Abstract
Stargardt disease (STGD1) is a form of inherited retinal dystrophy attributed to variants affecting function of the large ABCA4 gene and is arguably the most complex monogenic disease. Therapeutic trials in patients depend on identifying causal ABCA4 variants in trans, which is complicated by extreme allelic and clinical heterogeneity. We report the genetic architecture of STGD1 in the young genetically isolated population of Newfoundland, Canada. Population-based clinical recruitment over several decades yielded 29 STGD1 and STGD1-like families (15 multiplex, 14 singleton). Family interviews and public archival records reveal the vast majority of pedigree founders to be of English extraction. Full gene sequencing and haplotype analysis yielded a high solve rate (38/41 cases; 92.7%) for STGD1 and identified 16 causative STGD1 alleles, including a novel deletion (NM_000350.3: ABCA4 c.67-1delG). Several STGD1 alleles of European origin (including NM_000350.3: ABCA4 c.5714 + 5G>A and NM_000350.3: ABCA4 c.5461-10T>C) have drifted to a relatively high population frequency due to founder effect. We report on retinal disease progression in homozygous patients, providing valuable allele-specific insights. The least involved retinal disease is seen in patients homozygous for c.5714 + 5G>A variant, a so-called “mild” variant which is sufficient to precipitate a STGD1 phenotype in the absence of other pathogenic variants in the coding region and intron/exon boundaries of ABCA4. The most severe retinal disease is observed in cases with ABCA4 c.[5461-10T>C;5603A>T] complex allele. We discuss the advantages of determining genetic architecture in genetic isolates in order to begin to meet the grand challenge of human genetics.
Subject terms: Genetics research, Haplotypes
Introduction
In order to meet the grand challenge of human genetics, that is, to understand what causes disease and translate this knowledge to improve health outcomes, we need to know the number and population frequency of disease variants, the magnitude of their effects on phenotype and gene–gene and gene–environment interactions. In other words, we first need to understand genetic architecture [1]. For monogenic disorders, the genetic architecture is simplified as disease variants are, by definition, highly penetrant and environmental and gene–gene interactions are minimized. We believe that the very characteristics that make genetic isolates disproportionality valuable for disease mapping (i.e., reduced genetic heterogeneity, shared ancestry, environment, and lifestyle), along with their small size, also make them practical choices for studying genetic architecture.
Stargardt disease 1 (STGD1, [MIM: 248200]) is an autosomal recessive form of inherited juvenile macular degeneration with an estimated prevalence of 1 in 8000–10,000 [2–5] and usually presents as progressive bilateral central vision loss, often to the point of legal blindness. Although it has been 20 years since the ABCA4 gene was cloned [6], research is more intense than ever because the functional consequence of the >1000 ABCA4 variants associated with STGD1 [7] and a broad spectrum of other retinal dystrophies, including cone-rod dystrophy (arCRD) [8–11], retinitis pigmentosa (arRP) [8, 12–15], and age-related macular degeneration (AMD) [15–19], is largely unknown. Patients with ABCA4-related disease have pathogenic variants in the ABCA4 gene which encodes a large, 2273 amino acid photoreceptor-specific transporter involved in the removal of toxic retinoid compounds from photoreceptors [20]. ABCA4-related retinal dystrophies have a major impact on quality of life and clinical interventions including stem cell therapy, gene replacement therapy, and pharmacological agents are currently being developed [21]. In the case of STGD1, diagnostic screening is challenging. Direct sequencing of the entire open reading frame, the most comprehensive approach, reveals biallelic variants affecting function in 65–70% of patients [22]. The aggregate population frequency of ABCA4 alleles affecting function is high (1 in 20) [23]. The vast majority of patients are compound heterozygotes and may carry three or more causative alleles, making phenotypic attribution to specific ABCA4 variants difficult [24, 25]. Clinical heterogeneity both within and between families is thought to depend on the unique combination of ABCA4 variants in the genome of individual patients. Traditionally, ABCA4 variants frequent in the population were thought to be benign and variants incapable of precipitating STGD1 phenotype in the homozygous state, categorized as “mild” (hypomorphic) although recent studies are challenging this classification schema [7]. Phasing of ABCA4 variants, identifying two causal ABCA4 variants in trans in the germline of patients, is essential to confirm the clinical diagnosis, to provide prognostic and recurrence risk assessments and to determine eligibility for therapeutic trials [23, 26]. The utility of using whole exome/genome approaches for screening purposes in clinical settings is currently being explored [27, 28].
We set out to determine the genetic architecture of STGD1 in the well-defined genetic isolate of Newfoundland (NL), Canada. This is a prospective study, recruiting patients and their extended families over a period of 40 years, and includes serial clinical investigations and a comprehensive molecular approach. Although we had no expectations as to the frequency of STGD1 disease in NL, we anticipated reduced allelic heterogeneity, with the presence of one or more recurrent variants affecting function due to genetic drift. We also anticipated insights into genotype–phenotype correlations for specific ABCA4 alleles present in the homozygous state in STGD1 and STGD1-like patients.
Subjects and methods
Study population
According to archeological records, the Canadian province of NL, perched on the extreme northeast coast, has been inhabited for longer than any other place in North America. The first indigenous peoples (Maritime Archaic, Palaeoeskimo, and Beothuk) arrived 18,000 years ago, before the disappearance of the retreating Laurentide ice sheet from the last ice age. Recent mtDNA studies suggest that the relationships between these culturally distinct peoples predates their arrival by land across the continent of North America [29]. More recently, fisherfolk from ports in England and Ireland came to the island to prosecute the summer cod fishery. By 1775, English and Irish outports were established and 12,000 people overwintered on the island. By the late 19th century, hundreds of outports dotted the rugged, extensive 900-mile coastline, each isolated for long periods due to distance and rough seas. The island population expanded naturally (little immigration) so by the 1930s, more than 95% of residents were native born and the descendants of ~20,000 English and Irish settlers [30]. Both geographical and religious isolation (between Roman Catholic and Protestants) resulted in high kinship correlations, equivalent to that of the Bedouin of Kuwait in some communities [31]. The island population is a young genetic isolate (15–20 generations) and stratifies into three clusters, Protestant, Roman Catholic, and a small cluster of North American Indians [32].
We frequently observe unrelated patients on the island with rare monogenic diseases, including autosomal dominant, recessive, and X-linked disorders, that share the same pathogenic variant on an extended disease-associated haplotype (identical-by-descent) due to a common ancestor. Our most striking example is that of a particularly lethal form of sudden cardiac death due to Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC). We have uncovered hundreds of NM_024334.2: TMEM43 c.1073C>T; p.(S358L) carriers across numerous unrelated families, some pedigrees extending back nine generations, that all share the 2 Mb disease-associated haplotype due to a founder effect [33], the variant having been imported from Europe and dating back to the medieval period (400–700 AD) [34].
Patient recruitment
A population-wide recruitment effort on the island of NL (population 501,251) with the goal of identifying all patients with hereditary eye disease, including STGD1 and STGD1-like cases, began in 1978 with the establishment of an Ocular Genetics Clinic at the General Hospital in St. John’s and in select rural hospitals. A complete review of the provincial records of the Canadian National Institute for the Blind (CNIB) in the early 1980s determined the number and geographic distribution of registered CNIB clients with a possible hereditary form of blindness and also identified clients to be recruited to the study [35, 36]. STGD1 and STGD1-like patients from the island underwent comprehensive clinical examinations, including visual acuity, central and full visual fields, color vision testing (Ishihara and Farnsworth D-15), dark adaptation and/or full-field electroretinogram testing, retinal photographs and intravenous fluorescein. Extended family histories, including historical religious affiliations and community of origin, were obtained by family interviews and from the public archives. As STGD1 is a recessive disorder, we also noted consanguinity. All efforts were made to clinically reexamine patients in order to document the progression of retinal disease. This longitudinal study was approved by the institutional review board protocol # 02.116 (Human Research Ethics Board) and the Research Proposals Approval Committee of Eastern Health in St. John’s NL, Canada.
