Abstract
We investigated whether obsessive–compulsive (OC) symptoms from a population‐based sample could be analyzed to detect genetic variants influencing obsessive–compulsive disorder (OCD). We performed a genome‐wide association studies (GWAS) on the obsession (rumination and impulsions) and compulsion (checking, washing, and ordering/precision) subscales of an abbreviated version of the Padua Inventory (N = 8,267 with genome‐wide genotyping and phenotyping). The compulsion subscale showed a substantial and significant positive genetic correlation with an OCD case–control GWAS (r G = 0.61, p = .017) previously published by the Psychiatric Genomics Consortium (PGC‐OCD). The obsession subscale and the total Padua score showed no significant genetic correlations (r G = −0.02 and r G = 0.42, respectively). A meta‐analysis of the compulsive symptoms GWAS with the PGC‐OCD revealed no genome‐wide significant Single‐Nucleotide Polymorphisms (SNPs combined N = 17,992, indicating that the power is still low for individual SNP effects). A gene‐based association analysis, however, yielded two novel genes (WDR7 and ADCK1). The top 250 genes in the gene‐based test also showed a significant increase in enrichment for psychiatric and brain‐expressed genes. S‐Predixcan testing showed that for genes expressed in hippocampus, amygdala, and caudate nucleus significance increased in the meta‐analysis with compulsive symptoms compared to the original PGC‐OCD GWAS. Thus, the inclusion of dimensional symptom data in genome‐wide association on clinical case–control GWAS of OCD may be useful to find genes for OCD if the data are based on quantitative indices of compulsive behavior. SNP‐level power increases were limited, but aggregate, gene‐level analyses showed increased enrichment for brain‐expressed genes related to psychiatric disorders, and increased association with gene expression in brain tissues with known emotional, reward processing, memory, and fear‐formation functions.
Keywords: gene expression, genome‐wide association study (GWAS), obsessive–compulsive symptoms, OCD, Padua Inventory‐revised
1. INTRODUCTION
Obsessive–compulsive disorder (OCD) is characterized by recurrent, unwanted thoughts (obsessions) and/or repetitive behaviors (compulsions). The repetitive behaviors or mental acts (such as hand washing, ordering, and checking) are performed in response to an obsession or according to rules that must be applied rigidly. They are aimed at preventing or reducing the distress of a feared event or situation, a fear which at the same time is clearly unrealistic and/or excessive. OCD is associated with considerable suffering and markedly impairs individuals' social and occupational functioning. The lifetime population prevalence of OCD is estimated to be 2–3% (Kessler et al., 2005).
Genetic studies have firmly established that OCD has a significant heritable component. A family study has shown evidence for increased odds‐ratios in family members of OCD probands (Pauls, Alsobrook 2nd, Goodman, Rasmussen, & Leckman, 1995). However, family studies cannot exclude that the shared rearing environment between family members plays a role in the etiology of the disease, thus biasing heritability estimates. Twin studies may overcome this limitation; however, these studies of OCD diagnosis have been limited in sample size (van Grootheest, Cath, Beekman, & Boomsma, 2005). Twin studies examining obsessive–compulsive symptoms in the general population estimated its heritability to be around 40% (den Braber et al., 2016; Iervolino, Rijsdijk, Cherkas, Fullana, & Mataix‐Cols, 2011; van Grootheest et al., 2005; van Grootheest, Cath, Beekman, & Boomsma, 2007; Zilhão et al., 2015). Overall, these studies suggest that a modest, but significant proportion of the liability for OCD is heritable.
OCD is relatively underrepresented in genome‐wide association studies (GWAS), both in sample size and number. The largest case–control meta‐analysis of GWAS to date included ~2,800 cases, which falls well short of the >40,000 cases for many other psychiatric disorders (International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) and OCD Collaborative Genetics Association Studies (OCGAS) et al., 2018). No significant associations were reported, likely due to the sample size being smaller than required for studies of complex disorders. One solution to increase sample size is to include data from large databases of validated health questionnaires in individuals that have been genotyped, including nonclinical, population‐based samples with information on obsessive–compulsive symptoms. This requires that the scores on the questionnaire, or the scores on the subscales, reflect the underlying genetic liability and thus genetically correlate with the clinically established diagnosis of OCD. Since very large numbers of participants are a necessity to identify common genetic variants from GWAS, such self‐report symptom data may be crucial to increase the sample size of the existing case–control GWASs. For ADHD, it was recently demonstrated that this is a viable option, provided that the correct statistical meta‐analytic technique is used (Demontis et al., 2019). Here, we aim to use a similar approach for a GWAS of OCD. We will first run GWASs on OC symptom scores and subscales from the Padua Inventory and establish whether the genetic variants underlying OC symptoms are associated with those underlying OCD (International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) and OCD Collaborative Genetics Association Studies (OCGAS) et al., 2018), by estimating their genetic correlation. Secondly, we will meta‐analyze the GWASs on OCD and correlated OC symptom subscales, to examine whether this results in an increase in power to detect underlying genetic variants in a range of follow‐up analyses.
2. METHODS
2.1. Subjects
Twins and their family members (parents, children, siblings) registered at the Netherlands Twin Register (NTR; Boomsma et al., 2002) were included in the OC symptom analyses. Every 2–3 years subjects who participate in NTR studies receive self‐report surveys, which contain a variety of questionnaires related to health, personality, demographics, lifestyle, and psychiatric disorders. Data on OC symptoms were available for 20,528 subjects (N = 10,285 in the year 2002 and N = 15,803 in the year 2005, with N = 5,560 overlaps). Of these, N = 8,267 (64% female) had genotype data available and were of Dutch ancestry. Their mean age was 41.6 years (SD 15.4; age range between 18 and 80 years); Supporting Information Figure S1 shows the age histogram. Informed consent was obtained from all participants. The study was approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Centre, Amsterdam, an Institutional Review Board certified by the U.S. Office of Human Research Protections (IRB number IRB00002991 under Federal‐wide Assurance—FWA00017598; IRB/institute codes, NTR 03‐180).
