Summary
Enabling data reuse and knowledge discovery is increasingly critical in modern science, and requires an effort towards standardising data publication practices. This is particularly challenging in the plant phenotyping domain, due to its complexity and heterogeneity.
We have produced the MIAPPE 1.1 release, which enhances the existing MIAPPE standard in coverage, to support perennial plants, in structure, through an explicit data model, and in clarity, through definitions and examples.
We evaluated MIAPPE 1.1 by using it to express several heterogeneous phenotyping experiments in a range of different formats, to demonstrate its applicability and the interoperability between the various implementations. Furthermore, the extended coverage is demonstrated by the fact that one of the datasets could not have been described under MIAPPE 1.0.
MIAPPE 1.1 marks a major step towards enabling plant phenotyping data reusability, thanks to its extended coverage, and especially the formalisation of its data model, which facilitates its implementation in different formats. Community feedback has been critical to this development, and will be a key part of ensuring adoption of the standard.
Keywords: findability, interoperability, metadata, phenomics, plant phenotyping, reusability, standards
Introduction
The volume of data being generated in the life sciences demands good data management practices to enable reusability. While it is common practice to publish standardised sequencing data in public repositories, other data types are often only made available through scientific publications, and can be hard to find (Vines et al., 2014), interpret or reuse. In a survey with over a 1000 participants, more than half agreed that lack of access to data is a ‘major impediment to progress in science’ and ‘has restricted their ability to answer scientific questions’, and most pointed out that data may easily be misinterpreted due to its complexity or poor quality (Tenopir et al., 2011).
In the plant phenotyping domain, data reuse is both pressing and challenging (Spindel & McCouch, 2016; Tardieu et al., 2017; Ćwiek‐Kupczyńska, 2018). On the one hand, the development of automated high‐throughput and high‐resolution technologies has contributed to a scale‐up in the number, complexity and size of plant phenotyping datasets. This has been amplified by the increasing number of long‐term, highly multilocal phenotyping networks aiming to decipher the interaction between genotype and environment (Millet et al., 2019). Conversely, the reuse and meta‐analyses of phenotyping data are particularly challenging due to the heterogeneity of this domain that encompasses many types of experimental sites (field, glasshouse, controlled environment), plants (crops, forest trees), collected data (images, physical measurements, chemical assays, molecular biology assays), and experimental designs (factors being tested, timing, field layouts, etc.). Furthermore, plant phenotype hinges not only on the interaction between genotype and environment, but also developmental stage and epigenome status (King et al., 2010), which raises the challenges of integrating genotypic and phenotypic data (Pommier et al., 2019b).
A successful example of data reuse in this domain is the study by Hurtado‐Lopez et al. (2015), who reused field trial datasets and integrated them with quantitative trait locus (QTL) data to yield novel insights into genotype by environment (GxE) interactions in potato. Because the original experimental data followed no standardisation guidelines, the authors had to manually assemble detailed metadata during the preprocessing of the data from descriptions in unstructured text. To facilitate such studies, so that they may become the norm rather than the exception, it is essential that the scientific community adopt good data management and publication practices (Zamir, 2013).
The requirements for data reuse in science have been formalised in the FAIR data principles (Wilkinson et al., 2016). They state the criteria that scientific data must fulfil to be findable, accessible, interoperable and reusable by both humans and machines, which hinge on having rich, harmonised, machine‐readable, high‐quality metadata describing the data as explicitly and objectively as possible.
Four key components are needed from research communities to meet these requirements: metadata standards which list the fields required for interpreting the data from a given experimental domain; machine‐readable (meta)data exchange formats in which to express and share the (meta)data; ontologies or controlled vocabularies to describe (meta)data values and ensure that they are objective, consistent and unambiguous across datasets; and searchable data repositories with a well established protocol for machine access.
The need for a metadata standard was first recognised in the life sciences by the microarray community, who developed MIAME (Minimum Information About a Microarray Experiment) (Brazma et al., 2001). This was soon followed by similar standards for other domains (e.g. Field et al., 2008; Bustin et al., 2009; Lapatas et al., 2015) as can be seen on FAIRsharing (Sansone et al., 2019). In the plant phenotyping domain, the need for metadata to document experiments was initially addressed independently by the developers of phenotyping databases, such as BreedBase (BreedBase team, 2020), GnpIS (Steinbach et al., 2013), PIPPA (PIPPA team, 2020) and Plant Hybrid Information System (PHIS) (Neveu et al., 2019), which resulted in a multitude of implicit, often database‐specific standards. However, the need for an explicit consensus to enable interoperability between these databases brought this community together to develop MIAPPE (Minimum Information About a Plant Phenotyping Experiment), the first and so far only community metadata standard for the plant phenotyping domain (Krajewski et al., 2015).
MIAPPE had three guiding principles: to minimise the chance of a researcher missing important information in the documentation of an experiment; to support the annotation of content with community‐relevant vocabularies; and to promote a data format implementation. MIAPPE marked a critical step towards the FAIRness of plant phenotyping data, as concluded in a survey of c. 50 citations of this standard in publications and web portals (Krajewski & Ćwiek‐Kupczyńska, 2020). However, there were aspects to improve, such as the coverage, usability and clarity of the standard. In particular, MIAPPE lacked fields needed to capture experiments with woody plants, as it was conceived primarily with crop plants in mind, and it lacked an explicit data model, which left some researchers struggling to understand how to represent their experiments.
The microarray community was again among the first to produce a machine‐readable (meta)data exchange format in the form of MAGE‐Tab (MicroArray Gene Expression tabular) (Rayner et al., 2006), a standardised format for MIAME. This gave rise to the broader‐purpose ISA‐Tab (Investigation/Study/Assay tab‐delimited) format (Rocca‐Serra et al., 2010; Sansone et al., 2012), which was adopted by more domains, including plant phenotyping, with an ISA‐Tab implementation of MIAPPE (Ćwiek‐Kupczyńska et al., 2016).
The use of ontologies and controlled vocabularies in the life sciences dates back to Linnaeus's taxonomy, but they have witnessed a more recent boom after the creation of the Gene Ontology (The Gene Ontology Consortium, 2019), and currently number in the several hundred, as seen on BioPortal (Noy et al., 2009). For the plant phenotyping domain, there are a number of ontologies that cover different key aspects. The Crop Ontology (Shrestha et al., 2012) models plant traits and methods for assessing them in several species‐specific ontologies. It merits special reference, in that it aims at standardising the methods used by data producers for phenotyping and the way they are reported, rather than only at terminological standardisation. It therefore includes an implicit metadata standard, the trait‐method‐scale trio, which was incorporated into MIAPPE. The Planteome project (Cooper et al., 2018) developed three key ontologies: the Plant Trait Ontology (Arnaud et al., 2012) modelling species‐independent plant traits under a broader scope than the Crop Ontology and serving as a reference ontology for multispecies analyses; the Plant Ontology (Jaiswal et al., 2005) covering plant anatomical structures and development stages and enabling interplant comparisons; and the Plant Experimental Conditions Ontology (Cooper et al., 2018) describing plant treatments. In addition to these, relevant ontologies include: the Agronomy Ontology (Aubert et al., 2017) covering agronomic practices, techniques and variables; the Environment Ontology (Buttigieg et al., 2013) describing natural environments; and the Statistics Ontology (Statistics Ontology Project, 2020) devoted to statistical methods. All these ontologies and several others are indexed in AgroPortal (Jonquet et al., 2018) which serves as the reference repository and search service for plant‐related ontologies.
Finally, while searchable data repositories have long been the norm in the life sciences, especially concerning gene and protein data, only in the last decade has it become common practice to enable machine access to their data via application programming interfaces (APIs). Currently all major databases, such as GenBank (Benson et al., 2013) or UniProt (The UniProt Consortium, 2019), provide such access, but most smaller databases do not. This was the case for the plant phenotyping domain up until recently, with its numerous, independent and heterogeneous local databases. To address this problem and enable interoperability between databases, the plant community undertook the development of the Breeding API (BrAPI) (Selby et al., 2019), a common API for data search and retrieval that can be implemented by plant breeding databases irrespective of their internal data model. Like the databases it aims to connect, BrAPI also has an implicit (meta)data model that aims to reconcile the metadata available in existing databases, spanning organisational metadata, plant phenotypic (meta)data and genotypic (meta)data. BrAPI was initiated independently from MIAPPE, so while there is substantial overlap between the two resources, there are also a few key differences in their metadata fields, as well as differences in terminology.
