Abstract
The class of fungal indole alkaloids containing the bicyclo[2.2.2]diazaoctane ring is comprised of diverse molecules that display a range of biological activities. While much interest has been garnered due to their therapeutic potential, this class of molecules also displays unique chemical functionality, making them intriguing synthetic targets. Many elegant and intricate total syntheses have been developed to generate these alkaloids, but the selectivity required to produce these structures with high yield presents great barriers. Alternatively, if we can understand the molecular mechanisms behind how fungi make these complex molecules, we can leverage the power of nature to perform these chemical transformations. Here, we describe the various studies regarding the evolutionary development of enzymes involved in fungal indole alkaloid biosynthesis.
Keywords: biosynthesis, Diels-Alderase, natural products, non-ribosomal peptides, monooxygenase
1. Introduction to Fungal Indole Alkaloids
The fungal indole alkaloid class of natural products contains molecules with unique structural properties and a variety of biological activities. In particular, the subgroup typified by the bicyclo[2.2.2]diazaoctane ring has been extensively studied within our group. Initially, feeding studies using isotopically labeled precursors and biomimetic chemical syntheses provided insight into the biosynthetic pathways for these molecules. These efforts were later supplemented with genome sequencing and bioinformatics analyses,[1] greatly advancing our understanding of how these natural products are generated, yet we have only recently utilized our capabilities to address the knowledge gaps with detailed biochemical characterization. Multiple strategies including molecular and structural biology have brought our foundational understanding of these pathways to the forefront of the field.
The extent of our work has focused on the families of monoketopiperazines (MKP) such as the paraherquamides (Phq)[2, 3] and malbrancheamides (Mal),[4] and diketopiperazines (DKP) such as the stephacidins,[5] notoamides (Not),[6–8] and brevianamides (Bvn) (Figures 1–3).[9] These molecules display significant biological activities with potential therapeutic value such as anthelmintics,[10–13] vasodilators,[14] and anticancer agents.[15] All bear a similar core scaffold, but vary structurally based on key functionalities including oxidation, halogenation, and additional ring systems. Thus, our initial bioinformatic analyses have enabled us to link the construction of the common core to the homologous enzymes, while cluster-specific genes provide the basis for structural differences.[1] There is a clear evolutionary divergence between the MKP and DKP containing families within this class. Thus, installation of similar functional groups may be accomplished by different types of enzymes. Additionally, parallel work in pathways that do not contain the core bicycle reveal compelling biosynthetic branch points. Extensive characterization of the fumitremorgin,[16, 17] spirotryprostatin,[18] and aurachin[19, 20] families by a number of groups has provided insights into the homologous transformations within these varied bicyclo[2.2.2]diazaoctane ring containing metabolic systems.
Figure 1.
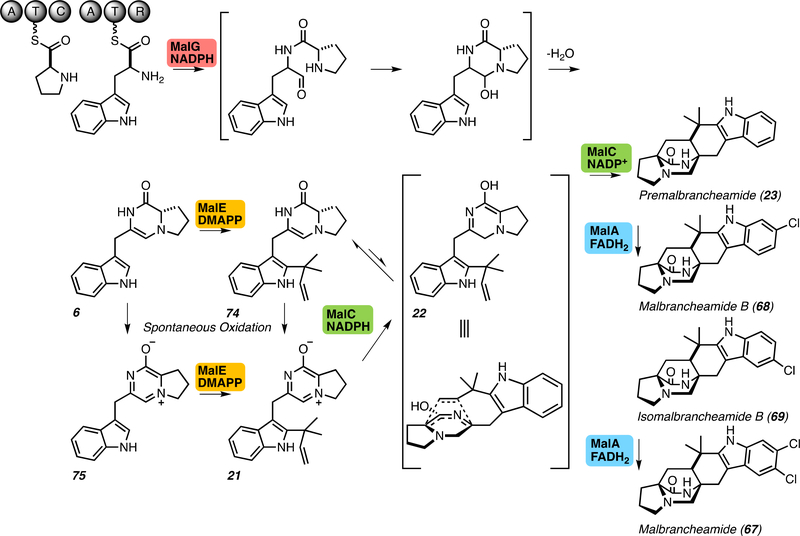
Malbrancheamide biosynthetic pathway. The pathway includes nonribosomal peptide synthetase (NRPS) MalG, prenyltransferase MalE, intramolecular Diels-Alderase MalC, and flavin-dependent halogenase MalA. MalG and MalC require nicotinamide adenine dinucleotide phosphate (NADPH) cofactor for catalysis, while MalA requires the reduced flavin adenine dinucleotide (FADH2) cofactor provided by a reductase partner. MalE prenylates the indole C2 position with a prenyl group provided by dimethylallyl pyrophosphate (DMAPP).
Figure 3.
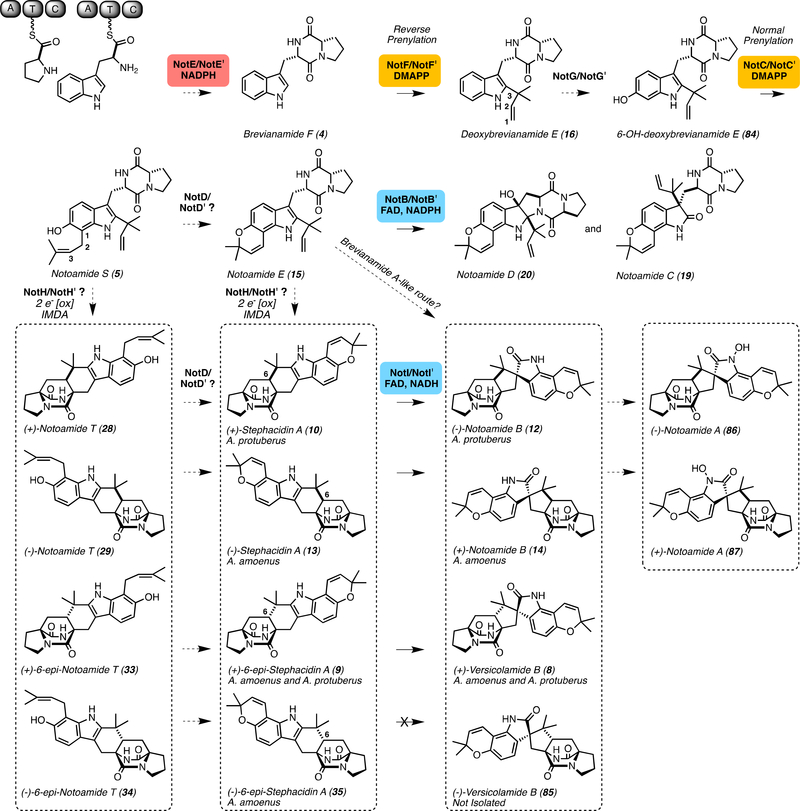
Notoamide biosynthetic pathway. The pathway includes nonribosomal peptide synthetase (NRPS) NotE, prenyltransferases NotF and NotC, putative hydroxylase NotG, a proposed but unidentified intramolecular Diels-Alderase, and flavin-dependent monooxygenases NotB and NotI. Prenyl group numbering distinguishes the reverse and normal prenyltransferases. NotE requires nicotinamide adenine dinucleotide phosphate (NADPH) cofactor for catalysis, while NotB and NotI require flavin adenine dinucleotide (FAD) and the reduced NAD(P)H cofactor. NotF catalyzes reverse prenylation of the indole C2 position and NotG normal prenylates the indole ring with prenyl groups provided by dimethylallyl pyrophosphate (DMAPP).
2. Distinct Nonribosomal Peptide Synthetases
The first step in the biosynthesis of these fungal indole alkaloids is catalyzed by the multidomain nonribosomal peptide synthetases (NRPSs). The architecture of the NRPS consists of adenylation (A), thiolation (T), condensation (C), and reductase (R) domains, where the first A domain in this class of compounds is selective for proline, distinguishing these systems from other fungal bimodular NRPS pathways. The domain construction of the terminal module determines the oxidation state of the offloaded dipeptide, and subsequently the core scaffold. The DKP containing molecules are produced by an NRPS with A-T-C-A-T-C domain organization, whereas the MKP containing molecules involve an NRPS with A-T-C-A-T-R organization. For example, the notoamide (DKP) biosynthetic pathway contains an NRPS with a terminal condensation domain (NotE),[21] while the malbrancheamide and paraherquamide (MKPs) biosynthetic pathways include NRPSs with terminal reductase domains.[1, 22] The different terminal domains and the coinciding mono- or diketopiperazine systems, indicate that the NRPSs offload distinct dipeptide intermediates for the subsequent intramolecular Diels-Alder (IMDA) reaction. Thus, it is now evident that the mechanism for the IMDA cyclization is based on the organization of the upstream NRPS.[1]
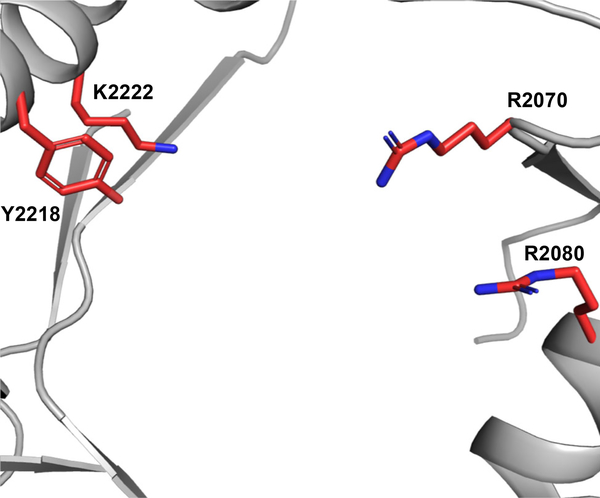
The mechanism for the MKP-producing NRPS has been identified based on recent investigations of the Phq and Mal systems by our group.[22] The distinguishing terminal reductase domain belongs to the family of short chain dehydrogenase reductase (SDR) nicotinamide adenine dinucleotide phosphate (NAD(P)H)-dependent oxidoreductases. These enzymes perform a hydride and proton transfer involving the nicotinamide cofactor and an active site tyrosine, among other conserved amino acids.[23, 24] The NRPS couples l-proline and l-tryptophan to produce an l-Pro-l-Trp aldehyde product through cyclization and dehydration (Figures 1 and 2). Under conditions that contain the NRPS alone, the linear aldehyde product spontaneously oxidizes to a cyclic aromatic zwitterionic species.[22] This catalytic mechanism requires the NADPH cofactor to provide the reducing equivalents, and both PhqB and MalG R-domains contain the canonical SDR tyrosine and lysine active site residues (Figure 4).[22]
Figure 2.
