Abstract
Plasmodiophora brassicae (Wor.) is an obligate intracellular plant pathogen affecting Brassicas worldwide. Identification of effector proteins is key to understanding the interaction between P. brassicae and its susceptible host plants. To date, there is very little information available on putative effector proteins secreted by P. brassicae during a secondary infection of susceptible host plants, resulting in root gall production. A bioinformatics pipeline approach to RNA‐Seq data from Arabidopsis thaliana (L.) Heynh. root tissues at 17, 20, and 24 d postinoculation (dpi) identified 32 small secreted P. brassicae proteins (SSPbPs) that were highly expressed over this secondary infection time frame. Functional signal peptides were confirmed for 31 of the SSPbPs, supporting the accuracy of the pipeline designed to identify secreted proteins. Expression profiles at 0, 2, 5, 7, 14, 21, and 28 dpi verified the involvement of some of the SSPbPs in secondary infection. For seven of the SSPbPs, a functional domain was identified using Blast2GO and 3D structure analysis and domain functionality was confirmed for SSPbP22, a kinase localized to the cytoplasm and nucleus.
Keywords: Brassica, clubroot, effectors, galls, kinase, RNA‐Seq
THE Rhizarian protist Plasmodiophora brassicae (Wor.), a soil‐borne pathogen of the Order Plasmodiophorales, is responsible for clubroot, one of the most devastating diseases affecting Brassica plants, with billion‐dollar losses worldwide (Burki et al. 2010; Dixon 2009). As an obligate biotrophic intracellular pathogen, P. brassicae depends on its host for propagation and only resting spores or short‐lived zoospores occur outside of plant tissues (Kageyama and Asano 2009). The life cycle of P. brassicae starts in the soil, where resting spores, which can remain viable for up to 20 yr (Hwang et al. 2012), germinate in response to the presence of plant hosts. During the secondary infection, susceptible plants develop galls that disrupt water and nutrient uptake, leading to wilting, stunting, and, in some instances, death of the infected plant (Dixon 2009).
Many details of the P. brassicae life cycle and the P. brassicae–Brassica host interaction are still unknown. In order to manipulate plant defenses and enable parasitic colonization, many eukaryotic biotrophic plant pathogens have evolved a repertoire of effector proteins that in some cases are responsible for the suppression of plant immunity or changes in plant morphology that can increase infection success (Dodds and Rathjen 2010; Jones and Dangl 2006; Sugio et al. 2011). The concept of an effector protein is very broad, but basically it can be considered as any small secreted protein that facilitates the pathogen infection process (Dodds and Rathjen 2010). Conserved pathogen‐associated molecular patterns (PAMPs) induce PAMP‐triggered immunity in a host (Dodds and Rathjen 2010). This is the first level of a host immune response that can be overcome by effector proteins produced by adapted pathogens (Dodds and Rathjen 2010).
Putative effectors have been identified in the genomes or transcriptomes of biotrophic plant pathogens, such as oomycetes and fungi, based on the presence of putative small secreted proteins and the expression profiles of these candidates (Duplessis et al. 2011; Hacquard et al. 2012). A similar strategy has been followed to identify putative secreted proteins in the genome of P. brassicae (Chen et al. 2017; Rolfe et al. 2016; Schwelm et al. 2015); however, none of these studies have been performed in the model plant Arabidopsis thaliana or have focused on describing the effector proteins involved during P. brassicae secondary infection and gall formation.
In this study, we used RNA‐Seq analysis to identify genes encoding small secreted P. brassicae proteins (SSPbPs) that were up‐regulated during infection of the model plant A. thaliana (L.) Heynh. at 17, 20, and 24 d postinoculation (dpi). We have identified and characterized 32 putative SSPbPs with a high probability of important roles in the P. brassicae infection process and subsequent clubroot development and progress.
Materials and Methods
Biological material and pathogen inoculation
Plasmodiophora brassicae pathotype P3 (Pb3) (Strelkov et al. 2006) was obtained from Dr. Gary Peng (AAFC—Saskatoon Research Centre), and Arabidopsis thaliana ecotype Columbia (Col.0, susceptible to clubroot disease) seeds were used in this study. Canola (Brassica napus, cultivar Westar) plants were used to generate resting spores. Canola plants, at the fourth leaf stage, were inoculated, and at 35 dpi fresh galls were collected. Galls were ground with a sterilized mortar and pestle in 5 ml 10% sucrose, and the liquid suspension was passed through eight‐layered cheesecloth. The filtered suspension was centrifuged at 10 g for 1 min to further remove root tissue debris. Supernatant was transferred to a fresh sterile tube and centrifuged at 1,900 g for 10 min to pellet the resting spores. The pellet was resuspended in 500 µl ddH2O, and spore density was determined using a haemocytometer.
Arabidopsis seeds were germinated in Sunshine Mix #3 soil (Sun Gro Horticulture Inc., Vancouver, BC, Canada) in a growth chamber (Conviron E8, CMP6050 control system) under a 16/8‐h light/dark cycle, 100 μmol (photons)/m2/s, and a constant 22 °C. Uniform 10‐d‐old seedlings were transferred to Sunshine Mix #4 soil (Sun Gro Horticulture Inc.) with four plants in each “square pot” of an 18‐pot sheet. Seedlings were allowed to acclimate in these pots for four additional days before 400 μl of 5 × 107 resting spores/ml was applied to each plant where the stem entered the soil. Control plants were inoculated with 400 μl of distilled water and grown in separate trays in the same growth chamber. For each time period, six pots (24 plants) were sampled. Plants were treated in blocks and sampled randomly, and each experiment was replicated three times. Pots were watered as required and fertilized once per week with 300 mg/L 20‐20‐20 Plant‐Prod® fertilizer (Master Plant‐Prod Inc., Brampton, ON, Canada).
Disease assessment
Disease index (DI) was determined for infected plants at the three time points, 17, 20, and 24 dpi as previously described by Siemens et al. (2002) to replicate the conditions used for the RNA‐Seq library generation. Root infection was assessed on a 0–4 scale: 0, no symptoms; 1, small galls mainly on lateral roots; 2, small galls on the main and lateral roots; 3, medium‐sized galls with possible negative effect on plant growth; and 4, severe galls on both main and lateral roots, deformed roots, and impaired growth. Three independent replicates of 24 plants were used to determine the DI of infected plants.
RNA isolation
To confirm the RNA‐Seq results, total RNA was extracted from infected and control root tissues at 17, 20, and 24 dpi using a TRIzol‐based extraction method (Chomczynski and Mackey 1995). To determine whether putative effector protein genes were expressed during primary infection (before 14 dpi), RNA was extracted from infected and mock‐infected Arabidopsis plants at 0, 2, 5, 7, 14, 21, and 28 dpi as described above.
RNA‐Seq data analysis
The raw RNA‐Seq library data were produced prior to this study (Irani et al. 2018). Reads were trimmed with Trimmomatic ver. 0.36. (Bolger et al. 2014), with minimum quality score 15, before alignment to the A. thaliana and P. brassicae Pbe3.h15 genomes using STAR 2.5 (Dobin and Gingeras 2015). Reads that did not map to Arabidopsis were used as queries in BlastX against the P. brassicae genome. Plasmodiophora brassicae sequences aligned against reads with an E‐value lower than 0.05 were selected for further analysis, alongside the sequences corresponding to the reads that directly mapped to the P. brassicae genome. Differentially expressed P. brassicae protein‐encoding genes with log2 fold changes ≥ 1 (Bergemann and Wilson 2011) were analyzed using SignalP v4.1 and TargetP v1.1 software (Emanuelsson et al. 2000; Peterson et al. 2011) to identify the differentially expressed P. brassicae secretome (Fig. 1). Predicted differentially expressed secreted proteins lacking a transmembrane domain, having a signal peptide prediction score ≥ 0.7, and a length of < 400 amino acids were analyzed using Blast2GO (Conesa et al. 2005), and we manually looked for cysteine residue number, RxLR motif, Pexel motif, and any functional motif present in the P. brassicae small secreted proteins as previously suggested (Pérez‐López et al. 2018). These proteins were further analyzed with ApoplastP (Sperschneider et al. 2017a) and Localizer (Sperschneider et al. 2017b). Heat maps were generated using the gplots package in R (Warnes et al. 2016).
Figure 1.
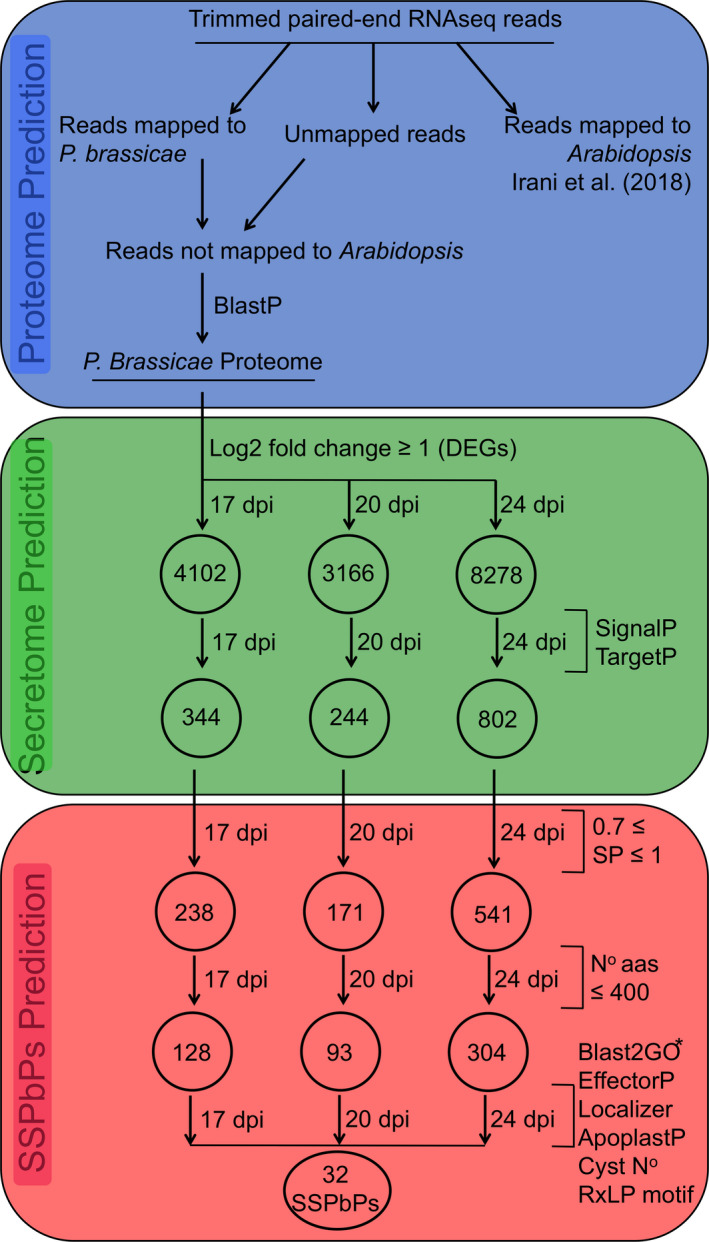
Bioinformatic pipeline for the identification of candidate small secreted Plasmodiophora brassicae proteins (SSPbPs) differentially expressed during secondary infection of Arabidopsis. The three main steps of the pipeline are as follows: in blue, P. brassicae transcribed proteome prediction; in green, differentially expressed transcriptome prediction; and in red, SSPbP prediction.