Full ABCA4 gene sequencing
So far as we know, all disease-causing variants for STGD1 reside in the ABCA4 gene. Of the 29 STGD1 or STGD1-like families enrolled in the study, five families were subsequently molecularly diagnosed with achromatopsia (S10, S12, S14, S15, and S20) and excluded from further investigation (Fig. 1). Bidirectional Sanger sequencing of the 50 exons and flanking intronic sequences of the ABCA4 gene was performed on 41 STGD1 cases from 24 families (Fig. 1). Genomic DNA was isolated from peripheral blood using a standard salting out procedure [37]. PCR amplification of the ABCA4 gene (GenBank NM_000350.2) was performed according to Azarian et al. [38]. except for exon 29 where primers were redesigned using Primer 3 software [39]. PCR products were size fractionated (1% agarose gels), stained (ethidium bromide or SYBR© Safe, Life Technologies, USA), documented (Kodak GEL 200; Mandel Scientific, Canada) and subjected to analysis according to standard protocols (ABI PRISM 3130XL or 3700 DNA Analyzer; Applied Biosystems, Foster City, CA; Mutation Surveyor Software, SoftGenetics LLC State College, PA 16803). Primer sequences are available upon request. Data from this study have been submitted to the public Leiden Open Variation Database.
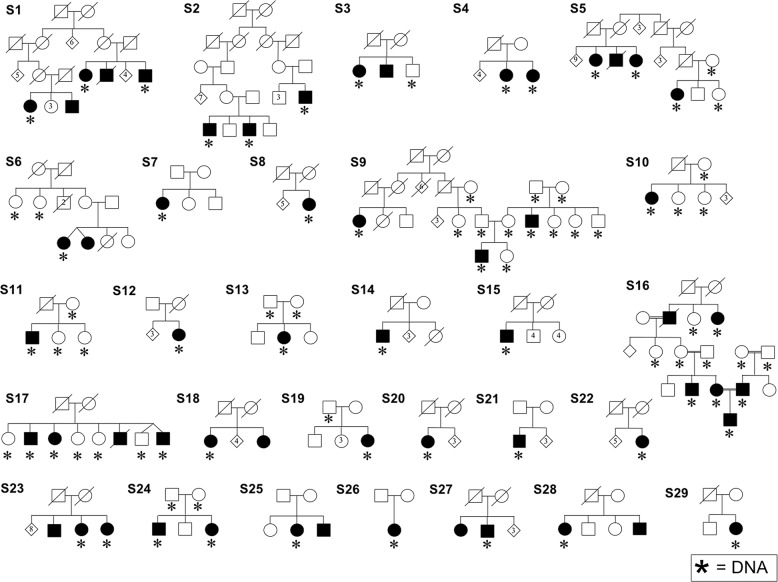
Fig. 1. Pedigrees of the 46 cases with Stargardt macular dystrophy or Stargardt-like disease recruited over three decades in Newfoundland.
Filled symbols represent relatives clinically diagnosed with Stargardt or Stargardt-like disease. An asterisk symbol indicates DNA was available for study.
Variant interpretation and STGD1-associated haplotypes
From the sequencing data, ABCA4 variants of interest were identified consisting of both novel and pathogenic or likely pathogenic variants as reported in the primary literature, public databases (e.g., Retinal Information Network, Retnet; Human Gene Mutation Database, HGMD; Leiden Open source Variant Database, LOVD, ClinVar), and in silico prediction software (e.g., Alamut). Variant classification based on the American College of Medical Genetics and Genomics criteria is provided (Supplementary Table 1). In each family, we genotyped variants of interest in parents and extended relatives when available to test for co-segregation with retinal disease consistent with a recessive mode of inheritance.
For autosomal recessive disorders and especially for STGD1, phasing is critical as two causal variants on the same chromosome (in cis) predict an unaffected carrier of a complex allele. To determine parent-of-origin for the purpose of phasing, intragenic single nucleotide polymorphisms (SNPs) that were identified in probands were also genotyped in extended family members to construct STGD1-associated haplotypes. To determine if recurrent pathogenic variants were due to founder effect, STGD1 haplotypes were compared between families. Recurrent variants may result from genomic hotspots (multiple events) or from single variants passed down through generations from a common ancestor. Despite the extreme allelic heterogeneity of ABCA4, we anticipated, due to the nature of the peopling of NL, to identify the same (recurrent) pathogenic variant in two or more unrelated patients due to a shared common ancestor and subsequent amplification in the population due to genetic drift.
Population frequencies of alleles and knowledge transfer to clinic
To provide a targeted gene panel to the local clinic, we designed a custom diagnostic panel including the 16 variants identified in this study using the iPLEX PRO Chemistry and the Agena MassARRAY platform. PCR primers were designed according to the manufacturer protocols (Design Assay Suite and Typer4 software; Agena Bioscience). We determined analytical validity (sensitivity, specificity, accuracy, and reproducibility) with a subset of molecularly characterized (“solved”) patients and partnered with the local genetics clinic to offer genetic counseling to families. Anonymized population control samples from NL and outbred populations (Canada, United Kingdom) were run on the custom panel and minor allele frequencies (MAFs) compared using Fisher’s Exact Test.
Results
The vast majority of STGD1 patients are of English extraction
Of the 29 STGD1 or STGD1-like families enrolled in the study, 41 cases were diagnosed with STGD1 (Fig. 1). Although the current population of NL is nearly equal parts English and Irish, the majority of STGD1 pedigree founders are of English extraction and trace their NL origins to a localized area within Conception Bay, a region of early English settlement in the northeastern part of the island (Supplementary Fig. 1) [40]. One singleton case (S11) originates on the French island of Saint Pierre, located 16 miles off the south coast of NL. Five families (S1, S2, S5, S9, S16) have two or more affected sibships; S9 represents two families connected by marriage and S16 is particularly notable with four consanguinity loops and an affected trio (Fig. 1).
Full gene sequencing and haplotype analysis achieves a high solve rate
Comprehensive molecular characterization of 41 STGD1 cases resulted in 38 solved and three unsolved cases (Families S5, S24), yielding a high case solve rate of 92.7%. Bidirectional Sanger sequencing yielded 16 pathogenic or likely pathogenic variants with an MAF of <0.01 in gnomAD browser (Table 1). We also identified a novel deletion, NM_000350.3: ABCA4 c.67-1delG, in three cases. This variant is located at the 3′ splice acceptor site of intron 1 and in silico analyses predict that it results in the removal of exon 2 from the mature ABCA4 transcript (MaxEnt [41], NNSPLICE [42], and Human Splice Finder [43]). Cascade screening and comprehensive haplotype analyses helped to confirm causality of the 16 ABCA4 alleles. Of the 41 STGD1 cases, 18 cases (43.9%) are homozygotes (Table 1). We found that 38/41 (92.7%) of STGD1 or STGD1-like cases had two pathogenic or likely pathogenic variants identified (Table 2). Most notably, the majority of solved cases (28/38; 73.7%) have at least one of two recurrent ABCA4 alleles (Table 2). We observed phenotypic heterogeneity, both within and between families; however, all diagnoses fell within the spectrum associated with ABCA4.
Table 1.
