds. Good correspondence in the pairwise coancestry matrices of the two runs indicates convergence of the MCMC samples to the posterior distribution. Without loss of generality, we used the first of these two runs in our main analysis.
Ancestry profiles of the French population and spatial pattern of genetic structure EEMS
We used ADMIXTURE v1.3 [29] to estimate mixture coefficients of each individual. We performed runs for values of K between 2 and 10, with fivefold cross-validation using the set of pruned SNPs, as described in the PCA analyses. To identify if cluster differences existed, we performed a one-way analysis of variance (ANOVA) on the admixture components, followed by post hoc pairwise comparisons.
We estimated an effective migration surface using the software EEMS [30]. We ran EEMS with slightly different grids to investigate how/whether these changes affected the results. Plots were generated in R using the “rEEMSplots” package according to instructions in the manual. For both datasets the full set of SNPs was included. To avoid difference that could be due to difference in coverage of the different geographic regions in the two datasets, we selected, in both datasets, the same number of individuals in each department before running EEMS. For more information on the specific pipeline, see Supplementary Data.
IBD-estimated population size
We estimated the recent effective population size with IBDNe [31]. IBDNe was run with the default parameters and a minimum IBD segment length of 4 cM (mincm = 4). We used the default settings to filter IBD segments from IBDseq v. r1206 software package [32]. Breaks and short gaps in IBD segments were removed with the merge-ibd-segments utility program. For IBD detection, we varied the minimum IBD segment length in centiMorgan units by the mincm parameter (mincm argument) from the default value, 2–8 cM. IBDNe analysis was applied on the whole SU.VI.MAX and 3C datasets as well as on the major subpopulations from FineSTRUCTURE clustering. Growth rates were calculated with the formula . We assumed a generation time of 30 years, as assumed in the original paper.
Principal component analysis (PCA) and FST in modern populations
Both PCA and FST analyses were carried out on a pruned set of SNPs in each dataset independently and using the smartpca tool in the EIGENSOFT program (v6.1.1) [25]. The pairwise FST matrices were estimated using the option ‘fsthiprecision = YES’ in smartpca. We calculated the mean FST between clusters inferred by FineSTRUCTURE as group labels. In each dataset, SNPs in strong LD were pruned out with PLINK in a two-step procedure. SNPs located in known regions of long range LD in European populations were excluded from the analysis [33]. Then, SNPs in strong LD were pruned out using the ‘indep-pairwise’ command in PLINK. The command was run with an LD r2 = 0.2, a window size of 50 SNPs and 5 SNPs to shift the window at each step. This led to a subset of 100,973 SNPs and 83,246 SNPs in the 3C and SU.VI.MAX datasets, respectively. To evaluate the geographic relevance of PCs, we tested for the significance of association between the latitude and longitude of each département and PCs coordinates (‘cor.test’ function in R) using a Spearman’s rank correlation coefficient.
Relation with neighboring European populations: 1000G and HGDP
We assembled SNP data matching either the SU.VI.MAX or the 3C genotype data (after quality control) with the European individuals from the 1000G phase 3 reference panel and from the Human Genome Diversity Panel data [34] (HGDP, Illumina HuHap 650k), to generate four genome-wide SNP datasets analyzed independently.
The 1000G reference panel served as donor populations when estimating ancestry proportions. First, in order to define a set of donor groups from 1000G Europe (EUR), we used the subset of unrelated and outbred individuals generated in the study of Gazal et al. [35]. Four European populations were considered: north west Europe (GBR, n = 85 and CEU, n = 94), Spain (IBS, n = 107) and Italy (TSI, n = 104). These 390 Europeans individuals were then combined with individuals from both datasets independently resulting in a set of 484,874 common SNPs with 3C and a set of 232,148 common SNPs with SU.VI.MAX. The filtered datasets (after pruning) included 1160 individuals genotyped on 100,851 SNPs in the 3C Study and 1804 individuals genotyped on 64,653 SNPs in SU.VI.MAX. We inferred European ancestry contributions in France using the novel haplotype-based estimation of ancestry implemented in SOURCEFIND [36]. SOURCEFIND has been shown to give a greater accuracy than the usual nonnegative least squares regression for inferring proportion of admixture but because it is recommended to use homogeneous donor groups, we ran FineSTRUCTURE on the four European populations defined above and selected the level of clustering describing the main features of the donor populations. These European donor groups served as reference in SOURCEFIND. We performed analysis of variance (ANOVA) on French admixture component per cluster group to identify whether cluster differences existed.
Additional analyses combining the European participants of the HGDP panel were carried out in order to estimate the contribution of the Basque population of our South-West clusters. A total of 160 European HGDP participants were included from eight populations: Adygei (n = 17), French-Basque (n = 24), French (n = 29), Italian (n = 13), Italian from Tuscany (n = 8), Sardinian (n = 28), Orcadian (n = 16), and Russian (n = 25). Using the same procedure for merging panels, the filtered datasets (after pruning) included 930 individuals and 93,938 SNPs in the 3C Study and 1574 individuals and 57,775 SNPs in SU.VI.MAX.
