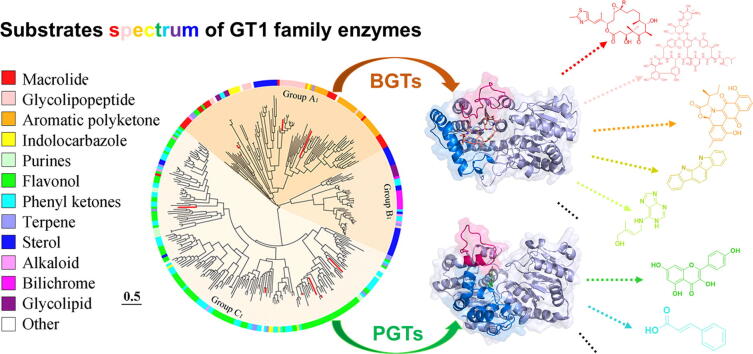
Graphical abstract
Keywords: Glycosyltransferase (GT), GT1 family, Phylogenetic distribution, Substrates spectrum, Sequence length disparity, Distinct binding motifs
Highlights
-
•
The phylogenetic distribution of GT1 family enzymes showed domain-dependent pattern.
-
•
The domain-dependent characterized GTs showed distinct substrates spectrum.
-
•
Two regions that discriminate the GT1 enzymes from different domains were identified.
Abstract
Glycosyltransferases (GTs) are responsible for transferring glycosyl moieties from activated sugar donors to certain acceptors, among which the GT1 family enzymes have been known for their outstanding glycosylation capacities toward diverse natural products, such as glycolipids, flavonoids and macrolides etc. However, there still lacks a systematic collation of this important family members. In this minireview, all the GT1 family sequences were phylogenetically analyzed, and the grouping of GT1 proteins exhibited a taxonomic life domain-dependent pattern, revealing many untapped clades of GTs. The further phylogenetic analysis of the characterized GTs facilitated the classification of substrates coverage of GT1 family enzymes from different life domains, whereby the GTs from bacteria can tolerate a wider spectrum of chemical skeletons as substrates, showing higher promiscuity than those from other domains. Furthermore, the sequence sizes of GTs from different domains were compared to understand their different substrates selectivity. Based on the multiple sequence alignments of 28 representative GT1 enzymes with crystal structures, two critical regions located in the N-terminal of GTs were identified, which were most variable among sequences from different taxonomic domains and essential for substrates binding and/or catalysis. The key roles of these two regions were validated by enumerating the influential residues that interacted with substrates in the representative structures from bacteria and plants. The atlas for GT1 family in terms of phylogeny, substrates selectivity, sequence length, and critical motifs provides the clues for the exploration of unknown GT1s and rational engineering of known enzymes, synthesizing novel promising glycoconjugates for pharmaceutical application.
1. Introduction
Glycosyltransferases (GTs), as natural biocatalysts, can catalyze the transfer of the glycosyl moieties from activated sugar donors to a diverse range of acceptors [1], [2]. Many acceptor molecules like proteins, nucleic acid, and secondary metabolites such as antibiotics, alkaloids and plant hormones are widely used for medicinal, agricultural, and industrial application [3], [4]. According to anchor atom for the glycosidic appendage in the acceptor substrates, GTs can be generally classified into O-, N-, S-, and C-GTs. The donor substrates are often activated form of sugar (mainly the NDP-pentoses and NDP-hexoses) mediated by high-energy phosphoester bond(s), and a diversity of (modified) sugar residues exhibit distinct effects on the physiological functions of glycoside products [5], [6]. According to the Carbohydrate-Active enZYmes Database (CAZy database, http://www.cazy.org/), GTs constitute a superfamily, classified into 110 families according to the sequence identity, conserved motifs, and stereochemistry of the glycosidic linkage, etc [7], [8], [9]. Structural characterization of the reported GT representatives exhibited two predominate topologies: GT-A and GT-B folds [10], [11], [12], but variant folds have also been reported, like GT-C fold combining topological elements between canonical GT-A and GT-B folds [13]. The GT-A fold possess a central β-sheet surrounded by α-helices and two tightly abutting β/α/β domains, and a highly conserved DxD motif (ASP-X-ASP) coordinates with a divalent cation [14]. The GT-B fold consists of two separate and flexibly linked Rossmann-like domains, and the catalytic cleft is located between these two domains [11], [15]. GT-B fold contains a relatively conserved donor binding motif mainly at the C-terminal domain, whereas the N-terminal domain is highly divergent, exhibiting the greater plasticity of topology [2], [10]. Thus, the GT-B structure shows flexible substrates binding pattern compared to the GT-A fold. In terms of reaction mechanism, the inverting GTs could utilize a direct SN2-like displacement so that the stereochemistry at C-1 of sugar substrates is inverted in the glycosylated products, while the retaining GTs could retain the original configuration of sugar substrates, and two consecutive SN2 reactions could be the mechanism: the sugar moiety first binds to the retaining GT through SN2 reaction which leads to an inverting configuration at sugar C1, after which, the sugar is transferred to the acceptor through another SN2 reaction so that sugar reverts to its initial conformation [11], [16]. Generally, each families of GTs possess unitary catalytic mechanism and structural fold. For instance, family GT1, GT9, GT10 and family GT3, GT4, GT5 are inverting and retaining enzymes, respectively, which adopt the GT-B fold. Family GT2, GT16, GT25 and Family GT6, GT8, GT24 are inverting and retaining enzymes, respectively, which adopt the GT-A fold [17].