Frequencies of the 16 pathogenic/likely pathogenic ABCA4 (NM_000350.3) variants identified in 24 families (41 cases) in population controls and unsolved retinal disease cases.
| Variant | Effect | Homo | Het | NL Cases MAFa | gnomAD MAFb | NL MAFc | Non-NL MAFd |
|---|---|---|---|---|---|---|---|
| c.67-1delG | Splice | 0 | 3 | 0.073 | ND | ND | 0.002 |
| c.634C>T | p.(Arg212Cys) | 0 | 1 | 0.024 | 0.00011 | ND | ND |
| c.2564G>A | p.(Trp855Ter) | 0 | 5 | 0.122 | 0.00002 | 0.00760 | 0.002 |
| c.2588G>C | p.(Gly863Ala) | 0 | 4 | 0.098 | 0.0043 | 0.00770 | 0.0061 |
| c.3322C>T | p.(Arg1108Cys) | 0 | 3 | 0.073 | 0.00013 | ND | 0.002 |
| c.3323G>A | p.(Arg1108His) | 0 | 1 | 0.024 | 0.0002 | ND | ND |
| c.4139C>T | p.(Pro1380Leu) | 1 | 2 | 0.098 | 0.00023 | 0.00190 | ND |
| c.4163T>C | p.(Leu1388Pro) | 2 | 4 | 0.195 | 0.000004 | ND | ND |
| c.4469G>A | p.(Cys1490Tyr) | 0 | 2 | 0.049 | 0.00006 | ND | ND |
| c.4537delC | p.(Gln1513Argfs13) | 0 | 1 | 0.024 | 0.00001 | ND | ND |
| c.4577C>T | p.(Thr1526Met) | 0 | 3 | 0.073 | 0.00006 | ND | ND |
| c.5461-10T>C | Exon skipping | 3 | 6 | 0.293 | 0.00021 | 0.0019 | ND |
| c.5603A>T | p.(Asn1868Ile) | 5 | 10 | 0.488 | 0.0393 | 0.0374 | NA |
| c.5714 + 5G>A | Splice | 7 | 15 | 0.707 | 0.00022 | 0.0163e | ND |
| c.6089G>A | p.(Arg2030Gln) | 0 | 1 | 0.024 | 0.00035 | ND | ND |
| c.6449G>A | p.(Cys2150Tyr) | 0 | 1 | 0.024 | 0.00035 | ND | ND |
Homo homozygous, Het heterozygous, NA not available, ND not detected.
aRatio of STGD1 cases with at least one copy of the pathogenic allele.
bhttps://gnomad.broadinstitute.org.
cMinor allele frequencies (MAF) calculated using a total of 562–1056 alleles.
dMAF calculated using a total of 982–996 alleles.
eStatistical significance (p = 0.00001; Fisher’s Exact Test).
Table 2.
Disease-causing and likely pathogenic alleles (simple and complex) in ABCA4 (NM_000350.3) and their phenotypic effect in 24 Stargardt families from an isolated population.
| Family member | Variants identified | Age | Age dx | VA now | VF | CV defect | ERG | Phenotype |
|---|---|---|---|---|---|---|---|---|
| S1:III-4 | c.[5714 + 5G>A];[4163T>C] | 90 | 20 | R-CF L-CF | Central scotoma | Mild | NA | Central choroidal sclerosis |
| S1:III-6 | c.[5714 + 5G>A];[4163T>C] | 75 | 45 | R-CF 3 ft L-HM | Central scotoma | Medium | NA | STGD |
| S1:V-1 | c.4577C>T(;)4163T>C | NA | NA | NA | NA | NA | NA | NA |
| S2:IV-5 | c.[5714 +5G>A];[67-1delG] | 35 | 17 | R-20/400 L-20/400 | Central scotoma | Mild | Rod-normal cone-slight dec | CRD |
| S2:V-1 | c.[5714 + 5G>A];[67-1delG] | 16 | 14 | R-20/400 L-20/400 | Central scotoma | Mild | Rod-normal | CRD |
| S2:V-3 | c.[5714 + 5G>A];[67-1delG] | 20 | 18 | R-20/300 L-20/300 | Central scotoma | Mild | Rod-dec cone-normal | CRD |
| S3:II-1 | c.4469G>A(;)6089G>A(;)5603A>T | NA | NA | NA | NA | NA | NA | NA |
| S4:II-2 | c.[5714 + 5G>A];[5714 + 5G>A] | 56 | 52 | R-20/60 L-20/40 | Central scotoma | NA | Rod-slight dec cone-slight dec | STGD |
| S4:II-3 | c.[5714 + 5G>A];[5714 + 5G>A] | 66 | 47 | R-20/80 L-CF | Central scotoma | Severe | Rod-slight dec cone-slight dec | STGD |
| S5:II-2 | c.[4163T>C];[4163T>C] | NA | NA | NA | NA | NA | NA | NA |
| S5:II-4 | c.[4163T>C];[4163T>C] | NA | NA | NA | NA | NA | NA | NA |
| S5:III-1 | c.[5714 + 5G>A];[4163T>C] | 52 | 26 | R-20/400 L-20/300 | Central scotoma | Normal | Rod-dec cone-Normal | Atrophic macula |
| S6:III-1 | c.[5714 + 5G>A];[3322C>T] | 63 | 23 | R-CF L-CF | NA | Mild | Rod-normal cone-normal | STGD |
| S7:II-1 | c.[5714 + 5G>A];[5714 + 5G>A] | 56 | 26 | R-20/400 L-20/200 | Central scotoma | Mild | Rod-normal cone-normal | STGD |
| S8:II-2 | c.4577C>T(;)5603A>T | NA | NA | NA | NA | NA | NA | NA |
| S9:III-1 | c.[5714 + 5G>A];[5461-10T>C;5603A>T] | 67 | 12 | R-CF L-CF | Central scotoma correct | NA | Rod-dec cone-dec | CRD |
| S9:III-8 | c.[5714 + 5G>A;2588G>C];[5714 + 5G>A] | 46 | 29 | R-20/60 L-20/20 | Normal | Normal | Rod-normal cone-normal | STGD |
| S9:IV-1 | c.[5714 + 5G>A;2588G>C]; [5461-10T>C;5603A>T] | 21 | 15 | R-20/200 L-20/300 | Central scotoma | Medium | NA | STGD |
| S11:II-1 | c.[2588G>C;5603A>T];[6449G>A] | NA | NA | NA | NA | NA | NA | NA |
| S13:II-2 | c.[2588G>C;5603A>T];[4537delC] | NA | NA | NA | NA | NA | NA | NA |
| S16:II-4 | c.[2564G>A];[5461-10T>C;5603A>T] | 71 | 11 | R-HM L-HM | Peripheral islands only | Severe | NA | CRD |
| S16:IV-2 | c.[5461-10T>C;5603A>T]; [5461-10T>C;5603A>T] | 38 | 8 | R-20/400 L-20/400 | Peripheral Islands only | Severe | Rod-dec cone-dec | CRD |
| S16:IV-3 | c.[5461-10T>C;5603A>T]; [5461-10T>C;5603A>T] | 39 | 12 | R-20/400 L-CF | Mid-peripheral islands only | Severe | Rod-dec cone-dec | CRD |
| S16:IV-4 | c.[5714 + 5G>A];[5461-10T>C;5603A>T] | 41 | 13 | R-20/200 L-20/400 | Central scotoma | NA | Rod-dec cone-normal | CRD |
