Abstract
Oxysterols are critical regulators of inflammation and cholesterol metabolism in cells. They are oxidation products of cholesterol and may be differentially metabolised in subcellular compartments and in biological fluids. New analytical methods are needed to improve our understanding of oxysterol trafficking and the molecular interplay between the cellular compartments required to maintain cholesterol/oxysterol homeostasis. Here we describe a method for isolation of oxysterols using solid phase extraction and quantification by liquid chromatography-mass spectrometry, applied to tissue, cells and mitochondria.
We analysed five monohydroxysterols; 24(S)-hydroxycholesterol, 25-hydroxycholesterol, 27-hydroxycholesterol, 7α-hydroxycholesterol, 7 ketocholesterol and three dihydroxysterols 7α-24(S)dihydroxycholesterol, 7α-25dihydroxycholesterol, 7α-27dihydroxycholesterol by LC-MS/MS following reverse phase chromatography. Our new method, using Triton and DMSO extraction, shows improved extraction efficiency and recovery of oxysterols from cellular matrix. We validated our method by reproducibly measuring oxysterols in mouse brain tissue and showed that mice fed a high fat diet had significantly lower levels of 24S/25diOHC, 27diOHC and 7ketoOHC. We measured oxysterols in mitochondria from peripheral blood mononuclear cells and highlight the importance of rapid cell isolation to minimise effects of handling and storage conditions on oxysterol composition in clinical samples. In addition, in vitro cell culture systems, of THP-1 monocytes and neuronal-like SH-SH5Y cells, showed mitochondrial-specific oxysterol metabolism and profiles were lineage specific. In summary, we describe a robust and reproducible method validated for improved recovery, quantitative linearity and detection, reproducibility and selectivity for cellular oxysterol analysis. This method enables subcellular oxysterol metabolism to be monitored and is versatile in its application to various biological and clinical samples.
Keywords: Oxysterol, Cholesterol, Subcellular, Metabolism, Monocytes, Neuroblastoma, Mitochondria, Blood, Liquid chromatography-mass spectrometry, Whole cell, Dihydroxycholesterol, Peripheral blood mononuclear cell, Brain oxysterol
Highlights
-
•
New method for extraction and quantification of mono and dihydroxysterols from mitochondria.
-
•
Method offers high sensitivity and selectivity allowing quantification of oxysterols at ≥5pg.μl−1.
-
•
Successful application to tissue, primary cells and cell lines.
-
•
Common oxysterol metabolites are present in blood and brain cell mitochondria.
1. Introduction
Oxysterols are biologically important molecules that regulate cell signalling, contribute to regulating cholesterol homeostasis and are essential for bile acid and steroid hormone biosynthesis [1]. Oxysterols are formed by oxidation and addition of hydroxyl groups onto cholesterol hydrocarbon rings and side chains [2,3]. Enzymatic catalysed additions of hydroxyl to the side chain by cytochrome P450 (CYP) enzymes generate mono- and di-hydroxysterols; 24(S)-hydroxycholesterol (24OHC), 25-hydroxycholesterol (25OHC), 27-hydroxycholesterol (27OHC), 20-hydroxycholesterol, 22-hydroxycholesterol, 7α-24(S)-dihydroxycholesterol (24SdiOHC), 7α-25-dihydroxycholesterol (25diOHC) and 7α-27-dihydroxycholesterol (27diOHC) [4]. Hydroxylation of the hydrocarbon rings occurs by oxygen free radical attack forming hydroxylated sterols 7α-hydroxycholesterol (7αOHC) and 7β-hydroxycholesterol, oxysterols with a ketone group 7-ketocholesterol (7ketoOHC), epoxy cholesterols [5β, 6β-epoxy cholesterol (5β, 6β-epox), 5α, 6α-epoxy cholesterol (5α, 6α-epox) and cholestan-3β, 5α, 6β-triol [5,6].
The cellular localisation of enzymes responsible for oxysterol generation is critical to local oxysterol homeostasis [2]. In addition, the redox states of subcellular compartments are different; the endoplasmic reticulum (ER) has a strong reducing environment to enable protein folding, whereas the mitochondria stores glutathione but is exposed to greater potential for oxidation from reactive oxygen species (ROS) leakage by the electron transport chain, increasing the potential for local cholesterol autoxidation [2]. Interaction between endosomes, ER, mitochondria, cell membrane, peroxisomes and lysosomes is necessary for cholesterol and oxysterol trafficking and sterol homeostasis intracellularly; for instance, cholesterol is transported to mitochondria from the ER either by direct membrane contact between ER and mitochondria, or mobilised in lipid droplets (cholesterol esters) [7,8]. In mitochondria, the steroidogenic acute regulatory protein (StAR) transports cholesterol from the outer to inner membrane and controls mitochondrial sterol trafficking [7,9]. Less is known about oxysterol trafficking between the subcellular compartments [2].
Changes to oxysterol levels are implicated in the pathogenesis of several diseases and targeting cholesterol and oxysterol dyshomeostasis may provide a platform for development of novel therapeutics. Dysregulation of cholesterol metabolism has been associated with metabolic diseases such as cancer, heart and liver diseases and motor neurone diseases [2,10,11].
Oxysterols are known to be cytotoxic at high concentration; accumulation of 7-ketoOHC in lysosomes triggered membrane destabilization and induced cell death in human monocytic U937 cells [12]. At a subcellular level, accumulation of 7-ketoOHC and 7β-hydroxycholesterol by cholesterol autoxidation caused redox imbalance and peroxisomal dysfunction in nerve cells [13,14]. Mitochondrial dysfunction and aberrant oxysterol metabolism have been recognized in the pathogenesis of diseases such as atherosclerosis, carcinogenesis and multiple neurodegenerative diseases including Alzheimer's disease [[15], [16], [17], [18], [19], [20]]. In vitro studies have shown that the addition of 7β-hydroxysitosterol and 7β-OHC induced loss of mitochondrial membrane potential, membrane permeabilisation, oxidative stress and apoptosis in human colon cancer, Caco-2 cells [21]. Again, high concentrations of 24OHC caused necroptosis in human neuroblastoma SH-SY5Y cells whereas sublethal concentrations of 24OHC induced protection against cytotoxic stress in these cells [22,23]. Similarly, accumulation of 25OHC has been shown to inhibit inflammasome activation in macrophages [24]. In neurodegenerative conditions, such as Alzheimer's disease, oxysterol levels are linked to neuroinflammation, mitochondrial oxidative damage and cell death [[25], [26], [27]]. Thus, altered oxysterol metabolism at both cellular and subcellular levels associates with altered cell function and the pathogenesis of several chronic diseases. To improve understanding of any contribution of oxysterols to disease and in particular, to explore the mitochondria-oxysterol association and role in disease, there is a need to reproducibly measure oxysterols in different subcellular compartments.
Oxysterols are ~10–1000 fold lower in abundance as compared to cholesterol in cells and biological fluids [2,28]. The structural and chemical properties of oxysterols are similar to each other and pose a challenge in detection and quantification using analytical techniques [2,29,30]. Analysis of oxysterols has been performed by both gas chromatography-mass spectrometry and high performance liquid chromatography-mass spectrometry (HPLC-MS) with the latter providing more sensitive targeted detection of sterols with the advanced technique of multiple reaction monitoring (MRM) or MS/MS that tracks precursor to product ion formation [[31], [32], [33]]. Oxysterol quantification using LC-MS/MS has emerged as a popular analytical method, developed and adopted by many research groups. A number of methods have been described to date for measuring oxysterols in human plasma, serum, and cerebrospinal fluid, and these methods involve derivatisation of oxysterol moieties for subsequent quantifications [[34], [35], [36]]. Dias et al., 2018, developed a multistep LC gradient for efficient direct MS detection and quantification of oxysterols from human plasma reducing the risk of artefactual oxidation during sample processing [29]. The application of LC-MS methods for cellular oxysterol analysis is an emerging field and has recently been applied in dietary intervention studies [29,37,38], for example Beck et al., 2018 isolated and quantified oxysterols from carp cell lines, showing an increase in cellular 25diOHC after exogenous supplementation with 25OHC.
Oxysterol content also differs according to the cell type, tissue and organism [2]. The different structural composition of discrete cells, tissue and biological fluids requires that the methods for isolation of lipids and oxysterols are tailored for the biological sample under study. To date, no comprehensive method for oxysterol quantification at cellular and subcellular levels has been reported. In this study, we describe the isolation of mitochondria from whole cells, followed by isolation of lipids and oxysterols from mitochondria. We adapted our LC-MS/MS method for characterisation of mono and dihydroxycholesterols from mitochondria.
Here we have compared our newly developed method to two previously published methods for extraction of lipids in mitochondrial isolates from tissues by Bird et al., 2013 [38] and for measuring free oxysterols in plasma by Dias et al., 2018 [29]. Our newly developed method shows improved recovery, selectivity and highly sensitive detection and quantification of oxysterols from discrete cellular compartments. Oxysterol distribution in whole cell and mitochondrial isolates were profiled in THP-1 monocytes, SH-SY5Y neuroblastoma cell and peripheral blood mononuclear cells (PBMC) isolated from whole blood of healthy individuals. Our analysis showed differential oxysterol distribution across cell types indicating cell and tissue specific oxysterol metabolism. Furthermore, measurement of mitochondrial specific oxysterol profile indicated local metabolism of oxysterols that was distinct from the whole cell and endoplasmic reticulum. This method enables quantification of oxysterols in mitochondria to elucidate compartment specific metabolism in clinical samples.
2. Materials and methods
2.1. Ethical statement for blood and mouse tissue samples
Blood samples were collected from healthy volunteers with ethics committee approval and with informed consent.
The mouse experimental procedures were conducted under the State Council of the People's Republic of China (Decree No. 2 of the State Science and Technology Commission) October 31, 1988. Amendment Regulations and approval of the local ethics committee for use effective March 1, 2017. The experimental procedures in the University of Surrey received ethical clearance under NASPA ethical review assessment under reference number NERA-2017-011-Bio. Effective date September 26, 2019.
2.2. PBMC isolation from blood
2.2.1. Blood samples
Blood samples were collected from healthy individuals and were kept 1 h at room temperature or 24, 48 and 72 h at 4 °C. Whole blood (10ml) was diluted 1:1 with RPMI + GlutaMAX, layered onto Histopaque-1077 (Sigma) and centrifuged at 1000g for layer separation. The PBMC layer was retained, washed and resuspended in 9ml STE buffer (250mM Sucrose, 5mM Tris and 2mM EGTA, pH7.4)/0.5% BSA (purchased from Sigma Aldrich)).
2.3. Cell culture
The human monocytic THP-1 cell line (ATCC TIB-202) was grown in RPMI 1640 medium supplemented with 10% heat inactivated fetal bovine serum (FBS) at 37 °C, 5% CO2 and 95% humidity. Reagents were purchased from Sigma Aldrich. SH-SY5Y cells were grown in Dulbecco's modified Eagle's medium/F-12 with GlutaMAX supplemented with 10% (v/v) foetal bovine serum, and penicillin (100U/ml) and streptomycin (100μg/ml). Cell cultures were grown to 6 × 15cm plates at 80–90% confluency and harvested through trypsinisation. The cell pellet was washed in DMEM/F12 media before resuspending in STE/0.5% BSA and proceeding with homogenisation and mitochondrial isolation as described above for blood samples.
