Abstract
Hypoxia is a hallmark of the tumour microenvironment with profound effects on tumour biology, influencing cancer progression, the development of metastasis and patient outcome. Hypoxia also contributes to genomic instability and mutation frequency by inhibiting DNA repair pathways. This review summarises the diverse mechanisms by which hypoxia affects DNA repair, including suppression of homology-directed repair, mismatch repair and base excision repair. We also discuss the effects of hypoxia mimetics and agents that induce hypoxia on DNA repair, and we highlight areas of potential clinical relevance as well as future directions.
Introduction
Hypoxia, or low oxygen content, is a hallmark of the tumour microenvironment, arising when tumour cell proliferation outpaces the development of sufficient vasculature. This leads to regions within the tumour with sparse or structurally and functionally abnormal vasculature (1). Hypoxia and anoxia are found in up to 60% of locally advanced solid tumours across a wide range of tumour types, including breast, uterine, cervical, head and neck, prostate, rectal, pancreatic, lung, brain, liver, renal cell, soft tissue sarcoma, non-Hodgkin’s lymphoma and melanoma (2). Tumour hypoxia can be both chronic, as is seen when oxygen diffusion is limited by spatial distribution of the vasculature, or acute, e.g. when disruptions in perfusion due to fluctuating vasoconstriction lead to transient hypoxic episodes on a timescale of minutes to hours (3). Furthermore, in tumours, dynamic changes in perfusion can lead to cycles of hypoxia and re-oxygenation, generating reactive oxygen species (3).
Hypoxia is a poor prognostic factor for patient outcome and is associated with invasive growth and metastasis (2). Furthermore, hypoxia can cause resistance to cancer therapeutics, including radiation and chemotherapy. In the case of radiation therapy, this is primarily due to reduced formation of free radicals, thereby lessening the DNA damage caused by radiation (4). Chemotherapy resistance is likely due partially to poor drug delivery and reduced cell proliferation (5). Many of the biological responses to hypoxia are mediated through hypoxia-inducible factors (HIFs), transcription factors whose expression is stabilised under conditions of low oxygen (6). HIF-dependent transcriptional changes affect several biological pathways, including apoptosis, proliferation, migration, metabolism and immortalisation, and may contribute to the therapeutic resistance and poor prognosis phenotypes observed in hypoxia (7).
Hypoxia contributes to genomic instability in tumours and is associated with high rates of mutation frequency, DNA over-replication, fragile site induction and microsatellite instability (8–12). As hypoxia does not generate DNA damage, this increased genomic instability can be attributed to decreased capacity of cellular DNA repair pathways. The effects of hypoxia on DNA repair are complex and multifaceted, and include regulation of several DNA repair pathways through transcriptional, translational, post-translational and epigenetic mechanisms. Ultimately, hypoxia decreases capacity for high-fidelity repair pathways, thereby increasing mutagenesis and genomic instability. In this review, we summarise the mechanisms by which hypoxia and hypoxia mimetics affect DNA repair pathways and highlight areas of potential clinical relevance as well as future directions.
Defining, measuring and modelling hypoxia
There is substantial variability in the physiologic oxygenation levels in normal human tissues, which can range from oxygen partial pressure or oxygen tension (pO2) levels of 23–70 mm Hg (13). Lower oxygenation levels are considered hypoxic; however, there does not appear to be a consensus on the quantitative definition of hypoxia. In general, at pO2 < 10 mm Hg, activation of HIFs (14) and resistance to radiation therapy (15) can be observed. Further, tumours with median pO2 <10 mm Hg show worse outcomes than those with higher median pO2 (2). Anoxia refers to a complete lack of oxygen, or pO2 = 0 mm Hg.
A variety of different techniques have been used to quantitatively measure hypoxia in tumours. Oxygen tension can be directly measured using an electrode (16). Indirect measurements of hypoxia include the use of bioreductive compounds, such as 2-nitroimidazole derivatives, which accumulate intracellularly under hypoxic conditions due to their propensity to covalently bind to thiol groups (17). These compounds can be imaged ex vivo using immunohistochemistry or flow cytometry as well as in vivo using positron emission tomography or magnetic resonance imaging (17). Efforts have also been made to quantify hypoxia on the basis of expression of hypoxia-induced proteins, including HIFs and carbonic anhydrase IX (CAIX) (18). Gene-expression-based hypoxia signatures have also been identified in a number of cancer types (19,20).
These techniques have been used in an effort to quantify relative oxygenation within and between different tumour types. Using mRNA-based hypoxia signatures to quantitatively compare hypoxia in >8000 human tumours across 19 different tumour types, a recent study found that head and neck, cervical and lung squamous cell tumours were the most hypoxic, whereas thyroid and prostate adenocarcinomas were the least hypoxic (21). However, prostate cancer has also been cited as having a relatively low median oxygen tension, consistent with a high degree of hypoxia, when compared with other tumour types (22). It is important to note that a direct comparison of relative levels of hypoxia between tumour types is limited by the large amount of variance found within tumour types. In fact, Bhandari et al. (21) found that >40% of the variance within hypoxia scores occurred within individual tumour types. In addition, the temporal and spatial heterogeneity of hypoxia within an individual tumour further complicates efforts to quantify and compare levels of hypoxia.
Although specialised chambers, hoods and incubators are commonly used by researchers to tightly control oxygen levels for both in vitro and in vivo experiments, these techniques do not mimic the spatial heterogeneity of hypoxia as it occurs in human tumours. However, recent advances in microfluidics and biomaterials have allowed for the development of novel model systems to induce hypoxia with a higher degree of spatial control (23,24).