| S16:V-1 | c.[5461-10T>C;5603A>T]; [5461-10T>C;5603A>T] | 14 | 8 | R-20/500 L-20/300 | Central scotoma | Mild | Rod-dec cone-dec | CRD |
| S17:II-2 | c.[5714 + 5G>A];[5714 + 5G>A] | 62 | 39 | R-20/200 L-20/50 | Central scotoma | Severe | Rod-dec cone-normal | STGD |
| S17:II-3 | c.[5714 + 5G>A];[5714 + 5G>A] | 63 | 46 | R-20/30 L-CF | Central scotoma | NA | NA | STGD |
| S17:II-8 | c.[5714 + 5G>A];[5714 + 5G>A] | 52 | 41 | R-20/400 L-20/40 | Central scotoma | NA | Rod-slight dec cone-normal | STGD |
| S18:II-1 | c.2564G>A(;)4139C>T | NA | NA | NA | NA | NA | NA | NA |
| S19:II-3 | c.3322C>T(;)3323G>A | NA | NA | NA | NA | NA | NA | NA |
| S21:II-1 | c.[4139C>T];[4139C>T] | NA | NA | NA | NA | NA | NA | NA |
| S22:II-2 | c.[5714 + 5G>A];[2564G>A] | 57 | 14 | R-20/400 L-CF | Peripheral Islands | Severe | Rod-dec cone-dec | CRD |
| S23:II-3 | c.[5461-10T>C;5603A>T]; [5603A>T] | 52 | 14 | R-20/200 L-20/200 | NA | NA | NA | NA |
| S23-II-4 | c.[5461-10T>C;5603A>T]; [5603A>T] | 51 | 20 | R-CF L- 20/400 | NA | NA | NA | NA |
| S24:II-1 | c.[2564G>A;5603A>T] | NA | NA | NA | NA | NA | NA | NA |
| S24:II-3 | c.[2564G>A;5603A>T] | NA | NA | NA | NA | NA | NA | NA |
| S25:II-2a | c.4139C>T(;)5714 + 5G>A | 36 | 34 | R-20/20 L-20/25 | Small central scotomas | normal | NA | STGD |
| S26:II-1a | c.4577C>T(;)5714 + 5G>A | 32 | 12 | R-20/50−1 L-20/40−1 | Central scotoma | NA | Rod-slight dec cone-slight dec | NA |
| S27:II-2 | c.[5714 + 5G>A];[3322C>T] | 50 | NA | R-20/80 L-20/150 | Small central scotomas | NA | Rod-slight dec cone-normal | STGD |
| S28:II-1a | c.4163T>C(;)4469G>A | NA | NA | NA | NA | NA | NA | NA |
| S29:II-2a | c.5714 + 5G>A(;)634C>T | 47 | 45 | L-20/20 R-20/20 | Normal | NA | NA | STGD |
Bold indicates recurrent founder alleles.
CF counting fingers, HM hand motion, STGD Stargardt disease, CRD cone-rod dystrophy, NA not available, dec decreased, VA visual acuity, VF visual fields, dx diagnosis, ft feet, R right, L left, CV central vision, ERG electroretinogram.
aABCA4 genetic testing performed at Asper Biogene (no phasing information available).
Recurrent c.5714 + 5G>A allele is due to a major founder effect
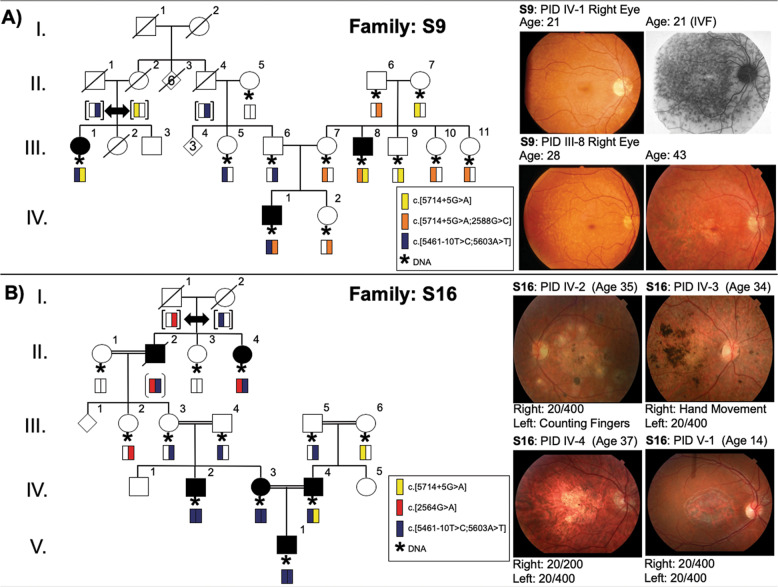
The most frequent ABCA4 allele in NL is c.5714 + 5G>A, surprisingly detected in 22/38 (57.9%) of solved cases across 13 families and represented the only pathogenic or likely pathogenic variant identified in all coding and exon/intron flanking regions within the gene. The c.5714 + 5G>A splice variant was first described in 1998 and is one of the most common STGD1 alleles currently reported in Northern Europe [8, 44, 45]. Most patients with the c.5714 + 5G>A allele shared the entire disease-associated haplotype (Fig. 2 and Table 2). Comparison of STGD1 haplotypes revealed patients in 13 families shared a common c.5714 + 5G>A haplotype (Fig. 2). In S9, two families connected by marriage, a c.5714G>A haplotype was introduced from the left side of the pedigree and a distinct c.5714 + 5G>A complex allele c.[2588G>C;5714 + 5G>A] was introduced on the right side of the pedigree (Fig. 3a). When compared with the 2019 ProgStar study [46], the c.5714 + 5G>A allele has a much higher frequency in NL cases (0.023 vs 0.707) and it is significantly increased (p = 0.00001) in the NL population compared with non-NL controls. The occurrence of two haplotypes with the c.5714 + 5G>A splice variant (Fig. 3a: yellow and orange haplotypes) suggests that it was introduced into the population at least twice and likely underwent natural expansion from the Conception Bay area, historically the most densely populated area of the province. We conclude that the c.5714 + 5G>A variant is recurrent in this population due to major founder effect. In seven homozygous cases, haplotype analysis revealed that the variant was the only pathogenic or likely pathogenic variant on the STGD1-associated haplotype. Furthermore, longitudinal observations show it is associated with a definite STGD1 phenotype with visual loss by middle age (Table 2).
Fig. 2. Examples of STGD1-associated haplotypes identified in Newfoundland.
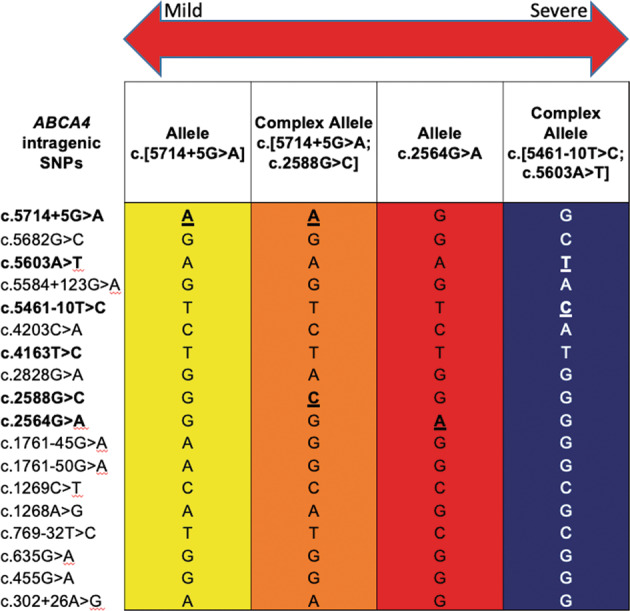
STGD1 haplotypes are read vertically using intragenic SNPs (5′ to 3′) residing in cis on chromosome 1p22.1 (pathogenic variants bolded). The first two haplotypes represent the recurrent NM_000350.3: ABCA4 c.5714 + 5G>A haplotype (yellow) and the ABCA4 c.[5714 + 5G>A;2588G>C] complex allele (orange), which are both associated with a milder phenotype. The ABCA4 c.2564G>A haplotype (red) is associated with a moderately severe phenotype. The blue haplotype denotes the recurrent ABCA4 c.[5461–10T>C;5603A>T] complex allele associated with the most severe phenotype.
