Summary
Loss of barley Mildew Resistance Locus O (MLO) is known to confer durable and robust resistance to powdery mildew (Blumeria graminis), a biotrophic fungal leaf pathogen. Based on the increased expression of MLO in mycorrhizal roots and its presence in a clade of the MLO family that is specific to mycorrhizal‐host species, we investigated the potential role of MLO in arbuscular mycorrhizal interactions.
Using mutants from barley (Hordeum vulgare), wheat (Triticum aestivum), and Medicago truncatula, we demonstrate a role for MLO in colonization by the arbuscular mycorrhizal fungus Rhizophagus irregularis.
Early mycorrhizal colonization was reduced in mlo mutants of barley, wheat, and M. truncatula, and this was accompanied by a pronounced decrease in the expression of many of the key genes required for intracellular accommodation of arbuscular mycorrhizal fungi.
These findings show that clade IV MLOs are involved in the establishment of symbiotic associations with beneficial fungi, a role that has been appropriated by powdery mildew.
Keywords: arbuscular mycorrhizal fungi, barley, Medicago truncatula, Mildew Resistance Locus O (MLO), powdery mildew
Short abstract
See also the Commentary on this article by Zuccaro & Langen, 227: 279–282.
Introduction
Plants have co‐evolved with microbes, which is evident in the elaborate strategies that plants use to both promote beneficial symbioses and to restrict pathogenesis. For example, to establish beneficial endosymbioses like arbuscular mycorrhization, plants have dedicated host signalling pathways involving hundreds of genes (Oldroyd, 2013; Bravo et al., 2016). Similarly, to keep apace with pathogens, components of the plant immune system are rapidly evolving and expanding (Chisholm et al., 2006; Han, 2019). Pathogens employ strategies to take advantage of host genes – so‐called ‘susceptibility factors’ – to cause disease (O'Connell & Panstruga, 2006; Boevink et al., 2016). One example of a susceptibility factor is the barley Mildew resistance locus O (MLO) gene – referred to here as MLO1 to distinguish it from other family members – which is required for successful colonization by powdery mildew fungi, Erysiphales (Jørgensen, 1992). However, the extent to which host susceptibility factors are required for beneficial symbioses is unknown.
MLO encodes a plasma membrane protein with seven transmembrane helices and a C‐terminal calmodulin‐binding domain (Devoto et al., 1999; Kim et al., 2002). Most research on MLO1 has been on its role in powdery mildew susceptibility. Barley lines with mutations in MLO1 have been successfully deployed to provide broad‐spectrum, durable resistance to powdery mildew since 1942 (Freisleben & Lein, 1942). Despite considerable progress on the characterization of the powdery mildew resistance phenotype and its underlying molecular mechanism, MLO1's biological role remains unclear (Kim et al., 2002; Piffanelli et al., 2002; Consonni et al., 2010). Notably, orthologues of MLO1 are only present in plant species that can host arbuscular mycorrhizal fungi (Bravo et al., 2016). The study of other MLO family members has revealed their involvement in diverse plant processes, including pollen tube reception (AtMLO7/NORTIA) and root thigmomorphogenesis (Kessler et al., 2010; Bidzinski et al., 2014). The ancestral function of MLO proteins is unknown, but their presence in green algae suggests that the so‐far characterized roles are derivative (Jiao et al., 2011). In Arabidopsis thaliana, loss of MLO family members belonging to a separate clade from barley MLO1 confers resistance to powdery mildew in a redundant fashion (Kusch et al., 2016; Kuhn et al., 2017). Interestingly, one of these can complement the Atmlo7 pollen tube reception phenotype, suggesting a conserved biochemical function of MLO proteins (Jones et al., 2017).
Most land plants can form an endosymbiosis with members of the Glomeromycotina, reflecting the ancient origins of this interaction (Remy et al., 1994; Spatafora et al., 2016). This mutualistic relationship supplies water and nutrients – particularly phosphate, but also nitrogen and zinc – to the host plant (Cooper & Tinker, 1978; Govindarajulu et al., 2005; Watts‐Williams & Cavagnaro, 2012). Several key components of the signalling pathway required for establishing mycorrhizal symbioses (the common SYM pathway) are conserved in green algae, suggesting that land plant ancestors were preadapted for symbiosis (Delaux et al., 2013 2015). Furthermore, MLO1 was previously suggested to be required for colonization by arbuscular mycorrhizal fungi (Ruiz‐Lozano et al., 1999). By contrast, a recent study found that a Hvmlo1 mutant had increased mycorrhizal colonization (Hilbert et al., 2019). These studies were limited to a single allele evaluated at a single time point. Another study of an MLO from a different clade found no evidence for a role in arbuscular mycorrhization of pea (Humphry et al., 2011).
Using time‐course analyses of mutants from barley, wheat, and Medicago truncatula and transcript profiling of Hvmlo1 mutant mycorrhizal roots, we reveal a clear role for MLO in the early stages of arbuscular mycorrhizal colonization in angiosperms. The findings suggest a role for clade IV MLOs in the establishment of beneficial fungal symbiosis, which has been exploited by pathogens such as powdery mildew.
Materials and Methods
Phylogenetic analyses
Sequences were aligned using Muscle (Edgar, 2004), and a maximum likelihood tree was constructed using Mega X following a Jones–Taylor–Thornton model with 100 bootstrap replications (Kumar et al., 2018). Protein sequences and references are in Supporting Information Methods S1.
Barley, wheat and M. truncatula mutant molecular analyses
Details of the barley (Hordeum vulgare L.) and wheat (Triticum aestivum L.) mlo mutants used are in Methods S2. All primers are listed in Methods S3. To identify homozygous M. truncatula Gaertn. mlo8 Tnt1‐insertion mutants, DNA was extracted (DNeasy 96 Plant Kit; Qiagen), and genotyping of segregating seedling populations was performed by PCR. To detect MtMLO8 cDNA, RNA was extracted from mycorrhizal M. truncatula roots (Plant RNeasy Kit; Qiagen). DNases were removed (Turbo DNA‐free; Ambion), and 150 ng of RNA was retrotranscribed (SuperScript IV reverse transcriptase; Invitrogen). cDNA was amplified by RT‐PCR (Phusion High Fidelity Polymerase kit; New England Biolabs, Ipswich, MA, USA).
