Summary
Organ size is a major agronomic trait that determines grain yield and biomass production in crops. However, the molecular mechanisms controlling organ size, especially in legumes, are poorly understood.
Using forward genetic approaches in a Tnt1 insertion mutant population of the model legume Medicago truncatula, we identified SMALL LEAF AND BUSHY1 (SLB1), which is required for the control of organ size and lateral branching.
Loss of function of SLB1 led to reduced leaf and flower size but increased lateral branch formation in M. truncatula. SLB1 encodes an F‐box protein, an orthologue of Arabidopsis thaliana STERILE APETALA (SAP), that forms part of an SKP1/Cullin/F‐box E3 ubiquitin ligase complex. Biochemical and genetic analyses revealed that SLB1 controls M. truncatula organ growth and lateral branching by modulating the stability of BIG SEEDS1 (BS1). Moreover, the overexpression of SLB1 increased seed and leaf size in both M. truncatula and soybean (Glycine max), indicating functional conservation.
Our findings revealed a novel mechanism by which SLB1 targets BS1 for degradation to regulate M. truncatula organ size and shoot branching, providing a new genetic tool for increasing seed yield and biomass production in crop and forage legumes.
Keywords: BIG SEEDS1, F‐box protein, lateral branching, Medicago truncatula, organ size, SCF complex, SMALL LEAF AND BUSHY1
Introduction
Organ size is an important parameter in the characterisation of organ morphology and function. How plants control organ size is not only an intriguing, fundamental question in developmental biology, but it is also a crucial issue for improving grain yield and biomass production in crops. However, the genetic and molecular mechanisms that determine final organ or organism size are still poorly understood in plants.
Plant organ growth is mainly controlled by two coordinated developmental processes: cell division and cell expansion (Horiguchi et al., 2006). Leaf development provides an excellent model system for analysing the coordination of these two important processes. During leaf development in eudicot plants, cells throughout the leaf primordia primarily undergo general proliferative cell division. Subsequently, a front of cell cycle arrest occurs in the apex‐to‐base direction. Immediately after the formation of this arrest front, there is a gradient of cell differentiation and expansion (Donnelly et al., 1999; Kazama et al., 2010; Andriankaja et al., 2012). Although most cells begin to differentiate and enlarge, meristematic cells divide continuously to generate specific cell types within each cell layer, such as meristemoid and procambium cells, which are required for the formation of stomatal stem cells and vascular cells, respectively (White, 2006; Bergmann & Sack, 2007; Pillitteri & Torii, 2012). In Arabidopsis thaliana, meristemoid cells generate c. 67% of all pavement cells in cotyledons and 48% in leaves (Geisler et al., 2000), implying that meristemoid cell division contributes significantly to final epidermal area in plants.
Molecular genetic studies in diverse plant species have revealed a number of key factors involved in regulating organ size by regulating cell division rate (Achard et al., 2009; Rojas et al., 2009; Eloy et al., 2011; Zhang et al., 2015), the duration of cell division (Hu et al., 2003; Palatnik et al., 2003; Kim & Kende, 2004; Horiguchi et al., 2005; Disch et al., 2006; Anastasiou et al., 2007; Dewitte et al., 2007; Li et al., 2008; Rodriguez et al., 2010; Li et al., 2012; Xia et al., 2013; Du et al., 2014), or cell expansion (Kim et al., 1998; Kim et al., 1999; Hu et al., 2006; Deprost et al., 2007; Kurepa et al., 2009; Sonoda et al., 2009; Xu & Li, 2011; Lu et al., 2014). Notably, recent studies on a series of organ and seed size mutants have revealed that the ubiquitin‐proteasome pathway plays an important role in plant organ size control. For example, the ubiquitin‐binding protein DA1 functions synergistically with the E3 ubiquitin ligases DA2 and BIG BROTHER (BB)/ENHANCER OF DA1 (EOD1) to control organ and seed size by limiting cell proliferation (Disch et al., 2006; Li et al., 2008; Xia et al., 2013). UBIQUITIN‐SPECIFIC PROTEASE15 (UBP15), encoded by SUPPRESSOR2 OF DA1 (SOD2), positively regulates seed size by promoting cell proliferation, and DA1 physically associates with UBP15 to modulate its stability (Du et al., 2014). In A. thaliana, STERILE APETALA (SAP)/SUPPRESSOR OF DA1 (SOD3) encodes an F‐box protein that functions as a component of the SKP1/Cullin/F‐box (SCF) E3 ubiquitin ligase complex (Wang et al., 2016). SAP interacts with PPDs (PEAPOD1 and PEAPOD2) and KIXs (kinase‐inducible domain interacting proteins KIX8 and KIX9) and targets the KIX–PPD repressor complex for degradation to regulate organ growth (Gonzalez et al., 2015; Wang et al., 2016; Li et al., 2018). Similarly, in Capsella rubella, decreasing SAP activity shortens the cell proliferation period and reduces the number of petal cells (Sicard et al., 2016), while disrupting the cucumber (Cucumis sativus L.) SAP orthologue LITTLELEAF (LL) reduces leaf, flower, and fruit size and seed weight but increases lateral branch formation (Yang et al., 2018), which is not observed in A. thaliana sap/sod3 mutants, pointing to both the conserved and diverse roles of SAP in controlling lateral organ growth among plant species.
Legumes comprise one of the largest monophyletic families, with c. 700 genera and 18 000 species (Dong et al., 2005). Legumes are second only to grasses in terms of economic and nutritional value (Graham & Vance, 2003). In addition to serving as important sources of protein and oils for the human diet, legumes are used as livestock forage and silage and as soil‐enhancing green manure (Graham & Vance, 2003) through fixing atmospheric nitrogen in association with rhizobial bacteria. However, due to an ancient genome duplication event, most important legume crops and forages, such as soybean (Glycine max) and alfalfa (Medicago sativa), have complex genomic structures (Shoemaker et al., 2006). Thus, the genetic and molecular mechanisms that determine organ size in legumes are largely elusive.
A key regulator of seed size and seed weight, BIG SEEDS1 (BS1), was recently identified in the diploid model legume plant Medicago truncatula (Ge et al., 2016). BS1 encodes a TIFY family transcriptional regulator related to tandemly repeated PPDs in A. thaliana that regulate the sizes of leaves and fruits but not seeds (White, 2006; Wang et al., 2016). Notably, loss of function of BS1 leads to enlarged seeds, as well as fruits and leaves, in M. truncatula, whereas the downregulation of BS1 orthologues in soybean significantly increases seed size, weight and quality (Ge et al., 2016), pointing to the great potential of using BS1 for legume crop improvement. Despite this progress, the role of BS1 in regulating organ size in legumes is still poorly understood.
In this study, we isolated and characterised the M. truncatula mutant small leaf and bushy1 (slb1), which exhibits smaller leaves and petals but more lateral branches than the wild‐type. SLB1 is an orthologue of A. thaliana SAP, encoding an F‐box protein that forms part of a SKP1/Cullin/F‐box E3 ubiquitin ligase complex. We demonstrate that SLB1 controls organ size and lateral branching by modulating the stability of BS1. Finally, overexpressing SLB1 increased seed and leaf size in both M. truncatula and soybean, suggesting that SLB1 represents a new genetic tool for increasing grain yield and biomass production in legume crops.
Materials and Methods
Plant materials and growth conditions
Medicago truncatula strain R108 and soybean cultivar Williams 82 were used for all experiments described in this study. slb1‐1 (NF11180), slb1‐2 (NF20634), and slb1‐3 (NF19156) were identified from a Tnt1 retrotransposon‐tagged mutant collection of M. truncatula R108. Plants were grown in a glasshouse under the following conditions: 24°C : 22°C, 16 h : 8 h, day : night photoperiod, and 60–70% relative humidity. The primers used to identify the Tnt1 insertions are listed in Supporting Information Table S1.
Morphological analysis
Projected area of leaves, petals and seeds were measured by scanning to generate digital images, followed by analysis using Olympus CellSens Standard 1.14 and imagej software (https://imagej.nih.gov/ij/). Cell numbers were calculated by dividing the leaf area by the average cell area.
Vector construction and plant transformation
To generate the constructs used for complementation, a 7878‐bp fragment was amplified from M. truncatula R108 gDNA using the primers gSLB1‐F and gSLB1‐R and ligated to the pCAMBIA2300 vector (after digestion with EcoRI and SmaI) using the In‐Fusion cloning system (Clontech, Mountain View, CA, USA), yielding gSLB1; this construct contained the 2400‐bp upstream sequence, the entire gene sequence and the 650‐bp downstream sequence of SLB1. The 2400‐bp SLB1 promoter was cloned into the pBGWFS7 vector using the Gateway system (Invitrogen) to generate the pSLB1:GUS (β‐glucuronidase) construct. CRISPR/Cas9 vector construction was performed as previously described (Meng et al., 2017). Briefly, the U6 promoter and single guide RNA (sgRNA) scaffold were amplified using the primer sets MtU6‐F1/MtU6‐R1 and BS1‐sgRNA‐F/R1, respectively. The U6 promoter and sgRNA scaffold were integrated by PCR using the primers MtU6‐F1/R1 and ligated into the linearised destination vector pFGC5941 digested with XbaI using the In‐Fusion cloning system. The green fluorescent protein (GFP)‐SLB1 fragment was amplified by fusing the coding sequence (CDS) of GFP with SLB1 using the primers GFP‐attB1‐F and SLB1‐attB2‐R, followed by cloning into the binary vector pEarlygate203 using the Gateway system to generate the 35S:GFP‐SLB1 construct. The constructs were introduced into Agrobacterium tumefaciens by chemical transformation. Agrobacterium tumefaciens strain AGL1 was used for M. truncatula transformation, and strain EHA105 was used for soybean transformation as described (Tadege et al., 2011; Chen et al., 2018). All primers used are listed in Table S1.