The way forward for enabling FAIR plant phenotyping data lies in bringing together all of the components described above. MIAPPE would be the cornerstone of such an architecture, specifying the metadata that is needed and connecting metadata fields to the ontologies recommended to fill them, as well as reconciling the several implicit metadata models of existing knowledge resources. BrAPI would serve as the means for federating the many independent plant phenotyping databases to enable findability and accessibility, and should enforce and validate the MIAPPE compliance of datasets. The MIAPPE ISA‐Tab implementation would support data publication and exchange. And potentially all of the ontologies listed above would play a role in describing the data and metadata of plant phenotyping experiments in a standardised and unambiguous way. However, it is clear that further development effort on these resources is needed to attain such a goal.
In this paper, we detail the efforts of an international consortium to enhance the MIAPPE standard towards enabling FAIR plant phenotyping data. We describe the following refinements: (i) the extension of MIAPPE to accommodate a wider range of use cases (including those relevant to perennial and woody plants); (ii) the specification of a data model underlying the standard, to facilitate its interpretation and usage; (iii) the formalisation of MIAPPE in a computer‐interpretable format (using the Web Ontology Language, OWL) to enable dataset validation and computational analysis; and (iv) the alignment of MIAPPE and BrAPI to enable the exposure of MIAPPE‐compliant datasets via BrAPI endpoints.
Materials and Methods
Development of MIAPPE 1.1
To take on the challenge of improving MIAPPE, the community gathered both life and computer scientists. The former drove the documentation and description of the standard, ensuring that the terms and definitions are meaningful and not purely technical. The latter took the initiative for the technical aspects, involving data formalisation, organisation, integration, sharing and interoperability. This ongoing partnership ensures that MIAPPE bridges the domains of life and data science and addresses the needs of both communities.
The development of MIAPPE 1.1 was carried out collaboratively using simple and efficient protocols and format (spreadsheet). Throughout the process, drafts were presented and discussed with the international community through consultations by emails, the MIAPPE consortium GitHub issue tracker (MIAPPE Contributors, 2020a) and during ‘bring your own data’ training sessions.
Like its predecessor, MIAPPE 1.1 is a metadata standard that formally organises the documenting of a phenotyping dataset, including environmental aspects. It primarily structures the metadata, imposing no constraints on the data itself (which may consist of images, other binary data, tabular files, etc.).
In comparison with MIAPPE 1.0, MIAPPE 1.1 introduces several new concepts while preserving most of those already present. The major change, however, is that it moved from a simple checklist to a fully formalised data model that makes explicit mandatory information, restrictions and expectations and thus represents a major improvement in clarity from MIAPPE 1.0.
The key changes in MIAPPE 1.1 fall into one of three categories, which are detailed in the following subsections: scope extension, interoperability and data model specification. In addition to these, the MIAPPE data model has been formalised in OWL as the Plant Phenotyping Experiment Ontology (PPEO).
Note: throughout the rest of this document, we use italics to denote MIAPPE concepts, <angle brackets> to denote ontology concepts, and “double quotes” for MIAPPE field value examples.
Scope extension
The scope of MIAPPE 1.0, mostly restricted to field crops, was extended in MIAPPE 1.1 to encompass woody plants, mainly by enabling the identification of plant materials by their geolocation coordinates, which are typically used to identify forest trees instead of plant identifiers (e.g. GenBank accession numbers) used in crop research. Two levels for plant material identification and description are available in MIAPPE 1.1: (i) its identification within the experiment (biological material); and (ii) its identification before the experiment (material source), allowing for individual plants, lots or progeny to be described and related to previously published or publicly accessible material. Furthermore, the preprocessing field (previously called pretreatments) can describe any type of action performed on the material source before it is used as the experimental biological material (for instance “tree transplantation” and “grafting”).
Interoperability
MIAPPE 1.1 incorporates several metadata standards and practices that cover parts of its scope, in order to ensure interoperability and avoid remodelling and redefining aspects that are already well established: the generic metadata fields (e.g. identifier, title, version, date) in MIAPPE 1.1 are largely based on the DataCite metadata model of Dublin Core (DataCite Metadata Working Group, 2014); the fields for biological material identification are based on the Multi‐Crop Passport Descriptors (mcpd) v.2.1 (Alercia et al., 2015); the observation unit concept and its fields were imported from BrAPI and GnpIS‐Ephesis (Pommier et al., 2019b); and the observed variable section is largely based on the data model of the Crop Ontology.
Additionally, to further foster interoperability, MIAPPE 1.1 includes precise definitions and examples for each of its fields, with recommendations for the use of controlled vocabularies, ontologies and ISO norms whenever appropriate. For example, the ISO 8601 norm is recommended for dates, Crop Ontology terms are recommended in the observed variable section, and Plant Ontology terms are recommended for characterising samples. These definitions and recommendations clarify the intended usage of MIAPPE 1.1 in a way that is accessible to biologists and breeders, while promoting compliance with the FAIR principles.
Data model specification
The specification of a data model was essential to clarify MIAPPE's structure, and improves its internal consistency. The construction of a formal model helped: (i) ascertain the roles and relationships of the MIAPPE 1.0 checklist's main categories and concepts; and (ii) extend those concepts to a broader range of experiments.
Objects in the MIAPPE 1.1 data model correspond to sections in the MIAPPE 1.1 checklist. A schematic view of the data model is presented in Fig. 1.
Fig. 1.
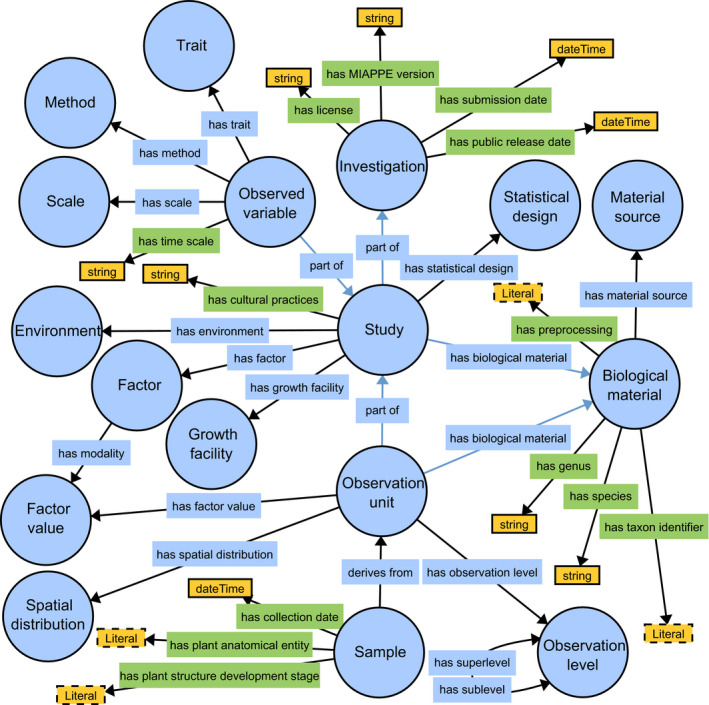
Subset of the Plant Phenotyping Experiment Ontology representing the MIAPPE data model. Generated using WebVOWL (http://editor.visualdataweb.org/) and edited manually. Circles indicate classes. Object properties are shown in blue rectangles, and data properties are shown in green rectangles. Yellow rectangles represent literals.
The MIAPPE data model is reconciled with the more generic data models underlying the ISA‐Tab exchange format (Rocca‐Serra et al., 2010) through key objects such as Investigation and Study (ISA) and specialised representations such as BrAPI (Selby et al., 2019) with entities such as Observation unit and Observation variable (BrAPI).
Data model formalisation (PPEO)
While the specification of the MIAPPE data model addresses the concern of improving the clarity of the standard for users, it does not address machine readability, which is important to enable validation and facilitate implementation at scale. The need for the latter led us to encode the MIAPPE standard in OWL as the PPEO (Pommier et al., 2020).
In PPEO, each MIAPPE section is encoded as an ontology class, with additional classes declared to group linked MIAPPE fields (e.g. <method> groups the linked fields method description and method accession number). Each MIAPPE field is encoded as an ontology data property, which specifies the type of value expected (e.g. <has collection date> for class <sample>, which must take a date–time value). The relations between classes are formalised through object properties (e.g. <has biological material> connects <observation unit> to <biological material>). Cardinality restrictions imposed by the data model are encoded as ontology restrictions on the corresponding classes (e.g. an <investigation> must have at least one <study>). Ontology usage recommendations are encoded as annotations. Finally, to facilitate the implementation of MIAPPE in various forms, PPEO includes labels expressing corresponding names of each class in different resources (BrAPI, ISA‐Tab).