Plant growth conditions and mycorrhization and powdery mildew assays
For mycorrhization experiments, barley, wheat, and M. truncatula were grown in pots containing 80–90% Terragreen/sand and 10–20% mycorrhizal inoculum (homogenized soil substrate containing Allium schoenoprasum roots colonized by Rhizophagus irregularis DAOM 197198). Full descriptions of seed sterilization, soils substrates, and growth conditions for mycorrhization and powdery mildew assays are in Methods S4.
Agrobacterium rhizogenes‐mediated gene transfer
Agrobacterium rhizogenes‐mediated gene transfer (Boisson‐Dernier et al., 2001), was performed using strain AR1193. The promoter‐GUS construct pMtMLO8:GUS was generated using the Gateway® system (Primrose & Twyman, 2013): entry vector pDONR207; destination vector 243_pKGW‐GGRR. The 1983 bp promoter region of MtMLO8 (Medtr3g115940) was amplified using primers with Gateway‐compatible end sequences (Methods S3). The complementation construct pMtMLO8:MtMLO8 was generated using the Golden Gate system (Engler et al., 2009) using the same MtMLO8 promoter region fused to the 1656 bp coding region. Domesticated DNA parts were synthesized by GeneArt® (Life Technologies, Carlsbad, CA, USA). pL2V‐50507 was used for the L2 backbone, with pLjUBIQUTIN:DsRed:t53S placed in position 1, and pMtMLO8:MLO8:t35S in position 2.
Fungal staining and quantification
Staining (ink, GUS, and WGA‐Alexa488) and visualization methods are summarized in Methods S5. Samples were blinded, and mycorrhizal colonization structures quantified using the gridline intersect method (Giovannetti & Mosse, 1980). Powdery mildew fungal structures were quantified by calculating the percentage host cell entry (the ratio of germinated spores : colony formation).
Gene expression analyses
RNAs were extracted from root tissue (Plant RNeasy Kit; Qiagen) and were treated with DNase (RNase‐Free DNase Set; Qiagen). Methods for RT‐qPCR and transcriptome analyses using RNA‐sequencing can be found in Methods S6.
Results
To assess a potential function for MLO1 in mycorrhization, we compared expression levels of barley MLO1 (HvMLO1) in mycorrhizal and nonmycorrhizal roots (Fig. 1a). At 22 d post‐inoculation (dpi) with R. irregularis, RT‐qPCR analysis indicated that HvMLO1 was induced in mycorrhizal roots. We then evaluated mycorrhizal phenotypes of barley Hvmlo1 mutants by quantification of mycorrhizal fungal colonization structures over a time‐course. Arbuscules and vesicles were quantified at 22, 29, and 36 dpi with R. irregularis (20% inoculum) in barley cv Ingrid wild‐type (WT), Hvmlo1‐1, and Hvmlo1‐5 roots (Fig. 1b; Gene structures in Methods S2). We found a reduction in arbuscule and vesicle occurrence in Hvmlo1‐1 and Hvmlo1‐5 compared to WT at 22 dpi. This difference was maintained in Hvmlo1‐5 at 29 dpi, whereas Hvmlo1‐1 showed a similar phenotype to WT. By 36 dpi, no significant difference in mycorrhization between the lines was detectable. WT roots were fully colonized by mycorrhizal fungi by 22 and 29 dpi, whereas the Hvmlo1 mutants established full colonization between 29 and 36 dpi. Overall, reduced mycorrhization was more evident at early time points, suggesting that mycorrhizal development in Hvmlo1 is delayed but later recovers to WT levels.
Figure 1.
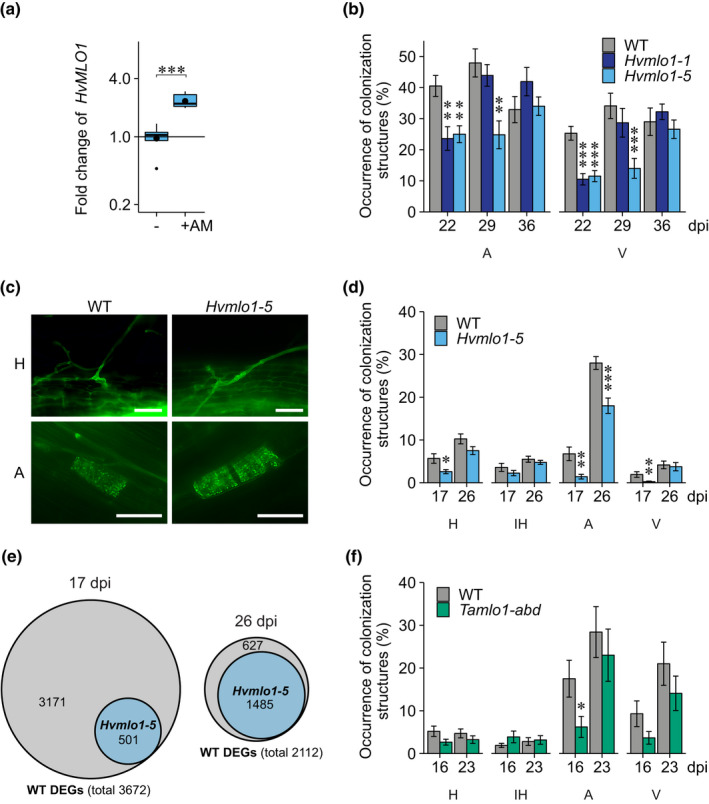
Mycorrhizal colonization in barley (Hordeum vulgare) and wheat (Triticum aestivum) mlo1 mutants. (a) Relative expression of HvMLO1 in wild‐type (WT) barley cv Ingrid roots without (−) and with Rhizophagus irregularis (+AM) at 22 d post inoculation (dpi). Relative expression levels were measured by RT‐qPCR and normalized to HvEF1alpha (barley). Statistical comparisons were made relative to nonmycorrhizal root samples. Boxplots represent means (large black dots), medians (black lines), 25–75% quartile (box), and upper/lower quartile ±1.5 × interquartile range (whiskers) of eight biological replicates (Student's t‐test: ***, P < 0.001). (b, d, f) Quantification of arbuscular mycorrhizal colonization structures in barley cv Ingrid and wheat cv KN1999 WT and mlo1 roots after inoculation with R. irregularis: hyphopodia (H), intraradical hyphae (IH), arbuscules (A), and vesicles (V). The binomial occurrences of mycorrhizal structures are shown as a percentage of the total number of root sections assessed. Statistical comparisons have been made to the WT. Values are the mean of 12 biological replicates ± 1 SEM (error bars) (General Linear Model with a logit link function; ANOVA; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (c) Appearance of hyphopodia (H) and arbuscules (A) in WT and Hvmlo1‐5 roots at 17 dpi with R. irregularis. Mycorrhizal fungal structures were stained using Alexa Fluor 488 wheat germ agglutinin. Photographs indicate representative examples from 40 observations. Bars, 50 μm. (e) Venn diagrams showing the number of differentially regulated genes (DEGs) in mycorrhizal roots relative to uninoculated roots in WT and the proportion of genes similarly responding in Hvmlo1‐5 using a cut‐off of fold change > 2; FDR‐corrected P‐value < 0.05.
