Abstract
Abstract
Meat tenderness is the most important criterion in food quality because it strongly influences the consumer’s satisfaction. Tenderness generally depends on connective tissue and sarcomere length of muscle. One of the effective methods for meat tenderizing is protease treatment. In this study, Manihot esculenta root was chosen as a protease source due to its skin blistering effect, suggesting the presence of strong proteolytic activity. The extraction of the crude protease was optimized by using response surface methodology (RSM) with four independent variables, which were pH (X1), CaCl2 (X2), Triton X-100 (X3) and 2-mercaptoethanol (X4). Based on the RSM model, all the independent variables were significant and the optimum extraction conditions were pH 9, 3.24 mM CaCl2, 4.12% Triton X-100 and 6.32 mM 2-mercaptoethanol. Tukey’s test results showed that the difference between the expected and experimental protease activity value was 0.05%. A reduction of meat firmness was observed when samples treated with enzyme were compared with a control by using a texture analyser. Electrophoretic patterns also showed extensive proteolysis and a reduction of intensity and number of the protein bands in the treated sample. SEM clearly revealed the degradation of muscle fibres and connective tissue of meat treated with crude protease.
Graphic abstract
Electronic supplementary material
The online version of this article (10.1007/s13197-020-04317-5) contains supplementary material, which is available to authorized users.
Keywords: Meat tenderization, Response surface methodology (RSM), Manihot esculenta, Texture analyzer, SDS-PAGE, Scanning electron microscopy (SEM)
Introduction
Tenderness is an essential parameter in meat quality and has been studied for many years. It plays a major role in marketability, which can influence the purchase decision of consumers. Meat tenderness generally depends upon sarcomere length, connective tissue and the extent of proteolytic degradation of muscles (Arshad et al. 2016). Therefore, one of the popular methods used by researchers around the world for meat tenderizing is treatment by a proteolytic enzyme called protease. Proteases, also called peptidases or proteinases, are enzymes that perform proteolysis, which is one of the most important biological reactions. Protease is able to hydrolyse the peptide bonds that link the amino acids together in the polypeptide chain forming the protein. Hence, it can degrade the muscle protein of meat to increase the tenderness (Sun et al. 2018).
Most of the protease industrial sources arise from animals and microorganisms. However, both of these sources can raise several issues, such as food adulteration as a religious issue. Therefore, plants such as Manihot esculenta from the Euphorbiaceae family have been identified as potential halal protease sources. In the Euphorbia plant, proteases are present in virtually every part, such as stem, fruit, flower, leaf, root gum and latex (Mahajan et al. 2016). The effect of skin blistering from its latex has suggested the presence of strong proteolytic activity of the plant. Previous studies show that serine proteases were found in the latex of the Euphorbiaceae family (Sobottka et al. 2014).
However, the protease extraction from plants is comparatively difficult due to the presence of a high concentration of phenolic compounds and phenol oxidase, which promote enzymatic browning, and subsequently result in enzyme inactivation (Jones and Saxena 2013). Therefore, researchers around the world have incorporated multiple variables such as reducing agents to overcome this problem. These reagents need to be optimized so that the highest response can be achieved. Optimization of these variables can be done using response surface methodology (RSM). RSM is an advanced method that is useful for studying the effect of several variables influencing the response by varying them simultaneously (Chu et al. 2015) and carrying out a lower number of experiments at less cost (Kumar et al. 2013).
In this study, the crude enzyme from Manihot esculenta was extracted and optimized by using RSM with four parameters, namely pH (50 mM phosphate buffer, pH 6, 50 mM Tris buffer, pH 8, 50 mM glycine–NaOH buffer, pH 10), CaCl2 (1, 5.5 and 10 mM), Triton X-100 [1% (v/v), 3% (v/v), 5% (v/v)] and 2-mercaptoethanol (1, 5.5 and 10 mM). Then the optimized crude protease was applied as a meat tenderizer by using three different methods, which were texture analyser, electrophoresis and scanning electron microscopy (SEM).
Materials and methods
Materials
The samples of Manihot esculenta root were obtained from a local Manihot esculenta orchard in Terengganu, Malaysia. The outer skin was peeled to obtain the flesh of root and cut into small pieces and then subjected to protease extraction.
Extraction of crude enzyme
Four independent variables were used in designing the experiment before protease extraction, as shown in Supplementary Table S1. Twenty g of Manihot esculenta was extracted in 100 mL of chilled appropriate buffer solution (50 mM phosphate buffer, pH 6, 50 mM Tris buffer, pH 8, 50 mM glycine–NaOH buffer, pH 10) that contained Triton X-100, CaCl2 and 2-mercaptoethanol using a Waring blender for 5 min. The crude enzyme was filtered using Whatman filter paper No. 3. Then, the filtrate was centrifuged for 30 min at 9910 rpm (Hermle Z 323 K, Hermle Labortechnik GmbH, Wehingen, Germany) at 4 °C. The supernatant was collected and stored at 4 °C for further analysis (Ahmad et al. 2019).
Protease activity
The protease activity was determined using the Folin–Ciocalteu method with some modification (Darwesh et al. 2019). A mixture of 0.9 mL of 50 mM phosphate buffer (pH 7.5), 5 mL of 0.65% (w/v) casein solution and 0.1 mL of crude enzyme was incubated at 37 °C for 10 min. Five mL of 110 mM trichloroacetic acid solution was added to terminate the reaction. Then, non-hydrolysed casein was filtered using a 0.25 µm polypropylene syringe filter. The mixture containing 2 mL of filtrate, 5 mL of 500 mM Na2CO3 and 1 mL of 0.5 mM Folin–Ciocalteu Reagent was incubated at 37 °C for 30 min. The absorbance of the standard and samples was determined at 660 nm by using Lambda 35 UV–Vis Spectrometer (Perkin Elmer, USA). The protease activity was determined based on the release of µmole tyrosine/min using casein as substrate and was calculated using Eq. 1 below:
| 1 |
where IU = µmole tyrosine/min; DF = dilution factor; Wt = weight of sample, g; 0.1 = volume of enzyme (mL) used.