Remarkably, the number of characterized enzymes of GT1 family is largest among the 110 GT families in the CAZy database, largely because of their excellent glycosylation capacities toward numerous valued small molecules [18], [19], [20], [21], [22]. The inverting GT1 family enzymes all adopt the GT-B fold and usually involve in negligent discrimination of acceptors due to the flexibility in N-terminal domain [23]. However, most GTs preferentially select one specific sugar donor because of a set of highly conserved amino acid (aa) residues in C-terminal domain, like “PSPG motif” in plants [24]. Similar motifs are also identified in GTs from other taxonomic life domains, such as bacteria and animals [25], [26]. GT1 family glycosyltransferases are found in almost all domains of life, but the members from disparate domains show diverse functions. For instance, the “antibiotic glycosylation” is commonplace in the microorganisms. The typical GTs glycosylating antibiotic are divergent in the phylogenetic classification [27], and show activities to various chemical scaffolds, including polyketides [28], macrolides [29], [30], [31] and glycolipopeptides [32], [33]. These GT1 enzymes are crucial for the biosynthesis of glycosylated natural products, and play a vital role in the last step of molecular assembly and the glycosylated products often exhibit stronger pharmaceutical activities than their precursors [34]. GT1 enzymes in plant kingdom also glycosylate massive bioactive natural products with low molecular weight, such as flavonoids, phenyl ketones, terpenoids and steroids [35], modulating the stability, solubility, and biological activity of the aglycons, and regulating the plant hormones or detoxifying the xenobiotics [36]. Moreover, the “substrate promiscuity” are ubiquitous for GT1 family enzymes from most life domains, and this enlightened glycosylation capacity prompts the application through enzymatic engineering [2], [34], [37].
So far, our understanding of GT1 family enzymes is still limited due to the small number of the resolved crystal structures [38]. In light of the importance of GT1 family glycosyltransferases, it is meaningful to have an overview on the classification and substrates recognition of GT1 family enzymes. In this study, all the GT1 family members deposited in the CAZy database were phylogenetically analyzed, whereby the GTs were grouped related to the life domains classification, and numerous unexploited GTs were revealed especially for those harbored in bacteria (BGTs). Furthermore, the substrates coverage of the characterized GTs from different taxonomic life domains were summarized, and BGTs were found to accept a broader spectrum of chemical skeletons as substrates. We also discussed two motifs that most discriminated the enzymes from bacteria or plants, which played a pivotal role in substrates binding and recognition. Our studies pave the way for the future exploration of uncharacterized GT1 proteins and/or rational engineering of known ones.
2. The occurrence frequency of GT1 in various organisms
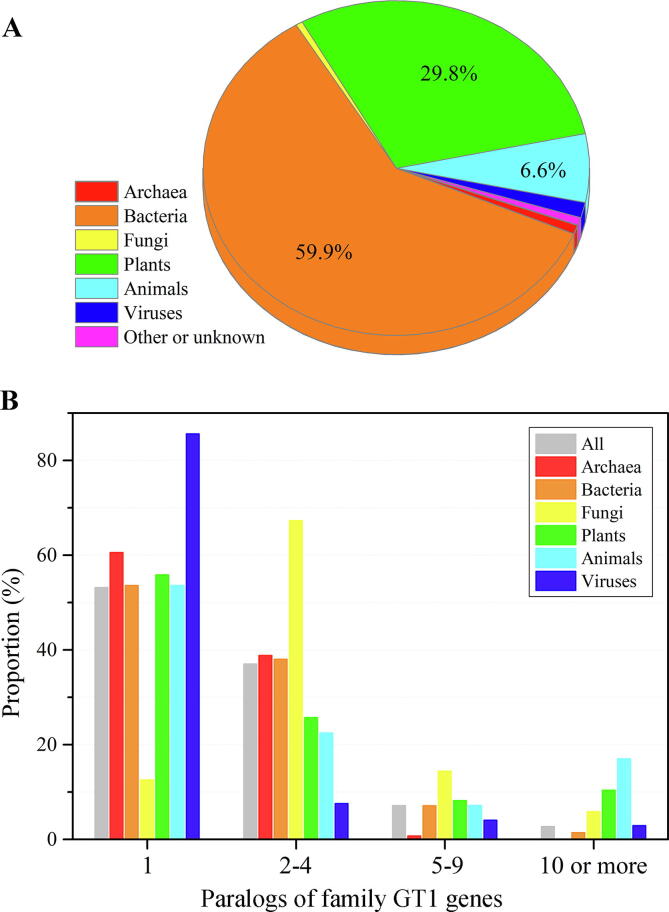
According to CAZy database, GT1 family contains more than 20,000 members, distributed in nearly 8,000 different life domains that cover almost all major life taxonomies such as bacteria, animals, plants, fungi, viruses, etc, though they display an uneven distribution. Nearly 60% of all the GT1 family members are originated from the bacteria (BGTs), followed by 30% from plants (PGTs) and approximately 7% from animals (AGTs) (Fig. 1A). Among the 8,000 life domains containing GT1 family, 81.2% and 9.1% are bacteria and plants, respectively, while every other domains account for less than 3%. It is not uncommon that a same GT1-containing host encode multiple paralogs of GT1 sequences. To be specific, 53.2% of the referred 8,000 life domains encode a single paralog of the GT1 genes, 37.0% encode 2–4 paralogs, 7.2% encode 5–9 paralogs, and 2.7% encoded at least 10 paralogs. Notably, 16.9% and 10.4% of animals and plants encode at least 10 paralogs of GT1 family, respectively, whereas only 1.4% of bacterial genomes can reach this number (Fig. 1B). Overall, these data revealed that the existing GT1 sequences are predominantly derived from bacteria, almost two times those from plants and animals, though it is likely that more bacterial genomes have been sequenced than plants and animals. However, multiple paralogs of the GT1 family genes are more likely to happen in the animals and plants genomes (Table S1).
