Summary
Epichloë festucae is an endophytic fungus that forms a symbiotic association with Lolium perenne. Here we analysed how the metabolome of the ryegrass apoplast changed upon infection of this host with sexual and asexual isolates of E. festucae.
A metabolite fingerprinting approach was used to analyse the metabolite composition of apoplastic wash fluid from uninfected and infected L. perenne. Metabolites enriched or depleted in one or both of these treatments were identified using a set of interactive tools. A genetic approach in combination with tandem MS was used to identify a novel product of a secondary metabolite gene cluster.
Metabolites likely to be present in the apoplast were identified using marvis in combination with the BioCyc and KEGG databases, and an in‐house Epichloë metabolite database. We were able to identify the known endophyte‐specific metabolites, peramine and epichloëcyclins, as well as a large number of unknown markers.
To determine whether these methods can be applied to the identification of novel Epichloë‐derived metabolites, we deleted a gene encoding a NRPS (lgsA) that is highly expressed in planta. Comparative MS analysis of apoplastic wash fluid from wild‐type‐ vs mutant‐infected plants identified a novel Leu/Ile glycoside metabolite present in the former.
Keywords: apoplast, endophyte, Epichloë, metabolome, symbiosis
Introduction
The fungal endophyte Epichloë festucae forms symbiotic associations with temperate grasses of the Festuca, Lolium and Koeleria genera (Leuchtmann et al., 1994). Hyphae grow in the intercellular (apoplastic) spaces of aerial tissues, systemically colonizing the host leaf sheath, leaf blade, and inflorescences (May et al., 2008; Scott et al., 2012). Hyphae attach to the host cell wall through an adhesive matrix and cease growing in the later stages of host development, though they remain metabolically active (Tan et al., 2001; Christensen & Voissey, 2007). Epichloë species growing in planta produce a wide variety of secondary metabolites (SMs) that protect the host against herbivores (Malinowski & Belesky, 2000; Clay & Schardl, 2002; Saikkonen et al., 2016). The most well characterized Epichloë SMs are the ergot alkaloids, lolines, indole‐diterpenes, and pyrrolopyrazines such as peramine (Young et al., 2005; Schardl et al., 2013; Berry et al., 2015). Ergot alkaloids and indole‐diterpenes are anti‐mammalian mycotoxins (Florea et al., 2016; Philippe, 2016), while lolines and peramine protect against insect herbivores (Rowan & Gaynor, 1986; Wilkinson et al., 2000; Tanaka et al., 2005; Schardl et al., 2007; Pan et al., 2014).
In addition to these well‐known SMs, Epichloë endophytes are known to produce ribosomally synthesized and post‐translationally modified peptides (RiPPs) derived from the secreted polypeptide GigA (Johnson et al., 2015). Imperfect 27‐amino‐acid repeats within the GigA sequence are cleaved to release multiple different cyclic RiPPs, which are collectively known as the ‘epichloëcyclins’. The spectrum of epichloëcyclins produced varies between different endophyte strains as a result of differences in encoded GigA sequences; for example, epichloëcyclins A–N have been characterized, but only A–E are present in plants infected with E. festucae strain Fl1 (Johnson et al., 2015). Epichloë species are also known to protect the host, to varying degrees, from a range of fungal pathogens (Gwinn & Gavin, 1992; Bonos et al., 2005; Clarke et al., 2006; Steinebrunner et al., 2008; Niones & Takemoto, 2014, 2015; Tian et al., 2017). In addition to protection from these biotic stresses, Epichloë endophytes also protect hosts from abiotic stresses such as drought (Arachevaleta et al., 1989; Hahn et al., 2007) although the physiological basis of this phenomenon is not known. Collectively, these and other studies have shown that Epichloë endophytes have an important biological role in both natural and agricultural ecosystems (Johnson et al., 2013).
There are many changes in the host transcriptome upon endophyte symbiosis establishment, including differential expression of genes involved in host hormone production, pathogenesis‐related gene expression, adaptation to biotic stresses, and host cell wall metabolism (Ambrose & Belanger, 2012; Dupont et al., 2015; Schmid et al., 2016; Dinkins et al., 2017, 2019). Given these major changes in the transcriptome it is likely that endophyte symbiosis results in metabolome reprogramming. While transcriptome studies have given us significant insights into how host and endophyte reprogramming might occur (Eaton et al., 2010, 2015; Chujo et al., 2019), these experiments provide very little insight into the exact metabolite changes that occur within the apoplastic environment where the two organisms interact. While some of the SMs synthesized by Epichloë are known to be mobilized throughout the host plant, presumably facilitating systemic host protection (Koulman et al., 2007), little is known about their exact composition in the apoplast. The aim of this study was to compare the composition of metabolites in the apoplast wash fluid of Epichloë‐infected vs mock‐infected plants by a nontargeted liquid chromatography‐coupled high‐resolution electrospray ionization mass spectroscopy (LC‐HR‐MS)‐based metabolome analysis. In particular, we set out to identify unknown compounds that could be novel symbiosis‐induced E. festucae or host‐synthesized metabolites that contribute to host protection.
Materials and Methods
Endophyte inoculations and growth conditions
Endophyte‐free and common‐toxic endophyte (CTE)‐infected seeds (Lolium perenne cv Samson) were germinated on 3% water agar. Endophyte‐free seedlings were inoculated with E. festucae strains as previously described (Latch & Christensen, 1985). Plants were grown in root trainers in an environmentally controlled growth room at 22°C with a 16 h : 8 h, light : dark photoperiod (c. 100 µmol m−2 s−1) and at 10 wk post‐inoculation, tested for the presence of the endophyte by immunoblotting (Tanaka et al., 2005).
Two‐week‐old seedlings of Triticum aestivum (cv Summit) were infected with Zymoseptoria tritici (WAI321) by syringe infiltration into the second leaf with spores at a concentration of 5 × 106 ml−1.
Escherichia coli cultures were grown in lysogeny broth (LB) or on LB agar supplemented with 100 μg ml−1 ampicillin (Miller, 1972). Epichloë festucae cultures were grown on 2.4% (w/v) potato‐dextrose (PD) agar or in PD broth (Moon et al., 1999, 2000).
Generation of gene deletion and complementation strains
Biological materials and primers can be found in Supporting Information Tables S1 and S2. Plasmid DNA was extracted using the High Pure Plasmid Isolation Kit (Roche). Fungal DNA was extracted as previously described (Byrd et al., 1990). Cloning and screening PCR reactions were performed using Q5 High‐Fidelity DNA and One‐Taq DNA polymerases (New England Biolabs (NEB), Ipswich, MA, USA) respectively. Products of PCR reactions were purified using the Wizard® SV Gel and PCR Clean‐Up system (Promega). Sequencing reactions were performed using the big‐dye terminator v.3.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA), and separated using an ABI3730 genetic analyser (Applied Biosystems). Sequence data wer assembled and analysed using macvector sequence assembly software, v.12.0.5 (MacVector Inc., Apex, NC, USA).
The lgsA deletion and complementation constructs were assembled using Gibson Assembly (Gibson et al., 2009). The lgsA (gene model EfM3.056230) (Schardl et al., 2013) replacement construct pKG39 was prepared by recombining a 2.6 kb pAN7‐1 vector backbone (amplified using primers pRS426F/pRS426R from a pAN7‐1 plasmid DNA template), a 1.4 kb hygromycin resistance cassette (primers hphF/hphR; pSF15.15 plasmid DNA template), and 1 and 2.3 kb sequence fragments that flank the lgsA gene in the Fl1 genome (primers KG167/168 and KG169/170, respectively; E. festucae Fl1 genomic DNA template). The ΔlgsA complementation construct pKG40 was prepared using a 5.5 kb pRS426 backbone (primers pRS426F/pRS426R; pRS426 plasmid DNA template) and a 4.2 kb genomic DNA fragment containing lgsA (primers KG167KG170; E. festucae Fl1 genomic DNA template). Plasmids were transformed into chemically competent E. coli DH5α cells and transformants selected using ampicillin (100 μg ml−1), and screened by restriction enzyme digestion. Candidate clones were verified by insert sequencing.
The lgsC (gene model EfM3.062310) deletion construct pCE57 was assembled using yeast recombinational cloning (Colot et al., 2006). For this, 5′ and 3′ sequences flanking the lgsC gene of 1432 and 967 bp respectively, were PCR‐amplified with Phusion® polymerase from genomic DNA template using primer sets pRS426‐cpsA‐F/cpsA‐hph‐R and hph‐cpsA‐F/cpsA‐pRS426‐R, which contained sequences that overlap with the yeast vector pRS426 and hph cassette. The PtrpC‐hph cassette was amplified with primers hph‐F/hphR. Yeast cells were transformed with the EcoRI/XhoI‐linearized pRS426 backbone together with the lgsC 5′ and 3′ flanks and the PtrpC‐hph cassette, as previously described (Gietz & Woods, 2002). Transformants were selected on media lacking uracil, and plasmid DNA isolated and transformed into E. coli to select for plasmids containing the in silico‐predicted sequence of pCE57.
Epichloë festucae protoplasts (Young et al., 2005) were prepared and transformed with 2–3 μg of target DNA, as previously described (Itoh et al., 1994). Epichloë festucae lgsA deletion strains were obtained using the Split Marker system (Rahnama et al., 2016). Two PCR fragments (primers KG171/SplitR and SplitF/KG172, pKG39 plasmid DNA template) were transformed into Fl1 protoplasts and transformants selected using hygromycin (150 μg ml−1). Epichloë festucae lgsC deletion strains were obtained by transforming a PCR‐amplified linear product from pCE57 using primers AR84/AR85. Transformants were subcultured three times onto selection medium and putative lgsA ‘knockouts’ were PCR‐screened using primer sets KG173/174 (1.5 kb knockout and 1.1 kb wild‐type (WT) products), and KG175/10 and KG11/176 (1.2 and 2.3 kb products for 5′ and 3′ flanks where correct integration has occurred). lgsC putative ‘knockouts’ were PCR‐screened using primer sets cps7/cps8 (4 kb knockout and 3.7 kb WT products), and cps7/TC44 and cps8/TC45 (1.56 kb and 2.04 kb products for 5′ and 3′ flanks where correct integration has occurred).
