Abstract
Background
Emerging biomarkers from medical imaging or molecular characterization of tumour biopsies open up for combining the two and exploiting their synergy in treatment planning of cancer patients. We generated a paired data set of imaging- and gene-based hypoxia biomarkers in cervical cancer, appraised the influence of intratumour heterogeneity in patient classification, and investigated the benefit of combining the methodologies in prediction of chemoradiotherapy failure.
Methods
Hypoxic fraction from dynamic contrast enhanced (DCE)-MR images and an expression signature of six hypoxia-responsive genes were assessed as imaging- and gene-based biomarker, respectively in 118 patients.
Findings
Dichotomous biomarker cutoff to yield similar hypoxia status by imaging and genes was defined in 41 patients, and the association was validated in the remaining 77 patients. The two biomarkers classified 75% of 118 patients with the same hypoxia status, and inconsistent classification was not related to imaging-defined intratumour heterogeneity in hypoxia. Gene-based hypoxia was independent on tumour cell fraction in the biopsies and showed minor heterogeneity across multiple samples in 9 tumours. Combining imaging- and gene-based classification gave a significantly better prediction of PFS than one biomarker alone. A combined dichotomous biomarker optimized in 77 patients showed a large separation in PFS between more and less hypoxic tumours, and separated the remaining 41 patients with different PFS. The combined biomarker showed prognostic value together with tumour stage in multivariate analysis.
Interpretation
Combining imaging- and gene-based biomarkers may enable more precise and informative assessment of hypoxia-related chemoradiotherapy resistance in cervical cancer.
Funding
Norwegian Cancer Society, South-Eastern Norway Regional Health Authority, and Norwegian Research Council.
Keywords: Prognostic biomarker, Medical imaging, Gene expression signature, Hypoxia, Intratumour heterogeneity, Cervical cancer
Research in context.
Evidence before this study: Many cancer patients die of metastasis and treatment resistance caused by low levels of oxygen (hypoxia) in their tumour. Better treatment strategies are required, and biomarkers assessing the tumour hypoxia status are needed for selection of patients to clinical trials and individual treatment regimes. Medical imaging and tumour biopsies are cornerstones in the diagnostic procedures, and both imaging parameters and biopsy-based gene expression signatures show promise as hypoxia biomarkers. The two methodologies have different strengths, and there is a need to understand how they relate to each other and should be combined to fully exploit their potential. Imaging provide information about the entire tumour, whereas biopsy-based biomarkers are derived only from a small part. Variations across the tumour volume, intratumour heterogeneity, is thus a factor potentially affecting hypoxia assessment. A PubMed search with the terms “hypoxia”, “cancer”, “biomarker”, “gene signature”, “imaging”, and “survival”, without restricting the search by date or language, found no studies comparing imaging- and gene-based hypoxia classification of patients in relation to intratumour heterogeneity and treatment resistance.
Added value of this study: We report the generation of a paired data set on imaging- and gene-based hypoxia biomarkers in 118 cervical cancer patients, enabling the comparison and combination of the two methodologies in hypoxia assessment. By using an imaging biomarker constructed from MR images with high spatial resolution compared to biopsy size, intratumour heterogeneity in imaging-defined hypoxia could be assessed on a scale of relevance for the gene-based biomarker. Our results revealed robust classification for both biomarkers, independent on intratumour heterogeneity. Measuring the gene-based biomarker in a small sample relative to the whole tumour volume thus seem to provide reliable hypoxia information. For 25% of the patients, the tumours were classified with a different hypoxia status using the imaging-based biomarker as compared to the gene-based biomarker. The two biomarkers therefore seem to contain different information, and our results showed that combining them for hypoxia classification, could improve the separation in progression-free survival for patients classified with a more or a less hypoxic tumour.
Implications of all the available evidence: Our study encourages implementation of a multifactorial biomarker in patients with cervical cancer, where imaging and gene expression signature are combined to assess hypoxia-related treatment resistance and thereby enable more information about the disease before treatment-decision. The approach is clinically feasible as the data are based on MR images and biopsies obtained during state-of-the-art diagnostics. Combining tumour phenotype information from both methodologies may be useful also for other cancer types as well as other tumour phenotypes than hypoxia.
Alt-text: Unlabelled box
1. Introduction
Advances in medical imaging and molecular characterization of tumours have shown promise for identifying treatment-resistant cancer and deciding therapy [1], [2], [3]. Incorporation of the two methodologies in the clinic may improve treatment-decision, and is an important step towards precision medicine [3,4]. Hypoxia is a major adverse feature of solid tumours, leading to metastases and resistance to radiotherapy, chemotherapy, and possibly molecular targeted drugs and immunotherapies [5,6]. Promising imaging- and gene-based hypoxia biomarkers have been proposed, including candidates derived from positron emission tomography (PET) and magnetic resonance (MR) images [7], and gene expression signatures recorded in tumour biopsies [8]. In particular, PET with the hypoxia tracer F-18-fluoromisonidazole (FMISO) has shown potential for targeted, local radiation dose escalation in head and neck cancer [9]. Also, dynamic contrast enhanced (DCE)-MR imaging (MRI) has shown benefit in monitoring effects of the hypoxia-modifying drug sorafenib in cervical cancer [10]. Moreover, gene expression signatures with predictive impact in hypoxia-modifying combination therapies of head and neck and bladder cancer have been presented [11,12] and some are evaluated in ongoing intervention trials (e.g. NCT01950689, NCT01880359, NCT02661152, NCT04275713). These developments constitute an excellent basis for combining imaging and molecular characterization to exploit advantages of each methodology in an extended treatment decision support system that includes hypoxia.
Imaging- and gene-based hypoxia biomarkers provide different information of value for treatment planning. Imaging can non-invasively visualize hypoxia in three dimensions prior to and during therapy, assess intratumour heterogeneity, and monitor therapy responses repeatedly [13]. Approaches based on MR and PET can be implemented without high costs and changes in the hospital's infrastructure, since these modalities are part of the state-of-the-art diagnostic procedures for many cancer types, including cervical cancer [14]. On the other hand, gene expression signatures capture the transcriptional state of cells and can inform about hypoxia-related resistance mechanisms at play in individual tumours [15]. This is of utmost importance for the choice of hypoxia-targeting drug amongst a large number of existing and upcoming agents for combination therapies [16]. A major obstacle is, however, that the information provided in a biopsy may be biased by the cellular composition of the sample and intratumour heterogeneity in hypoxia [17]. To exploit the potential synergy between imaging- and gene-based hypoxia biomarkers, a better understanding of how their information relates to each other is crucial.
A major challenge in studies comparing the two methodologies is a shortage of paired imaging and gene data in patient cohorts; existing reports are few and based on small cohorts [18]. We have proposed imaging- and gene-based hypoxia biomarkers for cervical cancer patients, derived from DCE-MR images and gene expression data, respectively [19], [20], [21], [22]. The biomarkers have shown prognostic impact in several independent cohorts [19,21,23], and although they both inform about hypoxia, their underlying biology differs. The imaging biomarkers depend on physiological features related to oxygen supply and consumption, such as blood perfusion, vascular density, and cell density [24], while the gene-based biomarkers measure expression of hypoxia responsive genes. In the present work, we generated a unique paired data set of two of the most promising imaging- and gene-based biomarkers for 118 cervical cancer patients [21,22]. We compared the performance of the two biomarkers in relation to the intratumour heterogeneity, and further investigated how they could be combined in prediction of treatment resistance.