Validation of RNA‐Seq data and expression profile
To confirm RNA‐Seq results, qRT‐PCR was performed for 24 of the SSPbP‐encoding genes identified in this study. Total RNA extracted from infected and control plants at 17, 20, and 24 dpi was quantified with the Qubit™ RNA BR Assay Kit (Thermo Fisher Scientific, Mississauga, ON, Canada), and 2 μg of total RNA was used to synthesize cDNA using the QuantiTect® Reverse Transcription Kit (Qiagen, Toronto, ON, Canada) following the manufacturer's recommendations. Real‐time qPCR was performed using SsoAdvanced™ Universal SYBR® Green Supermix (Bio‐Rad, Mississauga, ON, Canada) in a 20 μl final volume containing 300 nM of each primer and 2 μl of cDNA diluted 1:5 (v:v) in RNase‐free water. Amplification was carried out using a C1000 thermocycler base with a CFX96 Real‐Time System (Bio‐Rad), and reactions were quantified using Bio‐Rad CFX Manager software (v.3.1). Expression of the ELONGATION FACTOR‐LIKE (PbEFL) gene (SRX027224) was also determined (Burki et al. 2010), and transcript levels of P. brassicae genes were normalized against ACTIN2 (At3g18780) (Irani et al. 2018). Relative transcript levels were calculated using the 2−ΔΔCt method (Livak 1997; Livak and Schmittgen 2001). The primer list is presented in Table S2.
cDNAs for root tissue at 0, 2, 5, 7, 14, 21, and 28 dpi and the corresponding controls were diluted 1:80 (v:v) in RNase‐free water and used to detect the starting point of expression for 21 of the genes encoding putative effectors. PCR was performed using DreamTaq Green Master Mix (Thermo Scientific) in a 25 μl final volume containing 1 μM of each primer and 1 μl of diluted cDNA. Amplification was carried out using a SimpliAmp™ Thermal Cycler (Thermo Scientific), with an annealing temperature of 55 °C and 35 cycles. The cDNAs for the seven time points were also used to monitor expression of the Arabidopsis ELONGATION FACTOR gene (AtEF, AT2G18720) and the P. brassicae ELONGATION FACTOR‐LIKE gene (PbEFL, SRX027224) as controls.
Structural analysis of SSPbPs
Model structures for SSPbPs with putative functions were created using SWISS‐MODEL (Arnold et al. 2006; Kiefer et al. 2009), based on crystal structures in the PDB database with the highest structural similarity to each SSPbP. A model structure was also created for the closest related protein sequence identified through BlastP (protein–protein BLAST®). The models of SSPbP and its putative paralog were aligned using PyMOL (De Lano 2002).
Transmission electron microscopic analysis
For microscopic analysis, infected Arabidopsis roots were sampled at 20 and 24 dpi. Tissues were fixed overnight in a fresh solution of 2% glutaraldehyde at room temperature, washed three times in freshly prepared 0.1 M phosphate buffer pH 7.4 (PB) for 30 min, and postfixed in 1% osmium tetroxide (Sigma‐Aldrich, Oakville, ON, Canada) for 4 h with a final step of three washes with distilled water for 30 min each time. Tissues were dehydrated in a gradient of ethanol from 25% to 100% before incubation in propylene oxide (Alfa Aesar, Haverhill, MA) for 8 h. Dehydrated tissues were embedded in a 100% resin mold and incubated at 60 °C for polymerization. The mold was used to generate ultra‐thin sections (60 nm) with a Microstar diamond knife (Huntsville, TX) on a Reichert‐Jung microtome (Reichert microscopic service; Depew, NY) and captured onto single‐slot copper grids coated with Formvar or 100 naked mesh copper grids. Ultra‐thin sections were stained with 2% uranyl acetate (EMS) for 30 min in the dark, followed by Reynold's lead citrate solution for 10 min and observed with a Hitachi HT7700 Transmission Electron Microscope (TEM).
Signal peptide validation assay
For standard cloning, the terminal ends of the amplified DNA fragments coding the signal peptides of the 32 SSPbPs were digested by restriction enzymes and ligated with the vector using T4 DNA ligase (Fermentas Life Science, Burlington, ON, Canada). To validate the SSPbPXX signal peptide (24 aa), the 72 bp fragment was amplified by PCR using as template the cDNA obtained from infected Arabidopsis plants with designated primer pairs (Table S2). As a positive control, the LOW‐MOLECULAR‐WEIGHT CYSTEINE‐RICH 78 (LCR78, At1g19610) signal peptide was amplified (28 aa, 84 bp) by PCR using as template the cDNA obtained from uninfected Arabidopsis plants with designated primer pairs (Table S2). The amplified signal peptide fragments were introduced into pSUC2 (Prof. Sophien Kamoun, The Sainsbury Laboratory) linearized by EcoRI‐XhoI.
The yeast invertase assay was used to validate the SSPbPXX signal peptide (Jacobs et al. 1997), following the protocol previously described (Yin et al. 2018). Briefly, chemicompetent Saccharomyces cerevisiae yeast strain YTK12 (Dr. Hossein Borhan, AAFC—Saskatoon Research Centre) was transformed with pSUC2 and pSUC2 constructs using the lithium acetate method (Gietz and Schiestl 2007). After transformation, yeast cells were spread on YPDA plates and incubated at 30 °C for 2–3 d. Positive clones were confirmed through PCR with primer pair pSUC2F/pSUC2R (Table S2) and streaked on CMD‐W (0.67% yeast N base without amino acids, 0.075% tryptophan dropout supplement, 2% sucrose, 0.1% glucose, and 2% agar) plates and YPRAA (1% yeast extract, 2% peptone, 2% raffinose, and 2 μg/ml antimycin A) plates and incubated at 30 °C for 3–4 d. Invertase enzymatic activity was analyzed as previously described (Yu et al. 2019), with an active signal peptide producing a colorimetric change of 2,3,5‐triphenyltetrazolium chloride (TCC: Sigma‐Aldrich). As well as yeast transformed with pSUC2_SSPbPXXSN, empty yeast strain YTK12 was included as a negative control, along with YTK12 transformed with pSUC2, while YTK12 transformed with pSUC2_LCR78SP was the positive control.
Production of recombinant proteins
For protein expression, SSPbP22‐coding sequence with the attB1 and attB2 recombination sites at the 5′ and 3′ ends was amplified with the designated primer pairs (Table S2). The attB‐flanked DNA fragments were cloned into pDONR™/Zeo (Thermo Fisher Scientific) using BP Clonase™ (Thermo Fisher Scientific) following the manufacturer's recommendations, generating entry clones.
To obtain recombinant SSPbP22‐His, the coding sequence was cloned into pDEST17 using LR Clonase™ (Thermo Fisher Scientific) and the SSPbP22_pDONR/Zeo entry vector following the manufacturer's recommendations. The list of vectors generated in this study can be seen in Table S3.
For protein expression, SSPbP22‐His_pDEST17 was used to transform Escherichia coli BL21 (DE3) cells. Escherichia coli transformants were grown in 50 ml of LB medium supplemented with ampicillin (100 μg/ml) at 37 °C, 200 rpm for 3 h. Protein expression was induced by the addition of 20% arabinose to a final concentration of 0.02% at 37 °C, 200 rpm for a further 2 h. Cell pellets were lysed using a sonicator VirTis equipped with a microtip (VirSonic, Gardiner, NY). Pellets expressing SSPbP22‐His were resuspended in 8 ml of guanidinium lysis buffer, sonicated on ice using six 10‐s bursts at high intensity with a 10‐s cooling period between each burst. The resulting lysate was centrifuged at 3,000 g for 15 min. SSPbP22‐His was purified using Ni‐NTA agarose (Thermo Fisher Scientific) through hybrid conditions following the manufacturer's recommendations. Protein concentration was measured using the Qubit Protein Assay Kit (Thermo Fisher Scientific).
Kinase activity assay
For the in vitro kinase assay, recombinant SSPbP22 (20 pmol) was incubated at 28 °C for 30 min in a final volume of 20 μl containing 0.5 mg/ml myelin basic protein (Santa Cruz Biotechnology, Santa Cruz, CA) and 20 μM ATP. Kinase activity was measured as ADP production using an Universal Kinase Assay Kit (Abcam, Cambridge, U.K.), following the manufacturer's recommendations. A reaction without SSPbP22 was included as the negative control, together with a reaction containing 20 μM ADP as the positive control.
Dot‐blot analysis
The kinase reactions, with and without SSPbP22, together with Arabidopsis total protein extracted in PEB protein extraction buffer, following the manufacturer's recommendations (Agrisera, Vännäs, Sweden), were immobilized on 0.2‐μm Immun‐Blot® PVDF Membrane (Bio‐Rad) using a 96‐well Bio‐Dot® (Bio‐Rad). Phosphorylated proteins were visualized using 1:1,000 polyclonal anti‐Phospho‐(Ser/Thr) antibody (Abcam), followed by mouse anti‐rabbit‐HRP (Santa Cruz Biotechnology). Blocking was performed with 5% skim milk (BD Difco, Mississauga, ON, Canada). The chemiluminescent signal from the horseradish peroxidase (HPR) was observed using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) through a ChemiDoc™ Imaging System (Bio‐Rad).
Protein localization
To express SSPbP22‐GFP in plants, the coding sequence amplified with primers SSPbP22attB1nS/SSPbP22attB2nS (Table S2) was cloned under the control of the CaMV 35S promoter into pEarlyGate 103 using LR Clonase™ (Thermo Fisher Scientific) and the pDONZeo_SSPbP22nS entry vector, following the manufacturer's recommendations. Constructs generated are listed in Table S3.
Agrobacterium tumefaciens containing SSPbP22‐GFP was infiltrated at an OD600 0.3 into leaves of 4‐wk‐old Nicotiana benthamiana plants. Cells expressing the fluorescent protein fusion were observed using a Zeiss LSM‐880 confocal microscope with a PL APO 40×/1.3 water‐dipping objective at no more than 2 d postinfiltration. GFP fluorescence was detected using 488/500–530 nm excitation/emission wavelengths.