RNA extraction and gene expression analysis
Total RNA was extracted from various organs of M. truncatula plants using TRIzol Reagent (Invitrogen). cDNA was generated by reverse transcription with SuperScript (Invitrogen). Reverse transcription PCR (RT‐PCR) was performed using a 2× Taq PCR Master Mix (UPTECH) using MtActin as a control. Quantitative RT‐PCR was performed as previously described (Wang et al., 2017) with at least three biological and three technical replicates for both the samples and controls. All primers used are listed in Table S1.
Histochemical GUS staining
The GUS staining assay was performed as described (Niu et al., 2015), and images of the GUS staining patterns of tissues were collected with a digital camera mounted on an Olympus SZX‐16 stereoscope.
Subcellular localisation and confocal microscopy
To determine the subcellular localisation of SLB1, A. tumefaciens strain GV2260 containing 35S:GFP‐SLB1 and the nuclear marker plasmid mRFP‐AHL22 were simultaneously infiltrated into 4‐wk‐old Nicotiana benthamiana leaves (Xiao et al., 2009); pMDC32‐GFP was used as a control. P19 from tomato bushy stunt virus was used to inhibit transgene silencing. The fluorescence signals were observed 48–60 h after infiltration under a Zeiss LSM 700 confocal microscope.
Sequence alignment and phylogenetic analysis
Sequence alignment was performed using ClustalW (http://www.genome.jp/tools/clustalw/). Bootstrap values of 1000 permutations for the neighbour‐joining phylogenetic tree were generated using mega 6.0 software (http://www.megasoftware.net/).
Yeast two‐hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays
The Y2H assay was performed using the Matchmaker Gold System (Clontech). The coding sequence of SLB1 was cloned into the pGBKT7 vector, and the coding sequences of MtASK1, MtASK2, and BS1 were cloned into the pGADT7 vector using the Gateway system (Invitrogen). The bait and prey plasmids were cotransformed into yeast strain Y2H Gold (Clontech). For the auxotrophic assay, yeast colonies were inoculated onto SD/−Leu/−Trp (DDO) and SD/−Trp/−Leu/−His/−Ade (QDO) plates and incubated in the dark at 28°C for 3 d. All primers used are listed in Table S1.
The BiFC assay was conducted as described (Meng et al., 2019). Briefly, SLB1 was cloned into pEarlygate201‐YN, while MtASK1, MtASK2 and BS1 were cloned to pEarlygate202‐YC using the Gateway system (Invitrogen). SLB1‐nYFP, MtASK1‐cYFP, MtASK2‐cYFP and BS1‐cYFP were introduced into A. tumefaciens strain GV2260. Various combinations of transformed A. tumefaciens cells were simultaneously infiltrated into 4‐wk‐old N. benthamiana leaves. P19 from tomato bushy stunt virus was used to inhibit transgene silencing. Fluorescence signals were observed 48–60 h after infiltration under a Zeiss LSM 700 confocal microscope.
Co‐immunoprecipitation (Co‐IP) assay
A Co‐IP assay was performed as previously described with minor modifications (Meng et al., 2019). The CDSs of MtASK1, MtASK2, MtCUL1, and MtCUL2 were cloned into binary vector pGWB17 using the Gateway system (Invitrogen) to generate 35S:MtASK1‐Myc, 35S:MtASK2‐Myc, 35S:MtCUL1‐Myc, and 35S:MtCUL2‐Myc. A. tumefaciens strain GV2260 containing different combinations of 35S:GFP‐SLB1, 35S:MtASK1‐Myc, 35S:MtASK2‐Myc, 35S:MtCUL1‐Myc, 35S:MtCUL2‐Myc, and 35S:GFP constructs were infiltrated into N. benthamiana leaves. Total proteins were extracted from the samples with extraction buffer (50 mM Tris‐HCl, pH 7.5, 1 mM ethylene diamine tetraacetic acid (EDTA), 150 mM NaCl, 0.05–0.1% Tween 20, 10% glycerol, 1× protease inhibitor cocktail, and 1 mM phenylmethylsulfonyl fluoride (PMSF)). After protein extraction, 50 μl of anti‐GFP Magarose beads (Smart Life Sciences, Changzhou, Jiangsun, China) were added to the protein extract, and the mixture was incubated for 4 h at 4°C. The beads were washed at least four times with protein extraction buffer, and the proteins were eluted by adding SDS protein loading buffer and boiling the samples for 10 min. The clear supernatant was analysed by SDS‐PAGE and examined by immunoblot analysis using anti‐GFP (Abmart M20004S, 1/5000) and anti‐Myc (Abmart M20002L, 1/5000) antibodies.
Degradation assay
For the degradation assay, the CDS of BS1 and MtWD40‐1 were cloned into binary vector pEarlygate202 and pGWB17 using the Gateway system to generate 35S:Flag‐BS1 and 35S:MtWD40‐1‐Myc, respectively. N. benthamiana leaves transiently expressing 35S:Flag‐BS1 and 35S:GFP‐SLB1 or 35S:GFP or 35S:MtWD40‐1‐Myc were harvested and ground into a fine powder. Here, c. 1 g of N. benthamiana tissue was homogenised in 5 ml of native extraction buffer (50 mM Tris‐MES (2‐(N‐morpholino)ethanesulfonic acid), pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, 10 mM ATP, 0.1% NP‐40, and 1× protease inhibitor cocktail) and filtered through a 70‐μm nylon filter. The lysates were incubated at 30°C, and samples were taken at the indicated time points. The samples were centrifuged at 6000 g for 1 min and the supernatant removed. Loading buffer was adding to the precipitate, followed by boiling for 10 min. For the proteasome inhibitor assay, MG132 (200 μM) or dimethyl sulfoxide (as a control) was added to the lysates. Protein levels were analysed by immunoblotting with anti‐FLAG antibody (MBL M185‐7, 1/5000).
Pollen staining and in vitro pollen germination
Pollen staining was performed as previously described (Alexander, 1969). Flowers of wild‐type and slb1‐1 were collected, fixed in Carnoy's fixative for 2 h, and stained with Alexander's solution for 2 h at room temperature. The samples were destained in 10% glycerol for 45 min before observation.
For pollen germination tests, drops of pollen germination medium (10% sucrose, 0.015% boric acid) were added on a microscope slide, pollens from the mature anthers of wild‐type and slb1‐1 were sprinkled on the medium. The slide was placed on wet filter paper in moist plastic dish and incubated for 1.5 h at room temperature. The pollen tubes were imaged using an Olympus SZX‐16 stereoscope.
GenBank accession numbers
GenBank accession numbers are as follows: MN954689 for SLB1 (Medtr5g097060), KM668032 for BS1 (Medtr1g102900), XP_003612225.1 for MtASK1 (Medtr5g022710), XP_003612227.1 for MtASK2 (Medtr5g022730), XP_013463300.1 for MtCUL1 (Medtr2g437390), XP_013458250.1 for MtCUL2 (Medtr4g119413), XP_003602392.1 for MtWD40‐1 (Medtr3g092840).
Results
Identification and characterisation of M. truncatula slb1 mutants
To investigate the molecular mechanisms underlying the control of organ size in legumes, we identified a mutant with obviously reduced leaf size named small leaf and bushy1‐1 (slb1‐1) from a forward genetic screen of Tnt1 retrotransposon‐tagged M. truncatula genotype R108 plants (Fig. 1a–c) (Tadege et al., 2008; Yarce et al., 2013). By contrast with the wild‐type, the slb1‐1 mutant exhibits arrested leaf expansion in both the longitudinal and transverse directions, leading to reduced leaf size and heart‐shaped blade morphology (Fig. 1c,e). The slb1‐1 mutant also exhibited smaller flowers compared with the wild‐type (Figs 1d, S1). A close examination showed that slb1‐1 has a small carpel and retarded anthers, even though the dissected pollens could germinate normally, while the carpel of slb1‐1 exhibited aberrant development with reduced and abnormal ovules, resulting in sterility (Fig. S2). Further examination of leaf epidermal cells in slb1‐1 showed that the cell size was not obviously altered in this mutant, suggesting that its altered leaf size and shape are mainly caused by reduced cell proliferation (Fig. S3). Consistent with this notion, the transcript levels of the cell division markers Histone H4 and cyclin D were significantly reduced in slb1‐1 leaves, as determined by quantitative RT‐PCR (Fig. 1f). In addition, slb1‐1 has more lateral branches than the wild‐type due to accelerated axillary bud outgrowth rather than increased axillary bud formation (Fig. 1a,b,g,h).
Figure 1.