Fig. 1 represents a subset of the PPEO.
MIAPPE 1.1 overview
The MIAPPE sections, which correspond to objects in the data model, are the following:
Investigation – the entry point of each MIAPPE dataset. It contains several general metadata fields (e.g. title, description, submission/publication dates), including some critical for FAIRness (unique identifier, licence, MIAPPE version). One or more publications may be associated with the investigation.
Study – corresponds to one experiment and defines its location (which by definition must be single per study) and duration. It lists general fields documenting the experiment (e.g. experimental design, cultural practices, growth facility). Like the investigation, it contains a unique identifier field. An investigation must have one or more studies.
Person – contains contact details for each contributor of an entire investigation or an individual study, including the role of the person.
Data file – references a data file of the MIAPPE dataset (e.g. a tabular file containing the results of observations, an image file), which may be attached to the dataset (referenced by name) or available in an online repository (referenced by URL). A version and a description must be provided for each data file. A study may have any number of data files.
Biological material – identifies and describes the plant materials used in the studies. Plant materials must be identified through a biological material ID field, which can be institution‐specific or platform‐specific (e.g. seed lot number for annual plants, clone number for perennials or an experimental plant ID), and is recommended to follow the MCPD convention of holding institute identifier (FAO WIEWS code) plus a unique identifier of the individual plant material provided by that institute. They must also be identified through the organism field, which indicates the unique taxonomic identifier of the biological material in a standard such as the NCBI taxonomy. Optionally, they may be identified through the fields genus, species and infraspecific name, where textual names are expected (but should follow accepted standards). They may also be identified through geographical coordinates (i.e. latitude, longitude, altitude, and coordinates uncertainty), as is common for forest trees. The biological material preprocessing describes the biological material pretreatments, applied (e.g. to the seeds, or the tree cuttings) before the beginning of the experiment. Finally, the material source fields identify the origin or provenance of the biological material (e.g. gene bank accession, in situ material like an orchard, tree material provenance including forest wild site, laboratory‐specific populations). These fields include the material source ID (which follows the same recommendations as the biological material ID), the material source DOI (for referencing material sources listed in repositories), four geographical coordinates fields (same as for biological material), and finally a textual description. The biological material section thus covers a minimal subset of the MCPD standard used by gene banks, while also enabling interoperability and data linking through the use of identifiers (namely NCBI taxonomy identifiers) both between MIAPPE‐compliant datasets and with external datasets. Moreover, through the provision of external identifiers to resources detailing their biological material (e.g. DOIs, accessions to gene banks or genome archives) researchers can encompass additional information, such as extended MCPD information (e.g. synonyms, genealogy) and genotypic information. Last but not least, with these additions, MIAPPE 1.1 can handle cases such as forest tree clonal trials, where the plants identified solely through biological material coordinates in one study are used to generate new plant material for another study, in which their identification is done by specifying the location of the material source.
Environment – describes a management practice parameter (e.g. sowing density, rooting medium composition) that was kept constant throughout the study across all observation units (to be described later). It applies to the whole study and has only a type (parameter) and a value. There can be discrepancies between intended environmental settings (e.g. target temperature in a glasshouse) and actual measurements of environmental observed variables (e.g. hourly temperature measured with four sensors). A study may have any number of environments.
Experimental factor – describes a management practice that varied between observation units in a study, assessing the effect of which is the object of the study. Experimental factors can be biotic or abiotic (e.g. pest, disease interaction, cultural practice) and are characterised by a type, a description and a list of possible values. For instance, a “drought” experimental factor can discriminate “rainfed” and “irrigated” blocks, and a “nitrogen input level” can identify groups of plants under “high nitrogen input”, “low nitrogen input” and “no nitrogen inputs”. A study may have one or more experimental factors.
Event – describes a discrete occurrence at a specific time that affected the whole study or one or more observation units, which can be the application of a field/glasshouse practice (e.g. planting, fungicide application) or an unpredictable happening (e.g. rainfall, pathogen attack). Events allow a general traceability of the conditions/events, and have been adopted since their usefulness was successfully demonstrated in the PHIS (Neveu et al., 2019). Events include a type, ideally taken from an ontology such as the Crop Research Ontology (Shrestha, 2020) or the Agronomy Ontology (Aubert et al., 2017), a date and a description, but no dedicated field for categorical or numerical values. Events can be repeated through time (e.g. to capture repeating cultural practices, such as adding fertiliser) by duplicating the type and description while providing a new date.
Observation unit – is the experimentation object on which phenotypic and environmental parameters are measured and to which experimental factors are applied. It is characterised by a type or level, which can be a single “plant”, a group of plants (“pot”, “plot”, “block”), or the whole “study”. These types are hierarchical, meaning that we can have observation units and corresponding observations made from the study level down to the plant level. In some cases, an observation unit may contain no plant (e.g. raw plots after harvest or areas of a forest without tree), but can still be the object of environmental observations. Optionally, an observation unit can have a cross‐reference to an external database, such as BioSamples (Courtot et al., 2019). Also optionally, it can have one or more spatial distribution key‐value pairs that locate the observation unit in the experimental hierarchy (e.g. “block: 1”) or globally (e.g. “latitude: +43.619261”). A study should have one or more observation units.
Sample – represents subplant material that was physically collected from an observation unit and was stored and processed before observations are made on it (e.g. in molecular studies). When traceability of sample processing is not needed, subplant observations can be assigned directly to the corresponding plant‐level observation unit without the use of a sample as an intermediary. In such cases, the observed variable should describe that the observation is made on a plant part (e.g. “leaf chlorophyll content”, “grain protein content”) and include additional information on how the sampling was made in the textual description. The sample description field contains a free text description such as organism count, oxygenation, salinity or storage attributes. The plant anatomical entity and the plant structure development stage give more details on the sample properties at the time of sampling, which is specified with the field collection date. A sample must be derived from a single observation unit, but each observation unit may have any number of derived samples.
Observed variable – documents a phenotypic or environment parameter that was observed and recorded as part of the study. It follows the Crop Ontology model of representing variables as combinations of a trait, a method and a scale. Trait details the characteristic being observed/measured (e.g. “plant height”). Method describes the procedure used in the observation/measurement (e.g. “with a measuring tape, starting at ground level”). Scale indicates the unit or scale with which observations/measurements were recorded (e.g. “cm”). Observed variables, traits, methods and scales are each identified by name, and may have a reference to the corresponding ontology concept (ideally from the Crop Ontology). Observed variables also have an ID by which they are referenced in the data file. Methods can also have a description plus an additional reference, usually from the literature. The time scale indicates the unit of time (e.g. “date–time”, or “growing degree days”) used to timestamp observations of this observed variable.
Note that in MIAPPE 1.1, the description of environment aspects is broken into several sections so as to allow flexibility in capturing and representing environment information: environment (fixed parameters throughout the study), experimental factors (fixed set of values for the study which vary between observation units), observed variables (measured during the study) and potentially events (discrete occurrences such as heavy rain).
MIAPPE implementations
MIAPPE is a general specification that needs to be adopted and implemented by data repositories and exchange tools if it is to be easily usable. The MIAPPE 1.1 update encompasses four major implementations which are discussed in the following subsections: (i) an ISA file archive backed by an updated ISA‐Tab configuration, developed in collaboration with the ISA Framework team; (ii) a web service implementation through BrAPI, developed in close collaboration between the MIAPPE and BrAPI communities; (iii) a spreadsheet template developed and used as training material to introduce biologists to MIAPPE, which can also be used for simple metadata exchange; and (iv) finally, an RDF implementation based on the PPEO.
MIAPPE ISA‐Tab
The ISA Framework encompasses a model and a set of serialisations (TAB, JSON and RDF) to describe the experimental metadata with links to data files, code, articles and other digital objects. It comes with a suite of associated tools, is extensively used in the life sciences (The ISA Team, 2020), and among the endorsed resources of the ELIXIR Interoperability Platform.
MIAPPE 1.0 already included an ISA‐Tab implementation in the form of a configuration file. This implementation has been revised in view of the changes in MIAPPE 1.1, and the configuration file has been updated accordingly. This configuration (ISA‐Tab for Plant Phenotyping Contributors, 2020) can be used with the ISA Creator tool, to more easily produce MIAPPE‐compliant ISA‐Tab archives.
