Abstract
The genera Kernia and Acaulium comprise species commonly isolated from dung, soil, decaying meat and skin of animal. The taxonomy of these fungi has been controversial and relies mainly on morphological criteria. With the aim to clarify the taxonomy and phylogeny of these fungi, we studied all the available ex-type strains of a large set of species by means of morphological and molecular phylogenetic analyses. Phylogenetic analysis of the partial internal transcribed spacer region (ITS) and the partial 28S rDNA (LSU) showed that the genera Kernia and Acaulium were found to be separated in two distinct lineages in Microascaceae. Based on morphological characters and multilocus phylogenetic analysis of the ITS, LSU, translation elongation factor 1α and β-tubulin genes, the species in Kernia and Acaulium were well separated and two new combinations are introduced, i.e. Acaulium peruvianum and Acaulium retardatum, a new species of Kernia is described, namely Kernia anthracina. Descriptions of the phenotypic features and molecular phylogeny for identification are discussed for accepted species in two genera in this study.
Subject terms: Fungal systems biology, Microbiology, Fungi
Introduction
Kernia was erected by Nieuwland1 for a group of fungi with cleistothecial, with Kernia nitida (Saccardo) Nieuwland as type species firstly and subsequent species have all been characterized by fascicled hair-like ascocarp appendages and reddish-brown to brown ascospores. In 1971, the ascomycete genus Kernia is emended to revised concepts by Malloch2. Five species, K. bifurcotricha Saxena & Mukerji, K. hippocrepida Malloch & Cain, K. nitida (Sacc.) Nieuwland, K. hyalina Malloch & Cain and K. pachypleura Malloch & Cain, are included and two species, K, brachytricha (Ames) Benjamin and K. geniculotricha Seth, are placed in synonymy with K. nitida. Besides, K. bartlettii (Massee & Salmon) Benjamin, K. furcotricha Tandon & Bilgrami, and K. spirotricha Benjamin, are excluded from Kernia. And some new species and combinations were added to it later as K. ovata (Booth) Malloch & Cain, K. retardata Udagawa & T. Muroi, K. setadispersa, K. cauquensis, K. irregularis, K. peruviana Udagawa & Furuya, K. columnaris (H.J. Swart) Woudenb & Samson3–10. In recently, K. hyalina is excluded from Kernia by phylogenetic analysis based on a combined LSU and ITS sequence dataset and morphological characteristics11. Although 11 species are accepted in Kernia, many described species are of doubtful identity because their type materials are lost and their protologues are uninterpretable.
The genus Acaulium was established as the sexual morph and the type species is Acaulium albonigrescens Sopp.12, and this genus was considered as congeneric with Microascus13–15. Acaulium is characterised by annellidic conidiogenesis, guttulate conidia and mycelium forming abundant hyphal fascicles and has generally been considered a synonym of Scopulariopsis but recently was re-instated as an accepted genus of Microascaceae with three species as A. acremonium (Delacr.) Sandoval-Denis, Guarro & Gené, A. albonigrescens Sopp, Skr. Vidensk.-Selsk and A. caviariforme (Malloch & Hubart) Sandoval-Denis, Guarro & Gené16. Acaulium album, formerly known as Doratomyces putredinis, is transferred to Acaulium and redescribed by Woudenberg10 based on morphological, physiological and molecular phylogenetic analyses. In addition, A. pannemaniae Sandoval-Denis is introduced in this genus by morphological and phylogenetic analyses of LSU17. Five species are currently accepted at present10.
Kernia currently comprises species that are commonly isolated from the dung of animal2, 8–10, except two species K. retardata and K. peruviana8, 9, which isolated from soil. Acaulium species have been reported from a variety of environments such as skin of a horse, decaying meat, soil and so on10, 16. From beginning, the genera of Kernia and Acaulium have been controversial and rely mainly on morphological criteria. Recent molecular studies have demonstrated that the Microascaceae contains several closely related genera and difficult to separate morphologically16. Multilocus phylogenetic analysis have considerably improved our understanding of species concepts in many fungal groups18–25, but the studies for revising the genera of Kernia and Acaulium are relatively limited. Besides, during our investigation of intestinal fungi in animals in China, three particular Kernia isolates from the dung of marmot were isolated. The present work also aims to clarify the taxonomic position of these strains as putative new species using the genealogical concordance analysis24. We provide a multigene (ITS, LSU, TEF, TUB) phylogeny of Kernia and Acaulium and related fungi based on a large set of strains, which includes all available ex-type cultures and well-identified reference strains from international culture collections.
Results
Generic circumscription
DNA sequences determined in this study are deposited in GenBank, and accession numbers are listed in Table 1. To delineate generic boundaries, we conducted a phylogenetic analysis using the combined LSU and ITS datasets including 29 currently accepted species belonging to nine genera of Microascaceae and one species of the family Graphiaceae. Graphium penicillioides were selected as outgroup (Fig. 1). The final alignment consisted of 31 strains and contained 1,385 characters (LSU 796, ITS 589). Figure 1 shows the ML tree including ML bootstrap values (bs) and posterior probabilities (pp) values. The trees obtained from ML and Bayesian analyses of the individual loci and the combined analysis showed congruent topologies. The phylogenetic inferences (Fig. 1) showed that Kernia and Acaulium were monophyletic, the species of Kernia and Acaulium clustered into a single, well-supported lineage (bs = 100%/pp = 100%), respectively. Figure 2 is demonstrated that the colonies of K. peruviana CBS 320.91, K. retardata CBS 707.82 and A. album CBS 539.85 can form white to pale grey colonies with dense hyphal fascicles. Therefore, K. peruviana and K. retardata were identified as the Acaulium species in this study. However, other type Kernia species grow slowly and form compact, brown to dark brown colonies apparently different with the Acaulium species. Among with the K. geniculotricha and K. nitida were located in the same clade with the value of (bs = 94%/pp = 91%) and combined with morphological characters, they could be identified as the same species in this research. Three Kernia strains which were isolated from the dung of marmot, were clustered with the species of K. hippocrepida and have a well-supported value (bs = 100%/pp = 100%).
Table 1.