2. Materials and methods
2.1. Patient cohort and study design
Totally 118 patients with locally advanced carcinomas of the uterine cervix, prospectively recruited to our observational chemoradiotherapy trial at the Norwegian Radium Hospital from 2001 to 2007, were included (Supplementary Table S1). Treatment and follow-up were performed as described [20]. In short, external radiation of 50 Gy in 25 fractions was given to the tumour, parametria, and adjacent pelvic wall, while the remaining pelvis received 45 Gy. This was followed by brachytherapy of totally 25 Gy to the tumour in 5 fractions. Concurrent cisplatin (40 mg/m2) was given weekly in maximum six courses according to tolerance. Follow up was performed by standard procedures. When symptoms of relapse were noted, MRI of pelvis and retroperitoneum as well as X-ray of thorax were performed. The study was approved by the Regional Committee for Medical and Health Research Ethics in southern Norway (S-01129). All patients gave written informed consent.
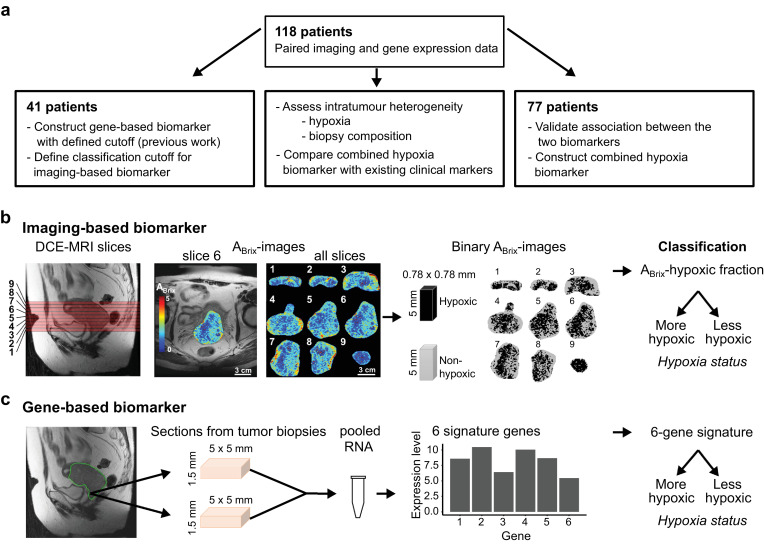
High quality paired DCE-MR images and gene expression data were available for all patients. The cohort constituted a subgroup of 41 patients which were used to construct and define the classification cutoff of the gene-based biomarker in previous work [21]. The classification cutoff of the imaging-based biomarker had not been determined before [22], and the same 41 patients were used to define this cutoff (Fig. 1a). The remaining 77 patients were used to validate the association between the two biomarkers and to construct a combined biomarker. The performance of this combined biomarker in relation to existing clinical markers was evaluated in the entire cohort of 118 patients (Fig. 1a). The entire cohort was also used to compare the performance of the individual biomarkers in relation to the intratumour heterogeneity.
Fig. 1.
Study design and hypoxia biomarkers. (a) Patients included at different stages of the study. Independent subgroups of the total cohort of 118 patients were used to determine biomarker cutoff for classification (n = 41) and to validate the association between the biomarkers and construct the combined biomarker (n = 77). The total cohort was used to assess the importance of intratumour heterogenity for biomarker performance and compare the combined biomarker with existing clinical markers. (b) Determination of the imaging-based biomarker from left to right, sagittal T2W-image of the pelvic showing localization of image slices numbered from lower to upper part of the tumour, example of axial ABrix-image of the tumour (slice 6) superimposed on T2W-image, ABrix-images of all slices covering the tumour, binary ABrix-images of the same slices showing voxels in hypoxic and non-hypoxic regions according to an ABrix threshold value of 1.56, and classification based on the hypoxic fraction of all slices combined. (c) Determination of the gene-based biomarker from left to right, sagittal T2W-image of the pelvic showing localization of the region accessible for biopsies in the lower part of tumour, the approximate size of the sections from 1–4 biopsies (median 2) taken from each tumour and pooled for RNA isolation, expression data of 6 signature genes, and classification based on the signature value.
2.2. Imaging-based biomarker
Diagnostic DCE-MR images for the imaging-based biomarker were acquired using a Signa Horizon LX-1.5T scanner (GE Medical Systems) with a pelvic-phased-array coil and a fast bolus injection of 0.1 mmol/kg body weight of Gd-DTPA (Magnevist®, Schering) [19,22] The dynamic T1-weighted series were acquired with a fast spoiled gradient recalled echo sequence and included 2–12 (median of seven) axial slices covering the whole tumour (Fig. 1b), with a slice thickness of 5 mm, slice gap of 1 mm, and in-plane resolution of 0.78 mm. Axial T2-weighted (T2W) images from a fast spin echo sequence were used for tumour delineation. The uptake of contrast agent; i.e. the relative signal intensity increase as a function of time after Gd-DTPA injection, was recorded for each voxel.
The ABrix-parameter in Brix pharmacokinetic model [24] was derived from the contrast uptake curves [19]. Fraction of voxels in hypoxic regions was calculated from the ABrix-values of all tumour voxels, and this ABrix-hypoxic fraction was used as biomarker (Fig. 1b). The biomarker can be visualized in binary ABrix-images and is thus a clinically usable imaging biomarker [22]. The ABrix threshold for hypoxia was 1.56. This threshold was established in a subgroup of our patients to reflect a hypoxia level associated with chemoradiotherapy resistance [22], and was considered to be valid for our extended cohort examined with the same MR machine. To define a cutoff of ABrix-hypoxic fraction for dichotomous classification of tumours, we considered the hypoxia status of the 41 patients used to construct the gene-based biomarker [21] (Fig. 1a). This hypoxia status from previous work was determined from the 20–30th percentile of the ABrix-histogram for the entire tumour, and is less feasible as imaging biomarker than the ABrix-hypoxic fraction applied in the present study. The tumours classified as more hypoxic and as less hypoxic had an ABrix-hypoxic fraction above and below 0.38, respectively, and this cutoff was used to classify tumours according to their hypoxia status with the imaging biomarker (Fig. 1b).
2.3. Gene-based biomarker
Gene expression profiles for the gene-based biomarker were generated based on pooled RNA from one to four biopsies (median of two) per tumour. The biopsies were taken from the lower, accessible region of the tumour the day after MRI (Fig. 1c), immediately snap frozen and stored at −80 °C. From each biopsy, 30 × 50 µm slices (approximately 5 × 5 × 1.5 mm) were used for RNA isolation. Assay methods for RNA isolation and gene expression measurement by Illumina BeadArray WG-6 v3 and HT-12 v4 (Illumina Inc.) were described previously [21]. To compare classification from different biopsies within tumours, two to four biopsies from nine patients (24 biopsies in total) were available for individual analyses. Expression data were derived for each of these biopsies, using Illumina HT-12 v4 BeadArray and the same procedure as for the pooled samples. Tumour cell fraction, defined as the percentage of tumour cells in a haematoxylin and eosin stained section from the central part of the biopsy, was available for all biopsies.
The gene-based biomarker was constructed by evaluating the log2-transformed expression value of 31 hypoxia responsive genes associated with ABrix in a cohort of 42 patients, as reported previously [21]. In short, based on the ability of the expression values to separate patients correctly according to an ABrix-defined hypoxia status, a six-gene signature (DDIT3, ERO1A, KCTD11, P4HA2, STC2, UPK1A) was identified and used as biomarker [21]. The six-gene signature provides a continuous output value calculated as a weighted sum of the expression level of all signature genes [21]. The signature has a predefined cutoff of zero, which was used for dichotomous classification of tumours or samples according to their hypoxia status as more or less hypoxic (Fig. 1c).