To further assess the mycorrhizal phenotype of Hvmlo1 during the early stages of colonization, we performed a detailed analysis of symbiotic structures (hyphopodia, intraradical hyphae, arbuscules and vesicles) at early time points using less R. irregularis inoculum (10% inoculum) (Fig. 1c,d). At 17 dpi, Hvmlo1‐5 showed a significant reduction in the occurrence of hyphopodia, arbuscules, and vesicles compared to WT. Similar to the previous experiment, by 26 dpi, Hvmlo1‐5 showed a significant reduction in only arbuscule occurrence compared to WT. A similar phenotype was observed between WT and Hvmlo1‐5 in a barley cv Pallas near‐isogenic line, in two separate experiments (Fig. S1a), suggesting the phenotype is stable across different genetic backgrounds. We observed no differences in the morphology of hyphopodia or arbuscules in Hvmlo1‐5 mutants (Fig. 1c).
To gain insight into the nature of the delayed colonization of Hvmlo1‐5, we carried out comparative transcriptomic analyses at 17 dpi and 26 dpi with R. irregularis (Figs 1e, S2). At 17 dpi, there were considerably fewer differentially regulated genes (DEGs) in Hvmlo1‐5 than WT, with 86% (3171/3672) of all WT DEGs not responding in Hvmlo1‐5. By 26 dpi, despite the recovery of observable colonization in the mutant, there was still a substantial (c. 30%) reduction of the number of DEGs in Hvmlo1‐5 relative to WT, suggesting persistent perturbation of the mycorrhizal interaction.
To further investigate the nature of the differences in Hvmlo1‐5, we examined the expression of potential barley orthologues of genes known to be involved in mycorrhizal symbiosis in other species. Amongst the genes induced more than two‐fold in WT – but not in Hvmlo1‐5 – at 17 dpi, were genes required for early colonization and components of the common signalling pathway required for nodulation and mycorrhization, VAPYRIN, NSP1, NSP2, DMI2, IPD3 and NOPE1 homologues (Catoira et al., 2000; Ané et al., 2002; Lévy et al., 2004; Messinese et al., 2007; Pumplin et al., 2010; Murray et al., 2011; Nadal et al., 2017) (Tables 1, S1, S2; Fig. S3). This suggests that MLO1 is required for timely or full‐activation of early processes of mycorrhizal colonization. Also, the expression of potential orthologues of several genes involved in arbuscule development and function were affected in Hvmlo1‐5, including RAM1, RAD1, RAM2, EXO70I, STR, STR2 AMT2‐3, AMT3‐4 and PT4 homologues (Table 1; Fig. S3) (Harrison et al., 2002; Zhang et al., 2010; Gobbato et al., 2012; Wang et al., 2012; Breuillin‐Sessoms et al., 2015; Xue et al., 2015; Zhang et al., 2015). Generally, these genes were induced to a lesser extent than WT at 17 dpi, and partly or fully recovered by 26 dpi, which mirrors the initial delay and later recovery of the colonization phenotype. Together, these phenotypic and transcriptomic results indicate a role for MLO1 in the early stage of mycorrhization.
Table 1.
Counts per million (CPM) values of potential barley (Hordeum vulgare (Hv)) orthologues of Medicago truncatula (Mt) genes previously described to be involved in mycorrhizal symbiosis in wild‐type (WT) and Hvmlo1‐5 nonmycorrhizal and mycorrhizal roots: significant fold changes between WT and Hvmlo1‐5 at corresponding time points and treatments are indicated.
| Mt gene | Mt identifier | Hv identifier | Mean normalized CPMs | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 17 dpi | 26 dpi | |||||||||
| Nonmycorrhizal | Mycorrhizal | Nonmycorrhizal | Mycorrhizal | |||||||
| WT | Hvmlo1‐5 | WT | Hvmlo1‐5 | WT | Hvmlo1‐5 | WT | Hvmlo1‐5 | |||
| VAPYRIN | Medtr6g027840 | HORVU4Hr1G002180 | 14.3 | 11.0 | 32.2 | 16.2 *** | 17.0 | 18.8 | 62.4 | 63.6 |
| NSP1 | Medtr8g020840 | HORVU2Hr1G104160 | 1.0 | 2.6 | 4.0 | 2.2 * | 6.0 | 6.9 | 8.9 | 8.6 |
| NSP2 | Medtr3g072710 | HORVU4Hr1G061310 | 2.7 | 3.2 | 11.2 | 2.5 *** | 4.5 | 5.8 | 47.2 | 57.3 |
| DMI1 | Medtr2g005870 | HORVU5Hr1G120340 | 28.3 | 33.3 | 44.2 | 33.2 ** | 61.1 | 62.1 | 80.6 | 74.6 |
| DMI2 | Medtr5g030920 | HORVU2Hr1G058820 | 11.1 | 17.6 | 72.8 | 29.2 *** | 57.0 | 71.3 | 181.0 | 195.0 |
| DMI3 | Medtr8g043970 | HORVU1Hr1G068660 | 14.0 | 17.0 | 23.8 | 14.9 ** | 38.4 | 35.2 | 52.3 | 48.6 |
| IPD3 | Medtr5g026850 | HORVU7Hr1G008420 | 12.6 | 19.8 | 56.4 | 22.9 *** | 54.7 | 56.7 | 158.8 | 138.8 |
| NOPE1 |
Medtr3g093270 Medtr3g093290 |
HORVU2Hr1G005470 | 9.1 | 10.1 | 53.5 | 18.5 *** | 18.8 | 15.9 | 121.8 | 149.8 |
| RAD1 | Medtr4g104020 | HORVU3Hr1G088780 | 0.3 | 0.5 | 7.6 | 0.4 *** | 1.5 | 1.6 | 16.9 | 15.6 |
| RAM2 | Medtr1g040500 | HORVU4Hr1G011110 | 5.3 | 4.7 | 30.6 | 5.5 *** | 7.7 | 8.5 | 77.7 | 68.1 |
| EXO70I | Medtr1g017910 | HORVU7Hr1G052100 | 0.1 | 0.1 | 11.6 | 2.2 *** | 0.1 | 0.1 | 34.0 | 37.4 |
| STR | Medtr8g107450 | HORVU5Hr1G060690 | 1.3 | 0.5 | 14.5 | 1.7 *** | 0.7 | 0.4 | 56.1 | 50.4 |
| STR2 | Medtr5g030910 | HORVU3Hr1G066220 | 0.0 | 0.0 | 26.2 | 2.5 *** | 0.1 | 0.2 | 83.3 | 84.2 |
| AMT2‐3 | Medtr8g074750 | HORVU3Hr1G082610 | 0.0 | 0.0 | 14.3 | 0.7 *** | 0.0 | 0.0 | 53.8 | 45.5 |
| AMT2‐4 | Medtr7g115050 | HORVU5Hr1G095030 | 0.0 | 0.0 | 3.0 | 0.5 *** | 0.0 | 0.0 | 9.8 | 12.0 |
| PT4 | Medtr8g074750 | HORVU6Hr1G058690 | 0.0 | 0.0 | 3.9 | 1.0 | 0.0 | 0.0 | 16.6 | 21.0 |
FDR‐corrected P‐value; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
dpi, days post inoculation.