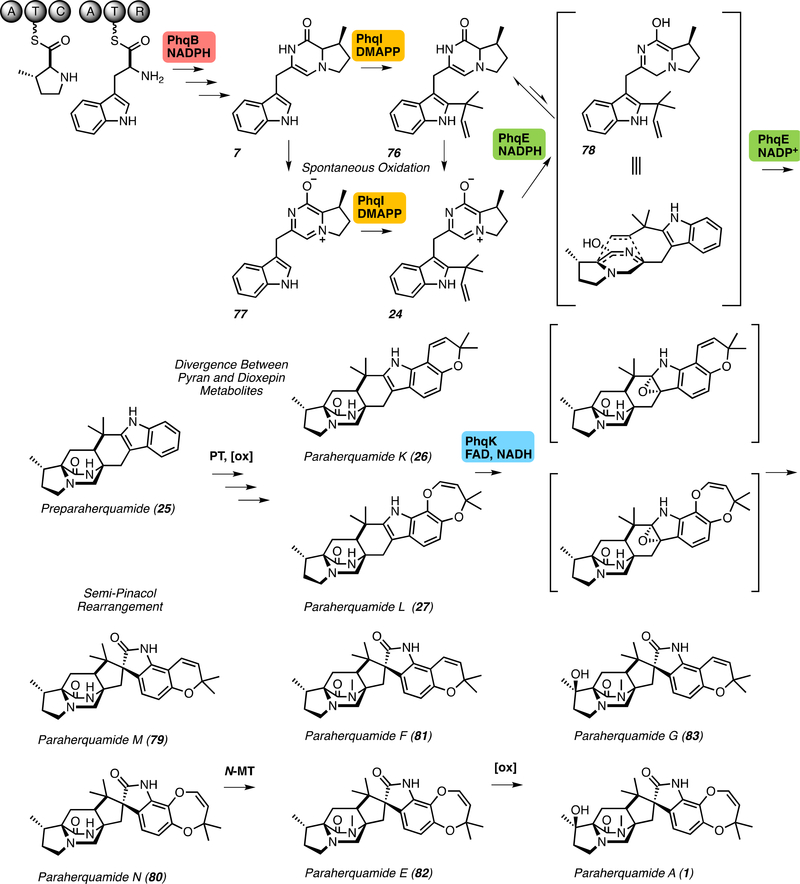
Paraherquamide biosynthetic pathway. The pathway includes nonribosomal peptide synthetase (NRPS) PhqB, prenyltransferase PhqI, intramolecular Diels-Alderase PhqE, and flavin-dependent monooxygenase PhqK. A putative prenyltransferase and oxidative enzyme are proposed to be involved in the formation of the pyran and dioxepin rings. PhqB and PhqE require nicotinamide adenine dinucleotide phosphate (NADPH) cofactor for catalysis, while PhqK requires flavin adenine dinucleotide (FAD) and the reduced nicotinamide adenine dinucleotide (NADH) cofactor. PhqI prenylates the indole C2 position with a prenyl group provided by dimethylallyl pyrophosphate (DMAPP).
Figure 4.
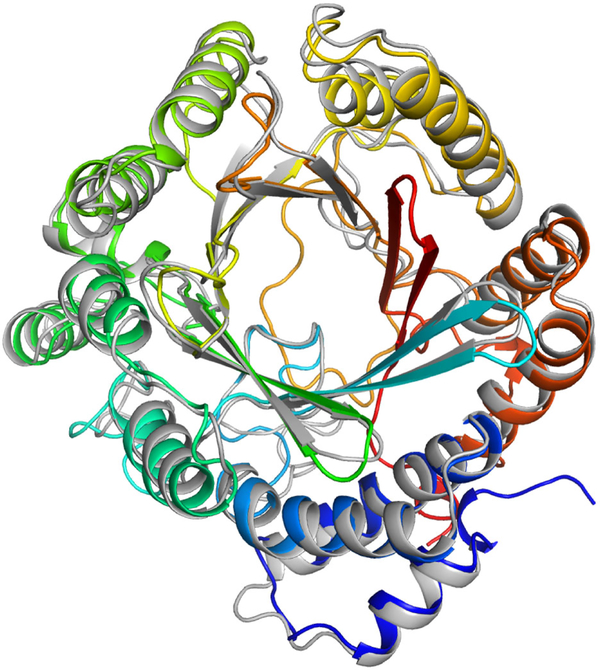
Nonribosomal peptide synthetase (NRPS) PhqB reductase domain X-ray crystal structure in complex with nicotinamide adenine dinucleotide phosphate (NADPH) with canonical short chain dehydrogenase reductase (SDR) catalytic amino acids displayed (PDB ID: 6NKI). Model generated using PyMol Molecular Graphics System.
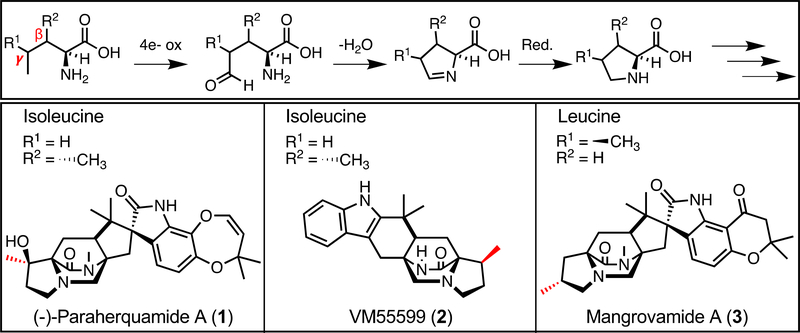
The key difference between the Mal and Phq NRPS systems is the incorporation of β-methyl proline into the dipeptide core within the paraherquamide pathway. Through isotopic feeding studies in Penicillium fellutanum (ATCC: 20841), it was determined that isoleucine is the biosynthetic precursor to β-methyl proline,[25] and that this nonproteinogenic amino acid is directly incorporated into paraherquamide A (1).[26] The enzyme responsible for this amino acid transformation is proposed to perform an oxidative cyclization of the nitrogen atom and Cδ aldehyde, followed by a selective reduction on the same face as the methyl (Figure 5).[25–27] In vitro validation of this reaction scheme was demonstrated for the biosynthesis of UCS1025A, a fungal polyketide alkaloid of a different class.[28] The subsequent hydroxylation observed on the proline moiety in the paraherquamides occurs later in the biosynthesis, following bicyclic ring formation (Figure 2).[29] Interestingly, Penicillium sp. IMI 332995 produces another β-methyl proline-containing compound VM55599 (2), which has the opposite stereochemistry of the bicyclic ring and the same stereochemistry of the methyl group,[25, 30] indicating that these Penicillium strains may produce a broad repertoire of β-methyl proline analogs. Additionally, the recently discovered mangrovamide A (3) contains a unique γ-methyl proline moiety.[31] We hypothesize that this functionality is generated by a leucine-selective enzyme (Figure 5) involving oxidation, cyclization, and reduction, similar to the respective transformations in the biosynthesis of echinocandin,[32] griselimycin,[33] and the nostopeptolides.[34]
Figure 5.
Proposed mechanism for the generation of the methyl proline-containing molecules and examples of molecules generated from isoleucine and leucine precursors.
3. Redundant Prenyltransferases
Prenyltransferases catalyze normal and reverse prenylation reactions in natural product biosynthetic pathways, where the bond is formed between the substrate and C1 of the isoprene unit in normal prenylation and between the substrate and C3 of the isoprene in reverse prenylation (Figure 3, deoxybrevianamide E (16) and notoamide S (5) numbering). We have found that most of the bicyclo[2.2.2]diazaoctane fungal indole alkaloid gene clusters contain an additional prenyltransferase relative to the number of prenyl units incorporated into the molecule. The initial reverse prenylation occurs at the indole C2 position of the NRPS dipeptide product,[22] while prenylation of the phenolic portion of the indole to form the pyran and dioxepin rings can occur before (Figure 3) or after (Figure 2) formation of the [2.2.2] fused bicycle. Precursor incorporation studies have provided insight on the origin of the prenyl groups. P. fellutanum, P. brevicompactum, and Aspergillus ustus all demonstrated incorporation of [13C2]-acetate into paraherquamide A, brevianamide A, and austamide, respectively, indicating a mevalonic acid pathway origin of the prenyl group. Additionally, the geminal dimethyl group stemming from the indole C2 position was found to be non-selectively assigned, thus the facial bias of the reverse prenylation in the biosynthesis of these molecules varies by pathway.[35]
Enzymatic analyses have provided key details for the prenylation reactions within these systems. Within notoamide biosynthesis in the marine-derived Aspergillus protuberus (formerly Aspergillus sp. MF297–2), NotF has been characterized as the reverse prenyltransferase or deoxybrevianamide E synthase (Figure 3).[21, 36, 37] It was hypothesized that brevianamide F (4) would be the substrate for NotF based on previous work focused on fumitremorgin biosynthesis,[16, 17] although in the case of the fumitremorgins, 4 underwent normal prenylation at the indole C2 position.[17] The notoamide biosynthetic pathway also contains a normal prenyltransferase (NotC) that is involved in the formation of notoamide S (5), indicating its significance in pyran ring formation.[21] Additionally, the NotF homolog BrePT from the terrestrial Aspergillus versicolor NRRL573) has been characterized.[38] Although these prenyltransferases have high sequence identity, BrePT displayed a broader substrate scope, while maintaining its indole C2 regioselectivity.
Our investigations of the MKP pathways have led to the characterization of the indole C2-reverse prenyltransferases MalE and PhqI (Figures 1 and 2).[22] We determined that the free-standing reduced dipeptide product was the favored substrate (6 and 7), rather than either the oxidized zwitterion or an NRPS-carrier protein tethered substrate. Additionally, free tryptophan was not prenylated, refuting the role of an early-stage modification. A presumed redundant prenyltransferase, MalB, displayed low activity with the reduced dipeptide (6) as substrate. Sequence alignments and structural comparison revealed that MalB is missing two integral strands of the central β barrel likely responsible for its attenuated activity (Figure 6).
Figure 6.
Comparison of MalE (rainbow) and MalB (gray) Phyre2[82] models displaying the missing Cterminus of MalB (red). Model generated using PyMol Molecular Graphics System.
4. Enantioselective Diels-Alderases
Initial proposals for the biosynthesis of the bicyclo[2.2.2]diazaoctane core involved an IMDA reaction to form the exo and endo products.[39] Early biosynthetic hypotheses assumed that both the MKP and DKP systems were formed through a common DKP intermediate, with the tryptophan carbonyl of the MKPs derived from a net four-electron reduction occurring after a putative Diels-Alder construction.[39] The search for the Diels-Alderase within this class of molecules was hindered by the inability to identify candidate enzymes through bioinformatic analysis. This was due to the unique identities of the respective Diels-Alderases that result from divergent evolutionary processes. Recent studies have shown that Diels-Alderase enzymes typically lose their original function and have evolved to control stereoselectivity for the cycloaddition reaction.[22, 40–46] Additionally, bound cofactors no longer serve a catalytic function (e.g. methyl transfer, redox), and only play a structural role in maintaining the proper active site conformation for stereochemical control.[40–46] Moreover, most Diels-Alderases have no identified catalytic residues, leading to the conclusion that these biocatalysts function by constraining the substrate in the proper orientation.[40–46] Within this class of bicyclo[2.2.2]diazaoctane ring containing molecules, the cycloaddition is proposed to be stereospecific based on the syn- or anti- configuration of the bridged bicycle relative to C6 (Figure 3, stephacidin A numbering).[39] The biosynthetic IMDA enzyme is proposed to have strict diastereo- and enantioselectivity, but some organisms have been found to produce both antipodes.[9, 47]
4.1. Isolation and Precursor Incorporation Studies
Within the notoamide (Not’) producing organism A. amoenus, both (−)-stephacidin A (13) and (+)-versicolamide B (8) have been isolated (Figure 3). The production of 8 suggests that the putative IMDA reaction leading to the major metabolites within the producing organism may suffer from stereochemical leakage with respect to facial selectivity of cycloaddition involving the reverse isoprene moiety (dienophile) anchored at the indole C2 position.[48] Interestingly, the organism must then produce (+)-6-epi-stephacidin A (9), but not (+)-stephacidin A (10), indicating the presence of multiple highly selective IMDAases or one semi-selective IMDAase.
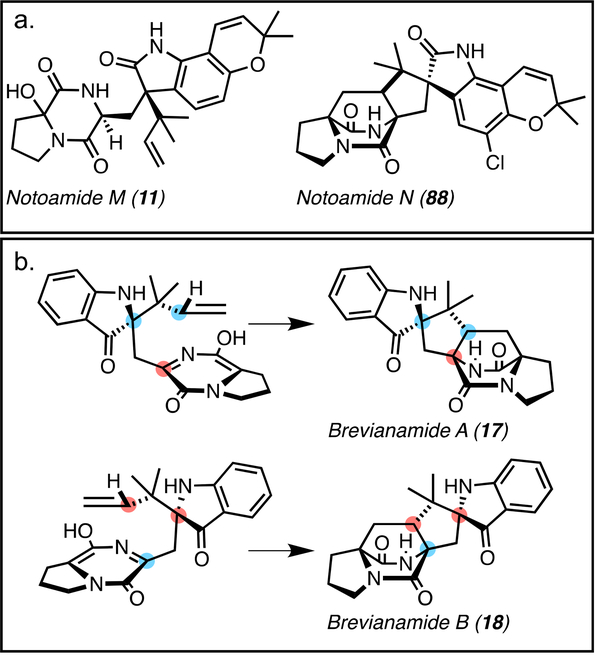
The marine-derived Aspergillus sp. (Not) also produces intermediates that may be relevant to the Diels-Alder reaction in this organism. The isolation of notoamide M (11), which is hydroxylated at C17, raised questions regarding the biosynthetic Diels-Alder reaction within the Not pathway (Figure 7a).[36] It was proposed that 11 would undergo a dehydration to form the azadiene intermediate for cycloaddition; alternatively, 11 could be an artifact due to addition of HO− to the transient azadiene species.