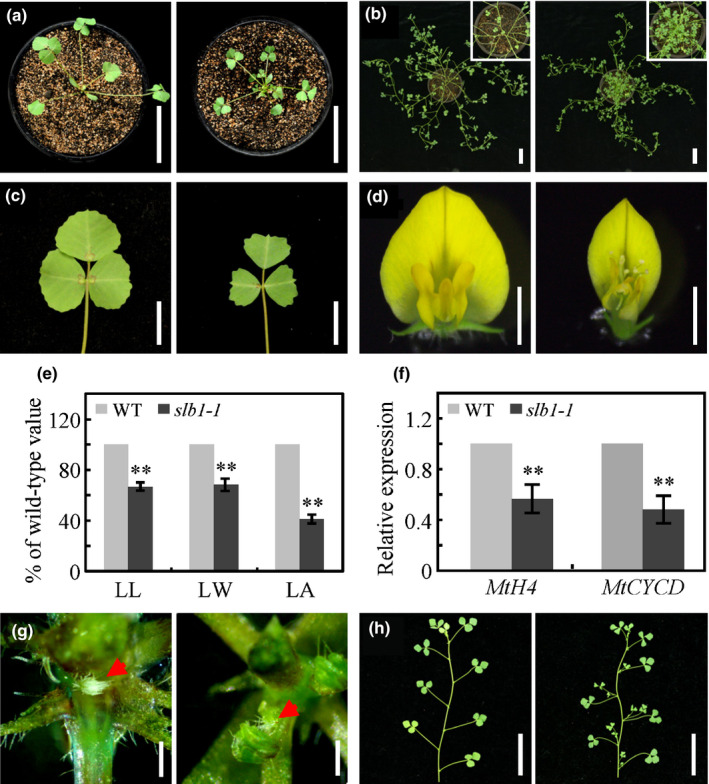
Morphological comparison of Medicago truncatula wild‐type vs slb1‐1 plants. (a) 3‐wk‐old wild‐type (WT, left) and slb1‐1 (right) seedlings. Bars, 5 cm. (b) 10‐wk‐old WT and slb1‐1 plants. The insets show close‐up views of the shoot base. Bars, 5 cm. (c, d) Phenotypes of leaves (c) and flowers (d) in WT (left) and slb1‐1 (right). Bars, 1 cm for leaves and 2 mm for flowers. (e) Leaf length (LL), leaf width (LW), and leaf area (LA) of WT and slb1‐1. Bars represent means ± SD (n = 16); asterisks indicate significant differences from the WT (**P < 0.01, Student's t‐test). (f) Transcript levels of MtH4 and MtCYCD in slb1‐1. Bars represent means ± SD (n = 3); asterisks indicate significant differences from the WT (**P < 0.01, Student's t‐test). (g) Comparison of axillary bud formation between WT (left) and slb1‐1 (right). Red arrows indicate axillary buds. Bars, 0.5 mm. (h) Dissected WT (left) and slb1‐1 (right) branches, showing accelerated bud elongation in slb1‐1. Bars, 5 cm.
Molecular cloning of the SLB1 gene
The slb1‐1 mutant was backcrossed with the wild‐type plants. All of the F1 progeny were wild‐type‐like, and in a segregating F2 population, the wild‐type‐like and mutant plants showed a segregation ratio of 3 : 1 (115 : 47), suggesting that the mutant phenotype was controlled by a single recessive gene. To identify the gene responsible for the pleiotropic defects in slb1‐1, the Tnt1 flanking sequence tags (FSTs) of slb1‐1 mutant were obtained from Medicago truncatula Mutant Database (https://medicago-mutant.noble.org/mutant/index.php) and analysed by PCR‐based genotyping in segregating populations (Tadege et al., 2008). The FST, NF11180A_high_3, which segregated with the mutant phenotype of slb1‐1, was analysed by blast searches against the M. truncatula genome at NCBI (http://www.ncbi.nlm.nih.gov/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) to obtain the full‐length sequence of SLB1. PCR and RT‐PCR were carried out to amplify the SLB1 genomic and coding sequences (CDS), respectively. The SLB1 gene contains two exons, with the Tnt1 retrotransposon inserted at the middle of exon 2 in slb1‐1 632 bp upstream of the translational stop codon (Fig. 2a). RT‐PCR revealed that the full‐length coding sequence of SLB1 was not transcribed in slb1‐1 (Fig. 2b). To confirm the notion that the slb1‐1 mutant phenotype is caused by disruption of SLB1, we obtained two additional Tnt1 insertion lines (NF20634 and NF19156) with the same phenotypes as described for slb1‐1 via phenotypic screening (Fig. S4). Analysis of flanking sequences showed that NF20634 and NF19156 contained Tnt1 insertions at different locations in exon 2 of SLB1; we therefore named these lines slb1‐2 and slb1‐3, respectively (Fig. 2a). RT‐PCR analysis revealed that the transcripts of SLB1 were abolished in these two mutants (Fig. 2b). The identity of SLB1 was further confirmed by genetic complementation. We introduced a construct including the 2.4‐kb promoter region, the entire SLB1 genomic DNA sequence, and the 0.65‐kb downstream region of SLB1 (gSLB1) into slb1‐1 plants by A. tumefaciens‐mediated transformation. The phenotypes of the complemented transgenic slb1‐1 plants (gSLB1/slb1‐1) were comparable with those of the wild‐type (Fig. 2c). Collectively, these data confirmed the notion that disrupting SLB1 function resulted in the altered organ size and increased branching of the slb1 mutants.
Figure 2.

Cloning of the Medicago truncatula SLB1 gene. (a) Schematic representation of the gene structure of SLB1 and the Tnt1 insertion sites in slb1‐1, slb1‐2 and slb1‐3. (b) RT‐PCR analysis of SLB1 expression in the wild‐type (WT) and various slb1 alleles. MtActin was used as the loading control. (c) Phenotype analysis of branches (top panel), leaves (middle panel), and flowers (lower panel) of WT, slb1‐1, and slb1‐1 plants complemented with gSLB1 (gSLB1/slb1‐1). Bars, 5 cm in the top panel, 1 cm in the middle panel, and 2 mm in the lower panel.
Expression analysis of SLB1 and subcellular localisation of SLB1
SLB1 was expressed at high levels in flowers and axillary buds, moderate levels in seeds and roots, and low levels in leaves, stems and fruits, as revealed by quantitative RT‐PCR (Fig. 3a). To gain a better spatial and temporal resolution, we analysed the expression of the GUS reporter gene fused to the 2.4‐kb SLB1 promoter fragment in transgenic M. truncatula plants. In the shoot apex, we found that GUS staining was observed in the shoot apical meristem, the floral meristem and emerging leaf primordia (Fig. 3b). During leaf development, higher GUS activity and transcript abundance of SLB1 were detected in younger leaves than older ones (Figs 3c,d, S5a). GUS staining was also observed at the pulvinus regions where leaflets were attached (Fig. 3d). Buds developing in the leaf axils showed high GUS staining, and GUS activity was observed in stipules (Fig. 3e). In very young flower buds, GUS staining was detected in most floral organs including sepals, petals, stamens, and carpels (Fig. 3f). As the flower develops further, GUS staining was observed at anthers, developing ovules as well as immature seeds (Fig. 3g–i). Further quantitative RT‐PCR analysis showed that SLB1 was highly expressed during the early stages of floral buds, but the levels were reduced at the later stages (Fig. S5b). The expression pattern of SLB1 supports its function in controlling leaf and flower size as well as lateral branching in M. truncatula.
Figure 3.