To investigate the conservation of function for this MLO, we used phylogenetic analysis to identify potential functional orthologues of HvMLO1 (Fig. 2). The analysis included mycorrhizal host species: basal angiosperm Amborella trichopoda; dicots Glycine max, M. truncatula, Pisum sativum, and Solanum lycopersicum; and monocots H. vulgare, Oryza sativa, and T. aestivum, as well as nonmycorrhizal host species: moss Physcomitrella patens; and dicots Arabidopsis thaliana, Beta vulgaris, Dianthus caryophyllus, and Lupinus angustifolius. As previously shown (Kusch et al., 2016), the phylogenetic analysis supported the view that embryophyte MLO proteins diverged into seven clades. HvMLO1 groups in clade IV, which is comprised only of species that are mycorrhizal‐hosts, while powdery mildew susceptibility factors identified in monocots and dicots are found in clades IV and V, respectively.
Figure 2.
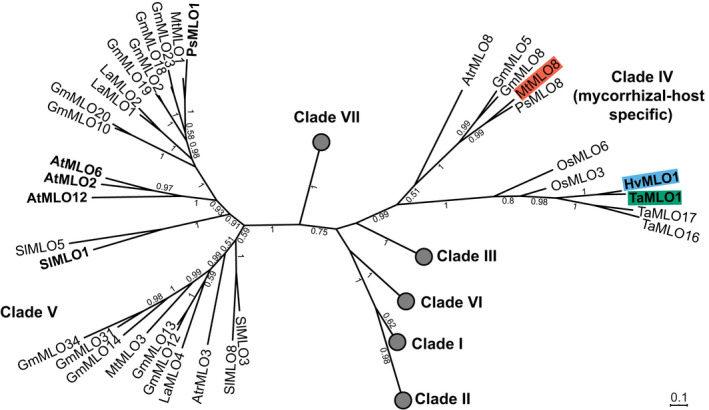
MLO phylogeny. A phylogenetic tree showing the relationship between MLO proteins from Amborella trichopoda (Atr), Arabidopsis thaliana (At), Beta vulgaris (Bv), Dianthus caryophyllus (Dc), Glycine max (Gm), Hordeum vulgare (Hv), Lupinus angustifolius (La), Medicago truncatula (Mt), Oryza sativa (Os), Pisum sativum (Ps), Physcomitrella patens (Pp), Solanum lycopersicum (Sl) and Triticum aestivum (Ta). The T. aestivum MLO protein dataset often included near‐identical homoeoalleles (A, B and D copies); therefore, for simplicity, only one homoeoallele was included in the phylogenetic analysis (A homoeoallele where possible). The tree was generated using Mega X from an amino acid alignment. The phylogenetic tree was calculated via the maximum likelihood method, using Jones–Taylor–Thornton modelling with 100 bootstrap replications. Bootstrap values > 0.5 are shown. The scale bar indicates the evolutionary distance based on the amino acid substitution rate. Circles indicate collapsed clades; known powdery mildew susceptibility factors are labelled in bold, and candidate MLO proteins for a role in mycorrhization are highlighted.
The wheat functional orthologue of HvMLO1, TaMLO1, is a powdery mildew host susceptibility factor (Wang et al., 2014). Tamlo1‐abd contains TALEN‐induced mutations in all three homoeoalleles (A, B, and D) (Wang et al., 2014). To assess the mycorrhizal phenotype of Tamlo1‐abd during the early stages of mycorrhization, we performed a detailed analysis of mycorrhizal structures (hyphopodia, intraradical hyphae, arbuscules, and vesicles) at early time points similar to those used for barley (Figs 1f, S1b). At 16 dpi with R. irregularis, Tamlo1‐abd showed a significant reduction in arbuscule occurrence compared to WT. However, consistent with our findings in barley, by 23 dpi Tamlo1‐abd colonization levels were similar to WT. Wheat mlo1 mutants exhibited the delayed mycorrhizal phenotype in two separate experiments, suggesting a conserved role for MLO1 in mycorrhization in cereals.
To determine whether MLO's role in mycorrhization extends to dicots, we investigated M. truncatula MtMLO8, the apparent orthologue of HvMLO1, which was previously found to be conserved exclusively in mycorrhizal host plants (Bravo et al., 2016). We first tested its expression in mycorrhizal roots, and we found that, like HvMLO1, MtMLO8 was induced relative to nonmycorrhizal roots (Figs 3a, S4a), consistent with data from several studies available in the public database (Benedito et al., 2008; Breakspear et al., 2014; Luginbuehl et al., 2017). To establish in which cell types MtMLO8 is expressed, we studied its expression in M. truncatula roots using the MtMLO8 promoter to drive GUS expression using the Agrobacterium rhizogenes mediated hairy root transformation system. MtMLO8 promoter activity was strongly associated with cortical cells containing arbuscules (Figs 3b, S4b).
Figure 3.