2.4. Subcellular fractionation and mitochondrial isolation
SH-SY5Y cells (107 cells) were trypsinised and collected by centrifugation at 1000g for 5 mins. PBMCs, THP-1 monocytes and SH-SY5Y cells were washed with phosphate buffer saline prior to subcellular fractionation and mitochondrial isolation using a method developed by Kappler et al., 2016 [39]. Cells were re-suspended in 9ml STE +0.5% BSA (250mM Sucrose, 5mM Tris and 2mM EGTA, pH7.4)/0.5% BSA) buffer and placed on ice prior to homogenisation using a loose fitted 40ml Wheaton Dounce tissue grinder containing a further 9ml of buffer. 1ml of the cell homogenate was collected separately for whole cell (WC) analysis with the remaining homogenate centrifuged at 1000g for 10 minutes at 4 °C, with pellets re-suspended and this step repeated. A crude mitochondrial pellet was obtained by centrifugation of the supernatant at 10,400g for 10 minutes at 4 °C. To gain a purer mitochondrial fraction this pellet was resuspended in 200μl STE, layered onto 25% Percoll gradient (25% Percoll (GE Healthcare), 25% 2xSTE and 50% STE + 0.5% BSA) and ultracentrifuged at 80,000g for 20 minutes at 4 °C. The resultant ER/mitochondria and enriched mitochondrial layers were extracted, washed with STE followed by PBS, resuspended in molecular grade water and stored at −80 °C. For PBMCs and SH-SY5Y cells, three fractions mitochondrial (M), ER/M and WC were analysed. The following THP-1 monocyte subtractions, WC, M, ER/M and WCR (remaining WC fraction pellet without M and ER/M, obtained after first centrifugation step of the homogenate), were used for method development and validations.
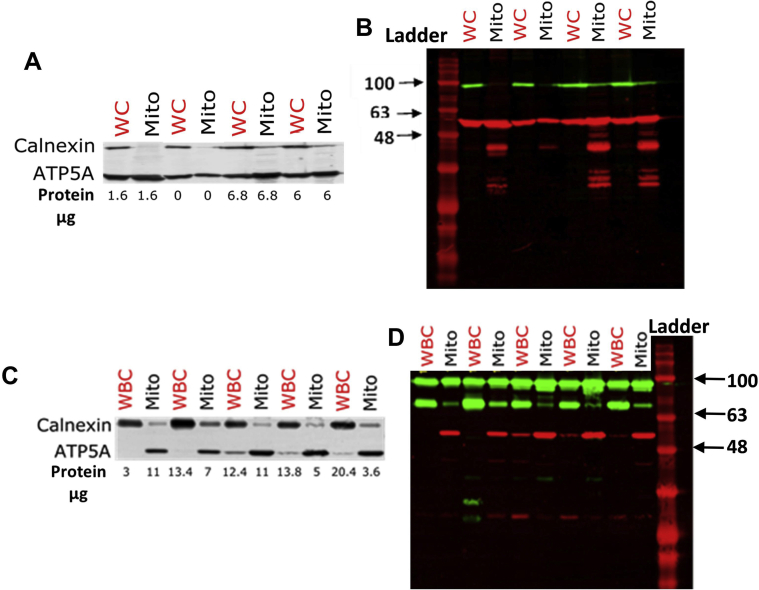
2.5. Western blot analysis of mitochondrial isolates
Protein quantification was performed using a Pierce bicinchoninic acid protein assay kit (ThermoFisher), with electrophoresis using SDS-PAGE and immunodetection using calnexin (Abcam, ab22595) and ATP5A (Abcam, ab14748) followed by fluorescent DyLight secondary antibodies (Invitrogen, SA5-10036 and 35519) and visualisation using a CLx imaging system scanner (LI-COR Bioscience), using Geneflow BLUeye protein ladder (245kDa) ladder (Supplementary Fig. 1).
2.6. High fat diet-fed mouse experiments and brain tissue sections
Four weeks old C57BL/6 male mice were maintained in Nanjing- China SPF animal facility on a 07.00–19.00 day/light cycle at 20–22 °C with food and water ad libitum. They were housed four in a cage and fed either control (normal) diet or high fat diet (HFD) ad libitum for four weeks. The body weight of the mice was taken every week. The animals in HFD treated group were fed a 60% fat, 20% carbohydrate and 20% protein per kcal% while the animals in the control group received a 10% fat, 70% carbohydrate and 20% protein per kcal%. At the end of their respective periods of feeding mice were culled through cervical dislocation for tissue collection. Isolated serum was aliquoted, snap frozen and stored at −80 °C freezer. Brains were removed, and with systematic sampling, part of the tissues were snap frozen and stored at −80 °C. Brain tissues (n = 5) from control and high fat diet (HFD) mice were sectioned into ~50mg hemispheres, weighed and used for lipid extraction.
2.7. Isolation of lipids from tissues, cell lines and blood PBMCs
Sample lysis: Mouse tissue (10–50mg) was weighed and re-suspended in 70μl of 0.1% Triton X-100, 40μl dimethylsulfoxide (DMSO) and 5 μl of butylatedhydroxytoluene (BHT) (90 mg ml−1) as an antioxidant. Tissue sections were disrupted using a handheld rotor-stator homogenizer TissueRuptor II for disruption of tissues sections. The homogeniser probe was cleaned with ethanol/water- 30 seconds cycle and tissue disruption was carried out for 30 seconds (until the material homogenised), followed by cleaning of the probe. Whole cells (THP-1 monocytes, neuroblastoma SHY-5Y cell lines and blood PBMC), mitochondria and endoplasmic reticulum/mitochondria fractions were re-suspended in lysis buffer 70μl of 0.1% Triton X-100, 40μl DMSO and 5 μl of BHT (90 mg ml−1).
Homogenised tissue and cellular fractions were sonicated in the lysis buffer for 20 minutes. Standards and quality controls were prepared by adding a range of concentrations of each of the 9 authentic oxysterols (0, 0.005, 0.010, 0.025, 0.050, 0.075, 0.1, 0.25, 0.5, 0.75 and 1ng.μl−1) within a mix into 30μl of whole cell lysate used as the matrix, followed by addition of lysis buffer and sonication for 20 minutes. 3 μl of 22SOHC-D7 (50ng.μl−1) external standard was added to the samples and standards and vortexed. Lipid extraction was carried out in fume hood following the method of Bird et al., 2013, with modifications adapted for this work. 190μl of LC-MS grade methanol (MEOH) and 380μl of dichloromethane (DCM) were added to the 110μl− 1 sample lysates and standards and vortexed for 20 seconds. 120μl of LC-MS grade water was added to the samples and standards, vortexed for 10 seconds and the tubes were allowed to stand at room temperature for 10 minutes. Samples and standards were centrifuged at 7168g, 8 °C for 10 minutes. The upper and interphase phase of extract was collected separately, dried and used for protein analysis. The lower DCM layer ~350 μl with the lipid extract was collected separately and dried under N2 gas using Turbovap LV purchased from Biotage. A fraction of the lipid extract (15μl) was collected separately and dried for cholesterol quantification.
2.8. Oxysterol extraction
The dried DCM lower phase was reconstituted in 500μl of MEOH and 1.5ml of LC-MS grade water containing 0.1% formic acid and mixed well. A vacuum manifold connected to a multichannel filtration apparatus was used for the extraction of oxysterols. Oasis HLB solid phase extraction (SPE) cartridges (Waters) were used for selective separation of oxysterols from the lipid extracts. Cartridges were fitted into the vacuum manifold and primed with 800μl MEOH and 600μl H2O + 0.1% formic acid. Samples were added slowly to the cartridges and the flow through was discarded. Samples were washed with 600μl H2O + 0.1% formic acid and 600μl hexane to remove polar non-binding analytes and excess cholesterol. Butyl acetate (1ml) was added to the cartridges and eluates were collected in a fresh tube with a controlled flow of 1 drop per second. Butyl acetate eluates were dried under N2 gas and reconstituted in 40μl of 40% MEOH containing 0.1% formic acid.
2.9. LC-MS/MS analysis
Standards (10μl) and samples (20μl) were injected into a NUCLEOSIL C18 column (100-5 125/2) fitted with a guard column for the liquid chromatography (LC) analysis. Our previously developed solvent composition in Dias et al., 2018 was used for separation of oxysterols. Solvent A is 70% MEOH, 10% water, 0.1% formic acid and solvent B is 90% isopropanol, 10% MEOH and 0.1% formic acid. A multistep solvent gradient was applied using ACQUITY Ultra performance liquid chromatography (UPLC) quaternary system purchased from Waters. The LC method used here was developed previously by Dias et al., 2018, and was set to 0 minutes 16% B- 7 minutes 16% B; 7 minutes 16% B-11 minutes 24% B; 11 minutes 24%B-25 minutes 100%B; 25 minutes 100%B-30 minutes 100% B; 30 minutes 100% B- 32 minutes 16% B and was held at 16% B up to 48 minutes. Mass spectrometry analysis was performed using a Xevo TQ-S Triple Quadrupole Mass Spectrometer (Waters) and operated with the positive electrospray ionisation (ESI) mode. Nitrogen gas is used for desolvation at 500 °C and a flow rate of 900 L/hour−1 and collision gas argon at a flow rate of 0.15 ml/minute−1 for collision. MRM transitions were set up using authentic oxysterol standards (Avanti Polar) lipids. Standards include- 24(S) hydroxycholesterol (700061P), 25-hydroxycholesterol (700019P), 27-hydroxycholesterol (700021P), 7α-hydroxycholesterol (700034P), 7 ketocholesterol (700015P), 7α,27-dihydroxycholesterol (700136P), 7α,24 (R/S)-dihydroxycholesterol (700119P), 7α,25-dihydroxycholesterol (700078P). The mass spectrometry method was set up with the MRM transitions for 9 analytes using the Intellistart feature in Xevo and are detailed in Table 1. The two best MRM transitions were identified for each analyte-the most abundant MRM as the quantifier and the second most abundant was used as the qualifier. Authentic oxysterols were dissolved in 50% methanol, 0.1% formic acid and working stocks were prepared in a range of 10ng.μl−1 to 50ng.μl−1. Analytes were infused directly into the MS system and transitions were determined manually by inspection of the chemistry of the analyte and optimisation of the ionisation parameters using the inbuilt Intellistart feature of the Xevo TQs system. Infusions were performed at 10–20μl min−1 and the cone voltage and collision energy were optimised in order to obtain the 5 best MRM transitions. 7α,24Sdihydroxychoelsterol (24SdiOHC) and 7α,25Sdihydroxychoelsterol (25diOHC) had identical MS/MS transitions and co-eluted with identical retention times. So, these two analytes were analysed together as 24S25diOHC for detection and quantification. For every batch of standards and samples, an unprocessed standard mixture and pooled samples were used as the quality control for the analyses.