Fig. 1.
The distribution statistics of GT1 enzymes from diverse life domains. (A) GT1 family exhibits an uneven distribution pattern in different domains of life. (B) The statistics of paralogs of family GT1 genes in different life domains.
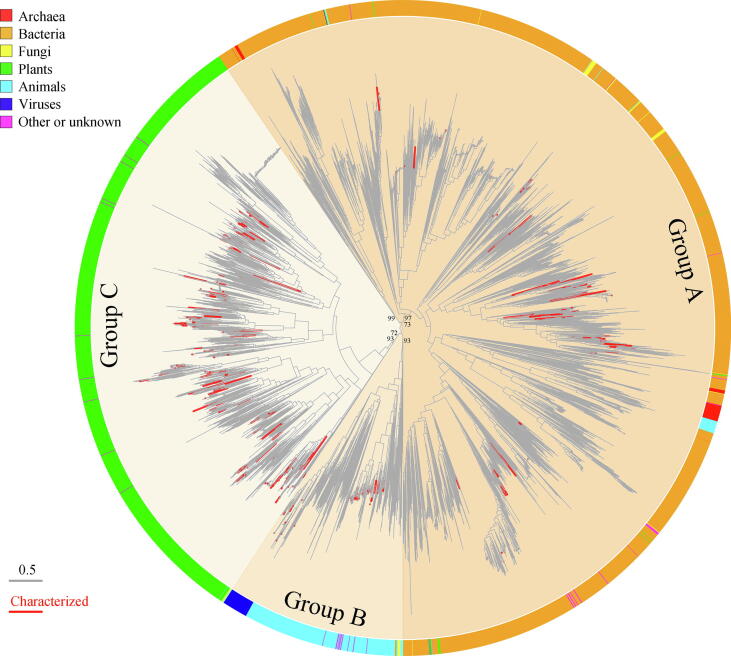
The phylogenetic tree analysis of the retrieved >20,000 GT1 protein sequences presented three major groups A, B and C (Fig. 2). The members residing in group A, B and C accounted for 59.5%, 9.2% and 31.3%, respectively, and they were separated early during the GT1 family evolution. Interestingly, the phylogenetic grouping was highly related to the life domains classification, whereby BGTs and PGTs accounted for more than 90% of group A and C, respectively, and the AGTs occupied over 80% of group B, which also included most of the virus-derived GTs (VGTs). The GTs from other life domains were much fewer, and unevenly distributed in the three groups. From the perspective of life domains, 99.8% of BGTs and 100% of archaea-derived GTs (ArGTs) belonged to group A, 91.5% of AGTs and 99.4% of VGTs belonged to group B, and 98.9% of PGTs belonged to group C. Intriguingly, our phylogenetic analysis of GT1 family reveals the gene horizontal transfer among the different life domains takes place. For example, group A is dominated by bacterial proteins, but also contains members from animals (1.1%), fungi (FGTs) (1.0%), or plants (0.6%). Actually, several members of the GT1 family or other GT families have been found to undergo horizontal gene transfer [39], [40], [41].
Fig. 2.
The phylogenetic tree of all the GT1 family members. The three groups A, B and C were distinguished by different background. The origin of the GT1 were color labeled in peripheral circle. The characterized GT1 enzymes in the branches were highlighted by red lines, and the functions of GT1 in many clades awaits characterization. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Only 1.6% of GT1 enzymes deposited in CAZy database have been characterized, whose natural substrates were identified. The proportion of the characterized GTs from plants (3.0%), animals (2.9%), and viruses (2.6%) were slightly higher than that from bacteria (0.7%), while no characterized ArGTs have been reported yet. Although a large number of GTs have been sequenced, the functions of the majority of GTs are still unknown. More remarkably, many clades of GTs in Fig. 2 remain untapped, which indicate a huge space for the discovery of novel members of GTs for the glycosylation of pharmaceutically important natural products.
3. Substrates spectrum of the characterized GT1 family members
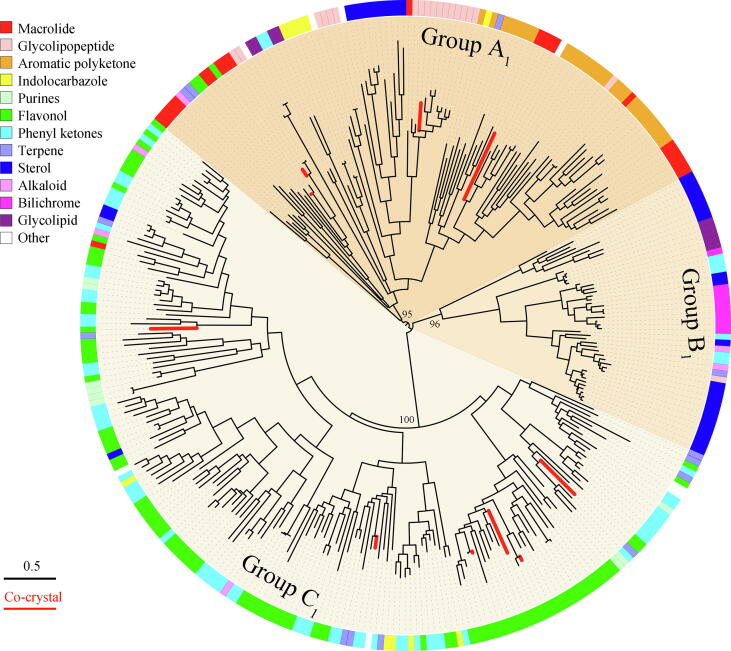
GT1 family glycosyltransferases can glycosylate numerous valuable molecules, however, there is still a lack of an overview of substrates spectrum of GTs from different taxonomic life domains. Herein, we phylogenetically analyzed the 317 characterized representatives in the GT1 family, and summarized their natural substrates (Fig. 3). Similar to the phylogenetic classification of all the GT1 family members, the phylogenetic grouping of the characterized GTs was also taxonomic life domain-dependent, separating into three major groups A1, B1 and C1, which were the subgroups of group A (bacteria), B (animals) and C (plants), respectively (Fig. S1). Over 97% of constituting characterized sequences in group A1 were from bacteria (BCGTs), and the animal-derived characterized GTs (ACGTs) accounted for more than 80% of group B1, while group C1 was merely composed of the plant-derived characterized GTs (PCGTs).