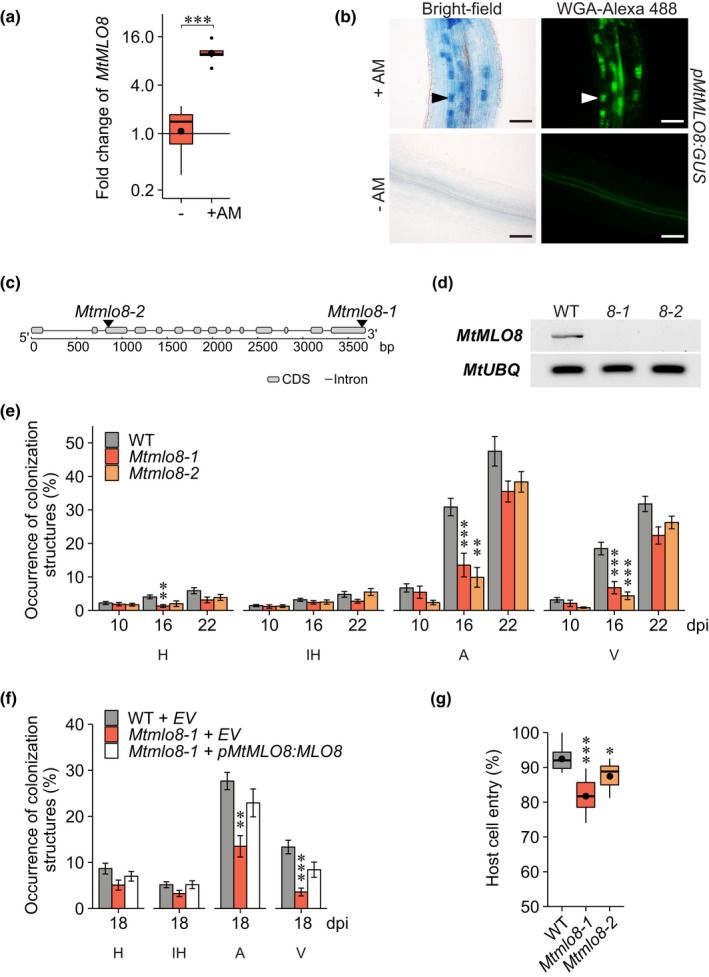
MtMLO8 is involved in mycorrhizal colonization of Medicago truncatula. (a) Relative expression of MtMLO8 in wild‐type (WT) M. truncatula cv R108 without (−) and with Rhizophagus irregularis (+AM) at 22 d post inoculation (dpi). Relative expression levels were measured by RT‐qPCR and normalized to the geometric mean of MtUBIQUITIN and MtPTB. Statistical comparisons were made relative to nonmycorrhizal root samples. Boxplots represent means (large black dots), medians (black lines), 25–75% quartile (box), and upper/lower quartile ±1.5 × interquartile range (whiskers) of eight biological replicates (Student's t‐test: ***, P < 0.001). (b) Activity of the MtMLO8 promoter in mycorrhizal (+AM) and nonmycorrhizal (−AM) WT roots at 21 dpi with R. irregularis, assessed using a promoter‐GUS fusion. Bright‐field and corresponding green fluorescence images of M. truncatula hairy roots expressing the β‐glucuronidase (GUS) gene under the control of the MtMLO8 promoter. Mycorrhizal fungal structures were visualized using Alexa Fluor 488 wheat germ agglutinin. Arrowheads indicate examples of cells containing arbuscules. Bars, 100 μm. (c) Gene structure of M. truncatula MtMLO8. Arrows indicate the Tobacco retrotransposon 1 (Tnt1) insertion sites in the Mtmlo8 mutants. (d) RT‐PCR was used to detect the accumulation of the MtMLO8 transcript in WT, Mtmlo8‐1, and Mtmlo8‐2 mutant roots. MtUBIQUITIN was used as a constitutive control. (e) Quantification of mycorrhizal structures in M. truncatula cv R108 WT, Mtmlo8‐1 and Mtmlo8‐2 roots. (f) Complementation of the Mtmlo8‐1 mutants and quantification of mycorrhizal structures in hairy roots. Mtmlo8‐1 was transformed with pMtMLO8:MtMLO8 or empty vector (EV) control, and WT were transformed with EV control. The binomial occurrences of mycorrhizal structures, hyphopodia (H), intraradical hyphae (IH), arbuscules (A), and vesicles (V), are shown as a percentage of the total number of root systems assessed. (g) Quantification of powdery mildew infection on WT, Mtmlo8‐1 and Mtmlo8‐2 leaves at 72 h post inoculation with Erysiphe pisi. The binomial occurrences of successful host cell entry (ratio of germinated spores: colony formation) are shown as a percentage of the total number of spores observed. For (e–g) statistical comparisons have been made to the WT. Bars represent the mean of 15 (e) and 10 (f) biological replicates ± 1 SEM (error bars) and boxplots (g) represent means (black dots), medians (black lines), 25–75% quartile (box), and upper/lower quartile ±1.5 × interquartile range (whiskers) of eight biological replicates (General Linear Model with a logit link function; ANOVA, post hoc pairwise; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
To investigate whether MtMLO8 functions during mycorrhization, seeds from two independent M. truncatula cv R108 lines carrying Tnt1 insertions in the coding sequence of these genes were obtained, and homozygous mutants were identified (Fig. 3c,d) (Tadege et al., 2008). We performed a detailed analysis of mycorrhizal structures (hyphopodia, intraradical hyphae, arbuscules, and vesicles) at multiple time points in WT, Mtmlo8‐1, and Mtmlo8‐2. Compared to WT, there was a significant reduction in arbuscules and vesicles at 16 dpi in both mutant alleles of Mtmlo8 (Fig. 3e), consistent with our results using the orthologous mutants in barley and wheat. By 22 dpi – and at later time points (Fig. S5a) – there was no difference between the lines. We observed no impairment in the morphology of mycorrhizal structures – hyphopodia and arbuscules – in Mtmlo8 mutants (Fig. S5b). To validate that mycorrhizal phenotype observed was due to mutations in MtMLO8, and not a consequence of additional Tnt1 insertions or other mutations in the mutant backgrounds, we expressed MtMLO8 from its native promoter in Mtmlo8‐1 mutant roots by transformation with Agrobacterium rhizogenes. When arbuscule occurrence in roots of WT plants transformed with the empty vector reached c. 30% – corresponding to when the Mtmlo8 phenotype could be observed – mycorrhizal structures (hyphopodia, intraradical hyphae, arbuscules, and vesicles) were quantified in the transgenic roots. The expression of MtMLO8 under the control of its native promoter successfully complemented the Mtmlo8‐1 mutant phenotype (Fig. 3f). In summary, these results suggest that mutations in MtMLO8 affect the early stages of mycorrhization and delay arbuscule development.