The overview of the mapping between MIAPPE 1.1 and ISA‐Tab sections is shown in Table 1. The Investigation and the Study express the same concepts in both MIAPPE and ISA‐Tab, and many of their fields are listed under the corresponding sections. There are also direct correspondences for experimental factors (Study Factors), biological material (Source), observation units (Samples) and samples (Extracts). The remaining MIAPPE‐specific fields are stored as ISA‐Tab Comments. ISA Protocols must include a protocol named “Growth” holding the MIAPPE cultural practices field and environment parameters, one protocol for the “Phenotyping” process, plus an optional list of protocols with Type “Event” to handle MIAPPE events, with specific occurrences listed in an external Events file. Finally, the ISA Sampling Protocol indicates the derivation of a MIAPPE sample from an observation unit. Each ISA‐Tab Assay represents one data file measured at one observation level. Observed variables are listed in the trait definition file, and referenced in the data files. The data files are formatted according to the common practices of the domain and contain references to that Variable ID, the measured values and times plus any information which researchers might deem useful.
Table 1.
Mapping between MIAPPE and ISA‐Tab sections.
| MIAPPE section | ISA‐Tab section | ISA‐Tab section specification |
|---|---|---|
| Investigation | Investigation/investigation publications | |
| Study | Study/study design descriptors/study protocols | |
| Person | Investigation contacts/study contacts | |
| Data file | Study | With comment fields |
| Biological material | Source | |
| Environment | Study protocols | Growth type protocol |
| Experimental factor | Study Factors | |
| Event | Study protocols | Event type protocols and external Events file |
| Observation unit | Sample | |
| Sample | Extract/study protocols | Sampling type protocol |
| Observed variable | Observed variable | In external trait definition file |
The table lists the MIAPPE sections with the ISA‐Tab sections holding their fields. MIAPPE‐exclusive fields have been added as comments in the corresponding sections. The detailed mapping can be found in Supporting Information Table S1, and in the MIAPPE repository (https://github.com/MIAPPE/MIAPPE/tree/master/MIAPPE_Checklist‐Data‐Model‐v1.1/MIAPPE_mapping).
Breeding API (BrAPI)
The Breeding API (BrAPI) (Selby et al., 2019) is a RESTful API developed by an international open‐source community for querying plant breeding data, already implemented by several databases, and selected by the European biological data infrastructure ELIXIR as the cornerstone of its plant data search service. It is therefore critical that BrAPI and MIAPPE be compatible.
The collaboration between the BrAPI and MIAPPE teams has aimed at ensuring the compatibility of the two schemas, and the latest brapi (v.1.3) covers most of MIAPPE. The main sections correspond to the same concepts and either share the same name or have direct correspondence, such as investigation (BrAPI Trial) and biological material (BrAPI Germplasm). Some MIAPPE sections are currently absent from BrAPI (e.g. environment, event) but have already been proposed as additions to BrAPI and are under consideration for the next major release. The mapping between MIAPPE and BrAPI is overviewed in Table 2.
Table 2.
Mapping between MIAPPE sections and BrAPI objects.
| MIAPPE | BrAPI object |
|---|---|
| Investigation | Trial |
| Study | Study |
| Person | Contact |
| Data file | Data link |
| Biological material | Germplasm |
| Environment | Environment parameter |
| Experimental factor | Treatment |
| Event | Events |
| Observation unit | Observation unit |
| Sample | Samples |
| Observed variable | Variable |
The table lists the MIAPPE sections with the BrAPI objects holding their fields (in the current and future versions). The detailed mapping for each field can be found on the MIAPPE GitHub repository and in Supporting Information Table S1, and in the MIAPPE repository (https://github.com/MIAPPE/MIAPPE/tree/master/MIAPPE_Checklist‐Data‐Model‐v1.1/MIAPPE_mapping).
Finally, BrAPI datasets can be exported as MIAPPE‐compliant ISA‐Tab archives using the BrAPI2ISA tool (BrAPI2ISA contributors, 2020).
Spreadsheet template
The spreadsheet template for MIAPPE (MIAPPE Contributors, 2020b) was developed mainly for training purposes, as a simpler alternative to ISA‐Tab. It is an explicit representation of MIAPPE, where each section has been placed in a separate worksheet. This template facilitates the understanding of the connections between documentation, data model and actual data. For training, it is important that the data model and one‐to‐many relationships (Fig. 1) be explicitly presented and comprehensively explained to the users (e.g. biologists or data managers).
RDF based on the PPEO
PPEO was conceived to enable the direct expression of MIAPPE datasets in RDF (W3C, 2020a), by instantiating the ontology. Moreover, because PPEO explicitly maps MIAPPE to its implementations, it should be straightforward to convert MIAPPE datasets expressed in any of them to RDF.
MIAPPE and BrAPI are also connected through PPEO, which not only maps the two resources, but also includes classes exclusive to BrAPI, such as the <observation> class (which is outside of the scope of MIAPPE, as it pertains to data). A proof of concept has demonstrated the feasibility of producing linked data through BrAPI using the JSON‐LD format (W3C, 2020b), with PPEO enabling the semantic mapping (see this dataset: Oury et al. (2020)).
Having MIAPPE datasets in RDF enables the use of a wide range of available tools for reasoning and analysis, and facilitates data integration (by enabling data linking and cross‐referencing at the semantic level) and validation.
Results
To evaluate the applicability of the standard and the functionality of its implementations, plant scientists were asked to describe their phenotyping experiments using MIAPPE 1.1. The datasets were provided by: the Instituto de Biologia Experimental e Tecnológica (iBET), Portugal; the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Germany; the Genetic and Genomic Information System (GnpIS) of the Institut National de la Recherche Agronomique, France; and the Vlaams Instituut voor Biotechnologie (VIB), Belgium. The datasets are summarised in Table 3, and described in detail in Supporting Information Notes S1. All of them are listed under Papoutsoglou et al. (2019), and their files can be retrieved through the repositories listed there (Baute et al., 2019a, 2019b, 2019c, 2020; Chaves et al, 2019a, 2019b; Junker, 2020; Junker & Li, 2020; Michotey, 2019, 2020; Oury et al., 2019a, 2019b; Pea et al., 2019a, 2019b).
Table 3.
Overview of some characteristics of the example datasets.
| Publication | Inácio et al. (2017) | Junker et al. (2015) | Li et al. (2019) | Oury et al. (2018) | Monclus et al. (2012) | Baute et al. (2015) | Dell'Acqua et al. (2015) | Baute et al. (2016) |
|---|---|---|---|---|---|---|---|---|
| Biological material | Natural population trees identified by geographical location; material source not identified Population | Mutant; multiple replicates RIL Population | Mutant and wild‐types | Genebank material | Clonal material with material source traceability; includes crosses and populations | RIL | Population | RIL |
| Throughput | Low | High | High | Low | Low | High | Low | High |
| Plant type | Forest tree | Model plant | Crop | Crop | Forest tree | Crop | Crop | Crop |
| Setting | Field; three locations | Automated glasshouse, controlled environment, four experimental factors | Automated glasshouse | Multiyear multilocal field network | Field | Glasshouse | Field | Glasshouse |
| Field Dataset | Glasshouse Cork oak | Arabidopsis | Barley | Wheat | Poplar | Maize | Maize | Maize |
The experiments on the table encompass different experimental settings, plant types and throughput. RIL, recombinant inbred line.
The datasets span model, crop and perennial plants in a variety of experimental settings, as well as various MIAPPE 1.1 implementations. They demonstrate the ability of MIAPPE to handle diverse experimental designs, including automated glasshouses (IPK and VIB datasets), field networks for crops (GnpIS) and forest trees (iBET and GnpIS) with multiple scales and repetitions. Perennial plant use cases feature time series data, that is several observations across time for the same observed variable on the same plant. Field networks (GnpIS wheat) demonstrate the use case of a multilocal and multiannual dataset where each location represents one study over several years. Several datasets demonstrate also the use of experimental factors such as cultural practices (nitrogen level in GnpIS wheat) or experimental questions (covered or uncovered plants in IPK Arabidopsis). The observed variables proved to be well suited for very diverse destructive and nondestructive measurements adapted to agronomic (e.g. yield, grain weight), morphological (e.g. plant height), stress (e.g. disease or game), molecular (e.g. protein content) and physiological (e.g. photosynthetic efficiency) data. The data types covered by the observed variables are mostly numeric or textual, but also include images (IPK barley). These observed variables were described using references to ontologies (Michotey et al., 2019; Pommier et al., 2019a; Michotey & Chaves, 2020) whenever possible, but ad hoc variables were also used in specific cases not covered by ontologies. One of the most challenging aspects addressed by MIAPPE and successfully demonstrated by the datasets is the documentation of the biological material. The datasets clearly demonstrate how to organise information for model plants (IPK Arabidopsis), mutants (IPK barley), recombinant inbred line and population (VIB maize), GenBank reference accessions (GnpIS wheat) and perennial plants including in situ material (iBET cork oak stands) or dedicated experimental locations acting as experimental tree fields with populations or crosses (GnpIS poplar).