2.2. Padua‐revised (abbreviated) OC symptoms
The OC symptom self‐report data were collected by the abbreviated Dutch translation of the Padua Inventory‐Revised (Burns, Keortge, Formea, & Sternberger, 1996; Cath, van Grootheest, Willemsen, van Oppen, & Boomsma, 2008; van Oppen, 1992). Supporting Information Table S1 shows the questions and subscales of the OC symptom scores. The Padua Inventory‐revised separates the worry, thought‐related items from the behavioral, compulsive items. Six items are included to measure symptoms of impulsive thoughts and rumination. The remaining six items measure behavioral symptoms of OCD, namely checking, washing, and precision (ordering, counting). Since the OC symptoms scale showed strong evidence for skew, we transformed the data with a square‐root transformation to minimize the right skew (Zilhão et al., 2015).
2.3. Genotyping and imputation for OC symptoms
A full description of genotyping, preimputation QC, and imputation of the NTR OC symptoms GWAS is provided in the Supporting Information methods. Several critical steps and parameters are presented here. Samples were removed if DNA sex did not match the expected phenotype, if the Plink heterozygosity F statistic was <−0.10 or >0.10, or if the genotyping call rate was <0.90. SNPs were removed if the minor allele frequency (MAF) <0.01, if the Hardy–Weinberg equilibrium p‐value <1 × 10–5, if call rate <0.95, or if the N Mendel errors >20. Palindromic AT/GC SNPs with an MAF range between 0.4 and 0.5 were removed to avoid possible strand alignment issues. These were applied to each genotyping platform that was used. After imputation, the datasets of each genotyping platform were merged and QC repeated. Ancestry outliers (non‐Dutch ancestry) were defined based on Principal Components Analysis (PCA) by projecting 10 PCs from 1000G reference Phase 3v5. We finally filtered on population based and sample MAF filtered at 0.03. Allele‐frequency differences between 1000G reference and sample over 0.20 were removed (10,260 SNPs).
2.4. GWAS on OC symptoms
We ran GWASs for the total score on the Padua scale, the obsessions scale, and the compulsions scale with ~4.5 M SNPs in a model with linear mixed effects correcting for population stratification and the genetic relatedness between family members, as implemented in GCTA (Yang, Lee, Goddard, & Visscher, 2011). Sex, age, age2, and 10 population stratification PCs were specified as fixed effects. We created two types of matrices to model genetic relatedness. The matrix covered the full genetic relatedness matrix including unrelated subjects. The second, family‐based matrix was created from the first by setting all relatedness values under 0.05 to zero. This models the additive genetic effects within and between families separately, thus correcting for both family dependence and ancestry dependence in the SNP effects. Both the full genetic relatedness matrix and the family matrix were used in the association analysis. We excluded information of each chromosome out of the relatedness estimations (leave‐one‐chromosome‐out method) so as to avoid adding information from the currently tested SNP in the residual of the linear mixed model. We inspected LD‐score regression intercept estimates to test for adequate control of the complex relatedness in the sample.
2.5. Meta‐analysis of PGC‐OCD‐EA and OC symptoms GWAS
We meta‐analyzed the results with the PGC‐OCD GWAS (International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) and OCD Collaborative Genetics Association Studies (OCGAS) et al., 2018) of 2,688 cases and 7,037 controls of European ancestry. We meta‐analyzed this (dichotomous) case–control PGC‐OCD GWAS with the (continuous) OC symptoms GWAS using the genome‐wide association meta‐analysis (GWAMA) method described in (Demontis et al., 2019) and implemented in R. The population prevalence was set to 0.01 with the actual number of cases and controls entered.
2.6. SNP heritability
SNP heritability for PGC‐OCD and OC‐compulsions were established with LD‐score regression (Bulik‐Sullivan, Loh, et al., 2015) to estimate the proportion of variance that could be explained by the aggregated effect of the SNPs. The method is based on the assumption that a regression of the phenotype on the SNP dosage includes the effects of all SNPs in LD with the tested SNP. On average, an SNP that tags many other SNPs will have a higher probability of tagging a causal variant than one that tags a few other SNPs. Accordingly, for highly polygenic traits, SNPs with a higher average LD score have stronger effect sizes than SNPs with lower LD scores. When regressing the effect size obtained from the GWAS against the LD score for each SNP, the slope of the regression line gives an estimate of the proportion of variance accounted for by all analyzed SNPs. Standard LD scores were used based on the Hapmap 3 reference panel, restricted to European populations.
2.7. Genetic correlation
We used cross‐trait LD‐score regression to estimate the genetic covariation between traits based on GWAS summary statistics (Bulik‐Sullivan, Finucane, et al., 2015). We chose LD‐score regression genetic correlation estimates as it was found to be reasonably robust and unbiased (Lee, McGue, Iacono, & Chow, 2018). In addition, LD‐score is often used in large genetic correlation studies (e.g., Brainstorm Consortium, 2018), which makes our results readily comparable. The alternative of calculating polygenic score based correlations are strongly biased toward zero for the current sample sizes and require an independent sample for testing. In LD‐score regression, the genetic covariance is estimated using the slope from the regression of the product of z‐scores from two GWAS studies on the LD score. The estimate obtained from this method represents the genetic correlation between the two traits based on all polygenic effects captured by SNPs. Standard LD scores were used as provided by Bulik‐Sullivan et al. (Bulik‐Sullivan, Loh, et al., 2015) based on the 1,000 genomes reference set, restricted to European populations.
2.8. Gene‐based test and enrichment analysis
We performed an MAGMA positional gene‐based test of association based SNP effects around genes with a 10kbp margin around the 3′ and 5′ UTR as implemented in FUMA (Watanabe, Taskesen, van Bochoven, & Posthuma, 2017). We performed enrichment tests by comparing the top 250 genes from the gene‐based tests to several types of annotated gene sets. Ten brain‐expression gene sets were selected in FUMA based on the GTEx v7 database. From these tissues, differentially expressed genes (DEGs) were defined as those showed significant up‐ or down‐regulation compared to the average expression in all other 52 tissues. Enrichment of these genes in the top 250 genes was determined using the hypergeometric test. In addition, we determined whether sets of GWAS catalog reported genes were overrepresented in the top 250 genes. Finally, we compared the significance of these tests between the original PGC‐OCD GWAS and the meta‐analysis OCD + compulsion symptoms in order to establish whether stronger enrichment could be obtained.