Candidate ‘knockouts’ were confirmed by Southern blot analysis. Genomic DNA restriction enzyme digests (NEB), separated by agarose gel electrophoresis, were transferred to positively charged nylon membranes (Roche) and fixed by UV light cross‐linking in a Cex‐800 UV light cross‐linker (Ultra‐Lum, Claremont, CA, USA) at 254 nm for 2 min. Digoxigenin‐dUTP (DIG) labelling, and hybridization of the pKG39 DNA probe, and nitroblue tetrazolium chloride and 5‐bromo‐4‐chloro‐3‐indolyl‐phosphate (NBT/BCIP) visualization, were performed using the DIG High Prime DNA Labelling and Detection Starter Kit I (Roche). HindIII generates hybridizing fragments of 8, 2.6 and 1.8 kb fragments in WT, and 10.9 and 1.8 kb fragments in ‘knockout’ strains. Complementation strains were generated by cotransforming pKG40 and pII99 plasmid DNA into ΔlgsA protoplasts using geneticin (200 μg ml−1) selection. Complementation strains were PCR‐screened using the primers KG171/172 (4.2 kb complementation product).
Microscopy analyses
Culture morphology was analysed using an Olympus IX71 inverted fluorescence microscope (Olympus Corp., Tokyo, Japan), with the filters set for capturing differential interference contrast. Infected pseudostem tissues for Confocal Laser‐Scanning Microscopy (CLSM) were stained as previously described (Becker et al., 2016, 2018). CLSM images were captured using a Leica SP5 DM6000B confocal microscope (488 nm argon and 561 nm DPSS laser, 40× oil immersion objective, NA = 1.3; Leica Microsystems, Wetzlar, Germany).
Apoplast extractions for metabolite analysis
To extract apoplastic fluid, L. perenne leaf and pseudostem tissues were rinsed and submerged in water, and vacuum‐infiltrated at c. 60 mbar for 3 × 10 min cycles. Tissues were lightly blotted dry to remove surface water and placed in 50 ml polypropylene Falcon tubes retrofitted with a plastic sieve wedged above the conical section of the tube. Tubes were centrifuged at 2000 g for 10 min to collect apoplastic wash fluid in the bottom of the tube, which was removed to a separate microtube and centrifuged at 17 000 g for 10 min to remove cellular debris, then analysed or stored at −20°C.
Apoplastic wash fluid from infected and uninfected seedlings of T. aestivum (cv Summit) were harvested at 2 wk post‐infection with Z. tritici (WAI321). The leaves were removed from the plant and submerged in sterile distilled water under vacuum for 5 min. The infiltrated leaves were then dried and placed into the barrel of a 30 ml syringe which in turn was inserted inside a sterile 50 ml centrifuge tube. The leaves were centrifuged at 5000 g for 5 min at 4°C after which the extracted apoplast wash fluid was quickly transferred to a 1.5 ml Eppendorf tube and frozen in liquid nitrogen before being freeze‐dried overnight and stored at −20°C (Solomon & Oliver, 2001).
Metabolite fingerprinting
Two‐phase extraction of apoplastic fluids
The two‐phase extraction method with methyl‐tert‐butyl ether (MTBE), methanol and water (Matyash et al., 2008) was adapted from a previous method (Feussner & Feussner, 2019) and modified for apoplastic wash fluid. Frozen apoplastic wash fluid (−80°C) was thawed on ice, vortexed and 200 µl was added to 150 µl of methanol in a glass vial followed by addition of 500 µl MTBE. The sample was vortexed and shaken for 1 h in the dark and then 120 µl of water was added. Samples were incubated for 10 min for phase separation and centrifuged for 15 min at 800 g. The upper (nonpolar) and lower (polar) phases were separately removed, transferred to new glass vials, and dried under a stream of nitrogen. The metabolites of the polar extraction phase were dissolved in 60 µl of methanol : acetonitrile : water (1 : 1 : 12, v/v/v). The metabolites of the nonpolar extraction phase were dissolved in 60 µl of 80% methanol. All samples were shaken for 15 min and centrifuged for 10 min. From each sample, 50 µl was transferred into a glass microvial, covered with argon and used for metabolite fingerprinting analysis.
Data acquisition
For metabolite fingerprinting analysis an ultraperformance liquid chromatography system coupled to the photo diode array detector eλ (Waters Corp., Milford, MA, USA) and the high‐resolution orthogonal time‐of‐flight MS (HR‐MS) LCT Premier (Waters) was used as previously described (Feussner & Feussner, 2019). Samples of the polar and the nonpolar extraction phases were analysed in the positive and in the negative electrospray ionization (ESI) mode, resulting in four datasets.
Data processing and data mining
Peak selection and alignment for each dataset was done with the software markerlynx xs for masslynx v.4.1 (Waters). This data deconvolution procedure resulted in four data matrixes of several thousand metabolite features (3258 and 1797 features for the polar extraction phase for positive and negative ESI, respectively; 1750 and 740 features for the nonpolar extraction phase for positive and negative ESI, respectively (Tables S3–S6)). The software suite marvis (Kaever et al., 2015; http://marvis.gobics.de/) was used for further data processing, such as ranking, filtering, adduct correction, merging of datasets as well as for data mining such as clustering and automated database search. Subsequently, ANOVA and Benjamini–Hochberg for multiple testing were applied with the tool marvis filter to rank the metabolite features of each of the four data matrices and filter them for a false discovery rates (FDR) of < 0.003. After adduct correction the data matrices were combined. This resulted in a dataset of 203 metabolite features (Tables S7, S8). Next, the tool marvis‐cluster was used for clustering and visualizing the intensity profiles of the selected features by means of one‐dimensional self‐organizing maps (1D‐SOMs). Finally, marvis‐pathway was applied to facilitate the annotation of metabolite features based on accurate mass information. The databases KEGG (http://www.kegg.jp; Tanabe & Kanehisa, 2012), BioCyc (http://biocyc.org; Caspi et al., 2012) and an in‐house database specific for SMs of Epichloë (Table S9) were used in combination with a framework for metabolite set enrichment analysis to annotate the metabolite features.
Verification of the chemical structure of marker compounds
To confirm or to elucidate the chemical identity of marker metabolites, LC‐HR‐MS/MS analyses were performed by UHPLC LC 1290 Infinity (Agilent Technologies, Santa Clara, CA, USA) coupled to the 6540 UHD accurate‐mass Q‐TOF LC‐MS instrument with Agilent Dual Jet Stream Technology as ESI source (Agilent Technologies).
Whole‐pseudostem extractions for polar metabolite analysis
Pseudostem tissues were snap‐frozen in liquid nitrogen, freeze‐dried, and 50 mg (DW) of each sample was placed into 2 ml impact‐resistant screw‐cap microtubes containing 3 × 3.2 mm diameter stainless steel beads and ground to a powder using a FastPrep® FP120 Cell Disrupter System (Thermo Scientific Savant, Waltham, MA, USA). Ground samples were mixed with 1 ml 50% (v/v) methanol and incubated at room temperature in the dark with end‐over‐end rotation at 40 rpm for 1 h. Cellular debris was removed by centrifugation at 17 000 g for 10 min, and the supernatant transferred to an amber glass high‐pressure liquid chromatography (HPLC) vial through a 13‐mm‐diameter, 0.45‐µm‐pore polytetrafluoroethylene syringe filter (Jet BioFil, Guangzhou, China). Extracts were then analysed or stored at −20°C.
Whole pseudostem and apoplastic fluid extracts generated for identification of the putative LgsA product were analysed using a Q Exactive Focus hybrid quadrupole‐orbitrap high‐resolution mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Each sample (5 µl injection) was separated on a 2.1 × 150 mm Zorbax Eclipse Plus C18 column with 1.8 µm particle size (Agilent Technologies) with a flow rate of 0.2 ml min−1 and a linear gradient profile using water with 0.1% formic acid as eluent A and acetonitrile with 0.1% formic acid as eluent B, with time 0 min (T 0) at 5% B, T 1 at 5% B, T 21 at 95% B, T 26 at 95% B, T 27 at 5% B, followed by equilibration to initial conditions over the following 6 min.
Samples (five replicates) were analysed by HPLC‐coupled HR positive ESI MS (LC‐MS) using a capture window of 133–2000 m/z, and the resulting mass spectra were compared using Compound Discoverer 2.1 (Thermo Scientific, Waltham, MA, USA) to identify features exhibiting highly differential profiles between sample conditions. Mass spectra from samples of mock‐inoculated (uninfected) plants were used as controls for baseline subtraction.
Bioinformatics
The E. festucae WT (Fl1/E894) genome is available at http://csbio‐l.csr.uky.edu/ef894‐2011/ (Schardl et al., 2013). The WT axenic culture and in planta transcriptome data used here are available from the Sequence Read Archive (SRA) under Bioproject PPRJNA447872 (Hassing et al., 2019).