While the datasets showcase the applicability of MIAPPE to diverse experimental settings, they by no means represent the full extent of its coverage. Additional settings that were contemplated in the conception of MIAPPE but are not covered by the examples include: high‐throughput phenotyping facilities with plants manipulated by conveyor belts, which produce large volumes of data with respect to the positions of plants and their development; precision agriculture field studies with drones and sensors capturing a wealth of data both about plant development and the environment; and cases where tracing the identity of plant materials is more complex. In the interest of demonstrating MIAPPE's coverage, Table 4 presents additional examples of settings and details their modelling in MIAPPE.
Table 4.
Modelling possibilities for complicated experiment details.
| No. | Scenario | MIAPPE modelling |
|---|---|---|
| (1) | Heterozygous parent genotypes are used to derive a crossing population exhibiting significant phenotypic segregation. Genotype tracing is necessary. | The cross of the parents is mentioned in the material source. Each of the progeny is treated as a biological material derived from the same material source, and is attributed a unique ID. |
| (2) | Each tree in a field is observed through several sensors, at the roots and near its top. | Observation unit levels: “plant”. Each tree is a single observation unit. Each sensor measures one or many observed variables, (e.g. “Canopy temperature”, “Cork thickness”, …) |
| (3) | A sensor is placed in the middle of the field. | An observation unit is created for the sensor. No plants have to be present for an observation unit to be valid, as long as that observation unit is used to produce measurements or express experimental factor values. |
| (4) | Multilocal, multiyear field phenotyping network |
Observation unit levels: “study”> “genotype”> “plot”. The whole network is an investigation. Each location is a study over several years. The biological material list is shared for the whole investigation. The list of observed variable definitions is also shared by all studies. The measured data and observations can be at the “plant” or “plot” level, or as a per‐genotype average within each study. Study‐level observations can be measurements from a meteorological station. |
| (5) | Time series of event or observation. |
Observation unit levels: any. Study type: any. Observed variables list the time scale they use. In the data file, a single observed variable is measured several times, each value being timestamped in julian days, growing degree days or any other time scale. The same applies with events with a given event type recorded several times at different time stamps. |
The table shows more specific scenarios that may be necessary to accommodate in MIAPPE, and the proposed modelling for them inside the standard.
Discussion
A global metadata standard is a key component for enabling FAIR data in any research domain, by providing a common framework under which researchers can describe their datasets with the necessary information for their interpretation, thus promoting interoperability and reusability. MIAPPE aims at serving such a role for the plant phenotyping community, and the first version of the standard took ample strides in that direction. In this work, we summarise the steps taken to extend and improve the usability of the standard.
The datasets presented in the Results demonstrate the broader applicability of MIAPPE 1.1, which was one of the main goals behind the update. The datasets span a variety of settings (e.g. woody, crop and model plants; glasshouses, single fields and field networks; single‐year and multiannual experiments) and include aspects that could not be modelled under MIAPPE 1.0 (the most critical being the identification of biological materials using geographical coordinates).
MIAPPE 1.1 also has improved in flexibility and usability compared to the previous version. It has clearer definitions, examples, and when applicable, ontology recommendations for all fields. It has an explicit data model available in schematic form and encoded in OWL as PPEO. It has fewer mandatory fields, since not all of them are applicable to all experiments. It allows different strategies for modelling aspects such as environmental parameters: under the environment section, as events or as observed variables. The improved biological material description and the new material source can now handle gene banks and experimental collections with either the bare minimum identification or very detailed information, including infraspecific description, provenance, complex processing or identification mechanism. We received evidence for the improved usability of MIAPPE from the community, during two open requests for feedback and in several training sessions. The specification of the data model and the enriched definitions and examples were highlighted as clear improvements.
While MIAPPE promotes interoperability and reusability, the other two FAIR principles (findability and accessibility) rely mainly on BrAPI, which enables data search and retrieval through machine access. However, for these two resources to be part of a common scheme for enabling FAIR plant phenotyping data, it is necessary to ensure that BrAPI calls adequately cover MIAPPE and enable searches by all key MIAPPE fields. The process of reconciling MIAPPE and BrAPI was undertaken in parallel with the MIAPPE 1.1 update, through a collaboration between the BrAPI team, the ELIXIR Interoperability platform and Plant Sciences community, the EMPHASIS Plant Phenotyping Infrastructure and the CGIAR. BrAPI will be fully MIAPPE 1.1 compliant once its (currently beta) 2.0 release is finalised.
This reconciliation and the interoperability between the various MIAPPE 1.1 implementations is demonstrated in our Results, as most datasets are available in two different implementations (including BrAPI), in many cases through automatic conversion. This is critical, as MIAPPE aims to support a wide range of users and applications, from data submission by life scientists to data exchange, validation and even reasoning by machines. Formats supporting all these applications must not only be available but also be interconvertible.
While, from a technical standpoint, we believe that the merits of MIAPPE 1.1 speak for themselves, we are well aware of the many hurdles ahead of getting any standard widely adopted by the community it seeks to serve. Indeed, there are several dozen standards currently deprecated in the FAIRsharing portal and surely many more have been lost to history.
One of the main factors behind wide adoption is having community engagement throughout the development process. Indeed, GO (The Gene Ontology Consortium, 2019) has been so successful because it emerged from the communities involved in gene function annotation for several model organisms and has remained open to input from the community throughout its history. The story behind MIAPPE is curiously similar, as its development gathered several researchers involved in plant phenotyping repositories, the process of updating it had extensive direct engagement with the community, and it remains open to community input through its GitHub repository (MIAPPE Contributors, 2020a). MIAPPE 1.1 therefore gathers as close to a community‐wide consensus as is possible to get and distil into a clear and well‐organised standard, especially considering the heterogeneity and complexity of the plant phenotyping domain, and the difficulty in reconciling the perspectives of its different subdomains and experiment types. We will further foster its adoption through constant efforts of outreach and dissemination, to gather new communities and ensure the long‐term usefulness of the standard.
Also critical for adoption is demand: when funders and/or publishers require compliance with a standard or data publication practice – such as depositing sequencing data in one of the public gene banks – it tends to be widely adopted. In the case of MIAPPE, the demand consists of the increasing pressure from funding agencies towards compliance with the FAIR data principles. Researchers working in plant phenotyping and seeking FAIR data solutions will be pointed towards MIAPPE thanks to its presence in the FAIRsharing portal (FAIRsharing.org: MIAPPE, 2020) and above all to the endorsement of ELIXIR, which led the MIAPPE 1.1 update and is helping shape the policies and lay the foundations needed for enacting the FAIR principles.
Equally critical is usability, as researchers tend to view the need for standardisation and reusability as a burden and often do as little effort as they can get away with when submitting a dataset, unless the process is virtually effortless. While MIAPPE's usability was improved with the 1.1 update, it is still missing an easy‐to‐use submission interface. For this reason, we are engaging with popular data management tools such as COPO (The COPO team, 2020) or FAIRDOM (Wolstencroft et al., 2017) to incorporate MIAPPE and thus enable user‐friendly MIAPPE‐compliant dataset submission.
Last but not least, in order to persist, a standard must constantly evolve to keep up with technical and scientific advances. In this regard, the 1.1 update demonstrates that MIAPPE is very much a living standard, and we are already starting the next phase of development of MIAPPE. It will concentrate on two main aspects: extending its coverage of environmental aspects (initiated by the EMPHASIS members of the MIAPPE community) and facilitating the recording of technical aspects of material and data processing (e.g. sensors, cameras, software, configurations, calibrations), which are becoming increasingly important. Within this scope, another possible improvement could be to establish a formal complementarity with the ICASA standard, which is used by the agronomic and modelling communities, and provides variables for agronomic management practices, treatments, environmental conditions and measurements of crop responses (White et al., 2013). There is overlap between these two standards – with MIAPPE dedicated to plant phenotyping as used by geneticists, biologists and some agronomists and ICASA to ‘any field experiment or crop production situation’ – but the scope of each extends far beyond that of the other and there are obvious complementarities between them. User feedback from these endeavours will steer further developments, by revealing areas where improvement is desired by the community.