Relation with founding ancient populations: hunter-gatherers, early farmers, and Steppe pastoralist populations
To understand the contribution of ancient populations to modern French populations, we merged each of the two datasets, 3C, and SU.VI.MAX, with publicly available ancient DNA samples [37–39]. Based on the supervised ADMIXTURE analysis performed by Mathieson et al. [40], we only used samples whose assignment probability to the corresponding group was >0.99 and, in the case of steppe pastoralists (SP), we only kept samples reported to belong to the Yamnaya complex, considered the source of the migration events spreading steppe-related ancestry across central and western Europe in the last 3500 years [41]. PCA was conducted by projecting the ancient DNA samples onto the principal components computed from modern samples, which included those in each of our datasets and 17 European populations from the Human Origins array [42]. PCA analysis was performed using smartpca software included in the EIGENSOFT package version 6.1.4 [25] with lsqproject option set to YES. Independent SNPs were selected as previously (See PCA section). The filtered datasets (after pruning) included 69,772 and 64,661 SNPs in 3C and SU.VI.MAX, respectively.
Outgroup f3-statistics of the form f3(Outgroup; Ancient, French cluster) were computed to measure the shared drift between a pair of populations, in our case between each of the possible combinations between ancient populations Western Hunter Gatherers (WHG), Early Farmers (EF), and Steppe Pastoralist (SP) and the six French clusters. D-statistics were computed to formally assess the excess of sharing between ancient populations and a specific modern French cluster, in relation to the remaining clusters. We used ADMIXTOOLS version 5.1 [43] to compute outgroup f3-statistics, D-statistics and assess statistical significance using the default parameters and the block jackknife approach implemented in the software package.
To obtain unbiased estimates of the WHG, EF, and SP mixture proportions contributing to modern French, we used the method initially introduced by Haak et al. [44] and currently implemented in qpAdm program within the ADMIXTOOLS version 5.1 package. Based on a matrix of f4-statistics computed from a set of outgroup populations (“right” populations) and a set of target and source populations (“left” populations), this method estimates mixture proportions contributing to the “target” without the need of a parameterized model describing the relationship between a set of test populations and a set of outgroup populations. P values reported refer to the probability of rejecting the null hypothesis, i.e., that the target population does not contain ancestry from another source that is differentially related to the right populations.
Results
ChromoPainter/FineSTRUCTURE analysis reveals consistent fine-scale genetic stratification within France
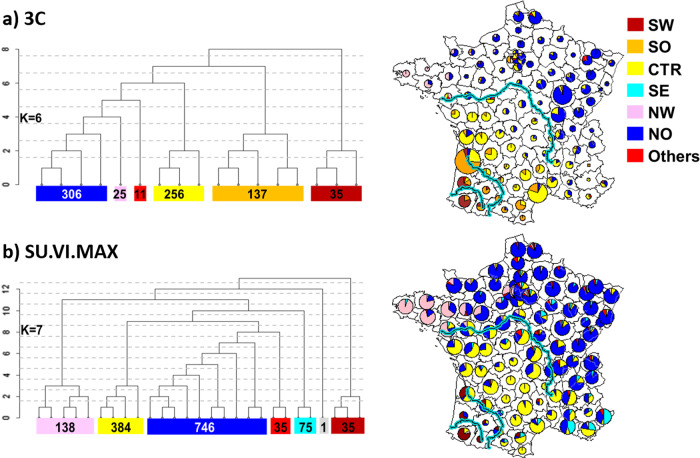
Results of FineSTRUCTURE analysis reveal fine-scale population patterns within France at a very fine level that are very consistent in the two datasets (Fig. 1). FineSTRUCTURE identified, respectively, 17 and 27 clusters in 3C and SU.VI.M.AX, demonstrating local population structure (Fig. S3). Even though the sampling distributions of individuals varies slightly between datasets both analyses show very concordant partitions with a broad correlation between clusters and geographic coordinates. The major axis of genetic differentiation runs from the south to the north of France.
Fig. 1. Clustering of the French individuals into six or seven clusters as inferred by FineSTRUCTURE analysis.
(a) 3C Study (770 individuals) and (b) SU.VI.MAX (1414 individuals). The tree structure is shown as well as a map of France with pie-charts giving, for each département, the proportion of individuals belonging to each cluster. Three main rivers of France (Loire, Garonne, and Adour from north to south) are drawn on the maps.