Figure 7.
a. Hydroxylated notoamide M (11) and halogenated notoamide N (88). These molecules exemplify some of the unique secondary metabolites in the notoamide family of natural products. b. Conformations of the azadiene en route to the major diastereomer brevianamide A (17) and the minor diastereomer brevianamide B (18).
The notoamides and stephacidins are part of the same family of bicyclo[2.2.2]diazaoctane alkaloids and are often co-isolated from a single strain. A unique aspect of these systems is that A. protuberus and A. amoenus produce antipodal versions of these molecules. While A. protuberus produces exo products (+)-stephacidin A (10)/(−)-notoamide B (12) and endo products (+)-6-epi-stephacidin A (9)/(+)-versicoloamide B (8), A. amoenus produces (−)-stephacidin A (13)/(+)-notoamide B (14), where in both systems the exo-metabolites are favored (Figure 3).[49] After isolation of notoamide E (15) from the marine-derived Aspergillus sp., doubly 13C-labeled 15 was prepared for feeding studies in an unidentified marine-derived Aspergillus sp.[37] Surprisingly, it was not incorporated into the [2.2.2]-bicyclic ring containing molecules, and no bicycle-containing structures were generated.[37] These data contrast with the brevianamide system in P. brevicompactum, where [3H]-deoxybrevianamide E (16) was incorporated into brevianamides A (17) and B (18), implying that the bicyclic ring was constructed from the reverse prenyl group and the DKP ring.[50] Alternatively, notoamide E was observed to be incorporated into notoamide C (19), notoamide D (20), and trace amounts of 3-epi-notoamide C in A. amoenus (Figure 3).[51] To highlight the differences between the two strains, larger amounts of 3-epi-notoamide C were produced within the marine strain,[37] while the formation of the bicyclo[2.2.2]diazaoctane ring was not completely abolished in the terrestrial strain as trace amounts of unlabeled 10 and 12 were produced.[51] The presence of 15 appears to suppress the formation of stephacidin A (10 and 13) and notoamide B (12 and 14), suggesting that this compound inhibits or diverts enzymatic machinery responsible for the production of the bicyclo[2.2.2]-diazaoctane-containing metabolites. These findings suggested a branch point in the biosynthetic pathway, just prior to the formation of 15, perhaps after the formation of 5 (Figure 3).[51]
Enzyme-controlled facial selectivity appears to be lost within the brevianamide biosynthetic Diels-Alder reaction in Penicillium brevicompactum.[50] It was proposed that the semi-pinacol rearrangement occurs on the 3-hydroxyindolenine, setting the R-absolute stereochemistry at the indoxyl quaternary center. Oxidation of the DKP subunit forms the azadiene of which the major rotamer would form 17 and the minor rotamer would form 18 (Figure 7b).[50] Therefore, the preponderance of 17 over 18 is likely due to intrinsic energy barriers in the cycloaddition reaction (non-enzyme catalyzed).[52] Through the utilization of frontier molecular orbital theory it was predicted that, for the energy levels of a relatively electron-rich diene (such as that in the DKP) to effectively interact with a dienophile, powerful electron-withdrawing groups would need to be present in the dienophile. Since the prenyl group is an isolated electron-neutral vinyl group, one would not expect the [4+2] cycloaddition in DKPs to be spontaneous without catalysis, but biomimetic syntheses[53] have shown that the azadiene undergoes a spontaneous IMDA to generate a mixture of brevianamides with enantiomorphic bicyclo[2.2.2] ring systems. While spontaneous, diastereocontrol of the IMDA may be impacted by upstream transformations such as the early-stage semi-pinacol rearrangement to generate the 3-spiro-ψ-indoxyl species.[52]
4.2. Enzymology
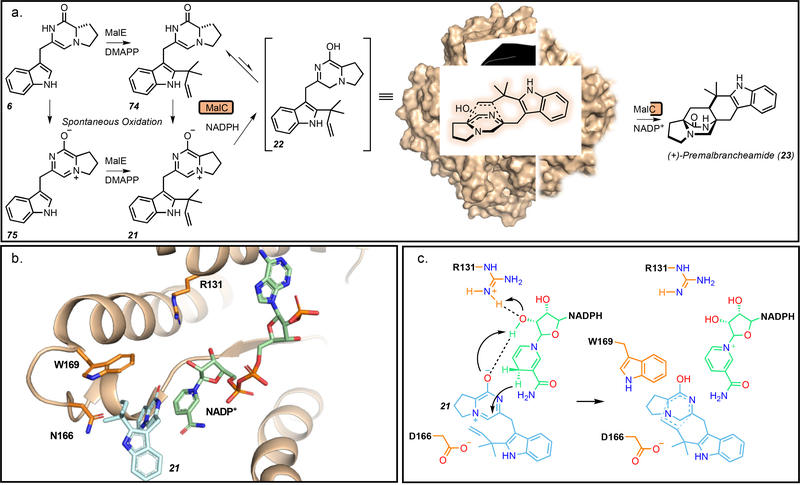
Recent work within our lab has provided key biochemical evidence to support the hypothesis of a biosynthetic Diels-Alder enzyme within MKP systems.[22] MalC was found to catalyze enantioselective cycloaddition as a bifunctional reductase/Diels-Alderase through NADPH-dependent reduction of the prenylated zwitterion 21 to generate the reactive azadiene 22, followed by an enzyme-controlled enantioselective cycloaddition reaction to generate exclusively (+)-premalbrancheamide (23). In the absence of the enzyme, chemical reduction of the prenylated zwitterion leads to racemic 23, thus the enzyme-dependent shift in enantiomeric excess and rate increase led to the conclusion that this reaction is enzyme catalyzed (Figures 1 and 8).
Figure 8.
a. Reactions performed by MalC. Reduction and stereochemical control are performed by MKP IMDAase MalC which “holds” the substrate in the proper conformation to generate (+)-premalbrancheamide (23). b. Active site of PhqE in complex with the prenyl zwitterion 21. c. Proposed mechanism involving donation of a proton from R131 and stabilization of the zwitterion by D166 followed by hydride transfer from the nicotinamide adenine dinucleotide phosphate (NADPH) cofactor to generate the unstable azadiene. The cofactor and W169 provide diastereo- and enantioselective control of the cycloaddition. Model generated using PyMol Molecular Graphics System.
The crystal structures of MalC and the homolog PhqE (from the paraherquamide pathway) provided further mechanistic insight. Their closest structural homologs are canonical SDRs, although the Diels-Alderases lack the essential catalytic residues for SDR activity (Tyr, Lys, Asn, Ser).[54] MalC Asp165 forms part of a conserved PDPGW motif and was shown to be required for reduction of the substrate prior to the Diels-Alder reaction. The structure showed that Asp165 has moved about 3Å closer to the substrate than the analogous amino acid in canonical SDRs, highlighting a larger change of the active site architecture. Based on mutagenesis and computational studies, a mechanism for the reduction and Diels-Alder selectivity was proposed. An arginine residue and the 2’-OH of the NADPH ribose initiate the reaction with NADPH acting as the hydride donor and Asp165 stabilizing the positive charge of the prenyl zwitterion to facilitate the formation of the reactive azadiene intermediate. Stereocontrol seemed to be primarily driven by active site shape complementarity including aromatic amino acids and the cofactor, which hold the substrate in the proper conformation (Figure 8).
The work on the Phq system was consistent with prior feeding studies and predictions, where the β-methyl-Pro-Trp dipeptide was formed by the NRPS.[25, 29] Disruption of phqE in Penicillium simplicissimum using the CRISPR-Cas9 system generated a mutant strain that accumulated the β-methyl-Pro-Trp prenyl zwitterion intermediate 24 (Figure 2).[22] With the addition of the PhqE crystal structures in complex with the substrate, the product, and the NADP+ cofactor, a more thorough mechanistic understanding was achieved. Preference for the NADPH cofactor is explained by an electrostatic interaction between Lys50 and the cofactor 2’-phosphate. Additionally, a relatively short distance between the nicotinamide C4 hydride donor and the presumed deoxy C5 acceptor of the product premalbrancheamide (23) indicated that reduction and cycloaddition are highly coordinated.
Amino acid sequence comparisons, structural data, and NADPH cofactor dependence support the hypothesis that the Diels-Alderases catalyzing formation of the MKP bicyclo[2.2.2]diazaoctane ring system evolved from an SDR progenitor. The active site pocket in the MKP systems clearly orients the substrate for proper enantiocontrol, while the evolution of the active site for its new function is evident. This spectacular display of protein evolution has led to an enzyme that maintained its reductase function without the canonical catalytic residues, while also providing stereocontrol for the Diels-Alder [4+2] cyclization. Moreover, our studies have revealed that the MKP and DKP pathways have evolved in a convergent manner based on the NRPS terminal domain identity, and redox chemistry leading to the dipeptide precursor ring system.
An alternative mechanism for the formation of the bicyclo[2.2.2]diazaoctane ring was discovered for the biosynthesis of brevianamide A.[52] While the previously discussed MKP pathways involved formation of the bicyclic core prior to spirocycle formation, brevianamide production employs a reversed sequence of biosynthetic steps where the oxidized indole intermediate is formed prior to a subsequent DKP oxidation and cyclization. The initial indole oxidation by BvnB and isomerization by BvnE is similar to the NotB-catalyzed formation of notoamides C and D (discussed in detail below). This is followed by a P450-catalyzed DKP oxidation (by BvnD) to the azadiene and spontaneous cyclization to the bicyclo[2.2.2]diazaoctane core.
5. Flavin Monooxygenases (FMOs) and Spirocycle Formation
The spiro-oxindole center is prevalent within members of this indole alkaloid family including paraherquamides,[2, 3] notoamides,[6–8] marcfortines,[55, 56] sclerotiamide,[57] mangrovamides,[31] and asperparalines,[58] among others (Figures 2, 3, 5, 7B). The stereochemistry of the spirocycle is proposed to be controlled by the facial selectivity of the initial indole 2,3-epoxidation, followed by its collapse to a 2-alkoxyindole intermediate, and a semi-pinacol rearrangement to generate the final spirocyclized product.[59]
5.1. Paraherquamides
Based on previous data,[29] it was hypothesized that the [2.2.2]bicyclic ring is formed with the non-oxidized tryptophanyl moiety and that oxidations of the indole ring to form both the dioxepin and the spiro-oxindole must occur after formation of preparaherquamide (25) (Figure 2).[60] To probe the timing of spirocyclization and dioxepin ring formation, triply deuterium-labelled 7-hydroxy-25 was introduced to a culture of Penicillium fellutanum; the results indicated that indole C7 oxidation is not the immediate step following the IMDA reaction, but still may occur prior to spiro-oxindole formation.[60] Recent gene disruption studies in Penicillium simplicissimum established that the pyran and dioxepin rings are both formed prior to spirocyclization, elucidating two key intermediates en route to paraherquamides A (1) and G (83).[61]
The genetic knock-out data in Penicillium simplicissimum have also revealed that the epoxidation and semi-pinacol rearrangement occur through two parallel pathways. The biosynthetic pathway diverges after the formation of 25 to form either a pyran or a dioxepin ring on the indole (see below for a discussion of the proposed mechanism for assembly of the heptacyclic molecules). The PhqK FMO accepts both paraherquamides K (26) and L (27) and performs a facially-selective epoxidation, with a controlled collapse of the epoxide to form the spiro-oxindole (Figure 2). Crystal structures revealed that substrate orientation in the PhqK active site determined the facial selectivity of epoxidation. Collapse of the presumed 2,3-indole epoxide intermediate is strongly influenced by the pyran/dioxepin oxygen, and the presence of a catalytic arginine which serves as a general acid. Based on Michaelis-Menten kinetic parameters, 27 appears to be the favored substrate, but both 26 and 27 can undergo semi-pinacol rearrangement to the respective spiro-oxindole product.