RNA‐Seq data
The RNA‐Seq data are available at the sequence read archive (SRA) with the ID SRP160427.
Results
Defining and identifying the small secreted protein (SSPbP) repertoire of Plasmodiophora brassicae during secondary infection
To identify candidate small secreted P. brassicae (Pb) proteins (SSPbPs), the RNA‐Seq data previously generated by our group were analyzed. For this, we constructed a bioinformatics pipeline based on the characteristics of fungi and oomycetes plant pathogen‐secreted proteins, along with the evolutionarily related pathogen, Plasmodium (Ellis and Dodds 2011; Marti et al. 2004). The pipeline was organized into three sections, (i) Pb proteome prediction, (ii) Pb differentially expressed secretome, and (iii) small secreted Pb proteins (Fig. 1). From all the reads mapping to P. brassicae Pbe3, we identified 4,102 differentially expressed, protein‐encoding genes at 17 dpi, 3,166 at 20 dpi, and 8,278 at 24 dpi (Fig. 1). Of these, we identified 344, 244, and 802 genes encoding proteins with a signal peptide and without a transmembrane domain at 17, 20, and 24 dpi, respectively (Fig. 1). To ensure that only predicted proteins with a high probability of being secreted were further analyzed, we only kept those with a SignalP v4.1 score equal to or higher than 0.7 (Fig. 1). Further restrictions were placed on the pipeline to select only those genes encoding proteins of equal to, or shorter than, 400 amino acids—that is, “small” proteins (Fig. 1). Finally, this subset of SSPbPs was analyzed using Blast2GO (Conesa et al. 2005) and BlastX (https://blast.ncbi.nlm.nih.gov/Blastx) for annotation, Localizer and ApoplastP (Sperschneider et al. 2017a,b) to localize the proteins in the plant cell, as well as manual curation to identify cysteine‐rich proteins and the presence of effector motifs such as RxLP and Pexel, resulting in 32 SSPbP candidates (Fig. 1).
Four of the 32 SSPbPs were identified as cysteine‐rich proteins (2.5% or higher Cys is considered “Cys‐rich”; Hacquard et al. 2012). SSPbP41, a 386 amino acid protein, has nine (2.6%) Cys residues. Fourteen (3.8%) of the 366 amino acids of SSPbP61 are Cys residues, while the 213 amino acid SSPbP01 contains eight (3.7%) Cys residues and the 200 amino acid SSPbP81 contains six (3%) Cys residues.
The putative SSPbP sequences were searched for the RxLR motif, arginine–any amino acid–leucine–arginine, a motif in the N‐terminal of some oomycete and fungal effectors responsible for secretion of the effector (Ellis and Dodds 2011). Only one of the 32 putative SSPbPs, SSPbP97, carries the RxLR motif, with it located toward the C‐terminus (Fig. 2A). A second conserved characteristic motif of effector proteins containing the RxLR motif, the N‐terminal located DEER (Asp‐Glu‐Glu‐Arg) motif (Wirthmueller et al. 2013), is not present in SSPbP97 (Fig. 2A).
Figure 2.
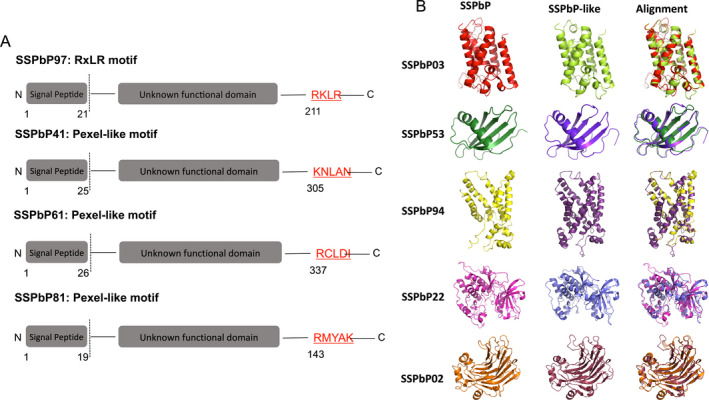
SSPbPs with motifs previously identified in effector proteins. A. SSPbPs with RxLR and Pexel‐like motifs. B. 3D model of SSPbPs with predicted functional domain. The identity of SSPbP‐like proteins presented in the figure and their alignment is described in the main text.
The Plasmodium export element Pexel has been identified in effector proteins from Plasmodium as key for the secretion of these effectors (Marti et al. 2004). This motif is a N‐terminal domain pentameric motif comprised of a positively charged, hydrophilic amino acid (Arg or Lys) in position one, a hydrophobic amino acid (Leu or Ile) in position three, and another less conserved amino acid (predominantly Asp, Glu, or Gln) in position 5, with a noncharged amino acid (Ser, Thr, Cys, Met, Asn, or Gln) in positions two and four. Although none of the putative SSPbPs identified in this study presented an exact Pexel motif, three of the proteins contain a Pexel‐like sequence: SSPbP61, RCLDI; SSPbP41, KNLAN; and SSPbP81, RMYAK (Fig. 2A).
An ankyrin domain was identified in three of the 32 SSPbPs (Table 1). Ankyrin domains have been identified as mediators of protein–protein interactions (Voronin and Kiseleva 2007), a key process involved in the interplay between effector–receptor interactions. SSPbP43 contains two ankyrin repeats between amino acids 206 and 304, while SSPbP71 presents three ankyrin repeats between amino acids 115 and 230 and SSPbP93 contains two ankyrin repeats between amino acids 223 and 308.
Table 1.
Predicted small secreted Plasmodiophora brassicae proteins (SSPbPs) with SP score > 0.7 and length < 400 aa
| SSPbP no. | Length (aa) | MW (kDa) | IPa | SPb score | Cleavage site (bp) | Cleavagec score | ID and/or motifs | Localizationd | Plasmodiophora brassicae (Pb) protein |
|---|---|---|---|---|---|---|---|---|---|
| SSPbP01 | 213 | 23.86 | 5.9 | 0.857 | 26 | 0.710 | HPe | C | PBRA_000804 |
| SSPbP02 | 398 | 43.9 | 5.79 | 0.827 | 22 | 0.723 | Glucanase | N | PBRA_002335 |
| SSPbP03 | 390 | 41.87 | 10.77 | 0.866 | 20 | 0.824 | Serine protease | Ch, M | PBRA_002618 |
| SSPbP04 | 123 | 12.95 | 9.49 | 0.869 | 28 | 0.682 | Cons. HPf | N | PBRA_005973 |
| SSPbP05 | 335 | 35.68 | 9.56 | 0.824 | 23 | 0.521 | SMCg | C | PBRA_002298 |
| SSPbP10 | 368 | 38.54 | 5.34 | 0.854 | 22 | 0.650 | Cons. HP | C | PBRA_007831 |
| SSPbP11 | 301 | 33.42 | 5.62 | 0.773 | 23 | 0.378 | HP | C | PBRA_004359 |
| SSPbP18 | 287 | 41.03 | 6.95 | 0.906 | 22 | 0.811 | HP | C | PBRA_001856 |
| SSPbP21 | 283 | 30.48 | 6.4 | 0.759 | 26 | 0.433 | SMC | C | PBRA_002100 |
| SSPbP22 | 375 | 41.95 | 5.21 | 0.778 | 28 | 0.567 | Kinase | C, N | PBRA_008980 |
| SSPbP31 | 266 | 27.73 | 7.02 | 0.727 | 18 | 0.351 | HP | N | PBRA_005440 |
| SSPbP32 | 204 | 22.01 | 6.85 | 0.779 | 30 | 0.632 | Cons. HP | A, Ch | PBRA_008250 |
| SSPbP41 | 386 | 40.00 | 4.99 | 0.906 | 24 | 0.913 | HP | A | PBRA_006112 |
| SSPbP42 | 206 | 21.51 | 7.81 | 0.751 | 18 | 0.438 | Cons. HP | N | PBRA_005499 |
| SSPbP43 | 311 | 33.68 | 5.02 | 0.860 | 26 | 0.747 | ANKh | C | PBRA_006649 |
| SSPbP44 | 96 | 9.65 | 5.23 | 0.798 | 21 | 0.566 | HP | A | PBRA_005716 |
| SSPbP51 | 395 | 42.34 | 7.92 | 0.916 | 22 | 0.832 | Cons. HP | Ch, M, N | PBRA_007344 |
| SSPbP52 | 206 | 28.39 | 4.96 | 0.840 | 17 | 0.536 | HP | N | PBRA_002122 |
| SSPbP53 | 140 | 14.55 | 4.55 | 0.819 | 24 | 0.596 | Cysteine protease inhibitor | A | PBRA_008207 |
| SSPbP54 | 248 | 27.4 | 8.54 | 0.776 | 20 | 0.792 | Cons. HP | M | PBRA_003240 |
| SSPbP61 | 366 | 40.06 | 7.31 | 0.755 | 25 | 0.318 | Kinase | N | PBRA_001067 |
| SSPbP62 | 242 | 26.55 | 4.96 | 0.751 | 22 | 0.372 | Cons. HP | C | PBRA_009168 |
| SSPbP71 | 389 | 42.44 | 7.02 | 0.776 | 31 | 0.649 | ANK | C | PBRA_001722 |
| SSPbP72 | 229 | 25.45 | 6.97 | 0.789 | 20 | 0.454 | Cons. HP | C | PBRA_008279 |
| SSPbP73 | 236 | 24.47 | 8.76 | 0.910 | 19 | 0.840 | Cons. HP | N | PBRA_001286 |
| SSPbP81 | 200 | 21.98 | 5.07 | 0.765 | 18 | 0.558 | Cyclin | C | PBRA_005813 |
| SSPbP86 | 227 | 25.47 | 6.12 | 0.871 | 23 | 0.622 | HP | C | PBRA_000257 |
| SSPbP91 | 154 | 15.68 | 9.17 | 0.789 | 20 | 0.789 | Cons. HP | A | PBRA_005502 |
| SSPbP92 | 385 | 41.71 | 8.96 | 0.746 | 26 | 0.362 | SMC | N | PBRA_002300 |
| SSPbP93 | 315 | 34.39 | 5.17 | 0.848 | 26 | 0.701 | ANK | C | PBRA_006655 |
| SSPbP94 | 307 | 33.32 | 8.46 | 0.745 | 21 | 0.260 | Translocase | N | PBRA_005298 |
| SSPbP97 | 233 | 25.23 | 5.76 | 0.832 | 20 | 0.622 | Cons. HP | C | PBRA_003620 |
| PbBSMTi | 377 | 45.21 | 4.92 | 0.728 | 21 | 0.505 | Methyl transferase | C | AFK13134 |
Isoelectric point of the SSPbP.
Signal peptide score, also known as D (discrimination) score, is the value used to distinguish proteins with signal peptides from proteins with no signal peptides.