Experimental design and statistical analysis
Four independent variables (pH, 2-mercaptoethanol, Triton X-100, CaCl2) were optimized using RSM (Design Expert, Stat-Ease, Version 11.1.0.1, Minneapolis, MN). A Faced-Centered Central Composite Design (FCCCD) was used in the optimization process with four variables at three levels with 30 runs.
Experimental data were fitted to the second-order regression equation:
| 2 |
where Y is the predicted response; b0 is the intercept; b1, b2, b3 and b4 are linear coefficients; b11, b22, b33, and b44 are squared coefficients; b12, b13, b14, b23, b24 and b34 are interaction coefficients; X1 = pH, X2 = CaCl2, X3 = Triton X-100 and X4 = 2-mecaptoethanol.
Validation of the model
The protease extraction conditions were numerically optimised for the maximum protease activity based on the regression analysis and the 3D surface plots of the independent variables. Tukey’s test was used to compare between the means by using JMP Pro 13 (SAS Institute Inc., USA). The experimental value of the response was measured under the optimal recommended conditions of the extraction and was compared with the predicted value by means of replicate determination (n = 3) in order to determine the validity of the model. Results were considered statistically significant at p ≤ 0.05.
Protein assay
The protein content of crude extract was determined using Bradford method (Gallagher and Wiley 2008). Bovine Serum Albumin (BSA) was used as a reference standard. A series of standard protein and 0.1 mL of crude enzyme was added into different test tubes. Two mL of Bradford reagent was added to all the test tubes and mixed. The tubes were incubated for 30 min at room temperature. The absorbance of blank, standards and sample were determined at 595 nm using Lambda 35, UV–Vis Spectrometer (Perkin Elmer, USA).
Specific activity
The specific activity of enzyme is the number of enzyme units per milligram of protein by the following equation (Sun et al. 2019):
| 3 |
Effectiveness of enzyme on meat tenderization
Texture analyzer
The texture was analysed using a CT3 texture analyser (Brookfield, Ametek Inc., USA), equipped with a general probe kit, which was a 60 mm wide knife edge and also analysed by methods with minor modification (Zhu et al. 2018). Ten rectangular (2 cm × 2 cm) shaped samples of raw beef were prepared. One sample was used as a control and other samples were treated with an enzyme with different volumes of the enzyme (0.3, 0.6, 0.9 mL). Each volume was repeated three times. The blade was pressed down at a constant speed of 2 mm s−1 through the samples. The shear force applied on each meat sample was recorded (Rawdkuen et al. 2013).
Electrophoresis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using the following method (Gallagher and Wiley 2008; Chanalia et al. 2018) with electrophoresis apparatus (Bio-Rad Laboratories Inc, CA, USA). Minced raw beef (0.8 g) was added to each test tube. One test tube was used as a control and 0.8 mL of the enzyme was added to another test tube. The test tube containing enzyme was incubated for 20 min. The test tube was then placed in a boiling water bath for 2 min to inactive the enzyme. Three mL of 6 M urea containing 2% (w/v) SDS was added to each test tube, including the control. The test tubes were mixed thoroughly and allowed to stand for 10 min. The sample and control were centrifuged at 5300 rpm for 10 min. One mL of supernatant for each test tube was thoroughly mixed with 1 mL of sample buffer. About 10 µL of sample was used for loading the gel. Electrophoresis was performed at constant voltage mode of 110 V until the tracking dye reached the lower end of the gel. The gel was removed and stained with Coomassie blue for 4–5 h. The gel was then destained by shaking it gently in destaining solution [10% (v/v) acetic acid: 25% (v/v) methanol: 65% (v/v) water) (Copeland 1994).
Scanning electron microscopy
The microstructure of the samples was determined using a scanning electron microscope (Zeiss Evo50, Germany) with some minor modification (Hu et al. 2018). Muscle specimens were prepared to the required size and washed with 0.1 M phosphate buffer. The sample was incubated with 1% (w/v) osmium tetroxide for 2 h at room temperature. The samples were then washed twice with distilled water before being dehydrated in ethanol with a serial concentration of 50%, 75%, 95% and 100% (v/v). The samples were incubated again in 100% (v/v) acetone after dehydration. They were then critical point dried (CPD) by using CO2 as a transition fluid. The dried samples were mounted on a bronze stub and sputter-coated with gold. The specimens were observed with an SEM with a magnification of 154× at an acceleration voltage of 10 kV.
Results and discussions
Optimization of variables by central composite rotatable design (CCRD)
According to statistical analysis, the optimum results could be obtained from the optimized model. All the 30 designated experiments in Supplementary Table S2 were conducted to obtain the protease activity for each run. The protease activity and test variables from Manihot esculenta were expressed by the following regression equation:
| 4 |
where Y is predicted response; X1, X2, X3, X4 are linear terms of pH, CaCl2, Triton X- 100 and 2-mercaptoethanol respectively; X21, X22, X23, X24 were quadratic terms of pH, CaCl2, Triton X-100 and 2-mercaptoethanol respectively; X1X2, X1X3, X1X4, X2X3, X2X4, X3X4 represent the interaction terms of the variables tested. Based on the regression equation above, the predicted response for each run can be determined.
Table 1 shows the ANOVA results for the statistical significance of the response surface quadratic polynomial model. The result shows that the model was significant due to the high F value (22.35) and low p value (< 0.0001). The values of p < 0.05 indicate the high significance of the corresponding coefficients (Wang et al. 2020). The lack of fit was obtained to measure the inadequacy of the model (Ameer et al. 2017). Based on the ANOVA results, the lack of fit was not significant with low F value (0.5559) and high p value (0.7992). Thus, it indicated that the model is adequate to describe the experimental data.
Table 1.