Results
Endophyte symbiosis alters the metabolite composition of the host apoplast
In nature, the SM composition of E. festucae‐infected host tissues varies considerably depending on the E. festucae strain used (Schardl et al., 2007, 2013; Young et al., 2009). We therefore included two E. festucae strains, Fl1 and CTE, in our analysis. The former is a commonly used laboratory strain that forms a stable association with L. perenne (Leuchtmann et al., 1994; Scott et al., 2012) while the latter is a naturally occurring asexual endophyte of L. perenne (Christensen et al., 1993). While a range of SMs are known to be synthesized in each of these associations, our knowledge of the biosynthetic capability of Fl1 is much more extensive because of the availability of a complete genome sequence and the relative ease with which mutants can be generated (Scott et al., 2012; Schardl et al., 2013; Winter et al., 2018). To determine how Fl1 and CTE strains affect the host apoplastic metabolome, we extracted apoplastic wash fluid from leaves of mock‐inoculated (Mock), Fl1‐infected, and CTE‐infected L. perenne associations by two‐phase extraction and analysed both extraction phases by a metabolite fingerprinting approach using LC‐HR‐MS in both positive and negative ESI‐modes (Feussner & Feussner, 2019). This analysis resulted in several thousand metabolite features (Tables S3–S6). Filtering features of the four datasets by FDR < 0.003 selected 203 metabolite features. As LC‐ESI‐MS analyses tend to form adducts, these 203 features may represent in total c. 60–80 metabolites, which show strong differences in their intensity profiles between treatments. Principal component analysis of this dataset showed a clear separation on principal component 1 (PC1) for the apoplast metabolome of mock‐treated and FI1‐infected ryegrass, while PC2 separates all three treatments (Fig. S1). Next, the intensity profiles of the selected features were clustered and visualized by means of 1D‐SOMs and organized into four clusters (Fig. 1a; Table S7). Cluster 1 contains 36 metabolite features that are specifically enriched in CTE‐infected samples, cluster 3 contains 106 features that are specifically enriched in Fl1‐infected samples, and cluster 4 contains 42 features that are enriched in both CTE and Fl1‐infected samples. Cluster 2 contains 19 metabolite features that are depleted in the apoplastic wash fluid of endophyte‐infected plants in comparison to the mock‐treated plants.
Fig. 1.
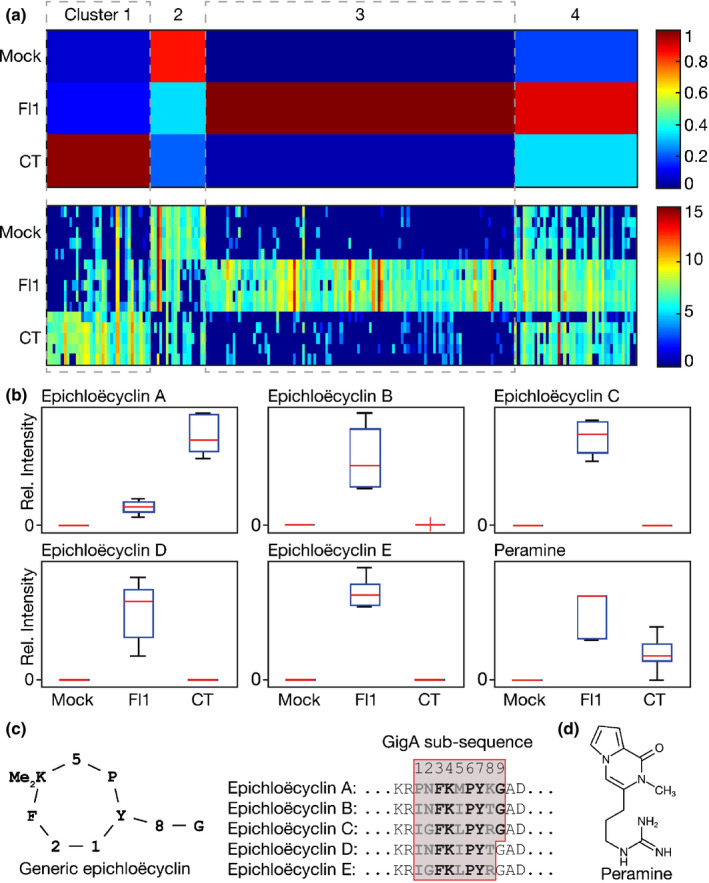
Metabolic fingerprinting of apoplastic wash fluid extracted from Epichloë festucae‐infected and uninfected Lolium perenne. Apoplastic wash fluid obtained from mock‐inoculated (M), Fl1‐infected (F) and common‐toxic endophyte (CTE)‐infected (C) L. perenne associations at 18 wk post‐inoculation were analysed by ultraperformance liquid chromatography‐electrospray‐ionization/quadrupole time‐of‐flight MS. (a) marvis one dimensional‐self organizing maps (1D‐SOM) intensity‐based clustering of the 203 features (false discovery rate < 0.003) detected in apoplastic wash fluid using ESI (blue low, red high). Five samples of M, F and C were analysed. Polar and nonpolar, positive and negative ESI data have been combined. (b) Metabolites confirmed to be in the apoplastic wash fluid samples by ultrahigh performance liquid chromatography‐quadrupole time‐of‐flight‐high resolution‐MS/MS and their respective intensities. The red mark of the box plot indicate the median. The bottom and top edges of the box represent the 25th and 75th percentiles, while the whiskers show the most extreme data points not considered outliers. (c) Generic representation of epichloëcyclin structure and GigA repeat sequences from which epichloëcyclin A–E are derived. (d) Structure of peramine.
To assign identities to these features, the accurate mass information was used for a metabolite set enrichment analysis supported by the software tool marvis‐pathway (Kaever et al., 2015). The databases BioCyc and KEGG, as well as an in‐house database, containing 117 SMs previously described for Epichloë (Table S9), were used to assign putative metabolite identities for these apoplastic features. The E. festucae metabolite peramine was identified in both CTE and Fl1‐infected samples, with a stronger accumulation in the Fl1‐infected samples (Fig. 1b,d). The identity of peramine was confirmed unequivocally by LC‐HR‐MS/MS analysis (Table S8; Fig. S2). A set of epichloëcyclins were detected as double‐charged ions ([M + 2H]2+) using positive‐mode ESI on Epichloë‐infected samples (Fig. 1b,c; Table S8). Epichloëcyclin A was detected weakly in Fl1‐infected samples and strongly in CTE‐infected samples, whereas epichloëcyclins B–E accumulated exclusively in Fl1‐infected samples (Figs 1b, S3, S4). Sequencing gigA from CTE, which is part of a four‐gene cluster (Eaton et al., 2010; Fig. S5a), revealed that this gene encodes a polypeptide that contains two identical epichloëcyclin A repeats (Fig. S5b). In addition to peramine and the epichloëcyclins, several other features representing so far unknown chemical structures were also present exclusively in the apoplastic wash fluid of endophyte‐infected associations (Tables S7, S8) including two putative peptides, which are represented by the CTE‐specific cluster 1 (Fig. S6). A putatively annotated Kojibiose‐related metabolite (322.0643 Da), a putatively annotated N‐(hydroxypentyl) acetamide (145.1101 Da) and a compound of 471.1952 Da were found in the FI1‐specific cluster 3 (Table S8). Such markers are of considerable interest as they represent potential novel bioactive fungal secondary metabolites.
Identification of a novel amino acid glycoside produced by the NRPS‐like protein LgsA
To determine whether we could connect a novel metabolite identified by the nontargeted apoplast metabolome analysis to an uncharacterized E. festucae gene clusters, we chose to delete a gene (model EfM3.056230) encoding a nonribosomal peptide synthetase (NRPS)‐like protein that is part of a putative SM gene cluster (Fig. 2; Eaton et al., 2015). The genes from this cluster are expressed at very low levels in axenic cultures of E. festucae Fl1, but are dramatically upregulated when E. festucae is growing in planta (Fig. 2; Table S10; Winter et al., 2018; Hassing et al., 2019). These genes were previously shown to be significantly downregulated in three L. perenne associations containing symbiotically defective E. festucae mutants (Eaton et al., 2015). We have provisionally named this cluster LGS (leucine/isoleucine glycoside synthesis) based on the identification of the product described below. The NRPS‐encoding gene (lgsA) from this cluster encodes the 1049 amino acid‐long protein LgsA which contains an N‐terminal thiolation (T) domain, a central condensation (C) domain, and a C‐terminal adenylylation (A) domain (Fig. S7). The presence of an A‐domain in LgsA suggests incorporation of an amino acid substrate into any product, but the domain structure of LgsA (T‐C‐A) is atypical compared with previously characterized NRPS and NRPS‐like proteins. The other genes in the LGS cluster are predicted to encode an ABC transporter (lgsD), a haloacid dehydrogenase‐like hydrolase (gene 1), a mitochondrial substrate carrier protein (gene 2), a NADP‐binding oxidoreductase (lgsB), a FAD‐dependent oxidoreductase (gene 3), and a glycosyl transferase (lgsC) (Table 1). These putative functions suggested a product containing both amino acid and sugar residues that may be exported into the apoplastic space.
Fig. 2.
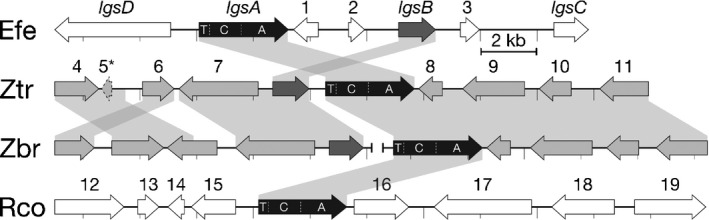
Microsyntenic comparison of LGS loci between Epichloë, Zymoseptoria and Ramularia spp. The Epichloë festucae LGS cluster genes with known or suspected functions are named lgsA–lgsC, and all other genes assigned numbers. Predicted functions of all labelled genes are listed in Table 1. Syntenic blocks are indicated by grey shading, and darker gene colouring indicates a higher degree of homology between species.