2.4. Heterogeneity and clustering of hypoxic regions
Intra- and inter-tumour heterogeneity in the biomarkers were estimated based on data for several samples per tumour, using random-effects one-way analysis-of-variance (ANOVA) models [25,26], where the total variance (T) is divided into the within-tumour (W) and between-tumour (B) variance. As a measure of the intratumour heterogeneity that is invariant to the measurement unit, W/T, where T = W + B, was used. Thus, a W/T > 0.5 indicates a larger intratumour than intertumour heterogeneity, whereas forW/T < 0.5 the heterogeneity within tumours is less than between tumours. When the biomarker value for each tumour is calculated based on the average of k samples instead of one, the within-tumour variance (Wk) is reduced by a factor 1/k, and thus the intratumour heterogeneity of the biomarker using these averages (Wk/T) is given by:
where W and B are the within- and between-tumour variance calculated using ANOVA with single sample values [26]. ANOVA was performed on continuous biomarker values, and for the imaging-based biomarker, logit-transformed data were used.
Clustering of hypoxic regions was identified from the variance in imaging-based hypoxic fraction of numerous virtual samples randomly collected within an image slice. Each sample included 12 voxels, mimicking the size of a biopsy used for the gene-based biomarker. The number of virtual samples per patient was set to 1/12 of the total number of voxels in the corresponding slice. The variation in hypoxic fraction for the virtual samples from a tumour depends on the spatial distribution of the hypoxic areas as well as the hypoxic fraction of the slice, with lower variation for low and high hypoxic fractions. Therefore, to identify tumours with more or less clustering of hypoxic regions, the virtual sample variation adjusted for hypoxic fraction of whole slice was used as follows: The standard deviation (SD) of the hypoxic fractions for the virtual samples was calculated for each tumour and plotted versus the hypoxic fraction of the whole slice. Then, a generalized additive model (GAM) was fitted to the data to separate tumours with more and less clustering, defined as having positive and negative residuals, respectively.
2.5. Hypoxia classification by imaging and genes combined
The imaging- and gene-based biomarkers were combined into a composite hypoxia measure for each patient to define a new classification cutoff that included both biomarkers. Based on a correlation plot of the continuous biomarker values, the optimal line for separation of the more and less hypoxic tumours to achieve the best association to progression-free survival (PFS) was determined:
where y and x are the values of the two biomarkers and y0 and x0 are the points where the line intersects the y- and x-axis, respectively. The optimal separation line and, hence, x0 and y0, were determined in an iterative procedure, where the log-rank test was used to test the difference in PFS between patients with more and less hypoxic tumours for numerous possible lines. The distance (d) from the point (x,y) to the separation line was calculated for each patient to classify the tumour as more or less hypoxic:
where less hypoxic tumours have d < 0 and more hypoxic tumours have d ≥ 0.
2.6. Statistics
Clinical endpoint was PFS for follow-up until five years, where time from diagnosis to disease-related death or first event of relapse was used and patients were censored as described [21]. Kaplan-Meier survival curves were compared using log-rank test. The univariate Cox proportional-hazards (PH) model was used to determine hazard ratios (HR), and Cox uni- and multivariate PH analyses were performed to evaluate prognostic significance. Assumptions of PHs were confirmed graphically using log-minus-log plots. Nested Cox models were compared with a likelihood ratio test.
Associations were estimated by Pearson's or Spearman's correlation and reported by the correlation coefficients r and rho, respectively. Differences between groups were assessed with Fisher's exact test, Wilcoxon rank-sum test or Kruskal-Wallis test, as appropriate. Significance level was 5%, and all tests were two-sided. All analyses were performed using R [27], version 3.6.0. HRs are presented with 95% confidence interval (CI).
2.7. Data availability
The gene expression data have been deposited to the NCBI's Gene Expression Omnibus (GEO) database: GSE146114.
3. Results
3.1. Relationship between imaging- and gene-based classification
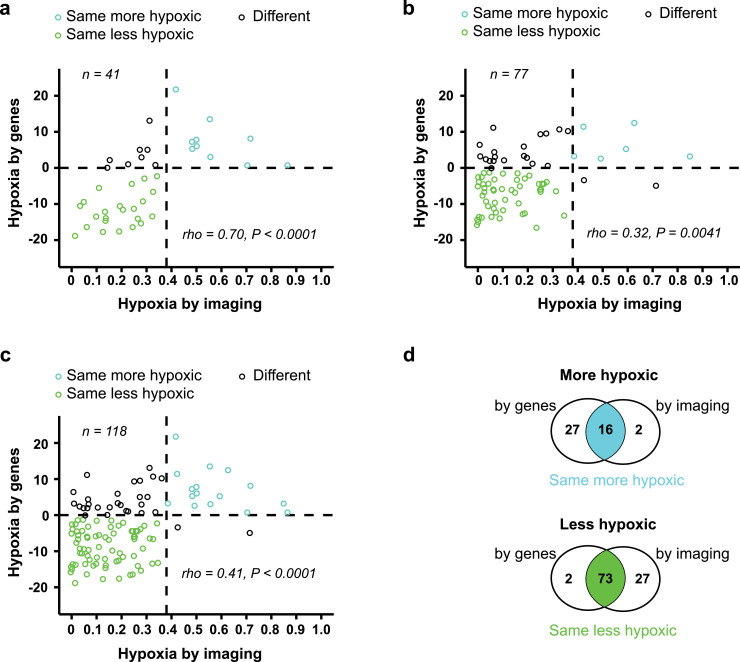
Classification by the two biomarkers was compared in correlation plots of the continuous biomarker values (Fig. 2). Based on the subgroup of 41 patients used to construct the gene-based biomarker in previous work [21], we first confirmed a strong correlation between imaging and genes for our imaging biomarker ABrix-hypoxic fraction, as well as a significant association in classification (P < 0.0001;,Fisher's exact test) (Fig 2a). Totally 80% of the tumours were classified with the same hypoxia status by imaging and genes in these patients. These associations were further validated in the 77 remaining patients. A positive correlation between the biomarker values was found (Fig. 2b), and a significant association in classifications (P = 0.01, Fisher's exact test,) with 73% of the tumours classified with the same hypoxia status.
Fig. 2.
Comparison of imaging-and gene-based hypoxia classification. Correlation plots of biomarker values, showing gene-defined versus imaging-defined hypoxia for 41 patients used in construction of the gene-based biomarker and to define cutoff of both biomarkers (a), 77 remaining patients (b) and all 118 patients (c). Dotted lines, classification cutoff of each biomarker, defining three groups according to the similarity in classification by imaging and genes. P-value and regression coefficient (rho) from Spearman correlation analysis are indicated. (d) Venn diagram showing the overlap of imaging- and gene-based classification of more and less hypoxic tumours for all 118 patients.
To compare the two biomarkers in the following analyses of intratumour heterogeneity, the two subgroups of patients were merged (Fig. 1a). Based on the correlation plot, three classification groups were defined; group 1 (n = 16, more hypoxic) and 2 (n = 73, less hypoxic) tumours had the same hypoxia status by both biomarkers, whereas group 3 tumours (n = 29) were classified with different hypoxia status (Fig. 2c-d). Although equal classification by the two biomarkers was generally achieved (75%), a significant group of tumours was thus differently classified. Most of these tumours; i.e., 27 out of 29, were more hypoxic by genes (Fig. 2c), and a higher number of tumours was classified as more hypoxic by genes (n = 43) than by imaging (n = 18).