Fig. 3. Progression of retinal phenotype associated with allele-specific haplotypes.
Bars below symbols represent STGD1-associated haplotypes in Fig. 2. a Family S9 shows retinal phenotypes of two different compound heterozygotes. IV-1 has four pathogenic variants arranged into two complex alleles: NM_000350.3: ABCA4 c.[5461–10T>C;5603A>T];[5714 + 5G>A;2588G>C] (orange and blue bars). III-8 has three pathogenic variants arranged into two alleles: ABCA4 c.[5714 + 5G>A;2588G>C];[5714 + 5G>A] (yellow and orange bars). b Family S16 shows severe retinal phenotypes of patients either homozygous (blue bars) or compound heterozygous (blue/red and blue/yellow bars) for the ABCA4 c.[5461–10T>C;5603A>T] complex allele. An asterisk symbol indicates DNA was available for study and symbol “[]” indicates inferred haplotype.
Recurrent c.5461-10T>C Allele also due to a Founder Effect
The c.5461-10T>C ABCA4 allele was detected in 9/38 (23.7%) of solved cases across three families (S9, S16, S23) and forms a complex allele with c.5603A>T in 9/9 (100%) of STGD1 cases (Table 2and Fig. 2). First reported in 1999 [10], this intronic splicing variant is the third most frequent ABCA4 allele identified in STGD1 patients of European or African descent and functional studies reveal it causes skipping of exon 39 (100%) or 39–40, reduction in full-length mRNA and reduced protein levels in homozygotes [44, 45, 47, 48]. The c.5461–10T>C variant in the ABCA4 gene has been reported previously in the homozygous state and in the presence of a second ABCA4 pathogenic variant, in association with STGD1 disease [45, 49], has been consistently clinically classified as a pathogenic variant and is a severe variant. Our nine cases included three homozygous patients and 9/9 (100%) of cases shared the STGD1 complex allele c.[5461–10T>C;5603A>T] (Fig. 2). Haplotype analysis revealed that ABCA4 c.5461–10T>C is identical-by-descent and recurrent due to founder effect. Haplotype analysis in S16 (with four consanguinity loops) demonstrated the affected trio to be a case of pseudodominance (Fig. 3b). The affected son (PID V-1) is homozygous for the recurrent ABCA4 c.[5461–10T>C;5603A>T] complex allele, inheriting one copy from his homozygous mother and a second copy from his father. When compared with the 2019 ProgStar study [46], the c.5461–10T>C variant is amplified in NL cases (0.048 vs 0.293). Although we observe the c.5461–10T>C allele in ethnically matched controls, the frequency is not significantly amplified in NL (p > 0.05; Table 1).
Recurrent c.5603A>T hypomorphic allele
The ABCA4 c.5603A>T; p.(Asn1868Ile) variant was the second most frequent allele observed in this study (Table 1) and there is evidence to support the variant being a hypomorphic allele as it is phenotypically characterized by late onset of symptoms (4th decade) and foveal sparing (85%) only when in trans with a deleterious mutation [7, 50]. Complex alleles involving the ABCA4 c.5603A>T; p.(Asn1868Ile) variant were detected in 13 STGD1 cases including nine solved cases with c.[5603A>T; 5461–10T>C], two solved cases with c.[5603A>T; 2588G>C]) with a third complex allele, c.[5603A>T;2564G>A], identified in two cases in a single unsolved family (Table 2). There is evidence supporting the ABCA4 c.2588G>C; p.(Gly863Ala) variant as being pathogenic producing less protein, reducing ATP binding and hydrolysis and retinal transfer [51]. In our study, the ABCA4 c.2588G>C; p.(Gly863Ala) variant causes disease only when in cis with ABCA4 c.5714 + 5G>A or c.5603A>T and may not act as a disease-associated hypomorphic variant, but rather as a modifier (Table 2).
Novel pathogenic variant/complex alleles
We identified a novel variant, ABCA4 c.67-1delG, in third cousins once removed in family S2, which seems likely to have been transmitted by one of the pedigree founders four generations back (Fig. 1 and Table 1). The fact that this novel ABCA4 variant was in trans with the recurrent c.5714 + 5G>A allele helped to confirm its pathogenicity (Table 2). As ABCA4 c.67-1delG was also identified in a non-NL control sample it is probably not a private variant; however, we cannot be certain as the identity of the non-NL control is unknown (Table 1). We also identified four complex alleles: c.[5461-10T>C;5603A>T], c.[5714 + 5G>A;2588G>C], c.[2588G>C;5603A>T], and c.[2564G>A;5603A>T] (Table 2).
Natural history of retinal disease attributable to specific STGD1 alleles
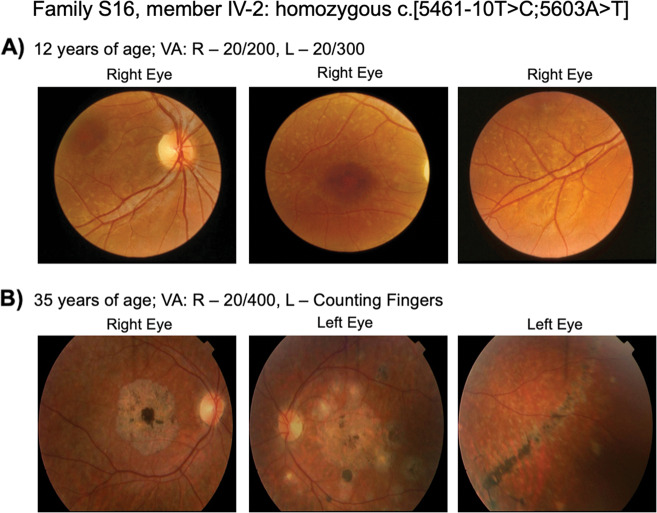
Homozygous STGD1 patients are rare [24] due to the extreme allelic heterogeneity in ABCA4 disease, making it difficult to determine accurate genotype–phenotype prognoses when counseling patients and their families. Prospective clinical examinations over several decades in 18/38 (47.4%) of solved cases, which are homozygous, conclusively reveal that ABCA4 c.5461-10T>C variant is the most severe STGD1 allele in this population (Table 2 and Fig. 2). Patients having one or two copies of the ABCA4 c.[5461-10T>C;5603A>T] allele are characterized by the earliest age of onset, diagnosed with STGD1 as young as 8 years of age, and by the second or third decade, all progressed to a severe arCRD or arRP phenotype with atrophic retina and sparse pigment clumping with reduced or extinguished rod and cone function on ERG testing (Figs. 3 and 4). For example, in family S16, the retinal pattern over a 23-year period has progressed to the point where it does not present as STGD1 disease (Fig. 4). In contrast, ABCA4 c.5714 + 5G>A cases experience a mild-moderate vision loss with homozygotes having a less severe and later onset of decrease in acuity than compound heterozygotes (Table 2).
Fig. 4. Progressive course of retinal disease in patients with the NM_000350.3: ABCA4 c.[5461–10T>C;5603A>T] haplotype.
Retinal photos of patient PID IV-2 (Family S16) showing phenotype at: a age 12 and b age 35. The patient presented early with typical STGD1 but by clinical follow-up in their 3rd decade, the retinal disease progressed to the point where the clinical presentation was more typical of cone-rod dystrophy or retinitis pigmentosa.