Table 1.
Putative functions of LGS cluster‐proximal genes in Epichloë, Zymoseptoria and Ramularia spp.
| Gene | Gene model(s) 1 | Putative function |
|---|---|---|
| lgsA | EfM3.056230, ZT1E4_G1886, TI39_contig403g00012, RCC_03904 | NRPS‐like, A‐T‐C domain structure |
| lgsB | EfM3.062330, ZT1E4_G1887, TI39_contig42g00014 | NADP‐binding oxidoreductase |
| lgsC | EfM3.062310 | Glycosyl transferase type 2 |
| lgsD | EfM3.056220 | ABC transporter type I |
| 1 | EfM3.056240 | Haloacid dehydrogenase‐like hydrolase |
| 2 | EfM3.062340 | Mitochondrial substrate carrier |
| 3 | EfM3.062320 | FAD‐binding oxidoreductase |
| 4 | ZT1E4_G1891, TI39_contig42g00011 | Transcription factor |
| 5 | ZT1E4_G1890, TI39_contig42g00012 | MFS sugar transporter |
| 6 | ZT1E4_G1889, TI39_contig42g00010 | Anhydro‐N‐acetylmuramic acid kinase |
| 7 | ZT1E4_G1888, TI39_contig42g00013 | DNA repair metallo‐beta‐lactamase |
| 8 | ZT1E4_G1885, TI39_contig403g00011 | Integral membrane protein, unknown function |
| 9 | ZT1E4_G1884, TI39_contig403g00010 | Nitrogen permease regulator 3 |
| 10 | ZT1E4_G1883, TI39_contig403g00009 | Mitochondrial substrate carrier |
| 11 | ZT1E4_G1882, TI39_contig403g00008 | Integral membrane protein, collagen binding/endopeptidase activity |
| 12 | RCC_03900 | Ubiquitin‐conjugating enzyme E2 |
| 13 | RCC_03901 | Integral membrane protein, SUR7/Rim9‐like |
| 14 | RCC_03902 | Unknown |
| 15 | RCC_03903 | Histidine phosphatase superfamily |
| 16 | RCC_03905 | Glycoside hydrolase family 47 |
| 17 | RCC_03906 | Sodium/calcium exchanger |
| 18 | RCC_03907 | Amino acid/polyamine transport |
| 19 | RCC_03908 | Unknown |
EfM3 gene models for Epichloë festucae are from the UKY genome database (http://www.endophyte.uky.edu). Gene models for Zymoseptoria tritici (ZT1E4), Zymoseptoria brevis (TI39), and R. collo‐cygni (RCC) are from the Ensembl Fungi database (http://fungi.ensembl.org/index.html).
To investigate the function of LgsA in E. festucae, a replacement construct (pKG39) was prepared and two overlapping PCR‐amplified linear fragments of this plasmid were introduced into the genome of E. festucae strain Fl1 by homologous recombination (Fig. S8). PCR screening of c. 20 hygromycin‐resistant transformants identified two candidates – ∆lgsA#2 and ∆lgsA#11 – that had PCR product patterns consistent with targeted replacement events. Southern blot analysis of genomic DNA digests from these transformants probed with a PCR fragment derived from pKG39 confirmed that both candidates contained a single‐copy integration of the replacement construct at the target lgsA gene locus, with no additional ectopic integrations (Fig. S8).
Macroscopic and microscopic analyses revealed that the culture morphologies of both ∆lgsA strains were indistinguishable from the WT (Fig. S9). To determine the host interaction phenotype, WT and ∆lgsA strains were inoculated into L. perenne seedlings and the phenotype of infected plants was examined at 8 wk post‐planting (Fig. S10). The whole plant and cellular phenotypes of plants infected with the ∆lgsA mutants were indistinguishable from WT‐infected plants (Fig. S10). These results were not unexpected given that the likely function of the product of this gene is the synthesis of a bioprotective metabolite, which is typically phenotypically neutral when plants are grown under laboratory conditions in the absence of biotic stress (Tanaka et al., 2005; Young et al., 2005; Saikia et al., 2012).
To examine the chemotype of these associations, whole‐pseudostem extracts and apoplastic wash fluid samples were extracted from L. perenne plants infected with WT, ∆lgsA mutants (both ∆lgsA#2 & ∆lgsA#11), or mock‐inoculated plants. Comparison of the LC‐MS data of pseudostem extract samples from Fl1‐infected and ∆lgsA‐infected plants did not identify any significantly different signal abundances. By contrast, comparison of apoplastic wash fluid samples identified a [M + H]+ 472.2022 feature in Fl1‐infected samples that was undetectable in all ∆lgsA apoplast wash fluid samples (Figs 3a,c, S11a). Targeted analysis revealed that this metabolite could also be detected in whole‐pseudostem extracts from WT‐infected plants, although signal intensity was an order of magnitude lower than from apoplastic wash fluid samples. The functional genetic link between lgsA and this metabolite of 471.1949 Da (neutral mass) was confirmed by demonstrating that metabolite production could be restored by reintroduction of a WT lgsA sequence into both ∆lgsA strains (Fig. 3a). The metabolite of 471.1949 Da was absent from apoplastic wash fluid samples from L. perenne infected with E. festucae var lolii CTE.
Fig. 3.
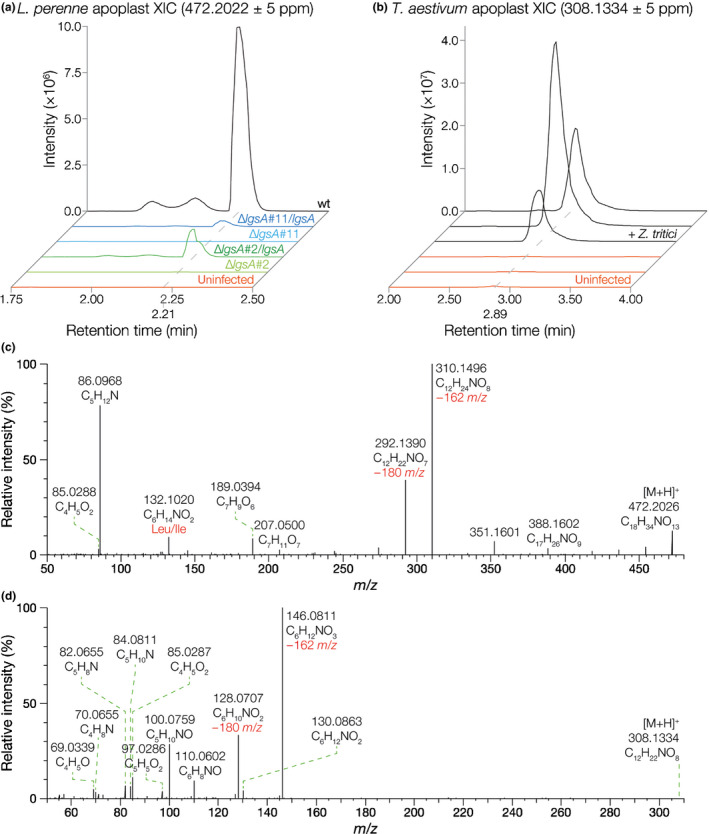
Identification of metabolites encoded by the Epichloë festucae and Zymoseptoria tritici LGS gene clusters. (a) Extracted ion chromatograms generated by LC‐high resolution‐MS (LC‐HR‐MS) analysis of apoplastic wash fluid samples extracted from uninfected, E. festucae ∆lgsA‐infected (two independent mutants), E. festucae ∆lgsA/lgsA‐infected (∆lgsA strains complemented with lgsA), and E. festucae wild‐type (WT)‐infected Lolium perenne plants. Three plants were analysed per association, with representative examples shown here. (b) Extracted ion chromatograms generated by LC‐HR‐MS analysis of apoplastic wash fluid samples extracted from uninfected and Z. tritici‐infected Triticum aestivum plants. Three plants were analysed per association, with results from all samples shown here. (c) High‐resolution accurate mass positive electrospray ionization high resolution‐MS/MS (ESI‐HR‐MS/MS) spectrum of the 472 m/z E. festucae LGS precursor ion fragmented by higher‐energy collisional dissociation (HCD) at 20% energy. (d) High‐resolution accurate mass positive ESI‐HR‐MS/MS spectrum of the 308 m/z putative Z. tritici LGS precursor ion fragmented by HCD at 20% energy.
Integration and comparison of the 13C (m/z 473.2056) and 18O (m/z 474.2064) isotopologue peak areas to the m/z 472.2022 base peak area established sum formula constraints of C18–21 and O13–14 for this LGS metabolite (Fig. S12a). Applying the nitrogen rule, the odd nominal mass of this compound (471 Da) suggests that its molecular formula contains an odd number of nitrogen atoms. Furthermore, the absence of a detectable 15N (m/z 473.1992) isotopologue suggests this compound contains only a single nitrogen atom. The metabolite was therefore assigned the elemental composition of C18H33NO13 (calculated mass, 471.1952 Da; observed mass, 471.1949 Da). LC‐HR‐MS/MS analysis suggested this LGS metabolite has a structure containing a leucine (Leu) or isoleucine (Ile) moiety shown by the diagnostic fragments of m/z 132.1020 (C6H14NO2) and m/z 86.0962 (C5H12N) (Fig. 3c; Table S8). Neutral loss of one hexose moiety (162.053 Da (C6H10O5) or 180.0636 Da (C6H12O6)) from the precursor ion of m/z 472.2026 led to fragments of m/z 310.1496 (C12H24NO8) and m/z 292.1390 (C12H22NO7), respectively. A further neutral loss of 178.0476 Da (C6H10O6) is then observed, probably resulting from loss of an oxidized hexose derivative, leaving the fragment of m/z 132.1020 (C6H14NO2), which represents the Leu/Ile‐moiety (Fig. 3c).