3.2. Heterogeneity in hypoxia across the tumour volume
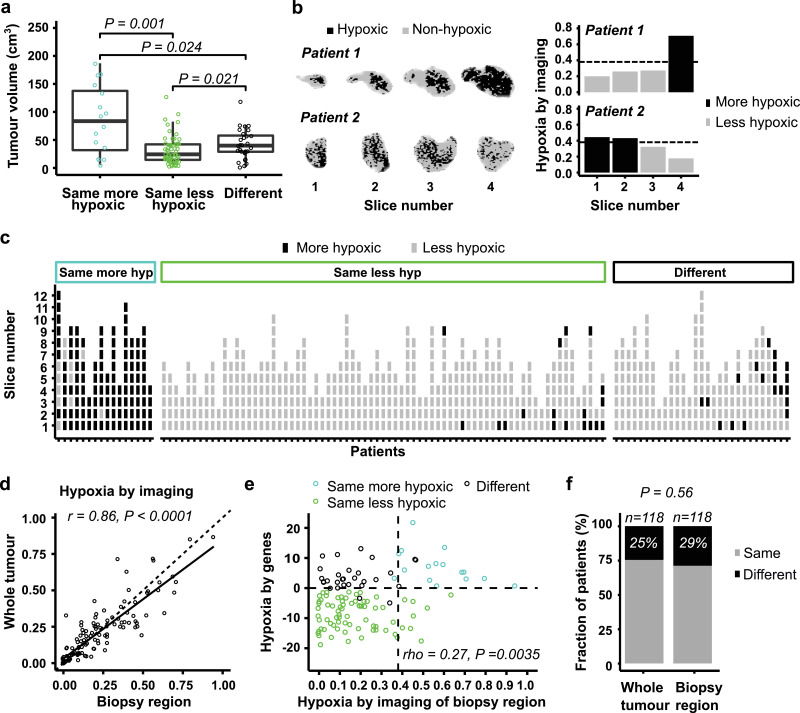
Inconsistent classification by the two biomarkers could be more common in large than small tumours, since the biopsies include only a small part of the tumour and may therefore have lower ability to record the hypoxia status correctly in these cases. In contrast to this hypothesis, the largest tumours were generally classified equally, as more hypoxic by both biomarkers (Fig. 3a). However, tumours classified differently had a higher volume than those defined as less hypoxic by both biomarkers (Fig. 3a). A detailed investigation of regional differences in hypoxia was therefore performed by first using the imaging data to assess the heterogeneity across image slices. The binary images revealed pronounced differences up to 0.54 in hypoxic fraction between slices of individual tumours, and the hypoxia status of each slice could differ (Fig. 3b-c; Supplementary Fig. S1). However, hypoxic fraction per slice showed a small intratumour heterogeneity with a W/T of 0.16. Moreover, most slices (93%) were classified with the same hypoxia status as the whole tumour (Fig. 3c).
Fig. 3.
Heterogeneity in hypoxia across image slices. (a) Tumour volume for the three classification groups defined in Fig. 2c. Jitter plot overlaid a boxplot, where the boxes extend from the first to third quartile with the median value indicated. P-values from Wilcoxon rank-sum test is indicated. (b) Binary images showing voxels in hypoxic and non-hypoxic regions of two patients with heterogeneous hypoxia status across image slices (left), and imaging-defined hypoxia (hypoxic fraction) of each slice (right). Dotted lines, classification cutoff (0.38). (c) Classification of individual image slices by the imaging-based biomarker. The patients are divided into three classification groups defined in Fig. 2c, and are sorted according to increasing biomarker value of the whole tumour for each group. (d) Correlation between imaging-defined hypoxia of the biopsy region and the whole tumour. Dotted line, 1:1 relationship; solid line, linear regression line. (e) Comparison of the imaging- and gene-based biomarker values using image slice 1 from the biopsy region. Dotted lines, classification cutoff of each biomarker. The three classification groups defined in Fig. 2c are indicated by colours. (f) Fraction of patients classified with same or different hypoxia status by imaging and genes, using images from the whole tumour or the biopsy region. Number of patients (n) and P-value from Fisher's exact test are indicated. (d, e) P-value and regression coefficient from Pearson (d) and Spearman (e) correlation analysis are indicated.
The image slice covering the biopsy region (i.e., slice 1) was selected to investigate how well the hypoxia status of this region reflected the status of the whole tumour. Imaging-defined hypoxia based on this slice showed a strong correlation with the value based on all slices (r = 0.86, P < 0·0001; Fig. 3d). Moreover, both a significant correlation between imaging- and gene-defined hypoxia (rho = 0.27, P = 0·0035; Fig. 3e) as well as the similarity in classification by imaging and genes (Fig. 3f) were retained, but not improved, when the imaging data were derived from the biopsy region rather than the whole tumour. These results indicate that discordant hypoxia classification by imaging and genes was not related to different hypoxia status of the biopsy region compared to the whole tumour. In particular, the hypoxia status of the biopsy region seemed to be representative for the whole tumour.
3.3. Heterogeneity in hypoxia within the biopsy region
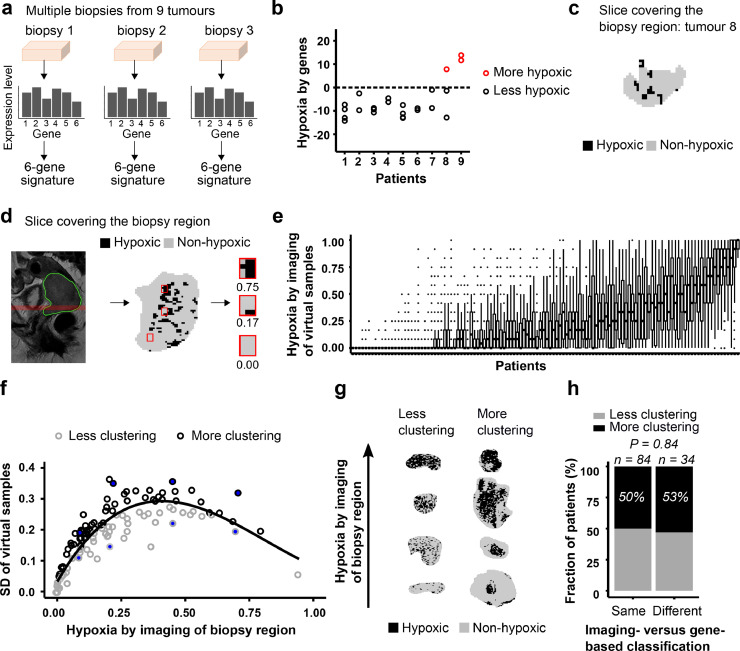
The heterogeneity in hypoxia was further investigated within the biopsy region. In a subgroup of nine patients, hypoxia was assessed by genes for multiple biopsies from each tumour (Fig. 4a). The intratumour heterogeneity in gene-defined hypoxia, determined as a W/T of 0.33, was smaller than the heterogeneity between tumours. By using two to four biopsies, the intratumour heterogeneity (Wk/T) was reduced to 0.20 (2 biopsies), 0.14 (3 biopsies) and 0.11 (4 biopsies). However, the number of biopsies used for patient classification in Fig. 2 (i.e., 1–4) was not associated with the similarity in imaging- and gene-based classification (Supplementary Fig. S2). The use of 2 or more biopsies therefore seemed not to result in more cases of equal classification. Moreover, for eight of nine tumours in our multiple biopsy experiment there was a complete concordance in classification amongst the biopsies (Fig. 4b). For the remaining tumour (no. 8, Fig. 4b), one biopsy was classified as more hypoxic and two as less hypoxic. The binary image of the slice covering the biopsy region (slice 1) of this tumour showed large spatial variation with clustering of hypoxic areas (Fig. 4c), which could imply that the biomarker value strongly depended on the exact location of the biopsy.