Cleavage score, or C‐score (cleavage), is the value that is used to determine whether a signal peptide is processed by the host cell and removed in the mature protein.
A = apoplast; C = cytoplasm; Ch = chloroplast; M = mitochondria; N = nucleus.
Hypothetical protein.
Conserved hypothetical protein.
Chromosome segregation protein.
Ankyrin domains.
Methyl transferase characterized by Ludwig‐Müller et al. (2015).
Three of the SSPbPs contain the conserved N‐terminal SMC domain from chromosome segregation protein that is involved in cell cycle progression (Jessberger 2002). However, in all cases the SMC domain is located downstream of the N‐terminal, between amino acids 28 and 177 in SSPbP05, amino acids 29 and 148 in SSPbP21, and amino acids 37 and 215 in SSPbP91 (Table 1). Nineteen (56.2%) of the 32 SSPbPs were identified as hypothetical proteins (Table 1). However, eleven of these were designated conserved hypothetical proteins (conserved between species, but with no designated function).
Using Localizer, 15 of the 32 SSPbPs were identified as cytoplasmic proteins, 11 contained nuclear localization signals (NLS), three were identified as mitochondrial, and three were identified as probably chloroplastic (Table 1). Using ApoplastP, SSPbP32, SSPbP41, SSPbP44, SSPbP53, and SSPbP91 were identified as apoplastic (Table 1). Interestingly, some SSPbPs shared more than one predicted location in the plant cell, for example, SSPbP22 predicted to be both nuclear and cytoplasmic, while SSPbP51 was predicted to be chloroplastic and mitochondrial, as well as nuclear (Table 1).
SSPbPs with predicted functions
Of the 32 predicted SSPbPs, a functional domain was identified in seven (Table 1). SSPbP81 was identified as a cyclin, with the functional domain between amino acids 61 and 153 (Table 1). This Blast2GO result was confirmed through structural alignment against the Beta vulgaris cyclin P3 (Genbank XP_010674760) (Fig. 2B).
From amino acid alignment with serine proteases from Oceanithermus profundus (Genbank WP_01345690) and Lachnospiraceae bacterium (Genbank WP_090036406), SSPbP03 was putatively identified as a serine protease (Table 1), with confirmation by 3D structural alignment (Fig. 2B).
SSPbP53 was identified as a cysteine protease inhibitor, with the highest amino acid similarity to a cystatin (Genbank CAD20980) from the dust mite Lepidoglyphus destructor (Fig. 2B).
SSPbP94 was identified as a translocase (Table 1), similar to an ATP/ADP mitochondrial carrier protein lacking a signal peptide (Genbank CUG91522) from the protozoan Bodo saltans (Fig. 2B). SSPbP94 contains an ADP/ATP translocase domain between amino acids 53 and 225; however, TargetP did not identify a mitochondrial localization signal or any transmembrane domain in the SSPbP94 sequence (Table 1).
SSPbP02, between amino acids 1 and 260, contains a predicted fungal 1,3(4)‐beta‐D‐glucanase (Genbank OAD01513) domain from the fungus Mucor circinelloides (Table 1 and Fig. 2B).
Both SSPbP22 and SSPbP61 were identified as kinases (Table 1), with SSPbP22 showing the highest amino acid similarity with a protein kinase (Genbank OQR91891) from the oomycete Achlya hypogyna, while SSPbP61 showed the highest amino acid similarity with a protein kinase (Genbank OHS94657) from the protist Tritrichomonas foetus. Although both SSPbPs were identified as kinases, the SWISS‐MODEL structure for SSPbP22 was created using the crystal structure of the human cAMP‐dependent protein kinase catalytic subunit alpha as template (Qin et al. 2018), while that for SSPbP61 used the crystal structure of the human tyrosine kinase fibroblast growth factor receptor 1 as template (Tsai et al. 2019). Alignment of the SWISS‐MODEL structure for SSPbP22 with that for the A. hypogyna protein kinase supported the kinase identity for SSPbP22 (Fig. 2B), while the SWISS‐MODEL alignment between SSPbP61 and the T. foetus protein kinase confirmed the kinase identity for SSPbP61 (Fig. S1).
Thirty‐one SSPbPs have a functional signal peptide
The secretory activity directed by the 32 SSPbP signal peptides (SPs) was tested individually. Thirty‐one of these SP sequences showed secretory activity in yeast as demonstrated by the growth assay on CMD‐W and YPRAA plates (presented for 8 in Fig. S2) and by the reduction of 2,3,5‐triphenyltetrazolium chloride (TTC) to insoluble red‐colored 1,3,5‐triphenylformazan (TPF) (Fig. 3). All the 31 SPs showed a similar red tone to that of the Arabidopsis‐secreted protein LOW‐MOLECULAR‐WEIGHT CYSTEINE‐RICH 78 (LCR78, AT1G19610) positive control (Hu et al. 2011). The only predicted SP that did not demonstrate invertase activity in the YTK12 yeast strain was from SSPbP31, which coincidently had the lowest SP score (0.727). Yeast transformed with pSUC_SSPbP31 was unable to grow on YPRAA or reduce TCC (Fig. S2 and Fig. 3). Untransformed YTK12 cells were unable to grow on any plate (Fig. S2), and while YTK12 + empty pSUC2 vector was able to grow on CMD‐W, it was unable to reduce TCC (Fig. 3).
Figure 3.

Validation of the signal peptide of SSPbPs through yeast invertase assay. Constructs containing the signal peptide sequence of SSPbPs (pSUC2_SSPbPsSP), LCR78 (pSUC2_LCR78SP), and empty vector (EV) pSUC2 in yeast YTK12 were plated on CMD‐W (Trp depleted), and the enzymatic activity of secreted invertase was determined using the reduction of TTC to the red‐colored formazan assay. NR, not transformed. Images are representative of three replicates using independent biological samples.
Disease index and SSPbP expression confirmation
The conditions used during the RNA‐Seq analysis were replicated in order to confirm the expression profiles previously obtained. The disease index (DI) at the three time points 17, 20, and 24 dpi correlated with that in our previous transcriptome study of clubroot infection (Irani et al. 2018). Consistent with previous studies (Irani et al. 2018; Malinowski et al. 2012), a swelling of the roots was first visible on most Arabidopsis roots between 16 and 18 dpi, with gall formation at 20 dpi and large galls producing severe wilting at 24 dpi (Fig. 4A). Secondary infection was further confirmed at 20 and 24 dpi when multiple secondary plasmodia were observed in infected host cells of Arabidopsis root tissue (Fig. 4B). These multinucleated secondary plasmodia increased the size of the infected cell when compared to noninfected host cells. At 21–24 dpi, when disease symptoms were fully developed, the cell wall of infected cells had broken down (Fig. 4C, D), facilitating plasmodia movement from cell to cell and subsequent gall formation (Donald et al. 2008; Riascos et al. 2011).
Figure 4.
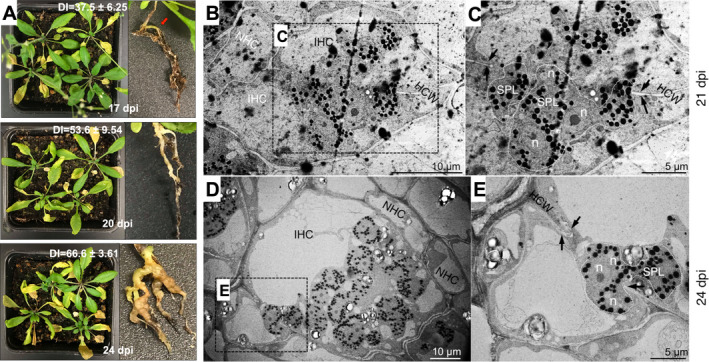
Secondary infection in Arabidopsis at the time points utilized in this study. A. Phenotype of shoots and roots of Arabidopsis plants infected with Plasmodiophora brassicae at 17, 20, and 24 dpi. Disease index mean ± standard error is indicated. n = 3. Red arrow indicates the first visible symptoms at 17 dpi. B–E. TEM micrographs of P. brassicae secondary plasmodia in Arabidopsis‐infected root cells at 21 and 24 dpi. B. Infected tissue at 21 dpi showing infected host cells (IHC), noninfected host cells (NHC), and secondary plasmodia (SPL) with numerous lipid droplets (black dots), C. Magnification of the dashed‐line square in B, with secondary plasmodia and arrows indicating thinning of the host cell wall (HCW). D. Infected tissue at 24 dpi showing IHC, NHC, and SPL with higher number of lipid droplets. E. Magnification of the dashed‐line square in D, with secondary plasmodia and arrows indicating broken HCW.
The RNA‐Seq data for the 32 predicted SSPbPs were used to generate a heat map of normalized gene expression at 17, 20, and 24 dpi, with the most strongly activated response occurring at 24 dpi (Fig. S3A). Although transcript levels for most of the SSPbPs were lowest at 17 dpi, eight genes (SSPbP03, SSPbP04, SSPbP11, SSPbP21, SSPbP51, SSPbP52, SSPbP72, and SSPbP81) showed higher transcript levels at 17 and 24 dpi, with reduced transcript levels at 20 dpi (Fig. S3A). RNA‐Seq data reported for Brassica rapa, B. napus, and B. oleracea at 35 dpi (Schwelm et al. 2015) were also heat mapped, confirming that the predicted SSPbPs in this study were also highly expressed in three other plant hosts after 24 dpi (Fig. S3A). To confirm the RNA‐Seq data, transcript levels for 24 randomly selected SSPbPs representing all of the proposed categories for the SSPbPs (hypothetical proteins, conserved hypothetical proteins, SSPbPs with ankyrin domains, SSPbPs with SMC domains, and the SSPbPs with predicted functional domain (Table 1)) were determined by qRT‐PCR (Fig. S3B). Transcript levels for each SSPbP gene, relative to P. brassicae ELONGATION FACTOR‐LIKE gene (PBRA_001540), confirmed the RNA‐Seq data (Fig. S3B).
Of the 32 SSPbPs, the expression profiles of 21 over the P. brassicae life cycle from 0 to 28 dpi, with 0, 2, 5, and 7 dpi representing primary infection and 14, 21, and 28 dpi representing secondary infection, were determined. All selected SSPbP‐encoding genes were expressed in planta, with 10 of the 21 SSPbPs highly expressed during both primary and secondary infection, suggesting that they are involved in all stages of the P. brassicae life cycle, including the development of primary plasmodia (Fig. 5).
Figure 5.