2.9. Expression analysis
To examine to what extent genes associated with OCD + compulsion symptoms meta‐analysis are expressed in the brain, we performed S‐Predixcan analysis on the meta‐analyzed GWAS results. S‐Predixcan is based on Predixcan (Gamazon et al., 2015). Predixcan uses RNAseq gene‐expression associations present in the GTEx database to build sparse elastic net models, one model for each pair of the 53 tissues and ~30,000 genes. Individual SNP data is then used to impute gene expression. These imputed gene expressions are then associated with the phenotype, resulting in tissue‐specific associations of gene‐expression with the phenotype. S‐Predixcan (Barbeira et al., 2018) is an extension of Predixcan and can be used with summary statistics only.
3. RESULTS
3.1. Genome‐wide association studies
Supporting Information Figure S2A–C shows the Manhattan plots for the obsessions, compulsions, and original PGC‐OCD GWASs. The respective Q–Q plots are shown in Supporting Information Figure S3A–C.
3.2. Compulsive symptoms show significant genetic correlation with OCD
Table 1 shows the results of the SNP heritability and genetic correlation analysis for the three GWASs using LD‐score regression with the original PGC‐OCD. The SNP heritability is consistent with previous results (den Braber et al., 2016). The compulsions subscale showed an almost significant heritability at 11.6% (z = 1.88, p = .06). The obsessions subscale showed lower heritability and did not approach significance. The compulsions composite scale (contamination/ordering/counting/checking symptoms) showed a substantial and significant correlation with the PGC‐OCD GWAS (r G = 0.61, p = .017). The obsessions subscale and the full‐scale GWAS did not show a significant r G with the PGC‐OCD GWAS.
Table 1.
SNP‐based heritability of the Padua Inventory full‐scale score GWAS, the compulsions and obsessions subscales, and their genetic correlation with the PGC‐OCD GWAS
| Heritability | Genetic correlation with PGC‐OCD | |||||
|---|---|---|---|---|---|---|
| h 2 | SE | r G | SE | z | p | |
| Compulsions | 0.116 | 0.062 | 0.61 | 0.255 | 2.378 | .017 |
| Obsessions | 0.058 | 0.065 | −0.02 | 0.322 | −0.068 | .946 |
| Full scale | 0.102 | 0.060 | 0.42 | 0.254 | 1.672 | .095 |
Notes: SNP‐based heritabilities and genetic correlations were estimated using LD‐score regression.
3.3. Meta‐analysis
Because only the compulsion subscale showed significant r G, this subscale was selected for meta‐analyzing with the PGC‐OCD GWAS. Supporting Information Figure S2D shows the Manhattan plot for the OCD + compulsion symptoms meta‐analysis. Figure S3D shows the associated Q–Q, and Figure 1 shows the comparison of the original PGC‐OCD Q–Q to the one for the OCD + compulsion symptoms meta‐analysis. The inflation median lambdas were comparable for the original and extended GWASs, λ = 1.032 and λ = 1.033, respectively, indicating a marginally higher lambda for the meta‐analysis. Lambdas for the upper 10% percentile were 1.0205 and 1.0405, respectively, indicating a difference in inflation for top SNP effects. LD score regression intercepts were near 1.0 (0.9889, SE = 0.0065 and 0.9937, SE = 0.0085, for the original PGC‐OCD and the OCD + compulsion symptoms meta‐analysis, respectively), indicating successful control of the ancestry effects.
Figure 1.
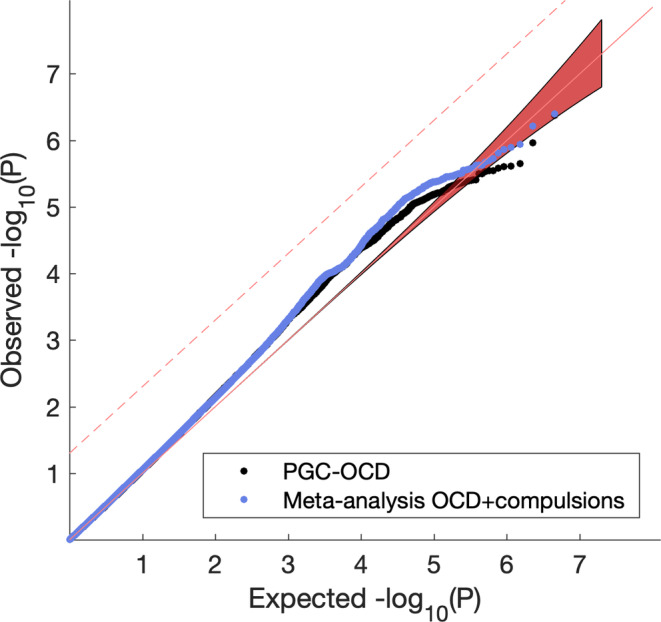
Q–Q plot of observed SNP p‐values against expected p‐values under the null. Black is the original PGC‐OCD GWAS, blue is the meta‐analysis of PGC‐OCD with compulsions. Dashed line is FDR q = 0.05 [Color figure can be viewed at wileyonlinelibrary.com]
3.4. Gene‐based tests
Supporting Information figure shows the gene‐based Manhattan plot, and Figure 2 shows the associated Q–Q for the gene‐based test results obtained using MAGMA (de Leeuw, Mooij, Heskes, & Posthuma, 2015). Four genes were significantly associated with OCD + compulsion symptoms meta‐analysis after correcting for multiple testing: KIT, GRID2, WDR7, and ADCK1 at FDR q = 0.05. Of these, WDR7 and ADCK1 are novel, whereas the other two confirmed findings from the original PGC‐OCD GWAS.
Figure 2.