5.2. Notoamides
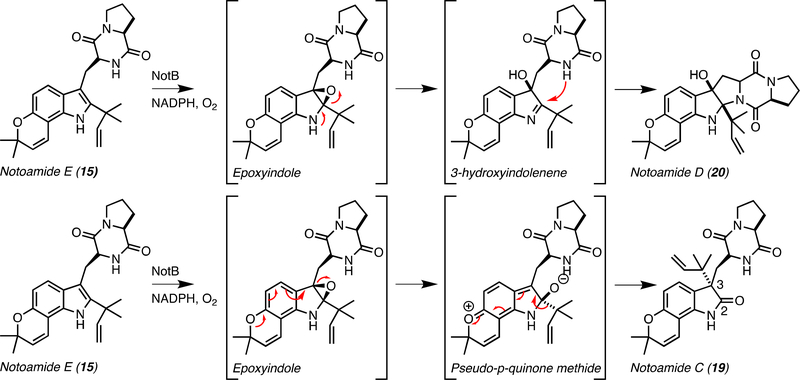
The oxidative conversion of notoamide E (15) was investigated through biomimetic oxaziridine-based syntheses in which notoamide C (19)[62] (48%), 3-epi-notoamide C[62] (28%), and minor amounts of notoamide D (20) and 2,3-epi-notoamide D were produced (Figure 3).[7] The electronic properties of the indole ring were found to influence the regiochemistry of oxidation of the C2–C3 indole bond, resulting in either the pyrroloindole or the oxindole.[7] While the synthetic method generated unequal product distribution, the organism produced almost equal amounts of 19 and 20, implying that the responsible biosynthetic enzyme can override the inherent reactivity of the molecule.[7]
Enzymatic characterization of the FMO NotB revealed that both 19 and 20 can be produced from 15, and it was proposed that both can be formed through β-epoxidation of the C2–C3 indole bond (Figure 9).[7] Ring-opening of the β−2,3-epoxyindole intermediate to the 3-hydroxyindolenine species followed by N-C ring closure from the diketopiperazinyl NH generates pyrroloindole 20 as the major product. The minor product 19 is derived from the pseudoquinone methide species and subsequent α-face migration of the prenyl group from C2 to C3. It was also proposed that an interaction between the indole C2 oxyanion intermediate and DKP N-H determines whether the pyran ring contributes to the breakdown of the epoxide (Figure 9). The N-H can stabilize the negatively charged intermediate, but this interaction does not occur when the epoxide collapses to the indole C3 position, thus the hydroxyl is formed immediately, producing 20.[7] The products resulting from α-face epoxidation were not observed in the enzymatic reaction, thus it was proposed that their isolation in the precursor incorporation study was an artifact of the excessive amounts of 15 within the culture medium.[62]
Figure 9.
Proposed mechanism for assembly of notoamides C (19) and D (20) by the flavin-dependent monooxygenase NotB.
Because 15 halted production of bicyclo[2.2.2]diazaoctane ring containing metabolites, it was proposed that an enzyme other than NotB was required to catalyze formation of the spiro-oxindole within this family.[7] Intriguingly, it was reported that A. protuberus produced (+)-stephacidin A (10) and (−)-notoamide B (12),[6] while A. amoenus produced the enantiomers (−)-stephacidin A (13) and (+)-notoamide B (14) (Figure 3).[48] Due to the antipodal relationship of the metabolites isolated from A. protuberus (marine) and A. amoenus (terrestrial), an R-selective monooxygenase was proposed to exist within the marine strain and an S-selective monooxygenase within the terrestrial strain.[48] At first, whether the alternative FMO would act before or after the IMDA remained unclear. It was considered that the breakdown of the indole 2,3-epoxide followed by semi-pinacol rearrangement of the isoprenyl group from C2 to C3 could produce intermediates that undergo oxidation and tautomerization to yield the azadiene intermediates for the IMDA.[36] However, the isolation of notoamide T (28/29) and stephacidin A (10/13),[63] as well as the precursor incorporation studies led to the biosynthetic hypothesis that the IMDA occurs prior to spirocyclization. Late-stage spiro-oxindole formation was probed by introducing doubly 13C-labeled racemic stephacidin A (10/13) to cultures of A. amoenus and A. protuberus; analysis of the metabolites revealed enantiospecific incorporation of intact 13 into 14 in A. amoenus and 10 into 12 in A. protuberus, and the unreacted 10 and 13 were re-isolated from the respective fungal extracts.[63] These data provided evidence of divergent flavoenzymes with opposite R/S-substrate selectivity in each strain.
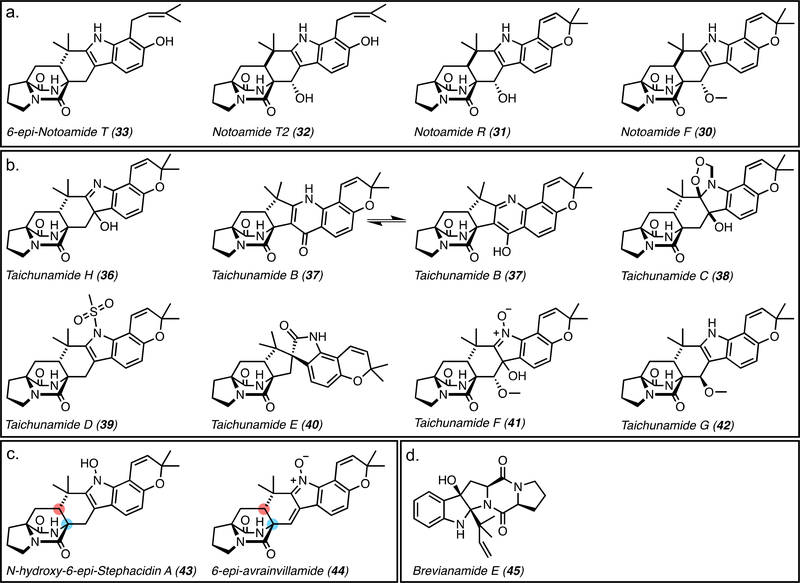
In feeding studies, racemic D,L-[13C]2-notoamide T (28 and 29) was incorporated into 10 and 14 in A. amoenus,[64] indicating that 28 and 29 are converted to 10 and 13, respectively, and 13 is rapidly converted to 14, while 10 accumulates as a shunt product. In A. protuberus D,L-[13C]2-notoamide T (28/29) was incorporated into 10/13, 12/14, notoamide F (30), notoamide R (31), and notoamide T2 (32) (all racemic) (Figures 3 and 10A).[64] This led to a contradictory notion that the gene products in A. protuberus do not discriminate between the two enantiomers of notoamide T (28/29) or stephacidin A (10/13) in the oxidative conversion to the final metabolites.[64] While growth conditions were consistent between the two experiments, it is possible that small changes such as substrate concentration induced higher reactivity (broader substrate scope) in the second incorporation study.
Figure 10.
a. Secondary metabolites with anti and syn configuration. The syn products (30-32) are all generated from notoamide T in vivo. b. Taichunamides H (Aspergillus versicolor HDN11–84) and B-G (Aspergillus taichungensis (IBT 19404) with anti- relative stereochemistry of the bicyclic ring. c. Anti- bridged bicycles produced by Aspergillus taichungensis ZHN-7–07. d. Brevianamide E (45), which is generated from deoxybreviamide E (16).
Additionally, the presence of 6-epi-isomers raised some ambiguity in the selectivity of the IMDAase (Figure 3). (+)-Versicolamide B (8) was isolated from A. amoenus and was the first member of this family to possess the anti-relative stereochemistry within the bicyclo[2.2.2]diazaoctane ring system (Figure 3).[48] Racemic 6-epi-notoamide T (33/34) was converted to 6-epi-stephacidin A (9/35) and (+)-versicolamide B (8) in A protuberus.[49] The isolation and incorporation of notoamide S (5) into notoamides C (19), D (20), (−)-stephacidin A (13), (+)-notoamide B (14), and (+)-versicolamide B (8) in A. amoenus[65, 66] indicated that the FMO can also accept (+)-6-epi-stephacidin A (9). The observation of an enantiomeric mixture of 6-epi-stephacidin A (9/35) enriched with the (−)-isomer from A. amoenus indicated that the enzymes upstream of the FMO were not selective. While the A. amoenus FMO transforms 9 to 8, there is not a suitable oxidase for the (−)-6-epi-stephacidin A (35) shunt metabolite.[49] It is intriguing that the closely related Aspergillus species have evolved enantiodivergent pathways to the stephacidins and notoamides, but converged on the production of (+)-versicolamide B (8).[49]
In vitro work with NotI (A. protuberus) and NotI’ (A. amoenus) demonstrated the conversion of 10 and 13 to 12 and 14, although a clear preference was observed for 13 (Figure 3).[67, 68] Both NotI and NotI’ also converted 9 to 8 but no reaction was observed with 35. This is consistent with the conversion observed in A. amoenus where 8 was produced and 35 was determined to be a shunt metabolite.[49] The ability to convert 10 may be an evolved trait from an ancestral enzyme previously selective for 13, as high reactivity for this substrate is retained in both NotI and NotI’.
The presence of both early-stage and late-stage FMOs in notoamide biosynthesis indicates that two routes may be employed to generate the bicyclic moiety within this pathway (Figure 3). The production of similar products by NotB and BvnB/BvnE have led us to believe that a P450 monooxygenase homologous to BvnD may be present in the notoamide biosynthetic gene cluster (NotH). In combination with the precursor incorporation studies, this work provides evidence that this biosynthetic system utilizes two seemingly divergent routes to converge on the same bicyclo[2.2.2]diazaoctane-containing notoamides, each with its own inherent selectivity to furnish the various notoamide natural products. If this prediction is ultimately determined to be the case, we can hypothesize that the notoamide producers might be the ancestral strain from which the other fungal indole alkaloid producers diverged to utilize just a single mechanism to generate the bicyclo[2.2.2]diazaoctane core.
5.3. Taichunamides
The isolation of fungal natural products with the anti- relative stereochemistry of the bicyclic ring generated questions regarding the driving force for selectivity of the IMDA reaction in these families. Taichunamide H (36), isolated from mangrove-derived fungus Aspergillus versicolor HDN11–84, and taichunamides B-G (37-42), isolated from the fungus Aspergillus taichungensis (IBT 19404), contain an anti-bicyclo[2.2.2]diazaoctane core (Figure 10B).[69, 70] They are proposed to share the common precursors notoamide S (5), (+)-6-epi-notoamide T (33), and (+)-6-epi-stephacidin A (9). Additionally, A. taichungensis ZHN-7–07 produces 9, N-hydroxy-6-epi-stephacidin A (43), and 6-epi-avrainvillamide (44), all with anti- relative configuration (Figure 10C).[71] This indicates that the taichunamide biosynthetic enzymes share some overlap or crosstalk with those from the notoamide biosynthetic pathway in these producing organisms, but in all cases they seem to be selective for the anti-form of the bicyclic ring configuration.