In both datasets, the coarsest level of genetic differentiation (i.e., the assignment into two clusters) separates the south-western regions from the rest of France (Figs. S4 and S5). Next levels of tree structures slightly differ between the two datasets but converge into a common geographic partition at k = 6 clusters in 3C and k = 7 in SU.VI.MAX (Fig. 1). The clusters are geographically stratified and labels were assigned to reflect geographic origin: the South-West (SW) for the dark-red cluster, the South (SO) for the orange cluster, the Center (CTR) for the yellow cluster, the North-West (NW) for the pink cluster, the North (NO) for the blue cluster and the South-East (SE) for the cyan cluster. In each dataset, one cluster (labeled “Others” and colored in red) included individuals geographically dispersed over France. Furthermore, one cluster identified in SU.VI.MAX included only one individual and was removed in further analysis so that k = 7 also resumed to six clusters in SU.VI.MAX. At this tree level of six clusters, individuals from the NO, NW, and CTR clusters are clearly separated in the two datasets. The SW cluster and part of the SO cluster in 3C match geographically the SW cluster identified in SU.VI.MAX while the SE subgroup was not detected in the 3C. This might be explained by differences in the geographic coverage between the two studies especially in the south of France. Indeed, SU.VI.MAX has a better coverage of the south-east, whereas 3C lacks data from this region and the reverse is true for the south-west. In the two datasets, two large clusters (CTR and NO) are found that cover most of the central and northern France. Notably, even at the finest level of differentiation (17 and 27 clusters in 3 C and SU.VI.MAX respectively), these clusters remain largely intact.
The broad-scale genetic structure of France in six clusters strikingly aligns with two major rivers of France, “La Garonne” and “La Loire” (Fig. 1). At a finer-scale, the “Adour” river divides the SW to the SO cluster in the 3C dataset. The mean FST between clusters inferred by FineSTRUCTURE (Tables S1 and S2) are small, confirming subtle differentiation. In both datasets, the strongest differentiation is between the SW cluster and all other regions. These FST values vary from 0.0016 with the SO cluster to 0.004 with the NW cluster in the 3C dataset and from 0.0009 with the CTR cluster to 0.0019 with the NW cluster in SU.VI.MAX. Finally, besides this subtle division, genetic differentiation within France is also due to isolation by distance as shown by the gradient exhibited on the values of the 1st component of the PCA (Fig. S6).
Different genetic ancestry profiles that could have been shaped by gene flow barriers
Results obtained by using ADMIXTURE corroborate the FineSTRUCTURE analysis with the SW cluster being the most different from the other groups (Figs. S7 and S8). At k = 2, the SW cluster shows a light blue component that is significantly less frequent in the other groups (ANOVA post hoc tests, p value < 10−6) (Fig. S9). In the 3C dataset, the proportion of light blue tends to decrease gradually from the south-western part of France to the center of France (CTR) to finally remain similar in the north of France (NO, NW, and Others). In SU.VI.MAX, the proportion of light blue component tends to discriminate the north from the south of France (Fig. S9). For k = 3, a third major component can be defined, the light green ancestry. In the 3C Study this component is predominant in the north of France (NW and NO clusters) and almost absent in the SW while in SU.VI.MAX this component is predominant in the SE and minimal in the extreme west of France (NW and SW). At k = 6, both datasets highlight the differentiation of the SW and the NW cluster from the others clusters.
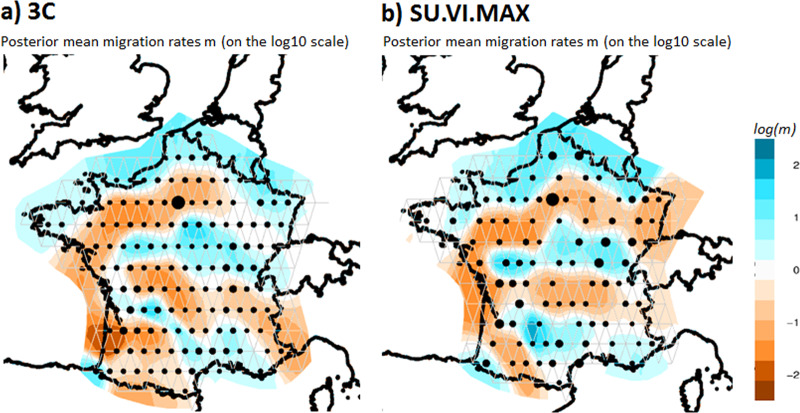
We performed EEMS analysis in order to identify gene flow barriers within France; i.e. areas of low migration. We varied the number of demes from 150 to 300 demes and selected a grid of 250 demes showing good concordance between datasets (Fig. S10). In both datasets, we identified a genetic barrier around the south-west region (Fig. 2). This barrier mirrors the first division in the FineSTRUCTURE. The plots also reveal a gene flow barrier around Bretagne in the north-west and in the central area of France, which covers the separation of the NO cluster. Finally, another barrier is also present on the south-east side that roughly corresponds to the location of the Alps at the border with Northern Italy.
Fig. 2. Estimated effective migration surfaces of France obtained from EEMS.
(a) 3 C and (b) SU.VI.MAX datasets. The color scale reveals low (blue) to high (orange) genetic barriers between populations localized on a grid of 250 demes. Each dot is proportional to the number of populations included.