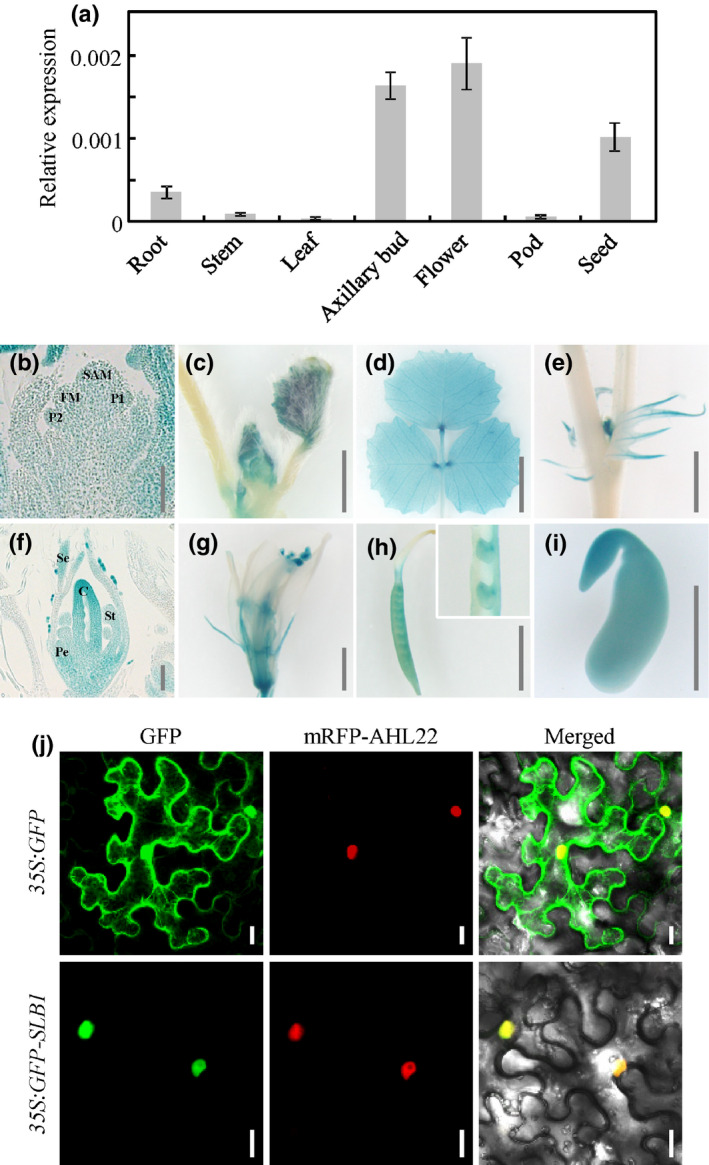
Expression pattern of SLB1 in Medicago truncatula and subcellular localisation of SLB1 protein. (a) SLB1 transcript levels in different tissues, as revealed by quantitative RT‐PCR. MtActin was used as an internal control. Bars represent means ± SD (n = 3). (b–i) GUS activity analysis in different tissues of wild‐type plants transformed with the pSLB1:GUS construct. GUS staining in shoot apical meristem (SAM), floral meristem (FM), and leaf primordia (P) (b), developing leaves (c, d), axillary bud (e), young floral bud (f), mature flower (g), carpel and dissected ovules (h), and immature seed (i). C, carpel; Pe, petal; Se, sepal; St, stamen. Bars, 100 μm in (b, f), 2 mm in (c, e, g–i), 5 mm in (d). (j) Subcellular localisation of GFP and GFP–SLB1 in tobacco leaf epidermal cells. The nuclear protein AHL22 was used as nuclear localisation marker. Bars, 20 μm.
To determine the subcellular localisation of SLB1, we fused the N terminus of SLB1 with GFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter and transformed the construct into tobacco (Nicotiana benthamiana) leaf epidermal cells by agro‐infiltration. By contrast with the GFP control, which was localised to both the cytoplasm and nuclei of epidermal cells, the GFP–SLB1 fusion protein was mainly localised to the nucleus (Fig. 3j).
SLB1 functions within an SCF complex
Blast searches against the NCBI database showed that SLB1 encodes an F‐box protein homologous to A. thaliana SAP. SLB1 and SAP share 43.7% amino acid sequence identity (using full‐length sequences), with strong conservation at the F‐box motif (Fig. S6). F‐box proteins have been shown to function as the substrate‐recruiting subunits of SKP1/Cullin/F‐box‐protein (SCF) complexes (Kipreos & Pagano, 2000; Cardozo & Pagano, 2004). Given that SLB1 contains the highly conserved F‐box motif, we asked whether SLB1 functions within an SCF complex in M. truncatula. We searched for potential SKP1 and Cullin‐related proteins in the Medicago truncatula Genome Database (http://www.medicagogenome.org) using the A. thaliana ASK1/2 and Cullin1 proteins as query sequences. We identified two SKP1‐like proteins (which we named MtASK1 and MtASK2) and two Cullin1‐like proteins (MtCUL1 and MtCUL2) (Fig. S7). SLB1 interacted with both MtASK1 and MtASK2 in an Y2H assay (Fig. 4a). These interactions were verified via a BiFC assay in N. benthamiana leaves using split YFP. Strong yellow fluorescence was clearly observed between SLB1 fused to the N‐terminal half of YFP (SLB1–nYFP) and MtASK1 or MtASK2 fused to the C‐terminal half of YFP (MtASK1–cYFP or MtASK2–cYFP), whereas no YFP fluorescent signals were detected in the negative control (combination of SLB1–nYFP and cYFP alone; Fig. 4b).
Figure 4.
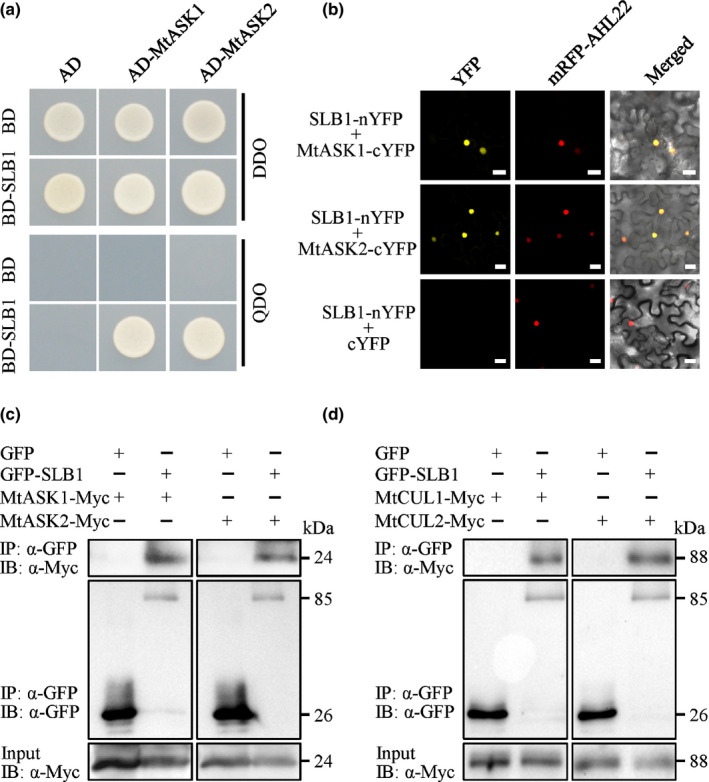
Medicago truncatula SLB1 physically associates with components of the SCF complex. (a) Interaction of SLB1 with MtASK1 and MtASK2 in an Y2H assay. Auxotrophic growth indicates the interaction of each protein. DDO and QDO indicate SD/−Trp/−Leu and SD/−Trp/−Leu/−His/−Ade, respectively. (b) Interaction of SLB1 with MtASK1 and MtASK2 in Nicotiana benthamiana leaf epidermal cells using a BiFC assay. AHL22 was used as a nuclear localisation marker. Bars, 20 μm. (c) SLB1 associates with MtASK1 and MtASK2 in N. benthamiana, as revealed in a Co‐IP assay. Total proteins were immunoprecipitated with anti‐GFP beads, and the immunoblots were probed with anti‐GFP and anti‐Myc antibodies. MtASK1‐Myc and MtASK2‐Myc were detected in the immunoprecipitated GFP–SLB1 complex. IP and IB indicate immunoprecipitation and immunoblot, respectively. (d) SLB1 associates with MtCUL1 and MtCUL2 in N. benthamiana, as revealed in a Co‐IP assay. Total proteins were immunoprecipitated with anti‐GFP beads, and the immunoblots were probed with anti‐GFP and anti‐Myc antibodies. MtCUL1‐Myc and MtCUL2‐Myc were detected in the immunoprecipitated GFP–SLB1 complex. IP and IB indicate immunoprecipitation and immunoblot, respectively.
To explore whether SLB1 physically associates with an SCF complex in planta, we performed Co‐IP analysis to detect the interactions of SLB1 with MtASK1, MtASK2, MtCUL1 and MtCUL2 in vivo. We transiently co‐expressed 35S:GFP‐SLB1 with 35S:MtASK1‐Myc or 35S:MtASK2‐Myc in N. benthamiana leaves. Transient coexpression of 35S:GFP with 35S:MtASK1‐Myc or 35S:MtASK2‐Myc was used as a negative control. We isolated total proteins from the samples, incubated them with anti‐GFP Magarose beads to immunoprecipitate GFP‐SLB1 and GFP, and used anti‐GFP and anti‐Myc antibodies to detect the immunoprecipitated proteins. MtASK1‐Myc and MtASK2‐Myc were captured in the immunoprecipitation complex containing GFP–SLB1, but not in the control, indicating that SLB1 physically associates with MtASK1 and MtASK2 in planta (Fig. 4c). When we transiently co‐expressed 35S:GFP‐SLB1 with 35S:MtCUL1‐Myc or 35S:MtCUL2‐Myc in N. benthamiana leaves, MtCUL1‐Myc and MtCUL2‐Myc were also detected in the immunoprecipitated GFP–SLB1 complex (Fig. 4d). These results suggested that SLB1 functions within an SCF ubiquitin ligase complex in M. truncatula.
SLB1 physically interacts with BS1 and targets it for degradation
SAP interacts with PPD proteins in A. thaliana and targets them for degradation to control organ size. Given that BS1 is a PPD orthologue in M. truncatula that plays a conserved role in controlling organ size in legumes, we asked whether SLB1 physically interacts with BS1 and targets it for degradation. Expression pattern analyses showed that BS1 transcripts were present in most tissues, including roots, stems, leaves, fruits and seeds and were highly expressed in flowers and axillary buds (Fig. S8a). Furthermore, the GFP–BS1 fusion protein primarily localised to the nucleus (Fig. S8b), indicating that both spatial expression and localisation of BS1 are associated with that of SLB1. Furthermore, the Y2H assay showed that SLB1 interacts with BS1 (Fig. 5a) and this interaction was verified in N. benthamiana leaves via a BiFC assay using split YFP (Fig. 5b).