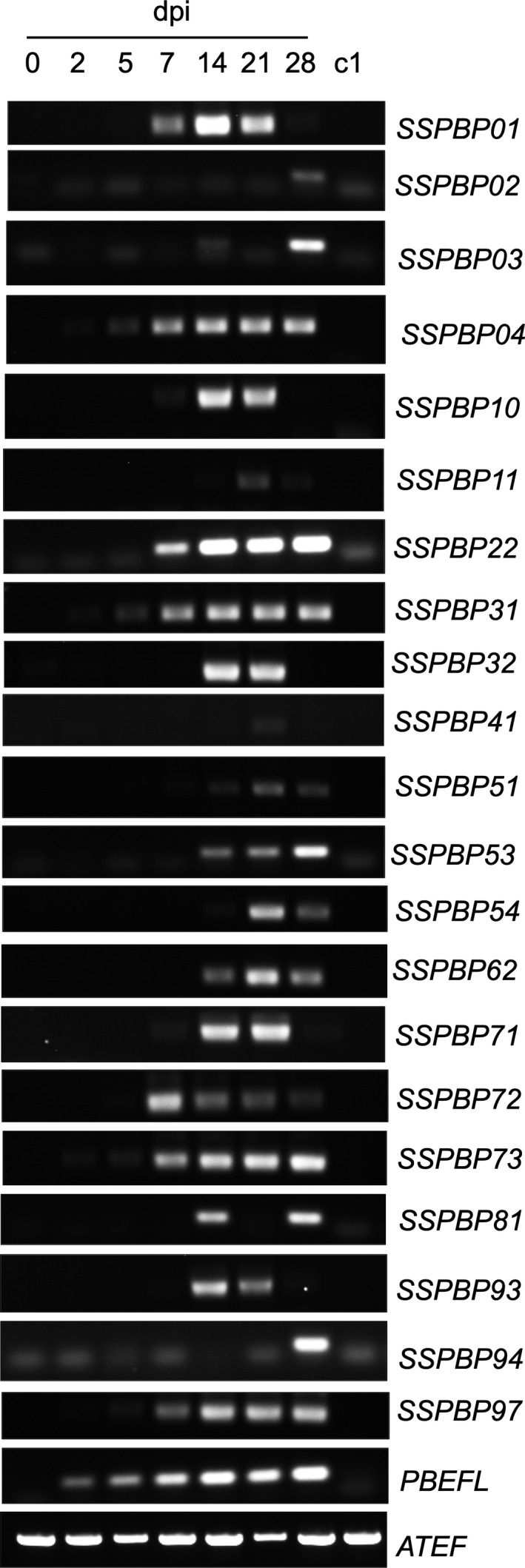
RT‐PCR amplification of 21 selected SSPbP‐encoding genes at 0, 2, 5, 7, 14, 21, and 28 dpi. c1 is cDNA from mock‐inoculated Arabidopsis root tissue at 7 dpi. Plasmodiophora brassicae ELONGATION FACTOR‐LIKE gene (PbEF1) was used as the pathogen internal control, with the Arabidopsis thaliana ELONGATION FACTOR gene (AtEF) as host control. Images are representative of three replicates using independent biological samples.
SSPbP22 is a cytosolic and nuclear kinase
To confirm whether SSPbP22 is an active kinase, as predicted using in silico analysis, the recombinant His tagged protein was obtained (Fig. 6A). The resulting His‐SSPbP22 was able to phosphorylate a MAP kinase substrate, as indicated by the production of ADP (Fig. 6B and Table S1) and with phosphorylated substrate determined by dot‐blot assay with Phospho p44/p42 MAPK (Fig. 6C).
Figure 6.
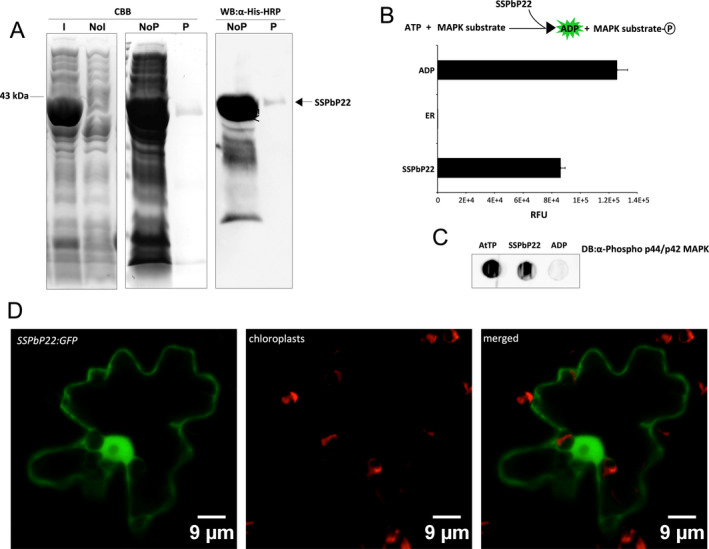
SSPbP22 kinase activity validation. A. Coomassie Brilliant Blue (CBB) gel of production of SSPbP22 (~43 kDa) in Escherichia coli BL21DE3 cells, induced (I) and noninduced (NoI) by arabinose and SSPbP22 nonpurified (NoP) and purified (P) using Ni‐NTA agarose. Purified and nonpurified SSPbP22 was also visualized through Western blot (WB) with anti‐His antibody. B. Kinase reaction scheme and kinase activity of SSPbP22. The reaction without kinase protein served as negative control. ADP served as positive control. RFU—relative fluorescence units. C. Dot blot detecting substrate phosphorylated by SSPbP22. Arabidopsis total protein was used as positive control. D. Confocal images showing SSPbP22‐GFP localized to the cytoplasm and the nucleus of N. benthamiana epidermal cells (Scale bars, 9 μm.)
The cytosolic and nuclear locations predicted for SSPbP22 (Table 1) were confirmed in N. benthamiana leaves, where SSPbP22‐GFP was observed to localize in both the cytosol and the nucleus (Fig. 6D).
Discussion
During infection, many pathogen‐secreted proteins are effectors involved in pathogen differentiation, communication, and development (Rovenich et al. 2014). Plasmodiophora brassicae is an obligate parasite, and although primary and secondary infections have been investigated (Chen et al. 2019; Feng et al. 2013), the molecular mechanisms responsible for infection and the effector proteins involved in secondary infection are still unknown. Unlike oomycete, fungal, and bacterial effectors that are readily identified based on the presence of characteristic motifs such as Y/F/WxC (Godfrey et al. 2010), LXLFLAK (Whisson et al. 2007), RxLR (Ellis and Dodds 2011), and the conserved Nudix box (GX5EX7REUXEEXGU) (Dong and Wang 2016), the identification of P. brassicae effectors is more problematic (Chen et al. 2019; Pérez‐López et al. 2018; Schwelm et al. 2015). Although Rolfe et al. (2016) presented data resulting from RNA‐Seq analysis of P. brassicae Canadian pathotypes, to date, our study is the only completely available RNA‐Seq data set (SRP160427) for a Canadian pathotype. Primarily, in this study, we have identified small secreted P. brassicae proteins (SSPbPs) differentially expressed during the interaction of the pathogen with the model plant A. thaliana (Fig. 5 and Fig. S3). The pipeline used in this study (Fig. 1) could be modified by other research groups by varying some parameters such as the SP score, amino acid length, or whether to keep proteins with transmembrane domains to narrow down or broaden subsequent searches of the RNA‐Seq data.
As a biotrophic pathogen, P. brassicae is able to avoid host recognition and cell death, and, in doing so, promotes the proliferation of host root tissues to produce a nutrient‐sink gall habitat. Many mechanisms involved in these processes are still unknown in P. brassicae, but effectors from well‐studied biotrophic plant pathogens such as the Basidiomycete rusts and the Ascomycete powdery mildews have been studied and characterized, and are the basis of disease resistance breeding strategies (Vleeshouwers and Oliver 2014). While in this study it might appear that the number of predicted small secreted proteins for P. brassicae (32 SSPbPs) is small, our findings correlate with the number of small secreted proteins (SSPs) predicted for the obligate parasite aster yellows phytoplasma (56 predicted SSPs) (Sugio et al. 2011), as well as with the number of effector candidates recently reported for P. brassicae during primary infection (Chen et al. 2019). In comparison with the biotrophic rust fungi Melampsora larici‐populina (Hacquard et al. 2012), or the smut pathogen Ustilago hordei (Ökmen et al. 2018) with hundreds of predicted secreted proteins, 32 SSPbPs is a small number, but there are some points that should be considered for P. brassicae: (i) This pathogen is an obligate parasite only able to survive in the plant cell or as a dormant resting spore in the soil, (ii) the life cycle is less complex than any currently known biotrophic fungal plant pathogen, and (iii) the RNA‐Seq data were generated during P. brassicae secondary infection in Arabidopsis.
A common characteristic of effectors such as Avr2, Avr4, Avr4E, and Avr9 of several plant pathogens, including Cladosporium fulvum (syn. Passalora fulva), an asexual extracellular fungal pathogen of tomato (Stergiopoulos and de Wit 2009), is a high number of Cys residues in their sequences. Four of the 32 predicted SSPbPs in this study were identified as Cys‐rich proteins. While in some biotrophic pathogens, such as M. larici‐populina, Cys‐rich proteins dominate the secretome with up to 63% of the SSPs containing more than four Cys residues, many well‐characterized effectors do not contain any Cys in their sequences (Hacquard et al. 2012).
The N‐terminal RxLR motif has been identified in effector proteins of oomycetes as necessary for translocation into the host cell (Ellis and Dodds 2011; Yaeno et al. 2011). The motif is required for an effector–phosphatidylinositol phosphate interaction within the host cell plasma membrane, thereby mediating entry into the host cell by the pathogen (Kale et al. 2010). Only SSPbP97 contains an RxLR‐like motif; however, unlike in oomycetes, the motif is located toward the C‐terminal of the protein. While RxLR motifs have been previously reported in putative secreted proteins in both the Canadian P. brassicae pathotype Pb3 and the European pathotype Pbe3, confirmation of these proteins as effectors and characterization of their functions have yet to be determined (Rolfe et al. 2016; Schwelm et al. 2015). The DEER motif, generally found in effector proteins containing the RxLR motif, is not found in SSPbP97, leading to the possibility that the RxLR‐like motif in SSPbP97 may not be involved in any interaction with the host cell plasma membrane. SSPbP97 was highly expressed between 7 and 28 dpi (Fig. 5), during secondary plasmodia colonization of cortical cells, galls being formed, and disruption of the host cell wall (Fig. 4), leading to resting spore dispersal into the soil (Kageyama and Asano 2009; Schuller and Ludwing‐Müller 2016). A similar high expression of SSPBP97 was also detected in Pb‐infected Brassicas at 35 dpi (Fig. S3) (Schwelm et al. 2015).