5.4. Brevianamides
The anti-relative stereochemistry in the brevianamides was proposed to be generated from the oxindole intermediate. [3H]-labeled deoxybrevianamide E (16) (Figure 3) was shown to be efficiently incorporated into brevianamides A (17), B (18) (Figure 7B), and E (45) (Figure 10D) in Penicillium brevicompactum. It was proposed that 45 may be a biosynthetic precursor to 17 and 18, but [3H]-labeled 45 was not incorporated into either, disproving its intermediacy within the pathway.[50] Thus, it is likely that oxidation of the indole occurs at C3 prior to the IMDA. An R-selective hydroxylation is proposed to generate the 3-hydroxyindolenine which can either cyclize to form 45 or undergo a semi-pinacol rearrangement to form the C2-prenylated intermediate. Oxidation to the azadiene would provide the proper substrate for the IMDA.[50, 52] The semi-pinacol rearrangement is proposed to proceed through general base catalysis to generate the indoxyl.48 In comparison, paraherquamide biosynthesis utilizes general acid catalysis to generate an oxindole moiety,[61] demonstrating that these systems have evolved divergent mechanisms for spirocycle formation.
5.5. Effect on the Diels-Alder Product
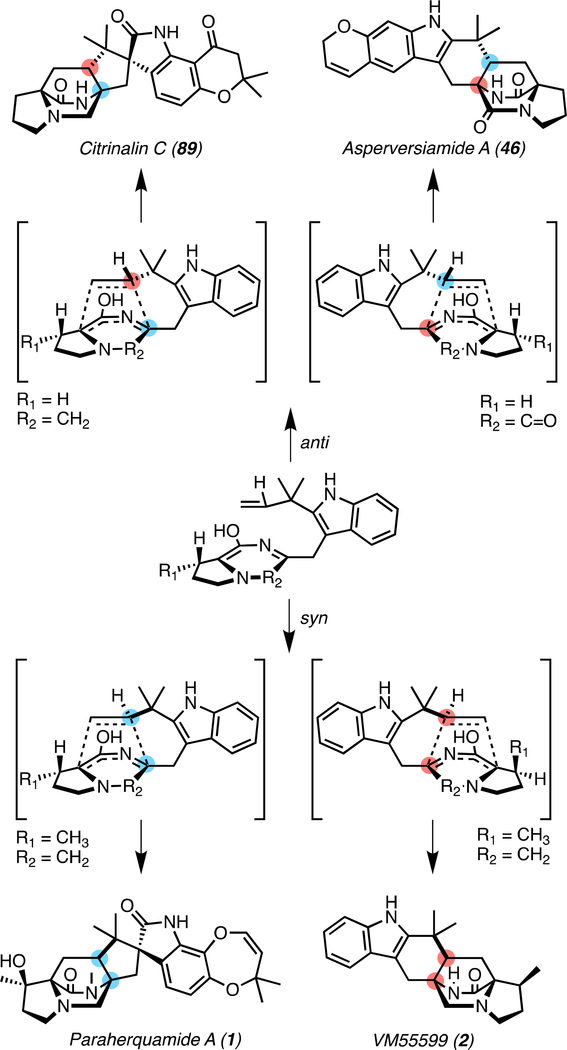
With the knowledge that the spiro-oxindole is differentially installed preceding or following the IMDA reaction in various strains, the implications of this distinction regarding other reactions within the biosynthetic sequence must be considered. Density functional theory computational studies have been performed to elucidate the stereochemical features of the cyclization step involved in paraherquamide A (1) and VM55599 (2) biosynthesis (Figure 11).[72] It was determined that the non-oxidized indole favors syn diastereoselectivity, which agrees well with precursor incorporation experiments by Williams et al.[35, 50] concluding that the syn cycloaddition could take place via the non-oxidized tryptophanyl moiety and that the oxidation of the indole ring occurs after the IMDA cycloaddition in paraherquamide biosynthesis.[72] In the computational study, anti-selectivity was found for the cycloadditions of the oxindole-based derivatives, indicating that the FMO reaction could precede the IMDA in cases where anti-bicyclo[2.2.2]diazaoctane rings are observed.[72] Contrary to this hypothesis, nonspirocyclized anti-MKP members of this family have also been isolated,[73–75] indicating that there may be various divergent routes to the syn/anti-bicyclo[2.2.2]diazaoctane ring. Monooxygenases have evolved to perform the presumed epoxidation and semi-pinacol rearrangement at different points within the biosynthesis, affecting the stereochemical outcome of the Diels-Alder reaction.[50] While this computational study presents a substrate-controlled mechanism for directing the IMDA, the enzyme active site conformation may provide a catalyst-controlled means to overcome the innate reactivity. With this in mind, IMDA enzymes may be selective for either the oxidized or non-oxidized substrate, and the orientation in which they bind may direct the reaction. If this is the case, there may have been a parallel evolution of the IMDA enzyme and the FMO in each of the biosynthetic gene clusters.
Figure 11.
Conformations conducive to syn or anti bicyclic ring formation. Examples from each subgroup of the fungal indole alkaloids are shown.
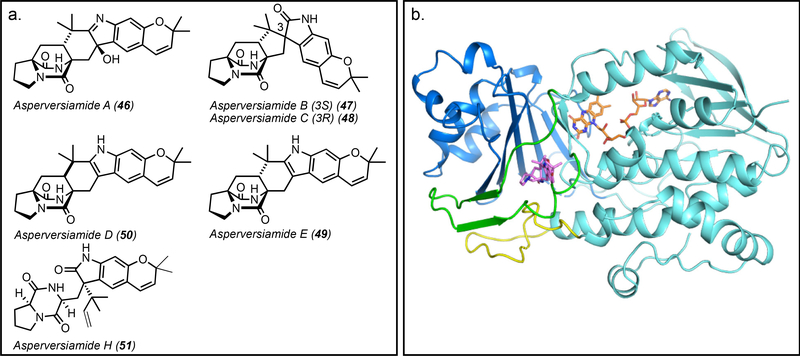
Interestingly, in the case of the asperversiamides, both syn and anti products have been isolated from the same organism, a marine-derived Aspergillus versicolor. While asperversiamides A-C (46-48) and E (49) contain the anti-bicyclo[2.2.2]diazaoctane ring, asperversiamide D (50) contains the syn-ring (Figure 12a). With the isolation of the prenylated precursor asperversiamide H (51), one can infer that the Diels-Alder reaction is the last step in the pathway to 46-49, following the oxindole formation, leading to an anti-configuration.[47] This system is unique in that it may contain two different monooxygenases since one would result in the semi-pinacol rearrangement and the other would generate a hydroxylated product.[47] Additionally, one FMO would act at an early stage, and the other at a late stage with various options for the IMDA. The IMDA enzyme may be non-selective and accept both substrates, or it may have an evolved active site that can override the inherent selectivity of the molecule. Among numerous possibilities, there may even be two different IMDA enzymes/mechanisms, serving to generate the diastereomeric products. On the other hand, recent work on the brevianamide biosynthetic pathway has revealed that the early-stage semi-pinacol rearrangement to generate the 3-spiro-ψ-indoxyl directs the subsequent IMDA reaction in a substrate-controlled manner.[22] This demonstrates another example of differentiation within this class of fungal indole alkaloids, where the pathways have evolved unique mechanisms to generate molecular diversity.
Figure 12.
a. Asperversiamides A-E (46-49) and H (51) isolated from a marine-derived Aspergillus versicolor. Asperversiamides A-C and E display anti- relative stereochemistry and asperversiamide D displays syn- relative stereochemistry. b. Domains of PhqK flavin-dependent monooxygenase (PDB ID: 6PVI). PhqK flavin adenine dinucleotide (FAD)-binding domain (cyan) and three insertions that form the substrate-binding domain (blue, green, yellow) are highlighted. The substrate is shown in magenta and the FAD cofactor is shown in orange. Model generated using PyMol Molecular Graphics System.
5.6. FMO Evolution
There are a variety of different types of FMOs that have diverged into two groups defined by whether they require a reductase partner.[76] For standalone enzymes, such as those involved in spirocyclization, the reactions with the electron donor and oxygen are catalyzed by a single protein. While some FMOs have two distinct cofactor binding domains, others (such as PhqK) contain a single nucleotide binding domain for the FAD, and have developed a NADPH binding groove on the surface of the protein. Over time, the flavin monooxygenases have undergone domain fusion events, eventually leading to the development of a new domain for substrate binding which serves to further differentiate the members of this enzyme class (Figure 12b).
5.7. Mechanism of Spiro-oxindole Formation
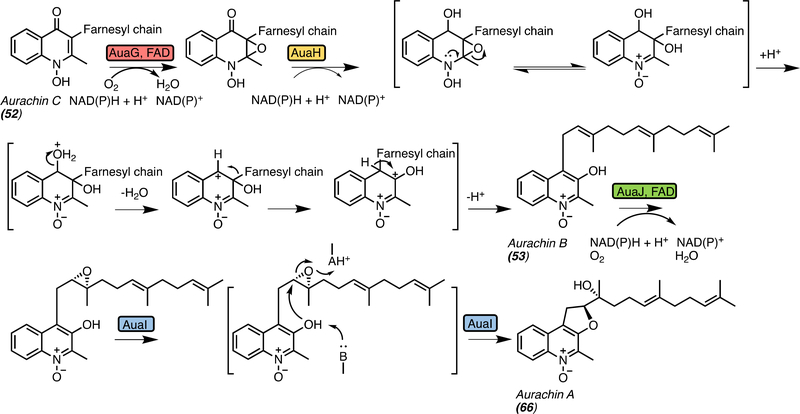
Mechanistic implications for spiro-oxindole formation can be gleaned from systems outside the bicyclo[2.2.2]diazaoctane ring containing family. The conversion of aurachin C (52) to B (53) is proposed to involve the migration of the prenyl group from position C3 to C4 through a semi-pinacol rearrangement,[20] similar to that proposed for the spirocylization in the fungal indole alkaloids (Figure 13). The proposed mechanism involves an epoxidation of the quinoline core of 52 by AuaG followed by AuaH-catalyzed ring opening of the epoxide and semi-pinacol rearrangement, while the proposed mechanism for the fungal indole alkaloids is initiated with an epoxidation of the indole C2=C3, and the breakdown of the epoxide leads to a spontaneous semi-pinacol rearrangement to generate the spirocycle. In the aurachin pathway two enzymes catalyze the semi-pinacol-type rearrangement where the main product of AuaG appears to be an energetically less-favored isomer that is immediately stabilized by reduction and subsequent aromatization catalyzed by AuaH reductase.[20] In contrast, the fungal indole alkaloid systems have evolved a single dual-function enzyme.
Figure 13.
Aurachin biosynthesis. AuaG and AuaH perform nicotinamide adenine dinucleotide phosphate (NAD(P)H)-dependent epoxidation and semi-pinacol rearrangement like the bifunctional PhqK, and AuaJ and AuaI catalyze epoxidation and cyclization similar to the pyran and dioxepin ring formation in fungal indole alkaloid biosynthesis. Both AuaG and AuaJ are flavin adenine dinucleotide (FAD)-dependent enzymes.
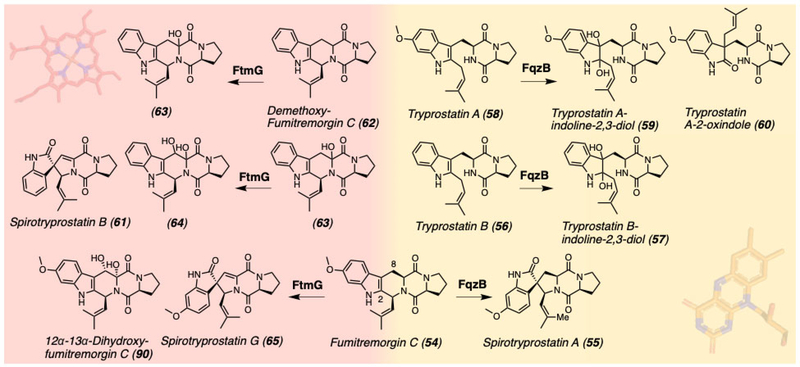
A detailed study of spiro-ring formation in spirotryprostatins derived from the fumitremorgin biosynthetic pathway has also shed light on this topic (Figure 14).[18] FqzB is an FAD-dependent monooxygenase from the unrelated fumiquinazoline biosynthetic pathway which catalyzes the spirocycle formation on fumitremorgin C (54) to generate spirotryprostatin A (55) via an epoxidation route (Figure 14).[18] The conversion of tryprostatin B (56) to tryprostatin B-indoline-2,3-diol (57) and conversion of tryprostatin A (58) to tryprostatin A-indoline-2,3-diol (59) and tryprostatin A-2-oxindole (60) indicates that FqzB catalyzes epoxidation of the C2-C3 olefin of the indole on 56 and 58. The formation of 60 indicates that the mechanism involves a semi-pinacol rearrangement of the epoxidized intermediate.[18]
Figure 14.