Founder effects amplify specific alleles but not STGD1 prevalence or carrier frequency
According to Hardy–Weinberg Law, we know that violating the first assumption of random mating can cause large deviations from the frequency of individuals homozygous for an autosomal recessive condition. After complete clinical ascertainment, we identified 41 NL residents with STGD1 or STGD1-like disease (41/501,251; q2 = 0.0000818) and calculate the prevalence to be 1 in 12,255, which is lower than the prevalence reported for other populations of 1 in 8000–10,000 [2–5, 22] and calculate the expected carrier frequency (2pq) to be 0.178. The actual carrier frequency (summation from Table 1) equals 0.0728, is also lower than expected. Using a custom diagnostic panel of 16 STGD1 pathogenic or likely pathogenic variants, five variants in the NL controls had a higher MAF compared with non-NL controls (Table 1) and the ABCA4 c.5714 + 5G>A allele is significantly higher (p = 0.00001) compared with other Northern European populations (Table 1). In this genetic isolate, it is likely that the reduced genetic heterogeneity (founder effect) combined with a high degree of kinship correlation, geographical, and religious isolation have precipitated the unprecedented rate of homozygous STGD1 patients observed in this study without a corresponding increase in STGD1 prevalence or population carrier frequency (Table 1).
Discussion
The genetic architecture of STGD1 in Newfoundland, Canada
STGD1 in this young genetic isolate is almost exclusively of English origin and characterized by drastically reduced allelic heterogeneity with the concomitant amplification of specific European alleles due to genetic drift. Sixteen ABCA4 (NM_000350.3) pathogenic variants, including a novel deletion, account for 92.7% of solved cases, with a disproportionate number of homozygous cases. The common European ABCA4 c.5714 + 5G>A splice variant resides on two distinct disease haplotypes, occurs in 58% (14/24) of families and the frequency is significantly amplified in the NL population. Public archival records, family interviews and comprehensive haplotype analysis reveal that the ABCA4 c.5714 + 5G>A allele was likely introduced by English settlers early in the peopling of the island population. Key genotype-specific insights in homozygous patients include the confirmation of the ABCA4 c.5714 + 5G>A allele as a true pathogenic but mild allele. Despite the presence of recurrent founder variants, the prevalence of STGD1 in NL is similar to that of outbred populations in North America and Europe.
Advantages of a population-based approach in genetic isolates
We believe the high solve rate (92.7%) is due to the triad of population-based clinical ascertainment, comprehensive molecular screening, and the presence of recurrent founder variants. To date, the vast majority of natural history studies on the progression of retinal diseases are retrospective from single centers serving outbred populations, resulting in calls for natural history studies of large cohorts of molecularly proven patients [22]. Our successful approach was serial retinal investigations over four decades on molecularly characterized cases and their extended families who share ancestry and environment, while leveraging key resources including public archives, community-based ophthalmologists and patient advocacy databases (e.g., the CNIB).
The design and execution of successful clinical trials depend on accurate predictions of clinical outcomes for specific alleles. In autosomal dominant disorders, this is relatively straightforward, as each affected person harbors a single disease allele, and the natural history can be gleaned from multiple affected individuals over several generations with the exact same disease allele. In recessive disorders, and particularly for STGD1 in outbred populations, the majority of affected individuals are compound heterozygotes. In the NL population, we have a rare opportunity to track the progression of retinal disease in seven patients homozygous for c.5714 + 5G>A (one of the most common European variants) often reported as a mild allele [52] with mild-to-moderate vision loss and late onset of decrease in acuity, as well as 15 compound heterozygous. Full gene sequencing and haplotype analysis confirmed that c.5714 + 5G>A was actually the only pathogenic or likely pathogenic variant on the STGD1-associated haplotype in seven homozygotes.
In this study, we relied on phasing to determine which alleles were in cis to identify the presence and composition of complex alleles, a particular challenge for ABCA4-related retinal diseases. Patients in our study who inherited two copies of ABCA4 c.5461–10T>C, the third most frequent variant associated with STGD1 patients of European or African descent [44, 45, 47] are the most severely affected, presenting with a classic STGD phenotype and rapidly progressing to RP or arCRD with severely reduced or extinguished cone and rod responses on electroretinogram testing as early as 8 years of age.
In genetic isolates, recurrent variants can unmask rare recessive alleles. In this study, the predominance of ABCA4 c.5714 + 5G>A served to unmask ABCA4 alleles in the region of early English settlement. This included a novel ABCA4 deletion pathogenic variant (c.67-1delG) which co-segregated with disease in three cases in a multiplex family and was inherited in trans with the founder ABCA4 c.5714 + 5G>A variant (c.[67-1delG];[5714 + 5G>A]), increasing our confidence that the novel variant affects function. We also uncovered a case of an affected trio with STGD1. Pseudodominant transmission (vertical inheritance of a recessive trait) in STGD1 occurs more often than might be expected, probably because of the relatively high carrier frequency of pathogenic ABCA4 variants in the general population. Huckfeldt et al. [53] recently reported a case of pseudodominant transmission where there were three alleles in the family and Lee et al. [25] reported four alleles in a family from the UK. In our study, the affected trio turned out to be a case where an affected son inherited two copies of the severe c.[5461–10T>C;5603A>T] allele, one from his homozygous mother and the other from his compound heterozygous father. Molecular characterization of this case is not only interesting from a genetic architecture lens, but also clinically significant as it drastically increases the recurrence risk for this family from 25 to 100%, as there are no normal ABCA4 alleles in the parental generation.
Prevalence of STGD1 in the NL population
At the population level, recurrent variants and increased homozygosity may lead one to assume that the disease prevalence of STGD1 in NL is comparatively relatively high. We find empirical evidence to the contrary. In genetically isolated populations, it is more likely that members will marry someone with the same variant and have homozygous children, resulting in a higher portion of homozygotes, concomitant with a lower number of disease alleles, a feature of recessive conditions in the Hutterites [54]. In the context of STGD1 disease with >1000 variants affecting function worldwide, we identified a mere 16 ABCA4 alleles accounting for all cases in this population, although we do observe a skewed geographical distribution of cases.
Limitations of a population-based approach in a genetic isolate
A major challenge of any STGD1 study is determining the actual age at onset of vision loss, especially for older participants (70–92 years old). As there were no ophthalmologists in rural NL three or more decades ago, age at diagnosis for many of our senior patients was unknown, and a lack of access to health care is still a feature of rural NL. Three cases across two families remain unsolved, representing a particular conundrum for genetic conditions as the lack of family history makes it impossible to determine the mode of inheritance. Retinal disease in these cases could be due to deep intronic or copy number variants in ABCA4 or due to variants in other genes. Although we did not search for deep intronic variants or large deletions, we tracked STGD1-associated haplotypes across multiple generations and established parental contributions, confirming homozygous cases are truly homozygous rather than a point mutation in trans with a deletion. Whole genome sequencing approaches may solve these cases, although first studies suggest that copy number variations (large deletions and insertions) at the ABCA4 locus are rare [23].
With respect to genetic architecture, we limited this study to STGD1 alleles identified in patients. To identify all disease-causing ABCA4 variants in a population would require a random sampling design and a whole exome/genome approach, with the caveat that discerning pathogenic variants that occur in unaffected people would be difficult for novel variants or variants of unknown significance. For example, the dearth of STGD1 and STGD1-like cases on the west coast of the island may mean there is a broader range of STGD1 alleles that have not been unmasked due to paucity of recurrent STGD1 alleles in this localized area. Although there are concerns that disease variants identified in genetic isolates are not relevant to outbred populations, this study reaffirms that the NL population represents a potent amplifier for a subset of Old-World variants. In this way, specific disease variants, expanded due to genetic drift, can be examined with phenotypic depth and breadth in the context of reduced environmental exposures.