IBD-derived demographic inferences reveal a rapid expansion over the last 150 generations
Demographic inferences based on IBD patterns in the two datasets were also very concordant. We observed a very rapid increase—four orders of magnitude—of the effective population size Ne in the last 150 generations (4500 years assuming a generation time of 30 years, Fig. S11). This is in accordance with previous observations [45] which report an explosive growth of human population in Europe in the last 100 generations. However, the increase of Ne was not constant over time and a rapid decrease of Ne was observed in both datasets between 12 and 22 generations ago, which corresponds to the time period between 1300 and 1700 AD. The population growth in the period preceding and the period following this decrease were also different. While Ne increased 2.5 orders of magnitude in the preceding period, which lasted 128 generations, it increased in the same order of magnitude in the following and much shorter period of the last 12 generations. In-between these two periods, we detected a bottleneck that could reflect the devastating Black Death (1347–1351 AD). This bottleneck seems to affect mainly the Northern part of France (Fig. S12) and this result is robust to change in the parameter related to the IBD segment length (Fig. S13).
Different contributions of Northern Europe and Basque heritage in the six French genetic clusters
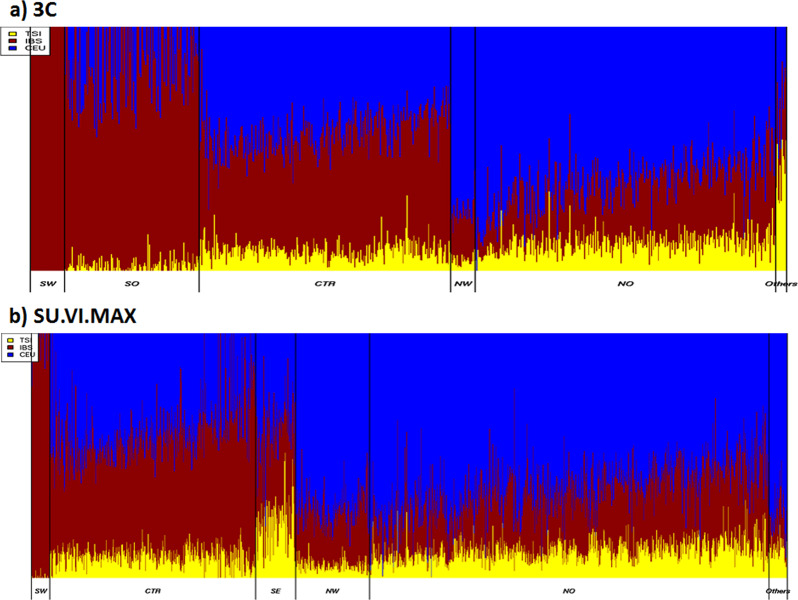
To study the relationship between the genetic clusters observed in France and neighboring European populations, we combined our two datasets with the 1000 G European dataset. As a first step, we ran FineSTRUCTURE on the 1000G European populations excluding Finland and found that they could be divided into three donor groups as CEU and GBR clustered together (north-west Europe) (Fig. S14). We estimated European ancestry contributions in France with SOURCEFIND and reported the total levels of ancestry proportions for each individual grouped by cluster (Fig. 3). We observed similar patterns of admixture between datasets. The proportion of each admixture component from neighboring European countries was significantly different between the six FineSTRUCTURE clusters in both the 3C and SU.VI.MAX datasets (ANOVA, p value < 10−16). As expected, the contribution from north-west Europe (CEU and GBR) was more marked in the north than in the south of France where, instead, the contribution from southern Europe was stronger. The overall contribution from north-west Europe was substantially higher in the NW than in the NO cluster (76% vs 64% in the 3C and 72% vs 63% in SU.VI.MAX). TSI was contributing to the SE cluster while IBS was mainly contributing to the SW cluster, which again was very coherent with the geographic places of birth of individuals. In both dataset, SW had the highest proportions of IBS component. Part of this IBS component could in fact reflect a Basque origin as shown on the PCA plot obtained when combining 3C, SU.VI.MAX, and HGDP European dataset (Fig. S15). This trend is even more pronounced in the 3C where few individuals are grouped together with Basque individuals in the first three dimensions. To explore the relationship between French clusters, data from 3C and SU.VI.MAX were combined and analysed together with PCA. Starting from a set of 216,865 common SNPs, the filtered datasets (after pruning) included 2184 individuals and 63,602 SNPs. We found a strong overlap between clusters except for the SW region, reflecting fewer Basque individuals in the SU.VI.MAX data than in the 3C data (Fig. S16). This SW region geographically corresponds to the “Aquitaine” region described by Julius Caesar in his “Commentarii de Bello Gallico”[1] (Fig. S1).
Fig. 3. Ancestry profiles from the three neighboring European populations inferred by SOURCEFIND.
(a) 3 C and (b) SU.VI.MAX individuals datasets. In each cluster, individuals are ordered according to the latitude of their reported birthplace.