Strains and sequence accession numbers included in this study.
| Species | Strain no. | Isolation source | Location | GenBank accession no. | |||
|---|---|---|---|---|---|---|---|
| ITS | TUB | LSU | TEF | ||||
| Acaulium acremonium | CBS 290.38T | Skin of a horse | Denmark | LM652456 | LN851108 | LN851001 | HG380362 |
| Acaulium albonigrescens | CBS 109.69T | Litter, treated | Japan | KY852469 | LN851111 | KY852480 | LN851058 |
| Acaulium album | CBS 539.85T | Hair in dung in pole cat | Netherlands | MN991960 | MN982419 | MN991968 | MN982411 |
| Acaulium caviariforme | CBS 536.87T | Decaying meat | Belgium | LM652392 | LN851112 | LN851005 | LN851059 |
| Acaulium pannemaniae | CBS 145025T | Soil | Netherlands | LS999990 | LS999993 | LS999991 | LS999992 |
| Acaulium peruvianum | CBS 320.91T | Soil | Peru | MN991959 | MN982418 | MN991966 | – |
| Acaulium retardatum | CBS 707.82T | From paddy soil | Japan | MN991961 | – | MN991969 | MN982412 |
| Cephalotrichum asperulum | CBS 582.71T | Soil | Argentina | LN850960 | LN851114 | LN851007 | LN851061 |
| Cephalotrichum brevistipitatum | CBS 157.57T | Solanum tuberosum | Netherlands | LN850984 | LN851138 | LN851031 | LN851084 |
| Cephalotrichum dendrocephalum | CBS 528.85T | Cultivated soil | Iraq | LN850966 | LN851120 | NG_059041 | LN851067 |
| Cephalotrichum microsporum | CBS 523.63T | Wheat field soil | Germany: | LN850967 | LN851121 | LN851014 | LN851068 |
| Gamsia columbina | CBS 233.66T | Sandy soil | Germany | LN850990 | LN851147 | LN851039 | LN851092 |
| Graphium penicillioides | CBS 102632T | Populus nigra | Czech Republic | KY852474 | – | KY852485 | – |
| Kernia anthracina | CGMCC 3.19001T | Dung of marmot | Beijing, China | MK773539 | MK773545 | MK773542 | MK773568 |
| Kernia anthracina | CGMCC 3.19002 | Dung of marmot | Beijing, China | MK773540 | MK773546 | MK773543 | MK773569 |
| Kernia anthracina | CGMCC 3.19003 | Dung of marmot | Beijing, China | MK773541 | MK773547 | MK773544 | MK773570 |
| Kernia columnaris | CBS 159.66T | Dung of hare | South Africa | MN991957 | MN982416 | MN991962 | MN982409 |
| Kernia geniculotricha | CBS 599.68T | On dung of Oryctolagus cuniculus | Germany | MN991956 | MN982414 | MN991964 | MN982408 |
| Kernia hippocrepida | CBS 774.70T | On dung of Erethizon dorsatus | Ontario, Canada | MN991954 | MN982413 | – | MN982406 |
| Kernia nitida | CBS 282.52T | Chrysolina sanguinolenta | France | MN991955 | MN982415 | MN991963 | MN982407 |
| Kernia pachypleura | CBS 776.70T | On dung of Loxodonta africana | Uganda | MN991958 | MN982417 | MN991965 | MN982410 |
| Microascus cirrosus | CBS 217.31T | Leaf of Prunus sp. | Italy | KX923838 | – | AF400860 | – |
| Microascus longirostris | CBS 196.61T | Wasp's nest | USA: Maine | LM652421 | LM652634 | LN851043 | LM652566 |
| Microascus senegalensis | CBS 277.74T | Mangrove soil | Senegal | KX923929 | – | AF400867 | – |
| Petriella musispora | CBS 745.69 | On rotten wood of Populus grandidentata | Ontario, Canada | MH859407 | – | AF027663 | – |
| Petriella setifera | CBS 390.75 | skin lesion in Tursiops truncatus | Netherlands | AY882353 | – | AF027664 | – |
| Petriellopsis africana | CBS 311.72T | Brown sandy soil | Namibia | AJ888425 | – | EF151331 | – |
| Pseudallescheria ellipsoidea | CBS 418.73T | Soil | Tajikistan | EF151323 | MH271617 | AF027671 | – |
| Wardomyces anomalus | CBS 299.61T | Air cell of egg | Canada: Ontario | LN850992 | LN851149 | LN851044 | LN851095 |
| Wardomyces inflatus | CBS 367.62T | Greenhouse soil | Belgium | LN850994 | LN851153 | AF400886 | LN851099 |
| Wardomyces pulvinatus | CBS 112.65T | Salt-marsh | England, UK | LN850997 | LN851156 | LN851051 | LN851102 |
CBS: CBS Fungal Biodiversity Centre, Utrecht, The Netherlands.
Sequences newly generated in this study are indicated in bold.
‘T’ represents type strain.
Figure 1.
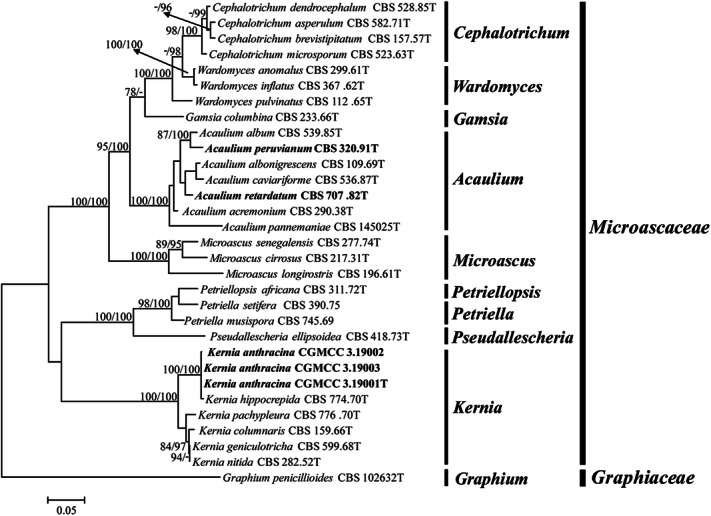
Maximum likelihood (ML) tree obtained from the combined LSU and ITS sequences of 31 representative taxa of Microascaceae and Graphiaceae. Numbers on the branches are ML bootstrap values (bs) above 75%, followed by Bayesian posterior probabilities (pp) above 95%. A dash (–) indicates support value lower than 75% bs or 95% pp. Branch lengths are proportional to distance. Ex-type strains are indicated with T. The tree was rooted to Graphium penicillioides (CBS 102632).
Figure 2.