Meeting the grand challenge of human genetics—were we successful?
How close did we come to determining the genetic architecture of STGD1 disease in this genetic isolate, and translating knowledge to the clinic? We took STGD1, arguably the most difficult monogenic disease, and over the course of four decades identified all prevalent cases and their extended families. We then used a comprehensive molecular approach and determined the underlying pathogenic variants and their phenotypic impact over the lifespan. Furthermore, we designed and validated a targeted diagnostic panel on a customizable platform and provided genetic counseling to all patients and their families in collaboration with the local clinic, enabling their eligibility for therapeutic trials.
Renewed calls for longitudinal studies emphasize the need for much more effort to be placed on phenotyping patients and family members in our current era of whole genome approaches [54, 55]. Although outside the scope of this project, we now have a unique opportunity in NL to look at the population level for all ABCA4 variants and to examine individual carriers for retinal phenotypes in order to capture age of onset, and mild manifestations of the same variants in the absence of ascertainment bias. In this way, the exact molecular effect of the >1000 ABCA4 variants associated with STGD1 and a broad spectrum of other retinal dystrophies (arCRD, arRP, and AMD) may be better understood. A focus on monogenic diseases in isolated populations with manageable population size, reduced heterogeneity, and local expertize, provides unique opportunities for meeting the grand challenge of understanding the causes of genetic disease and translating the information to the clinic.
Web resources
Ensembl Genome Browser, http://www.ensembl.org/index.html
Genome Aggregation Database, https://gnomad.broadinstitute.org
Human Gene Mutation Database (HGMD) http://www.hgmd.cf.ac.uk/ac/
Leiden Open Variation Database (LOVD): ABCA4 Locus database: http://www.lovd.nl/ABCA4 (Individual IDs #00233811 - #00233851)
OMIM, http://www.omim.org/
RetNet – Retinal Information Network, https://sph.uth.edu/retnet/home.htm
UCSC Genome Browser, http://genome.ucsc.edu
Supplementary information
Acknowledgements
We are truly grateful to the patients and their families who participated in this study for many years; to Dr Gordon J. Johnston for early clinical studies; to technical staff and undergraduate students who contributed over the years, and thanks to Dr Kathy Hodgkinson and the peer reviewers for improving the paper. We are thankful to reviewers and the editor for constructive comments. This work was supported by The Janeway Children’s Hospital Foundation, Canada Foundation for Innovation (New Investigator Award no. 9384); Leaders Opportunity Fund no. 13120), Genome Canada (Atlantic Medical Genetics and Genomics Initiative), Atlantic Canada Opportunities Agency—Atlantic Innovation Fund (ACOA-AIF), Research and Development Corporation (RDC), Faculty of Medicine, Memorial University, and the Government of NL.
Author contributions
The project was conceived by JSG and TLY. Patients/charts were examined/reviewed by JSG, DB, and JW. Experiments performed by HR, DG, JH and AS. The data were analyzed by DDO, HR, NA, JSG, JAP, TB, JH and TLY. The paper was written by JSG, DDO, and TLY.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jane S. Green, Darren D. O’Rielly
Supplementary information
The online version of this article (10.1038/s41431-020-0581-4) contains supplementary material, which is available to authorized users.
References
- 1.Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet. 2018;19:110–24. doi: 10.1038/nrg.2017.101. [DOI] [PubMed] [Google Scholar]
- 2.Lambertus S, van Huet RA, Bax NM, Hoefsloot LH, Cremers FP, Boon CJ, et al. Early-onset stargardt disease: phenotypic and genotypic characteristics. Ophthalmology. 2015;122:335–44. doi: 10.1016/j.ophtha.2014.08.032. [DOI] [PubMed] [Google Scholar]
- 3.Burke TR, Tsang SH. Allelic and phenotypic heterogeneity in ABCA4 mutations. Ophthalmic Genet. 2011;32:165–74. doi: 10.3109/13816810.2011.565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michaelides M, Hunt DM, Moore AT. The genetics of inherited macular dystrophies. J Med Genet. 2003;40:641–50. doi: 10.1136/jmg.40.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujinami K, Lois N, Davidson AE, Mackay DS, Hogg CR, Stone EM, et al. A longitudinal study of stargardt disease: clinical and electrophysiologic assessment, progression, and genotype correlations. Am J Ophthalmol. 2013;155:1075–88. doi: 10.1016/j.ajo.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Allikmets R. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;17:122. doi: 10.1038/ng0997-122a. [DOI] [PubMed] [Google Scholar]
- 7.Zernant J, Lee W, Collison FT, Fishman GA, Sergeev YV, Schuerch K, et al. Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J Med Genet. 2017;54:404–12. doi: 10.1136/jmedgenet-2017-104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cremers FP, van de Pol DJ, van Driel M, den Hollander AI, van Haren FJ, Knoers NV, et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum Mol Genet. 1998;7:355–62. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- 9.Kitiratschky VB, Grau T, Bernd A, Zrenner E, Jagle H, Renner AB, et al. ABCA4 gene analysis in patients with autosomal recessive cone and cone rod dystrophies. Eur J Hum Genet. 2008;16:812–9. doi: 10.1038/ejhg.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, et al. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet. 2000;67:960–6. doi: 10.1086/303079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yzer S, van den Born LI, Zonneveld MN, Lopez I, Ayyagari R, Teye-Botchway L, et al. Molecular and phenotypic analysis of a family with autosomal recessive cone-rod dystrophy and Stargardt disease. Mol Vis. 2007;13:1568–72. [PubMed] [Google Scholar]
- 12.Martinez-Mir A, Paloma E, Allikmets R, Ayuso C, del Rio T, Dean M, et al. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet. 1998;18:11–2. doi: 10.1038/ng0198-11. [DOI] [PubMed] [Google Scholar]
- 13.Papaioannou M, Ocaka L, Bessant D, Lois N, Bird A, Payne A, et al. An analysis of ABCR mutations in British patients with recessive retinal dystrophies. Invest Ophthalmol Vis Sci. 2000;41:16–9. [PubMed] [Google Scholar]
- 14.Riveiro-Alvarez R, Vallespin E, Wilke R, Garcia-Sandoval B, Cantalapiedra D, Aguirre-Lamban J, et al. Molecular analysis of ABCA4 and CRB1 genes in a Spanish family segregating both Stargardt disease and autosomal recessive retinitis pigmentosa. Mol Vis. 2008;14:262–7. [PMC free article] [PubMed] [Google Scholar]
- 15.Shroyer NF, Lewis RA, Yatsenko AN, Lupski JR. Null missense ABCR (ABCA4) mutations in a family with stargardt disease and retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2001;42:2757–61. [PubMed] [Google Scholar]
- 16.Allikmets R. Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. Am J Hum Genet. 2000;67:487–91. doi: 10.1086/303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277:1805–7. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- 18.Shroyer NF, Lewis RA, Allikmets R, Singh N, Dean M, Leppert M, et al. The rod photoreceptor ATP-binding cassette transporter gene, ABCR, and retinal disease: from monogenic to multifactorial. Vis Res. 1999;39:2537–44. doi: 10.1016/s0042-6989(99)00037-1. [DOI] [PubMed] [Google Scholar]
- 19.Souied EH, Ducroq D, Gerber S, Ghazi I, Rozet JM, Perrault I, et al. Age-related macular degeneration in grandparents of patients with Stargardt disease: genetic study. Am J Ophthalmol. 1999;128:173–8. doi: 10.1016/s0002-9394(99)00145-2. [DOI] [PubMed] [Google Scholar]