Fig. 4.
Heterogeneity in hypoxia within biopsy region. (a) Determination of gene-based biomarker for multiple biopsies from 9 tumours. (b) Gene-based biomarker value for 2–4 biopsies from each of 9 tumours (24 samples). Dotted line, classification cutoff. (c) Binary image of the biopsy region for tumour 8 in (b). (d) Illustration of the simulation experiment. Sagittal T2W-image of the pelvic with indication of selected image slice covering the biopsy region (left), binary image of selected slice showing voxels in hypoxic and non-hypoxic regions and location of 3 virtual samples, each of 12 voxels (middle), binary image and biomarker value (hypoxic fraction) of the 3 virtual samples (right). (e), Imaging-based biomarker value of the virtual samples for each patient sorted according to increasing biomarker value of the biopsy region. The boxes extend from the first to third quartile with the median value indicated. (f), Standard deviation (SD) of the imaging-based biomarker value of the virtual samples versus biomarker value of the biopsy region. Line, generalized additive model (GAM) fitted to the data to separate tumours with more (above the line) or less (below the line) clustering of hypoxic regions. Filled circles, tumours displayed in (g). (g) Binary images showing less or more clustering of hypoxic regions for 8 tumours indicated in (f). (h), Fraction of patients with less or more clustering of hypoxic regions for patients with same or different hypoxia status by imaging (biopsy region) and genes. Number of patients (n) and P-value from Fisher's exact test are indicated.
To address this hypothesis, we performed a simulation experiment to assess the spatial variation in hypoxia within the biopsy region, using the imaging data of slice 1. We randomly sampled numerous areas of biopsy size, each of 12 voxels, within the slice and determined the hypoxic fraction for each of these virtual samples (Fig. 4d–e). The virtual samples showed a large intratumour heterogeneity, with a W/T of 0.50, indicating high spatial variation in imaging-defined hypoxia across the biopsy region. Tumours with less or more clustering of hypoxic regions were identified by comparing the variation in the imaging-defined hypoxia of the virtual samples for each tumour, correcting for the whole-slice hypoxic fraction (Fig. 4f). Considerable difference in the degree of clustering was found across the tumours, in agreement with a visual inspection of the image slices (Fig. 4g; Supplementary Fig. S3-S4). Clustering was, however, not associated with inconsistent classification by imaging and genes (Fig. 4h). Overall, the gene-based biomarker seemed to be reproducible across biopsies, and spatial variation in hypoxia within the biopsy region could not explain differences in classification by the two biomarkers.
3.4. Heterogeneity in biopsy composition and gene-based hypoxia
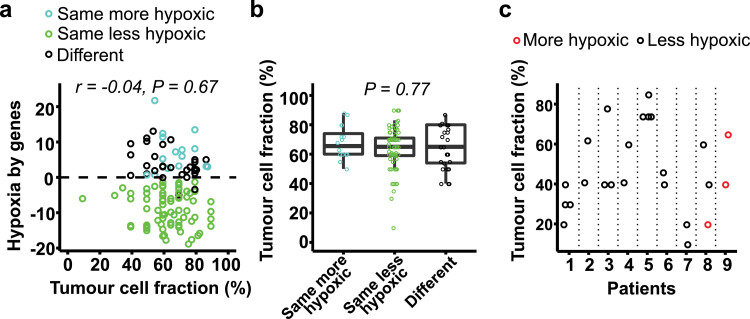
Tumour cell fraction was 50% or higher in most biopsies underlying the gene-based biomarker, but showed a broad range from 10% to 90% (median of 65%). No correlation between this fraction and gene-defined hypoxia was found (Fig. 5a). In particular, both more and less hypoxic tumours were identified at tumour cell fractions up to 90% and down to 40%. All three tumours with a fraction below 40% were classified as less hypoxic by genes, but these were also less hypoxic by imaging (Fig. 5a). Moreover, there was no significant difference in this fraction between any of the classification groups (Fig. 5b). A more detailed analysis was also performed, using data from our multiple biopsy experiment (Fig. 4a). Although the fraction could differ up to 40% for some biopsies from the same tumour (Fig. 5c, tumours no. 3 and 8), they were generally classified with the same hypoxia status. For tumour no. 8, the only biopsy out of three that was classified as more hypoxic had the lowest tumour cell fraction of 20%. Altogether, the gene-based classification seemed to be independent on biopsy composition.
Fig. 5.
Tumour cell fraction and gene-based classification. (a) Gene-based biomarker value versus tumour cell fraction in biopsy (n = 118). Mean fraction of multiple biopsies used for classification of the tumour is shown. The colour indicates the three classification groups defined in Fig. 2c. Dotted line, classification cutoff. P-value and regression coefficient (r) from Pearson correlation analysis are indicated. (b) Tumour cell fraction of the three classification groups. Jitter plot overlaid a boxplot, where the boxes extend from the first to third quartile with the median value indicated. P-value from Kruskal-Wallis test is indicated. (c) Tumour cell fraction and hypoxia status by the gene-based biomarker of individual biopsies from 9 tumours presented in Fig. 4b.
3.5. Construction of a combined imaging- and gene-based biomarker
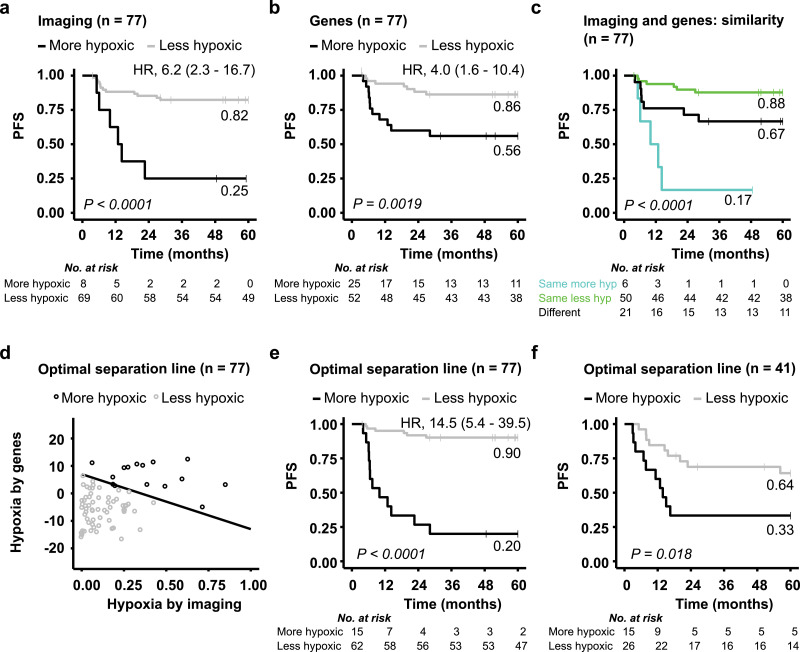
The above results indicated considerable robustness of the two biomarkers, where different classification of some tumours seemed not to be caused by intratumour heterogeneity. It was therefore likely that the biomarkers provided complementary information that combined could lead to more precise prediction of treatment outcome. To address this hypothesis, we considered the 77 patients who were previously not used to construct the gene-based biomarker (Fig. 1a). Both biomarkers showed prognostic impact in this subgroup of patients, with a significantly lower PFS probability for patients with a more hypoxic than a less hypoxic tumour (Fig. 6a-b). Moreover, the lowest PFS probability of 0.17 was achieved for patients with more hypoxia by both biomarkers (group 1 tumours) (Fig. 6c). While patients with less hypoxia by both biomarkers (group 2) showed the highest PFS probability of 0.88, those with more hypoxia by one biomarker only (group 3) had an intermediate PFS probability of 0.67 (Fig. 6c). Some of the latter patients thus experienced relapse and should have been included amongst the high-risk group 1 patients. A Cox PH model with the two dichotomous biomarkers as covariates showed a significant contribution of both of them in the model (P < 0.05). Moreover, this model fitted the PFS data significantly better than a model with either imaging or genes only (likelihood ratio test, P = 0.03 and P = 0.01, respectively). Altogether, this supported that the two biomarkers combined would yield a better outcome prediction than only one biomarker.