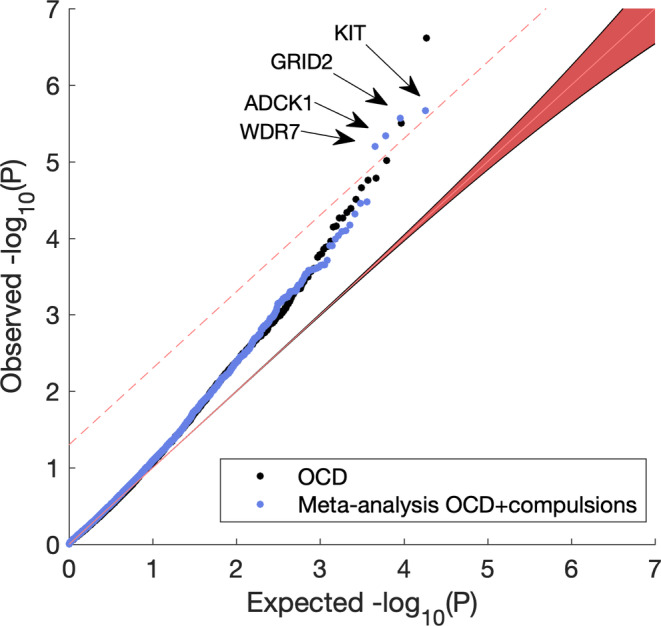
Q–Q plot of observed against expected p‐values under the null of the MAGMA gene‐based test. Dashed line is FDR q = 0.05. The meta‐analysis OCD + compulsion symptoms (blue) resulted in four significant discoveries, KIT, GRID2, WDR7, and ADCK1. The latter two are novel findings compared to the original PGC‐OCD GWAS (black) [Color figure can be viewed at wileyonlinelibrary.com]
3.5. Altered enrichment of brain‐expressed genes
We performed an enrichment test of the top 250 genes from the MAGMA positional gene‐based analysis, scanned for the enrichment of brain‐expressed genes from the GTEx database (v7) and for genes reported in GWAS catalog (https://www.ebi.ac.uk/gwas/) associated with psychiatric traits.
Figure 3 shows the significance of enrichment of tissue‐specific genes in original PGC‐OCD before (left, in black) and after meta‐analyzing with compulsive symptoms (right, in orange) for all neural tissues excluding cerebellum and spinal cord. Whole blood and two brain‐unrelated tissues (spleen, stomach) were added for reference. Brain‐tissue DEGs were significantly enriched in the PGC‐OCD GWAS, with the expressed genes in the anterior cingulated cortex (ACC), nucleus accumbens (NAcc), and amygdala as top significant tissues. ACC, amygdala, frontal cortex, and hippocampus showed strong increases in significance, indicating that more brain‐expressed DEGs were present in the top 250 genes of the meta‐analysis compared to the original PGC‐OCD. Other brain tissues showed marginal change (caudate, putamen, NAcc, hypothalamus). Other tissues (cortex and substantia nigra) showed a decrease in effect.
Figure 3.
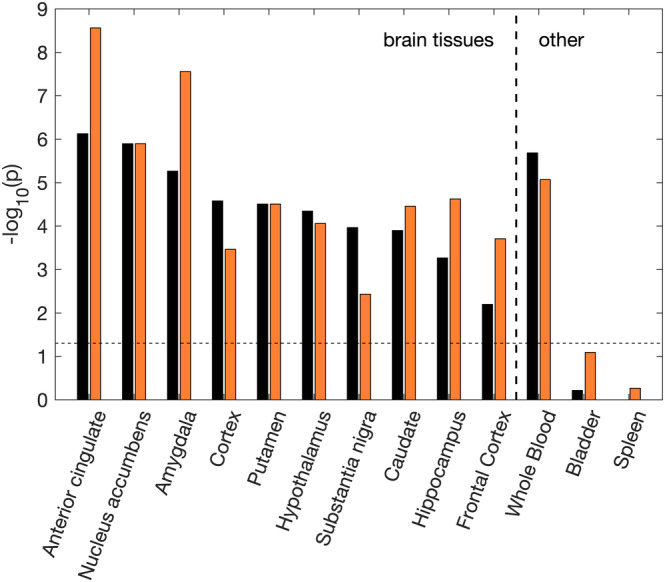
Enrichment results for the PGC‐OCD (black) and the OCD + compulsion symptoms meta‐analysis (orange). Enrichment of tissue‐specific differentially expressed genes (DEGs) was determined using the hypergeometric test. The y‐axis shows the Bonferroni corrected significance of the test as –log 10(p). The dashed line indicates the significance threshold (p = .05). Significance strongly increased for some brain tissues (over 1 point increase for ACC, amygdala, hippocampus and frontal cortex), and decreased for others (over 1 point decrease for cortex, substantia nigra) [Color figure can be viewed at wileyonlinelibrary.com]
3.6. Enrichment of psychiatric and behavioral gene sets
Gene sets from the GWAS catalog are available in FUMA for enrichment analysis. Both the original PGC‐OCD GWAS and the meta‐analysis with compulsive symptoms showed a large set of significant results after Bonferroni correction, including many psychiatric/behavioral traits. Figure 4 shows the −log10(p) of the enrichment tests of these traits. Schizophrenia genes were strongly enriched, and significantly increased over 3 orders of magnitude for the meta‐analysis. Significance increased also for “Tourette's or OCD” and “Autism or Schizophrenia” genes. The schizophrenia genes that contributed to the enrichment effect included the KIT gene, as well as genes on region 3p21 (ITIH4, TMEM112). Genes in this region have been related to bipolar disorder and schizophrenia as well as brain functional activity (Smit et al., 2018).
Figure 4.