To assess whether, like HvMLO1 and TaMLO1, MtMLO8 might also have a role in powdery mildew susceptibility, we inoculated WT and Mtmlo8 mutant leaves with an isolate of powdery mildew that can infect M. truncatula, Erysiphe pisi. We observed a small but statistically significant reduction in the percentage of successful host cell entry in Mtmlo8 compared to WT (Fig. 3g). This result indicates that MtMLO8 is required not only for early mycorrhizal colonization but also for powdery mildew colonization.
Discussion
Since MLO's function in facilitating powdery mildew infection is disadvantageous to the host, it follows that it must also fulfil some other beneficial role that explains its conservation throughout evolutionary history. The phylogenetic analysis, mutant phenotypes, and gene expression studies presented here point to a symbiotic role for members of the MLO clade IV and suggest that – at least in part – selection to maintain this gene is related to its function in mycorrhizal interactions. Recent evidence also suggests a positive role for MLO1 in supporting endophytic interactions with Serendipita indica in barley (Hilbert et al., 2019). A common thread between the established roles of MLO in fungal interactions, pollen‐tube reception, and thigmotropism is touch‐sensing. Indeed, MLO proteins have been reported to accumulate at the site of powdery mildew penetration (Bhat et al., 2005), as well as at the contact point between the pollen tube and synergid cell (Kessler et al., 2010). These responses could result from a generic response to mechanical stimuli. Notably, a role for mechanosensing has been proposed for the receptor kinase Feronia (Hamant & Haswell, 2017), which was shown to be required for proper re‐localization of MLO7/NORTIA to the site of pollen tube contact in the synergid cell (Kessler et al., 2010; Jones et al., 2017). However, other cues may activate MLO, for example, the biotrophic fungi Blumeria graminis, R. irregularis, and Serendipita indica produce effectors that could target MLO (Kloppholz et al., 2011; Akum et al., 2015; Requena et al., 2018; Pennington et al., 2019; Saur et al., 2019). Regardless of how it is activated, it will be of interest to study the localization of MLO1 or its orthologues during hyphopodium formation.
The mycorrhizal phenotype of the mlo mutants examined was not severe, but may be expected to have measurable consequences on fitness in a natural setting with competition for limited resources. Furthermore, global gene expression was still affected at the later time point, so a detailed study of growth and nutrient uptake characteristics of the mutant may show differences. Also, the increased expression of other MLOs during mycorrhization in M. truncatula (Fig. S3a) suggests that – as for other mlo phenotypes such as powdery mildew resistance (Kuhn et al., 2017) and synergid cell reception (Jones & Kessler, 2017) – MLOs act redundantly, with different MLOs potentially contributing unequally across different cell types and developmental stages. It would be interesting to test whether MtMLO3 – which was highly induced during mycorrhization – and MLO genes with high epidermal expression (Breakspear et al., 2014), function together with MtMLO8 during arbuscular mycorrhizal colonization. The powdery mildew phenotype of Mtmlo8 was relatively minor. Additional MLO genes may contribute to powdery mildew susceptibility – as is the case in Arabidopsis thaliana – for example, MtMLO1, the apparent orthologue of PsMLO1, which is involved in powdery mildew susceptibility in pea (Humphry et al., 2011; Pavan et al., 2011). Notably, the narrow time window for mycorrhizal phenotype definition and potential functional redundancy with gene family members could explain why MLO has not so far been identified in mutagenesis screens.
Despite having yield penalties and potentially enhancing susceptibility to necrotrophic pathogens, mutants for MLO have been deployed in agriculture for decades (Brown, 2002; McGrann et al., 2014). Understanding the role of MLO in symbiotic interactions with mycorrhizal fungi is therefore vital information for sustainable agriculture (Martin et al., 2018). Our detailed phenotypic and transcriptomic analyses in both cereals and M. truncatula provide the basis for further elucidation of MLO function in mycorrhization and how plants balance this beneficial association with susceptibility to parasitic powdery mildews. Since mycorrhizal fungi and powdery mildew respectively infect root and shoot, it may be possible to generate genotypes that could fully support mycorrhiza while remaining nonhosts for powdery mildew.
Author contributions
CNJ performed experimental work and data analyses. MC, JDM and CJR supervised and co‐directed the research. CNJ, JDM, CJR and MC wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Mycorrhization in barley (Hordeum vulgare) cv Pallas and wheat (Triticum aestivum) cv KN199 wild‐type (WT) and mlo mutants.
Fig. S2 Proportion of differentially expressed genes (DEGs) in barley (Hordeum vulgare) cv Ingrid wild‐type (WT) and Hvmlo1‐5 mutants in mycorrhizal roots relative to uninoculated roots.
Fig. S3 Relative expression of potential barley (Hordeum vulgare) orthologues of genes known to be involved in mycorrhizal symbiosis.
Fig. S4 Medicago truncatula MLO gene expression.
Fig. S5 Mycorrhization in Medicago truncatula wild‐type (WT) and Mtmlo8 mutants.
Methods S1 MLO protein sequences.
Methods S2 Gene structure of barley (Hordeum vulgare) and wheat (Triticum aestivum) MLO1.
Methods S3 Primer sequences.
Methods S4 Sterilization methods and growth conditions.
Methods S5 Staining and visualization methods.
Methods S6 Gene expression analyses.
Table S1 Normalized read counts for RNA‐seq samples obtained from barley (Hordeum vulgare) cv Ingrid wild‐type and Hvmlo1‐5 roots at 17 dpi and 26 dpi with and without Rhizophagus irregularis.
Table S2 RNA‐seq analysis – Fold changes (log2) of RNA‐seq samples obtained from barley (Hordeum vulgare) cv Ingrid wild‐type and Hvmlo1‐5 roots at 17 dpi and 26 dpi with and without Rhizophagus irregularis.