Analysis of variance for the evaluation of the quadratic model
| Source of variation | Sum of square | Degree of freedom | Mean square | F value | p value |
|---|---|---|---|---|---|
| Model | 134.50 | 14 | 9.61 | 22.35 | < 0.0001 |
| Residual | 6.45 | 15 | 0.4299 | ||
| Lack of fit | 3.39 | 10 | 0.3395 | 0.5559 | 0.7992 |
| Pure error | 3.05 | 5 | 0.6107 | ||
| Total | 140.95 | 29 |
R2 = 0.9543, Adjusted R2 = 0.9116, Predicted R2 = 0.8313, Adeq precision = 13.8786, C.V.% = 10.75, PRESS = 23.78
The coefficient of determination (R2) gives a measure of how much variability in the observed response can be explained by the experimental parameters and their extraction. The value of R2 was 0.9543, which is a relatively high value because 95.43% of the experimental variability can be explained by the model while only 4.57% was not explained by the model. The adjusted R2 value corrects the R2 value for the sample size and for the number of terms in the model. Based on the result, the adjusted R2 was 0.9116, which is quite high and indicates a high correlation between the observed and predicted values. The difference between the predicted R2 value (0.8313) and adjusted R2 value (0.9116) was less than 0.2. Therefore, it shows that the predicted R2 is in reasonable agreement with the adjusted R2, at the same time indicating a highly significant model. When expressed in percentages, the value of predicted R2 was 83.13%, which indicates that the variables in the response can be clarified precisely by this model (Ahmad et al. 2019).
The adeq precision is used to measure the signal to noise ratio and determines whether the model can be used to navigate the design space. If the value is greater than 4, it is desirable and shows an adequate signal. In this study, the value of adeq precision (13.8786) was greater than 4, which indicated an adequate signal and this model can be used to navigate the design space. The coefficient of variation (C.V.) and prediction residual errors sum square (PRESS) value were 10.75% and 23.78, respectively. The value of C.V. was considered a low value and indicated the model was precise and reliable (Joardar et al. 2018). The smaller the value of PRESS, the better the model can fit each point in the design.
By using the normal probability plot graph in Supplementary Fig. S1(a), the significance of the variables on protease activity was determined by comparing the magnitude of the effects with standard errors. As can be seen on the probability plot graph, the experimental data were aligned with a straight line. This indicated that the data fitted well. Hence, all the independent variables (pH, CaCl2, Triton X-100, 2-mercaptoethanol) had significant effect on protease activity (p < 0.05). Both Supplementary Fig. S1(a) and S1(b) are important to check the fitted model and to determine whether the model is adequate or not (Vasiee et al. 2016; Prabhu and Karthikeyan 2018). Figure S1(b) shows the plot of residuals versus the predicted response. As can be seen, the residuals scatter randomly within the red line on the display, suggesting that the variance of the original observation is constant for all values of response. Both of the plots are acceptable; it can conclude that the model is adequate to explain the protease activity.
Based on Supplementary Fig. S2(a), protease activity was increased significantly approaching optimum pH (pH 9) and decreased when pH was above 9. A buffer was required to maintain the stability of the enzyme and reduced the denaturation of protease activity by controlling the pH value. Normally, the natural pH inside plant cells ranges between 6 and 7 (Ahmad et al. 2012). However, in this study, the optimum pH for protease extraction from Manihot esculenta was 9. A similar result was found in a previous study, the optimum pH for serine protease from Euphorbia neriifolia Lin., which belongs to the same family as Manihot esculenta, was pH 9.5 (Yadav et al. 2012).
Supplementary Fig. S2(b) showed that the protease activity was increased slowly approaching the optimum level of CaCl2 (3.24 mM) and decreased when the concentration of CaCl2 was further increased. Metal ion can enhance the protease activity due to the ionic binding formed between metal ion and protein, subsequently made a protease become compact and stable (Krishnan and Murugan 2015; Zhang et al. 2019). The best metal ion was Ca2+ because it had the largest ionic radius (0.099 nm) among divalent metal cations (Mn2+, Zn2+, Mg2+ and Cu2+) where the activation of protease activity increased as the ionic radius was increased (Ahmad et al. 2012).
As can be seen in Supplementary Fig. S2(c), the protease activity increases sharply when approaching the optimum level of Triton X-100 (4.19%) and decreased slightly when it reached the highest level of Triton X-100 (5%). Triton X-100 is an ionic detergent use in protease extraction to disrupt the membranes and essential for solubilizing membrane proteins. Triton X-100 is suitable to use in extraction because it is less harmful compared with other detergents such as SDS (Krishnan and Murugan 2015). The protease activity was increased rapidly as the concentration of 2-mercaptoethanol approaching the optimum level (6.32 mM) and decreased slowly when it over the optimum level [Supplementary Fig. S2(d)]. The 2-mercaptoethanol was used as a reducing agent and antioxidant to protect against denaturation and enhanced the protease activity (Matkawala et al. 2019). The 2-mercaptoethanol in the extraction buffer are essential to maintain the reduced state of sulfhydryl groups in the enzyme and to reduce oxidation of other components such as phenolic compounds.
The p value is used as a tool to decide the significance of each of the coefficients to understand the interaction between the variables (Dinarvand et al. 2017). Based on Table 2, all the independent variables, pH (X1), CaCl2 (X2), Triton X-100 (X3) and 2-mercaptoethanol (X4) were significant (p < 0.05). Based on the t-value, the independent variables for protease activity were arranged in the following descending order, Triton X-100 > pH > 2-mercaptoethanol > CaCl2. Triton X-100 shows the strongest effect on protease activity compared with other variables while CaCl2 shows the weakest effect on protease activity.
Table 2.