Different contributions of three ancient populations (WHG, EF, SP) in the six French genetic clusters
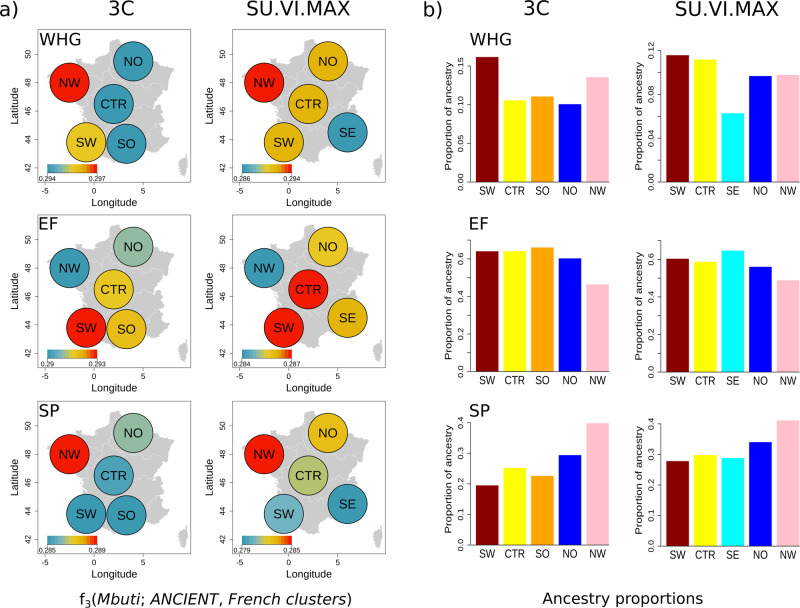
Ancient DNA studies suggest that modern European genetic diversity was shaped by the dispersal of three main ancestral populations: hunter-gatherers, Neolithic farmers, and post-Neolithic SP [3, 37, 44]. We used the outgroup f3-statistics of the form f3(Outgroup; Ancient, French cluster) to test whether the contributions of these populations vary across the identified French clusters. The NW cluster was found to have the largest level of shared drift with both WHG and SP (Fig. 4a), which is compatible with its distribution on the PCA (Fig. S17), and the lowest levels of shared drift with EF. Contrarily, the SE cluster exhibits the lowest levels of shared ancestry with WHG and SP. The highest levels of shared drift with EF are observed in the CTR and SW clusters. Furthermore, negative values of D-statistics of the form D(Mbuti, WHG/SP; NW, remaining clusters) suggest that NW shares more ancestry with WHG and SP than any other French cluster (|Z| > 4, excluding for the D(Mbuti, WHG; NW, SW) in the 3C dataset) (Fig. S18). On the other hand, positive values for D-statistics of the form D(Mbuti, EF; NW, remaining clusters) indicate that the NW cluster has a dearth of sharing with EF in comparison with most of the other populations (|Z| > 4, excepting for the D(Mbuti, EF; NW, NO) = 0.002, Z = 3.064 in the 3C dataset).
Fig. 4. Relationship between French clusters and three ancient populations: western hunter-gatherers (WHG), early Neolithic farmers (EF), and Steppe pastoralists (SP).
(a) Outgroup f3-statistics of the form f3(Mbuti; Ancient population, French clusters) plotted onto the map of France. (b) Ancestry proportions estimated with qpAdm [43] assuming a model where each French cluster results from the mixture of WHG, EF, and SP-related ancestry. Maps were generated using the R statistical package along with ‘maptools’ library.
Interestingly, D-statistics of the form D(Mbuti, EF; SW, remaining clusters) suggest that the SW cluster has increased sharing with EF in comparison with all the other clusters in the 3C dataset (D = −0.0029, −0.0045, −0.0065, and −0.0025, Z = −4.667, −7.179, −7.185, and −4.200 when the French clusters = CTR, NO, NW, and SO, respectively) and with the NW cluster in SU.VI.MAX (D = −0.0038, Z = −5.961). Given that the SW cluster is genetically closer to Basques (Fig. S15) our results are consistent with recently reported evidence for high levels of shared drift between Basques and EF [39, 46].
To estimate the proportions of ancestry related to each ancestral population we modeled the French clusters as a mixture of WHG, EF, and SP-related ancestry. The results show that the French genetic landscape is predominantly of EF-related ancestry (ancestry proportions vary between 46.5 and 66.2%, Fig. 4b), which was found to follow a north-south cline (Fig. S17) with the lowest values found in the NW (<50%). Nevertheless, ancestry proportion estimates also point to a substantial contribution (19.6–41.2%) from SP or SP-related populations to the France clusters mainly in the NW (Fig. S19), for which we observed the largest values (39.9% and 41.2% in 3C and SU.VI.MAX, respectively). WHG were estimated to have contributed less to the ancestry of the French populations with estimated proportions varying between 6.3 and 16.2% with the largest values observed in the SW consistently across the two datasets (16.2% and 11.6% in 3 C and SU.VI.MAX, respectively). In general ancestry proportion estimates are compatible with the outgroup f3-statistics with the exception that, in contrast with the outgroup f3-statistics, ancestry proportions point to larger contributions of WHG to the SW and not to the NW cluster (Figs. 4b and S19). However, the three-way models seem not to fit well the data (P values vary between 0.0021–0.0416 and 0–0.0156 for SU.VI.MAX and 3C clusters, respectively). One possible explanation for this could be ancestry heterogeneity within the source populations, which we expect to be limited as we included only ancient samples with an assignment probability >0.99 to each of the ancestral populations. Furthermore, the f4 statistics matrix seems to point to the presence of a small Eastern HG-like ancestry in our modern populations and/or more recent migration from North Africa within some of the clusters (data not shown).