Fig. 3.
Phylogenetic tree analysis of the characterized GTs from the GT1 family. The three groups A1, B1 and C1 were color labelled with different background according to those in Fig. 2. The substrate specificities of the characterized GT1 family were depicted by different colors in peripheral circle. The characterized GT1 enzymes with co-crystal structures reported were highlighted by red lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The phylogenetically separated three groups of characterized GTs showed intriguingly distinct substrates spectrum. The GTs in group A1 possessed a wide range of substrates selectivity, which accepted almost all the listed types of compounds in Fig. 3 as substrates, including high-molecular weight compounds like glycolipopeptides and low-molecular weight compounds like purines. Moreover, even the closely located branches of GTs were capable of glycosylating disparate chemical skeletons, indicating the potential substrates promiscuity of the BCGTs (Fig. 3). In contrast, the PCGTs had a comparatively narrow spectrum of substrates selectivity, showing preference on phenyl ketones or flavonoids, and the ACGTs in group B1 were more inclined to mediate sterols, bilichromes or alkaloid compounds glycosylation. However, it was noteworthy that no strict catalytic boundary existed among the three groups of glycosyltransferases, and one certain substrate type could be catalyzed by GTs from different groups. For example, all the groups A1, B1 and C1 of GTs could tolerate sterols as natural substrates; flavonoids could be catalyzed by GTs in group A1, B1. On the other hand, some chemical scaffolds were only recognized by the GTs from a single group, wherein only BCGTs rather than ACGTs and PCGTs have been hitherto reported to catalyze macrolides and purines.
Although the “promiscuity” is common among GT1 family enzymes [5], [34], [42], [43], BCGTs seem to have evolved a higher level of “promiscuity” than ACGTs and PCGTs. The Streptomyces antibioticus glycosyltransferase OleD has been reported to have activity on strikingly different architectures, including alkaloids, flavonoids, macrolides, and sterols [22], [44], [45]; Likewise, the enzyme BsGT-1 from Bacillus subtilis was able to glycosylate a series of compounds, such as phenyl ketones, flavonoids, terpenes, and macrolides [46], [47]. The plant-derived glycosyltransferases show limited substrate promiscuity. Although the permissive C-glycosyltransferase GcCGT tolerated different types of NDP-sugar as donors, the sugar acceptors were restricted to the structurally related phenyl ketones or flavonoids [48]; the plant-derived glycosyltransferase UGT74AN1 accepted a broad range of chemical skeletons to form O-, N-, and S-glycosides, but it was more keen on the glycosylation of steroids [21].
The glycosylation is important for tuning the physiochemical properties of natural products, and it may also execute potential important biological functions for organisms, which form driven forces for organisms to orchestrate the glycosyltransferases to efficiently mediate this reaction. The taxonomic life domain-dependent pattern of substrates spectrum for GT1 enzymes revealed in Fig. 3 can well reflect the adaptive evolution of this family. To be specific, the flavonoids or phenyl ketones are ubiquitous in plants, which play important roles for their survival [35], [49], and thus it is the plants that have evolved a strategy to modify them. Steriods are oftentimes materials for the production of hormones and bilichrome type compounds that are essential for blood, and thus ACGTs have evolved to accept these catalogs of chemicals. Bacteria produce a plethora of structurally diverse compounds for various purposes, such as antibiotics and siderophores, which probably require a larger number of GTs to handle them (Fig. 1A). Especially, bacteria contain comparatively less paralogs of the GT1 family genes (Fig. 1B), which could contribute to evolve a higher promiscuity of GT1 enzymes for bacteria to cope with the complicated challenges. For instance, one of the important roles of bacterial GTs is “natural antidote” to reduce threats from toxic products produced by themselves [22], or their competitors [50].