Coefficient estimates by the quadratic model
| Terms | Coefficient estimate | Sum of squares | DF | Mean square | F value | p value | t-value |
|---|---|---|---|---|---|---|---|
| X1 | 0.6294 | 7.13 | 1 | 7.13 | 16.59 | 0.0010 | 4.073 |
| X2 | − 0.3517 | 2.23 | 1 | 2.23 | 5.18 | 0.0380 | − 2.276 |
| X3 | 1.08 | 20.82 | 1 | 20.82 | 48.44 | < 0.0001 | 6.960 |
| X4 | 0.6206 | 6.93 | 1 | 6.93 | 16.12 | 0.0011 | 4.015 |
| X21 | − 1.11 | 3.18 | 1 | 3.18 | 7.39 | 0.0158 | − 2.719 |
| X22 | − 0.6176 | 0.9884 | 1 | 0.9884 | 2.30 | 0.1502 | − 1.516 |
| X23 | − 1.10 | 3.15 | 1 | 3.15 | 7.33 | 0.0162 | − 2.707 |
| X24 | − 1.03 | 2.74 | 1 | 2.74 | 6.36 | 0.0234 | − 2.523 |
| X1X2 | − 0.5125 | 4.20 | 1 | 4.20 | 9.78 | 0.0069 | − 3.127 |
| X1X3 | 0.5638 | 5.09 | 1 | 5.09 | 11.83 | 0.0037 | 3.439 |
| X1X4 | − 0.3575 | 2.04 | 1 | 2.04 | 4.76 | 0.0455 | − 2.181 |
| X2X3 | 0.0400 | 0.0256 | 1 | 0.0256 | 0.0596 | 0.8105 | 0.244 |
| X2X4 | − 0.1287 | 0.2652 | 1 | 0.2652 | 0.6170 | 0.4444 | − 0.785 |
| X3X4 | − 0.2100 | 0.7056 | 1 | 0.7056 | 1.64 | 0.2196 | − 1.281 |
There were six interactions between the variables that were significant (p < 0.05), namely X21, X23, X24, X1X2, X1X3 and X1X4 and the other interactions (X22, X2X3, X2X4, X3X4) were not significant. It shows that all the significant interactions give a stronger effect on protease activity compared with not significant variables, which give a weak effect on protease activity. This explanation can be supported based on the t-values in Table 2. The t-value of significant interactions was higher than with not significant variables. The higher the t-value, the stronger the effect of the variables on protease activity (Ahmad et al. 2019). Based on the t-values in Table 2, pH.Triton X-100 was the strongest interaction effect on the protease activity followed by pH.CaCl2 > pH.2-mercaptoethanol > Triton X-100.2-mercaptoethanol > CaCl2.2-mercaptoethanol > CaCl2.Triton X-100.
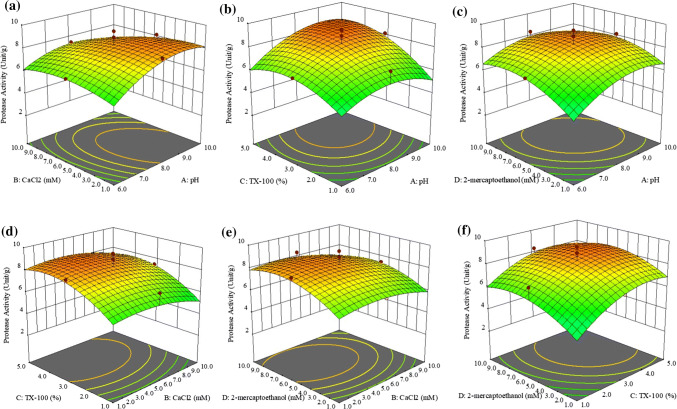
Based on Fig. 1a, the optimum protease activity was recorded at pH 9 and 3.24 mM of CaCl2. It shows that the protease activity of Manihot esculenta increased rapidly when approaching optimum pH while for CaCl2, protease activity increases slowly when it approaches 3.24 mM. Therefore, pH was more effective in increasing the protease activity because it had a higher (4.073) t-value than CaCl2 (− 2.276) and prevented the protease from denaturing. A similar result was obtained in the previous study that been conducted by Ahmad et al. (2012, 2019) where pH has a stronger effect on protease activity than CaCl2.
Fig. 1.
The effects of a pH.CaCl2, b pH.Triton X-100, c pH.2-mercaptoethanol, d CaCl2. Triton X-100, e CaCl2.2-mercaptoethanol, f Triton X-100.2-mercaptoethanol time on protease activity (Unit/g) in 3D Plot
The protease activity was more sensitive to changes of Triton X-100 than pH (Fig. 1b). As can be seen, the protease activity increased only slightly approaching optimum pH, while for Triton X-100, it increased rapidly. It shows that Triton X-100 has a stronger effect on protease activity than pH. This result was supported by the higher t-value for Triton X-100 (6.960) than pH, which has a lower t-value (4.073). The enzyme was highly stable in the presence of a non-ionic surfactant such as Triton X-100 (Amid et al. 2014). It means that Triton X-100 has a stronger effect in increasing the protease activity. This result can be supported by a previous study conducted by Ahmad et al. (2012) where Triton X-100 gives greater influence on protease activity compared to pH.
Changes in both pH and 2-mercaptoethanol have a strong impact on protease activity. Based on Fig. 1c, the increases in protease activity approaching optimum pH and 2-mercaptoethanol were quite similar. However, the protease activity gradually decreased when pH and 2-mercaptoethanol exceeded optimal conditions. When comparing the t-values for both variables, the t-value of pH (4.073) was only slightly higher than for 2-mercaptoethanol (4.015). Based on the previous study, the reducing agent (2-mercaptoethanol) has a stronger effect on protease activity than did pH (Ahmad et al. 2019).
CaCl2 had a lower influence than Triton X-100 as protease activity only increased slightly on approaching the optimum value of CaCl2 compared with Triton X-100, which was increased significantly (Fig. 1d). This can be justified based on the t-values of both variables, where CaCl2 (− 2.276) had a lower t-value than Triton X-100 (6.960). According to the Bhange et al. (2016), surfactants like Triton X-100 show twofold increases in protease activity. Thus, it showed that Triton X-100 has a stronger impact on protease activity. Triton X-100 also has a greater influence on protease activity than CaCl2 (Ahmad et al. 2012).