Figure 5.
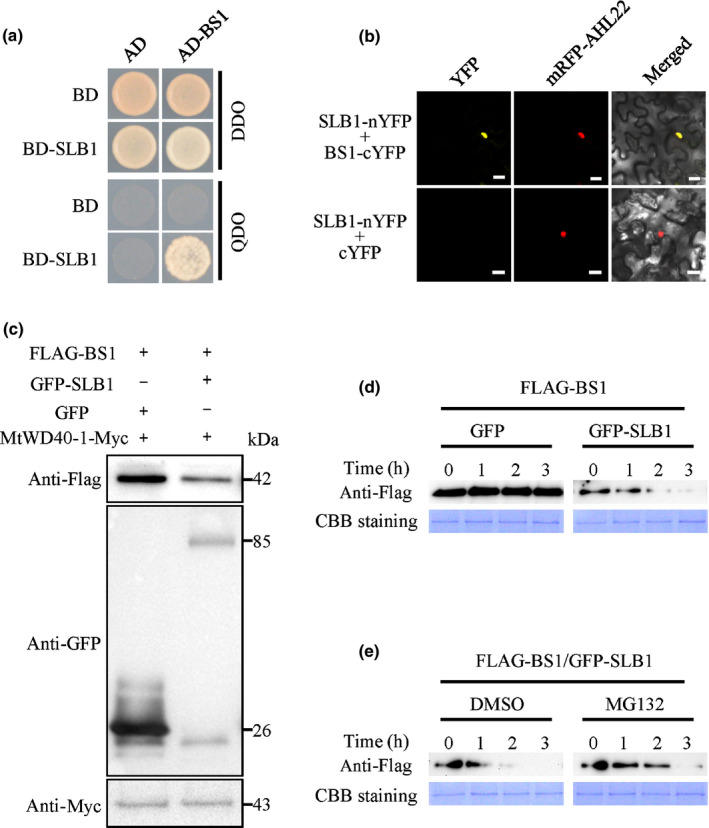
Medicago truncatula SLB1 interacts with and targets BS1 for degradation. (a) Interaction between SLB1 and BS1 in an Y2H assay. Auxotrophic growth indicates the interaction of each protein. DDO and QDO indicate SD/−Trp/−Leu and SD/−Trp/−Leu/−His/−Ade, respectively. (b) Interaction between SLB1 and BS1 in Nicotiana benthamiana leaf epidermal cells using a BiFC assay. AHL22 was used as a nuclear localisation marker. Bars, 20 μm. (c) SLB1 promotes the degradation of BS1 in vivo. Immunoblotting analysis of total protein corresponding to agro‐infiltrated N. benthamiana leaves with the indicated plasmids. The abundance of FLAG−BS1 was detected using anti‐FLAG antibody, and that of GFP−SLB1 was detected using anti‐GFP antibody. MtWD40‐1‐Myc detection using anti‐Myc antibody served as a loading control. (d) SLB1 promotes the degradation of BS1 in an in vitro protein degradation assay. Protein samples from tobacco leaves coexpressing FLAG–BS1 and GFP–SLB1 or GFP were incubated at 30°C for the indicated times. The abundance of FLAG–BS1 was detected using anti‐FLAG antibody. Coomassie Brilliant Blue (CBB) staining served as a loading control. (e) BS1 degradation is inhibited by MG132. Protein samples from tobacco leaves coexpressing FLAG–BS1 and GFP–SLB1 were treated with MG132 or dimethyl sulfoxide (DMSO, as control) at 30°C for the indicated times. The accumulation of FLAG‐BS1 protein was detected by immunoblotting with anti‐FLAG antibody. CBB staining served as a loading control.
To investigate whether SLB1 modulates the stability of BS1 protein, we transiently expressed FLAG‐BS1 with GFP‐SLB1 or GFP in N. benthamiana leaves. Compared with coexpression with GFP, the stability of BS1 was reduced by coexpression with SLB1 (Fig. 5c). In vitro degradation experiments indicated that SLB1 promotes the degradation of BS1, which was suppressed by treatment with MG132, a 26S proteasome‐specific inhibitor (Fig. 5d,e). These results indicated that SLB1 promotes the degradation of BS1 via the ubiquitin‐26S proteasome‐dependent pathway.
SLB1 genetically interacts with BS1 to control organ size and lateral branching
As SLB1 associates with BS1 and regulates its stability, we investigated whether BS1 functions with SLB1 in a common pathway to control organ size and lateral branching in M. truncatula. Compared with the wild‐type, plants with disrupted expression of BS1 in the R108 background, as generated by CRISPR/Cas9 (BS1‐CR), displayed an arrested axillary bud outgrowth, but exhibited obviously increased leaf and flower size (Figs 6a–g, S9), which is consistent with the phenotype of the previously reported bs1‐1 mutant in the cv Jemalong A17 background (Ge et al., 2016). By contrast, disrupting BS1 in the slb1 mutant background not only suppressed the reduced leaf and flower size phenotype, and the branching phenotype of slb1‐1, but also significantly increased the size of the leaves and flowers compared with wild‐type R108, which is similar to the phenotype of the bs1 single mutant (Fig. 6a–g). These results clearly indicated that BS1 is epistatic to SLB1 with respect to the control of organ size and lateral branching in M. truncatula.
Figure 6.
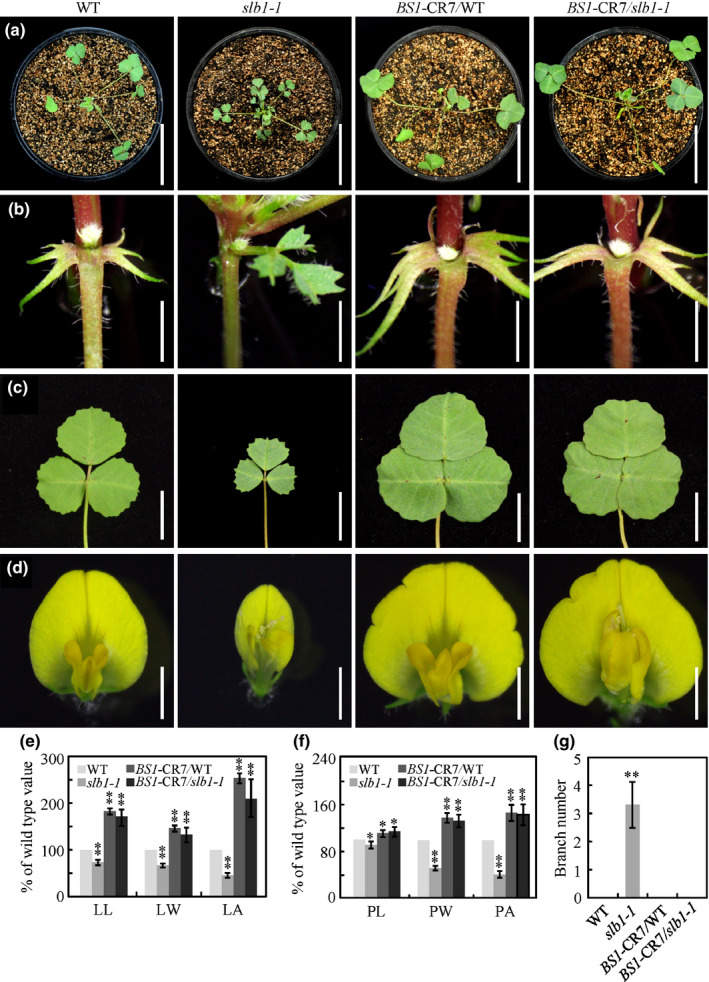
SLB1 genetically interacts with BS1 to control Medicago truncatula organ size and lateral branching. (a–d) Phenotypic analyses of branches (a), axillary buds (b), leaves (c), and flowers (d) in the wild‐type (WT), slb1‐1, BS1‐CR7/WT and BS1‐CR7/slb1‐1. Bars, 5 cm in (a), 2 mm in (b), 1 cm in (c) and 2 mm in (d). (e) Leaf length (LL), leaf width (LW), leaf area (LA) of WT, slb1‐1, BS1‐CR7/WT and BS1‐CR7/slb1‐1 plants. Bars represent means ± SD (n = 15); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test). (f) Vexillum petal length (PL), vexillum petal width (PW), vexillum petal area (PA) of WT, slb1‐1, BS1‐CR7/WT and BS1‐CR7/slb1‐1 plants. Bars represent means ± SD (n = 15); asterisks indicate significant differences from the WT (*P < 0.05, **P < 0.01, Dunnett's test). (g) Branch number of 3‐wk‐old WT, slb1‐1, BS1‐CR7/WT and BS1‐CR7/slb1‐1 seedlings. Branch indicates axillary bud length >5 mm. Bars represent means ± SD (n = 10); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test).