4. The disparity in the sequence length of GT1 proteins from different taxonomic life domains
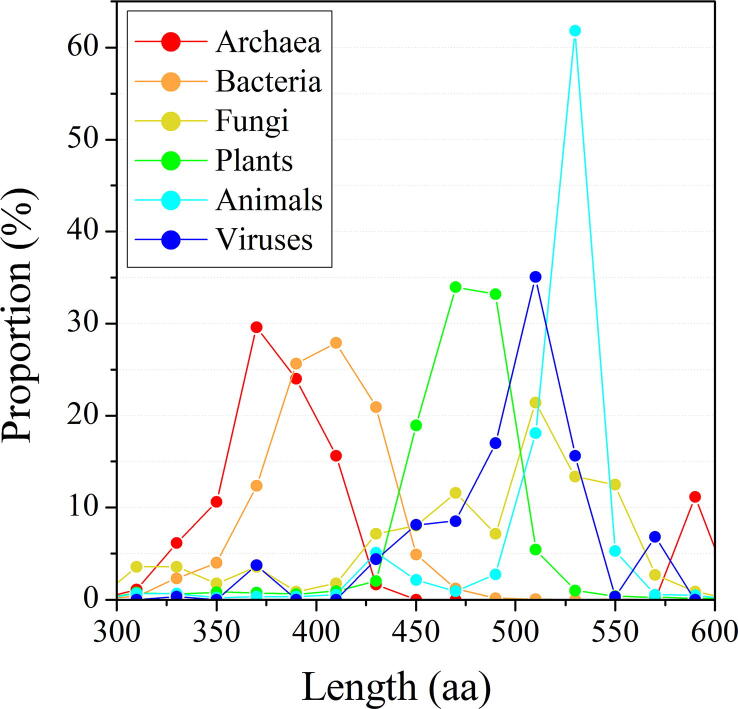
To investigate the structural differences of the GTs from different life domains, we first compared their lengths of primary sequences. The proportion of proteins of different lengths from each domain of life was detailed in Fig. 4, which clearly distinguished the frequencies and variations. For example, the length variation of AGTs peaked in the range of 425–575 aa was much smaller than that of FGTs peaked in the range of 500–550 aa. The lengths of ArGTs and BGTs (mainly belonging to Group A) were relatively shorter, of which the average lengths were 400.6 ± 69.8 and 402.0 ± 27.9 residues, respectively. The PGTs and FGTs (mainly belonging to Group C) had the medium average lengths, with 470.3 ± 31.7 and 479.1 ± 68.1 residues, respectively, while the AGTs and VGTs (mainly belonging to Group B) were comparatively longer, of which the average lengths were 494.7 ± 42.0 and 514.3 ± 38.0 residues, respectively (Table 1). Hence, the GTs from different life domains showed distinct amino acid sequence lengths and distribution pattern. Notably, this feature may be not applicable to other GT-B families. For example, the GT10 family members from bacteria have no significant difference in protein length compared to the members from eukaryote (Fig. S2).
Fig. 4.
The proportion of proteins of different lengths from each life domains. Every 20 amino acid residues were binned as one scatter. The frequency and variation in the protein sequence length was represented by the peak height and width.
Table 1.
Comparison of the average length (aa) of GTs from different life domains.
| GTs from different life domains | Length (aa) of whole Protein | Length (aa) of Region A | Length (aa) of Region B |
|---|---|---|---|
| 17 BGTs with crystal structures | 400.8 ± 22.0 | 38.06 ± 9.66 | 43.29 ± 14.03 |
| 11 PGTs with crystal structures | 460.7 ± 14.9 | 24.36 ± 2.77 | 63.00 ± 13.39 |
| All archaea-derived GTs (ArGTs) | 400.6 ± 69.8 | 37.11 ± 7.56 | 21.68 ± 6.76 |
| All bacteria-derived GTs (BGTs) | 402.0 ± 27.9 | 39.85 ± 8.03 | 41.45 ± 14.62 |
| All plant-derived GTs (PGTs) | 470.3 ± 31.7 | 40.89 ± 14.02 | 49.45 ± 13.33 |
| All fungi-derived GTs (FGTs) | 479.1 ± 68.1 | 27.77 ± 6.70 | 64.48 ± 10.15 |
| All animal-derived GTs (AGTs) | 494.7 ± 42.0 | 47.31 ± 7.02 | 73.13 ± 10.48 |
| All virus-derived GTs (VGTs) | 514.3 ± 38.0 | 45.89 ± 3.52 | 75.48 ± 5.31 |
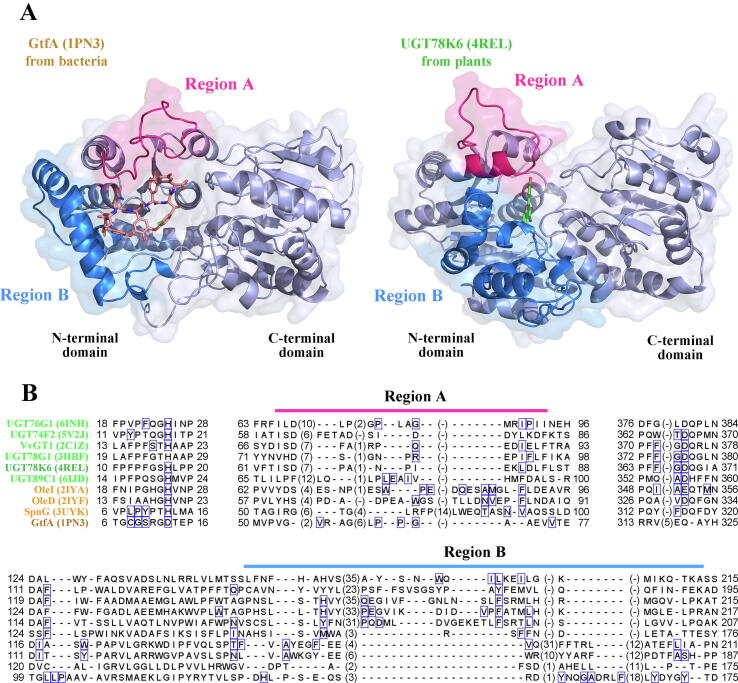
When we wrote this manuscript, 92 structures resolved from 35 GT1 family proteins were deposited in the PDB database. Seven proteins were excluded due to the defect in the requisite information (i.e. lack of UniProt ID, NMR-resolved structures, or unavailable structures), and the remaining 28 representative enzymes with complete crystal structures were subjected for further structural analysis, involving 17 GTs from bacteria and 11 GTs from plants. These proteins showed highly similar topology of GT-B fold consisting of two distinct β/α/β Rossmann domains [11]. GT-B fold normally proceeded flexible domain movement with a fickle substrate-binding pattern, and the compressed-domain global motion was often associated with critical regions [10]. The structure-based multiple sequence alignment of the 28 GTs indicated the structural differences between the 17 BGTs and 11 PGTs were mainly concentrated in two regions (Region A and Region B) located at the N-terminal, wherein a loop and a partial α-helix structure existed, respectively. For the Region A, the average lengths in the structures of 17 BGTs and 11 PGTs were 38.1 ± 9.7 aa and 24.4 ± 2.8 aa, respectively. For the Region B, the average lengths of 17 BGTs and 11 PGTs were 43.3 ± 14.0 aa and 63.0 ± 13.4 aa, respectively. Two typical crystal structures GtfA (PDB number: 1PN3) from bacterium Amycolatopsis orientalis and UGT78K6 (PDB number: 4REL) from plant Clitoria ternatea were showcased in Fig. 5A. The Region A of GtfA formed a disordered long loop, while the loop in the Region A of UGT78K6 was relatively shorter and fitted with a part of α-helix. The loop enhances the plasticity of GTs, allowing a more flexible and inclusive substrate-binding pattern, whereas the helical structures increases rigidity of GTs. The Region B of GtfA consisted of an α-helix and a loop, and both of the two parts were significantly shorter than the counterparts of UGT78K6. Moreover, the Region B of GtfA exhibited structural eversion, expanding the entrance of the catalytic cavity to facilitate the substrates binding, while the Region B of UGT78K6 displayed structural inversion, limiting the entry of the substrates. These structural features could contribute to the promiscuity differences between BGTs and PGTs: BGTs exhibit a wider spectrum of substrates recognition.