Fig. 6.
Combined imaging- and gene-based biomarker. Kaplan-Meier curves for progression-free survival (PFS) of 77 patients with a more or less hypoxic tumour classified by imaging (a) or genes (b), and for the three classification groups defined in Fig. 2c (c). (d) Correlation plot of biomarker values for the same 77 patients, showing gene-defined versus imaging-defined hypoxia. The optimal line for classifying patients with a more (above the line) and less (below the line) hypoxic tumour to achieve the strongest association to PFS is shown. (e, f) Kaplan-Meier curves for PFS of patients classified into the two groups defined in (d) for the 77 patients used to define the line (e), and the 41 remaining patients (f). P-values from log-rank test, number of patients at risk, 60-month recurrence probability and HR with 95% CIs are indicated in the Kaplan-Meier plots.
A combined dichotomous biomarker was constructed, aiming to identify patients with relapse in the intermediate group as high-risk patients. In the correlation plot of the two continuous biomarker values, a line was identified that divided the patients into two groups; one group with more hypoxia and another with less hypoxia, to yield the strongest association to PFS (Fig. 6d). Numerous lines were tested by changing the intersections with the two biomarker axes (Supplementary Fig. S5). A 10-fold cross-validation was applied, resulting in the same optimal line in each of the ten analyses. By using this line as classification cutoff, some patients originally defined as more hypoxic by one biomarker only, was moved to the more hypoxic group. This yielded a large separation in PFS of 0.70 between the more and less hypoxic group with a 60 months survival probability of 0.20 and 0.90, respectively (Fig 6e). Moreover, HR increased from 6.2 and 4.0 for the imaging- and gene-based biomarkers, respectively, to 14.5 for the combined biomarker (Fig. 6a, b, e). Our strategy to combine the two biomarkers therefore further increased the prognostic impact of hypoxia classification. The optimal line identified for the 77 patients also separated the 41 remaining patients into two groups with different PFS probability (Fig. 6f), demonstrating robustness in this approach.
3.6. Performance of the combined biomarker
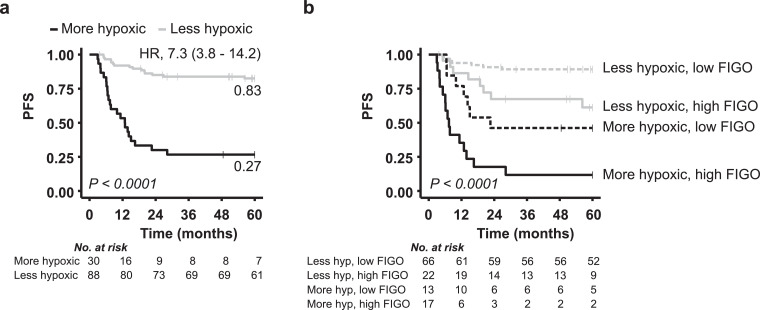
To evaluate the performance of the combined biomarker, the two subgroups of 41 and 77 patients were merged. A strong prognostic impact of the biomarker with a HR of 7.3 was found for this large cohort of 118 patients (Fig. 7a). The biomarker was further compared with the existing clinical markers pelvic lymph node status, tumour stage, and tumour volume, where the patients were classified into groups based on positive or negative lymph node status, median tumour volume or high (3, 4A) or low (1B, 2) tumour stage according to FIGO (Supplementary Table S1). In univariate Cox PH analyses, hypoxia status and tumour stage showed the strongest association to PFS with HRs above 4.5 (Table 1). Moreover, the significance of hypoxia status was retained in multivariable Cox PH analysis together with tumour stage (Table 1). Hence, although tumour stage is a strong prognostic factor in cervical cancer, hypoxia status appeared to have independent prognostic impact that may be of clinical value. This is demonstrated by the survival curves in Fig. 7b, showing considerable difference in PFS probability for patients with a more and less hypoxic tumour both in cases of a high and a low tumour stage.
Fig. 7.
Prognostic value of hypoxia classification by the combined biomarker. Kaplan-Meier curves for progression-free survival (PFS) of 118 patients classified with a more or less hypoxic tumour (a) and for the same classification groups stratified for a low and a high tumour stage according to FIGO (b). P-values from log-rank test, number of patients at risk, 60-month recurrence probability and HR with 95% CIs (a) are indicated.
Table 1.
Cox PH regression analyses based on 118 patients.
| Univariate analyses |
Multivariate analysisd |
|||||
|---|---|---|---|---|---|---|
| p | HR | 95% CI | p | HR | 95% CI | |
| Pelvic lymph node status | 0.051 | 1.9 | 1.0–3.7 | N.S. | - | - |
| Tumour stagea | < 0.001 | 4.6 | 2.3–8.9 | < 0.001 | 3.4 | 1.7–6.7 |
| Tumour volumeb | 0.033 | 2.1 | 1.1–4.1 | N.S. | – | – |
| Hypoxiac | < 0.001 | 7.3 | 3.7–14.2 | < 0.001 | 5.8 | 3.0–11.5 |
Abbreviations: HR, hazard ratio; CI, confidence interval; FIGO, Federation International de Gynecologie et d'Obstetrique; N.S., non-significant.
Patients were divided into two groups based on a FIGO stage of 1B-2B and 3A-4A.
Patients were divided into two groups based on the median tumour volume of 31.1 cm3.
Patients were classified with a more or less hypoxic tumour by a composite hypoxia measure based on imaging and genes combined.
In the multivariate analysis, the same result was obtained for forward and backward selection.
4. Discussion
The present study addresses an unmet need in the diagnostics of cancer patients; to better understand how an imaging- and a gene-based biomarker relate to each other and should be combined in an extended treatment decision support system. Construction of a unique, paired data set for a decent number of patients enabled reliable comparison of two hypoxia biomarkers and evaluation of their prognostic potential in a combined setting. Moreover, by utilizing an imaging biomarker constructed from MR images with high spatial resolution compared to biopsy size, intratumour heterogeneity in hypoxia could be assessed on a scale of relevance for the gene-based biomarker. Although considerable heterogeneity was found, this seemed to have no major influence on the performance of the two biomarkers, and a synergy in prediction of treatment resistance was demonstrated by combining their hypoxia information.