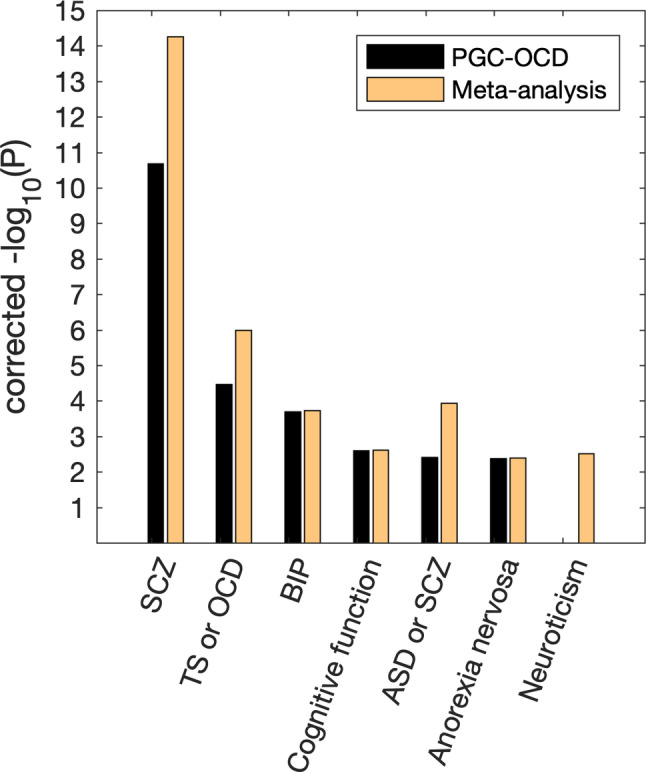
Significance of the enrichment of genes reported in psychiatric/behavioral GWASs, as found in the top 250 genes in the PGC‐OCD GWAS and the current meta‐analysis. Y‐axis shows Bonferroni corrected –log 10(p). Either equal significance or a strong increase in the effect was observed when meta‐analyzing OCD with compulsive symptoms. ASD, autism spectrum disorders; BIP, bipolar disorder; SCZ, schizophrenia; TS, Tourette's syndrome [Color figure can be viewed at wileyonlinelibrary.com]
3.7. S‐Predixcan gene‐expression associations
Imputed gene expression was not significantly associated with the OCD + compulsion symptoms combined phenotype in any of the 10 brain tissues. The expression of RP11‐446E9 in Anterior Cingulate Cortex almost reached significance after Bonferroni correction for the number of genes tested within this tissue (p = .06). Comparing the results from the PGC‐OCD GWAS to the meta‐analysis, Figure 5 shows the difference in log(p‐values) for genes that fall under a range of p‐value thresholds, with values >0 indicating that the meta‐analysis has stronger effects. The figure highlights that increased significance is not present for lower threshold values, but they are for the stronger associations.
Figure 5.
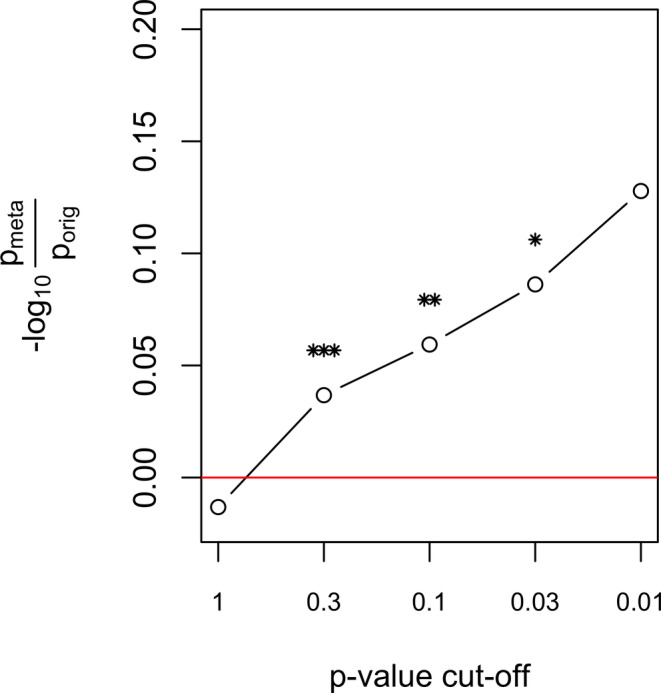
S‐Predixcan resulted in stronger p‐values after meta‐analyzing for the association of a gene's tissue expression with the phenotype. The figure shows the ratio of the p‐values of the meta‐analysis to the original GWAS (x‐axis), log‐transformed. This was performed for genes reaching a specific threshold (x‐axis) in the original GWAS. The difference in log p‐values for threshold 0.3–0.01, indicating stronger effects in the meta‐analysis. It also suggests that adding compulsive symptoms only strengthens top genetic expression effects in OCD. The average –log 10(p) difference across 10 brain tissues is shown. *p < .05, **p < .01, ***p < .001 [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
We found a substantial and significant genetic correlation between the existing OCD case–control GWAS and our GWAS of compulsion symptoms based on the abbreviated Padua Inventory. For this subscale—which holds questions on behavioral symptoms related to checking behavior, precision (ordering and counting), and contamination fear/washing behavior—the genetic correlation was estimated at r G = 0.61, p = .017. The obsessions subscale, on the other hand, was not significantly genetically correlated with the PGC‐OCD GWAS. This may reflect the observation that the majority of OCD cases have washing and checking symptoms, thus biasing the OCD GWAS toward these symptoms. The remainder of the Padua Inventory items involves questions on thoughts and worries that may be less specific to OCD. Adding all items scales in equal proportions to the GWAS (i.e., all available Padua item scores) reduced the genetic correlation substantially. Therefore, to optimally add symptom scale analyses to complement case–control GWAS, it might be prudent to include only scores on the questions interrogating the behavioral component of OCD. Note, however, that recent work has suggested that the absence of genome‐wide genetic correlation does not preclude the absence of any correlated SNP‐effect, merely that the summed effect across all SNP effects is zero (Frei et al., 2019). Such partial overlap could still play a role between the obsessions subscale and the OCD GWAS. Future investigations could establish whether positively and negatively correlated SNP effects are indeed present between obsessive symptoms and OCD.