Fig. 5.
Comparison of the structures and sequence lengths of Region A and Region B in BGTs and PGTs. (A) The representative crystal structures of the bacterial protein GtfA and botanical protein UGT78K6. The major differences among GtfA and UGT78K6 lied in the Region A and Region B that were depicted in red and blue, respectively. (B) The multiple sequence alignment of the 10 GT sequences for which co-crystal structures were known, including 4 BGTs labelled in orange color and 6 PGTs labelled in green color. The Region A and B were labeled with lines and the number in the brackets represented the amino acids omitted. The influential amino acid residues that interacted with substrates in the co-crystals were marked with blue boxes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As the disparity in the sequences and structures of GT1 family proteins from different life domains may be mainly located in the referred two regions, we further calculated the approximate lengths of Region A and Region B in GT1 family proteins, based on the referenced crystal structures and multiple sequence alignment. As shown in Table 1, the sequence lengths of Region A and Region B in GT1 family proteins indeed differed in a taxonomic life domain-dependent manner. The length differentiation in these two regions may be critical to the structural diversity and substrate recognition of GTs from different life domains.
5. The influential residues in the Region A and B of co-crystal structures of BGTs and PGTs
To analyze the interactions of the region A and B with substrates, we collected 10 co-crystal structures of the GT1 family, including 4 BGTs and 6 PGTs. The loop regions of region A and B turned out to be the loop N3 and N5, which had variable conformations and flexible interactions with substrates. Moreover, the α-helixes adjacent to N3 and N5 often formed an internal hydrophobic cavity, influencing substrates selectivity and promiscuity [10]. The multiple sequence alignment of the 10 GT sequences was performed and the influential amino acid residues, which formed hydrogen bonds or hydrophobic interactions with acceptors in the N-terminal were marked (Fig. 5B). We found that the quantity of influential residues distributing in Region A and B were distinct between BGTs and PGTs due to the length difference in these two regions. The Region A of 4 BGTs had more influential residues than that of 6 PGTs, while the Region B of 6 PGTs contained more influential residues than BGTs. Therefore, the lengths of Region A and B, along with the distribution of the influential residues in these two regions, may be pivotal for the discrimination of substrate selectivity of GTs from different life domains, which requires further experimental verification.
6. Summary and outlook
Enzyme-catalyzed glycosylation is believed to be an effective approach to improve the physiochemical properties of compounds, and thus the discovery of novel glycosyltransferases is of utmost importance. Though a large number of GT1 family enzymes have been revealed owing to the development of Next-Generation Sequencing technologies, the functions of the majority remain uncharacterized. Our study showed that the phylogenetic grouping of the GT1 enzymes were taxonomic life domain-dependent, and that GTs finely evolve and orchestrate the substrates spectrum dependent of the life domains that they are originated from. In addition, the phylogenetic analysis also illuminated a large number of GTs that remain yet unexplored, which could be potentially applied to glycosylate a broader range of chemical scaffolds, especially for the BGTs that have evolved the most diversity and promiscuity. We also made to compare the sequence lengths of GTs from various life domains, and accordingly identified two critical motifs in the N-terminal domain of GT1 enzymes through structure-based multiple sequence alignment, which most discriminated GTs of different origin and probably play a pivotal role in the substrate recognition and/or catalysis. All in all, the sketchy knowledge in terms of phylogenetic distribution, substrates spectrum, sequence length, and critical structural motifs of GT1 family glycosyltransferases from different life domains, will be beneficial for the rational exploitation of this important family of enzymes, to promote the efficient enzymatic synthesis of biologically important glycosylated compounds.