DNA damage response
Coordination of the cellular response to DNA damage relies largely on the activity of a family of phosphoinositide 3-kinase-related kinases, which includes ataxia-telangiectasia-mutated kinase (ATM), ataxia telangiectasia and Rad3-related protein (ATR) and DNA-dependent protein kinase (DNA-PK). These kinases are activated via post-translational modifications in response to DNA damage [specifically double-strand breaks (ATM and DNA-PK) and replication stress (ATR)], where they subsequently phosphorylate a number of substrates, thereby coordinating the cellular response to DNA damage, including DNA repair, cell cycle control and apoptosis (25). In the absence of DNA damage, hypoxia has been shown to activate these kinases through post-translational modifications, ultimately protecting cells from hypoxia-induced replication stress and re-oxygenation-induced DNA damage.
Hypoxia induces ATM-dependent signalling, including Chk2 phosphorylation (26). This signalling cascade is likely mediated through hypoxia-induced replication stress in the context of a heterochromatin-like state (27). ATM-induced Chk2 phosphorylation subsequently causes G2 cell-cycle arrest upon re-oxygenation (28). Hypoxia-induced ATM and Chk2 activation protect cells against apoptosis after exposure to both hypoxia and re-oxygenation, as cells lacking ATM or Chk2 show reduced clonogenic survival and increased apoptosis after exposure to hypoxia as well as hypoxia/re-oxygenation (26,28). The protective effect of ATM under hypoxia conditions likely relies on activation of the mammalian target of rapamycin (mTOR) pathway, as treatment with the mTOR inhibitor rapamycin reduces the induction of apoptosis after exposure to hypoxia in ATM-deficient cells (29).
In addition, hypoxia induces replication arrest, thereby activating ATR-dependent signalling, including induction of Chk1 phosphorylation (30). Hypoxia-dependent ATR/Chk1 activation subsequently causes induction of γH2AX and phosphorylation of p53 (31). Furthermore, activation of both ATR and Chk1 protects against apoptosis after re-oxygenation, as knockdown of either ATR or Chk1 decreases cellular survival and increases apoptosis after exposure to hypoxia/re-oxygenation (32).
It has also been reported that hypoxia induces activation of DNA-PK via phosphorylation of Ser2056 of the catalytic subunit (33), through a mechanism relying on histone acetylation. Hypoxia-induced DNA-PK activation was also shown to regulate HIF-1 expression (33). However, although the primary consequence of DNA-PK activation is repair of double-strand breaks (DSBs) through the non-homologous end joining (NHEJ) pathway, hypoxia is not associated with an increase in nuclear localisation of NHEJ factors, such as XRCC4 (33), and thus its impact on NHEJ, as further discussed later, remains unclear.
Homology-directed repair
Homology-directed repair (HDR) represents a high-fidelity mechanism by which cells repair DSBs. This pathway, which is limited to the S and G2 phases of the cell cycle, uses a homologous sequence to serve as a template for repair (34). Hypoxia suppresses HDR through several mechanisms, including transcriptional, translational and epigenetic repression of the expression of key repair factors, which may ultimately lead to increased mutagenesis and genomic instability.
Hypoxia reduces the transcription of key HDR factors BRCA1 and RAD51 through the induction of nuclear E2F4/p130 complexes, which bind to E2F consensus sites in the BRCA1 and RAD51 promoters, suppressing gene expression (35,36). The induction of these repressive transcription regulatory complexes is associated with hypophosphorylation of p130 (36). It has been proposed that p130 hypophosphorylation is mediated by protein phosphatase 2, which is activated under hypoxic conditions (37). Expression of the Fanconi’s anaemia protein FANCD2, which plays a role in HDR, is also suppressed by a similar mechanism under hypoxic conditions (38).
Translational repression of several key HDR factors has also been documented under hypoxic conditions. Under hypoxic conditions that only mildly affect global translation efficiency, dramatic suppression of RAD51, RAD51B, RAD51C, RAD51D, RAD54, XRCC3, BRCA1 and BRCA2 protein expression and mRNA polysomal fractions was observed, suggesting that hypoxia reduces the translational efficiency of specific HDR genes (39). MicroRNAs (miRNAs) also mediate translational repression of HDR factors under hypoxic conditions. The miRNAs miR210 and miR373 are both induced by hypoxic conditions, and have been shown to suppress RAD52 and RAD23B expression, respectively, by targeting the 3′-untranslated region of these genes (40). In addition, miR-155 is induced by hypoxia (41,42) and its overexpression increases mutation frequency (43). miR-155 has been shown to both suppress RAD51 expression and induce a functional HDR deficit (44). Regulation of HDR also occurs at the epigenetic level, as hypoxia promotes epigenetic silencing of the BRCA1 and RAD51 via the lysine-specific histone demethylase LSD1 (45). Hypoxia also induces expression of the Polycomb protein EZH2, leading to epigenetic suppression of RAD51 expression (46).
A correlation between decreased HDR factor expression and hypoxia has been documented in both mouse tumour models and in the clinical setting. RAD51 and EF5, a marker of hypoxia, was noted in colorectal tumour xenografts (47). In addition, in a series of clinical breast cancer resection specimens, expression of the hypoxia marker CAIX has been shown to inversely correlate with BRCA1 expression as detected by quantitative immunofluorescence (48).
Regulation of HDR through hypoxia may also occur through the production of the hypoxia-induced metabolite S-2-hydroxyglutarate, which has recently been shown to functionally inhibit HDR (49). Although the mechanism of this repression has yet to be elucidated, it may be similar to the mechanism by which the oncometabolite R-2-hydroxyglutarate suppresses HDR, namely suppression of the α-ketoglutarate-dependent lysine demethylase KDM4A and KDM4B (49).