Based on the genetic correlation results, we ran the meta‐analysis with the PCG‐OCD GWAS and the GWAS of compulsive symptoms. This meta‐analysis increased the sensitivity of the GWAS to find genes in gene‐based and gene‐enrichment analyses. We observed two novel discoveries at FDR p = .05 (ADCK1 and WDR7) in addition to the KIT and GRID2 genes that were previously identified (International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) and OCD Collaborative Genetics Association Studies (OCGAS) et al., 2018). SNPs in the WDR7 region have shown trend associations with Tourette's syndrome/OCD (p = 4 × 10−6; Yu et al., 2015), but note that this report included data from the current GWAS. In addition, SNPs near WDR7 have been suggestively associated with alcohol dependence (p = 8 × 10−6; Edwards et al., 2012). The ADCK1 gene is a novel finding. An SNP near ADCK1 (rs740658277) has been reported in relation to schizophrenia, schizophrenia symptom severity, and response to paliperidone (p = 7 × 10−7; Li et al., 2017). Note that these associations near the WDR7 and ADCK1 genes to psychiatric liabilities are only suggestive, indicating that future research must confirm that these variants are part of the genetic overlap between OCD and other psychiatric disorders.
More substantial power increases were found in the enrichment analysis using gene sets from GTEx and GWAS reports summarized in the GWAS catalog. The OCD GWAS showed highly significant enrichment of genes expressed in the anterior cingulate cortex and nucleus accumbens. The involvement of these tissues is consistent with their putative role in OCD, reward processing, and as contributing substrates in the cortico‐striato‐thalamo‐cortical circuitry known to be affected in OCD (Denys et al., 2010; Hibar et al., 2018; van den Heuvel et al., 2016). The effects were increased by meta‐analyzing the GWAS with the compulsive symptoms GWAS. In addition, the meta‐analysis showed increased enrichment of amygdala DEGs, again consistent with the role of this subcortical structure in fear learning and OCD (van den Heuvel et al., 2004).
Gene‐set analysis revealed strong increases in the GWAS catalog reported genes for traits known to be related to OCD. The original PGC‐OCD GWAS showed significant enrichment of genes linked to schizophrenia, consistent with the known genetic overlap between the disorders (Brainstorm Consortium, 2018; Bulik‐Sullivan, Finucane, et al., 2015; Martin, Taylor, & Lichtenstein, 2018). This effect increased by over three orders of magnitude in significance after meta‐analyzing with the compulsive symptoms GWAS. Likewise, the significance of the enrichment of “Tourette's or OCD” genes increased by over one order of magnitude. The increased enrichment indicates that the top 250 genes were increasingly selective for psychiatric traits known to be genetically correlated with OCD. This indicates that the expression of compulsive behavior is selectively associated with these genes.
The increased enrichments in several psychiatric and brain‐expression gene sets were observed without a notable difference in the magnitude of SNP effects between the original OCD GWAS and the meta‐analysis. The fact that these power increases were minor after adding compulsive symptoms is likely a consequence of the small size of the compulsion symptoms dataset (N < 10 k). Even so, the power increases on an SNP‐aggregate level observed here suggest that a larger OCD symptoms GWAS could be useful for obtaining increased SNP effects. Moreover, such a power increase is relatively easy to obtain, requiring a relatively short questionnaire with just six compulsion‐symptom items from the Padua Inventory.
To summarize, we observed significant SNP‐based genetic correlations between the PGC‐OCD GWAS and a GWAS of compulsive symptoms in a general population sample. This provided evidence that compulsion symptoms substantially overlap with the genetic liability for clinical diagnosis of OCD, and serves recent calls for doing genome‐wide symptom scale analyses to create insight into psychiatric disorder etiology (Davis, 2019). We showed that meta‐analyzing the OCD case–control and compulsions symptoms GWAS results have added value in the gene‐based and gene‐enrichment analyses. This included additional significant genes in the gene‐based test and subsequent enrichment analysis of brain‐expressed genes. These results bode well for larger population‐based samples to be merged with clinical samples to increase power for finding the genetic mechanisms underlying OCD.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
D.J.A.S., D.C., and D.I.B. conceived and designed the analyses, and drafted the manuscript, D.J.A.S., N.R.Z., A.d.B., and J.‐J.H. performed the analyses, H.F.I. provided analysis tools, D.D., E.J.C.d.G., and K.J.H.V. revised the manuscript.
Supporting information
Data S1 Supplementary methods.
Table S1 Padua Inventory (abbreviated) questionnaire items.
Figure S1 Age distribution histogram.
Figure S2. Manhattan plots for the Padua Inventory obsessions subscale (A), compulsions subscale (B), the original PGC‐OCD (C), and the meta‐analysis of OCD + compulsions (D).
Figure S3. Q–Q plots for the Padua Inventory obsessions subscale (A), compulsions subscale (B), the original PGC‐OCD (C), and the meta‐analysis of OCD + compulsions (D).
Figure S4. Manhattan plot for the MAGMA gene‐based test of association for the meta‐analysis OCD + compulsions GWAS.
ACKNOWLEDGMENTS
We are indebted to the Psychiatric Genomics Consortium Tourette's Syndrome/Obsessive–Compulsive Disorder workgroup (PGC TS/OCD), in particular, Dongmei Yu and Carol Mathews, for making the GWAS summary statistics available for download. We thank the twins and their family members who participate in the studies of the NTR. This study was supported by the Biobanking and Biomolecular Resources Research Infrastructure BBMRI‐NL (NWO 184.021.007 and 184.033.111); FP7‐ People‐2012‐ITN, project: TS‐EUROTRAIN, grant number 316978; ZonMW (Addiction) 31160008; and European Research Council (ERC—230374); NWO‐Groot 480‐15‐001/674: Netherlands Twin Registry Repository; and Tourette Syndrome Association USA 2008: the genetic epidemiology of tics. Genotyping was realized by grants Rutgers University Cell and DNA Repository (NIMH U24 MH068457‐06), the National Institutes of Health (NIH—R01 D0042157‐01A1, R01 MH58799‐03, MH081802, DA018673, R01 DK092127‐04, Grand Opportunity grants 1RC2 MH089951, and 1RC2 MH089995); the Avera Institute for Human Genetics, Sioux Falls, South Dakota (USA). D.I.B. acknowledges the Royal Netherlands Academy of Science Professor Award (PAH/6635). K.J.H.V. is supported by the Foundation Volksbond Rotterdam.