Optimum protease activity of Manihot esculenta was observed at 3.24 mM of CaCl2 and 6.32 mM of 2-mercaptoethanol (Fig. 1e). Both variables affected the protease activity significantly. According to Ahmad et al. (2019). 2-mercaptoethanol was more important than CaCl2, as indicated by the t-value in Table 2. The t-values of 2-mercaptoethanol and CaCl2 were 4.015 and − 2.276, respectively. Besides, 2-mercaptoethanol had a stronger effect when it acted individually compared with working together with CaCl2.
Figure 1f shows that protease activity increased minimally when approaching the optimum value of 2-mercaptoethanol (6.32 mM) compared with Triton X-100. Thus, 2-mercaptoethanol had a weaker effect on protease activity than Triton X-100 as justified by the t-values of both variables. The 2-mercaptoethanol (4.015) had a lower t-value than Triton X-100 (6.960). In addition, Triton X-100 has a greater influence on protease activity, as reported in the previous study the t-value of Triton X-100 was the highest compared with other variables (Ahmad et al. 2019).
Optimization and validation of the model
Supplementary Table S2 also shows the values of predicted and actual (experimental) protease activity of each run. The correlation shows a high degree of similarity which indicates the accuracy and applicability of RSM for the optimum protease extraction Manihot esculant.a The optimum protease activity obtained from the RSM model was 6.3017 units/mL, calculated based on Eq. 4, of which x1 (pH) is 9.04 Unit/g, x2 (CaCl2 concentration) is 3.24 mM, x3 (Triton X-100) is 4.19% (v/v), and x4 (2-mercaptoethanol) is 6.32 mM. The verification test was conducted at the optimum level of variables and the average protease activity obtained was 8.99 Unit/g. A comparison of the calculated (9.04 Unit/g) and experimental (8.99 Unit/g) protease activity values showed satisfactory agreement within 95% confidence interval. Furthermore, this data was supported by the Tukey test, which is insignificant difference at 95%, p = 0.05. It is desirable that optimum extraction conditions should yield the highest protease activity. The protein concentration of the crude extract obtained under the optimum conditions (experimental) and predicted 1.251 mg/mL and 1.206 mg/mL The predicted resulted is based on Fibriana and Upaichit (2015) findings, where the protein content of Euphorbia maculata was 1.206 mg/mL and its belongs to similar family of Manihot esculanta. Tuckey test shows that there are not significant difference (96%) between the predicted and experimental results. While, the specific activity of the crude protease obtained was 0.1201 Unit/mg. Specific activity is important to measure the purity of enzyme (Foustoukos 2014).
Meat tenderization
Texture analyser
Based on Fig. 2, the shear forces for control is significantly different (p < 0.05) than the treated samples (0.3, 0.6 and 0.9 mL) at 1.95 ± 0.057, 1.66 ± 0.012, 0.97 ± 0.049 and 0.46 ± 0.011 N, respectively. The values were lower in all treated samples than for the control without crude protease addition. The value of shear force was significantly decreased (p < 0.05) as the amount of protease used in the meat was increased (Sun et al. 2018). By adding 0.3, 0.6 and 0.9 mL of crude protease, the shear force was significantly decreased (p < 0.05) by 14%, more than 50% and 76%, respectively when compared with the control. Similar results were also obtained when using papain protease in meat tenderizer (Zhang et al. 2017). By knowing the value of shear force for each sample and control, the level of firmness for each sample can be determined (Ketnawa and Rawdkuen 2011). Based on the shear force, the treated samples (0.9 mL) had the lowest firmness compared with other samples because they had lower shear force. Therefore, it does not require a lot of force to break down the myofibrillar protein compared with other samples. The action of the proteolytic enzyme is the main factor contributing to the reduction of meat firmness. The meat firmness is related to the acceptable meat quality for the consumer.
Fig. 2.
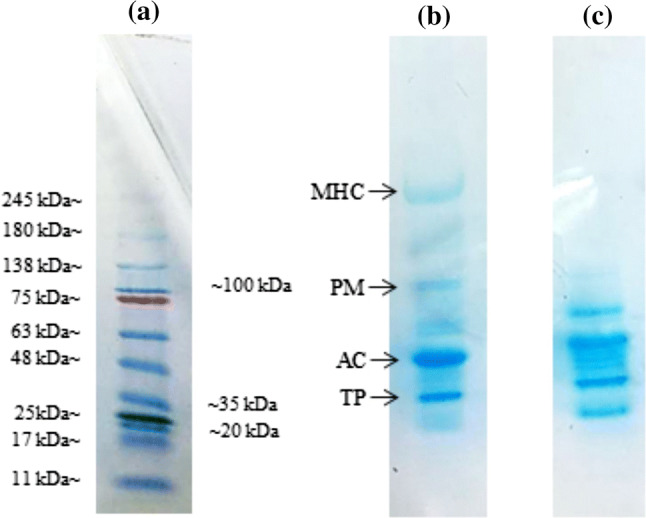
The electrophoretic analysis of a Standard ladder protein, b control, and c after protease treatment
Electrophoresis (SDS-PAGE)
The SDS-PAGE pattern is shown in Fig. 3a. Based on the SDS-PAGE pattern, there are four main muscle proteins that can be observed, namely, myosin heavy chain, paramyosin, actin and troponin. The calibration curve graph between log MV and migration distance of the band was used to calculate the molecular weight of bands that appeared on the gel (Supplementary Fig. S3). It can be seen that the most intense band was actin (40 kDa) and the second most intense was troponin (25 kDa). The molecular weights of actin and troponin are 43 kDa (Dutta et al. 2019; Yang et al. 2013) and 19–30 kDa (Lana and Zolla 2016), respectively. The highest molecular weight was 237 kDa and it was assumed that the band belongs to the myosin heavy chain based on the previous study done by Ghowsi (2012) and Zhang et al. (2019). It was reported that the molecular weight for myosin heavy chain was 220 kDa. The second highest molecular weight belongs to paramyosin (106 kDa) (Onopiuk et al. 2017). The analysis of the main myofibrillar proteins in meat showed the highest level of myosin heavy chain (MHC) followed by actin and troponin (Vigoreaux 2006). It shows quite a similar pattern to the result obtained here.