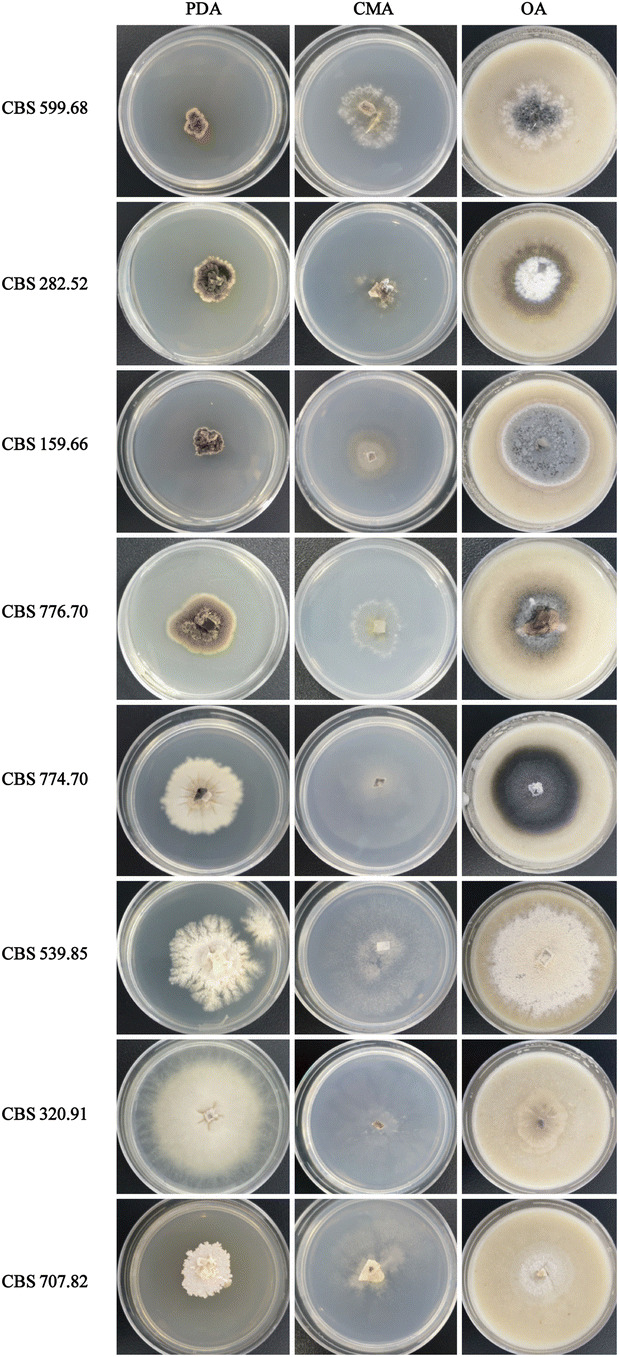
The colony morphology of the species in Kernia and Acaulium was growing on PDA, CMA and OA after 35 days, respectively.
Phylogeny of two genera and genealogical concordance analysis
The second dataset was composed of 19 taxa (including the outgroup) with the following four loci combined: ITS (1–434), LSU (435–1,163), TEF (1,164–2,024), TUB (2,025–2,401). The phylogenic tree was constructed and the branch support values (≥ 50%) and the Bayesian posterior probabilities from Bayesian analyses (≥ 95%) were indicated (Fig. 3). The phylogenetic tree grouped 19 strains into three clades comprising Kernia (bs = 100%/pp = 100%) and Acaulium (bs = 98%/pp = 100%) subclades with high bootstrap values. Coupled with morphological characteristics (Fig. 2), two new combination species A. peruvianum and A. retardatum are proposed. Besides, Three Kernia strains were clustered with the species of K. hippocrepida in independent group and have a well-supported value (bs = 100%/pp = 100%).
Figure 3.
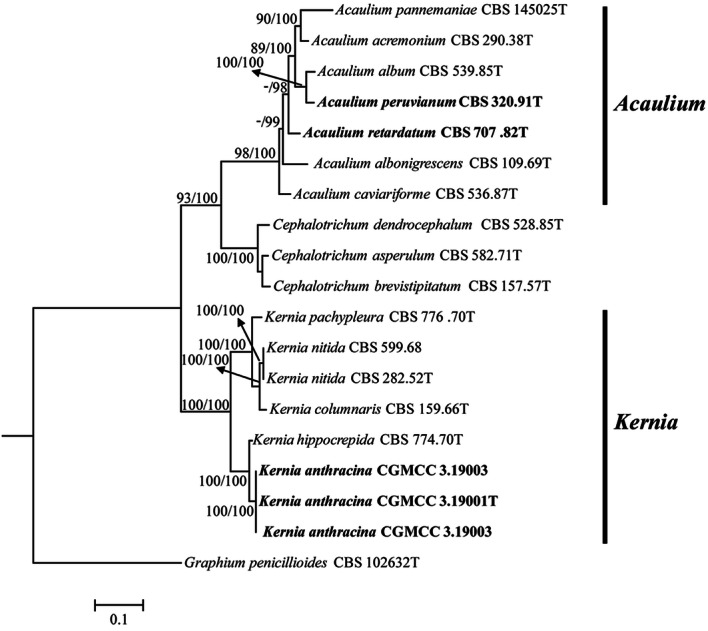
Maximum likelihood (ML) tree obtained from the combined LSU, ITS, TEF and TUB sequences of 19 representative taxa included most of species in the genera of Kernia and Acaulium. Numbers on the branches are ML bootstrap values (bs) above 75% and Bayesian posterior probabilities (pp) above 95%. A dash (–) indicates support value lower than 75% bs or 95% pp. Branch lengths are proportional to distance. Ex-type strains are indicated with T. The tree was rooted to Graphium penicillioides (CBS 102632).
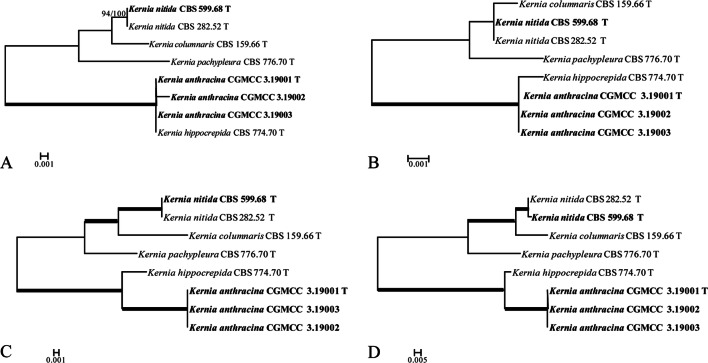
BLAST searches of GenBank using the ITS sequences of three Kernia strains isolated from the dung of marmot revealed that three strains showed 94.7% similarity to K. nitida CBS 282.52T (KY852476). Multiple sequence alignment and sequence polymorphism analysis were further carried out in the ITS region of three Kernia strains. Compared with K. hippocrepida CBS 774.70T, had only two variable positions that is transition and indel in the ITS region. However, sequence polymorphism analysis in the TUB region indicated that strain CGMCC 3.19001T have 35 variable positions, including 12 transitions, 7 transversions, and 16 indels, indicating a lower similarity (93.2%) to K. hippocrepida CBS 774.70T (Supplementary Table 1). In genealogical concordance analysis, K. hippocrepida and K. anthracina strains were divided into different statistically supported subclades in TUB and TEF tree with a high bootstrap value (bs = 100%/pp = 100%) (Fig. 4). In addition, K. geniculotricha CBS 599.68T and K. nitida CBS 282.52T were clustered together and have a high similarity (99%) from four different gene tree.
Figure 4.
Delimitation of Kernia anthracina and its closely related species based on the separate analyses of four loci: ITS (A), LSU (B), TEF (C) and TUB (D). ML bootstrap values (bs) above 97% and Bayesian posterior probabilities (pp) above 99% are represented as bold lines. The TUB and TEF tree divides K. anthracina and K. hippocrepida isolates into different clades while the ITS loci place these isolates on a single branch. Genealogical concordance is seen in four trees, which supports K. anthracina and K. hippocrepida as distinct species.