Acknowledgements
The wheat Tamlo1‐abd was kindly supplied by Caixia Gao, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China. Erysiphe pisi (Ep) CJ001 spores kindly supplied by Professor Diego Rubiales, Institute for Sustainable Agriculture, CSIC, Córdoba, Spain. The authors would like to thank Burkhard Steuernagel for aligning our RNA‐sequencing data and advice with analyses; James Brown for advice on statistics; Alina‐Andrada Igna for her preliminary experiments in M. truncatula; and Erin Zess for feedback during the editing of this manuscript. The authors want to thank past and present members of the Ridout, Murray, and Charpentier laboratories for help and advice. JDM was supported by the National Key R&D Programme of China (2016YFA0500500), the Biotechnology and Biological Sciences Research Council (grant no. BB/L010305/1 [David Phillips Fellowship]). CNJ was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) Norwich Research Park Bioscience Doctoral Training Grants BB/J014524/1 and BB/M011216/1. MC is supported by the BBSRC grant BB/P007112/1. CJR is supported by BBSRC grant BB/P012574/1.
See also the Commentary on this article by Zuccaro & Langen, 227: 279–282.
Contributor Information
Jeremy D. Murray, Email: jeremy.murray@jic.ac.uk.
Christopher J. Ridout, Email: christopher.ridout@jic.ac.uk.
References
- Akum FN, Steinbrenner J, Biedenkopf D, Imani J, Kogel K‐H. 2015. The Piriformospora indica effector PIIN_08944 promotes the mutualistic Sebacinalean symbiosis. Frontiers in Plant Science 6: 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané J‐M, Lévy J, Thoquet P, Kulikova O, de Billy F, Penmetsa V, Kim D‐J, Debellé F, Rosenberg C, Cook DR et al 2002. Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod Factor transduction, nodulation, and mycorrhization. Molecular Plant–Microbe Interactions 15: 1108–1118. [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres‐Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T. 2008. A gene expression atlas of the model legume Medicago truncatula . The Plant Journal 55: 504–513. [DOI] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze‐Lefert P, Panstruga R. 2005. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proceedings of the National Academy of Sciences, USA 102: 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidzinski P, Noir S, Shahi S, Reinstaedler A, Gratkowska DM, Panstruga R. 2014. Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana MILDEW LOCUS O genes. Plant, Cell & Environment 37: 2738–2753. [DOI] [PubMed] [Google Scholar]
- Boevink PC, McLellan H, Gilroy EM, Naqvi S, He Q, Yang L, Wang X, Turnbull D, Armstrong MR, Tian Z. 2016. Oomycetes seek help from the plant: Phytophthora infestans effectors target host susceptibility factors. Molecular Plant 9: 636–638. [DOI] [PubMed] [Google Scholar]
- Boisson‐Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. 2001. Agrobacterium rhizogenes‐transformed roots of Medicago truncatula for the study of nitrogen‐fixing and endomycorrhizal symbiotic associations. Molecular Plant–Microbe Interactions 14: 695–700. [DOI] [PubMed] [Google Scholar]
- Bravo A, York T, Pumplin N, Mueller LA, Harrison MJ. 2016. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nature Plants 2: 15208. [DOI] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE. 2014. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. The Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuillin‐Sessoms F, Floss DS, Gomez SK, Pumplin N, Ding Y, Levesque‐Tremblay V, Noar RD, Daniels DA, Bravo A, Eaglesham JB et al 2015. Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the Ammonium Transporter 2 family protein AMT2;3. The Plant Cell 27: 1352–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JK. 2002. Yield penalties of disease resistance in crops. Current Opinion in Plant Biology 5: 339–344. [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet E‐P, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J. 2000. Four genes of Medicago truncatula controlling components of a Nod Factor transduction pathway. The Plant Cell 12: 1647–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. 2006. Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Consonni C, Bednarek P, Humphry M, Francocci F, Ferrari S, Harzen A, Loren Ver, van Themaat E, Panstruga R. 2010. Tryptophan‐derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiology 152: 1544–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KM, Tinker P. 1978. Translocation and transfer of nutrients in vesicular‐arbuscular mycorrhizas: ii. Uptake and translocation of phosphorus, zinc and sulphur. New Phytologist 81: 43–52. [Google Scholar]
- Delaux P‐M, Radhakrishnan GV, Jayaraman D, Cheema J, Malbreil M, Volkening JD, Sekimoto H, Nishiyama T, Melkonian M, Pokorny L. 2015. Algal ancestor of land plants was preadapted for symbiosis. Proceedings of the National Academy of Sciences, USA 112: 13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P‐M, Séjalon‐Delmas N, Bécard G, Ané J‐M. 2013. Evolution of the plant–microbe symbiotic ‘toolkit’. Trends in Plant Science 18: 298–304. [DOI] [PubMed] [Google Scholar]
- Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze‐Lefert P. 1999. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. Journal of Biological Chemistry 274: 34993–35004. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden Gate shuffling: a one‐pot DNA shuffling method based on Type IIs restriction enzymes. PLoS ONE 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisleben R, Lein A. 1942. Über die Auffindung einer mehltauresistenten Mutante nach Röntgenbestrahlung einer anfälligen reinen Linie von Sommergerste. Naturwissenschaften 30: 608–608. [Google Scholar]
- Giovannetti M, Mosse B. 1980. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist 84: 489–500. [Google Scholar]
- Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P. 2012. A GRAS‐type transcription factor with a specific function in mycorrhizal signaling. Current Biology 22: 2236–2241. [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bücking H, Lammers PJ, Shachar‐Hill Y. 2005. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435: 819–823. [DOI] [PubMed] [Google Scholar]
- Hamant O, Haswell ES. 2017. Life behind the wall: sensing mechanical cues in plants. BMC Biology 15: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GZ. 2019. Origin and evolution of the plant immune system. New Phytologist 222: 70–83. [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. 2002. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. The Plant Cell 14: 2413–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert M, Novero M, Rovenich H, Mari S, Grimm C, Bonfante P, Zuccaro A. 2019. MLO differentially regulates barley root colonization by beneficial endophytic and mycorrhizal fungi. Frontiers in Plant Science 10: 1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry M, Reinstädler A, Ivanov S, Bisseling T, Panstruga R. 2011. Durable broad‐spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss‐of‐function mutations in PsMLO1 . Molecular Plant Pathology 12: 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS et al 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Jones DS, Kessler SA. 2017. Cell type‐dependent localization of MLO proteins. Plant Signaling & Behavior 12: 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DS, Yuan J, Smith BE, Willoughby AC, Kumimoto EL, Kessler SA. 2017. MILDEW RESISTANCE LOCUS O function in pollen tube reception is linked to its oligomerization and subcellular distribution. Plant Physiology 175: 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen IH. 1992. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63: 141–152. [Google Scholar]