Non-homologous end joining
Although hypoxia has a clear effect on HDR, its effects on the other major pathway of double-strand break repair, NHEJ are less well defined. NHEJ, which is responsible for repairing the majority of DSB breaks in human cells (50), is required to maintain genomic stability. However, this rapid and relatively error-prone pathway of repair, which relies on DNA end processing and ligation, can also introduce insertion and deletion mutations (50), thereby increasing genomic instability.
Several mRNA-based studies seem to indicate that hypoxia causes a downregulation of NHEJ factor expression. A microarray analysis of cultured breast cancer cells showed significant downregulation of NHEJ factor mRNA expression under hypoxia (51). Furthermore, inverse correlations have been observed between RNA-based hypoxia (HIF-2α) signatures and gene expression in several DNA repair pathways, including NHEJ (52).
However, whether the reported downregulation of NHEJ factor mRNA translates to reduced expression of NHEJ proteins is unclear. Although hypoxia has been shown to decrease mRNA expression of the NHEJ factor Ku70 in prostate cancer cells, these same conditions did not cause a reduction in Ku70 protein expression (53). Furthermore, various studies have reported conflicting effects of hypoxia on NHEJ factor protein expression, including no change (35), upregulation (54,55) and downregulation (52). In addition, functional assays indicate that hypoxia does not suppress, and may even enhance, NHEJ capacity (35,54). Augmented NHEJ capacity would represent an increase in the use of an error-prone repair pathway, which would contribute to the mutagenesis and genomic instability observed under hypoxic conditions.
Although the exact effects of hypoxia on NHEJ remain to be elucidated, it is likely that multiple mechanisms underlie the observed changes in repair factor expression and functional repair. Furthermore, the impact of hypoxia on NHEJ may be affected by the nature of the hypoxic conditions as well as cell-type specificities, and additional work is needed to more clearly define these effects.
Mismatch repair
Mismatch repair (MMR), which removes mismatched bases from newly synthesised DNA, is particularly important for maintenance of genomic stability (56), as cells with deficiencies in MMR develop a mutator phenotype, or an increase in mutation rates, which can lead to the development of cancer (56). Hypoxia suppresses MMR through multiple mechanisms, including decreased gene expression of the MMR factors MLH1 and MSH2 (57,58). Under hypoxic conditions, transient transcriptional repression of MLH1 and MSH2 occurs due to a shift in promoter occupancy from activating c-Myc/Max to repressive Mnt/Max and Mad1/Max complexes, respectively (58). Transcriptional repression of MSH2 also occurs due to displacement of the transcriptional activator c-Myc from Sp1 by HIF-1α (59). Finally, hypoxia also induces the expression of transcriptional repressors differentiated embryo chondrocyte 1 and 2, which suppress MLH1 transcription by binding to E-box-like motifs in the MLH1 promoter (60).
Together these mechanisms repress MMR factor expression under hypoxic conditions. Hypoxia has been correlated with low protein expression of MSH2 and MLH1 in mouse xenograft models (61,62). These results have also been validated in a clinical setting, as HIF-1α expression has been found to inversely correlate with MSH2 expression in sporadic human colon cancer (59). Hypoxia also regulates MMR through miRNA-mediated suppression of MMR gene expression. The hypoxia-induced miRNA miR-155 has been shown to suppress MLH1, MSH2 and MSH6 expression (43,63) and to induce microsatellite instability, a hallmark of MMR deficiency (63).
Epigenetic regulation also underlies the suppression of MMR by hypoxia. This is evidenced by the fact that the histone deacetylase inhibitor, trichostatin A, prevents downregulation of MLH1 under hypoxic conditions (57,60,64). Furthermore, hypoxia increases markers of epigenetic repression, including H2K9 (65) and H3K9 (62) methylation, and decreases markers of activation, including H3K9 acetylation and H3K4 methylation (62), at the MLH1 promoter. Hypoxia-mediated H3K4 demethylation occurs via the action of the H3K4 demethylases LSD1 and PLU-1 (62). Prolonged hypoxia also induces stable silencing of the MLH1 promoter through a mechanism requiring LSD1 (62).
Nucleotide excision repair
Nucleotide excision repair (NER) is the major pathway of DNA repair for removal of bulky DNA lesions caused by ultraviolet (UV) irradiation, environmental mutagens and specific cancer therapeutics (66). The effects of hypoxia on NER are unclear. Some evidence indicates that hypoxia suppresses NER. For example, severe hypoxia has been shown to cause reduced NER capacity and UV irradiation-induced hypermutability (67). However, moderate hypoxia has been shown to increase, and not decrease, NER capacity (68). Many NER genes contain hypoxia response elements in their promoters (69), however, it is unclear whether hypoxia downregulates mRNA expression of these genes. Furthermore, protein levels of NER genes, such as XPA, XPB, XPD and XPG, remain unchanged after hypoxia exposure (57,67). The expression of one NER factor, ERCC1, is downregulated at both the mRNA and protein level after hypoxia exposure (70). Ultimately, further work is needed to more conclusively determine the effects of hypoxia on NER, including a more comprehensive study of dose and time responses.
Base excision repair
Base excision repair (BER) is essential for maintaining genomic integrity after oxidative damage (71). Hypoxia has been shown to decrease expression of several BER factors, including OGG1, MYH, POLB, APE1, RPA and PCNA (72). Hypoxia also downregulates the expression of ASCIZ/ATMIN (73), a zinc-finger protein that may play a role in BER (74,75). Furthermore, hypoxia causes a reduction in functional BER as measured by MYH- and OGG1-specific glycosylase assays and an increase in accumulated residual base damage after hydrogen peroxide exposure (72).