Taxonomy
Based on the results of the above multilocus sequence analysis and a morphological analysis, the species of the genera Acaulium and Kernia have been reassessed accordingly. Their current circumscription is revised and several new taxa and combinations are proposed as follows:
Acaulium peruvianum (Udagawa & Furuya) L. Su comb. nov. Fig. 5.
Figure 5.
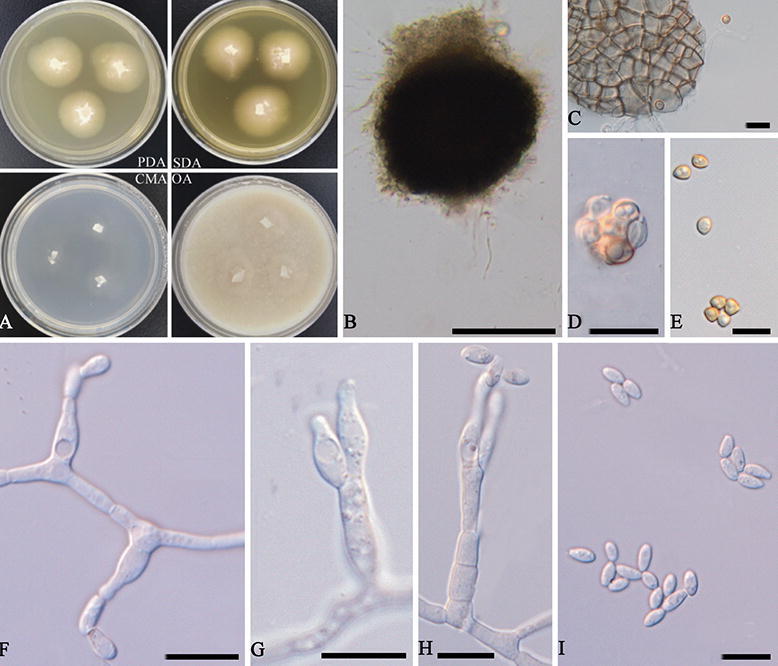
Acaulium peruvianum (ex-type CBS 320.91). (A) Colonies on different media after 10 days at 20 °C. (B) Ascoma. (C) Ascomatal peridium. (D,E) Ascospores. (F–H) Conidiophores and conidiogenous cells. (I) Conidia. Scale bars: (B) = 100 μm; (C–I) = 10 μm.
MycoBank: MB 834193.
Basionym: Kernia peruviana Udagawa & Furuya, Mycotaxon 33: 295. 1988.
Hyphae hyaline to subhyaline, smooth-walled, 1–4 μm (x̅ = 2.7 μm) wide. Conidiophores often arising from the substratum or from the aerial mycelium, branched or unbranched, septate, smooth, cylindrical, 9–25 × 2–4 μm (x̅ = 14.0 × 3.0 μm). Conidiogenous cells solitary or more commonly united into synnemata, percurrent in conidiophores or produced on hyphae in laterally, flask-shaped, subhyaline and smooth-walled, 6–11 × 2–3.5 μm (x̅ = 8.9 × 2.7 μm). Conidia ellipsoidal to fusiform, with a truncate base and rounded or bluntly pointed apex, subhyaline, smooth and slightly thick-walled, 3.5–7 × 1–3 μm (x̅ = 5.0 × 2.2 μm). Sexual morph observed. Cleistothecia superficial, non-ostiolate, dark brown to black, globose, 119–160 μm (x̅ = 143.9 μm) diam., glabrous at maturity except for a few hyphal attachments. Asci 8-spored, globose to ovoid, evanescent. Ascospores irregularly arranged, pale yellowish brown to brown, broadly ovoid to fusiform, 3–5 × 2–4 μm (x̅ = 4.0 × 2.8 μm).
Colonies on PDA reaching 17 mm diameter after 10 days at 20 °C, planar, finely felty with tufts of mycelium in center, white to cream. On SDA reaching 21 mm diameter, planar, finely felty with tufts of mycelium in center, white to cream. On CMA reaching 16 mm diameter, planar, subhyaline. On OA reaching 15 mm diameter, planar to low convex, white to creamcoloured centre, margin discrete.
Specimens examined. Peru, Tamshiyacu, near Iquitos, T. Akiyama, from soil, 1987, S. Udagawa (culture ex-type CBS 320.91 = NHL 2,985).
Notes. This species was originally placed in Kernia based on morphological features of the well developed sexual morph9. In our phylogenetic analysis, the ex-type culture of Acaulium peruvianum grouped with high statistical support with species of A. album. A. peruvianum is morphologically different with A. album, A. peruvianum produces sexual and asexual morphs in culture, besides, most of conidiogenous cells directly produced from hyphae. However, A. album has abundant monoverticillate, irregularly biverticillate and terverticillate, or reduced to single conidiogenous cells10. Conidiogenous cells of A. album are smaller (6–8.5 × 2.5–3 μm) than A. peruvianum.
Acaulium retardatum (Udagawa & T. Muroi) L. Su comb. nov. Fig. 6.
Figure 6.
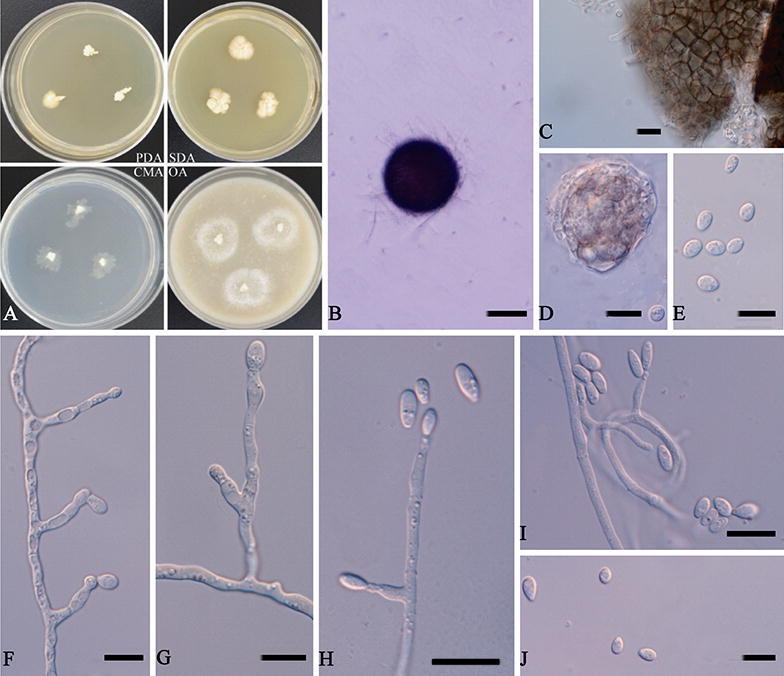
Acaulium retardatum (ex-type CBS 707.82). (A) Colonies on different media after 10 days at 20 °C. (B) Ascoma. (C) Ascomatal peridium. (D) Ascus. (E) Ascospores. (F–I) Conidiophores and conidiogenous cells. (J) Conidia. Scale bars: (B) = 100 μm; (C–J) = 10 μm.