Table 1.
MRM transitions for oxysterols.
| Analyte | Quantifier transition | Qualifier transition | CV | CE | Retention time (min) |
|---|---|---|---|---|---|
| 7α hydroxycholesterol | 367.35 -> 147.42 | 367.35 -> 159.12 | 94 | 22, 22 | 18, 20.70 |
| 22(S)-hydroxycholesterol-d7 | 392.19 -> 159.11 | 392.19 -> 105.4 | 2 | 24, 44 | 15.15 |
| 7α,24(S)dihydroxycholesterol | 383.11 -> 81.05 | 383.11 -> 105.03 | 38 | 30, 42 | 6.08, 10.20 |
| 7α,25dihydroxycholesterol | 383.31 -> 81.05 | 383.31 -> 95.09 | 12 | 30, 30 | 6.04, 10.62 |
| 7α,27dihydroxycholesterol | 401.43 -> 159.05 | 401.43 -> 81.06 | 40 | 24, 34 | 6.83, 11.97 |
| 24(S)hydroxycholesterol | 367.35 -> 95.03 | 367.35 -> 147.09 | 12 | 30, 24 | 15.98 |
| 25 hydroxycholesterol | 367.35 -> 147.09 | 367.35 -> 158.54 | 40 | 24,18 | 16.34 |
| 27 hydroxycholesterol | 385.35 -> 81.06 | 385.35 -> 95.09 | 48 | 30,26 | 17.05 |
| 7-ketocholesterol | 401.43 -> 95.05 | 401.43 -> 109.48 | 44 | 36, 34 | 18.75 |
2.10. Cholesterol and protein quantification
Cholesterol content in the lipid extract was quantified using the Invitrogen™ Molecular Probes™ Amplex™ Red Cholesterol Assay Kit (10236962) with 96 well plate assay set up that detects free and esterified cholesterol released from hydrolysis of cholesteryl esters. Protein was quantified using Pierce Bichinchonic acid assay kit (ThermoFisher) following the manufacturer's protocol. Fluorescence (excitation-525 nm, emission- 585 nm) for Amplex™ Red cholesterol assay kit and absorbance (562 nm) for BCA assay kit were measured using SpectraMax i3x microplate reader and SoftMax Pro Software (Molecular Devices). The lower limit of quantitation (LLOQ) for cholesterol using the cholesterol assay kit and for protein were verified to be 0.08ng.μl−1 and 5ng.μl−1 respectively.
2.11. Oxysterol quantification and data analysis
Mass spectrometry data was processed using MassLynx and TargetLynx software (Waters). Oxysterols were identified by comparing its retention time, exact mass, and MS/MS spectra to that of its authentic standard. To check the extraction efficiency and recovery of oxysterols from the cellular matrix, authentic standards were used. Authentic standards were mixed with cellular lysate prior to oxysterol extraction and LC-MS/MS analysis. A blank sample consisting of cell lysate and no authentic standard added was analysed in parallel to correct for interference from endogenous analytes. Peak Detection and integration was performed with ApexTrack integration feature built in MassLynx. ApexTrack integration automatically determines appropriate integration parameters for peak width and threshold. Peak areas were extracted for the quantifier MRM transition for each analyte. Standard curves were generated using the authentic oxysterols with concentrations 0, 0.005, 0.010, 0.025, 0.050, 0.075, 0.1, 0.25, 0.5, 0.75 and 1 ng μl−1. Linear regression analysis was done to derive the equation for the line of best-fit and the concentration of oxysterols in samples was calculated from this equation. The limit of quantification (LOQ) was determined as per the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) guidelines of LLOQ defined as the lowest point on a calibration curve accepted based on an analytical precision cut-off of a 20% relative standard deviation. The lowest calibration curve point with co-efficient of variation <20% was evaluated from six independent measurements (two replicates for each) done on different days was set as the LLOQ. Measurements below lower limit of quantification (LLOQ) were assigned LLOQ/2 values, following the models developed by Keizer et al., 2015 and Beal, 2001 to incorporate concentration data below the limit of quantification [40,41]. Graphs and statistical analysis including two-tailed student's t-tests, ANOVA analysis and regression analysis were performed using Graphpad prism 8.0 and in Microsoft excel. Principal component analysis (PCA) was performed using IBM SPSS statistical analysis software.
3. Results
3.1. Extraction and characterisation of oxysterols from whole cell and mitochondria
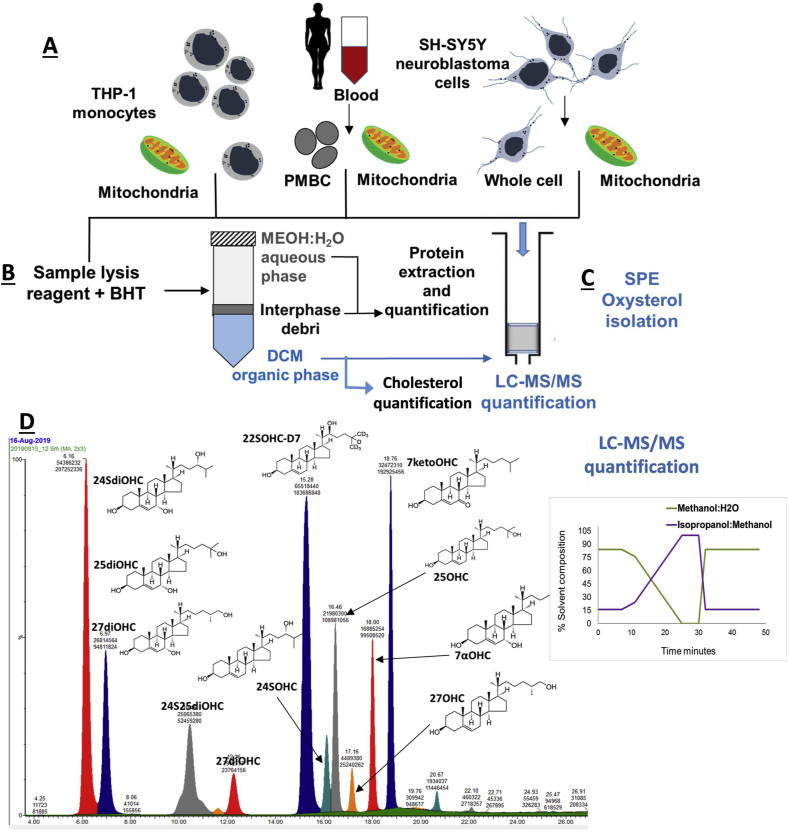
Cellular/subcellular fractions were generated from THP-1 monocytes, PBMC and SH-SH5Y neuroblastoma cells for investigation (Fig. 1A). Fraction enrichment was confirmed by western blotting using calnexin (ER) and ATP5A (mitochondrial) markers (Fig. S1). Next, we isolated lipids and oxysterol pools from unfractionated whole cells and subcellular fractions (Fig. 1B and C) with authentic standards incorporated into whole cell (WC) lysate to mimic the effect of a cellular matrix, which may impair extraction efficiency. Eight authentic oxysterol standards were used (Table 1) and 22S-hydroxycholesterol-d7 (22SOHCD7) as the external standard for setting up the LC-MS/MS quantitation methodology (Fig. 1D).
Fig. 1.
Methodology-work flow for isolation of lipids and oxysterols from cellular and subcellular compartments. Three cell types - THP-1 monocytes, peripheral blood mononuclear cells (PBMC) and SH-SY5Y neuroblastoma cells-were used for this analysis. The work flow involves stage A - isolation of mitochondria from whole cells, stage B - sample lysis and lipid extraction-cholesterol and protein measurements, stage C - oxysterol isolation using solid phase extraction (SPE) followed by LC-MS/MS profiling using a multistep gradient of solvent A - 70% methanol (MeOH) + 30% H2O + 0.1% formic acid and solvent B - 90% isopropanol (IPA) + 10% MeOH and 0.1% formic acid.
The first extraction method M1, which had been established for human plasma [29], involved protein precipitation with methanol followed by oxysterol isolation using solid phase extraction (SPE) with polymeric reversed-phase sorbent. We developed a MS/MS method using a Waters Triple quad mass spectrometer to detect both mono and dihydroxycholesterols using the MRMs in Table 1. MRMs were set up by direct infusion of authentic standard into the mass spectrometer followed by optimisation of cone voltage and collision energy to obtain two best transitions for each of the nine oxysterols.
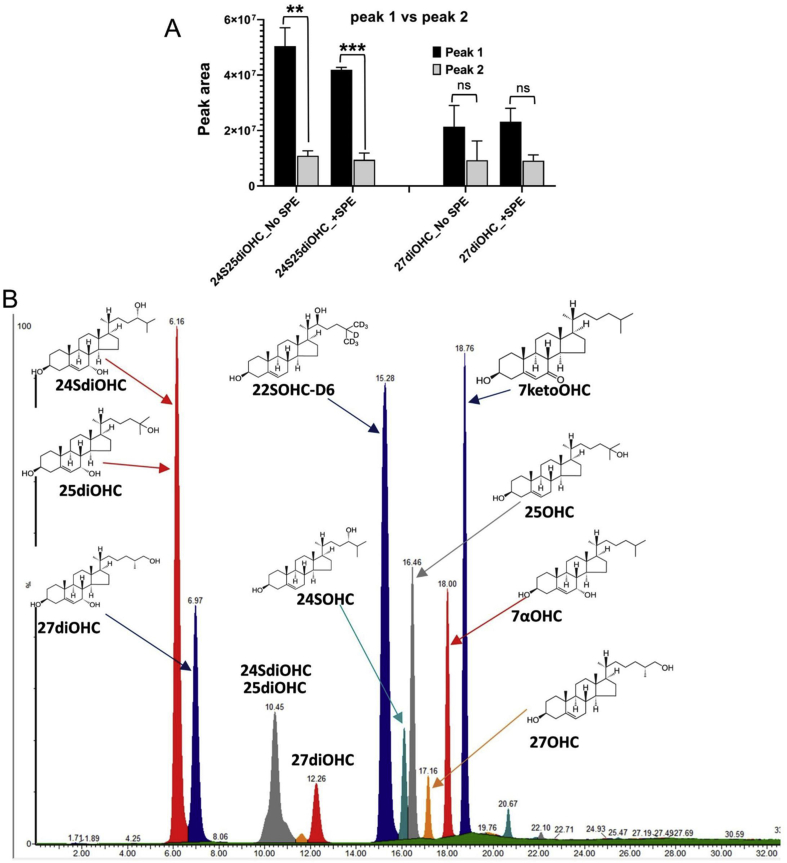
We employed our previously developed multistep gradient LC method using a C18 reverse phase (RP) column [29] for chromatographic separation of oxysterols (Fig. 1D). The three dihydroxysterols analysed by our LC-MS/MS produced two chromatographic peaks each due to their isomeric features (Supplementary Figs. S2A and B). The two-peak feature was also observed with the 7α-dihydroxycholesterols reported by others using LC analysis [42,43]. The first chromatographic peak was the dominant one for the dihydroxycholesterols and as used in peak area analysis (Fig. S2A). The second peaks (peak 2) for 24S25diOHC and 27diOHC were 3/7th and 5/9th of the first peak respectively. We chose the first dominant peak for quantification because of the significantly larger peak area as compared to the second peak. This choice avoids the risk of losing quantification because of the non-detectability of the second peaks in samples with relatively low concentration of the dihydroxycholesterols.