Spirotryprostatin and fumitremorgin biosynthetic pathway crosstalk involving cytochrome P450 FtmG and flavin-dependent monooxygenase FqzB.
Within the same organism, a cytochrome P450 FtmG from the fumitremorgin biosynthetic pathway catalyzes the spiro-ring formation observed in spirotryprostatin B (61).[18] FtmG converts dimethoxy-fumitremorgin C (62) into 61 as well as mono-hydroxyl (63) and diol (64) forms of dimethoxy-fumitremorgin C.[18] Based on the intermediate structures, a reaction mechanism was proposed involving a P450 heme-catalyzed initial radical formation and subsequent two rounds of hydroxylation of 62.[18] The production of 61 by FtmG is proposed to occur through radical migration from C8 to C2 and subsequent hydroxylation at C2 can set up the molecule for a semi-pinacol rearrangement involving a concomitant spiro-ring formation. Additionally, FtmG crosses into the fumitremorgin biosynthetic pathway to convert fumitremorgin C to spirotryprostatin G (65) (Figure 14).[18]
A. fumigatus maintains two orthogonal spiro-ring formation systems in its secondary metabolite biosynthetic pathway: an FAD-dependent route (for the formation of 55) catalyzed by the FMO FqzB, and a radical route (for the formation of 61 and 65) catalyzed by the P450 FtmG.[18] This study highlights the versatile role of oxygenating enzymes in the biosynthesis of structurally complex natural products and indicates that cross-talk of different biosynthetic pathways promotes product diversification in natural product biosynthesis.[18]
6. Dioxepin and Pyran Ring Formation
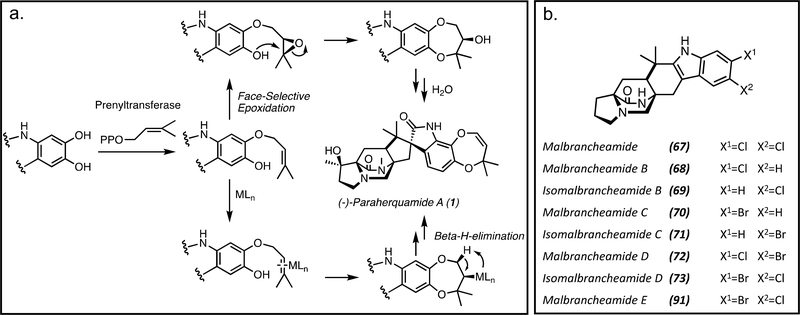
Within P. fellutanum, isoprene units from the mevalonic acid (mevalonate) pathway were incorporated into the dioxepin ring of paraherquamide A (1).[35] It was proposed that the formation of the dioxepin is due to a facially selective epoxidation of the olefin followed by a stereospecific ring-opening of the epoxide and dehydration.[35] Another alternative for the dioxepin formation would be a face-selective complex with a transition metalloprotein to the olefinic π-system followed by stereospecific intramolecular nucleophilic addition and reductive elimination to the enol ether (Figure 15a).[35] The hydroxylation and epoxidation are both proposed to be performed by currently unidentified oxidative enzymes. This fungal system contrasts with the non-heme iron-dependent enzyme deguelin cyclase found in plants.[77, 78]
Figure 15.
a. Proposed mechanisms for dioxepin ring formation involving an oxidative enzyme with a metal cofactor (MLn) and a prenyltransferase. b. Halogenated malbrancheamides. These molecules are generated by the flavin-dependent halogenases MalA and MalA’ in Malbranchea aurantiaca and Malbranchea graminicola, respectively.
Within the notoamide-producing strain A. amoenus, [13C]2-notoamide T (28/29) was incorporated into (+)-stephacidin A (10) and (+)-notoamide B (14) (Figure 3).[64] This indicates that both 28 and 29 were accepted by the pyran ring forming enzyme to generate 10 and 13, respectively. Thus, the enzyme responsible for pyran formation must have evolved to accept both enantiomers.
The asperversiamides have a unique connectivity with the isoprenyl substituent located on the indole C5 to form the pyrano-[3,2-f]indole moiety as opposed to the angularly fused pyrano[2,3-g]indole (Figures 11 and 12a).[47] The prenyltransferase responsible for this transformation must have C5 selectivity as opposed to typically observed indole C7 prenylation. Consequently, the downstream enzymes, such as those responsible for catalyzing pyran ring formation must have evolved in parallel to perform their function on the new metabolite.
1.2.6.1. Mechanism of Pyran and Dioxepin Ring Formation
Similar biosynthetic enzymes involved in aurachin production in the myxobacterium Stigmatella aurantiaca Sg a15 have provided insight into the potential mechanism to form the pyran and dioxepin moieties. This involved the 3,4-migration of the farnesyl chain to produce A-type aurachins, which was proposed to proceed via 2,3-indole epoxidation (AuaG) and subsequent reduction by AuaH as described above (Figure 13).[19] AuaJ is proposed to perform an epoxidation that is followed by AuaI-catalyzed hydrolysis to form aurachin A (66), which functions similarly to the pyran and dioxepin ring forming enzymes within fungal indole alkaloid biosynthesis.[19] In this case, two enzymes (AuaG and AuaH) perform the proposed function of PhqK and NotI in paraherquamide and notoamide biosynthesis, respectively. PhqK and NotI have evolved as dual-function enzymes to perform the epoxidation and selective semi-pinacol rearrangement in their respective pathways. It is possible that the epoxidation and cyclization to form the pyran and dioxepin rings may also be mediated by a single fungal enzyme, but the mechanism remains unclear (Figure 15a).
7. Late-Stage Halogenation
Halogenation of the fungal bicyclo[2.2.2.] indole alkaloids appears to be fairly specific to the Malbranchea strains M. aurantiaca[4] and M. graminicola.[78] The halogenases MalA (M. aurantiaca) and MalA’ (M. graminicola) are responsible for the iterative chlorination to produce the natural product malbrancheamide (67).[4, 78, 80] These halogenases catalyze chlorination and bromination reactions to generate monochlorinated malbrancheamide B (68) and isomalbrancheamide B (69), monobrominated malbrancheamide C (70) and isomalbrancheamide C (71), mixed halogen malbrancheamide D (72) and isomalbrancheamide D (73), and the dibrominated malbrancheamide E[61] (Figure 15b). The monobrominated molecules had previously been isolated from M. graminicola, and in vitro work with MalA and MalA’ demonstrated that both strains were capable of generating the brominated and chlorinated molecules. The MalA/A’ study included the first computational analysis of the flavin-dependent halogenase (FDH) family, and demonstrated a modified mechanism for fungal FDHs. Additionally, it was determined that MalA is stereospecific, distinguishing a single antipodal natural substrate. Thus, when incubated with racemic premalbrancheamide, only (+)-premalbrancheamide (23) was converted to (+)-malbrancheamide (67).[22]
The notoamide family also contains a halogenated secondary metabolite, notoamide N (88), which contains an indole C5 chlorination (Figure 7a).[36] Based on what we have observed with the production of spiromalbramide,[79] and in reactions with NotI/PhqK, we can hypothesize that the pyran ring formation and the halogenation of the [2.2.2] bicyclic product would occur prior to the spirocyclization. The molecule could be halogenated first through a similar mechanism to MalA, then the pyran ring would be formed, followed by spirocyclization. However, the corresponding halogenating enzyme has yet to be identified.
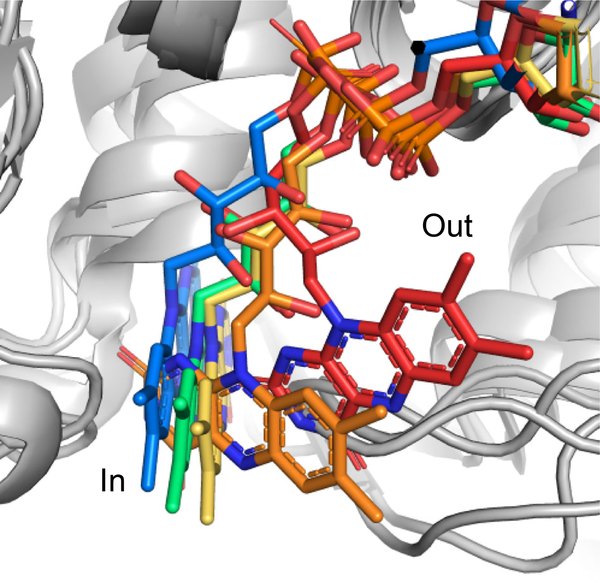
The spiro-oxindole-forming enzymes and FDHs are both part of the overarching FMO family. While the enzymes that catalyze spiro-oxindole formation are typically discrete proteins, the FDHs require an FAD reductase partner to initiate catalysis. The stand-alone enzymes have a highly dynamic FAD cofactor that moves between “in” and “out” conformations as it is reduced and oxidized, respectively. After a structure-based evolutionary analysis of FMOs, it was clear that the halogenases have evolved to bind the FAD in a conformation that resembles the “in” conformation of the stand-alone enzymes. Along this evolutionary trajectory, the enzyme also refashioned the FAD redox chemistry to produce hypohalous acid and direct this halogenating agent through the production of a putative lysine haloamine intermediate[81] (Figure 16).
Figure 16.
Structural comparison of the flavin cofactor in flavin-dependent monooxygenases (FMO)s. Some FMOs catalyze oxidation reactions and others catalyze halogenation reactions. PhqK (red), PqsL FMO, PDB: 6fho (orange), MalA’ halogenase, PDB: 5wgr (yellow), PltA halogenase, PDB: 5dbj (green), and decarboxylative hydroxylase, PDB: 5eow (blue). The discrete conformations captured in these structures indicate the ancestral relationship of these flavin-dependent enzymes. Model generated using PyMol Molecular Graphics System.
8. Discussion
Fungal indole alkaloids that contain the bicyclo[2.2.2]diazaoctane ring system are a unique family of molecules and corresponding biosynthetic enzymes. The NRPSs bear terminal condensation or reductase domains, which lead to the formation of DKP or MKP bicyclic ring systems, respectively. Additionally, the incorporation of β-methyl proline and γ-methyl proline has been accommodated through enzymes that have evolved for assembly from isoleucine or leucine. Following the NRPS reaction, a selective reverse prenylation of the indole C2 position occurs.
The IMDA enzyme performs a central role to form the bicyclic core common to this class of molecules. Initially annotated as SDRs, the MKP IMDAases MalC and PhqE perform an initial reduction prior to controlling the selective [4+2] cycloaddition reaction. This is an example of the only characterized Diels-Alder enzyme that retains its ancestral functionality.[44, 82] Interestingly, DFT calculations suggest that the syn-/anti- diastereospecificity of the IMDA reaction can be substrate-driven based on the oxidation state of the indole ring, where presence of the FMO-derived spiro-oxindole moiety reverses the intrinsic diastereospecificity. Natural examples of pre- and post-IMDA spirocyclization have been identified in these pathways. Lastly, halogenation is performed by flavin-dependent halogenases (MalA/MalA’), which have only been identified in the mal gene clusters. In each of the aforementioned diversification mechanisms, a unique family of molecules with intriguing bioactivities has been generated. This pool of biosynthetic knowledge forms the basis to explore the utility of these biocatalysts to generate and diversify small molecules and expand the potential of this class of broadly bioactive natural products.