Author contributions
EAP, DF, DA, EA, INA, IC, FC, GC, BVC, HC‐K, BD, RF, KG, AJ, GJK, PK, ML, M‐AL, CM, MO, RO, HP, RR‐G, ŽR, JCR, PR‐S, S‐AS, US, FT, CU, BU, RGFV, SW, PJK, CMM, A‐FA‐B and CP contributed to the development of MIAPPE 1.1, participating in the discussions and responding to the public request for comments. A‐FA‐B, BD, CMM, CP, DA, DF, EAP, FC, GC, HC‐K, IC and PR‐S contributed to the MIAPPE implementations. A‐FA‐B, AJ, BVC, BD, CM, CMM, CP, DA, FC, IC, ML and US contributed to the evaluation of the new version of MIAPPE. EAP coordinated the drafting of the manuscript, with contributions from A‐FAB, CMM, CP, DF and IC. A‐FA‐B, CMM, CP and PJK coordinated the research. All authors read, provided feedback and approved the final manuscript.
Supporting information
Notes S1 Summaries of the datasets used to evaluate MIAPPE 1.1.
Table S1 Detailed mapping between MIAPPE, ISA‐Tab and BrAPI fields.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was based on extensive reviews from and interactions with the broader MIAPPE community. We are grateful for all feedback. This work was supported by the European Union's Horizon 2020 programme through the ELIXIR‐EXCELERATE project, Grant Agreement no. 676559, the EMPHASIS‐Prep project, Grant Agreement no. 739514, and the research infrastructure for life science data ELIXIR. It was also funded by: the National Research Institute for Agriculture, Food and Environment (INRAE); the French Infrastructure en Biologie Santé ‘Phenome‐FPPN’ supported by the French National Research Agency (ANR‐11‐INBS‐0012); the Portuguese Fundação para a Ciência e Tecnologia through project BioData.pt (Grant no. 22231, co‐financed by FEDER); the German‐Plant‐Phenotyping Network (DPPN) and the German Network for Bioinformatics Infrastructure (de.NBI), both funded by the German Federal Ministry of Education and Research (BMBF) (project identification number: 031A053, 031A536A,C). Philippe Rocca‐Serra and Susanna‐Assunta Sansone are funded by UK Research Councils (BB/L024101/1, BB/L005069/1), Wellcome Trust (212930/Z/18/Z, 208381/A/17/Z), European Union (H2020‐EU.3.1, 634107, H2020‐EU.1.4.1.3, 654241, H2020‐EU.1.4.1.1, 676559), IMI (116060) and NIH Data Common Fund. In‐kind contribution was made by the Crop Ontology project team supported by the Integrated Breeding Platform, by Planteome (NSF award IOS:1340112), and by the CGIAR Platform on Big Data for Agriculture. Some developments were supported by the RDA Europe project funded by the European Commission. Inês Chaves acknowledges DL 57/2016/CP1351/CT0003. Paweł Krajewski and Hanna Ćwiek‐Kupczyńska acknowledge funding from EPPN2020 grant agreement no. 731013 and National Science Centre grant 2016/21/N/ST6/02358. Evangelia A. Papoutsoglou acknowledges a grant from WUR Plant Breeding. The authors declare that they have no conflicts of interest.
Contributor Information
Evangelia A. Papoutsoglou, Email: evangelia.papoutsoglou@wur.nl.
Cyril Pommier, Email: cyril.pommier@inrae.fr.
References
- Alercia A, Diulgheroff S, Mackay M. 2015. FAO/Bioversity multi‐crop passport descriptors v.2.1 [MCPD V.2.1] ‐ December 2015. Bioversity International 11 p. [WWW document] URL https://hdl.handle.net/10568/69166 [accessed 10 February 2020]. [Google Scholar]
- Arnaud E, Cooper L, Shrestha R, Menda N, Nelson RT, Matteis L, Skofic M, Bastow R, Jaiswal P, Mueller LA et al 2012. Towards a reference plant trait ontology for modeling knowledge of plant traits and phenotypes. Proceedings of the International Conference on Knowledge Engineering and Ontology Development 2012: 220–225. [Google Scholar]
- Aubert C, Buttigieg PL, Laporte M‐A, Devare M, Arnaud E. 2017. CGIAR agronomy ontology. [WWW document] URL http://purl.obolibrary.org/obo/agro.owl [accessed 10 February 2020]. Licensed under CC‐BY‐4.0. [Google Scholar]
- Baute J, De Block J, Inzé D. 2019a. Zea mays biparental RIL population ‐ growth chamber phenotyping data [Data set]. Zenodo. doi: 10.5281/zenodo.3553692. [DOI] [Google Scholar]
- Baute J, De Block J, Inzé D. 2019b. Zea mays MAGIC RIL population ‐ growth chamber phenotyping data [Data set]. [WWW document] URL doi: 10.5281/zenodo.3553768. [DOI] [Google Scholar]
- Baute J, De Block J, Inzé D. 2019c. Zea mays MAGIC RIL population ‐ growth chamber phenotyping data (PIPPA BrAPI endpoint) [Data set]. [WWW document] URL https://pippa.psb.ugent.be/BrAPIPPA/brapi/v1/trials/2 [accessed 10 February 2020]. [Google Scholar]
- Baute J, De Block J, Inzé D. 2020. Zea mays biparental RIL population ‐ growth chamber phenotyping data (PIPPA BrAPI endpoint) [Data set]. [WWW document] URL https://pippa.psb.ugent.be/BrAPIPPA/brapi/v1/trials/1 [accessed 10 February 2020]. [Google Scholar]
- Baute J, Herman D, Coppens F, De Block J, Slabbinck B, Dell’Acqua M, Pè ME, Maere S, Nelissen H, Inzé D. 2015. Correlation analysis of the transcriptome of growing leaves with mature leaf parameters in a maize RIL population. Genome Biology 16: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baute J, Herman D, Coppens F, De Block J, Slabbinck B, Dell'Acqua M, Pè ME, Maere S, Nelissen H, Inzé D. 2016. Combined large‐scale phenotyping and transcriptomics in maize reveals a robust growth regulatory network. Plant Physiology 170: 1848–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch‐Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Research 41: D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BrAPI2ISA Contributors . 2020. BrAPI2ISA. [WWW document] URL https://github.com/elixir‐europe/plant‐brapi‐to‐isa [accessed 10 February 2020]. [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC et al 2001. Minimum information about a microarray experiment (MIAME) – toward standards for microarray data. Nature Genetics 29: 365–371. [DOI] [PubMed] [Google Scholar]
- BreedBase Team . 2020. BreedBase. [WWW document] URL https://breedbase.org/ [accessed 10 February 2020]. [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL et al 2009. The MIQE guidelines: Minimum information for publication of quantitative real‐time PCR experiments. Clinical Chemistry 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Buttigieg PL, Morrison N, Smith B, Mungall CJ, Lewis SE. 2013. The environment ontology: contextualising biological and biomedical entities. Journal of Biomedical Semantics 4: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Miguel CM, Faria D, Costa BV. 2019a. Enabling reusability of plant phenomic datasets with MIAPPE 1.1 ‐ supplementary dataset iBET [Data set] . Portail Data INRAE. doi: 10.15454/AH6U4A. [DOI]
- Chaves I, Miguel CM, Faria D, Costa BV. 2019b. Enabling reusability of plant phenomic datasets with MIAPPE 1.1 ‐ supplementary dataset iBET (PHENO BrAPI endpoint) [Data set]. [WWW document] URL https://brapi.biodata.pt/brapi/v1/trials/2 [accessed 10 February 2020]. [Google Scholar]
- Cooper L, Meier A, Laporte M‐A, Elser JL, Mungall C, Sinn BT, Cavaliere D, Carbon S, Dunn NA, Smith B et al 2018. The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics. Nucleic Acids Research 46: D1168–D1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot M, Cherubin L, Faulconbridge A, Vaughan D, Green M, Richardson D, Harrison P, Whetzel PL, Parkinson H, Burdett T. 2019. BioSamples database: an updated sample metadata hub. Nucleic Acids Research 47: D1172–D1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ćwiek‐Kupczyńska H. 2018. Striving for semantics of plant phenotyping data. Semantics, Analytics, Visualization SAVE‐SD 2017, SAVE‐SD 2018: 161–169.