MycoBank: MB 834194.
Basionym: Kernia retardata Udagawa & T. Muroi, Trans. Mycol. Soc. 22(1): 18. 1981.
Hyphae hyaline to subhyaline, thin- and smooth-walled, 1–4 μm (x̅ = 2.6 μm) wide. Conidiophores branched or unbranched, septate, cylindrical. Conidiogenous cells flask-shaped to nearly cylindrical, subhyaline and smooth-walled, terminal or lateral in hyphae or hyphae coil, 8.5–23 × 2–5 μm (x̅ = 13.3 × 3.0 μm). Conidia ellipsoidal to fusiform, with a truncate base, subhyaline, smooth and slightly thick-walled, 4–10.5 × 3–6 μm (x̅ = 7.1 × 4.9 μm). Sexual morph observed. Cleistothecia superficial, non-ostiolate, dark brown to black, globose, 106.5–154 μm (x̅ = 129.1 μm) diam., glabrous at maturity except for a few hyphal attachments. Asci 8-spored, globose to subglobose, evanescent. Ascospores irregularly arranged, grey to pale yellow, broadly ovoid to ellipsoidal, 4–8 × 3–5 μm (x̅ = 5.6 × 3.7 μm).
Colonies on PDA reaching 5 mm diameter after 10 days at 20 °C, slow growing, raised centrally, with flat and irregular margin, white. On SDA reaching 9 mm diameter, moderately growing, raised centrally, aerial mycelium absent or sparse, white to cream. On CMA reaching 9 mm diameter, moderately growing, planar, white, margin discrete. On OA reaching 15 mm diameter, planar, white.
Specimens examined. Japan, Nishinasuno-machi, Nasu-gun, Tochigi, Udagawa, S, from rice-field soil, 1988, S. Udagawa (culture ex-type CBS 707.82 = NHL 2,879).
Notes. This species was originally placed in Kernia based on sexual morphological features8. The phylogenetic analysis shows that the ex-type culture of Acaulium retardatum grouped with statistical support with species of A. albonigrescens and A. caviariforme. A. retardatum is morphologically similar to A. caviariforme; both species produce sexual and asexual morphs in culture. However, A. caviariforme has fusiform, pale orange to copper-red ascospores, and brown, obovoid to ellipsoidal conidia16; ascospores of A. retardatum are smaller, broadly ovoid to ellipsoidal.
Kernia anthracina L. Su, H. Zhu & C. Qin, sp. nov. Fig. 7.
Figure 7.
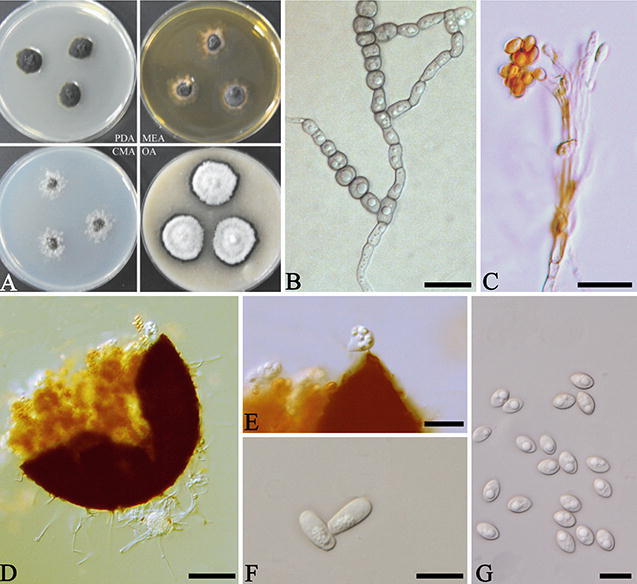
Kernia anthracina (CGMCC 3.19001). (A) Colonies on different media after 20 days at 30 °C. (B, C) Hyphae and conidiophores. (D) Cleistothecia. (E) Ascus. (F) Conidia. (G) Ascospores. Scale bars: (B,F, G) = 10 μm, (C–E) = 20 μm.
MycoBank: MB 830661.
Etymology: Referring to the coal-black colony.
Holotype: HMAS 255463.
Hyphae septate, branched, catenate, hyaline to subhyaline, mostly 2–4.5 μm (x̅ = 3.1 μm) wide. Conidiophores, with scopulariopsis-like branching pattern, produce acrospores. Conidia formed in slimy heads at the apex of the scopulariopsis-like branch, broadly clavate to ellipsoid with a slightly apiculate base, smooth to finely roughened, 6.5–14 × 1–5 μm (x̅ = 10.8 × 3.2 μm). Cleistothecia abundant in CMA, gregarious, superficial, non-ostiolate, glabrous at maturity, black, globose to subglobose, 55–106 μm (x̅ = 77.9 μm) diameter; peridium with a textura intricata. Asci ampulliform. Ascospores irregularly arranged, pale yellowish brown to straw coloured, ovoid to fusiform or ellipsoidal, 5–8 × 3–6 μm (x̅ = 6.9 × 4.8 μm). Optimal growth temperatures are 25–30 °C, no growth at 40 °C.
Colonies on PDA reaching 6 mm diameter after 20 days at 30 °C, 4 mm on SDA, 5 mm on CMA and 13 mm on OA. Colonies on PDA, black in obverse, compact, reverse black, raised centrally, aerial mycelium absent or sparse.
Specimens examined. China, Beijing, Fangshan District, in north center for experimental animal resources, Institute of medical laboratory animal science, Chinese academy of medical sciences, 116°13′ E, 39°48′ N, 58 m above sea level, from fresh dung samples of healthy Marmota monax, 7 December 2017, collected and isolated by L. Su (HOLOTYPE: HMAS 255463, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; dried culture of ex-type CGMCC 3.19001T on PDA), living cultures, CGMCC 3.19001, CGMCC 3.19002, CGMCC 3.19003.
Notes. Strains of K. anthracina have the typical features of Kernia such as compact growth, nonostiolate, ascomata, and ovoid to ellipsoidal ascospores28. K. anthracina not only has a scopulariopsis-like asexual morph, but is also supported by phylogenetic tree based on the combined four genes dataset (Fig. 3) and genealogical concordance analysis (Fig. 4). Although K. anthracina is closed to K. hippocrepida CBS 774.70T with ITS sequences, they apparently differs from morphological characters as K. hippocrepida produced reniform ascospores and conidiophores produced from coiled or irregularly twisted (Fig. 8A–D) while K. anthracina production of scopulariopsis-like conidiophores and ovoid to ellipsoidal ascospores.
Figure 8.