3.2. Extraction efficiency and recovery of oxysterols from cellular matrix
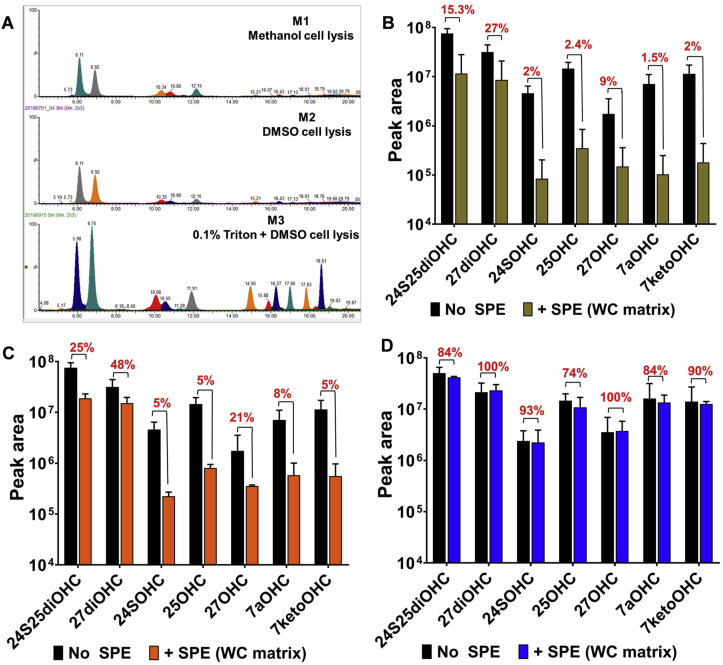
Method M1 (our previously reported method for plasma) led to poor chromatographic detection of oxysterol authentic standards within the cellular matrix (Fig. 2A). The maximum recoveries for dihydroxysterols and monohydroxysterols from the cellular matrix using M1 were 27% and 9% respectively relative to the neat authentic standards analysed without cellular matrix and SPE extraction (Fig. 2B). We next explored method M2 for lipid extraction from rat liver tissues published in Bird et al., 2013 [38]. This method involved isolation of lipid pools following cell lysis in dimethylsulfoxide (DMSO), followed extraction of lipids by methanol (MeOH):dichloromethane (DCM) (1:2) solvent extraction. We extracted oxysterols from the total lipids using a solid phase extraction (SPE) separation technique as described in Dias et al., 2018. M2 also gave poor recovery of oxysterol standards as seen from chromatograms (Fig. 2C). M2 gave a maximum of 48% for dihydroxysterols and 21% for monohydroxysterols (Fig. 2C). We speculated the poor recovery of oxysterols was due to the incomplete cell lysis and poor lipid extraction. Method M3 yielded well resolved oxysterol peaks and improved chromatographic separation (Fig. 2A). We used 0.1% Triton X-100 and DMSO as detergents and BHT as an antioxidant during cell lysis. LC-MS/MS analysis showed enhanced recovery of both mono and dihydroxysterols standards in WC matrix using the Triton cell lysis method (M3) (Fig. 2A and D, Fig. S2B). The recoveries of both mono and dihydroxycholesterols ranged from 74 to 100%.
Fig. 2.
Comparison of oxysterol recovery and matrix effects from three methods. (A) Three methods; M1 (methanol (MeOH) cell lysis and SPE oxysterol extraction, M2 dimethylsulfoxide (DMSO) cell lysis, methanol:dichloromethane (MeOH:DCM) lipid isolation and SPE oxysterol extraction, M3 (0.1% Triton X-100 + DMSO cell lysis, MeOH:DCM lipid extraction and SPE oxysterol extraction) were compared. Authentic standards were prepared in 40% methanol 0.1% formic acid (without cellular matrix and no SPE extraction) and used as the control (black bars). M3 improved chromatographic resolution of 10ng authentic oxysterol standards mixed with whole cell lysate as the matrix and isolated using the methodology described in Fig. 1 (B), (C) and (D) compares the recovery (ratio of +SPE peak area/No SPE peak area) expressed as % for the authentic standards in matrix using extraction methods M1 [29], M2 [38] and M3-developed in this work. M3 showed >70% recovery for all nine oxysterols. Measurements were done using standards at 1ng.μl-1 and are mean ± S.D. of 3–4 independent replicates.
3.3. Method validation for subcellular oxysterol quantitation
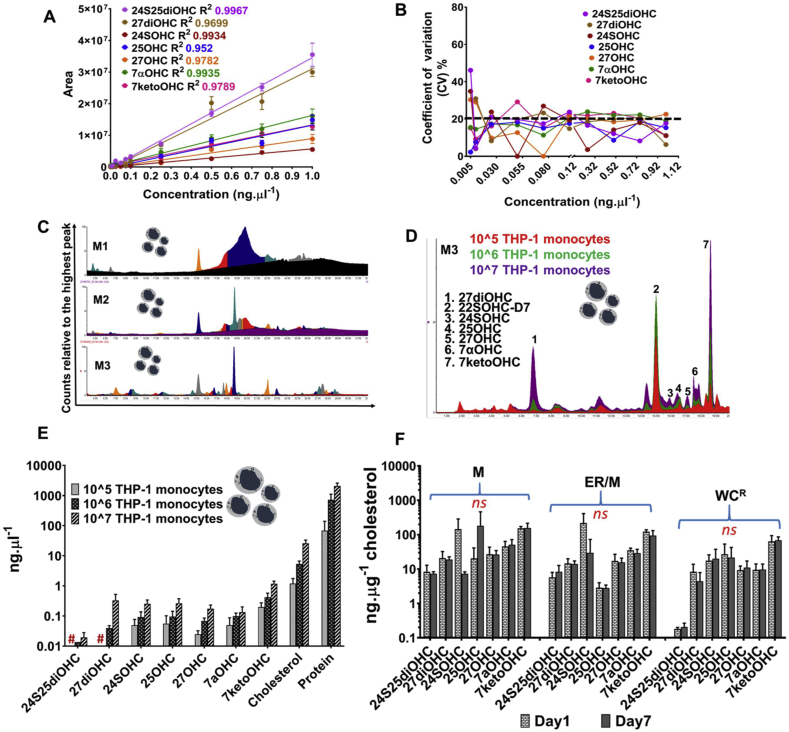
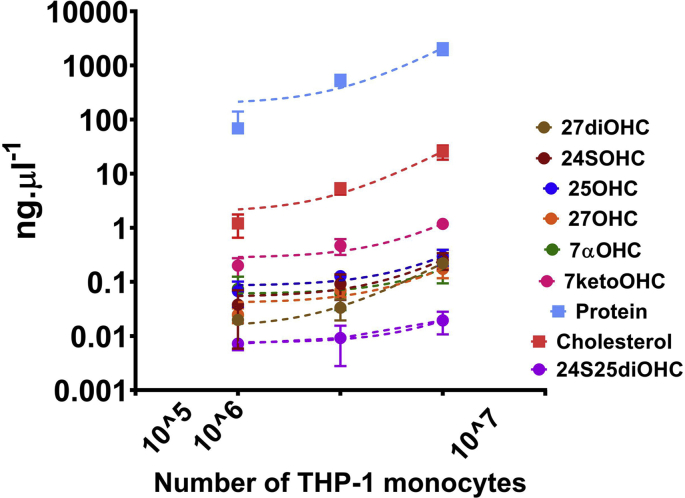
We used method M3 to generate standard curves with authentic oxysterols mixed in WC lysates in concentrations ranging from 5 pg. μl−1 to 1 ng μl−1. Regression analysis produced an R2 ≥ 0.9 for all oxysterols confirming linearity between concentration of an analyte and its peak area (Fig. 3A). The co-efficient of variation (CV) between the standard concentrations measured intra and inter days was calculated. The lower limit quantification (LLOQ) was set up with the lowest standard measurements that had a CV of <20% across intra- and inter-day measurements (Fig. 3B). Precision of the standard curves were monitored with quality control (QC) samples consisting of eight oxysterols that showed a CV of ≤20%. THP-1 monocytes were used to set up the quantitation and for reproducibility analysis. Fig. 3C compares the chromatographic resolution of oxysterols from THP-1 monocytes using the three methods. Method M3 showed the highest extraction efficiency of oxysterols. We then checked the linearity of the LC-MS/MS method by quantifying oxysterols from THP-1 monocytes with varying cell numbers. We used 105, 106 and 107 THP-1 cells as the starting material for extraction and quantification of oxysterols, cholesterol and protein (Fig. 3D and E, Supplementary Fig. S3). There were linear relationships between the number of monocytes used as the starting material, and monohydroxycholesterol, cholesterol and protein concentrations, implying 105 monocytes was the lower acceptable limit of starting material for detection of monohydroxycholesterols (Fig. 3E). However, 105 monocytes were insufficient for detection of dihydroxycholesterols (Fig. 3E). The limit of starting material for quantification of the three dihydroxycholesterols analysed here was 106 monocytes.
Fig. 3.
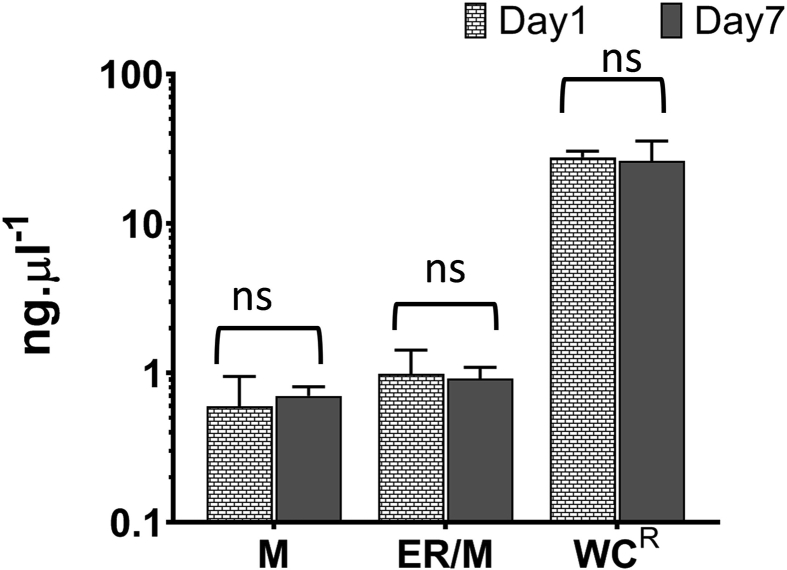
Method validation for oxysterol quantification in THP1 subcellular fractions. (A) Standard curve and R2 goodness-of-fit of the curve for oxysterol standards produced using method M3. (B) Co-efficient of variation of standard curve and identification of lower limit of quantification (LLOQ) and upper limit of quantification (ULOQ). LC injections for the standards were 10μl and the values are mean ± S.D. of 3–4 independent extractions and LC-MS/MS analysis. (C) Chromatographic resolution of oxysterols extracted from THP-1 monocytes using the methods M1, M2 and M3. (D) Chromatography of mono and dihydroxycholesterols from monocytes ranging from 105 to 107. (E) Quantification of oxysterols, cholesterol and protein in THP-1 monocytes and their linearity with number of cells used for extraction and LC-MS/MS analysis. # indicates the amount detected was close to the LLOQ and are unreliable measurements. (F) Reproducibility analysis of oxysterol extraction and quantitation. Data shown are mean ± S.D. of biological replicates (n = 3–5). Student's t-test (cut off *, p ≤ 0.05) was used to check statistically significant differences in the measurements (absolute and log transformed); ns denotes not significant.