- 20.Quazi F, Lenevich S, Molday RS. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat Commun. 2012;3:925. doi: 10.1038/ncomms1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith J, Ward D, Michaelides M, Moore AT, Simpson S. New and emerging technologies for the treatment of inherited retinal diseases: a horizon scanning review. Eye. 2015;29:1131–40. doi: 10.1038/eye.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanna P, Strauss RW, Fujinami K, Michaelides M. Stargardt disease: clinical features, molecular genetics, animal models and therapeutic options. Br J Ophthalmol. 2017;101:25–30. doi: 10.1136/bjophthalmol-2016-308823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zernant J, Xie YA, Ayuso C, Riveiro-Alvarez R, Lopez-Martinez MA, Simonelli F, et al. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum Mol Genet. 2014;23:6797–806. doi: 10.1093/hmg/ddu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujinami K, Sergouniotis PI, Davidson AE, Mackay DS, Tsunoda K, Tsubota K, et al. The clinical effect of homozygous ABCA4 alleles in 18 patients. Ophthalmology. 2013;120:2324–31. doi: 10.1016/j.ophtha.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Lee W, Xie Y, Zernant J, Yuan B, Bearelly S, Tsang SH, et al. Complex inheritance of ABCA4 disease: four mutations in a family with multiple macular phenotypes. Hum Genet. 2016;135:9–19. doi: 10.1007/s00439-015-1605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalkara D, Goureau O, Marazova K, Sahel JA. Let there be light: gene and cell therapy for blindness. Hum Gene Ther. 2016;27:134–47. doi: 10.1089/hum.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strom SP, Gao YQ, Martinez A, Ortube C, Chen Z, Nelson SF, et al. Molecular diagnosis of putative Stargardt disease probands by exome sequencing. BMC Med Genet. 2012;13:67. doi: 10.1186/1471-2350-13-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin W, Xiao X, Li S, Jia X, Guo X, Zhang Q. Identification of genetic defects in 33 probands with Stargardt disease by WES-based bioinformatics gene panel analysis. PLoS ONE. 2015;10:e0132635. doi: 10.1371/journal.pone.0132635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duggan AT, Harris AJT, Marciniak S, Marshall I, Kuch M, Kitchen A, et al. Genetic discontinuity between the Maritime Archaic and Beothuk populations in Newfoundland, Canada. Curr Biol. 2017;27:3149–56. doi: 10.1016/j.cub.2017.08.053. [DOI] [PubMed] [Google Scholar]
- 30.Mannion JJ. Memorial University of Newfoundland. Institute of Social and Economic Research.: The Peopling of Newfoundland: essays in historical geography. St. John’s, Nfld.: Institute of Social and Economic Research, Memorial University of Newfoundland; 1977. [Google Scholar]
- 31.Bear JC, Nemec TF, Kennedy JC, Marshall WH, Power AA, Kolonel VM, et al. Inbreeding in outport Newfoundland. Am J Med Genet. 1988;29:649–60. doi: 10.1002/ajmg.1320290324. [DOI] [PubMed] [Google Scholar]
- 32.Zhai G, Zhou J, Woods MO, Green JS, Parfrey P, Rahman P, et al. Genetic structure of the Newfoundland and Labrador population: founder effects modulate variability. Eur J Hum Genet. 2016;24:1063–70. doi: 10.1038/ejhg.2015.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merner ND, Hodgkinson KA, Haywood AF, Connors S, French VM, Drenckhahn JD, et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet. 2008;82:809–21. doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milting H, Klauke B, Christensen AH, Musebeck J, Walhorn V, Grannemann S, et al. The TMEM43 Newfoundland mutation p.S358L causing ARVC-5 was imported from Europe and increases the stiffness of the cell nucleus. Eur Heart J. 2015;36:872–81. doi: 10.1093/eurheartj/ehu077. [DOI] [PubMed] [Google Scholar]
- 35.Green JS, Bear JC, Johnson GJ. The burden of genetically determined eye disease. Br J Ophthalmol. 1986;70:696–9. doi: 10.1136/bjo.70.9.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green JS, Johnson GJ. Hereditary diseases as causes of blindness in Newfoundland: preliminary report. Can J Ophthalmol. 1983;18:281–4. [PubMed] [Google Scholar]
- 37.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azarian SM, Megarity CF, Weng J, Horvath DH, Travis GH. The human photoreceptor rim protein gene (ABCR): genomic structure and primer set information for mutation analysis. Hum Genet. 1998;102:699–705. doi: 10.1007/s004390050765. [DOI] [PubMed] [Google Scholar]
- 39.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 40.Manion JJ. The Peopleing of Newfoundland: Essays in Historical Geography. Memorial University of Newfoundland, St. John’s, Newfoundland. Canada: Institute of Scoial and Economic Research; 1977. [Google Scholar]
- 41.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 42.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 43.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sangermano R, Khan M, Cornelis SS, Richelle V, Albert S, Garanto A, et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018;28:100–10. doi: 10.1101/gr.226621.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sangermano R, Bax NM, Bauwens M, van den Born LI, De Baere E, Garanto A, et al. Photoreceptor progenitor mRNA analysis reveals exon skipping resulting from the ABCA4 c.5461-10T->C mutation in Stargardt disease. Ophthalmology. 2016;123:1375–85. doi: 10.1016/j.ophtha.2016.01.053. [DOI] [PubMed] [Google Scholar]
- 46.Fujinami K, Strauss RW, Chiang JP, Audo IS, Bernstein PS, Birch DG, et al. Detailed genetic characteristics of an international large cohort of patients with Stargardt disease: ProgStar study report 8. Br J Ophthalmol. 2018;103:390–7. [DOI] [PMC free article] [PubMed]
- 47.Aukrust I, Jansson RW, Bredrup C, Rusaas HE, Berland S, Jorgensen A et al. The intronic ABCA4 c.5461-10T>C variant, frequently seen in patients with Stargardt disease, causes splice defects and reduced ABCA4 protein level. Acta Ophthalmol. 2016;95:240–6. [DOI] [PubMed]
- 48.Aukrust I, Jansson RW, Bredrup C, Rusaas HE, Berland S, Jorgensen A, et al. The intronic ABCA4 c.5461-10T>C variant, frequently seen in patients with Stargardt disease, causes splice defects and reduced ABCA4 protein level. Acta Ophthalmol. 2017;95:240–6. doi: 10.1111/aos.13273. [DOI] [PubMed] [Google Scholar]
- 49.Burke TR, Allikmets R, Smith RT, Gouras P, Tsang SH. Loss of peripapillary sparing in non-group I Stargardt disease. Exp Eye Res. 2010;91:592–600. doi: 10.1016/j.exer.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Runhart EH, Sangermano R, Cornelis SS, Verheij J, Plomp AS, Boon CJF, et al. The common ABCA4 variant p.Asn1868Ile shows nonpenetrance and variable expression of Stargardt disease when present in trans with severe variants. Investig Ophthalmol Vis Sci. 2018;59:3220–31. doi: 10.1167/iovs.18-23881. [DOI] [PubMed] [Google Scholar]
- 51.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–5. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 52.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huckfeldt RM, East JS, Stone EM, Sohn EH. Phenotypic variation in a family with pseudodominant Stargardt disease. JAMA Ophthalmol. 2016;134:580–3. doi: 10.1001/jamaophthalmol.2015.5471. [DOI] [PubMed] [Google Scholar]
- 54.Chong JX, Ouwenga R, Anderson RL, Waggoner DJ, Ober C. A population-based study of autosomal-recessive disease-causing mutations in a founder population. Am J Hum Genet. 2012;91:608–20. doi: 10.1016/j.ajhg.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boycott KM, Rath A, Chong JX, Hartley T, Alkuraya FS, Baynam G, et al. International cooperation to enable the diagnosis of all rare genetic diseases. Am J Hum Genet. 2017;100:695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.