The imaging data suggested large regional differences in hypoxia both across the entire tumour volume and, at a smaller level, within the biopsy region. This is consistent with oxygen tension (pO2) measurements by electrodes, showing broad pO2 distributions of individual cervix tumours and a large variation in pO2 along electrode tracks of 5–10 mm [28], [29], [30], [31]. Although this heterogeneity has caused concern for the application of hypoxia biomarkers derived from only a limited part of the tumour [32], our study showed good agreement between the hypoxia status of the biopsy region and the whole tumour. Moreover, consistent classification by imaging and genes was found for almost all of the largest tumours, and for tumours with severe clustering of hypoxic regions. It therefore seems to be a minor hurdle for the gene-based biomarker that only the lower part of the tumour is accessible for biopsies and the tumours are large compared to biopsy size. However, it should be emphasized that heterogeneity in hypoxia may have led to different classification by the two biomarkers in a few cases.
The gene-based biomarker showed a low intratumour heterogeneity, consistent with studies reporting lower heterogeneity for most multigene signatures than for the individual signature genes [33]. When using up to four biopsies, the heterogeneity of the continuous biomarker value was reduced to around the 0.15 limit suggested for a biomarker to represent the tumour with satisfactory accuracy [34]. However, for dichotomous hypoxia classification, our biomarker seemed to be robust also when based on a single biopsy. Moreover, the cellular composition of the biopsies appeared to play no major role for the biomarker output, in accordance with a previous report on head and neck cancer [35]. It is therefore likely that the biomarker captured a molecular hypoxia phenotype characteristic of the tumour, independent of the number or composition of biopsies used. Expression of hypoxia responsive genes, as measured by the gene-based biomarker, is influenced by persistent genetic alterations such as DNA methylation, mutations, and copy number changes, in addition to an instant stimulatory effect on transcription by reduced oxygen concentration. One could therefore speculate that this biomarker reflects a phenotype providing tolerance of tumour cells to hypoxic stress, and thereby capability to tackle fluctuations in hypoxia, while the imaging biomarker provides an instant measure of the balance between oxygen consumption and supply in the tissue.
The use of both biomarkers significantly improved risk classification of the patients, probably by reflecting different hypoxia phenotypes related to tumour aggressiveness, in addition to reducing small effects of intratumour heterogeneity, technical uncertainties, and changes in hypoxia status during the time period between MRI and collection of biopsies [13,26]. Our new classification cutoff included information from both biomarkers and was directly transferrable to another subgroup of patients. This approach to combine biomarkers could therefore be of interest for a clinical implementation, and might also be exploited for other phenotypes than hypoxia. Our biomarker has a considerable potential of being adopted since it measures an important biological feature, for which several molecular targeted drugs are available or in pipeline [16]. Moreover, it enabled improved outcome prediction compared to existing clinical markers. However, before implementation, the biomarker must fulfil several criteria for translation [36,37].
To pass the first translational gap, the biomarker needs to be developed into a robust medical tool [37]. Technical and clinical validation has already been shown for our gene-based biomarker, by demonstrating transferability from a microarray assay to a more feasible RT-qPCR assay and prognostic significance in two independent patient cohorts [21]. Transferability of our imaging biomarker across MRI machines remains to be demonstrated. A previous study by Loncaster and colleagues [23], showing prognostic significance of an ABrix-defined parameter from another MRI machine than ours, encourages further work to address this challenging step. Moreover, it is reasonable to believe that an imaging biomarker assessing hypoxic fraction, like ours, would be more robust than the histogram-based biomarker considered previously [23]. For further assessment and cutoff validation of the two biomarkers combined, generation of paired imaging and gene-based data sets for large patient cohorts is needed and highly warranted. Finally, in an economic perspective, our combined biomarker is beneficial as the data are based on MRI and biopsies obtained during state-of-the-art diagnostics. This will facilitate passage of the second translational gap; to be integrated into routine patient care [37].
In conclusion, our study encourages implementation of a multifactorial biomarker based on imaging and genes to assess tumour hypoxia more precisely and extensively in cervical cancer. Moreover, our findings motivate for investigating such combination also in other cancer types, using imaging modalities suitable for the specific cancer type, e.g. FMISO in head and neck cancer [9] and diffusion weighted MRI in prostate cancer [38]. The combined biomarker may lead to a better trial design, both by providing more accurate identification of patients with treatment resistant disease and by proposing the most relevant hypoxia targeting drugs for intervention. This might be of utmost importance in cervical cancer since patients are already on the toxicity limit with the standard treatment and should not enter clinical trials without an expected benefit [39,40].
Author contributions
Conception and design: C.S. Fjeldbo and H. Lyng. Acquisition of data: C.S. Fjeldbo, T. Hompland, T. Hillestad, E.-K. Aarnes, G.B. Kristensen, and H. Lyng. Data analysis: C.S. Fjeldbo, T. Hompland, T. Hillestad, C.-C. Günther. Data interpretation: C.S. Fjeldbo, T. Hompland, T. Hillestad, C.-C. Günther, E. Malinen, and H. Lyng. Manuscript draft and editing: C. S. Fjeldbo and H. Lyng. All authors reviewed the manuscript and approved the final version. Study supervision: H. Lyng.
Declaration of Competing Interest
HL is registered as inventor of a patent application covering the clinical use of the hypoxia gene signature (WO2013/124,738).
Funding Sources
The work was supported by grants from The Norwegian Cancer Society (Grant No 107438 and 182451), The South-Eastern Norway Regional Health Authority (Grant No 2015020), and The Norwegian Research Council (ELIXIR Norway). The funders had no role in study design, data collection and analysis, decision to publish, or writing of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102841.