ABBREVIATIONS
- A
adenylation domain
- Asn
asparagine
- C
condensation domain
- DKP
diketopiperazine
- FAD
flavin adenine dinucleotide
- FDH
flavin-dependent halogenase
- FMO
flavin-dependent monooxygenase
- IMDA
intramolecular Diels-Alder
- Lys
lysine
- MKP
monoketopiperazine
- NADPH
nicotinamide adenine dinucleotide phosphate
- NRPS
nonribosomal peptide synthetase
- Pro
proline
- R
reductase domain
- SDR
short chain dehydrogenase
- Ser
serine
- T
thiolation domain
- Trp
tryptophan
- Tyr
tyrosine
REFERENCES
- 1.Li S, Srinivasan K, Tran H, Yu FA, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, Williams RM & Sherman DH (2012) Comparative analysis of the biosynthetic systems for fungal bicyclo[2.2.2]diazaoctane indole alkaloids: the (+)/(−)-notoamide, paraherquamide and malbrancheamide pathways, Medchemcomm. 3, 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamazaki M, Okuyama E, Kobayashi M & Inoue H (1981) The structure of paraherquamide, a toxic metabolite from Penicillium paraherquei, Tetrahedron Lett. 22, 135–136. [Google Scholar]
- 3.López-Gresa MP, González MC, Ciavatta L, Ayala I, Moya P & Primo J (2006) Insecticidal activity of paraherquamides, including paraherquamide H and paraherquamide I, two new alkaloids isolated from Penicillium cluniae, J Agr Food Chem. 54, 2921–2925. [DOI] [PubMed] [Google Scholar]
- 4.Martínez-Luis S, Rodríguez R, Acevedo L, González MC, Lira-Rocha A & Mata R (2006) Malbrancheamide, a new calmodulin inhibitor from the fungus Malbranchea aurantiaca, Tetrahedron. 62, 1817–1822. [Google Scholar]
- 5.Qian-Cutrone JF, Huang S, Shu YZ, Vyas D, Fairchild C, Menendez A, Krampitz K, Dalterio R, Klohr SE & Gao Q (2002) Stephacidin A and B: Two structurally novel, selective inhibitors of the testosterone-dependent prostate LNCaP cells, J Am Chem Soc. 124, 14556–14557. [DOI] [PubMed] [Google Scholar]
- 6.Kato H, Yoshida T, Tokue T, Nojiri Y, Hirota H, Ohta T, Williams RM & Tsukamoto S (2007) Notoamides A-D: Prenylated indole alkaloids isolated from a marine-derived fungus, Aspergillus sp., Angew Chem Int Edit. 46, 2254–2256. [DOI] [PubMed] [Google Scholar]
- 7.Grubbs AW, Artman GD, Tsukamoto S & Williams RM (2007) A concise total synthesis of the notoamides C and D, Angew Chem Int Edit. 46, 2257–2261. [DOI] [PubMed] [Google Scholar]
- 8.Tsukamoto S, Kato H, Samizo M, Nojiri Y, Onuki H, Hirota H & Ohta T (2008) Notoamides F-K, prenylated indole alkaloids isolated from a marine-derived Aspergillus sp., J Nat Prod. 71, 2064–2067. [DOI] [PubMed] [Google Scholar]
- 9.Birch AJ & Wright JJ (1969) Brevianamides - a new class of fungal alkaloid, Chem Commun, 644–645. [Google Scholar]
- 10.Blizzard TA, Marino G, Mrozik H, Fisher MH, Hoogsteen K & Springer JP (1989) Chemical modification of paraherquamide. 1. Unusual reactions and absolute stereochemistry, J Org Chem. 54, 2657–2663. [Google Scholar]
- 11.Blizzard TA, Mrozik H, Fisher MH & Schaeffer JM (1990) Chemical modification of paraherquamide. 2. Replacement of the C-14 methyl group, J Org Chem. 55, 2256–2259. [Google Scholar]
- 12.Blizzard TA, Marino G, Mrozik H, Schaeffer JM & Fisher MH (1989) Chemical Modification of Paraherquamide .3. Vinyl Ether Modified Analogs, Abstr Pap Am Chem S. 198, 42–Orgn. [Google Scholar]
- 13.Blizzard TA, Margiatto G, Mrozik H, Schaeffer JM & Fisher MH (1991) Chemical modification of paraherquamide. 4. 1-N-substituted analogs, Tetrahedron Lett. 32, 2441–2444. [Google Scholar]
- 14.Madariaga-Mazon A, Hernandez-Abreu O, Estrada-Soto S & Mata R (2015) Insights on the vasorelaxant mode of action of malbrancheamide, J Pharm Pharmacol. 67, 551–558. [DOI] [PubMed] [Google Scholar]
- 15.Artman GD, Grubbs AW & Williams RM (2007) Concise, asymmetric, stereocontrolled total synthesis of stephacidins A, B and notoamide B, J Am Chem Soc. 129, 6336–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiya S, Grundmann A, Li SM & Turner G (2006) The fumitremorgin gene cluster of Aspergillus fumigatus: Identification of a gene encoding brevianamide F synthetase, Chembiochem. 7, 1062–1069. [DOI] [PubMed] [Google Scholar]
- 17.Grundmann A & Li SM (2005) Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus, Microbiol-Sgm. 151, 2199–2207. [DOI] [PubMed] [Google Scholar]
- 18.Tsunematsu Y, Ishikawa N, Wakana D, Goda Y, Noguchi H, Moriya H, Hotta K & Watanabe K (2013) Distinct mechanisms for spiro-carbon formation reveal biosynthetic pathway crosstalk, Nat Chem Biol. 9, 818–825. [DOI] [PubMed] [Google Scholar]
- 19.Pistorius D, Li YY, Sandmanny A & Müller R (2011) Completing the puzzle of aurachin biosynthesis in Stigmatella aurantiaca Sg a15, Mol Biosyst. 7, 3308–3315. [DOI] [PubMed] [Google Scholar]
- 20.Katsuyama Y, Harmrolfs K, Pistorius D, Li YY & Müller R (2012) A semipinacol rearrangement directed by an enzymatic system featuring dual-function FAD-dependent monooxygenase, Angew Chem Int Edit. 51, 9437–9440. [DOI] [PubMed] [Google Scholar]
- 21.Ding YS, de Wet JR, Cavalcoli J, Li SY, Greshock TJ, Miller KA, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, Williams RM & Sherman DH (2010) Genome-based characterization of two prenylation steps in the assembly of the stephacidin and notoamide anticancer agents in a marine-derived Aspergillus sp., J Am Chem Soc. 132, 12733–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dan Q, Newmister SA, Klas KR, Fraley AE, A., M. T., Somoza AD, Sunderhaus JD, Ye Y, Shende VV, Yu F, Sander JN, Brown WC, Zhao L, Paton RS, Houk KN, Smith JL, Sherman DH & Williams RM (2019) Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase, Nat Chem. 11, 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavanagh KL, Jörnvall H, Persson B & Oppermann U (2008) The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes, Cell Mol Life Sci. 65, 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallberg Y, Oppermann U & Persson B (2010) Classification of the short-chain dehydrogenase superfamily using hidden Markov models, FEBS J. 277, 2375–2386. [DOI] [PubMed] [Google Scholar]
- 25.Stocking EM, Sanz-Cervera JF, Williams RM & Unkefer CJ (1996) Studies on the biosynthesis of paraherquamide A. Origin of the beta-methylproline ring, J Am Chem Soc. 118, 7008–7009. [DOI] [PubMed] [Google Scholar]
- 26.Stocking EM, Sanz-Cervera JF, Unkefer CJ & Williams RM (2001) Studies on the biosynthesis of paraherquamide. Construction of the amino acid framework, Tetrahedron. 57, 5303–5320. [Google Scholar]
- 27.Stocking EM, Martinez RA, Silks LA, Sanz-Cervera JF & Williams RM (2001) Studies on the biosynthesis of paraherquamide: Concerning the mechanism of the oxidative cyclization of L-isoleucine to beta-methylproline, J Am Chem Soc. 123, 3391–3392. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Tang M-C, Tang S, Gao S, Soliman S, Hang L, Xu W, Ye T, Watanabe K & Tang Y (2018) Genome mining and assembly-line biosynthesis of the UCS1025A pyrrolizidinone family of fungal indole alkaloids, J Am Chem Soc. 140, 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stocking EM, Sanz-Cervera JF & Williams RM (2001) Studies on the biosynthesis of paraherquamide: Synthesis and incorporation of a hexacyclic indole derivative as an advanced metabolite, Angew Chem Int Edit. 40, 1296–1298. [DOI] [PubMed] [Google Scholar]
- 30.Blanchflower SE, Banks RM, Everett JR & Reading C (1993) Further novel metabolites of the paraherquamide family, J Antibiot. 46, 1355–1363. [DOI] [PubMed] [Google Scholar]
- 31.Yang B, Dong JD, Lin XP, Zhou XF, Zhang YY & Liu YH (2014) New prenylated indole alkaloids from fungus Penicillium sp derived of mangrove soil sample, Tetrahedron. 70, 3859–3863. [Google Scholar]
- 32.Jiang W, Cacho RA, Chiou G, Garg NK, Tang Y & Walsh CT (2013) EcdGHK Are three tailoring iron oxygenases for amino acid building blocks of the echinocandin scaffold, J Am Chem Soc. 135, 4457–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukat P, Katsuyama Y, Wenzel S, Binz T, König C, Blankenfeldt W, Brönstrup M & Müller R (2017) Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity, Chem Sci. 8, 7521–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luesch H, Hoffmann D, Hevel JM, Becker JE, Golakoti T & Moore RE (2003) Biosynthesis of 4-methylproline in cyanobacteria: Cloning of nosE and nosF genes and biochemical characterization of the encoded dehydrogenase and reductase activities, J Org Chem. 68, 83–91. [DOI] [PubMed] [Google Scholar]
- 35.Stocking EM, Williams RM & Sanz-Cervera JF (2000) Reverse prenyl transferases exhibit poor facial discrimination in the biosynthesis of paraherquamide A, brevianamide A, and austamide, J Am Chem Soc. 122, 9089–9098. [Google Scholar]
- 36.Tsukamoto S, Kawabata T, Kato H, Greshock TJ, Hirota H, Ohta T & Williams RM (2009) Isolation of antipodal (−)-versicolamide B and notoamides L-N from a marine-derived Aspergillus sp., Org Lett. 11, 1297–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsukamoto S, Kato H, Greshock TJ, Hirota H, Ohta T & Williams RM (2009) Isolation of notoamide E, a key precursor in the biosynthesis of prenylated indole alkaloids in a marine-derived fungus, Aspergillus sp., J Am Chem Soc. 131, 3834–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin SQ, Yu X, Wang Q, Liu XQ & Li SM (2013) Identification of a brevianamide F reverse prenyltransferase BrePT from Aspergillus versicolor with a broad substrate specificity towards tryptophan-containing cyclic dipeptides, Appl Microbiol Biot. 97, 1649–1660. [DOI] [PubMed] [Google Scholar]
- 39.Porter AEA & Sammes PG (1970) A Diels-Alder reaction of possible biosynthetic importance, J Chem Soc Chem Comm, 1103. [Google Scholar]
- 40.Kim HJ, Ruszczycky MW, Choi SH, Liu YN & Liu HW (2011) Enzyme catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A, Nature. 473, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson GA, Zhang ZG, Tietz JI, Mitchell DA & van der Donk WA (2015) In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin, J Am Chem Soc. 137, 16012–16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wever WJ, Bogart JW, Baccile JA, Chan AN, Schroeder FC & Bowers AA (2015) Chemoenzymatic synthesis of thiazolyl peptide natural products featuring an enzyme-catalyzed formal [4+2] cycloaddition, J Am Chem Soc. 137, 3494–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian ZH, Sun P, Yan Y, Wu ZH, Zheng QF, Zhou SX, Zhang H, Yu FT, Jia XY, Chen DD, Mandi A, Kurtan T & Liu W (2015) An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins, Nat Chem Biol. 11, 259–U91. [DOI] [PubMed] [Google Scholar]