Discussion
In this paper, we studied the genetic structure of France using data from two independent cohorts of individuals born in different regions of France and whose places of birth could be geolocalized. Modern France has a strategic location at the western part of Europe and on migration routes between the south and north of Europe. Studying its genetic structure is thus of major interest to gain insight on the peopling of Europe. To date however, no exhaustive study had been conducted on the French genetic make-up and our work was intended to fill this gap.
The French genomes were found to map at their expected position in between north-west Europe (GBR and CEU), Italian and Spanish individuals from the 1000 Genomes Project. Within France, correlations were detected between genetic data and geographical information on the individual’s place of birth. Correlation changes gradually over space reflecting isolation by distance. Although we found this close correlation between allele frequency distributions and population geographic distributions, the distribution of genetic diversity within France was also found compatible with the presence of clusters likely explained by local barriers to gene flow.
An important division separates Northern from Southern France. It may coincide with the von Wartburg line, which divides France into “Langue d’Oïl” part (influenced by Germanic speaking) and “Langue d’Oc” part (closer to Roman speaking)—Fig S20. This border has changed through centuries and our north-south limit is close to the limit as it was estimated in the IXth century [47, 48]. This border also follows the Loire River, which has long been a political and cultural border between kingdoms/counties in the north and in the south (Fig. 1).
Regions with strong cultural particularities tend to separate. This is for example the case for Aquitaine in the South-West which has long represented a civilization on its own. The Brittany region is also detected as a separate entity in both datasets. This could be explained both by its position at the end of the continent where it forms a peninsula and, by its history since Brittany has been an independent political entity (Kingdom and, later, duchy of Bretagne), with stable borders, for a long time [49].
The extreme South-Western regions show the highest differentiation with the neighboring clusters. This is particularly strong in 3C dataset, where we even observe an additional cluster. This cluster is likely due to a higher proportion of possibly Basque individuals in 3C, which overlap with HGDP Basque defined individuals. FST between the south-west and the other French clusters was markedly higher than the FST between remaining French clusters. In 3C these values are comparable with what we observed between the Italian and the north-west of Europe (FST = 0.0035). We observed similar trends in SU.VI.MAX even though the level of differentiation with the SW was weaker.
We also observe that the broad-scale genetic structure of France strikingly aligns with two major rivers of France “La Garonne” and “La Loire” (Fig. 1). At a finer-scale, the “Adour” river partition the SW to the SO cluster in the 3C dataset. This alignment suggests moderate genetic isolation between riversides and might reflects isolation by resistance in which specific observed features such as river barriers (or altitude) influence connectivity. The Loire is a large, fierce river. Historically, at each slightly high flood it exceeded its banks and covered its valley over a width of several kilometers, leaving behind it, when it withdrew, mud and swamps. Such wide gap presents a serious obstacle to any kind of relations. The slope, even at the flattest part, is not lower than 0.2 m/km, three time more than for Seine River (Fig. S2). Similarly, the Garonne River also displays one of the fastest flow rates and is characterized by low number of fords and bridges: at least since Strabo, Garonne was a border between the Aquitans and Celtic Gauls [50].
While historical, cultural and political borders seem to have shaped the genetic structure of modern-days France, exhibiting visible clusters, the population is quite homogeneous with low FST values between clusters ranging from 2 × 10−4 up to 3 × 10−3. We find that each cluster is genetically close to the closest neighboring European country, which is in line with continuous gene flow at the European level. However, we observe that Brittany is substantially closer to the population from north-west Europe than to the north of France, in spite of both being equally geographically close. Migration of Britons in what was at the time Armorica (and is now Brittany) may explain this closeness. These migrations may have been quite constant during centuries although a two waves model is generally assumed. A first wave would have occurred in the IVth century when soldiers from British Isles were sent to Armorica, whereas the second wave consisted of Britons escaping the Anglo-Saxon invasions [51]. Additional analyses, on larger datasets may be required to discriminate between these various models.
Trying to understand the process that shaped the modern-day French genetic diversity, we assessed the impact of the three main ancestral populations who seem to have shaped modern European genetic diversity. We observed clear differentiation across clusters, aligning with what we know of the geographical expansion of these populations: early neolithic farmers migration arrived through the south-east of France, from Anatolia, and steppe pastoralist (SP) from the northern plains of Eurasia. While neolithic farmersʼ genome is predominant in modern France population, the SP contribution is higher in the north of France (NO and NW clusters).
The highest level of SP population in the NW cluster may be either due to its position, at an extreme of the possible Neolithic expansion or to later, and more recent migration from northern Europe with high SP proportion, i.e., Celtic and/or Anglo Saxon. Exploration of more recent rare genetic variation will allow a better disentangling of these two hypotheses. Interestingly, the south-western populations seem to harbor highest proportions of early farmers and hunter-gatherer ancestry. Therefore, the genetic structure of the French population was initially shaped by these Stone and Bronze Age events of population migrations and mixture, and then finely-tuned by natural geographical boundaries.