Translesion synthesis
Translesion synthesis describes a process by which cells use specialised polymerases, such as DNA polymerases ι, κ and ζ, to bypass DNA lesions in an effort to avoid replication fork stalling (76). Although this process can be essential for cellular survival after DNA damage, it is also error-prone and can lead to both mutagenesis and carcinogenesis. There is evidence suggesting that hypoxia may increase the use of this error-prone DNA repair pathway. For example, DNA polymerase ι expression is induced by hypoxia through HIF-1α-dependent mechanism (77). Furthermore, hypoxia decreases expression of polymerase δ, a high-fidelity DNA polymerase, via induction of miR-155 (43), and it has been hypothesised that this reduction may lead to a compensatory increase in the activity of more error-prone translesion polymerases, such as DNA polymerase ι. In addition, cells lacking the catalytic subunit of DNA polymerase ζ show decreased survival and increased chromosomal breaks after irradiation under hypoxic conditions (78), suggesting that translesion synthesis may play an important role in cellular responses to radiation in hypoxia. However, the exact effects of hypoxia on translesion synthesis remain to be elucidated.
Hypoxia mimetics
Several lines of evidence indicate that hypoxia mimetics induce similar impairments in DNA repair to hypoxia. For example, the iron chelator desferrioxamine mimics the effects of hypoxia by stabilising HIF-1α through inactivation of the iron-dependent proline hydroxylases that ordinarily inactive HIF (6). Desferrioxamine has been shown to suppress gene expression of both MMR and HDR factors (35,38,57). Nickel similarly inhibits the HIF proline hydroxylases (79) and induces transcriptional repression of HDR as well as MMR factors (80).
Inactivating mutations in the von Hippel-Lindau (VHL) protein mimic hypoxia by preventing the degradation of HIF-1α. It is estimated that between 60 and 80% of sporadic clear cell renal cell carcinoma (ccRCC), the most common form of kidney cancer, display VHL mutations (81). In ccRCC, VHL deficiency confers impairments in HDR via reduced expression of HDR factors BRCA1 and RAD51 (82). Similar to hypoxia, VHL deficiency also reduces FANCD2 and MLH1 expression (82).
We have also recently reported that the small-molecule inhibitor, cediranib, which was developed as a vascular endothelial growth factor receptor inhibitor (83), mimics the effects of hypoxia on HDR. Like hypoxia, treatment with cediranib suppresses expression of key HDR factors through a pathway mediated by E2F4 and p130, inducing a functional HDR deficit (84). The effects of cediranib on HDR have been attributed to inhibition of platelet-derived growth factor receptor (PDGFRs), as other small-molecule inhibitors of PDGFRs as well as small interfering RNA targeting PDGFRβ also impair HDR.
Clinical implications and future directions
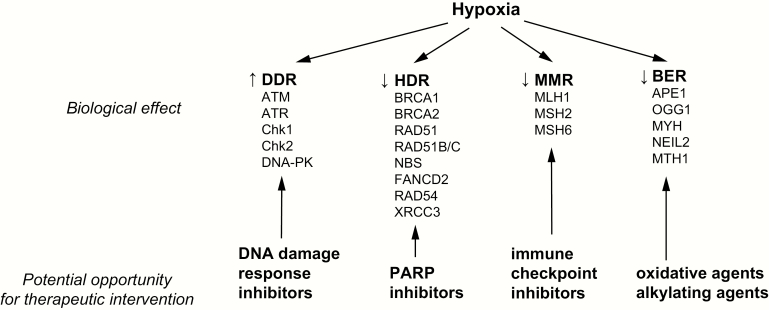
Although the suppression of DNA repair caused by hypoxia may lead to increased genomic instability and mutagenesis, it may also create opportunities for potential therapeutic targeting (Figure 1). For example, hypoxia-induced activation of DNA damage response (DDR) factors, such as ATM, ATR, Chk1 and Chk2, has been shown to protect cells against apoptosis under hypoxia and re-oxygenation. Therefore, the use of small-molecule inhibitors to such factors may re-sensitise hypoxic cells, inducing cellular death. For example, hypoxia sensitises cancer cells in culture to the ATR inhibitor VE-821 in a dose-dependent fashion (85). Small-molecule inhibitors targeting the DDR pathways are currently in clinical trials (86). Hypoxia-induced suppression of BER also creates an opportunity for potential therapeutic targeting, as it sensitises cells to oxidative and alkylating agents (72).
Figure 1.
Effects of hypoxia on DNA repair pathways with potential opportunities for therapeutic interventions.
Although acute hypoxia can cause resistance to radiation therapy by reducing the formation of free radicals, there is evidence that chronic hypoxia and re-oxygenation can confer radiosensitivity, likely due to suppression of HDR (39,87). The functional decrease in HDR caused by hypoxia also confers increased sensitivity to DNA crosslinking agents, such as mitomycin C and cisplatin (39,88).
Furthermore, by suppressing HDR, hypoxia also renders tumour cells sensitive to poly (ADP-ribose) polymerase (PARP) inhibitors, inducing a synthetic vulnerability in the tumour microenvironment. Evidence supporting this hypothesis includes the increased sensitivity of cancer cells grown in hypoxic culture conditions to PARP inhibition (89). Furthermore, treatment of tumour xenografts with the PARP inhibitor, ABT888, increased markers of DNA damage and apoptosis in hypoxic regions of tumours (20). Hypoxia mimetics similarly increase tumour cell sensitivity to PARP inhibition (82) and ionising radiation (80). The concept of a hypoxia-induced synthetic lethality is particularly intriguing given the ability of antiangiogenic therapeutic compounds, such as bevacizumab and cediranib, to promote tumour hypoxia (90–93). Phase 1 clinical trials have found the combination of the PARP inhibitor olaparib with antiangiogenics to be tolerable (94,95), and a phase 3 clinical trial evaluating the efficacy of combination bevacizumab and olaparib maintenance therapy is currently underway (PAOLA-1; NCT02477644). Interestingly, the combination of cediranib with olaparib has already been shown to improve progression-free survival in ovarian cancer in a recent clinical trial (96,97). The combination of cediranib and olaparib may be a particular effective strategy given the additional direct effect of cediranib on HDR, as described earlier. Indeed, cediranib has been shown to sensitise cancer cells to olaparib both in culture as well as in xenografts in vivo (84,98).