- Ćwiek‐Kupczyńska H, Altmann T, Arend D, Arnaud E, Chen D, Cornut G, Fiorani F, Frohmberg W, Junker A, Klukas C et al 2016. Measures for interoperability of phenotypic data: minimum information requirements and formatting. Plant Methods 12: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DataCite Metadata Working Group . 2014. DataCite metadata schema documentation for the publication and citation of research data. DataCite e.V. Version 3.1. doi: 10.5438/0010. [DOI]
- Dell'Acqua M, Gatti DM, Pea G, Cattonaro F, Coppens F, Magris G, Hlaing AL, Aung HH, Nelissen H, Baute J et al 2015. Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays . Genome Biology 16: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAIRsharing.org: MIAPPE . 2020. Minimum information about plant phenotyping experiment. [WWW document] doi: 10.25504/FAIRsharing.nd9ce9 [accessed 10 February 2020]. [DOI] [Google Scholar]
- Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV et al 2008. The minimum information about a genome sequence MIGS specification. Nature Biotechnology 26: 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado‐Lopez P, Tessema B, Schnabel S, Maliepaard C, Van der Linden C, Eilers P, Jansen J, van Eeuwijk F, Visser R. 2015. Understanding the genetic basis of potato development using a multi‐trait QTL analysis. Euphytica 204: 229–241. [Google Scholar]
- Inácio V, Barros PM, Costa A, Roussado C, Gonçalves E, Costa R, Graça J, Oliveira MM, Morais‐Cecílio L. 2017. Differential DNA methylation patterns are related to phellogen origin and quality of Quercus suber cork. PLoS ONE 12: e0169018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISA‐Tab for Plant Phenotyping Contributors . 2020. ISA‐Tab for plant phenotyping. [WWW document] URL https://github.com/MIAPPE/ISA‐Tab‐for‐plant‐phenotyping [accessed 10 February 2020]. [Google Scholar]
- Jaiswal P, Avraham S, Ilic K, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M et al 2005. Plant ontology (PO): a controlled vocabulary of plant structures and growth stages. Comparative and Functional Genomics 6: 388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonquet C, Toulet A, Arnaud E, Aubin S, Yeumo ED, Emonet V, Graybeal J, Laporte M‐A, Musen MA, Pesce V et al 2018. AgroPortal: a vocabulary and ontology repository for agronomy. Computers and Electronics in Agriculture 144: 126–143. [Google Scholar]
- Junker A.2020. Raw images files from quantitative monitoring of 484 Arabidopsis thaliana plants using high‐throughput plant phenotyping (MIAPPE 1.1 update) [Data set]. e!DAL ‐ Plant Genomics and Phenomics Research Data Repository (PGP), IPK Gatersleben, Seeland OT Gatersleben, Germany. doi: 10.5447/IPK/2020/3. [DOI]
- Junker A, Li M.2020. Phenotypic assessment of growth and coloration dynamics as well as photosynthetic efficiency parameters in barley HvASL mutants and wild type plants [Data set]. e!DAL ‐ Plant Genomics and Phenomics Research Data Repository (PGP), IPK Gatersleben, Seeland OT Gatersleben, Germany. doi: 10.5447/IPK/2020/4. [DOI]
- Junker A, Muraya MM, Weigelt‐Fischer K, Arana‐Ceballos F, Klukas C, Melchinger AE, Meyer RC, Riewe D, Altmann T. 2015. Optimizing experimental procedures for quantitative evaluation of crop plant performance in high throughput phenotyping systems. Frontiers in Plant Science 5: 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A, Weigelt‐Fischer K, Altmann T, Klukas C. 2020. Raw images files from quantitative monitoring of 484 Arabidopsis thaliana plants using high‐throughput plant phenotyping [Data set]. e!DAL ‐ Plant Genomics and Phenomics Research Data Repository (PGP), IPK Gatersleben, Seeland OT Gatersleben, Corrensstraße 3, 06466, Germany. doi: 10.5447/IPK/2016/7. [DOI]
- King GJ, Amoah S, Kurup S. 2010. Exploring and exploiting epigenetic variation in crops. Genome 53: 856–868. [DOI] [PubMed] [Google Scholar]
- Krajewski P, Chen D, Ćwiek H, van Dijk AD, Fiorani F, Kersey P, Klukas C, Lange M, Markiewicz A, Nap JP et al 2015. Towards recommendations for metadata and data handling in plant phenotyping. Journal of Experimental Botany 66: 5417–5427. [DOI] [PubMed] [Google Scholar]
- Krajewski P, Ćwiek‐Kupczyńska H. 2020. MIAPPE impact. [WWW document] URL https://www.miappe.org/publications/#impact [accessed 10 February 2020]. [Google Scholar]
- Lapatas V, Stefanidakis M, Jimenez RC, Via A, Schneider MV. 2015. Data integration in biological research: an overview. Journal of Biological Research‐Thessaloniki 22: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hensel G, Melzer M, Junker A, Tschiersch H, Arend D, Kumlehn J, Börner T, Stein N. 2019. Mutation of the ALBOSTRIANS ohnologous gene HvCMF3 impairs chloroplast development and thylakoid architecture in barley due to reduced plastid translation. bioRxiv. doi: 10.1101/756833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIAPPE Contributors . 2020a. MIAPPE Github repository. [WWW document] URL https://github.com/MIAPPE/MIAPPE [accessed 10 February 2020]. [Google Scholar]
- MIAPPE Contributors . 2020b. MIAPPE v1.1 training spreadsheet. [WWW document] URL https://github.com/MIAPPE/MIAPPE/tree/master/MIAPPE_Checklist‐Data‐Model‐v1.1/MIAPPE_templates [accessed 10 February 2020]. [Google Scholar]
- Michotey C. 2019. Integrating genome annotation and QTL position to identify candidate genes for productivity, architecture and water‐use efficiency in Populus spp ‐ supplementary dataset [Data set]. Portail Data INRAE. doi: 10.15454/EASUQV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michotey C. 2020. Integrating genome annotation and QTL position to identify candidate genes for productivity, architecture and water‐use efficiency in Populus spp ‐ Supplementary dataset (GnpIS BrAPI endpoint). [WWW document] URL https://urgi.versailles.inra.fr/faidare/brapi/v1/trials/aHR0cDovL2R4LmRvaS5vcmcvMTAuMTE4Ni8xNDcxLTIyMjktMTItMTcz [accessed 10 February 2020]. [Google Scholar]
- Michotey C, Chaves I. 2020. Woody plant ontology. [WWW document] URL http://www.cropontology.org/ontology/CO_357/Woody%20Plant%20Ontology [accessed 10 February 2020]. [Google Scholar]
- Michotey C, Chaves I, Anger C, Jorge V, Ehrenmann F, Jean F, Opgenoorth L. 2019. Woody plant ontology. Portail Data INRAE, V1. doi: 10.15454/JB2WCE [DOI] [Google Scholar]
- Millet EJ, Kruijer W, Coupel‐Ledru A, Prado SA, Cabrera‐Bosquet L, Lacube S, Charcosset A, Welcker C, van Eeuwijk F, Tardieu F. 2019. Genomic prediction of maize yield across European environmental conditions. Nature Genetics 51: 952–956. [DOI] [PubMed] [Google Scholar]
- Monclus R, Leplé J‐C, Bastien C, Bert P‐F, Villar M, Marron N, Brignolas F, Jorge V. 2012. Integrating genome annotation and QTL position to identify candidate genes for productivity, architecture and water‐use efficiency in Populus spp. BMC Plant Biology 12: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu P, Tireau A, Hilgert N, Nègre V, Mineau‐Cesari J, Brichet N, Chapuis R, Sanchez I, Pommier C, Charnomordic B et al 2019. Dealing with multi‐source and multi‐scale information in plant phenomics: the ontology‐driven Phenotyping Hybrid Information System. New Phytologist 221: 588–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, Jonquet C, Rubin DL, Storey M‐A, Chute CG et al 2009. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research 37: W170–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oury F‐X, Heumez E, Rolland B, Auzanneau J, Bérard P, Brancourt‐Hulmel M, Charrier X, Chiron H, Depatureaux C, Falchetto L et al 2018. Winter wheat (Triticum aestivum L) phenotypic data from the multiannual, multilocal field trials of the INRA Small Grain Cereals Network. Portail Data INRAE, V5. doi: 10.15454/1.4489666216568333E12. [DOI] [Google Scholar]