Appendix. Supplementary materials
References
- 1.Thorwarth D. Functional imaging for radiotherapy treatment planning: current status and future directions-a review. Br J Radiol. 2015;88(1051) doi: 10.1259/bjr.20150056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yaromina A., Krause M., Baumann M. Individualization of cancer treatment from radiotherapy perspective. Mol Oncol. 2012;6(2):211–221. doi: 10.1016/j.molonc.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambin P., van Stiphout R.G., Starmans M.H. Predicting outcomes in radiation oncology–multifactorial decision support systems. Nat Rev Clin Oncol. 2013;10(1):27–40. doi: 10.1038/nrclinonc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaffray D.A., Das S., Jacobs P.M., Jeraj R., Lambin P. How advances in imaging will affect precision radiation oncology. Int J Radiat Oncol Biol Phys. 2018;101(2):292–298. doi: 10.1016/j.ijrobp.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 5.Ye Y., Hu Q., Chen H. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat Metab. 2019;1(4):431–444. doi: 10.1038/s42255-019-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overgaard J. Hypoxic radiosensitization: adored and ignored. J Clin Oncol. 2007;25(26):4066–4074. doi: 10.1200/JCO.2007.12.7878. [DOI] [PubMed] [Google Scholar]
- 7.Horsman M.R., Mortensen L.S., Petersen J.B., Busk M., Overgaard J. Imaging hypoxia to improve radiotherapy outcome. Nat Rev Clin Oncol. 2012;9(12):674–687. doi: 10.1038/nrclinonc.2012.171. [DOI] [PubMed] [Google Scholar]
- 8.Yang L., West C.M. Hypoxia gene expression signatures as predictive biomarkers for personalising radiotherapy. Br J Radiol. 2018 doi: 10.1259/bjr.20180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welz S., Monnich D., Pfannenberg C. Prognostic value of dynamic hypoxia PET in head and neck cancer: results from a planned interim analysis of a randomized phase II hypoxia-image guided dose escalation trial. Radiother Oncol. 2017;124(3):526–532. doi: 10.1016/j.radonc.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Milosevic M.F., Townsley C.A., Chaudary N. Sorafenib increases tumor hypoxia in cervical cancer patients treated with radiation therapy: results of a phase 1 clinical study. Int J Radiat Oncol Biol Phys. 2016;94(1):111–117. doi: 10.1016/j.ijrobp.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Toustrup K., Sorensen B.S., Lassen P. Gene expression classifier predicts for hypoxic modification of radiotherapy with nimorazole in squamous cell carcinomas of the head and neck. Radiother Oncol. 2012;102(1):122–129. doi: 10.1016/j.radonc.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Taylor J., Eustace A. A gene signature for selecting benefit from hypoxia modification of radiotherapy for high-risk bladder cancer patients. Clin Cancer Res. 2017;23(16):4761–4768. doi: 10.1158/1078-0432.CCR-17-0038. [DOI] [PubMed] [Google Scholar]
- 13.O'Connor J.P., Rose C.J., Waterton J.C., Carano R.A., Parker G.J., Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21(2):249–257. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyng H., Malinen E. Hypoxia in cervical cancer: from biology to imaging. Clin Transl Imaging. 2017;5(4):373–388. doi: 10.1007/s40336-017-0238-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaccia A.J. Molecular radiobiology: the state of the art. J Clin Oncol. 2014;32(26):2871–2878. doi: 10.1200/JCO.2014.57.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rey S., Schito L., Koritzinsky M., Wouters B.G. Molecular targeting of hypoxia in radiotherapy. Adv Drug Deliv Rev. 2017;109:45–62. doi: 10.1016/j.addr.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Seoane J., De Mattos-Arruda L. The challenge of intratumour heterogeneity in precision medicine. J Intern Med. 2014;276(1):41–51. doi: 10.1111/joim.12240. [DOI] [PubMed] [Google Scholar]
- 18.Lock S., Linge A., Seidlitz A. Repeat FMISO-PET imaging weakly correlates with hypoxia-associated gene expressions for locally advanced HNSCC treated by primary radiochemotherapy. Radiother Oncol. 2019;135:43–50. doi: 10.1016/j.radonc.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Andersen E.K., Hole K.H., Lund K.V. Pharmacokinetic parameters derived from dynamic contrast enhanced MRI of cervical cancers predict chemoradiotherapy outcome. Radiother Oncol. 2013;107(1):117–122. doi: 10.1016/j.radonc.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Halle C., Andersen E., Lando M. Hypoxia-induced gene expression in chemoradioresistant cervical cancer revealed by dynamic contrast-enhanced MRI. Cancer Res. 2012;72(20):5285–5295. doi: 10.1158/0008-5472.CAN-12-1085. [DOI] [PubMed] [Google Scholar]
- 21.Fjeldbo C.S., Julin C.H., Lando M. Integrative analysis of DCE-MRI and gene expression profiles in construction of a gene classifier for assessment of hypoxia-related risk of chemoradiotherapy failure in cervical cancer. Clin Cancer Res. 2016;22(16):4067–4076. doi: 10.1158/1078-0432.CCR-15-2322. [DOI] [PubMed] [Google Scholar]
- 22.Li A., Andersen E., Lervåg C. Dynamic contrast enhanced magnetic resonance imaging for hypoxia mapping and potential for brachytherapy targeting. Phys Imaging Radiat Oncol. 2017;2:1–6. [Google Scholar]
- 23.Loncaster J.A., Carrington B.M., Sykes J.R. Prediction of radiotherapy outcome using dynamic contrast enhanced MRI of carcinoma of the cervix. Int J Radiat Oncol Biol Phys. 2002;54(3):759–767. doi: 10.1016/s0360-3016(02)02972-3. [DOI] [PubMed] [Google Scholar]
- 24.Tofts P.S. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7(1):91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 25.Rosner B. 8th edition. Cengage Learning; Boston, MA: 2016. Fundamentals of biostatistics; p. 927. xix. [Google Scholar]
- 26.Pintilie M., Iakovlev V., Fyles A., Hedley D., Milosevic M., Hill R.P. Heterogeneity and power in clinical biomarker studies. J Clin Oncol. 2009;27(9):1517–1521. doi: 10.1200/JCO.2008.18.7393. [DOI] [PubMed] [Google Scholar]
- 27.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: a language and environment for statistical computing. [Google Scholar]
- 28.Sundfor K., Lyng H., Kongsgard U.L., Trope C., Rofstad E.K. Polarographic measurement of pO2 in cervix carcinoma. Gynecol Oncol. 1997;64(2):230–236. doi: 10.1006/gyno.1996.4571. [DOI] [PubMed] [Google Scholar]
- 29.Hockel M., Knoop C., Schlenger K. Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiother Oncol. 1993;26(1):45–50. doi: 10.1016/0167-8140(93)90025-4. [DOI] [PubMed] [Google Scholar]
- 30.Brizel D.M., Rosner G.L., Prosnitz L.R., Dewhirst M.W. Patterns and variability of tumor oxygenation in human soft tissue sarcomas, cervical carcinomas, and lymph node metastases. Int J Radiat Oncol Biol Phys. 1995;32(4):1121–1125. doi: 10.1016/0360-3016(95)00106-9. [DOI] [PubMed] [Google Scholar]
- 31.Lyng H., Sundfor K., Rofstad E.K. Oxygen tension in human tumours measured with polarographic needle electrodes and its relationship to vascular density, necrosis and hypoxia. Radiother Oncol. 1997;44(2):163–169. doi: 10.1016/s0167-8140(97)01920-8. [DOI] [PubMed] [Google Scholar]
- 32.Le Q.T., Courter D. Clinical biomarkers for hypoxia targeting. Cancer Metastasis Rev. 2008;27(3):351–362. doi: 10.1007/s10555-008-9144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukovic J., Han K., Pintilie M. Intratumoral heterogeneity and hypoxia gene expression signatures: is a single biopsy adequate. Clin Transl Radiat Oncol. 2019;19:110–115. doi: 10.1016/j.ctro.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachtiary B., Boutros P.C., Pintilie M. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12(19):5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 35.Toustrup K., Sorensen B.S., Metwally M.A. Validation of a 15-gene hypoxia classifier in head and neck cancer for prospective use in clinical trials. Acta Oncol. 2016;55(9–10):1091–1098. doi: 10.3109/0284186X.2016.1167959. [DOI] [PubMed] [Google Scholar]
- 36.de Gramont A., Watson S., Ellis L.M. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol. 2015;12(4):197–212. doi: 10.1038/nrclinonc.2014.202. [DOI] [PubMed] [Google Scholar]
- 37.O'Connor J.P., Aboagye E.O., Adams J.E. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14(3):169–186. doi: 10.1038/nrclinonc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hompland T., Hole K.H., Ragnum H.B. Combined MR imaging of oxygen consumption and supply reveals tumor hypoxia and aggressiveness in prostate cancer patients. Cancer Res. 2018;78(16):4774–4785. doi: 10.1158/0008-5472.CAN-17-3806. [DOI] [PubMed] [Google Scholar]
- 39.Barbera L., Thomas G. Management of early and locally advanced cervical cancer. Semin Oncol. 2009;36(2):155–169. doi: 10.1053/j.seminoncol.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Trifiletti D.M., Tyler Watkins W., Duska L., Libby B.B., Showalter T.N. Severe gastrointestinal complications in the era of image-guided high-dose-rate intracavitary brachytherapy for cervical cancer. Clin Ther. 2015;37(1):49–60. doi: 10.1016/j.clinthera.2014.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression data have been deposited to the NCBI's Gene Expression Omnibus (GEO) database: GSE146114.