We tested the reproducibility of oxysterol extraction and quantification in subcellular fractions - mitochondria (M), endoplasmic reticulum/mitochondria (ER/M) and remaining cell lysate (WCR) (Fig. 3F). Cellular fractions from 107 THP-1 monocytes were isolated on the same day and stored for two different lengths of time at −80 °C, from one to seven days prior to oxysterol extraction and LC-MS/MS analysis. The amounts of oxysterol were normalised to the cholesterol quantified for each subfraction (Fig. 1). Cholesterol quantitation was performed using fluorometric cholesterol red assay and the LLOQ was verified to be 0.08 ng μl−1. There were no statistically significant differences between the measurements of oxysterols and cholesterol after seven days of storage at −80 °C (Fig. 3F, Supplementary Fig. S4). The amounts of 25OHC and 7ketoOHC, the sterols that can also be generated by cholesterol autoxidation, did not vary significantly between Day 1 and Day 7 indicating negligible autoxidation under the storage conditions for fractionated cells in the presence of BHT and at −80 °C. The data from Day 1 and Day 7 also confirms the inter day precision of oxysterol extraction and LC-MS/MS analysis of samples fractionated and stored for up to one week.
3.4. Application of the method to characterise tissue oxysterols
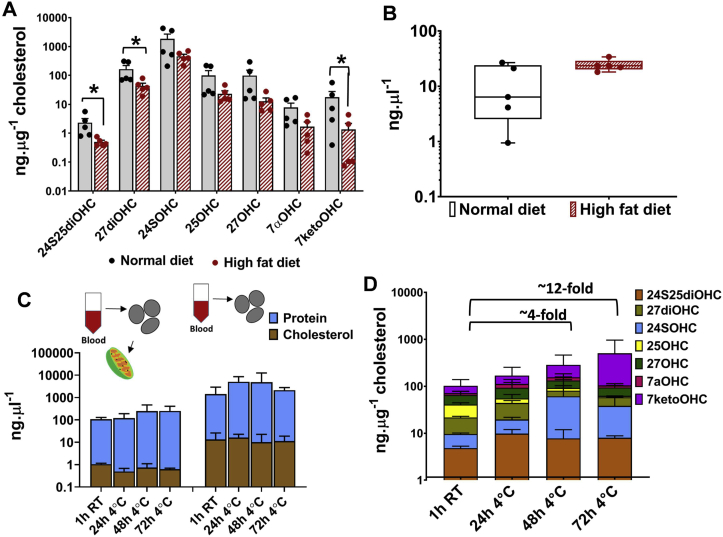
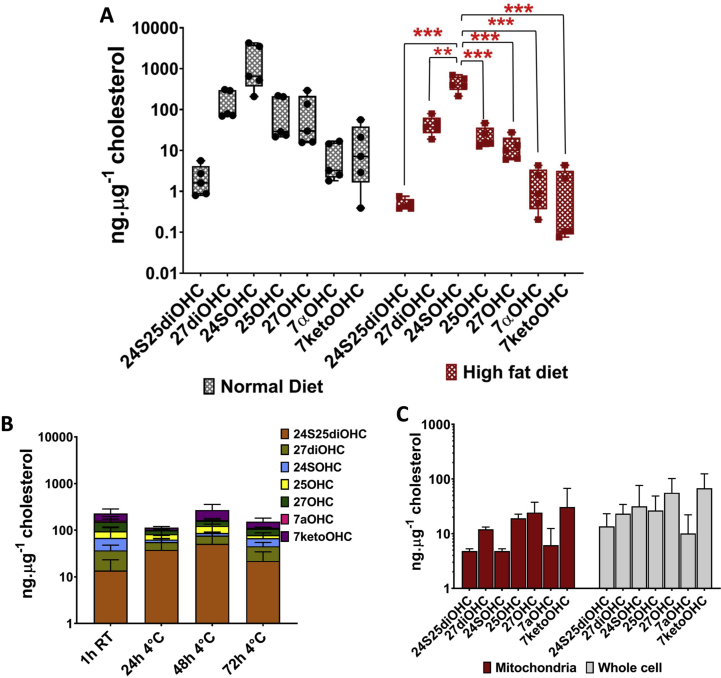
We applied our optimised method M3 for oxysterol extraction and quantitation in tissues including mouse brain and whole blood (Fig. 4). We quantified oxysterols normalised to cholesterol in brains from two groups of mice, one fed on normal chow and one on high fat diet to validate the method (Fig. 4A). We show 24SOHC as the most abundant oxysterol in the brain tissues of both groups of mice, consistent with previous observations [43,44] (Supplementary Fig. S5A). Our data shows 27OHC, 25OHC and 27diOHC as the next most abundant oxysterols, with levels at least 10-fold lower concentrations than 24SOHC (Supplementary Fig. S5A). The monohydroxysterol, 24SOHC, was also the most abundant sterol in high fat diet mice although the corrected levels of 24SOHC were reduced four fold from 6108.6 ± 2535 ng μg−1 cholesterol in normal diet fed mice to 1512 ± 251 ng μg−1 cholesterol in high fat diet mice (Fig. 4A). The brain levels of all oxysterols were lower in high fat diet fed mice with significant decreases in levels of 24S25diOHC, 27diOHC and 7ketoOHC (Fig. 4A). This was not explained simply by the correction used for the elevated cholesterol levels in high fed diet mouse brain (twofold higher than that of the normal diet fed mice) (Fig. 4B). The analysis here established the versatility and applicability of the method to extract and quantify oxysterol from tissue sections and the biologically relevant and reproducible oxysterol distributions obtained from mouse brain.
Fig. 4.
Oxysterol profiles in tissues. (A) Oxysterol profiles in brain tissues of mice fed with normal chow diet vs. high fat diet. Statistical significance was calculated using paired t-test (two-tailed) with α = 0.05; *, P < 0.05. (B) Cholesterol levels in brain tissues of mice fed with normal chow diet vs. high fat diet. Values are mean ± S. D (n = 5). (C) Cholesterol and protein levels in the mitochondria and whole peripheral blood mononuclear (PBMC). (D) Changes in oxysterol levels of mitochondria isolated from blood stored under various conditions. Values are mean ± S.D. (n = 2 to 5). ANOVA analysis and statistical test using Holm-Sidak method, with α = 0.05 without assuming a consistent S. D were performed to check for statistically significant differences in mitochondrial oxysterols isolated from blood stored under the four storage periods- 1 hour/room temperature, 24 hour/4 °C, 48 hour/4 °C and 72 hour/4 °C.
We applied our method to measure oxysterol profiles in whole blood PBMCs and their mitochondrial subfractions to validate the applicability of this method to clinical samples. Blood samples were collected from healthy individuals and were stored under four different conditions- 1h at room temperature, 24h at 4 °C, 48h at 4 °C and 72h at 4 °C prior to PBMC and subsequent mitochondrial isolation. This analysis was used to determine optimal storage conditions for blood samples and to determine whether the various storage and handling conditions introduced variations in the subsequent subcellular oxysterol measurements. Oxysterols were extracted from whole PBMCs and mitochondria harvested from blood stored under the four aforementioned conditions and compared to analyse the effect of storage times on oxysterols. Fig. 4C shows that the protein and cholesterol content in mitochondria and whole PBMC were not affected by storage, however, we observed a trend for increase in the 7ketoOHC profiles in the mitochondrial fraction with the increase in blood storage period (up to 12-fold; Fig. 4D), suggesting that cholesterol autoxidation occurs in mitochondria fractionated from blood leukocytes with increased storage times. This trend of 7ketoOHC accumulation was not observed in whole PBMCs isolated from stored blood (Supplementary Fig. S5B). Also, we did not observe such change in the protein and cholesterol measurements, indicative of the small contribution of oxysterols to total cholesterol.
The analysis here shows variations in subcellular oxysterol levels as a consequence of the experimental conditions used prior to oxysterol extraction, and that these variations were captured by the quantitation method developed in this study. The analysis highlights the importance of a standardised storage condition for analysis of clinical samples to minimise autoxidation. The analysis also showed differential oxysterol metabolism in brain and blood tissues. Unlike brain tissues, for which 24SOHC was the abundant sterol, in PBMC mitochondrial fractions and PBMCs, 27OHC and 7ketoOHC were the most abundant oxysterols indicating differences in oxysterol metabolism between tissues (Supplementary Fig. S5C).