Hypoxia has been shown to induce a longstanding deficiency in MMR by stable silencing of MMR promoters. Cancers with MMR appear to be particularly responsive to immune checkpoint inhibitors (99,100). Thus, it is possible that cancers with substantial hypoxic fractions may be more responsive to immune checkpoint inhibitors due to their acquired MMR deficiency. Hypoxia also upregulates PDL1 expression in a HIF-1α-dependent fashion (101,102), providing another reason that immune checkpoint inhibitor therapy may be particularly effective in highly hypoxic tumours regardless of changes in MMR status. However, to date no studies have evaluated the relative cytotoxic effects of immune checkpoint inhibitor therapy on normoxic and hypoxic fractions of tumours in vivo.
A preponderance of literature demonstrates that hypoxia profoundly impairs genomic stability by inhibiting a number of high-fidelity DNA repair pathways, including HDR, MMR and BER. Compounds or genetic mutations that mimic or induce hypoxia have comparable effects on DNA repair. Hypoxia also activates DDR pathways, which appears to serve a protective role. However, the effects of hypoxia on other DNA repair pathways, including NHEJ, NER and translesion synthesis remain incompletely elucidated. Furthermore, many of the studies on the role of hypoxia on DNA repair have been performed only under in vitro conditions; therefore, extending these findings to physiologic conditions using tumour model systems remains an important future direction. Further studies in a more physiologic model system will also allow for the study of the role of the tumour microenvironment, including contributions from stromal and immune cells, in regulating the effects of hypoxia. This work will lead to a more comprehensive understanding of the impacts of hypoxia on tumour biology and how DNA repair and mutagenesis is regulated. It may also ultimately contribute to the development of new therapeutic paradigms to target a population of relatively treatment-resistant cancer cells.
Funding
This work was supported by National Institutes of Health (NIH) grants R01ES005775 and R35CA197574 to P.M.G. and by NIH grant F30CA221065 to A.R.K. Support for this research was also provided by the NIH Medical Scientist Training Program Training Grant T32GM007205.
Conflict of interest statement: None declared.
References
- 1. Multhoff G., Radons J. and Vaupel P (2014) Critical role of aberrant angiogenesis in the development of tumor hypoxia and associated radioresistance. Cancers (Basel)., 6, 813–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaupel P. and Mayer A (2007) Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev., 26, 225–239. [DOI] [PubMed] [Google Scholar]
- 3. Vaupel P. and Mayer A (2014) Hypoxia in tumors: pathogenesis-related classification, characterization of hypoxia subtypes, and associated biological and clinical implications. Adv. Exp. Med. Biol., 812, 19–24. [DOI] [PubMed] [Google Scholar]
- 4. Rockwell S., Dobrucki I. T., Kim E. Y., Marrison S. T. and Vu V. T (2009) Hypoxia and radiation therapy: past history, ongoing research, and future promise. Curr. Mol. Med., 9, 442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaupel P., Thews O. and Hoeckel M (2001) Treatment resistance of solid tumors: role of hypoxia and anemia. Med. Oncol., 18, 243–259. [DOI] [PubMed] [Google Scholar]
- 6. Schofield C. J. and Ratcliffe P. J (2004) Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol., 5, 343–354. [DOI] [PubMed] [Google Scholar]
- 7. Harris A. L. (2002) Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer, 2, 38–47. [DOI] [PubMed] [Google Scholar]
- 8. Reynolds T. Y., Rockwell S. and Glazer P. M (1996) Genetic instability induced by the tumor microenvironment. Cancer Res., 56, 5754–5757. [PubMed] [Google Scholar]
- 9. Yuan J. and Glazer P. M (1998) Mutagenesis induced by the tumor microenvironment. Mutat. Res., 400, 439–446. [DOI] [PubMed] [Google Scholar]
- 10. Rice G. C., Hoy C. and Schimke R. T (1986) Transient hypoxia enhances the frequency of dihydrofolate reductase gene amplification in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. U. S. A., 83, 5978–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Young S. D., Marshall R. S. and Hill R. P (1988) Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc. Natl. Acad. Sci. U. S. A., 85, 9533–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coquelle A., Toledo F., Stern S., Bieth A. and Debatisse M (1998) A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol. Cell, 2, 259–265. [DOI] [PubMed] [Google Scholar]
- 13. Carreau A., El Hafny-Rahbi B., Matejuk A., Grillon C. and Kieda C (2011) Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med., 15, 1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bracken C. P., Fedele A. O., Linke S., Balrak W., Lisy K., Whitelaw M. L. and Peet D. J (2006) Cell-specific regulation of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J. Biol. Chem., 281, 22575–22585. [DOI] [PubMed] [Google Scholar]
- 15. Höckel M., Schlenger K., Mitze M., Schäffer U. and Vaupel P (1996) Hypoxia and radiation response in human tumors. Semin. Radiat. Oncol., 6, 3–9. [DOI] [PubMed] [Google Scholar]
- 16. Vaupel P., Schlenger K., Knoop C. and Höckel M (1991) Oxygenation of human tumors: evaluation of tissue oxygen distribution in breast cancers by computerized O2 tension measurements. Cancer Res., 51, 3316–3322. [PubMed] [Google Scholar]
- 17. Kizaka-Kondoh S. and Konse-Nagasawa H (2009) Significance of nitroimidazole compounds and hypoxia-inducible factor-1 for imaging tumor hypoxia. Cancer Sci., 100, 1366–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hutchison G. J., Valentine H. R., Loncaster J. A., et al. (2004) Hypoxia-inducible factor 1alpha expression as an intrinsic marker of hypoxia: correlation with tumor oxygen, pimonidazole measurements, and outcome in locally advanced carcinoma of the cervix. Clin. Cancer Res., 10, 8405–8412. [DOI] [PubMed] [Google Scholar]