In Plasmodium falciparum, the Pexel motif is a targeting motif directing translocation of effectors across the parasitophorous vacuolar membrane (Marti et al. 2004). Three of the identified SSPbPs carry a Pexel‐like motif. A plant pathogen effector protein sharing a highly similar motif with Plasmodium spp. has been previously identified as the Plasmodium‐like virulence effector HgGLAND18, a suppressor of plant innate immunity from the soybean cyst nematode Heterodera glycines (Noon et al., 2016).
Based on the structures and predicted domains in the 32 SSPbPs, more than 25% of them appear to be implicated in the regulation of the cell cycle. The ankyrin repeat (ANK), a 33‐residue motif that is usually organized in tandem and mediates protein–protein interactions, is involved in many cellular functions, such as cell cycle, in eukaryotes (Voronin and Kiseleva 2007). Deletion of the ANK repeats resulted in defects in intracellular proliferation of the pathogen in mammalian cells (Price et al. 2010). Eukaryotic‐like genes containing ankyrin repeats (ANK) have been identified in Legionella pneumophila, bacteria that have co‐evolved with protozoa (Price et al. 2010). The three ANK‐containing SSPbPs (Table 1) may be involved in plant cell proliferation and/or P. brassicae life‐cycle transitions. The SSPbPs with N‐terminal domains from chromosome segregation protein (SMC) (Table 1) may also share a similar function, aiding in pathogen proliferation and/or colonization of the host cell. SMC proteins act together with other proteins in a range of chromosomal transitions, including chromosome condensation, sister‐chromatid cohesion, recombination, DNA repair, and epigenetic silencing of gene expression (Jessberger 2002). It is foreseeable that these processes require tight control during P. brassicae infection, especially during secondary infection and gall formation such that viable resting spores are produced. Schwelm et al. (2015) identified more than 200 ANK‐containing proteins in the P. brassicae genome. Further complicating matters during secondary infection, an increase in meristematic activity of the host tissues also occurs (Kageyama and Asano 2009; Malinowski et al. 2012).
Cell cycle progression is driven by cyclin‐dependent kinases (CDKs) and associated cyclins that regulate CDK activity and confer substrate specificity (Bulankova et al. 2010). SSPbP81 was identified as a cyclin (Table 1), and SSPbP22 and SSPbP61 were identified as kinases, possible counterparts in cyclin–kinase interactions. In many plant pathogens, a variety of kinases have been identified as effectors (Franceschetti et al. 2017; Lo Presti et al. 2015). The influence of plant pathogens on host cell cycle has been identified as a strong evolutionary force resulting in plants with high numbers of CDK inhibitors (CKIs) in their genomes (Fox and Duronio 2013). CKIs negatively control cell cycle progression, contribute to cell cycle checkpoints, and can promote a modified cell cycle resulting in endoreduplication and increased ploidy (Fox and Duronio 2013). CKIs contribute to the innate immunity of Arabidopsis (Hamdoun et al. 2016). To date, there is no record of any plant pathogen cyclin influencing a host cell cycle; however, taking into account the impact of P. brassicae on host meristematic activity and the reprogramming of the host cell during gall formation, the role of SSPbP81 as an effector cyclin protein cannot be ignored.
SSPbP53 is identified as a cysteine protease inhibitor (Table 1). A number of previously identified plant pathogen effectors, such as Avr2 from the fungal plant pathogen Cladosporium fulvum (Song et al. 2009), EPIC1 and EPIC2B from the oomycete Phytophthora infestans (Song et al. 2009), and Pit2 from Ustilago maydis (Mueller et al. 2013), inhibit plant apoplastic cysteine proteases, an interaction that appears key for establishing compatibility (Lo Presti et al. 2015). As a putative cysteine protease inhibitor, SSPbP53 may play an important role in P. brassicae pathogenicity. The apoplastic space has been identified as a major “battleground” for plant–pathogen interactions (Wang and Wang 2018), and besides SSPbP53, another four SSPbPs with predicted apoplastic localizations (Table 1) were identified.
Serine proteases are widely distributed in all organisms, carrying out diverse functions such as digestion, immune response, and defense (Yousef et al. 2003). The role of serine proteases in pathogenicity has been well established in the human pathogenic protozoans P. falciparum (Roggwiller et al. 1996), Trypanosoma cruzi (Burleigh et al. 1997), and Entamoeba spp. (Makioka et al. 2009). Although their exact mode of action is generally unknown, there are many reports of serine proteases also having key roles in the pathogenicity of plant pathogens, such as the fungal pathogen Colletotrichum coccodes (Redman and Rodriguez 2002), the oomycete Phytophthora infestans (Hamill et al. 2006), and the bacterium responsible for disease ring rot of potato, Clavibacter michiganensis subsp. sepedonicus (Nissinen et al. 2009). The P. brassicae serine protease Pro1 (Genbank accession number GU082362), identified as a member of the S28 family of proteases, was previously characterized as important in stimulating resting spore germination (Feng et al. 2010).
While ADP/ATP translocases have not previously been defined as effector proteins, they do play key roles in pathogen survival in the host cell (Vahling et al. 2010). The production of ADP/ATP translocases is a common and characteristic feature of many obligate parasites with a reduced genome including the plant pathogen “Candidatus Liberibacter asiaticus” and in some obligate intracellular bacteria belonging to Chlamydiales and Rickettsiales (Jain et al. 2017; Merfa et al. 2019; Schmitz‐Esser et al. 2004, 2008; Vahling et al. 2010). A common feature of ADP/ATP translocases is a signal peptide for secretion and no transmembrane domain (Schmitz‐Esser et al. 2004): SSPbP94 was identified as such. Plasmodium falciparum utilizes an ADP/ATP exchange system to facilitate host–parasite metabolic interactions and, besides the ATP transporter located in the plasma membrane, also contains an ADP/ATP transporter (AdT) in mitochondria, key for oxidative phosphorylation (Choi and Mikkelsen 1990; Kanaani and Ginsburg 1989). SSPbP94 does not contain a mitochondrial target signal, suggesting that it is exported and might play a role in scavenging ATP from infected host cells, although it does not contain a recognized transmembrane domain.
Plant cell wall‐degrading enzymes, secreted by biotrophic plant‐pathogenic fungi, facilitate their penetration of host plant cells (Gibson et al. 2011), and some, identified as effectors, are specifically required for penetration (Lo Presti et al. 2015). The biotrophic plant pathogens Ustilago maydis, Blumeria graminis, and Puccinia graminis all secrete endo‐1,3(4)‐beta‐glucanase, among other cell wall‐degrading enzymes (Gibson et al. 2011), responsible for the endohydrolysis of (1‐3)‐ or (1‐4)‐linkages in beta‐d‐glucans within the plant or the pathogen cell wall (Sova et al. 1970). Highly expressed at 24 dpi, when host cell walls are being broken down and secondary plasmodia are establishing, SSPbP02, identified as a putative endo‐1,3(4)‐beta‐glucanase, may have a crucial role in secondary infection of root cortical cells and subsequent gall formation.
Conclusion
Specific interactions between well‐characterized host target proteins and pathogen effectors have been identified in a number of host–pathogen interactions (Chen et al. 2017; Salcedo et al. 2017); however, for P. brassicae this study is one of the first to characterize effector proteins with potential roles in clubroot pathogenesis. RNA‐Seq data, over a time course of secondary infection, were validated through qRT‐PCR, and a complete in silico prediction of small secreted P. brassicae proteins (SSPbPs) was carried out. The pipeline designed by our group and applied in this study was successful in identifying a number of putative effectors, for which expression and signal peptide activity were experimentally validated, along with the structural domain identified of one of the candidates and its subcellular localization. Subcellular localization of the other candidate effector proteins, together with identification of the interacting plant proteins, is the next step toward confirming the functions of the SSPbPs reported in this study as P. brassicae effector proteins. Our efforts are fully focusing on this challenging and extremely important task.
Author Contribution
EPL, JT, CDT, MW, AJK, YW, MMH, and PCBS designed the research; EPL, MMH, JT, and MW performed the research; EPL and MW analyzed the data; EPL, CDT, AJK, YW, MMH, and PCBS wrote and/or edited the manuscript.
Supporting information
Figure S1. 3D model of SSPbP61, SSPbP61‐like and their respective alignment.
Figure S2. Signal peptide validation of eight randomly selected SSPbPs and SSPbP31, effector candidate with lower SP score (Table 1).
Figure S3. Expression of SSPbPs identified in this study.
Table S1. Raw data of SSPbP22 kinase activity assay.
Table S2. Primers designed and used in this study.
Table S3. Plasmids constructions generated in this study.
Acknowledgments
The authors thank Tim Dumonceaux from AAFC—Saskatoon Research Centre for the support offered during the qPCR analysis and to Arne Schwelm from the Swedish University of Agricultural Sciences for the critical revision of the manuscript. This work was supported by Saskatchewan Agriculture Development Fund (SADF) and the Saskatchewan Canola Development Commission (SaskCanola) funding to PCBS, YW, and CDT.
Literature Cited
- Arnold, K. , Bordoli, L. , Kopp, J. & Schwede, T. 2006. The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics, 22:195–201. [DOI] [PubMed] [Google Scholar]
- Bergemann, T. L. & Wilson, J. 2011. Proportion statistics to detect differentially expressed genes: a comparison with log‐ratio statistics. BMC Bioinformatics, 12:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. & Usadel, B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulankova, P. , Riehs‐Kearnan, N. , Nowack, M. K. , Schnittger, A. & Riha, K. 2010. Meiotic progression in Arabidopsis is governed by complex regulatory interactions between SMG7, TDM1, and the meiosis I‐specific cyclin TAM. Plant Cell, 22:3791–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki, F. , Kudryavtsev, A. , Matz, M. V. , Aglyamova, G. V. , Bulman, S. , Fiers, M. , Keeling, P. J. & Pawlowski, J. 2010. Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol. Biol., 10:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh, B. A. , Caler, E. V. , Webster, P. & Andrews, N. W. 1997. A cytosolic serine endopeptidase from Trypanosoma cruzi is required for the generation of Ca2+ signalling in mammalian cells. J. Cell Biol., 136:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Li, Y. , Yan, R. , Xu, L. , Ren, L. , Liu, F. , Zeng, L. , Yang, H. , Chi, P. , Wang, X. , Chen, K. , Ma, D. & Fang, X. 2019. Identification and characterization of Plasmodiophora brassicae primary infection effector candidates that suppress or induce cell death in host and nonhost plants. Phytopathology, 109(10):1689–1697. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Upadhyaya, N. M. , Ortiz, D. , Sperschneider, J. , Li, F. , Bouton, C. , Breen, S. , Dong, C. , Xu, B. , Zhang, X. , Mago, R. , Newell, K. , Xia, X. , Bernoux, M. , Taylor, J. M. , Steffenson, B. , Jin, Y. , Zhang, P. , Kanyuka, K. , Figueroa, M. , Ellis, J. G. , Park, R. F. & Dodds, P. N. 2017. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science, 358:1607–1610. [DOI] [PubMed] [Google Scholar]
- Choi, I. & Mikkelsen, R. B. 1990. Plasmodium falciparum: ATP/ADP transport across the parasitophorous vacuolar and plasma membranes. Exp. Parasitol., 71:452–462. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. & Mackey, K. 1995. Short technical report. Modification of the TRIZOL reagent procedure for isolation of RNA from polysaccharide‐and proteoglycan‐rich sources. Biotechniques, 19(6):942–945. [PubMed] [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J. M. , Terol, J. , Talón, M. & Robles, M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- De Lano, W. L. 2002. Pymol: An open‐source molecular graphics tool. CCP4 Newsletter On Protein Crystallography, 40: 82–92.