Studying the evolution of the French population size based on genetic data, we observe a very rapid increase in the last generations. This observation is in line with what has been seen in European populations [45]. This “Demographic Transition”, seem to be the result of a reduction in death rate, due to improvements in agriculture (Agricultural Revolution of the XVIIIth century), the mitigation of famines through the improvement of road and financial networks, allowing the delivery and conservation of food [52]. Moreover, with the climatic situation becoming more favorable, poor harvests and epidemics became less frequent [53]. We also observe, in the preceding period, a depression during a period spanning from 12 to 22 generations ago. This may correspond to a period spanning from 1300 to 1700 AD. This period was characterized by a deep depression in population size due to a long series of plague events. While the population size in kingdom of France was estimated to be 20 Million in 1348, it dropped down to 12 Million in 1400, followed by an uneven trajectory to recover the 20 Million at the end of the reign of Louis XIVth (1715 AD) [54].
However, the decrease we observe in the genetic data does seem to affect mainly the Northern part of France, and for instance is mainly observed in the NO cluster. We see no reason for this trend based on historical records (Fig. S21) except perhaps the last plague epidemics in 1666–1670 that was limited to the NO of France. Alternatively, a more diffuse population in the SO (which is in general hilly or mountainous) may explain a lower impact of these dramatic episodes. Plague is expected to have had a very strong impact on the population demography in the past as some epidemics led to substantial reduction in the population sizes [55]. However, we could not detect in our data any footprint of the Justinian plague (541–767 AD) although, according to historical records it had a major impact on the population at that time. This may be due to difficulty to estimate population changes in ancient times, deeper than 50–100 generations, especially in presence of more recent bottleneck and given our reduced sample sizes in some of the groups and IBD resolution power. We expect that increasing sample size especially for the FineSTRUCTURE subgroups with small sample sizes will help getting more detailed information farther in the past.
The strength of our studies lies on concordant observations from two datasets where ascertainment scheme and genotyping strategy were different. But our study also highlights how clustering and spatial genetic pattern analyses can be sensitive to these sampling schemes [56]. Indeed, the concordance between the two studies constantly improved as we forced the 3C sampling towards a geographically uniform distribution, one individual per birth town/village, closer to the geographically homogeneous selection of SU.VI.MAX: this was true for both the ChromoPainter/FineSTRUCTURE clusters and for the migration barriers evidenced using EEMS.
Identification of genetic structure is important to guide future studies of association both for common, but more importantly, for rare variants [57]. In the near future, interrogating the demographic history of France from genetic data will bring more precise results, thanks to whole genome sequencing that, along with new methods, could allow us to perform more detailed demographic inference.
Supplementary information
Acknowledgements
Part of this work was supported by the French National Research Agency (FROGH: ANR-16-CE12–0033) and the European Union via the Marie Skłodowska-Curie actions (PRESTIGE-2017–4–0018). We are most grateful to the Bioinformatics Core Facility of Nantes (BiRD, Biogenouest) for its technical support.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
On behalf of the 3C study: Philippe Amouyel, Jean-François Dartigues, Christophe Tzourio
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Aude Saint Pierre, Joanna Giemza, Isabel Alves, Emmanuelle Génin, Christian Dina
Change history
3/5/2020
An amendment to this article has been published and can be accessed via a link at the top of the article.
Change history
3/11/2020
A Correction to this paper has been published: 10.1038/s41431-020-0604-1
Contributor Information
Aude Saint Pierre, Email: aude.saintpierre@univ-brest.fr.
Christian Dina, Email: christian.dina@univ-nantes.fr.
Supplementary information
The online version of this article (10.1038/s41431-020-0584-1) contains supplementary material, which is available to authorized users.
References
- 1.Caesar CJ. De bello Gallico Commentarius Primus. p. 58–60 BCE.
- 2.Lazaridis I. The evolutionary history of human populations in Europe. Curr Opin Genet Dev. 2018;53:21–7. doi: 10.1016/j.gde.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–13. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornette J. Collection Histoire de France. Paris, France: Belin; 2009–12.
- 5.Cornette J. Atlas Histoire de France (481–2005). Paris, France: Belin; 2016.