- Kessler SA, Shimosato‐Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. 2010. Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971. [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze‐Lefert P. 2002. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451. [DOI] [PubMed] [Google Scholar]
- Kloppholz S, Kuhn H, Requena N. 2011. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Current Biology 21: 1204–1209. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Lorek J, Kwaaitaal M, Consonni C, Becker K, Micali C, Loren Ver, van Themaat E, Bednarek P, Raaymakers TM et al 2017. Key components of different plant defense pathways are dispensable for powdery mildew resistance of the Arabidopsis mlo2 mlo6 mlo12 triple mutant. Frontiers in Plant Science 8: 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch S, Pesch L, Panstruga R. 2016. Comprehensive phylogenetic analysis sheds light on the diversity and origin of the MLO family of integral membrane proteins. Genome Biology and Evolution 8: 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet E‐P, Ané J‐M, Lauber E, Bisseling T et al 2004. A putative Ca2+ and calmodulin‐dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ. 2017. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178. [DOI] [PubMed] [Google Scholar]
- Martin FM, Harrison MJ, Lennon S, Lindahl B, Öpik M, Polle A, Requena N, Selosse MA. 2018. Cross‐scale integration of mycorrhizal function. New Phytologist 220: 941–946. [DOI] [PubMed] [Google Scholar]
- McGrann GR, Stavrinides A, Russell J, Corbitt MM, Booth A, Chartrain L, Thomas WT, Brown JK. 2014. A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. Journal of Experimental Botany 65: 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E, Mun J‐H, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono J‐J, Cook DR. 2007. A novel nuclear protein interacts with the symbiotic DMI3 calcium‐ and calmodulin‐dependent protein kinase of Medicago truncatula . Molecular Plant–Microbe Interactions 20: 912–921. [DOI] [PubMed] [Google Scholar]
- Murray JD, Muni RRD, Torres‐Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J. 2011. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula . The Plant Journal 65: 244–252. [DOI] [PubMed] [Google Scholar]
- Nadal M, Sawers R, Naseem S, Bassin B, Kulicke C, Sharman A, An G, An K, Ahern KR, Romag A. 2017. An N‐acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nature Plants 3: 17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RJ, Panstruga R. 2006. Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytologist 171: 699–718. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews Microbiology 11: 252. [DOI] [PubMed] [Google Scholar]
- Pavan S, Schiavulli A, Appiano M, Marcotrigiano AR, Cillo F, Visser RG, Bai Y, Lotti C, Ricciardi L. 2011. Pea powdery mildew er1 resistance is associated to loss‐of‐function mutations at a MLO homologous locus. Theoretical and Applied Genetics 123: 1425–1431. [DOI] [PubMed] [Google Scholar]
- Pennington HG, Jones R, Kwon S, Bonciani G, Thieron H, Chandler T, Luong P, Morgan SN, Przydacz M, Bozkurt T. 2019. The fungal ribonuclease‐like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathology 15: e1007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze‐Lefert P. 2002. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiology 129: 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primrose SB, Twyman R. 2013. Principles of gene manipulation and genomics. Oxford, UK: John Wiley & Sons. [Google Scholar]
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. 2010. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. The Plant Journal 61: 482–494. [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H. 1994. Four hundred‐million‐year‐old vesicular arbuscular mycorrhizae. Proceedings of the National Academy of Sciences, USA 91: 11841–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena N, Voss S, Betz R, Heidt S, Corradi N. 2018. RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Frontiers in Microbiology 9: 2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Lozano JM, Gianinazzi S, Gianinazzi‐Pearson V. 1999. Genes involved in resistance to powdery mildew in barley differentially modulate root colonization by the mycorrhizal fungus Glomus mosseae . Mycorrhiza 9: 237–240. [Google Scholar]
- Saur IML, Bauer S, Lu X, Schulze‐Lefert P. 2019. A cell death assay in barley and wheat protoplasts for identification and validation of matching pathogen AVR effector and plant NLR immune receptors. Plant Methods 15: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A. 2016. A phylum‐level phylogenetic classification of zygomycete fungi based on genome‐scale data. Mycologia 108: 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M et al 2008. Large‐scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula . The Plant Journal 54: 335–347. [DOI] [PubMed] [Google Scholar]
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE. 2012. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Current Biology 22: 2242–2246. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J‐L. 2014. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology 32: 947–951. [DOI] [PubMed] [Google Scholar]
- Watts‐Williams SJ, Cavagnaro TR. 2012. Arbuscular mycorrhizas modify tomato responses to soil zinc and phosphorus addition. Biology and Fertility of Soils 48: 285–294. [Google Scholar]
- Xue L, Cui H, Buer B, Vijayakumar V, Delaux P‐M, Junkermann S, Bucher M. 2015. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus . Plant Physiology 167: 854–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Blaylock LA, Harrison MJ. 2010. Two Medicago truncatula half‐ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. The Plant Cell 22: 1483–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Pumplin N, Ivanov S, Harrison MJ. 2015. EXO70I is required for development of a sub‐domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Current Biology 25: 2189–2195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Mycorrhization in barley (Hordeum vulgare) cv Pallas and wheat (Triticum aestivum) cv KN199 wild‐type (WT) and mlo mutants.
Fig. S2 Proportion of differentially expressed genes (DEGs) in barley (Hordeum vulgare) cv Ingrid wild‐type (WT) and Hvmlo1‐5 mutants in mycorrhizal roots relative to uninoculated roots.
Fig. S3 Relative expression of potential barley (Hordeum vulgare) orthologues of genes known to be involved in mycorrhizal symbiosis.
Fig. S4 Medicago truncatula MLO gene expression.
Fig. S5 Mycorrhization in Medicago truncatula wild‐type (WT) and Mtmlo8 mutants.
Methods S1 MLO protein sequences.
Methods S2 Gene structure of barley (Hordeum vulgare) and wheat (Triticum aestivum) MLO1.
Methods S3 Primer sequences.
Methods S4 Sterilization methods and growth conditions.
Methods S5 Staining and visualization methods.
Methods S6 Gene expression analyses.
Table S1 Normalized read counts for RNA‐seq samples obtained from barley (Hordeum vulgare) cv Ingrid wild‐type and Hvmlo1‐5 roots at 17 dpi and 26 dpi with and without Rhizophagus irregularis.
Table S2 RNA‐seq analysis – Fold changes (log2) of RNA‐seq samples obtained from barley (Hordeum vulgare) cv Ingrid wild‐type and Hvmlo1‐5 roots at 17 dpi and 26 dpi with and without Rhizophagus irregularis.