3.5. Application of the method to profile mitochondrial oxysterols of cell lines
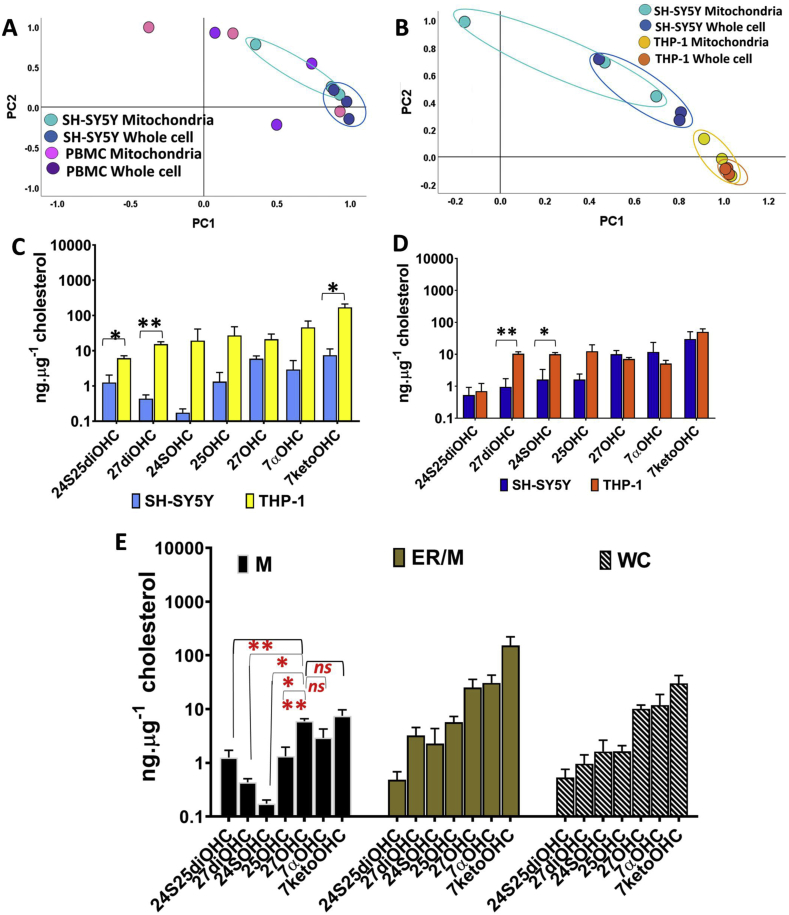
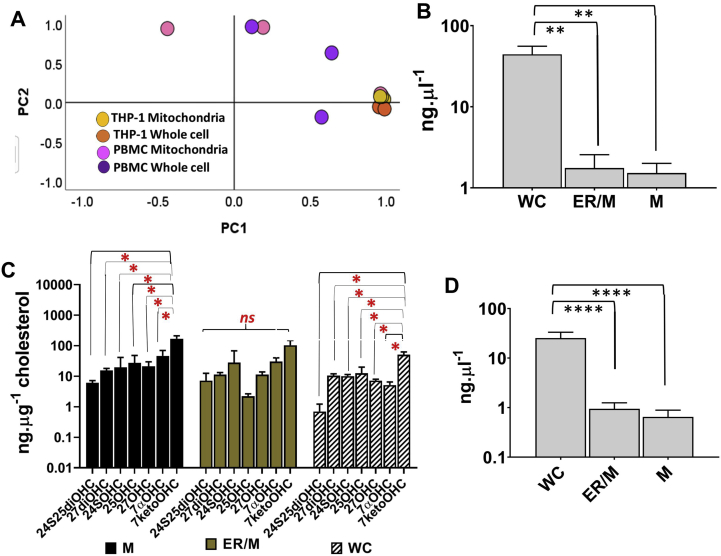
The optimised method M3 was applied to measure oxysterol profiles in mitochondria, ER/mitochondrial and whole cell fractions of SH-SY5Y cells. We compared the mitochondrial and whole cell oxysterol profiles of SH-SY5Y to that of THP-1 monocytic cell lines and PBMCs in order to capture cell lineage specific differences. Fig. 5A and B shows principal component analysis (PCA) plots of SH-SY5Y vs. PBMC and SH-SY5Y vs. THP-1 oxysterols respectively. The oxysterol component profiles of SH-SY5Y were grouped distinctly to that of PBMC and THP-1s respectively. The differences were pronounced in the mitochondrial and whole cell profiles of SH-SY5Y vs. THP-1 (Fig. 5B). In particular, the levels of 24S25diOHC, 27diOHC and 7ketoOHC were higher in THP-1 mitochondrial fractions than the SH-SY5Y mitochondrial fractions (Fig. 5C). In whole THP-1 monocytes, 27diOHC and 24SOHC were higher than that measured in SH-SY5Y cells (Fig. 5D). These analysis shows cell specific oxysterol metabolism with distinct mitochondrial profiles in the two cell types. We compared THP-1 profiles to that of PBMCs by PCA and show distinct grouping of the mitochondrial and whole cell oxysterol profiles from THP-1 monocytes (Fig. S6A).
Fig. 5.
Oxysterol profiles in different cell types. (A) Principal component analysis (PCA) of mitochondrial and whole cell oxysterol measurements of SH-SY5Y neuroblastoma cells vs. blood PBMCs. (B) PCA of mitochondrial and whole cell oxysterol measurements of SH-SY5Y vs. THP-1 monocytes. (C) Mitochondrial oxysterol levels and (D) Whole cell oxysterol levels in SH-SY5Y and THP-1. E, Compartment specific oxysterol profiles in SH-SY5Y. Values are mean ± S.D. (n = 3). Values are mean ± S.D. (n = 3). Statistical significance calculated by unpaired t-test Welch's correction (two-tailed) on log data, ns, not significant, *p < 0.05, **P < 0.005, ***P < 0.0005.
We next explored compartment specific oxysterol metabolism from whole cell (WC), mitochondria (M) and endoplasmic reticulum/mitochondria (ER/M) fractions of SH-SY5Y cells. The levels of total cholesterol were lower in M and ER/M fractions and oxysterol distributions were distinct in the subcellular compartments (Figs. S6B and 5E). For instance, 27OHC concentrations were significantly higher than the other enzymatically derived mono- and di-hydroxycholesterols in mitochondria, while 7ketocholesterol was the dominant oxysterol in ER/M and WC fractions. 24SOHC was the least abundant oxysterol in mitochondria, while 24S25diOHC were the least abundant in ER/M and WC. The overall mitochondrial oxysterol concentrations were lower than in ER/M and WC fractions.
Compartment-specific oxysterol profiles were also determined in THP-1 monocytes and 7ketoOHC was the predominant oxysterol in M and WC fractions (Fig. S6C). Unlike in SH-SY5Y cells, where the overall mitochondrial oxysterol concentrations were lower than in ER/M and WC fractions, the oxysterol levels in the three compartments of THP-1 monocytes were comparable albeit the levels of cholesterol in M and ER/M fractions were over tenfold lower than that in the WC (Fig. S6D). These analyses confirm that oxysterol profiles are distinct between mitochondria, endoplasmic reticulum/mitochondria and whole cells, confirming compartment specific oxysterol metabolism in cells.
4. Discussion
Recent advances in high throughput analytical techniques have enabled measurement of oxysterols in various biological systems [16,32,45]. Our newly developed method allowed simultaneous detection of mono and dihydroxycholesterols across mouse brain tissues, cell lines and blood samples, providing a versatile method that can be applied to various biological samples.
Extraction efficiency and recovery of oxysterols from cells and tissues without artefactual oxidation is critical for quantitation, because endogenous sterols are present in trace amounts in cells and tissues (Dias et al., 2018). Here we used authenticated standards to confirm the extraction efficiency/recovery, which was ≥74% for the nine sterols including the external standard. The use of surrogate heavy stable isotope-labelled standards offers an additional advantage to check and correct for the chromatographic shifts in the peak width and retention times for endogenous analytes. They may be evaluated to replace the external deuterated standard and used for quantitation if they have identical recovery and matrix effects as the analyte of interest. Our previously developed method (Dias et al., 2018) was successfully employed to quantify monohydroxycholesterols in plasma [29]. Although this method allowed >80% recovery of sterols from a plasma matrix, it failed to provide similar recovery of sterols from a cell matrix, confirming the essential requirement to tailor methods for optimal oxysterol extraction efficiency in cells and tissues. Eukaryotic cells are highly compartmentalised systems with cholesterol and oxysterol metabolism localised in specific subcellular compartments [2,46]. Therefore, complete cell lysis and release of oxysterol pools from the cellular niche has required a new rigorous method, as developed in this study, using Triton X-100 for efficient extraction.
Autoxidation of cholesterol during sample processing is a critical factor to account for during sterol analysis [29,47]. Use of antioxidants and also an initial separation step to remove cholesterol is adopted to minimise risk of autoxidation artefacts. Use of SPE columns for cholesterol removal is a widely accepted oxysterol selective technique that was also included in our method [29,30]. A previous study reported the use of SPE silica columns to cause a 1–3% cholesterol autooxidation to 5/6 (α/β)-epoxycholesterol [32]; however, the effect of such oxidation and biological interference was shown to be minimal in our previous study [29]. Also, our current method did not result in any change to oxysterol levels particularly in 7ketoOHC that could be generated by autoxidation from cholesterol during sample storage, handling and extraction process if the cells were stored intact. The extraction and quantitation performed on two different days for THP-1 cells did not lead to any significant variation in subcellular oxysterol distribution.
However, blood stored under four different conditions (time and temperature) prior to PBMC harvest and cellular fractionation, introduced biological variation, including increased cholesterol autoxidation to 7ketoOHC particularly in mitochondria. The effects of processing and handling on clinical specimens have already been recognized and evaluated for serum and plasma samples where standardized procedures are advised to avoid confounding factors and bias for validation studies and biomarker discoveries [48]. Following from our analysis, we stress the practice of uniform clinical sample handling and minimal blood storage time to avoid autoxidation artefacts that may arise e.g. from mitochondrial oxidative stress during storage that was measured here as mitochondrial 7ketoOHC and which was not present in whole cell PBMCs.
The choice of solvent compositions for HPLC sterol analysis affects the signal intensity. Acetonitrile as a mobile phase has negative effects on the electrospray ionisation process and reduction in signal intensity of oxysterols due to adduct formations [32]. Solvent compositions of methanol/water are preferably and widely used for LC oxysterol analyses [3,32,49]. In our method we used a multistep gradient composed of methanol/water/isopropanol that allowed baseline separation of the monohydroxycholesterols and in particular, separation of the two critical oxysterols- 24SOHC and 25OHC [29]. A limitation of existing LC methods is the co-elution of the two dihydroxycholesterols- 24SdiOHC and 25diOHC. These two are difficult to resolve by LC and also by MS/MS because of their identical MRM transitions. The two dihydroxycholesterols could not be separately resolved on the C-18 reverse phase, because of the weaker interaction between these analytes and the column bed. In the future, the baseline resolution of these dihydroxycholesterols may be improved by new alternate column chemistry.
We evaluated the biological validity of our method for oxysterol analysis by measuring oxysterol profiles from mouse brain sections. In alignment with other studies by Griffiths and Wang, 2009 [31], our method also identified 24SOHC as the most abundant oxysterol in mouse brain. Our results also showed significant decrease in 24S25diOHC, 27diOHC and 7ketoOHC levels after an increase in dietary fat intake. Lower concentrations of dihydroxysterols may be indicative of cholesterol accumulation in the ER, leading to ER stress and impaired activity of specific ER cholesterol metabolising enzymes such as Cyp7b1 [50]. Previous studies have described the association between increased dietary cholesterol and dementia [51], and others have shown a decrease in 24SOHC and increase an in 27OHC and 7ketoOHC in post-mortem brains of patients with advanced dementia [52]. A different study evaluated the effects of high fat diet on hypothalamic oxysterols in C57BL/6 mice, showing high plasma cholesterol, no significant modifications 24SOHC and 27OHC and a decrease in the levels of 7ketoOHC consistent with our oxysterol measurements from brain tissues of high fat diet fed mice [53]. Guillemot-Legris et al., 2015 also showed significant modifications in hepatic, plasma and adipose tissue oxysterols, 4βhydroxycholesterol, 25OHC and 27OHC in a time course dependent manner. Sozen et al., 2018 also evaluated the effects of high cholesterol diet on fatty acid, oxysterol and scavenger receptor levels in heart tissues of rabbits, and showed both cholesterol and oxysterols including 4βhydroxycholesterol, 25OHC, 27OHC and 7ketoOHC levels were elevated in heart tissues of high cholesterol fed animals [54]. Taken together the findings from this study and the previous research suggests that the high fat dietary effects on oxysterol accumulation are tissue dependent.