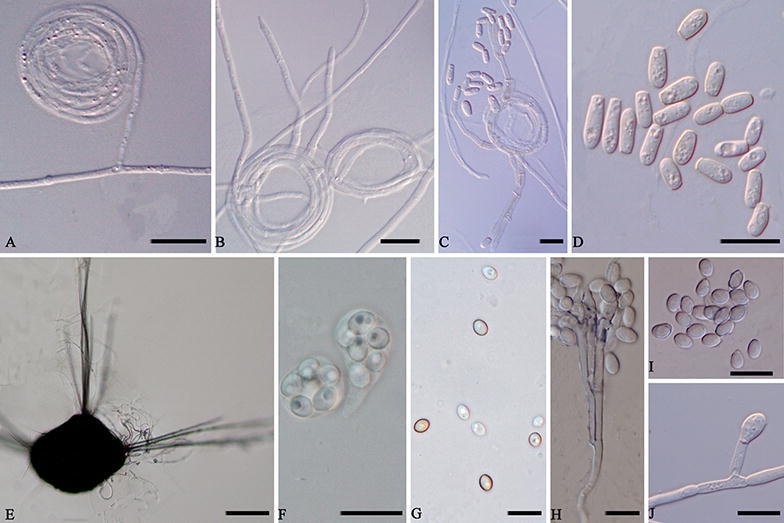
Morphology characters of the two species. Kernia hippocrepida (CBS 774.70). (A,B) Hyphae coil. (C) Conidiophores. (D) Conidia. Kernia nitida (CBS 599.68). (E) Cleistothecia. (F) Ascus. (G) Ascospores. (H) Conidiophores and conidiogenous cells. (I,J) Conidia. Scale bars: (A–D,F–J) = 10 μm; (E) = 100 μm.
Kernia nitida (Saccardo) Nieuwland. Amer. Midland Natur. 4: 379. 1916. Figure 8E–J.
Basionym: Magnusia nitida Saccardo. Michelia, 1: 123. 1878.
Synonym: Kernia geniculotricha Seth. Acta Bot. Neerl. 17: 481. 1968.
Description and illustrations: Seth (1968).
Specimens examined. Germany, near Hamburg, dung of rabbit, 1968, H.K. Seth (culture ex-type CBS 599.68 = ATCC 18529).
Notes. Although Malloch and Cain28 placed the species of K. geniculotricha as the synonym of K. nitida only based on numerous drawings and aquarels which are thus proposed, the isolate CBS 599.68 studied and proposed here conforms to the morphological characteristics of descriptions and phylogeny of K. nitida. The isolate of CBS 599.68 forms compact colonies on PDA, simple or branched, hyaline to light brown conidiophores, and bearing a cluster of annellophores directly at the apex or repeatedly and compactly branching to form a dense penicillus, 10–20.5 × 1–3.5 μm (x̅ = 14.4 × 2.0 μm), produced in clusters of two to three at the tips of the conidiophores or metulae, rarely solitary, flaskshaped to nearly cylindrical, 7–15 × 1–4 μm (x̅ = 9.7 × 2.5 μm); conidia ovoid to ellipsoidal, 3–6.5 × 2.5–4.5 μm (x̅ = 5.1 × 3.2 μm). Besides, The isolate has a sexual morph characterised by abundant cleistothecia on CMA, gregarious, superficial, non-ostiolate, black, opaque, ovoid, 143–323.5 μm (x̅ = 203.9 μm) diam., hairs emerging as two opposing or triangle symmetrical on the cleistothecium, dark brown to black. Asci 8-spored, globose to ampulliform, 8–18 μm (x̅ = 11.2 μm) diam, evanescent. Ascospores irregularly arranged, pale brown, smooth, broadly ovoid to globose, 3.5–6 × 2.5–5.5 μm (x̅ = 4.9 × 3.6 μm). In addition, K. geniculotricha is a well-circumscribed species described from on dung of Oryctolagus cuniculus in Germany. All these characters are similar to the species of K. nitida. Combined with phylogeny and genealogical concordance analysis, we identified the K. geniculotricha as the synonym of K. nitida.
Identification keys
According to the morphological features, identification keys were constructed for the different genera including all the phylogenetic species recognised in this study (Supplementary information 1).
Discussion
The family Microascaceae was established by Luttrell26, comprising saprobic and plant pathogenic species. Some species of Microascaceae are opportunistic pathogens and show intrinsic resistance to antifungal agents11, 20, 27. Recent molecular studies have demonstrated that the Microascaceae contains several closely related genera that are difficult to separate morphologically11 including Microascus, Scedosporium and Scopulariopsis. Recently, three of the most debated genera of the family, Microascus, Scopulariopsis and Pithoascus were revised by morphology and multigene phylogeny11, 16. As a result, several taxa were excluded from these genera and appeared as a new lineage within the Microascaceae as Acaulium11.
In this study we have reviewed the taxonomic circumscription of species in the genera of Kernia and Acaulium, traditionally referred to as sexual and asexual morphs, respectively, and two genera using a polyphasic approach based on the genealogical concordance analysis, phylogeny and morphological data. These results show that Kernia and Acaulium constitute two phylogenetically distant lineages, combining the results of phenotypic data, delineate the accepted species of the two new combination species, proposing a new species, which are clarifying the identity of species as Kernia nitida and K. geniculotricha, reclassifying the white synnematous species as Acaulium peruvianum and A. retardatum, and describing a new species K. anthracina isolated from the dung of marmot. The species of Kernia are mainly isolated from dung of various animals, while the species of Acaulium have a worldwide distribution and are mainly isolated from dung, litter, soil, skin of a horse and decaying meat4–10, 16.
Sandoval-Denis et al.11 was firstly attempts to clarify phylogenetically the relationships among the different genera of the Microascaceae by the use of partial LSU and ITS sequences. Subsequently, Microascaceae was revised by Sandoval-Denis et al.16 based on morphological, physiological and molecular phylogenetic analyses using DNA sequence data of four loci (ITS, LSU, TEF and TUB). These studies demonstrated that several genera of Microascaceae raised questions concerning correct positions of several members of the family and their generic circumscriptions, suggesting a possible subdivision of Microascus and Scopulariopsis into several smaller genera as Kernia and Acaulium. Our results based on the phylogenetic reconstructions of two loci (LSU, ITS) indicated that Kernia and Acaulium fall into two groups (Fig. 1). Besides, it also shows that Kernia and Acaulium species can be well separate by phylogentic analysis of four loci (LSU, ITS, TEF and TUB). As known that Acaulium is characterised by the formation of pale colonies with dense hyphal fascicles and the presence of abundant oil drops in the mycelium, conidia and ascospores, showing a guttulate appearance16. The new combination species A. peruvianum and A. retardatum clustered in Acaulium group, in which the species produce white and pale grey colonies, and have a wide isolation source. A new species was clustered in Kernia group, which form compact dark brown or black colonies, and mainly isolated from dung (Figs. 2, 3). In addition, species delineation was also assessed in the genus of Kernia as the closed species of K. anthracina and K. hippocrepida under the genealogical concordance analysis using DNA sequence data of four loci (Fig. 4).