- 19. Buffa F. M., Harris A. L., West C. M. and Miller C. J (2010) Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer, 102, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winter S. C., Buffa F. M., Silva P., et al. (2007) Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res., 67, 3441–3449. [DOI] [PubMed] [Google Scholar]
- 21. Bhandari V., Hoey C., Liu L. Y., et al. (2019) Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet., 51, 308–318. [DOI] [PubMed] [Google Scholar]
- 22. Brown J. M. and Wilson W. R (2004) Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer, 4, 437–447. [DOI] [PubMed] [Google Scholar]
- 23. Brennan M. D., Rexius-Hall M. L., Elgass L. J. and Eddington D. T (2014) Oxygen control with microfluidics. Lab Chip, 14, 4305–4318. [DOI] [PubMed] [Google Scholar]
- 24. Park K. M. and Gerecht S (2014) Hypoxia-inducible hydrogels. Nat. Commun., 5, 4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blackford A. N. and Jackson S. P (2017) ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell, 66, 801–817. [DOI] [PubMed] [Google Scholar]
- 26. Gibson S. L., Bindra R. S. and Glazer P. M (2005) Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer Res., 65, 10734–10741. [DOI] [PubMed] [Google Scholar]
- 27. Olcina M. M., Foskolou I. P., Anbalagan S., Senra J. M., Pires I. M., Jiang Y., Ryan A. J. and Hammond E. M (2013) Replication stress and chromatin context link ATM activation to a role in DNA replication. Mol. Cell, 52, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Freiberg R. A., Hammond E. M., Dorie M. J., Welford S. M. and Giaccia A. J (2006) DNA damage during reoxygenation elicits a Chk2-dependent checkpoint response. Mol. Cell. Biol., 26, 1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cam H., Easton J. B., High A. and Houghton P. J (2010) mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol. Cell, 40, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hammond E. M., Denko N. C., Dorie M. J., Abraham R. T. and Giaccia A. J (2002) Hypoxia links ATR and p53 through replication arrest. Mol. Cell. Biol., 22, 1834–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammond E. M., Dorie M. J. and Giaccia A. J (2003) ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J. Biol. Chem., 278, 12207–12213. [DOI] [PubMed] [Google Scholar]
- 32. Hammond E. M., Dorie M. J. and Giaccia A. J (2004) Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer Res., 64, 6556–6562. [DOI] [PubMed] [Google Scholar]
- 33. Bouquet F., Ousset M., Biard D., Fallone F., Dauvillier S., Frit P., Salles B. and Muller C (2011) A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. J. Cell Sci., 124, 1943–1951. [DOI] [PubMed] [Google Scholar]
- 34. Goldstein M. and Kastan M. B (2015) The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med., 66, 129–143. [DOI] [PubMed] [Google Scholar]
- 35. Bindra R. S., Gibson S. L., Meng A., Westermark U., Jasin M., Pierce A. J., Bristow R. G., Classon M. K. and Glazer P. M (2005) Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res., 65, 11597–11604. [DOI] [PubMed] [Google Scholar]
- 36. Bindra R. S. and Glazer P. M (2007) Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene, 26, 2048–2057. [DOI] [PubMed] [Google Scholar]
- 37. Di Conza G., Trusso Cafarello S., Loroch S., et al. (2017) The mTOR and PP2A pathways regulate PHD2 phosphorylation to fine-tune HIF1α levels and colorectal cancer cell survival under hypoxia. Cell Rep., 18, 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scanlon S. E. and Glazer P. M (2014) Hypoxic stress facilitates acute activation and chronic downregulation of Fanconi anemia proteins. Mol. Cancer Res., 12, 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan N., Koritzinsky M., Zhao H., Bindra R., Glazer P. M., Powell S., Belmaaza A., Wouters B. and Bristow R. G (2008) Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res., 68, 605–614. [DOI] [PubMed] [Google Scholar]
- 40. Crosby M. E., Kulshreshtha R., Ivan M. and Glazer P. M (2009) MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res., 69, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bruning U., Cerone L., Neufeld Z., et al. (2011) MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol. Cell. Biol., 31, 4087–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Babar I. A., Czochor J., Steinmetz A., Weidhaas J. B., Glazer P. M. and Slack F. J (2011) Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol. Ther., 12, 908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Czochor J. R., Sulkowski P. and Glazer P. M (2016) mir-155 overexpression promotes genomic instability by reducing high-fidelity polymerase delta expression and activating error-prone DSB repair. Mol. Cancer Res., 14, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gasparini P., Lovat F., Fassan M., et al. (2014) Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. U. S. A., 111, 4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu Y., Chu A., Turker M. S. and Glazer P. M (2011) Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol. Cell. Biol., 31, 3339–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang C. J., Yang J. Y., Xia W., et al. (2011) EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell, 19, 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chan N., Pires I. M., Bencokova Z., et al. (2010) Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res., 70, 8045–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Neumeister V. M., Sullivan C. A., Lindner R., et al. (2012) Hypoxia-induced protein CAIX is associated with somatic loss of BRCA1 protein and pathway activity in triple negative breast cancer. Breast Cancer Res. Treat., 136, 67–75. [DOI] [PubMed] [Google Scholar]