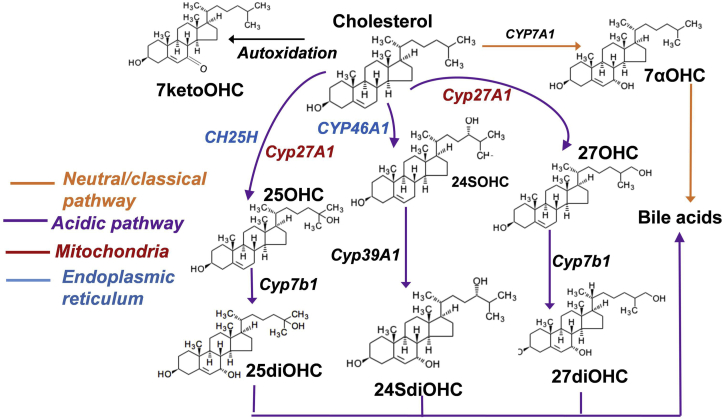
Biosynthesis and metabolism of oxysterols is distinctive within subcellular compartments with Cyp27A1 (mitochondrial specific sterol 27 hydroxylase) functions in the acidic pathway of bile acid synthesis and catalyses 27 hydroxylation of cholesterol to produce 27OHC (Fig. 6) [55,56] and is expressed exclusively in mitochondria. However, whereas Cyp7b1, Cyp46A1 and Cyp39A1 are more prevalent in the ER [2,56,57] (Fig. 6). We measured compartment specific oxysterol profiles in SH-SY5Y and THP-1 cell lines, of neural and myeloid origin respectively [58,59], and showed markedly different oxysterol distribution between two cell models, particularly in the mitochondrial subfractions of the cells. A previous study adopted a radioisotopically labelled approach to trace oxysterol formation in THP1 cells [60]. After addition of labelled cholesterol, between 10-20% was converted to oxysterols within 24 hours. Here, we also measured a similar proportion of oxysterol to cholesterol in THP-1 monocytes but not in SH-SY5Y cells. Considering both THP-1 and SH-SY5Y are cancer cells and that they have a dominant glycolytic metabolism [60,61], our analyses shows that independent of this, oxysterol metabolism is significantly distinct in the mitochondrial fractions of these cells.
Fig. 6.
Cholesterol-oxysterol metabolic pathway. Cholesterol is converted to bile acids by classical and acidic pathway. Enzymes specific to mitochondria and endoplasmic reticulum are colour coded. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
After the autoxidised oxysterol 7ketoOHC, 27OHC was the abundant enzymatically produced oxysterol in SH-SY5Y mitochondrial fractions confirming the mitochondrial activity of sterol 27-hydroxylase, and this is consistent with previous findings [2,56,57]. The levels of cholesterol were lower in the endoplasmic reticulum and mitochondrial fractions than the whole cells of both THP-1 monocytes and SH-SY5Y cells. Plasma membrane contains majority of cholesterol at cellular levels and this explains the minimal amount of cholesterol measured in THP-1 and SH-SY5Y subfractions and that majority of cholesterol is retained in the membrane fractions of these cells [61,62]. Despite the minimal cholesterol levels in the mitochondrial fractions, the oxysterol levels were comparable to that of the whole cells, particularly in the THP-1 monocytes. This highlights mitochondria as one of the primary cellular compartment housing significant oxysterol metabolism at the cellular level.
In summary, we have developed a robust, a highly sensitive and selective method for extraction and quantification of oxysterols from mitochondria. We show the application of our method to identify mitochondrial specific oxysterol metabolism in different cell lines including SH-SY5Y, PBMC and THP-1. The method offers versatility in its application to tissue sections, cell lines and blood leukocytes for profiling the oxysterol distribution and in particular, to study cholesterol/oxysterol metabolism in the context of disease.
Declaration of competing interest
None.
Acknowledgements
K Borah and HR Griffiths acknowledge INClusilver funded by the European Union, grant number H2020–INNOSUP‐2017‐2017 731349; NeutroCure funded by the European Union, grant number H2020-FETOPEN-01-2018-2019-2020 861878 and Faculty Research Support Fund (FRSF) fund from the University of Surrey 2019–2020. K Borah also acknowledges support of training grant 2019 Ref T022 from VALIDATE network. I Ampong, D Gao and HR Griffiths acknowledge funding from BBSRC (China Partnering Award BB/M028100/2. D Gao acknowledges funding from the National Natural Science Foundation of China (NFSC) (Grant No. 81873665). IHKD acknowledges funding from Alzheimer's research UK midlands network grant 2019. AH Crosby and EL Baple acknowledge support from the Hereditary Spastic Paraplegia Support Group and The Diamond Jubilee Doctoral Scholarship Fund.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101595.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
References
- 1.Luu W., Sharpe L.J., Capell-Hattam I., Gelissen I.C., Brown A.J. Oxysterols: old tale, new twists. Annu. Rev. Pharmacol. Toxicol. 2016;56:447–467. doi: 10.1146/annurev-pharmtox-010715-103233. [DOI] [PubMed] [Google Scholar]
- 2.Dias I.H., Borah K., Amin B., Griffiths H.R., Sassi K., Lizard G., Iriondo A., Martinez-Lage P. Localisation of oxysterols at the sub-cellular level and in biological fluids. J. Steroid Biochem. Mol. Biol. 2019;193 doi: 10.1016/j.jsbmb.2019.105426. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths W.J., Wang Y. Oxysterol research: a brief review. Biochem. Soc. Trans. 2019;47:517–526. doi: 10.1042/BST20180135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fakheri R.J., Javitt N.B. 27-Hydroxycholesterol, does it exist? On the nomenclature and stereochemistry of 26-hydroxylated sterols. Steroids. 2012;77:575–577. doi: 10.1016/j.steroids.2012.02.006. 2012. [DOI] [PubMed] [Google Scholar]
- 5.Björkhem I., Björkhem I. Vol. 110. 2002. (Do oxysterols control cholesterol homeostasis ? Find the latest version : do oxysterols control cholesterol homeostasis ?). 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björkhem I., Heverin M., Leoni V., Meaney S., Diczfalusy U. Oxysterols and Alzheimer's disease. Acta Neurol. Scand. Suppl. 2006;185:43–49. doi: 10.1111/j.1600-0404.2006.00684.x. [DOI] [PubMed] [Google Scholar]
- 7.Jefcoate C. High-flux mitochondrial cholesterol trafficking, a specialized function of the adrenal cortex. J. Clin. Invest. 2002;110:881–890. doi: 10.1172/JCI0216771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scharwey M., Tatsuta T., Langer T. Mitochondrial lipid transport at a glance. J. Cell Sci. 2013;126:5317–5323. doi: 10.1242/jcs.134130. [DOI] [PubMed] [Google Scholar]
- 9.English G. Mitochondrial cholesterol trafficking: impact on inflammatory mediators. Biosci. Horizons. 2010;3:1–9. doi: 10.1093/biohorizons/hzq002. [DOI] [Google Scholar]
- 10.Neuschwander-Tetri B.A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–788. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- 11.Rickman O.J., Baple E.L., Crosby A.H. Lipid metabolic pathways converge in motor neuron degenerative diseases. Brain. 2019:1–15. doi: 10.1093/brain/awz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vejux A., Namsi A., Nury T., Moreau T., Lizard G. Biomarkers of amyotrophic lateral sclerosis: current status and interest of oxysterols and phytosterols. Front. Mol. Neurosci. 2018;11:1–13. doi: 10.3389/fnmol.2018.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baarine M., Andréoletti P., Athias A., Nury T., Zarrouk A., Ragot K., Vejux A., Riedinger J.M., Kattan Z., Bessede G., Trompier D., Savary S., Cherkaoui-Malki M., Lizard G. Evidence of oxidative stress in very long chain fatty acid–treated oligodendrocytes and potentialization of ROS production using RNA interference directed knockdown of ABCD1 and ACOX1 peroxisomal proteins. Neuroscience. 2012;213:1–18. doi: 10.1016/j.neuroscience.2012.03.058. [DOI] [PubMed] [Google Scholar]
- 14.Mutemberezi V., Guillemot-Legris O., Muccioli G.G. Oxysterols: from cholesterol metabolites to key mediators. Prog. Lipid Res. 2016;64:152–169. doi: 10.1016/j.plipres.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Soderblom C., Stadler J., Jupille H., Blackstone C., Shupliakov O., Hanna M.C. Targeted disruption of the Mast syndrome gene SPG21 in mice impairs hind limb function and alters axon branching in cultured cortical neurons. Neurogenetics. 2010;11:369–378. doi: 10.1007/s10048-010-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanaswamy R., Iyer V., Khare P., Lou Bodziak M., Badgett D., Zivadinov R., Weinstock-Guttman B., Rideout T.C., Ramanathan M., Browne R.W. Simultaneous determination of oxysterols, cholesterol and 25-hydroxy-vitamin D3 in human plasma by LC-UV-MS. PloS One. 2015;10:1–15. doi: 10.1371/journal.pone.0123771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X., Sidhu R., Porter F.D., Yanjanin N.M., Speak A.O., Te Vruchte D.T., Platt F.M., Fujiwara H., Scherrer D.E., Zhang J., Dietzen D.J., Schaffer J.E., Ory D.S. A sensitive and specific LC-MS/MS method for rapid diagnosis of Niemann-Pick C1 disease from human plasma. J. Lipid Res. 2011;52:1435–1445. doi: 10.1194/jlr.D015735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellanti F., Villani R., Tamborra R., Blonda M., Iannelli G., di Bello G., Facciorusso A., Poli G., Iuliano L., Avolio C., Vendemiale G., Serviddio G. Synergistic interaction of fatty acids and oxysterols impairs mitochondrial function and limits liver adaptation during NAFLD progression. Redox Biol. 2018;15:86–96. doi: 10.1016/j.redox.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musso G., Gambino R., Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog. Lipid Res. 2013;52:175–191. doi: 10.1016/j.plipres.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Serviddio G., Blonda M., Bellanti F., Villani R., Iuliano L., Vendemiale G. Oxysterols and redox signaling in the pathogenesis of non-alcoholic fatty liver disease. Free Radic. Res. 2013;47:881–893. doi: 10.3109/10715762.2013.835048. [DOI] [PubMed] [Google Scholar]
- 21.Roussi S., Gossé F., Aoudé-Werner D., Zhang X., Marchioni E., Geoffroy P., Miesch M., Raul F. Mitochondrial perturbation, oxidative stress and lysosomal destabilization are involved in 7β-hydroxysitosterol and 7β-hydroxycholesterol triggered apoptosis in human colon cancer cells. Apoptosis. 2006;12:87. doi: 10.1007/s10495-006-0485-y. [DOI] [PubMed] [Google Scholar]
- 22.Kölsch H., Ludwig M., Lütjohann D., Rao M.L. Neurotoxicity of 24-hydroxycholesterol, an important cholesterol elimination product of the brain, may be prevented by vitamin E and estradiol-17β. J. Neural. Transm. 2001;108:475–488. doi: 10.1007/s007020170068. [DOI] [PubMed] [Google Scholar]