Overexpression of SLB1 results in increased leaf and seed size in M. truncatula and soybean
Seed size is a major agronomic trait of crop plants. Given that SLB1 and BS1 function in a common genetic pathway, we wondered whether SLB1 could be used to modify seed size in legume crops. To test this idea, we introduced the full‐length SLB1 CDS into M. truncatula as well as the major legume crop soybean, under the control of the 35S promoter via A. tumefaciens‐mediated transformation. Both the SLB1‐overexpressing transgenic M. truncatula and soybean plants showed significantly increased organ size, including leaves, fruits and seeds, compared with those of wild‐type plants (Fig. 7a–e,g–k). To quantitatively measure these improvements, we evaluated total above ground biomass yield of 9‐wk‐old wild‐type and SLB1 overexpressing M. truncatula plants. The average fresh weight of 35S:GFP‐SLB1#5 and 35S:GFP‐SLB1#7 transgenic M. truncatula had a 1.35‐fold and 1.32‐fold increase, respectively, in total biomass compared with the wild‐type, while the total amount of dry biomass yield of 35S:GFP‐SLB1#5 and 35S:GFP‐SLB1#7 had increased by over 1.43‐fold and 1.37‐fold per plant, respectively (Fig. S10). Moreover, we evaluated the seed size and seed weight of SLB1‐overexpressing M. truncatula and soybean plants after maturity. The average seed size and seed weight of the SLB1‐overexpressing M. truncatula plants increased by 1.58‐fold and 1.22‐fold, respectively, compared with the wild‐type (Fig. 7e,f), while the seed size and seed weight increased by over 1.49‐fold and 2.08‐fold, respectively, in SLB1‐overexpressing transgenic soybean (Fig. 7k,l). These results indicated that SLB1 plays a conserved role in controlling leaf and seed size in these plants, suggesting that it could be used to increase grain and biomass yield in legume crops.
Figure 7.
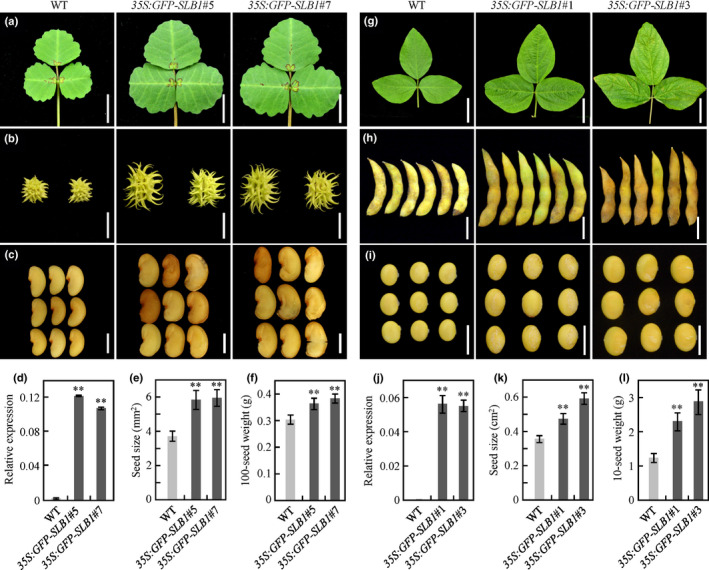
SLB1 overexpression increases leaf, fruit and seed size in Medicago truncatula and soybean. (a–c) Phenotypes of leaves (a), fruits (b), and seeds (c) of wild‐type (WT) and SLB1‐overexpressing M. truncatula. Bars, 1 cm in (a) and (b) and 2 mm in (c). (d) Transcript levels of SLB1 in transgenic M. truncatula plants, as revealed by quantitative RT‐PCR. MtActin was used as an internal control. Bars represent means ± SD (n = 3); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test). (e) Comparison of seed size in WT and SLB1‐overexpressing M. truncatula plants. Bars represent means ± SD (n = 15); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test). (f) Comparison of 100‐seed weight of WT and SLB1‐overexpressing M. truncatula plants. Bars represent means ± SD (n = 3); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test). (g–i) Phenotypes of leaves (g), fruits (h), and seeds (i) of WT and SLB1‐overexpressing soybean. Bars, 5 cm in (g), 2 cm in (h), and 1 cm in (i). (j) Transcript levels of SLB1 in transgenic soybean plants, as revealed by quantitative RT‐PCR. GmActin11 was used as an internal control. Bars represent means ± SD (n = 3); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test). (k) Comparison of seed size in WT and SLB1‐overexpressing soybean. Bars represent means ± SD (n = 15); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test). (l) Comparison of 10‐seed weight of WT and SLB1‐overexpressing soybean. Bars represent means ± SD (n = 5); asterisks indicate significant differences from the WT (**P < 0.01, Dunnett's test).
Discussion
Functional conservation and divergence of M. truncatula SLB1 and A. thaliana SAP in controlling organ growth
In the model eudicot A. thaliana, SAP was initially identified as a floral regulator, as the loss‐of‐function sap mutant exhibited severe defects in inflorescence, flower and ovule development, leading to sterile flowers with reduced petal size (Byzova et al., 1999). Another allelic mutant of SAP, sod3‐1, showed reduced leaf, flower and fruit size due to decreased cell numbers, indicating that SAP also functions as a master regulator of organ size (Wang et al., 2016). Similarly, in this study, we demonstrated that disrupting SLB1 in M. truncatula leads to reduced leaf and flower size as well as poor fertility with abnormal ovules (Figs 1c–e, S1, S2), suggesting that SLB1 and SAP share a conserved role in regulating vegetative and reproductive organ development. Nevertheless, the slb1 mutant produces more branches than the wild‐type (Fig. 1a,b), which was not observed in A. thaliana sap or sod3 mutants. A genetic complementation experiment confirmed that this additional branching phenotype is caused by the disruption of SLB1 function (Fig. 2c), indicating that SLB1 is also involved in regulating branch development in M. truncatula. Notably, recent studies in cucumber showed that disrupting Leaf length (LL), an orthologue of SAP, leads to small organ size but the formation of multiple lateral branches (Yang et al., 2018), suggesting that SLB1 and LL have pleiotropic effects on branching in both M. truncatula and cucumber, although the regulation of organ size appears to be a conserved function of SAP/LL/SLB1 in diverse plant species. These results are in agreement with previous phylogenetic analysis showing that SLB1 and LL group together, but distinct from the closely related SAP (Yang et al., 2018), implying a functional divergence of SAP orthologues in M. truncatula and cucumber.
SAP controls organ growth in A. thaliana by targeting PPD and KIX for degradation (Wang et al., 2016; Li et al., 2018). Our results indicated that SLB1 can function within an SCF complex (Fig. 4) and physically interact with BS1, a PPD homologue in M. truncatula, and modulate its stability (Fig. 5), suggesting that SLB1 uses a repressor module similar to PPD‐KIX‐TPL to control organ size. Supporting this scenario, disrupting BS1 using CRISPR/Cas9 suppressed the leaf and flower phenotypes of slb1 (Figs 6, S9). However, by contrast with the ppd mutant, which only partially suppressed the organ growth phenotypes of A. thaliana sod3‐1, disrupting BS1 in the slb1 mutant background not only recovered the organ growth phenotype but also resulted in enlarged leaves and flowers (Fig. 6c–f). These findings suggested that SLB1 controls organ size in M. truncatula primarily by modulating the stability of BS1. Given the different regulatory modules present in M. truncatula vs A. thaliana, the identification and characterisation of M. truncatula KIX genes should shed light on the roles of the SAP‐PPD‐KIX regulatory module in diverse plant systems.
SLB1 controls M. truncatula lateral branching by modulating the stability of BS1
F‐box proteins play a variety of roles in regulating phytohormone signalling and stress responses during plant development. For example, the F‐box proteins TRANSPORT INHIBITOR RESPONSE1 (TIR1) and CORONATINE‐INSENSITIVE PROTEIN1 (COI1) are involved in auxin and jasmonic acid signalling, respectively (Dharmasiri et al., 2005; Kepinski & Leyser, 2005; Sheard et al., 2010). Studies of a series of branching mutants, including more axillary growth (max) in Arabidopsis, ramosus (rms) mutants in pea, and dwarf (d) mutants in rice have revealed that the F‐box proteins MAX2/RMS4/D3 are responsible for perceiving and transducing strigolactone (SL) signals to regulate shoot branching (Stirnberg et al., 2002; Ishikawa et al., 2005; Johnson et al., 2006; Stirnberg et al., 2007). In this study, we demonstrated that the F‐box protein SLB1 acts as a novel regulator of lateral branching by modulating the stability of BS1 in M. truncatula. BS1 was previously shown to control organ size in M. truncatula, including seed, fruit and leaf size, via a regulatory module that targets primary cell proliferation (Ge et al., 2016). The disruption of BS1 fully restored the branching phenotype of the slb1 mutant (Fig. 6b,g), suggesting that BS1 has an epistatic effect on lateral branching. Thus, our findings define a novel genetic and molecular mechanism of the F‐box protein SLB1 and the organ size regulator BS1 in controlling lateral branching in M. truncatula.