CRediT authorship contribution statement
Peng Zhang: Conceptualization, Methodology, Investigation, Writing - original draft, Validation. Zheng Zhang: Conceptualization, Methodology, Resources, Investigation, Visualization, Validation. Lijuan Zhang: Resources, Validation. Jingjing Wang: Data curation, Validation. Changsheng Wu: Conceptualization, Supervision, Writing - review & editing, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was financially supported by the National Key R&D Program of China (2019YFA0905700), and National Natural Science Foundation of China (NSFC) (Nos. 31900042 and 81973215) to C.S.W.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.06.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gloster T.M. Advances in understanding glycosyltransferases from a structural perspective. Curr Opin Struct Biol. 2014;28:131–141. doi: 10.1016/j.sbi.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard S., Thorson J.S. Enzymatic tools for engineering natural product glycosylation. Curr Opin Chem Biol. 2006;10(3):263–271. doi: 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Bowles D., Isayenkova J., Lim E.K., Poppenberger B. Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol. 2005;8(3):254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Marschall E., Cryle M.J., Tailhades J. Biological, chemical, and biochemical strategies for modifying glycopeptide antibiotics. J Biol Chem. 2019;294(49):18769–18783. doi: 10.1074/jbc.REV119.006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gantt R.W., Peltier-Pain P., Singh S., Zhou M., Thorson J.S. Broadening the scope of glycosyltransferase-catalyzed sugar nucleotide synthesis. Proc Natl Acad Sci USA. 2013;110(19):7648–7653. doi: 10.1073/pnas.1220220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y., Yao M., Wang Y., Ding M., Zha J., Xiao W. Advances in engineering UDP-sugar supply for recombinant biosynthesis of glycosides in microbes. Biotechnol Adv. 2020;107538 doi: 10.1016/j.biotechadv.2020.107538. [DOI] [PubMed] [Google Scholar]
- 7.Coutinho P.M., Deleury E., Davies G.J., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328(2):307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 8.Campbell J.A., Davies G.J., Bulone V., Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326(Pt 3):929. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombard Vincent, Golaconda Ramulu Hemalatha, Drula Elodie, Coutinho Pedro M., Henrissat Bernard. The carbohydrate-active enzymes database (CAZy) in 2013. Nucl Acids Res. 2014;42(D1):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang A., Singh S., Phillips G.N., Jr., Thorson J.S. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr Opin Biotechnol. 2011;22(6):800–808. doi: 10.1016/j.copbio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lairson L.L., Henrissat B., Davies G.J., Withers S.G. Glycosyltransferases: structures, functions, and mechanisms. Ann Rev Biochem Ann Rev Palo Alto. 2008:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 12.Brockhausen I. Crossroads between bacterial and mammalian glycosyltransferases. Front Immunol. 2014;5:492. doi: 10.3389/fimmu.2014.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrus D., Kellokumpu S., Glumoff T. Crystal structures of eukaryote glycosyltransferases reveal biologically relevant enzyme homooligomers. Cell Mol Life Sci CMLS. 2018;75(5):833–848. doi: 10.1007/s00018-017-2659-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ünligil U.M., Rini J.M. Glycosyltransferase structure and mechanism. Curr Opin Struct Biol. 2000;10(5):510–517. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 15.Vrielink A., Ruger W., Driessen H.P., Freemont P.S. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994;13(15):3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang D.M., Liu J.H., Wu H., Wang B.B., Zhu H.J., Qiao J.J. Glycosyltransferases: mechanisms and applications in natural product development. Chem Soc Rev. 2015;44(22):8350–8374. doi: 10.1039/c5cs00600g. [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12(7):1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulichak A.M., Lu W., Losey H.C., Walsh C.T., Garavito R.M. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43(18):5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C., Griffith B.R., Fu Q., Albermann C., Fu X., Lee I.-K. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313(5791):1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 20.Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B.G., Tarling C.A. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25(6):1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen C., Huang W., Zhu X.-L., Li X.-S., Zhang F., Jiang R.-W. UGT74AN1, a permissive glycosyltransferase from asclepias curassavica for the regiospecific steroid 3-O-glycosylation. Org Lett. 2018;20(3):534–537. doi: 10.1021/acs.orglett.7b03619. [DOI] [PubMed] [Google Scholar]
- 22.Bolam D.N., Roberts S., Proctor M.R., Turkenburg J.P., Dodson E.J., Martinez-Fleites C. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc Natl Acad Sci USA. 2007;104(13):5336–5341. doi: 10.1073/pnas.0607897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palcic M.M. Glycosyltransferases as biocatalysts. Curr Opin Chem Biol. 2011;15(2):226–233. doi: 10.1016/j.cbpa.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Paquette S., Møller B.L., Bak S. On the origin of family 1 plant glycosyltransferases. Phytochemistry. 2003;62(3):399–413. doi: 10.1016/s0031-9422(02)00558-7. [DOI] [PubMed] [Google Scholar]