- 23.Yamanaka K., Saito Y., Yamamori T., Urano Y., Noguchi N. 24(S)-hydroxycholesterol induces neuronal cell death through necroptosis, a form of programmed necrosis. J. Biol. Chem. 2011;286:24666–24673. doi: 10.1074/jbc.M111.236273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang E.V., McDonald J.G., Russell D.W., Cyster J.G. Oxysterol restraint of cholesterol synthesis prevents AIM2 inflammasome activation. Cell. 2017;171:1057–1071. doi: 10.1016/j.cell.2017.09.029. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamba P., Testa G., Gargiulo S., Staurenghi E., Poli G., Leonarduzzi G. Oxidized cholesterol as the driving force behind the development of Alzheimer's disease. Front. Aging Neurosci. 2015;7:1–21. doi: 10.3389/fnagi.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johri A., Beal M.F. Johri, mitochondrial dysfunction in neurodegenerative diseases. J. Pharmacol. Exp. Therapeut. 2012;342:619–630. doi: 10.1124/jpet.112.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y., Chen M., Jiang J. Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion. 2019;49:35–45. doi: 10.1016/j.mito.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 28.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dias I.H.K., Milic I., Lip G.Y.H., Devitt A., Polidori M.C., Griffiths H.R. Simvastatin reduces circulating oxysterol levels in men with hypercholesterolaemia. Redox Biol. 2018;16:139–145. doi: 10.1016/j.redox.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths W.J., Abdel-Khalik J., Crick P.J., Yutuc E., Wang Y. New methods for analysis of oxysterols and related compounds by LC–MS. J. Steroid Biochem. Mol. Biol. 2016;162:4–26. doi: 10.1016/j.jsbmb.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 31.Griffiths W.J., Wang Y. Analysis of neurosterols by GC–MS and LC–MS/MS. J. Chromatogr. B. 2009;877:2778–2805. doi: 10.1016/j.jchromb.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 32.McDonald J.G., Thompson B.M., McCrum E.C., Russell D.W. Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:76–6879. doi: 10.1016/S0076-6879(07)32006-5. [DOI] [PubMed] [Google Scholar]
- 33.Dzeletovic S., Breuer O., Lund E., Diczfalusy U. Determination of cholesterol oxidation products in human plasma by isotope dilution-mass spectrometry. Anal. Biochem. 1995;225:73–80. doi: 10.1006/abio.1995.1110. [DOI] [PubMed] [Google Scholar]
- 34.Honda A., Yamashita K., Hara T., Ikegami T., Miyazaki T., Shirai M., Xu G., Numazawa M., Matsuzaki Y. Highly sensitive quantification of key regulatory oxysterols in biological samples by LC-ESI–MS/MS. J. Lipid Res. 2009;50:350–357. doi: 10.1194/jlr.D800040-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Sidhu R., Jiang H., Farhat N.Y., Carrillo-Carrasco N., Woolery M., Ottinger E., Porter F.D., Schaffer J.E., Ory D.S., Jiang X. A validated LC–MS/MS assay for quantification of 24(S)-hydroxycholesterol in plasma and cerebrospinal fluid. J. Lipid Res. 2015;56:1222–1233. doi: 10.1194/jlr.D058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crick P.J., William B.T., Abdel-Khalik J., Matthews I., Clayton P.T., Morris A.A., Bigger B.W., Zerbinati C., Tritapepe L., Iuliano L., Wang Y., Griffiths W.J. Quantitative charge-tags for sterol and oxysterol analysis. Clin. Chem. 2015;61:400–411. doi: 10.1373/clinchem.2014.231332. [DOI] [PubMed] [Google Scholar]
- 37.Beck A., Jordan L.K., Herlitze S., Amtmann A., Christian J., Brogden G., Adamek M., Naim H.Y., Maria Becker A. Quantification of sterols from carp cell lines by using HPLC-MS. Sep. Sci. Plus. 2018;1:11–21. doi: 10.1002/sscp.201700021. [DOI] [Google Scholar]
- 38.Bird S.S., Marur V.R., Stavrovskaya I.G., Kristal B.S. Qualitative characterization of the rat liver mitochondrial lipidome using LC-MS profiling and high energy collisional dissociation (HCD) all ion fragmentation. Metabolomics. 2013;9:67–83. doi: 10.1007/s11306-012-0400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kappler L., Li J., Häring H.U., Weigert C., Lehmann R., Xu G., Hoene M. Purity matters: a workflow for the valid high-resolution lipid profiling of mitochondria from cell culture samples. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keizer R.J., Jansen R.S., Rosing H., Thijssen B., Beijnen J.H., Schellens J.H.M., Huitema A.D.R. Incorporation of concentration data below the limit of quantification in population pharmacokinetic analyses. Pharmacol. Res. Perspect. 2015;3:1–15. doi: 10.1002/prp2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beal S.L. Ways to fit a PK model with some data below the quantification limit. J. Pharmacokinet. Pharmacodyn. 2001;28:481–504. doi: 10.1023/A:1012299115260. [DOI] [PubMed] [Google Scholar]
- 42.Meljon A., Theofilopoulos S., Shackleton C.H.L., Watson G.L., Javitt N.B., Knölker H.J., Saini R., Arenas E., Wang Y., Griffiths W.J. Analysis of bioactive oxysterols in newborn mouse brain by LC/MS. J. Lipid Res. 2012;53:2469–2483. doi: 10.1194/jlr.D028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meljon A., Crick P.J., Yutuc E., Yau J.L., Seckl J.R., Theofilopoulos S., Arenas E., Wang Y., Griffiths W.J. Mining for Oxysterols in Cyp7b1−/− Mouse Brain and Plasma: Relevance to Spastic Paraplegia Type 5. Biomolecules. 2019;vol. 9 doi: 10.3390/biom9040149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saeed A.A., Genové G., Li T., Lütjohann D., Olin M., Mast N., Pikuleva I.A., Crick P., Wang Y., Griffiths W., Betsholtz C., Björkhem I. Effects of a disrupted blood-brain barrier on cholesterol homeostasis in the brain. J. Biol. Chem. 2014;289:23712–23722. doi: 10.1074/jbc.M114.556159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valverde-Som L., Carrasco-Pancorbo A., Sierra S., Santana S., Ruiz-Samblás C., Navas N., Burgos J.S., Cuadros-Rodríguez L. Separation and Determination of Some of the Main Cholesterol-Related Compounds in Blood by Gas Chromatography-Mass Spectrometry (Selected Ion Monitoring Mode) Separations. 2018;vol. 5 doi: 10.3390/separations5010017. [DOI] [Google Scholar]
- 46.Brown A.J., Mander E.L., Gelissen I.C., Kritharides L., Dean R.T., Jessup W. Cholesterol and oxysterol metabolism and subcellular distribution in macrophage foam cells: accumulation of oxidized esters in lysosomes. J. Lipid Res. 2000;41:226–236. [PubMed] [Google Scholar]
- 47.Helmschrodt C., Becker S., Schröter J., Hecht M., Aust G., Thiery J., Ceglarek U. Fast LC-MS/MS analysis of free oxysterols derived from reactive oxygen species in human plasma and carotid plaque. Clin. Chim. Acta. 2013;425:3–8. doi: 10.1016/j.cca.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 48.Tuck M.K., Chan D.W., Chia D., Godwin A.K., Grizzle W.E., Krueger K.E., Rom W., Sanda M., Sorbara L., Stass S., Brenner D.E. Standard operating procedures for serum and plasma collection. J. Proteome Res. 2010;8:113–117. doi: 10.1021/pr800545q.Standard. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mutemberezi V., Buisseret B., Masquelier J., Guillemot-Legris O., Alhouayek M., Muccioli G.G. Oxysterol levels and metabolism in the course of neuroinflammation: insights from in vitro and in vivo models. J. Neuroinflammation. 2018;15:1–16. doi: 10.1186/s12974-018-1114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Widenmaier S.B., Snyder N.A., Nguyen T.B., Arduini A., Lee G.Y., Arruda A.P., Saksi J., Bartelt A., Hotamisligil G.S. NRF1 is an ER membrane sensor that is central to cholesterol homeostasis. Cell. 2017;171 doi: 10.1016/j.cell.2017.10.003. 1094.e15-1109.e15. [DOI] [PubMed] [Google Scholar]
- 51.Dias I.H.K., Polidori M.C., Li L., Weber D., Stahl W., Nelles G., Grune T. Plasma levels of HDL and carotenoids are lower in dementia patients with vascular comorbidities. J. Alzheim. Dis. 2014;40:399–408. doi: 10.3233/JAD-131964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Testa G., Staurenghi E., Zerbinati C., Gargiulo S., Iuliano L., Giaccone G., Fantò F., Poli G., Leonarduzzi G., Gamba P. Changes in brain oxysterols at different stages of Alzheimer's disease: their involvement in neuroinflammation. Redox Biol. 2016;10:24–33. doi: 10.1016/j.redox.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillemot-Legris O., Mutemberezi V., Cani P.D., Muccioli G.G. Obesity is associated with changes in oxysterol metabolism and levels in mice liver, hypothalamus, adipose tissue and plasma. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep19694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sozen E., Yazgan B., Sahin A., Ince U., Ozer N.K. High cholesterol diet-induced changes in oxysterol and scavenger receptor levels in heart tissue. Oxid. Med. Cell. Longev. 2018:8520746. doi: 10.1155/2018/8520746. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mutational Analysis of CYP27A1 Assessment of 27-hydroxylation of cholesterol and 25-hydroxylation of vitamin D. Metabolism. 2007;56:1248–1255. doi: 10.1016/j.metabol.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X., Hylemon P., Pandak W.M., Ren S. Enzyme activity assay for cholesterol 27-hydroxylase in mitochondria. J. Lipid Res. 2006;47:1507–1512. doi: 10.1194/jlr.M600117-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Honda A., Miyazaki T., Ikegami T., Iwamoto J., Maeda T., Hirayama T., Saito Y., Teramoto T., Matsuzaki Y. Cholesterol 25-hydroxylation activity of CYP3A. J. Lipid Res. 2011;52:1509–1516. doi: 10.1194/jlr.M014084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider L., Giordano S., Zelickson B.R., Johnson M., Benavides G., Ouyang X., Fineberg N., Darley-usmar V.M. Differentiation of SH cells. 2012;51:2007–2017. doi: 10.1016/j.freeradbiomed.2011.08.030.Differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raulien N., Friedrich K., Strobel S., Rubner S., Baumann S., von Bergen M., Körner A., Krueger M., Rossol M., Wagner U. Fatty acid oxidation compensates for lipopolysaccharide-induced Warburg effect in glucose-deprived monocytes. Front. Immunol. 2017;8:1–12. doi: 10.3389/fimmu.2017.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y., Arnal-Levron M., Lagarde M., Moulin P., Luquain-Costaz C., Delton I. THP1 macrophages oxidized cholesterol, generating 7-derivative oxysterols specifically released by HDL. Steroids. 2015;99:212–218. doi: 10.1016/j.steroids.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 61.Thelen A.M., Zoncu R. Emerging roles for the lysosome in lipid metabolism. Trends Cell Biol. 2017;27:833–850. doi: 10.1016/j.tcb.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.