Smit DJA, Cath D, Zilhão NR, et al. Genetic meta‐analysis of obsessive–compulsive disorder and self‐report compulsive symptoms. Am J Med Genet Part B. 2020;183B:208–216. 10.1002/ajmg.b.32777
Funding information FP7 People: Marie‐Curie Actions, Grant/Award Number: 316978; H2020 European Research Council, Grant/Award Number: 230374; National Institute of Mental Health, Grant/Award Number: MH068457‐06; National Institutes of Health, Grant/Award Numbers: 1RC2 MH089951, 1RC2 MH089995, D0042157‐01A1, MH58799‐03, MH081802, DA018673, DK092127‐04; Nederlandse Organisatie voor Wetenschappelijk Onderzoek, Grant/Award Numbers: 184.021.007, 184.033.111, 480‐15‐001/674; ZonMw, Grant/Award Number: 31160008
REFERENCES
- Barbeira, A.N. , Dickinson, S.P. , Bonazzola, R., Zheng, J. , Wheeler, H.E. , Torres, J.M. , … Im, H. K. (2018). Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun, 9, 1–20. 10.1101/045260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma, D. I. , Vink, J. M. , van Beijsterveldt, T. C. E. M. , Geus, E. J. C. d. , Beem, A. L. , Mulder, E. J. C. M. , … van Baal, G. C. M. (2002). Netherlands Twin Register: A focus on longitudinal research. Twin Research and Human Genetics, 5(5), 401–406. 10.1375/twin.5.5.401 [DOI] [PubMed] [Google Scholar]
- Brainstorm Consortium . (2018). Analysis of shared heritability in common disorders of the brain. Science (New York, N.Y.), 360(6395), eaap8757. 10.1126/science.aap8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik‐Sullivan, B. K. , Finucane, H. K. , Anttila, V. , Gusev, A. , Day, F. R. , Loh, P.‐R. , … Neale, B. M. (2015). An atlas of genetic correlations across human diseases and traits. Nature Genetics, 47(11), 1236–1241. 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik‐Sullivan, B. K. , Loh, P.‐R. , Finucane, H. K. , Ripke, S. , Yang, J. , Schizophrenia Working Group of the Psychiatric Genomics Consortium , … Neale, B. M. (2015). LD score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nature Genetics, 47(3), 291–295. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, G. L. , Keortge, S. G. , Formea, G. M. , & Sternberger, L. G. (1996). Revision of the Padua Inventory of obsessive compulsive disorder symptoms: Distinctions between worry, obsessions, and compulsions. Behaviour Research and Therapy, 34(2), 163–173. 10.1016/0005-7967(95)00035-6 [DOI] [PubMed] [Google Scholar]
- Cath, D. C. , van Grootheest, D. S. , Willemsen, G. , van Oppen, P. , & Boomsma, D. I. (2008). Environmental factors in obsessive–compulsive behavior: Evidence from discordant and concordant monozygotic twins. Behavior Genetics, 38(2), 108–120. 10.1007/s10519-007-9185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L. K. (2019). Common knowledge: Shared genetics in psychiatry. Nature Neuroscience, 22(3), 331–332. 10.1038/s41593-019-0346-y [DOI] [PubMed] [Google Scholar]
- de Leeuw, C. A. , Mooij, J. M. , Heskes, T. , & Posthuma, D. (2015). MAGMA: Generalized gene‐set analysis of GWAS data. PLoS Computational Biology, 11(4), e1004219. 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis, D. , Walters, R. K. , Martin, J. , Mattheisen, M. , Als, T. D. , Agerbo, E. , … Neale, B. M. (2019). Discovery of the first genome‐wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genetics, 51(1), 63–75. 10.1038/s41588-018-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Braber, A. , Zilhão, N. R. , Fedko, I. O. , Hottenga, J.‐J. , Pool, R. , Smit, D. J. A. , … Boomsma, D. I. (2016). Obsessive–compulsive symptoms in a large population‐based twin‐family sample are predicted by clinically based polygenic scores and by genome‐wide SNPs. Translational Psychiatry, 6(2), e731. 10.1038/tp.2015.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denys, D. , Mantione, M. , Figee, M. , van den Munckhof, P. , Koerselman, F. , Westenberg, H. , … Schuurman, R. (2010). Deep brain stimulation of the nucleus accumbens for treatment‐refractory obsessive–compulsive disorder. Archives of General Psychiatry, 67(10), 1061–1068. 10.1001/archgenpsychiatry.2010.122 [DOI] [PubMed] [Google Scholar]
- Edwards, A. C. , Aliev, F. , Bierut, L. J. , Bucholz, K. K. , Edenberg, H. , Hesselbrock, V. , … Dick, D. M. (2012). Genome‐wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatric Genetics, 22(1), 31–41. 10.1097/YPG.0b013e32834acd07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei, O. , Holland, D. , Smeland, O. B. , Shadrin, A. A. , Fan, C. C. , Maeland, S. , … Dale, A. M. (2019). Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nature Communications, 10(1), 1–11. 10.1038/s41467-019-10310-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon, E. R. , Wheeler, H. E. , Shah, K. P. , Mozaffari, S. V. , Aquino‐Michaels, K. , Carroll, R. J. , … Im, H. K. (2015). A gene‐based association method for mapping traits using reference transcriptome data. Nature Genetics, 47(9), 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P. , Cheung, J. W. , Medland, S. E. , Mufford, M. S. , Jahanshad, N. , Dalvie, S. , … Enhancing Neuro Imaging Genetics through Meta Analysis (ENIGMA) Consortium and International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) . (2018). Significant concordance of genetic variation that increases both the risk for obsessive–compulsive disorder and the volumes of the nucleus accumbens and putamen. The British Journal of Psychiatry: The Journal of Mental Science, 213(1), 430–436. 10.1192/bjp.2018.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iervolino, A. C. , Rijsdijk, F. V. , Cherkas, L. , Fullana, M. A. , & Mataix‐Cols, D. (2011). A multivariate twin study of obsessive–compulsive symptom dimensions. Archives of General Psychiatry, 68(6), 637–644. 10.1001/archgenpsychiatry.2011.54 [DOI] [PubMed] [Google Scholar]