The absence of clear diagnostic morphological characters can be used to identify species which belonging to the Kernia and Acaulium species2, 11, 16. The species of ‘K. geniculotricha’ and K. nitida have been identified two different species according to morphologically characters, but molecular data can easily identified them as the same species, using any of the four genes studied here. Some species as A. peruvianum and A. retardatum isolates were initially identified as K. peruviana, K. retardate at CBS based on their morphology. Combined the molecular data, the group of species that would previously have been included in Kernia and Acaulium are easily recognized. Our phylogeny demonstrates that, although Kernia and Acaulium share similar morphological and ecological traits, they are in fact genetically distant. The phylogenetic data is supported by relevant morphological differences, such as the color of colonies, the shape of ascospores or conspicuously hairy ascomata2, 8–10.
The new species, K. anthracina and K. hippocrepida, are very similar in ITS but easily distinguished by TUB and TEF sequences. All Kernia species can be well separated with TUB and TEF partial gene sequences. Based on ITS alone, K. anthracina and K. hippocrepida cannot be distinguished (Fig. 4), but morphology and TUB and TEF sequences clearly differentiate them. The lack of the isotype herbarium specimens examined here prevented us from conclusively characterizing five of the other described species K. bifurcotricha, K. setadispersa, K. cauquensis, K. irregularis, K. ovata, leaving them as nomena dubia. From our study, we found that it is easy to identify the species of Kernia and Acaulium by polyphasic approach.
The delimitation of the two genera in this study contributes to an integrated phylogeny of the family Microascaceae. The two monophyletic genera currently accepted are statistically supported in the four-locus phylogeny (Fig. 3). There are seven species included in Acaulium by our revision, while ten species in Kernia. It is regret that some species of Kernia absent holotype material and unavailable for these species. Therefore, further studies are needed to establish a comprehensive modern classification of the Kernia and to give better insight into the evolutionary relationships among the species in the genus.
Materials and methods
Eight Kernia and Acaulium ex-type strains were obtained from the CBS culture collection (CBS) housed at the Westerdijk Fungal Biodiversity Institute (WI), Utrecht, the Netherlands. More isolates potentially related to the obtained Acaulium strains were selected based on a preliminary phylogenetic analysis of LSU + ITS sequences from GenBank, as well as several cultures of the Kernia, which isolated from the feces of Marmota monax25, maintained in China General Microbiological Culture Collection Center (CGMCC) in China. All the strains used in this study are listed in Table 1. The strains were incubated on different media such as Potato dextrose agar (PDA), Malt extract agar (MEA), Sabouraud Dextrose Agar (SDA), Corn meal agar (CMA), and Oatmeal agar (OA) (Becton, Dickinson & Co.) at 20 °C. Colony morphology and microscopic characteristics were examined, measured and photographed after incubation for 10 days with the methods of Su et al.25. Means and standard deviations (SD) were calculated from at least 50 measurements. The ex-type living cultures were deposited in the China General Microbiological Culture Collection Center (CGMCC). The dried culture and microscope slide were deposited in Herbarium Mycologicum, Academia Sinica, Beijing, China (HMAS). Nomenclatural novelties and descriptions were registered in MycoBank (https://www.MycoBank.org).
DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from mycelia grown on PDA or OA plates using the protocol of Guo et al.28. Primers ITS1 and ITS4 were used to amplify the ITS region of the nuclear rRNA gene29, primers LROR/LR5 primers were used for the partial 28S rDNA (LSU)30, primers 983F and 2218R31 for the elongation factor 1-α gene (TEF), and primers Bt2a and Bt2b32 for the partial β-tubulin gene (TUB). PCR was performed in a 25 μL reaction volume containing 1.0 μL DNA template, 1.0 μL of each forward and reverse primers, 12.5 μL 2 × MasterMix (Tiangen Biotech Co. Ltd., Beijing, China) and 10.5 μL ddH2O with the following cycling parameters: 94 °C for 40 s; 35 cycles at 94 °C for 40 s, annealing temperature specific for the gene amplified (52 °C for LSU, 55 °C for TEF and ITS, 58 °C for TUB) for 60 s and 72 °C for 120 s; and a final extension at 72 °C for 10 min. The PCR products were sequenced by Beijing Sunbiotech Co. Ltd. (Beijing, China). Sequences were compared with accessions in the GenBank database via a BLASTn search to determine the most likely taxonomic designation.
Phylogenetic analysis
Sequence data of the four loci were aligned with Clustal X33. Reference sequences were retrieved from GenBank and the accession numbers indicated in Table 1. Manual editing of sequences was performed in MEGA634. The concatenated sequences (LSU + ITS) or (LSU + TUB + TEF + ITS) were assembled using SeaView35 and alignments were deposited in TreeBASE (www. treebase.org, submission no.: S25764). The combined dataset of two or four loci was analyzed phylogenetically using Bayesian MCMC36 and Maximum Likelihood37, respectively. For the Bayesian analyses, the models of evolution were estimated by using MrModeltest 2.338. Posterior probabilities (PP)39,40 were determined by Markov Chain Monte Carlo sampling (MCMC), Six simultaneous Markov chains were run for 2,000,000 generations and trees were sampled every 100th generation (resulting in 20,000 total trees). The first 4,000 trees represented the burn-in phase of the analyses and were discarded and the remaining 16,000 trees were used for calculating PP in the majority rule consensus tree. For the ML analysis in RAxML37, the GTRGAMMA model was used for all partitions, in accordance with recommendations in the RAxML manual against the use of invariant sites. Analyses were performed using the CIPRES web portal41. Trees were visualised in TreeView 1.6.642.
Supplementary information
Supplementary information 1 (DOCX 14 kb)
Supplementary information 2 (DOCX 13 kb)
Acknowledgements
The authors are indebted to CBS culture collection (CBS) housed at the Westerdijk Fungal Biodiversity Institute (WI), Utrecht, the Netherlands for providing ex-type strains and their useful suggestions. This study was supported by the Chinese Academy of Medical Sciences Innovation Fund for Medical Science (no. 2017-12M-2-005, 2016-12M-2-006), National Natural Science Foundation of China (no. 31700022) and National Mega projects of China for Major Infectious Diseases (2017ZX10304402).
Author contributions
L.S. performed all the experimental work, culturing the samples, isolating in pure culture the fungi and performing their phenotypic characterization, as well as the DNA extraction and purification, gene sequencing and data processing for phylogenetic analysis, being one of the major contributors of this manuscript. H.Z., and C.Q., supervised all steps of the experimental work by L.S., and reviewing of the draft several times. Y.N. help buy type strains from CBS, gave useful suggestions to write the manuscript and reviewed the draft several times. Y.G. and X.D. contributed actively in the identification and taxonomy of the fungal strains, and reviewed the draft several times. J.G. and L.Z. gave useful suggestions to write the manuscript and reviewed several times the draft. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/10/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
is available for this paper at 10.1038/s41598-020-67347-1.