Analysis of apoplastic wash fluid from Zymoseptoria tritici‐infected wheat plants for LGS‐like metabolites
To determine whether proteins similar to LgsA are encoded by genes in other fungi, a tblastn search was performed against the NCBI database using the E. festucae LgsA protein sequence as query. This analysis identified matches to NRPS‐like proteins in Zymoseptoria brevis (60% amino acid identity), Z. tritici (60%), and Ramularia collo‐cygni (68%) (Fig. S7) that share the same unusual T‐C‐A domain structure as LgsA. Homologues to other putative LGS cluster genes were either absent or located at different loci in these Zymoseptoria and Ramularia spp., with the exception of the putative NADP‐binding oxidoreductase (lgsB), which was located immediately adjacent to the lgsA homologues of both Zymoseptoria species (Fig. 2).
We therefore compared apoplastic wash fluid samples from wheat plants infected with Z. tritici to uninfected wheat plants by LC‐HR‐MS. These analyses showed that Z. tritici‐infected wheat apoplastic wash fluid samples do not contain the 471 Da metabolite produced by the E. festucae LGS cluster. Data‐dependent LC‐HR‐MS/MS spectra generated from the Z. tritici‐infected wheat apoplast samples were therefore filtered to identify metabolites exhibiting a neutral loss of 162.0523 Da (C6H10O5/hexose‐moiety) or 178.0476 Da (C6H10O6/hexuronic acid‐moiety), which are characteristic features of the Epichoë LGS product. Candidates exhibiting these neutral losses were eliminated if they were also present in the LC‐HR‐MS spectra of uninfected wheat apoplastic wash fluid samples, exhibited a substantially later retention time compared with the 2.21 min E. festucae LGS metabolite, or exhibited elemental compositions or LC‐HR‐MS/MS spectrum features that were not consistent with the properties expected of an LGS‐like product. This left a single [M + H]+ 308.1334 feature with a retention time of 2.89 min as the sole remaining candidate for a Z. tritici LGS cluster product in this dataset, corresponding to a metabolite with an elemental composition of C12H21NO8 (calculated mass, 307.1267 Da; observed mass, 307.1261 Da; Figs 3b,d, S12b). Analysis of the LC‐HR‐MS/MS spectrum for this compound shows that the product ion of m/z 146.0811 remaining after the loss of the hexose‐moiety as well as the corresponding fragments of m/z 128.0707 and 100.0759 could derive from a six‐carbon amino acid, although this does not appear to be a Leu/Ile‐moiety (Figs 3d, S11b). Given that the substrate‐specifying ‘A‐domain code’ (Stachelhaus et al., 1999) of the Z. tritici LgsA homologue (DGLMYAVILK) has diverged somewhat from that of E. festucae LgsA (DALLYGIMAK) it is possible that these two proteins bind different amino acid substrates.
LgsC is required for biosynthesis of the E. festucae LGS product
Given that the E. festucae LGS cluster has a gene (lgsC; EfM3.062310) encoding a putative glycosyl transferase that is absent from the Z. tritici cluster, we hypothesized that this gene could encode an enzyme catalysing the conjugation of the second monosaccharide present in the 471 Da E. festucae LGS product. We therefore prepared a replacement construct (pCE57) for lgsC, and transformed the linear product of this construct into protoplasts of E. festucae. A PCR screen of hygromycin‐resistant transformants identified four transformants (ΔlgsC#6‐6, #7‐6, #11‐4 and #12‐6) in which the lgsC gene was deleted, with this result confirmed by Southern blot analysis (Fig. S13). Seedlings of L. perenne were infected with E. festucae WT, ΔlgsC#6‐6 and ΔlgsC#12‐6 and, as expected, L. perenne plants infected with the ∆lgsC mutant exhibited the same host interaction phenotype as WT‐infected plants (Fig. S14). Whole‐pseudostem extracts and apoplastic wash fluid samples were harvested from three mature plants of each association and analysed by LC‐HR‐MS. Apoplastic wash fluid samples from the ∆lgsC mutants lacked the 471 Da LGS product, demonstrating that LgsC is required for LGS biosynthesis in E. festucae (Fig. 4). LC‐HR‐MS analysis of apoplastic fluid and pseudostem extracts targeting metabolites with the sum formulas C12H23NO8, C12H21NO8, C12H23NO7, and C12H21NO7 revealed that ∆lgsC‐infected material does not appear to accumulate any of these hypothesized monoglycosylated LGS intermediates. However, given that the 471 Da E. festucae LGS product itself was barely detectable in pseudostem extracts, it is possible that a monoglycosylated intermediate that is not exported into the apoplast is present at low levels.
Fig. 4.
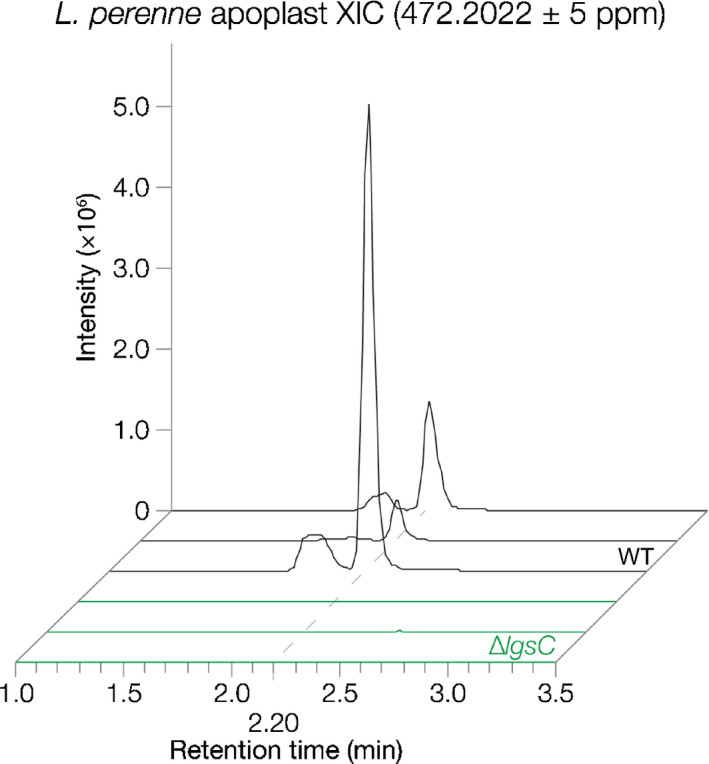
LgsC is required for synthesis of the Epichloë festucae LGS product. Extracted ion chromatograms generated by LC‐high resolution‐MS analysis of apoplastic wash fluid samples extracted from Lolium perenne plants infected with E. festucae wild‐type (WT) or ∆lgsC strains (two independent mutants). Three plants were analysed per association, with representative examples shown here.
Discussion
Establishment of a mutualistic symbiotic association between E. festucae and L. perenne results in a dramatic change to both the host and fungal transcriptome (Dupont et al., 2015; Winter et al., 2018), leading to a reprogramming of plant host and mycosymbiont metabolism. While there are many changes in primary metabolism (Rasmussen et al., 2008; Zhang et al., 2009; Dupont et al., 2015), it is the changes to fungal secondary metabolism that best define these associations as many compounds within this group have known bioprotective roles in natural ecosystems (Schardl et al., 2012). The preferential expression of genes encoding these alkaloid biosynthetic enzymes in the host plant makes it challenging to identify novel endophyte‐induced metabolites given the complexity of the leaf extract metabolome. However, by focusing on the apoplast, where the endophyte grows, the metabolite complexity is reduced, thereby enabling the identification of novel products. Here we describe for the first time a nontargeted analysis of the L. perenne apoplast metabolome and how it changes upon establishment of a symbiotic interaction with E. festucae.
The metabolite differences between the apoplast metabolome of mock‐infected vs E. festucae‐infected ryegrass is surprisingly uncomplex, with just 203 metabolite features identified distinguishing these two physiological states. This is in contrast to more complex metabolome changes reported for the plant fungal pathogen Verticillium longisporum (Floerl et al., 2012). Of these features, 184 were shown to be enriched in the endophyte‐infected apoplastic fluid wash with the remaining 19 features corresponding to depletions in endophyte‐infected apoplastic fluid wash. As expected, fungal metabolites known to be preferentially accumulated in the host are among those enriched upon establishment of endophyte symbiosis, including peramine (Tanaka et al., 2005), and epichloëcyclins (Johnson et al., 2015). No lolines were detected in this analysis, as the two E. festucae strains used lack this biosynthetic capability (Schardl et al., 2013).
The epichloëcyclins were among the most abundant metabolites synthesized in response to endophyte symbiosis. These are RiPPs generated through proteolytic cleavage and cyclization of linear peptide repeats that comprise the pro‐peptide product of gigA. This gene is one of the most highly expressed in the endophyte–grass symbiotic interaction and is part of a four‐gene cluster (Eaton et al., 2010). This cluster also includes kexB, which encodes a kexin presumably required for cleavage of the repeats from the pro‐peptide; a gene encoding a protein with a DUF3328 domain, which is also found in UstYa/UstYb, which are proteins required for cyclization of the ustiloxin precursors in Aspergillus flavus (Nagano et al., 2016; Ye et al., 2016); and a gene encoding a hypothetic protein of unknown function. The cyclic nonapeptides epichloëcyclin A, B and C, and the cyclic octapeptides epichloëcylin B and C (Johnson et al., 2015), were found in the apoplastic wash fluid of ryegrass infected with strain Fl1, whereas only epichloëcyclin A was found in apoplastic wash fluid from CTE‐infected plants. These results are in agreement with the repeat structures of the corresponding GigA‐derived peptides from these E. festucae strains.