Fig. 3.
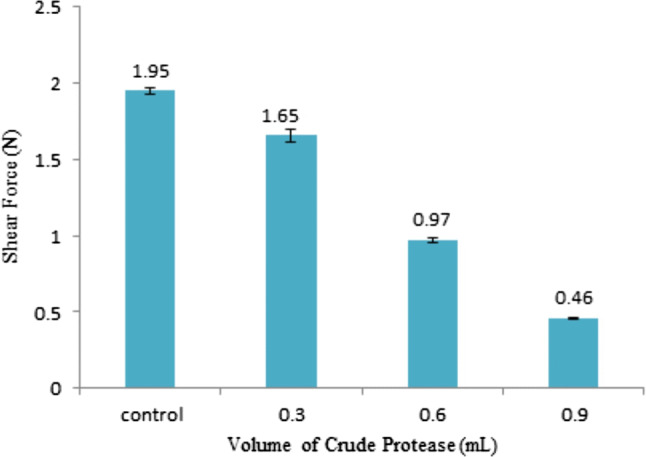
The firmness (Shear Force, N) of muscle samples treated with different volumes of Ambarella crude extract (mL)
When comparing the control and enzyme-treated samples, there was increased proteolysis of muscle protein by observing the reduction of number and intensity of bands (Ketnawa and Rawdkuen 2011; Ahmad et al. 2019). The intensity of the MHC band was clearly decreased as the band disappeared after being treated with the enzyme. It shows that the MHC was markedly degraded into lower molecular weight products as shown at the bottom of the gels. The paramyosin also was degraded as the intensity of the paramyosin band was reduced. As reported by Wada et al. (2002), the plant proteinases affected the structure of the myofibrillar protein and caused the degradation. Degradation of myofibrillar protein occurs when the protein interacted with the catalytic triad of serine, histidine and aspartic acid that are located at the active site of serine protease as expected to be found in Manihot esculenta (Weston et al. 2002). The tenderness was determined based on the degradation of muscle protein. Because the enzyme affected MHC by fragmenting them into smaller molecular weight protein bands, it shows that the enzyme from Manihot esculenta potentially has high proteolytic activity, making the meat more tender. The observation of fragments of MHC is important because it gives a significant correlation to meat tenderness (Zhang et al. 2019).
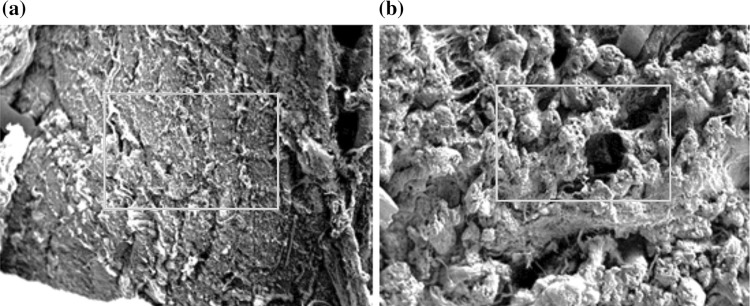
Scanning electron microscopy (SEM)
Connective tissue plays an important role in determining the eating quality of meat. Figure 4 shows the microstructure of control and sample treated with crude protease. Magnification was 154× for control and sample, at an acceleration voltage of 10 kV. Based on Fig. 4, the control had compacted structure and was closely bound to each other. The broken muscle fibres can be observed in the enzyme-treated sample. Besides, there was a loss of muscle fibres’ interaction and less attached fibres. As can be seen, there was interfibrillar space and big gaps were revealed between the muscle fibres. This is because of the degradation of collagen and sarcolemma surrounding the muscle fibres. Collagen is abundant in connective tissues, so, the degradation of collagen is a big contributing factor in meat tenderness (Mikołajczak et al. 2019). The degradation of muscle fibers can be proven by observing the Fig. 4. It shows significant differences of structure between the control and sample as the fibres were broken and the cell membranes were degraded. Another factor of meat tenderization is disruption of connective tissue structure (Purslow 2018). Based on the previous result in terms of texture properties and SDS-PAGE, the microstructure of enzyme-treated sample showed a strong correlation. In a previous study, the effect of papain, pineapple and ginger juice on the microstructure of goose meat was examined using SEM and it was found that the muscle was ruptured in the sample treated with proteases due to the action of exogenous protease (Gao et al. 2011).
Fig. 4.