- Oury F‐X, Heumez E, Rolland B, Auzanneau J, Bérard P, Brancourt‐Hulmel M, Charrier X, Chiron H, Depatureaux C, Falchetto L et al 2020. Winter wheat (Triticum aestivum L) phenotypic data from the multiannual, multilocal field trials of the INRA Small Grain Cereals Network. [WWW document] URL https://urgi.versailles.inra.fr/ephesis/ephesis/viewer.do#dataResults/trialSetIds=8 [accessed 10 February 2020]. [Google Scholar]
- Oury F‐X, Pommier C, Charmet G. 2019a. Enabling reusability of plant phenomic datasets with MIAPPE 1.1 ‐ supplementary dataset [Data set]. Portail Data INRAE. doi: 10.15454/1AFKZ2. [DOI] [Google Scholar]
- Oury F‐X, Pommier C, Charmet G. 2019b. Enabling reusability of plant phenomic datasets with MIAPPE 1.1 ‐ supplementary dataset INRA Wheat (GnpIS BrAPI endpoint) [Data set]. [WWW document] URL https://urgi.versailles.inra.fr/faidare/brapi/v1/trials/dXJuOlVSR0kvdHJpYWwvNw== [accessed 10 February 2020]. [Google Scholar]
- Papoutsoglou EA, Faria D, Arend D, Arnaud E, Athanasiadis IN, Chaves I, Coppens F, Cornut G, Costa BV, Ćwiek‐Kupczyńska H et al 2019. Enabling reusability of plant phenomic datasets with MIAPPE 1.1 ‐ supplementary dataset. Portail Data INRAE. doi: 10.15454/1YXVZV [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pea G, Hlaing AL, Aung HH, Pè ME. 2019a. Zea mays MAGIC RIL population ‐ field trial phenotyping data [Data set]. Zenodo. doi: 10.5281/zenodo.3553749. [DOI] [Google Scholar]
- Pea G, Hlaing AL, Aung HH, Pè ME. 2019b. Zea mays MAGIC RIL population ‐ field trial phenotyping data (PIPPA BrAPI endpoint) [Data set]. [WWW document] URL https://pippa.psb.ugent.be/BrAPIPPA/brapi/v1/trials/3 [accessed 10 February 2020]. [Google Scholar]
- PIPPA Team . 2020. PIPPA ‐ PSB interface for plant phenotype analysis. [WWW document] URL https://pippa.psb.ugent.be/ [accessed 10 February 2020]. [Google Scholar]
- Pommier C, Ćwiek‐Kupczyńska H, Faria D, Papoutsoglou E, Bruno CBV, Laporte M‐A, Chaves I, Neveu P, Cornut G, Ruiz M et al 2020. Plant phenotype experiment ontology (PPEO). [WWW document] URL http://purl.org/ppeo [accessed 10 February 2020]. [Google Scholar]
- Pommier C, Letellier T, Pietragalla J, Laporte MA, Arnaud E, Le Gouis J, Shrestha R. 2019a. Wheat crop ontology. Portail Data INRAE, V1. doi: 10.15454/3EDMCP [DOI] [Google Scholar]
- Pommier C, Michotey C, Cornut G, Roumet P, Duchêne E, Flores R, Lebreton A, Alaux M, Durand S, Kimmel E et al 2019b. Applying FAIR principles to plant phenotypic data management in GnpIS. Plant Phenomics 2019: 1671403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner TF, Rocca‐Serra P, Spellman PT, Causton HC, Farne A, Holloway E, Irizarry RA, Liu J, Maier DS, Miller M et al 2006. A simple spreadsheet‐based, MIAME‐supportive format for microarray data: MAGE‐TAB. BMC Bioinformatics 7: 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca‐Serra P, Brandizi M, Maguire E, Sklyar N, Taylor C, Begley K, Field D, Harris S, Hide W, Hofmann O et al 2010. ISA software suite: supporting standards‐compliant experimental annotation and enabling curation at the community level. Bioinformatics 26: 2354–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone S‐A, McQuilton P, Rocca‐Serra P, Gonzalez‐Beltran A, Izzo M, Lister AL, Thurston M. 2019. FAIRsharing as a community approach to standards, repositories and policies. Nature Biotechnology 37: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone S‐A, Rocca‐Serra P, Field D, Maguire E, Taylor C, Hofmann O, Fang H, Neumann S, Tong W, Amaral‐Zettler L et al 2012. Toward interoperable bioscience data. Nature Genetics 44: 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby P, Abbeloos R, Backlund JE, Basterrechea Salido M, Bauchet G, Benites‐Alfaro OE, Birkett C, Calaminos VC, Carceller P, Cornut G et al 2019. BrAPI – an application programming interface for plant breeding applications. Bioinformatics 35: 4147–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R. 2020. IBP crop research ontology CO_715. [WWW document] URL www.cropontology.org/ontology/CO_715/Crop%20Research [accessed 10 February 2020]. [Google Scholar]
- Shrestha R, Matteis L, Skofic M, Portugal A, McLaren G, Hyman G, Arnaud E. 2012. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Frontiers in Physiology 3: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindel JE, McCouch SR. 2016. When more is better: how data sharing would accelerate genomic selection of crop plants. New Phytologist 212: 814–826. [DOI] [PubMed] [Google Scholar]
- Statistics Ontology Project . 2020. Statistics Ontology (STATO). [WWW document] URL http://stato‐ontology.org/ [accessed 10 February 2020]. [Google Scholar]
- Steinbach D, Alaux M, Amselem J, Choisne N, Durand S, Flores R, Keliet A‐O, Kimmel E, Lapalu N, Luyten I et al 2013. GnpIS: an information system to integrate genetic and genomic data from plants and fungi. Database 2013: bat058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Cabrera‐Bosquet L, Pridmore T, Bennett M. 2017. Plant phenomics, from sensors to knowledge. Current Biology 27: R770–R783. [DOI] [PubMed] [Google Scholar]
- Tenopir C, Allard S, Douglass K, Aydinoglu AU, Wu L, Read E, Manoff M, Frame M. 2011. Data sharing by scientists: practices and perceptions. PLoS ONE 6: e21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The COPO team . 2020. COPO – collaborative open plant omics. [WWW document] URL https://copo‐project.org/ [accessed 10 February 2020]. [Google Scholar]
- The Gene Ontology Consortium . 2019. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Research 47: D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ISA Team . 2020. ISA commons. [WWW document] URL https://www.isacommons.org/ [accessed 10 February 2020]. [Google Scholar]
- The UniProt Consortium . 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research 47: D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vines TH, Albert AY, Andrew RL, Débarre F, Bock DG, Franklin MT, Gilbert KJ, Moore J‐S, Renaut S, Rennison DJ. 2014. The availability of research data declines rapidly with article age. Current Biology 24: 94–97. [DOI] [PubMed] [Google Scholar]
- W3C . 2020a. RDF 1.1 concepts and abstract syntax. [WWW document] URL https://www.w3.org/TR/rdf11‐concepts/ [accessed 10 February 2020]. [Google Scholar]
- W3C . 2020b. JSON‐LD 1.0. [WWW document] URL https://www.w3.org/TR/json‐ld/ [accessed 10 February 2020]. [Google Scholar]
- White JW, Hunt L, Boote KJ, Jones JW, Koo J, Kim S, Porter CH, Wilkens PW, Hoogenboom G. 2013. Integrated description of agricultural field experiments and production: the ICASA Version 2.0 data standards. Computers and Electronics in Agriculture 96: 1–12. [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J‐W, da Silva Santos LB, Bourne PE et al 2016. The FAIR guiding principles for scientific data management and stewardship. Scientific Data 3: 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstencroft K, Krebs O, Snoep JL, Stanford NJ, Bacall F, Golebiewski M, Kuzyakiv R, Nguyen Q, Owen S, Soiland‐Reyes S et al 2017. FAIRDOMHub: a repository and collaboration environment for sharing systems biology research. Nucleic Acids Research 45: D404–D407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir D. 2013. Where have all the crop phenotypes gone? PLoS Biology 11: e1001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Notes S1 Summaries of the datasets used to evaluate MIAPPE 1.1.
Table S1 Detailed mapping between MIAPPE, ISA‐Tab and BrAPI fields.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