- 25.Quiros L.M., Aguirrezabalaga I., Olano C., Mendez C., Salas J.A. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol Microbiol. 1998;28(6):1177–1185. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- 26.Kapitonov D., Yu R.K. Conserved domains of glycosyltransferases. Glycobiology. 1999;9(10):961–978. doi: 10.1093/glycob/9.10.961. [DOI] [PubMed] [Google Scholar]
- 27.Liang D., Qiao J. Phylogenetic analysis of antibiotic glycosyltransferases. J Mol Evol. 2007;64(3):342–353. doi: 10.1007/s00239-006-0110-2. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmeister D., Wilkinson B., Foster G., Sidebottom P.J., Ichinose K., Bechthold A. Engineered urdamycin glycosyltransferases are broadened and altered in substrate specificity. Chem Biol. 2002;9(3):287–295. doi: 10.1016/s1074-5521(02)00114-x. [DOI] [PubMed] [Google Scholar]
- 29.Kuo M.S., Chirby D.G., Argoudelis A.D., Cialdella J.I., Coats J.H., Marshall V.P. microbial glycosylation of erythromycin-A. Antimicrob Agents Chemother. 1989;33(12):2089–2091. doi: 10.1128/aac.33.12.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez C., Olano C., Mendez C., Salas J.A. Characterization of a Streptomyces antibioticus gene cluster encoding a glycosyltransferase involved in oleandomycin inactivation. Gene. 1993;134(1):139–140. doi: 10.1016/0378-1119(93)90189-a. [DOI] [PubMed] [Google Scholar]
- 31.Brautaset T., Sekurova O.N., Sletta H., Ellingsen T.E., Strøm A.R., Valla S. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol. 2000;7(6):395–403. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 32.Mulichak A.M., Losey H.C., Lu W., Wawrzak Z., Walsh C.T., Garavito R.M. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci USA. 2003;100(16):9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulichak A.M., Losey H.C., Walsh C.T., Garavito R.M. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure (London, England : 1993) 2001;9(7):547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 34.Luzhetskyy A., Mendez C., Salas J.A., Bechthold A. Glycosyltransferases, important tools for drug design. Curr Top Med Chem. 2008;8(8):680–709. doi: 10.2174/156802608784221514. [DOI] [PubMed] [Google Scholar]
- 35.Osmani S.A., Bak S., Moller B.L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry. 2009;70(3):325–347. doi: 10.1016/j.phytochem.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Yonekura-Sakakibara K., Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66(1):182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- 37.Thibodeaux C.J., Melancon C.E., 3rd, Liu H.W. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed Engl. 2008;47(51):9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meech R., Hu D.G., McKinnon R.A., Mubarokah S.N., Haines A.Z., Nair P.C. The UDP-Glycosyltransferase (UGT) superfamily: new members, new functions, and novel paradigms. Physiol Rev. 2019;99(2):1153–1222. doi: 10.1152/physrev.00058.2017. [DOI] [PubMed] [Google Scholar]
- 39.Ahn S.J., Dermauw W., Wybouw N., Heckel D.G., Van Leeuwen T. Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochem Mol Biol. 2014;50:43–57. doi: 10.1016/j.ibmb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Brew K., Tumbale P., Acharya K.R. Family 6 glycosyltransferases in vertebrates and bacteria: inactivation and horizontal gene transfer may enhance mutualism between vertebrates and bacteria. J Biol Chem. 2010;285(48):37121–37127. doi: 10.1074/jbc.R110.176248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes A.L. Origin of Ecdysosteroid UDP-glycosyltransferases of Baculoviruses through Horizontal Gene Transfer from Lepidoptera. Coevolution. 2013;1(1):1–7. doi: 10.1080/23256214.2013.858497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moremen K.W., Haltiwanger R.S. Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat Chem Biol. 2019;15(9):853–864. doi: 10.1038/s41589-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams G.J., Gantt R.W., Thorson J.S. The impact of enzyme engineering upon natural product glycodiversification. Curr Opin Chem Biol. 2008;12(5):556–564. doi: 10.1016/j.cbpa.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams G.J., Goff R.D., Zhang C., Thorson J.S. Optimizing glycosyltransferase specificity via “hot spot” saturation mutagenesis presents a catalyst for novobiocin glycorandomization. Chem Biol. 2008;15(4):393–401. doi: 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou M., Hamza A., Zhan C.G., Thorson J.S. Assessing the regioselectivity of OleD-catalyzed glycosylation with a diverse set of acceptors. J Nat Prod. 2013;76(2):279–286. doi: 10.1021/np300890h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai L., Li J., Yao P., Zhu Y., Men Y., Zeng Y. Exploiting the aglycon promiscuity of glycosyltransferase Bs-YjiC from Bacillus subtilis and its application in synthesis of glycosides. J Biotechnol. 2017;248:69–76. doi: 10.1016/j.jbiotec.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 47.Zhang P., Zhang Z., Li Z.F., Chen Q., Li Y.Y., Gong Y. Phylogeny-guided characterization of glycosyltransferases for epothilone glycosylation. Microb Biotechnol. 2019;12(4):763–774. doi: 10.1111/1751-7915.13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang M., Li F.-D., Li K., Wang Z.-L., Wang Y.-X., He J.-B. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J Am Chem Soc. 2020 doi: 10.1021/jacs.9b12211. [DOI] [PubMed] [Google Scholar]
- 49.Shao H., He X., Achnine L., Blount J.W., Dixon R.A., Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17(11):3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C., Liu X., Zhang P., Wang Y., Li Z., Li X. Bacillus licheniformis escapes from Myxococcus xanthus predation by deactivating myxovirescin A through enzymatic glucosylation. Environ Microbiol. 2019;21(12):4755–4772. doi: 10.1111/1462-2920.14817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.