Regarding the alkaloids, both E. festucae strains have a functional copy of peramine synthetase, perA, and accumulate peramine in the apoplastic space as expected, given the known systemic distribution of this metabolite within the plant (Koulman et al., 2007). While metabolites with masses that match indole‐diterpenes known to be synthesized by these strains were detected (Saikia et al., 2012), the low signal intensity precluded their confirmation by LC‐HR‐MS/MS. The absence of ergot alkaloids, which are known to be synthesized by these strains in planta (Chujo & Scott, 2014), is probably a result of their lack of transport into the apoplast.
As anticipated, there were many metabolites of unknown structure, highlighting the utility of this type of analysis to identify novel endophyte‐bioprotective metabolites. We therefore employed a combined genetics/metabolomics approach using the well‐developed genetic system of E. festucae strain Fl1 (Scott et al., 2012, 2018) to demonstrate the utility of this apoplastic wash fluid method for identification of putative novel fungal metabolites. For this analysis we chose to delete the NRPS‐encoding gene lgsA, which is a component of a putative seven‐gene SM cluster that is downregulated in L. perenne associations with three different symbiotically defective E. festucae mutants (Eaton et al., 2015). All seven genes were found here to be significantly upregulated in the Fl1 in planta transcriptome compared with axenic culture. As expected for a gene encoding a putative bioprotective metabolite (Tanaka et al., 2005; Young et al., 2005; Saikia et al., 2012), the axenic culture and plant interaction phenotypes of the E. festucae ΔlgsA mutant were indistinguishable from the WT. A 471 Da metabolite that was absent in apoplastic wash fluid samples from plants infected with the ΔlgsA mutants was identified by LC‐HR‐MS in extracts of apoplastic wash fluid from L. perenne plants infected with WT E. festucae Fl1. The failure to identify this compound in an equivalent comparative analysis of whole‐pseudostem extracts of these same plants highlights the power of working with the much less complex apoplastic metabolome. LC‐HR‐MS/MS analysis suggests this LGS metabolite has a structure containing a Leu/Ile moiety linked to a glycoside constituent. Given the relatively small quantities of apoplastic wash fluid obtained using the method described here, definitive structural determination will probably require development of a high‐volume extraction method or an alternative approach such as overexpressing the cluster genes in a heterologous fungal host to obtain sufficient product for NMR analysis (Van de Bittner et al., 2018; van Dolleweerd et al., 2018). The absence of this 471 Da metabolite in the apoplastic fluid of L. perenne with CTE suggests that the genes are absent, mutated, or not expressed in this strain. However, the presence of the LGS cluster across several Epichloë species suggests this is a widespread SM of this genus (Eaton et al., 2015).
A comparative synteny analysis of the genomes of other filamentous fungi identified homologues to the NRPS‐encoding lgsA gene in the genomes of the cereal pathogens R. collo‐cygni, Z. brevis, and Z. tritici, with conserved homologues to the putative NADP‐binding oxidoreductase‐encoding gene lgsB located immediately adjacent in the Zymoseptoria genomes. LC‐HR‐MS analysis of extracts of apoplastic wash fluid samples from wheat infected with WT Z. tritici compared with apoplastic wash fluid from uninfected wheat plants identified a 307 Da metabolite that appears to have some structural similarities to the 471 Da metabolite, but with only a single glycosyl constituent. This structure is consistent with the absence of a conserved homologue to the putative glycosyl transferase‐encoding gene lgsC in these species. However, more substantial evidence, such as analysis of a Z. tritici ∆lgsA mutant, will be required to prove the hypothesized link between the Z. tritici LGS cluster and this 307 Da metabolite.
While the accessory gene content of a secondary metabolism cluster can vary considerably between fungal species, the genes encoding synthesis of the basic structural motif for a class of secondary metabolites are typically conserved (Schardl et al., 2013). The conservation of lgsA and lgsB between the LGS clusters of Zymoseptoria and Epichloë spp. therefore suggests that LgsA and LgsB catalyse the first reactions in LGS biosynthesis. The presence of a Leu/Ile product ion in the LC‐HR‐MS/MS spectrum of the E. festucae LGS product suggests that the A‐domain of LgsA binds one of these amino acids as substrate. However, NRPS T‐domains typically accept activated amino acyl substrates from an upstream A‐domain (Weissman, 2015). Given the unusual T‐C‐A structure of LgsA, it is unclear if the LgsA T‐domain could accept an activated Leu/Ile substrate from the downstream A‐domain. An alternative hypothesis would be that the LgsA T‐domain is loaded with an oxidized hexose derivative provided by LgsB. The LgsA C‐domain would then catalyse condensation between the amino acid and sugar moieties, which would be linked by an ester or amide bond depending on whether the T‐domain is loaded with the Leu/Ile or sugar constituent, respectively. LgsC would add the glycosyl group observed in E. festucae, either before or after the LgsA‐catalysed condensation event. The final LGS pathway product would then be exported into the apoplast by a transporter, presumably the putative ABC transporter encoded by lgsD in E. festucae. While not specifically analysed here, other co‐clustered genes in Epichloë and Zymoseptoria spp. may also encode proteins that contribute to LGS biosynthesis in their respective species.
While we do not know what the biological functions are for the compounds identified here, we make two observations: first, the gene clusters that encode the enzymes for the synthesis of these novel SMs appear to be limited to filamentous fungi that infect monocots of the subfamily Pooideae, and second, this gene cluster is dispensable for a mutualistic symbiotic interaction between E. festucae and L. perenne. Whether these products have a role in bioprotection, virulence or some other interaction role remains to be determined. It will be interesting to analyse the host interaction phenotype of a Z. tritici ΔlgsA mutant.
In conclusion, we describe for the first time how the metabolome of L. perenne changes upon establishment of symbiosis with either asexual or sexual isolates of E. festucae. Interestingly, the changes are not as dramatic as observed for fungal pathogen infection by V. longisporum, though the total number of studies in this field are still rather limited. Given the lower complexity of the apoplast metabolome in comparison to the metabolome of green tissues we have demonstrated here the power of a combined genetics/metabolomics approach to identify new metabolites in host apoplastic wash fluids. It will be of considerable interest to determine the structures of other unknown metabolites in this and other fungal–plant associations, and to identify the biological function of these metabolites in these symbioses.
Author contributions
KAG, KF, DB, IF and BS planned and designed the research. KAG, KF, DB, CJE, AR and PS performed the experiments. KAG, KF, DB, CHM, IF and BS analysed the data. KAG, DB and BS wrote the manuscript. KAG, DB and KF contributed equally to this work.
Supporting information
Fig. S1 Principal component analysis (PCA) of 203 metabolite features.
Fig. S2 Extracted ion chromatogram of peramine.
Fig. S3 Extracted ion chromatogram of epichloëcyclins A–E.
Fig. S4 Confirmation of the identity of epichloëcyclins A–E.
Fig. S5 gigA gene cluster organization and sequences.
Fig. S6 Extracted ion chromatogram of a putative peptide.
Fig. S7 LgsA predicted domain structure and multiple sequence alignment.
Fig. S8 lgsA deletion and complementation construct design and strain screening.
Fig. S9 Culture morphology of WT and ∆lgsA strains.
Fig. S10 Host interaction and cellular phenotypes of L. perenne infected with WT and ∆lgsA strains.
Fig. S11 ESI‐HR‐MS/MS fragmentation of 472 m/z metabolite in negative mode.
Fig. S12 472 and 308 metabolite isotope analysis.
Fig. S13 Strategy for deletion of E. festucae lgsC, Southern blot and PCR analysis.
Fig. S14 Host interaction phenotype of L. perenne infected with WT and ∆lgsC strains.
Table S1 Biological material.
Table S2 Primers used in this study.
Table S3 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the polar extraction phase, analysed in positive ESI‐mode.
Table S4 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the polar extraction phase, analysed in negative ESI mode.
Table S5 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the nonpolar extraction phase, analysed in positive ESI mode.
Table S6 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the nonpolar extraction phase, analysed in negative ESI mode.
Table S7 Data matrix of 203 high quality metabolite features (false discovery rate < 0.003) obtained by metabolite fingerprinting (UPLC‐ESI‐TOF‐MS analysis) of apoplastic wash fluids from mock‐treated, FI1‐ and CTE‐infected L. perenne.
Table S8 Infection markers identified by metabolite fingerprinting (UPLC‐ESI‐TOF‐MS analysis) and verified by UHPLC‐ESI‐QTOF‐HR‐MS/MS analysis or coelution.
Table S9 Epichloë metabolite database.
Table S10 Differences in expression of lgs cluster genes in planta compared with axenic culture.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This research was supported by a grant from the Tertiary Education Commission to the Bio‐Protection Research Centre and by Massey University. BS was supported by an Alexander von Humboldt Research Award. IF was supported by the Deutsche Forschungsgemeinschaft (ZUK 45/2010). The authors thank Matthew Savoian, Panimalar Vijayan and Niki Minards (Manawatu Microscopy and Imaging Centre) for technical assistance, Nazanin Noorifar (Massey University) for assistance with plant inoculations, David Lun (Massey University) for assistance with MS, Richard Johnson (AgResearch) for provision of E. festucae lgsA sequences, Benedikt Ni (University of Goettingen) for compiling the Epichloë metabolite database and Christopher Schardl (University of Kentucky) for provision of Clavicipitaceae genome sequences.