SEM micrographs of beef in a control and b treatment with Manihot esculenta protease
Conclusion
The results obtained in this experiment indicate that the crude protease from Manihot esculenta can be used as a meat tenderizer. Based on the statistical analysis of RSM, the optimum variables of extraction that obtained for pH, CaCl2, Triton X-100 and 2-mercaptoethanol were pH 9, 3.24 mM, 4.19% and 6.32 mM respectively. SDS-PAGE, SEM and texture analysis supports the effectiveness of the crude protease acts a meat tenderizer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The project was funded by the Fundamental Research Grant Scheme (Grant No. FRGS16-058-0557).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmad MN, Liew SL, Yarmo MA, Said M. Optimization of protease extraction from Horse Mango (Mangifera foetida Lour) kernels by a response surface methodology. Biosci Biotechnol Biochem. 2012;76(8):1438–1444. doi: 10.1271/bbb.120073. [DOI] [PubMed] [Google Scholar]
- Ahmad MN, Mat Noh NA, Abdullah EN, Yarmo MA, Mat Piah MB, Ku Bulat KH. Optimization of a protease extraction using a statistical approach for the production of an alternative meat tenderizer from Spondias cytherea roots. J Food Process Preserv. 2019;43(11):1–14. doi: 10.1111/jfpp.14192. [DOI] [Google Scholar]
- Ameer K, Seong-Woo B, Jo Y, Hyun-Gyu L, Ameer A, Joong-Ho K. Optimization of microwave-assisted extraction of total extract stevioside and rebaudioside-A from Stevia rebaudiana (Bertoni) leaves using response surface methodology (RSM) and artificial neural network (ANN) modelling. Food Chem. 2017;229:198–207. doi: 10.1016/j.foodchem.2017.01.121. [DOI] [PubMed] [Google Scholar]
- Amid M, Abd Manap MY, Zohdi NK. Purification and characterization of alkaline-thermostable protease enzyme from Pitaya (Hylocereus polyrhizus) waste: a potential low cost of the enzyme. Biomed Res Int. 2014;1:1–8. doi: 10.1155/2014/259238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad MS, Kwan JH, Imran M, Sohaib M, Aslam A, Nawaz I, Amjad Z, Khan U, Javed M. Plant and bacterial proteases: a key towards improving meat tenderization, a Mini review. Cogent Food Agric. 2016;2(1):1–10. doi: 10.1080/23311932.2016.1261780. [DOI] [Google Scholar]
- Bhange K, Chaturvedi V, Bhatt R. Simultaneous production of detergent stable keratinolytic protease, amylase, and biosurfactant by Bacillus subtilis PF1 using agro industrial waste. Biotechnol Rep. 2016;10:94–104. doi: 10.1016/j.btre.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanalia P, Gandhi D, Attri P, Dhanda S. Extraction, purification and characterization of low molecular weight Proline iminopeptidase from probiotic L. plantarum for meat tenderization. Int J Biol Macromol. 2018;109:651–663. doi: 10.1016/j.ijbiomac.2017.12.092. [DOI] [PubMed] [Google Scholar]
- Chu B, Shi Y, Li Z, Tian H, Li W. Optimization of gentiside extraction from Gentiana rigescens Franch. ex Hemsl. by response surface methodology. J Anal Methods Chem. 2015;2015(1):1–8. doi: 10.1155/2015/819067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA. Methods of protein analysis: a practical guide to laboratory protocols. USA: Springer; 1994. [Google Scholar]
- Darwesh OM, El-Hawary AS, El Kelany US, El-Sherbiny GM. Nematicidal activity of thermostable alkaline protease produced by Saccaromonospora viridis strain Hw G550. Biotechnol Rep (Amst) 2019;24:e00386. doi: 10.1016/j.btre.2019.e00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarvand M, Rezaee M, Foroughi M. Optimizing culture conditions for production of intra and extracellular inulinase and invertase from Aspergillus niger ATCC 20611 by response surface methodlogy (RSM) Braz J Microbiol. 2017;48(3):427–441. doi: 10.1016/j.bjm.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta P, Jijumon AS, Mazumder M, Dileep D, Mukhopadhyay AK, Gourinath S, Maiti S. Presence of actin binding motif in VgrG-1 toxin of Vibrio cholera reveals the molecular mechanism of actin cross-linking. Int J Biol Macromol. 2019;13:775–785. doi: 10.1016/j.ijbiomac.2019.04.026. [DOI] [PubMed] [Google Scholar]
- Fibriana F, Upaichit A. Proteases from latex of Euphorbia spp. and its application on milk clot formation. J Biol Biol Educ. 2015;7(2):92–99. doi: 10.15294/biosaintifika.v7i2.3951. [DOI] [Google Scholar]
- Foustoukos D, et al. Specific activity. In: Amils R, et al., editors. Encyclopedia of astrobiology. Berlin: Springer; 2014. [Google Scholar]
- Gallagher SR, Wiley EA. Current protocols essential laboratory techniques. New York: Wiley; 2008. [Google Scholar]
- Gao H, Zhou G, Zeng J, Ma H, Pan R, Yu X. Tenderization of goose meat by papain, pineapple juice and ginger juice treatment. Adv Mater Res. 2011;236–238(1):2349–2352. doi: 10.4028/www.scientific.net/AMR.236-238.2349. [DOI] [Google Scholar]
- Ghowsi K. Electrophoresis. In: Pighin DG, editor. Electrophoresis as a useful tools in studying the quality of meat products. 1. Rijeka: Intech; 2012. pp. 117–136. [Google Scholar]
- Hu J, Ge S, Huang C, Cheung PCK, Lin L, Zhang Y, Zheng B, Lin S, Huang X. Tenderization effect of whelk meat using ultrasonic treatment. Food Sci Nutr. 2018;6(7):1848–1857. doi: 10.1002/fsn3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joardar N, Mukherjee S, Babu SPS. Thioredoxin reductase from the bovine filarial parasite Setaria cervi: studies on its localization and optimization of the extraction. Int J Biol Macromol. 2018;107:2375–2384. doi: 10.1016/j.ijbiomac.2017.10.114. [DOI] [PubMed] [Google Scholar]
- Jones AMP, Saxena PK. Inhibition of phenylpropanoid biosynthesis in Artemisia annua L.: a novel approach to reduce oxidative browning in plant tissue culture. PLoS ONE. 2013;8:1–13. doi: 10.1371/journal.pone.0076802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketnawa S, Rawdkuen S. Application of bromelain extract for muscle foods tenderization. Food Nutr Sci. 2011;2(5):393–401. doi: 10.4236/fns.2011.25055. [DOI] [Google Scholar]