- 49. Sulkowski P. L., Corso C. D., Robinson N. D., et al. (2017) 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med, 9, eaal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chang H. H. Y., Pannunzio N. R., Adachi N. and Lieber M. R (2017) Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol., 18, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fanale D., Bazan V., Caruso S., Castiglia M., Bronte G., Rolfo C., Cicero G. and Russo A (2013) Hypoxia and human genome stability: downregulation of BRCA2 expression in breast cancer cell lines. Biomed Res. Int., 2013, 746858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jongen J. M. J., van der Waals L. M., Trumpi K., Laoukili J., Peters N. A., Schenning-van Schelven S. J., Govaert K. M., Borel Rinkes I. H. M. and Kranenburg O (2017) Downregulation of DNA repair proteins and increased DNA damage in hypoxic colon cancer cells is a therapeutically exploitable vulnerability. Oncotarget, 8, 86296–86311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meng A. X., Jalali F., Cuddihy A., Chan N., Bindra R. S., Glazer P. M. and Bristow R. G (2005) Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother. Oncol., 76, 168–176. [DOI] [PubMed] [Google Scholar]
- 54. Madan E., Gogna R. and Pati U (2012) p53 Ser15 phosphorylation disrupts the p53-RPA70 complex and induces RPA70-mediated DNA repair in hypoxia. Biochem. J., 443, 811–820. [DOI] [PubMed] [Google Scholar]
- 55. Ren Y., Hao P., Dutta B., Cheow E. S., Sim K. H., Gan C. S., Lim S. K. and Sze S. K (2013) Hypoxia modulates A431 cellular pathways association to tumor radioresistance and enhanced migration revealed by comprehensive proteomic and functional studies. Mol. Cell. Proteomics, 12, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiricny J. (2006) The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol., 7, 335–346. [DOI] [PubMed] [Google Scholar]
- 57. Mihaylova V. T., Bindra R. S., Yuan J., et al. (2003) Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell. Biol., 23, 3265–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bindra R. S. and Glazer P. M (2007) Co-repression of mismatch repair gene expression by hypoxia in cancer cells: role of the Myc/Max network. Cancer Lett., 252, 93–103. [DOI] [PubMed] [Google Scholar]
- 59. Koshiji M., To K. K., Hammer S., Kumamoto K., Harris A. L., Modrich P. and Huang L. E (2005) HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol. Cell, 17, 793–803. [DOI] [PubMed] [Google Scholar]
- 60. Nakamura H., Tanimoto K., Hiyama K., et al. (2008) Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors, DEC1 and DEC2. Oncogene, 27, 4200–4209. [DOI] [PubMed] [Google Scholar]
- 61. Shahrzad S., Quayle L., Stone C., Plumb C., Shirasawa S., Rak J. W. and Coomber B. L (2005) Ischemia-induced K-ras mutations in human colorectal cancer cells: role of microenvironmental regulation of MSH2 expression. Cancer Res., 65, 8134–8141. [DOI] [PubMed] [Google Scholar]
- 62. Lu Y., Wajapeyee N., Turker M. S. and Glazer P. M (2014) Silencing of the DNA mismatch repair gene MLH1 induced by hypoxic stress in a pathway dependent on the histone demethylase LSD1. Cell Rep., 8, 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Valeri N., Gasparini P., Fabbri M., et al. (2010) Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. U. S. A., 107, 6982–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rodríguez-Jiménez F. J., Moreno-Manzano V., Lucas-Dominguez R. and Sánchez-Puelles J. M (2008) Hypoxia causes downregulation of mismatch repair system and genomic instability in stem cells. Stem Cells, 26, 2052–2062. [DOI] [PubMed] [Google Scholar]
- 65. Chen H., Yan Y., Davidson T. L., Shinkai Y. and Costa M (2006) Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res., 66, 9009–9016. [DOI] [PubMed] [Google Scholar]
- 66. Schärer O. D. (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol., 5, a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yuan J., Narayanan L., Rockwell S. and Glazer P. M (2000) Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res., 60, 4372–4376. [PubMed] [Google Scholar]
- 68. Liu Y., Bernauer A. M., Yingling C. M. and Belinsky S. A (2012) HIF1α regulated expression of XPA contributes to cisplatin resistance in lung cancer. Carcinogenesis, 33, 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rezvani H. R., Mahfouf W., Ali N., et al. (2010) Hypoxia-inducible factor-1alpha regulates the expression of nucleotide excision repair proteins in keratinocytes. Nucleic Acids Res., 38, 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dudás J., Schartinger V. H., Romani A., Schweigl G., Kordsmeyer K., Marta P. I., Url C., Kral F. and Riechelmann H (2014) Cell cycle association and hypoxia regulation of excision repair cross complementation group 1 protein (ERCC1) in tumor cells of head and neck cancer. Tumour Biol., 35, 7807–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. David S. S., O’Shea V. L. and Kundu S (2007) Base-excision repair of oxidative DNA damage. Nature, 447, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chan N., Ali M., McCallum G. P., Kumareswaran R., Koritzinsky M., Wouters B. G., Wells P. G., Gallinger S. and Bristow R. G (2014) Hypoxia provokes base excision repair changes and a repair-deficient, mutator phenotype in colorectal cancer cells. Mol. Cancer Res., 12, 1407–1415. [DOI] [PubMed] [Google Scholar]
- 73. Leszczynska K. B., Göttgens E. L., Biasoli D., Olcina M. M., Ient J., Anbalagan S., Bernhardt S., Giaccia A. J. and Hammond E. M (2016) Mechanisms and consequences of ATMIN repression in hypoxic conditions: roles for p53 and HIF-1. Sci. Rep., 6, 21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Oka H., Sakai W., Sonoda E., et al. (2008) DNA damage response protein ASCIZ links base excision repair with immunoglobulin gene conversion. Biochem. Biophys. Res. Commun., 371, 225–229. [DOI] [PubMed] [Google Scholar]