References
- Ambrose KV, Belanger FC. 2012. SOLiD‐SAGE of endophyte‐infected red fescue reveals numerous effects on host transcriptome and an abundance of highly expressed fungal secreted proteins. PLoS ONE 7: e53214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arachevaleta M, Bacon CW, Hoveland CS, Radcliffe DE. 1989. Effect of the tall fescue endophyte on plant response to environmental stress. Agronomy Journal 81: 83–90. [Google Scholar]
- Becker M, Becker Y, Green K, Scott B. 2016. The endophytic symbiont Epichloë festucae establishes an epiphyllous net on the surface of Lolium perenne leaves by development of an expressorium, an appressorium‐like leaf exit structure. New Phytologist 211: 240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker Y, Green K, Scott B, Becker M. 2018. Artificial inoculation of Epichloë festucae into Lolium perenne, and visualization of endophytic and epiphyllous fungal growth. Bio‐protocol 8: e2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Takach JE, Schardl CL, Charlton ND, Scott B, Young CA. 2015. Disparate independent genetic events disrupt the secondary metabolism gene perA in certain symbiotic Epichloë species. Applied and Environmental Microbiology 81: 2797–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonos SA, Wilson MM, Meyer WA, Funk RC. 2005. Suppression of red thread in fine fescues through endophyte‐mediated resistance. Applied Turfgrass Science 10: 1094. [Google Scholar]
- Byrd AD, Schardl CL, Songlin PJ, Mogen KL, Siegel MR. 1990. The β‐tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne). Current Genetics 18: 347–354. [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA et al 2012. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Research 40: D742–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen MJ, Leuchtmann A, Rowan DD, Tapper BA. 1993. Taxonomy of Acremonium endophytes of tall fescue (Festuca arundinacea), meadow fescue (F. pratensis) and perennial rye‐grass (Lolium perenne). Mycological Research 97: 1083–1092. [Google Scholar]
- Christensen MJ, Voissey CR. 2007. The biology of the endophyte/grass partnership In: Popay AJ, Thom ER, eds. Proceedings of the 6th International Symposium on Fungal Endophytes of Grasses. Grasslands research and practice series no. 13. Christchurch, New Zealand: New Zealand Grassland Association, 123–133. [Google Scholar]
- Chujo T, Lukito Y, Eaton CJ, Dupont PY, Johnson LJ, Winter D, Cox MP, Scott B. 2019. Complex epigenetic regulation of alkaloid biosynthesis and host interaction by heterochromatin protein I in a fungal endophyte–plant symbiosis. Fungal Genetics and Biology 125: 71–83. [DOI] [PubMed] [Google Scholar]
- Chujo T, Scott B. 2014. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte‐plant symbiosis. Molecular Microbiology 92: 413–434. [DOI] [PubMed] [Google Scholar]
- Clarke BB, White JFJ, Hurley RH, Torres MS, Sun S, Huff DR. 2006. Endophyte‐mediated suppression of dollar spot disease in fine fescues. Plant Disease 90: 994–998. [DOI] [PubMed] [Google Scholar]
- Clay K, Schardl C. 2002. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. The American Naturalist 160: S99–S127. [DOI] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. 2006. A high‐throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proceedings of the National Academy of Sciences, USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins RD, Nagabhyru P, Graham MA, Boykin D, Schardl CL. 2017. Transcriptome response of Lolium arundinaceum to its fungal endophyte Epichloë coenophiala . New Phytologist 213: 324–337. [DOI] [PubMed] [Google Scholar]
- Dinkins RD, Nagabhyru P, Young CA, West CP, Schardl CL. 2019. Transcriptome analysis and differential expression in tall fescue harboring different endophyte strains in response to water deficit. The Plant Genome 12: 180071. [DOI] [PubMed] [Google Scholar]
- van Dolleweerd CJ, Kessans SA, Van de Bittner KC, Bustamante LY, Bundela R, Scott B, Nicholson MJ, Parker EJ. 2018. MIDAS: A modular DNA assembly system for synthetic biology. ACS Synthetic Biology 7: 1018–1029. [DOI] [PubMed] [Google Scholar]
- Dupont PY, Eaton CJ, Wargent JJ, Fechtner S, Solomon P, Schmid J, Day RC, Scott B, Cox MP. 2015. Fungal endophyte infection of ryegrass reprograms host metabolism and alters development. New Phytologist 208: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton CJ, Cox MP, Ambrose B, Becker M, Hesse U, Schardl CL, Scott B. 2010. Disruption of signaling in a fungal‐grass symbiosis leads to pathogenesis. Plant Physiology 153: 1780–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton CJ, Dupont P‐Y, Solomon PS, Clayton W, Scott B, Cox MP. 2015. A core gene set describes the molecular basis of mutualism and antagonism in Epichloë species. Molecular Plant–Microbe Interactions 28: 218–231. [DOI] [PubMed] [Google Scholar]
- Feussner K, Feussner I. 2019. Comprehensive LC‐MS‐based metabolite fingerprinting approach for plant and fungal‐derived samples In: D'Alessandro A, ed. High‐throughput metabolomics: methods and protocols. New York, NY, USA: Springer, 167–185. [DOI] [PubMed] [Google Scholar]
- Floerl S, Majcherczyk A, Possienke M, Feussner K, Tappe H, Gatz C, Feussner I, Kues U, Polle A. 2012. Verticillium longisporum infection affects the leaf apoplastic proteome, metabolome, and cell wall properties in Arabidopsis thaliana . PLoS ONE 7: e31435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea S, Phillips TD, Panaccione DG, Farman ML, Schardl CL. 2016. Chromosome‐end knockoff strategy to reshape alkaloid profiles of a fungal endophyte. G3: Genes, Genomes, Genetics 6: 2601–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single‐stranded carrier DNA/polyethylene glycol method. Methods in Enzymology 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Gwinn KD, Gavin AM. 1992. Relationship between endophyte infestation level of tall fescue seed lots and Rhizoctonia zeae seedling disease. Plant Disease 76: 911–914. [Google Scholar]
- Hahn H, McManus MT, Warnstorff K, Monahan BJ, Young CA, Davies E, Scott B. 2007. Neotyphodium fungal endophytes confer physiological protection to perennial ryegrass (Lolium perenne L.) subjected to water deficit. Environmental and Experimental Botany 63: 183–199. [Google Scholar]
- Hassing B, Winter D, Becker Y, Mesarich CH, Eaton CJ, Scott B. 2019. Analysis of Epichloë festucae small secreted proteins in the interaction with Lolium perenne . PLoS ONE 14: e0209463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Johnson R, Scott B. 1994. Integrative transformation of the mycotoxin‐producing fungus, Penicillium paxilli . Current Genetics 25: 508–513. [DOI] [PubMed] [Google Scholar]
- Johnson LJ, De Bonth ACM, Briggs LR, Caradus JR, Finch SC, Fleetwood DJ, Fletcher LR, Hume DE, Johnson RD, Popay AJ et al 2013. The exploitation of epichloae endophytes for agricultural benefit. Fungal Diversity 60: 171–188. [Google Scholar]
- Johnson RD, Lane GA, Koulman A, Cao M, Fraser K, Fleetwood DJ, Voisey CR, Dyer JM, Pratt J, Christensen M et al 2015. A novel family of cyclic oligopeptides derived from ribosomal peptide synthesis of an in planta‐induced gene, gigA, in Epichloë endophytes of grasses. Fungal Genetics and Biology 85: 14–24. [DOI] [PubMed] [Google Scholar]
- Kaever A, Landesfeind M, Feussner K, Mosblech A, Heilmann I, Morgenstern B, Feussner I, Meinicke P. 2015. MarVis‐Pathway: integrative and exploratory pathway analysis of non‐targeted metabolomics data. Metabolomics 11: 764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulman A, Lane GA, Christensen MJ, Fraser K, Tapper BA. 2007. Peramine and other fungal alkaloids are exuded in the guttation fluid of endophyte‐infected grasses. Phytochemistry 68: 355–360. [DOI] [PubMed] [Google Scholar]
- Latch GCM, Christensen MJ. 1985. Artificial infection of grasses with endophytes. Annals of Applied Biology 107: 17–24. [Google Scholar]
- Leuchtmann A, Schardl CL, Siegel MR. 1994. Sexual compatibility and taxonomy of a new species of Epichloë symbiotic with fine fescue grasses. Mycologia 86: 802–812. [Google Scholar]
- Malinowski DP, Belesky DP. 2000. Adaptations of endophyte‐infected cool‐season grasses to environmental stresses: mechanisms of drought and mineral stress tolerance. Crop Science 40: 923–940. [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. 2008. Lipid extraction by methyl‐tert‐butyl ether for high‐throughput lipidomics. Journal of Lipid Research 49: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KJ, Bryant MK, Zhang X, Ambrose B, Scott B. 2008. Patterns of expression of a lolitrem biosynthetic gene in the Epichloë festucae‐perennial ryegrass symbiosis. Molecular Plant–Microbe Interactions 21: 188–197. [DOI] [PubMed] [Google Scholar]
- Miller JH. 1972. Experiments in molecular genetics. New York, NY, USA: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Moon CD, Scott B, Schardl CL, Christensen MJ. 2000. The evolutionary origins of Epichloë endophytes from annual ryegrasses. Mycologia 92: 1103–1118. [Google Scholar]
- Moon CD, Tapper BA, Scott B. 1999. Identification of Epichloë endophytes in planta by a microsatellite‐based PCR fingerprinting assay with automated analysis. Applied and Environmental Microbiology 65: 1268–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano N, Umemura M, Izumikawa M, Kawano J, Ishii T, Kikuchi M, Tomii K, Kumagai T, Yoshimi A, Machida M et al 2016. Class of cyclic ribosomal peptide synthetic genes in filamentous fungi. Fungal Genetics and Biology 86: 58–70. [DOI] [PubMed] [Google Scholar]