- Dixon, G. R. 2009. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul., 28(3):194–202. [Google Scholar]
- Dobin, A. & Gingeras, T. R. 2015. Mapping RNA‐seq reads with STAR. Curr. Protoc. Bioinformatics, 51:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P. N. & Rathjen, J. P. 2010. Plant immunity: towards an integrated view of plant– pathogen interactions. Nat. Rev. Gen., 11(8):539–548. [DOI] [PubMed] [Google Scholar]
- Donald, E. C. , Jaudzems, G. & Porter, I. J. 2008. Pathology of cortical invasion by Plasmodiophora brassicae in clubroot resistant and susceptible Brassica oleracea hosts. Plant. Pathol., 57:201–209. [Google Scholar]
- Dong, S. & Wang, Y. 2016. Nudix effectors: a common weapon in the arsenal of plant pathogens. PLoS Pathog., 12(8):e1005704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis, S. , Cuomo, C. A. , Lin, Y.‐C. , Aerts, A. , Tisserant, E. , Veneault‐Fourrey, C. , Joly, D. L. , Hacquard, S. , Amselem, J. , Cantarel, B. L. , Chiu, R. , Coutinho, P. M. , Feau, N. , Field, M. , Frey, P. , Gelhaye, E. , Goldberg, J. , Grabherr, M. G. , Kodira, C. D. , Kohler, A. , Kües, U. , Lindquist, E. A. , Lucas, S. M. , Mago, R. , Mauceli, E. , Morin, E. , Murat, C. , Pangilinan, J. L. , Park, R. , Pearson, M. , Quesneville, H. , Rouhier, N. , Sakthikumar, S. , Salamov, A. A. , Schmutz, J. , Selles, B. , Shapiro, H. , Tanguay, P. , Tuskan, G. A. , Henrissat, B. , Van de Peer, Y. , Rouzé, P. , Ellis, J. G. , Dodds, P. N. , Schein, J. E. , Zhong, S. , Hamelin, R. C. , Grigoriev, I. V. , Szabo, L. J. & Martin, F. 2011. Obligate biotrophy features unravelled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA, 108:9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J. G. & Dodds, P. N. 2011. Showdown at the RXLR motif: serious differences of opinion in how effector proteins from filamentous eukaryotic pathogens enter plant cells. Proc. Natl. Acad. Sci. USA, 108(35):14381–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O. , Nielsen, H. , Brunak, S. & von Heijne, G. 2000. Predicting subcellular localization of proteins based on their N‐terminal amino acid sequence. J. Mol. Biol., 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Feng, J. , Hwang, R. U. , Hwang, S.‐F. , Strelkov, S. E. , Gossen, B. D. , Zhou, Q.‐X. & Peng, G. 2010. Molecular characterization of a serine protease Pro1 from Plasmodiophora brassicae that stimulates resting spore germination. Plant. Pathol., 11(4):503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J. , Hwang, S.‐F. & Strelkov, S. E. 2013. Studies into primary and secondary infection processes by Plasmodiophora brassicae on canola. Plant. Pathol., 62:177–183. [Google Scholar]
- Fox, D. T. & Duronio, R. J. 2013. Endoreplication and polyploidy: insights into development and disease. Development, 140:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschetti, M. , Maqbool, A. , Jiménez‐ Dalmaroni, M. J. , Pennington, H. G. , Kamoun, S. & Banfield, M. J. 2017. Effectors of filamentous plant pathogens: commonalities amid diversity. Microbiol. Mol. Biol. Rev., 81:e00066‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D. M. , King, B. C. , Hayes, M. L. & Bergstrom, G. C. 2011. Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Curr. Opin. Microbiol., 14:264–270. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D. & Schiest, R. H. 2007. High‐efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc., 2:31–34. [DOI] [PubMed] [Google Scholar]
- Godfrey, D. , Böhlenius, H. , Pedersen, C. , Zhang, Z. , Emmersen, J. & Thordal‐Christensen, H. 2010. Powdery mildew fungal effector candidates share N‐terminal Y/F/WxC‐motif. BMC Genom., 11:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard, S. , Joly, D. L. , Lin, Y.‐C. , Tisserant, E. , Feau, N. , Delaruelle, C. , Legue, V. , Kohler, A. , Tanguay, P. , Petre, B. , Frey, P. , Van de Peer, Y. , Rouzé, P. , Martin, F. , Hamelin, R. C. & Duplessis, S. 2012. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici‐populina (poplar leaf rust). Mol. Plant Microbe Interact., 25:279–293. [DOI] [PubMed] [Google Scholar]
- Hamdoun, S. , Zhang, C. , Gill, M. , Kumar, N. , Churchman, M. , Larkin, J. C. , Kwon, A. & Lu, H. 2016. Differential roles of two homologous cyclin‐dependent kinase inhibitor genes in regulating cell cycle and innate immunity in Arabidopsis. Plant Physiol., 170:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill, J. A. , Selby, C. & Cooke, L. R. 2006. Aggressiveness of Phytophthora infestans isolates correlates with their proteolytic activity. Acta Hort., 725:673–678. [Google Scholar]
- Hu, T. T. , Pattyn, P. , Bakker, E. G. , Cao, J. , Cheng, J.‐F. , Clark, R. M. , Fahlgren, N. , Fawcett, J. A. , Grimwood, J. , Gundlach, H. , Haberer, G. , Hollister, J. D. , Ossowski, S. , Ottilar, R. P. , Salamov, A. A. , Schneeberger, K. , Spannagl, M. , Wang, X. & Guo, Y.‐L. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet., 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, S. F. , Strelkov, S. E. , Feng, J. , Gossen, B. D. & Howard, R. J. 2012. Plasmodiophora brassicae: a review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol., 13:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani, S. , Trost, B. , Waldner, M. , Nayidu, N. , Tu, J. , Kusalik, A. J. , Todd, C. D. , Wei, Y. & Bonham‐Smith, P. C. 2018. Transcriptome analysis of response to Plasmodiophora brassicae infection in the Arabidopsis shoot and root. BMC Genom., 19:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, K. A. , Collins‐Racie, L. A. , Colbert, M. , Duckett, M. , Golden‐Fleet, M. , Kelleher, K. , Kriz, R. , LaVallie, E. R. , Merberg, D. , Spaulding, V. , Stover, J. , Williamson, M. J. & McCoy, J. M. 1997. A genetic selection for isolating cDNAs encoding secreted proteins. Gene, 198:289–296. [DOI] [PubMed] [Google Scholar]
- Jain, M. , Munoz‐Bodnar, A. & Gabriel, D. W. 2017. Concomitant loss of the glyoxalase system and glycolysis makes the uncultured pathogen “Candidatus Liberibacter asiaticus” an energy scavenger. Appl. Environ. Microbiol., 83(23):e01670‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger, R. 2002. The many functions of smc proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol., 3:767–778. [DOI] [PubMed] [Google Scholar]
- Jones, J. D. G. & Dangl, J. L. 2006. The plant immune system. Nature, 444:323–329. [DOI] [PubMed] [Google Scholar]
- Kageyama, K. & Asano, T. 2009. Life cycle of Plasmodiophora brassicae . J. Plant Growth Regul., 28(3):203–211. [Google Scholar]
- Kale, S. D. , Gu, B. , Capelluto, D. G. , Dou, D. , Feldman, E. , Rumore, A. , Arredondo, F. D. , Hanlon, R. , Fudal, I. , Rouxel, T. , Lawrence, C. B. , Shan, W. & Tyler, B. M. 2010. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell, 142:284–295. [DOI] [PubMed] [Google Scholar]
- Kanaani, J. & Ginsburg, H. 1989. Metabolic interconnection between the human malarial parasite Plasmodium falciparum and its host erythrocyte. Regulation of ATP levels by means of an adenylate translocator and adenylate kinase. J. Biol. Chem., 264:3194–3199. [PubMed] [Google Scholar]
- Kiefer, F. , Arnold, K. , Kunzli, M. , Bordoli, L. & Schwede, T. 2009. The SWISS‐MODEL repository and associated resources. Nucleic Acid Res., 37:387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. 1997. ABI Prism 7700 sequence detection system. User Bulletin no. 2. PE Applied Biosystems, AB website, bulletin reference. 4303859B 777802‐002.