The development of shoot branching generally comprises two distinct steps: the formation of the axillary meristems in the leaf axils and the outgrowth of axillary buds (Shimizu‐Sato & Mori, 2001). In some plant species, the outgrowth of axillary buds can be suppressed by the primary shoot, a phenomenon known as apical dominance (Sachs & Thimann, 1964; Cline, 1991). The phytohormones auxin and cytokinin have long been implicated in the process, in which auxin inhibited the outgrowth of axillary buds by affecting the supply of cytokinin to axillary buds (Eklof et al., 2000; Li & Bangerth, 2003; Nordstrom et al., 2004; Tanaka et al., 2006). Notably, many flowering plants with sterility issues exhibited increased branching, which is thought to be an indirect effect of the auxin export levels from growing meristems vs fruits (Hensel et al., 1994; Ware et al., 2019). The observation of sterility and increased branching in the slb1 mutant suggests that the SLB1 regulation of lateral bud outgrowth may be connected to auxin transport. Moreover, recent studies with branching mutants in several plant species have demonstrated that SLs, a group of terpenoid lactones, are newly identified phytohormone that repress shoot branching (Gomez‐Roldan et al., 2008; Umehara et al., 2008). The slb1 mutant showed defects in axillary bud dormancy, leading to accelerated lateral branch growth, while disrupting BS1 restores the axillary bud dormancy of slb1, indicating that the SLB1‐BS1 regulatory module involves in the regulation of M. truncatula apical dominance, probably via affecting phytohormone crosstalk network.
It is worth noting that the slb1 mutant shows a decreased cell proliferation in most lateral organs while an accelerated lateral bud outgrowth, suggesting the cell proliferation might be enhanced in lateral buds by involving additional regulators epistatic to SLB1. The TCP (TEOSINTE BRANCHED1/CYCLOIDEA/PCF) family genes may be interesting candidates for investigation for possible roles in SLB1–BS1‐mediated control of shoot branching in M. truncatula. For example, the BRANCHED1 (BRC1) gene is thought to integrate various environmental and hormonal signals, including auxin, cytokinin and SL signalling, to regulate bud growth (Aguilar‐Martinez et al., 2007; Braun et al., 2012; Dun et al., 2012; Barbier et al., 2019). Further elucidating the actions of the SLB1–BS1 regulatory module and investigating their interactions with phytohormones, including auxin, cytokinin and SLs and other branching regulators, like BRC1, will enlighten our understanding of the genetic network underlying this fundamental process.
Increasing seed yield by manipulating SLB1 expression in legume crops
Organ size is an important agronomic trait that influences crop yields (Li et al., 2019). Soybean, a major crop worldwide, provides up to 69% of proteins and 30% of oils in the human diet (Lam et al., 2010; Zhou et al., 2015). To meet the needs of the rapidly increasing human population, soybean breeders are faced with the challenge of designing a high‐efficiency breeding strategy for developing soybean varieties with higher yields and improved quality (Masuda & Goldsmith, 2009; Ray et al., 2013). In fact, several laboratories are currently focused on increasing seed size via genetic engineering to improve soybean yields. Downregulating the BS1 orthologues, GmBS1 and GmBS2, using artificial microRNA was successfully used to modify seed size, thereby increasing soybean yields (Ge et al., 2016). However, this type of knockout strategy is often limited due to the redundancy of gene targets, especially in legumes with complex genomes. As overexpressing SLB1 resulted in enlarged seed and leaf phenotypes similar to those produced by downregulating BS1 orthologues (Fig. 7), SLB1 represents an alternative target for genetic manipulation of seed size, perhaps serving as a new tool for significantly improving seed yield in soybean and other legume crops.
Author contributions
PY, LN and HL conceived and designed the research. PY, QM, HW and DF performed the experiments. XW, YP and JW contributed analytical tools. PY, MT, LN and HL analysed the data and wrote the manuscript. LN and HL contributed equally to this work. This work has not been submitted elsewhere for publication, and all authors reviewed the manuscript.
Supporting information
Fig. S1 Comparison of vexillum petal size of M. truncatula wild‐type and slb1‐1.
Fig. S2 The M. truncatula slb1 mutant shows defects in floral organ development.
Fig. S3 Disruption of SLB1 leads to significant decreases in leaf size, primarily by affecting cell proliferation in M. truncatula.
Fig. S4 Phenotypes of the M. truncatula wild‐type and slb1 alleles.
Fig. S5 Transcript abundance of SLB1 in M. truncatula leaves and flowers at different developmental stages.
Fig. S6 Sequence alignment of M. truncatula SLB1 and Arabidopsis SAP.
Fig. S7 Phylogenetic analysis of ASK1/2‐like and CUL1‐like family proteins in M. truncatula.
Fig. S8 Expression pattern of BS1 in M. truncatula and subcellular localisation of BS1.
Fig. S9 Targeted mutagenesis of M. truncatula BS1 using the CRISPR/Cas9 system.
Fig. S10 SLB1 overexpression improves biomass yield in M. truncatula.
Table S1 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was supported by grants from the National Transgenic Science and Technology Program (2019ZX08010‐002), Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS) (No. CAAS‐ZDXT2019004), and Central Public‐interest Scientific Institution Basal Research Fund (No. 1610392020005). Development of M. truncatula Tnt1 insertion lines was supported by the US National Science Foundation (DBI 0703285 and IOS‐1127155) and Noble Research Institute LLC. The authors declare no competing financial interests.
Contributor Information
Lifang Niu, Email: niulifang@caas.cn.
Hao Lin, Email: linhao@caas.cn.
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. 2009. Gibberellin signaling controls cell proliferation rate in Arabidopsis . Current Biology 19: 1188–1193. [DOI] [PubMed] [Google Scholar]
- Aguilar‐Martínez JA, Poza‐Carrión C, Cubas P. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MP. 1969. Differential staining of aborted and nonaborted pollen. Stain Technology 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M. 2007. Control of plant organ size by KLUH/CYP78A5‐dependent intercellular signaling. Developmental Cell 13: 843–856. [DOI] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Muhlenbock P, Skirycz A, Gonzalez N, Beemster GT et al 2012. Exit from proliferation during leaf development in Arabidopsis thaliana: a not‐so‐gradual process. Developmental Cell 22: 64–78. [DOI] [PubMed] [Google Scholar]
- Barbier FF, Dun EA, Kerr SC, Chabikwa TG, Beveridge CA. 2019. An update on the signals controlling shoot branching. Trends in Plant Science 24: 220–236. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. 2007. Stomatal development. Annual Review of Plant Biology 58: 163–181. [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot J‐P, Boutet‐Mercey S, Dalmais M, Antoniadi I, Li X, Maia‐Grondard A, Le Signor C, Bouteiller N et al 2012. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiology 158: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byzova MV, Franken J, Aarts MG, de Almeida‐Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC. 1999. Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes & Development 13: 1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nature Reviews Molecular Cell Biology 5: 739–751. [DOI] [PubMed] [Google Scholar]
- Chen L, Cai Y, Liu X, Yao W, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W. 2018. Improvement of soybean Agrobacterium‐mediated transformation efficiency by adding glutamine and asparagine into the culture media. International Journal of Molecular Sciences 19: 3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG. 1991. Apical dominance. Botanical Review 57: 318–358. [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C. 2007. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Reports 8: 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V et al 2007. Arabidopsis CYCD3 D‐type cyclins link cell proliferation and endocycles and are rate‐limiting for cytokinin responses. Proceedings of the National Academy of Sciences, USA 104: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F‐box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. 2006. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage‐dependent manner. Current Biology 16: 272–279. [DOI] [PubMed] [Google Scholar]
- Dong ZC, Zhao Z, Liu CW, Luo JH, Yang J, Huang WH, Hu XH, Wang TL, Luo D. 2005. Floral patterning in Lotus japonicus . Plant Physiology 137: 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. 1999. Cell cycling and cell enlargement in developing leaves of Arabidopsis . Developmental Biology 215: 407–419. [DOI] [PubMed] [Google Scholar]
- Du L, Li N, Chen L, Xu Y, Li Y, Zhang Y, Li C, Li Y. 2014. The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin‐specific protease UBP15/SOD2 in Arabidopsis . Plant Cell 26: 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. 2012. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiology 158: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklof S, Astot C, Sitbon F, Moritz T, Olsson O, Sandberg G. 2000. Transgenic tobacco plants co‐expressing Agrobacterium iaa and ipt genes have wild‐type hormone levels but display both auxin‐ and cytokinin‐overproducing phenotypes. The Plant Journal 23: 279–284. [DOI] [PubMed] [Google Scholar]
- Eloy NB, de Freitas Lima M, Van Damme D, Vanhaeren H, Gonzalez N, De Milde L, Hemerly AS, Beemster GT, Inze D, Ferreira PC. 2011. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. The Plant Journal 68: 351–363. [DOI] [PubMed] [Google Scholar]
- Ge L, Yu J, Wang H, Luth D, Bai G, Wang K, Chen R. 2016. Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proceedings of the National Academy of Sciences, USA 113: 12414–12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD. 2000. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Roldan V, Fermas S, Brewer PB, Puech‐Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC et al 2008. Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Pauwels L, Baekelandt A, De Milde L, Van Leene J, Besbrugge N, Heyndrickx KS, Cuellar Perez A, Durand AN, De Clercq R et al 2015. A repressor protein complex regulates leaf growth in Arabidopsis. Plant Cell 27: 2273–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP. 2003. Legumes: importance and constraints to greater use. Plant Physiology 131: 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB. 1994. The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis . Plant Physiology 106: 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. 2006. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana . Journal of Plant Research 119: 37–42. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. 2005. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana . The Plant Journal 43: 68–78. [DOI] [PubMed] [Google Scholar]