- 44.Ohashi M, Liu F, Hai Y, Chen MB, Tang MC, Yang ZY, Sato M, Watanabe K, Houk KN & Tang Y (2017) SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis, Nature. 549, 502–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Tang MC, Tang SB, Gao S, Soliman S, Hang L, Xu W, Ye T, Watanabe K & Tang Y (2018) Genome mining and assembly-line biosynthesis of the UCS1025A pyrrolizidinone family of fungal alkaloids, J Am Chem Soc. 140, 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato N, Nogawa T, Takita R, Kinugasa K, Kanai M, Uchiyama M, Osada H & Takahashi S (2018) Control of the stereochemical course of [4+2] cycloaddition during trans-decalin formation by Fsa2-family enzymes, Angew Chem Int Edit. 57, 9754–9758. [DOI] [PubMed] [Google Scholar]
- 47.Li HQ, Sun WG, Deng MY, Zhou Q, Wang JP, Liu JJ, Chen CM, Qi CX, Luo ZW, Xue YB, Zhu HC & Zhang YH (2018) Asperversiamides, linearly fused prenylated indole alkaloids from the marine-derived fungus Aspergillus versicolor, J Org Chem. 83, 8483–8492. [DOI] [PubMed] [Google Scholar]
- 48.Greshock TJ, Grubbs AW, Jiao P, Wicklow DT, Gloer JB & Williams RM (2008) Isolation, structure elucidation, and biomimetic total synthesis of versicolamide B, and the isolation of antipodal (−)-stephacidin A and (+)-notoamide B from Aspergillus versicolor NRRL 35600, Angew Chem Int Edit. 47, 3573–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato H, Nakahara T, Sugimoto K, Matsuo K, Kagiyama I, Frisvad JC, Sherman DH, Williams RM & Tsukamoto S (2015) Isolation of notoamide S and enantiomeric 6-epi-stephacidin A from the fungus Aspergillus amoenus: Biogenetic implications, Org Lett. 17, 700–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanz-Cervera JF, Glinka T & Williams RM (1993) Biosynthesis of the brevianamides - Quest for a biosynthetic Diels-Alder cyclization, J Am Chem Soc. 115, 347–348. [Google Scholar]
- 51.Finefield JM, Greshock TJ, Sherman DH, Tsukamoto S & Williams RM (2011) Notoamide E: Biosynthetic incorporation into notoamides C and D in cultures of Aspergillus versicolor NRRL 35600, Tetrahedron Lett. 52, 1987–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye Y, Du L, Zhang X, Newmister SA, Zhang W, Mu S, Minami A, McCauley M, Alegre-Requena JV, Fraley AE, Androver-Castellano ML, Shende VV, Oikawa H, Kato H, Tsukamoto S, Paton R, Williams RM, Sherman DH & Li S (2019) Cofactor-independent Pinacolase directs non-Diels-Alderase biogenesis of the brevianamides, ChemRxiv. DOI: 10.26434/chemrxiv.9122009 [DOI] [Google Scholar]
- 53.Williams RM, Sanz-Cervera JF, Sancenon F, Marco JA & Halligan KM (1998) Biomimetic Diels-Alder cyclizations for the construction of the brevianamide, paraherquamide, sclerotamide, asperparaline and VM55599 ring systems, Bioorgan Med Chem. 6, 1233–1241. [DOI] [PubMed] [Google Scholar]
- 54.Jornvall H, Persson B, Krook M, Atrian S, Gonzalezduarte R, Jeffery J & Ghosh D (1995) Short-chain dehydrogenases reductases (Sdr), Biochemistry. 34, 6003–6013. [DOI] [PubMed] [Google Scholar]
- 55.Polonsky J, Merrien MA, Prange T, Pascard C & Moreau S (1980) Isolation and structure (X-ray analysis) of marcfortine A, a new alkaloid from Penicillium roqueforti, JCS Chem Comm, 601–602. [Google Scholar]
- 56.Prangé T, Billion MA, Vuilhorgne M, Pascard C, Polonsky J & Moreau S (1981) Structures of marcfortine B and marcfortine C (X-ray analysis), alkaloids from Penicillium roqueforti, Tetrahedron Lett. 22, 1977–1980. [Google Scholar]
- 57.Whyte AC & Gloer JB (1996) Sclerotiamide: a new member of the paraherquamide class with potent antiinsectan activity from the sclerotia of Aspergillus sclerotiorum, J Nat Prod. 59, 1093–1095. [DOI] [PubMed] [Google Scholar]
- 58.Hayashi H, Nishimoto Y & Nozaki H (1997) Asperparaline A, a new paralytic alkaloid from Aspergillus japonicus JV-23, Tetrahedron Lett. 38, 5655–5658. [DOI] [PubMed] [Google Scholar]
- 59.Greshock TJ, Grubbs AW, Tsukamoto S & Williams RM (2007) A concise, biomimetic total synthesis of stephacidin A and notoamide B, Angew Chem Int Edit. 46, 2262–2265. [DOI] [PubMed] [Google Scholar]
- 60.Sommer K & Williams RM (2009) Studies on paraherquamide biosynthesis: synthesis of deuterium-labeled 7-hydroxy-pre-paraherquamide, a putative precursor of paraherquamides A, E, and F, Tetrahedron. 65, 3246–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraley AE, Caddell Haatveit K, Ye Y, Kelly SP, Newmister SA, Yu F, Williams RM, Smith JL, Houk KN & Sherman DH (2020) Molecular basis for spirocycle formation in the paraherquamide biosynthetic pathway, J Am Chem Soc. DOI: 10.1021/jacs.9b09070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li SY, Finefield JM, Sunderhaus JD, McAfoos TJ, Williams RM & Sherman DH (2012) Biochemical characterization of NotB as an FAD-dependent oxidase in the biosynthesis of notoamide indole alkaloids, J Am Chem Soc. 134, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finefield JM, Kato H, Greshook TJ, Sherman DH, Tsukamoto S & Williams RM (2011) Biosynthetic Studies of the notoamides: Isotopic synthesis of stephacidin A and incorporation into notoamide B and sclerotiamide, Org Lett. 13, 3802–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sunderhaus JD, McAfoos TJ, Finefield JM, Kato H, Li SY, Tsukamoto S, Sherman DH & Williams RM (2013) Synthesis and bioconversions of notoamide T: A biosynthetic precursor to stephacidin A and notoamide B, Org Lett. 15, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McAfoos TJ, Li SY, Tsukamoto S, Sherman DH & Williams RM (2010) Studies on the biosynthesis of the stephacidins and notoamides. Total synthesis of notoamide S, Heterocycles. 82, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li SY, Finefield JM, Sunderhaus JD, McAfoos TJ, Williams RM & Sherman DH (2012) Biochemical characterization of NotB as an FAD-dependent oxidase in the biosynthesis of Nntoamide indole alkaloids, J Am Chem Soc. 134, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klas KR, Kato H, Frisvad JC, Yu FA, Newmister SA, Fraley AE, Sherman DH, Tsukamoto S & Williams RM (2018) Structural and stereochemical diversity in prenylated indole alkaloids containing the bicyclo [2.2.2]diazaoctane ring system from marine and terrestrial fungi, Nat Prod Rep. 35, 532–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fraley AE, Tran HT, Tripathi A, Newmister SA, Kelly SP, Kato H, Tsukamoto S, Li S, Williams RM, Sherman DH (2019) Flavin-dependent monooxygenases NotI and NotI’ mediate spiro-oxindole formation in biosynthesis of the notoamides, ChemRxiv. DOI: 10.26434/chemrxiv.11302604.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kagiyama I, Kato H, Nehira T, Frisvad JC, Sherman DH, Williams RM & Tsukamoto S (2016) Taichunamides: Prenylated indole alkaloids from Aspergillus taichungensis (IBT 19404), Angew Chem Int Edit. 55, 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li F, Zhang ZZ, Zhang GJ, Che Q, Zhu TJ, Gu QQ & Li DH (2018) Determination of taichunamide H and structural revision of taichunamide A, Org Lett. 20, 1138–1141. [DOI] [PubMed] [Google Scholar]
- 71.Cai SX, Luan YP, Kong XL, Zhu TJ, Gu QQ & Li DH (2013) Isolation and photoinduced conversion of 6-epi-stephacidin from Aspergillus taichungensis, Org Lett. 15, 2168–2171. [DOI] [PubMed] [Google Scholar]
- 72.Domingo LR, Zaragoza RJ & Williams RM (2003) Studies on the biosynthesis of paraherquamide A and VM99955. A theoretical study of intramolecular Diels-Alder cycloaddition, J Org Chem. 68, 2895–2902. [DOI] [PubMed] [Google Scholar]
- 73.Hu XL, Bian XQ, Wu X, Li JY, Hua HM, Pei YH, Han AH & Bai J (2014) Penioxalamine A, a novel prenylated spiro-oxindole alkaloid from Penicillium oxalicum TW01–1, Tetrahedron Lett. 55, 3864–3867. [Google Scholar]
- 74.Lin ZJ, Wen JN, Zhu TJ, Fang YC, Gu QQ & Zhu WM (2008) Chrysogenamide A from an endophytic fungus associated with Cistanche deserticola and its neuroprotective effect on SH-SY5Y cells, J Antibiot. 61, 81–85. [DOI] [PubMed] [Google Scholar]
- 75.Li CW, Wu CJ, Cui CB, Xu LL, Cao F & Zhu HJ (2016) Penicimutamides A-C: Rare carbamate-containing alkaloids from a mutant of the marine-derived Penicillium purpurogenum G59, Rsc Adv. 6, 73383–73387. [Google Scholar]
- 76.Mascotti ML, Ayub MJ, Furnham N, Thornton JM & Laskowski RA (2016) Chopping and changing: the evolution of the flavin-dependent monooxygenases, J Mol Biol. 428, 3131–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhandari P, Crombie L, Harper MF, Rossiter JT, Sanders M & Whiting DA (1992) Mechanism and stereochemistry of the enzyme-catalyzed formation of a 2,2-dimethylchromene ring from a prenylated phenol - Conversion of rot-2’-enonic acid into deguelin by deguelin cyclase, J Chem Soc Perkin Trans. 1, 1685–1697. [Google Scholar]
- 78.Crombie L, Rossiter JT, Vanbruggen N & Whiting DA (1992) Deguelin cyclase, a prenyl to chromen transforming enzyme from Tephrosia vogellii, Phytochemistry. 31, 451–461. [Google Scholar]
- 79.Watts KR, Loveridge ST, Tenney K, Media J, Valeriote FA & Crews P (2011) Utilizing DART mass spectrometry to pinpoint halogenated metabolites from a marine invertebrate-derived fungus, J Org Chem. 76, 6201–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fraley AE, Garcia-Borrás M, Tripathi A, Khare D, Mercado-Marin EV, Tran H, Dan Q, Webb GP, Watts KR, Crews P, Sarpong R, Williams RM, Smith JL, Houk KN & Sherman DH (2017) Function and structure of MalA/MalA’, iterative halogenases for late-stage C-H functionalization of indole alkaloids, J Am Chem Soc. 139, 12060–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yeh E, Blasiak LC, Koglin A, Drennan CL & Walsh CT (2007) Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases, Biochemistry. 46, 1284–1292. [DOI] [PubMed] [Google Scholar]
- 82.Zheng QF, Gong YK, Guo YJ, Zhao ZX, Wu ZH, Zhou ZX, Chen DD, Pan LF & Liu W (2018) Structural insights into a flavin-dependent [4+2] cyclase that catalyzes trans-decalin formation in pyrroindomycin biosynthesis, Cell Chem Biol. 25, 718–727. [DOI] [PubMed] [Google Scholar]
- 83.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJ (2015) The Phyre2 web portal for protein modelling, prediction and analysis, Nat Protoc. 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]