- 75. Jurado S., Smyth I., van Denderen B., et al. (2010) Dual functions of ASCIZ in the DNA base damage response and pulmonary organogenesis. PLoS Genet., 6, e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Goodman M. F. and Woodgate R (2013) Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol., 5, a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ito A., Koshikawa N., Mochizuki S., Omura K. and Takenaga K (2006) Hypoxia-inducible factor-1 mediates the expression of DNA polymerase iota in human tumor cells. Biochem. Biophys. Res. Commun., 351, 306–311. [DOI] [PubMed] [Google Scholar]
- 78. Shimizu N., Ooka M., Takagi T., Takeda S. and Hirota K (2015) Distinct DNA damage spectra induced by ionizing radiation in normoxic and hypoxic cells. Radiat. Res., 184, 442–448. [DOI] [PubMed] [Google Scholar]
- 79. Davidson T. L., Chen H., Di Toro D. M., D’Angelo G. and Costa M (2006) Soluble nickel inhibits HIF-prolyl-hydroxylases creating persistent hypoxic signaling in A549 cells. Mol. Carcinog., 45, 479–489. [DOI] [PubMed] [Google Scholar]
- 80. Scanlon S. E., Scanlon C. D., Hegan D. C., Sulkowski P. L. and Glazer P. M (2017) Nickel induces transcriptional down-regulation of DNA repair pathways in tumorigenic and non-tumorigenic lung cells. Carcinogenesis, 38, 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Robinson C. M. and Ohh M (2014) The multifaceted von Hippel-Lindau tumour suppressor protein. FEBS Lett., 588, 2704–2711. [DOI] [PubMed] [Google Scholar]
- 82. Scanlon S. E., Hegan D. C., Sulkowski P. L. and Glazer P. M (2018) Suppression of homology-dependent DNA double-strand break repair induces PARP inhibitor sensitivity in VHL-deficient human renal cell carcinoma. Oncotarget, 9, 4647–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wedge S. R., Kendrew J., Hennequin L. F., et al. (2005) AZD2171: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res., 65, 4389–4400. [DOI] [PubMed] [Google Scholar]
- 84. Kaplan A. R., Gueble S E., Liu Y., et al. (2019) Cediranib suppresses homology-directed DNA repair through down-regulation of BRCA1/2 and RAD51. Sci Transl Med, 11, eaav4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pires I. M., Olcina M. M., Anbalagan S., Pollard J. R., Reaper P. M., Charlton P. A., McKenna W. G. and Hammond E. M (2012) Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br. J. Cancer, 107, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pilié P. G., Tang C., Mills G. B. and Yap T. A (2019) State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol., 16, 81–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kumareswaran R., Ludkovski O., Meng A., Sykes J., Pintilie M. and Bristow R. G (2012) Chronic hypoxia compromises repair of DNA double-strand breaks to drive genetic instability. J. Cell Sci., 125, 189–199. [DOI] [PubMed] [Google Scholar]
- 88. Strese S., Fryknäs M., Larsson R. and Gullbo J (2013) Effects of hypoxia on human cancer cell line chemosensitivity. BMC Cancer, 13, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hegan D. C., Lu Y., Stachelek G. C., Crosby M. E., Bindra R. S. and Glazer P. M (2010) Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc. Natl. Acad. Sci. U. S. A., 107, 2201–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Grkovski M., Emmas S. A. and Carlin S. D (2017) 18F-Fluoromisonidazole kinetic modeling for characterization of tumor perfusion and hypoxia in response to antiangiogenic therapy. J. Nucl. Med., 58, 1567–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jiang Y., Allen D., Kersemans V., Devery A. M., Bokobza S. M., Smart S. and Ryan A. J (2015) Acute vascular response to cediranib treatment in human non-small-cell lung cancer xenografts with different tumour stromal architecture. Lung Cancer, 90, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Heijmen L., Ter Voert E. G., Punt C. J., et al. (2014) Monitoring hypoxia and vasculature during bevacizumab treatment in a murine colorectal cancer model. Contrast Media Mol. Imaging, 9, 237–245. [DOI] [PubMed] [Google Scholar]
- 93. Presta L. G., Chen H., O’Connor S. J., Chisholm V., Meng Y. G., Krummen L., Winkler M. and Ferrara N (1997) Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res., 57, 4593–4599. [PubMed] [Google Scholar]
- 94. Liu J. F., Tolaney S. M., Birrer M., et al. (2013) A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur. J. Cancer, 49, 2972–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dean E., Middleton M. R., Pwint T., Swaisland H., Carmichael J., Goodege-Kunwar P. and Ranson M (2012) Phase I study to assess the safety and tolerability of olaparib in combination with bevacizumab in patients with advanced solid tumours. Br. J. Cancer, 106, 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu J. F., Barry W. T., Birrer M., et al. (2014) Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet. Oncol., 15, 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu J. F., Barry W.T., Birrer M., et al. (2019) Overall survival and updated progression-free survival outcomes in a randomized phase 2 study of combination cediranib and olaparib versus olaparib in relapsed platinum-sensitive ovarian cancer. Ann. Oncol., 30, 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lin Z. P., Zhu Y. L., Lo Y. C., et al. (2018) Combination of triapine, olaparib, and cediranib suppresses progression of BRCA-wild type and PARP inhibitor-resistant epithelial ovarian cancer. PLoS One, 13, e0207399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Overman M. J., McDermott R., Leach J. L., et al. (2017) Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet. Oncol., 18, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Le D. T., Uram J. N., Wang H., et al. (2015) PD-1 Blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med., 372, 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Noman M. Z., Desantis G., Janji B., Hasmim M., Karray S., Dessen P., Bronte V. and Chouaib S (2014) PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med., 211, 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Barsoum I. B., Smallwood C. A., Siemens D. R. and Graham C. H (2014) A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res., 74, 665–674. [DOI] [PubMed] [Google Scholar]