- Livak, K. J. & Schmittgen, T. D. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods, 25:402–408. [DOI] [PubMed] [Google Scholar]
- Lo Presti, L. , Lanver, D. , Schweizer, G. , Tanaka, S. , Liang, L. , Tollot, M. , Zuccaro, A. , Reissmann, S. & Kahmann, R. 2015. Fungal effectors and plant susceptibility. Ann. Rev. Plant Biol., 66:513–545. [DOI] [PubMed] [Google Scholar]
- Ludwig‐Müller, J. , Jülken, S. , Geib, K. , Richter, F. , Mithöfer, A. , Šola, I. , Rusak, G. , Keenan, S. & Bulman, S. 2015. A novel methyltransferase from the intracellular pathogen Plasmodiophora brassicae methylates salicylic acid. Mol. Plant Pathol., 16(4):349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makioka, A. , Kumagai, M. , Kobayashi, S. & Takeuchi, T. 2009. Involvement of serine proteases in the excystation and metacystic development of Entamoeba invadens . Parasitol. Res., 105:977–987. [DOI] [PubMed] [Google Scholar]
- Malinowski, R. , Smith, J. A. , Fleming, A. J. , Scholes, J. D. & Rolfe, S. A. 2012. Gall formation in clubroot‐infected Arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. Plant J., 71:226–238. [DOI] [PubMed] [Google Scholar]
- Marti, M. , Good, R. T. , Rug, M. , Knuepfer, E. & Cowman, A. F. 2004. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science, 306:1930–1933. [DOI] [PubMed] [Google Scholar]
- Merfa, M. V. , Pérez‐López, E. , Naranjo, E. , Jain, M. , Gabriel, D. W. & De La Fuente, L. 2019. Progress and obstacles in culturing ‘Candidatus Liberibacter asiaticus’, the bacterium associated with Huanglongbing. Phytopathology, 109(7):1092–1101. [DOI] [PubMed] [Google Scholar]
- Mueller, A. N. , Ziemann, S. , Treitschke, S. , Aßmann, D. & Doehlemann, G. 2013. Compatibility in the Ustilago maydis–maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog., 9:e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissinen, R. , Xia, Y. , Mattinen, L. , Ishimaru, C. A. , Knudson, D. L. , Knudson, S. E. , Metzler, M. & Pirhonen, M. 2009. The putative secreted serine protease Chp‐7 is required for full virulence and induction of a non‐host hypersensitive response by Clavibacter michiganensis subsp. sepedonicus . Mol. Plant Microbe Interact., 22:809–819. [DOI] [PubMed] [Google Scholar]
- Noon, J. B. , Qi, M. , Sill, D. N. , Muppirala, U. , Eves‐van, D. , Akker, S. , Maier, T. R. , Dobbs, D. , Mitchum, M. G. , Hewezi, T. & Baum, T. J. 2016. A Plasmodium‐like virulence effector of the soybean cyst nematode suppresses plant innate immunity. New Phytol., 212:444–460. [DOI] [PubMed] [Google Scholar]
- Ökmen, B. , Mathow, D. , Hof, A. , Lahrmann, U. , Aßmann, D. & Doehlemann, G. 2018. Mining the effector repertoire of the biotrophic fungal pathogen Ustilago hordei during host and non‐host infection. Mol. Plant Pathol., 19(12):2603–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐López, E. , Waldner, M. , Hossein, M. , Kusalik, A. J. , Wei, Y. , Bonham‐Smith, P. C. & Todd, C. D. 2018. Identification of Plasmodiophora brassicae effectors – a challenging goal. Virulence, 9(1):1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, T. M. , Brunak, S. , von Heijne, G. & Nielsen, H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8:785–786. [DOI] [PubMed] [Google Scholar]
- Price, C. T. D. , Al‐Khodor, S. , Al‐Quadan, T. & Kwaik, Y. A. 2010. Indispensable role for the eukaryotic‐like ankyrin domains of the Ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect. Immun., 78(5):2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, L. , Sankaran, B. , Aminzai, S. , Casteel, D. & Kim, C. 2018. Structural basis for selective inhibition of human PKG Iα by the balanol‐like compound N46. J. Biol. Chem., 293(28):10985–10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman, R. S. & Rodriguez, R. J. 2002. Characterization and isolation of an extracellular serine protease from the tomato pathogen Colletotrichum coccodes, and its role in pathogenicity. Mycol. Res., 106:1427–1434. [Google Scholar]
- Riascos, D. , Ortiz, E. , Quintero, D. , Montoya, L. & Hoyos‐Carvajal, L. 2011. Histopathological and morphological alterations caused by Plasmodiophora brassicae in Brassica oleracea L. Agron. Colomb., 29:57–61. [Google Scholar]
- Roggwiller, E. , Bétoulle, M. R. M. , Blisnick, T. & Breton, C. B. 1996. A role for erythrocyte band degradation by the parasite gp76 serine protease in the formation of the parasitophorous vacuole during invasion of erythrocytes by Plasmodium falciparum . Mol. Biochem. Parasit., 82:13–24. [DOI] [PubMed] [Google Scholar]
- Rolfe, S. A. , Strelkov, S. E. , Links, M. G. , Clarke, W. E. , Robinson, S. J. , Djavaheri, M. , Malinowski, R. , Haddadi, P. , Kagale, S. , Parkin, I. A. P. , Taheri, A. & Borhan, M. H. 2016. The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host Brassica spp. BMC Genom., 17:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovenich, H. , Boshoven, J. C. & Thomma, B. P. H. J. 2014. Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol., 20:96–103. [DOI] [PubMed] [Google Scholar]
- Salcedo, A. , Rutter, W. , Wang, S. , Akhunova, A. , Bolus, S. , Chao, S. , Anderson, N. , De Soto, M. F. , Rouse, M. & Szabo, L. 2017. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science, 358:1604–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz‐Esser, S. , Haferkamp, I. , Knab, S. , Penz, T. , Ast, M. , Kohl, C. , Wagner, M. & Horn, M. 2008. Lawsonia intracellularis contains a gene encoding a functional rickettsia‐like ATP/ADP translocase for host exploitation. J. Bacteriol., 190(17):5746–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz‐Esser, S. , Linka, N. , Collingro, A. , Beier, C. L. , Neuhaus, H. E. , Wagner, M. & Horn, M. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J. Bacteriol., 186(3):683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller, A. & Ludwing‐Müller, J. 2016. Histological methods to detect the clubroot pathogen Plasmodiophora brassicae during its complex life cycle. Plant. Pathol., 65:1223–1237. [Google Scholar]
- Schwelm, A. , Fogelqvist, J. , Knaust, A. , Jülke, S. , Lilja, T. , Bonilla‐Rosso, G. , Karlsson, M. , Shevchenko, A. , Dhandapani, V. , Choi, S. R. , Kim, H. G. , Park, J. Y. , Lim, Y. P. , Ludwig‐Müller, J. & Dixelius, C. 2015. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep., 5:11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens, J. , Nagel, M. , Ludwig‐Müller, J. & Sacristan, M. D. 2002. The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: parameters for disease quantification and screening of mutant lines. J. Phytopathol., 150:592‐05. [Google Scholar]
- Song, J. , Wina, J. , Tian, M. , Schornack, S. , Kaschani, F. , Ilyas, M. , van der Hoorn, R. A. L. & Kamoun, S. 2009. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA, 106(5):1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sova, V. V. , Elyakova, L. A. & Vaskovsky, V. E. 1970. Purification and some properties of β‐1,3‐glucan glucanohydrolase from the crystalline style of bivalvia, Spisula sachalinensis. Biochim. Biophys. Acta, 212(1):111–115. [DOI] [PubMed] [Google Scholar]
- Sperschneider, J. , Catanzariti, A.‐M. , DeBoer, K. , Petre, B. , Gardiner, D. M. , Singh, K. B. , Dodds, P. N. & Taylor, J. M. 2017b. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep., 7:44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Dodds, P. N. , Singh, K. B. & Taylor, J. M. 2017a. ApoplastP: prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol., 217(4):1764–1778. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. & de Wit, P. J. G. M. 2009. Fungal effector proteins. Annu. Rev. Phytopathol., 47:233–263. [DOI] [PubMed] [Google Scholar]
- Strelkov, S. E. , Tewari, J. P. & Smith‐Degenhardt, E. 2006. Characterization of Plasmodiophora brassicae populations from Alberta, Canada. Can. J. Plant Pathol., 28:467–474. [Google Scholar]
- Sugio, A. , Kingdom, H. N. , MacLean, A. M. , Grieve, V. M. & Hogenhout, S. A. 2011. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA, 108(48):E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K.‐C. , Chang, C.‐D. , Cheng, M.‐H. , Lin, T.‐Y. , Lo, Y.‐N. , Yang, T.‐W. , Chang, F.‐L. , Chiang, C.‐E. , Lee, Y.‐C. & Yen, Y. 2019. Chicken‐derived humanized antibody targeting a novel epitope F2pep of fibroblast growth factor receptor 2: potential cancer therapeutic agent. ACS Omega, 4:2387–2397. [Google Scholar]
- Vahling, C. M. , Duan, Y. P. & Lin, H. 2010. Characterization of an ATP translocase identified in the destructive plant pathogen “Candidatus Liberibacter asiaticus”. J. Bacteriol., 192:834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V. G. A. A. & Oliver, R. P. 2014. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact., 27(3):196–206. [DOI] [PubMed] [Google Scholar]
- Voronin, D. A. & Kiseleva, E. V. 2007. Functional role of proteins containing ankyrin repeats. Tsitologiia, 49(12):989–999. [PubMed] [Google Scholar]
- Wang, Y. & Wang, Y. 2018. Trick or treat: microbial pathogens evolved apoplastic effectors modulating plant susceptibility to infection. Mol. Plant Microbe Interact., 31:6–12. [DOI] [PubMed] [Google Scholar]
- Warnes, G. R. , Bolker, B. , Bonebakker, L. , Gentleman, R. , Huber, W. , Liaw, A. , Lumley, T. , Maechler, M. , Magnusson, A. , Moeller, S. , Schwartz, M. & Venables, B. 2016. gplots: Various R programming tools for plotting data. R package version 3.0.1. https://CRAN.R-project.org/package=gplots
- Whisson, S. C. , Boevnik, P. C. , Moleleki, L. , Avrova, A. O. , Morales, J. G. , Girloy, E. M. , Armstrong, M. R. , Grouffaud, S. , van West, P. , Chapman, S. , Hein, I. , Toth, I. K. , Pritchard, L. & Birch, P. R. J. 2007. A translocation signal for delivery of oomycete effector proteins into host plant cell. Nature, 450:115–118. [DOI] [PubMed] [Google Scholar]
- Wirthmueller, L. , Maqbool, A. & Banfield, M. J. 2013. On the front line: structural insights into plant‐pathogen interactions. Nat. Rev. Microbiol., 11:761–776. [DOI] [PubMed] [Google Scholar]
- Yaeno, T. , Li, H. , Chaparro‐Garcia, A. , Schornack, S. , Koshiba, S. , Watanabe, S. , Kigawa, T. , Kamoun, S. & Shirasu, K. 2011. Phosphatidyl inositol monophosphate‐binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc. Natl. Acad. Sci. USA, 108:14682–14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W. , Wang, Y. , Chen, T. , Lin, Y. & Luo, C. 2018. Functional evaluation of the signal peptides of secreted proteins. Bio‐Protocol, 8:e2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef, G. M. , Kopolovic, A. D. , Elliott, M. B. & Diamandis, E. P. 2003. Genomic overview of serine proteases. Biochem. Biophys. Res. Commun., 305:28–36. [DOI] [PubMed] [Google Scholar]
- Yu, F. , Wang, S. , Zhang, W. , Tang, J. , Wang, H. , Yu, L. , Zhang, X. , Fei, F. & Li, J. 2019. Genome‑wide identification of genes encoding putative secreted E3 ubiquitin ligases and functional characterization of PbRING1 in the biotrophic protist Plasmodiophora brassicae . Curr. Genet., 65:1355–1365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. 3D model of SSPbP61, SSPbP61‐like and their respective alignment.
Figure S2. Signal peptide validation of eight randomly selected SSPbPs and SSPbP31, effector candidate with lower SP score (Table 1).
Figure S3. Expression of SSPbPs identified in this study.
Table S1. Raw data of SSPbP22 kinase activity assay.
Table S2. Primers designed and used in this study.
Table S3. Plasmids constructions generated in this study.