References
- 1.Nieuwland JA. Critical notes on new and old genera of plants. VIII. Am. Midl. Nat. 1916;4:379–386. doi: 10.2307/2992735. [DOI] [Google Scholar]
- 2.Malloch D, Cain RF. The genus Kernia. Can. J. Bot. 1971;49:855–867. doi: 10.1139/b71-126. [DOI] [Google Scholar]
- 3.Lodha BC. Studies on coprophilous fungi IV. Some cleistothecial Ascomycetes. J. India Bot. Soc. 1971;50:196–208. [Google Scholar]
- 4.Malloch D, Cain RF. The genus Thielavia. Mycologia. 1973;65:1055–1077. doi: 10.1080/00275514.1973.12019527. [DOI] [Google Scholar]
- 5.Locquin-Linard M. A propos des genres non ostiolés placés dans la famille des Microascaceae (Ascomycètes). Création d’un nouveau genre: Enterocarpus. Rev. Mycol. 1977;41:509–523. [Google Scholar]
- 6.Locquin-Linard M. Kernia setadispersa, nouvelle espèce de la famille des Microascaceae (Ascomycètes) Cryptogamie Mycol. 1980;1:29–32. [Google Scholar]
- 7.Calviello BO. Contribucion al estudio de Ascomycetes argentinos. II. Una nueva especie de “Kernia (Microascales)”. Revta Mus. Argent. Ciencias Nat. Bernardino Rivadavia Bot. 1979;5:239–243. [Google Scholar]
- 8.Udagawa S, Muroi T. Notes on some Japanese Ascomycetes XVI. Trans. Mycol. Soc. Japan. 1981;22:11–26. [Google Scholar]
- 9.Udagawa S, Furuya K. Emericellopsis sphaerospora and Kernia peruviana, two new soil-borne cleistothecial Ascomycetes. Mycotaxon. 1988;33:291–301. [Google Scholar]
- 10.Woudenberg JHC, et al. Cephalotrichum and related synnematous fungi with notes on species from the built environment. Stud. Mycol. 2017;88:137–159. doi: 10.1016/j.simyco.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandoval-Denis M, et al. Redefining Microascus, Scopulariopsis and allied genera. Persoonia. 2016;36:1–36. doi: 10.3767/003158516X688027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sopp, O. J. Monographie der Pilzgruppe Penicillium mit besonderer Berücksichtigung der in Norwegen gefunden Arten. Videnskaps Selskapets Skrifter 1. Matematisk-Natur. Klasse11, 1–207 (1912).
- 13.Curzi M. Una nuova specie di Microascus. Bolletino della Stazione di Patologia Vegetale di Roma. 1930;10:302–309. [Google Scholar]
- 14.Barron GL, Cain RF, Gilman JC. The genus Microascus. Can. J. Bot. 1961;39:1609–1631. doi: 10.1139/b61-143. [DOI] [Google Scholar]
- 15.Morton FJ, Smith G. The genera Scopulariopsis Bainier, Microascus Zukal, and Doratomyces Corda. Mycol. Pap. 1963;8:1–96. [Google Scholar]
- 16.Sandoval-Denis M, et al. Phylogeny and taxonomic revision of Microascaceae with emphasis on synnematous fungi. Stud. Mycol. 2016;83:193–233. doi: 10.1016/j.simyco.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crous PW, et al. Fungal Planet description sheets 785–867. Persoonia. 2018;41:333. doi: 10.3767/persoonia.2018.41.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lackner M, de Hoog GS. Parascedosporium and its relatives: Phylogeny and ecological trends. IMA Fungus. 2011;21:39–48. doi: 10.5598/imafungus.2011.02.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summerbell RC, et al. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 2011;68:139–162. doi: 10.3114/sim.2011.68.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lackner M, et al. Proposed nomenclature for Pseudallescheria scedosporium and related genera. Fungal Divers. 2014;67:1–10. doi: 10.1007/s13225-014-0295-4. [DOI] [Google Scholar]
- 21.Samson RA, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su L, Deng H, Niu YC. Phialemoniopsis endophytica sp. nov., a new species of endophytic fungi from Luffa cylindrical in Henan, China. Mycol. Prog. 2016;15:48. doi: 10.1007/s11557-016-1189-5. [DOI] [Google Scholar]
- 23.Su L, Deng H, Niu Y. Phylogenetic analysis of Plectosphaerella species based on multi-locus DNA sequences and description of P. sinensis sp. nov. Mycol. Prog. 2017;16:823–829. doi: 10.1007/s11557-017-1319-8. [DOI] [Google Scholar]
- 24.Su L, Niu YC. Multi-locus phylogenetic analysis of Talaromyces species isolated from cucurbit plants in China and description of two new species T. cucurbitiradicus and T. endophyticus. Mycologia. 2018;110:375–386. doi: 10.1080/00275514.2018.1432221. [DOI] [PubMed] [Google Scholar]
- 25.Su L, et al. Lecanicillium coprophilum (Cordycipitaceae, Hypocreales), a new species of fungus from the feces of Marmota monax in China. Phytotaxa. 2019;387:55–62. doi: 10.11646/phytotaxa.387.1.4. [DOI] [Google Scholar]
- 26.Malloch D. New concepts in the Microascaceae illustrated by two new species. Mycologia. 1970;62:727–740. doi: 10.1080/00275514.1970.12019019. [DOI] [Google Scholar]
- 27.Sandoval-Denis M, et al. Scopulariopsis, a poorly known opportunistic fungus: spectrum of species in clinical samples and in vitro responses to antifungal drugs. J. Clin. Microbiol. 2013;51:3937–3943. doi: 10.1128/JCM.01927-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo LD, Hyde KD, Liew ECY. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000;147:617–630. doi: 10.1046/j.1469-8137.2000.00716.x. [DOI] [PubMed] [Google Scholar]
- 29.White TJ, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand MA, Sninsky DH, Innis JJ, White TJ, et al., editors. PCR protocols: A guide to methods and applications. London: Academic; 1990. pp. 315–322. [Google Scholar]
- 30.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/JB.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 32.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995;61:1323–1330. doi: 10.0000/PMID7747954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JD, et al. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 36.Altekar G, et al. Parallel metropolis-coupled markov chain monte carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nylander JAA, et al. Accounting for phylogenetic uncertainty in biogeography: A Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus) Syst. Biol. 2008;57:257–268. doi: 10.2307/20143140. [DOI] [PubMed] [Google Scholar]
- 39.Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 40.Zhaxybayeva O, Gogarten JP. Bootstrap, bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002;3:1–15. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Gateway computing environments workshop, p 1–8 (2010).
- 42.Page RDM. Treeview: An application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information 1 (DOCX 14 kb)
Supplementary information 2 (DOCX 13 kb)