- Krishnan VGM, Murugan K. Purification, characterization and kinetics of protease inhibitor from fruits of Solanum aculeatissimum Jacq. Food Sci Hum Wellness. 2015;4(3):97–107. doi: 10.1016/j.fshw.2015.06.003. [DOI] [Google Scholar]
- Kumar SNA, Ritesh SK, Sharmila G, Muthukumaran C. Extraction optimization and characterization of water soluble red purple pigment from floral bracts of Bougainvillea Glabra. Arab J Chem. 2013;10(2):S2145–S2150. doi: 10.1016/j.arabjc.2013.07.047. [DOI] [Google Scholar]
- Lana A, Zolla L. Proteolysis in meat tenderization from the point of view of each single protein: a proteomic perspective. J Proteomics. 2016;16(147):85–97. doi: 10.1016/j.jprot.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Mahajan RT, Khan JV, Khan TA. Evaluation of proteolytic activity of some Euporbian garden plants. Int J Life Sci Sci Res. 2016;2(4):355–360. doi: 10.21276/ijlssr.2016.2.4.7. [DOI] [Google Scholar]
- Matkawala F, Nighojkar S, Kumar A, Nighojkar A. A novel thio-dependent serine protease from Neocosmospora sp. N1. Heliyon. 2019;5:e022246. doi: 10.1016/j.heliyon.2019.e02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikołajczak B, Iwańska E, Spychaja A, Danyluka B, Montowska M, Grześ B, Banach JK, Żywica R, Pospiec E. An analysis of the influence of various tenderising treatments on the tenderness of meat from Polish Holstein-Friesian bulls and the course of changes in collagen. Meat Sci. 2019;158:107906. doi: 10.1016/j.meatsci.2019.107906. [DOI] [PubMed] [Google Scholar]
- Onopiuk A, Pottorak A, Sun DW, Wierzbicka A. Effects of selected myofibrillar protein activities on beef tenderization process based on electrophoretic analysis. J Food Process Eng. 2017;41(1):1–8. doi: 10.1111/jfpe.12596. [DOI] [Google Scholar]
- Prabhu MV, Karthikeyan R. Comparative studies on modelling and optimization of hydrodynamic parameters on inverse fluidized bed reactor using ANN-GA and RSM. Alex Eng J. 2018;57:3019–3032. doi: 10.1016/j.aej.2018.05.002. [DOI] [Google Scholar]
- Purslow PP. Contribution of collagen and connective tissue to cooked meat toughness; some paradigms reviewed. Meat Sci. 2018;144:127–134. doi: 10.1016/j.meatsci.2018.03.026. [DOI] [PubMed] [Google Scholar]
- Rawdkuen S, Jaimakreu M, Benjakul S. Physicochemicl properties and tenderness of meat samples using proteolytic extract from Calotropis procera latex. Food Chem. 2013;136(215):909–916. doi: 10.1016/j.foodchem.2012.08.077. [DOI] [PubMed] [Google Scholar]
- Sobottka AM, Tonial F, Sytwala S, Melzig M. Proteinase activity in latex of three plants of the family Euphorbiaceae. Braz J Pharm Sci. 2014;50(3):559–565. doi: 10.1590/S1984-82502014000300015. [DOI] [Google Scholar]
- Sun Q, Fusheng F, Geng F, Luo Y, Gong S, Jiang Z. A novel aspartic protease from Rhizomucor miehei expressed in Pichia pastoris and its application on meat tenderization and preparation of turtle peptides. Food Chem. 2018;245:570–577. doi: 10.1016/j.foodchem.2017.10.113. [DOI] [PubMed] [Google Scholar]
- Sun F, Hu Y, Chen Q, Kong B, Liu Q. Purification and biochemical characteristics of the extracellular protease from Pediococcus pentosaceus isolated from Harbin dry sausages. Meat Sci. 2019;156:156–165. doi: 10.1016/j.meatsci.2019.05.030. [DOI] [PubMed] [Google Scholar]
- Vasiee A, Behbahani BA, Yazdi FT, Moradi S. Optimization of the production conditions of the lipase produced by Bacillus cereus from rice flour through Plackett–Burman Design (PBD) and response surface methodology (RSM) Microb Pathog. 2016;101:36–43. doi: 10.1016/j.micpath.2016.10.020. [DOI] [PubMed] [Google Scholar]
- Vigoreaux JO. Nature’s versatile engine: insect flight muscle inside and out. New York: Springer; 2006. [Google Scholar]
- Wada M, Suzuki T, Yaguti Y, Hasegawa T. The effects of pressure treatment with Kiwi fruit protease on adult cattle semitendinosus muscle. Food Chem. 2002;78(2):167–171. doi: 10.1016/S0308-8146(01)00395-8. [DOI] [Google Scholar]
- Wang W, Chena F, Zheng F. Russell B (2020) Optimization of synthesis of carbohydrates and 1-phenyl-3-methyl-5pyrazolone (PMP) by response surface methodology (RSM) for improved carbohydrate detection. Food Chem. 2020;309:125686. doi: 10.1016/j.foodchem.2019.125686. [DOI] [PubMed] [Google Scholar]
- Weston AR, Rogers Pas RW, Althen TG. The role of collagen in meat tenderness. Prof Anim Sci. 2002;18(2):107–111. doi: 10.15232/S1080-7446(15)31497-2. [DOI] [Google Scholar]
- Yadav RP, Patel AK, Jagannadham MV. Neriifolin S, a dimeric serine protease from Euphorbia neriifolia Linn.: purification and biochemical characterisation. Food Chem. 2012;132(1):1296–1304. doi: 10.1016/j.foodchem.2011.11.107. [DOI] [PubMed] [Google Scholar]
- Yang J, Zou L, Hu Z, Chen W, Zhang J, Zhu J, Fang X, Yuan W, Hu X, Hu F, Rao X. Identification and characterization of a 43 kDa actin protein involved in the DENV-2 binding of ECV304 cells. Microb Infect. 2013;15:310–318. doi: 10.1016/j.micinf.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Zhang B, Sun Q, Hai-Jie L, Shao-Zhen L, Zheng-Qiang J. Characterication of actinidin from Chinese kiwifruit cultivars and its applications in meat tenderization and production of angiotension I-converting enzyme (ACE) inhibitory peptides. LWT-Food Sci Technol. 2017;78:1–7. doi: 10.1016/j.lwt.2016.12.012. [DOI] [Google Scholar]
- Zhang L, Wang W, Zhou F, Zheng Y, Wang X. Tenderness and histochemistry of muscle tissues from Eriocheir sinensis. Food Biosci. 2019 doi: 10.1016/j.fbio.2019.100479. [DOI] [Google Scholar]
- Zhu X, Kaur L, Staincliffe M, Boland M. Actinidin pretretatment and sous vide cooking of beef brisket: effects on meat microstructure, texture and in vitro protein digestibility. Meat Sci. 2018;145:256–265. doi: 10.1016/j.meatsci.2018.06.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.