- Hu Y, Poh HM, Chua NH. 2006. The Arabidopsis ARGOS‐LIKE gene regulates cell expansion during organ growth. The Plant Journal 47: 1–9. [DOI] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH. 2003. The Arabidopsis auxin‐inducible gene ARGOS controls lateral organ size. The Plant Cell 15: 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. 2005. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant and Cell Physiology 46: 79–86. [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogne K, Beveridge CA, Rameau C. 2006. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long‐distance signals. Plant Physiology 142: 1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T, Ichihashi Y, Murata S, Tsukaya H. 2010. The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5‐dependent mobile growth factor in developing leaves of Arabidopsis thaliana . Plant and Cell Physiology 51: 1046–1054. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F‐box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Saito Y, Uchimiya H. 1999. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis . Proceedings of the National Academy of Sciences, USA 96: 9433–9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. 1998. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P‐450 family that is required for the regulated polar elongation of leaf cells. Genes & Development 12: 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kende H. 2004. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis . Proceedings of the National Academy of Sciences, USA 101: 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, Pagano M. 2000. The F‐box protein family. Genome Biology 1: REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Wang S, Li Y, Zaitlin D, Pierce AJ, Smalle JA. 2009. Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiology 150: 178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong F‐L, Li MW, He W, Qin N, Wang B et al 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genetics 42: 1053–1059. [DOI] [PubMed] [Google Scholar]
- Li C, Bangerth F. 2003. Stimulatory effect of cytokinins and interaction with IAA on the release of lateral buds of pea plants from apical dominance. Journal of Plant Physiology 160: 1059–1063. [DOI] [PubMed] [Google Scholar]
- Li N, Liu Z, Wang Z, Ru L, Gonzalez N, Baekelandt A, Pauwels L, Goossens A, Xu R, Zhu Z et al 2018. STERILE APETALA modulates the stability of a repressor protein complex to control organ size in Arabidopsis thaliana . PLoS Genetics 14: e1007218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xu R, Li Y. 2019. Molecular networks of seed size control in plants. Annual Review of Plant Biology 70: 435–463. [DOI] [PubMed] [Google Scholar]
- Li S, Liu Y, Zheng L, Chen L, Li N, Corke F, Lu Y, Fu X, Zhu Z, Bevan MW et al 2012. The plant‐specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana . New Phytologist 194: 690–703. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW. 2008. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana . Genes & Development 22: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wang T, Persson S, Mueller‐Roeber B, Schippers JH. 2014. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nature Communications 5: 3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Goldsmith PD. 2009. World soybean production: area harvested, yield, and long‐term projections. International Food and Agribusiness Management Review 12: 143–162. [Google Scholar]
- Meng Y, Hou Y, Wang H, Ji R, Liu B, Wen J, Niu L, Lin H. 2017. Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula . Plant Cell Reports 36: 371–374. [DOI] [PubMed] [Google Scholar]
- Meng Y, Wang Z, Wang YQ, Wang C, Zhu B, Liu H, Ji W, Wen J, Chu C, Tadege M et al 2019. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40‐1 to regulate carotenoid‐derived flower pigmentation in Medicago truncatula . Plant Cell 31: 2751–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Lin H, Zhang F, Watira TW, Li G, Tang Y, Wen J, Ratet P, Mysore KS, Tadege M. 2015. LOOSE FLOWER, a WUSCHEL‐like Homeobox gene, is required for lateral fusion of floral organs in Medicago truncatula . The Plant Journal 81: 480–492. [DOI] [PubMed] [Google Scholar]
- Nordstrom A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G. 2004. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin‐cytokinin‐regulated development. Proceedings of the National Academy of Sciences, USA 101: 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU. 2012. Mechanisms of stomatal development. Annual Review of Plant Biology 63: 591–614. [DOI] [PubMed] [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA. 2013. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8: e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. 2010. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CA, Eloy NB, Lima Mde F, Rodrigues RL, Franco LO, Himanen K, Beemster GT, Hemerly AS, Ferreira PC. 2009. Overexpression of the Arabidopsis anaphase promoting complex subunit CDC27a increases growth rate and organ size. Plant Molecular Biology 71: 307–318. [DOI] [PubMed] [Google Scholar]
- Sachs T, Thimann KV. 1964. Release of lateral buds from apical dominance. Nature 201: 939–940. [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben‐Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J et al 2010. Jasmonate perception by inositol‐phosphate‐potentiated COI1‐JAZ co‐receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu‐Sato S, Mori H. 2001. Control of outgrowth and dormancy in axillary buds. Plant Physiology 127: 1405–1413. [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter J, Doyle JJ. 2006. Paleopolyploidy and gene duplication in soybean and other legumes. Current Opinion in Plant Biology 9: 104–109. [DOI] [PubMed] [Google Scholar]
- Sicard A, Kappel C, Lee YW, Wozniak NJ, Marona C, Stinchcombe JR, Wright SI, Lenhard M. 2016. Standing genetic variation in a tissue‐specific enhancer underlies selfing‐syndrome evolution in Capsella . Proceedings of the National Academy of Sciences, USA 113: 13911–13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda Y, Sako K, Maki Y, Yamazaki N, Yamamoto H, Ikeda A, Yamaguchi J. 2009. Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. The Plant Journal 60: 68–78. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. 2007. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. The Plant Journal 50: 80–94. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis . Development 129: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Tadege M, Lin H, Bedair M, Berbel A, Wen J, Rojas CM, Niu L, Tang Y, Sumner L, Ratet P et al 2011. STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris . Plant Cell 23: 2125–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M et al 2008. Large‐scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula . The Plant Journal 54: 335–347. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. 2006. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. The Plant Journal 45: 1028–1036. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda‐Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K et al 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- Wang H, Niu L, Fu C, Meng Y, Sang D, Yin P, Wu J, Tang Y, Lu T, Wang ZY et al 2017. Overexpression of the WOX gene STENOFOLIA improves biomass yield and sugar release in transgenic grasses and display altered cytokinin homeostasis. PLoS Genetics 13: e1006649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li N, Jiang S, Gonzalez N, Huang X, Wang Y, Inze D, Li Y. 2016. SCF(SAP) controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana . Nature Communications 7: 11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware A, Walker CH, Simura J, Ljung K, Bishopp A, Wilson ZA, Bennett T. 2019. Auxin export from proximal fruits drives arrest in competent inflorescence meristems. bioRxiv: 542662. [DOI] [PubMed] [Google Scholar]
- White DW. 2006. PEAPOD regulates lamina size and curvature in Arabidopsis . Proceedings of the National Academy of Sciences, USA 103: 13238–13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Li N, Dumenil J, Li J, Kamenski A, Bevan MW, Gao F, Li Y. 2013. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis . Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Chen F, Yu X, Lin C, Fu YF. 2009. Over‐expression of an AT‐hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana . Plant Molecular Biology 71: 39–50. [DOI] [PubMed] [Google Scholar]
- Xu R, Li Y. 2011. Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana . Development 138: 4545–4554. [DOI] [PubMed] [Google Scholar]
- Yang L, Liu H, Zhao J, Pan Y, Cheng S, Lietzow CD, Wen C, Zhang X, Weng Y. 2018. LITTLELEAF (LL) encodes a WD40 repeat domain‐containing protein associated with organ size variation in cucumber. The Plant Journal 95: 834–847. [DOI] [PubMed] [Google Scholar]
- Yarce JC, Lee HK, Tadege M, Ratet P, Mysore KS. 2013. Forward genetics screening of Medicago truncatula Tnt1 insertion lines. Methods in Molecular Biology 1069: 93–100. [DOI] [PubMed] [Google Scholar]
- Zhang XR, Qin Z, Zhang X, Hu Y. 2015. Arabidopsis SMALL ORGAN 4, a homolog of yeast NOP53, regulates cell proliferation rate during organ growth. Journal of Integrative Plant Biology 57: 810–818. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y et al 2015. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nature Biotechnology 33: 408–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Comparison of vexillum petal size of M. truncatula wild‐type and slb1‐1.
Fig. S2 The M. truncatula slb1 mutant shows defects in floral organ development.
Fig. S3 Disruption of SLB1 leads to significant decreases in leaf size, primarily by affecting cell proliferation in M. truncatula.
Fig. S4 Phenotypes of the M. truncatula wild‐type and slb1 alleles.
Fig. S5 Transcript abundance of SLB1 in M. truncatula leaves and flowers at different developmental stages.
Fig. S6 Sequence alignment of M. truncatula SLB1 and Arabidopsis SAP.
Fig. S7 Phylogenetic analysis of ASK1/2‐like and CUL1‐like family proteins in M. truncatula.
Fig. S8 Expression pattern of BS1 in M. truncatula and subcellular localisation of BS1.
Fig. S9 Targeted mutagenesis of M. truncatula BS1 using the CRISPR/Cas9 system.
Fig. S10 SLB1 overexpression improves biomass yield in M. truncatula.
Table S1 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