- 6.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–7. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science. 2002;298:2381–5. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 8.Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, et al. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am J Hum Genet. 2000;66:979–88. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David A, Peter D. The International HapMap Consortium. A haplotype map of the human genome. Nature 2005;437:1299–320. [DOI] [PMC free article] [PubMed]
- 10.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, Fabianova E, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008;16:1413–29. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- 12.Humphreys K, Grankvist A, Leu M, Hall P, Liu J, Ripatti S, et al. The genetic structure of the Swedish population. PLoS ONE. 2011;6:e22547. doi: 10.1371/journal.pone.0022547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdellaoui A, Hottenga JJ, de Knijff P, Nivard MG, Xiao X, Scheet P, et al. Population structure, migration, and diversifying selection in the Netherlands. Eur J Hum Genet. 2013;21:1277–85. doi: 10.1038/ejhg.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert E, O’Reilly S, Merrigan M, McGettigan D, Molloy AM, Brody LC, et al. Author correction: the Irish DNA atlas: revealing fine-scale population structure and history within Ireland. Sci Rep. 2018;8:7208. doi: 10.1038/s41598-018-24846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, et al. The fine-scale genetic structure of the British population. Nature. 2015;519:309–14. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bycroft C, Fernandez-Rozadilla C, Ruiz-Ponte C, Quintela I, Carracedo A, Donnelly P, et al. Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula. Nat Commun. 2019;10:551. doi: 10.1038/s41467-018-08272-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karakachoff M, Duforet-Frebourg N, Simonet F, Le Scouarnec S, Pellen N, Lecointe S, et al. Fine-scale human genetic structure in Western France. Eur J Hum Genet. 2015;23:831–6. doi: 10.1038/ejhg.2014.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hercberg S, Galan P, Preziosi P, Bertrais S, Mennen L, Malvy D, et al. The SU.VI.MAX Study: a randomized, placebo-controlled trial of the health effects of antioxidant vitamins and minerals. Arch Intern Med. 2004;164:2335–42. doi: 10.1001/archinte.164.21.2335. [DOI] [PubMed] [Google Scholar]
- 19.3C Study Group. Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology 2003;22:316–25. [DOI] [PubMed]
- 20.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 21.Haworth S, Mitchell R, Corbin L, Wade KH, Dudding T, Budu-Aggrey A, et al. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat Commun. 2019;10:333. doi: 10.1038/s41467-018-08219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–73. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 26.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 28.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petkova D, Novembre J, Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 2016;48:94–100. doi: 10.1038/ng.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browning SR, Browning BL. Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am J Hum Genet. 2015;97:404–18. doi: 10.1016/j.ajhg.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browning BL, Browning SR. Detecting identity by descent and estimating genotype error rates in sequence data. Am J Hum Genet. 2013;93:840–51. doi: 10.1016/j.ajhg.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–5. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 35.Gazal S, Sahbatou M, Babron MC, Genin E, Leutenegger AL. High level of inbreeding in final phase of 1000 Genomes Project. Sci Rep. 2015;5:17453. doi: 10.1038/srep17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chacon-Duque JC, Adhikari K, Fuentes-Guajardo M, Mendoza-Revilla J, Acuna-Alonzo V, Barquera R, et al. Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nat Commun. 2018;9:5388. doi: 10.1038/s41467-018-07748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathieson I, Reich D. Differences in the rare variant spectrum among human populations. PLoS Genet. 2017;13:e1006581. doi: 10.1371/journal.pgen.1006581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olalde I, Mallick S, Patterson N, Rohland N, Villalba-Mouco V, Silva M, et al. The genomic history of the Iberian Peninsula over the past 8000 years. Science. 2019;363:1230–4. doi: 10.1126/science.aav4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathieson I, Alpaslan-Roodenberg S, Posth C, Szecsenyi-Nagy A, Rohland N, Mallick S, et al. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anthony DW. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. Princeton, USA: Princeton University Press; 2007.
- 42.Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–24. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient admixture in human history. Genetics. 2012;192:1065–93. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, et al. Massive migration from the Steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–11. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–3. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiang CWK, Marcus JH, Sidore C, Biddanda A, Al-Asadi H, Zoledziewska M, et al. Genomic history of the Sardinian population. Nat Genet. 2018;50:1426–34. doi: 10.1038/s41588-018-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wartburg W von. Les origines des peuples romans. Paris, France: Presses Universitaires de France; 1941.
- 48.Chaurand J. Nouvelle histoire de la langue française. Paris, France: Seuil; 2012.
- 49.Leprohon R. Vie et mort des Bretons sous Louis XIV. Brasparts, France: Beltan; 1984.
- 50.Lasserre F. Strabon, Geographie. Paris, France: Belles Lettres; 1974.
- 51.Fleuriot L. Les Origines de la Bretagne: L'émigration. Lausanne, Switzerland: Payot; 1999.
- 52.Lévy ML. La transition démographique en Occident. Institut National des Études Démographiques 1979.
- 53.Cornette J. Absolutisme et Lumières 1652-783. Paris, France: Hachette; 2008.
- 54.Dupâquier J. Collection Histoire de la population française. Presses Universitaires de France: Paris, France; 1995.
- 55.Biget JL, Bove B, Cornette J. Le temps de la Guerre de Cent ans (1328-453). Belin: Paris, France; 2009.
- 56.Lawson DJ, van Dorp L, Falush D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat Commun. 2018;9:3258. doi: 10.1038/s41467-018-05257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Persyn E, Redon R, Bellanger L, Dina C. The impact of a fine-scale population stratification on rare variant association test results. PLoS ONE. 2018;13:e0207677. doi: 10.1371/journal.pone.0207677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.