- International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF‐GC) and OCD Collaborative Genetics Association Studies (OCGAS) , Arnold, P. D. , Askland, K. D. , Barlassina, C. , Bellodi, L. , Bienvenu, O. J. , … Zai, G. (2018). Revealing the complex genetic architecture of obsessive–compulsive disorder using meta‐analysis. Molecular Psychiatry, 23(5), 1181–1188. 10.1038/mp.2017.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, R. C. , Berglund, P. , Demler, O. , Jin, R. , Merikangas, K. R. , & Walters, E. E. (2005). Lifetime prevalence and age‐of‐onset distributions of DSM‐IV disorders in the National Comorbidity Survey Replication. Archives of General Psychiatry, 62(6), 593–602. 10.1001/archpsyc.62.6.593 [DOI] [PubMed] [Google Scholar]
- Lee, J. J. , McGue, M. , Iacono, W. G. , & Chow, C. C. (2018). The accuracy of LD score regression as an estimator of confounding and genetic correlations in genome‐wide association studies. Genetic Epidemiology, 42(8), 783–795. 10.1002/gepi.22161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Wineinger, N. E. , Fu, D.‐J. , Libiger, O. , Alphs, L. , Savitz, A. , … Schork, N. J. (2017). Genome‐wide association study of paliperidone efficacy. Pharmacogenetics and Genomics, 27(1), 7–18. 10.1097/FPC.0000000000000250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J. , Taylor, M. J. , & Lichtenstein, P. (2018). Assessing the evidence for shared genetic risks across psychiatric disorders and traits. Psychological Medicine, 48(11), 1759–1774. 10.1017/S0033291717003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls, D. L. , Alsobrook, J. P. , Goodman, W. , Rasmussen, S. , & Leckman, J. F. (1995). A family study of obsessive–compulsive disorder. The American Journal of Psychiatry, 152(1), 76–84. 10.1176/ajp.152.1.76 [DOI] [PubMed] [Google Scholar]
- Smit, D.J.A. , Wright, M.J. , Meyers, J.L. , Martin, N.G. , Ho, Y.Y.W. , Malone, S.M. , … Boomsma, D.I. 2018. Genome‐wide association analysis links multiple psychiatric liability genes to oscillatory brain activity. Human Brain Mapping,39,4183–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel, O. A. , van Wingen, G. , Soriano‐Mas, C. , Alonso, P. , Chamberlain, S. R. , Nakamae, T. , … Veltman, D. J. (2016). Brain circuitry of compulsivity. European Neuropsychopharmacology: The Journal of the European College of Neuropsychopharmacology, 26(5), 810–827. 10.1016/j.euroneuro.2015.12.005 [DOI] [PubMed] [Google Scholar]
- van den Heuvel, O. A. , Veltman, D. J. , Groenewegen, H. J. , Dolan, R. J. , Cath, D. C. , Boellaard, R. , … van Dyck, R. (2004). Amygdala activity in obsessive–compulsive disorder with contamination fear: A study with oxygen‐15 water positron emission tomography. Psychiatry Research: Neuroimaging, 132(3), 225–237. 10.1016/j.pscychresns.2004.06.007 [DOI] [PubMed] [Google Scholar]
- van Grootheest, D. S. , Cath, D. C. , Beekman, A. T. , & Boomsma, D. I. (2005). Twin studies on obsessive–compulsive disorder: A review. Twin Research and Human Genetics, 8(5), 450–458. 10.1375/twin.8.5.450 [DOI] [PubMed] [Google Scholar]
- van Grootheest, D. S. , Cath, D. C. , Beekman, A. T. , & Boomsma, D. I. (2007). Genetic and environmental influences on obsessive–compulsive symptoms in adults: A population‐based twin‐family study. Psychological Medicine, 37(11), 1635–1644. 10.1017/S0033291707000980 [DOI] [PubMed] [Google Scholar]
- van Oppen, P. (1992). Obsessions and compulsions: Dimensional structure, reliability, convergent and divergent validity of the Padua inventory. Behaviour Research and Therapy, 30(6), 631–637. 10.1016/0005-7967(92)90008-5 [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Taskesen, E. , van Bochoven, A. , & Posthuma, D. (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8(1), 1826 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Lee, S. H. , Goddard, M. E. , & Visscher, P. M. (2011). GCTA: A tool for genome‐wide complex trait analysis. The American Journal of Human Genetics, 88(1), 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D. , Mathews, C. A. , Scharf, J. M. , Neale, B. M. , Davis, L. K. , Gamazon, E. R. , … Pauls, D. L. (2015). Cross‐disorder genome‐wide analyses suggest a complex genetic relationship between Tourette's syndrome and OCD. The American Journal of Psychiatry, 172(1), 82–93. 10.1176/appi.ajp.2014.13101306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilhão, N. R. , Smit, D. J. A. , den Braber, A. , Dolan, C. V. , Willemsen, G. , Boomsma, D. I. , & Cath, D. C. (2015). Genetic and environmental Contributions to stability in adult obsessive compulsive behavior. Twin Research and Human Genetics, 18(1), 52–60. 10.1017/thg.2014.77 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Supplementary methods.
Table S1 Padua Inventory (abbreviated) questionnaire items.
Figure S1 Age distribution histogram.
Figure S2. Manhattan plots for the Padua Inventory obsessions subscale (A), compulsions subscale (B), the original PGC‐OCD (C), and the meta‐analysis of OCD + compulsions (D).
Figure S3. Q–Q plots for the Padua Inventory obsessions subscale (A), compulsions subscale (B), the original PGC‐OCD (C), and the meta‐analysis of OCD + compulsions (D).
Figure S4. Manhattan plot for the MAGMA gene‐based test of association for the meta‐analysis OCD + compulsions GWAS.