- Niones JT, Takemoto D. 2014. An isolate of Epichloë festucae, an endophytic fungus of temperate grasses, has growth inhibitory activity against selected grass pathogens. Journal of General Plant Pathology 80: 337–347. [Google Scholar]
- Niones JT, Takemoto D. 2015. VibA, a homologue of a transcription factor for fungal heterokaryon incompatibility, is involved in antifungal compound production in the plant‐symbiotic fungus Epichloë festucae . Eukaryotic Cell 14: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Bhardwaj M, Faulkner JR, Nagabhyru P, Charlton ND, Higashi RM, Miller AF, Young CA, Grossman RB, Schardl CL. 2014. Ether bridge formation in loline alkaloid biosynthesis. Phytochemistry 98: 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe G. 2016. Lolitrem B and indole diterpene alkaloids produced by endophytic fungi of the genus Epichloë and their toxic effects in livestock. Toxins 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnama M, Forester N, Ariyawansa KGSU, Voisey CR, Johnson LJ, Johnson RD, Fleetwood DJ. 2016. Efficient targeted mutagenesis in Epichloë festucae using a split marker system. Journal of Microbiological Methods 134: 62–65. [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Parsons AJ, Fraser K, Xue H, Newman JA. 2008. Metabolic profiles of Lolium perenne are differentially affected by nitrogen supply, carbohydrate content, and fungal endophyte infection. Plant Physiology 146: 1440–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan DD, Gaynor DL. 1986. Isolation of feeding deterrents against Argentine stem weevil from ryegrass infected with the endophyte Acremonium loliae . Journal of Chemical Ecology 12: 647–658. [DOI] [PubMed] [Google Scholar]
- Saikia S, Takemoto D, Tapper BA, Lane GA, Fraser K, Scott B. 2012. Functional analysis of an indole‐diterpene gene cluster for lolitrem B biosynthesis in the grass endosymbiont Epichloë festucae . FEBS Letters 586: 2563–2569. [DOI] [PubMed] [Google Scholar]
- Saikkonen K, Young CA, Helander M, Schardl CL. 2016. Endophytic Epichloë species and their grass hosts: from evolution to applications. Plant Molecular Biology 90: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Grossman RB, Nagabhyru P, Faulkner JR, Mallik UP. 2007. Loline alkaloids: Currencies of mutualism. Phytochemistry 68: 980–996. [DOI] [PubMed] [Google Scholar]
- Schardl CL, Young CA, Faulkner JR, Florea S, Pan J. 2012. Chemotypic diversity of epichloae, fungal symbionts of grasses. Fungal Ecology 5: 331–344. [Google Scholar]
- Schardl CL, Young CA, Hesse U, Amyotte SG, Andreeva K, Calie PJ, Fleetwood DJ, Haws DC, Moore N, Oeser B et al 2013. Plant‐symbiotic fungi as chemical engineers: multi‐genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genetics 9: e1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J, Day R, Zhang N, Dupont PY, Cox MP, Schardl CL, Minards N, Truglio M, Moore N, Harris DR et al 2016. Host tissue environment directs activities of an Epichloë endophyte, while it induces systemic hormone and defense responses in its native perennial ryegrass host. Molecular Plant–Microbe Interactions 30: 138–149. [DOI] [PubMed] [Google Scholar]
- Scott B, Becker Y, Becker M, Cartwright G. 2012. Morphogenesis, growth and development of the grass symbiont Epichloë festucae In: Martin JP, Di Pietro A, eds. Morphogenesis and pathogenicity in fungi. Heidelberg, Germany: Springer‐Verlag, 243–264. [Google Scholar]
- Scott B, Green K, Berry D. 2018. The fine balance between mutualism and antagonism in the Epichloë festucae–grass symbiotic interaction. Current Opinion in Plant Biology 44: 32–38. [DOI] [PubMed] [Google Scholar]
- Solomon PS, Oliver RP. 2001. The nitrogen content of the tomato leaf apoplast increases during infection by Cladosporium fulvum . Planta 213: 241–249. [DOI] [PubMed] [Google Scholar]
- Stachelhaus T, Mootz HD, Marahiel MA. 1999. The specificity‐conferring code of adenylation domains in nonribosomal peptide synthetases. Chemistry & Biology 6: 493–505. [DOI] [PubMed] [Google Scholar]
- Steinebrunner F, Schiestl FP, Leuchtmann A. 2008. Ecological role of volatiles produced by Epichloë: differences in antifungal toxicity. FEMS Microbiology Ecology 64: 307–316. [DOI] [PubMed] [Google Scholar]
- Tan YY, Spiering MJ, Scott V, Lane GA, Christensen MJ, Schmid J. 2001. In planta regulation of extension of an endophytic fungus and maintenance of high metabolic rates in its mycelium in the absence of apical extension. Applied and Environmental Microbiology 67: 5377–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Kanehisa M. 2012. Using the KEGG database resource. Current Protocols in Bioinformatics 38: 1.12.1–1.12.43. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B. 2005. A symbiosis expressed non‐ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Molecular Microbiology 57: 1036–1050. [DOI] [PubMed] [Google Scholar]
- Tian Z, Wang R, Ambrose KV, Clarke BB, Belanger FC. 2017. The Epichloë festucae antifungal protein has activity against the plant pathogen Sclerotinia homoeocarpa, the causal agent of dollar spot disease. Scientific Reports 7: 5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Bittner KC, Nicholson MJ, Bustamante LY, Kessans SA, Ram A, van Dolleweerd CJ, Scott B, Parker EJ. 2018. Heterologous biosynthesis of nodulisporic acid F. Journal of the American Chemical Society 140: 582–585. [DOI] [PubMed] [Google Scholar]
- Weissman KJ. 2015. The structural biology of biosynthetic megaenzymes. Nature Chemical Biology 11: 660–670. [DOI] [PubMed] [Google Scholar]
- Wilkinson HH, Siegel MR, Blankenship JD, Mallory AC, Bush LP, Schardl CL. 2000. Contribution of fungal loline alkaloids to protection from aphids in a grass‐endophyte mutualism. Molecular Plant–Microbe Interactions 13: 1027–1033. [DOI] [PubMed] [Google Scholar]
- Winter DJ, Ganley ARD, Young CA, Liachko I, Schardl CL, Dupont PY, Berry D, Ram A, Scott B, Cox MP. 2018. Repeat elements organise 3D genome structure and mediate transcription in the filamentous fungus Epichloë festucae . PLoS Genetics 14: e1007467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Minami A, Igarashi Y, Izumikawa M, Umemura M, Nagano N, Machida M, Kawahara T, Shin‐Ya K, Gomi K et al 2016. Unveiling the biosynthetic pathway of the ribosomally synthesized and post‐translationally modified peptide ustiloxin B in filamentous fungi. Angewandte Chemie International Edition 55: 8072–8075. [DOI] [PubMed] [Google Scholar]
- Young CA, Bryant MK, Christensen MJ, Tapper BA, Bryan GT, Scott B. 2005. Molecular cloning and genetic analysis of a symbiosis‐expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Molecular Genetics and Genomics 274: 13–29. [DOI] [PubMed] [Google Scholar]
- Young CA, Tapper BA, May K, Moon CD, Schardl CL, Scott B. 2009. Indole‐diterpene biosynthetic capability of Epichloë endophytes as predicted by ltm gene analysis. Applied and Environmental Microbiology 75: 2200–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DX, Nagabhyru P, Schardl CL. 2009. Regulation of a chemical defense against herbivory produced by symbiotic fungi in grass plants. Plant Physiology 150: 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Principal component analysis (PCA) of 203 metabolite features.
Fig. S2 Extracted ion chromatogram of peramine.
Fig. S3 Extracted ion chromatogram of epichloëcyclins A–E.
Fig. S4 Confirmation of the identity of epichloëcyclins A–E.
Fig. S5 gigA gene cluster organization and sequences.
Fig. S6 Extracted ion chromatogram of a putative peptide.
Fig. S7 LgsA predicted domain structure and multiple sequence alignment.
Fig. S8 lgsA deletion and complementation construct design and strain screening.
Fig. S9 Culture morphology of WT and ∆lgsA strains.
Fig. S10 Host interaction and cellular phenotypes of L. perenne infected with WT and ∆lgsA strains.
Fig. S11 ESI‐HR‐MS/MS fragmentation of 472 m/z metabolite in negative mode.
Fig. S12 472 and 308 metabolite isotope analysis.
Fig. S13 Strategy for deletion of E. festucae lgsC, Southern blot and PCR analysis.
Fig. S14 Host interaction phenotype of L. perenne infected with WT and ∆lgsC strains.
Table S1 Biological material.
Table S2 Primers used in this study.
Table S3 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the polar extraction phase, analysed in positive ESI‐mode.
Table S4 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the polar extraction phase, analysed in negative ESI mode.
Table S5 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the nonpolar extraction phase, analysed in positive ESI mode.
Table S6 Data matrix of raw data obtained by UPLC‐ESI‐TOF‐MS‐based metabolite fingerprinting analysis of the nonpolar extraction phase, analysed in negative ESI mode.
Table S7 Data matrix of 203 high quality metabolite features (false discovery rate < 0.003) obtained by metabolite fingerprinting (UPLC‐ESI‐TOF‐MS analysis) of apoplastic wash fluids from mock‐treated, FI1‐ and CTE‐infected L. perenne.
Table S8 Infection markers identified by metabolite fingerprinting (UPLC‐ESI‐TOF‐MS analysis) and verified by UHPLC‐ESI‐QTOF‐HR‐MS/MS analysis or coelution.
Table S9 Epichloë metabolite database.
Table S10 Differences in expression of lgs cluster genes in planta compared with axenic culture.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


